Abstract
Ninety percent of clinical drug development fails despite implementation of many successful strategies, which raised the question whether certain aspects in target validation and drug optimization are overlooked? Current drug optimization overly emphasizes potency/specificity using structure‒activity-relationship (SAR) but overlooks tissue exposure/selectivity in disease/normal tissues using structure‒tissue exposure/selectivity–relationship (STR), which may mislead the drug candidate selection and impact the balance of clinical dose/efficacy/toxicity. We propose structure‒tissue exposure/selectivity–activity relationship (STAR) to improve drug optimization, which classifies drug candidates based on drug's potency/selectivity, tissue exposure/selectivity, and required dose for balancing clinical efficacy/toxicity. Class I drugs have high specificity/potency and high tissue exposure/selectivity, which needs low dose to achieve superior clinical efficacy/safety with high success rate. Class II drugs have high specificity/potency and low tissue exposure/selectivity, which requires high dose to achieve clinical efficacy with high toxicity and needs to be cautiously evaluated. Class III drugs have relatively low (adequate) specificity/potency but high tissue exposure/selectivity, which requires low dose to achieve clinical efficacy with manageable toxicity but are often overlooked. Class IV drugs have low specificity/potency and low tissue exposure/selectivity, which achieves inadequate efficacy/safety, and should be terminated early. STAR may improve drug optimization and clinical studies for the success of clinical drug development.
Key words: Drug development, Drug optimization, Clinical trial, Structure‒tissue exposure/selectivity relationship (STR), Structure‒tissue exposure/selectivity–activity relationship (STAR)
Graphical abstract
Structure‒tissue exposure/selectivity–activity relationship (STAR) selects drug candidates and balances clinical dose/efficacy/toxicity.
1. Why 90% of clinical drug development fails?
Drug discovery and development is a long, costly, and high-risk process that takes over 10–15 years with an average cost of over $1–2 billion for each new drug to be approved for clinical use1. For any pharmaceutical company or academic institution, it is a big achievement to advance a drug candidate to phase I clinical trial after drug candidates are rigorously optimized at preclinical stage. However, nine out of ten drug candidates after they have entered clinical studies would fail during phase I, II, III clinical trials and drug approval2,3. It is also worth noting that the 90% failure rate is for the drug candidates that are already advanced to phase I clinical trial, which does not include the drug candidates in the preclinical stages. If drug candidates in the preclinical stage are also counted, the failure rate of drug discovery/development is even higher than 90%.
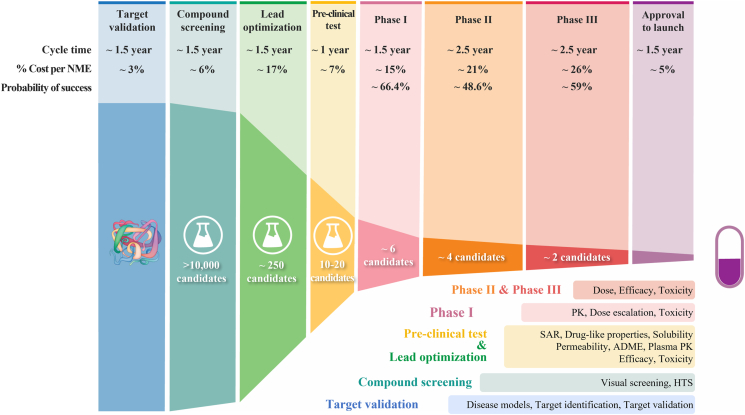
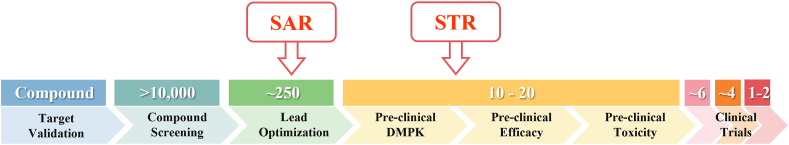
Analyses of clinical trial data from 2010 to 2017 show four possible reasons attributed to the 90% clinical failures of drug development: lack of clinical efficacy (40%–50%), unmanageable toxicity (30%), poor drug-like properties (10%–15%), and lack of commercial needs and poor strategic planning (10%)2,4. Generally, drug development follows a classical process (Fig. 1), which includes vigorous genetic and genomic target validation, high-throughput screening (HTS) of drug candidate molecules, rigorously drug optimization for activity and drug-like properties, preclinical efficacy and toxicity testing, and biomarker-guided selection of patients and optimal clinical trial designs. In the past few decades, each step of the above drug development has been rigorously optimized and validated, while many successful strategies have been rightfully implemented in the drug development process to select the best drug candidates for clinical studies. Despite this validated effort, the overall success rate of clinical drug development remains low at 10%–15%5, 6, 7. Such persistent high failure rate raises several questions: Why 90% of clinical drug development fails despite implementation of many successful strategies in the past several decades? Did we overlook certain aspects of drug development process leading to high failure? How can we improve the success rate of clinical drug development?
Figure 1.
The process of drug discovery and development, and the failure rate at each step.
2. What are the successful strategies to improve each aspect of drug development process in the past decades?
2.1. Select best lead drug candidate to achieve adequate clinical efficacy
Since 40%–50% of clinical failure of drug development is due to lack of clinical efficacy, tremendous effort has been devoted to improving drug efficacy in preclinical and clinical studies. In the target validation process, the disease targets have been rigorously confirmed using genetic, genomic, and proteomic studies in cell lines, tissues, preclinical models, and human disease models5,7, 8, 9, 10, 11, 12. However, true validation of any a new molecular target in human disease is challenging before a drug can be successfully developed since biological discrepancy among in vitro, in disease animal model, and human disease may hinder the true validation of the molecular target's function8, 9, 10, 11, 12. This discrepancy makes the development of the first-in-class drugs difficult. In the drug screening process, both virtual computation screening and HTS of chemical libraries have often been conducted to select the best scaffold and eliminate non-specific binding to the targets6,7,12,13. The artificial intelligence (AI) and machine learning computational tools further improve the computation-aided drug design process14,15. HTS using protein-based biochemical assays, cell-based phenotypical assays, or organism-based assays improve the efficiency and specificity of the hits6,7. During drug optimization process, the lead compounds are extensively optimized through structure–activity relationship (SAR) to achieve high affinity and specificity to their molecular targets (with Ki or IC50 at low nmol/L or pmol/L range) and to limit off-target effect16, 17, 18, 19, 20. However, the validation of the pharmacological effect of a drug molecule is a result of drug's inhibition of its intended molecular target may also be challenging since the pharmacological effect (efficacy and toxicity) of the molecule may be due to the inhibition of some unknow molecular targets12. Interestingly, some drugs have been successfully developed even though their targets are different from their intended targets as reported previously12. In such case, however, the drug optimization using SAR to an intended target may mislead the optimization effort. During preclinical testing, the compounds are usually tested and optimized to show excellent efficacy in preclinical animal disease models. Many animal models have been established to mimic human disease conditions, although finding best animal models to fully recapitulate the disease phenotype or pathophysiology of human disease remains a challenge5. During clinical study process, clinical trial design and dose regimen selections from phase I to phase III have been rigorously optimized. In addition, genomics and genetic biomarkers have also been extensively used to select patients for clinical trials to improve the success rate of drug development21. It worth noting that most of drug discovery effort (before clinical studies) have been rightly devoted in these studies to select the best lead drug candidates with validated molecular targets related to human diseases. However, high proportion of many “perfect” drug candidates still failed during clinical phase I, II, and III studies. Although those above drug discovery efforts are well justified, overemphasis of these above effort, which is the current practice in drug discovery, may be helpful, but may not solve the persist high failure rate of clinical drug development.
2.2. Select best lead drug candidate to minimize clinical toxicity
Various strategies have been used in toxicology studies to minimize drug development failure due to unmanageable clinical toxicity22,23. Drug's toxicity could be resulted from either off-target or on-target inhibition of the molecular targets. In order to reduce off-target toxicity, drug screening against other targets is usually performed24,25. For instance, development any kinase inhibitor often needs to be screened against other several hundred kinases. The selectivity of disease related kinase target vs. other kinase targets are often calculated by the ratios of their IC50, in which at least 10-fold selectivity is preferred. On the other hand, if a drug candidate has on-target toxicity, which is caused by the inhibition of the disease related target, the solution is limited where titration of dose regimen may be helpful. Further, drug candidates are often evaluated if they would inhibit several known toxicity targets in the major vital organs. For instance, in vitro and in vivo hERG assays are usually performed as a predictive marker of cardiotoxicity (lethal torsade de pointe arrhythmia) of the lead compounds26. Finally, drug candidates may also cause chemical-induced toxicity without clear known targets. Toxicogenomics is often used for early assessment of potential toxicity14,27. Structure of drug candidates may need to be further modified for minimizing drug‒protein adducts or drug‒DNA adducts for reducing potential organ toxicity28. In vitro/in vivo animal studies are always conducted for potential genotoxicity and carcinogenicity22. In practice, acute and chronic toxicity of the drug candidates, which mimics clinical dose regimen, are always examined in three species of animals23. Since there may be difference between animal models and humans, during clinical trials, the dose regimen is often optimized using various approaches to maintain the therapeutic windows with adequate efficacy and manageable toxicity23. However, regardless drug candidates have off-target or on-target toxicity to a molecular target, the accumulation of drug candidates in the vital organs or blood cells is one of the major factors for toxicity. Unfortunately, there is no well-developed strategy to optimize drug candidates to reduce tissue accumulation in the major vital organs to minimize the potential toxicity.
2.3. Select best lead drug candidate with optimal drug-like properties
Poor drug-like properties contributed to the 30%–40% drug development failures in the 1990s; but they only account for 10%–15% of drug development failures today29. This improvement benefits from rigorous selection criteria for drug-like properties during drug optimization, including solubility, permeability, protein binding, metabolic stability, and in vivo pharmacokinetics, such as bioavailability (F), drug exposure (AUC), Cmax, t1/2, clearance CL, and volume distribution V30. Certain cut-off values of these drug-like properties have been used as criteria to select the best lead compounds. The “rule of 5” have been considered in chemical structure design: (1) less than 5 H-bond donors; (2) molecular weight less than 500; (3) cLogP less than 5; (4) less than 10 H-bond acceptors31. A polar surface area of less than 140 A2 is also desired32. In vitro permeability of more than 2 × 10−6–3 × 10−6 cm/s is preferred for better oral absorption33. Pre-formulation and formulation studies are conducted to increase drug solubility and improve oral bioavailability based on the biopharmaceutical classification system34,35. In vitro microsomal stability t1/2 > 45–60 min is preferred. Meanwhile, preclinical pharmacokinetics are rigorously performed to select lead compounds with suitable drug-like properties with bioavailability F > 30%, half-life t1/2 > 4–6 h, and clearance CL < 25% hepatic blood flow Q36,37.
It is important to note that clinical drug development failure due to poor drug-like properties has been significantly improved in the past 20 years29, which suggests the right strategy in drug-like property optimization. However, overall success rate of clinical drug development has not been significantly improved and remained at low of 10%–15%. However, although all in vitro and in vivo assessment of drug-like properties are well developed and well justified, the current selection criteria using plasma pharmacokinetics, which may or may not guide the correct selection of lead drug candidates to be advanced to clinical trials. Drug candidates with better plasma exposure are often selected to advance to clinical studies, while drug candidates with low exposure in the plasma are often eliminated without further development38,39. However, the clinical dose/efficacy/toxicity of drug candidates is determined by the effective drug exposure/selectivity in the disease-targeted organs vs. normal organs, but current drug optimization process in drug candidate selection has not fully utilized a criteria to assess drug exposure/selectivity in the disease-targeted organs vs. normal organs, which may have misled drug candidate selection40. The assumption, which the drug plasma exposure may be used as an indicator of therapeutic exposure in disease-targeted tissue, is based on the “free drug hypothesis” to select lead drug candidate. This hypothesis believes that only free unbound drug from plasma (not plasma protein bound drug) can distribute to disease-targeted tissues to interact with its molecular target; while free drug exposure in the plasma would be similar to the disease-targeted tissues at steady state41,42; and thus drug exposure in the plasma can be used to predict the pharmacodynamic effect of the drug candidate. However, this “free drug” hypothesis may only apply to a limited class of drug candidates but not applicable to many other compounds since many factors can cause an asymmetric free drug distribution between plasma and tissue42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64. Therefore, drug exposure in the plasma, without knowing the exposure in disease-targeted tissue/normal tissues, may mislead the selection of drug candidate to clinical trials51, 52, 53,65.
2.4. Optimize strategic planning in drug development
Poor strategic planning, which may include change in therapeutic focus, company mergers, or poor clinical study conduct, accounts for 10% of drug development failures2. The merging of pharmaceutical companies may increase the number of duplications of drugs that is forced to terminate2,4. All pharmaceutical companies have developed meticulous development plan with a detailed roadmap and milestones to advance new compounds from the lab through each stage of development. A multidisciplinary project team of experienced experts often work together in strategic planning with the help of various business models and analytic tools66. Artificial Intelligence (AI) has brought state-of-the-art analytical tools that enable pharmaceutical companies to predict patients’ needs and market trends in a more efficient and cost-effective way14.
3. What are the overlooked aspects in drug discovery/optimization leading to high failure rate of clinical drug development?
3.1. True target validation, which confirms the molecular target is the cause of human disease and drug's intended target, is still challenging for the success of clinical drug development
Two types of target validations need to be rigorously investigated for any drug discovery and development program8,9,16, 17, 18, 19, 20. One type of target validation is to confirm a molecular target is indeed a cause for human disease. Although this type of target validation is extensively studied using various genetic or genomic methodology in in vitro cell lines, in animal disease models, and in human disease models, the biological discrepancy of a molecular target remains between in vitro and in vivo or between disease in animal and human10. In reality, a molecular target would not be fully validated until a successful drug is developed, which poses the challenges for first-in-class drug discovery and development. Another type of target validation is to confirm if a molecular target is the intended target of the drug molecules, which is usually confirmed by the specificity/affinity to bind the molecular target through SAR studies16, 17, 18, 19, 20. The pharmacological effect of the drug molecules can be compared to the effect of genetic alteration of the target using siRNA knockdown or CRISPR gene editing11,12. However, validation of drug target is still challenging since drug's pharmacological effect (efficacy and toxicity) may be resulted from inhibition of unknown molecular targets that may be different from their intended targets12, which subsequently impact the drug optimization process as described below. Interestingly, some drugs have been successfully developed even though their targets are different from their intended targets as reported previously12. Indeed, many challenges still remain for these two types of target validation, which may cause high failure rate of clinical drug development as discussed previously8,9. However, even if the targets are truly validated, many drug candidates still have high failure rate in clinical phase I, II, and III trials, which highlight the importance of drug candidate optimization process as described below.
3.2. Current drug optimization may have overemphasized one aspect but overlooked others that may mislead drug candidate selection and impact the balance of clinical dose/efficacy/toxicity
3.2.1. Optimization of two aspects of lead drug candidates is required in the current drug optimization process
During drug optimization, two major aspects of the compounds are rigorously optimized: (1) the potency and specificity of the lead compounds to inhibit the molecular target are rigorously optimized through SAR, where low Ki or IC50 at low nmol/L or even pmol/L range is desired to achieve better efficacy and decrease off-target effect; and (2) drug-like properties of the lead compounds are also extensively optimized using certain cut-off values as acceptable criteria for drug solubility, permeability, stability, protein binding, and plasma PK parameters. Despite significant efforts to improve each aspect of the drug development process in the past few decades, the success rate of drug development remains at 10%–15%5, 6, 7. The persistent high failure rate raises questions as to whether some aspects of drug optimization are overlooked despite the validated molecular targets.
3.2.2. The balance of drug exposure/selectivity in disease-targeted tissues vs. healthy tissues is overlooked in drug optimization that may mislead drug candidate selection and tip the balance of clinical dose/efficacy/toxicity
In clinical drug development, a delicate balance needs to be achieved among clinical dose, efficacy, and toxicity to optimize the benefit/risk ratios in patients. An ideal drug candidate would have high potency and specificity to inhibit its molecular target without off-target effect, high drug exposure in disease-targeted tissues to achieve adequate efficacy at an optimal dose (ideally at low doses), and minimal drug exposure in healthy tissues to avoid toxicity at optimal doses (even at high doses). However, the current drug optimization process may have overly emphasized the potency/specific using SAR studies but overlooked the balance of drug exposure/selectivity in disease-targeted tissues vs. healthy tissues, which may have misled the drug candidate selection, tipped the balance among clinical dose/efficacy/toxicity, and caused high clinical failure rate.
The drug candidates, which are selected using current drug optimization criteria, usually show good efficacy in vitro and in animal models if an adequate dose is given. However, once a drug candidate is selected and advanced to clinical trials, one of the most important questions is whether patient could tolerate the toxicity when the adequate efficacy is achieved. The clinical drug development failure due to the lack of efficacy often does not mean the drug candidates do not work, but it is most likely because these drugs could not show satisfactory efficacy in the disease-targeted organs even at maximal tolerable dose (MTD) that already showed toxicity in healthy organs. These drugs would surely show efficacy at disease-targeted organs at a dose above MTD, but patient cannot tolerate the higher dose in the healthy organs. On other hand, the clinical drug development failure due to the unmanageable toxicity is most likely due to the fact that it showed toxicity at normal healthy organs even at low dose before the drug can achieve any efficacy in disease-targeted organs. Therefore, the success/failure of clinical drug development depends on a delicate balance among clinical dose, efficacy in disease-targeted organs, and toxicity in normal healthy organs. Therefore, it is important to test whether the drug candidates can reach adequate exposure in disease-targeted organs without too much drug exposure in healthy normal organs. However, these aspects are not considered during drug optimization process, which may mislead lead drug candidate selection and impact the balance of clinical dose/efficacy/toxicity. This may be one of the overlooked attributes in drug optimization process, which contribute to the high failure rate of clinical drug development despite validated molecular targets.
Fortunately, during the optimization of drugs targeting central nerve system (CNS), the drug exposure in the brain or drug's ability to cross blood‒brain barrier (BBB) are always a selection criteria for drug candidate selection52,54, 55, 56, 57,67, in addition to the optimization of SAR and drug-like properties. It is well accepted that if a drug candidate has difficulty penetrating BBB to reach brain tissue, it will not achieve adequate efficacy, which shall be terminated early in drug development.
However, drug optimization in other therapeutic areas rarely adapt a criterion to ensure drug exposure in the disease-targeted organs vs. normal organs. For anticancer drug discovery, target engagement is indeed often assessed in xenograft model, or occasionally in human tumor resection58, 59, 60, 61, 62,68, 69, 70, 71. However, how structure modifications of the drug molecule may alter drug exposure in the tumors vs. normal tissues is rarely assessed with an assumption that high plasma drug exposure is adequate to achieve high tumor exposure. In clinical trials of anticancer drugs, the target engagement is often monitored in peripheral blood mononuclear cells (PBMC) for clinical dose selection since PBMC can be easily obtained in clinical studies. However, target engagement in PBMC is known to be a poor surrogate biomarker that is easily saturated at low doses while target engagement in tumor tissues is barely adequate. It is widely accepted that dose optimization based on plasma drug exposure for target engagement in PBMC is only a reference and starting point for dose optimization in solid tumors and eventual doses that achieve maximal tumor inhibition or regression are much higher than the dose that yields maximal target engagement in blood. Once dose escalation is performed in clinical studies, the suboptimal balance of clinical dose/efficacy/toxicity may lead to clinical development failures.
For instance, analysis of U.S. Food and Drug Administration (FDA)-approved EGFR inhibitors (gefinitib, lapatinib, erlotinib, and vandetanib) showed that similar pharmacophore with certain modifications have distinct indications to treat very different cancer types with various anticancer efficacy72, 73, 74, 75. However, their SAR and drug-like properties of these EGFR inhibitors cannot fully explain their distinct clinical anticancer efficacies and toxicities. The structure modifications may significantly impact their drug exposure in different tumor types that may significantly contribute to their anticancer efficacy (unpublished data), in addition to SAR and drug-like properties. Similarly, several successful BTK inhibitors, such as ibrutinib, acalabrutinib, and zanubrutinib showed distinct efficacy/toxicity profiles76,77, while spebrutinib failed at early clinical trial78. In addition to the drug specificity/potency and drug-like properties, their tissue exposure in the disease-targeted organs vs. normal organs may also contribute their efficacy/toxicity, which needs further studies.
In addition, analysis of FDA-approved or clinical failed selective estrogen receptor modulators (SERMs) also showed slight structure modifications may alter the balance among clinical dose, clinical efficacy and toxicity, which may impact its success in clinical development79, 80, 81, 82, 83. For instance, more than 600 clinical trials have been conducted for SERMs clinical development, where 11 SERMs have been approved and many SERMs have been failed in clinical studies81, 82, 83. Some of SERMs have very similar structures with only very slight modifications, yet they have shown distinct efficacy in various indications to treat breast cancer, osteoporosis, and menopausal symptoms. Their SAR and drug-like properties cannot fully explain the difference among their clinical dose, efficacy, and toxicity79, 80, 81, 82, 83. The previous study showed that slight structure modifications may significantly alter drug exposure and drug selectivity in different tissues (such as tumor, fat pad, and bone), which impacts their clinical dose, efficacy and toxicity40.
Further, the most recent example of antiviral drug remdesivir for treatment of COVID-19 also demonstrates the importance of drug exposure in the disease-targeted tissue vs. normal tissues for the delicate balance of dose/efficacy/toxicity. Remdesivir only showed very limited clinical efficacy for treatment of COVID-19 despite good in vitro activity against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)84,85. At a dose of 100 mg, the exposure/selectivity of remdesivir and its active metabolites may be too low to achieve outstanding efficacy in the lung to kill the SARS-CoV-2 virus, but too high in the kidney to cause toxicity86. In addition, many methods have been used to estimate drug concentrations of anti-infective agents in the lung for their studying their efficacy on disease modulation37,53.
These examples clearly demonstrate that drug candidate selection and the delicate balance for clinical dose/efficacy/toxicity are not only determined by SAR and drug-like properties, but also impacted by the balance of drug exposure in disease-targeted tissues vs. normal healthy organs, which is often overlooked in drug optimization and clinical studies.
4. How to improve drug optimization to select better drug candidates, balance clinical dose/efficacy/toxicity, and improve success rate of clinical drug development?
The success of clinical drug development will be determined by best drug candidate selection and the delicate balance among clinical dose/efficacy/toxicity, in addition to true target validation. The balance of clinical dose/efficacy/toxicity of a drug candidate in human trials is not only determined by its potency/specificity to inhibit its molecular targets (through SAR studies), but also by its exposure/selectivity in disease-targeted organs vs. normal organs (through structure‒tissue exposure/selectivity relationship, STR).
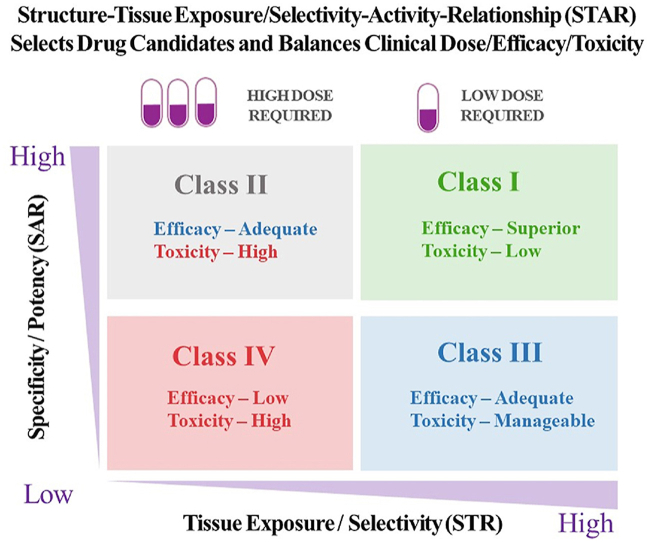
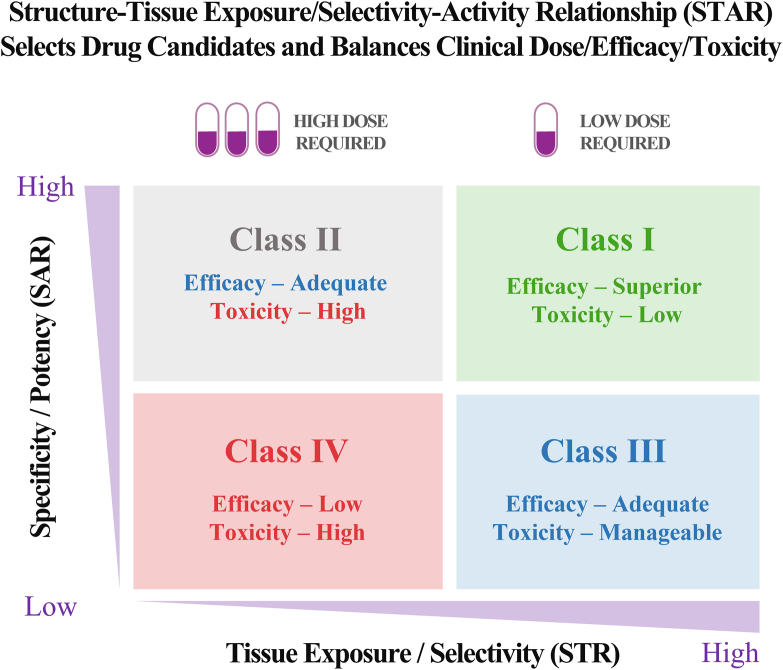
We propose a structure‒tissue exposure/selectivity–activity relationship (STAR) system to improve drug optimization process. STAR system classifies drug candidates into four different classes (I‒IV) based on three aspects: drug potency/selectivity to inhibit the molecular target using SAR studies with IC50 or Ki (high or low); drug tissue exposure/selectivity using STR studies (high or low); dose requirement for balancing clinical efficacy/toxicity (high or low). The four different classes of drug candidates (classes I‒IV) require different strategies to select lead drug candidates, optimize clinical doses, and balance clinical efficacy/toxicity. Successful application of STAR will improve the efficiency of drug optimization and clinical studies for four different classes of drug candidates to improve the success rate of clinical drug development (Fig. 2).
Figure 2.
Structure‒tissue selectivity/exposure–activity relationship (STAR) selects better drug candidates and balances clinical dose/efficacy/toxicity to improve drug optimization for successful clinical drug development.
In this STAR system, SAR explores structural modifications that alter the compound's binding affinity and specificity to the molecular target. Drug potency and specificity can be optimized using SAR to obtain compounds with low Ki or IC50 at low nmol/L or pmol/L range for better efficacy and lower toxicity6,7,87. SAR studies have been well established and validated in the current drug optimization process. In addition, STR studies structural modifications that changes drug exposure/selectivity in disease-targeted tissues vs. normal healthy organs, which not only impacts lead compound selection during drug optimization process, but also determines the balance among clinical dose/efficacy/toxicity in human trials40,88. However, STR studies have not been well established in drug optimization process and clinical trial design. Further, the balance of clinical efficacy/toxicity is always dose-dependent. A delicate balance among clinical dose/efficacy/toxicity always needs to be achieved for a successful clinical drug development to optimize the benefit/risk ratios in patients.
4.1. Class I
Class I drug candidates have high specificity/potency against their molecular targets (Ki < several hundred nmol/L by SAR studies) and high exposure/selectivity in disease-targeted tissue (by STR studies). A low dose is required for class I drug candidates to achieve adequate efficacy. Toxicity is low in healthy organs, benefiting from high disease tissue selectivity and low tissue exposure/selectivity in normal organs. Class I drugs may have either high or low drug exposure in the plasma. Class I drug candidates are most desirable and will have the highest success rate in clinical trials with balanced clinical dose/efficacy/toxicity. For instance, the following successful approved drugs are likely to be the class I drugs, such as antiviral drug sofosbuvir (NS5B polymerase inhibitor), cholesterol treatment drug atorvastatin (HMG-CoA reductase inhibitor), and erectile dysfunction treatment drug sildenafil (PDE5 inhibitor), and anticancer agent acalabrutinib (BTK inhibitor). It is worth noting that some of class I drugs may also have high exposure/selectivity in both disease-targeted tissues and some normal organs (related to toxicity) compared to drug exposure in the plasma, which may have both high efficacy and high incidence of adverse events. The following drugs are likely to be these types of examples, such as anticancer agents vandetanib (VEGFR/RET/EGFR inhibitor), tamoxifen (SERM), pomalidomide (E3 ligase inhibitor), and doxorubicin (TOPO I inhibitor).
4.2. Class II
Class II drug candidates have high specificity/potency against their molecular targets (Ki < several hundred nmol/L by SAR studies) and low tissue exposure/selectivity in disease-targeted tissue (by STR studies). To achieve adequate efficacy in disease-targeted tissues, high doses are often required for class II drug candidates due to their low exposure/selectivity in disease-targeted organs. However, toxicity may become unmanageable because of the high dose and low tissue exposure/selectivity in diseased tissues (but high tissue exposure/selectivity in normal organs).
Currently, most drug optimization efforts are focused on this class II compounds to improve their specificity/potency (Ki). The lead compounds are often optimized to have low nmol/L or even pmol/L IC50 or Ki. During lead drug candidate selection, compounds with high plasma exposure and better plasma PK parameters are often selected for clinical studies39,89. However, in vivo drug concentration in the plasma often needs to be achieved at μmol/L for adequate efficacy, especially for anticancer drugs. One would wonder why there is a large discrepancy between in vitro IC50 and in vivo EC50. One of the important factors may be due to the low drug exposure/selectivity in the disease-targeted tissues. In such cases, high drug exposure in plasma may mislead drug candidate selection and poor optimization of clinical dose/efficacy/toxicity. Since drug exposure in the plasma is determined by both elimination (clearance) and distribution (drug in different organs) processes90, drug candidates with high plasma exposure may be attributed either to low elimination or low tissue distribution. A drug with high plasma exposure but low tissue exposure in the disease-targeted tissues may not be preferred, because low target tissue exposure may result in inadequate efficacy. In such cases, a high dose is often used to achieve adequate efficacy, but such a high dose may lead to high toxicity in other vital organs91. If a drug candidate has lower tissue selectivity and exposure in the disease-targeted organs but higher tissue selectivity and exposure to other vital healthy organs, it may have a narrow therapeutic window and may not be able to reach its therapeutic concentration due to high toxicity. Further, if a drug candidate has low selectivity and exposure in both disease-targeted and healthy vital organs, it may not have safety concerns even with high drug plasma exposure in phase I studies, but it may not reach the desired efficacy in phase II/III studies. Unfortunately, we often observed such lead compounds, which have high potency/specificity, low tissue exposure/selectivity, high plasma exposure, good safety profile, advertised in phase I studies with high hope, but are failed in later phase III studies due to lack of efficacy. Therefore, class II drug candidates should be cautiously evaluated for their indications, dose optimization, and balance of clinical efficacy and toxicity. Several drugs are likely to be class II drugs, such as anticancer agents ibrutinib (successful BTK inhibitor), spebrutinib (failed BTK inhibitor), and fedritinib (successful JAK2 inhibitor). Antiviral drug remdesivir (RdRp inhibitor) is also likely to be class II drugs if its antiviral activity is considered to be high (IC50 at low nmol/L in Calu-3 cells). However, if remdesivir's antiviral activity is considered to be low (IC50 at μmol/L in Vera cells), it is likely to be classified as the class IV drugs as described below.
4.3. Class III
Class III drug candidates have low but adequate specificity/potency against their molecular targets (several hundred nmol/L < Ki < 1 μmol/L by SAR studies) but have high tissue exposure/selectivity in disease-targeted tissues (by STR studies). A low to medium dose is required for class III drug candidates to achieve adequate efficacy. Toxicity would be manageable because of low to medium doses, high exposure/selectivity in disease-targeted orans and low exposure/selectivity in normal organs. Currently, class III compounds may have low exposure in the plasma and are likely to be overlooked. They are often terminated at the early stage of the drug optimization process because of low plasma drug exposure. However, low plasma exposure may also be due to high tissue distribution, especially when drug candidates have good in vivo stability and relatively reasonable bioavailability. Such compounds may have high target tissue exposure, which is beneficial for achieving better efficacy at low doses, while low doses may also minimize toxicity in healthy organs. These class III drug candidates may balance clinical dose/efficacy/toxicity, which may improve the success rate from phase I to phase III clinical trials. However, a large proportion of these types of compounds may have been mistakenly terminated before they could be advanced to clinical trials. It is worth noting that the specificity and potency of class III compounds still need IC50 < 1 μmol/L, since compounds with IC50 > several μmol/L may not achieve adequate efficacy regardless of tissue exposure/selectivity. There are very few drugs on the markets that belong to class III drugs since many class III drug candidates may have been terminated in the early drug discovery process. The anticancer drug Thalidomide (an E3 ligase inhibitor) is likely to be class III drugs.
4.4. Class IV
Class IV drug candidates have low specificity/potency against their molecular targets (Ki > 1 to several μmol/L by SAR studies) and low exposure/selectivity in disease-target tissues (by STR studies). High doses are often required for class IV drug candidates to achieve some level of efficacy but still insufficient. Toxicity would become unmanageable because of the high dose and low tissue exposure/selectivity in disease-targeted organ but high tissue exposure/selectivity in normal organs. Class IV drug candidates are the most undesirable and should be terminated at an early stage of the drug optimization process. It is worth noting that drug candidates with low potency/specificity (Ki > one to several μmol/L) should be evaluated with caution during the advancement to clinical trials, regardless of tissue exposure/selectivity. Most of class IV drug candidates are likely failed in the clinical development with very limited successful examples.
It is worth noting that the drug examples in each classes (I–IV) are estimation based on their plasm PK and clinical dose/efficacy/toxicity without experimental data since STR is not currently used as a drug optimization process. More detailed studies are required to accurately assign these drugs into four different classes (I‒IV) based on their tissue exposure/selectivity in the future as described below.
It is important to note that the true target validation is equally important for successful clinical drug development. In addition, the current successful strategies are valid and well justified to overcome the four possible reasons for 90% of clinical development failure. The overlooked STAR is one of the major factors, but not the only one, that needs to be considered for the 90% failure rate of clinical drug development. Application of STAR will improve the efficiency of drug optimization and clinical studies for four different classes of drug candidates to improve the success rate. However, application STAR alone would not guarantee 90%–100% success rate of clinical drug development, but it may significantly improve the success rate. Improvement of success rate from 10%‒15% to 30%–40% would have significant impact in overall drug development.
5. How to implement STAR in drug optimization process and future perspectives
5.1. Measurement of SAR and STR
SAR studies have been widely applied in drug optimization processes using computation-aided drug design and in vitro screening by measuring the IC50 or Ki (nmol/L) of the compound binding to its molecular targets based-protein assays or cell-based assays. This has been widely reviewed previously16, 17, 18, 19, 20 and is not included in this commentary. Target validation has also been extensively reviewed previously8, 9, 10, 11, 12 and is not included in this commentary.
However, STR studies are rarely used as a drug optimization criterion in the current drug optimization to select the lead drug candidates and to balance clinical dose/efficacy/toxicity. The cut-off values to define high or low tissue exposure/selectivity have not been defined that requires further research. Experimentally, STR can be measured using drug tissue exposure (AUC), partition coefficient Kp, tissue selectivity over all organs (%), and AUC ratio of disease-targeted tissues vs. normal vital organs related to toxicity. It is important to note that tissue exposure/selectivity concept is different from volume of distribution (V) in traditional pharmacokinetics. Tissue exposure/selectivity of a drug in each organ will be determined by absorption/bioavailability, distribution, protein/tissue binding, metabolism of the drugs in each organ. The tissue exposure/selectivity of each organ will be different from another organ. Traditional volume of distribution is a mathematical term to describe how easy a drug molecule can distribute into tissues in general, but it cannot distinguish which tissue is high or low.
5.2. Tissue exposure/selectivity measurement for STR
Drug tissue exposure (AUC) describes the amount of total drug (bound and free) accumulated in certain tissues which is determined by drug exposure (AUC) in the plasma and tissue using the partition coefficient (Kp)92 as shown in Eq. (1):
| (1) |
where drug exposure in the plasma and tissue can be calculated using total drug concentration vs. time curve, and Kp values can be calculated by total drug concentrations in tissues vs. plasma (Ctissue/Cplasma) or total drug AUC in tissue vs. plasma (AUCtissue/AUCplasma)92.
Drug tissue selectivity affects the therapeutic window of drugs to balance the dose, efficacy, and toxicity. It can be described using the two equations as shown in Eqs. (2), (3), depending on various experimental purposes and conditions. Drug tissue selectivity can be described by the proportion of total drug concentration or exposure in certain tissues related to efficacy or toxicity among the total drug concentration or exposure to all the tissues as shown in Eq. (2):
| (2) |
where the sum of Ctissue or AUCtissue is the total drug concentration or AUC in the tissues. Eq. (2) is suitable for drug candidates with unknown toxicity profiles. The measurement of drug selectivity in all major tissues can provide information on the potential toxicity of drug candidates. However, the measurement of total drug tissue exposure in all major tissues is time-consuming and labor-intensive; thus, it is only suitable for screening from a small number of drug candidates.
Therefore, drug tissue selectivity can also be described by the ratios of total drug concentration or exposure in disease-targeted tissues related to drug efficacy to that in healthy vital organs related to toxicity as shown in Eq. (3):
| (3) |
where Ceff or AUCeff is the drug concentration or AUC in tissue related to efficacy, and Ctox or AUCtox is the drug concentration or AUC in tissue related to toxicity. Eq. (3) can be used when major or vital toxicity of drug candidates is clearly demonstrated and related to one or two tissues. Eq. (3) does not require the measurement of all tissue concentrations, which is less time consuming and less labor intensive.
5.3. Measurement of total drug concentration/exposure vs. free drug concentration/exposure for STR
It is debatable if the concentration or exposure of total drugs or free drugs should be used for above calculations for lead drug candidate selection according to the traditional “free drug hypothesis” of small molecules. “Free drug hypothesis” suggests that the total drug concentration in the plasma includes an equilibrium between free unbound drugs and plasma protein-bound drugs41,93; only free unbound drugs (but not protein-bound drugs) could distribute to other tissues, while the protein-bound drugs serve as reservoir in the plasma to release the free drugs41,93; the total drug concentrations in the tissues also include the free unbound drugs and bound drugs; only free unbound drugs can interact with their molecular targets for pharmacological functions41,93; at steady state, the free drug concentration in tissue is similar or equal to the free drug concentration in plasma41,42, and thus, drug concentration or exposure of in plasma can be used to predict the pharmacologic effects in the targeted tissues.
Although it is correct that only free drugs could engage their molecular targets for pharmacological actions while bound drugs could serve as a reservoir to release free drugs for pharmacological actions, it is incorrect to assume only free drugs can distribute to other tissues based on the following reasons: (1) The assumption of only free drugs for distribution into tissues completely overlooks the trafficking of plasma proteins themselves (such as albumin) from systemic circulation to tissues. In fact, albumin is actively transported from plasma to extra-cellular matrix, and to tissues through FcRn and other active transport process45, which carries fatty acids or drugs to different tissues. Previous study found that albumin-bound small molecules (tyrosine kinase inhibitors) interacting with albumin-binding proteins on vascular and in tissues, such as gp18, gp30, gp60/albondin, and secreted protein acidic and cysteine-rich (SPARC), which mediates tissue accumulation of these small molecules in normal tissues, which is associated with their toxicity46. Protein-bound drugs may also contribute higher drug accumulation in tumor tissues than that in the normal tissues40. (2) “The free drug hypothesis” is incorrect in the following situations since an asymmetric free drug distribution between plasma and tissue occurs: the uptake and efflux drug transporters involved in drug uptake and extrusion (e.g., liver and brain)42, 43, 44,49; ionizable drugs affected by the pH gradient and “lysosomal trapping” effect42,50; drugs covalently binding to the target42; prodrugs, and protein degraders42. It is well known that free drug hypothesis does not apply to antibody–drug conjugates, nanomedicine, nucleic acids42; and drug locally administrated (e.g., inhalation)37,48,94, which are not included in this review.
Therefore, the “free drug hypothesis” is only true in some limited cases when drugs are passive diffused from plasma to its target and not rapidly cleared, but it is incorrect in many other situations42. Free drug fraction is a very important parameter in both plasma and tissues in the drug optimization process since it is indeed true that only free drug could bind to its molecular target for pharmacological function. However, overemphasis of free drug fraction in the plasma by only measuring plasma protein binding during drug optimization process may be misleading. We propose to use the total drug exposure/selectivity in disease-targeted organs vs. normal organs since equilibria and transport of both free unbound drugs and protein-bound drugs are presented in normal tissues and disease-targeted tissues51, 52, 53. Previous preclinical and clinical studies also directly used total tissue exposure or Kp (total drug in tissue/plasma ratio) to screen drug candidates and evaluate dose dependent efficacy/toxicity54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64. The clinical development group from AstraZeneca developed translational Pharmacokinetics and Pharmacodynamic (PK/PD) model, which suggests total drug levels were preferred over free drug level for clinical PK/PD relationships48,64. The free drug concentration in the tissues may still be very useful, which is very difficult to measure. Its use for lead drug candidate selection may still be debatable. Several methods are available to determine free unbound drugs (fu) in the tissues, among which equilibrium dialysis is the most commonly used method95. However, most of the current methods to determine fu use tissue homogenates, which destroys all subcellular structures and cannot truly represent free drug concentration in the tissues at the site of action42,52.
5.4. Developing in vitro high-throughput screening to study STAR
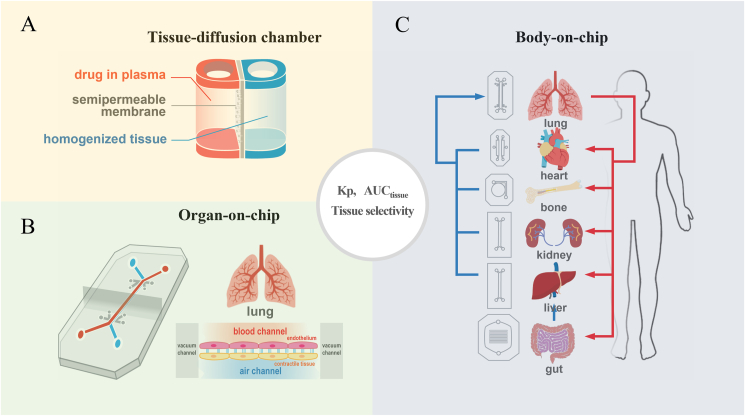
High throughput study of SAR has been successfully implemented in drug optimization process8,10,16, 17, 18, 19, 20, and thus it will not be reviewed here. However, high throughput STR methodology is still lacking. In the future, it would be ideal to develop in vitro screening tools to screen STR, in addition to SAR studies, during the early stage of the optimization process. The diffusion chamber may provide an easy and high-throughput method for screening tissue exposure and selectivity of drug candidates (Fig. 3A). The amount of drug diffusion from the plasma chamber (mimicking drug in systemic circulation) into the homogenized tissue chamber (mimicking different tissues, such as the liver, kidney, heart, etc.) can be measured over time. The plasma partition coefficient (Kp) can be calculated using either total drug concentration or AUC ratios of tissue chambers/plasma chambers over different time points. Meanwhile, the total drug exposure and tissue selectivity of the drugs in different tissues can be calculated using Equations (1), (2), (3), which can be confirmed by correlation with in vivo tissue exposure and selectivity in animal models.
Figure 3.
In vitro high-throughput screening tool to study the structure–tissue selectivity/exposure–activity relationship (STR). (A) Tissue diffusion chamber may provide an easy and high throughput screening to study structure‒tissue exposure/selectivity relationship (STR). (B) Single-organ chips, and (C) Body-on-a-Chip integrates multiple organ units to study STR in different organs.
The emergence of organ-on-a-chip technologies may also provide a better HTS for STR (Fig. 3B and C). Organon-on-a-chip technologies may incorporate human physiological conditions that contribute to drug tissue exposure/selectivity. Various single-organ chips provide tissue barriers with multicellular vascular or epithelial interfaces of organs (e.g., blood vessel networks, the lung, and the gut), which determines drug penetration in certain tissues. Some organs-on-a-chip may also provide tissue-level organization, such as parenchymal cells (e.g., the liver, heart, skeletal muscle, and tumors), which contribute to the binding of the drug to specific tissues96,97. Multi-organ chips or “body-on-a-chip” could be developed to integrate multiple organ units that mimic the entire human body and determine the STR in tissues of different organs98.
5.5. Developing artificial intelligent (AI)-aided computation model to evaluate STAR
AI-aided computational tools have been successfully used in 3D protein structure prediction of molecular targets, in design of drug molecules to inhibit the molecular target, and in study of SAR of drug molecule interaction with its targets15,99, 100, 101. Although there have been some attempts to use AI and machine learning technology to utilize structural information of the compounds in predicting drug-like properties and plasma PK102,103, these new technologies have not been used to successfully study STR. In the future, it is ideal to use AI and machine learning for structure representations (molecule graph notation, linear notation, or chemical descriptors) to predict STR in lead compound selection and clinical trial design and balance the dose–efficacy–toxicity. For instance, physicochemical descriptors of lead compounds can be computationally analyzed using cheminformatics. These descriptors may include, but are not limited to, structural keys, hashed fingerprints; autocorrelation, charge, logP, protein binding, refractivity, compositional, topological, and connectivity descriptors, composite descriptors from MOE (Molecular Operating Environment), and Kappa shape indices. AI-based computation mode can be used to analyze the relationship between chemical descriptors and tissue exposure, tissue/plasma partition coefficient, and tissue selectivity for a selected set of compounds with in vitro and in vivo STR data40. Once these information of adequate number of compounds have been established by AI-aided computation modeling, for any newly designed compounds, the prediction of SAR and STR will be performed using AI-based computation analysis before synthesis, which may reduce effort during drug optimization. In addition, much needed information for AI-based computation modeling for the FDA-approved drug has already included in the NDA application package in FDA database, such as the physicochemical properties, 14C mass balance studies in different tissues, clinical efficacies, and preclinical and clinical toxicities. If these data can be collected, AI-aided computation analysis can be performed to predict STR in relationship to clinical dose, efficacy, adverse events, and toxicity. However, this effort requires collaboration among academia, the FDA, and pharmaceutical industry to develop and validate these AI-aided computational tools.
Traditionally, physiologically based pharmacokinetic (PBPK) modeling has been used to predict total drug tissue exposure from total drug concentration in the blood (not only in the plasma), which can be scaled from animal models to humans. However, PBPK modeling relies on extensive tissue concentration data from preclinical animal models, which are labor intensive. If the in vitro screening method using a diffusion chamber or organs-on-a-chip can be used to predict tissue exposure/selectivity, PBPK can be established based on the in vitro screening data, which may improve the predictive power and practical application of the STR for lead compound selection and clinical trial design. PBPK modeling incorporates all physiological parameters in each organ to predict the drug concentration over time in all tissues37,104. Presently, PBPK models have been integrated into the drug development process and regulatory submissions with the main purpose of qualitatively and quantitatively predicting drug–drug interactions and scaling PK properties to humans to support initial dose selection in pediatric and first-in-human trials37,105. However, the prediction of a clinical tissue PK profile using PBPK modeling is challenging for the following reasons: (1) PBPK models constructed based on compound-specific preclinical data are usually unable to be refined and updated with clinical observation where tissue exposure in humans is difficult to detect; and (2) slight structure modification of the compounds may dramatically alter tissue exposure/selectivity in different organs, where traditional PBPK cannot be practically performed in real-time decision making of lead compound selection or clinical trial design. Some preliminary efforts have used PBPK modeling for the translation of drug's PK in the lung between animal species and humans for 12 inhaled bronchodilator drugs, which were used to study the dose-dependent clinical effect of the drugs on lung function37,64.
5.6. Preclinical animal models to study STAR
Currently, several lead compounds selected based on their SAR studies are routinely tested in disease animal model for efficacy. It is feasible to test 5–10 compounds for one molecular target, which requires no further review in this commentary. However, preclinical animal test of compounds for STR studies require the measurement of drug exposure/selectivity in all tissues that are labor intensive. Therefore, it is not feasible to test hundreds of compounds in STR studies. Therefore, it is important to decide which step of the drug optimization process the STR studies should be implemented. Here, we propose a feasible early stage drug optimization strategy using STAR to select lead compounds for clinical studies (Fig. 4). In general, approximately 200–400 compounds are synthesized for SAR studies during the drug optimization process for a single drug target. After screening the in vitro activity for specificity and potency of these compounds, several dozens of compounds would undergo in vitro ADME screening (solubility, permeability, stability, and protein binding) to select a few compounds for in vivo pharmacokinetic studies based on plasma PK profiles. Several compounds with acceptable plasma PK profiles should be further tested for STR studies to determine their tissue exposure and selectivity in preclinical animal models, in addition to efficacy testing. The in vitro and in vivo activities (by SAR), in vitro ADME, in vivo plasma PK, and in vivo tissue exposure and selectivity (by STR) should be considered to select the best lead compound for clinical studies, which may improve the success rate of drug development.
Figure 4.
Implementation of structure‒tissue selectivity/exposure–activity relationship (STAR) for lead compound selection in the current drug optimization process.
5.7. Developing non-invasive imaging technologies in human to evaluate STAR in clinical trials
Ideally, in vivo imaging technology would be tremendously useful for studying STAR in human clinical trials to improve the success rate of drug development. Although imaging modalities have been used in clinical trials for drug development, currently there is no suitable imaging technology to study STAR in clinical trials. Dynamic position emission tomography (PET) imaging may be used to visualize radioactivity-labeled compounds for target engagement in vivo or observe compound tissue exposure, both of which can be used to study STR. For instance, quantitative assessment of target engagement provides a potential method to visualize drug distribution and delivers direct evidence for dose selection to achieve adequate drug exposure in the target organ to ensure pharmacological activity88. PET imaging has been used in the clinic to assess target engagement in tissues using radiolabeled neurokinin 1 (NK1) receptor antagonists, which help in decision-making in clinical trials. Clinical PET imaging studied receptor occupancy using [18F] SPARQ, which showed that high NK1 receptor occupancy by the antagonist did not translate to therapeutic efficacy. The PET imaging study confirmed that the lack of efficacy is due to the invalid hypothesis of pharmacological mechanism, but not due to the inadequate drug exposure in the brain, which led to the decision to stop pursuing NK1 antagonist for anti-depression or anxiety treatment106,107. Rather, the adequate drug exposure in the brain of aprepitant, a selective NK1 antagonist, was later approved for prevention of acute and delayed chemotherapy-induced nausea and vomiting108. The PET imaging study helped to optimize the dose to exhibit full central nervous system target engagement, thereby achieving adequate efficacy and minimize drug–drug interactions in treating cancer patients. However, the most commonly used positron emitting radioisotopes decay fast with a relatively short half-life (e.g., 20 min for C-11 and 110 min for F-18), which only allows for short-term detection of drug tissue exposure/selectivity and target engagement109. Although F-18 labeled compounds in PET imaging have a slightly longer half-life (110 min), F-18 labeling of compounds may alter drug exposure and selectivity in different organs, as shown in the previous study40. The C-11 labeling of the compound may have minimal effect on drug tissue exposure/selectivity, but it only has an 11 min half-life. C-14-labeled compounds are widely used in animal models and humans for mass balance studies. The distribution studies of C-14-labeled compounds in animal models may provide useful information in lead compound selection based on tissue exposure/selectivity, but it is only used in humans for mass balance studies without knowing tissue exposure/selectivity110. Other in vivo imaging methods, such as magnetic resonance imaging, are only able to visualize human anatomy without the capability to visualize drug molecules in the body with adequate sensitivity and specificity. Mass spectrometry imaging (MSI) is currently used only to visualize drug molecules in ex vivo tissue sections, while the hand-held probe of MSI is in the development process; therefore, it is not yet capable of detecting or imagining drug molecules in the human body. Clearly, more imaging modalities for studying STAR are desired for selecting drug candidates and balancing clinical dose/efficacy/toxicity in the future.
6. Conclusions
In the past few decades, 90% of clinical drug developments have failed during clinical phase I, II, and III clinical studies and drug approval due to four possible reasons: lack of clinical efficacy, unmanageable toxicity, poor drug-like properties, and lack of commercial needs and poor strategic planning. Although many successful strategies are correctly implemented to overcome the four possible reasons of 90% of clinical development failures, the success rate of clinical drug development remains at 10%–15% in the past few decades. This high failure rate raises the question of whether certain aspects of drug development are overlooked? On the one hand, true target validation, which confirms the molecular target is the cause of human disease and drug's intended target, is still challenging for the success of clinical drug development. On the other hand, current drug optimization may have overemphasized one aspect but overlooked others that may mislead drug candidate selection and unbalance clinical dose/efficacy/toxicity.
In clinical drug development, in order to achieve a delicate balance among clinical dose, efficacy, and toxicity to optimize the benefit/risk ratios in patients, an ideal drug candidate would have high potency and specificity to inhibit its molecular target without off-target effect, high drug exposure in disease-targeted tissues to achieve adequate efficacy at an optimal dose (ideally at low doses), and minimal drug exposure in healthy tissues to avoid toxicity at optimal doses (even at high doses). The delicate balance of clinical dose/efficacy/toxicity of drug candidates in clinical trials is not only determined by their potency/specificity in inhibiting its molecular targets (through SAR studies), but also by their tissue exposure/selectivity in disease-targeted tissues and normal tissues (through STR studies). However, current drug optimization process overly emphasized drug's potency/specificity through SAR studies but overlooked drug's tissue exposure/selectivity in disease-targeted tissues vs. normal tissues through STR studies, which may have misled the drug candidate selection, impacted clinical dose optimization, and tipped the balance of clinical efficacy and toxicity.
We propose a STAR system to improve the drug optimization process, which classifies drug candidates into four different classes based on three aspects: drug potency/specificity (high or low), drug tissue exposure/selectivity (high or low) and required dose for balancing clinical efficacy/toxicity (high or low). The four different classes of drug candidates (classes I‒IV) require different strategies to select lead drug candidates, optimize clinical doses, and balance clinical efficacy/toxicity. In this STAR system, class I drug candidates have high specificity/potency and high exposure/selectivity, which requires low dose to achieve balanced efficacy/safety and are most desirable with a high success rate. Class II drug candidates have high specificity/potency and low tissue exposure/selectivity, which needs a high dose to achieve adequate efficacy but may have unmanageable toxicity. Class II drug candidates need to be cautiously evaluated to balance clinical dose/efficacy/toxicity. Class III drug candidates have relatively low but adequate specificity/potency but high tissue exposure/selectivity, which may require a low to medium dose to achieve adequate efficacy with manageable toxicity. The class III drug candidates may have a high clinical success rate but are often overlooked due to poor plasma drug exposure at an early stage of drug discovery. Class IV drug candidates have low specificity/potency and low exposure/selectivity, which often requires high dose and shows inadequate efficacy with high toxicity and should be terminated as early as possible. In the future, the STAR system can be improved using AI-aided computation modeling, in vitro screening, in vivo testing, and non-invasive imaging technology. Application of STAR will improve the efficiency of drug optimization and clinical studies for four different classes of drug candidates to improve the success rate of clinical drug development.
Author contributions
Duxin Sun conceived the idea and wrote the manuscript. Wei Gao wrote the manuscript. Hongxiang Hu revised manuscript and designed the figures. Simon Zhou revised the idea and manuscript. All of the authors have read and approved the final manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare. Simon Zhou is an employee of Bristol Meyer Squibb Company.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences
References
- 1.Hinkson I.V., Madej B., Stahlberg E.A. Accelerating therapeutics for opportunities in medicine: a paradigm shift in drug discovery. Front Pharmacol. 2020;11:770. doi: 10.3389/fphar.2020.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dowden H., Munro J. Trends in clinical success rates and therapeutic focus. Nat Rev Drug `Discov. 2019;18:495–496. doi: 10.1038/d41573-019-00074-z. [DOI] [PubMed] [Google Scholar]
- 3.Takebe T., Imai R., Ono S. The current status of drug discovery and development as originated in United States academia: the influence of industrial and academic collaboration on drug discovery and development. Clinical and translational science. 2018;11:597–606. doi: 10.1111/cts.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison R.K. Phase II and phase III failures: 2013‒2015. Nat Rev Drug Discov. 2016;15:817–818. doi: 10.1038/nrd.2016.184. [DOI] [PubMed] [Google Scholar]
- 5.Nielsch U., Fuhrmann U., Jaroch S. Springer Nature; New York: 2016. New approaches to drug discovery. [Google Scholar]
- 6.Hughes J.P., Rees S., Kalindjian S.B., Philpott K.L. Principles of early drug discovery. Br J Pharmacol. 2011;162:1239–1249. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiriiri G.K., Njogu P.M., Mwangi A.N. Exploring different approaches to improve the success of drug discovery and development projects: a review. Futur J Pharm Sci. 2020;6:27. [Google Scholar]
- 8.Emmerich C.H., Gamboa L.M., Hofmann M.C.J., Bonin-Andresen M., Arbach O., Schendel P., et al. Improving target assessment in biomedical research: the GOT-IT recommendations. Nat Rev Drug Discov. 2021;20:64–81. doi: 10.1038/s41573-020-0087-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis K.D., Aghaeepour N., Ahn A.H., Angst M.S., Borsook D., Brenton A., et al. Discovery and validation of biomarkers to aid the development of safe and effective pain therapeutics: challenges and opportunities. Nat Rev Neurol. 2020;16:381–400. doi: 10.1038/s41582-020-0362-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moffat J.G., Vincent F., Lee J.A., Eder J., Prunotto M. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat Rev Drug Discov. 2017;16:531–543. doi: 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- 11.Fellmann C., Gowen B.G., Lin P.C., Doudna J.A., Corn J.E. Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat Rev Drug Discov. 2017;16:89–100. doi: 10.1038/nrd.2016.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin A., Giuliano C.J., Palladino A., John K.M., Abramowicz C., Yuan M.L., et al. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci Transl Med. 2019;11 doi: 10.1126/scitranslmed.aaw8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green D.V. Virtual screening of chemical libraries for drug discovery. Expet Opin Drug Discov. 2008;3:1011–1026. doi: 10.1517/17460441.3.9.1011. [DOI] [PubMed] [Google Scholar]
- 14.Mak K.K., Pichika M.R. Artificial intelligence in drug development: present status and future prospects. Drug Discov Today. 2019;24:773–780. doi: 10.1016/j.drudis.2018.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Mao J., Akhtar J., Zhang X., Sun L., Guan S., Li X., et al. Comprehensive strategies of machine-learning-based quantitative structure‒activity relationship models. iScience. 2021;24:103052. doi: 10.1016/j.isci.2021.103052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holdgate G.A., Meek T.D., Grimley R.L. Mechanistic enzymology in drug discovery: a fresh perspective. Nat Rev Drug Discov. 2018;17:115–132. doi: 10.1038/nrd.2017.219. [DOI] [PubMed] [Google Scholar]
- 17.Adhikari N., Amin S.A., Trivedi P., Jha T., Ghosh B. HDAC3 is a potential validated target for cancer: an overview on the benzamide-based selective HDAC3 inhibitors through comparative SAR/QSAR/QAAR approaches. Eur J Med Chem. 2018;157:1127–1142. doi: 10.1016/j.ejmech.2018.08.081. [DOI] [PubMed] [Google Scholar]
- 18.Anighoro A. Underappreciated chemical interactions in protein–ligand complexes. Methods Mol Biol. 2020;2114:75–86. doi: 10.1007/978-1-0716-0282-9_5. [DOI] [PubMed] [Google Scholar]
- 19.Johnstone S., Albert J.S. Pharmacological property optimization for allosteric ligands: a medicinal chemistry perspective. Bioorg Med Chem Lett. 2017;27:2239–2258. doi: 10.1016/j.bmcl.2017.03.084. [DOI] [PubMed] [Google Scholar]
- 20.Sun Z.G., Yang Y.A., Zhang Z.G., Zhu H.L. Optimization techniques for novel c-Met kinase inhibitors. Expet Opin Drug Discov. 2019;14:59–69. doi: 10.1080/17460441.2019.1551355. [DOI] [PubMed] [Google Scholar]
- 21.Malone E.R., Oliva M., Sabatini P.J.B., Stockley T.L., Siu L.L. Molecular profiling for precision cancer therapies. Genome Med. 2020;12:8. doi: 10.1186/s13073-019-0703-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guidance for industry and review staff recommended approaches to integration of genetic toxicology study results US department of health and human services food and drug administration center for drug evaluation and research (CDER) https://wwwfdagov/media/72266/download Available from:
- 23.Guidance for Industry M3(R2) nonclinical safety studies for the conduct of human clinical trials and marketing authorization for pharmaceuticals US department of health and human services food and drug administration center for drug evaluation and research (CDER) and center for biologics evaluation and research (CBER) https://www.fda.gov/media/71542/download Available from:
- 24.Ravikumar B., Aittokallio T. Improving the efficacy-safety balance of polypharmacology in multi-target drug discovery. Expet Opin Drug Discov. 2018;13:179–192. doi: 10.1080/17460441.2018.1413089. [DOI] [PubMed] [Google Scholar]
- 25.Van Vleet T.R., Liguori M.J., Lynch J.J., 3rd, Rao M., Warder S. Screening strategies and methods for better off-target liability prediction and identification of small-molecule pharmaceuticals. SLAS Discov. 2019;24:1–24. doi: 10.1177/2472555218799713. [DOI] [PubMed] [Google Scholar]
- 26.Kramer J., Obejero-Paz C.A., Myatt G., Kuryshev Y.A., Bruening-Wright A., Verducci J.S., et al. MICE models: superior to the HERG model in predicting Torsade de Pointes. Sci Rep. 2013;3:2100. doi: 10.1038/srep02100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vahle J.L., Anderson U., Blomme E.A.G., Hoflack J.C., Stiehl D.P. Use of toxicogenomics in drug safety evaluation: current status and an industry perspective. Regul Toxicol Pharmacol. 2018;96:18–29. doi: 10.1016/j.yrtph.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 28.Evans D.C., Watt A.P., Nicoll-Griffith D.A., Baillie T.A. Drug–protein adducts: an industry perspective on minimizing the potential for drug bioactivation in drug discovery and development. Chem Res Toxicol. 2004;17:3–16. doi: 10.1021/tx034170b. [DOI] [PubMed] [Google Scholar]
- 29.Kola I., Landis J. Can the pharmaceutical industry reduce attrition rates? Nat Rev Drug Discov. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 30.Kerns E.H., Di L. In: Drug-like properties concepts, structure design and methods from ADME to toxicity optimization. Di L., Kerns E.H., editors. Academic Press; San Diego: 2008. Chapter 2—advantages of good drug-like properties; pp. 6–16. [Google Scholar]
- 31.Lipinski C.A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1:337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 32.Balimane P.V., Han Y.H., Chong S. Current industrial practices of assessing permeability and P-glycoprotein interaction. AAPS J. 2006;8:E1–E13. doi: 10.1208/aapsj080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun D., Lennernas H., Welage L.S., Barnett J.L., Landowski C.P., Foster D., et al. Comparison of human duodenum and Caco-2 gene expression profiles for 12,000 gene sequences tags and correlation with permeability of 26 drugs. Pharm Res. 2002;19:1400–1416. doi: 10.1023/a:1020483911355. [DOI] [PubMed] [Google Scholar]
- 34.Sun D., Yu L.X., Hussain M.A., Wall D.A., Smith R.L., Amidon G.L. In vitro testing of drug absorption for drug 'developability' assessment: forming an interface between in vitro preclinical data and clinical outcome. Curr Opin Drug Discov Dev. 2004;7:75–85. [PubMed] [Google Scholar]
- 35.Amidon G.L., Lennernas H., Shah V.P., Crison J.R. A theoretical basis for a biopharmaceutic drug classification: the correlation of in vitro drug product dissolution and in vivo bioavailability. Pharm Res. 1995;12:413–420. doi: 10.1023/a:1016212804288. [DOI] [PubMed] [Google Scholar]
- 36.Kerns E.H., Di L. In: Drug-like properties concepts, structure design and methods from ADME to toxicity optimization. Di L., Kerns E.H., editors. Academic Press; San Diego: 2008. Chapter 19 - pharmacokinetics; pp. 228–241. Kerns EH, Di L. [Google Scholar]
- 37.Davies M., Jones R.D.O., Grime K., Jansson-Lofmark R., Fretland A.J., Winiwarter S., et al. Improving the accuracy of predicted human pharmacokinetics: lessons learned from the AstraZeneca drug pipeline over two decades. Trends Pharmacol Sci. 2020;41:390–408. doi: 10.1016/j.tips.2020.03.004. [DOI] [PubMed] [Google Scholar]
- 38.Di L., Kerns E.H., Carter G.T. Drug-like property concepts in pharmaceutical design. Curr Pharmaceut Des. 2009;15:2184–2194. doi: 10.2174/138161209788682479. [DOI] [PubMed] [Google Scholar]
- 39.Yusof I., Segall M.D. Considering the impact drug-like properties have on the chance of success. Drug Discov Today. 2013;18:659–666. doi: 10.1016/j.drudis.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 40.Wei Gao H.H., Lipeng Dai, He Miao, Yuan Hebao, Zhang Huixia, Liao Jinhui, et al. Structure‒tissue exposure/selectivity relationship (STR) may correlate with clinical efficacy/safety. Acta Pharm Sin B. 2022 in press. [Google Scholar]
- 41.Bohnert T., Gan L.S. Plasma protein binding: from discovery to development. J Pharmacol Sci. 2013;102:2953–2994. doi: 10.1002/jps.23614. [DOI] [PubMed] [Google Scholar]
- 42.Zhang D., Hop C., Patilea-Vrana G., Gampa G., Seneviratne H.K., Unadkat J.D., et al. Drug concentration asymmetry in tissues and plasma for small molecule-related therapeutic modalities. Drug Metab Dispos. 2019;47:1122–1135. doi: 10.1124/dmd.119.086744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shitara Y., Horie T., Sugiyama Y. Transporters as a determinant of drug clearance and tissue distribution. Eur J Pharmaceut Sci. 2006;27:425–446. doi: 10.1016/j.ejps.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Giacomini K.M., Huang S.M., Tweedie D.J., Benet L.Z., Brouwer K.L.R., Chu X., et al. Membrane transporters in drug development. Nat Rev Drug Discov. 2010;9:215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abdallah M., Mullertz O.O., Styles I.K., Morsdorf A., Quinn J.F., Whittaker M.R., et al. Lymphatic targeting by albumin-hitchhiking: applications and optimisation. J Contr Release. 2020;327:117–128. doi: 10.1016/j.jconrel.2020.07.046. [DOI] [PubMed] [Google Scholar]
- 46.Ghinea N. Anti-angiogenic therapy: albumin-binding proteins could mediate mechanisms underlying the accumulation of small molecule receptor tyrosine kinase inhibitors in normal tissues with potential harmful effects on health. Diseases. 2021;9:28. doi: 10.3390/diseases9020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poulin P., Burczynski F.J., Haddad S. The role of extracellular binding proteins in the cellular uptake of drugs: impact on quantitative in vitro-to-in vivo extrapolations of toxicity and efficacy in physiologically based pharmacokinetic-pharmacodynamic research. J Pharmacol Sci. 2016;105:497–508. doi: 10.1002/jps.24571. [DOI] [PubMed] [Google Scholar]
- 48.Hendrickx R., Lamm Bergström E., Janzén D.L.I., Fridén M., Eriksson U., Grime K., et al. Translational model to predict pulmonary pharmacokinetics and efficacy in man for inhaled bronchodilators. CPT Pharmacometrics Syst Pharmacol. 2018;7:147–157. doi: 10.1002/psp4.12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamazaki M., Suzuki H., Sugiyama Y. Recent advances in carrier-mediated hepatic uptake and biliary excretion of xenobiotics. Pharm Res. 1996;13:497–513. doi: 10.1023/a:1016077517241. [DOI] [PubMed] [Google Scholar]
- 50.Poulin P. Drug Distribution to human tissues: prediction and examination of the basic assumption in in vivo pharmacokinetics–pharmacodynamics (PK/PD) research. J Pharmacol Sci. 2015;104:2110–2118. doi: 10.1002/jps.24427. [DOI] [PubMed] [Google Scholar]
- 51.Hammarlund-Udenaes M. Active-site concentrations of chemicals—are they a better predictor of effect than plasma/organ/tissue concentrations? Basic Clin Pharmacol Toxicol. 2010;106:215–220. doi: 10.1111/j.1742-7843.2009.00517.x. [DOI] [PubMed] [Google Scholar]
- 52.Gonzalez D., Schmidt S., Derendorf H. Importance of relating efficacy measures to unbound drug concentrations for anti-infective agents. Clin Microbiol Rev. 2013;26:274–288. doi: 10.1128/CMR.00092-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rizk M.L., Zou L., Savic R.M., Dooley K.E. Importance of drug pharmacokinetics at the site of action. Clin Transl Sci. 2017;10:133–142. doi: 10.1111/cts.12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Charvériat M., Lafon V., Mouthon F., Zimmer L. Innovative approaches in CNS drug discovery. Therapie. 2021;76:101–109. doi: 10.1016/j.therap.2020.12.006. [DOI] [PubMed] [Google Scholar]
- 55.Ghosh K.K., Padmanabhan P., Yang C.T., Ng D.C.E., Palanivel M., Mishra S., et al. Positron emission tomographic imaging in drug discovery. Drug Discov Today. 2022;27:280–291. doi: 10.1016/j.drudis.2021.07.025. [DOI] [PubMed] [Google Scholar]
- 56.Patel N.C. Methods to optimize CNS exposure of drug candidates. Bioorg Med Chem Lett. 2020;30:127503. doi: 10.1016/j.bmcl.2020.127503. [DOI] [PubMed] [Google Scholar]
- 57.Tonge P.J. Drug-target kinetics in drug discovery. ACS Chem Neurosci. 2018;9:29–39. doi: 10.1021/acschemneuro.7b00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dai Q., Wilhelm S., Ding D., Syed A.M., Sindhwani S., Zhang Y., et al. Quantifying the ligand-coated nanoparticle delivery to cancer cells in solid tumors. ACS Nano. 2018;12:8423–8435. doi: 10.1021/acsnano.8b03900. [DOI] [PubMed] [Google Scholar]
- 59.Northfelt D.W., Martin F.J., Working P., Volberding P.A., Russell J., Newman M., et al. Doxorubicin encapsulated in liposomes containing surface-bound polyethylene glycol: pharmacokinetics, tumor localization, and safety in patients with AIDS-related Kaposi's sarcoma. J Clin Pharmacol. 1996;36:55–63. doi: 10.1002/j.1552-4604.1996.tb04152.x. [DOI] [PubMed] [Google Scholar]
- 60.Harrington K.J., Mohammadtaghi S., Uster P.S., Glass D., Peters A.M., Vile R.G., et al. Effective targeting of solid tumors in patients with locally advanced cancers by radiolabeled pegylated liposomes. Clin Cancer Res. 2001;7:243–254. [PubMed] [Google Scholar]
- 61.Arrieta O., Medina L.A., Estrada-Lobato E., Ramirez-Tirado L.A., Mendoza-Garcia V.O., de la Garza-Salazar J. High liposomal doxorubicin tumour tissue distribution, as determined by radiopharmaceutical labelling with 99mTc-LD, is associated with the response and survival of patients with unresectable pleural mesothelioma treated with a combination of liposomal doxorubicin and cisplatin. Cancer Chemother Pharmacol. 2014;74:211–215. doi: 10.1007/s00280-014-2477-x. [DOI] [PubMed] [Google Scholar]
- 62.Luan X., Yuan H., Song Y., Hu H., Wen B., He M., et al. Reappraisal of anticancer nanomedicine design criteria in three types of preclinical cancer models for better clinical translation. Biomaterials. 2021;275:120910. doi: 10.1016/j.biomaterials.2021.120910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun D., Zhou S., Gao W. What went wrong with anticancer nanomedicine design and how to make it right. ACS Nano. 2020;14:12281–12290. doi: 10.1021/acsnano.9b09713. [DOI] [PubMed] [Google Scholar]
- 64.Kuepfer L., Niederalt C., Wendl T., Schlender J.F., Willmann S., Lippert J., et al. Applied concepts in PBPK modeling: how to build a PBPK/PD model. CPT Pharmacometrics Syst Pharmacol. 2016;5:516–531. doi: 10.1002/psp4.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muller P.Y., Milton M.N. The determination and interpretation of the therapeutic index in drug development. Nat Rev Drug Discov. 2012;11:751–761. doi: 10.1038/nrd3801. [DOI] [PubMed] [Google Scholar]
- 66.Müller K. The science and business of drug discovery: demystifying the jargon. By Edward D. Zanders. ChemMedChem. 2012;7:533–534. [Google Scholar]
- 67.Raevsky O.A., Mukhametov A., Grigorev V.Y., Ustyugov A., Tsay S.C., Jih-Ru Hwu R., et al. Applications of multi-target computer-aided methodologies in molecular design of CNS drugs. Curr Med Chem. 2018;25:5293–5314. doi: 10.2174/0929867324666170920154111. [DOI] [PubMed] [Google Scholar]
- 68.Babic I., Kesari S., Nurmemmedov E. Cellular target engagement: a new paradigm in drug discovery. Future Med Chem. 2018;10:1641–1644. doi: 10.4155/fmc-2018-0139. [DOI] [PubMed] [Google Scholar]
- 69.Barlaam B., Casella R., Cidado J., Cook C., De Savi C., Dishington A., et al. Discovery of AZD4573, a potent and selective inhibitor of CDK9 that enables short duration of target engagement for the treatment of hematological malignancies. J Med Chem. 2020;63:15564–15590. doi: 10.1021/acs.jmedchem.0c01754. [DOI] [PubMed] [Google Scholar]
- 70.Guo W.H., Qi X., Yu X., Liu Y., Chung C.I., Bai F., et al. Enhancing intracellular accumulation and target engagement of PROTACs with reversible covalent chemistry. Nat Commun. 2020;11:4268. doi: 10.1038/s41467-020-17997-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McNulty D.E., Bonnette W.G., Qi H., Wang L., Ho T.F., Waszkiewicz A., et al. A high-throughput dose–response cellular thermal shift assay for rapid screening of drug target engagement in living cells, exemplified using SMYD3 and Ido1. SLAS Discov. 2018;23:34–46. doi: 10.1177/2472555217732014. [DOI] [PubMed] [Google Scholar]
- 72.Brassard M., Rondeau G. Role of vandetanib in the management of medullary thyroid cancer. Biologics. 2012;6:59–66. doi: 10.2147/BTT.S24220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Inoue A., Kobayashi K., Maemondo M., Sugawara S., Oizumi S., Isobe H., et al. Updated overall survival results from a randomized phase III trial comparing gefitinib with carboplatin–paclitaxel for chemo-naive non-small cell lung cancer with sensitive EGFR gene mutations (NEJ002) Ann Oncol. 2013;24:54–59. doi: 10.1093/annonc/mds214. [DOI] [PubMed] [Google Scholar]
- 74.Kim A., Jang M.H., Lee S.J., Bae Y.K. Mutations of the epidermal growth factor receptor gene in triple-negative breast cancer. J Breast Cancer. 2017;20:150–159. doi: 10.4048/jbc.2017.20.2.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cohen P., Cross D., Janne P.A. Kinase drug discovery 20 years after imatinib: progress and future directions. Nat Rev Drug Discov. 2021;20:551–569. doi: 10.1038/s41573-021-00195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Byrd J.C., Hillmen P., Ghia P., Kater A.P., Chanan-Khan A., Furman R.R., et al. Acalabrutinib versus ibrutinib in previously treated chronic lymphocytic leukemia: results of the first randomized phase III trial. J Clin Oncol. 2021;39:3441–3452. doi: 10.1200/JCO.21.01210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sharman J.P., Egyed M., Jurczak W., Skarbnik A., Pagel J.M., Flinn I.W., et al. Acalabrutinib with or without obinutuzumab versus chlorambucil and obinutuzmab for treatment-naive chronic lymphocytic leukaemia (ELEVATE TN): a randomised, controlled, phase 3 trial. Lancet. 2020;395:1278–1291. doi: 10.1016/S0140-6736(20)30262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y., Ramírez-Valle F., Xue Y., Ventura J.I., Gouedard O., Mei J., et al. Population pharmacokinetics and exposure response assessment of CC-292, a potent BTK Inhibitor, in patients with chronic lymphocytic leukemia. J Clin Pharmacol. 2017;57:1279–1289. doi: 10.1002/jcph.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.U.S. Food and Drug Administration . Initial U.S. Approval; 1977. SOLTAMOX® (tamoxifen citrate) oral solution.https://www.accessdata.fda.gov/drugsatfda_docs/label/2005/021807lbl.pdf Available from: [Google Scholar]
- 80.U.S. Food and Drug Administration . Initial U.S. Approval; 1997. EVISTA (raloxifene hydrochloride) tablet for oral use.https://www.accessdata.fda.gov/drugsatfda_docs/label/2007/022042lbl.pdf Available from: [Google Scholar]
- 81.Martinkovich S., Shah D., Planey S.L., Arnott J.A. Selective estrogen receptor modulators: tissue specificity and clinical utility. Clin Interv Aging. 2014;9:1437–1452. doi: 10.2147/CIA.S66690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Komm B.S., Mirkin S. An overview of current and emerging SERMs. J Steroid Biochem Mol Biol. 2014;143:207–222. doi: 10.1016/j.jsbmb.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 83.Maximov P.Y., Lee T.M., Jordan V.C. The discovery and development of selective estrogen receptor modulators (SERMs) for clinical practice. Curr Clin Pharmacol. 2013;8:135–155. doi: 10.2174/1574884711308020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., et al. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc Natl Acad Sci U S A. 2020;117:6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sun D. Remdesivir for treatment of COVID-19: combination of pulmonary and IV administration may offer aditional benefit. AAPS J. 2020;22:77. doi: 10.1208/s12248-020-00459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wassermann A.M., Wawer M., Bajorath J. Activity landscape representations for structure‒activity relationship analysis. J Med Chem. 2010;53:8209–8223. doi: 10.1021/jm100933w. [DOI] [PubMed] [Google Scholar]
- 88.Campbell B.R., Gonzalez Trotter D., Hines C.D., Li W., Patel M., Zhang W., et al. In vivo imaging in pharmaceutical development and its impact on the 3Rs. ILAR J. 2016;57:212–220. doi: 10.1093/ilar/ilw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Di L.K.E. Academic Press; 2015. Drug-like properties: concepts, structure design and methods from ADME to toxicity optimization. [Google Scholar]
- 90.Saha N. In: Pharmaceutical medicine and translational clinical research. Vohora D., Singh G., editors. Academic Press; Boston: 2018. Clinical pharmacokinetics and drug interactions; pp. 81–106. [Google Scholar]
- 91.Koyfman S.A., Agrawal M., Garrett-Mayer E., Krohmal B., Wolf E., Emanuel E.J., et al. Risks and benefits associated with novel phase 1 oncology trial designs. Cancer. 2007;110:1115–1124. doi: 10.1002/cncr.22878. [DOI] [PubMed] [Google Scholar]
- 92.Schmitt W. General approach for the calculation of tissue to plasma partition coefficients. Toxicol Vitro. 2008;22:457–467. doi: 10.1016/j.tiv.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 93.Deitchman A.N., Singh R.S.P., Derendorf H. Nonlinear protein binding: not what you think. J Pharmacol Sci. 2018;107:1754–1760. doi: 10.1016/j.xphs.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boger E., Evans N., Chappell M., Lundqvist A., Ewing P., Wigenborg A., et al. Systems pharmacology approach for prediction of pulmonary and systemic pharmacokinetics and receptor occupancy of inhaled drugs. CPT Pharmacometrics Syst Pharmacol. 2016;5:201–210. doi: 10.1002/psp4.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Di L., Umland J.P., Trapa P.E., Maurer T.S. Impact of recovery on fraction unbound using equilibrium dialysis. J Pharmacol Sci. 2012;101:1327–1335. doi: 10.1002/jps.23013. [DOI] [PubMed] [Google Scholar]
- 96.Zhang B., Korolj A., Lai B.F.L., Radisic M. Advances in organ-on-a-chip engineering. Nat Rev Mater. 2018;3:257–278. [Google Scholar]
- 97.Bhatia S.N., Ingber D.E. Microfluidic organs-on-chips. Nat Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 98.Herland A., Maoz B.M., Das D., Somayaji M.R., Prantil-Baun R., Novak R., et al. Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nat Biomed. 2020;4:421–436. doi: 10.1038/s41551-019-0498-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Carracedo-Reboredo P., Linares-Blanco J., Rodriguez-Fernandez N., Cedron F., Novoa F.J., Carballal A., et al. A review on machine learning approaches and trends in drug discovery. Comput Struct Biotechnol J. 2021;19:4538–4558. doi: 10.1016/j.csbj.2021.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vamathevan J., Clark D., Czodrowski P., Dunham I., Ferran E., Lee G., et al. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov. 2019;18:463–477. doi: 10.1038/s41573-019-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jiménez-Luna J., Grisoni F., Schneider G. Drug discovery with explainable artificial intelligence. Nat Mach Intell. 2020;2:573–584. [Google Scholar]
- 103.David L., Thakkar A., Mercado R., Engkvist O. Molecular representations in AI-driven drug discovery: a review and practical guide. J Cheminf. 2020;12:56. doi: 10.1186/s13321-020-00460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhuang X., Lu C. PBPK modeling and simulation in drug research and development. Acta Pharm Sin B. 2016;6:430–440. doi: 10.1016/j.apsb.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang X., Yang Y., Grimstein M., Fan J., Grillo J.A., Huang S.M., et al. Application of PBPK modeling and simulation for regulatory decision making and its impact on US prescribing information: an update on the 2018-2019 submissions to the US FDA's Office of Clinical Pharmacology. J Clin Pharmacol. 2020;60(Suppl 1):S160–S178. doi: 10.1002/jcph.1767. [DOI] [PubMed] [Google Scholar]
- 106.Keller M., Montgomery S., Ball W., Morrison M., Snavely D., Liu G., et al. Lack of efficacy of the substance p (neurokinin1 receptor) antagonist aprepitant in the treatment of major depressive disorder. Biol Psychiatr. 2006;59:216–223. doi: 10.1016/j.biopsych.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 107.Michelson D., Hargreaves R., Alexander R., Ceesay P., Hietala J., Lines C., et al. Lack of efficacy of L-759274, a novel neurokinin 1 (substance P) receptor antagonist, for the treatment of generalized anxiety disorder. Int J Neuropsychopharmacol. 2013;16:1–11. doi: 10.1017/S1461145712000065. [DOI] [PubMed] [Google Scholar]
- 108.Hargreaves R., Ferreira J.C., Hughes D., Brands J., Hale J., Mattson B., et al. Development of aprepitant, the first neurokinin-1 receptor antagonist for the prevention of chemotherapy-induced nausea and vomiting. Ann N Y Acad Sci. 2011;1222:40–48. doi: 10.1111/j.1749-6632.2011.05961.x. [DOI] [PubMed] [Google Scholar]
- 109.Matthews P.M., Rabiner E.A., Passchier J., Gunn R.N. Positron emission tomography molecular imaging for drug development. Br J Clin Pharmacol. 2012;73:175–186. doi: 10.1111/j.1365-2125.2011.04085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Penner N., Klunk L.J., Prakash C. Human radiolabeled mass balance studies: objectives, utilities and limitations. Biopharm Drug Dispos. 2009;30:185–203. doi: 10.1002/bdd.661. [DOI] [PubMed] [Google Scholar]