Abstract
Vogt-Koyanagi-Harada disease (VKH) is a rare autoimmune disease characterized by diffuse and bilateral uveitis, alopecia, tinnitus, hearing loss, vitiligo and headache. The transcriptional expression pattern of peripheral blood mononuclear cells (PBMC) in VKH remains largely unknown. In this study, mRNA sequencing was conducted in PBMC from VKH patients with active uveitis before treatment (n = 7), the same patients after prednisone combined with cyclosporine treatment (n = 7) and healthy control subjects strictly matched with gender and age (n = 7). We found 118 differentially expressed genes (DEGs) between VKH patients and healthy control subjects, and 21 DEGs between VKH patients before and after treatment. TRIB1 was selected as a potential biomarker to monitor the development of VKH according to the mRNA sequencing. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed to predict the possible biological functions and signaling pathways of DEGs. Neutrophil degranulation, peptidase regulator activity, secretory granule membrane, cellular response to peptide, growth factor binding and cell projection membrane were enriched as GO annotations of DEGs. Arachidonic acid metabolism and mitogen-activated protein kinase (MAPK) signaling pathway were potential signaling pathways involved in pathogenesis and drug response of VKH. A protein–protein interaction (PPI) network was constructed by STRING, and colony stimulating factor 1 receptor (CSF1R) was identified as the hubgene of all DEGs by Cytoscape. The cell type presumed to contribute to the aberrant expression of DEGs was analyzed with the use of publicly available single-cell sequencing data of PBMC from a healthy donor and single-cell sequencing dataset of monocytes from VKH patients. Our findings may help to decipher the underlying cellular and molecular pathogenesis of VKH and may lead novel therapeutic applications.
Keywords: Vogt-Koyanagi-Harada disease, uveitis, mRNA sequencing, Peripheral blood mononuclear cells
Introduction
VKH is a rare autoimmune disease,1 and various melanin-containing organs are affected by this systemic disorder.2 A rapidly decreasing visual acuity with blurred vision accompanied with headache, neck and back stiffness, hearing loss, tinnitus, vitiligo and alopecia are the major symptoms of VKH.3 Cataract, glaucoma, choroidal neovascular membranes, subretinal fibrosis may occur as complications in half of the patients with VKH.4 A definite etiology of VKH is not clear, whereby HLA-DRB1∗0405 carriers,5 single nucleotide polymorphism loci of different genotypes,6 vaccine injection,7 viral infections8,9 and recognition of melanin-related antigens2 have all been implicated in the etiology of VKH. Diagnosis of VKH mainly depends on clinical manifestations supplemented with auxiliary examinations like fluorescein angiography, angiography with indocyanine green, and optical coherence tomography.10 To date, no biomarkers are available for the diagnosis of VKH, although elevated carbonic anhydrase 2 (CA2) and ras-related protein Rap-1b (RAP1B) in exosomes isolated from plasma have been proposed as biomarkers in early stage VKH.11 The therapy of VKH often requires long-term oral corticosteroids12 combined with cyclosporine.13 The long-term use of corticosteroids and cyclosporine may lead to adverse side effects.14,15 In addition, some VKH patients responds poorly to the corticosteroid-cyclosporine therapy, which necessitates the search for novel drugs.16 Untimely or inappropriate treatment can lead to blindness in VKH. Therefore, elucidating the detailed cellular and molecular mechanisms underlying the pathogenesis and finding potential therapeutic targets for this chronical and relapsing disease is urgently needed.
Quantitative measurement of mRNA, a high throughput technology, for the detection of genes with good sensitivity and reproducibility,17 may contribute to the diagnosis, drug discovery18,19 and surveillance of disease.20 Aberrant mRNA expression profiles of PBMC have been described in several autoimmune diseases such as rheumatoid arthritis, inflammatory bowel disease,21 psoriasis22 and multiple sclerosis.23 Abnormal expression of suppressor of cytokine signaling 1 (SOCS1),24 interleukin 23 (IL-23),25 nucleotide-binding oligomerization domain-containing protein 1 (NOD1), NOD226 in PBMC have been reported in VKH.
To date, a comprehensive mRNA sequencing of PBMC in VKH has not yet been reported and was therefore the subject of the study represented here. In this study we compared the mRNA profile of PBMC in healthy control subjects and in VKH patients before and after treatment. We found several DEGs, which are potentially involved in the pathogenesis of VKH and which might be engaged in drug responses. GO annotation, KEGG enrichment, PPI network and analysis of single-cell sequencing data of PBMC from a healthy donor and single-cell sequencing data of monocytes from VKH patients were performed to deepen our understanding of these DEGs.
Materials and methods
Subjects
Diagnosis of VKH was based on the revised diagnostic criteria for VKH27 in combination with those reported earlier by our group.28 VKH patients with cells and flare in the anterior chamber and vitreous, keratic precipitates and diffuse choroiditis29 and who did not receive any treatment for more than 4 months before this attack were defined as VKH patient with active uveitis before treatment.
This study was approved by the Institutional Ethical Committee of The First Affiliated Hospital of Chongqing Medical University (#2018–048). Informed consents were obtained before the collection of samples. In this project, 7 VKH patients with active uveitis before treatment were recruited, their blood samples were also collected after the therapy with cyclosporine and prednisone for 8 weeks. Exclusions to the enrollment included diabetes mellitus, hypertension, dyslipidaemia, history of other autoimmune disease, immunodeficiency and other disorders. Healthy controls (n = 7) were strictly age- and gender-matched with the VKH patients.
Preparation of PBMC
Venous blood was collected in tubes containing ethylene diamine tetraacetic acid (EDTA), then kept on ice, and PBMC were isolated by gradient centrifugation within 1 h after collection. PBMC were isolated by Ficoll (TBD Science, Tianjin, China) density-gradient centrifugation (600 g for 30 min at 18 °C) according to the instruction manual, cells were suspended in ice-cold phosphate buffered solution and centrifuged at 300 g for 5 min at 4 °C twice. Purified PBMC were stored in liquid nitrogen until use. VKH PBMC samples used for RT-qPCR were collected previously by our lab.
mRNA sequencing and identification of DEGs
RNA extraction, library construction and mRNA sequencing were performed by Novogene (Beijing, China). Briefly, total RNAs were extracted by TRIzol, mRNAs were purified by poly-T oligo-attached magnetic beads. Sequencing libraries were generated by NEBNext Ultra RNA Library Prep Kit (NEB, USA) for Illumina, then sequenced on an Illumina Novaseq platform and 150 bp paired-end reads were generated. Differently expressed genes (DEGs) between two groups were analyzed by DESeq2 R package.30 P-values were adjusted by Benjamini and Hochberg's approach for controlling the false discovery rate. Genes with an adjusted P-value (padj) < 0.05 identified by DESeq2 were considered as DEGs. Hierarchical clustering analyses were performed on Fragments Per Kilobase per Million (FPKM) values and Z-score was used for row normalization. The color of each grid represents the normalized value of gene expression.
Extraction of RNA and reverse-transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using the TRIzol reagent (Invitrogen, USA). The concentration of total RNA was measured with a NanoPhotometer (IMPLEN, German). Reverse transcription was performed using Go-Taq polymerase (Promega, USA). A SYBR Green qPCR Master Mix (MCE, USA) for RT-qPCR was used with Applied Biosystems 7500 (Applied Biosystems, USA). β-Actin was used as internal control. Relative expression of target gene was determined by 2-△△t. The forward primer of β-actin was CATGTACGTTGCTATCCAGGC and reverse primer of β-actin was CTCCTTAATGTCACGCACGAT. The forward primer of SGK1 was GAACACAACAGCACAACAT and reverse primer of SGK1 was CCATACAGCATCTCATACAAG. The forward primer of TRIB1 was CCTTCTGGTTGGACGATAC and reverse primer of TRIB1 was CAAGAGGCTGCGAATGAG.
GO and KEGG enrichment analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEGs were performed with the cluster Profiler R package.31 Terms or pathways with a P-value less than 0.05 were identified as statistically enriched.
Protein–protein interaction
The interactions between differentially expressed genes were analyzed using the STRING database.32 The following settings were used: full network, evidence, all active interaction sources, medium confidence (0.400) and hide disconnected nodes in the network. The hub genes were calculated and visualized by cytoHubba in Cytoscape and ranked by degree.33
Single-cell RNA sequencing
The data of the PBMC single-cell sequencing data set from a healthy donor was downloaded from https://support.10xgenomics.com/single-cell-gene-expression/datasets, and then analyzed by the Seurat R package (3.2.0).34 Genes expressed in less than 3 cells were filtered out. Cells with 200–4000 detected genes and expressing <20% mitochondrial genes were retained for subsequent analysis. Data were normalized using the NormalizeData function. PCA dimensional reduction was performed by the RunPCA function. Twelve clusters were identified with the use of the FindNeighbors and FindClusters function at a resolution of 0.4. Using this approach we extracted the five clusters we focused on with canonical markers.35
Data of the single-cell sequencing of monocytes from healthy controls and VKH patients were downloaded from GSE148020, the processing workflow, parameters and marker genes were the same as they published.36 We extracted the monocytes, integrated the monocytes of one group and identified the different expressed genes in monocytes between VKH patients and healthy volunteers by the FindMarkers function with threshold settings of Fold Change >1.5 and padj <0.05.
Statistical analysis
The statistical significance of DEGs from the bulk sample was determined with the DESeq2 R package, and the statistical significance of DEGs from the single-cell sample was determined with the FindMarkers function, whereby a padj <0.05 was considered as statistically significant. The statistical analyses, as well as the diagrams, were performed with SPSS software and GraphPad Prism 7 software. The Shapiro–Wilk test was applied to test normality. Nonparametric test was used to examine the statistical relationship between two non-normally distributed data, and parametric test was used to examine the statistical relationship between two normal distributed data, P value less than 0.05 was considered as statistically significant. Detailed statistical methods were described in figure legends.
Results
Identification of DEGs
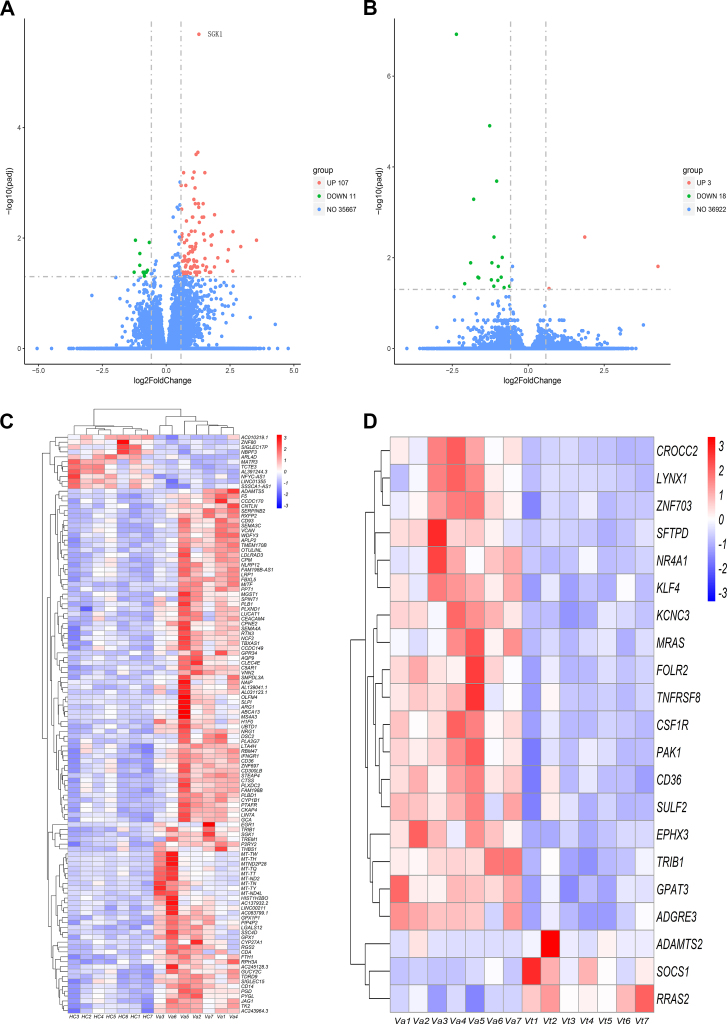
The basic information of the included VKH patients is shown in Table 1. The discovery of DEGs between two groups was identified by the R-package DESeq2 with threshold settings of Fold Change (FC) > 1.5 and padj < 0.05. DEGs were graphed as volcano plots. In total, 118 altered genes (107 up-regulated and 11 down-regulated) were detected when comparing healthy control subjects and VKH patients with active uveitis before treatment (Fig. 1A). Another 21 differentially expressed genes (3 up-regulated and 18 down-regulated) were found between VKH patients before and after therapy with cyclosporine and prednisone (Fig. 1B). DEGs with a similar expression pattern within a group were visualized by hierarchal clustering heatmap analysis (Fig. 1C, D).
Table 1.
Basic information of VKH patients.
| Sample Name | Age | Gender | Headache | Stiffness of neck and back | Tinnitus | Reduced hearing | Alopecia | Poliosis | Vitiligo | Active uveitis stage |
|---|---|---|---|---|---|---|---|---|---|---|
| Va1 | 30 | M | + | – | – | – | – | + | – | + |
| Va2 | 33 | M | + | + | + | – | – | – | – | + |
| Va3 | 46 | M | + | – | – | – | + | – | – | + |
| Va4 | 54 | M | + | – | – | + | – | – | + | + |
| Va5 | 61 | M | + | – | – | – | + | – | – | + |
| Va6 | 64 | M | – | + | – | + | – | – | – | + |
| Va7 | 22 | M | + | – | + | – | – | – | + | + |
Va, VKH patients with active uveitis before treatment. M, male.
Figure 1.
Visualization of DEGs. (A, B) Volcano plot of DEGs. Green and red dots represent downregulated and upregulated genes respectively with the criterion of FC > 1.5 and padj<0.05. Blue dots are genes not meeting the inclusion criterion of DEG. (C, D) Hierarchical clustering heatmap of DEGs. The color of each grid represents the normalized value of gene expression, the redder the color, the higher the expression level; the bluer the color, the lower the expression level. HC (n = 7), Va (n = 7), Vt (n = 7). HC, healthy control. Va, VKH patients with active uveitis before treatment. Vt, VKH patients after the therapy of cyclosporine and prednisone. The same number after Va and Vt represents the same patient.
Validation of some important DEGs
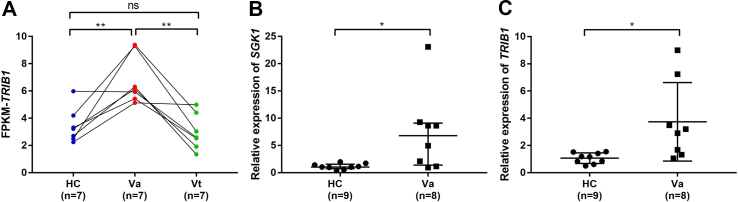
The mRNA-sequence analysis showed that the so called serum glucocorticoid regulated kinase 1 (SGK1) was upregulated in VKH patients with active uveitis before treatment when compared with healthy control subjects with the minimum padj value. The expression of TRIB1 was increased in VKH patients with active uveitis before treatment and was downregulated in VKH patients after treatment according to the mRNA sequencing (Fig. 2A), which indicates that TRIB1 is a potential biomarker to monitor the development of VKH. Upregulated mRNA expression of SGK1 and TRIB1 were further confirmed by RT-qPCR in another cohort of VKH patients with active uveitis before treatment and healthy control subjects (Fig. 2B, C).
Figure 2.
Expression of DEGs. (A) The expression of TRIB1 according to the mRNA sequencing. The statistical significance between healthy control subjects and VKH patients with active uveitis before treatment was examined by independent Mann Whitney test (P = 0.004). The statistical significance between VKH patients before and after treatment was examined by Wilcoxon matched-pairs signed rank test (P = 0.016). The statistical significance between healthy control subjects and VKH patients after treatment was examined by independent Mann Whitney test (P = 0.535). (B) The expression of SGK1 by RT-qRCR. The bars indicate median with interquartile range. The statistical significance between healthy control subjects and VKH patients with active uveitis before treatment was examined by independent Mann Whitney test (P = 0.011). (C) The expression of TRIB1 by RT-qRCR. The bars indicate median with standard deviation. The statistical significance between healthy control subjects and VKH patients with active uveitis before treatment was examined by t test (P = 0.035). ns, no statistical significance. HC, healthy control. Va, VKH patients with active uveitis before treatment. Vt, VKH patients after prednisone combined with cyclosporine treatment.
Functional analysis of DEGs
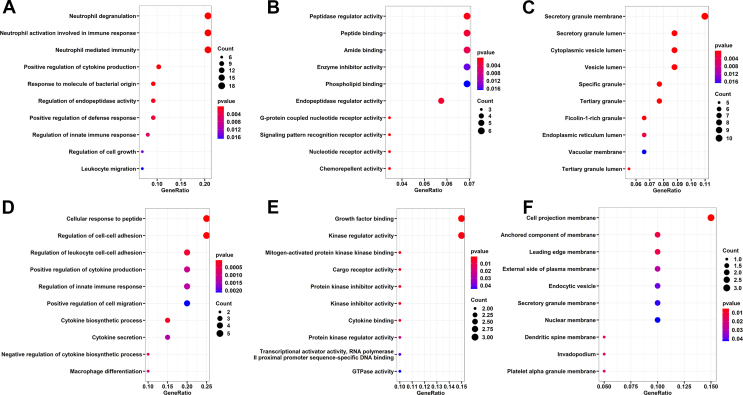
To ascertain the potential biological functions of the identified DEGs, we annotated them by Gene Ontology (GO) analysis. GO analysis includes three items: Cellular Component (CC), Molecular Function (MF) and Biological Process (BP).
The DEGs identified by comparing healthy controls with VKH patients with active uveitis before treatment revealed that neutrophil degranulation, neutrophil activation involved in immune response, response to molecule of bacterial origin, regulation of innate immune response and leukocyte migration were enriched in the BP category (Fig. 3A). Peptidase regulator activity, peptide binding, enzyme inhibitor activity and nucleotide receptor activity were enriched in the MF category (Fig. 3B). Secretory granule membrane, secretory granule lumen and cytoplasmic vesicle lumen were enriched in the CC category (Fig. 3C).
Figure 3.
Bubble diagram of enriched GO items of DEGs. The enriched GO terms of DEGs between healthy control and VKH patients with active uveitis before treatment. (A–C) BP, MF and CC enriched GO items of DEGs between VKH patients before and after treatment. (D–F) BP, MF and CC enriched GO items of DEGs between VKH patients before and after treatment. BP, Biological Process. MF, Molecular Function. CC, Cellular Component.
The DEGs found by comparing the same group of VKH patients before and after therapy with cyclosporine and prednisone included enriched growth factor binding, kinase regulator activity and mitogen-activated protein kinase (Fig. 3D). Cellular response to peptide, regulation of cell–cell adhesion, regulation of leukocyte cell–cell adhesion and positive regulation of cytokine production were enriched in the BP category (Fig. 3E). Cell projection membrane, anchored component of membrane, endocytic vesicle, and secretory granule membrane were enriched in the MF category (Fig. 3F).
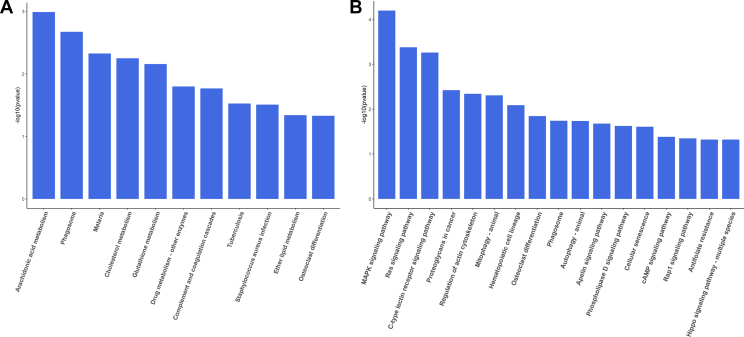
Pathway enrichment analysis of DEGs by KEGG
To explore the potential signaling pathway involved in the pathogenesis and treatment of VKH, we analyze the KEGG pathway enrichment. Arachidonic acid metabolism, phagosome, malaria, cholesterol metabolism, and glutathione metabolism pathways were enriched when comparing healthy control subjects and VKH patients with active uveitis before treatment (Fig. 4A). The MAPK signaling pathway, Ras signaling pathway, C-type lectin receptor signaling pathway and regulation of actin cytoskeleton pathways were enriched when comparing the same group of VKH patients before and after treatment (Fig. 4B).
Figure 4.
KEGG pathway enrichment analysis of DEGs. The enriched KEGG pathways of DEGs were revealed by bar graph. (A) The enriched KEGG pathways enriched for DEGs between healthy control and VKH patients with active uveitis before treatment. (B) The enriched KEGG pathways enriched for DEGs between VKH patients before and after treatment.
PPI network construction and identification of hubgene
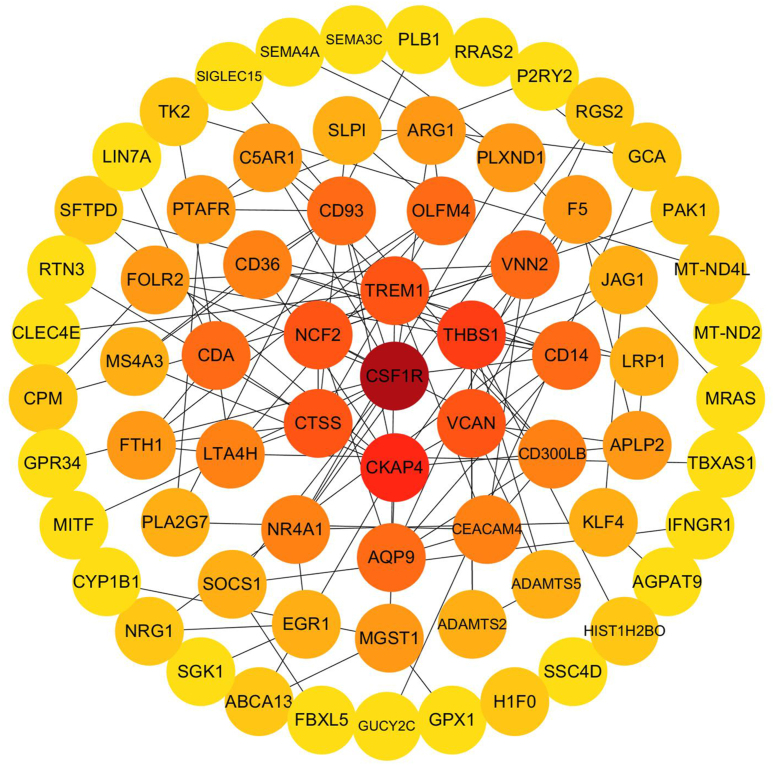
String, a well-known tool to assess the protein–protein interaction, was used to delineate the interactions between the identified DEGs. A PPI network was produced by String and drawn using the Cytoscape software. CSF1R was found to be hub gene as calculated by cytoHubba in Cytoscape (Fig. 5).
Figure 5.
Construction of PPI network among all DEGs. Each circle represents a differently expressed gene. The rank of the connection was sorted by color from deep red to light yellow.
Combined analysis of DEGs with single-cell sequencing
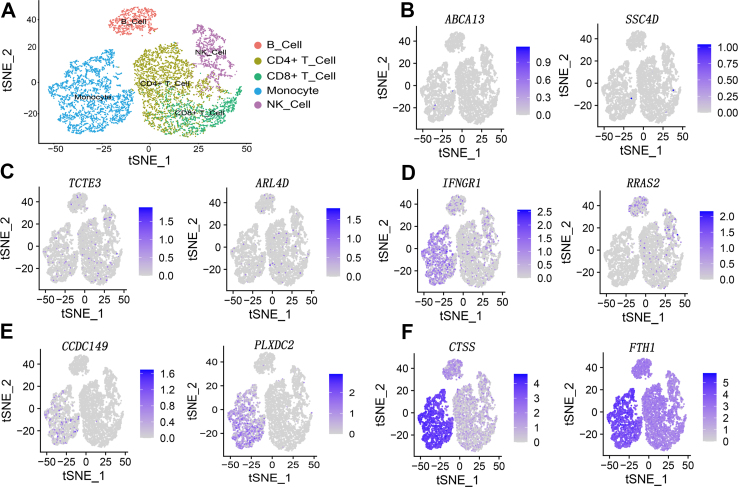
Since PBMC mainly consists of T cells, B cells, NK cells and monocytes, we extracted CD4+ T cell, CD8+ T cell, NK cell, B cell and monocyte from the single-cell sequencing dataset of PBMC (Fig. 6A). Then, we examined the distribution of our DEGs in these clusters. We found that there were mainly 5 types of distribution features in these DEGs. Type 1 included genes with almost no obvious expression in all clusters (Fig. 6B). Type 2 included genes that were sporadically expressed in very few cells in all clusters (Fig. 6C). Type 3 included genes expressed in part of the cells with a preference to certain clusters (Fig. 6D). Type 4 included genes that were mainly expressed in only one certain cluster (Fig. 6E). Type 5 included genes that were expressed in most cells among all clusters (Fig. 6F).
Figure 6.
Analysis of DEGs from single-cell sequencing data. (A) Pseudocolored t-distributed stochastic neighbor embedding (tSNE) plots of CD4+T cells (mustard), CD8+ T cells (green), NK cells (purple), B cells (red) and monocytes (blue). Each dot represents a single cell. (B–F) Representative distribution of DEGs across the five types of cells with the use of a single-cell sequencing data set from a healthy donor. The expression intensities are shown in different shades of purple, gray represents low expression, dark purple represents high expression. The name of the DEG is labeled on the top of the image.
Interestingly, we found that approximately 60% DEGs (healthy controls vs. VKH patients with active uveitis) were expressed only in monocytes according to the single-cell sequencing of the PBMC from a healthy donor (i.e., type 4). We wondered whether the expression changes of these DEGs could be attributed to monocytes. Therefore, we identified the DEGs of monocytes from healthy individuals and VKH patients with the use of single-cell sequencing of monocytes published earlier by others.36 However, the DEGs of PBMC shown in our study and DEGs of monocytes exhibited no intersection.
Discussion
In this study, where we used mRNA sequencing of PBMC, we identified 118 DEGs when comparing healthy control subjects with VKH patients, and 21 DEGs between patients before and after therapy with prednisone and cyclosporine. Bioinformatics analysis was performed to predict the biological role of DEGs, and two datasets of public available single-cell sequencing were used to infer the cell-specific origin of these DEGs. Our data suggest that peripheral immune cells may also participate in the pathogenesis of VKH as well as in the response to treatment.
When comparing DEGs from healthy controls with those obtained in VKH patients with active uveitis, several genes showed an evident expression change in VKH. SGK1 was upregulated in VKH, which is in agreement with earlier studies showing that SGK1 is also increased in PBMC from ulcerative colitis patients.37 SGK1, a serine/threonine kinase, is a sensor of multiple environmental signals.38 It has been reported that cytosolic Ca2+, reactive oxygen species, phosphatidylinositide-3-kinase, cyclic AMP, stress-activated protein kinase-5, stress-activated protein kinase-2, protein kinase C signaling could regulate the transcription of SGK1. Glucocorticoid receptor, nuclear factor kappa-B, reticuloendotheliosis viral oncogene homolog, activating transcription factor 6 have been shown to bind the promotor of SGK1.39 It is evident that the regulation of SGK1 is quite complex and which mechanisms are exactly operational in VKH is unclear and deserves further study.
The arachidonic acid metabolism pathway is the most obvious enriched KEGG pathway of the DEGs when comparing healthy control subjects and VKH patients with active uveitis before treatment. Arachidonic acid can be metabolized by enzymes to a spectrum of bioactive mediators that includes prostanoids, leukotrienes, epoxyeicosatrienoic acids, dihydroxyeicosatetraenoic acid, eicosatetraenoic acids, and lipoxins, which are strongly correlated with inflammation. Arachidonic acid not only plays an important role in cardiovascular biology and carcinogenesis, but also in autoimmune inflammatory diseases, such as asthma and arthritis.40 This enrichment indicates that activation of arachidonic acid metabolism pathway closely associates with the inflammation of VKH with active uveitis. Pathway analysis showed that MAPK, Ras and C-type lectin receptor signaling pathways were enriched when studying VKH patients longitudinally before and after treatment, suggesting that these pathways are involved in the process of disease remission. Suppressors of cytokine signaling 1 (SOCS1) was upregulated in treated VKH patients as compared to its expression before treatment. SOCS1 has been reported to exert an anti-inflammatory effect through the regulation of reactive oxygen species (ROS). Downregulated SOCS1 is associated with an induction of pro-inflammatory cytokines such as TNF-α, IL-1β and IL-6.41 We could not detect a difference when comparing SOCS1 in active uveitis stage VKH versus controls either by mRNA sequencing or RT-qPCR, which is not in agreement with earlier studies showing that SOCS1 was expressed at a 4.92-fold higher level in PBMC of VKH patient with active uveitis when compared with normal controls.24 Interestingly, the latter cohort of VKH patients with active uveitis had a history of corticosteroids use. Based on these results, it seems that upregulated SOCS1 emerges after the use of corticosteroids.
Another hitherto not yet described factor in VKH, included the enzyme tribble pseudokinase 1 (TRIB1). TRIB1 was upregulated in our VKH patients with active uveitis when compared to healthy controls and decreased again after treatment. It has been reported that TRIB1 plays a role in inflammatory signaling pathways.42 TRIB1 controls the polarization of macrophages,42 and the mRNA level of TRIB1 in PBMC is correlated with chronic antibody-mediated allograft failure.43 GO analysis showed that TRIB1 is associated with the response to molecule of bacterial origin, and enzyme inhibitory activity. Our data suggest that TRIB1 may act as an immune regulator in VKH and that it might serve as a potential biomarker to monitor disease activity and response to treatment.
Aberrant infiltrations of mononuclear immune cells in VKH lesions have been observed. Infiltration of T cells, B cells and macrophages in the choroid,44 3:1 ratio of CD4+ T cells and CD8+ T cells in vitiligo skin,45 abnormal frequency of lymphocytes in aqueous humor46 and cerebrospinal fluid47 have been reported in VKH. Based on these limited studies, it can be concluded that multiple types of mononuclear immune cell are involved in the pathological progress of VKH. However, the disease-specific cell type in VKH needs to be identified. Single-cell sequencing is a novel method for the disease researches and exploration of immune cell heterogeneity, which permits the detection of sequence at a single-cell resolution, it facilitates the identification of disease-associated cell type and the identification of categories in heterogeneous samples.48, 49, 50 To our knowledge, single-cell sequencing of PBMC has not yet been reported in VKH. In this study, the single-cell sequencing of PBMC from a healthy donor and single-cell sequencing in monocytes of VKH patients with corresponding normal control were used to further investigate the detail of DEGs identified by our bulk sample sequencing. We found that over half of our DEGs (VKH patients with active uveitis vs. healthy control subjects) were constitutively expressed in monocytes according to the single-cell sequencing of PBMC from a healthy donor. The expression of gene and the regulation of gene expression exhibit cell-type-specific properties,51,52 from which we hypothesized that monocytes significantly contribute to our identified DEGs. In addition, the most connected hubgene of our PPI is CSF1R, which is a known macrophage marker.53 However, we found no intersection between DEGs of monocytes and DEGs of PBMC. We speculate that this may be due to the following reasons. Firstly, there are differences in sequencing method, sequencing processes, the assembly of sequencing data and data processing between bulk sample sequencing and single cell sequencing, and therefore, comparing sequencing data obtained from different methods may produce biases. Secondly, the aberrant expression changes seen in PBMC were elicited by other non-monocyte clusters in VKH. Thirdly, we cannot exclude the possibility that there is a change in the frequency of monocytes in PBMC. Interestingly, CD14, a marker of monocyte,54 is upregulated in VKH patients with active uveitis before treatment when compared with healthy subjects. Fourthly, VKH is a rare disease, and the collection of sufficient samples from different untreated active patients is problematic. Our patient sample size is therefore relatively small. There were no females in the sequencing cohort, since most female VKH patients objected to donate blood samples for research purposes. However, men and women are equally affected by VKH in the Chinese Han population,55 and the same expression pattern of DEGs are observed in females by RT-qPCR as shown in Figure 2. Despite these limitations, we believe that elucidating which cell type contributes to the aberrant expression alterations detected in our study will help the discovery of disease-associated cell type.
Conclusions
This study is the first to our knowledge, which uses mRNA sequencing in PBMC from healthy control subjects and VKH patients with active uveitis before and after treatment. The differences in the mRNA expression profile among the tested groups provide evidence that peripheral immune cells are involved in the pathogenesis and treatment response of VKH. The data on mRNA expression of TRIB1 suggests that this gene may function as a biomarker in the management of VKH. Our data suggest that arachidonic acid metabolism may play an important role during the inflammatory events occurring during the development of VKH, and that treatment with prednisone combined with cyclosporine may target the MAPK signaling pathway. We speculate that monocyte, an ignored cell type in most previous VKH studies, plays an active role during VKH. We hope that our studies will help to clarify the underlying pathogenesis of VKH and lead to the development of novel drugs for the treatment of this rare disease.
Funding
This study was financially supported by National Natural Science Foundation Key Program, China (No. 81930023), Natural Science Foundation Major International (Regional) Joint Research Project, China (No. 81720108009), Chongqing Outstanding Scientists Project (2019), Chongqing Key Laboratory of Ophthalmology, China (No. CSTC, 2008CA5003), Chongqing Science & Technology Platform and Base Construction Program, China (No. cstc2014pt-sy10002) and the Chongqing Chief Medical Scientist Project, China (2018).
Conflict of interests
No conflict of interest declared.
Acknowledgements
We thank Zongren Xu and Qinfeng Ma for their help in this project.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Lavezzo M.M., Sakata V.M., Morita C., et al. Vogt-Koyanagi-Harada disease: review of a rare autoimmune disease targeting antigens of melanocytes. Orphanet J Rare Dis. 2016;11:29. doi: 10.1186/s13023-016-0412-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugita S., Sagawa K., Mochizuki M., Shichijo S., Itoh K. Melanocyte lysis by cytotoxic T lymphocytes recognizing the MART-1 melanoma antigen in HLA-A2 patients with Vogt-Koyanagi-Harada disease. Int Immunol. 1996;8(5):799–803. doi: 10.1093/intimm/8.5.799. [DOI] [PubMed] [Google Scholar]
- 3.Du L., Kijlstra A., Yang P., Vogt-Koyanagi-Harada disease: novel insights into pathophysiology, diagnosis and treatment. Prog Retin Eye Res. 2016;52:84–111. doi: 10.1016/j.preteyeres.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Read R.W., Rechodouni A., Butani N., et al. Complications and prognostic factors in Vogt-Koyanagi-Harada disease. Am J Ophthalmol. 2001;131(5):599–606. doi: 10.1016/s0002-9394(01)00937-0. [DOI] [PubMed] [Google Scholar]
- 5.Shi T., Lv W., Zhang L., Chen J., Chen H. Association of HLA-DR4/HLA-DRB1∗04 with Vogt-Koyanagi-Harada disease: a systematic review and meta-analysis. Sci Rep. 2014;4:6887. doi: 10.1038/srep06887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hou S., Du L., Lei B., et al. Genome-wide association analysis of Vogt-Koyanagi-Harada syndrome identifies two new susceptibility loci at 1p31.2 and 10q21.3. Nat Genet. 2014;46(9):1007–1011. doi: 10.1038/ng.3061. [DOI] [PubMed] [Google Scholar]
- 7.Kim M. Vogt-Koyanagi-Harada Syndrome following influenza vaccination. Indian J Ophthalmol. 2016;64(1):98. doi: 10.4103/0301-4738.178141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassili S.S., Peyman G.A., Gebhardt B.M., Daun M., Ganiban G.J., Rifai A. Detection of Epstein-Barr virus DNA by polymerase chain reaction in the vitreous from a patient with Vogt-Koyanagi-Harada syndrome. Retina. 1996;16(2):160–161. doi: 10.1097/00006982-199616020-00013. [DOI] [PubMed] [Google Scholar]
- 9.Yoshino N., Kawamura A., Ishii A., et al. Vogt-Koyanagi-Harada disease associated with influenza A virus infection. Intern Med. 2018;57(11):1661–1665. doi: 10.2169/internalmedicine.9819-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakata V.M., da Silva F.T., Hirata C.E., de Carvalho J.F., Yamamoto J.H. Diagnosis and classification of Vogt-Koyanagi-Harada disease. Autoimmun Rev. 2014;13(4–5):550–555. doi: 10.1016/j.autrev.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 11.Zheng H., Yang F., Ea V., et al. Proteomics profiling of plasma exosomes in VKH patients. Curr Mol Med. 2021;21(8):675–689. doi: 10.2174/1566524020666200719021653. [DOI] [PubMed] [Google Scholar]
- 12.Lai T.Y., Chan R.P., Chan C.K., Lam D.S. Effects of the duration of initial oral corticosteroid treatment on the recurrence of inflammation in Vogt-Koyanagi-Harada disease. Eye (Lond) 2009;23(3):543–548. doi: 10.1038/eye.2008.89. [DOI] [PubMed] [Google Scholar]
- 13.Cuchacovich M., Solanes F., Díaz G., et al. Comparison of the clinical efficacy of two different immunosuppressive regimens in patients with chronic vogt-koyanagi-harada disease. Ocul Immunol Inflamm. 2010;18(3):200–207. doi: 10.3109/09273941003587541. [DOI] [PubMed] [Google Scholar]
- 14.Liu D., Ahmet A., Ward L., et al. A practical guide to the monitoring and management of the complications of systemic corticosteroid therapy. Allergy Asthma Clin Immunol. 2013;9(1):30. doi: 10.1186/1710-1492-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naesens M., Kuypers D.R., Sarwal M. Calcineurin inhibitor nephrotoxicity. Clin J Am Soc Nephrol. 2009;4(2):481–508. doi: 10.2215/CJN.04800908. [DOI] [PubMed] [Google Scholar]
- 16.Chang R., Chen L., Su G., et al. Identification of Ribosomal Protein S4, Y-Linked 1 as a cyclosporin A plus corticosteroid resistance gene. J Autoimmun. 2020;112:102465. doi: 10.1016/j.jaut.2020.102465. [DOI] [PubMed] [Google Scholar]
- 17.Sumitomo S., Nagafuchi Y., Tsuchida Y., et al. Transcriptome analysis of peripheral blood from patients with rheumatoid arthritis: a systematic review. Inflamm Regen. 2018;38:21. doi: 10.1186/s41232-018-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith S.L., Plant D., Eyre S., Barton A. The potential use of expression profiling: implications for predicting treatment response in rheumatoid arthritis. Ann Rheum Dis. 2013;72(7):1118–1124. doi: 10.1136/annrheumdis-2012-202743. [DOI] [PubMed] [Google Scholar]
- 19.Pascual V., Chaussabel D., Banchereau J. A genomic approach to human autoimmune diseases. Annu Rev Immunol. 2010;28:535–571. doi: 10.1146/annurev-immunol-030409-101221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pham M.X., Teuteberg J.J., Kfoury A.G., et al. Gene-expression profiling for rejection surveillance after cardiac transplantation. N Engl J Med. 2010;362(20):1890–1900. doi: 10.1056/NEJMoa0912965. [DOI] [PubMed] [Google Scholar]
- 21.Mesko B., Poliska S., Szegedi A., et al. Peripheral blood gene expression patterns discriminate among chronic inflammatory diseases and healthy controls and identify novel targets. BMC Med Genomics. 2010;3:15. doi: 10.1186/1755-8794-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koczan D., Guthke R., Thiesen H.J., et al. Gene expression profiling of peripheral blood mononuclear leukocytes from psoriasis patients identifies new immune regulatory molecules. Eur J Dermatol. 2005;15(4):251–257. [PubMed] [Google Scholar]
- 23.Bomprezzi R., Ringnér M., Kim S., et al. Gene expression profile in multiple sclerosis patients and healthy controls: identifying pathways relevant to disease. Hum Mol Genet. 2003;12(17):2191–2199. doi: 10.1093/hmg/ddg221. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y.X., Wang H., Peng X.Y., Zhou Z.C. Expression of suppressor of cytokine signaling in peripheral blood monocular cells of patients with Vogt-Koyanagi-Harada disease. Zhonghua Yan Ke Za Zhi. 2009;45(11):1015–1019. [PubMed] [Google Scholar]
- 25.Chi W., Yang P., Li B., et al. IL-23 promotes CD4+ T cells to produce IL-17 in Vogt-Koyanagi-Harada disease. J Allergy Clin Immunol. 2007;119(5):1218–1224. doi: 10.1016/j.jaci.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 26.Deng B., Ye Z., Li L., et al. Higher expression of NOD1 and NOD2 is associated with Vogt-Koyanagi-Harada (VKH) syndrome but not Behcet's disease (BD) Curr Mol Med. 2016;16(4):424–435. doi: 10.2174/1566524016666160316153038. [DOI] [PubMed] [Google Scholar]
- 27.Read R.W., Holland G.N., Rao N.A., et al. Revised diagnostic criteria for Vogt-Koyanagi-Harada disease: report of an international committee on nomenclature. Am J Ophthalmol. 2001;131(5):647–652. doi: 10.1016/s0002-9394(01)00925-4. [DOI] [PubMed] [Google Scholar]
- 28.Yang P., Zhong Y., Du L., et al. Development and evaluation of diagnostic criteria for Vogt-Koyanagi-Harada disease. JAMA Ophthalmol. 2018;136(9):1025–1031. doi: 10.1001/jamaophthalmol.2018.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C., Wang L., Hu J., Li H., Kijlstra A., Yang P. Increased expression of IL-23 receptor (IL-23R) in Vogt-Koyanagi-Harada (VKH) disease. Curr Eye Res. 2018;43(11):1369–1373. doi: 10.1080/02713683.2018.1485952. [DOI] [PubMed] [Google Scholar]
- 30.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szklarczyk D., Franceschini A., Kuhn M., et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39(Database issue):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stuart T., Butler A., Hoffman P., et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du Y., Huang Q., Arisdakessian C., Garmire L.X. Evaluation of STAR and Kallisto on single cell RNA-Seq data alignment. G3 (Bethesda) 2020;10(5):1775–1783. doi: 10.1534/g3.120.401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Y., Hu Y., Xiao Y., et al. Genetic landscape and autoimmunity of monocytes in developing Vogt-Koyanagi-Harada disease. Proc Natl Acad Sci U S A. 2020;117(41):25712–25721. doi: 10.1073/pnas.2002476117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spagnuolo R., Dattilo V., D'Antona L., et al. Deregulation of SGK1 in ulcerative colitis: a paradoxical relationship between immune cells and colonic epithelial cells. Inflamm Bowel Dis. 2018;24(9):1967–1977. doi: 10.1093/ibd/izy158. [DOI] [PubMed] [Google Scholar]
- 38.Yang Y.H., Istomine R., Alvarez F., et al. Salt sensing by serum/glucocorticoid-regulated kinase 1 promotes Th17-like inflammatory adaptation of Foxp3(+) regulatory T cells. Cell Rep. 2020;30(5):1515–1529. doi: 10.1016/j.celrep.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lang F., Rajaxavier J., Singh Y., Brucker S.Y., Salker M.S. The enigmatic role of serum & glucocorticoid inducible kinase 1 in the endometrium. Front Cell Dev Biol. 2020;8:556543. doi: 10.3389/fcell.2020.556543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang B., Wu L., Chen J., et al. Metabolism pathways of arachidonic acids: mechanisms and potential therapeutic targets. Signal Transduct Target Ther. 2021;6(1):94. doi: 10.1038/s41392-020-00443-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim G.Y., Jeong H., Yoon H.Y., et al. Anti-inflammatory mechanisms of suppressors of cytokine signaling target ROS via NRF-2/thioredoxin induction and inflammasome activation in macrophages. BMB Rep. 2020;53(12):640–645. doi: 10.5483/BMBRep.2020.53.12.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnston J., Basatvat S., Ilyas Z., Francis S., Kiss-Toth E. Tribbles in inflammation. Biochem Soc Trans. 2015;43(5):1069–1074. doi: 10.1042/BST20150095. [DOI] [PubMed] [Google Scholar]
- 43.Ashton-Chess J., Giral M., Mengel M., et al. Tribbles-1 as a novel biomarker of chronic antibody-mediated rejection. J Am Soc Nephrol. 2008;19(6):1116–1127. doi: 10.1681/ASN.2007101056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakamoto T., Murata T., Inomata H. Class II major histocompatibility complex on melanocytes of Vogt-Koyanagi-Harada disease. Arch Ophthalmol. 1991;109(9):1270–1274. doi: 10.1001/archopht.1991.01080090096030. [DOI] [PubMed] [Google Scholar]
- 45.Okada T., Sakamoto T., Ishibashi T., Inomata H. Vitiligo in Vogt-Koyanagi-Harada disease: immunohistological analysis of inflammatory site. Graefes Arch Clin Exp Ophthalmol. 1996;234(6):359–363. doi: 10.1007/BF00190711. [DOI] [PubMed] [Google Scholar]
- 46.Ohta K., Yoshimura N. Expression of Fas antigen on helper T lymphocytes in Vogt-Koyanagi-Harada disease. Graefes Arch Clin Exp Ophthalmol. 1998;236(6):434–439. doi: 10.1007/s004170050102. [DOI] [PubMed] [Google Scholar]
- 47.Norose K., Yano A., Aosai F., Segawa K. Immunologic analysis of cerebrospinal fluid lymphocytes in Vogt-Koyanagi-Harada disease. Invest Ophthalmol Vis Sci. 1990;31(7):1210–1216. [PubMed] [Google Scholar]
- 48.Potter S.S. Single-cell RNA sequencing for the study of development, physiology and disease. Nat Rev Nephrol. 2018;14(8):479–492. doi: 10.1038/s41581-018-0021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papalexi E., Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18(1):35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 50.Stubbington M.J.T., Rozenblatt-Rosen O., Regev A., Teichmann S.A. Single-cell transcriptomics to explore the immune system in health and disease. Science. 2017;358(6359):58–63. doi: 10.1126/science.aan6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chadwick L.H. The NIH roadmap epigenomics Program data resource. Epigenomics. 2012;4(3):317–324. doi: 10.2217/epi.12.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grubert F., Srivas R., Spacek D.V., et al. Landscape of cohesin-mediated chromatin loops in the human genome. Nature. 2020;583(7818):737–743. doi: 10.1038/s41586-020-2151-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kato S., Okamura R., Kumaki Y., et al. Expression of TIM3/VISTA checkpoints and the CD68 macrophage-associated marker correlates with anti-PD1/PDL1 resistance: implications of immunogram heterogeneity. Oncoimmunology. 2020;9(1):1708065. doi: 10.1080/2162402X.2019.1708065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ong S.M., Teng K., Newell E., et al. A novel, five-marker alternative to CD16-CD14 gating to identify the three human monocyte subsets. Front Immunol. 2019;10:1761. doi: 10.3389/fimmu.2019.01761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang P., Ren Y., Li B., Fang W., Meng Q., Kijlstra A. Clinical characteristics of Vogt-Koyanagi-Harada syndrome in Chinese patients. Ophthalmology. 2007;114(3):606–614. doi: 10.1016/j.ophtha.2006.07.040. [DOI] [PubMed] [Google Scholar]