Abstract
The present review focuses on the roles and underlying mechanisms of action of hepatic nuclear factor-1 (HNF-1) in lipid metabolism and the development of lipid metabolism disorders. HNF-1 is a transcriptional regulator that can form homodimers, and the HNF-1α and HNF-1β isomers can form heterodimers. Both homo- and heterodimers recognize and bind to specific cis-acting elements in gene promoters to transactivate transcription and to coordinate the expression of target lipid-related genes, thereby influencing the homeostasis of lipid metabolism. HNF-1 was shown to restrain lipid anabolism, including synthesis, absorption, and storage, by inhibiting the expression of lipogenesis-related genes, such as peroxisome proliferator-activated receptor γ (PPARγ) and sterol regulatory element-binding protein-1/2 (SREBP-1/2). Moreover, HNF-1 enhances the expression of various genes, such as proprotein convertase subtilisin/kexin type 9 (PCSK9), glutathione peroxidase 1 (GPx1), and suppressor of cytokine signaling-3 (SOCS-3) and negatively regulates signal transducer and activator of transcription (STAT) to facilitate lipid catabolism in hepatocytes. HNF-1 reduces hepatocellular lipid decomposition, which alleviates the progression of nonalcoholic fatty liver disease (NAFLD). HNF-1 impairs preadipocyte differentiation to reduce the number of adipocytes, stunting the development of obesity. Furthermore, HNF-1 reduces free cholesterol levels in the plasma to inhibit aortic lipid deposition and lipid plaque formation, relieving dyslipidemia and preventing the development of atherosclerotic cardiovascular disease (ASCVD). In summary, HNF-1 transcriptionally regulates lipid-related genes to manipulate intracorporeal balance of lipid metabolism and to suppress the development of lipid metabolism disorders.
Keywords: ASCVD, HNF-1, Lipid metabolism, NAFLD, Obesity
Abbreviations
- HNF-1
hepatic nuclear factor-1
- PPARγ
peroxisome proliferator-activated receptor-γ
- ACC
acetyl-CoA carboxylase
- FAS
fatty acid synthetase
- L-FABP
liver fatty acid-binding protein
- SOCS-3
suppressor of cytokine signaling-3
- STAT
signal transducer and activator of transcription
- SigR1
sigma receptor 1
- DPP4
dipeptidyl peptidase 4
- NOX1
nicotinamide adenine dinucleotide phosphate oxidase 1
- PCSK9
proprotein convertase subtilisin/kexin type 9
- Angptl8
angiopoietin-like protein8
- SREBP-1/2
sterol regulatory element-binding transcription factor-1/2
- GPx1
glutathione peroxidase 1
- HL
hepatic lipase
- LPL
lipoprotein lipase
- A1254
aroclor 1254
- NAFLD
nonalcoholic fatty liver disease
- ASCVD
atherosclerotic cardiovascular disease
- NLS
nuclear localization signal
- ADI/II/III
activation domain I/II/III
- TFIID
transcription factor II D
- LDL-c
low density lipoprotein-cholesterol
- HDL-c
high density lipoprotein-cholesterol
- TG
triglycerides
- apoA/B
apolipoprotein A/B
- SNP
single nucleotide polymorphism
Introduction
Lipid homeostasis is critical to normal cellular and systemic functions and is fundamental for the maintenance of normal physiological functions of the organisms. An imbalance between lipid anabolism and catabolism can result in an increase in aberrant lipid deposition and lipid metabolic syndromes, such as nonalcoholic fatty liver disease (NAFLD),1,2 obesity,3 and atherosclerotic cardiovascular diseases (ASCVDs).4 The incidence rate of lipid metabolism disorders has continued to increase in recent decades, and these disorders have become the greatest threat to human health. Therefore, unravelling the regulatory mechanisms underlying lipid homeostasis may create a unique therapeutic window for treatment of lipid metabolism disorders by correction of lipid metabolism.
Hepatic nuclear factor-1 (HNF-1) is a protein important for development that was originally considered to be restricted to hepatocytes. HNF-1 has been conserved throughout evolution and is the promoters of the albumin gene and other hepatic-specific promoters in various species, including Xenopus, mice, rats, and humans; HNF-1 is critical for the initiation and maintenance of transcription of specific genes in vivo.5 A growing number of studies have highlighted the role of HNF-1 in various aspects of lipid metabolism.6, 7, 8 Abnormal expression and function of HNF-1 predispose patients to disturbances in lipid metabolism and elevate their risk for developing lipid disorder-based diseases.9,10
Although HNF-1 participates in the regulation of lipid metabolism, the understanding of specific regulatory mechanisms is limited by a lack of cohesive information and is superficial. For instance, as a key regulator of homeostasis of lipid metabolism, HNF-1 targets biological effectors of lipid metabolism, and aberrant expression and/or function of HNF-1 disrupts lipid homeostasis, resulting in the onset and progression of lipid metabolism disorders. The role of HNF-1 in extrahepatic lipid metabolism, especially in the cardiovascular system, has rarely been reported. The lack of exploration into these uncharted areas greatly limits the application of HNF-1 in basic research and clinical therapy.
To understand the roles of HNF-1 in the regulation of lipid metabolism and the development of lipid dysmetabolism, we provide a brief description of biochemical, structural, and biological properties of HNF-1, assess the effects of HNF-1 on lipid metabolism and related target genes implicated in lipid-regulating pathways, and finally summarize the roles of HNF-1 in lipid metabolism disorders. The present review emphasizes the significance of HNF-1 in the regulation of lipid metabolism and provides insight into the value of HNF-1 for the prevention and treatment of lipid metabolism disorders.
What is HNF-1?
HNF-1 is a homodimeric or heterodimeric transcriptional regulator formed by the HNF-1α and HNF-1β isoforms.11 The HNF-1α and HNF-1β genes are located on chromosomes 12 and 17, respectively.9 The HNF-1 mRNA precursor undergoes modification and splicing, which produces a mature mRNA containing 9 directly connected exons. This mature mRNA enters the cytoplasm through the nuclear pores to direct polypeptide synthesis on ribosomes. The peptide chain is folded into a coiled α-helix within the endoplasmic reticulum, and a dimer-polypeptide precursor is processed into a mature dimeric protein in the Golgi apparatus.12 Subcellular localization analysis indicated that HNF-1 is a nuclear protein with a nuclear localization signal (NLS) formed by 8 amino acids (aa 197-205). Nuclear importin acts as an HNF-1 receptor that recognizes and binds the NLS sequence of HNF-1; subsequently, the importin–HNF-1 complex is transported into the nucleus by an energy-dissipating mechanism to perform its biological functions.13
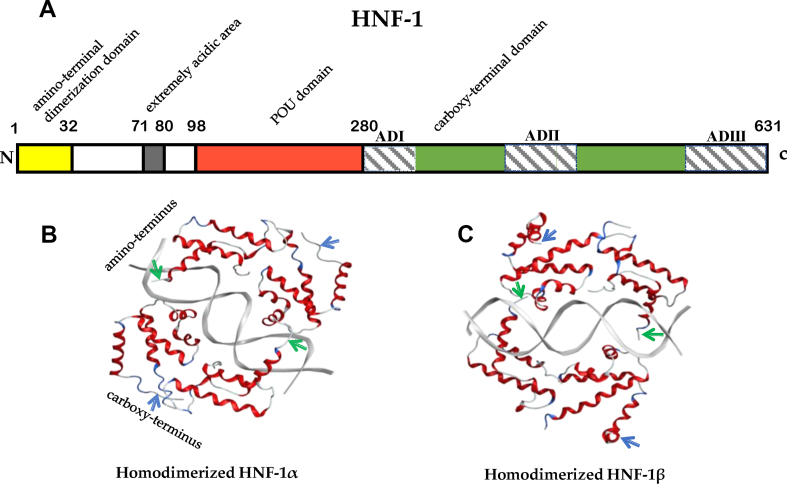
Human HNF-1 is a polypeptide composed of 631 amino acid residues. The first 280 amino acid residues of the polypeptide chain form an amino-terminal dimerization domain (aa 1-32), an extremely acidic area (aa 71-80), and a POU domain (aa 98-280).14 The dimerization domains are rich in α-helices that form an X-type four-helix bundle and are crucially important for homodimerization and heterodimerization of the HNF-1α and HNF-1β proteins.15 The POU domain includes the DNA-binding region with high homology among species. The DNA-binding regions of HNF-1 dimers bind to the conserved DNA sequence 5′GTTAATNATAAC3'.16,17 The conserved crimp structure of the DNA-binding region is stabilized by charged residues and hydrophobic bonds formed by the dimerization domain.16 The extremely acidic area and carboxy-terminal domain (aa 281-631) of the polypeptide chain are responsible for transcriptional activation effects of HNF-1.16 Three different transactivation domains are located in the carboxy terminus: activation domain I (ADI) includes amino acids 546-628 and contains abundant serine residues (26%); ADII includes amino acids 281-318; and ADIII includes amino acids 440-506 and contains 46.2% proline and serine residues.18 C-terminal amino acid residues include 48 amino acid residues within ADI and ADIII that are essential for transcriptional activation. The C-terminal region of HNF-1α includes 170 amino acid residues highly enriched in proline and provides 85% of the transactivation potential of HNF-1α.19 Moreover, HNF-1α can directly interact with three activation sites in the promoters to transactivate transcription of the target genes, and HNF-1β inactivates transcription of the target genes because it lacks ADI and ADIII (Fig. 1).16
Figure 1.
The structures of the HNF-1 protein. (A) A scheme of the structure of the HNF-1 protein. The polypeptide chain of human HNF-1 contains 631 amino acid residues, and the first 280 amino acid residues form an amino-terminal dimerization domain (aa 1-32), an extremely acidic area (aa 71-80), and a POU domain (aa 98-280). The carboxy-terminal domain (aa 281-631) is responsible for transcriptional activation by HNF-1 and contains three transactivation domains: activation domain I (ADI), including amino acids 546-628; ADII, including amino acids 281-318; and ADIII, including amino acids 440-506. (B) Three-dimensional structures of the HNF-1α homodimer. (C) Three-dimensional structures of the HNF-1β homodimer.  , amino-terminus;
, amino-terminus;  , carboxy-terminus.
, carboxy-terminus.
HNF-1 has been shown to recognize and bind with specific activation sites to enhance the expression of the target genes.17 Activation sites include the cis-acting CAAT box element in the promoter of the target genes and are critical for the activation of transcription in response to HNF-1. The activation sites of HNF-1 are located in the -84/-68 promoter region of the angiopoietin-like 8 (Angptl8) gene in human liver cells, in the upstream 100-200 amino acid-encoding base pair (bp) promoter region of the human albumin gene, and in the first 200 bp of the human CRP promoter (APRE2 and APREL regions).14,17,20 HNF-1 interacts with the activation sites of the target genes through its C-terminus, especially the proline residues in ADIII, which bind to the CAAT box of a promoter to transactivate the transcription of the target genes. The transactivation activity of HNF-1 mainly depends on the synergistic effect of ADI and ADIII, with ADII being nonfunctional.18 Additionally, the extremely acidic region of the HNF-1 protein has a conserved sequence rich in aspartic acid residues that forms a negatively charged β-fold that facilitates the assembly of the transcriptional initiation complex by interacting with transcription factor II D (TFIID) and initiating the transcription of the target genes.20
Accumulating evidence indicates that HNF-1 transcripts are expressed in the intestine, pancreas, kidney, and liver.21 HNF-1 participates in the regulation of cholesterol metabolism and fatty acid uptake and synthesis and influences homeostasis of lipid metabolism in hepatocytes and adipocytes.4,22,23 Mutations in HNF-1α can lead to enhanced adipogenesis in the liver; downregulation of HNF-1β causes liver steatosis, and a lack of HNF-1 leads to lipid dysmetabolism in the liver, pancreas, small intestine, and adipose tissue.24, 25, 26, 27 Mechanistic studies demonstrated that HNF-1 transcriptionally controls lipid-related genes ubiquitously involved in lipid metabolism and participates in the onset and progression of a variety of lipid metabolism disorders.26,28
HNF-1 in lipid metabolism
Epidemiological investigations have demonstrated associations between HNF-1 and lipid metabolism. Genome-wide association studies determined that the levels of plasma low-density lipoprotein-cholesterol (LDL-c) and triglycerides (TG) were higher and the levels of high-density lipoprotein-cholesterol (HDL-c) were lower in 19,840 individuals (and in 20,623 additional individuals) with HNF-1α deficiency due to mutations.29, 30, 31 Analysis of associations between common polymorphisms of the HNF-1α gene and plasma LDL-c and apolipoprotein B (apoB) levels indicated that deletions of the HNF-1α gene resulted in elevated levels of serum lipids in healthy young European Americans and African Americans.4,22 The results of polymorphism analysis indicated that HNF-1α phenotypes influence the serum HDL-c levels of 356 Japanese men; specifically, the subjects with the isoleucine phenotype had low serum HDL-c levels, and the subjects with the leucine phenotype had high serum HDL-c levels.32 Two single nucleotide polymorphisms (SNPs) of HNF-1α (rs2259820T and rs2464196A alleles) are significantly associated with higher levels of serum apoA1 and consistently reduced risk of coronary artery disease in the Chinese population.33 This epidemiological evidence indicates an important role of HNF-1 in the regulation of intracorporeal lipid metabolism.
HNF-1 directly participates in the regulation of lipid metabolism in vitro. Hepatic HNF-1α transcriptionally upregulated proprotein convertase subtilisin/kexin type 9 (PCSK9), which reduced LDL endocytosis and cholesterol uptake by mediating the degradation of LDL receptors and increasing the plasma LDL-c levels.34,35 Shende's group demonstrated that HNF-1α knockdown decreased circulating levels of PCSK9 protein and increased the intracellular accumulation of cholesterol in HepG2 hepatocytes and in primary hepatocytes of C57BL/6J mice; however, HNF-1β knockdown did not cause these alterations in vitro.34,36 Hu et al demonstrated that the loss of HNF-1α function resulted in a sharp accumulation of lipid droplets and promoted intracellular cholesterol accumulation in HepG2 hepatocytes transfected with HNF-1α siRNA.36, 37, 38 Wade and colleagues showed that HNF-1α overexpression stimulated apoA transcription, resulting in higher levels of lipoprotein A and lipid droplet accumulation in HepG2 cells.39 Furthermore, Wang's group confirmed these findings by showing that HNF-1β downregulation dramatically enhanced the differentiation of 3T3-L1 preadipocytes transfected with HNF-1β shRNA lentivirus and increased lipid droplet formation.40 These findings indicated that HNF-1 plays an important role in the regulation of intracellular lipid metabolism.
HNF-1 manipulates lipid metabolism processes. Shende and colleagues demonstrated that liver-specific knockdown of HNF-1α, but not HNF-1β, reduced the serum level of PCSK9, leading to a lower level of circulating LDL-c in C57BL/6J mice infected with shHNF-1α adenovirus.34 Taro et al demonstrated that HNF-1α deficiency promoted the expression of the enzymes of fatty acid synthesis and FAT genes, accelerating hepatic lipid accumulation and elevating serum levels of TG in HNF-1α-null mice with mutations generated by the Cre-loxP deletion system.41 Sandra Rebouissou et al showed that loss-of-function mutations of HNF-1α caused an aberrant promotion of lipogenesis in human hepatocellular adenomas.8,36,42 Additionally, Shih's group showed that HNF-1α knockout diminished the expression of bile acid transporters and enhanced the activity of cholesterol acyl transferase to increase the rate of synthesis of bile acids and cholesterol in the liver; HNF-1α deletion in Tcf1−/− mice was characterized by reduced hepatic lipase (HL) activity that impaired HDL catabolism.22,36,43 These results emphasize that HNF-1 is involved in intracorporeal aspects of lipid metabolism.
Pathways of HNF-1 in lipid metabolism regulation
HNF-1 is an important transcription factor that modulates the expression of lipid-related genes involved in lipid metabolism. HNF-1 can recognize and bind with specific cis-acting elements within gene promoters to transactivate the transcription and coordinate the expression of target lipid-related genes. HNF-1 is predominantly expressed in hepatic tissue and is involved in various aspects of hepatic lipid metabolism, including lipid uptake and biosynthesis, lipid transport and intracellular lipid storage, and oxidation of fatty acids. HNF-1 also influences lipid synthesis and absorption in adipocytes and modulates the expression of the genes related to adipocyte differentiation and fatty acid metabolism. HNF-1 controls the homeostasis of normal lipid metabolism via multiple regulatory pathways under physiological conditions (Table 1).
Table 1.
Regulatory pathways of HNF-1 in lipid metabolism.
| HNF-1 intervention | Animal/cell model | Targeted factors | Binding site | Lipid metabolism | References |
|---|---|---|---|---|---|
| HNF-1α↓ | HepG2 hepatocytes | miR-122↓ | --- | Lipid synthesis and absorption↑ | Hu et al (2020)36 Shende et al (2015)34 |
| HNF-1α-null mice | L-FABP↓ | 343-328 bp and 107-93 bp upstream of L-FABP promoter | Adipogenesis↑ | Akiyama et al (2000)41 Gordon et al (1985)47 Graham et al (2016)48 |
|
| LO2 hepatocytes | SOCS-3↓ | --- | Lipid synthesis and absorption↑ | Tan et al (2019)6 | |
| LO2 hepatocytes | STAT↑ | --- | Lipid synthesis and absorption↑ | Tan et al (2019)6 | |
| HepG2 and Huh7 cells | SigR1↑ | --- | Adipogenesis↑ | Villemain et al (2020)49 | |
| normolipidemic mice | PCSK9↓ | 380 bp upstream of promoter | LDLR degradation↓ | Shende et al (2015)34 Li et al (2015)35 |
|
| HNF-1α↑ | HNF-1α-null mice | PPARγ↓ | --- | Adipogenesis↓ | Lefterova et al (2009)45 Patitucci et al (2017)46 |
| HNF-1α/HNF-1β | C57BL mice | Angptl8↑ | 84-68 bp upstream of Angptl8 promoter | Lipid degradation↓ | Watanabe et al (2020)17 Wang et al (2013)50 |
| HNF-1β↓ | C57BL/6J mice | DPP4↑ | --- | Lipid oxidation↓ | Long et al (2017)1 Seidah et al (2003)2 |
| C57BL/6J mice | NOX1↑ | --- | Lipid oxidation↓ | Long et al (2017)1 Seidah et al (2003)2 |
|
| 3T3-L1 preadipocytes | PPARγ↑ | --- | Adipogenesis↑ | Wang et al (2017)40 Patitucci et al (2017)46 Wang et al (2020)51 |
|
| HNF-1β↑ | AML-12 hepatocytes | SREBP-1↓ | --- | Lipid synthesis and absorption↓ | Hu et al (2020)36 Wu et al (2017)44 |
| AML-12 hepatocytes | ACC↓ | --- | Lipid synthesis and absorption↓ | Wu et al (2017)44 | |
| AML-12 hepatocytes | L-FABP↓ | Lipid synthesis and absorption ↓ | Wu et al (2017)44 | ||
| AML-12 hepatocytes | FAS↓ | --- | Lipid synthesis and absorption ↓ | Wu et al (2017)43 | |
| C57BL/6J mice | GPx1↑ | --- | Lipid oxidation↑ | Long et al (2017)1 Seidah et al (2003)2 Wu et al (2017)44 |
Remarks: PPARγ, peroxisome proliferator-activated receptor-γ; ACC, acetyl-CoA carboxylase; FAS, fatty acid synthetase; L-FABP, liver fatty acid-binding protein; SOCS-3, suppressor of cytokine signaling-3; STAT, signal transducer and activator of transcription; SigR1, sigma receptor 1; DPP4, dipeptidyl peptidase 4; NOX1, nicotinamide adenine dinucleotide phosphate oxidase 1; PCSK9, proprotein convertase subtilisin/kexin type 9; Angptl8, angiopoietin-like 8; SREBP-1, sterol regulatory element binding transcription factor-1; GPx1, glutathione peroxidase 1. ↑, increased; ↓, decreased. ---, not determined.
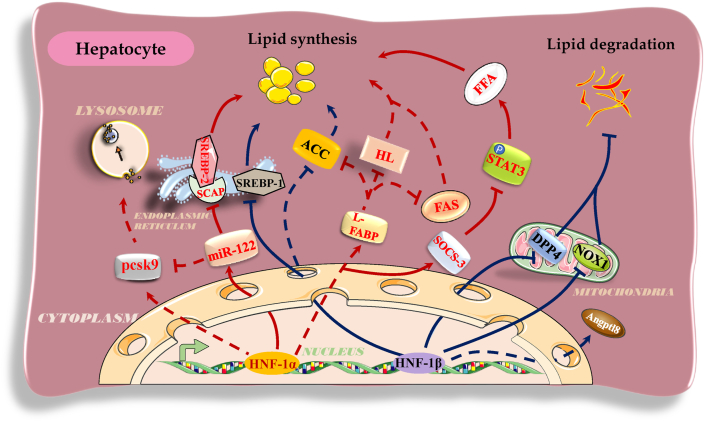
HNF-1 plays a pivotal role in lipid anabolism, including lipid synthesis, absorption, and storage. For example, HNF-1α directly upregulated the transcription of miR-122 to enhance miR-122-inhibited SCAP expression and to interfere with the maturation of SREBP-2, resulting in a decrease in lipid biosynthesis and lipid uptake by HepG2 hepatocytes.34,36 Similarly, overexpression of HNF-1β significantly decreased the expression of SREBP-1 and lipid-related synthases, such as liver fatty acid-binding protein (L-FABP), acetyl-CoA carboxylase (ACC), and fatty acid synthetase (FAS), leading to the suppression of lipid synthesis and absorption by AML-12 hepatocytes.44 HNF-1α overexpression transcriptionally repressed peroxisome proliferator-activated receptor γ (PPARγ) to abolish PPARγ-induced adipocyte differentiation in HNF-1α-null mice.45,46 HNF-1α binding to the regions 343-328 bp and 107-93 bp upstream of the L-FABP promoter increased the transcription of L-FABP mRNA to repress the expression of HL, ACC, and FAS, ultimately decreasing hepatic lipid generation and reducing serum TG levels in HNF-1α-null mice.41,47,48 HNF-1α promoted the expression of suppressor of cytokine signaling-3 (SOCS-3) and negatively regulated the signal transducer and activator of transcription (STAT) pathway by reducing the rate of STAT3 phosphorylation, resulting in inhibition of lipid anabolism or accelerated lipolysis and preventing free fatty acid-induced steatosis in LO2 hepatocytes.6 HNF-1α restrained the expression of sigma receptor 1 (SigR1) to reduce lipid droplet formation and storage capacity of neutral lipids in HepG2 and Huh7 cells, eventually preventing hepatic steatosis in hepatocellular adenoma.49 Dipeptidyl peptidase 4 (DPP4)/nicotinamide adenine dinucleotide phosphate oxidase 1 (NOX1) enhances the production of TG and the formation of lipid droplets in hepatocytes. HNF-1β binding to the promoters of DPP4 and NOX1 weakened the expression of these genes to restore hepatic TG homeostasis and alleviate liver injury and insulin resistance in C57BL/6J mice.1,2
HNF-1 plays an important role in lipid catabolism by accelerating lysosomal degradation and activating mitochondrial beta oxidation. Direct binding of HNF-1α to the region 380 bp upstream of the PCSK9 promoter increased hepatic and circulating PCSK9 levels, ultimately increasing the lysosomal degradation rate of hepatocellular LDLR and serum LDL-c level in normolipidemic mice.34,35 Additionally, hepatic HNF-1α indirectly inhibited the expression of PCSK9 by increasing miR-122 expression,36 indicating that this interaction may form a feedback pathway through which HNF-1α accurately controls the expression of PCSK9 and stabilizes the balance of lipid metabolism and plasma LDL-c levels. Direct binding of HNF-1α/HNF-1β to the regions 84 and 68 bp upstream of the Angptl8 promoter elevated the expression of this gene to subsequently inhibit the activity of lipoprotein lipase (LPL) and decreased the clearance rate of TG fatty acids in C57BL mice, ultimately severely disrupting TG metabolism.17,50 HNF-1 improves mitochondrial function to facilitate oxidative phosphorylation of fatty acids. For example, HNF-1β downregulated the transcription of PPARγ and of the downstream target genes regulated by aroclor 1254 (A1254), which disrupted mitochondrial function, accelerating the decomposition of TG, free fatty acids, and cholesterol via mitochondrial oxidative phosphorylation and ultimately suppressing the differentiation of 3T3-L1 cells into adipocytes.40,51 HNF-1β improved mitochondrial antioxidant potential and beta oxidation of fatty acids by promoting the transcription of glutathione peroxidase 1 (GPx1), leading to enhanced catabolism of cholesterol and TG and a reduction in the plasma lipid levels in C57BL/6J mice. HNF-1β inhibited the expression of both DPP4 and NOX1, induced the formation of a high superoxide environment, and resulted in a decrease in the glutathione content in the mitochondria, ultimately facilitating mitochondrial oxidative decomposition of lipids to maintain hepatic TG homeostasis and to prevent liver steatosis in C57BL/6J mice.1,2,44 These pathways are involved in the effects of HNF-1 to cooperatively maintain homeostasis of lipid metabolism under normal conditions. However, aberrant expression and function of HNF-1 under pathophysiological conditions disturb the balance of lipid metabolism and cause lipid metabolism diseases, such as NAFLD, obesity, and ASCVD.1,10,34,52, 53, 54
HNF-1 in lipid disorders
HNF-1 prevents the occurrence of NAFLD
NAFLD is an acquired dysfunctional metabolic syndrome mainly characterized by hepatic storage of excessive lipids and hepatocellular fatty degeneration resulting from the disorders of lipid metabolism in the liver.55 HNF-1 abated hepatocellular lipid synthesis by silencing the expression of sterol regulatory element-binding protein-1/2 (SREBP-1/2) and reduced the amount of free fatty acids and cholesterol internalized from extrahepatic tissues, preventing hepatic fat deposition and the development of NAFLD.36,44 HNF-1 countered STAT phosphorylation and the expression of DPP4/NOX1, ameliorating the oxidative function of mitochondria and hepatocellular lipid decomposition, which hindered hepatic lipidosis and the progression of NAFLD.1,2,6,56,57 Moreover, the deletion of HNF-1α/HNF-1β accelerated the synthesis of fatty acids and hepatic fat accumulation to boost the development of NAFLD in C57BL/6J mice. HNF-1β overexpression remarkably decreased hepatic lipid accumulation in both HF diet-fed mice and db/db mice.1,58 Inactivating HNF-1α mutations in exon 4 of the HNF-1α allele were reported to dramatically promote hepatocellular steatosis in a 60-year-old woman with hepatocellular adenoma.59 These data confirm that HNF-1 is a protective factor and potential target for NAFLD treatment (Fig. 2).
Figure 2.
The regulatory roles and pathways of HNF-1 in hepatocellular lipid metabolism and NAFLD. Abbreviations: PCSK9, proprotein convertase subtilisin/kexin type 9; miR-122, microRNA-122; SREBP-1/2, sterol regulatory element-binding protein-1/2; SCAP, SREBP cleavage-activating protein; ACC, acetyl-CoA carboxylase; L-FABP, liver fatty acid-binding protein; HL, hepatic lipase; FAS, fatty acid synthetase; SOCS-3, suppressor of cytokine signaling-3; STAT3, signal transducer and activator of transcription 3; FFA, free fatty acid; DPP4, dipeptidyl peptidase 4; NOX1, nicotinamide adenine dinucleotide phosphate oxidase 1; Angptl8, angiopoietin-like 8; and NAFLD, nonalcoholic fatty liver disease. Solid lines indicate regulatory pathways known to be associated with NAFLD, and dotted lines indicate regulatory pathways with unconfirmed associations with NAFLD. →, promotion; −, inhibition.
HNF-1 stunts the development of obesity
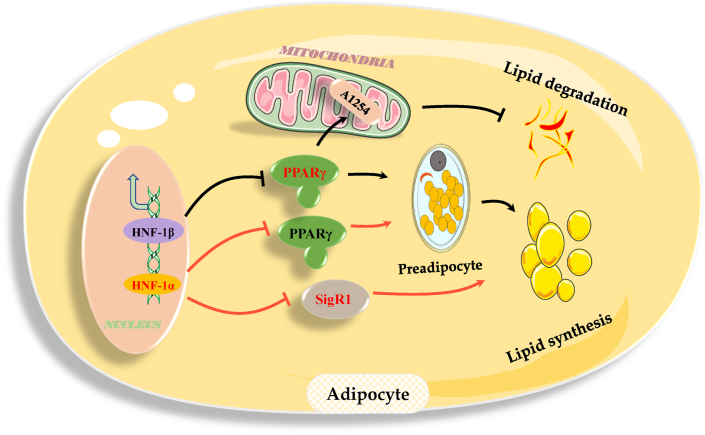
Obesity is due to dysregulation of the balanced control of body fat and energy metabolism that leads to abnormal distribution of body fat and surplus accumulation of lipids in white adipose tissue.60 HNF-1 impaired preadipocyte differentiation to reduce the number of adipocytes by inhibiting the expression of PPARγ.46 Moreover, HNF-1 repressed the expression of SigR1 to restrain the formation of lipid droplets in adipocytes, impeding adipogenesis and adipocyte hypertrophy.49 HNF-1β enhanced the oxidative function of mitochondria and facilitated adipolysis by restraining A1254-influenced expression, stunting the development of obesity.51 Additionally, HNF-1 suppressed excessive accumulation of lipids, such as TG, in white adipose tissue and significantly reduced obesity, which impairs physical health.40,45 HNF-1β downregulation governed adipose differentiation in C57B6/J mice.61 The rs7957197 SNP of HNF-1α was associated with significant progression of obesity in a randomized controlled trial of dietary intervention.10 Approximately 20% of Caucasian patients with HNF-1α mutations are obese or overweight.62 Recent findings indicated that HNF-1 has a promising therapeutic potential for limiting and/or reducing overweight and obesity (Fig. 3).
Figure 3.
The regulatory roles and pathways of HNF-1 in lipid metabolism in adipocytes and obesity. Abbreviations: PPARγ, peroxisome proliferator-activated receptor-γ; SigR1, sigma receptor 1; A1254, aroclor 1254-regulated processes. Solid lines indicate regulatory pathways known to be associated with obesity, and dotted lines indicate regulatory pathways with unconfirmed associations with obesity. →, promotion; −, inhibition.
HNF-1 affects the progression of ASCVD
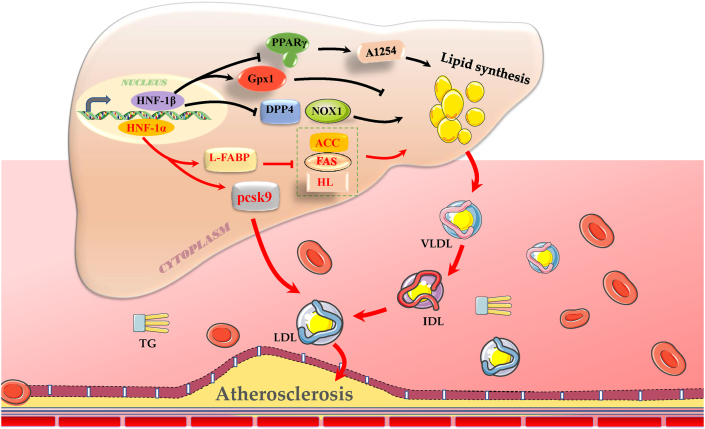
ASCVD is primarily ascribed to arterial atherosclerotic lesions, which are generally caused by atherogenic hyperlipidemia and lipoprotein disorders and lead to subintimal lipid deposition and the formation of atherosclerotic plaque.63,64 HNF-1 increased the expression of L-FABP and GPx1, facilitating hepatocellular decomposition of circulating lipids in mitochondria, decreasing the levels of lipids in the plasma, relieving dyslipidemia, and reducing the development of ASCVD.17,34,41,44,50 HNF-1 depressed the expression of PPARγ and DPP4/NOX1 to decrease lipid biosynthesis and accelerate lipid degradation, inhibiting aortic lipid deposition and lipid plaque formation and suppressing the occurrence of cardiovascular disease.1,39,46,65 The inhibition of HNF-1 resulted in a decrease in PCSK9 expression and circulating LDL-c levels, which ameliorated the development of ASCVD.66, 67, 68 The rs1183910 SNP of HNF-1α is associated with elevated serum levels of total cholesterol, LDL-c, and apoB in 60,283 individuals in the general Danish population.69 The HNF-1α G319S mutant genotype is associated with higher concentrations of plasma HDL-c and apoA1 in Oji-Cree subjects.31 In European Americans and African Americans, young adults carrying the rs1169288 and/or rs2464196 SNPs of HNF-1α have a higher risk of subclinical coronary atherosclerosis.4 These findings indicated that HNF-1 can influence the progression of atherosclerosis and is a potential contributor to a paradigm of ASCVD prevention (Fig. 4).
Figure 4.
The regulatory roles and pathways of HNF-1 in hepatic lipid metabolism and ASCVD. PPARγ, peroxisome proliferator-activated receptor γ; A1254, aroclor 1254-regulated processes; GPx1, glutathione peroxidase 1; DPP4, dipeptidyl peptidase 4; NOX1, nicotinamide adenine dinucleotide phosphate oxidase 1; L-FABP, liver fatty acid-binding protein; ACC, acetyl-CoA carboxylase; FAS, fatty acid synthetase; HL, hepatic lipase; LDL, low density lipoprotein; TG, triglyceride; VLDL, very low density lipoproteins; IDL, intermediate density lipoprotein; and ASCVD, atherosclerotic cardiovascular disease. Solid lines indicate regulatory pathways known to be associated with ASCVD, and dotted lines indicate regulatory pathways with unconfirmed associations with ASCVD. →, promotion; −, inhibition.
Conclusions, limitations, and prospects
HNF-1 is an essential transcription factor participating in lipid metabolism regulation and is a potential target for the treatment of lipid metabolism disorders. HNF-1 recognizes and binds to specific cis-acting elements in the gene promoters to induce the transcription and expression of the target genes and to influence the processes of lipid metabolism. HNF-1 inhibits the expression of lipogenesis-related genes, such as PPARγ, SREBP-1/2, and SigR1, restraining lipid anabolism processes, including synthesis, absorption, and storage. Moreover, HNF-1 induces various genes, such as PCSK9, GPx1, and SOCS-3/STAT, facilitating lipid catabolism in hepatocytes. Furthermore, abnormal expression of HNF-1 was shown to cause the development of lipid metabolism disorders, including NAFLD, obesity, and ASCVD.
Despite the proven effects of HNF-1 on lipid metabolism, many challenges should be addressed in the future. HNF-1 has been found to combine with cis-acting elements, such as CAAT boxes, to manipulate gene expression and lipid metabolism. Considering that HNF-1 can upregulate or downregulate gene expression under various conditions, it is unclear which cis-acting elements bind HNF-1α/β in addition to the CAAT box to trigger various biological functions. The specific mechanisms of HNF-1-induced transactivation of transcription of the target genes subsequent to HNF-1 binding to cis-acting elements remain unclear. Unlike the HNF-1α isoform, HNF-1β lacks the transactivation ADI and ADIII within the C-terminus, which may explain the distinct activity of HNF-1β in controlling the expression of the target genes. Furthermore, it is unknown whether HNF-1α and HNF-1β work synergistically or individually to regulate the transcription of downstream genes. The anomalous expression and function of HNF-1 are strongly associated with lipid dysmetabolism. However, it needs to be determined whether the abnormal expression of HNF-1 leads to disordered lipid metabolism or lipid dysmetabolism induces the abnormal HNF-1 expression that in turn aggravates lipid metabolism disorders. Lipid disorders are risk factors for atherogenesis, and a few reports have implicated HNF-1 in the development of atherosclerosis, especially in the regulation of lipid metabolism in macrophages under pro-atherosclerotic conditions.
HNF-1 is widely recognized as a promising therapeutic target that has broad application prospects for diagnosis and treatment of the disorders of lipid metabolism, stimulating ongoing in-depth investigations. For instance, tissue specificity of HNF-1α/β expression and lipid metabolism disorders in which HNF-1α/β are predominantly involved should be comprehensively analyzed to facilitate possible dual applications of HNF-1α/β as diagnostic markers and as therapeutic targets in the clinic. A thorough understanding of the molecular mechanisms underlying the regulatory effects of HNF-1 on lipid metabolism will provide the foundation for clinical interventions involving HNF-1 in the therapy of specific disorders of lipid metabolism. Additionally, the downstream molecules targeted by HNF-1α/β and the underlying mechanism controlling HNF-1α/β expression await further investigation, which will help to design novel synthetic molecules and to use new tactics to combat lipid metabolism disorders. Moreover, recent documents have reported that a new redox regulator PCB-153 downregulating HNF-1β expression caused the lipid metabolic disorder in hepatocytes and adipocytes,44 and a novel synthetic chemical 7030B-C5 competing with HNF-1α to bind with the target genes to exert lipid-lowering and anti-atherosclerotic effects in atherosclerosis-prone animals,66 which further substantiates the therapeutic value of HNF-1 as a potential target for lipid metabolism disorders.
Conflict of interests
The authors declare no potential conflicts of interest with respect to the authorship and publication of this article.
Funding
This work was financially supported by the National Natural Sciences Foundation of China (No. 81770460), the Natural Science Foundation of Guangxi Zhuang Autonomous Region, China (No. 2019JJA140728), Scientific Research Foundation for the Excellent Youth of the Education Department of Hunan Province (No. 18B264), Aid Program (No. 2017KJ268), and Key Lab for Clinical Anatomy & Reproductive Medicine (No. 2017KJ182) of the Science and Technology Bureau of Hengyang City, China.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Shan Li, Email: 791816918@qq.com.
Yuncheng Lv, Email: anthony0723@163.com.
References
- 1.Long Z., Cao M., Su S., et al. Inhibition of hepatocyte nuclear factor 1b induces hepatic steatosis through DPP4/NOX1-mediated regulation of superoxide. Free Radic Biol Med. 2017;113:71–83. doi: 10.1016/j.freeradbiomed.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seidah N.G., Benjannet S., Wickham L., et al. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc Natl Acad Sci U S A. 2003;100(3):928–933. doi: 10.1073/pnas.0335507100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hegele R.A., Cao H., Harris S.B., Zinman B., Hanley A.J., Anderson C.M. Gender, obesity, hepatic nuclear factor-1alpha G319S and the age-of-onset of type 2 diabetes in Canadian Oji-Cree. Int J Obes Relat Metab Disord. 2000;24(8):1062–1064. doi: 10.1038/sj.ijo.0801258. [DOI] [PubMed] [Google Scholar]
- 4.Reiner A.P., Gross M.D., Carlson C.S., et al. Common coding variants of the HNF1A gene are associated with multiple cardiovascular risk phenotypes in community-based samples of younger and older European-American adults: the Coronary Artery Risk Development in Young Adults Study and the Cardiovascular Health Study. Circ Cardiovasc Genet. 2009;2(3):244–254. doi: 10.1161/CIRCGENETICS.108.839506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frain M., Swart G., Monaci P., et al. The liver-specific transcription factor LF-B1 contains a highly diverged homeobox DNA binding domain. Cell. 1989;59(1):145–157. doi: 10.1016/0092-8674(89)90877-5. [DOI] [PubMed] [Google Scholar]
- 6.Tan J., Xu J., Wei G., et al. HNF1α controls liver lipid metabolism and insulin resistance via negatively regulating the SOCS-3-STAT3 signaling pathway. J Diabetes Res. 2019;2019:5483946. doi: 10.1155/2019/5483946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong B., Wu M., Li H., et al. Strong induction of PCSK9 gene expression through HNF1alpha and SREBP2: mechanism for the resistance to LDL-cholesterol lowering effect of statins in dyslipidemic hamsters. J Lipid Res. 2010;51(6):1486–1495. doi: 10.1194/jlr.M003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rebouissou S., Imbeaud S., Balabaud C., et al. HNF1alpha inactivation promotes lipogenesis in human hepatocellular adenoma independently of SREBP-1 and carbohydrate-response element-binding protein (ChREBP) activation. J Biol Chem. 2007;282(19):14437–14446. doi: 10.1074/jbc.M610725200. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi Y., Wang W., Ninomiya T., Nagano H., Ohta K., Itoh H. Liver enriched transcription factors and differentiation of hepatocellular carcinoma. Mol Pathol. 1999;52(1):19–24. doi: 10.1136/mp.52.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang T., Wang T., Heianza Y., et al. HNF1A variant, energy-reduced diets and insulin resistance improvement during weight loss: the POUNDS Lost trial and DIRECT. Diabetes Obes Metab. 2018;20(6):1445–1452. doi: 10.1111/dom.13250. [DOI] [PubMed] [Google Scholar]
- 11.Dohda T., Kaneoka H., Inayoshi Y., Kamihira M., Miyake K., Iijima S. Transcriptional coactivators CBP and p300 cooperatively enhance HNF-1alpha-mediated expression of the albumin gene in hepatocytes. J Biochem. 2004;136(3):313–319. doi: 10.1093/jb/mvh123. [DOI] [PubMed] [Google Scholar]
- 12.Bach I., Mattei M.G., Cereghini S., Yaniv M. Two members of an HNF1 homeoprotein family are expressed in human liver. Nucleic Acids Res. 1991;19(13):3553–3559. doi: 10.1093/nar/19.13.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bach I., Galcheva-Gargova Z., Mattei M.G., et al. Cloning of human hepatic nuclear factor 1 (HNF1) and chromosomal localization of its gene in man and mouse. Genomics. 1990;8(1):155–164. doi: 10.1016/0888-7543(90)90238-p. [DOI] [PubMed] [Google Scholar]
- 14.Courtois G., Baumhueter S., Crabtree G.R. Purified hepatocyte nuclear factor 1 interacts with a family of hepatocyte-specific promoters. Proc Natl Acad Sci U S A. 1988;85(21):7937–7941. doi: 10.1073/pnas.85.21.7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rose R.B., Endrizzi J.A., Cronk J.D., Holton J., Alber T. High-resolution structure of the HNF-1alpha dimerization domain. Biochemistry. 2000;39(49):15062–15070. doi: 10.1021/bi001996t. [DOI] [PubMed] [Google Scholar]
- 16.Mendel D.B., Hansen L.P., Graves M.K., Conley P.B., Crabtree G.R. HNF-1 alpha and HNF-1 beta (vHNF-1) share dimerization and homeo domains, but not activation domains, and form heterodimers in vitro. Genes Dev. 1991;5(6):1042–1056. doi: 10.1101/gad.5.6.1042. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe T., Ozawa A., Masuda S., et al. Transcriptional regulation of the Angptl8 gene by hepatocyte nuclear factor-1 in the murine liver. Sci Rep. 2020;10(1):9999. doi: 10.1038/s41598-020-66570-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toniatti C., Monaci P., Nicosia A., Cortese R., Ciliberto G. A bipartite activation domain is responsible for the activity of transcription factor HNF1/LFB1 in cells of hepatic and nonhepatic origin. DNA Cell Biol. 1993;12(3):199–208. doi: 10.1089/dna.1993.12.199. [DOI] [PubMed] [Google Scholar]
- 19.Bach I., Yaniv M. More potent transcriptional activators or a transdominant inhibitor of the HNF1 homeoprotein family are generated by alternative RNA processing. EMBO J. 1993;12(11):4229–4242. doi: 10.1002/j.1460-2075.1993.tb06107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toniatti C., Demartis A., Monaci P., Nicosia A., Ciliberto G. Synergistic trans-activation of the human C-reactive protein promoter by transcription factor HNF-1 binding at two distinct sites. EMBO J. 1990;9(13):4467–4475. doi: 10.1002/j.1460-2075.1990.tb07897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cereghini S., Ott M.O., Power S., Maury M. Expression patterns of vHNF1 and HNF1 homeoproteins in early postimplantation embryos suggest distinct and sequential developmental roles. Development. 1992;116(3):783–797. doi: 10.1242/dev.116.3.783. [DOI] [PubMed] [Google Scholar]
- 22.Shih D.Q., Bussen M., Sehayek E., et al. Hepatocyte nuclear factor-1alpha is an essential regulator of bile acid and plasma cholesterol metabolism. Nat Genet. 2001;27(4):375–382. doi: 10.1038/86871. [DOI] [PubMed] [Google Scholar]
- 23.Garruti G., Wang H.H., Bonfrate L., de Bari O., Wang D.Q., Portincasa P. A pleiotropic role for the orphan nuclear receptor small heterodimer partner in lipid homeostasis and metabolic pathways. J Lipids. 2012;2012:304292. doi: 10.1155/2012/304292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pontoglio M., Barra J., Hadchouel M., et al. Hepatocyte nuclear factor 1 inactivation results in hepatic dysfunction, phenylketonuria, and renal Fanconi syndrome. Cell. 1996;84(4):575–585. doi: 10.1016/s0092-8674(00)81033-8. [DOI] [PubMed] [Google Scholar]
- 25.Pontoglio M. Hepatocyte nuclear factor 1, a transcription factor at the crossroads of glucose homeostasis. J Am Soc Nephrol. 2000;11(Suppl 16):S140–S143. [PubMed] [Google Scholar]
- 26.Yamagata K., Oda N., Kaisaki P.J., et al. Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3) Nature. 1996;384(6608):455–458. doi: 10.1038/384455a0. [DOI] [PubMed] [Google Scholar]
- 27.Pontoglio M., Faust D.M., Doyen A., Yaniv M., Weiss M.C. Hepatocyte nuclear factor 1alpha gene inactivation impairs chromatin remodeling and demethylation of the phenylalanine hydroxylase gene. Mol Cell Biol. 1997;17(9):4948–4956. doi: 10.1128/mcb.17.9.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jump D.B., Botolin D., Wang Y., Xu J., Christian B., Demeure O. Fatty acid regulation of hepatic gene transcription. J Nutr. 2005;135(11):2503–2506. doi: 10.1093/jn/135.11.2503. [DOI] [PubMed] [Google Scholar]
- 29.Kathiresan S., Willer C.J., Peloso G.M., et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41(1):56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Triggs-Raine B.L., Kirkpatrick R.D., Kelly S.L., et al. HNF-1alpha G319S, a transactivation-deficient mutant, is associated with altered dynamics of diabetes onset in an Oji-Cree community. Proc Natl Acad Sci U S A. 2002;99(7):4614–4619. doi: 10.1073/pnas.062059799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hegele R.A., Cao H., Harris S.B., Hanley A.J., Zinman B., Connelly P.W. The private hepatocyte nuclear factor-1alpha G319S variant is associated with plasma lipoprotein variation in Canadian Oji-Cree. Arterioscler Thromb Vasc Biol. 2000;20(1):217–222. doi: 10.1161/01.atv.20.1.217. [DOI] [PubMed] [Google Scholar]
- 32.Babaya N., Ikegami H., Fujisawa T., et al. Association of I27L polymorphism of hepatocyte nuclear factor-1 alpha gene with high-density lipoprotein cholesterol level. J Clin Endocrinol Metab. 2003;88(6):2548–2551. doi: 10.1210/jc.2002-021891. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Y.J., Yin R.X., Hong S.C., Yang Q., Cao X.L., Chen W.X. Association of the HNF1A polymorphisms and serum lipid traits, the risk of coronary artery disease and ischemic stroke. J Gene Med. 2017;19(1–2):e2941. doi: 10.1002/jgm.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shende V.R., Wu M., Singh A.B., Dong B., Kan C.F., Liu J. Reduction of circulating PCSK9 and LDL-C levels by liver-specific knockdown of HNF1α in normolipidemic mice. J Lipid Res. 2015;56(4):801–809. doi: 10.1194/jlr.M052969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z., Liu Q. Hepatitis C virus regulates proprotein convertase subtilisin/kexin type 9 promoter activity. Biochem Biophys Res Commun. 2018;496(4):1229–1235. doi: 10.1016/j.bbrc.2018.01.176. [DOI] [PubMed] [Google Scholar]
- 36.Hu M., Huang X., Han X., Ji L. Loss of HNF1α function contributes to hepatocyte proliferation and abnormal cholesterol metabolism via downregulating miR-122: a novel mechanism of MODY3. Diabetes Metab Syndr Obes. 2020;13:627–639. doi: 10.2147/DMSO.S236915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X., Hassan W., Zhao J., et al. The impact of hepatocyte nuclear factor-1α on liver malignancies and cell stemness with metabolic consequences. Stem Cell Res Ther. 2019;10(1):315. doi: 10.1186/s13287-019-1438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu H.T., Lu F.H., Ou H.Y., et al. The role of hepassocin in the development of non-alcoholic fatty liver disease. J Hepatol. 2013;59(5):1065–1072. doi: 10.1016/j.jhep.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 39.Wade D.P., Lindahl G.E., Lawn R.M. Apolipoprotein(a) gene transcription is regulated by liver-enriched trans-acting factor hepatocyte nuclear factor 1 alpha. J Biol Chem. 1994;269(31):19757–19765. [PubMed] [Google Scholar]
- 40.Wang X., Wu H., Yu W., et al. Hepatocyte nuclear factor 1b is a novel negative regulator of white adipocyte differentiation. Cell Death Differ. 2017;24(9):1588–1597. doi: 10.1038/cdd.2017.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akiyama T.E., Ward J.M., Gonzalez F.J. Regulation of the liver fatty acid-binding protein gene by hepatocyte nuclear factor 1alpha (HNF1alpha). Alterations in fatty acid homeostasis in HNF1alpha-deficient mice. J Biol Chem. 2000;275(35):27117–27122. doi: 10.1074/jbc.M004388200. [DOI] [PubMed] [Google Scholar]
- 42.Odom D.T., Zizlsperger N., Gordon D.B., et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303(5662):1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiang J.Y., Kimmel R., Stroup D. Regulation of cholesterol 7alpha-hydroxylase gene (CYP7A1) transcription by the liver orphan receptor (LXRalpha) Gene. 2001;262(1–2):257–265. doi: 10.1016/s0378-1119(00)00518-7. [DOI] [PubMed] [Google Scholar]
- 44.Wu H., Yu W., Meng F., et al. Polychlorinated biphenyls-153 induces metabolic dysfunction through activation of ROS/NF-κB signaling via downregulation of HNF1b. Redox Biol. 2017;12:300–310. doi: 10.1016/j.redox.2017.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lefterova M.I., Lazar M.A. New developments in adipogenesis. Trends Endocrinol Metab. 2009;20(3):107–114. doi: 10.1016/j.tem.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 46.Patitucci C., Couchy G., Bagattin A., et al. Hepatocyte nuclear factor 1α suppresses steatosis-associated liver cancer by inhibiting PPARγ transcription. J Clin Invest. 2017;127(5):1873–1888. doi: 10.1172/JCI90327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gordon J.I., Elshourbagy N., Lowe J.B., Liao W.S., Alpers D.H., Taylor J.M. Tissue specific expression and developmental regulation of two genes coding for rat fatty acid binding proteins. J Biol Chem. 1985;260(4):1995–1998. [PubMed] [Google Scholar]
- 48.Graham R.P., Terracciano L.M., Meves A., et al. Hepatic adenomas with synchronous or metachronous fibrolamellar carcinomas: both are characterized by LFABP loss. Mod Pathol. 2016;29(6):607–615. doi: 10.1038/modpathol.2016.59. [DOI] [PubMed] [Google Scholar]
- 49.Villemain L., Prigent S., Abou-Lovergne A., et al. Sigma 1 receptor is overexpressed in hepatocellular adenoma: involvement of ERα and HNF1α. Cancers (Basel) 2020;12(8):2213. doi: 10.3390/cancers12082213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y., Quagliarini F., Gusarova V., et al. Mice lacking ANGPTL8 (Betatrophin) manifest disrupted triglyceride metabolism without impaired glucose homeostasis. Proc Natl Acad Sci U S A. 2013;110(40):16109–16114. doi: 10.1073/pnas.1315292110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang W., Zhao M., Zhao Y., Shen W., Yin S. PDGFRα/β-PI3K-Akt pathway response to the interplay of mitochondrial dysfunction and DNA damage in Aroclor 1254-exposed porcine granulosa cells. Environ Pollut. 2020;263(Pt A):114534. doi: 10.1016/j.envpol.2020.114534. [DOI] [PubMed] [Google Scholar]
- 52.Xiong X., Wang X., Lu Y., et al. Hepatic steatosis exacerbated by endoplasmic reticulum stress-mediated downregulation of FXR in aging mice. J Hepatol. 2014;60(4):847–854. doi: 10.1016/j.jhep.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 53.Kornfeld J.W., Baitzel C., Könner A.C., et al. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature. 2013;494(7435):111–115. doi: 10.1038/nature11793. [DOI] [PubMed] [Google Scholar]
- 54.Lake A.D., Chaput A.L., Novak P., Cherrington N.J., Smith C.L. Transcription factor binding site enrichment analysis predicts drivers of altered gene expression in nonalcoholic steatohepatitis. Biochem Pharmacol. 2016;122:62–71. doi: 10.1016/j.bcp.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rom O., Xu G., Guo Y., et al. Nitro-fatty acids protect against steatosis and fibrosis during development of nonalcoholic fatty liver disease in mice. EBioMedicine. 2019;41:62–72. doi: 10.1016/j.ebiom.2019.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Willebrords J., Cogliati B., Pereira I.V.A., et al. Inhibition of connexin hemichannels alleviates non-alcoholic steatohepatitis in mice. Sci Rep. 2017;7(1):8268. doi: 10.1038/s41598-017-08583-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhuo M.Q., Luo Z., Xu Y.H., Li D.D., Pan Y.X., Wu K. Functional analysis of promoters from three subtypes of the PI3K family and their roles in the regulation of lipid metabolism by insulin in yellow catfish Pelteobagrus fulvidraco. Int J Mol Sci. 2018;19(1):265. doi: 10.3390/ijms19010265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ni Q., Ding K., Wang K.Q., et al. Deletion of HNF1α in hepatocytes results in fatty liver-related hepatocellular carcinoma in mice. FEBS Lett. 2017;591(13):1947–1957. doi: 10.1002/1873-3468.12689. [DOI] [PubMed] [Google Scholar]
- 59.Sakellariou S., Morgan Y., Heaton N., Portmann B., Quaglia A., Tobal K. New monoallelic (partial tandem duplication) mutation of HNF1a gene in steatotic hepatocellular adenoma. Eur J Gastroenterol Hepatol. 2011;23(7):623–627. doi: 10.1097/MEG.0b013e328347964d. [DOI] [PubMed] [Google Scholar]
- 60.Li H., Li J., Qu Z., et al. Intrauterine exposure to low-dose DBP in the mice induces obesity in offspring via suppression of UCP1 mediated ER stress. Sci Rep. 2020;10(1):16360. doi: 10.1038/s41598-020-73477-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Su S., Wu G., Cheng X., et al. Oleanolic acid attenuates PCBs-induced adiposity and insulin resistance via HNF1b-mediated regulation of redox and PPARγ signaling. Free Radic Biol Med. 2018;124:122–134. doi: 10.1016/j.freeradbiomed.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 62.Tonooka N., Tomura H., Takahashi Y., et al. High frequency of mutations in the HNF-1alpha gene in non-obese patients with diabetes of youth in Japanese and identification of a case of digenic inheritance. Diabetologia. 2002;45(12):1709–1712. doi: 10.1007/s00125-002-0978-3. [DOI] [PubMed] [Google Scholar]
- 63.Parwin A., Najmi A.K., Ismail M.V., Kaundal M., Akhtar M. Protective effects of alendronate in Triton X-100-induced hyperlipidemia in rats. Turk J Gastroenterol. 2019;30(6):557–564. doi: 10.5152/tjg.2019.18076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gole H.K., Tharp D.L., Bowles D.K. Upregulation of intermediate-conductance Ca2+-activated K+ channels (KCNN4) in porcine coronary smooth muscle requires NADPH oxidase 5 (NOX5) PLoS One. 2014;9(8):e105337. doi: 10.1371/journal.pone.0105337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paik Y.H., Iwaisako K., Seki E., et al. The nicotinamide adenine dinucleotide phosphate oxidase (NOX) homologues NOX1 and NOX2/gp91(phox) mediate hepatic fibrosis in mice. Hepatology. 2011;53(5):1730–1741. doi: 10.1002/hep.24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X., Chen X., Zhang X., et al. A small-molecule inhibitor of PCSK9 transcription ameliorates atherosclerosis through the modulation of FoxO1/3 and HNF1α. EBioMedicine. 2020;52:102650. doi: 10.1016/j.ebiom.2020.102650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Momtazi A.A., Banach M., Pirro M., Katsiki N., Sahebkar A. Regulation of PCSK9 by nutraceuticals. Pharmacol Res. 2017;120:157–169. doi: 10.1016/j.phrs.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 68.Dong B., Singh A.B., Shende V.R., Liu J. Hepatic HNF1 transcription factors control the induction of PCSK9 mediated by rosuvastatin in normolipidemic hamsters. Int J Mol Med. 2017;39(3):749–756. doi: 10.3892/ijmm.2017.2879. [DOI] [PubMed] [Google Scholar]
- 69.Allin K.H., Nordestgaard B.G. Pleiotropic effects of HNF1A rs1183910 in a population-based study of 60,283 individuals. Diabetologia. 2014;57(4):729–737. doi: 10.1007/s00125-013-3156-x. [DOI] [PubMed] [Google Scholar]