Abstract
The autophagy adaptor protein SQSTM1/p62 is overexpressed in breast cancer and has been identified as a metastasis-related protein. However, the mechanism by which SQSTM1/p62 contributes to breast cancer progression and tumor microenvironment remains unclear. This study revealed that silencing SQSTM1/p62 expression suppressed breast cancer progression via regulating cell proliferation and reshaping the tumor microenvironment (TME). Here, we found that SQSTM1/p62 was overexpressed in multiple human cancer tissue types and that was correlated with poor patient overall survival (OS) and disease-free survival (DFS). Moreover, we found that short-hairpin RNA (shRNA)-mediated knockdown of p62 expression significantly inhibited cell proliferation, migration, and invasion, and promoted cell death in vitro, as well as suppressed breast cancer growth and lung metastasis in vivo. In addition, flow cytometry analysis of splenocytes and tumor infiltrating lymphocytes (TILs) indicated that the numbers of CD8α+ interferon (IFN)-γ+ cells (CTLs) and CD4+IFN-γ+ (Th1) cells were increased while those of CD4+IL-4+ (Th2) cells, tumor-associated macrophages (TAMs) and myeloid-derived suppressor cells (MDSCs) were decreased. RT-PCR analyses showed that the gene expression of Th1/Th2 cytokines changed in the tumor microenvironment. Silencing SQSTM1/p62 suppressed tumor cell lung metastasis. Together, our results provide strong evidence that silencing tumor cell SQSTM1/p62 inhibited tumor growth and metastasis through cell cycle arrest and TME regulation. This finding provides a novel molecular therapeutic strategy for breast cancer progression and metastasis treatment.
Keywords: Breast cancer, Cell cycle, Metastasis, SQSTM1/p62, Tumor microenvironment
Introduction
Breast cancer is the most common malignancy in women, produces a serious economic burden and is associated with poor clinical outcome.1,2 Triple-negative breast cancer has been recognized as a major global public health threat because it is resistant to commonly used chemotherapeutic drugs and immunotherapies.3 Although surgery is an effective treatment for breast cancer, increased circulating tumor cell-mediated metastasis enhances the risks of tumor recurrence and therapy failure.4 Therefore, it is essential to understand tumorigenesis and develop novel therapy combinations for breast cancer.
SQSTM1/p62, which was first identified as an autophagy adaptor consisting of the 4 structural domains PB1/TRAF6 binding domain (TB)/LC3-interacting region (LIR)/ubiquitin-associated (UBA) domain, participates in tumor cell autophagy and apoptosis.5 The LIR and UBA domains enable SQSTM1/p62 to function as an autophagy adaptor. In addition, SQSTM1/p62 directly enhances inflammatory gene expression through its TB via the nuclear factor kappa-B (NF-κB) pathway and activates the Nrf2-Keap1-denpented antioxidant response through its KIR region.6,7 Recent studies have demonstrated that SQSTM1/p62 not only plays an important role in autophagy regulation but also triggers tumorigenesis and metastasis in vivo. SQSTM1/p62 directly interacts with the vitamin D receptor on hepatic stellate cells, which negatively regulates liver inflammation and fibrosis and induces hepatocellular carcinoma (HCC) development.8 High expression of SQSTM1/p62 is associated with invasive phenotypes in breast cancer and relatively poor clinical outcomes, which depend on the interaction between SQSTM1/p62 and vimentin-mediated metastasis.9 Overexpression of SQSTM1/p62 in breast cancer induces cancer cell self-renewal via MYC expression, promoting cell growth and proliferation, which are dependent on the gene expression of let-7a and let-7b.10 Additionally, phosphorylation of p62 at serine 349 by PKC-δ is enhanced with VPS34 production, which suggests that p-p62 is associated with tumor growth and development.11 SQSTM1/p62-dependent selective autophagy recruits the LC3 protein and other cargoes to the autophagosome to maintain cellular homeostasis, which is correlated with flightless-1 (Flil) phosphorylation in clinical breast cancer samples.12 Thus, SQSTM1/p62 facilitates breast cancer tumorigenesis and metastasis.
The tumor microenvironment (TME), which consists of tumor cells, immune cells, fibroblasts, cytokines, etc., significantly influences tumorigenesis and the response to tumor immunotherapy, inducing poor clinical outcomes. Effector CD8+ cytotoxic T lymphocytes (CTLs) whose survival is dependent on the cytokine interleukin (IL)-2 perform a potent role in antitumor immunity via Interferon (IFN)-γ and Granzyme B production.13 Enhancement of T helper 1 (Th1) CD4+ T cell polarization in the TME provides a potential antitumor response in breast cancer via Xanthohumol (XN)-induced expression of T-bet and phosphorylation of signal transducer and activator of transcription 4 (STAT4).14 In addition to Th1 cells, tumor-specific CD4+ Th9 cells eradicate established murine tumors and establish a long-lasting antitumor response via Pu.1-Traf 6-NF-κB and eomes-granzyme pathway activation.15 Conversely, in the TME, myeloid-derived suppressor cells (MDSCs) mediate immune suppression via the expression of the cytokines TGF-β and IL-10 and suppression of the activity of effector T cells.16 Hypoxia downregulates the expression of STAT3 in MDSCs, resulting in the differentiation of tumor-associated macrophages (TAMs) and the inhibition of dendritic cell (DC) functions.17 Interestingly, SQSTM1/p62 also plays a critical role in TME regulation-induced cancer progression.5 Knocking out SQSTM1/p62 in fibroblasts induces the production of the inflammatory cytokines IL-6 and TGF-β in cancer-associated fibroblasts (CAFs), which promote tumor development.18 In clinical colorectal carcinoma (CRC) tissue, SQSTM1/p62 expression increases FOXP3+ regulatory T cell infiltrate density in the TME.19 Overall, the regulation of the TME balance plays an important role in the antitumor response.
Herein, we demonstrated the negative roles of SQSTM1/p62 in breast cancer progression and metastasis. A triple-negative murine breast cancer cell (4T1) xenograft model was used in this study to evaluate the function of SQSTM1/p62. Knocking down SQSTM1/p62 expression in tumor cells activated autophagy and induced cell cycle arrest via p27 upregulation in vitro. In addition, silencing the expression of SQSTM1/p62 inhibited tumor growth, altered cytokine production profiles and regulated immune cell infiltration into the TME in vivo. A p62-short-hairpin RNA (shRNA)-4T1 stable cell line showed obviously reduced metastasis to lung tissue. Thus, our study reported a novel mechanism involving SQSTM1/p62 in tumorigenesis and metastasis via cell cycle arrest and immune cell profile changes in the TME, which provides insight into the regulation of breast cancer progression by autophagy and provides potential drug targets for regulating tumor development.
Materials and methods
Ethics statement, mice, and cell lines
Six-to eight-week-old female BALB/c mice (16–18 g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd (Beijing, PR China), maintained under specific pathogen-free (SPF) conditions and raised in the central animal care services of the Institute of the Medical Biology Chinese Academy of Medical Sciences (IMBCAMS) & Perking Union Medical College (PUMC). Animal experiments were approved by the Ethics Committee of Animal Care and Welfare of IMBCAMS and PUMC (Permit Number: SYXK (dian) 2010–0007). All efforts were made to minimize animal suffering. The 4T1 triple-negative murine breast cancer cell line, which is a 6-thioguanine resistant cell line screened from the 401.4 tumor strain, was purchased from the cell bank of the Chinese Academy of Sciences. 293T cells were also purchased from the cell bank of the Chinese Academy of Sciences. The cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin and 1% nonessential amino acids.
Stable cell line construction and microscopy
A lentiviral expression system was employed for stable cell line construction, and the target gene sequence was cloned into the plasmid psi-LVRU6GP and verified by sequencing. The recombinant plasmid p62-shRNA-psi-LVRU6GP was co-transfected into 293T cells with the packaging plasmids pH1 and pH2, which were purchased from Invitrogen (Thermo Fisher Scientific, USA) at a ratio of 3:1:1, respectively. The day before transfection, 293T cells were harvested and seeded into petri dishes. The next day, the 293T cells were washed with fresh PBS and changed into FBS- and antibiotic-free Opti-MEM. The three sterile plasmids (40 μg, 3:1:1) were mixed gently together with 80 μL polyplus transfection™ (jetPRIME®, BioLegend, USA) in 800 μL Opti-MEM for 30 min at room temperature. Then, the mixture medium was transferred to the 293T cells, and the medium was replaced with complete RPMI 1640 medium 6 h later. The supernatants were harvested by centrifugation at 2000 × g for 10 min at 48 h and 72 h after transfection and then filtered through a 0.45-μm filter. Supernatants containing 10 μg/mL polybrene were added to 4T1 cells, and 48 h later, GFP-positive 4T1 cells were observed and imaged under a fluorescence microscope (Nikon, Japan). Puromycin was employed to kill non-infected 4T1 cells. Stable cell lines were identified with fluorescence microscopy, RT-PCR, and Western blot assays.
Tumor challenge and metastasis studies
To establish a tumor cell-grafted model, 1 × 106 cells mixed with Basement Membrane Matrix (BD Bioscience, San Jose, CA, USA) were injected subcutaneous (s.c.) into the right flank of each mouse to establish the p62-shRNA 4T1 group, NC-4T1 group and 4T1 model group (n = 6–8). To establish the 4T1 orthotopic (o.t.) model, 1 × 106 cells were inoculated into the second breast fat pad. The mice were monitored three times per week for tumor growth using a slide caliper as described previously,20 and the mice were sacrificed at experiment endpoints. To generate a murine lung metastasis model, mice were intravenously (i.v.) injected with 100 μL cell suspension containing a total of 1 × 105 cells to establish the p62-shRNA 4T1 group, NC-4T1 group and 4T1 model group (n = 8), and the mouse survival rate was monitored daily until day 24. All of the animal studies were performed twice, and one of the studies is shown in the figures.
Immunoblot analysis and antibodies
For SQSTM1/p62 and p27 kip1 immunoblot assays, p62-shRNA-, NC- and mock-4T1 cells were lysed with RIPA buffer containing a protease inhibitor at 4 °C for 30 min in a 1.5-mL tube. The supernatants were harvested by centrifugation at 12,000 × g at 4 °C for 10 min. A Bradford kit was employed for total protein quantitation, and then a 5 × loading buffer was added to the samples for immunoblot analysis. Ten micrograms of protein/well per samples were separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes. After blocking, the membranes were incubated with primary anti-mouse antibodies (1:1000 for p62 CST39749 and 1:1000 for p27 CST36985, CST, USA; ab207612, 1:2000 for Beclin-1 and 1:10,000 for β-actin ab6267, Abcam, UK) diluted in 0.5% BSA buffer at 4 °C overnight. Then, the membranes were washed 5 times with Tris-buffered saline Tween (TBST) buffer and incubated with secondary antibodies (horseradish peroxidase (HRP)-conjugated goat anti-rabbit and anti-mouse IgG antibodies, ab6721, 1 mg/mL, 1:5,000, Abcam, UK) for 60 min at room temperature. After the incubation, the membranes were washed 5 times, and the protein bands were imaged with enhanced chemiluminescence (ECL; Thermo Fisher Scientific, USA) plus a western blotting detection kit according to the manufacturer's instructions. ImageJ was used to quantify the grayscale values of the Western blot bands.
Real-time quantitative PCR
Total RNA was isolated from cells and tissues using TRIzol (RNAiso, Takara) and purified by the chloroform-phenol extraction method. cDNA was reverse transcribed with the SureScript First-stand cDNA Synthesis Kit (GeneCopoeia) and then detected by the All-in-One™ miRNA qRT-PCR Detection Kit (GeneCopoeia). Real-time quantitative PCR (RT-qPCR) was performed on a Bio-Rad CFX-96 Touch Real-Time Detection systems, all data were normalized to GAPDH. Primer sequences are listed in Table S1.
Flow cytometry assay
For flow cytometry analysis of immune cell infiltration, tumor and splenic lymphocytes were separated with a mouse lymphocyte separation kit (BioLegend, USA) and then washed with fresh RPMI 1640 medium. After PMA and ionomycin stimulation, the cells were washed with a staining buffer three times. The flow cytometry protocol was previously reported.20 Briefly, FITC-labeled anti-mouse CD8α and APC-labeled anti-mouse IFN-γ antibodies were used to stain CTLs, FITC-labeled anti-mouse CD4 and APC-labeled anti-mouse IFN-γ antibodies were used to stain Th1 cells, FITC-labeled anti-mouse CD4 and APC-labeled anti-mouse IL-4 antibodies were used to stain Th2, PE-labeled anti-mouse Gr-1 and APC-labeled anti-mouse CD11b antibodies were used to stain MDSCs, PE-labeled anti-mouse Ly6G and APC-labeled anti-mouse CD11b antibodies were used to stain neutrophils in BALF and PE-labeled anti-mouse F4/80 and APC-labeled anti-mouse CD11b antibodies were used to stain macrophages. All flow cytometry antibodies used in this research were purchased from BioLegend, USA. The cells were stained with premixed antibodies at 4 °C in the dark for 1 h. After washing, the cells were resuspended in 200 μL staining buffer and analyzed by flow cytometry (CytoFLEX LX, Beckman, USA), and the data were analyzed using CytExpert 2.0 software.
Cell cycle flow cytometry assay
For a cell cycle assay, stable cell lines were harvested by centrifugation at 1000×g at 4 °C for 10 min. Then, the cells were fixed with 75% absorbent ethanol at −20 °C overnight. The next day, the cells were washed three times with precooled PBS and stained with a cell cycle detection kit (BD, Cell cycle staining Kit, USA). The cells were incubated at 4 °C for 15 min, washed three times with a binding buffer, resuspended in 100 μL PBS and analyzed by flow cytometry (CytoFLEX LX, Beckman, USA), and all data were analyzed using CytExpert 2.0 software.
ELISA
Serum IL-6, IL-10, IL-1β, IL-4, IL-13, TNF-α, IFN-γ, and IL-12 levels in tumor-bearing mice were measured by mouse IL-6, IL-10, IL-1β, IL-4, IL-13, TNF-α, IFN-γ, and IL-12 Quantitative ELISA kits, respectively (R&D Systems) according to the manufacturer's protocol.
Preparation of tissue samples for histopathological analysis
Lung tissue samples were obtained, frozen in liquid nitrogen, embedded in optimal cutting temperature compound (OCT) and sectioned (5 μm thickness). The sections were stained with Hematoxylin and Eosin (H & E) and examined. To count the number of metastatic lung nodules, whole-lung tissue samples were stained with Bouin's buffer, and the left lobe of the lung was sectioned and stained with H & E staining.
Statistical analysis
GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, CA, USA) was used for statistical analyses. All data are given as the mean ± SEM. Statistical analyses were performed using a two-tailed Student's t-test for two-group comparisons, one-way ANOVA for multi group comparisons, and a Kaplan-Meier nonparametric log-rank test for survival rate comparisons. P ≤ 0.05 was considered significant. ∗ indicates P < 0.05, ∗∗ indicates P < 0.01, ∗∗∗ indicates P < 0.001, ∗∗∗∗ indicates P < 0.0001, and ns indicates nonsignificant.
Results
Overexpression of SQSTM1/p62 reduces cancer patient survival
To evaluate the potential relationship between SQSTM1/p62 and tumorigenesis, we analyzed SQSTM1/p62 gene expression across 33 cancer types in the GEPIA.2 databank (http://gepia2.cancer-pku.cn), and the data were shown as log2 (TPM+1) values. The expression of SQSTM1/p62 in cancer tissues was significantly increased than that in normal tissues (Fig. 1A). High expression of SQSTM1/p62 was strongly associated with poor cancer patient overall survival (OS) and disease-free survival (DFS), indicating that SQSTM1/p62 overexpression induced poor clinical outcomes (Fig. 1B) and the expression of SQSTM1 were increased on patient pathological stage (F = 5.13, P = 0.000106) (Fig. 1C). In addition, we used the STRING network tool (https://version11.string-db.org/) to construct a PPI network of SQSTM1, the SQSTM1 module contains 11 nodes and 54 edges (PPI enrichment P: < 1.0e-16) (Fig. 1D). The top 10 terms of GO and KEGG functional enrichment assay were listed as blown (Fig. 1E). To further confirm the role of SQSTM1 in prognostic values, we performed IHC to detect the protein expression of SQSTM1 in breast cancer patients using the IHC data from Human Protein atlas project (https://www.proteinatlas.org) (Fig. 1F). The IHC results reveal the high expression of SQSTM1 in breast cancer patients, among the 10 cases of BRCA tissues examined for SQSTM1 staining, 2 cases obtain high expression, 7 cases obtain medium expression, and 1 case obtains low expression. In line with these results, we performed a real-time PCR assay for SQSTM1/p62 gene expression in clinical breast cancer samples, and SQSTM1/p62 expression in tumor tissue was significantly higher than that in normal tissue (Fig. 1G). Thus, our results demonstrated that overexpression of SQSTM1/p62 induced poor clinical outcomes across pan-cancer types including breast cancer, indicating the poor clinical outcome.
Figure 1.
Overexpression of SQSTM1/p62 across pan-cancer types is associated with poor clinical outcome. (A) Boxplot analyses of the expression of SQSTM1/p62 across pan-cancer types in matched TCGA normal and GTEx data. (B) Overall survival (OS) and disease-free survival (DFS) curves of the high and low SQSTM1/p62 expression groups of related cancer patients. (C) Violin plots show SQSTM1 expression with patient pathological stage. (D) Module analysis of protein–protein interactions network for SQSTM1. (E) Histogram analysis of Biological process, Cellular component, Molecular function, and KEGG pathways for SQSTM1. (F) Representative images of SQSTM1 in breast cancer tissues via IHC staining. (G) Histogram analysis of SQSTM1/p62 gene expression in breast cancer tissues measured by real-time PCR (n = 15, T = 20). ∗, P < 0.05; the paired Student's t-test was employed for the RT-PCR assay and one-way ANOVA for the multi groups comparison. Kaplan-Meler survival curves show the correlations between SQSTM1 expression with OS and DFS times, and the log-rank was used for curve comparisons. Data are presented as the mean ± SEM.
SQSTM1/p62 regulates cell cycle arrest and promotes cell death in vitro
To identify the function of SQSTM1/p62 in breast cancer progression, we generated a p62-knockdown 4T1 stable cell line and a negative control (shNC) 4T1-GFP stable cell line using a lentiviral-based system (Fig. S1). We found that knocking down of SQSTM1/p62 significantly lead to decrease tumor cell proliferation in 4T1 breast cancer cells via a CCK-8 assay (Fig. 2A). In addition, the release of LDH in shp62 was higher compared to shNC and mock 4T1 cells, which was enhanced after starving treatment (Fig. 2B). We found that silencing p62 expression also enhanced cell death in vitro, which was consisted with the LDH release assay (Fig. 2C, D). Furthermore, we treated shp62 cells with IFN-γ to simulate the tumor microenvironment, more extracellular ATP was detected in the medium (Fig. 2E). We hypothesized that knocking down of SQSTM1/p62 expression in 4T1 cells directly affect cell proliferation due to the regulatory activation of cell cycle arrest. Silencing the expression of SQSTM1/p62 in 4T1 cells induced 75.95% of the cells to arrest in the G0/G1 phase, and this proportion of arrested cells was higher than that observed for shNC- or mock-4T1 cells (Fig. 2F). In addition, the percentages of S-phase and G2/M-phase cells in the shp62 group were significantly decreased (Fig. 2G). Western blot analysis of cell lysates of shp62, and shNC- revealed upregulation of the expression of p27, which negatively regulates the cell cycle by arresting the progression of cells from G1 phase into S phase (Fig. 2H). Strikingly, the level of LC3 was substantially increased in p62-shRNA 4T1 cells, which suggested that knocking down SQSTM1/p62 expression in 4T1 cells also induced autophagy in vitro. In line with these results, we co-cultured stable tumor cell lines with bone marrow-derived dendritic cells (BMDCs). The results revealed that shp62 promoted tumor cell death which could induce DC maturation (Fig. 2I). Overall, we identified that silencing SQSTM1/p62 in 4T1 cells induced cell cycle arrest and promoted cell death in vitro.
Figure 2.
Knockdown of SQSTM1/p62 delays the cell cycle and promotes cell death. (A) Proliferation curve of stable cell lines by the CCK-8 assay. shp62, shNC and mock 4T1 cells were shown here. (B) Histogram analysis of LDH release. The stable cell line was treated with- or without starvation for 24 h. (C) Representative flow cytometry images of cell apoptosis analyzed by Anneix V/7-AAD staining kit. (D) Histogram analysis of percentages of cell death. (E) Histogram analysis of concentration of ATP release in the cell culture. shp62 cells were treated with 100 µm 3-MA, 10 µM Rapa, and 2 ng/mL IFN-γ for 24 h, and the cell culture was harvested for testing. (F) Representative plots of flow cytometry analysis of the cell cycle. Silencing the expression of SQSTM1/p62 in 4T1 cells arrested the cell cycle in the G0/G1 phase. (G) Histogram analysis of the percentages of 4T1 cells in the different cell cycle phases in each group. (H) Immunoblotting assay showing that knocking down SQSTM1/p62 expression promotes p27 upregulation in vitro. (I) Histogram analysis of the activation of DCs co-cultured with shp62, shNC and mock 4T1 cells. ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001; and ns non-significance as determined by one-way ANOVA assay for comparison. Data are presented as the mean ± SEM.
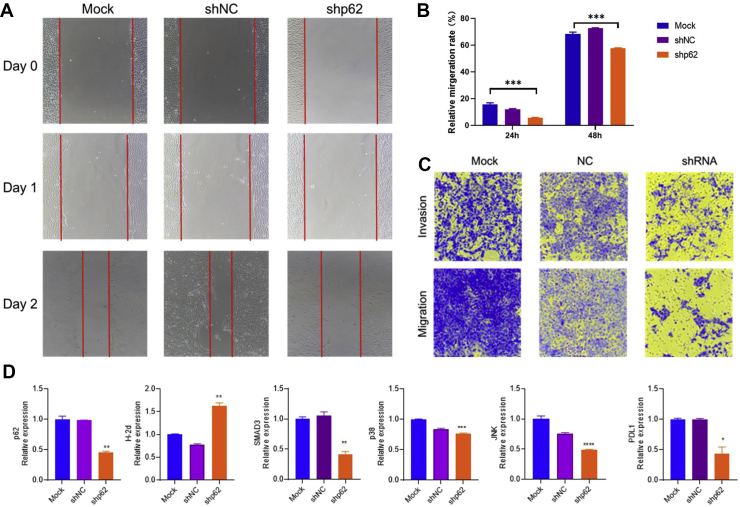
SQSTM1/p62 fosters the migration of breast cancer in vitro
Since SQSTM1/p62 was associated with the proliferation in breast cancer cells, we further investigated the effect of SQSTM1/p62 on migration and invasion in vitro. Silencing 4T1 SQSTM1/p62 expression significantly decreased the migration rate in vitro via a wound-healing assay (Fig. 3A, B). Additionally, the shp62 stable cell showed the attenuated migration and invasion capabilities compared to shNC and mock 4T1 cells (Fig. 3C). Meanwhile, real-time PCR analysis suggested that knocking down SQSTM1/p62 suppressed p38, JNK, and SMAD3 expression levels. Strikingly, we also found that the immune checkpoint blockage PDL1 gene was inhibited in the shp62 stable cell line, which may lead to considerable antitumor response feedback in vivo (Fig. 3D). In total, knocking down p62 in breast cancer cells suppressed migration and invasion, as well as decreased PDL1 expression.
Figure 3.
Knock down of SQSTM1/p62 suppresses the migration and invasion of breast cancer cell in vitro. (A) Representative images of wound-healing assay. The results of mock, shNC, and shp62 stable cell line at day 0, 1, and 2 were showed here. (B) The quantification of migration rate in stable cell line at different time point. The results of 24 h and 48 h were shown here. (C) Representative images of migration and invasion assay. (D) The results of gene expression level by RT-PCR assay. The gene p62, p38, SMAD3, JNK and PDL1 were shown here. One-way ANOVA assay was employed for the comparison among three groups. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗ P < 0.0001.
Silencing SQSTM1/p62 expression significantly suppresses breast cancer growth in a 4T1-grafted model
We next evaluated the impact on tumor growth in vivo. Six-to eight-week-old mice were subcutaneously (s.c.) injected with shNC-, p62-shRNA- or Ctrl-4T1 cells at a dose of 1 × 106 cells per mouse, and tumor growth was monitored. Obviously, compared to the shNC group and Ctrl-4T1 group, the p62-shRNA group showed specifically inhibited tumor growth in vivo (Fig. 4A). Tumor growth was identical between the shNC group and Ctrl-4T1 group, indicating that silencing SQSTM1/p62 in 4T1 cells significantly delayed tumor growth. The tumor growth curve of each mouse was shown separately (Fig. 4B). The mice were sacrificed on day 20 after tumor cell inoculation, and tumor tissue samples were harvested and imaged. As shown in Figure 4C, the tumors in the p62-shRNA group were significantly smaller than those in shNC and Ctrl-4T1 group. Furthermore, we monitored the weight of the tumor tissue, spleen, and lung of each mouse in the individual groups (Fig. 4D–F). Consistent with the tumor growth curves and tumor sizes, the weights of the tumor, spleen, and lung tissues in the p62-shRNA group were the lightest, while there were no differences between the shNC group and Ctrl-4T1 group. In addition, we also employed a more convincing orthotopic (o.t.) implantation model in vivo, and the results showed that p62-shRNA tumors growth were slowest, tumor size, and weight, and spleen weight were lighter than that in shNC and Ctrl groups (Fig. 4G–L). We also knocked down the SQSTM1/p62 expression in TC-1 cell line. TC-1, transformed from C57BL/c (H-2b) lung epithelial cell by co-transfecting with HPV16 E6, E7 and Ras was broadly used as an HPV tumor model. Interestingly, p62-shRNA-TC-1 tumor grew slower than that in NC- and TC-1 model groups in vivo and the weight of spleen and tumor were also lighter (Fig. S2). Taken together, the above data demonstrate that SQSTM1/p62 is required for breast cancer tumorigenesis.
Figure 4.
SQSTM1/p62 controls progression of grafted murine breast cancer 4T1 tumors in vivo. (A)In vivo tumor growth curves. shp62-, shNC- and CTRL-4T1 cells were s. c. inoculated into the right flank of BALB/c mice (n = 6–8 per group) at a dose of 1 × 106 cells. (B) Tumor growth progression over time for the individual mice in each group. The animals were euthanized at experiment endpoint. (C) Representative images of tumor tissues in each group (n = 6). (D–F) Histogram analyses of the weights of the tumors, spleens and lungs in each group (n = 6). (G) Tumor growth curves of shp62-,shNC- and 4T1 CTRL group by orthotopic implantation in vivo (n = 6 per group). (H–J) Histogram analyses of the weights of the tumors, spleens and lungs in each group (n = 6). (K–L) Representative images of tumor tissues in each group (n = 6) with in vitro bright field vision and bioluminescence IVIS® biophotonic imaging system. ∗P < 0.01; ∗∗P < 0.001; and ns non-significance as determined by one-way ANOVA for the tumor, spleen and lung weight graphs and by two-way ANOVA for the tumor growth curves. Data are presented as the mean ± SEM.
Silencing SQSTM1/p62 regulates systemic immune cell infiltration into the spleen and tumor microenvironment
Activated CD8α+ cells and some CD4+ cells that produce IFN-γ, granzyme B and other components are effective killers of cancerous cells.13,21 However, other immunosuppressive cells such as MDSCs and M2 macrophages help tumor cells escape host immune surveillance in the TME.22 To examine whether silencing SQSTM1/p62 affects immune cell infiltration, we analyzed the immunocyte profiles of spleen. Flow cytometry analysis showed that there was no significant difference in the number of splenic CD8α+IFN-γ+ CTLs between the shNC group and Ctrl group, while knocking down SQSTM1/p62 expression significantly induced CTL accumulation in the spleen (P < 0.01). In line with this result, we found that CD4+IFN-γ+ Th1 cells exhibited an identical tendency (P < 0.001). Interestingly, CD4+IL-4+ Th2 cell numbers were significantly decreased in the shNC group compared with the Ctrl group, while silencing SQSTM1/p62 reduced Th2 cell infiltration into the spleen (P < 0.05). Analysis of the myeloid compartment showed decreased fractions of MDSCs and macrophages in the shp62 group. Conversely, there was no difference between the shNC and CTRL groups (P > 0.05) (Fig. 5A). Similarly, we build the orthotopic model and analyzed the tumor infiltrating lymphocytes (TILs) and splenocytes at the endpoint. We found that the percent of CD8+ T cells and CTLs were increased and the percent of MDSCs were decreased (Fig. 5B, C). Altogether, our results suggest that silencing SQSTM1/p62 increased antitumor lymphocytes accumulation and reduced immunosuppressive myeloid cell frequencies during tumor progression.
Figure 5.
Silencing the expression of SQSTM1/p62 in 4T1 cells enhances the immunogenicity of the tumor microenvironment during tumor development. (A) Histogram analyses of the percentages of splenocytes in s. c. model. The results of CD8α+IFN-γ+ CTLs, CD4+IFN-γ+ Th1 cells, CD4+IL-4+ Th2 cells, CD11b+Gr-1+ MDSCs, and CD11b+F4/80+ macrophages were shown here. (B) Histogram analyses of the percentages of splenocytes in o. t. model. The results of CD8 T cells, CTLs, MDSCs, and macrophages were shown here. (C) Histogram analyses of the percentages of tumor infiltration lymphocytes (TILs) in o. t. model. The results of CD8 T cells, CTLs, and MDSCs were listed here.∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗P < 0.0001; and ns non-significance as determined by one-way ANOVA for the flow cytometry analyses. n = 6 per group. Data are presented as the mean ± SEM.
Knocking down SQSTM1/p62 expression changes the expression of inflammatory cytokines and chemokines in the TME
To explore the mechanism by which SQSTM1/p62 impacts immune suppression, we further demonstrated the expression of inflammatory cytokines and chemokines in the TME, and found results consistent with the effects of SQSTM1/p62 on tumor suppression. Cytokine analyses of serum samples from tumor-bearing mice showed enhancement of the production of the antitumor cytokines IFN-γ, TNF-α and IL-12. In addition, the levels of the Th2 cytokines IL-4, IL-10 and IL-13 were significantly reduced in the p62-shRNA group, and those of the inflammatory cytokines IL-1β and IL-6 were also reduced via ELISA assay (Fig. 6A). Meanwhile, real-time PCR analysis suggested that cytokine profiles were changed in tumor tissues. Surprisingly, we found that the level of the pro-inflammatory cytokine IL-1α/β was increased in tumor tissue samples (Fig. 6B, C). In line with the serum cytokine results, the tumor tissue results showed that the expression of the Th2 cytokines IL-4 and IL-13 was significantly decreased (Fig. 6E–G). IL-2 is one of the most broadly biologically active cytokines produced by activated CD4+ and CD8+ T cells.21,23,24 Our results showed that the gene expression of IL-2 was significantly enhanced in the p62-shRNA group (Fig. 6D), which was in line with the results of the flow cytometry assay. Furthermore, p38-MAPK signal transduction pathway activation induced breast cancer invasion and metastasis. Silencing SQSTM1/p62 expression specifically inhibited p38 activation in tumor tissue samples (Fig. 6H). Together, these results suggest that silencing SQSTM1/p62 expression changes cytokine production profiles in the serum and gene expression profiles in tumor tissue. Silencing SQSTM1/p62 resulted in effector CTL and Th1 cell infiltration and decreased recruitment of Th2 cells, MDSCs and macrophages to the spleen (Fig. 5). To identify the mechanism by which SQSTM1/p62 impacts immune cell infiltration, we evaluated multiple chemokine expression changes in the TME. Real-time PCR results showed that CCL2, CCL5 and CXCL9 were three of the most significantly altered chemokines in tumor tissue samples (Fig. 6I). The level of the chemokine CCL2, which is also known as MCP-1 and displays chemotactic activity for monocytes, was significantly decreased (Fig. 6L). We also found that the level of CCL3 (MIP-2) was decreased (Fig. 6M). Moreover, the expression of genes involved in the CCL5-CXCL9 axis was significantly increased by silencing SQSTM1/p62 (Fig. 6J, K). Consistent with these results, the expression of chemokines changed due to silencing SQSTM1/p62 in 4T1 cells. Collectively, these results indicate that knocking down SQSTM1/p62 expression affects cytokine production and expression of chemokines.
Figure 6.
Knocking down SQSTM1/p62 reshapes cytokines production. (A) Heat map analysis of the serum inflammatory cytokine production fold changes via ELISA assay in each group. (B–H) Gene expression profiles of tumor tissue samples from mice analyzed using real-time PCR. IL-1α/1β, IL-2/6/13, IFN-γ and p38 gene expression was normalized to GAPDH expression. (I) Gene expression profiles of chemokines in tumor tissue samples analyzed using real-time PCR. Gene expression was normalized to GAPDH expression. (J–M) Histogram analyses of the gene expression levels of CCL1-5 and CXCL1/9. One out of three representative experiments is shown in the graphs, and each group contains two mice with three duplicate wells. Data are presented as the mean ± SEM. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; and ns non-significance as determined by one-way ANOVA for RT-PCR analysis.
p62-shRNA specifically inhibits tumor cells dissemination
Bone and lung metastases of breast cancer are important causes of cancer-related death.3 To evaluate the effect of SQSTM1/p62 silencing on cancer cell metastasis, we intravenously injected p62-shRNA-, shNC- or 4T1 cells at a dose of 1 × 105 cells per mouse and monitored survival. Nearly 80% of the mice in the p62-shRNA group survived beyond day 21, while less than 50% of the mice in the shNC–or 4T1 Ctrl group survived. Knocking down SQSTM1/p62 expression in tumor cells significantly enhanced the mouse survival rate (Fig. 7A). As expected, metastatic nodules were observed on the surface of the lungs in the shNC and Ctrl groups, and almost no metastatic nodules were observed in the p62-shRNA group (Fig. 7B). Consistent with these results, the number of metastatic nodules was decreased in the p62-shRNA group (Fig. 7C). H & E staining showed that immune cell infiltration into the lung tissue was significantly higher in the shNC and 4T1 Ctrl groups than in the p62-shRNA group (Fig. 7D). In line with these results, the number of metastatic nodules per microscopic field of view was also decreased in the p62-shRNA group compared with the shNC and 4T1 Ctrl groups (Fig. 7E). It has been reported that neutrophils enhance cancer cell metastasis via neutrophil extracellular trap (NET) formation.25 We evaluated the composition of immune cells in the lung broncho alveolar lavage fluid (BALF) via cytospin (Fig. 7F). Quantity counts showed that the number of tumor-promoting neutrophils was decreased in the p62-shRNA group (Fig. 7G). In addition, a flow cytometry assay showed that CD11b+ Ly6G+ neutrophil number was also significantly reduced in the BALF (Fig. 7H, I). Thus, these results demonstrated that knocking down SQSTM1/p62 expression in 4T1 cells inhibited lung metastasis and reduced neutrophil infiltration into the BALF.
Figure 7.
Knocking down the expression of SQSTM1/p62 in 4T1 cells decreases breast cancer metastasis. (A) Survival curves of the treated mice (n = 8) in each group. The animals were sacrificed at day 24 after i.v. injection of tumor cells at a dose of 1 × 105 cells per mouse. (B) Representative images of the lungs of the mice in each group (n = 3–5) (C) Histogram analyses of the metastatic lung nodule numbers of each group. The dots in the graph represent the number of macroscopic tumor nodules in the lungs of each mouse. (D) Representative H & E-stained left lung sections from each group. (E) Histogram analyses of lung macro-mets (no. nodules per microscopic field) in each section. (F) Representative image of cells in the BALF stained with Giemsa buffer by cytospin. (G) Histogram analyses of the percentage of infiltrating lung neutrophils per microscopic field (n = 16). (H) Representative flow cytometry plots of the cells in the BALF. The CD11b+ Ly6G+ cells detected by flow cytometry are shown as neutrophils. (I) Histogram analyses of the number of neutrophils in the BALF. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001; ∗∗∗∗P < 0.0001; and ns non-significance as determined by one-way ANOVA for the metastatic lung nodules and infiltrating neutrophils and by Kaplan-Meier log-rank test for the survival curves. Data are presented as the mean ± SEM.
Discussion
Autophagy is a double-edged sword for tumor progression and metastasis. The understanding of the autophagy pathway during tumorigenesis has rapidly advanced over recent years. For example, TME autophagy-mediated TNF and IL-6 pathway activation by stressed tumor cells rapidly induces tumor growth in a Drosophila melanogaster malignant tumor model.26 Previously, bacterial infection-mediated cell autophagy was shown to specifically inhibit chemotherapeutic drug-induced apoptosis. CAFs have been implicated as promotors of tumor progression, and new research suggests that autophagy is also upregulated in head and neck squamous cell carcinoma (HNSSC)-associated CAFs.27 Conversely, multiple autophagy inhibitors that focus on inhibiting lysosome formation, such as chloroquine (CQ) and hydroxy chloroquine (HCQ), and the functional autophagy regulators VPS34, ULK1, and ATG4B have been applied in clinical studies.28,29 Our study reveals that the autophagy adaptor SQSTM1/p62 is overexpressed in multiple tumor types from The Cancer Genome Atlas (TCGA) database, which has a poor effect on cancer patient OS and DFS. We also found that SQSTM1/p62 was highly expressed in breast cancer tumor tissue samples (Fig. 1).
A previous study reported that SQSTM1/p62 was necessary for HCC tumorigenesis, which was dependent on gene activation of NRF2 and mTORC1 and retraining HCC cell death.30 After screening by shRNA-mediated knockdown of SQSTM1/p62 expression in the triple-negative murine breast cancer cell line 4T1, we found that silencing SQSTM1/p62 not only inhibited cell proliferation but also promoted cell death in vitro (Fig. 2). Interestingly, the shp62 stable cell line growth slower in vitro and we found that p27 expression was increased in p62-shRNA 4T1 cells, suggesting that knocking down SQSTM1/p62 expression may induce cell cycle arrest in vitro. In line with this result, we used a cell cycle kit for detection, and the flow cytometry results suggested that silencing SQSTM1/p62 induced cell cycle arrest in cancer cells (Fig. 2), which is consistent with the results of a previous study showing that curcumin affects neural stem cell autophagy by modulating SQSTM1/p62.31 Moreover, we revealed that the migration and invasion capabilities were also reduced due to the inhabitation of SQSTM1/p62, which were also observed in osteosarcoma (Fig. 3). Furthermore, TGF-β1-mediated cell cycle arrest has been shown to increase the number of G0/G1-phase cells and decrease the proliferation of ovarian cancer cells in vitro and in vivo.32 We found that shp62 suppressed TGF-β pathway-related genes p38, JNK, and SMAD3 expression. Then, we found that stable knockdown p62 significantly suppressed tumor growth both in subcutaneous and orthotopic implantation model (Fig. 4). However, there were no differences between the shNC and model groups (Fig. 4). Additionally, tumor volumes and weights and spleen weights were significantly decreased by SQSTM1/p62 knockdown, but there was no change in lung weight (Fig. 4). Thus, our results demonstrated that knocking down the expression of SQSTM1/p62 in breast cancer cells inhibited tumorigenesis in vivo and induced cell cycle arrest via p27 upregulation in vitro.
Our previous studies demonstrated that TGF-β1- and HPV E7-presented chimeric virus-like particles significantly suppressed tumor growth through TME regulation.20,33 SQSTM1/p62 also participates in regulating the TME via TAMs, which promote tumor growth. Endogenous and exogenous stimuli induce mitochondrial damage in TAMs that results in the production of mitochondrial DNA (mtDNA) and reactive oxygen species (ROS), which directly activate NLRP3 inflammasome-mediated pyroptosis along with production of the tumor-promoting cytokines IL-1β and IL-18.34 In addition, SQSTM1/p62 has been reported to control this damage. Importantly, SQSTM1/p62-expressing CRCs regulate immunosuppressive FOXP3+ regulatory T cells in the TME.19 Knocking down SQSTM1/p62 expression in human stromal cells significantly decreases IL-6 and VCAM-1 levels in the TME, which results in tumor cell growth.35 Our finding that SQSTM1/p62 deficiency changed splenocytes immune cell profiles offers insights into the mechanism underlying SQSTM1/p62-mediated TME regulation. The numbers of antitumor CTLs and Th1 cells increased, while the numbers of tumor-promoting Th2 cells, TAMs and TANs decreased (Fig. 5). Additionally, TILs and splenocytes profiles regulation showed the same tendency in convincing orthotopic implantation model. The dendritic cell was one of the most important antigen presentation cells, whose maturation can promote T cell differentiation and proliferation. Here, we found that knockdown of p62 in 4T1 cells not only enhanced MHC I (H-2Kd) gene expression but also promoted the co-cultured DC maturation in vitro. The oncogenic transcription factor Twist1 has been identified as one of the key downstream effectors of SQSTM1/p62 whose degradation regulates tumorigenesis,36 induces CCL2 expression and recruits macrophages to promote angiogenesis.37 Further studies are required to identify how Twist1, a SQSTM1/p62 downstream signal, regulates immune cell infiltration into the TME during tumorigenesis. Additionally, TME cytokine production plays important roles in tumorigenesis and immunotherapy.22 We found that the levels of the Th1 cytokines IL-12, TNF-α and IFN-γ were increased, while those of the Th2 cytokines IL-4, IL-10 and IL-13 were decreased (Fig. 6). Altogether, our results suggested that knocking down SQSTM1/p62 expression in 4T1 cells affected immune cell infiltration and induced cytokine changes in the TME that inhibited tumor growth.
Chemokines facilitate immune cell infiltration into the TME to drive tumor progression or inhibition. Expression of CCL2/3/4/5 and CXCL9/10 is associated with the presence of tumor infiltrating lymphocytes (TILs) and Th1 immunity activation in melanoma.38 In breast cancer, high expression of CXCR4, CCR7 and CXCL12 has a critical role in determining the metastatic phenotype.39 In addition, the CCL5/CCR5 axis recruits immunosuppressive cells into the TME and promotes tumor progression in specific subtypes of breast cancer.40 High expression of CCL2 and CCL5 in estrogen receptor (ER)-positive breast cancers increases TAM infiltration and angiogenesis in the TME, which reveals potential therapeutic targets for ER (+) cancers.41 CCL5 also activates CCR3, which is highly expressed on CD4+ T cells, and the CCL5/CCR3 axis pathway enhances breast cancer metastasis, which is beneficial for Th2 polarization of CD4+ T cells.42 We observed that the CCL5/CXCL9 axis was activated in SQSTM1/p62-deficient tumors (Fig. 6), which was consistent with the CTL influx into the TME and a previous work.43 Further studies are required to establish how knocking down SQSTM1/p62 expression mediates CCL5 expression during tumorigenesis. We also found that the immune checkpoint blockage PDL1 expression was reduced in shp62 cells (Fig. 3D), which may turn the 4T1 cells from ‘cold’ tumor to ‘hot’ tumor and sensitive tumor cells to killer T cells. Furthermore, cancer metastasis and drug resistance are urgent problems in breast cancer immunotherapy. We found that silencing SQSTM1/p62 in tumor cells decreased lung metastasis and neutrophil influx in vivo (Fig. 7).
Conclusion
In summary, we demonstrated that knock down SQSTM1/p62 expression in breast cancer inhibited tumor progression and metastasis in vivo due to the regulation of immune cell infiltration into the TME, enhanced cell death, and cell cycle arrest via overexpression of p27. In addition to current tumor immunotherapies and chemotherapies to control cancer progression and metastasis, new strategies that regulate SQSTM1/p62 activation may also contribute to preventing tumorigenesis and metastasis. How SQSTM1/p62 regulates immune cell infiltration into the TME via chemokine expression and controls the cell cycle gene expression network during cancer tumorigenesis and metastasis are important questions that need to be answered in the future. In short, our results highlight the important functions of SQSTM1/p62 in breast cancer tumorigenesis and metastasis and present potential therapeutic targets for drug development to control cancer.
Ethics declaration
Animal experiments were approved by the Ethics Committee of Animal Care and Welfare of IMBCAMS and PUMC (Permit Number: SYXK (dian) 2010–0007).
Consent for publication
No identifying patient details are contained within this manuscript.
Availability of data and materials
All of the data and materials in this paper are available when requested.
Author contributions
JL Q, JR H, CB L and YB M conceptualized, designed, supervised this study and take responsibility for data integrity and accuracy of analysis; JR H, JL Q, acquired, analyzed and interpreted the data; SM J, X Y and HM B provide materials and technical support; JR H and JL Q wrote this manuscript; YB M revised and edited the manuscript. All authors read and approved the final manuscript.
Conflict of interests
The authors declare that they have no conflicts of interest.
Funding
This work was financially supported by the CAMS Initiative for Innovative Medicine (No. 2016-I2M-1-019), 2017-I2M-3- 022, the Fundamental Research Funds for the Central Universities of China (No. 3332019162), and the funds for IMBCAMS PhD Innovation (No. 2018018001), and the Foundation for Studying Abroad from the China Scholarship Council (No. 201808110121, 201906210477).
Acknowledgements
We thank Professor Jiankun Yu and Suping Dai from the Institute of Medical Biology, Chinese Academy of Medical Sciences (IMBCAMS).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2021.03.008.
List of Abbreviations
- ELISA
Enzyme-linked immunosorbent assay
- WB
Western Blotting
- H&E
Hematoxylin-eosin staining
- OS
Overall survival
- DFS
Disease-free survival
- shRNA
Short-hairpin RNA
- CTLs
Cytotoxic lymphocyte
- TAMs
Tumor-associated macrophages
- TANs
Tumor-associated neutrophils
- MDSCs
Myeloid-derived suppressor cells
- DC
Dendritic cell
- TME
Tumor microenvironment
- CAFs
Cancer-associated fibroblasts
- TB
PB1/TRAF6 binding domain
- LIR
LC3-interacting region
- UBA
Ubiquitin-associated domain
- NF-κB
Nuclear factor kappa-B
- HCC
Hepatocellular carcinoma
- IFN
Interferon
- IL
Interleukin
Appendix ASupplementary data
The following are the Supplementary data to this article:
References
- 1.Momenimovahed Z., Salehiniya H. Epidemiological characteristics of and risk factors for breast cancer in the world. Breast Cancer (Dove Med Press) 2019;11:151–164. doi: 10.2147/BCTT.S176070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan L., Strasser-Weippl K., Li J.J., et al. Breast cancer in China. Lancet Oncol. 2014;15(7):e279–e289. doi: 10.1016/S1470-2045(13)70567-9. [DOI] [PubMed] [Google Scholar]
- 3.Waks A.G., Winer E.P. Breast cancer treatment: a review. JAMA. 2019;321(3):288–300. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- 4.Weigelt B., Peterse J.L., van 't Veer L.J. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5(8):591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 5.Moscat J., Karin M., Diaz-Meco M.T. p62 in cancer: signaling adaptor beyond autophagy. Cell. 2016;167(3):606–609. doi: 10.1016/j.cell.2016.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saito T., Ichimura Y., Taguchi K., et al. p62/Sqstm1 promotes malignancy of HCV-positive hepatocellular carcinoma through Nrf2-dependent metabolic reprogramming. Nat Commun. 2016;7:12030. doi: 10.1038/ncomms12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng Z., Lim J., Wang Q., et al. ALS-FTLD-linked mutations of SQSTM1/p62 disrupt selective autophagy and NFE2L2/NRF2 anti-oxidative stress pathway. Autophagy. 2020;16(5):917–931. doi: 10.1080/15548627.2019.1644076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duran A., Hernandez E.D., Reina-Campos M., et al. p62/SQSTM1 by binding to vitamin D receptor inhibits hepatic stellate cell activity, fibrosis, and liver cancer. Cancer Cell. 2016;30(4):595–609. doi: 10.1016/j.ccell.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S.S., Xu L.Z., Zhou W., et al. p62/SQSTM1 interacts with vimentin to enhance breast cancer metastasis. Carcinogenesis. 2017;38(11):1092–1103. doi: 10.1093/carcin/bgx099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu L.Z., Li S.S., Zhou W., et al. p62/SQSTM1 enhances breast cancer stem-like properties by stabilizing MYC mRNA. Oncogene. 2017;36(3):304–317. doi: 10.1038/onc.2016.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang X., Bao Y., Liu H., et al. VPS34 stimulation of p62 phosphorylation for cancer progression. Oncogene. 2017;36(50):6850–6862. doi: 10.1038/onc.2017.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou F., Jiang X., Teng L., Yang J., Ding J., He C. Necroptosis may be a novel mechanism for cardiomyocyte death in acute myocarditis. Mol Cell Biochem. 2018;442(1–2):11–18. doi: 10.1007/s11010-017-3188-5. [DOI] [PubMed] [Google Scholar]
- 13.Klebanoff C.A., Gattinoni L., Palmer D.C., et al. Determinants of successful CD8+ T-cell adoptive immunotherapy for large established tumors in mice. Clin Canc Res. 2011;17(16):5343–5352. doi: 10.1158/1078-0432.CCR-11-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W., Pan Y., Gou P., et al. Effect of xanthohumol on Th1/Th2 balance in a breast cancer mouse model. Oncol Rep. 2018;39(1):280–288. doi: 10.3892/or.2017.6094. [DOI] [PubMed] [Google Scholar]
- 15.Lu Y., Wang Q., Xue G., et al. Th9 cells represent a unique subset of CD4+ T cells endowed with the ability to eradicate advanced tumors. Cancer Cell. 2018;33(6):1048–1060. doi: 10.1016/j.ccell.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Umansky V., Blattner C., Gebhardt C., Utikal J. The role of myeloid-derived suppressor cells (MDSC) in cancer progression. Vaccines. 2016;4(4):36. doi: 10.3390/vaccines4040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabrilovich D.I. Myeloid-derived suppressor cells. Cancer Immunol Res. 2017;5(1):3–8. doi: 10.1158/2326-6066.CIR-16-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valencia T., Kim J.Y., Abu-Baker S., et al. Metabolic reprogramming of stromal fibroblasts through p62-mTORC1 signaling promotes inflammation and tumorigenesis. Cancer Cell. 2014;26(1):121–135. doi: 10.1016/j.ccr.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosumi K., Masugi Y., Yang J., et al. Tumor SQSTM1 (p62) expression and T cells in colorectal cancer. OncoImmunology. 2017;6(3):e1284720. doi: 10.1080/2162402X.2017.1284720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu X., Li Y., Huang W., et al. Combined immunization against TGF-β1 enhances HPV16 E7-specific vaccine-elicited antitumour immunity in mice with grafted TC-1 tumours. Artif Cells Nanomed Biotechnol. 2018;46(sup2):1199–1209. doi: 10.1080/21691401.2018.1482306. [DOI] [PubMed] [Google Scholar]
- 21.Read K.A., Powell M.D., McDonald P.W., Oestreich K.J. IL-2, IL-7, and IL-15: multistage regulators of CD4(+) T helper cell differentiation. Exp Hematol. 2016;44(9):799–808. doi: 10.1016/j.exphem.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 22.Tang H., Qiao J., Fu Y.X. Immunotherapy and tumor microenvironment. Cancer Lett. 2016;370(1):85–90. doi: 10.1016/j.canlet.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyman O., Kolios A.G., Raeber M.E. Modulation of T cell responses by IL-2 and IL-2 complexes. Clin Exp Rheumatol. 2015;33(4 Suppl 92):S54–S57. [PubMed] [Google Scholar]
- 24.Grimm E.A., Owen-Schaub L. The IL-2 mediated amplification of cellular cytotoxicity. J Cell Biochem. 1991;45(4):335–339. doi: 10.1002/jcb.240450405. [DOI] [PubMed] [Google Scholar]
- 25.Park J., Wysocki R.W., Amoozgar Z., et al. Cancer cells induce metastasis-supporting neutrophil extracellular DNA traps. Sci Transl Med. 2016;8(361):e361ra138. doi: 10.1126/scitranslmed.aag1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katheder N.S., Khezri R., O'Farrell F., et al. Microenvironmental autophagy promotes tumour growth. Nature. 2017;541(7637):417–420. doi: 10.1038/nature20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.New J., Arnold L., Ananth M., et al. Secretory autophagy in cancer-associated fibroblasts promotes head and neck cancer progression and offers a novel therapeutic target. Cancer Res. 2017;77(23):6679–6691. doi: 10.1158/0008-5472.CAN-17-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy J.M.M., Towers C.G., Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17(9):528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chude C.I., Amaravadi R.K. Targeting autophagy in cancer: update on clinical trials and novel inhibitors. Int J Mol Sci. 2017;18(6):1279. doi: 10.3390/ijms18061279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umemura A., He F., Taniguchi K., et al. p62, upregulated during preneoplasia, induces hepatocellular carcinogenesis by maintaining survival of stressed HCC-initiating cells. Cancer Cell. 2016;29(6):935–948. doi: 10.1016/j.ccell.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J.L., Wang J.J., Cai Z.N., Xu C.J. The effect of curcumin on the differentiation, apoptosis and cell cycle of neural stem cells is mediated through inhibiting autophagy by the modulation of Atg 7 and p62. Int J Mol Med. 2018;42(5):2481–2488. doi: 10.3892/ijmm.2018.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bu S., Zhang Q., Wang Q., Lai D. Human amniotic epithelial cells inhibit growth of epithelial ovarian cancer cells via TGFbeta1-mediated cell cycle arrest. Int J Oncol. 2017;51(5):1405–1414. doi: 10.3892/ijo.2017.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu X., Li Y., Long Q., et al. Chimeric HBcAg virus-like particles presenting a HPV 16 E7 epitope significantly suppressed tumor progression through preventive or therapeutic immunization in a TC-1-grafted mouse model. Int J Nanomed. 2016;11:2417–2429. doi: 10.2147/IJN.S102467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong Z., Umemura A., Sanchez-Lopez E., et al. NF-kappaB restricts inflammasome activation via elimination of damaged mitochondria. Cell. 2016;164(5):896–910. doi: 10.1016/j.cell.2015.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiruma Y., Honjo T., Jelinek D.F., et al. Increased signaling through p62 in the marrow microenvironment increases myeloma cell growth and osteoclast formation. Blood. 2009;113(20):4894–4902. doi: 10.1182/blood-2008-08-173948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiang L., Zhao B., Ming M., et al. Regulation of cell proliferation and migration by p62 through stabilization of Twist 1. Proc Natl Acad Sci U S A. 2014;111(25):9241–9246. doi: 10.1073/pnas.1322913111. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Low-Marchelli J.M., Ardi V.C., Vizcarra E.A., van Rooijen N., Quigley J.P., Yang J. Twist 1 induces CCL2 and recruits macrophages to promote angiogenesis. Cancer Res. 2013;73(2):662–671. doi: 10.1158/0008-5472.CAN-12-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harlin H., Meng Y., Peterson A.C., et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009;69(7):3077–3085. doi: 10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller A., Homey B., Soto H., et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410(6824):50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 40.Velasco-Velázquez M., Xolalpa W., Pestell R.G. The potential to target CCL5/CCR5 in breast cancer. Expert Opin Ther Targets. 2014;18(11):1265–1275. doi: 10.1517/14728222.2014.949238. [DOI] [PubMed] [Google Scholar]
- 41.Svensson S., Abrahamsson A., Rodriguez G.V., et al. CCL2 and CCL5 are novel therapeutic targets for estrogen-dependent breast cancer. Clin Cancer Res. 2015;21(16):3794–3805. doi: 10.1158/1078-0432.CCR-15-0204. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Q., Qin J., Zhong L., et al. CCL5-Mediated Th2 immune polarization promotes metastasis in luminal breast cancer. Cancer Res. 2015;75(20):4312–4321. doi: 10.1158/0008-5472.CAN-14-3590. [DOI] [PubMed] [Google Scholar]
- 43.Dangaj D., Bruand M., Grimm A.J., et al. Cooperation between constitutive and inducible chemokines enables T cell engraftment and immune attack in solid tumors. Cancer Cell. 2019;35(6) doi: 10.1016/j.ccell.2019.05.004. 885–900.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data and materials in this paper are available when requested.