Abstract
In the past, contradictory statements have been made about the age of cancer genes. While phylostratigraphic studies suggest that cancer genes emerged during the transitional period from unicellularians (UC) to early metazoans (EM), life cycle studies suggest that they arose earlier. This controversy could not be resolved. Phylostratigraphic methods use data from somatic tumor gene collections containing or lacking polyploidy genes (PGCC genes) and compare them to genes from evolutionary node taxa. I analyze whether the selected taxa are suitable to resolve the above contradiction or not. Both cancer and amoebae life cycles have a reproductive asexual germline that produces germline stem cells (GSCs) and somatic cell lines that cannot. When the germline loses its reproductive function, the soma-to-germ transition forms a new reproductive germline. The reproductive polyploidy of cancer is homologous to the reproductive polyploidy of unicellular cysts. PGCCs repair DNA defects, reorganize the involved genome architecture and produce new GSCs. The present study refutes the dogma of the early metazoan origin of cancer. Cancer has a unicellular life cycle that was adopted by early metazoans to rescue themselves from evolutionary dead ends. Early metazoans controlled the unicellular life cycle through suppressor and anti-suppressor genes that could suspend or reactivate it. They are the archetypes of tumor suppressor genes and oncogenes. Cells of mammalians and humans that reach a similar impasse as early metazoans can reactivate the conserved life cycle of unicellularians.
Keywords: Cancer, Entamoeba, Gene age, Life cycle, Polyploidy, CSCs, EMT, PGCC
Abbreviations
- UC
unicellularians
- EM
early metazoans
- MC
multicellularians
- MM
mammalians
- MGRS
multinucleate genome repair structures (cancer); multinucleate genome repair syncytia (Entamoeba)
- PGCC
polyploid giant cancer cell
- aCLS
autonomous cyst-like structure
- GSCs
germline stem cells
- G + S
germ and soma
- GST
germ to soma transition
- SGT
soma to germ transition
- EMT
epithelial–mesenchymal transition
- MET
mesenchymal–epithelial transition
- GRN
gene regulatory network
- DDR
DNA damaged repair
- SSB
single-strand break
- DSB
double-strand break
- EAS
early surface antigens
Introduction
“Deep homology” is a recent concept in developmental biology and medicine and has its roots in evolution. Robert Kreisman noted in 2010 that all species are thought to have had the same ancestor hundreds of millions of years ago and that we all still share common, deep homologous genes.1 The same gene clusters (gene modules) that worked together millions of years ago still work together today. However, such gene modules are much easier to identify in less complex organisms than in human diseases or cancer. McGary et al studied more than 200,000 gene–phenotype associations in humans and lower animals and found matches in several species.2 Accordingly, scientists use a yeast model to study the angiogenesis of cancer and a worm model for breast cancer. All of these orthologous phenotypes reveal evolutionarily conserved gene networks. However, evolutionary models of human disease and cancer in their entirety are rare.
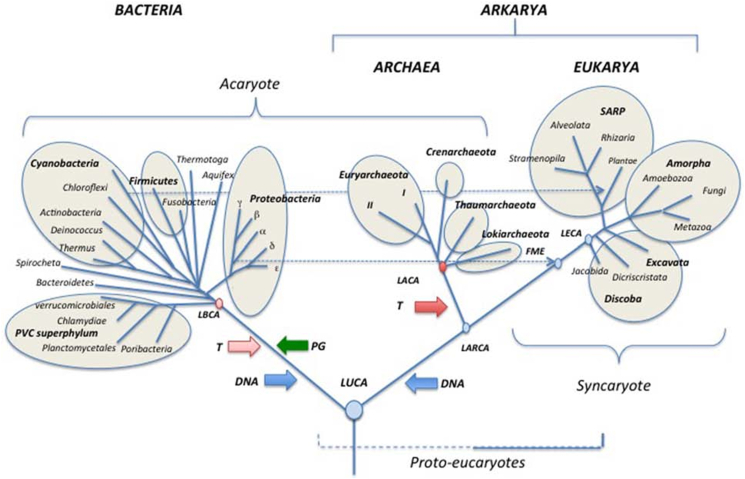
Over the last decade, I have studied the close similarity between the life cycle of Entamoeba and that of cancer. I found that both life cycles have many similarities and have summarized these in the “amoeba model” of cancer.3,4 Unfortunately, little is known about the ancestral gene module adopted by amoebozoans and metazoan cancers. However, there is no doubt that both cell systems share the same genetic machinery that goes back to the common unicellular ancestor. Not surprisingly, the life cycle of both systems consists of a reproductive germline and a somatic cell line (G + S life cycle). This is due to the fact that Amoebozoa (including Entamoeba) and Metazoa (including cancer) are the last branched sister clades of the phylogenetic tree of life. Both belong to the group Amorpha.5 (Fig. 1).
Figure 1.
Schematic universal tree updated from Forterre (2015) after Woese et al (1990). Amorpha is the last assemblage descended from the common ancestor LECA (Eukaria domain). It contains the three sister clades Amoebozoa, Metazoa, and Fungi. https://doi.org/10.3389/fmicb.2015.00717; CC BY 2015.
I disagree with the common dogma of the origin of cancer by early metazoans and argue that cancer is an expression of a pre-existing life cycle that was adopted by unicellular organisms, but also by early metazoans from their common ancestor to escape from evolutionary dead ends. I assume that early metazoans already controlled the adopted unicellular life cycle by suppressor and anti-suppressor genes and that these are the archetypes of tumor suppressors and oncogenes.
After branching, the unicellular G + S life cycles of Amoebozoa and early Metazoa cycles evolved separately. They optimized their genetic machinery in response to the environment. Entamoeba and metazoan cancers6 evolved to the parasitic lifestyle that optimally exploited host resources and niches. However, both systems retained the basic structure of the ancestral cell system and passed it on to subsequent generations without fundamental changing. This explains the deep evolutionary homology in terms of germline reproductive polyploidy, stemness, stem cell phenotypes, and stem cell repair, and suggests a more coherent evolutionary designation for participants in the life cycle of cancer.
Using phylostratigraphic methods and analyzing data from somatic tumor gene collections that contain or do not contain polyploidy genes from PGCCs and taxa with evolutionary nodes, the present work shows that cancer polyploidy is homologous to the deep polyploidy of the unicellular ancestors that evolved stemness and the first archaeal stem cells. The mechanisms of stem cell formation by reproductive polyploidization were adopted by protists such as Entamoeba as well as metazoan cancers.
Little is known about the evolutionary origin of cancer stem cell (CSC) and their archetypes, so much of this work is concerned with the evolutionary origin of CSCs. I argue that one of the crucial events in the evolution of pre-metazoans was the development of the inner cyst cell into a stem cell progenitor phenotype and founder stemness genes. The evolution of CSC archetypes traces back to the common ancestor that transformed the simple protective cyst of the ancestral flagellate into a reproductive cyst capable of stemness. Its haploid daughter cells transferred stem cell potential into the amoeboid stem cells that differentiate the germline. The germline of the common ancestor evolved to carry stemness.
This review begins with a brief introduction to the evolutionary “deep homology” and considers cancer evolution from the perspective of deep homology. It explains the cancer life cycle using the “amoeba model” and shows that CSCs are products of their unicellular life cycle of cancer. It concludes that the origin of the ancestral inherited life cycle is found in the polyploid reproductive cysts of single-celled organisms.
It can be assumed that cancer gene archetypes evolved in the pre-metazoan era in the form of a Gaussian bell diagram. Many G + S founder genes are cancer gene archetypes that arose early in evolution when simple protective cysts evolved into reproductive cysts that became polyploid. Last but not least, this work addresses the question of why metazoans did not completely abandon the unicellular G + S cycle. I suggest that unstable early metazoans that carried the G + S cycle for safety reasons suppressed it only to the extent that reactivation remained possible when evolutionary dead ends arose. This was beneficial for the whole-cell community as well as for individual cells. In the early era of metazoans (EM), new EM suppressor genes were integrated into the EM regulatory network, but also UC genes that were repurposed as anti-suppressor EM genes. This regulatory hub gene module allowed metazoans to control the G + S cycle and retain it as useful.
Deep homology, the ancestral gene module and cancer evolution
According to Shubin et al, “deep homology” means an ancient similarity of patterning mechanisms and a historical continuity through which regulatory circuits inherited from a common ancestor have been passed on in evolution. This means that new functions and phenotypes have not emerged from scratch; they have evolved by exploiting regulatory circuits that were already established in early eukaryotes.7 As Wagner added, individual structures that do not appear to be homologous at first consideration may have evolved from the genetic machinery of a common ancestor, using it as a pattern for homologous evolutionary programs.8 In recent years, deep homology approaches have evolved more and more towards a better understanding of the molecular and genetic mechanisms and function, moving away from the simple comparative morphology.
Tschopp and Tabin understand deep homology as the conservation of gene expression during evolution and speak from historical continuity, historical homology, and organizational hierarchy.9 They consider the homologies between the individual cell types of a given organ as the core of the modern concept of homology and assume that phenotypes result from complex gene regulatory networks (GRN) and gene modules rather than from single individual genes.10,11 According to the “kernel concept” of Davidson and Erwin regarding the hierarchy in the regulation of genes, kernels are sub-units of GRN that exhibit deep evolutionary preservation and are refractory to regulatory rewiring and underlie stability while at the base of the GRN hierarchy are so-called gene differentiation batteries that direct terminal cell differentiation.12
The evolution of clade genomes began with the evolutionary branching 750–800 million years ago13 and continues until today. Over time, ancestral genes have taken on additional or entirely new functions, adapted to analogous microenvironmental pressures in the tissue they parasitize, and passed these changes on to subsequent generations of cells, according to the Darwinian model of evolution. Recent work by Vendramin et al shows that the Darwinian model alone is insufficient to fully explain cancer evolution, as “single catastrophic events such as whole-genome duplication (WGD) position cancer evolution beyond Darwin”.14 It confirms Shubin's assumption that innovations have a history that goes far back deep in time.15 However, I think, WGD is not a catastrophic event, rather, it is a reproductive process inherited from the common ancestor.3,4
Cancer evolution through germline clones and subclones is certainly beyond Darwin. It is largely related to deregulated processes of soma-to-germ transition (SGT, EMT) caused by various environmental stress factors. Environmental stress leads to altered and incomplete second CSCs generations and multiclonal germline stem cells (GSCs).4 Cancer evolution is in contrast to the previously assumed linear sequential evolution of clonal cell division. Gradual changes in CSC progenitors provide cancer with a strong selective advantage, allowing earlier CSCs lineages to be displaced.14, 15, 16, 17
The life cycle of cancer explained by the “amoeba model”
The great similarity between the unicellular life cycles of Entamoeba and cancer contradicts the molecular phylostratigraphic theory for the early metazoan origin of cancer.18, 19, 20, 21 The deep relationship between the two cell systems, resulting from the temporally close branching of Amoebozoa and Metazoa (Amorpha group), is why the “amoeba model” is a uniquely valuable support for understanding the biology of cancer from the evolutionary perspective.4,22
The G + S life cycle of Entamoeba is closer to the common ancestor and more original than any other life cycle of unicellularians. Both cell systems – amoeba and cancer – use the deep homologous G + S gene module evolved by the common ancestors. Both systems have striking homologies: (i) A reproductive asexual germline capable of forming germline stem cells (GSCs, referred to as CSCs in cancer) and a somatic cell line without reproductive GSC function; (ii) Germ and soma cells that proliferate through asymmetric and symmetric cell cycles and can interconvert by transitioning from germ to soma (GST) and from soma to germ (SGT); both processes are referred to as MET and EMT in cancer; (iii) Oxygen-sensitive germlines that irreversibly lose their reproductive function due to irreparable DNA damage caused by excess oxygen; (iv) DNA damage repair (DDR) mechanisms to repair DNA replication and polyploidization defects and maintain genomic integrity of nascent GSCs/CSCs; (v) DNA DSB repair mechanisms via MGRS and PGCC structures, with or without homologous cell fusion4; It is also conceivable that the release of excess DNA material, as occurs in Entamoeba by MGRS or non-reproductive polyploidization/depolyploidization processes, results in circulating cancer DNA fragments.
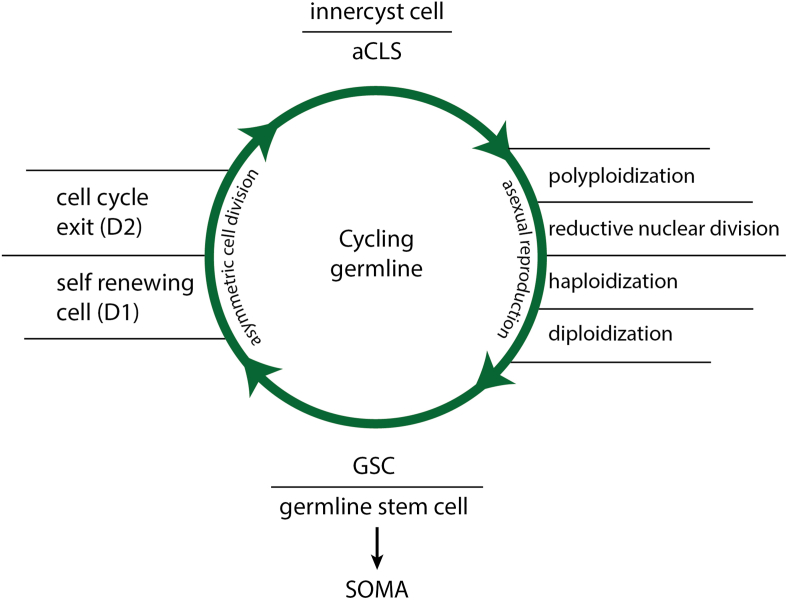
In Entamoeba, asexual reproductive cycles begin with the polyploidization of the inner cyst cell, which gives rise to eight haploid daughter cells that develop into diploid stem cells (Fig. 2). I refer to these “nascent” stem cells produced by the germline as GSCs. These GSCs differentiate into the reproductive germline that proliferates by asymmetric cell division. This type of cell division gives rise to non-identical daughter cells: one of them is the self-renewing SR daughter cell (D1 cell) and the other the non-proliferating sister cell (D2 cell) committed for reproductive polyploidization and GSC formation. Each D1/SR cell continues the normal cell cycle while D2 cells leave it to transform into a reproductive cyst. The inner cyst cell changes its structure and undergoes several whole cell cycle rounds (WGDs) without cytokinesis and cell division.4 The closed asexual circuit of Entamoeba's germline includes cysts, GSCs, self-renewing germline cells (D1), and cyst precursor cells (D2). In cancer, the term GSCs used for nascent CSCs has the advantage of clearly indicating the origin of primary CSCs.
Figure 2.
Amoeba model: the deep homologous germline of cancer (aCLS cycle). Under normoxic conditions, the cancer germline performs reproductive polyploid cycles to GSCs, which are the nascent cancer stem cells (CSCs).
Cancer stem cells are products of the G + S life cycle
Cancer stem cells are products of the G + S life cycle and are not derived from adult HSCs or de-differentiated cells as previously thought. They are products of the unicellular G + S life cycle adopted from early metazoans. Cancer is ubiquitous in metazoans,6 including low metazoans such as Holozoa and other lower animals. For 700–800 million years, metazoan cancers have had reproductive germlines that performed polyploid aCLS cycles via encystment (reproductive germline cycles) and produced GSCs. Nascent CSCs are GSCs of unicellular origin and, like Entamoeba GSCs, are defined by the asymmetric cell cycle, asymmetric cell division, and differentiation ability. During the evolution of cancer, the germline was involved in the formation of different classes of CSCs which are: (i) the nascent GSCs (primary CSCs), (ii) the SGT/EMT-induced CSCs (secondary CSCs), and (iii) the MGRS/PGCC-induced CSCs (tertiary, repair CSCs).4 They correspond to the GSC classes and mechanisms of GSC production observed in Entamoeba.
In cancer, the reproductive germline cycle begins with the committed D2 cell (precursor cell) that polyploidizes within a cell envelope thinner than the cyst wall. The cancer germline undergoes development similar to the Entamoeba's germline.4 There is a deep homology to the mammalian GSCs. The amoeba model substantiates the statement of Nayernia, who hypothesizes that mammalian (somatic) stem cells are the direct descendants of the germline.23,24 According to this hypothesis, the germline is the “mother line” of all somatic stem cell lineages in the adult body and the daughter GSCs are the only stem cells capable of passing genetic information from generation to generation.23,25
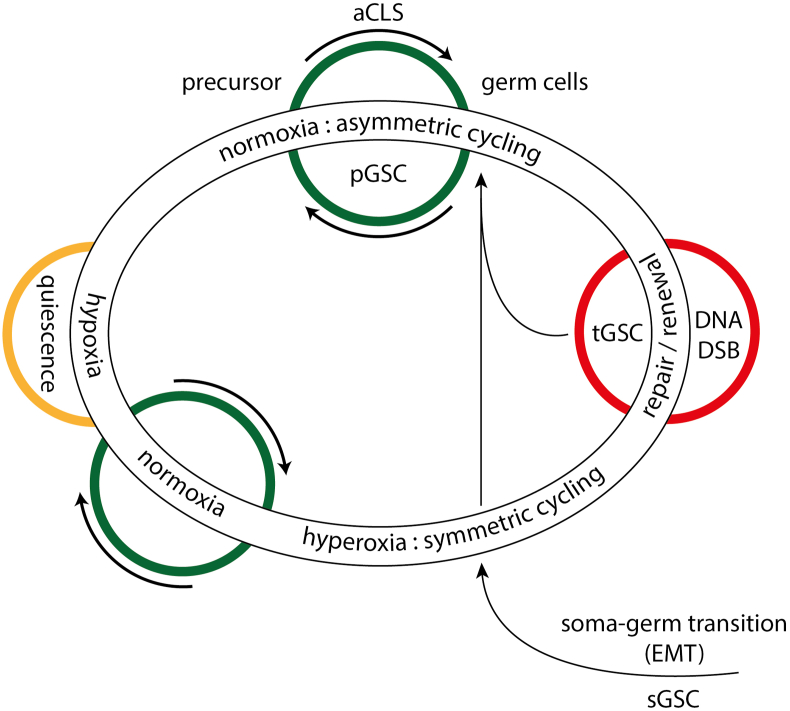
The germline of amoebae and cancer is oxygen-sensitive. Under conditions of excess-oxygen it loses its reproductive function and cannot produce nascent CSCs/GSCs (Fig. 3). As a result, soma cells are forced to form new reproductive germline clones that begin with the production of secondary sCSC/sGSC.4 When soma-to-germ transition (SGT/EMT) cannot be initiated, defective germline cells repair DNA defects and reorganize the genome architecture of the germline through multinucleate genome repair structures (MGRSs) or polyploid giant cancer cells (PGCCs) with or without cell fusion.
Figure 3.
Cancer germline evolution. The germline proliferates under both normoxic and hyperoxic living conditions. Under normoxic conditions and asymmetric cell division, the germline undergoes repetitive aCLS cycles by polyploidization (see Fig. 1). aCLS polyploidization is similar to the polyploidization that occurs in inner cyst cells of Entamoeba, which gives rise to multiple daughter cells and forms GSCs. CSCs are the homologous GSCs of cancer. The nascent GSCs/CSCs differentiate new germlines, clones, and somatic populations. Hyperoxia causes DNA DSB defects: The germline loses its reproductive function and stemness and ceases polyploidization and GSC/CSC production. The loss of function triggers soma-to-germ transition (SGT/EMT) and leads to new germlines or clones, and new sCSCs. Damaged germline cells can repair DNA defects and reorganize the genome integrity through multinucleated germline repair structures (MGRSs) or autonomous PGCCs not induced by therapeutics or radiation. pGSC, sGSC, tGSC: primary, secondary and tertiary CSCs. From: Niculescu.4
In the amoeba model, MGRS/PGCC polyploidization does not activate ancient developmental programs, as previously claimed.26,27 This occurs during the transition from the weakened multicellular life cycle of metazoans to the unicellular G + S life cycle of cancer. During cancer development, polyploidization occurs not only in MGRS/PGCC structures induced by chemotherapeutic agents or irradiation, but also in the early stages of cancer, when excess oxygen damages the germline. The damaged germline cells have lost their reproductive function and cannot form new CSCs. To repair DNA defects and remodel genome architecture, oxygen-damaged germline cells fused and formed autonomous MGRS/PGCC syncytia that repair DNA DSB damage and produce new effective CSCs capable to differentiating new reproductive germline clones. I disagree with the notion that polyploidy in cancer is an important driver of epigenetic change and that “atavistic” changes go hand in hand with cancer polyploidy.26 Rather, the “amoeba model” confirms the assumption that cancer “defects” (such as reproductive polyploidization) are mostly adaptive tools of survival programs that evolve during the evolution of life and are inherent to cancer.27
The deep homologous origin of the G + S life cycle of cancer lies in the reproductive cyst of unicellularians
Crucial to the evolution of unicellularians (UCs) was the transition from simple protective cysts to reproductive cysts capable of polyploidy. Cyclic polyploidy is the result of the evolution to asymmetric cell division.4 According to Schussnig, the archetype of the last eukaryotic common ancestor LECA was a mononuclear and motile “monad cell” that could form cysts as a defense against harmful living conditions.28 Within this protective envelope, the flagellar cell reduced its morphology to an amoeboid cell shape, which left the cyst wall after a resting phase. This inner cyst transformation occurred with successive regression of the flagellar phenotype. While early eukaryotic cysts were non-reproductive structures, later eukaryotic cysts did not cease replication and performed multiple rounds of WGDs without cytokinesis and cell division. Such polyploid cysts gave rise to more than two daughter cells. Polyploidization, initially random and facultative, became systemic due to better progeny survival and an evolutionary advantage.
The transition from protective to reproductive cysts had great evolutionary potential. In addition to phenotype transition, the reproductive function was transferred from the flagellar to the amoeboid cell type and its daughter cells. These germ cells were generated by polyploid cycles within the reproductive cyst, and the flagellar cells express only the somatic genome. The best evidence for this is provided by Proleptomonas, a free-living, mononuclear flagellate with a reproductive cyst 6–9 μm in diameter that belongs to the Cercozoa.29 Unlike other flagellates that divide vegetatively by symmetrical cell division. Proleptomonas reproduce only within the reproductive cyst and, like Entamoeba, produces eight daughter germ cells that form the germline. Early amoeboflagellate ancestors have laid the groundwork for germ and soma life cycles. The flagellate cell transferred its totipotency to the innercyst cell and its amoeboid daughter cells, which gradually took on stemness and differentiation capabilities, in contrast to the flagellate phenotype, which expressed only soma traits. Both phenotypes retained the capacity for inter-conversion and could transit from germ-to-soma (GST) and from soma-to-germ cells (SGT). The amoeboflagellate ancestor transferred the deep homologous G + S gene module to the branching taxa (Fig. 1) that adopted either the entire gene module, or only parts of it in active or inactive forms, or discarded them altogether. With rhizopodial transformation, pre-metazoans expanded the possibilities for colonial life,29 cell fusion, and lateral gene transfer (LGT). LGTs enable the asexual exchange of genetic information between different genomes.30
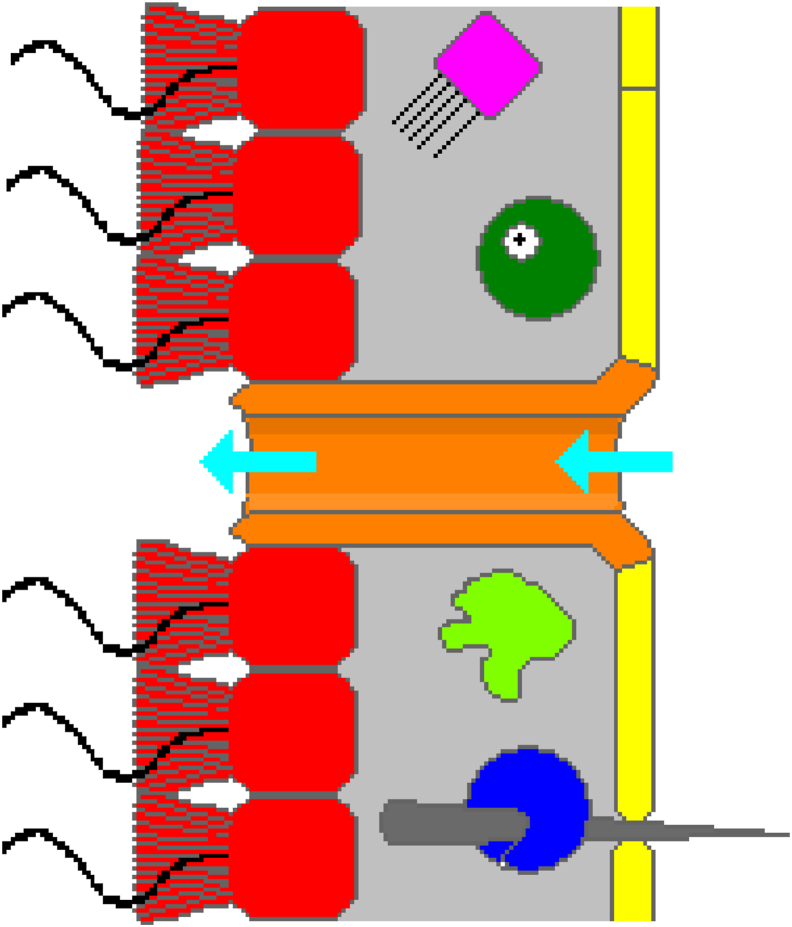
Accordingly, it can be assumed that the common ancestor of Entamoeba, Metazoa and cancer was a highly evolved amoeboflagellate with a relevant deep G + S module including genes for stemness and cell differentiation, polyploidy as well as GST and SGT processes. Sponges (Porifera), which belong to the oldest metazoans, support this hypothesis. Stem cells and cell differentiation in modern sponges arise from totipotent amebocytes (archeocytes) (Fig. 4) and not from flagellated choanocytes, which have only soma function and differentiate themselves from amebocytes. Nevertheless, SGT processes are also known in sponges, where soma choanocytes transform into totipotent amoebocytes.31
Figure 4.
Sponges architecture: choanocytes (red) and amoebocytes/archeocytes (green). From: Ruppert EE, Fox RS, Barnes RD. Invertebrate Zoology, ed. Brooks/Cole. p. 82. ISBN 978-0-03-025982-1 (https://en.wikipedia.org/wiki/Sponge).
Phylostratigraphic errors: choanoflagellates are not the taxa closest to metazoans
Conventional wisdom holds that choanoflagellates are the unicellularians that would be closest to metazoans. But this seems to be less and less true. Choanoflagellates and Amoebozoa are two groups to Metazoa. As revealed by Carr et al, little is known about choanoflagellate evolution.32 Unlike Amoebozoans, Choanoflagellates show a low propensity to form cysts and dual life cycles and there is no evidence that choanoflagellates have an exclusive relationship with Metazoa.33 Molecular data reject suggestions that Metazoa are descended from a true choanoflagellate ancestor. Much more is conceivable that both groups are descended from a common protistan ancestor. From this ancestor, metazoans evolved into truly multicellular organisms, whereas the choanoflagellates have maintained a predominantly solitary existence.
Recent molecular studies showed that the transcriptome of choanocytes bears little resemblance to the transcriptome of choanoflagellates.31 This finding clearly refutes the widely held view that metazoans evolved from a unicellular ancestor surrounded by an apical cilium, and a microvillar collar structurally similar to modern choanoflagellates.34, 35, 36 Brunet and King hypothesized that the first metazoans evolved from a colonial state of unicellular organisms with a long history of “simple multicellularity” that captured nutrients and bacteria with a flagellum surrounded by a microvillar collar and had a different genome than the genome of the colonial assemblage.35 The genome differences between choanoflagellates and simple metazoans such as sponges and their choanocytes are the result of the distinct genomes that arose in the colonial assemblage.
In contrast, amoebozoans show more evolutionary homology with Metazoa than with choanoflagellates. According to Kang et al, the last common ancestor of all amoebozoan species could spread propagules from a sporocarp-like fruiting body,37 a feature adopted by reproductive cysts and MGRSs and PGCCs of amoebae and cancer.4 Amoebozoa have well-developed life cycles and germ cells that form reproductive cysts, in contrast to the trophic somatic cells.38,39 Amoebozoans live in environments with varying oxygen levels and are best adapted to lower and higher O2 pressures. Most amoebozoans alternate between the trophozoite stage and one or more different stages.39 Cell fusion, followed by nuclear division, results in a large syncytial cell that forms spore-bearing structures.40 Many of these abilities are common to Entamoeba and cancer and are not observed in choanoflagellates.3,4
Conclusively, the genome of choanoflagellates is not the best reference for comparative studies between premetazoan ancestors, early metazoans, and cancer. Unlike Entamoebae, the life cycle of choanoflagellates is rudimentary and evolution into a reproductive germline is doubtful. Some choanoflagellates can form cysts when transferred to fresh media, but not much is known about them or whether encystation has a reproductive function. It is known that environmental changes, including the presence of oxygen-consuming bacteria, lead to oxygen reduction and produce the swarming life form, the role of which is not fully understood. By and large, Amoebozoa are evolutionarily and physiologically closer to the early metazoans than to vestigial choanoflagellates. Entamoeba for example has stem cells very similar to the stem cells of sponges. The totipotent stem cells of sponges that give rise to choanocytes are amoeboid cells called amebocytes or archaeocytes (Fig. 4). Choanoflagellates are only rudimentarily developed and are not suitable as phylostratigraphic reference taxa.
The dogma of the early metazoan origin of cancer
In 2007, Domazet-Lošo and Tauz introduced phylostratigraphic analysis to study human genome history and the evolutionary origin of genes related to human genome diseases, including cancer.18,19 Central to phylostratigraphic research is the assumption that new protein families that emerge at evolutionary nodes – such as the transition to multicellularity – are the major innovations for life initiated by founder genes. The aim of the phylostratigraphic analyses was to find out whether there is a link between the founder genes of multicellularity and cancer genes.
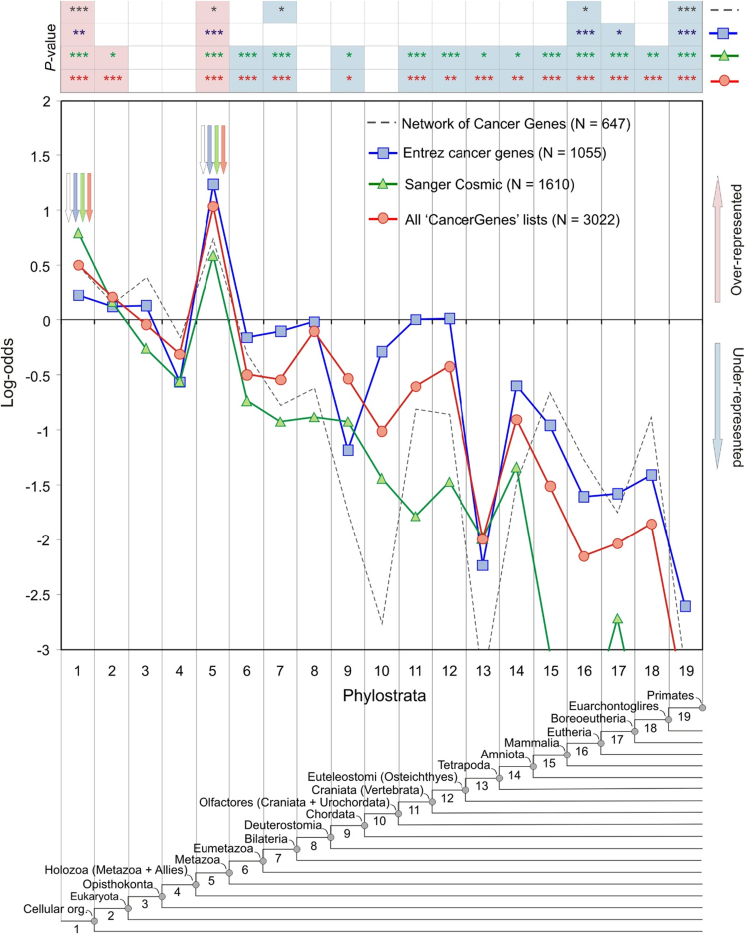
The phylostratigraphic map of human genes of Domazet-Lošo and Tauz has 19 phylostrata (ps)19 (Fig. 5). The first three layers harbor archaea and bacteria (ps1), lower eukaryotes (ps2), and fungi (ps3), while on the fourth phylostratum researchers placed the less adequate choanoflagellates, a decision that led to misinterpretation. Moreover, the analyzed gene material was derived from somatic cell material of tumors and metastases and was accordingly poor in CSC gene material, which could have provided more information on the age of the cancer's germline genome. Tumors and metastases are poorly populated with CSCs. The number of functional CSCs can decrease progressively and rapidly in tumors.41 After about 100 cell doublings and a spontaneous extinction probability of 95%, the metastatic effiiciency decreases to about 0.05% (loss of function).
Figure 5.
Statistical analysis of the cancer datasets using the 19-phylostrata map. From: Domazet-Lošo and Tautz18,19 (http://www.biomedcentral.com/1741-7007/8/66). CC BY 3.0.
Based on this map, the researchers found a significant number of cancer protein domains and cancer-like genes mainly at the level of ps1 (Archaea and Eukarya), followed by a decrease on ps2 and ps3 (primitive Eukaryota and Fungi) and a complete missing in choanoflagellates (ps4). A second significant peak occurs in the early metazoans of ps5 (Porifera) (Fig. 5). The occurrence of cancer-like genes was significantly reduced at the ps6- and ps7-levels (Eumetazoa and Bilateria) and at all levels beyond the emergence of vertebrates (ps11 and up).18,19
The thinking at the time was influenced by Weinberg, who assumed that cancer is a consequence of multicellular evolution.42 However, the peak observed in ps1 predates the emergence of multicellularity. Domazet-Lošo and Tauz believe that the deep peak indicates primitive protein domains associated with so-called cancer “caretaker” genes that arose during the formation of the first cells to ensure the stability of the genome. In contrast, the overexpressed ps5-peak would be due to the so-called “gatekeeper” genes, and contains oncogenes- and tumor suppressor-like genes.18,19 In the ontogenetic sequence of tumor progression, mutations in caretakers precede mutations in gatekeepers.
In more recent phylostratigraphic studies, Trigos et al reduced the number of phylostrata to 16 and grouped all human genes into three super-classes: 6719 unicellular ones (UC) originating from the phylostrata ps1–ps3, 7939 arising from the early metazoan (EM) of the phylostrata ps4–ps9, and 2660 genes arising from the mammalian phylostrata ps10–ps16 (MM).20,21 Accordingly, ancient genes related to cancer (ps1–ps3) are termed UC genes; they include genes of Opisthokonta (flagellated protozoa) and choanoflagellates, but not amoebozoans. The authors suggest that UC genes support basic cellular functions such as cell cycle, mitosis, proliferation and metabolic processes. In contrast, genes related to cancer (ps4–ps9), are referred to as EM genes; they represent genes with more complex cellular functions that arose with the transition to multicellularity.
Phylostratigraphic polyploidy studies reject the dogma of the EM origin of cancer
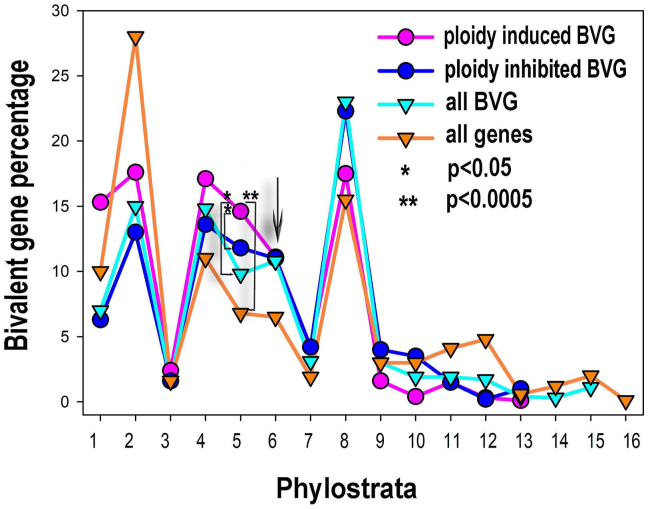
In an attempt to determine the evolutionary age of cancer polyploidy genes, the research group around Erenpreisa and Anatskaya analyzed the occurrence of polyploidy genes in the UC, EM, and MM phylostrata.26, 27 For this study, the researchers used transcriptomic data from tumors and tissue containing polyploid PGCCs. These PGCCs are multinucleate genome repair structures (MGRSs) capable of repairing defective germline cells and reorganizing the germline genome architecture.4 PGCCs generate new invasive CSCs capable of metastases.
The results in Figure 6 show that genes related to cancer polyploidy arose quite early in the evolutionary history. Both ploidy-related and bivalent ploidy-regulated genes (BVGs) belong to ps2 and occur at a time when primitive eukaryotes have switched from simple protective cysts to reproductive polyploid cysts. In contrast to the previous results of Domazet-Lošo and Tauz, the distribution of BVG genes show the expected Gaussian bell curve, ranging from ps1 to ps7.18,19 It is unfortunately interrupted by the misselected choanoflagellates and opistochonts on ps3. The curve flattens with the appearance of metazoans and finds his lowest point in the phylostrata representing Bilateralia and Chordata (ps6/ps7).
Figure 6.
The percentual proportions of gene origins and distribution of bivalent genes (BVG) in the phylostratic tree of life (strata 1–16) and the effect of polyploidy on it and authors statement. The upregulation of bi-valent genes by polyploidy includes strata 1, 2 (unicellularians), stratum 4 (metazoa) and, prominently, stratum 5 (eumetazoa—the appearance of embryo, germ layer, and gastrulation). The phylostrata are as follows: 1—cellular organisms, 2—Eukaryota, 3—Opisthokonta, 4—Metazoa, 5—Eumetazoa, 6—Bilateria, 7—Chordata,8—Euteleostomi, 9—Amniota, 10—Mammalia, 11—Ttheria, 12—Eutheria, 13—Euarchontoglires, 14—Catarrhini, 15—Homininae. From: Anatskaya et al26 CC BY 4.0.
It is clear that the work of Anatskaya et al traces cancer-related polyploidy genes to a time when the unicellular ancestor was developing reproductive polyploid cysts,27 and paving the way to stemness, cell differentiation, and G + S life cycles, overturning the dogma of the EM origin of cancer. The relationship between the reproductive polyploidy of cancer and the reproductive ploidy of ancient cysts is a clear supportive argument for cancer's unicellular origin. However, I disagree with the idea of the authors that polyploidy could “activate the recapitulation of evolutionary developmental programs.” In my opinion, cancer polyploidy is firstly (i) a reproductive mechanism of intact germlines and clones to generate nascent CSCs (cyclic reproductive polyploidy) and secondly (ii) a repair and regeneration mechanism by which therapy-induced PGCCs renew damaged germline cells and CSCs pools. In both cases, polyploidization takes place within the G + S life cycle. Polyploidization is not the driver that switch human cells into the unicellular G + S lifestyle; it is part of the already reactivated unicellular G + S life cycle and a distinct feature of unicellular lifestyle.
Suppression mechanisms of G + S life cycles evolved in early metazoans
Multicellularity could not prevail unless the unicellular G + S life cycle could be prevented. Mechanisms blocking the unicellular life cycle were required to maintain multicellularity. In this sense, early metazoans evolved gene regulatory networks (GRNs) that consisted partly of newly evolved EM genes, but in large part the UC genes were repurposed as anti-suppressor genes (oncogene archetypes). The goal of the antisuppressor genes was to shut down the suppressor networks (tumor suppressor archetypes) and restore the unicellular G + S life cycle. The transition period from UC to EM was the era of constant alternation between unstable EM states and reactivated UC states.
In tumors, there is much molecular evidence of ancient UC and EM genes or gene modules that are upregulated or downregulated during tumor development.20,21,27,43 Phylostratigraphic research has shown that the UC subnetwork of the human genome is connected to the multicellular subnetwork (MC) through “hub” genes that, in healthy cells, control and regulate the equilibrium between UC and MC genes.21,27,43 This hub gene network corresponds to the suppressors and anti-suppressor genes emerging in the EM era. According to phylostratigraphic analyses, anti-suppressor genes, in metazoans and humans, disrupt the principles of UC/MC cooperation, especially in advanced neoplasmic stages and metastases. They inactivate the MC genes, and upregulate the UC genes.43
Genes of UC and MC origin are involved in numerous processes of healthy human cells. In the somatic tumor cells molecular researchers identified a set of deeply homologous genes as regulators of the coexpression of the UC and MC subnetworks.21 Among this hub gene network, researchers found three UC genes from the phylostratum ps1 (ACTG1, RCC2, PKN2), five UC genes from ps3 and ps4 (PLEC, TLN1, VASP, DSP, CTTN), and four EM genes from ps5 (ILK, CTNNA1, CTNND1, and PKP3). Because of their central role in the human gene network, these 12 genes have fundamental vulnerability and play critical roles in cancer processes associated with genomic instability, late tumorigenesis, and metastasis.21,43 However, it is not sure that hub genes play a crucial role in cancer initiation and the conversion from a normal, healthy cell into malignant cells as claimed by previous authors.44, 45, 46
In tumors, Trigos et al found 12,812 downstream target genes and 1370 regulator genes and hypothesize that regulatory genes control a variety of target genes.21 Over half (56.42%) of the GRN genes are EM genes, while only 37.88% and 5.69% are UC and MM genes, respectively. The authors looked for genes with key regulatory roles controlling at least 10 downstream targets and designated them as master regulators. They found that 65.12% of the master regulators are EM genes, while only 28.49% and 6.40% are UC and MM genes respectively, and consider that the most important master regulators in the network are EM genes.
Genetic and non-genetic CSC heterogeneity arises from deregulated EMT processes
Posttherapeutic changes in the tumor microenvironment such as cell senescence and other environmental changes can alter both the phenotype and function of CSCs. However, CSCs may also lose their function prior to treatments if exposed to unfavorable growth factors such as hyperoxia (Fig. 3). Hyperoxic-grown germlines suffer DNA DSB defects. They signal their loss of function to soma cells, which respond by soma-to-germ transition (SGT/EMT) and generate new functional germline clones. This is a process of germline renewal inherited from unicellular ancestors. The “amoeba model” provides an evolutionary explanation for cancer cell theories such as “cellular plasticity” and “clonal evolution”, which state that nonstem cells and stem cells can interconvert and that genomic and genetic alteration accumulate over time, leading to more CSC heterogeneity and increased aggressiveness.47, 48, 49, 50
In the amoeba model, somatic cells arise (i) either from stem cell differentiation or (ii) from germ-to-soma transition (GST). Somatic cells have epigenetically silenced the germline genome but can reactivate it when the germline signals DNA damage and loss of reproductive function. SGT is also one of the causes of increased pathogenicity in parasitic Entamoeba. In cancer, germline signaling induces SGT/EMT that forms secondary germlines and clones. EMT-generated CSCs are usually more pathogenic, leading to metastases.
SGT/EMT processes experience both epigenetic and genetic alteration. As a result, genetically distinct CSC clones coexist in different tumors.51, 52, 53, 54, 55 They are differentially radiation sensitive and exhibit increased resistance to cancer therapies. Therapeutics can lead to adaptive responses and acquired radiation resistance and even more invasive tumors. Therapeutics preferentially kill non-stem cells and enrich CSC density in tumors from less than 1% to more than 80%.51,56,57 CSC clones with genomic alteration protect tumors from therapeutic effects. Second, the ability to repair DNA damage after irradiation, particularly severe DNA DSB damage, is higher in CSCs than in their nontumor counterparts.58 CSCs preferentially repairs DNA damage through ancient mechanisms of homologous recombination (HR) and MGRS/PGCC pathway.59,60
Acquired resistance to anti-cancer therapy usually results from nongenetic alteration such as DNA methylation, histone modifications, and other epigenetic mechanisms that alter gene expression without changing the genetic profile. Such mechanisms act more rapidly than genetic mutations.61, 62, 63 Although patterns of DNA methylation are generally faithfully transmitted to daughter cells, they can also be erased resulting in a large reduction in total DNA methylation.64
Cancer stem cells generated by germline have three distinct superclasses, for which I have proposed the designations (i) nascent CSCs or primary GSCs; they arise from repetitive aCLS cycling and cyclic reproductive polyploidization; (ii) EMT-generated CSCs (secondary CSCs) that arise when the damaged germline signals the loss of reproductive function to soma cells, and (iii) PGCC-generated CSCs (tertiary CSCs), arising from repair and genome reorganization processes that occur in both native PGCCs (MGRS) and therapy-induced PGCCs3,4 (Fig. 3). PGCCs have mechanisms of DNA restructuring and natural genetic engineering capacities necessary to rearrange genomic components and its system architecture. All CSCs are produced from the germline. Late EMT enables tumors to become more malignant and metastatic. Initiation, maintenance, and inhibition of EMT are regulated by small molecules of antiproliferative agents.65 External signaling molecules and intracellular signaling molecules activate various EMT transcription factors and mesenchymal EMT markers.
EMT processes involve the transition from specific epithelial markers (soma cell markers) to mesenchymal markers (CSC markers). Several transcription factors and other biomarkers are also involved in the orchestration of EMT that includes concomitant deletion, upregulation, and gene overexpression. Numerous cancer researchers agree that CSC formation is associated with EMT activation and that EMT leads to stem-like features and phenotypes that promote tumorigenesis and accelerated metastasis.66, 67, 68 Some of these CSCs express both soma and germline markers simultaneously. They are considered to be alternative CSC-states or CSC-like states, conditioned by partial EMT processes. Such intermediate EMT stages are better known as hybrid, early hybrid, and late hybrid EMT stages. The EMT-related CSCs are commonly referred to as “stem-like phenotype,” “stem- and EMT-based gene expression,” or “stem-like state” leading sometimes to confusion. Regardless of their origin, these are all cells with stemness function that have emerged from an earlier germline or a later germline with more or less invasive and metastatic potential.
In recent years, EMT has been increasingly understood as a common regulator of CSC phenotype, CSC production,69 and drug resistance. All in all, EMT is a critical mechanism leading to more malignancy and metastasis.70, 71, 72, 73, 74 It allows flexible gene expression between activation and repression.68 In contrast to the SGT processes in Entamoeba, cancer EMT are considerably deregulated by the immunosuppressive tumor microenvironment.75
Conclusions and perspectives
In recent years, the prevailing somatic mutation theory (SMT) of tumorigenesis has come under increasing pressure and several alternatives have been proposed76 and three main theories of cancer stem cell origin and CSC development have gained popularity. Each of them targets different phases of CSC evolution. One is the somewhat older “CSC model” theory, which proposes a unidirectional development model. This theory states, among other viewpoints, that primary CSCs divide asymmetrically to form a CSC pool and switch to symmetric cell cycling to increase the number of CSCs.48 The “clonal evolution model” assumes an accumulation of genomic and genetic alterations over time, leading to an increase in CSC heterogeneity and aggressiveness, while the “cellular plasticity model” assumes that non-CSCs and CSCs can interconvert. These theories are not mutually exclusive, but they address only part of the question. In contrast, the “amoeba model” of cancer, which I propose in the present paper, considers the evolution of cancer stem cells from the perspective of the “deep homology” of G + S gene module and its transition from a free-living cell system to a parasitic lifestyle which also occurs in Entamoeba.77 The “amoeba–cancer model” combines all three theories mentioned above into a more comprehensive theory of evolution. It distinguishes between “healthy” germline cells, which proliferate through asymmetric cell cycles and express stemness potential, and damaged germline cells, which lose its stem potential (loss of function) and proliferate through symmetric cell cycles.
Cancer ancestors were unicellularians evolving over millions and millions of years into sophisticated G + S life cycles. The origin of the deep homologous G + S gene module is in Ur-flagellates that could transform their protective cysts into reproductive cysts. The progeny of reproductive cysts evolved into stem cells, which differentiate germ and soma cell lines and cells that are capable of phenotype conversion. Damaged germline cells that lost the reproductive potential have learned to repair DNA defects and reorganize the genome architecture through MGRSs and/or PGCCs rearrangements. As colonial unicellular organisms or more advanced early metazoans began to evolve the integrated multicellular mode of life, evolution retained the old unicellular G + S life cycle as a stable protective measure for the maintenance of life. The extension of the GRN by new EM genes and repurposed UC genes took over the control and surveillance function over the ancient G + S gene module and its reactivation in case of evolutionary dead-ends. The extended GRN emerged suppressor and antisuppressor genes as archetypes of tumor suppressor genes (TSGs) and oncogenes. In parallel with the evolution of metazoans, the G + S gene module evolved the more complex cancer G + S life cycle contained in the human genome. From an evolutionary point of view, the G + S life cycle of unicellularians was not abandoned by metazoans, it was maintained as an “ultima ratio” in the genome of all metazoan and human cells.
The present work is one more analysis of the close relationship between the germline of cancer and the nascent cancer stem cells (nCSCs). I distinguish between three classes of CSCs: (i) nascent CSCs or primary CSCs are germline stem cells (GSCs) that arise from reproductive germline cycles; (ii) EMT-induced CSCs and EMT-induced CSC-like cells arise from somatic cells that transit into new germline cells, and clones through SGT/EMT processes; and (iii) high metastatic MGRS/PGCC-induced CSCs. These tertiary CSCs are the result of DNA repair and genome reorganization processes. Some SGT/EMT processes may be incomplete, defective, or deregulated, resulting in a broad spectrum of heterogenous CSCs and CSC-like cells that are either pure germline cells or hybrid cells with mixed germline and soma hallmarks.76
Non-genetic sources of phenotypic variation may play an important role in tumor initiation, tumor progression, and evolution.61
Recent research does not provide a definitive answer to the question of what cancer is. Is it an “atavism,” a “genetic disease,” or simply an ancient life cycle adopted by early metazoans to avoid evolutionary dead ends? It is now clear that cancer is not an atavism, as has been erroneously claimed in the literature in recent years. According to the definition by Wagner, an atavism is a morphophysiological trait controlled by an ancient gene regulatory network (GRN) that survives periods of disuse and can be reactivated and reused later lineages, even if it was not used in the immediate ancestors. Cancer is not an atavism because it was reactivated and reused continuously in the last 800 million years (Ma) by all intermediate ancestors of humans, mammalians, and vertebrates.8
To summarize: What we perceive as an extremely unpleasant and complicated human disease (cancer) is a rescue mechanism of a sick, weakened metazoan cell that cannot continue its multicellular life and find itself in the same dead-end situation as their early metazoan ancestors 800 million years ago. This rescue mechanism, which reactivates the framework of the deep unicellular G + S life cycle, leads to a kind of aggressive pseudo-parasitism within the multicellular host organism and eventually to its death, which parasitic Entamoeba also do. However, Entamoeba eliminates stem cell precursors via cyst excretion, whereas cancer, as an ancient cell system closed in the host organism, maintains all nascent CSC generations that further potentiate the disease. In a previous paper, I proposed that severely threatened cells are pulling old DNA repair mechanisms out of Pandora's tool box for unicellular organisms, inadvertently restarting the dormant G + S life cycle,22 doing more harm than good. In many countries around the world, nearly one in two people will develop cancer once in their lifetime. A large variety of metazoans are also affected by cancer.
Correctly understanding the origins and mechanisms of using the “amoeba model” can speed progress towards preventing or curing cancer. Interestingly, there is older evidence that Entamoeba cells change their surface antigen architecture when transferred from the normoxic intestine lumen (<5.7% O2 content) to the more oxygenated tissue (>5.7% O2).78 We have seen in the present work that tissue hyperoxia damages the oxygen-sensitive germline cells but not somatic cells, which tolerate oxygen and form tumors. It is further conceivable that the GST process also causes an increase in tumor surface antigens (somatic antigens). Similarely, Entamoeba has about 20 different somatic surface antigens,79 some of which – like cysteine peptidases (lysosomal cysteine peptidases, cysteine cathepsins) and serine-rich proteins (anchored serine proteases)80,81 – are potentially deep homologous antigen-sites and archetypes of tumor surface antigens. It will be of interest to learn more about the deep homology of early surface antigens (EAS) in pre-tumoral or early tumoral CSCs and somatic cancer cells; also what changes in EAS architecture occur during germ to soma conversion and the transition from hypoxia to hyperoxia. If deep homologous EAS could be discovered, they could also be targets for vaccines. More about the homologous life cycle of Entamoeba and the amoeba-cancer model occurs separately.82
Conflict of interests
The author declares no conflict of interests.
Acknowledgements
I express my gratitude to Dr. Dennis Thomas from the Cold Spring Harbor Laboratory, USA (native English speaker) for reading the manuscript and improving my English. Special thanks to Mr. Gregor Jaruga for the graphics.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Kreisman R. Deep homology opens up new possibilities for gene research. 2010. https://www.robertkreisman.com/medicalmalpractice-lawyer/deep_homology_opens_up_new_pos/ preprint at.
- 2.McGary K.L., Park T.J., Woods J.O., Cha H.J., Wallingford J.B., Marcotte E.M. Systematic discovery of non obvious human disease models through orthologous phenotypes. Proc Natl Acad Sci U S A. 2010;107(14):6544–6549. doi: 10.1073/pnas.0910200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niculescu V.F. aCLS cancers: genomic and epigenetic changes transform the cell of origin of cancer into a tumorigenic pathogen of unicellular organization and lifestyle. Gene. 2020;726:144174. doi: 10.1016/j.gene.2019.144174. [DOI] [PubMed] [Google Scholar]
- 4.Niculescu V.F. Germline evolution in cancer as explained by the germ and soma theory of dual cell systems. J Clin Anat Pathol. 2021;6(1):113. [Google Scholar]
- 5.Forterre P. The universal tree of life: an update. Front Microbiol. 2015;6:717. doi: 10.3389/fmicb.2015.00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albuquerque T.A.F., Drummond do Val L., Doherty A., de Magalhães J.P. From humans to hydra: patterns of cancer across the tree of life. Biol Rev Camb Philos Soc. 2018;93(3):1715–1734. doi: 10.1111/brv.12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shubin N., Tabin C., Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457(7231):818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- 8.Wagner G.P. Princeton Univerity Press; New Jersey, USA: 2014. Homology, Genes and Evolutionary Inovation. [Google Scholar]
- 9.Tschopp P., Tabin C.J. Deep homology in the age of next-generation sequencing. Philos Trans R Soc Lond B Biol Sci. 2017;372(1713):20150475. doi: 10.1098/rstb.2015.0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nachman M.W., Hoekstra H.E., D'Agostino S.L. The genetic basis of adaptive melanism in pocket mice. Proc Natl Acad Sci U S A. 2003;100(9):5268–5273. doi: 10.1073/pnas.0431157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wittkopp P.J., Carroll S.B., Kopp A. Evolution in black and white: genetic control of pigment patterns in Drosophila. Trends Genet. 2003;19(9):495–504. doi: 10.1016/S0168-9525(03)00194-X. [DOI] [PubMed] [Google Scholar]
- 12.Davidson E.H., Erwin D.H. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311(5762):796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 13.Erwin D.H. Early metazoan life: divergence, environment and ecology. Philos Trans R Soc Lond B Biol Sci. 2015;370(1684):20150036. doi: 10.1098/rstb.2015.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vendramin R., Litchfield K., Swanton C. Cancer evolution: Darwin and beyond. EMBO J. 2021;40(18):e108389. doi: 10.15252/embj.2021108389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shubin N. Vintage Books, Penguin Random House, LLC; New York: 2020. Some Assembly Required: Decoding Four Billion Years of Life, from Ancient Fossils to DNA. [Google Scholar]
- 16.Johnson N. Nothing in biology begins when you think it does. Evol Educ Outreach. 2020;13(1):24. [Google Scholar]
- 17.Davis A., Gao R., Navin N. Tumor evolution: linear, branching, neutral or punctuated? Biochim Biophys Acta Rev Cancer. 2017;1867(2):151–161. doi: 10.1016/j.bbcan.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Domazet-Loso T., Brajković J., Tautz D. A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet. 2007;23(11):533–539. doi: 10.1016/j.tig.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 19.Domazet-Loso T., Tautz D. Phylostratigraphic tracking of cancer genes suggests a link to the emergence of multicellularity in Metazoa. BMC Biol. 2010;8:66. doi: 10.1186/1741-7007-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trigos A.S., Pearson R.B., Papenfuss A.T., Goode D.L. Altered interactions between unicellular and multicellular genes drive hallmarks of transformation in a diverse range of solid tumors. Proc Natl Acad Sci U S A. 2017;114(24):6406–6411. doi: 10.1073/pnas.1617743114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trigos A.S., Pearson R.B., Papenfuss A.T., Goode D.L. Somatic mutations in early metazoan genes disrupt regulatory links between unicellular and multicellular genes in cancer. Elife. 2019;8:e40947. doi: 10.7554/eLife.40947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niculescu V.F. Is an ancient genome repair mechanism the Trojan Horse of cancer? Nov Appro in Can Study. 2021;5(5) NACS.000625. [Google Scholar]
- 23.Nayernia K. Germ cells, origin of somatic stem cells? Cell Res. 2008;18:S26. [Google Scholar]
- 24.Nayernia K., Lee J.H., Engel W., et al. From stem cells to germ cells and from germ cells to stem cells. Int J Reprod Biomed. 2007;5(2):41–44. [Google Scholar]
- 25.Solana J. Closing the circle of germline and stem cells: the Primordial Stem Cell hypothesis. Evodevo. 2013;4(1):2. doi: 10.1186/2041-9139-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anatskaya O.V., Vinogradov A.E., Vainshelbaum N.M., Giuliani A., Erenpreisa J. Phylostratic shift of whole-genome duplications in normal mammalian tissues towards unicellularity is driven by developmental bivalent genes and reveals a link to cancer. Int J Mol Sci. 2020;21(22):8759. doi: 10.3390/ijms21228759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erenpreisa J., Salmina K., Anatskaya O., Cragg M.S. Paradoxes of cancer: survival at the brink. Semin Cancer Biol. 2022;81:119–131. doi: 10.1016/j.semcancer.2020.12.009. [DOI] [PubMed] [Google Scholar]
- 28.Schussnig B. Verlag Ferdinand Berger & Söhne Ges.m.b.H.; Horn, Austria: 1948. Stammesgeschichtlicher Formenwandel und Gestaltungstypen im Reich der Pilze. [Google Scholar]
- 29.Mylnikov A.P., Karpov S.A. Review of diversity and taxonomy of cercomonads. Protistology. 2004;3:201–221. [Google Scholar]
- 30.Keeling P.J., Palmer J.D. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9(8):605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 31.Sogabe S., Hatleberg W.L., Kocot K.M., et al. Pluripotency and the origin of animal multicellularity. Nature. 2019;570(7762):519–522. doi: 10.1038/s41586-019-1290-4. [DOI] [PubMed] [Google Scholar]
- 32.Carr M., Leadbeater B.S., Hassan R., Nelson M., Baldauf S.L. Molecular phylogeny of choanoflagellates, the sister group to Metazoa. Proc Natl Acad Sci U S A. 2008;105(43):16641–16646. doi: 10.1073/pnas.0801667105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maldonado M. Choanoflagellates, choanocytes, and animal multicellularity. Invertebr Biol. 2004;123(1):1–22. [Google Scholar]
- 34.Cavalier-Smith T. Origin of animal multicellularity: precursors, causes, consequences-the choanoflagellate/sponge transition, neurogenesis and the Cambrian explosion. Philos Trans R Soc Lond B Biol Sci. 2017;372(1713):20150476. doi: 10.1098/rstb.2015.0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brunet T., King N. The origin of animal multicellularity and cell differentiation. Dev Cell. 2017;43(2):124–140. doi: 10.1016/j.devcel.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen C. Six major steps in animal evolution: are we derived sponge larvae? Evol Dev. 2008;10(2):241–257. doi: 10.1111/j.1525-142X.2008.00231.x. [DOI] [PubMed] [Google Scholar]
- 37.Kang S., Tice A.K., Spiegel F.W., et al. Between a pod and a hard test: the deep evolution of amoebae. Mol Biol Evol. 2017;34(9):2258–2270. doi: 10.1093/molbev/msx162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daniels E.W., Pappas G.D. Reproduction of nuclei in Pelomyxa palustris. Cell Biol Int. 1994;18(8):805–812. doi: 10.1006/cbir.1994.1113. [DOI] [PubMed] [Google Scholar]
- 39.Schilde C., Schaap P. In: Dictyostelium discoideum Protocols Second Edition (Methods in Molecular Biology) Ludwig Eichinger L., Rivero F., editors. Springer Science+Business Media, LLC; NY: New York: 2013. The Amoebozoa. [Google Scholar]
- 40.Stephenson S.L., Stempen H. Myxomycetes: a handbook of slime molds. Mycologia. 1995;87(3):424. [Google Scholar]
- 41.Kendal W.S. vol. 3. Springer Netherlands; Dordrecht: 2011. Gain and loss of cancer stem cells: effect on metastatic efficiency and treatment response; pp. 231–240. (Stem Cells and Cancer Stem Cells). [Google Scholar]
- 42.Weinberg R.A. Garland Science; Oxford: 2008. The Biology of Cancer. [Google Scholar]
- 43.Louka A., Takan I., Pavlopoulou A., Georgakilas A.G. Bioinformatic approaches to the investigation of the atavistic genes implicated in cancer. Front Biosci (Landmark Ed) 2021;26(8):279–311. doi: 10.52586/4944. [DOI] [PubMed] [Google Scholar]
- 44.Martincorena I., Raine K.M., Gerstung M., et al. Universal patterns of selection in cancer and somatic tissues. Cell. 2017;171(5):1029–1041. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller D.G. On the nature of susceptibility to cancer. The presidential address. Cancer. 1980;46(6):1307–1318. doi: 10.1002/1097-0142(19800915)46:6<1307::aid-cncr2820460602>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 46.Schinzel A.C., Hahn W.C. Oncogenic transformation and experimental models of human cancer. Front Biosci. 2008;13:71–84. doi: 10.2741/2661. [DOI] [PubMed] [Google Scholar]
- 47.Stratton M.R., Campbell P.J., Futreal P.A. The cancer genome. Nature. 2009;458(7239):719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walcher L., Kistenmacher A.K., Suo H., et al. Cancer stem cells-origins and biomarkers: perspectives for targeted personalized therapies. Front Immunol. 2020;11:1280. doi: 10.3389/fimmu.2020.01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reya T., Morrison S.J., Clarke M.F., Weissman I.L. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 50.Rich J.N. Cancer stem cells: understanding tumor hierarchy and heterogeneity. Medicine (Baltimore) 2016;95(1 Suppl 1):S2–S7. doi: 10.1097/MD.0000000000004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arnold C.R., Mangesius J., Skvortsova I.I., Ganswindt U. The role of cancer stem cells in radiation resistance. Front Oncol. 2020;10:164. doi: 10.3389/fonc.2020.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Batlle E., Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124–1134. doi: 10.1038/nm.4409. [DOI] [PubMed] [Google Scholar]
- 53.Bradshaw A., Wickremsekera A., Tan S.T., Peng L., Davis P.F., Itinteang T. Cancer stem cell hierarchy in glioblastoma multiforme. Front Surg. 2016;3:21. doi: 10.3389/fsurg.2016.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Atashzar M.R., Baharlou R., Karami J., et al. Cancer stem cells: a review from origin to therapeutic implications. J Cell Physiol. 2020;235(2):790–803. doi: 10.1002/jcp.29044. [DOI] [PubMed] [Google Scholar]
- 55.Prasetyanti P.R., Medema J.P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol Cancer. 2017;16(1):41. doi: 10.1186/s12943-017-0600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kreso A., O'Brien C.A., van Galen P., et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339(6119):543–548. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martins-Neves S.R., Cleton-Jansen A.M., Gomes C.M.F. Therapy-induced enrichment of cancer stem-like cells in solid human tumors: where do we stand? Pharmacol Res. 2018;137:193–204. doi: 10.1016/j.phrs.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 58.Rycaj K., Tang D.G. Cancer stem cells and radioresistance. Int J Radiat Biol. 2014;90(8):615–621. doi: 10.3109/09553002.2014.892227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mladenov E., Magin S., Soni A., Iliakis G. DNA double-strand-break repair in higher eukaryotes and its role in genomic instability and cancer: cell cycle and proliferation-dependent regulation. Semin Cancer Biol. 2016;37–38:51–64. doi: 10.1016/j.semcancer.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 60.Her J., Bunting S.F. How cells ensure correct repair of DNA double-strand breaks. J Biol Chem. 2018;293(27):10502–10511. doi: 10.1074/jbc.TM118.000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gunnarsson E.B., De S., Leder K., Foo J. Understanding the role of phenotypic switching in cancer drug resistance. J Theor Biol. 2020;490:110162. doi: 10.1016/j.jtbi.2020.110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brock A., Chang H., Huang S. Non-genetic heterogeneity -- a mutation-independent driving force for the somatic evolution of tumours. Nat Rev Genet. 2009;10(5):336–342. doi: 10.1038/nrg2556. [DOI] [PubMed] [Google Scholar]
- 63.Brown R., Curry E., Magnani L., Wilhelm-Benartzi C.S., Borley J. Poised epigenetic states and acquired drug resistance in cancer. Nat Rev Cancer. 2014;14(11):747–753. doi: 10.1038/nrc3819. [DOI] [PubMed] [Google Scholar]
- 64.Tycko B. Epigenetic gene silencing in cancer. J Clin Invest. 2000;105(4):401–407. doi: 10.1172/JCI9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ribatti D., Tamma R., Annese T. Epithelial-mesenchymal transition in cancer: a historical overview. Transl Oncol. 2020;13(6):100773. doi: 10.1016/j.tranon.2020.100773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ye X., Weinberg R.A. Epithelial-mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol. 2015;25(11):675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.May C.D., Sphyris N., Evans K.W., Werden S.J., Guo W., Mani S.A. Epithelial-mesenchymal transition and cancer stem cells: a dangerously dynamic duo in breast cancer progression. Breast Cancer Res. 2011;13(1):202. doi: 10.1186/bcr2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scheel C., Weinberg R.A. Phenotypic plasticity and epithelial-mesenchymal transitions in cancer and normal stem cells? Int J Cancer. 2011;129(10):2310–2314. doi: 10.1002/ijc.26311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Farmer P., Bonnefoi H., Anderle P., et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15(1):68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- 70.Lin Y.T., Wu K.J. Epigenetic regulation of epithelial-mesenchymal transition: focusing on hypoxia and TGF-β signaling. J Biomed Sci. 2020;27(1):39. doi: 10.1186/s12929-020-00632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lu W., Kang Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev Cell. 2019;49(3):361–374. doi: 10.1016/j.devcel.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nieto M.A., Huang R.Y., Jackson R.A., Thiery J.P. EMT: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 73.Dongre A., Weinberg R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20(2):69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 74.Tsai J.H., Donaher J.L., Murphy D.A., Chau S., Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22(6):725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Terry S., Savagner P., Ortiz-Cuaran S., et al. New insights into the role of EMT in tumor immune escape. Mol Oncol. 2017;11(7):824–846. doi: 10.1002/1878-0261.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Baker S.G. The case for a cancer paradox initiative. Carcinogenesis. 2021;42(8):1023–1025. doi: 10.1093/carcin/bgab052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Niculescu V.F. 2022. The germ and soma life cycle of Entamoeba and its stem cells as an evolutionary model for cancer life cycle. [Google Scholar]
- 78.Bhattacharya A., Ghildyal R., Prasad J., Bhattacharya S., Diamond L.S. Modulation of a surface antigen of Entamoeba histolytica in response to bacteria. Infect Immun. 1992;60(4):1711–1713. doi: 10.1128/iai.60.4.1711-1713.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Biller L., Matthiesen J., Kühne V., et al. The cell surface proteome of Entamoeba histolytica. Mol Cell Proteomics. 2014;13(1):132–144. doi: 10.1074/mcp.M113.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martin C.E., List K. Cell surface-anchored serine proteases in cancer progression and metastasis. Cancer Metastasis Rev. 2019;38(3):357–387. doi: 10.1007/s10555-019-09811-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pišlar A., Perišić Nanut M., Kos J. Lysosomal cysteine peptidases – molecules signaling tumor cell death and survival. Semin Cancer Biol. 2015;35:168–179. doi: 10.1016/j.semcancer.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 82.Niculescu V.F. Hyperoxia-mediated genome alteration in the germline of Entamoeba and the amoeba-cancer model. Genes Dis. 2022 Submitted for publication. [Google Scholar]