Abstract
A set of PCR primers was designed and validated for specific detection and quantification of Prevotella ruminicola, Prevotella albensis, Prevotella bryantii, Fibrobacter succinogenes, Selenomonas ruminantium-Mitsuokella multiacida, Streptococcus bovis, Ruminococcus flavefaciens, Ruminobacter amylophilus, Eubacterium ruminantium, Treponema bryantii, Succinivibrio dextrinosolvens, and Anaerovibrio lipolytica. By using these primers and the real-time PCR technique, the corresponding species in the rumens of cows for which the diet was switched from hay to grain were quantitatively monitored. The dynamics of two fibrolytic bacteria, F. succinogenes and R. flavefaciens, were in agreement with those of earlier, culture-based experiments. The quantity of F. succinogenes DNA, predominant in animals on the hay diet, fell 20-fold on the third day of the switch to a grain diet and further declined on day 28, with a 57-fold reduction in DNA. The R. flavefaciens DNA concentration on day 3 declined to approximately 10% of its initial value in animals on the hay diet and remained at this level on day 28. During the transition period (day 3), the quantities of two ruminal prevotella DNAs increased considerably: that of P. ruminicola increased 7-fold and that of P. bryantii increased 263-fold. On day 28, the quantity of P. ruminicola DNA decreased 3-fold, while P. bryantii DNA was still elevated 10-fold in comparison with the level found in animals on the initial hay diet. The DNA specific for another xylanolytic bacterium, E. ruminantium, dropped 14-fold during the diet switch and was maintained at this level on day 28. The concentration of a rumen spirochete, T. bryantii, decreased less profoundly and stabilized with a sevenfold decline by day 28. The variations in A. lipolytica DNA were not statistically significant. After an initial slight increase in S. dextrinosolvens DNA on day 3, this DNA was not detected at the end of the experiment. S. bovis DNA displayed a 67-fold increase during the transition period on day 3. However, on day 28, it actually declined in comparison with the level in animals on the hay ration. The amount of S. ruminantium-M. multiacida DNA also increased eightfold following the diet switch, but stabilized with only a twofold increase on day 28. The real-time PCR technique also uncovered differential amplification of rumen bacterial templates with the set of universal bacterial primers. This observation may explain why some predominant rumen bacteria have not been detected in PCR-generated 16S ribosomal DNA libraries.
The complex symbiotic microbiota of the rumen is responsible for breakdown of plant fiber, an ability the ruminal host animals lack. This microbiota is highly responsive to changes in diet, age, antibiotic use, and the health of the host animal and varies according to geographical location, season, and feeding regimen (reviewed in references 4, 12, 21, and 31). In the early days of rumen microbiology, these changes were monitored by cultivation-based techniques, but limitations inherent in the technique and the development of more sensitive and accurate molecular detection methods have brought new developments into the field. One of the first examples of this sort was reported by Stahl and coworkers (29), who used species- and group-specific 16S rRNA-targeted probes for enumeration of Fibrobacter (Bacteroides) succinogenes and Lachnospira multiparus in rumens treated with antibiotic. A genomic DNA fragment from Prevotella brevis (formerly Bacteroides ruminicola subsp. brevis B14) was used to monitor this strain upon introduction into the rumen (1). Then the 16S rRNA-targeting probe approach was used for quantification of Fibrobacter succinogenes (3, 36), ruminococci (14, 15, 36), Clostridium proteoclasticum (25), Butyrivibrio fibrisolvens (10, 13), and Methanomicrobium mobile (39).
The detection methods described above are based on sequences from cultivable strains of rumen microorganisms with known metabolic properties derived from laboratory experiments. However, because of cultivation limitations, the real diversity of rumen microorganisms may be substantially underestimated, and recently, several works describing the molecular diversity of rumen bacteria based on the retrieval and sequencing of SSU ribosomal DNA (rDNA) sequences have been published (32, 33, 38). Indeed, in these descriptions of ruminal bacterial diversity, the overwhelming majority of the retrieved sequences had very limited or no similarity to the previously recognized cultivable species and genera, and some sequences even had no clear phylum allocation. These experiments also demonstrated that clear bias toward the overrepresentation of easy-to-cultivate bacteria such as the Cytophaga-Flexibacter-Bacteroides group exists with cultivation-based diversity studies and enumeration. On the contrary, molecular analyses suggest that the numerically prevalent species, even under different diet conditions, are made up of bacteria belonging to the low-G+C, gram-positive bacterial phylum (32, 33). Molecular analyses reiterated the necessity of probe-directed isolation to saturate this molecular diversity with cultivable isolates, and our recent work has successfully demonstrated the feasibility of this approach (24). The availability of isolates representing the main groups of general diversity offers clear advantages with the possibility of assigning the functional role in the rumen based on the metabolic or physiological properties of pure cultures. Thus, the development of molecular detection of rumen bacteria, the functionality of which in the rumen is deduced from pure culture studies, remains an important subject for study. At present, however, only a few detection probes or PCR primers for rumen bacteria are available. In this study, we designed and validated PCR primers for detection of 13 cultivable rumen bacterial species: Prevotella ruminicola, Prevotella albensis, Prevotella bryantii, Fibrobacter succinogenes, Selenomonas ruminantium-Mitsuokella multiacida, Streptococcus bovis, Ruminococcus flavefaciens, Ruminobacter amylophilus, Eubacterium ruminantium, Treponema bryantii, Succinivibrio dextrinosolvens, and Anaerovibrio lipolytica. This primer set was used for detection and quantification of the corresponding species in the rumen under diet change conditions by a real-time PCR approach. In the second part of this work, we demonstrated the applicability of the real-time PCR approach to yet another problem of molecular ecology: confirmation of the phenomenon of differential amplification of DNA templates from pure cultures of rumen bacteria. This observation may have implications for analysis of PCR-generated libraries from other microbial ecosystems as well.
MATERIALS AND METHODS
Bacterial cultures and growth medium.
The following strains of rumen bacteria were used as reference strains: A. lipolytica (strain ATCC 32374T), E. ruminantium (strain ATCC 17233T), Escherichia coli (strain INVαF′; Invitrogen, Carlsbad, Calif.), Fibrobacter intestinalis (strain ATCC 43854T), F. succinogenes (strain ATCC 19169T), Megasphaera elsdenii (strain JCM1772T), M. multiacida (strain ATCC 27723T), P. ruminicola (strain ATCC 19189T), P. albensis (strain M384T), P. bryantii (strain B14T), R. amylophilus (strain ATCC 29744T), R. albus (strain ATCC 27210T), R. flavefaciens (strain ATCC 19208T), S. ruminantium (strain JCM6582T), S. bovis (strain JCM5802T), S. equinus (strain JCM7879T), S. dextrinosolvens (strain ATCC 19716T), T. bryantii (strain ATCC 33254T), and Wolinella succinogenes (strain ATCC 29543T). P. bryantii and P. albensis strains were kindly provided by H. J. Flint (Rowett Research Institute, Aberdeen, United Kingdom). Strains were cultured in medium 10 (5).
Sampling.
Samples were obtained from eight fistulated dry Holstein cows (with an average body weight of 560 ± 15 kg) kept at the National Institute of Animal Health, Tsukuba, Japan. Before the experiment, animals were maintained on a basal diet consisting of 3.5 kg of hay, 1 kg of hay cube, and 1.5 kg of concentrate fed twice a day. The composition of the concentrate was 24% ground wheat, 20% corn, 20% wheat bran, 10% soybean meal, 10% linseed meal, 6% gluten feed, 5% rice bran, 4% calcium carbonate, 0.5% dicalcium phosphate, and 0.5% microelements and vitamins. On day 0, five animals were switched to a high-grain diet for 4 weeks. This regimen consisted of two feedings: one in the morning (0.5 kg of hay, 2.4 kg of concentrate, and 3.6 kg of barley) and one in the evening (2.4 kg of concentrate and 3.6 kg of barley). In essence, the sampling procedure was performed as described previously (33). Briefly, the representative rumen content samples were obtained via fistula before the morning feeding. The samples were squeezed though two layers of cheesecloth and immediately used for DNA extraction. The rumen contents from days 0, 3, and 28 were used for this set of experiments.
DNA extraction.
Total genomic DNA from the rumen bacterial strains was extracted by the achromopeptidase method of Ezaki and coworkers (6, 7). Briefly, a 10-ml bacterial culture was pelleted by centrifugation (5,000 × g, 5 min) and resuspended in 3 ml of 5 mM EDTA (pH 8.0). The cells were treated with achromopeptidase (final concentration of 1 mg/ml) and lysed by adding 300 μl of 20% sodium dodecyl sulfate. After incubation at 55°C for 30 min, 3 ml of Tris-EDTA (TE)-buffered phenol (pH 8.0) was added. It was centrifuged at 10,000 × g for 10 min at room temperature. The supernatant was transferred to a fresh tube, extracted with buffered phenol, and isopropanol precipitated. Nucleic acids were dissolved in TE buffer (pH 8.0) and treated with DNase-free RNase. Samples were reextracted with the buffered phenol, ethanol precipitated, and dissolved in TE buffer. In further purification, the Qiagen Genomic-tip system (Qiagen, Tokyo, Japan) was used in accordance with the manufacturer's recommendations. The eluted DNA was precipitated with isopropanol, washed with 70% ethanol, dried, dissolved in sterile TE buffer, and stored at 4°C.
Total DNA from rumen fluid was extracted as described by Whitford and coworkers (38) with a Mini Bead-Beater (Biospec Products, Bartlesville, Okla.) for cell lysis. To minimize animal-to-animal variations, the aliquots of rumen fluid from five animals were mixed before DNA extraction. Four milliliters of rumen fluid was used for DNA extraction as described previously (33), but with a minor modification incorporating an additional DNA purification step. This was carried out with the Qiagen Genomic-tip system similarly to the procedure with the bacterial strains.
DNA concentrations were measured at 260 nm with a Beckman DU7500 spectrophotometer (Fullerton, Calif.). The DNA used for these experiments possessed an A260/A280 ratio higher than 1.8.
Design and synthesis of PCR primers.
The primers designed to detect the targeted species are listed in Table 1. The 16S rDNA sequences of targeted species were downloaded from GenBank and incorporated into our previously described alignments (24, 32, 33). Sequence regions specific for a given species (with >97% similarity) were searched online against GenBank sequences with the BLAST family of programs (18) to ensure the specificity of primers. These primers were also tested for the requirements imposed by real-time quantitative PCR (see below). The oligonucleotides were synthesized by Hokkaido System Science (Sapporo, Japan).
TABLE 1.
PCR primers for detection of rumen bacteria
| Target bacterium | Strain | Primer
|
Annealing temp (C) | Product size (bp) | |
|---|---|---|---|---|---|
| Forward | Reverse | ||||
| Prevotella ruminicola | ATCC 19189T | GGTTATCTTGAGTGAGTT | CTGATGGCAACTAAAGAA | 53 | 485 |
| Prevotella bryantii | B14T | ACTGCAGCGCGAACTGTCAGA | ACCTTACGGTGGCAGTGTCTC | 68 | 540 |
| Prevotella albensis | M384T | CAGACGGCATCAGACGAGGAG | ATGCAGCACCTTCACAGGAGC | 68 | 861 |
| Fibrobacter succinogenes | ATCC 19169T | GGTATGGGATGAGCTTGC | GCCTGCCCCTGAACTATC | 62 | 445 |
| Ruminobacter amylophilus | ATCC 29744T | CAACCAGTCGCATTCAGA | CACTACTCATGGCAACAT | 57 | 642 |
| Selenomonas ruminantium- Mitsuokella multiacida | JCM6582T | TGCTAATACCGAATGTTG | TCCTGCACTCAAGAAAGA | 53 | 513 |
| Streptococcus bovis | JCM5802T | CTAATACCGCATAACAGCAT | AGAAACTTCCTATCTCTAGG | 57 | 869 |
| Treponema bryantii | ATCC 33254T | AGTCGAGCGGTAAGATTG | CAAAGCGTTTCTCTCACT | 57 | 421 |
| Eubacterium ruminantium | ATCC 17233T | GCTTCTGAAGAATCATTTGAAG | TCGTGCCTCAGTGTCAGTGT | 57 | 671 |
| Anaerovibrio lipolytica | ATCC 33276T | TGGGTGTTAGAAATGGATTC | CTCTCCTGCACTCAAGAATT | 57 | 597 |
| Succinivibrio dextrinosolvens | ATCC 19716T | TGGGAAGCTACCTGATAGAG | CCTTCAGAGAGGTTCTCACT | 57 | 854 |
| Ruminococcus flavefaciens | ATCC 19208T | GGACGATAATGACGGTACTT | GCAATCYGAACTGGGACAAT | 62 | 835 |
Conventional PCR.
PCR was performed with the Takara Ex Taq PCR kit (Takara Shuzo, Osaka, Japan) and TaqStart antibody (Clontech, Palo Alto, Calif.). The PCR was conducted with a PE480 thermal cycler (Perkin-Elmer, Norwalk, Conn.). The amplification conditions were one cycle at 95°C for 3 min of denaturation, 35 cycles of 95°C for 30 s, various annealing temperatures (described in Table 1) for 30 s, and extension at 72°C for 1 min. A total of 25 μl of PCR mixture contained 300 nM each primer, 12.5 ng of purified template DNA, 1× Ex Taq reaction buffer, 200 μM each deoxynucleoside triphosphate (dNTP mixture), 1.25 U of Ex Taq DNA polymerase, and 220 ng of TaqStart antibody. The PCR products were separated by electrophoresis on agarose gels and stained with ethidium bromide.
Sequence and phylogenetic analyses of amplicons.
The specificity of amplifications was confirmed by sequencing and phylogenetic analysis. After detection of species-specific PCR amplicons in total rumen DNA, the PCR products were cloned into the TA cloning kit (Invitrogen, Carlsbad, Calif.), and the transformants were randomly picked up. The recombinant plasmids were extracted by the alkaline lysis miniprep method (2). Cycle sequencing was performed with the ThermoSequenase kit purchased from Amersham (Tokyo, Japan). The sequencing reaction products were read on a LI-COR M4000L automated DNA sequencer (Lincoln, Neb.). The obtained sequences were queried online by using the BLAST service at the National Center for Biotechnology Information (18). For phylogenetic confirmation of species-specific amplification, these sequences were incorporated into our previous alignments (24, 32, 33), and the phylogenetic positioning was done by constructing neighbor-joining trees (26) with the ClustalW program, version 1.74 (34).
Real-time PCR.
The quantification of each bacterial species DNA in total rumen DNA was performed with a LightCycler system (Roche, Mannheim, Germany). The FastStart DNA Master SYBR Green I was used for PCR amplification. The efficiency of PCR amplification was checked for various MgCl2 concentrations and annealing temperatures. The optimal amplification conditions for each primer pair were achieved with 5 mM (final concentration) MgCl2, 0.5 μM each primer, and the annealing temperatures shown in Table 1. The reaction mixture in 20 μl of the final volume contained 5 mM MgCl2, 2 μl of the 10× Mastermix (including FastStart enzyme, FastStart Taq DNA polymerase, reaction buffer, dNTP mixture, MgCl2, and SYBR Green I dye), 30 ng of template DNA, and 0.5 μM each primer. Amplification involved one cycle at 95°C for 10 min for initial denaturation and then 45 cycles of 95°C for 15 s followed by annealing at the temperatures shown in Table 1 for 5 s and then at 72°C for 30 s. Detection of the fluorescent product was set at the last step of each cycle. To determine the specificity of amplification, analysis of product melting was performed after each amplification. The melting curve was obtained by slow heating with a 0.1°C/s increment from 65°C to 95°C, with fluorescence collection at 0.1°C intervals. Additional specificity analyses included product size verification by gel electrophoresis of samples after the PCR run and melting point determination analysis. Dilutions of purified genomic DNA from control strains were used to construct species-specific calibration curves. These calibration curves were used for calculation of the species-specific DNA concentration in total rumen DNA preparations (in millimoles of 16S rDNA per milligram of total rumen DNA). The values and standard deviations presented in Table 4 are the repeated determinations of the same sample obtained with a DNA mix from the pooled rumen fluid of five animals (see the description of the sampling procedure). Total rumen DNA samples from days 0, 3, and 28 of our diet shift experiment (33) were used to monitor the dynamics of ruminal bacteria.
TABLE 4.
Quantification of rumen bacteria by real-time PCR during diet shift
| Bacteria targeted | DNA concna
|
||
|---|---|---|---|
| Day 0 | Day 3 | Day 28 | |
| P. ruminicola | 29.45 ± 3.6 | 205.2 ± 38.4 | 10.94 ± 1.9 |
| P. bryantii | 0.284 ± 0.1 | 74.77 ± 14.9 | 2.865 ± 0.5 |
| F. succinogenes | 12.32 ± 0.8 | 0.597 ± 0.0 | 0.215 ± 0.0 |
| S. ruminantium- M. multiacida | 7.114 ± 1.3 | 60.40 ± 6.6 | 16.92 ± 2.7 |
| S. bovis | 0.232 ± 0.1 | 15.63 ± 2.8 | 0.104 ± 0.0 |
| T. bryantii | 0.072 ± 0.0 | 0.043 ± 0.0 | 0.011 ± 0.0 |
| E. ruminantium | 0.585 ± 0.1 | 0.042 ± 0.0 | 0.048 ± 0.0 |
| A. lipolytica | 0.137 ± 0.1 | 0.440 ± 0.2 | 0.095 ± 0.0 |
| S. dextrinosolvens | 0.468 ± 0.1 | 0.733 ± 0.1 | NDb |
| R. flavefaciens | 2.891 ± 0.7 | 0.260 ± 0.0 | 0.324 ± 0.1 |
Day 0, before experiment, animals maintained on basal hay diet; day 3, animals fed high-grain diet for 3 days; day 28, animals fed high-grain diet for 28 days. The DNA concentration was measured as millimoles of 16S rDNA per milligram of total rumen DNA ± standard deviation (n = 4).
ND, not detected.
Nucleotide sequence accession number.
The 16S rDNA sequence data reported in this paper have been deposited in the GenBank database under accession no. AB056162 to AB056214.
RESULTS
Design and validation of species-specific primers.
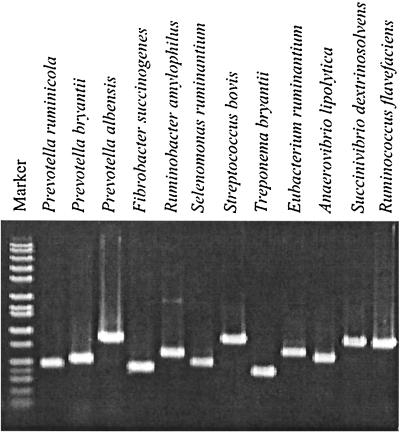
The primers were designed to satisfy the specificity within the range of sequences belonging to the same species. Cultivable and in vitro-retrieved sequences were used for alignments, and these alignments were searched for the regions that are conserved within a given bacterial species, but different from other species clusters on the phylogenetic tree. The primer sequences were then tested against online nucleotide databases to ensure their specificity. The resulting primer set (Table 1) produced PCR products of the expected size with test strains (Fig. 1). These primers were rigorously verified with DNA of 19 rumen bacterial species and E. coli (Table 2). The majority of PCR procedures were highly specific for the target species, except for S. bovis, which cross-reacted with S. equinus (Table 2), which is not surprising, since these strains are likely members of the same species (8, 37). The S. ruminantium-M. multiacida primer set amplified the targeted sequences from these two species of bacteria as expected (Table 2).
FIG. 1.
Amplification of control rumen bacterial DNA (strains are listed in Materials and Methods) with the primer set detailed in Table 1. A DNA size marker is in the extreme left lane.
TABLE 2.
Validation of PCR primers with control templates
| PCR primer seta | Validation result for:
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. ruminicola | P. bryantii | P. albensis | P. brevis | F. succinogenes | F. intestinalis | R. amylophilus | S. ruminantium | M. multiacida | S. bovis | S. equinus | T. bryantii | E. ruminantium | A. lipolytica | S. dextrinosolvens | R. flavefaciens | R. albus | M. elsdenii | W. succinogenes | E. coli | |
| P. ruminicola | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| P. bryantii | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| P. albensis | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| F. succinogenes | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| R. amylophilus | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − |
| S. ruminantium- M. multiacida | − | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − | − | − | − | − |
| S. bovis | − | − | − | − | − | − | − | − | − | + | + | − | − | − | − | − | − | − | − | − |
| T. bryantii | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − | − |
| E. ruminantium | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − | − |
| A. lipolytica | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − | − |
| S. dextrinosolvens | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − | − |
| R. flavefaciens | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | − | − | − | − |
See Table 1 for the corresponding primer set.
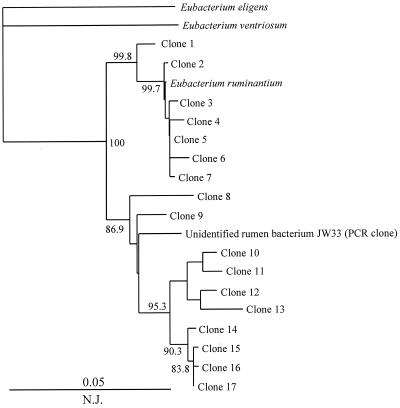
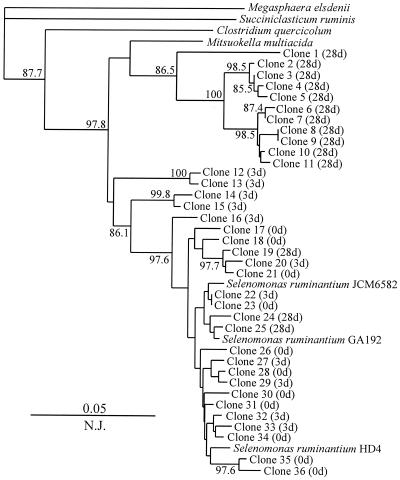
To verify the specificity of primers for analysis of complex microbial communities, total DNA was extracted from the rumen content, and amplifications were performed with each primer pair. Mini-libraries were constructed from each amplification reaction, and 12 to 36 randomly chosen clones from each of the 12 libraries were sequenced. The similarity values within each species-specific library were calculated (Table 3). In most cases, the similarity values within the species-specific libraries were well within the species definition range proposed on the basis of 16S rRNA similarity analysis (28). Despite the cross-reactivity of S. bovis primers with S. equinus (Table 2), the sequences retrieved with this set of primers formed a coherent group with a similarity value of 98.7% ± 0.5% (Table 3). It is necessary to note that during the database search, the S. bovis sequence displayed ≥97% similarity to other streptococcal sequences (S. infantarius, S. waius, S. gallolyticus, S. caprinus, S. intestinalis, and S. alactolyticus). Thus, on the basis of 16S rDNA sequences, these species are very similar, and other, less-conserved sequences are necessary for species-specific discrimination. Two primer sets, targeting E. ruminantium and S. ruminantium-M. multiacida, amplified sequences that had a lower degree of similarity (Table 3). Phylogenetic analysis of E. ruminantium-related sequences revealed two clusters: one grouping with the type strain and the other grouping with the in vitro-retrieved sequence of an unidentified rumen bacterium, JW33 (38) (Fig. 2). The latter cluster (with clones 8 to 17) has no cultivable representatives, and the availability of such a strain may help to define the new taxonomic boundaries within the Eubacterium species of the rumen and design specific primers for that group. At present, the E. ruminantium primer pair based on the type strain sequence detects the representatives of both these clusters. The S. ruminantium-M. multiacida primer set targets two species of rumen bacteria (Table 2), and the sequences amplified with this set of primers are clustered with M. multiacida and S. ruminantium sequences (Fig. 3).
TABLE 3.
Sequence analysis of clone libraries generated by species-specific primers from total rumen DNA samples
| Primer set | No. of clones sequenced | % Similarity (mean ± SD) |
|---|---|---|
| P. ruminicola | 24 | 98.5 ± 0.7 |
| P. bryantii | 14 | 98.6 ± 1.1 |
| P. albensis | 14 | 98.7 ± 0.5 |
| F. succinogenes | 25 | 97.6 ± 1.5 |
| R. amylophilus | 19 | 99.1 ± 0.4 |
| S. ruminantium-M. multiacida | 36 | 94.9 ± 4.1 |
| T. bryantii | 24 | 97.5 ± 1.7 |
| S. bovis | 12 | 98.7 ± 0.5 |
| E. ruminantium | 17 | 96.8 ± 2.3 |
| A. lipolytica | 21 | 97.6 ± 1.2 |
| S. dextrinosolvens | 21 | 98.1 ± 1.1 |
| R. flavefaciens | 25 | 98.2 ± 0.9 |
FIG. 2.
Phylogenetic placement of 16S rDNA sequences generated by an E. ruminantium primer set from total rumen DNA (day 3 of high-grain diet). The numbers represent the confidence levels (percentage) generated from 1,000 bootstrap trees. The scale bar is in fixed nucleotide substitutions per sequence position. Sequences 670 bp long were used in this analysis.
FIG. 3.
Phylogenetic placement of 16S rDNA sequences generated by an S. ruminatium-M. multiacida primer set from total rumen DNA on days 0 (0d), 3 (3d), and 28 (28d) of the switch to a high-grain diet. The numbers represent the confidence levels (percentage) generated from 1,000 bootstrap trees. The scale bar is in fixed nucleotide substitutions per sequence position. Sequences 513 bp long were used in this analysis.
Quantification of rumen bacteria during the diet switch.
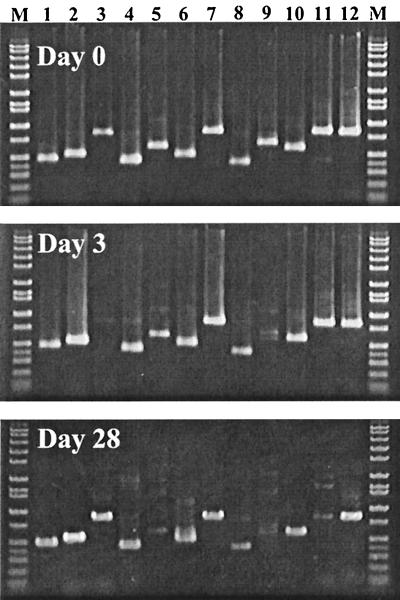
In the preliminary detection experiment with this primer set, we attempted to monitor at days 0, 3, and 28 of the experiment the presence of these bacterial species in the rumens of Holsteins for which the diet had been switched from hay to grain. At day 0, all targeted species were detected with our primer set (Fig. 4). At day 3, PCR signals of P. albensis and E. ruminantium were not detectable. At day 28, the intensity of the P. albensis signal was restored, but that of E. ruminantium was still not detectable. Also, the signals of S. dextrinosolvens and R. amylophilus were very weak in this sample (Fig. 4). All PCR products were of the expected size and were not contaminating by-products, which is essential for real-time PCR analysis.
FIG. 4.
Qualitative PCR detection of 12 bacteria in the rumens of cows for which the diet had been changed from hay to grain. Day 0, before the experiment, animals maintained on basal hay diet; day 3, animals fed a high-grain diet for 3 days; day 28, animals fed a high-grain diet for 28 days. Lanes: 1, P. ruminantium; 2, P. bryantii; 3, P. albensis; 4, F. succinogenes; 5, R. amylophilus; 6, S. ruminantium-M. multiacida; 7, S. bovis; 8, T. bryantii; 9, E. ruminantium; 10, A. lipolytica; 11, S. dextrinosolvens; and 12, R. flavefaciens. Lane M, DNA size marker.
The quantification of rumen bacteria during the diet switch was done with a LightCycler system under the optimized conditions described in Materials and Methods. The results of quantification are shown in Table 4. DNA of two bacteria, P. albensis and R. amylophilus, produced inconsistent amplification results with total DNA of the rumen contents, and these two species were omitted from analysis. The quantity of a fibrolytic bacterium, F. succinogenes, which was one of the major bacteria on the hay diet, fell 20-fold on the third day of the switch to a grain diet, and a further decline was observed on day 28, with a 57-fold reduction in F. succinogenes DNA (Table 4). A less dramatic reduction in DNA quantity was observed with the other fibrolytic bacterium, R. flavefaciens: the quantity of its DNA on day 3 declined to approximately 10% of its initial value in animals on the hay diet and remained at this level at day 28. During the transition period (day 3), the quantity of two ruminal prevotella DNAs increased considerably: that of P. ruminicola increased 7-fold, and that of P. bryantii increased 263-fold (Table 4). However, upon reaching the grain-adapted condition (day 28), the quantity of P. ruminicola DNA decreased 3-fold, while P. bryantii maintained the elevated level, outnumbering 10-fold the value found with the hay diet at the steady state (Table 4). The relative numbers of another xylanolytic bacterium, E. ruminantium, dropped 14-fold during the diet switch and were maintained at this level at the next measurement on day 28. The concentration of a rumen spirochete, Treponema bryantii, decreased less profoundly over the period of diet switch and stabilized with a sevenfold decline by day 28. The variations in A. lipolytica DNA were not statistically significant and therefore were not affected by diet change. After an initial slight increase in S. dextrinosolvens DNA on day 3, the DNA was not detected at the end of the experiment (Table 4). S. bovis was very responsive to the diet change and displayed a 67-fold increase during the transition period on day 3 (Table 4). However, the later measurement of the second grain diet steady state actually showed a twofold decline in comparison with the result for the hay diet at the steady state. The concentration of S. ruminantium-M. multiacida DNA also increased eightfold following the diet switch, but stabilized with only a twofold increase at the second steady state (Table 4). Sequencing the library of S. ruminantium-M. multiacida clones showed, however, that the contributions of these two species to the observed fluctuations are not equal: the transition period on day 3 is dominated by S. ruminantium, while the grain diet steady state is dominated by M. multiacida, with very few inclusions of S. ruminatium (Fig. 3).
Differential amplification of bacterial templates with universal bacterial primers.
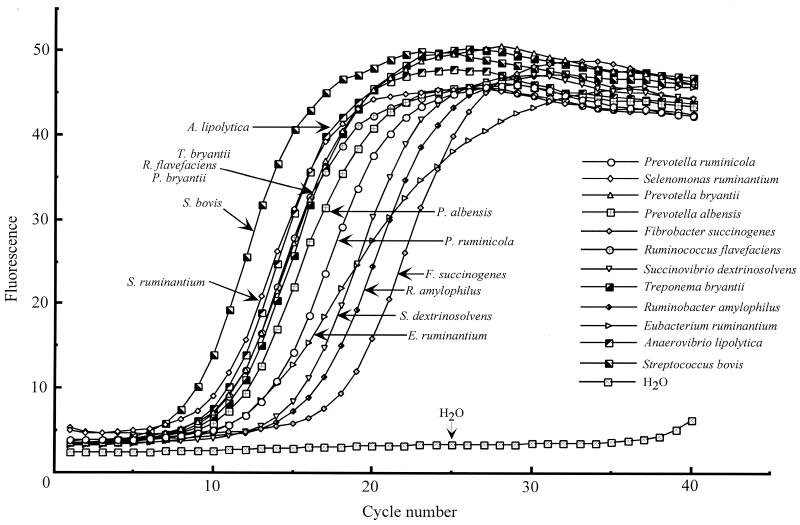
Another advantage offered by the real-time PCR approach is that the kinetics of amplification can be observed directly, thus allowing visual comparison of behaviors of several DNA templates under identical conditions. One problem associated with PCR-generated libraries is differential amplification of templates from a mix of environmental DNA, which distorts the real species distribution in a system (35). In molecular analysis of the rumen ecosystem, for example, several PCR-generated libraries reported by us and others (32, 33, 38) produced a total of 365 16S rDNA sequences, but the Fibrobacter-related sequence was encountered only once. This is in apparent contradiction to the results of the present study, and we attempted to test the amplification kinetics of individual rumen bacterial DNA templates (including F. succinogenes) under identical PCR conditions with the universal bacterial primer set 27f and 1525r (17). In these experiments, equal amounts (30 ng) of highly purified chromosomal DNAs were added to identical PCR mixes prepared from the master mix, but the kinetics of individual amplification varied widely (Fig. 5). For example, the threshold fluorescence for S. bovis was only 6.7 cycles, while the consistent fluorescence increase for the F. succinogenes template was extrapolated to occur only after the 15th cycle (Fig. 5). Other templates had intermediate cycle thresholds between these two extremes. The amplification kinetics of E. ruminantium DNA were different from those of the others and were actually nonexponential (Fig. 5), suggesting less-efficient annealing or extension of this template.
FIG. 5.
Differential amplification of rumen bacterial DNA templates with universal bacterial primers 27f and 1525r (17). Real-time PCR amplification was conducted essentially as described in Materials and Methods with 30 ng of each bacterial DNA template. PCR cycling was performed as follows: 95°C for 10 min of initial denaturation, then 40 cycles of 95°C for 15 s, 60°C for 5 s, and 72°C for 1 min. The fluorescence was captured at the end of the extension phase. The threshold fluorescence values were calculated with the LightCycler software and were as follows: S. bovis, 6.736 cycles; S. ruminantium, 8.375 cycles; A. lipolytica, 8.412 cycles; P. bryantii, 8.758 cycles; R. flavefaciens, 8.821 cycles; T. bryantii, 9.071 cycles; P. albensis, 9.592 cycles; P. ruminicola, 10.98 cycles; E. ruminantium, 10.28 cycles; S. dextrinosolvens, 12.59 cycles; R. amylophilus, 13.39 cycles; and F. succinogenes, 15.85 cycles.
DISCUSSION
The extreme complexity of the rumen microbiota has been uncovered in numerous publications employing isolation of pure cultures and description of their physiology. Based on this information, the putative contribution of these isolates to the overall metabolism and function of the rumen could be suggested. However, the next step—confirmation of the putative functionality by monitoring various functional groups by a cultivation-based approach—is very time- and labor-consuming, and the results, which are based on phenotypic characteristics, are not precise or conclusive. During the last decade, the use of molecular probes has become a popular approach in different fields of microbial ecology, and rumen microbiology is not exceptional in this regard. However, taking into consideration the enormous complexity of the rumen, the number of molecular probe tools specifically designed for monitoring the specific groups of microorganisms in this ecosystem is very limited. These few available molecular probes essentially target only seven bacterial species and one archeon (see the introduction). In this work, we designed and validated PCR primers for detection of 13 additional species of rumen bacteria. These primers were used in conjunction with real-time PCR, allowing accurate quantification of a target in a mix of total community DNA.
PCR primers for detection of 13 species of rumen bacteria were designed to satisfy the specificity and usability requirements of real-time PCR quantification. In the case of S. ruminantium and M. multiacida, it was not possible to design species-specific primers that would have sufficiently high melting temperatures for use in the LightCycler system. Therefore, this primer pair targets both species, but, if necessary, discrimination is still possible after sequence and phylogenetic analyses of the libraries produced with these primers (Fig. 3). Libraries obtained after amplification with E. ruminantium primers demonstrated a slightly higher level of diversity (96.8% of sequence similarity) than the ≥97% similarity sufficient for the integration into the same species nomination (28). Sequence and phylogenetic analyses (Fig. 2) demonstrated the existence of two clusters suggesting the necessity of additional species nominations. The second cluster, however, is represented exclusively by PCR-retrieved sequences, and further isolation and phenotypic characterization are necessary for new taxonomic designation. On the contrary, the 16S rDNA sequences of streptococci appeared to be less diverse, with many sequences having ≥97% sequence similarity, but which nevertheless have been elevated to the species level. An on-line similarity search demonstrated, for example, that S. bovis has 99% similarity to S. caprinus and 98% similarity to S. infantarius, S. waius, and S. alactolyticus, as well as being 97% similar to S. gallolyticus and S. intestinalis. Thus, the possibilities for designing only S. bovis-specific primers were limited, and our primer pair reacted with streptococci of other ecological origin, e.g., with S. infantarius and S. equinus. The close similarity of 16S rDNA sequences within the Streptococcus bovis-Streptococcus equinus complex has also been established in other investigations (8, 37), which suggests that less-conserved sequences, such as the intergenic region between 16S and 23S rDNAs, may be necessary for more accurate discrimination between them. In the present study, we assumed that the streptococci of other ecological origin are unlikely to be present in the rumen, but this requires further research. Another aspect of primer design was determining the primers' appropriateness for the LightCycler (or other real-time PCR) system. P. albensis and R. amylophilus primer pairs performed well with pure control cultures in both conventional and real-time PCR systems. Also, these two primer sets performed well in conventional PCR with total community DNA (Fig. 4). However, the results of their application in real-time PCR with total rumen DNA samples were inconsistent, supposedly because of very fast ramps between the denaturing and annealing temperatures and because of a short incubation time (5 s) during the annealing phase.
After validation, these primers were used to monitor and to quantify 11 rumen bacterial species during the diet shift from forage to grain. The dynamics of truly fibrolytic rumen bacteria were in good correlation with the diet change. The quantities of R. flavefaciens and F. succinogenes DNAs in total rumen DNA, which were arbitrarily taken as 100% on a hay diet, dropped correspondingly to more than 10- and 20-fold those found in animals on a grain diet. The R. flavefaciens data are also in good agreement with data from our previous experiments, in which we analyzed the 16S rDNA clone libraries produced from the same rumen DNA samples (33). In these experiments, R. flavefaciens-related sequences comprised 5.88% of all retrieved sequences from animals on the hay diet, but were undetectable (with a detection limit of ∼2%) for those on the grain diet (33). However, only a single F. succinogenes-related sequence has been found in several clone libraries reported to date (32, 33, 38), while our quantitative data do suggest that F. succinogenes may reach at least the same quantities as R. flavefaciens populations in animals on the hay diet. The clone libraries reported in these studies (32, 33, 38) were constructed with the universal bacterial primers, and we hypothesized that DNA of F. succinogenes may be amplified less efficiently than other bacterial templates present in a mix. Real-time PCR with various rumen bacterial templates and the universal bacterial primer set 27f and 1525r (17) confirmed that, under otherwise identical amplification conditions, this particular template has a prolonged lag phase compared with those of other templates (Fig. 5), which may be the reason for its underrepresentation in several clone libraries reported (32, 33, 38). Because the real-time PCR procedure uses the calibration curve obtained from pure cultures, more accurate quantification is possible in comparison with the PCR-generated clone libraries. Several factors that may contribute to differential amplification have been discussed (35). Presently recognized contributors are (i) genome size and rrn gene copy number, (ii) choice of primers and number of cycles, (iii) annealing efficiency and specificity of primers, (iv) G+C content, (v) DNA concentration, and (vi) DNA-associated molecules. In the case of F. succinogenes, this is definitely not a gene copy effect. In a previous work (20), we established that F. succinogenes possesses at least three rRNA operons, whereas an online search with the “fastest” S. bovis template checked against the genome of the taxonomically similar S. equi (http://www. sanger.ac.uk/Projects/S_equi/blast_server.shtml) produced only one high-scoring hit, suggesting this group may possess a single rRNA operon. Also, the efficiency of annealing and extension seems not to be a factor, as judged by the exponential increase in its fluorescence comparable to those of the other templates (Fig. 5). Poor annealing or extension appears to be a problem with the other bacterial template, E. ruminantium, which displays slower and nonexponential fluorescence kinetics compared with those of the other templates (Fig. 5). Since the difference in F. succinogenes amplification is largely attributed to the beginning of PCR cycling, the problem may be associated with the original DNA template, perhaps due to DNA-associated molecules.
Ruminal prevotella are known to possess oligosaccharolytic and xylanolytic activities and to occupy the ecological niches of the second line degraders (9). Comparative quantification of P. ruminicola on a hay diet suggested that this population is the most numerous among the populations studied. On a grain diet, the P. ruminicola count declines, but it still remains one of the predominant populations. The other representative of the genus, P. bryantii, demonstrated the opposite kinetics, suggesting its role in starch degradation. Both of these species demonstrated a tremendous increase during the transition period on day 3, and this observation correlates with our previous data from the clone libraries (33).
The saccharolytic spirochete T. bryantii has been shown to be associated with the fibrolytic bacteria of the rumen and, albeit not possessing any fibrolytic activity, could enhance fiber degradation in a coculture with fibrolytic bacteria (16, 30). In our experiment, the quantification of this bacterial DNA demonstrated kinetics similar to those of two fibrolytic bacteria, F. succinogenes and R. flavefaciens. The dynamic of two taxonomically different xylanolytic bacteria, E. ruminantium (belonging to low-G+C, gram-positive bacteria) and S. dextrinosolvens (belonging to the gamma subclass of Proteobacteria) also followed a similar decline during the diet switch, with the latter species not detectable on day 28. Based on the rate of lipolysis by pure cultures, A. lipolytica has been suggested to be an organism that may play an important role in the lipolytic activity of the rumen (23). However, no statistically significant changes were detected in the A. lipolytica DNA concentration during the shift to a grain diet containing increased amounts of lipids.
In our previous analysis of clone libraries generated from the rumen microbiota during the diet switch, we detected the numerical prevalence of low-G+C, gram-positive bacteria belonging to the Selenomonas-Succiniclasticum-Megasphaera group in Clostridium cluster IX in grain diet microbiota (33). The simultaneous quantification of two species in this group, S. ruminantium and M. multiacida, is in agreement with our earlier findings and demonstrates that these two bacteria represent the most numerous group in animals on a grain diet (Table 4). An amylolytic bacterium, S. bovis, has been considered as a major culprit in the development of lactic acidosis (19, 22), and selective inactivation of this bacterium by immunization results in reduced symptoms of lactic acidosis (11, 27). However, the absolute numbers of this bacterium seem to be low, and it was undetectable in our previous clone libraries with a detection limit of ∼2% (33). With the more sensitive approach implemented in this work, we were able to monitor its dynamics. Similarly with other amylolytic bacteria of the rumen, such as the prevotellas, S. bovis responded to the grain diet switch with a tremendous 67-fold increase. However, surprisingly, on the grain-adapted system, the numbers were twofold lower than on the hay diet. This suggests that, besides the amylolytic activity, this bacterium may possess other functional activities important for rumen digestion of plant polysaccharides.
To our knowledge, this is the first demonstration of the applicability of real-time PCR for quantification of bacterial species in a complex microbial ecosystem. Previous applications of this technique have been limited to detection and quantification of specific transcripts and, in clinical and veterinary microbiology, detection and quantification of pathogens, contaminants, and antibiotic resistance genes. We demonstrated that, with the availability of calibration strains, their dynamics could be accurately monitored in a complex mix, such as rumen content. DNA calibration curves could be based on the actual cell numbers, thus linking the cultivation and molecular detection methods. The approach implemented in this work can be applied to other microbial systems as well. In addition, the set of primers developed during this study not only is suitable for quantification purposes, but can also be used for rapid preliminary identification of other bacterial strains isolated from the rumen.
ACKNOWLEDGMENTS
The Laboratory of Rumen Microbiology was supported by grants from NIAI, Ministry of Agriculture, Forestry and Fisheries of Japan.
REFERENCES
- 1.Attwood G T, Lockington R A, Xue G P, Brooker J D. Use of a unique gene sequence as a probe to enumerate a strain of Bacteroides ruminicola introduced into the rumen. Appl Environ Microbiol. 1988;54:534–539. doi: 10.1128/aem.54.2.534-539.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birnboim H C, Doly J. A rapid alkaline extraction procedure for recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briesacher S L, May T, Grigsby K N, Kerley M S, Anthony R V, Paterson J A. Use of DNA probes to monitor nutritional effects on ruminal prokaryotes and Fibrobacter succinogenes S85. J Anim Sci. 1992;70:289–295. doi: 10.2527/1992.701289x. [DOI] [PubMed] [Google Scholar]
- 4.Bryant M P. Bacterial species of the rumen. Bacteriol Rev. 1959;23:125–153. doi: 10.1128/br.23.3.125-153.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldwell D R, Bryant M P. Medium without rumen fluid for nonselective enumeration and isolation of rumen bacteria. Appl Microbiol. 1966;14:794–801. doi: 10.1128/am.14.5.794-801.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ezaki T, Suzuki S. Achromopeptidase for lysis of anaerobic gram-positive cocci. J Clin Microbiol. 1982;16:844–846. doi: 10.1128/jcm.16.5.844-846.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ezaki T, Hashimoto Y, Takeuchi N, Yamamoto H, Liu S-L, Miura H, Matsui K, Yabuuchi E. Simple genetic methods to identify viridans group streptococci by colorimetric dot hybridization and fluorometric hybridization in microdilution wells. J Clin Microbiol. 1988;26:1708–1713. doi: 10.1128/jcm.26.9.1708-1713.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farrow J A E, Kruze J, Phillips B A, Bramley A J, Collins M D. Taxonomic studies on Streptococcus bovis and Streptococcus equinus: description of Streptococcus alactolyticus sp. nov. and Streptococcus saccharolyticus sp. nov. Syst Appl Microbiol. 1984;5:467–482. [Google Scholar]
- 9.Flint H J. The rumen microbial ecosystem—some recent developments. Trends Microbiol. 1997;5:483–488. doi: 10.1016/S0966-842X(97)01159-1. [DOI] [PubMed] [Google Scholar]
- 10.Forster R J, Gong J, Teather R M. Group-specific 16S rRNA hybridization probes for determinative and community structure studies of Butyrivibrio fibrisolvens in the rumen. Appl Environ Microbiol. 1997;63:1256–1260. doi: 10.1128/aem.63.4.1256-1260.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gill H S, Shu Q, Leng R A. Immunization with Streptococcus bovis protects against lactic acidosis in sheep. Vaccine. 2000;18:2541–2548. doi: 10.1016/s0264-410x(00)00017-7. [DOI] [PubMed] [Google Scholar]
- 12.Hungate R E. The rumen and its microbes. New York, N.Y: Academic Press; 1966. [Google Scholar]
- 13.Kobayashi Y, Foster R J, Teather R M. Development of a competitive polymerase chain reaction assay for the ruminal bacterium Butyrivibrio fibrisolvens OB156 and its use for tracking an OB156-derived recombinant. FEMS Microbiol Lett. 2000;188:185–190. doi: 10.1111/j.1574-6968.2000.tb09191.x. [DOI] [PubMed] [Google Scholar]
- 14.Krause D O, Dalrymple B P, Smith W J, Mackie R I, McSweeney C S. 16S rDNA sequencing of Ruminococcus albus and Ruminococcus flavefaciens: design of a signature probe and its application in adult sheep. Microbiology. 1999;145:1797–1807. doi: 10.1099/13500872-145-7-1797. [DOI] [PubMed] [Google Scholar]
- 15.Krause D O, Smith W J, Ryan F M, Mackie R I, McSweeney C S. Use of 16S-rRNA based techniques to investigate the ecological succession of microbial populations in the immature lamb rumen: tracking of a specific strain of inoculated Ruminococcus and interactions with other microbial populations in vivo. Microb Ecol. 1999;38:365–376. doi: 10.1007/s002489901006. [DOI] [PubMed] [Google Scholar]
- 16.Kudo H, Cheng K J, Costerton J W. Interactions between Treponema bryantii and cellulolytic bacteria in the in vitro degradation of straw cellulose. Can J Microbiol. 1987;33:244–248. doi: 10.1139/m87-041. [DOI] [PubMed] [Google Scholar]
- 17.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- 18.Madden T L, Tatusov R L, Zhang J. Application of network BLAST server. Methods Enzymol. 1996;266:131–141. doi: 10.1016/s0076-6879(96)66011-x. [DOI] [PubMed] [Google Scholar]
- 19.Nocey J E. Bovine acidosis: implications on laminitis. J Dairy Sci. 1995;80:1005–1028. doi: 10.3168/jds.S0022-0302(97)76026-0. [DOI] [PubMed] [Google Scholar]
- 20.Ogata K, Aminov R I, Nagamine T, Sugiura M, Tajima K, Mitsumori M, Sekizaki T, Kudo H, Minato H, Benno Y. Construction of a Fibrobacter succinogenes genomic map and demonstration of diversity at the genomic level. Curr Microbiol. 1997;35:22–27. doi: 10.1007/s002849900205. [DOI] [PubMed] [Google Scholar]
- 21.Ogimoto K, Imai S. Atlas of rumen microbiology. Tokyo, Japan: Japan Scientific Society Press; 1981. [Google Scholar]
- 22.Owens F N, Secrist D S, Hil W J, Gill D R. Acidosis in cattle: a review. J Anim Sci. 1998;76:275–286. doi: 10.2527/1998.761275x. [DOI] [PubMed] [Google Scholar]
- 23.Prins R A, Lankhorst A, van der Meer P, Van Nevel C J. Some characteristics of Anaerovibrio lipolytica, a rumen lipolytic organism. Antonie Leeuwenhoek. 1975;41:1–11. doi: 10.1007/BF02565031. [DOI] [PubMed] [Google Scholar]
- 24.Ramsak A, Peterka M, Tajima K, Martin J C, Wood J, Johnston M E, Aminov R I, Flint H J, Avgustin G. Unravelling the genetic diversity of ruminal bacteria belonging to the CFB phylum. FEMS Microbiol Ecol. 2000;33:69–79. doi: 10.1111/j.1574-6941.2000.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 25.Reilly K, Attwood G T. Detection of Clostridium proteoclasticum and closely related strains in the rumen by competitive PCR. Appl Environ Microbiol. 1998;64:907–913. doi: 10.1128/aem.64.3.907-913.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito N, Nei M. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 27.Shu Q, Gill H S, Leng R A, Rowe J B. Immunization with a Streptococcus bovis vaccine administered by different routes against lactic acidosis in sheep. Vet J. 2000;159:262–269. doi: 10.1053/tvjl.1999.0400. [DOI] [PubMed] [Google Scholar]
- 28.Stackebrandt E, Goebel B M. Taxonomic notice: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 29.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanton T B, Canale-Parola E. Treponema bryantii sp. nov., a rumen spirochete that interacts with cellulolytic bacteria. Arch Microbiol. 1980;127:145–156. doi: 10.1007/BF00428018. [DOI] [PubMed] [Google Scholar]
- 31.Stewart C S, Bryant M P. The rumen bacteria. In: Hobson PN, editor. The rumen microbial ecosystem. London, United Kingdom: Elsevier Applied Science; 1988. pp. 21–76. [Google Scholar]
- 32.Tajima K, Aminov R I, Nagamine T, Ogata K, Nakamura M, Matsui H, Benno Y. Rumen bacterial diversity as determined by sequence analysis of 16S rDNA libraries. FEMS Microbiol Ecol. 1999;29:159–169. [Google Scholar]
- 33.Tajima K, Arai S, Ogata K, Nagamine T, Matsui H, Nakamura M, Aminov R I, Benno Y. Rumen bacterial community transition during adaptation to high-grain diet. Anaerobe. 2000;6:273–284. [Google Scholar]
- 34.Thompson J D, Higgins D G, Gibson T J. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Wintzingerode V, Gobel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 36.Weimer P J, Waghorn G C, Odt C L, Mertens D R. Effect of diet on populations of three species of ruminal cellulolytic bacteria in lactating dairy cows. J Dairy Sci. 1999;82:122–134. doi: 10.3168/jds.S0022-0302(99)75216-1. [DOI] [PubMed] [Google Scholar]
- 37.Whitehead T R, Cotta M A. Development of molecular methods for identification of Streptococcus bovis from human and ruminal origins. FEMS Microbiol Lett. 2000;182:237–240. doi: 10.1111/j.1574-6968.2000.tb08901.x. [DOI] [PubMed] [Google Scholar]
- 38.Whitford M F, Foster R J, Beard C E, Gong J, Teather R M. Phylogenetic analysis of rumen bacteria by comparative sequence analysis of cloned 16S rRNA genes. Anaerobe. 1998;4:153–163. doi: 10.1006/anae.1998.0155. [DOI] [PubMed] [Google Scholar]
- 39.Yanagita K, Kamagata Y, Kawaharasaki M, Suzuki T, Nakamura Y, Minato H. Phylogenetic analysis of methanogens in sheep rumen ecosystem and detection of Methanomicrobium mobile by fluorescence in situ hybridization. 2000. [DOI] [PubMed] [Google Scholar]