Abstract
Aberrant activation of oncogenic signaling pathways in tumors can promote resistance to the antitumor immune response. However, single blockade of these pathways is usually ineffective because of the complex crosstalk and feedback among oncogenic signaling pathways. The enhanced toxicity of free small molecule inhibitor combinations is considered an insurmountable barrier to their clinical applications. To circumvent this issue, we rationally designed an effective tumor microenvironment-activatable prodrug nanomicelle (PNM) for cancer therapy. PNM was engineered by integrating the PI3K/mTOR inhibitor PF-04691502 (PF) and the broad spectrum CDK inhibitor flavopiridol (Flav) into a single nanoplatform, which showed tumor-specific accumulation, activation and deep penetration in response to the high glutathione (GSH) tumoral microenvironment. The codelivery of PF and Flav could trigger gasdermin E (GSDME)-based immunogenic pyroptosis of tumor cells to elicit a robust antitumor immune response. Furthermore, the combination of PNM-induced immunogenic pyroptosis with anti-programmed cell death-1 (αPD-1) immunotherapy further boosted the antitumor effect and prolonged the survival time of mice. Collectively, these results indicated that the pyroptosis-induced nanoplatform codelivery of PI3K/mTOR and CDK inhibitors can reprogram the immunosuppressive tumor microenvironment and efficiently improve checkpoint blockade cancer immunotherapy.
Key words: Pyroptosis, Prodrug, Stimuli-responsive, Immunotherapy, Targeted therapy, Tumor microenvironment
Graphical abstract
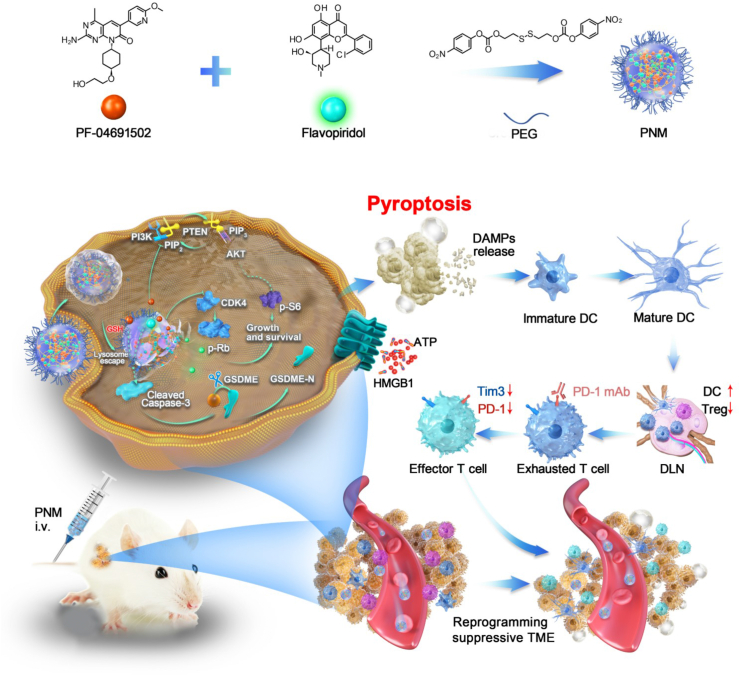
A GSH-activable prodrug nanomicelle co-delivering PI3K/mTOR inhibitor PF-04691502 and broad spectrum CDK inhibitor flavopiridol for potentiating cancer immunotherapy is developed.
1. Introduction
The success of immune checkpoint blockade (ICB) therapy in patients with a wide variety of malignancies has revolutionized the traditional treatment plan in clinical oncology practice1. Despite the encouraging results, only a small fraction of patients shows durable responses; most patients show nonresponse to ICB therapy, especially in certain solid tumors2. One of the major factors of tumor-intrinsic resistance to ICB therapy is the aberrant activation of multiple canonical oncogenic signaling pathways, contributing to the formation of an immunosuppressive tumor microenvironment3, 4, 5. Therefore, developing synergistic strategies targeting dual or multiple oncogenic signaling pathways may potentially reshape antitumor immune responses and enhance the response rate of ICB therapy.
Oncogenic signaling is an important component of tumor immune evasion, of which phosphoinositide 3-kinase (PI3K) and cyclin-dependent kinase (CDK) signaling are the most potent modulators of tumorigenesis and immune evasion6,7. PI3K was confirmed as one of the most frequently mutated genes in most solid tumors, including breast cancer8,9. Activation of the PI3K/Akt/mTOR pathway in tumor cells inhibits the cytotoxic effects of tumor-specific T cells and decreases T cell trafficking into tumors, which promotes resistance to T cell mediated immunotherapy10. PI3K/mTOR inhibitors can propagate T cell-mediated antitumor immunity by inducing MHC-I and MHC-II expression in tumor cells to improve antigen presentation11. The results of single-cell transcriptomic analysis indicated that the CDK4/6-driven signature was associated with immune exclusion and poor response to immune checkpoint blockade12. Furthermore, activation of the CDK-Rb signaling pathway usually compromises the therapeutic effect of PI3K signaling inhibitors13. Several studies have shown that the combination of PI3K and CDK4/6 inhibitors could overcome intrinsic and adaptive resistance, leading to tumor regression14,15. Therefore, combined targeting of oncogenic signaling pathway-PI3K/mTOR and CDK may effectively lead to tumor regression and modulate immunosuppressive tumor microenvironment.
Pyroptosis, a kind of gasdermin-induced programmed cell death that is characterized by cell swelling with big bubbles, pore formation and cell content release, has been considered an immune-stimulatory form of cell death that can activate antitumor immunity and enhance the antitumor effect of ICB therapy16, 17, 18, 19. Existing studies have indicated that molecular inhibitors targeting oncogenic signaling can elicit gasdermin E (GSDME)-induced pyroptotic tumor cell death20,21. Activation of the traditional apoptotic marker cleaved caspase-3 can also function as a pyroptosis regulator to selectively cleave GSDME to convert apoptosis into pyroptosis, which depends on the expression levels of GSDME in cells22. However, the pyroptosis induction ability of targeted anticancer agents has been substantially underestimated. Both PI3K/mTOR and CDK inhibitors can promote tumor cell death by inducing the activation of the pyroptosis regulator caspase-323. Consequently, we hypothesized that small molecule-targeted drug synergistically inhibiting PI3K/mTOR and CDK can induce pyroptosis in tumor cells, thereby enhancing the antitumor efficacy of ICB therapy.
However, the activation of the PI3K/mTOR and CDK signaling pathways is also found in normal tissues, which might contribute to systemic toxicity and reduce the bioavailability of molecular inhibitors24,25. Furthermore, adjacent normal tissue and hematopoietic cells are prone to cytotoxic agent-induced pyroptosis because of the widespread expression of gasdermins in normal tissues26. Therefore, how to improve the tumor targeting ability and reduce the side effects of small molecular inhibitors is an insurmountable barrier to their clinical application27. Stimuli-responsive prodrug nanomedicine is an important drug delivery system for achieving tumor-specific therapeutics, minimizing systemic toxicity due to the exposure of healthy tissue to cytotoxic agents28, 29, 30, 31, 32, 33, 34, 35, 36. Furthermore, a nanoparticle-based cancer therapy platform could be designed to modulate the immunosuppressive tumor microenvironment and reduce untoward off-target side effects37, 38, 39, 40, 41. Significant inherent differences between the tumor microenvironment and normal tissues can be readily exploited to design a suitable responsive drug release system42, 43, 44, 45, 46, 47, 48, 49, 50.
In this study, tumor microenvironment (TME)-activatable prodrug nanomicelle (PNM) was constructed by integrating the PI3K/mTOR inhibitor PF-04691502 (PF) and the broad spectrum CDK inhibitor flavopiridol (Flav) into a single nanoplatform through a GSH-responsive crosslinker (DBHD). The designed PNM showed tumor-specific accumulation, activation, release and deep penetration in response to the high glutathione (GSH) tumoral microenvironment, efficiently inhibiting tumor growth, alleviating myelosuppression and relieving the immunosuppressive tumor microenvironment (ITM). Furthermore, PNM could trigger GSDME-mediated immunogenic pyroptosis of tumor cells to elicit a robust antitumor immune response (Scheme 1). Moreover, synthetic PNM improved the antitumor effect and prolonged the survival time of anti-programmed cell death-1 (αPD-1) ICB therapy. In conclusion, we designed a GSH-responsive PNM-enabled horizontal blockade of the PI3K/mTOR and CDK pathways, which would be a reliable and effective strategy to robustly elicit pyroptotic cell death and improve antitumor immunotherapy.
Scheme 1.
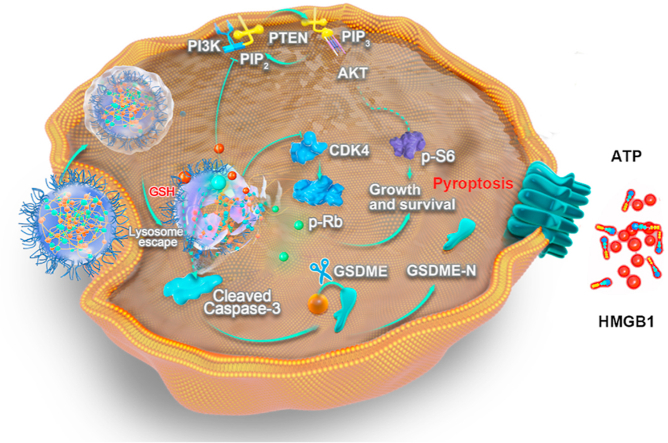
Schematic illustration of the targeted anticancer PNM inducing pyroptosis and improving antitumor immunity. Flav and PF can form stable prodrug nanomicelle (termed PNM) through the cross-linking of DBHD, which can be released in a high-GSH tumor microenvironment. After inducing pyroptosis of tumor cells, PNM could effectively elicit an antitumor immune response and improve the antitumor effect of αPD-1 immunotherapy.
2. Materials and methods
2.1. Materials and instruments
PF-04691502 (PF) (HY-15177) and Flavopiridol (Flav) (HY-10005) were purchased from MCE (NJ, USA). MPEG-NH2 was purchased from Xian Ruixi Biological Technology Co., Ltd. (Xian, China). RPMI-1640 medium, fetal bovine serum (FBS) and penicillin/streptomycin (PS) were acquired from ThermoFisher Scientific (Waltham, MA, USA). The in vivo anti-mouse PD-1 mAb (CD279) was obtained from BioXcell (West Lebanon, NH, USA). The antibodies used in this study were as follows: p-AKTS473 [#4060, cell signaling technology (CST), Boston, MA, USA], p-S6S235/236 (#4858, CST), CDK4 (GTX102993, GeneTex, San Antonio, TX, USA), p-RbS807/S811 (#8516, CST), CD8 (#98941, CST), Ki-67 (#12202, CST), Cleaved caspase-3 (#9664, CST), granzyme B (#44153, CST), Foxp3 (#12653, CST), GSDME (ab215191, Abcam, Cambridge, UK), HMGB1 (#6893, CST), FVD (eBioscience, San Diego, CA, USA), anti-CD45-APC-CY7 (BD Biosciences, Franklin Lakes, NJ, USA), anti-CD3-PE (BD Biosciences), anti-CD4-FITC (eBioscience), anti-CD8-PerCP (Biolegend, San Diego, CA, USA), anti-PD-1-BV421 (Biolegend), anti-Tim3-APC(Biolegend), anti-CD11b-FITC (Biolegend), anti-CD11c-FITC (eBioscience), anti-Ly6C-APC (eBioscience), anti-Ly6G-PE (eBioscience), anti-CD80-PE (eBioscience), anti-CD86-APC (eBioscience) and anti-Foxp3-PerCP (eBioscience). All other chemicals were purchased from commercial suppliers and used as received without further purification. The hydrodynamic diameter and surface potential were determined by a size analyzer Nano ZS90 (Malvern, UK). Morphological examination was performed with a JEOL 100CX II TEM (JEOL, Japan). The absorption spectra of free drugs and the PNM were measured by UV absorption spectroscopy (UV-1800, Shimadzu, Japan).
2.2. Cell culture
The 4T1 cells were cultured in RPMI-1640 medium (HyClone) with 10% fetal bovine serum (FBS) and 1% antibiotics containing penicillin and streptomycin at 37 °C in a 5% CO2 incubator.
2.3. Animals
Female BALB/c mice (6–8 weeks) were purchased from the Hubei Provincial Center for Disease Prevention and Control (Wuhan, China). All mice were housed in the specific pathogen-free, light-cycled, and temperature-controlled facility of the Hospital of Stomatology, Wuhan University. All animal experiments were approved and supervised by the Laboratory Animal Ethical Committee of Hospital of Stomatology, Wuhan University (S07919050G).
2.4. Preparation of the PNM
The synthesis of DBHD was carried out according to previous studies51. Typically, 9 μmol/L of Flav and 9 μmol/L of PF were dissolved in 1.5 mL DMSO in a reaction tube. After 30 min of stirring, 6 μmol/L of DBHD and 30 μmol/L TEA were dispersed in 0.15 mL DMSO dropwise into the reaction tube via a syringe under stirring. Then, the reaction tube was transferred to a 55 °C oil bath for 12 h in the dark, 6 μmol/L MPEG-NH2 was added to react with the excess cross-linker DBHD for end-capping and surface modification, and the reaction was continued for 12 h. Finally, PNM was obtained through dialysis to remove unreacted substances and DMSO in the reaction system. The potential and particle size of the synthesized PNM in different media were measured separately. Moreover, the stability of the particle size during the treatment period was measured. The Flav and PF loading efficiency and encapsulation efficiency were determined by high-performance liquid chromatography (HPLC) and calculated by the following Eqs. (1), (2):
| Loading efficiency (LE, %) = (Weight of Flav/PF in the PNM/Weight of PNM) × 100 | (1) |
| Encapsulation efficiency (EE, %) = (Weight of Flav/PF in the PNM/Weight of Flav/PF in feeding) × 100 | (2) |
2.5. In vitro drug release
The dialysis method was applied to study the responsive breakage of PNM in different mediums. The medium consisted of either (1) pH 7.4, 0.01 mol/L PBS buffer, (2) pH 7.4, 0.01 mol/L PBS buffer with 10 mmol/L GSH. PNM solutions (1.0 mL, 0.1 mg/mL) were transferred into dialysis bags (MW 3500) with 30 mL of such medium. At different time points, 1 mL buffer was removed and added to the same volume of the corresponding fresh buffer solution. The release of drugs in different media was measured using HPLC.
2.6. GSH consumption
ThiolTracker Violet (405 nm excitation, 525 nm emission) was used as the GSH consumption probe. First, 4T1 cells were seeded into 12-well plates at a density of 1 × 105 cells per well and incubated for 12 h. After all the cells were attached to the plate, fresh medium containing PNM (PF = 0.6 μg/mL, Flav = 0.6 μg/mL) was added and incubated at 37 °C for 4 h, while the group without treatment was set as the control. Then, the culture medium was removed, and ThiolTracker Violet (Thermo Fisher) (20 μmol/L) was added to incubate the cells. After 0.5 h of incubation, the cells were washed with PBS three times and observed by confocal laser scanning microscope (CLSM) (Olympus, Japan).
2.7. MTT assays
First, 4T1 cells (1 × 104 cells/well) were seeded in 96-well plates and cultured for 12 h. Then, the culture medium was removed, and fresh medium with PF (0, 0.2, 0.4, 0.6, 0.8 or 1 μg/mL), Flav (0, 0.2, 0.4, 0.6, 0.8 or 1 μg/mL), PF + Flav and PNM (equivalent dose of PF and Flav) was added. After incubation for 24 h, the medium was replaced with fresh medium containing 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (0.5 mg/mL). After 4 h of incubation, 100 μL DMSO was added to each well, and cell viability was evaluated by testing the absorbance at 570 nm.
2.8. Penetration in 3D tumor spheroids
DIR-loaded PNM was synthesized to observe the distribution of PNM in tumor cells and in vivo. After DBHD and TEA were added to the reaction tube and the reaction was continued for 12 h in a 55 °C oil bath, 30 μL DIR (10 mg/mL dissolved in DMSO) was added to the reaction tube, and the reaction was continued for 12 h. After that, the MPEG-NH2 was added to the mixture solution and the reaction was continued for 12 h. Next, DIR-loaded-PNM was obtained through dialysis. Multicellular spheroid (MCS)-based 4T1 cells were prepared according to our previous report51. Briefly, after the 4T1 MCS was formed, the medium was removed and fresh culture medium containing PNM-DIR was added to the MCS and incubated for one, four and 24 h. Images were obtained by CLSM.
2.9. Cellular uptake performance of the PNM
First, 4T1 cells (1 × 105 cells/well) were seeded in 12-well plates and cultured for 12 h at 37 °C. The medium was removed and washed with PBS three times, and then fresh medium containing PNM-DIR (PF = 0.4 μg/mL, Flav = 0.4 μg/mL) was added to culture the cells for 10 min, 30 min, 1 h, 2 h and 4 h. The cell uptake rate was evaluated by flow cytometry.
2.10. Lysosomes colocalization assay
First, 4T1 cells (1.2 × 105 cells/well) were seeded in 8-well plates and cultured for 12 h at 37 °C. The medium was removed and washed with PBS three times, and then fresh medium containing PNM-DIR (PF = 0.4 μg/mL, Flav = 0.4 μg/mL) was added. After incubation for 1, 4, 6, 8 and 12 h, the cells were stained with LysoTracker Green DND-99 and MitoTracker Green FM for 45 min, and the colocalization conditions were observed by CLSM.
2.11. Near-infrared fluorescence (NIFR) imaging and of pharmacokinetic profiles DIR-loaded PNM
Since the drug does not have fluorescence that can be excited, DIR was selected as a fluorescence probe to observe the distribution of the drug in tumor-bearing mice. DIR-loaded PNM and free DIR were injected into tumor-bearing mice (DIR, 1 μg/g per mouse) through the tail vein, and in vivo fluorescence images at 12, 36, 60 and 84 h were obtained by an IVIS Lumina imaging station (PerkinElmer, MA, USA). The main organs (heart, liver, spleen, lung and kidney) and tumors of mice were collected after the mice were euthanized, and measured by an imaging system.
To determine the pharmacokinetic profiles of PNM, 8 weeks old SD rats were divided into two groups: free Flav and PNM (Flav = 2.5 mg/kg, n = 3). After injection of free Flav or PNM, we separately collected 0.5 mL blood samples from the rats at 0.5, 3, 7, 24 and 48 h, and then the blood samples were centrifuged for 10 min at the speed of 3000 rpm to acquire serum. After precipitating the protein with methanol, the concentration of Flav in free Flav group and PNM group were detected by the HPLC-MS/MS (AB SCIEX, MA, USA).
2.12. Intercellular delivery of PNM-DIR
Intercellular delivery of PNM-DIR was utilized to better explain the deep infiltration effect of PNM52. 4T1 cells were seeded on coverslips (A, B, C in different wells) in 6-well plates, and then cells in coverslip A were separately treated with PNM-DIR or free-DIR (DIR = 0.9 μg/mL) for 8 h. The pretreated cells (in coverslips A) were moved and washed with PBS for 3 times and then cocultured with fresh cells (in coverslip B) for 24 h in fresh medium. The cells (in coverslip B) were withdrawn and cocultured with fresh cells (in coverslip C) for another 24 h in fresh medium. Then, the cells (A, B, C) were fixed with formalin for 15 min and washed with PBS for three times. At last, the slides were sealed by anti-quenching mounting medium with DAPI and observed with CLSM.
2.13. Western blotting analysis and cell supernatant collection
First, 4T1 cells were treated with PBS, PF (0.6 μg/mL), Flav (0.6 μg/mL), PF + Flav and PNM (equivalent dose of PF and Flav) for 24 h. After incubation, the cells were collected and lysed in lysis buffer for 20 min on ice, and then centrifuged at 12,000×g at 4 °C for 5 min. The concentrations of proteins were measured by a BCA Protein Assay Kit (Beyotime Biotechnology, Shanghai, China). After denaturation, equal amounts of proteins were separated by 10% SDS-PAGE gel electrophoresis and transferred to polyvinylidene fluoride (PVDF) membranes (Roche Diagnostics GmbH, Mannheim, Germany). The membranes were then blocked with 5% skimmed milk for 1 h and incubated with primary antibodies at 4 °C for 12 h. After incubation with the appropriate secondary antibody, the membrane was treated with chemiluminescent substrates, visualized using a chemiluminescence Odyssey Imaging System (Odyssey, Bad Homburg, Germany) to show the expression levels of proteins. Cell supernatants were harvested in the FBS free medium to avoid distortion of SDS-PAGE. After centrifugation to remove cell debris, cell supernatants were concentrated 10-fold using Amicon Ultra 10 kDa filter (Sigma-Aldrich, MO, USA). Concentrated supernatants were mixed with loading buffer (Bio-Rad, CA, USA) and analyzed by Western blotting.
2.14. Antitumor effects of the PNM in vivo
To evaluate the antitumor effect of PNM, 4T1 cells (1 × 106 cells) were subcutaneously inoculated into the right flank of each BALB/c mouse to establish tumors. After seven days, when the average tumor volume reached 80–100 mm3, the mice were randomly divided into five groups: PBS, PF (5 mg/kg, i.p.), Flav (5 mg/kg, i.p.), PF + Flav (equivalent dose of PF and Flav, i.p.) and PNM (equivalent dose of PF and Flav, i.v.). The prodrugs were injected every four days for four cycles. The tumor volume and body weight were recorded every two days, and the tumor volume was calculated by the following Eq. (3):
| V (mm3) = 1/2 × (Tumor length) × (Tumor width)2 | (3) |
2.15. Flow cytometric analysis
Flow cytometry was applied to characterize the change in immune cell populations in the tumor, draining lymph nodes (DLNs) and spleen, as previously reported53. Excised tumors and spleens were processed by a gentle MACS Dissociator (Miltenyl Biotec, Teterow, Germany) to obtain a single cell suspension, while the lymph nodes were ground through a filter to obtain a single cell suspension. Then, the cell suspension from the tumor was separated from Lymphoprep (STEMCELL Technologies Inc., #07801, Vancouver, Canada) to obtain mononuclear cells (including monocytes and lymphocytes). The single cell suspensions were stained with antibodies according to the manufacturer's protocols. The gating strategy for flow cytometry is shown in Supporting Information Fig. S1.
2.16. In vivo combination of αPD-1 with the PNM
To evaluate the synergistic antitumor effect of PNM with αPD-1 immunotherapy, 4T1 cells (1 × 106 cells) were subcutaneously inoculated into the right flank of each BALB/c mouse to establish tumors. After 8 days, when the tumor volume was notable, the mice were randomly divided into four groups: PBS, PNM (PF or Flav = 5 mg/kg, i.v.), αPD-1 (5 mg/kg, i.p.) and PNM+αPD-1 (equivalent dose of PNM and αPD-1). The tumor volume and body weight were recorded every two days until mouse death or the tumor volume reached 2500 mm3.
2.17. Immunohistochemistry and immunofluorescence
Paraffin-embedded excised tumors were cut into 4 μm thick sections for the following experiments. First, the sections were deparaffined and rehydrated after baking at 60 °C for 2 h. Then, the sections were placed in a pressure cooker with 0.01 mol/L citric acid buffer solution (pH 6.0), and antigens were retrieved by pressure cooking.
For immunohistochemical staining, 3% endogenous peroxidase blocker was added to the tissues to quench endogenous peroxidase and 10% goat serum was added to the sections to block non-specific binding. Then, the sections were incubated with primary antibodies. Thereafter, a secondary biotinylated IgG solution and an avidin-biotin-peroxidase reagents were incubated with the sections, and then, 3,3′-diaminobenzidine tetrachloride was added to detect the binding of these antibodies with their specific antigens. Finally, the slides were counterstained with hematoxylin for 1–2 min.
For immunofluorescence staining, 10% goat serum was added to the sections to block nonspecific binding for 2 h. Then, the sections were incubated with primary antibodies against CD11c (#97585, CST), HMGB1 (#6893, CST) and CRT (#97585, CST) from different species at 4 °C overnight. Next, the sections were incubated with secondary antibodies against DyLight 488 or DyLight 594 for another 2 h. After washing with PBS three times in the dark, the sections were sealed with anti-quenching fluorescent sealing tablets with DAPI. All sections were scanned with a 3D HISTECH Pannoramic Midi scanner (3D HISTECH Ltd., Budapest, Hungary).
2.18. Statistical analysis
GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, USA) was utilized to analyze the data. Unpaired t test was applied to analyze statistically significant differences between two groups. One-way analysis of variance (ANOVA) followed by Tukey's multiple comparisons and two-way ANOVA followed by Tukey's multiple comparisons was used to analyze statistically significant differences between more than two groups. The results are presented as the mean ± standard error of the mean (SEM) and statistical significance was determined as P < 0.05 (∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001).
3. Results and discussion
3.1. Synthesis and characterization of PNM
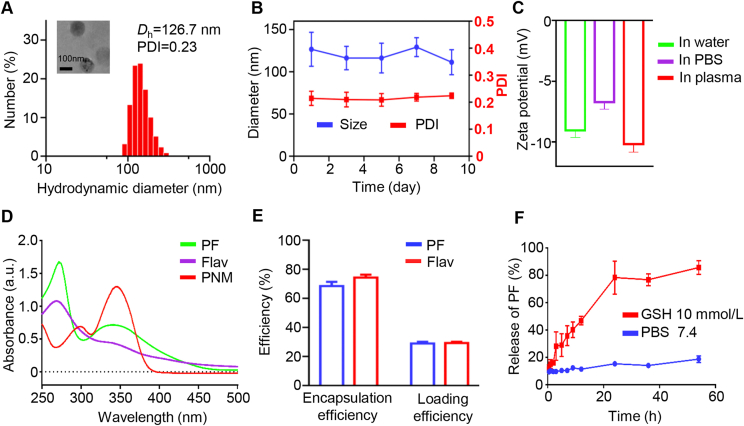
First, to prepare tumor microenvironment-responsive PNM, we synthesized GSH-responsive crosslinker of DBHD according to our previous report, which can be used to deliver drug molecules with amino and hydroxyl groups51. Then, we used the GSH-responsive crosslinker to crosslink the amino and hydroxyl group-containing molecules PF and Flav to form a stable prodrug nanomicelle (termed PNM). The size and shape of the resultant PNM were obtained by dynamic light scattering (DLS) and transmission electron microscopy (TEM). As shown in Fig. 1A and Supporting Information Fig. S2, TEM images and DLS clearly revealed that the PNM exhibited a spherical morphology and an average homogenous hydrodynamic diameter of 126.7 nm, which was favorable for its enrichment in tumors through blood circulation54. The polydispersity index (PDI), hydrodynamic diameter and zeta potential of the PNM showed a rational change within 9 days, suggesting that the PNM was stable and convenient for storage and animal inoculation (Fig. 1B and Supporting Information Fig. S3). Then, the zeta potential of the PNM in water, PBS and plasma was detected by DLS, and the results showed that there was no obvious change in different media, further illustrating its stability (Fig. 1C), which was beneficial for avoiding phagocytosis by macrophages and prolonging circulation. Furthermore, the successful synthesis of the PNM could be confirmed by UV absorption spectroscopy (Fig. 1D). The drug loading and encapsulation efficiency were measured by high-performance liquid chromatography (HPLC), while the resultant PNM displayed excellent PF and Flav loading efficiency (PF = 28.92 ± 2.11%, Flav = 29.33 ± 1.42%) and encapsulation efficiency (PF = 68.47 ± 5.00%, Flav = 74.26 ± 3.57%) (Fig. 1E). All of these results indicated the high stability and high drug loading of the PNM, which was conducive to its long-term storage and large-scale preparation.
Figure 1.
Physiochemical characterization of the PNM. (A) The average size of the PNM by DLS and TEM. (B) The long-term stability of the PNM in size and PDI. (C) The zeta potential of the PNM in water, PBS and plasma. (D) The UV absorption spectrum of PF, Flav and PNM. (E) The encapsulation efficiency and loading efficiency of the PNM. (F) The release profiles of PF in 10 mmol/L GSH and PBS. Data are shown as the mean ± SD (n = 3).
3.2. Drug release and GSH consumption
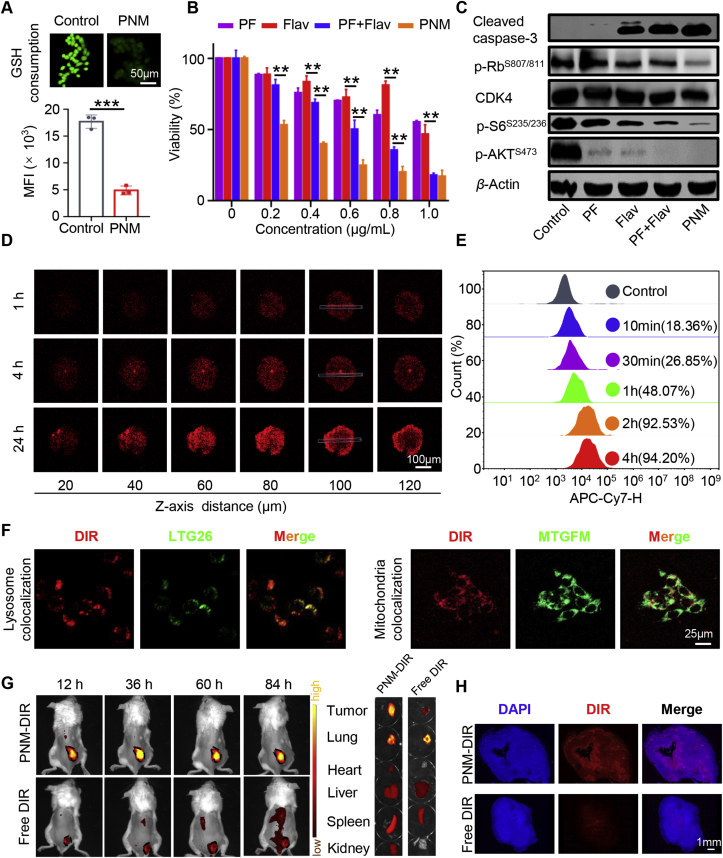
The responsive release of the PNM can avoid leakage in normal tissues and improve its biological safety. According to our design, the disulfide bonds distributed in the PNM can be activated by high concentrations of GSH in tumor cells, which can be transformed from stabilization to instability and eventually lead to the release of two active drugs. Therefore, we further examined GSH-triggered drug release from the PNM by simulating its release behavior in vitro. As shown in Fig. 1F, the release of PF increased to about 80% at 24 h in the presence of 10 mmol/L GSH, indicating the collapse of the nanomicelle in tumor cells. Such a high releasing property could help achieve optimal drug-induced inhibition in tumor sites and avoid unwanted systemic toxicity. The dense disulfide bonds in the PNM could react with GSH to destroy the redox equilibrium state in tumor cells to maximize the cytotoxic effect of the prodrug55. To verify the consumption of GSH in tumor cells, we used ThiolTracker Violet to observe the consumption of GSH in tumor cells. As shown in Fig. 2A, the green fluorescence of cells treated with the PNM was darker, while bright green fluorescence can be observed in untreated cells, indicating the consumption of GSH, which can promote tumor cell death by disrupting the redox balance.
Figure 2.
In vitro biological performance and in vivo biodistribution. (A) Confocal images showing 4T1 cells treated with PNM and then stained with ThiolTracker Violet. (B) MTT assay-derived curves of PF, Flav, PF + Flav and PNM in 4T1 cells after 24 h of treatment. (C) The relative protein expression levels of cleaved caspase-3, p-AKTS473, p-S6S234/235, CDK4 and p-RbS807/S811 in 4T1 cells after treatment with PF, Flav, PF + Flav and PNM for 24 h. (D) In vitro tumor permeability results for the 4T1 MCSs treated with PNM-DIR for 24 h. (E) Flow cytometric profiles of the cells treated with PNM-DIR for 10 min, 30 min, 1 h, 2 h and 4 h. (F) Confocal images showing the cells treated with PNM-DIR for 4 h and then stained with LTG26 and MTGFM. (G) IVIS spectrum images of the 4T1-bearing mice at different times upon injection of PNM-DIR or free DIR via tail vein injection. (H) Fluorescence of DIR in the tumor sections of the PNM-DIR or free DIR-treated mice at 84 h. Data are shown as the mean ± SD (n = 3) (∗∗P < 0.01; ∗∗∗P < 0.001).
3.3. Cell cytotoxicity and tumor penetration of the PNM in vitro
We selected 4T1 cells for the following research due to the overactivation of the PI3K/mTOR and CDK4/6 signaling pathways in breast cancer56. As shown in Fig. 2B, as expected, the combination of the two free drugs could cause more tumor cell death compared with single free drug, while the PNM showed stronger cytotoxicity on tumor cells than the other forms of drugs, possibly due to nanoparticle-accelerated or nanoparticle-augmented intracellular transport, improved water solubility and controlled release. Specifically, the cell viability with the PNM at a concentration of PF = 0.6 μg/mL just reached about 40%, while the other groups reached about 70%, 75% and 50%, respectively. And based on the cell cytotoxicity experiment, we have calculated synergy coefficient (CI) of PF and Flav by using CalcuSyn (version 1.1.1). As shown in Supporting Information Fig. S4, when fraction affected (Fa) > 0.3704, the CI < 1, which indicated that PF and Flav have a synergistic effect. Moreover, Western blotting assays were applied to verify the combined inhibitory effect of the PNM on the PI3K/mTOR and CDK4/6 signaling pathways in 4T1 cell. As shown in Fig. 2C and Supporting Information Figs. S5A–E, although PF reduced the expression of p-AKTS473 and p-S6S234/235, the expression of p-RbS807/S811 increased in the PF treatment group, indicating compensatory activation of the CDK-Rb signaling pathway when the PI3K signaling pathway was inhibited. And the combination of PF and Flav significantly inhibited the expression of p-AKTS473, p-S6S234/235 and p-RbS807/S811, indicating the rational design of combination therapy to overcome the adaptive resistance of targeted therapy. Furthermore, the expression level of cleaved caspase-3 increased in the combination group and the PNM group, suggesting that the rational combination of PF and Flav activated the caspase-associated signaling pathway, which could efficiently induce tumor cell death.
Complex delivery barriers in solid tumors severely impede drug penetration in tumor tissues57,58. The 4T1 MCS is often used to simulate solid tumors in vitro because of its three-dimensional characteristics; thus, MCS was established to observe the tissue penetration capability of the PNM. The fluorescence intensity in the MCS gradually increased with time, and the red fluorescence was distributed throughout the spheroids, including the edge and center, after 24 h of incubation (Fig. 2D). The quantification of dynamic fluorescence intensity in 100 μm sections also showed the time dependence of penetration through spheroids (Supporting Information Fig. S6A), indicating the excellent tumor penetration property of the PNM which will help improve the therapeutic effect of the prodrug on solid tumors. Furthermore, intercellular delivery of PNM-DIR was applied to better explain the excellent tumor penetration property of the PNM. As shown in Supporting Information Figs. S7A and B, the PNM-DIR-treated cells (coverslip A) showed widely distributed fluorescence and the infected cells (coverslips B and C) still exhibited obvious red fluorescence, indicating the PNM-DIR have the repeated infection properties, which was favorable for the deep penetration property of PNM in tumor.
3.4. Cellular uptake of the PNM by 4T1 cells in vitro
PNM labeled with DIR was first synthesized and measured by flow cytometry (FCM) and CLSM to evaluate the cellular uptake ability of the PNM in vitro. As shown in Fig. 2E, after 2 h of incubation with PNM-DIR, the percentage of positive cells increased to 92.53%, and the mean fluorescence intensity in the cells gradually increased, which indicated a gradual accumulation process of the PNM in tumor cells (Supporting Information Fig. S6B). After internalization by tumor cells, the PNM can be transported into lysosomes and further transported to other organelles. If the PNM cannot be transferred to other parts in the lysosome, it would result in low delivery efficiency and poor therapeutic effects. Therefore, a lysosome colocalization assay was applied to explore the behavior of the PNM in tumor cells, which shown that the colocalization coefficient of PNM and LysoTracker gradually decreases, indicating that PNM could escape from the lysosome. After incubation with PNM-DIR for 4 h, DIR fluorescence could be observed in mitochondria of cells (Fig. 2F, Supporting Information Figs. S6C, S6D, S8A and S8B), also indicating that the PNM can successfully escape to other parts of the cell from lysosomes.
3.5. Biodistribution and therapeutic effect of the PNM in vivo
Although the PI3K/mTOR signaling pathway is a major oncogenic driver implicated in 30%–40% of primary breast tumors59, the activation of PI3K is also required for T cell activation and DC maturation60, indicating that an inhibitor should ideally be preferentially targeted to tumor cells to avoid the unwanted effect on immune cells. Therefore, the tumor microenvironment-responsive ability of the PNM is important for reduced toxicity and enhanced therapeutic efficacy. To visualize the tumor-targeting ability and real-time distribution of the PNM, we labeled the PNM with fluorescent molecule (DIR), and the IVIS spectrum was used to investigate the distribution of the PNM in 4T1 tumor-bearing mice. As shown in Fig. 2G and Supporting Information Fig. S9A, fluorescence images showed that PNM accumulation in tumors achieved the highest level at 36 h post-injection, whereas free DIR in 4T1 tumors showed a weaker fluorescent signal. At 84 h post-injection, fluorescence images of organs (heart, liver, spleen, lung, kidney, tumor) showed that the PNM-DIR mainly accumulated in tumors, whereas the fluorescent signal of free DIR mainly accumulated in the lung (Fig. 2G and Supporting Information Fig. S9B). Analysis of tumor tissues also showed that PNM had deeper penetration than free DIR (Fig. 2H). These results indicated that the prepared PNM had the ability to enhance tumor aggregation and revealed its potential as an ideal nanocarrier for the specific delivery of therapeutic molecules in vivo. The pharmacokinetic profile of PNM was investigated by intravenously injecting the PNM or free Flav. And as shown in Supporting Information Fig. S10, the Flav in PNM exhibit higher concentration and longer blood circulation in compared with free Flav, consistent with the biodistribution behavior of PNM.
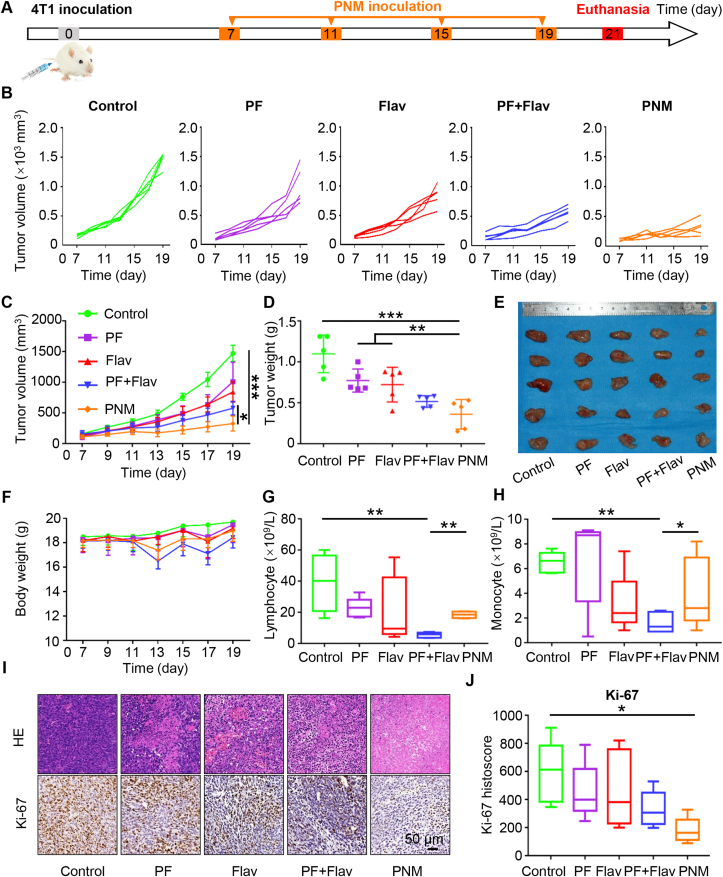
Encouraged by the superior tumor-targeting ability, we further evaluated the in vivo antitumor efficacy of the PNM in 4T1 tumor-bearing mice. The treatment schedule is displayed in Fig. 3A, and the mice were randomly divided into five groups (PBS, PF, Flav, PF + Flav and PNM) when the xenograft tumor was notable. As shown in Fig. 3B and C, PF or Flav alone exhibited little tumor inhibition, while the PF + Flav group showed more obvious tumor growth suppression than the PBS-treated group, confirming the synergistically enhanced therapeutic performance of combined therapy in comparison with PF or Flav alone therapy. Moreover, the tumor growth inhibition of the PNM was higher than that of the free drug combination, partially attributed to an effective accumulation of PNM at the tumor sites via the enhanced penetration and retention (EPR) effect and tumor microenvironment responsive release. A representative photograph and the corresponding weight of each excised tumor also validated the outstanding antitumor performance of the PNM (Fig. 3D and E). Furthermore, the mice in the PF + Flav group showed more obvious fluctuations in body weight during therapy in comparison with those in the PNM group (Fig. 3F), indicating that the prepared PNM can reduce the systemic toxicity caused by combination therapy. At the end of antitumor studies, we separately collected the blood serum and whole blood of different groups of mice for blood biochemical analysis and blood cell analysis. The concentrations of ALT, AST, CREA and UREA in blood and hematoxylin and eosin (H&E) staining in normal tissue (heart, liver, spleen, lung and kidney) showed no significant differences among the different groups of mice, indicating that free drug in combination or PNM did not cause significant damage (Supporting Information Figs. S11A‒D and S12). However, the blood cell analysis showed that free drug in combination significantly reduced the number of lymphocytes, monocytes, white blood cells and granulocytes compared with those of the PNM group, indicating that synthesis of PNM reduced the myelosuppression caused by the free targeted drug combination (Fig. 3G and H, Supporting Information Figs. S11E and F). Reduced myelosuppression could improve patients’ tolerance to small molecular inhibitors, which prolonged the drug administration time and strengthened the therapeutic effect. Furthermore, the synthesis of PNM reduced the inhibition of lymphocytes in peripheral blood and enhanced the inhibition of cancer cells, which would be a reliable and effective rationale for combination strategies of PI3K/mTOR and CDK inhibitors.
Figure 3.
In vivo therapeutic effect of PNM in 4T1 tumor-bearing mice. (A) Schedule of establishment of the 4T1 tumor-bearing mouse model and different treatments (n = 5). Individual (B) and average (C) tumor growth curves of the mice treated with PBS, PF, Flav, PF + Flav and PNM. Tumor weight (D) and tumor image (E) of excised tumors. (F) Body weight variation curves of the mice with different treatment schedules. Changes in lymphocyte (G) and monocytes (H) in the blood of the mice with different treatment schedules. (I) Representative pictures of HE staining and Ki-67 staining in tumor sections of different groups (scale bar = 50 μm). (J) The Ki-67 histoscore. Data are shown as the mean ± SEM (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001).
To further evaluate the therapeutic effect of PNM, we performed immunohistochemistry and H&E staining on the tumor tissue. As shown in Fig. 3I, evident deformation and shrinkage of the nuclei and destruction of membrane integrity were observed in the PNM group compared with the intact membrane and plump tumor cell nuclei in the control group (Fig. 3I). Then, we explored the expression levels of p-AKTS473, p-S6S235/236, CDK4 and p-RbS807/S811 in different groups by immunohistochemistry. As shown in Supporting Information Fig. S13, the expression of p-AKTS473, p-S6S235/236 and p-RbS807/S811 in the PNM group was significantly reduced compared with that of the group treated with free drug in combination, indicating the enhanced inhibition of the PI3K/mTOR and CDK signaling pathways in the PNM group. Thus, this treatment could efficiently inhibit tumor cell growth and survival, and reduce the activation of the PI3K/AKT/HIF-1α pathway. Furthermore, the expression of HIF-1α and Ki-67 in the PNM group was lower than that in the group treated with the free drug in combination (Fig. 3I and J and Supporting Information Figs. S14A and B), indicating the enhanced inhibition of cell proliferation and relieved tumor hypoxia, which would be helpful for T cell infiltration and improved antitumor immunity. Representative immunohistochemistry of cleaved caspase-3 also indicated that cell death in the PNM group was increased compared with that in the group treated with the free drug in combination (Supporting Information Figs. S14A and B).
3.6. PF combination with flav activated GSDME-dependent pyroptosis
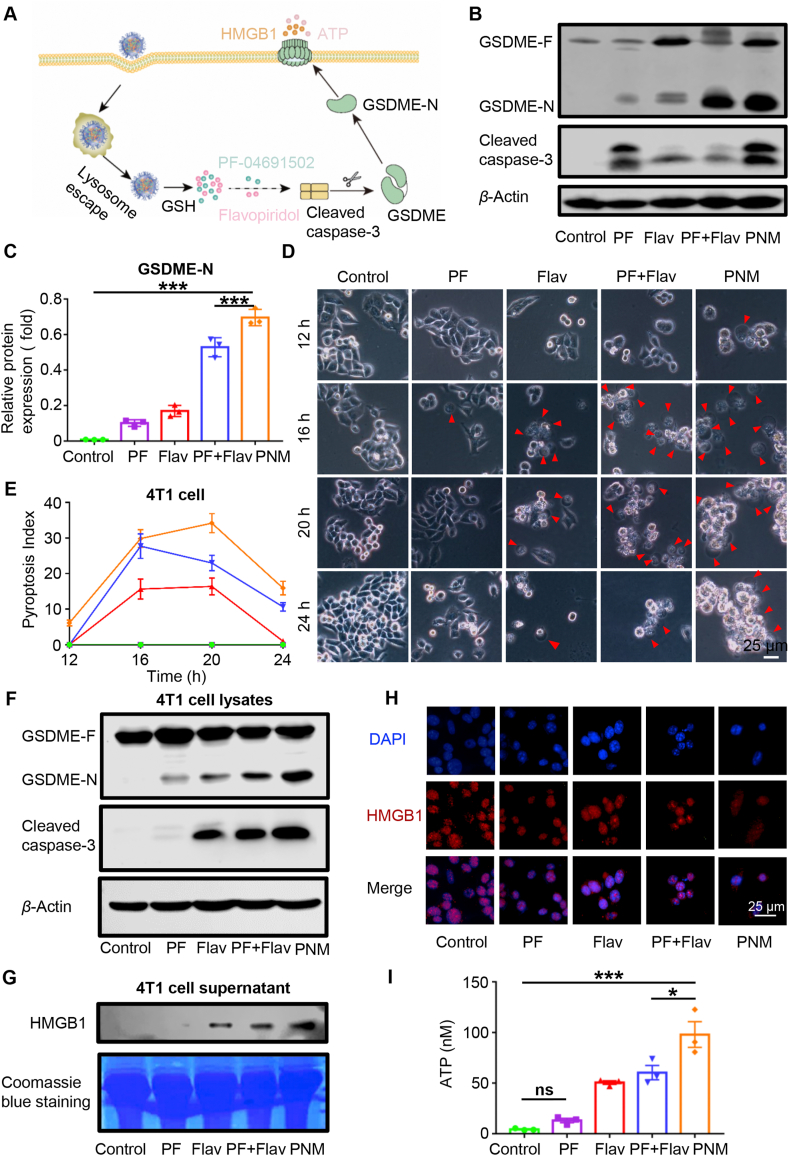
Pyroptosis is considered an immune-stimulatory form of cell death that can activate antitumor immunity16,17. Existing studies have indicated that molecular-targeted therapies can elicit concurrent apoptotic and GSDME-dependent pyroptotic tumor cell death20,21. Activation of the traditional apoptotic marker cleaved caspase-3 selectively cleaved GSDME to convert apoptosis into pyroptotic cell death61. Our research also indicated that the combination of PF and Flav could induce enhanced expression of the pyroptosis regulator cleaved caspase-3. Therefore, we explored whether PF in combination with Flav could activate GSDME-mediated pyroptosis (Fig. 4A). First, we used the collected tumor tissue for Western blotting to explore the activation of GSDME, which could cause pyroptosis of tumor cells. As shown in Fig. 4B and C, PF and Flav individually induced the expression of the cleaved 32 kDa GSDME fragment (GSDME-N) but to a lesser extent than the combination and PNM treatments. The cytomembrane translocation of calreticulin (CRT), an “eat me” signal, could promote phagocytosis by dendritic cells (DCs), thereby facilitating their tumor antigen presentation and recruitment of cytotoxic T cells62. The release of high-mobility group protein B1 (HMGB1) is known to promote antitumor immunity by binding to toll-like receptor four on DCs63. Then, we detected the expression of CRT and HMGB1 in tumor sections by immunohistochemistry. As shown in Supporting Information Figs. S15A and B, the cytomembrane expression of CRT was significantly increased in the PNM treatment group and might promote the recruitment of antigen presenting cells in tumors. The nuclear expression of HMGB1 in the PNM treatment group was significantly reduced, which indicated the release of HMGB1 from nucleus. These results indicated that the combination of PF and Flav could promote the activation of antitumor immunity by inducing immune-stimulatory cell death.
Figure 4.
PNM induction of pyroptosis in tumor cells. (A) Mechanistic illustration of PNM-induced pyroptosis in tumor cells. (B–C) GSDME-N and cleaved caspase-3 protein expression in different groups of tumor tissues by Western blotting. (D) Representative images of cells with different treatment schedules (scale bar = 25 μm). (E) The pyroptosis index (PyI) curve in different treatment groups. (F) GSDME-N and cleaved caspase-3 protein expression in 4T1 cell lysates with different treatment groups by Western blotting. (G) The HMGB1 protein expression in 4T1 cell supernatants from different treatment groups by Western blotting. (H) Fluorescence images of HMGB1 in 4T1 cells with different treatment schedules. (I) The concentration of ATP in 4T1 cell supernatant with different treatment groups at 24 h. Data are shown as the mean ± SEM (n = 3) (∗P < 0.05; ∗∗∗P < 0.001).
Given the diversity of cell components in the tumor microenvironment, the 4T1 cell line was utilized for in vitro experiments to identify the pyroptosis-inducing ability of PF and Flav in cancer cells. Pyroptosis is a kind of gasdermin-mediated programmed cell death, that features transmembrane pore formation and cellular swelling with big bubbles, causing the release of cell contents16. Therefore, we first observed the changes in cell morphology at different times in different treatment groups. Here, we introduced a pyroptosis index (PyI)64, calculated by Eq. (4):
| PyI (%) = (Pyroptotic cells with big bubbles/Total cells per snap shot) × 100 | (4) |
This method can better evaluate and quantify the pyroptosis-induced ability of the prodrug. As shown in Fig. 4D and E, the proportion of pyroptotic cell swelling with big bubbles gradually increased and separately reached a peak at 20 h (PyI = 16), 16 h (PyI = 27.7) and 20 h (PyI = 34) in the PF, PF + Flav and PNM groups. Furthermore, pyroptotic cells appeared earliest and were more abundant in the PNM treatment group compared with the other groups. Further, in order to better evaluate the morphology of pyroptosis, we have used the SEM to observe the cell treated with PNM, and the cell swelling with big bubble and pore formation was displayed (Supporting Information Fig. S16). Then, cell lysates were used for Western blotting to detect the expression of GSDME-N and cleaved caspase-3. We found that PF + Flav and PNM treatment significantly increased the expression of cleaved caspase-3 and GSDME-N (Fig. 4F), which is responsible for pore formation in the cytomembrane65. Thereafter, we detected the release of damage-associated molecular patterns (DAMPs), including HMGB1, CRT and ATP. As shown in Fig. 4G and H, Western blotting analysis and immunofluorescence of HMGB1 indicated that the release of HMGB1 in the cell supernatant in the combination and PNM treatment groups significantly increased compared with that in the free drug treatment group. Consistent with the results in tumor sections, we also found the cytomembrane translocation of CRT in the combination and PNM treatment groups (Supporting Information Fig. S17A). As shown in Fig. 4I and Supporting Information Fig. S17B, the PNM treatment showed a significantly increased concentration of ATP in the cell supernatant, which could mediate a “find me” signal in the tumor microenvironment to promote effective antitumor immune responses66. These data suggest that the combination of PF and Flav could induce GSDME-mediated pyroptosis in tumor cells to promote the release of DAMPs, which might improve the tumor microenvironment and elicit an effective antitumor immune response.
3.7. Immune modulation of the PNM in tumor microenvironment
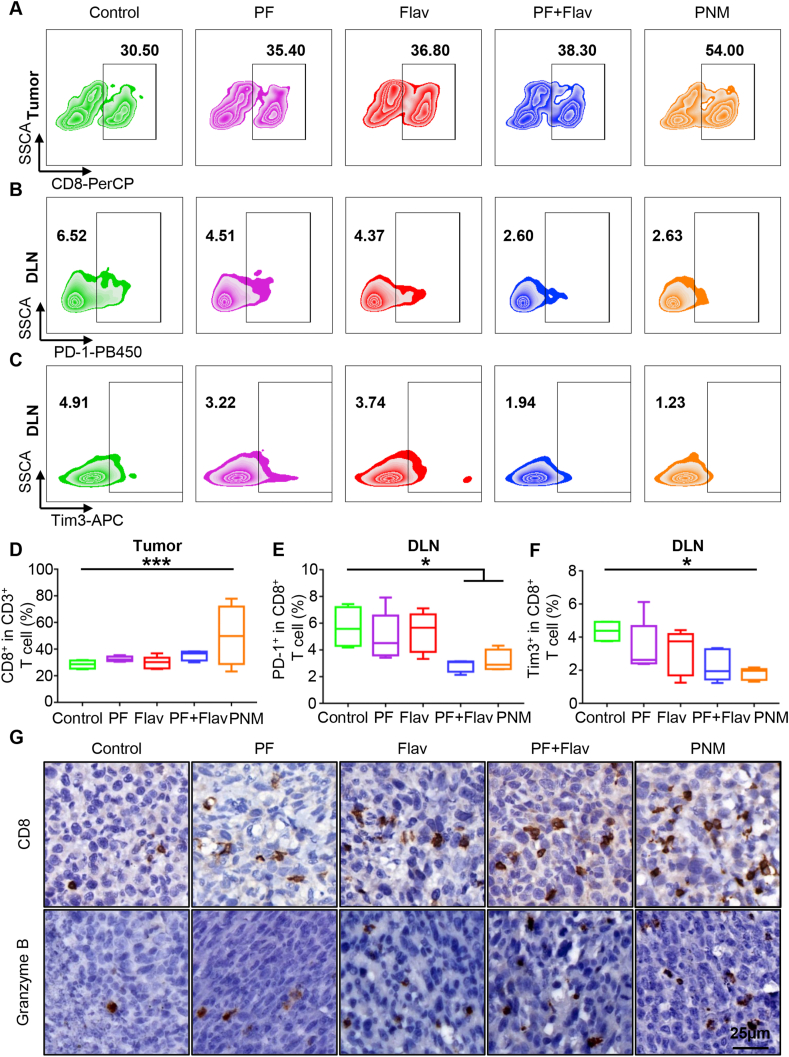
Inspired by the efficient oncogenic signaling inhibition and robust pyroptosis-induced ability of the PNM, we further explored the influence of PNM treatment on the antitumor immune response by analyzing the change of immune cell population in the tumor, DLNs and spleen. Within tumor tissues, the PNM induced a 50.15-±11.3% CD8+ T cell infiltration ratio, which was 1.75-, 1.5-, 1.68- and 1.4-fold higher than that of the PBS, PF, Flav and PF + Flav groups, respectively (Fig. 5A and D). The enhanced tumor infiltration of CD8+ T cells in the PNM treatment group was also verified by CD8 immunohistochemical examination of tumor sections (Fig. 5G). However, there was no increase in CD8+ T cells in spleen, indicating a limited effect on the systemic immune response (Supporting Information Figs. S18A and B). Furthermore, we also performed granzyme B staining in tumor sections, which is responsible for cytotoxic T cell-medicated tumor killing, and the result demonstrated that more granzyme B-positive cells could be observed in tumor sections of the PNM treatment group, indicating increased effector CD8+ T cells in tumors (Fig. 5G). These findings indicated that PNM treatment cumulatively triggered adaptive antitumor immunity in vivo, which was conducive to tumor control and potentiated T cell-based ICB therapy.
Figure 5.
Effects of PNM on T cell infiltration and exhaustion status. (A–C) Representative flow cytometric plots of CD8+ T cells in tumors and Tim3+ T and PD-1+ T cells in DLNs in each group. (D–F) Quantification of CD8+ T cells in tumors and Tim3+ T and PD-1+ T cells in DLNs in different groups (n = 5). (G) Representative immunohistochemical images of CD8 and granzyme B in tumor sections in different groups (scale bar = 25 μm). Data are shown as the mean ± SEM (∗P < 0.05; ∗∗∗P < 0.001).
Exhausted T cells, featuring sustained high expression of immune checkpoint molecules, including PD-1 and TIM3, are dysfunctional T cell populations, that exhibit impaired effector function and contribute to the immune evasion of tumors1. Within the tumor tissues, although increased infiltration of CD8+ T cells was identified, we found that PNM treatment did not induce a significant change in the PD1+CD8+ T and TIM3+CD8+ T cell populations, which suggested a limited effect on reversing T cell exhaustion of the PNM and might provide a rationale for combining PNM with ICB therapy (Supporting Information Figs. S18A and B). Afterwards, although PD1+CD8+ T and TIM3+CD8+ T cell populations took a small fraction in DLNs compared with that in tumors, they were significantly reduced in the DLNs of the PNM treatment group compared with the control group (Fig. 5B, C, 5E and 5F), showing that the fragile immunosuppressive microenvironment in the DLNs is more likely to be reshaped.
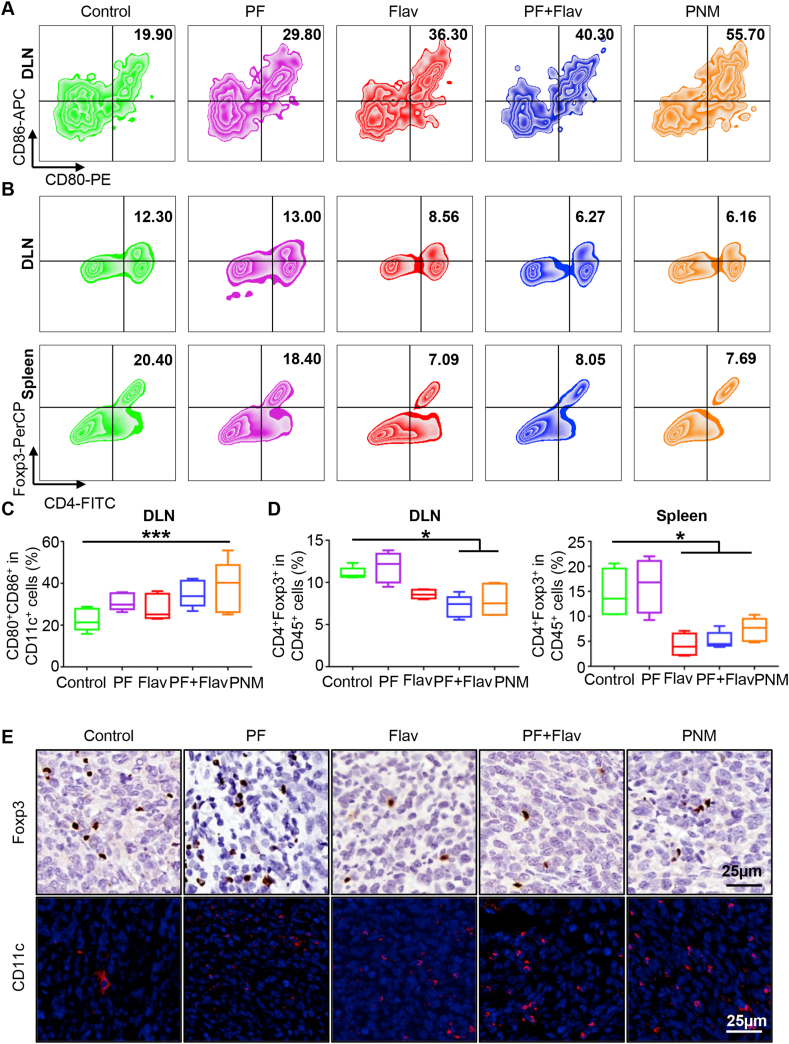
Mature DCs play a crucial role in antigen presentation to T lymphocytes, thus promoting intratumoral infiltration of CD8+ T cells. As shown in Fig. 6A and C, PNM-mediated targeted therapy robustly facilitated 38.06 ± 5.54% DC maturation (CD80+CD86+/CD11c+) compared with 22.56 ± 2.37% mature DCs in the DLNs in the control group, which indicated the initiation of an adaptive immune response and improved antigen presentation in the DLNs in the PNM group. The increase of CD11c immunofluorescence in tumor sections in the PNM treatment group (Fig. 6E) also demonstrated the increased infiltration of DCs in tumors. The inhibition of CDK oncogenic signaling could relieve the immunosuppressive microenvironment by reducing the number of Tregs11. Herein, we observed that the PNM group showed a reduced proportion of Treg (CD4+Foxp3+ T cell in CD45+ cell) in the DLNs, spleen and tumor from 11.08 ± 0.31%, 14.53 ± 2.47% and 15.58 ± 1.8% to 7.90 ± 0.83%, 7.35% ± 1.03% and 11.18 ± 2.44% respectively, which indicated that the immunosuppressive microenvironment could be alleviated by PNM treatment (Fig. 6B and D, Supporting Information Figs. S19A and B). The immunohistochemistry in tumor sections also showed that the PNM treatment could downregulate the expression of Foxp3, which could regulate the development of Tregs and function as a biomarker of Tregs in histology (Fig. 6E). Furthermore, we detected the impact of PNM treatment on another important immunosuppressive cell population myeloid-derived suppressor cells (MDSCs), which play a prominent role in immunosuppression in tumors. We found that the PNM treatment group had a reduced proportion of polymorphonuclear (PMN)-MDSCs (Ly6Ghigh Ly6C−/low in CD11b+) from 28.42 ± 3.44% to 16.19 ± 3.48% in tumors, which might be beneficial for relieving the immunosuppressive status in the tumor microenvironment (Figs. S19A and B). Overall, our research indicated that the designed PNM could significantly inhibit immunosuppressive cell populations, and efficiently promote DC maturation and infiltration of T cells in tumors.
Figure 6.
PNM promotes DC maturation and relieves the immunosuppressive tumor microenvironment. (A and B) Representative flow cytometric plots of CD80+CD86+ cells in DLNs and CD4+Foxp3+ Treg cells in the DLNs and spleen in each group. (C and D) Quantification of CD80+CD86+ cells and CD4+Foxp3+ Treg cells in different group (n = 5). (E) Representative immunohistochemical images of Foxp3 and immunofluorescence of CD11c in tumor sections in different groups (scale bar = 25 μm). Data are shown as the mean ± SEM (∗P < 0.05; ∗∗∗P < 0.001).
3.8. The combination of PNM and αPD-1 improved the antitumor efficacy
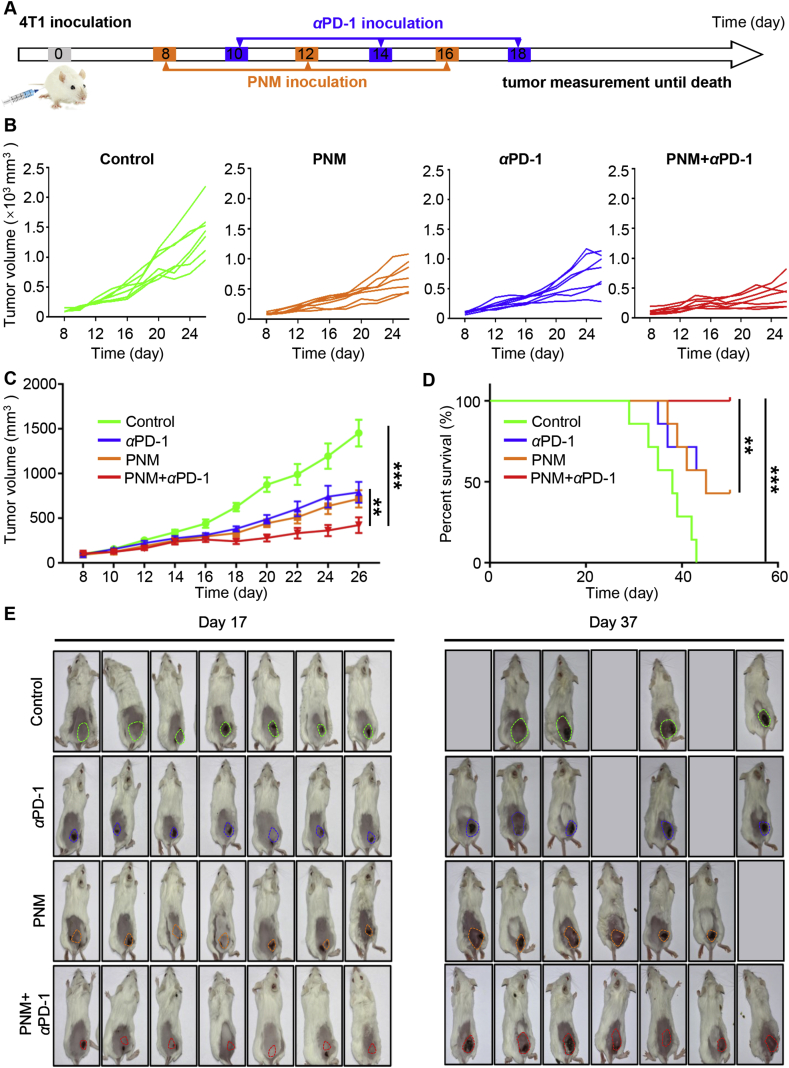
Motivated by the improved tumor microenvironment after PNM treatment, we further explored its ability to potentiate T cell-based checkpoint blockade therapy. The treatment schedule is displayed in Fig. 7A, while PBS, αPD-1, PNM and PNM+αPD-1 were administered for three cycles in 4T1 tumor-bearing mice, respectively. The tumor volume was calculated every two days by a digital caliper until mouse death or the tumor volume reached 2500 mm3.The tumor growth curve indicated that the combination of the PNM with αPD-1 delayed transplanted tumor growth in comparison with PNM and αPD-1 individual treatment (Fig. 7B and C). Furthermore, we did not find significant body weight fluctuation in the PNM+αPD-1 treatment group (Supporting Information Fig. S20), indicating the safety of the combination therapy. The survival curve indicated that the median survival time of the tumor-bearing mice in the control group was 36 days, while the mice in the PNM+αPD-1 treatment group survived until the end of the experiment (50 days) (Fig. 7D). The pictures of the mice in the different groups on Days 17 and 37 also showed that PNM+αPD-1 treatment delayed tumor growth and prolonged the median survival compared with PNM or αPD-1 treatment alone (Fig. 7E). These founding identified that engineered PNM could improve the therapeutic effect of αPD-1 and provide a rationale for the combination of targeted PNM with ICB therapy in cancer.
Figure 7.
Improved antitumor effect of PNM in combination with αPD-1. (A) Schedule of 4T1 tumor establishment and PNM+αPD-1 treatment (n = 7). Individual (B) and average (C) tumor growth curve of mice in different treatment groups (n = seven mice per group). (D) The survival curve of mice in different treatment groups (n = seven mice per group). (E) Photographs of tumors at days 17 and 37 after the first injection are shown. Data are shown as the mean ± SEM (∗∗P < 0.01; ∗∗∗P < 0.001).
4. Conclusions
In conclusion, we report an effective tumor microenvironment-activatable prodrug nanomicelle (PNM) that shows tumor-specific accumulation, activation, release and deep penetration. For the first time, we found that PNM codelivery of PI3K/mTOR and CDK inhibitors can synergistically trigger GSDME-mediated immunogenic pyroptosis of tumor cells to improve tumor immunogenicity and DC maturation. We also introduced an innovative concept pyroptosis index (PyI) to better evaluate and quantify the pyroptosis-inducing ability. This work also identified the synergistic antitumor effect of targeted therapy inhibiting the PI3K and CDK signaling pathways with αPD-1 immunotherapy and may provide inspiration for overcoming ICB resistance.
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (82072996, 81874131, 51703187), the National Key Research and Development Program (2017YFSF090107, China) and the Hubei Province Natural Science Funds for Distinguished Young Scholar (2017CFA062, China), Innovative research team of high-level local universities in Shanghai (ZLCX20180500, China), Chongqing Talents of Exceptional Young Talents Project (CQYC202005029 and cstc2021ycjh-bgzxm0061, China) and the Venture & Innovation Support Program for Chongqing Overseas Returnees (cx2021017). The authors also want to thank Bifeng Yuan and Yajing Feng from Wuhan university for their excellent technical assistance on pharmacokinetic detection and Shuyan Liang and Zhixin Qiu from Wuhan Biobank Co., Ltd. for their excellent technical assistance on flow cytometry.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.02.024.
Contributor Information
Zhigang Xu, Email: zgxu@swu.edu.cn.
Zhijun Sun, Email: sunzj@whu.edu.cn.
Author contributions
Zhijun Sun and Zhigang Xu designed and conceptualized this study. Qichao Yang, Xianbin Ma, Tian Zhang, Yao Xiao and Leilei Yang performed the in vitro and in vivo experiments. Shaochen Yang, Mengyun Liang, Shuo Wang and Zhizhong Wu collected and analyzed the data. Qichao Yang and Xianbin Ma drafted the manuscript. All of the authors have read and approved the final manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Postow M.A., Callahan M.K., Wolchok J.D. Immune checkpoint blockade in cancer therapy. J Clin Oncol. 2015;33:1974–1982. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chow L.Q.M., Haddad R., Gupta S., Mahipal A., Mehra R., Tahara M., et al. Antitumor activity of pembrolizumab in biomarker-unselected patients with recurrent and/or metastatic head and neck squamous cell carcinoma: results from the phase Ib KEYNOTE-012 expansion cohort. J Clin Oncol. 2016;34:3838–3845. doi: 10.1200/JCO.2016.68.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalbasi A., Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol. 2020;20:25–39. doi: 10.1038/s41577-019-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wellenstein M.D., De Visser K.E. Cancer-cell-intrinsic mechanisms shaping the tumor immune landscape. Immunity. 2018;48:399–416. doi: 10.1016/j.immuni.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Spranger S., Gajewski T.F. Impact of oncogenic pathways on evasion of antitumour immune responses. Nat Rev Cancer. 2018;18:139–147. doi: 10.1038/nrc.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fruman D.A., Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13:140–156. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otto T., Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer. 2017;17:93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kandoth C., Mclellan M.D., Vandin F., Ye K., Niu B.F., Lu C., et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie Q., Chi S., Fang Y., Sun Y., Meng L., Ding J., et al. PI3Kalpha inhibitor impairs AKT phosphorylation and synergizes with novel angiogenesis inhibitor AL3810 in human hepatocellular carcinoma. Signal Transduct Target Ther. 2021;6:130. doi: 10.1038/s41392-021-00522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George S., Miao D., Demetri G.D., Adeegbe D., Rodig S.J., Shukla S., et al. Loss of PTEN is associated with resistance to anti-PD-1 checkpoint blockade therapy in metastatic uterine leiomyosarcoma. Immunity. 2017;46:197–204. doi: 10.1016/j.immuni.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petroni G., Buque A., Zitvogel L., Kroemer G., Galluzzi L. Immunomodulation by targeted anticancer agents. Cancer Cell. 2021;39:310–345. doi: 10.1016/j.ccell.2020.11.009. [DOI] [PubMed] [Google Scholar]
- 12.Jerby-Arnon L., Shah P., Cuoco M.S., Rodman C., Su M.J., Melms J.C., et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell. 2018;175:984–997. doi: 10.1016/j.cell.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vora S.R., Juric D., Kim N., Mino-Kenudson M., Huynh T., Costa C., et al. CDK4/6 inhibitors sensitize PIK3CA mutant breast cancer to PI3K inhibitors. Cancer Cell. 2014;26:136–149. doi: 10.1016/j.ccr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zainal N.S., Lee B.K.B., Wong Z.W., Chin I.S., Yee P.S., Gan C.P., et al. Effects of palbociclib on oral squamous cell carcinoma and the role of PIK3CA in conferring resistance. Cancer Biol Med. 2019;16:264–275. doi: 10.20892/j.issn.2095-3941.2018.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Li X., Liu X., Chen Y., Yang C., Tan C., et al. Simultaneous inhibition of PI3Kalpha and CDK4/6 synergistically suppresses KRAS-mutated non-small cell lung cancer. Cancer Biol Med. 2019;16:66–83. doi: 10.20892/j.issn.2095-3941.2018.0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu D., Wang S., Yu G., Chen X. Cell death mediated by the pyroptosis pathway with the aid of nanotechnology: prospects for cancer therapy. Angew Chem Int Ed Engl. 2020;60:8018–8034. doi: 10.1002/anie.202010281. [DOI] [PubMed] [Google Scholar]
- 17.Li J., Anraku Y., Kataoka K. Self-boosting catalytic nanoreactors integrated with triggerable crosslinking membrane networks for initiation of immunogenic cell death by pyroptosis. Angew Chem Int Ed Engl. 2020;59:13526–13530. doi: 10.1002/anie.202004180. [DOI] [PubMed] [Google Scholar]
- 18.Wang Q., Wang Y., Ding J., Wang C., Zhou X., Gao W., et al. A bioorthogonal system reveals antitumour immune function of pyroptosis. Nature. 2020;579:421–426. doi: 10.1038/s41586-020-2079-1. [DOI] [PubMed] [Google Scholar]
- 19.Zhao P., Wang M., Chen M., Chen Z., Peng X., Zhou F., et al. Programming cell pyroptosis with biomimetic nanoparticles for solid tumor immunotherapy. Biomaterials. 2020;254:120142. doi: 10.1016/j.biomaterials.2020.120142. [DOI] [PubMed] [Google Scholar]
- 20.Lu H., Zhang S., Wu J., Chen M., Cai M.C., Fu Y., et al. Molecular targeted therapies elicit concurrent apoptotic and GSDME-dependent pyroptotic tumor cell death. Clin Cancer Res. 2018;24:6066–6077. doi: 10.1158/1078-0432.CCR-18-1478. [DOI] [PubMed] [Google Scholar]
- 21.Erkes D.A., Cai W., Sanchez I.M., Purwin T.J., Rogers C., Field C.O., et al. Mutant BRAF and MEK inhibitors regulate the tumor immune microenvironment via pyroptosis. Cancer Discov. 2020;10:254–269. doi: 10.1158/2159-8290.CD-19-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogers C., Fernandes-Alnemri T., Mayes L., Alnemri D., Cingolani G., Alnemri E.S. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun. 2017;8:14128. doi: 10.1038/ncomms14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blunt M.D., Carter M.J., Larrayoz M., Smith L.D., Aguilar-Hernandez M., Cox K.L., et al. The PI3K/mTOR inhibitor PF-04691502 induces apoptosis and inhibits microenvironmental signaling in CLL and the emicro-TCL1 mouse model. Blood. 2015;125:4032–4041. doi: 10.1182/blood-2014-11-610329. [DOI] [PubMed] [Google Scholar]
- 24.Swanton C. Cell-cycle targeted therapies. Lancet Oncol. 2004;5:27–36. doi: 10.1016/s1470-2045(03)01321-4. [DOI] [PubMed] [Google Scholar]
- 25.Hanker A.B., Kaklamani V., Arteaga C.L. Challenges for the clinical development of PI3K inhibitors: strategies to improve their impact in solid tumors. Cancer Discov. 2019;9:482–491. doi: 10.1158/2159-8290.CD-18-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y., Gao W., Shi X., Ding J., Liu W., He H., et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547:99–103. doi: 10.1038/nature22393. [DOI] [PubMed] [Google Scholar]
- 27.Sun T., Zhang G., Wang Q., Chen Q., Chen X., Lu Y., et al. A targeting theranostics nanomedicine as an alternative approach for hyperthermia perfusion. Biomaterials. 2018;183:268–279. doi: 10.1016/j.biomaterials.2018.04.016. [DOI] [PubMed] [Google Scholar]
- 28.Tao W., Wang J., Parak W.J., Farokhzad O.C., Shi J. Nanobuffering of pH-responsive polymers: a known but sometimes overlooked phenomenon and its biological applications. ACS Nano. 2019;13:4876–4882. doi: 10.1021/acsnano.9b01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nie W., Wu G., Zhang J., Huang L.L., Ding J., Jiang A., et al. Responsive exosome nano-bioconjugates for synergistic cancer therapy. Angew Chem Int Ed Engl. 2020;59:2018–2022. doi: 10.1002/anie.201912524. [DOI] [PubMed] [Google Scholar]
- 30.Hou B., Zhou L., Wang H., Saeed M., Wang D., Xu Z., et al. Engineering stimuli-activatable boolean logic prodrug nanoparticles for combination cancer immunotherapy. Adv Mater. 2020;32 doi: 10.1002/adma.201907210. [DOI] [PubMed] [Google Scholar]
- 31.Hickey J.W., Vicente F.P., Howard G.P., Mao H.Q., Schneck J.P. Biologically inspired design of nanoparticle artificial antigen-presenting cells for immunomodulation. Nano Lett. 2017;17:7045–7054. doi: 10.1021/acs.nanolett.7b03734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo L., Qi Y., Zhong H., Jiang S., Zhang H., Cai H., et al. GSH-sensitive polymeric prodrug: synthesis and loading with photosensitizers as nanoscale chemo-photodynamic anti-cancer nanomedicine. Acta Pharm Sin B. 2022;12:424–436. doi: 10.1016/j.apsb.2021.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma X., Yang S., Zhang T., Wang S., Yang Q., Xiao Y., et al. Bioresponsive immune-booster-based prodrug nanogel for cancer immunotherapy. Acta Pharm Sin B. 2022;12:451–466. doi: 10.1016/j.apsb.2021.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R.E., Costanza F., Niu Y., Wu H., Hu Y., Hang W., et al. Development of self-immolative dendrimers for drug delivery and sensing. J Contr Release. 2012;159:154–163. doi: 10.1016/j.jconrel.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 35.Wang Q., Guo X., Chen Y., Wu Z., Zhou Y., Sadaf S., et al. Theranostics system caged in human serum albumin as a therapy for breast tumors. J Mater Chem B. 2020;8:6877–6885. doi: 10.1039/d0tb00377h. [DOI] [PubMed] [Google Scholar]
- 36.Jiang W., Wang J.L., Yang J.B., He Z.W., Hou Z.H., Luo Y.L., et al. Acidity-triggered TAT-presenting nanocarriers augment tumor retention and nuclear translocation of drugs. Nano Res. 2018;11:5716–5734. [Google Scholar]
- 37.Zheng P., Ding B., Jiang Z., Xu W., Li G., Ding J., et al. Ultrasound-augmented mitochondrial calcium ion overload by calcium nanomodulator to induce immunogenic cell death. Nano Lett. 2021;21:2088–2093. doi: 10.1021/acs.nanolett.0c04778. [DOI] [PubMed] [Google Scholar]
- 38.Sun Y., Feng X., Wan C., Lovell J.F., Jin H., Ding J. Role of nanoparticle-mediated immunogenic cell death in cancer immunotherapy. Asian J Pharm Sci. 2021;16:129–132. doi: 10.1016/j.ajps.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng X., Xu W., Liu J., Li D., Li G., Ding J., et al. Polypeptide nanoformulation-induced immunogenic cell death and remission of immunosuppression for enhanced chemoimmunotherapy. Sci Bull. 2021;66:362–373. doi: 10.1016/j.scib.2020.07.013. [DOI] [PubMed] [Google Scholar]
- 40.Liu J., Li Z., Zhao D., Feng X., Wang C., Li D., et al. Immunogenic cell death-inducing chemotherapeutic nanoformulations potentiate combination chemoimmunotherapy. Mater Des. 2021;202:109465. [Google Scholar]
- 41.Feng X., Xu W., Li Z., Song W., Ding J., Chen X. Immunomodulatory nanosystems. Adv Sci. 2019;6:1900101. doi: 10.1002/advs.201900101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bai S., Yang L.L., Wang Y.J., Zhang T., Fu L.Q., Yang S.C., et al. Prodrug-based versatile nanomedicine for enhancing cancer immunotherapy by increasing immunogenic cell death. Small. 2020;16:2000214. doi: 10.1002/smll.202000214. [DOI] [PubMed] [Google Scholar]
- 43.Shen F.Y., Feng L.Z., Zhu Y.J., Tao D.L., Xu J., Peng R., et al. Oxaliplatin-/NLG919 prodrugs-constructed liposomes for effective chemoimmunotherapy of colorectal cancer. Biomaterials. 2020;255:120190. doi: 10.1016/j.biomaterials.2020.120190. [DOI] [PubMed] [Google Scholar]
- 44.Chu C.C., Lin H.R., Liu H., Wang X.Y., Wang J.Q., Zhang P.F., et al. Tumor microenvironment-triggered supramolecular system as an in situ nanotheranostic generator for cancer phototherapy. Adv Mater. 2017;29:1605928. doi: 10.1002/adma.201605928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu P., Wang X., Wang Z., Ma W., Guo J., Chen J., et al. Light-activatable prodrug and AIEgen copolymer nanoparticle for dual-drug monitoring and combination therapy. ACS Appl Mater Interfaces. 2019;11:18691–18700. doi: 10.1021/acsami.9b02346. [DOI] [PubMed] [Google Scholar]
- 46.Liu J., Huang Y., Kumar A., Tan A., Jin S., Mozhi A., et al. pH-Sensitive nano-systems for drug delivery in cancer therapy. Biotechnol Adv. 2014;32:693–710. doi: 10.1016/j.biotechadv.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 47.Yang Z., Chen Q., Chen J., Dong Z., Zhang R., Liu J., et al. Tumor-pH-responsive dissociable albumin-tamoxifen nanocomplexes enabling efficient tumor penetration and hypoxia relief for enhanced cancer photodynamic therapy. Small. 2018;14 doi: 10.1002/smll.201803262. [DOI] [PubMed] [Google Scholar]
- 48.Yang N., Xiao W.Y., Song X.J., Wang W.J., Dong X.C. Recent advances in tumor microenvironment hydrogen peroxide-responsive materials for cancer photodynamic therapy. Nano-Micro Lett. 2020;12:15. doi: 10.1007/s40820-019-0347-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang Y., Zhang L., Peng C., Zhang S., Chen S., Qian X., et al. Tumor microenvironments self-activated nanoscale metal organic frameworks for ferroptosis based cancer chemodynamic/photothermal/chemo therapy. Acta Pharm Sin B. 2021;11:3231–3243. doi: 10.1016/j.apsb.2021.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He M., Yu L., Yang Y., Zou B., Ma W., Yu M., et al. Delivery of triptolide with reduction-sensitive polymer nanoparticles for liver cancer therapy on patient-derived xenografts models. Chin Chem Lett. 2020;31:3178–3182. [Google Scholar]
- 51.Ma X., Zhang T., Qiu W., Liang M., Gao Y., Xue P., et al. Bioresponsive prodrug nanogel-based polycondensate strategy deepens tumor penetration and potentiates oxidative stress. Chem Eng J. 2021;420:127657. [Google Scholar]
- 52.Ju C.Y., Mo R., Xue J.W., Zhang L., Zhao Z.K., Xue L.J., et al. Sequential intra-intercellular nanoparticle delivery system for deep tumor penetration. Angew Chem Int Ed Engl. 2014;53:6253–6258. doi: 10.1002/anie.201311227. [DOI] [PubMed] [Google Scholar]
- 53.Xie W., Deng W.W., Zan M., Rao L., Yu G.T., Zhu D.M., et al. Cancer cell membrane camouflaged nanoparticles to realize starvation therapy together with checkpoint blockades for enhancing cancer therapy. ACS Nano. 2019;13:2849–2857. doi: 10.1021/acsnano.8b03788. [DOI] [PubMed] [Google Scholar]
- 54.Sun Q., Ojha T., Kiessling F., Lammers T., Shi Y. Enhancing tumor penetration of nanomedicines. Biomacromolecules. 2017;18:1449–1459. doi: 10.1021/acs.biomac.7b00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei D., Yu Y., Zhang X., Wang Y., Chen H., Zhao Y., et al. Breaking the intracellular redox balance with diselenium nanoparticles for maximizing chemotherapy efficacy on patient-derived xenograft models. ACS Nano. 2020;14:16984–16996. doi: 10.1021/acsnano.0c06190. [DOI] [PubMed] [Google Scholar]
- 56.Michaloglou C., Crafter C., Siersbaek R., Delpuech O., Curwen J.O., Carnevalli L.S., et al. Combined inhibition of mTOR and CDK4/6 is required for optimal blockade of E2F function and long-term growth inhibition in estrogen receptor-positive breast cancer. Mol Cancer Therapeut. 2018;17:908–920. doi: 10.1158/1535-7163.MCT-17-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chauhan V.P., Stylianopoulos T., Boucher Y., Jain R.K. Delivery of molecular and nanoscale medicine to tumors: transport barriers and strategies. Annu Rev Chem Biomol Eng. 2011;2:281–298. doi: 10.1146/annurev-chembioeng-061010-114300. [DOI] [PubMed] [Google Scholar]
- 58.Wang T.T., Wang D.G., Liu J.P., Feng B., Zhou F.Y., Zhang H.W., et al. Acidity-triggered ligand-presenting nanoparticles to overcome sequential drug delivery barriers to tumors. Nano Lett. 2017;17:5429–5436. doi: 10.1021/acs.nanolett.7b02031. [DOI] [PubMed] [Google Scholar]
- 59.Gonzalez-Angulo A.M., Ferrer-Lozano J., Stemke-Hale K., Sahin A., Liu S., Barrera J.A., et al. PI3K pathway mutations and PTEN levels in primary and metastatic breast cancer. Mol Cancer Therapeut. 2011;10:1093–1101. doi: 10.1158/1535-7163.MCT-10-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ladygina N., Gottipati S., Ngo K., Castro G., Ma J.Y., Banie H., et al. PI3Kgamma kinase activity is required for optimal T-cell activation and differentiation. Eur J Immunol. 2013;43:3183–3196. doi: 10.1002/eji.201343812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Broz P., Pelegrin P., Shao F. The gasdermins, a protein family executing cell death and inflammation. Nat Rev Immunol. 2020;20:143–157. doi: 10.1038/s41577-019-0228-2. [DOI] [PubMed] [Google Scholar]
- 62.Obeid M., Tesniere A., Ghiringhelli F., Fimia G.M., Apetoh L., Perfettini J.L., et al. Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat Med. 2007;13:54–61. doi: 10.1038/nm1523. [DOI] [PubMed] [Google Scholar]
- 63.Akira S., Takeda K., Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 64.Xiao Y., Zhang T., Ma X., Yang Q.C., Yang L.L., Yang S.C., et al. Microenvironment-responsive prodrug-induced pyroptosis boosts cancer immunotherapy. Adv Sci. 2021;8:2101840. doi: 10.1002/advs.202101840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z., Zhang Y., Xia S., Kong Q., Li S., Liu X., et al. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature. 2020;579:415–420. doi: 10.1038/s41586-020-2071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Martins I., Tesniere A., Kepp O., Michaud M., Schlemmer F., Senovilla L., et al. Chemotherapy induces ATP release from tumor cells. Cell Cycle. 2009;8:3723–3728. doi: 10.4161/cc.8.22.10026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.