Abstract
Many immunocompromised patients mount suboptimal humoral immunity after SARS-CoV-2 mRNA vaccination. Here, we assessed the single-cell profile of SARS-CoV-2-specific T cells post-mRNA vaccination in healthy individuals and patients with various forms of immunodeficiencies. Impaired vaccine-induced cell-mediated immunity was observed in many immunocompromised patients, particularly in solid-organ transplant and chronic lymphocytic leukemia patients. Notably, individuals with an inherited lack of mature B cells, i.e., X-linked agammaglobulinemia (XLA) displayed highly functional spike-specific T cell responses. Single-cell RNA-sequencing further revealed that mRNA vaccination induced a broad functional spectrum of spike-specific CD4+ and CD8+ T cells in healthy individuals and patients with XLA. These responses were founded on polyclonal repertoires of CD4+ T cells and robust expansions of oligoclonal effector-memory CD45RA+ CD8+ T cells with stem-like characteristics. Collectively, our data provide the functional continuum of SARS-CoV-2-specific T cell responses post-mRNA vaccination, highlighting that cell-mediated immunity is of variable functional quality across immunodeficiency syndromes.
Keywords: SARS-CoV-2, COVID-19, T cells, mRNA vaccine
Many immunocompromised individuals respond poorly to mRNA vaccination and are at higher risk of developing severe COVID-19. Gao and Cai et al. provide the single-cell landscape of SARS-CoV-2-specific T cells after mRNA vaccination and observe that the functional quality of memory T cell responses varies considerably across different immunodeficiency syndromes.
Introduction
SARS-CoV-2 mRNA vaccination has proven highly effective for reducing the incidence of symptomatic and severe COVID-19 (Polack et al., 2020). The generation of neutralizing antibodies (nAbs) has been identified as a strong correlate of protection following mRNA vaccination (Khoury et al., 2021). Nevertheless, current vaccines are based on the initial Wuhan SARS-CoV-2 sequence, whereas variants of concern (including B.1.617.2 and B.1.1.529) are known to partly subvert nAbs (Planas et al., 2021, 2022) and, to a lesser extent, T cell recognition (Gao et al., 2022). This has galvanized the need to gain a complete picture of adaptive immune responses that might form additional layers of protection from severe COVID-19. T cells serve as a critical component in establishing effective humoral and cell-mediated immunity in most infections. Both CD4+ and CD8+ T cell responses are induced following SARS-CoV-2 vaccination (Sahin et al., 2020) and have previously been shown to form long-lasting immunity following both yellow fever vaccination (Fuertes Marraco et al., 2015) and SARS-CoV-1 natural infection (Le Bert et al., 2020; Ng et al., 2016). Following the first dose of SARS-CoV-2 mRNA vaccination, spike-specific T cell responses arise before the emergence of nAbs and may contribute to the early protective efficacy observed already after one dose (Kalimuddin et al., 2021). Additionally, CD4+ and CD8+ T cells are necessary to generate protective antibody responses and control SARS-CoV-2 replication in the respiratory tract in murine and non-human primate models (Israelow et al., 2021; McMahan et al., 2021). These data highlight the need for effective vaccines to induce a complementary humoral and cell-mediated SARS-CoV-2-specific immune response to establish protection.
Although the mRNA-based BNT162b2 (Pfizer/BioNTech) and mRNA-1273 (Moderna) vaccines have proven to be highly effective in the general population (Baden et al., 2021; Polack et al., 2020), previous studies on immunocompromised populations have observed weaker or absent humoral immune responses after vaccination (Embi et al., 2021; Pimpinelli et al., 2021; Rahav et al., 2021). Individuals with primary or secondary immunodeficiencies (e.g., human immunodeficiency virus [HIV], hematological malignancy, and immunosuppressive treatments) also exhibit increased morbidity and mortality rates compared with immunocompetent individuals following SARS-CoV-2 infection (Shields et al., 2021). Nevertheless, the spectrum of immunodeficiencies most likely renders into differential responses following vaccination and whether immunodeficiency affects the maintenance of long-term memory responses is unknown. A previous immunological study in a cohort of multiple sclerosis (MS) patients undergoing anti-CD20 treatment has demonstrated that robust cell-mediated immune responses can arise in the relative absence of humoral immunity post-vaccination (Apostolidis et al., 2021). The possibility that different immunodeficient conditions might elicit differential humoral and cell-mediated immunity is of utmost interest, given that these states might offer insight into immune correlates of protection from severe COVID-19 in the scenario of breakthrough infections.
Our understanding of T cell responses following mRNA vaccination is relatively scarce compared with humoral immunity. Previous studies using conventional flow-cytometry analysis have indicated that mRNA vaccines rapidly mobilize an early CD8+ T cell response after the first dose (prime) when both spike-specific CD4+ T cells and nAbs are hardly detectable in the circulation (Oberhardt et al., 2021). Following a second dose, a robust spike-specific CD4+ T cell response is induced, and this is generally of a higher magnitude than the CD8+ T cell response (Guerrera et al., 2021; Mateus et al., 2021). Through conventional flow analysis, mRNA vaccination has been shown to polarize the CD4+ T cell response primarily into a T helper 1 (Th1) (Sahin et al., 2020) and a T follicular helper (Tfh) cell (Painter et al., 2021) phenotype. Longitudinal analysis has further demonstrated that polyfunctional spike-specific CD4+ and CD8+ T cells, based on interferon-γ (IFN-γ), tumor necrosis factor (TNF), interleukin-2 (IL-2), and CD107a expression of stem cell memory (TSCM: CD45RA+CCR7+CD95+) and central memory (TCM: CD45RA−CCR7+) phenotypes, persist up to 6 months after mRNA vaccination (Guerrera et al., 2021). Simple classifications of spike-specific and other antigen-specific CD4+ and CD8+ T cells into helper and memory subsets have traditionally been based on a few key chemokine receptors or cytokines. However, most antigen-experienced T cells are part of a continuum of cell states and phenotypes that creates a heterogeneous pool of clonotypes beyond classical Th and memory classifications. Through the use of next-generation secretome and single-cell sequencing technologies, it is possible to generate a deeper profile of the functional landscape of SARS-CoV-2-specific T cells and bring nuance to the definition of fixed CD4+ and CD8+ T cell archetypes (Kiner et al., 2021).
To fill this knowledge gap, we investigated the functional profile of SARS-CoV-2-specific T cells following mRNA-based BNT162b2 vaccination in healthy donors and across a spectrum of immunodeficiencies. Using conventional flow cytometry and secretome analysis, we show that many immunocompromised patient groups exhibit non-coordinated humoral and cell-mediated immune responses, which does not preclude the detection of SARS-CoV-2-specific T cell responses at 6 months post-vaccination. Additionally, our data indicate that certain immunocompromised patients, e.g., those with X-linked agammaglobulinemia (XLA), still elicit functional T cell responses despite the lack of mature B cells and antibody responses post-vaccination. Through a combined activation-induced sorting pipeline, we defined the single-cell functional landscape of spike-specific CD4+ and CD8+ T cell clonotypes in healthy individuals and those with an inherent lack of mature B cells—beyond traditional Th and memory cell characteristics. Our findings define the functional nature of spike-specific CD4+ and CD8+ T cell clonotypes following mRNA vaccination and demonstrate highly variable humoral and cell-mediated immunity across the range of immunodeficiency disorders.
Results
The magnitude of SARS-CoV-2-specific T cell responses after mRNA vaccination differs across different immunocompromised states
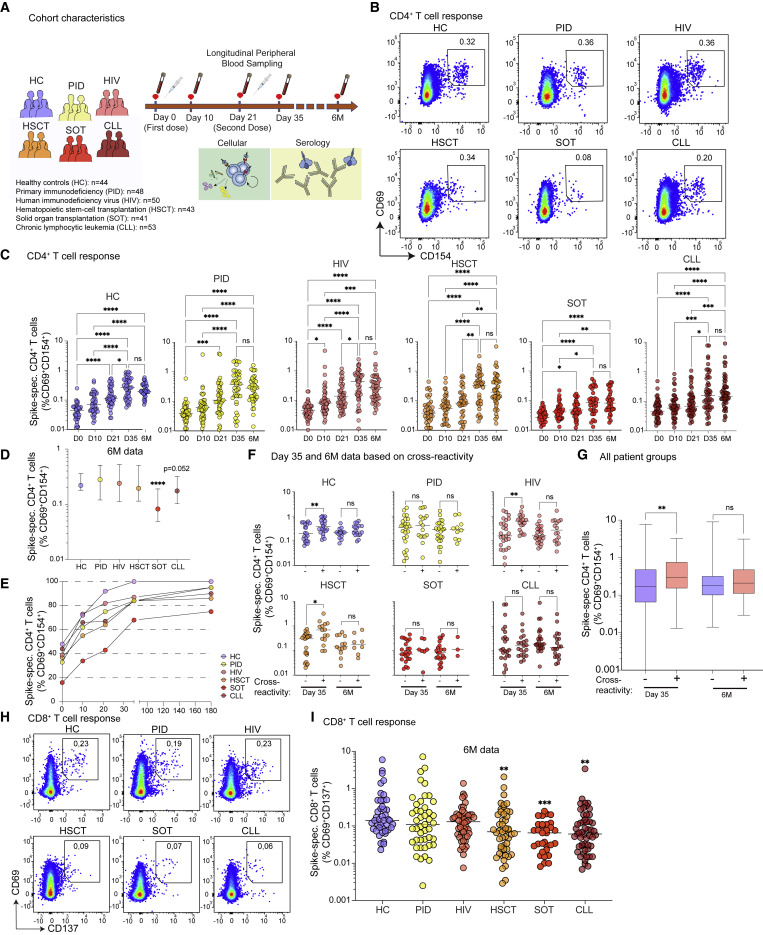
To examine the impact of immunodeficiency on cell-mediated immunity to SARS-CoV-2 vaccination, we initiated a prospective open-label clinical trial (COVAXID, EudraCT, no. 2021-000175-37) to investigate the immunogenicity of the BNT162b2 vaccine across a broad spectrum of immunocompromised patients and healthy controls (HCs) (Bergman et al., 2021). In this clinical trial, we also collected peripheral blood mononuclear cells (PBMCs) from a subset of patients, including HCs (n = 44) and individuals with (1) primary immunodeficiency (PID) (n = 48), (2) HIV infection (n = 50), (3) post hematopoietic stem cell transplantation (HSCT) (n = 43), (4) post solid-organ transplantation (SOT) (n = 41), and (5) chronic lymphocytic leukemia (CLL) (n = 53) (Figure 1 A; Table S1). All study participants had negative PCR tests at baseline and no previous documented COVID-19. Plasma and PBMCs were analyzed at five time points during vaccination (days 0, 14, 21, and 35 and 6 months) (Figure 1A).
Figure 1.
Longitudinal T cell responses after SARS-CoV-2 mRNA vaccination in healthy controls and immunocompromised patients
(A) Schematic of the longitudinal study design, involving six cohorts of healthy and immunocompromised patient groups across five time points.
(B and C) (B) Representative flow plots at day 35 (n = 279 independent experiments) showing CD69 and CD154 expression after spike-peptide pool stimulation on memory CD4+ T cells (above), and (C) plots of their frequencies over time (below).
(D) Spike-specific CD4+ T cell frequencies at 6 months.
(E) Spike-specific CD4+ T cell frequencies across all time points.
(F) Spike-specific CD4+ T cell frequencies at ay 35 and 6 months based on the presence or absence of pre-existing day 0 responses.
(G) Spike-specific CD4+ T cell frequencies at day 35 and 6 months based on the presence or absence of pre-existing day 0 responses with data combined from all individuals.
(H) Representative flow plots at 6 months (n = 279 independent experiments) showing CD69 and CD137 expression after spike-peptide pool stimulation of memory CD8+ T cells.
(I) Spike-specific (CD69+CD137+) CD8+ T cell frequencies at 6 months.
Graphs show median ± interquartile range (IQR) (D and G) or median values (C, F, and I). (C, D, and I) Kruskal-Wallis test with Dunn’s post-test. (F and G) Mann-Whitney test. ∗ p< 0.05, ∗∗ p< 0.01, ∗∗∗ p< 0.001, ns, not significant. See also Figures S1 and S2.
We first tracked the longitudinal evolution of total T cell responses in all groups before and post-vaccination using IFN-γ ELISpot assay after overlapping SARS-CoV-2 spike-peptide pool stimulations. IFN-γ+ T cell responses were undetectable in most donors prior to vaccination but increased after the first dose and continued to increase after the second dose (day 35) in all study groups (Figure S1A). On day 35, the CLL and SOT cohort showed the lowest fraction of detectable IFN-γ+ responses (Figure S1A), where the magnitude of responses was significantly lower overall compared with HCs (Figure S1B).
To differentiate between CD4+ and CD8+ T cell responses, we next used spike-peptide pool stimulations and assessed activation-induced markers (AIMs), namely CD69 together with CD40L (CD154) for detection of vaccine (spike)-specific CD4+ T cells and CD69 together with 4-1BB (CD137) to identify spike-specific CD8+ T cells (Figure S1C). Most HCs responded rapidly to the first mRNA vaccine dose (day 10), and all showed detectable spike-specific CD4+ T cells after two doses (day 35 and 6 months) (Figures 1B and 1C). Younger individuals (18–39 years) showed a trend toward higher frequencies of spike-specific CD4+ T cells at day 35 but similar frequencies with elderly patient groups at 6 months (Figure S2A). The PID cohort responded well to the vaccine, particularly after the second vaccine dose (Figures 1B and 1C). All subjects with monogenic and other PIDs, followed by CD4 lymphopenia, common variable immunodeficiency (CVID), and XLA, had detectable spike-specific CD4+ T cell responses at day 35 and 6 months (Figure S2B). Most HIV-infected patients generated positive CD4+ T cell responses regardless of their infection stage (<300 cells/mm3: 100%; >300 cells/mm3: 83%) at day 35, which were maintained up to 6 months after vaccination (Figures 1B, 1C, and S2C). Most individuals undergoing HSCT displayed positive CD4+ T cell responses at day 35 and 6 months (Figures 1B and 1C), although the time after HSCT influenced the response (Figure S2D). The SOT group showed a significant increase in CD4+ T cell responses only after the second vaccine dose (Figures 1B and 1C). Particularly patients treated with mycophenolate mofetil (MMF) soon after transplantation had a notably poor CD4+ T cell response (Figure S2E). Similarly, CLL patients produced significantly increased CD4+ T cell responses only after the second vaccine dose (Figures 1B and 1C), where particularly patients with ongoing ibrutinib (a BTK inhibitor) treatment generated lower CD4+ T cell responses (Figure S2F). At 6 months, the frequencies of spike-specific CD4+ T cells were comparable between HC, PID, HIV, and HSCT, whereas SOT and CLL individuals exhibited significantly lower responses compared with HCs (Figure 1D). Overall, all HCs generated detectable spike-specific CD4+ T cell responses at day 35 and 6 months, while more than one patient from each immunodeficiency group had undetectable responses (Figure 1E).
Pre-existing immunity correlates with increased SARS-CoV-2-specific CD4+ T cell responses early after mRNA vaccination
Several studies have reported the existence of cross-reactive (pre-existing) T cells that recognize SARS-CoV-2 in peripheral blood and tissue (Braun et al., 2020; Grifoni et al., 2020; Niessl et al., 2021; Schulien et al., 2021; Sekine et al., 2020). Similar to these previous reports, we detected pre-existing CD4+ T cells in 10%–40% of each patient group (Figure 1E). Notably, the SOT group, which responded poorly to the vaccine (Figure 1D), also displayed the lowest frequencies of pre-existing CD4+ T cells (day 0) (Figure 1E). Individuals with pre-existing responses subsequently generated higher frequencies of CD4+ T cell responses after the second vaccine dose at day 35, but not 6 months (Figure 1F). These data align with previous reports (Loyal et al., 2021). Still, here we also observed that pre-existing CD4+ T cells were associated with improved spike-specific CD4+ T cell responses across separate groups of immunodeficient patients (Figures 1F and 1G).
Immunocompromised patient groups generate variable magnitudes of SARS-CoV-2-specific CD8+ T cell responses
We next assessed spike-specific CD8+ T cells and found that these responses were induced post-mRNA vaccination (Figures 1H and S1C) but at lower frequencies than corresponding spike-specific CD4+ T cells. Nevertheless, similar trends were observed, where the frequencies of spike-specific CD8+ T cells were comparable between HC, PID, and HIV and lower in HSCT, SOT, and CLL individuals at 6 months (Figure 1I). In addition, we confirmed that the frequencies of IFN-γ+ CD8+ T cells were comparable between HC, PID, HIV, and HSCT and lower in SOT and CLL individuals at day 35 (Figure S1D).
Finally, we tested whether T cell responses against the Delta (B.1.617.2) variant were affected in individuals with PID or HCs. We observed no frequency differences between Delta and wild-type spike-specific CD4+ and CD8+ T cells (Figure S2G). These data indicate that cell-mediated cross-recognition of Delta is not impacted to a high degree during immunodeficient states.
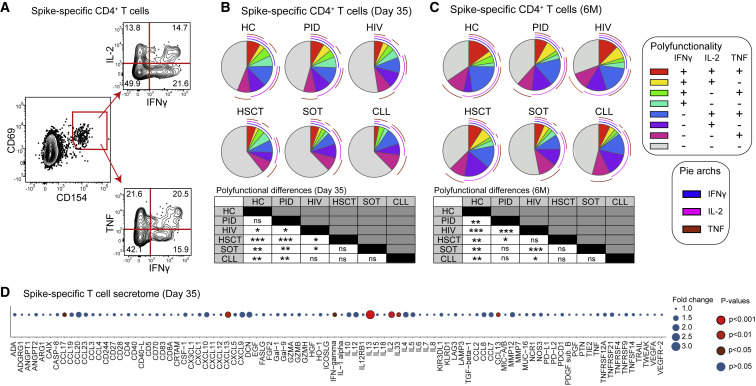
Immunodeficiency syndromes differentially impact the functional quality of SARS-CoV-2-specific T cells post-mRNA vaccination
To assess the functional features of spike-specific (CD69+CD154+) CD4+ T cells, we combined the AIM assay with a polyfunctional assessment of IFN-γ, TNF, and IL-2 at day 35 and 6 months post-vaccination (Figure 2 A). Few spike-specific CD4+ T cells expressed all cytokines together, and single or dual cytokine expression patterns differed between cohorts (Figures 2B and 2C). More specifically, the HC and PID groups experienced the highest spike-specific CD4+ T cell polyfunctionality, where IFN-γ was the least frequently expressed cytokine, particularly in the HIV, HSCT, SOT, and CLL groups at day 35 (Figures 2B and S3A). Increased polyfunctional characteristics of spike-specific CD4+ T cells were observed at 6 months post-vaccination, where HCs showed a diverse polyfunctional profile compared with all immunodeficient groups (Figures 2C and S3A).
Figure 2.
Functional profile of spike-specific T cells
(A) Representative flow gating strategy (n = 279 independent experiments) for IFN-γ, IL-2, and TNF expression within the spike-specific CD4+ T cell population.
(B) Comparison of the co-expression pattern for IFN-γ, IL-2, and TNF molecules at day 35.
(C) Comparison of the co-expression pattern for IFN-γ, IL-2, and TNF molecules at 6 months.
(D) Bubble plot of the fold change over unstimulated background for secreted proteins after spike-peptide pool stimulation of PBMCs from day 35.
Graphs show median values (B–D). (B and C) Permutation test. (D) Mann-Whitney test between stimulated and unstimulated conditions. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns, not significant. See also Figure S3.
Many spike-specific CD4+ T cells did not express IFN-γ, TNF, or IL-2 across the patient groups. We, therefore, expanded our functional T cell analyses by measuring the secretome of spike-peptide-stimulated PBMCs at days 0 and 35 with the Olink platform. On day 35, there was a marked increase in fold change of secreted molecules compared with the background cross-reactive secretome at day 0 (Figure 2D). This consisted of multiple cytokines and chemokines that distinguish Tfh, Th1, and Th2 polarized cells, including IL-13, IL-2, CXCL13, IL-4, and IFN-γ (Figure 2D). Additionally, CCL13 and CCL17 were significantly elevated compared with DMSO controls after spike stimulations, whereas other chemokines such as CCL20, CXCL19, and CXCL10 showed elevated tendencies (Figure 2D). To investigate the relative magnitude of protein secretion after spike stimulation, we compared the fold-change differences between the various patient groups and HCs. We detected a similar protein signature across all groups (Figure S3B). Collectively, our findings demonstrate that SARS-CoV-2 mRNA vaccination induces a functionally diverse T cell response in HCs and certain immunocompromised patient groups.
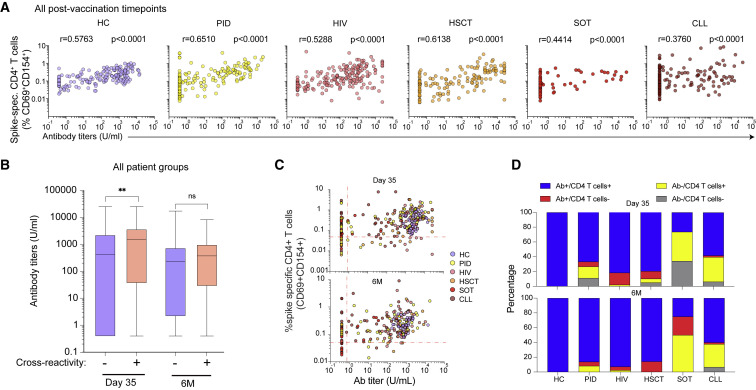
Healthy and immunocompromised individuals show discordant correlations between CD4+ T cell and antibody responses
CD4+ T cell help is essential for developing germinal center reactions and forming high-affinity antibodies (Crotty, 2011). We confirmed that the frequencies of spike-specific CD4+ T cells and antibody titers were positively correlated in all patient groups (Figure 3 A). Notably, the CLL and SOT groups showed reduced antibody responses and displayed a poor correlation coefficient with spike-specific CD4+ T cells compared with the other groups (Figure 3A). Extending to our previous observations about the impact of pre-existing responses, we observed that HCs with pre-existing CD4+ T cells produced higher antibody titers at day 35. This trend was not replicated in the immunocompromised groups and at a later point (6 months) across all cohorts (Figure S3C). Nevertheless, after pooling all groups together, we found that individuals with pre-existing CD4+ T cells subsequently generated higher antibody titers at day 35 (Figure 3B).
Figure 3.
Relationship between spike-specific antibody and CD4+ T cell responses after mRNA vaccination
(A) Correlative analysis of spike-specific antibody and CD4+ T cell responses combined from all post-vaccination time points (days 10, 21, and 35 and 6 months).
(B) Distribution of spike-specific antibody titers based on the presence or absence of pre-existing (day 0) CD4+ T cell responses.
(C) Scatter plot of spike-specific antibody titers and CD4+ T cell response frequencies at day 35. Dashed lines represent the limit of detectable responses (antibody threshold = 0.8 U/mL, CD4+ threshold = 0.05%, after background subtraction).
(D) The proportion of individuals with detectable antibody and/or spike-specific CD4+ T cell responses at day 35 and 6 months.
Graphs show median ± IQR (B). (A) Spearman correlation. (B) Mann-Whitney test. ∗ p < 0.05, ∗∗ p < 0.01, ns = not significant. See also Figure S3.
We further examined discordant cell-mediated and humoral responses by comparing the proportion of individuals with detectable or undetectable spike-specific CD4+ T cell and antibody responses at day 35 and 6 months post-vaccination across all cohorts. Only individuals within the HC group generated detectable spike-specific antibody and CD4+ T cell responses, with variable humoral and cell-mediated immunity in the immunodeficient groups (Figure S3C). The CLL and SOT groups had the largest proportion of antibody-deficient vaccine responses at day 35 and 6 months (Figures 3C and 3D). Additionally, a lower fraction of detectable CD4+ T cell responses when compared with humoral responses were observed in the HIV and HSCT groups at both day 35 and 6 months post-vaccination and in the SOT group at 6 months (Figures 3C and 3D). The PID group experienced heterogeneous humoral and cell-mediated response patterns depending on underlying conditions (Figures 3C and 3D). Notably, a fraction of individuals generated spike-specific CD4+ T cell responses in the absence of detectable antibody responses, as exemplified by the XLA patients within the PID group who intrinsically lack mature B cells and the capacity to produce antibodies at day 35 (Figure S3D; Table S2). These data suggest that despite undetectable antibody responses, spike-specific T cell immunity may be efficiently formed after mRNA vaccination.
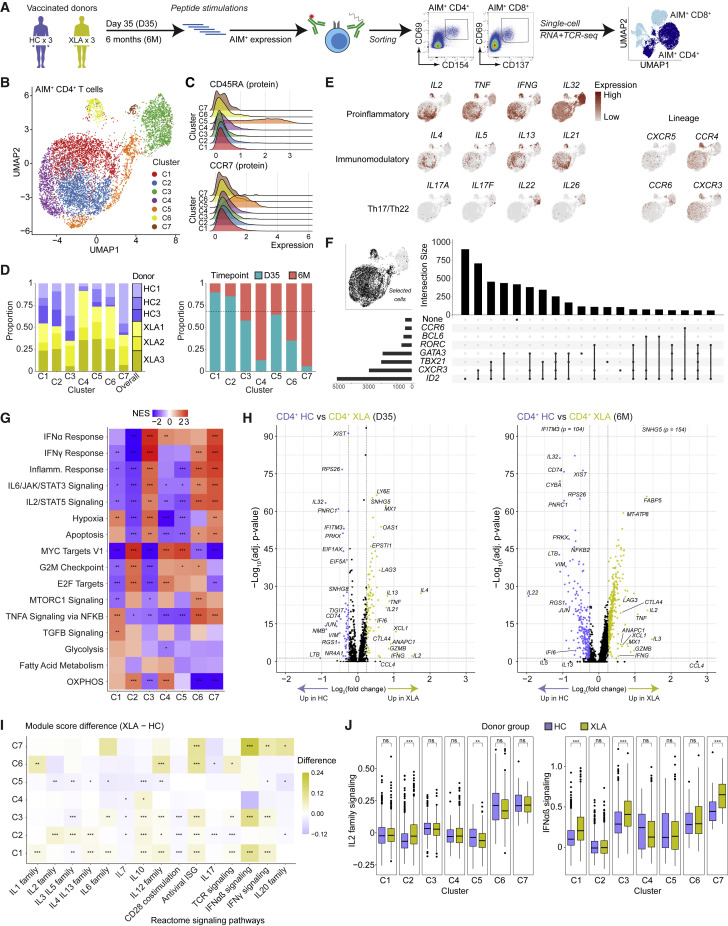
Longitudinal single-cell transcriptomics provides a high-resolution map of spike-specific T cells post-vaccination
Our observation that CD4+ T cell responses could be generated in the absence of mature B cells prompted us to investigate the functional capacity of spike-specific T cell responses in the HC and XLA groups at day 35 (n = 6) and at 6 months (n = 5) post-vaccination. In this light, we used the 10X Genomics platform with 5′ V(D)J chemistry and a panel of oligo-conjugated antibodies (CD4, CD8, CCR7, CD45RA, and CXCR5) (Figure 4 A) to perform single-cell sequencing on sorted spike-specific CD4+ (CD69+CD154+) and CD8+ (CD69+CD137+) T cells (n = 7,401 and n = 3,773 cells after quality control, respectively) (Table S3). Samples from the HC and XLA groups were stained, hashed, sorted (Figure S4A), and then sequenced. Cells with high mitochondrial content, low or high library sizes, co-expression of hashing antibodies, or co-expression of CD4 and CD8 were removed (Figure S4B). Louvain clustering and Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction identified ten subsets consisting primarily of conventional CD4+ and CD8+ T cell phenotypes (Figures S4C and S4D). Localized regions in each cluster corresponded to a cell’s donor or time point of origin (Figure S4E), but cells from multiple donors contributed to each cluster (Figure S4F). Within the CD8+ compartment, we detected CD8-expressing unconventional (including γδ T and NK cell) populations based on gene, T cell receptor sequencing, and protein signatures (Figures S4D, S4G, and S4H). For subsequent analyses in the CD8+ compartment, we filtered based on marker gene expression and TCR identity to remove γδ T, NK, NKT, and MAIT cell subsets (see STAR Methods).
Figure 4.
Single-cell transcriptome landscape of spike-specific CD4+ T cells in HCs and patients with XLA post-mRNA vaccination
(A) Schematic workflow for single-cell sequencing of spike-specific T cells.
(B) UMAP visualization of CD4+ T cell clusters (C1–C7) identified from HC and XLA patients at the day 35 and 6 months.
(C) Ridge plots of CD45RA and CCR7 protein expression across CD4+ T cell clusters.
(D) Bar plots of the composition of each cluster according to donor or time point of origin. The dotted line represents the overall distribution of cells between day 35 and 6 months.
(E) UMAP visualizations colored by gene expression intensity.
(F) UpSet plot of polarizing helper subset marker co-expression.
(G) Heatmap of normalized enrichment scores calculated using GSEA for Hallmark gene sets for each cluster. NES, normalized enrichment score.
(H) Volcano plot of differentially expressed genes between CD4+ T cells from HC and XLA donors at the day 35 and 6-month time points. Genes with very low p values consisting only of sex-chromosome-linked genes were removed for visualization purposes.
(I) Heatmap showing the difference in median module scores between XLA and HC donors across clusters. Curated gene sets were obtained from the Reactome pathway database.
(J) Distribution of module scores for signaling pathways from the Reactome pathway database.
Graphs show median ± IQR (J). (G) GSEA permutation test. (I and J) Mann-Whitney test. (G, H, I, and J) ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ns, not significant.
See also Figures S4 and S5.
mRNA vaccination generates a spectrum of functional CD4+ T cell subsets in both HC and XLA donors
To evaluate the molecular phenotypes of spike-specific CD4+ T cells, we selected only CD4+ T cells by oligo-conjugated antibody staining. Seven clusters were identified (C1–C7) (Figure 4B), with distinct cytokine expression and memory phenotypes. Most clusters and donors were of a central- or effector-memory phenotype (TCM/EM: CD45RA−CCR7+/−) (Figures 4C, S5A, and S5B), except for a TSCM cluster (C5: CD45RA+CCR7+) (Figure 4C; Table S4), which also expressed markers of memory cells (FAS and IL2RB) at frequencies similar to other clusters (Figure S5C). The 6-month time point populations from XLA donors were concentrated in cluster C4 (Figure 4D), with the remaining 6-month cells distributed throughout the other clusters (Figure S5D). Notably, the most highly enriched genes in C3 were LTB, S100A4, and IL32 (Table S4) and were distinguished by a unique Th17 or Th22 cell cytokine profile, including IL22 and IL26 (Figure 4E). The remaining clusters were highly polyfunctional in their cytokine profile and expressed proinflammatory cytokines (IL2, IFNG, and TNF) but also detectable transcripts of immunomodulatory and Tfh- and Th2 cell-lineage cytokines (IL4, IL5, IL13, and IL21) in a continuous spectrum (Figure 4E). Inspection of lineage marker expression showed evidence for Th1- and Th2 cell- lineages (CXCR3 and CCR4) (Figure 4E), whereas intracellular marker expression confirmed a stronger polarization of the Th1 cell lineage based on ID2 expression (Figure 4F). Notably, Th1 polarized cells also co-expressed non-Th1 cell-lineage markers, including GATA3 and RORC (Figure 4F), thus forming a spectrum of transcriptional states. Next, using the marker genes for each cluster, we applied gene set enrichment analysis (GSEA), which identified distinct pathway enrichment for IFN and inflammatory responses (in clusters C3, C6, and C7), which was contrasted by enrichment for cell cycling signatures (Myc targets and G2M checkpoint) (in clusters C2, C4, and C5), and TNFA and TGFB signaling in cluster C1 (Figure 4G).
Next, we directly investigated how XLA-immunodeficiency may impact CD4+ T cell responses following mRNA vaccination compared with HCs. Patients with XLA demonstrated signatures enriched for functional Th1 or Th2 cell responses (IL2, IFNG, and IL4) (Figure 4H). Notably, these signatures were evident at both the day 35 and 6 month time points. Additionally, module scoring showed higher expression for Reactome pathways associated with IL signaling by patients with XLA (Figure 4I). These differences were particularly pronounced for IL-2 family signaling and IFNαβ signaling for XLA donors in the largest clusters of C1 and C3, respectively (Figure 4J). Collectively, these data indicate that patients with XLA generated robust spike-specific CD4+ T cells with similar to greater functionality than HCs.
XLA patients experience polyfunctional effector CD8+ T cell responses post-vaccination
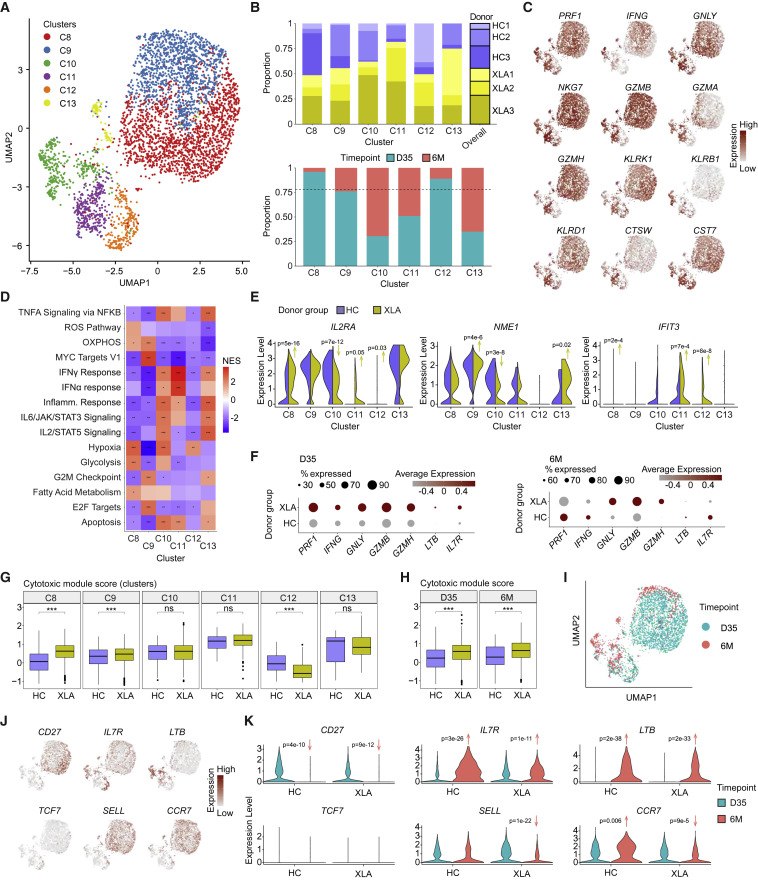
We further investigated conventional spike-specific CD8+ T cell responses. Six clusters (C8–C13) were identified using Louvain clustering visualized by UMAP dimensionality reduction (Figure 5 A; Table S5), using cells from XLA and HC donors at the day 35 and 6 month time points (Figure 5B). Most of each patient's response was dominated by the TEM and effector-memory CD45RA+ (TEMRA) phenotypes with increasing proportions of the latter at the 6-month time point, particularly among patients with XLA (Figures S5E and S5F). To assess the functional capacity of the CD8+ T cell response, we analyzed the transcriptional expression of a set of 12 effector-associated molecules (Zhang et al., 2020), which showed highly polyfunctional responses across all clusters (Figure 5C). Using GSEA (Hallmark pathways), the CD8+ T cell clusters with the greatest functional responses were clusters C10, C11, and C13, enriched for IFN, inflammatory, and IL signaling, respectively (Figure 5D).
Figure 5.
Single-cell analysis reveals the functional spectrum of spike-specific CD8+ T cell responses in HCs and XLA patients
(A) UMAP visualization of CD8+ T cell clusters (C8–C13) identified from HC and XLA donors at day 35 and 6 months. Day 35.
(B) Bar plots of the composition of each cluster according to donor or time point of origin. The dotted line represents the overall distribution of cells between day 35 and 6 months.
(C) UMAP visualizations for expression of a curated cytotoxic gene signature set colored by gene expression intensity.
(D) Heatmap of normalized enrichment scores calculated using GSEA for Hallmark gene sets for each cluster. NES, normalized enrichment score.
(E) Violin plots of gene expression for markers of activation.
(F) Dot plot of gene expression between all CD8+ T cells from HC and XLA donors at day 35 and 6 months.
(G) Distribution of module scores for a cytotoxic gene signature across clusters.
(H) Distribution of module scores for a cytotoxic gene signature across time points.
(I) UMAP visualization for the time point of origin of cells.
(J) UMAP visualizations for expression of memory gene signatures colored by gene expression intensity.
(K) Violin plots of memory gene signatures split by time point and donor group of origin.
Graphs show median ± IQR (G and H). (D) GSEA permutation test. (E, G, H and K) Mann-Whitney test. (D, G, and H) ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001, ns, not significant. See also Figure S5.
We next compared CD8+ T cell responses between HCs and patients with XLA by investigating their phenotypic composition, effector responses, and activation status. Differential gene expression between the two patient groups confirmed that patients with XLA generated highly responsive CD8+ T cells with comparable or higher expression for early activation molecules (IL2RA, NME1, and IFIT3) (Figure 5E; Szabo et al., 2019) and increased expression of effector transcripts (including PRF1, IFNG, GNLY, GZMB, and GZMH) (Figure 5F). We next scored the cytotoxic potential of patients with XLA and HCs by calculating the module scores for a set of cytotoxic genes (Figure 5C), which showed comparable or significantly higher cytotoxicity from XLA-patient-derived cells in most clusters (Figure 5G), and over time (Figure 5H).
We furthermore compared the transcriptome profile of spike-specific CD8+ T cells between day 35 and 6 months. CD8+ T cells at the two different time points were localized to distinct regions of the UMAP visualization (Figure 5I) and characterized by significantly higher transcript expression of the memory markers IL7R and LTB at 6 months (Figures 5J and 5K). XLA donors showed decreased SELL and CCR7 transcript expression at 6 months (Figure 5K). Overall, our results demonstrate that mRNA vaccination induces polyfunctional and persistent spike-specific CD8+ T cell responses in both HCs and patients with XLA, where the latter show robust cytotoxic capacity following anamnestic responses.
Clonality provides evidence for diverse recruitment and expansion of spike-specific T cells following vaccination
Our sequencing approach allowed us to characterize the clonal composition of total spike-specific CD4+ and CD8+ T cells between XLA and HC donors over time (Table S6). In the CD4+ T cell compartment, Shannon’s entropy diversity was not significantly different between the day 35 and 6 month time points and did not segregate by donor group (Figure S6A). Compositionally, the CD4+ T cell repertoire was polyclonal, with few clones observed more than once (Figures S6B and S6C). The repertoire diversity in the CD8+ T cell compartment was also highly similar over time (Figure S6D). However, the CD8+ T cell repertoire was composed of multiple expanded clones, where the largest twenty clones typically accounted for half of the total population (Figure S6E). Furthermore, most clones were observed at least twice, with a single clone detected up to 107 times (donor HC3 at day 35) (Figure S6F). Collectively, mRNA vaccination mounts polyclonal repertoires of CD4+ T cells and robust expansions of oligoclonal CD8+ T repertoires in both XLA and HC donors.
Persisting CD8+ T cell clones display a TEMRA phenotype with stem-like traits
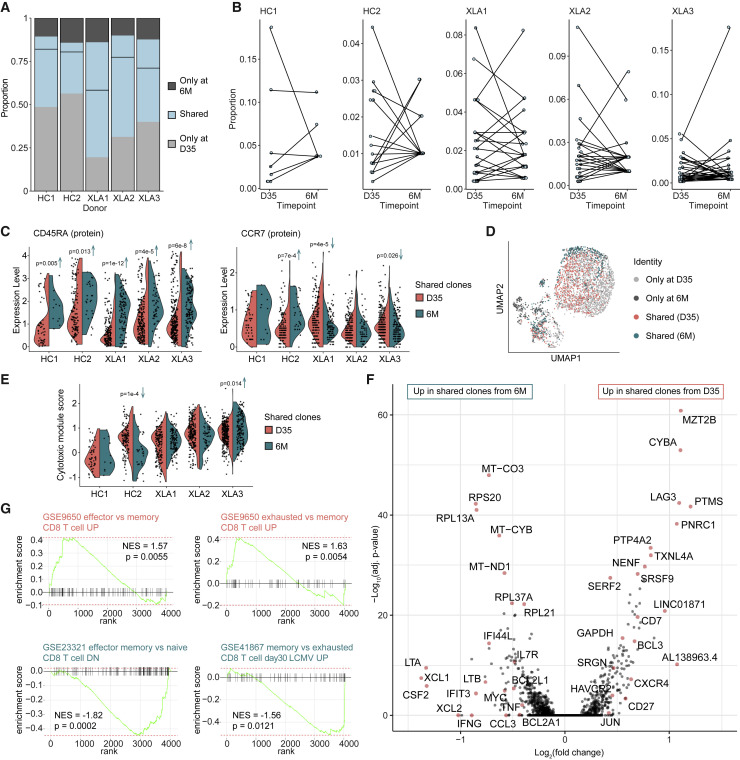
The presence of expanded CD8+ T cell clones in both donor groups presented an opportunity to characterize the subset of identical clones identified at both day 35 and 6 months (Figure 6 A). The largest clone at day 35 was no longer the predominant clone at 6 months (Figure 6B), suggesting that early clonal expansion does not necessarily predict the persistent spike-specific clonal landscape. Next, we compared protein expression of memory markers of shared clones at the day 35 and 6 month time points. We found increased CD45RA but unchanged CCR7 expression at the 6-month time point, indicating a transition into the TEMRA phenotype (Figure 6C). Cells with shared clonotypes from both time points collectively shared phenotypic characteristics such as a tendency for co-localization in the same UMAP clusters, particularly cluster C3 (Figure 6D) and equivalent cytotoxic scores in four of the five donors (Figure 6E). Despite this fact, we observed an upregulation of specific functional (IFNG, TNF, XCL1, XCL2, LTB, and LTA) and memory transcripts (IL7R, MYC, and BCL2L1) at 6 months (Figure 6F), indicative of a polarized stem-like cytotoxic state. GSEA confirmed enriched effector and exhausted (activated) transcriptional signatures at day 35 compared with 6 months after vaccination. In contrast, the shared clones from the 6-month time point showed increased features of naive-like and memory traits (Figure 6G), despite the TEMRA phenotype. In summary, our analyses demonstrate that both HC and XLA donors successfully generate memory CD8+ T cell clonotypes that persist, to a high degree, as TEMRA cells with combined cytotoxic and stem-like characteristics.
Figure 6.
Phenotypic evolution of shared CD8+ T cell clones at 6 months post-vaccination
(A) Stacked bar plot of the proportion of the CD8+ T cell repertoire with paired clonotype (CDR3) identity from each donor observed at only one or both time points. The dividing line represents the contribution of clonotypes from each time point for each donor.
(B) Dot plot of the frequency of only cells with clonotype identity observed in both time points. Identical clonotypes are connected.
(C) Violin plots of the protein expression of CD45RA and CCR7 in cells with clonotype identity observed in both time points.
(D) UMAP visualization of the distribution of cells with clonotype identity observed in only two or both time points.
(E) Violin plot of the cytotoxic module score in cells with clonotype identity observed in both time points.
(F) Volcano plot of differentially expressed cells' genes from day 35 and 6 months with shared clonotypes.
(G) GSEA of C7 immunologic signature gene sets using the ranked set of differentially expressed genes between shared clonotypes from day 35 and 6 months.
(C, E, and F) Mann-Whitney test. (G) GSEA permutation test. See also Figure S6.
Discussion
Coordinated humoral and cell-mediated immunity is critical for protection from viral disease. In contrast to antibody responses, however, the T cell arm remains relatively understudied after COVID-19 mRNA vaccination—particularly in patients with immunodeficiencies. Given that many of these individuals mount suboptimal B cell responses (Apostolidis et al., 2021; Bange et al., 2021), it remains essential to understand if these individuals can still generate effective cell-mediated immunity. These studies in immunocompromised states might be important and set the stage for future work identifying immune correlates of severe COVID-19 in breakthrough infections. In the present study, we addressed the impact of a broad range of immunodeficiencies on the longitudinal induction of T cell responses post-mRNA vaccination. We found that CLL and SOT patients notably elicited poor non-coordinated humoral and cell-mediated immune responses following two vaccination doses. In contrast, several immunocompromised patients, including patients with an inherited lack of mature B cells, mounted spike-specific T cells in the absence of antibody responses. Single-cell transcriptome analysis confirmed that individuals lacking B cells elicited robust clonotypic expansion of polyfunctional spike-specific CD4+ and CD8+ T cells following mRNA vaccination. These data provide a single-cell framework of anamnestic T cell responses following mRNA vaccination in a broad group of immunocompromised patients and indicate that individuals can mount highly functional T cell responses in the genetic absence of mature B cells.
Immunodeficient patients are disproportionally affected by severe COVID-19 (Fung and Babik, 2021) as well as relapsing and chronic SARS-CoV-2 infection (Brown et al., 2022). As such, booster vaccinations have been prioritized for specific immunocompromised patient groups, where ongoing boosters might be needed in the future (e.g., for transplanted patients and those with certain hematologic malignancies). The findings from our study support this strategy, given that these patients, as well as many with CLL, show poor antibody and T cell responses following two-dose mRNA vaccine regimens. Our observation that T cell responses in specific immunocompromised cohorts mirror the dynamics of healthy individuals over at least 6 months is promising given that other studies have shown waning protection (Naaber et al., 2021; Tartof et al., 2021), which may be rescued by booster vaccinations in healthy adults and result in lower rates of infection (Bar-On et al., 2021; Barda et al., 2021). Although SOT and CLL patients generally had suboptimal adaptive immunity compared to the other groups, we found that a fraction of these patients had detectable T cell responses in the absence of antibodies. As such, it will be of interest to further understand if detectable SARS-CoV-2-specific T cell responses, as described here, after two doses will be beneficial and serve as an immune correlate of improved antibody response in future booster studies of transplanted and other immunocompromised patient groups.
Previous studies have identified pre-existing T cells to SARS-CoV-2 in unexposed individuals (Braun et al., 2020; Grifoni et al., 2020; Niessl et al., 2021; Schulien et al., 2021). Recently, studies have also observed a correlation between pre-existing CD4+ T cells and enhanced immune responses following SARS-CoV-2 infection and mRNA vaccination (Loyal et al., 2021; Mateus et al., 2020). We confirm these findings in HCs and demonstrate that pre-existing CD4+ T cells to the spike antigen are associated with increased cell-mediated and humoral immunity across multiple immunodeficient states. We found that patients following SOT had the lowest frequency of pre-existing spike-reactive CD4+ T cells compared with all other groups, which partly could explain the lower frequency of T cell responders in this group compared with the different cohorts. Whether the presence of pre-existing CD4+ T cells is only an association or an actual link to increased B cell activation remains to be determined. Nevertheless, given that high numbers of virus-specific CD4+ T cells and cognate Th functions promote germinal center reactions and increased B cell proliferation, it seems plausible that cross-reactive responses might enhance antibody responses. Further studies are needed to mechanistically determine if cross-reactive CD4+ T cells can provide helper functions to improve antibody responses after mRNA vaccination.
In contrast to healthy individuals, we found that all immunocompromised patient groups had a certain degree of undetectable T cell or antibody responses. All patients with XLA belonged to this category with detectable T cell responses, except for one, despite the lack of antibody responses following mRNA vaccination. As such, these patients serve as a natural model to mechanistically dissect the human vaccine-induced T cell response in the absence of functional B cells. Our single-cell transcriptome data demonstrated that patients with XLA successfully generated CD4+ and CD8+ T cell responses of increased cytotoxic potential compared with healthy individuals. Furthermore, CD8+ T cell clonotypes were characterized by a TEMRA phenotype with memory characteristics at 6 months, suggesting the potential for long-term maintenance. These results were unexpected but in line with recent reports in MS patients treated with anti-CD20 (rituximab) (Apostolidis et al., 2021), highlighting future possibilities to generate neoantigen-based mRNA vaccination strategies in patients with B cell deficiencies. Previous case reports of patients with XLA have suggested that these patients can develop severe COVID-19. Nevertheless, they can resolve infections (Soresina et al., 2020), potentially through augmented T cell responses. The potential mechanism for increased cytotoxic CD8+ T cell signatures in B cell deficiencies remains unknown. Still, it could be a consequence of a lack of antibody-mediated antigen clearance or diminished creation of immune complexes on dendritic cells (Li et al., 2014). Future studies of patients with XLA and other B cell deficiencies will be excellent models to fully understand how well vaccine-induced T cell responses can mediate protection from severe COVID-19.
The T cell receptor repertoire is relatively fixed following vaccination; therefore, future protection against emerging variants depends on recruiting sufficiently broad repertoires. Our study used a peptide pool covering the entire spike antigen, allowing us to identify the total spike-specific CD4+ and CD8+ T cell repertoire using combined single-cell transcriptome and clonotype analysis. Our results demonstrate that mRNA vaccination mounts a polyclonal repertoire of spike-specific CD4+ T cells and a robust expansion of a relatively narrow cytolytic CD8+ T cell clonal repertoire. These data may reflect a greater tendency for peptide stimulations to favor the detection of spike-specific CD4+ T cells, as previous studies using multimer-based assays have shown high frequencies of SARS-CoV-2-specific CD8+ T cells after infection (Saini et al., 2021). Nevertheless, through our combined approach with flow cytometry and single-cell sequencing, we could map the total spike-specific CD4+ and CD8+ T cell response—beyond traditional Th subsets or memory populations using conventional flow cytometry. Our single-cell data demonstrate that mRNA vaccination mounts a robust CD4+ T cell response, segregated into continuous functional spectrums instead of discrete T helper subsets. For instance, the Th1 cell-biased response, as previously reported (Sahin et al., 2020), is also enriched for Th2 and Tfh cell signatures, whereas Th17 or Th22 cell-associated molecules are present in a unique cluster expressing IL32, LTB, and other memory markers. These results align with recent single-cell reports on antigen-specific CD4+ T cells, showing a polarized continuity of transcriptional gradients rather than fixed Th archetypes (Kiner et al., 2021). The spike-specific CD8+ T cell response also exhibited a profile of continuums with overlapping functions in different clusters that were highly heterogeneous between individuals and split between resting and highly cytotoxic responses. Our analysis of T cell responses at 6 months revealed a high degree of clonotypic maintenance in the CD8+ T cell populations, as has been reported after infection (Adamo et al., 2022), and the development of a TEMRA CD8+ T cell phenotype characterized by increased expression of memory transcripts, including IL7R and LTB.
In summary, we here provide a profile of the total spike-specific CD4+ and CD8+ T cell responses following mRNA vaccination in healthy individuals and patients with different immunocompromising disorders. We show that mRNA vaccines induce a broad spectrum of highly active antiviral T cell clonotypes that aid in explaining how these vaccines generate protective responses against severe COVID-19 in healthy individuals. Our data also provide evidence of a highly heterogeneous mRNA vaccine response in immunocompromised patient groups, where patients with an inherited lack of mature B cells can still induce highly functional T cell responses in the absence of antibodies. Nevertheless, many immunodeficient patients show persistently reduced cell-mediated immunity and may need future booster doses and prophylactic anti-SARS-CoV-2 monoclonal antibodies.
Limitations of the study
Our study contains several limitations. The study was designed to follow individuals over time to understand how various primary and secondary immunodeficiencies may differently impact immune responses to mRNA vaccination. As such, our cohort is highly heterogeneous, with differences in sex and age that may affect our results. Furthermore, this is a human study, and we cannot mechanistically study if T cell responses in the absence of antibodies can protect patients with B cell deficiencies from severe COVID-19. Future studies using T cell-based vaccines might be able to answer those questions. Finally, we have not assessed cross-reactive SARS-CoV-2-specific cell-mediated immune responses to Omicron. However, previous studies in immunocompetent individuals have found that established SARS-CoV-2-specific T cell responses can efficiently cross-recognize the Omicron variant (Gao et al., 2022; Keeton et al., 2022; Liu et al., 2022; Tarke et al., 2022), suggesting that similar results might be seen in immunocompromised patients.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-human CCR6 BUV737 (clone 11A9) | BD Biosciences | Cat# 612780; RRID:AB_2870109 |
| Anti-human CCR7 APC-Cy7 (clone G043H7) | BioLegend | Cat# 353212; RRID:AB_10916390 |
| Anti-human CD3 BUV805 (clone UCHT1) | BD Biosciences | Cat# 612895; RRID:AB_2870183 |
| Anti-human CD4 FITC (clone SK3) | BD Biosciences | Cat# 566911; RRID:AB_2739682 |
| Anti-human CD4 BUV496 (clone SK3) | BD Biosciences | Cat# 612936; RRID:AB_2870220 |
| Anti-human CD8 BV711 clone (RPA-T8) | BioLegend | Cat# 301044; RRID:AB_2562906 |
| Anti-human CD8 BUV395 (clone RPA-T8) | BD Biosciences | Cat# 563795; RRID:AB_2722501 |
| Anti-human CD14 BV510 (clone M5E2) | BioLegend | Cat# 301842; RRID:AB_2561946 |
| Anti-human CD19 BV510 (clone HIB19) | BioLegend | Cat# 302242; RRID:AB_2561668 |
| Anti-human CD45RA BV570 (clone HI100) | BioLegend | Cat# 304132; RRID:AB_2563813 |
| Anti-human CD69 BV650 (clone FN50) | BioLegend | Cat# 310934; RRID:AB_2563158 |
| Anti-human CD69 BUV563 (clone FN50) | BD Biosciences | Cat# 748764; RRID:AB_2873167 |
| Anti-human CD107A BV785 (clone H4A3) | BioLegend | Cat# 328644; RRID:AB_2565968 |
| Anti-human CD137 (4-1BB) PE-Cy7 (clone 4B4-1) | BioLegend | Cat# 309818; RRID:AB_2207741 |
| Anti-human CD154 (CD40L) BV421 (clone 24-31) | BioLegend | Cat# 310824; RRID:AB_2562721 |
| Anti-human CD194 (CCR4) BV605 (clone 1G1) | BD Biosciences | Cat# 562906; RRID:AB_2737882 |
| Anti-human CXCR3 BV750 (clone 1C6) | BD Biosciences | Cat# 746895; RRID:AB_2871692 |
| Anti-human CXCR5 BB515 (clone RF8B2) | BD Biosciences | Cat# 564624; RRID:AB_2738871 |
| Anti-human IFN-ɣ PE (clone B27) | BD Biosciences | Cat# 506507; RRID:AB_315440 |
| Anti-human IL-2 PE-Dazzle594 (clone MQ1-17H12) | BioLegend | Cat# 500344; RRID:AB_2564091 |
| Anti-human Ki67 AF647 (clone B56) | BD Biosciences | Cat# 558615; RRID:AB_647130 |
| Anti-human CD38 APC-R700 (clone HIT2) | BD Biosciences | Cat# 564979; RRID:AB_2744373 |
| Anti-human granzyme B BB790 (clone GB11) | BD Biosciences | Custom conjugate |
| Anti-human PD-1 BV711 (clone EH12.2H7) | BioLegend | Cat# 329928; RRID:AB_2562911 |
| Anti-human CD40 primary antibody (clone HB14) | Miltenyi Biotec | Cat# 130-108-041; RRID:AB_2660897 |
| Anti-human TNF-ɑ BV650 (clone Mab11) | BD Biosciences | Cat# 502936; RRID:AB_2563884 |
| TotalSeqC Anti-human CCR7 (clone G043H7) | BioLegend | Cat# 353251; RRID:AB_2800943 |
| TotalSeqC Anti-human CXCR5 (clone J252D4) | BioLegend | Cat# 356939; RRID:AB_2800968 |
| TotalSeqC Anti-human CD4 (clone RPA-T4) | BioLegend | Cat# 300567; RRID:AB_2800725 |
| TotalSeqC Anti-human CD8 (clone SK1) | BioLegend | Cat# 344753; RRID:AB_2800922 |
| TotalSeqC Anti-human CD45RA (clone HI100) | BioLegend | Cat# 304163; RRID:AB_2800764 |
| TotalSeq-C0251 anti-human Hashtag 1 | BioLegend | Cat# 394661; RRID:AB_2801031 |
| TotalSeq-C0252 anti-human Hashtag 2 | BioLegend | Cat# 394663; RRID:AB_2801032 |
| TotalSeq-C0253 anti-human Hashtag 3 | BioLegend | Cat# 394665; RRID:AB_2801033 |
| TotalSeq-C0254 anti-human Hashtag 4 | BioLegend | Cat# 394667; RRID:AB_2801034 |
| Co-Stimulatory Antibodies (CD28/CD49d) | BD Biosciences | Cat# 347690; RRID:AB_647457 |
| Biological samples | ||
| Cryopreserved peripheral blood mononuclear cells | Bergman et al., 2021 | EudraCT no. 2021-000175-37 |
| Chemicals, peptides, and recombinant proteins | ||
| BD Golgi Stop (with Monensin) | BD Biosciences | Cat# 554724; RRID:AB_2869012 |
| Brefeldin A | BioLegend | Cat# 420601 |
| Brilliant Stain Buffer Plus | BD Biosciences | Cat# 566385; RRID:AB_2869761 |
| DNAse | Sigma-Aldrich | Cat# 4716728001 |
| FoxP3/Transcription Factor Buffer Set | ThermoFisher Scientific | Cat# 00-5523-00 |
| LIVE/DEAD Fixable Aqua Dead Cell stain Kit | ThermoFisher Scientific | Cat# L34957 |
| Paraformaldehyde | Biotium | Cat# 22023 |
| RPMI-1640 without L-Glutamine | Cytiva | Cat# SH30096.01 |
| SARS-CoV-2 Spike Peptide pools | Peptides & Elephants | UniProt: P0DTC2 |
| Fetal bovine serum | Sigma-Aldrich | Cat# F7524 |
| Penicillin-streptomycin | Cytiva | Cat# SV30010 |
| L-glutamine | Sigma-Aldrich | Cat# 59202C |
| Critical commercial assays | ||
| ELISpot Pro: Human IFN-γ kit | Mabtech | Cat# 3420-2APT-2 |
| Elecsys Anti- SARS-CoV-2 S | Roche diagnostics | Cat# 09 289 267 190 |
| Immuno-Oncology Olink panel | Olink | RRID:SCR_003899 |
| Chromium Next GEM Single Cell 5’ Library & Gel Bead Kit v1.1 | 10X Genomics | Cat# PN-1000165 |
| Chromium Next GEM Chip G Single Cell Kit | 10X Genomics | Cat# PN-1000120 |
| Single Index Kit T Set A | 10X Genomics | Cat# PN-1000213 |
| Single Index Kit N Set A | 10X Genomics | Cat# PN-1000212 |
| Chromium Single Cell V(D)J Enrichment Kit, Human T Cell | 10X Genomics | Cat# PN-1000005 |
| Deposited data | ||
| Processed single cell RNA-seq data | This paper | ArrayExpress: E-MTAB-11845 |
| Software and algorithms | ||
| FlowJo Version 10 | BD Biosciences | RRID: SCR_008520 |
| R (programming language) | RRID: SCR_001905 | |
| SPICE | Roederer et al., 2011 | RRID: SCR_016603 |
| Seurat R package | Hao et al., 2021 | RRID:SCR_016341 |
| ggplot2 R package | RRID:SCR_014601 | |
| Cell Ranger | 10X Genomics | RRID:SCR_017344 |
| Other | ||
| Molecular Signatures Database (mSigDB) v7.5.1 | Liberzon et al., 2011, Liberzon et al., 2015 | RRID:SCR_016863 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Marcus Buggert (marcus.buggert@ki.se).
Materials availability
This study did not generate any new unique reagents.
Experimental model and subject details
Study design
The objective of this study was to characterize the SARS-CoV-2-specific T cells response following mRNA COVID-19 vaccination in healthy and immunocompromised individuals.
Human subjects and ethics
In this longitudinal study, healthy controls (n=44) and patients with primary immunodeficiency (PID, n=48), human immunodeficiency virus infection (HIV, n=50), hematopoietic stem cell transplantation (HSCT, n=43), solid organ transplantation (SOT, n=41), chronic lymphocytic leukemia (CLL, n=53) were recruited between March and June of 2021. Detailed patient characteristics have been described elsewhere for the Covaxid study (Bergman et al., 2021). Plasma and PBMCs were collected immediately before the first vaccine dose (Day 0), ten days after the first vaccine dose (Day 10), immediately before the second vaccine dose (Day 21), fourteen days after the second vaccine dose (Day 35) and six months after the first vaccine dose (6 months). The trial was registered at EudraCT (no. 2021-000175-37) and clinicaltrials.gov (no. 2021-000175-37). The study was approved by the Swedish Medical Product Agency (ID 5.1-2021-5881) and the Swedish Ethical Review Authority (ID 2021-00451). PBMCs were isolated via standard density gradient centrifugation and cryopreserved in fetal bovine serum (FBS) containing 10% dimethyl sulfoxide (DMSO). Plasma samples were collected from all individuals and stored at −80°C.
Peptides
Peptide pool stimulations used 316 overlapping peptides (15-mers with 11 aa overlap) (Peptides&Elephants) from the entire SARS-CoV-2 spike glycoprotein (UniProt: P0DTC2). The spike peptide pool was reconstituted in DMSO (10–20 mg/ml), diluted to 100 μg/ml in PBS, aliquoted, and stored at −20°C. Peptides for the VOC analysis spanned the entire SARS-CoV-2 spike protein and corresponded to the ancestral Wuhan (wild type) sequence or the B.1.617.2 (Delta) variant. Peptides were 15-mers overlapping by 10 amino acids and were synthesized as crude material (TC Peptide Lab, San Diego, CA), reconstituted in DMSO, and diluted in PBS as described above.
Method details
IFN-γ ELISpot assay
Cryopreserved PBMCs were resuspended in complete medium (RPMI 1640 containing 10% FBS, 1% penicillin-streptomycin, and 1% L-glutamine) and rested for 3-4 hours prior to assay. All experiments were performed in duplicates with 2.5 x 105 PBMCs per well supplemented with purified anti-CD28 and CD49d (347690, BD Biosciences) at 0.15 μg/mL and stimulated with either spike glycoprotein peptide pool (0.5 μg/ml) or negative control containing PBS with DMSO (<1%). The assay was performed using pre-coated plates and reagents from a human IFN-γ ELISpotPRO kit (3420-2APT-2, Mabtech). PBMCs were stimulated for 20 hours in assay plates incubated at 37°C and 5% CO2. Spots were counted using the automated IRIS ELISpot reader (Mabtech) and Apex software (Mabtech) using default settings. Visible artifacts were removed by masking. Results were expressed as spot forming units (SFUs) per million PBMCs, calculated by subtracting the mean negative control wells from the mean of spike peptide pool stimulation wells. A positive response after subtraction was determined by the median of all negative control wells + 2 standard deviations (>37 SFU/106 PBMCs). Results were excluded from analysis if the mean of duplicate negative control wells were >100 SFUs/106 PBMCs.
Serum protein quantification using proximity extension assay
Culture supernatants from Spike and negative control stimulations from the ELISpot assays were harvested immediately after the 20-hour stimulation (above) and stored at -80°C until sent for analysis. Ninety-two proteins from the Immuno-Oncology Olink panel were assessed using proximity extension assay technology (Olink AB) on 20 individuals from each group at Day 0 and a subset of each cohort group (13 PID, 9 HIV, 8 HSCT, 13 SOT, 6 CLL, 11 HC) at Day 35. One sample from Day 0 did not pass quality control and was excluded from the analysis.
Activation-induced marker (AIM) assay
Cryopreserved PBMCs were thawed quickly, resuspended in complete medium in the presence of DNase I (10 U/ml; Sigma-Aldrich), and rested at 1×106 cells/well in 96-well U-bottom plates (Corning) for 3 hours at 37°C. The medium was then supplemented with anti-CXCR5–BB515 (clone RF8B2; BD Biosciences), followed 15 min later by the spike peptide pool (0.5 μg/ml), and a further 1 hour later by brefeldin A (1 μg/ml; Sigma-Aldrich) and monensin (0.7 μg/ml; BD Biosciences). Negative control wells contained equivalent DMSO. After 9 hours, cells were washed in PBS supplemented with 2% FBS and 2 mM EDTA (FACS buffer) and stained with other chemokine receptors for 10 min at 37°C. Additional surface stains were performed for 30 min at room temperature in the presence of Brilliant Stain Buffer Plus (BD Biosciences). Viable cells were stained by exclusion using a LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Thermo Fisher Scientific). Cells were then washed in FACS buffer and fixed/permeabilized using a FoxP3 Transcription Factor Staining Buffer Set (Thermo Fisher Scientific). Intracellular stains were performed for 30 min at room temperature. Stained cells were washed in FACS buffer, fixed in PBS containing 1% paraformaldehyde (PFA; Biotium), and acquired using a FACSymphony A5 (BD Biosciences). Flow cytometry reagents are listed in STAR Methods key resources table. To address T cell responses against overlapping spike peptides from the WT and Delta variant, we conducted similar AIM measurements for surface molecules (CD69, CD154, and 41-BB) described below in the cell sorting section.
Serology assay
Serum samples were analyzed using the quantitative Elecsys® Anti-SARS-CoV-2 S assay (Roche Diagnostics) on the Cobas 8000 e801pro to detect antibodies to the SARS-CoV-2 spike protein receptor-binding domain (RBD). The detectable range is between 0.40 and 250 U/mL, with a cut-off for positive results at ≥ 0.80 U/ml. Positive samples with antibody titers of >250 U/mL were re-tested following a 1/10 dilution and, in applicable cases, a 1/100 dilution which increased the upper detection level to 25,000 U/ml (Bergman et al., 2021).
Cell sorting
Cryopreserved PBMCs were thawed quickly, resuspended in complete medium in the presence of DNase I (10 U/ml; Sigma-Aldrich), and rested at 3-5× 106 cells/well in 24-well plates (Corning) for 3 hours at 37°C. The medium was then supplemented with CD40 block (clone HB14; Miltenyi Biotec), followed 15 minutes later by the spike peptide pool (0.5 μg/ml). Negative control wells contained equivalent DMSO. After 12 hours, cells were washed in PBS, followed by viable staining with LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Thermo Fisher Scientific) for 10 minutes at room temperature. Cells were washed in PBS supplemented with 2% FBS, 2 mM EDTA (FACS buffer), and stained with two TotalSeq-C antibodies (anti-CCR7 clone G043H7 and anti-CXCR5 clone J252D4) (BioLegend) for 10 min at 37°C. Immediately after without washing, immunofluorescent-antibody staining (CD4, CD8, CD40L, CD69, CD14, CD19, 4-1BB), anti-human Hashtag antibody staining (clone LNH-94/2M2, BioLegend), and remaining TotalSeq-C antibody stainings (anti-CD45RA clone HI100, anti-CD4 clone RPA-T4, anti-CD8 clone SK1) (BioLegend) were performed for 30 minutes at room temperature in the presence of Brilliant Stain Buffer Plus (BD Biosciences). Stained cells were then washed, resuspended in FACS buffer, and sorted using an MA900 Multi-Application Cell Sorter (Sony Biotechnology) (Figure S4A). Cells from HC and XLA individuals within the same batch were pooled together, generating three individual sample sets in total.
Single-cell RNA-sequencing
Sorted cells were immediately loaded onto a Chromium Single Cell Chip (10x Genomics) with a target capture rate of ∼5000 single cells. Captured mRNA and molecules from antibody-derived tags (ADTs) and hashtag oligos (HTOs) were converted to cDNA and pooled for Illumina sequencing according to the manufacturer’s instructions for Chromium Next GEM Single Cell V(D)J 5’ v1.1 library preparation. Two library pools were prepared, one containing all the gene expression products and a second library containing enriched TCR products and TotalSeq/hashtag products. Each library pool was sequenced on separate lanes of a single NovaSeq6000 SP100 flow cell with an 8-base index read and a 26-base read 1 containing barcodes and unique molecular identifiers (UMIs) and a 98-base read 2 containing transcript sequences to a depth of approximately 50,000 to 90,000 reads per cellular barcode.
Data processing
Sequencing read alignment to reference genome (version GRCh38), gene expression quantification, and TCR reconstruction was performed using the multi command in the Cellranger pipeline (10x Genomics, v6.1.1). Relevant barcode sequences were provided for the alignment and quantification of HTOs and ADTs. Expression data (gene, protein, and hashtag) were combined using the R package Seurat (v4.0.3) (Hao et al., 2021) with the Read10X function. Cells were removed if they contained more than 7% of reads aligned to mitochondrial genes or expressed fewer than 700 genes or more than 5700 genes. Transcript expression was normalized using the LogNormalize method implemented in Seurat’s NormalizeData function. Expression for ADTs and HTOs were transformed using the CLR (centered log-ratio) method. HTO demultiplexing was performed using Seurat’s HTODemux function. Cells were grouped into either the CD4 or CD8 compartments based on higher ADT expression. TCR data was imported using the import_vdj command from the djvdj R package. For cells with more than one reconstructed alpha or beta chain, additional chains were discarded to retain only a single alpha and a single beta chain with the highest expression. Cells were considered to belong to a common clonotype if their CDR3 amino acid sequences matched precisely.
Identification of unconventional subsets expressing CD8
We observed that some cells stained positive for the CD8 protein but were not conventional CD8+ T cells. Clustering analysis identified two transcriptional clusters of γδ T cells and NK cells (Figure S4F). Unconventional CD8 expressing subsets were removed from CD8+ T cell analyses according to the following criteria: NK cells (NK cluster), MAIT cells (TRAJ33 paired with TRAV1-2 and TRBV20-1 or TRAV1-2 and TRBV6), γδ T cells (TRDV1 + or TRGV3 + or TRGV9 +) and invariant NKT cells (TRAV10+ and TRAJ18+).
Clustering and subset identification
Gene expression values were scaled and centered using the ScaleData function (Seurat), and highly variable genes were selected using the Seurat vst algorithm. Clusters were identified using the FindNeighbors function with 15 dimensions and the FindClusters function with the Louvain algorithm. Dimensionality reduction was performed using the UMAP algorithm, and clusters were named based on the top differentially expressed genes from a Wilcox rank-sum test.
Differential expression
Marker genes for each UMAP cluster and pairwise differential expression analyses were calculated using the FindAllMarkers and FindMarkers functions (Seurat) with logfc.threshold=0.01. Pairwise comparisons for individual markers between two groups were compared by the Mann-Whitney test implemented in the wilcox.test R function.
Gene set enrichment
Differentially expressed genes were ranked according to average log2 fold change values and used as input for gene set enrichment analysis (GSEA) with the fgsea command and fgsea R package (v1.18) with 5000 permutations. Hallmark and C7 immunologic signature gene sets were downloaded from the MSigDB.
Module scoring
Gene module scoring was performed using the AddModuleScore function (Seurat). Gene sets classified under the Reactome terms ‘Signaling by Interleukins’ and ‘Adaptive Immune System’ were downloaded from MSigDB. A manually curated gene set for CD8+ T cell cytotoxicity was sourced from a recent study of total peripheral responses during COVID-19 (Zhang et al., 2020).
TCR analysis
TCR analyses were performed using the scRepertoire R package (v1.3.2) (Borcherding et al., 2020) and custom R commands. The proportion of repertoire space occupied by each clone was calculated using the clonalProportion function. Shannon’s entropy was calculated using the following formula: -Σ(clonotype frequency ∗ log2(clonotype frequency)).
Coexpression visualizations
Coexpression analyses visualized using UpSet plots were produced using the UpSetR R package (v1.4) (Conway et al., 2017).
Data analysis and statistics
Flow cytometry data were analyzed using FlowJo software version 10.7.1 (FlowJo LLC). The gating strategy is shown in Figure S1. Spike-specific T cell responses of >0.05% after background reductions were considered positive. Only responses assigned as positive based on these criteria were included in downstream analyses to limit the impact of background noise. Stimulation indices were only included if the calculations were based on >10 cells in each marker+ population. Statistical analyses were performed using Prism version 9 for macOS (GraphPad Software Inc.). Significance between two paired groups was assessed using the Wilcoxon signed-rank test, and significance between two unpaired groups was assessed using the Mann-Whitney test. Significance among three or more unpaired groups or among paired groups with missing values was assessed using the Kruskal-Wallis test with Dunn’s post-test. Categorical variables were compared using Fisher’s exact test.
Acknowledgments
This study has been funded by grants from the SciLifeLab National COVID-19 Research Program and financed by the Knut and Alice Wallenberg Foundation, the Swedish Research Council, Nordstjernan AB, Region Stockholm (Clinical research position-, ALF-, and CIMEDgrants), and a graduate student fellowship from Karolinska Institutet. In addition, the Swedish patient organizations of Primary Immunodeficiencies (PIO) and Hematology (Swedish Blood Cancer Foundation) provided grants. M.B. was supported by the Swedish Research Council, the Knut and Alice Wallenberg Foundation, the European Research Council, Karolinska Institutet, the Swedish Society of Medicine, the Swedish Cancer Society, the Swedish Childhood Cancer Fund, the Åke Wibergs Stiftelse, and the Jonas Söderquist Stiftelse. This work has partly been supported by NIH contract 75N9301900065 (A.S. and D. Weiskopf). J.N. was supported by an EMBO postdoctoral fellowship (ALTF 1062-2020). The authors acknowledge support from the National Genomics Infrastructure in Stockholm funded by Science for Life Laboratory, the Knut and Alice Wallenberg Foundation and the Swedish Research Council, and SNIC/Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure.
Author contributions
Conceptualization, Y.G., C.C., J.K.S., C.I.E.S., P.B., H.-G.L., S.A., and M.B.; sample collection, O.R.-B., J.L., P.C., A.C., O.B., L.H., S. Mielke, P.N., M.A., A.P.-P., C.B., T.K., T.P., T.R.M., J.R.M., M.S., K.H., J.V., G.S., A.Ö., F.H., M.S.C., P.L., C.I.E.S., P.B., H.-G.L., S.A., and M.B.; investigation, Y.G., C.C., D. Wullimann., J.L., O.R.-B., G.B., S. Muschiol, S.A., and M.B.; formal analysis, Y.G., C.C., D. Wullimann, and S. Muschiol; visualization, Y.G., C.C., D. Wullimann, T.S., and M.B.; resources, A.G., D. Weiskopf, A.S., J.K.S., H.-G.L., S.A., and M.B.; funding acquisition, H.-G.L., S. Mielke, K.L., S.A., and M.B.; supervision, J.K.S., H.-G.L., S.A., and M.B.; writing—original draft, Y.G., C.C., and M.B.; writing—review and editing, C.C., J.N., and M.B.
Declaration of interests
M.B. is a consultant for Oxford Immunotec. A.S. is a consultant for Gritstone, Flow Pharma, Arcturus, Immunoscape, CellCarta, Oxford Immunotech, and Avalia. A.S. has filed for patent protection for various aspects of T cell epitope and vaccine design work.
Published: July 19, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2022.07.005.
Supplemental information
Data and code availability
The processed scRNA-seq data reported in this paper are available in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession number ArrayExpress: E-MTAB-11845.
All codes supporting the current study are available from the corresponding author on request.
References
- Adamo S., Michler J., Zurbuchen Y., Cervia C., Taeschler P., Raeber M.E., Baghai Sain S.B., Nilsson J., Moor A.E., Boyman O. Signature of long-lived memory CD8+ T cells in acute SARS-CoV-2 infection. Nature. 2022;602:148–155. doi: 10.1038/s41586-021-04280-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolidis S.A., Kakara M., Painter M.M., Goel R.R., Mathew D., Lenzi K., Rezk A., Patterson K.R., Espinoza D.A., Kadri J.C., et al. Cellular and humoral immune responses following SARS-CoV-2 mRNA vaccination in patients with multiple sclerosis on anti-CD20 therapy. Nat. Med. 2021;27:1990–2001. doi: 10.1038/s41591-021-01507-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bange E.M., Han N.A., Wileyto P., Kim J.Y., Gouma S., Robinson J., Greenplate A.R., Hwee M.A., Porterfield F., Owoyemi O., et al. CD8+ T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 2021;27:1280–1289. doi: 10.1038/s41591-021-01386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barda N., Dagan N., Cohen C., Hernán M.A., Lipsitch M., Kohane I.S., Reis B.Y., Balicer R.D. Effectiveness of a third dose of the BNT162b2 mRNA COVID-19 vaccine for preventing severe outcomes in Israel: an observational study. Lancet. 2021;398:2093–2100. doi: 10.1016/S0140-6736(21)02249-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-On Y.M., Goldberg Y., Mandel M., Bodenheimer O., Freedman L., Kalkstein N., Mizrahi B., Alroy-Preis S., Ash N., Milo R., et al. Protection of BNT162b2 vaccine booster against Covid-19 in Israel. N. Engl. J. Med. 2021;385:1393–1400. doi: 10.1056/NEJMoa2114255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman P., Blennow O., Hansson L., Mielke S., Nowak P., Chen P., Söderdahl G., Österborg A., Smith C.I.E., Wullimann D., et al. Safety and efficacy of the mRNA BNT162b2 vaccine against SARS-CoV-2 in five groups of immunocompromised patients and healthy controls in a prospective open-label clinical trial. EBiomedicine. 2021;74:103705. doi: 10.1016/j.ebiom.2021.103705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borcherding N., Bormann N.L., Kraus G. scRepertoire: an R-based toolkit for single-cell immune receptor analysis. F1000Res. 2020;9:47. doi: 10.12688/f1000research.22139.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Brown L.K., Moran E., Goodman A., Baxendale H., Bermingham W., Buckland M., Abdulkhaliq I., Jarvis H., Hunter M., Karanam S., et al. Treatment of chronic or relapsing COVID-19 in immunodeficiency. J. Allergy Clin. Immunol. 2022;149:557–561.e1. doi: 10.1016/j.jaci.2021.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway J.R., Lex A., Gehlenborg N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics. 2017;33:2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. Follicular helper CD4 T cells (TFH) Annu. Rev. Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- Embi P.J., Levy M.E., Naleway A.L., Patel P., Gaglani M., Natarajan K., Dascomb K., Ong T.C., Klein N.P., Liao I.C., et al. Effectiveness of 2-dose vaccination with mRNA COVID-19 vaccines Against COVID-19-associated hospitalizations Among immunocompromised adults - nine states, January–September 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:1553–1559. doi: 10.15585/mmwr.mm7044e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuertes Marraco S.A., Soneson C., Cagnon L., Gannon P.O., Allard M., Abed Maillard S., Montandon N., Rufer N., Waldvogel S., Delorenzi M., Speiser D.E. Long-lasting stem cell-like memory CD8+ T cells with a naive-like profile upon yellow fever vaccination. Sci. Transl. Med. 2015;7:282ra48. doi: 10.1126/scitranslmed.aaa3700. [DOI] [PubMed] [Google Scholar]
- Fung M., Babik J.M. COVID-19 in Immunocompromised Hosts: What We Know So Far. Clin. Infect. Dis. 2021;72:340–350. doi: 10.1093/cid/ciaa863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Cai C., Grifoni A., Müller T.R., Niessl J., Olofsson A., Humbert M., Hansson L., Österborg A., Bergman P., et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat. Med. 2022;28:472–476. doi: 10.1038/s41591-022-01700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrera G., Picozza M., D'Orso S., Placido R., Pirronello M., Verdiani A., Termine A., Fabrizio C., Giannessi F., Sambucci M., et al. BNT162b2 vaccination induces durable SARS-CoV-2 specific T cells with a stem cell memory phenotype. Sci Immunol. 2021;5:eabl5344. doi: 10.1126/sciimmunol.abl5344. [DOI] [PubMed] [Google Scholar]
- Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelow B., Mao T., Klein J., Song E., Menasche B., Omer S.B., Iwasaki A. Adaptive immune determinants of viral clearance and protection in mouse models of SARS-CoV-2. Sci. Immunol. 2021;6:eabl4509. doi: 10.1126/sciimmunol.abl4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalimuddin S., Tham C.Y.L., Qui M., de Alwis R., Sim J.X.Y., Lim J.M.E., Tan H.C., Syenina A., Zhang S.L., Le Bert N., et al. Early T cell and binding antibody responses are associated with COVID-19 RNA vaccine efficacy onset. Medicine. 2021;2:682–688.e4. doi: 10.1016/j.medj.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeton R., Tincho M.B., Ngomti A., Baguma R., Benede N., Suzuki A., Khan K., Cele S., Bernstein M., Karim F., et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature. 2022;603:488–492. doi: 10.1038/s41586-022-04460-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury D.S., Cromer D., Reynaldi A., Schlub T.E., Wheatley A.K., Juno J.A., Subbarao K., Kent S.J., Triccas J.A., Davenport M.P. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 2021;27:1205–1211. doi: 10.1038/s41591-021-01377-8. [DOI] [PubMed] [Google Scholar]
- Kiner E., Willie E., Vijaykumar B., Chowdhary K., Schmutz H., Chandler J., Schnell A., Thakore P.I., LeGros G., Mostafavi S., et al. Gut CD4(+) T cell phenotypes are a continuum molded by microbes, not by TH archetypes. Nat. Immunol. 2021;22:216–228. doi: 10.1038/s41590-020-00836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Li F., Smith P., Ravetch J.V. Inhibitory Fcgamma receptor is required for the maintenance of tolerance through distinct mechanisms. J. Immunol. 2014;192:3021–3028. doi: 10.4049/jimmunol.1302934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon Arthur, Birger Chet, Thorvaldsdóttir Helga, Ghandi Mahmoud, Mesirov Jill P., Tamayo Pablo. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Systems. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon Arthur, Subramanian Aravind, Pinchback Reid, Thorvaldsdóttir Helga, Tamayo Pablo, Mesirov Jill P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Chandrashekar A., Sellers D., Barrett J., Jacob-Dolan C., Lifton M., McMahan K., Sciacca M., VanWyk H., Wu C., et al. Vaccines elicit highly conserved cellular immunity to SARS-CoV-2 Omicron. Nature. 2022;603:493–496. doi: 10.1038/s41586-022-04465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyal L., Braun J., Henze L., Kruse B., Dingeldey M., Reimer U., Kern F., Schwarz T., Mangold M., Unger C., et al. Cross-reactive CD4(+) T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science. 2021;374:eabh1823. doi: 10.1126/science.abh1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Dan J.M., Zhang Z., Rydyznski Moderbacher C., Lammers M., Goodwin B., Sette A., Crotty S., Weiskopf D. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science. 2021;374:eabj9853. doi: 10.1126/science.abj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naaber P., Tserel L., Kangro K., Sepp E., Jürjenson V., Adamson A., Haljasmägi L., Rumm A.P., Maruste R., Kärner J., et al. Dynamics of antibody response to BNT162b2 vaccine after six months: a longitudinal prospective study. Lancet Reg Health Eur. 2021;10:100208. doi: 10.1016/j.lanepe.2021.100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng O.W., Chia A., Tan A.T., Jadi R.S., Leong H.N., Bertoletti A., Tan Y.J. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessl J., Sekine T., Lange J., Konya V., Forkel M., Maric J., Rao A., Mazzurana L., Kokkinou E., Weigel W., et al. Identification of resident memory CD8(+) T cells with functional specificity for SARS-CoV-2 in unexposed oropharyngeal lymphoid tissue. Sci. Immunol. 2021;6:eabk0894. doi: 10.1126/sciimmunol.abk0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhardt V., Luxenburger H., Kemming J., Schulien I., Ciminski K., Giese S., Csernalabics B., Lang-Meli J., Janowska I., Staniek J., et al. Rapid and stable mobilization of CD8(+) T cells by SARS-CoV-2 mRNA vaccine. Nature. 2021;597:268–273. doi: 10.1038/s41586-021-03841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter M.M., Mathew D., Goel R.R., Apostolidis S.A., Pattekar A., Kuthuru O., Baxter A.E., Herati R.S., Oldridge D.A., Gouma S., et al. Rapid induction of antigen-specific CD4(+) T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity. 2021;54:2133–2142.e3. doi: 10.1016/j.immuni.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpinelli F., Marchesi F., Piaggio G., Giannarelli D., Papa E., Falcucci P., Pontone M., Di Martino S., Laquintana V., La Malfa A., et al. Fifth-week immunogenicity and safety of anti-SARS-CoV-2 BNT162b2 vaccine in patients with multiple myeloma and myeloproliferative malignancies on active treatment: preliminary data from a single institution. J. Hematol. Oncol. 2021;14:81. doi: 10.1186/s13045-021-01090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.H., Porrot F., Staropoli I., Lemoine F., et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahav G., Lustig Y., Lavee J., Ohad Benjamini, Magen H., Hod T., Noga Shem-Tov, Shmueli E.S., Drorit Merkel, Ben-Ari Z., et al. BNT162b2 mRNA COVID-19 vaccination in immunocompromised patients: a prospective cohort study. EClinicalmedicine. 2021;41:101158. doi: 10.1016/j.eclinm.2021.101158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer Mario, Nozzi Joshua L., Nason Martha C. SPICE: Exploration and analysis of post-cytometric complex multivariate datasets. Cytometry Part A. 2011;79:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- Saini S.K., Hersby D.S., Tamhane T., Povlsen H.R., Amaya Hernandez S.P., Nielsen M., Gang A.O., Hadrup S.R. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8(+) T cell activation in COVID-19 patients. Sci. Immunol. 2021;6:eabf7550. doi: 10.1126/sciimmunol.abf7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., Sagar D.F., Salvat Lago M., Decker A., et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8(+) T cells. Nat. Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields A.M., Burns S.O., Savic S., Richter A.G., Consortium U.P.C. COVID-19 in patients with primary and secondary immunodeficiency: the United Kingdom experience. J. Allergy Clin. Immunol. 2021;147:870–875.e871. doi: 10.1016/j.jaci.2020.12.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soresina A., Moratto D., Chiarini M., Paolillo C., Baresi G., Focà E., Bezzi M., Baronio B., Giacomelli M., Badolato R. Two X-linked agammaglobulinemia patients develop pneumonia as COVID-19 manifestation but recover. Pediatr. Allergy Immunol. 2020;31:565–569. doi: 10.1111/pai.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo P.A., Levitin H.M., Miron M., Snyder M.E., Senda T., Yuan J., Cheng Y.L., Bush E.C., Dogra P., Thapa P., et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nat. Commun. 2019;10:4706. doi: 10.1038/s41467-019-12464-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Coelho C.H., Zhang Z., Dan J.M., Yu E.D., Methot N., Bloom N.I., Goodwin B., Phillips E., Mallal S., et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell. 2022;185:847–859.e11. doi: 10.1016/j.cell.2022.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartof S.Y., Slezak J.M., Fischer H., Hong V., Ackerson B.K., Ranasinghe O.N., Frankland T.B., Ogun O.A., Zamparo J.M., Gray S., et al. Effectiveness of mRNA BNT162b2 COVID-19 vaccine up to 6 months in a large integrated health system in the USA: a retrospective cohort study. Lancet. 2021;398:1407–1416. doi: 10.1016/S0140-6736(21)02183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Y., Wang X.M., Xing X., Xu Z., Zhang C., Song J.W., Fan X., Xia P., Fu J.L., Wang S.Y., et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020;21:1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The processed scRNA-seq data reported in this paper are available in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession number ArrayExpress: E-MTAB-11845.
All codes supporting the current study are available from the corresponding author on request.