Abstract
Forkhead box M1 (FOXM1) is a proliferative transcription factor and plays a vital role in many cancers. However, the function and molecular mechanism of FOXM1 in esophageal squamous cell carcinoma (ESCC) remain poorly understood. Hence, we aim to clarify the molecular basis of FOXM1-mediated ESCC progression. In this study, bioinformatics analysis showed that FOXM1 was mainly involved in key signal pathways, including cell proliferation, cell cycle, and homologous recombination in ESCC, and predicted that CDC6 might be a potential regulatory target gene of FOXM1. The results revealed that FOXM1 and CDC6 were significantly overexpressed in ESCC tissue and cell line, and their expression was positively correlated. Further studies showed that FOXM1 directly transcriptionally activated CDC6 by binding to its promoter region in ESCC cells. Moreover, FOXM1 mediated ESCC cell proliferation by regulating CDC6 expression, which may be related to promoting G1-S phase transition of cell cycle. Taken together, FOXM1-CDC6 axis mediates ESCC malignant proliferation and may serve as a potential biological target for ESCC treatment.
Keywords: FOXM1, CDC6, esophageal squamous cell carcinoma, tumor proliferation
Introduction
Esophageal carcinoma is a common malignancy of the upper digestive tract, with >570,000 new cases globally in 2018 (Bray et al., 2018). Esophageal squamous cell carcinoma (ESCC) represents the predominant subtype of esophageal carcinoma, accounting for about 84% of all cases (Arnold et al., 2020). Owing to the limitation of clinical methods for early diagnosis and treatment, ESCC still has a poor 5-year survival rate (Lin et al., 2014). Thus, a better understanding of the molecular pathogenesis of ESCC is an urgent research objective.
Forkhead box M1 (FOXM1), a Forkhead box transcription factor, is overexpressed in a variety of solid human tumors (Borhani and Gartel, 2020). Abnormally upregulated FOXM1 is associated with tumorigenesis and poor patient prognosis (Bach et al., 2018; Nandi et al., 2018). Activated FOXM1 leads to multiple cancer phenotypes, including but not limited to regulating cell proliferation, DNA damage repair, cell invasion, angiogenesis, and chemoresistance (Liao et al., 2018). A meta-analysis showed that FOXM1 is a major predictor of cancer poor prognosis (Gentles et al., 2015).
As FOXM1 affects various characteristics of human cancers, it has been identified as a key target in cancer therapies (Halasi and Gartel, 2013). Therefore, a closer look at direct FOXM1 targets as well as the signaling pathways involving FOXM1 will contribute to developing new targeted cancer treatment.
Currently, the molecular mechanisms underlying FOXM1 regulation in ESCC are poorly understood. Through analyzing ESCC data sets of GEO and TGCA, we found that CDC6 may be a novel transcriptional target of FOXM1. It is well known that FOXM1 promotes cellular proliferation and cell cycle progression, mainly through regulating genes related to cell cycle (Petrovic et al., 2008). Moreover, CDC6 is a key regulator of initiation of DNA replication in eukaryotic cells and plays important roles in the activation and maintenance of cell cycle checkpoints (Gonzalez et al., 2017). We thus hypothesized that the pro-oncogenic functions of FOXM1 might be linked to its transcriptional regulation of CDC6 in ESCC.
In this study, we further explored the biological function and potential mechanism of FOXM1 in ESCC mediated by CDC6. Our study provides valuable clues for developing more effective therapeutic strategies for the treatment of ESCC.
Materials and Methods
Bioinformatics analysis of ESCC data
Two ESCC RNA-seq data sets were downloaded from TCGA database (https://portal.gdc.cancer.gov/) and GEO database (GSE130078; https://www.ncbi.nlm.nih.gov/geo/). Differential gene expression analysis was performed with the R package edgeR at an adjusted p-value cutoff of <0.001. The Ensembl gene ID was converted to gene symbol using biomaRt R package (Durinck et al., 2009). Multiscale embedded gene coexpression network analysis (MEGENA) is a novel coexpression network analysis method that can effectively construct a fast planar filtered network (Song and Zhang, 2015). MEGENA analysis, using MEGENA v1 3.7 R package, was performed on 447 differentially expressed genes (DEGs) of GSE130078 data set.
We utilized a multiomics data analysis tool (omicsbean; www.omicsbean-cancer.com) to perform Kyoto Encyclopedia of Gene and Genome (KEGG) and protein–protein interaction (PPI) analysis. The threshold for KEGG significance was padjusted < 0.05, and the PPI network model used a confidence cutoff of 800. Pearson correlation analysis between genes was analyzed using the cBioPortal online platform (http://cbioportal.org/). In addition, we also downloaded four ESCC chip data from GEO database, including GSE53625, GSE23400, GSE161533, and GSE44021. The downloaded data were used for mRNA expression level analysis of FOXM1 and CDC6, and significance testing was performed using the Wilcoxon Rank Sum test.
Cell culture
KYSE450, KYSE510, and HET-1A cells were purchased from the American Type Culture Collection. All cell lines were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS) and were maintained at 37°C in a humidified 5% CO2 incubator.
Small-interfering RNAs, plasmids, and transfection
We used small-interfering RNA (siRNA) for FOXM1 knockdown, and the sequence information of siRNAs is shown in Supplementary Table S1. The coding sequence (CDS) of FOXM1 and CDC6 genes was cloned into the vector pcDNA3.1(+). Plasmids or siRNAs were transfected into ESCC cells using lipofectamine 3000 transfection reagent (Invitrogen).
RNA extraction and real-time PCR
Total RNA was extracted using NucleoZOL (Macherey-Nagel). Complementary DNA was synthesized using a PrimeScriptl RT reagent Kit (Takara). Real-time PCR was performed using UltraSYBR Mixture (High ROX, CWBIO). The relative mRNA expression levels were normalized to endogenous GAPDH. Relative expression was calculated using the 2−ΔΔCt method. Primer sequences were listed in Supplementary Table S2.
Western blot
Cells were lysed in radio-immunoprecipitation assay (RIPA) buffer (Beyotime, China) containing protease inhibitor cocktail (Beyotime). Total proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). The membrane was incubated with the primary antibodies, including FOXM1, CDC6, and GAPDH, overnight at 4°C. Blot was detected with secondary antibody conjugated horseradish peroxidase (HRP). The relative level of protein expression was normalized to GAPDH for each sample. Antibodies used in this study are shown as follows: Primary antibodies were FOXM1 (ab207298; Abcam), CDC6 (BS4036; Bioworld), and GAPDH (60004-1-Ig; Proteintech). Secondary antibodies included goat anti-rabbit IgG-HRP (SA00001-2; Proteintech) and goat anti-mouse IgG-HRP (SA00001-1; Proteintech).
Immunohistochemistry
This study was approved by the Ethics Committees of Fujian Provincial Hospital and abided by the Declaration of Helsinki (K2018-12-045). A total of 18 paired ESCC and adjacent normal tissue sections were obtained from the department of pathology, Fujian Provincial Hospital. Detailed information of ESCC patients used for immunohistochemistry are shown in Supplementary Table S3. The paraffin sections were dewaxed and rehydrated using xylene and a series of grades of alcohol. Tissue slides were boiled in citric acid repair solution (Maixin) in a high-pressure cooker for 10 min for antigen retrieval.
The endogenous peroxidase was blocked with peroxidase blocking reagent (Maixin), and nonspecific binding of antibodies was prevented through incubation with animal nonimmune serum for 15 min at room temperature. After removing the blocking buffer, tissue slides were incubated in the primary antibody at 4°C overnight. Subsequently, tissue slides were incubated with second antibody at 37°C for 15 min and then stained with 3,3-Diaminobenzidine (DAB) and counterstained with hematoxylin. The slides were placed onto a microscope (Leica) to observe or acquire images. Two experienced pathologists blinded to the clinical data scored the immunohistochemistry results.
CCK-8 assay
Cell viability was examined by CCK-8 assay (Beyotime). In brief, cells were seeded in 96-well plates (2 × 104 cells/mL). After cell treatment, 10 μL of CCK-8 was added and incubated for 2 h at 37°C. Finally, the absorbance under 450 nm was measured by a microplate reader.
Colony formation assay
Cells were seeded into six-well plates (3 × 103 cells/well) and incubated at 37°C and 5% CO2 for 2 weeks. Cells were fixed with 4% polyformaldehyde and subsequently stained with 1% crystal violet.
EdU assay
EdU assay was performed using a BeyoClick™ EdU-488 kit (Beyotime). Cells were cultured with EdU working solution for 2 h, and then fixed with 4% paraformaldehyde for 15 min. After being washed for three times, plates were incubated with 0.3% Triton X-100 for cell permeabilization. Then, cells were incubated with click reaction solution for 30 min in a dark environment. After Hoechst 33342 nuclear staining was performed, cell images were captured by a fluorescence microscope and analyzed by ImageJ software.
Flow cytometry analysis
Cell cycle status was detected using a cell cycle and apoptosis analysis kit (Beyotime). Cells were collected and fixed with 70% alcohol for 2 h at 4°C. The collected cells were stained using propidium iodide. Then, the stained cells were analyzed by flow cytometer.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were carried out by EpiQuik™ ChIP Kit (P-2002; Epigentek). In brief, cells were cross-linked with 1% formaldehyde. The cross-linked chromatin was then sonicated to fragments ranging from 300 to 500 bp. The DNA fragments were immunoprecipitated with the indicated FOXM1 antibody. Normal rabbit or mouse IgG was used as a negative control. Input DNA was extracted from the sample before immunoprecipitation. The immunoprecipitated DNA and input DNA were analyzed by PCR and agarose gel electrophoresis. The primers were used to amplify the predictive binding sites (BSs) of CDC6 promoter regions are shown in Supplementary Table S4.
Dual-luciferase reporter assay
KYSE510 cells were transfected with pGL3-CDC6 promoter (or truncated CDC6 promotors), PRL-TK, pcDNA3.1(+), or pcDNA3.1(+)-FOXM1 using Lipo3000 reagent (Invitrogen) for 48 h. The luciferase and Renilla activity in cell lysates were then measured using Duo-Lite Luciferase Assay System Kit (Vazyme). The fluorescence intensity was measured using a SpectraMax Paradigm multimode microplate reader (SpectraMax iD5). Firefly luciferase activity was normalized to renilla luciferase activity to correct for the differences in transfection efficiency.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 8 software. Each experiment was performed in duplicate at least three times. Data were presented as the means ± standard deviation, and p-value ≤0.05 was considered to be statistically significant. Continuous data were analyzed using Student's t-test. Fisher's exact test was applied to analyze FOXM1 and CDC6 expression based on immunohistochemistry scores.
Results
Functional analyses of FOXM1 using a bioinformatics approach
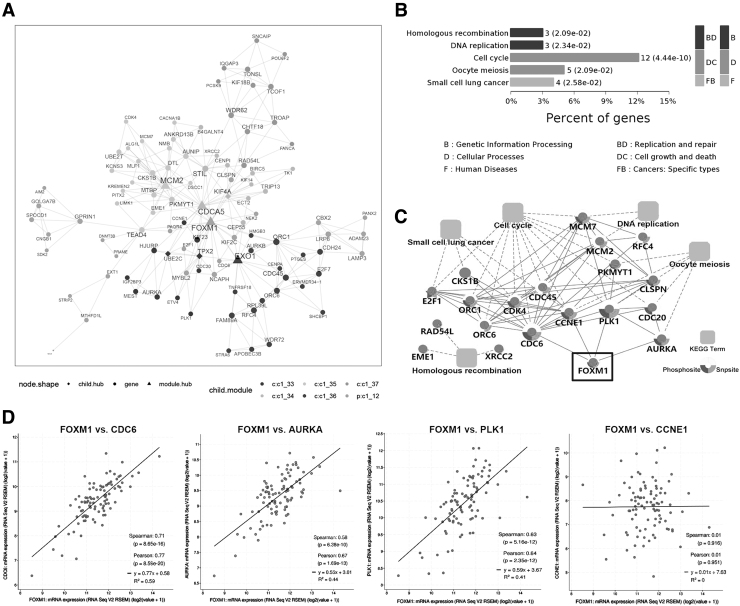
Differential gene expression analysis identified 1808 DEGs (upregulated: 1009, downregulated: 799) in the GSE130078 data set, and 1798 DEGs (upregulated: 857, downregulated: 941) in the TGCA_ESCC data set. A total of 447 overlapped DEGs, including 121 upregulated and 326 downregulated genes, were identified by Venn analysis (Supplementary Fig. S1). MEGENA analysis revealed a total of 30 differentially expressed modules (Supplementary Fig. S2, FDR <0.05). Among them, c1_12 module contained 101 genes, and FOXM1 was a hub gene of this module (Fig. 1A).
FIG. 1.
Functional analyses of FOXM1 using bioinformatics approach. (A) The coexpression network of c1_12 module in which FOXM1 is a hub gene was constructed using the overlapping differentially expressed genes as input to the MEGENA R package. Triangles represent hub network genes, whereas circles represent non-hub network genes. Node colors represent distinct subnetwork clusters, and node size is directly proportional to node degree. (B) KEGG enrichment analysis of C1_12 module genes. (C) PPI networks were constructed based on PPIs in STRING database and significant KEGG enrichment pathways. (D) Correlation analysis between FOXM1 and CDC6 mRNA expression in the TCGA-ESCC data set. ESCC, esophageal squamous cell carcinoma; FOXM1, Forkhead box M1; KEGG, Kyoto Encyclopedia of Gene and Genome; MEGENA, multiscale embedded gene coexpression network analysis; PPI, protein–protein interaction.
Next, we performed KEGG pathway analysis of C1_12 module genes, and the results showed that five signaling pathways were significantly enriched (padjusted <0.05), including cell cycle, DNA replication, oocyte meiosis, homologous recombination, and small cell lung cancer (Fig. 1B). Interestingly, these enrichment pathways were highly consistent with the main biological functions of FOXM1 as a proliferation stimulator.
PPI network suggested that FOXM1 might act through interacting with CDC6, CCNE1, PLK1, and AURKA (Fig. 1C). Furthermore, we found that the mRNA expression levels of FOXM1 and CDC6, PLK1, and AURKA were positively correlated in 95 ESCC patients, with FOXM1 and CDC6 having the largest Pearson correlation coefficient (Fig. 1D). In addition, CDC6 is a key gene for the initiation of DNA replication in the cell cycle (Gonzalez et al., 2017). Based on the aforementioned analysis, we believe that CDC6 may be a new key target gene of FOXM1 in ESCC.
FOXM1 and CDC6 overexpression in ESCC tissues and cell lines
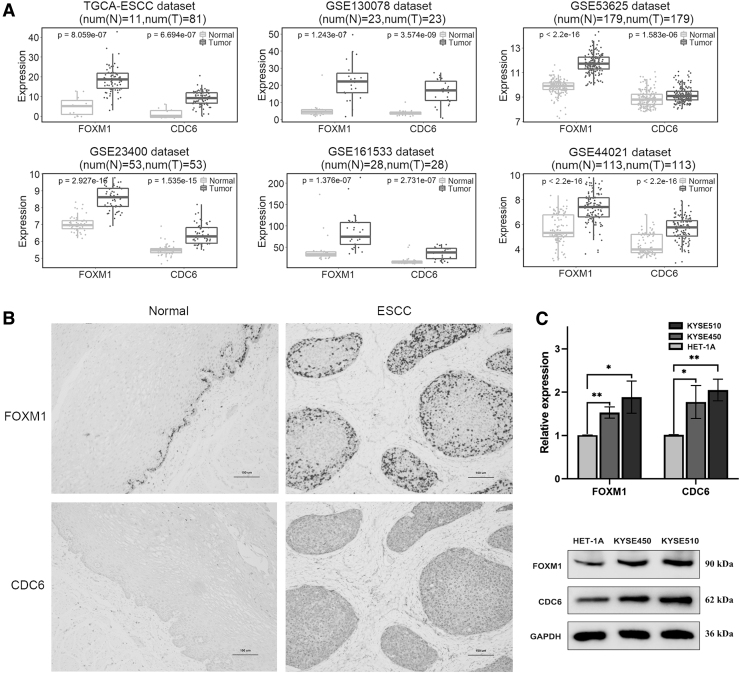
To preliminarily explore the expression of FOXM1 and CDC6 in ESCC, we used public data sets from TCGA and GEO to evaluate FOXM1 and CDC6 mRNA expression in ESCC and adjacent normal tissue samples. The results showed that FOXM1 and CDC6 mRNA expressions were significantly upregulated in ESCC group compared with adjacent tissue group (Fig. 2A). We further detected FOXM1 and CDC6 expression in both adjacent normal esophageal squamous epithelium and ESCC tissues by immunohistochemistry (Fig. 2B). FOXM1 positive staining was mainly observed in the nucleus and partly in the cytoplasm, which was brown.
FIG. 2.
Expression of FOXM1 and CDC6 in ESCC tissues and cell lines. (A) FOXM1 and CDC6 mRNA expression in ESCC and adjacent normal tissue samples from TCGA and GEO data sets. (B) Immunohistochemical staining images of FOXM1 and CDC6 expression in a representative ESCC and matched adjacent normal tissue samples. (C) mRNA and protein levels of FOXM1 and CDC6 were detected in ESCC cell lines (KYSE450 and KYSE510) and normal cell line (HET-1A) by qRT-PCR and western blot, respectively. *p ≤ 0.05; **p ≤ 0.01. qRT-PCR, quantitative real time PCR.
The positive expression level of FOXM1 (15/18, 83.33%) was significantly higher in ESCC tissues than in adjacent noncancerous tissues (4/18, 22.22%) (p < 0.001). CDC6 positive staining appeared in yellow or brownish yellow granules and was predominantly localized to the nucleus and cytoplasm area. The positive expression level of CDC6 in ESCC tissues (11/18, 61.11%) was also significantly higher than that in adjacent noncancerous tissues (3/18, 16.67%) (p < 0.01). Correlation analysis showed that the co-positivity was 61.11% (11/18), whereas the co-negativity was 22.22% (4/18), and the correlation coefficient was 0.670 (cor.test, p < 0.01).
Finally, we detected the mRNA and protein expression levels of FOXM1 and CDC6 in ESCC cell lines (KYSE450 and KYSE510) and normal cell lines (HET-1A) by quantitative real time PCR (qRT-PCR) and western blot, respectively. Both mRNA and protein levels of FOXM1 and CDC6 were significantly upregulated in ESCC cell lines compared with HET-1A cells (Fig. 2C). These results showed that FOXM1 and CDC6 were significantly highly expressed in ESCC, and that the expression levels of FOXM1 and CDC6 are positively correlated.
FOXM1 positively regulates CDC6 expression through binding to the CDC6 promoter
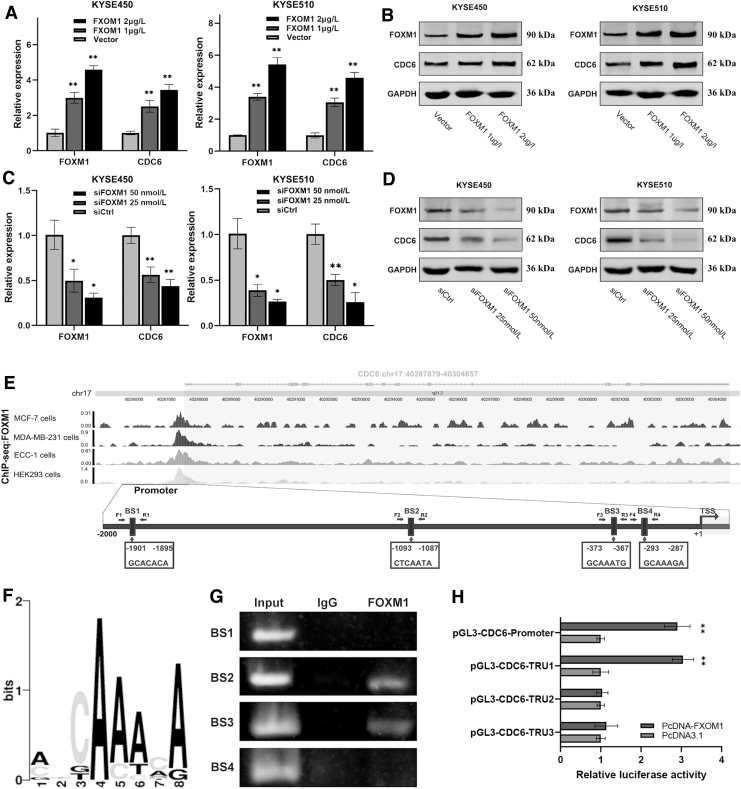
To determine whether FOXM1 regulates CDC6 expression in ESCC, we gradient transfected ESCC cell lines with FOXM1 overexpression vector or siRNA, respectively. Real-time PCR and western blot showed that the mRNA and protein expression levels of CDC6 increased in gradient after gradient overexpression of FOXM1 (Fig. 3A, B). Conversely, after specifically interfering FOXM1 with siRNA, the mRNA and protein levels of CDC6 decreased in gradient (Fig. 3C, D). These results suggested FOXM1 positively regulates CDC6 expression.
FIG. 3.
FOXM1 positively regulates CDC6 expression through binding to the CDC6 promoter. KYSE450 and KYSE510 cells were transfected with FOXM1 overexpression vector or siFOXM1 in gradient concentration, and then the mRNA (A, C) and protein (B, D) expression levels of FOXM1 and CDC6 were detected by qRT-PCR and western blot, respectively. (E) ChIP-seq binding peaks of FOXM1 within CDC6 promoter were visualized using the WashU browser from the Cistrome project, and schematic illustration showing the four potential FOXM1 BSs in CDC6 promoter. (F) The core motif of FOXM1 BS sequence. (G) The binding of FOXM1 to CDC6 promoter was examined by ChIP-PCR. ChIP-PCR products in the input, IgG, and FOXM1 groups were analyzed by agarose gel electrophoresis. (H) Luciferase constructs for wild-type or three truncated CDC6 promoter was transfected into KYSE510 cells along with FOXM1 overexpression plasmid or control vector. Luciferase activities were measured, and renilla luciferase activity normalized to firefly luciferase activity and plotted as fold change in luciferase activity relative to the control (n = 3). *p ≤ 0.05; **p ≤ 0.01. BS, binding site; ChIP, chromatin immunoprecipitation.
To explore whether FOXM1 regulates CDC6 through direct or indirect transcriptional activation, we used the cistrome data browser based on ChIP-seq data of FOXM1, and multiple binding peaks were found in the CDC6 promoter sequence (Fig. 3E). We used online software LASAGNA-Search 2.0 and sequence conservation analysis to determine that the CDC6 promoter region contained four putative FOXM1-BSs (BS1, -1901 to -1895; BS2, -1093 to -1087; BS3, -373 to -367; and BS4, -293 to -287) (Fig. 3E, F). To verify these potential BSs, we performed a ChIP-PCR assay. We found that FOXM1 is bound to the DNA fragment from BS2 and BS3 BSs in the CDC6 promoter region (Fig. 3G). Next, we performed a luciferase assay to test whether the BSs of FOXM1 in the CDC6 promoter affect its transcriptional activity.
The full-length promoter sequence of CDC6 (from −2000 bp upstream to 100bp downstream of transcription start site [TSS]) and three truncated CDC6 promoter sequences were connected into pGL3-basic vector, respectively, and the recombinant vectors were verified by sequencing and restriction enzyme digestion (Supplementary Fig. S3). Luciferase reporter assay showed that the luciferase activity was significantly increased in CDC6-promoter and CDC6-TRU1 groups, but the luciferase activity of the other two groups did not change significantly (Fig. 3H). The aforementioned results revealed that the BS2 BS is necessary for FOXM1 transcriptional activation, and the truncated promoter sequence containing BS3 and BS4 sites cannot activate FOXM1 transcriptional activation. So, we reasoned that CDC6 is a direct target of FOXM1, which binds to the BS2 site of the CDC6 promoter.
FOXM1 mediates malignant proliferation of ESCC cells through CDC6
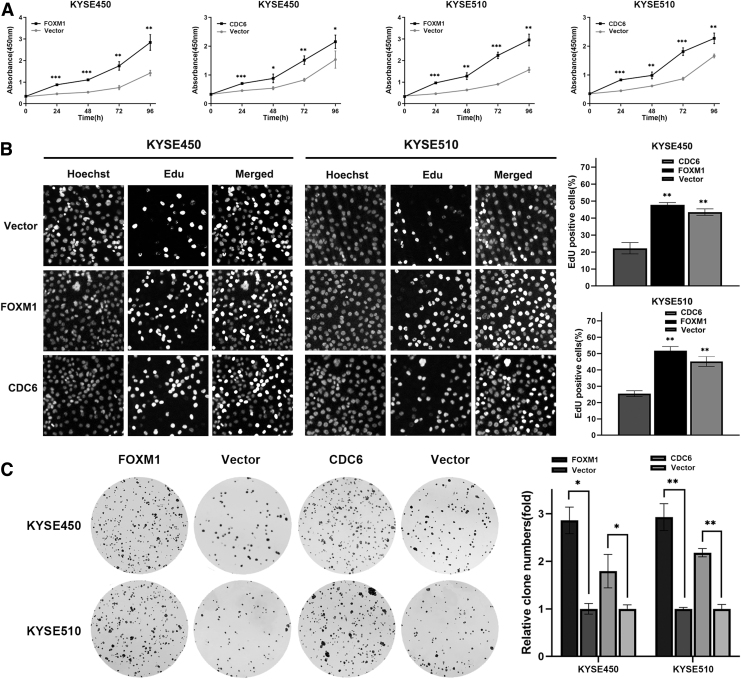
To explore the effects of FOXM1 and CDC6 on malignant proliferation of ESCC cells, CCK-8, EdU, and plate colony formation experiments were carried out. CCK-8 assay demonstrated that cell proliferative capacity increased with culture time in all groups, and the FOXM1 or CDC6 overexpression group had a significantly higher cell proliferative capacity when compared with the vector group (Fig. 4A). Then, to eliminate the adverse effects of cell death and other factors on the results of CCK-8, cell proliferation was further examined through EdU assay.
FIG. 4.
FOXM1 and CDC6 promote ESCC cell proliferation. (A) FOXM1 or CDC6 overexpression increases the cell viability of ESCC cells, assessed with CCK-8 assay. (B) FOXM1 or CDC6 overexpression increases the proliferation rate of ESCC cells as shown by EdU staining. (C) FOXM1 or CDC6 overexpression increases colony formation of ESCC cells. Data represent the mean ± SD of three independent experiments. Statistical analyses were performed by Student's t-test. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001. SD, standard deviation.
EdU analysis indicated the percentage of EdU-positive cells was significantly higher in the FOXM1 or CDC6 overexpression group, indicating that DNA replication activity was enhanced and cell proliferation rate was accelerated in ESCC cells (Fig. 4B). Moreover, the results of plate cloning showed that compared with the control group, the FOXM1 or CDC6 overexpression group increased clone size and clone formation number in ESCC cells (Fig. 4C). These results suggested that overexpression of FOXM1 or CDC6 promoted malignant proliferation of ESCC cells.
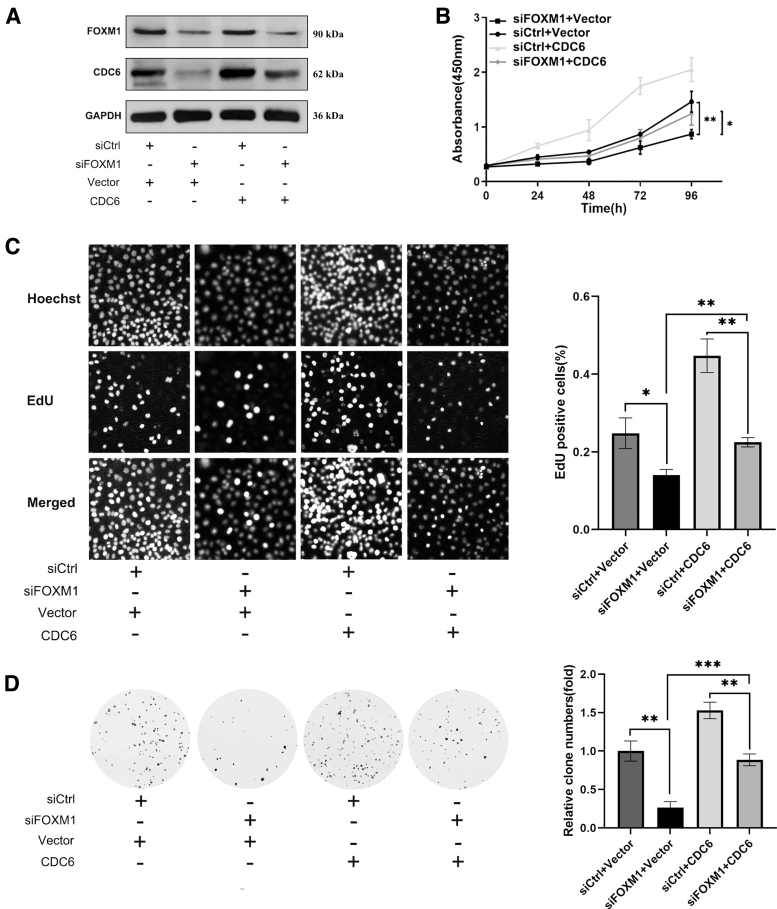
Since CDC6 is a transcriptional target gene of FOXM1, FOXM1 promotes malignant cell proliferation in many cancers. Therefore, we carried out interference and rescue experiments to confirm whether FOXM1 promotes ESCC proliferation by promoting CDC6 expression. CDC6 protein was forcibly overexpressed in FOXM1 knockdown ESCC cells (Fig. 5A), and a series of cell proliferation phenotypic changes were identified in ESCC cells. CCK-8 results indicated that compared with the control group (vector+siCtrl), FOXM1 knockdown group significantly reduced ESCC cell proliferation (Fig. 5B).
FIG. 5.
FOXM1 mediates malignant proliferation of ESCC cells through CDC6. (A) ESCC cells were transfected with FOXM1 knockdown (siFOXM1) and CDC6 overexpression (CDC6) vectors and the protein expression levels of FOXM1 and CDC6 were detected by western blot. The CCK-8 (B), EdU (C), and colony formation (D) assays were evaluated in ESCC cells transfected with siFOXM1 and CDC6. Data represent the mean ± SD of three independent experiments. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
However, after overexpression of CDC6 on the basis of FOXM1 knockdown, cell proliferative capacity of ESCC cells was partially enhanced. EdU incorporation assay yielded similar results (Fig. 5C). Colony formation result showed that CDC6 overexpression could partially abrogate clone formation ability suppressed by FOXM1 knockdown (Fig. 5D). These findings demonstrate that the promoting effect of FOXM1 on the malignant proliferation of ESCC depends on promoting CDC6 expression.
FOXM1 promotes ESCC cell proliferation by driving G1-S phase transition
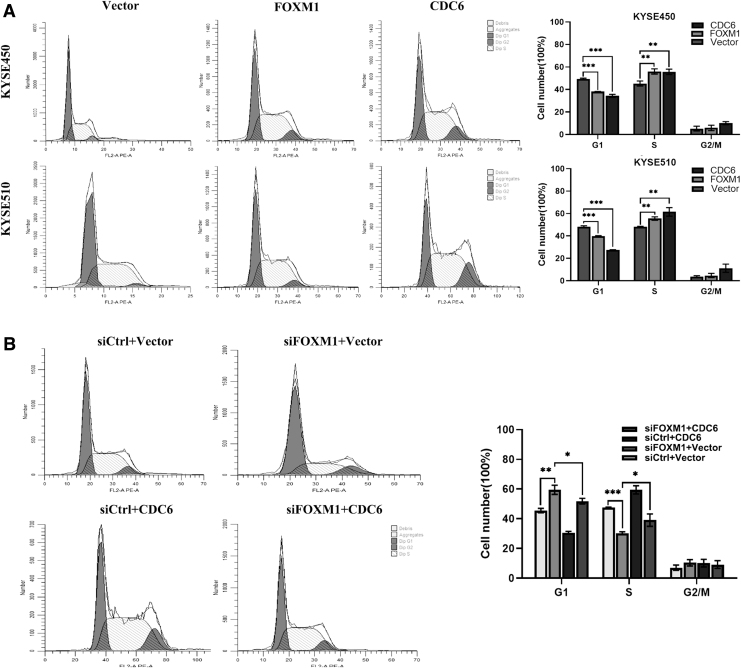
Given the essential role of CDC6 in the initiation of DNA replication, we were interested in determining whether FOXM1 mediated ESCC cell proliferation through the effect of CDC6 on cell cycle. Flow cytometry was used to examine the cell cycle distribution in ESCC cells. Interestingly, after overexpression of FOXM1 or CDC6 protein, the proportion of cells in the S phase significantly increased, the proportion of cells in the G1 phase significantly decreased, whereas no significant change in proportion was observed in the G2-M phase (Fig. 6A).
FIG. 6.
FOXM1 promotes ESCC cell proliferation by driving G1-S phase transition. (A) Cell cycle distribution was analyzed by flow cytometry after FOXM1 or CDC6 overexpression. (B) Flow cytometry analysis was used to determine cell cycle distribution (left) after interference and rescue experiment in KYSE510 cells. The proportion of cells in each phase of the cell cycle was plotted in histograms. Data represent the mean ± SD of three independent experiments. Statistical analyses were performed by Student's t-test. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
These data indicated that both FOXM1 and CDC6 promoted the G1-S cell cycle progression in ESCC cells. Therefore, we guessed that FOXM1 accelerated cell cycle progression by regulating CDC6 expression, thereby promoting cell proliferation. To verify this hypothesis, we performed interference and rescue experiment in KYSE510 cells. The results showed that FOXM1 knockdown inhibited ESCC cell transition from the G1 to S phase, and could be partially reversed by CDC6 overexpression (Fig. 6B). Based on the aforementioned results, we concluded that FOXM1 mediated ESCC cell proliferation depending on upregulating CDC6 expression and promoting G1-S cycle transition.
Discussion
FOXM1 is a wide range of cancer-promoting factor, which is overexpressed in many human cancers (Liao et al., 2018; Borhani and Gartel, 2020). The pan-cancer high expression pattern of FOXM1 suggests that most cancers may share common genetic characteristics that can drive FOXM1 expression, indicating the great potential of FOXM1 as a cancer therapeutic target. FOXM1 expression were strictly regulated by multiple signaling pathways.
Generally, FOXM1 expression was inhibited by growth inhibition signals, differentiation signals, and tumor suppressors (such as ERβ, P53, and KLF4), and activated by growth signals (such as HGF, GF, and EGF) (Kalathil et al., 2020). Moreover, FOXM1 was also a key downstream effector of PI3K/AKT, MAPK/ERK, and Raf/MEK/MAPK signaling pathways (Puig et al., 2017; Li et al., 2021). This is in line with the results that FOXM1 was significantly highly expressed in 33 kinds of cancers through pan-cancer analysis (Supplementary Fig. S4). These results indicated that FOXM1 was likely a crucial downstream regulatory gene of a variety of cancer-related pathways.
As a transcription factor, FOXM1 acts its biological function mainly by promoting the expression of downstream target genes. Therefore, gene coexpression network analysis is an effective method to identify the functional coexpression gene modules related to FOXM1. MEGENA analysis is considered to be a coexpression network analysis method better than weighted correlation network analysis (WGCNA), which can reveal more biologically meaningful coexpression gene clusters and new key regulators of cancer biological processes (Song and Zhang, 2015). In this study, we found c1_12 module in which FOXM1 as a hub gene through MEGENA analysis, and KEGG pathway analysis of genes in this module showed the cell cycle pathway was the most significantly enriched term.
Furthermore, the PPI network analysis indicated that FOXM1 had predicted interactions with these genes, including CDC6, CCNE1, PLK1, and AURKA in ESCC. Interestingly, it has been reported that MELK promoted FOXM1 phosphorylation and phosphorylated FOXM1 then activated downstream targets PLK1 in ESCC cells (Chen et al., 2020). Moreover, further correlation analysis suggested that CDC6 may be a key target gene for FOXM1. Therefore, the interaction between FOXM1 and CDC6 in ESCC needs to be further explored.
CDC6 is an essential regulator of DNA replication. CDC6, together with ORC and CDTl, promotes the sequential recruitment and assembly of two MCM2-7 complexes and make them become a stable double hexamer (pre-replication complex [pre-RC]) on double-stranded DNA (Ticau et al., 2015). Abnormal cell cycle regulation, including the regulation of DNA replication initiation, is the main reason for the uncontrolled proliferation of malignant tumors. CDC6 is abnormally highly expressed in various human tumors, including pancreatic cancer, non-small cell lung cancer, cervical cancer, and bladder cancer (Lim and Townsend, 2020).
Multiple studies have shown that abnormal CDC6 expression is closely related to the proliferation of tumor cells. For example, high expression of CDC6 promotes cell proliferation in epithelial ovarian cancer and is associated with poor prognosis (Deng et al., 2016). In contrast, silencing CDC6 significantly inhibited the proliferation and promoted apoptosis of tongue squamous cell carcinoma cells (Feng et al., 2013). In this study, we found that CDC6 were abnormally overexpressed in ESCC tissues and cell lines. More importantly, upregulated CDC6 promoted cell proliferation in ESCC, suggesting its oncogenic effect.
Continuous malignant proliferation is one of the most important characteristics of cancer phenotype (Hanahan, 2022). FOXM1 mainly maintains cell proliferation by regulating the transcription of cell cycle-related gene (Halasi and Gartel, 2013). FOXM1 has been reported to modulate key regulators of cell cycle, thus resulting in G1/S and G2/M transitions of cell cycle (Krishnan et al., 2017; Bu et al., 2020). In addition, FOXM1 is involved in regulating the expression of centromeric proteins CENPA, CENPB, and CENPF, and also plays an important role in chromosome separation and spindle assembly (Shirakawa et al., 2017; Nilsson et al., 2020).
In this study, we identified for the first time that CDC6 was a direct target of FOXM1, and FOXM1 regulated its translation in ESCC. In addition, FOXM1 participated in cell proliferation and cell cycle progression regulation by modulation of CDC6 expression.
Owing to the important role in tumor progression, FOXM1 has been widely considered as a potential key target for human cancer treatment. In recent years, several investigators have developed a variety of FOXM1 targeted drug strategies for tumor treatment, including RNA interference, proteasome inhibitor, SUMO protein modification (Huang et al., 2015; Borhani and Gartel, 2020). Studies have shown that silencing FOXM1 by RNAi can eliminate estrogen stimulated breast cancer cell proliferation and inhibit acquired tamoxifen resistance (Halasi and Gartel, 2013). In a recent study, NB-5 was identified as a FOXM1 inhibitor by screening small molecule libraries, which effectively inhibited cell proliferation in breast cancer (Ziegler et al., 2019).
Moreover, SUMO modification of FOXM1 weakens its activity and triggering the mitotic delay of breast cancer cells (Myatt et al., 2014). However, these targeted drugs are also accompanied by severe side effects or drug resistance, which is largely related to the insufficient understanding of FOXM1 regulatory mechanism (Gormally et al., 2014). Therefore, exploring FOXM1 regulatory mechanism in ESCC will help FOXM1 and its target genes to become the key therapeutic targets for ESCC treatment. Our study showed that FOXM1 mediated cell proliferation and tumor growth by promoting CDC6 expression in ESCC, suggesting that FOXM1-CDC6 axis may be a potential target for ESCC treatment.
Conclusion
In summary, our study showed that FOXM1 and CDC6 are aberrantly overexpressed in ESCC. FOXM1 positively regulates CDC6 expression through binding to the CDC6 promoter. Furthermore, FOXM1 mediates malignant proliferation of ESCC cells through regulating CDC6 expression, and then driving G1-S phase transition of the cell cycle. Our study reveals a novel mechanism by which FOXM1 acts its oncogenic role in the progression of ESCC, suggesting FOXM1-CDC6 axis is a potential therapeutic target for ESCC treatment.
Supplementary Material
Authors' Contributions
Investigation, data curation, formal analysis, software analysis, and writing by X.C. Investigation, data curation, and writing by J.C. Investigation, data curation, and formal analysis by X.Y. Conceptualization, resources, funding acquisition, and supervision by G.L. Project administration and supervision by T.C.
Disclosure Statement
No competing financial interests exist.
Funding Information
This study was supported by the Medical Innovation Foundation, Fujian Province (Grant No. 2018-CX-3). This study is also supported by the National Natural Science Foundation of China (Grant No. 61872218).
Supplementary Material
References
- Arnold, M., Ferlay, J., Berge, M.I., and Soerjomataram, I. (2020). Global burden of oesophageal and gastric cancer by histology and subsite in 2018. Gut 69, 1564–1571. [DOI] [PubMed] [Google Scholar]
- Bach, D.H., Long, N.P., Luu, T.T., Anh, N.H., Kwon, S.W., and Lee, S.K. (2018). The dominant role of forkhead box proteins in cancer. Int J Mol Sci 19, 3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borhani, S., and Gartel, A.L. (2020). FOXM1: a potential therapeutic target in human solid cancers. Expert Opin Ther Targets 24, 205–217. [DOI] [PubMed] [Google Scholar]
- Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R.L., Torre, L.A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68, 394–424. [DOI] [PubMed] [Google Scholar]
- Bu, H., Li, Y., Jin, C., Yu, H., Wang, X., Chen, J., et al. (2020). Overexpression of PRC1 indicates a poor prognosis in ovarian cancer. Int J Oncol 56, 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., Wei, Q., Bi, S., and Xie, S. (2020). Maternal embryonic leucine zipper kinase promotes tumor growth and metastasis via stimulating FOXM1 signaling in esophageal squamous cell carcinoma. Front Oncol 10, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, Y., Jiang, L., Wang, Y., Xi, Q., Zhong, J., Liu, J., et al. (2016). High expression of CDC6 is associated with accelerated cell proliferation and poor prognosis of epithelial ovarian cancer. Pathol Res Pract 212, 239–246. [DOI] [PubMed] [Google Scholar]
- Durinck, S., Spellman, P.T., Birney, E., and Huber, W. (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 4, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, C.J., Lu, X.W, Luo, D.Y., Li, H.J., and Guo, J.B. (2013). Knockdown of Cdc6 inhibits proliferation of tongue squamous cell carcinoma Tca8113 cells. Technol Cancer Res Treat 12, 173–181. [DOI] [PubMed] [Google Scholar]
- Gentles, A.J., Newman, A.M., Liu, C.L., Bratman, S.V., Feng, W., Kim, D., et al. (2015). The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med 21, 938–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, S., Klatt, P., Delgado, S., Conde, E., Lopez, R.F., Sanchez, C.M., et al. (2017). Retraction: oncogenic activity of Cdc6 through repression of the INK4/ARF locus. Nature 547, 246. [DOI] [PubMed] [Google Scholar]
- Gormally, M.V., Dexheimer, T.S., Marsico, G., Sanders, D.A., Lowe, C., Matak, V.D., et al. (2014). Suppression of the FOXM1 transcriptional programme via novel small molecule inhibition. Nat Commun 5, 5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halasi, M., and Gartel, A.L. (2013). Targeting FOXM1 in cancer. Biochem Pharmacol 85, 644–652. [DOI] [PubMed] [Google Scholar]
- Hanahan, D. (2022). Hallmarks of cancer: new dimensions. Cancer Discov 12, 31–46. [DOI] [PubMed] [Google Scholar]
- Huang, C.J., Wu, D., Khan, F.A., and Huo, L.J. (2015). DeSUMOylation: an important therapeutic target and protein regulatory event. DNA Cell Biol 34, 652–660. [DOI] [PubMed] [Google Scholar]
- Kalathil, D., John, S., and Nair, A.S. (2020). FOXM1 and cancer: faulty cellular signaling derails homeostasis. Front Oncol 10, 626836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan, A., Dhanya, K., Babu, P.S., Jagadeeshan, S., Prasad, M., and Nair, S.A. (2017). Oncogenic actions of SKP2 involves deregulation of CDK1 turnover mediated by FOXM1. J Cell Biochem 118, 797–807. [DOI] [PubMed] [Google Scholar]
- Li, Y., Wang, Y., Zhang, W., Wang, X., Chen, L., and Wang, S. (2021). BKM120 sensitizes BRCA-proficient triple negative breast cancer cells to olaparib through regulating FOXM1 and Exo1 expression. Sci Rep 11, 4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, G.B., Li, X.Z., Zeng, S., Liu, C., Yang, S.M., Yang, L., et al. (2018). Regulation of the master regulator FOXM1 in cancer. Cell Commun Signal 16, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, N., and Townsend, P.A. (2020). Cdc6 as a novel target in cancer: oncogenic potential, senescence and subcellular localisation. Int J Cancer 147, 1528–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, D.C., Hao, J.J., Nagata, Y., Xu, L., Shang, L., Meng, X., et al. (2014). Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet 46, 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myatt, S.S., Kongsema, M., Man, C.Y., Kelly, D.J., Gomes, A.R., Khongkow, P., et al. (2014). SUMOylation inhibits FOXM1 activity and delays mitotic transition. Oncogene 33, 4316–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi, D., Cheema, P.S., Jaiswal, N., and Nag, A. (2018). FoxM1: repurposing an oncogene as a biomarker. Semin Cancer Biol 52, 74–84. [DOI] [PubMed] [Google Scholar]
- Nilsson, M.B., Sun, H., Robichaux, J., Pfeifer, M., Mcdermott, U., Travers, J., et al. (2020). A YAP/FOXM1 axis mediates EMT-associated EGFR inhibitor resistance and increased expression of spindle assembly checkpoint components. Sci Transl Med 12, 559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic, V., Costa, R.H., Lau, L.F., Raychaudhuri, P., and Tyner, A.L. (2008). FoxM1 regulates growth factor-induced expression of kinase-interacting stathmin (KIS) to promote cell cycle progression. J Biol Chem 283, 453–460. [DOI] [PubMed] [Google Scholar]
- Puig, B.A., Vinyals, A., Ferreres, J.R., Aguilera, P., Cabré E., Tell, M.G., et al. (2017). AURKA overexpression is driven by FOXM1 and MAPK/ERK activation in melanoma cells harboring BRAF or NRAS mutations: Impact on melanoma prognosis and therapy. J Invest Dermatol 137, 1297–1310. [DOI] [PubMed] [Google Scholar]
- Shirakawa, J., Fernandez, M., Takatani Ti., El, O.A., Jungtrakoon, P., Okawa, E.R., et al. (2017). Insulin signaling regulates the FoxM1/PLK1/CENP-A pathway to promote adaptive pancreatic β cell proliferation. Cell Metab 25, 868–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W.M., and Zhang, B. (2015). Multiscale embedded gene co-expression network analysis. PLoS Comput Biol 11, e1004574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticau, S., Friedman, L.J., Ivica, N.A., Gelles, J., and Bell, S.P. (2015). Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. Cell 161, 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler, Y., Laws, M.J., Sanabria, G.V., Kim, S.H., Dey, P., Smith, B.P., et al. (2019). Suppression of FOXM1 activities and breast cancer growth in vitro and in vivo by a new class of compounds. NPJ Breast Cancer 5, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.