Abstract
Although COVID-19 vaccines have been developed, multiple pathogenic coronavirus species exist, urging on development of multispecies coronavirus vaccines. Here we develop prototype lipid nanoparticle (LNP)-mRNA vaccine candidates against SARS-CoV-2 Delta, SARS-CoV, and MERS-CoV, and we test how multiplexing LNP-mRNAs can induce effective immune responses in animal models. Triplex and duplex LNP-mRNA vaccinations induce antigen-specific antibody responses against SARS-CoV-2, SARS-CoV, and MERS-CoV. Single-cell RNA sequencing profiles the global systemic immune repertoires and respective transcriptome signatures of vaccinated animals, revealing a systemic increase in activated B cells and differential gene expression across major adaptive immune cells. Sequential vaccination shows potent antibody responses against all three species, significantly stronger than simultaneous vaccination in mixture. These data demonstrate the feasibility, antibody responses, and single-cell immune profiles of multispecies coronavirus vaccination. The direct comparison between simultaneous and sequential vaccination offers insights into optimization of vaccination schedules to provide broad and potent antibody immunity against three major pathogenic coronavirus species.
Keywords: mRNA vaccine, multispecies coronavirus vaccine, multiplexed vaccination, sequential vaccination, SARS-CoV-2, SARS-CoV, MERS-CoV, cross-reactivity, systems immunology, single-cell profiling
Graphical abstract
Peng et al. demonstrate the efficacy of mRNA vaccine candidates targeting three major pathogenic coronavirus species (SARS-CoV-2, SARS-CoV, and MERS-CoV) and find that vaccination schedules and antigen immunogenicity influence the magnitude of antibody responses. Single-cell profiling reveals immune population and transcriptomics changes associated with multiplexed mRNA vaccination against these coronaviruses.
Introduction
Coronaviridae is a large family of viral species constantly evolving (V’Kovski et al., 2021). Coronaviruses are genetically diverse RNA viruses that exhibit broad host range among mammals, where the infections cause a wide range of diseases, ranging from the common cold to severe illnesses and death (V’Kovski et al., 2021) (Hu et al., 2021). Multiple zoonotic coronavirus species evolved to infect humans and became highly contagious, pathogenic, and even fatal, leading to epidemics worldwide (V’Kovski et al., 2021). To date, seven known coronavirus species have evolved to infect humans (V’Kovski et al., 2021). There are three known highly pathogenic human coronavirus species to date, severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), all of which can cause severe respiratory or multiorgan diseases and can be fatal (Zhu et al., 2020). There are also thousands of potentially highly pathogenic coronaviruses circulating in animal reservoirs globally (Alluwaimi et al., 2020; Cui et al., 2019; Hu et al., 2021; Latif and Mukaratirwa, 2020). SARS-CoV-2 is the pathogen that causes coronavirus disease 2019 (COVID-19) (V’Kovski et al., 2021), an ongoing multiwave worldwide pandemic (Cohn et al., 2021) that has claimed over 5 million lives to date. SARS-CoV and MERS-CoV emerged in humans in 2002 (Peiris et al., 2003) and 2012 (Zaki et al., 2012), and have high case fatality rates (∼10% for SARS-CoV and ∼35% for MERS-CoV, relative to ∼1% for SARS-CoV-2) (Abdelrahman et al., 2020). Thus, it is important to develop effective vaccines against these highly pathogenic coronavirus species.
Before the COVID-19 pandemic, no effective vaccine had been approved to prevent the spread of coronaviruses. Previous SARS and MERS vaccine devolvement (Bosaeed et al., 2021; Folegatti et al., 2020; Li et al., 2020; Pallesen et al., 2017; Su et al., 2021), although at earlier stages, together with global efforts, led to rapid development of multiple COVID-19 vaccines against SARS-CoV-2 (Tregoning et al., 2021). The most prominent and efficacious vaccine belongs to the lipid nanoparticle (LNP) mRNA vaccine category, with the first two emergency use approvals issued to Moderna and Pfizer-BioNTech mRNA vaccines (Baden et al., 2021; Polack et al., 2020). Although successful vaccines against SARS-CoV-2 have been developed to control COVID-19, no effective vaccines exist that can counter multiple pathogenic coronavirus species including SARS-CoV and MERS-CoV. Thus, it is important to develop multispecies coronavirus vaccines, not only to help fight the ongoing pandemic but also to prevent re-emergence of these previously existent dangerous pathogens, as well as to gain insights to prepare for future zoonotic pathogenic coronavirus outbreaks.
The successs of LNP-mRNA vaccine against COVID-19 led to the natural hypothesis of multiplexed vaccination against multiple coronavirus species. Certain prior studies in other virus families such as influenza, herpes simplex virus (HSV) and cytomegalovirus (CMV) demonstrated initial feasibility of using two or more mRNA vaccine constructs in mixture (Awasthi et al., 2019; Freyn et al., 2020; John et al., 2018). These earlier foundational studies tested the concept of mRNA vaccine using multiple antigens in CMV, influenza, and HSV-2. However, it is still important to test them in the context of coronaviruses, which are the cause of the current pandemic. Our studies directly test multispecies coronavirus vaccines against the pathogenic circulating coronaviruses that have emerged to infect humans.
Moreover, the immunogenicity of multispecies coronavirus vaccines needs to be studied, for example LNP-mRNA vaccines against MERS-CoV, SARS-CoV-2, and SARS-CoV. Two recent studies used a chimeric mRNA, or a protein-based antigen complex, against two or more coronaviruses (Cohen et al., 2021; Martinez et al., 2021). The Martinez study generated chimeric vaccine constructs only for SARS-CoV, SARS-CoV-2, and other non-pathogenic species (e.g., HKU3-1 and RsSHC014, but not MERS-CoV). However, chimeric spikes would not be able to capture all full-length spikes, losing part of the critical antigenic regions (e.g., S1 or S2) for one species or the other. The two highly successful approved mRNA vaccines used full-length spike (Baden et al., 2021; Polack et al., 2020). Finally, the optimal vaccination schedule involving a multiplexed vaccination needs to be explored; for example, whether vaccination by administering all mRNAs simultaneously would be effective and whether spacing out the different mRNA vaccine shots would lead them to perform better.
To gain initial answers to some of these questions, we directly generated species-specific LNP-mRNA vaccine candidates and tested them either alone or in combination in vivo. We generated LNP-mRNAs specifically encoding the HexaPro engineered full-length spikes of SARS-CoV-2 Delta variant, SARS-CoV, and MERS-CoV, and systematically studied their immune responses in animal models.
Results
Design and biophysical characterization of triplex coronavirus vaccine against SARS-CoV-2, SARS-CoV, and MERS-CoV
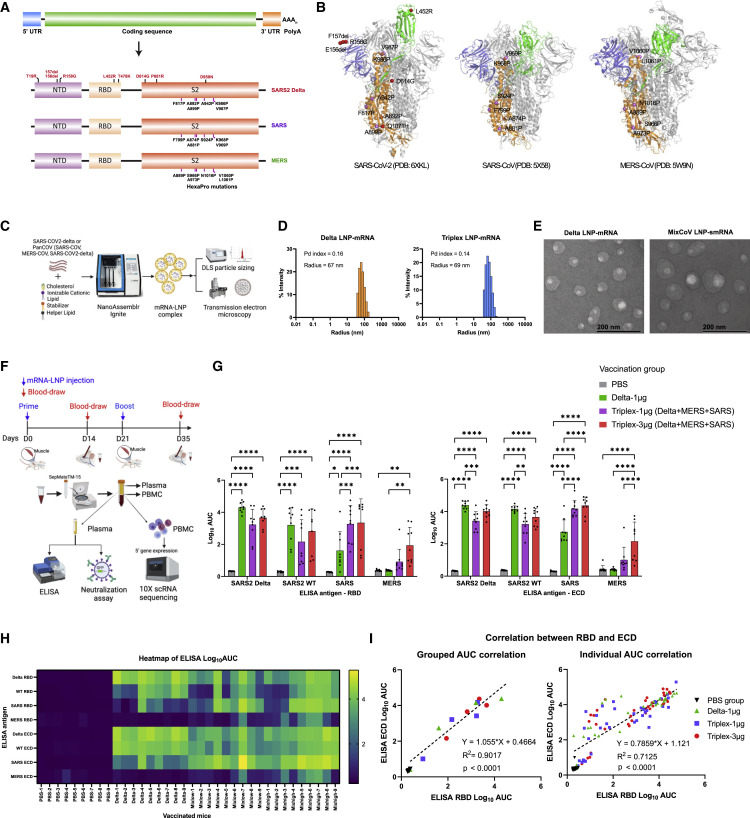
We first designed vaccine candidate constructs encoding full-length spike mRNA of SARS-CoV-2 (hereafter labeled SARS2) Delta variant (Delta), SARS-CoV (SARS), and MERS-CoV (MERS) (Figures 1A, 1B, and S1A). Each construct contains a 5′ untranslated region (UTR), an open reading frame (ORF), a 3′ UTR, and a polyA signal. The ORFs encode full-length spikes of defined species (SARS2, SARS, and MERS), in which six additional proline mutations (HexaPro) were introduced in the S2 domain of the respective species (Figures 1A and 1B), based on the homologous amino acid positions of SARS-CoV-2, in order to improve expression and stable prefusion state of spikes (Hsieh et al., 2020). The Delta construct ORF encodes the spike of SARS-CoV-2 Delta variant, which has nine mutations (T19R, 156del, 157del, R158G, L452R, T478K, D614G, P681R, and D950N) as compared with the original wild-type (WT; WA-1 or WA1) virus (Figures 1A and 1B). We tested each of these mRNA constructs and showed that they all successfully generate functional protein upon introduction into mammalian cells, as evident by surface binding to the cognate human receptors, hACE2 for SARS-CoV and SARS-CoV-2 and hDPP4 for MERS, respectively (Figures S1B and S1C).
Figure 1.
Antibody responses induced by Triplex LNP-mRNA vaccination against SARS-CoV-2 Delta, SARS-CoV, and MERS-CoV in vivo
(A) Schematics of mRNA vaccine construct design against pathogenic human coronavirus species. Each construct has regulatory elements (5′ UTR, 3′ UTR, and polyA) and spike ORF. The domain structures as well as engineered mutations of translated spike proteins of SARS-CoV-2 Delta variant (Delta), SARS-CoV (SARS), and MERS-CoV (MERS) are shown.
(B) Engineered mutations in spike protein structures of SARS-CoV-2 Delta, SARS-CoV, and MERS-CoV. The N-terminal domain (NTD, blue), receptor binding domain (RBD, green), and S2 subunit (orange) of one protomer along with homologous HexaPro mutations (pink) and Delta variant mutations (red) are highlighted in the spike trimer structures.
(C) Schematics of characterization of LNP-mRNA vaccine formulations. Assembly procedure of LNP-mRNA vaccine on NanoAssemblr Ignite and downstream biophysical characterization assays.
(D) Histogram displaying radius distribution of LNP-mRNA formulations of SARS-CoV-2 Delta and a Triplex (Delta + SARS + MERS) (abbreviated as Triplex-CoV or MixCoV), measured by dynamic light scattering. The polydispersity index and mean radius of each LNP sample are shown at the top left corner.
(E) Transmission electron microscopy images of Delta and Triplex-CoV LNP-mRNAs.
(F) Schematics of vaccination schedule of the Triplex LNP-mRNA formulations, as well as downstream assays to evaluate the antibody responses and other immunological profiles.
(G) Binding antibody titers of plasma samples from mice administered with PBS or different LNP-mRNAs (n = 9 mice from one independent experiment) against RBD or ectodomain (ECD) of SARS-CoV-2 wild-type (WT, Wuhan/WA-1), Delta variant, SARS, and MERS spikes. The binding antibody titers were quantified by area under the curve of log10-transformed titration curve (log10 AUC) in Figure S2. The mice were intramuscularly injected with two doses (×2, 2 weeks apart) of PBS, 1 μg of SARS-CoV-2 Delta variant LNP-mRNA (delta), and 1 μg or 3 μg equal-mass mixture of Delta, SARS, and MERS LNP-mRNA (Triplex-CoV).
(H) Overall heatmap of antibody titers of individual mice (one column represents one mouse, n = 9) against eight spike antigens in ELISA (one row represents one antigen).
(I) Correlation of antibody titers against RBD (y value) and ECD (x value) of same coronavirus spike, by individual mouse, or by averaged group (n = 9 mice × 4 antigens).
Statistical information is provided in STAR Methods. See also Figures S1 and S6.
To multiplex these constructs, we prepared equal-mass mixture of spike mRNA of Delta, SARS, and MERS, which were then encapsulated by LNPs on a microfluidics instrument, to generate a triplex LNP-mRNA formulation of vaccine candidate (termed as Triplex or MixCoV, interchangeable aliases) (Figure 1C). We also generated a Delta singlet LNP-mRNA for testing in parallel. The size and homogeneity of assembled LNPs were evaluated by dynamic light scattering and transmission electron microscopy (Figures 1D and 1E). The Delta LNP-mRNA and Triplex LNP-mRNA showed monodispersed size distribution with average radius of 70 ± 3.8 nm and 71 ± 3.6 nm, and polydispersity indices of 0.160 and 0.157, respectively. To evaluate the immunogenicity of Delta and Triplex LNP-mRNA vaccines, C57BL/6Ncr (B6) mice were immunized intramuscularly with two doses (prime and boost) of 1 μg Delta LNP-mRNA, and 1 μg or 3 μg (total mRNA mass) Triplex LNP-mRNA, three weeks apart (Figure 1F). The peripheral blood mononuclear cells (PBMCs) and plasma were collected 2 weeks post boost. The mice humoral response including binding and neutralizing antibody response against spike antigens were examined by ELISA and neutralization assays using collected plasma samples. Single-cell RNA sequencing (scRNA-seq) was performed to profile the systemic immune repertoires and their respective transcriptomics in vaccinated animals (Figure 1F).
Immune responses to triplex coronavirus LNP-mRNA vaccination against SARS2, SARS, and MERS
Compared with the PBS control group, the 1 μg Delta LNP-mRNA, and 1 μg and 3 μg Triplex LNP-mRNA all elicited potent antibody response, as seen in the high post-boost binding antibody titers against both receptor binding domain (RBD) and ectodomain (ECD) of Delta, WT, and SARS spikes (Figures 1G, 1H, and S1). Among the three vaccination groups, only 3 μg Triplex LNP-mRNA significantly boosted mouse immunity to MERS antigens (Figures 1G, 1H, and S1). As the Delta and Triplex vaccines used the Delta variant as spike antigen, their responses to Delta ELISA antigen were found to be slightly higher than the WT antigen response (Figures 1G and S1). Despite of the lack of SARS spike antigen in the vaccine, the Delta LNP-mRNA induced antibodies that cross-react with SARS spike but not MERS spike (Figures 1G and S1), consistent with the respective degree of homology between these species (Figure S1A). The titers are at similarly high level between the 1 μg and 3 μg Triplex groups for SARS and SARS2 spikes (Figure 1G), while there is a trend of dose-dependent increase although statistically insignificant (Figure S1). Compared with those of MixCoV 1 μg or 3 μg groups, SARS binding antibody titer in the 1 μg Delta LNP-mRNA group was significantly lower. A dose-dependent increased trend of antibody titers was observed for MERS spike in the two Triplex vaccination groups (Figure 1G). Within the Triplex groups, it is worth noting that antibody titer against MERS was 10- to 20-fold lower than that against SARS-CoV or SARS-CoV-2. Considering Delta spike mRNA at the same dose, mice in the 1 μg Delta and 3 μg Triplex (that also contains 1 μg Delta mRNA) groups showed similar titers of antibodies against SARS2 WT and Delta spikes, although an insignificant trend of lower titers was observed in the 3 μg Triplex mice (Figures 1G and S1). Both ECD and RBD ELISA antigen panels showed highly correlated results among four spike types used (Figure 1I). In addition, a subset of animals showed relatively higher titer to ECD than RBD (Figure 1I, off-the-diagonal data points), potentially due to the additional antibody reactivity outside RBD in those animals.
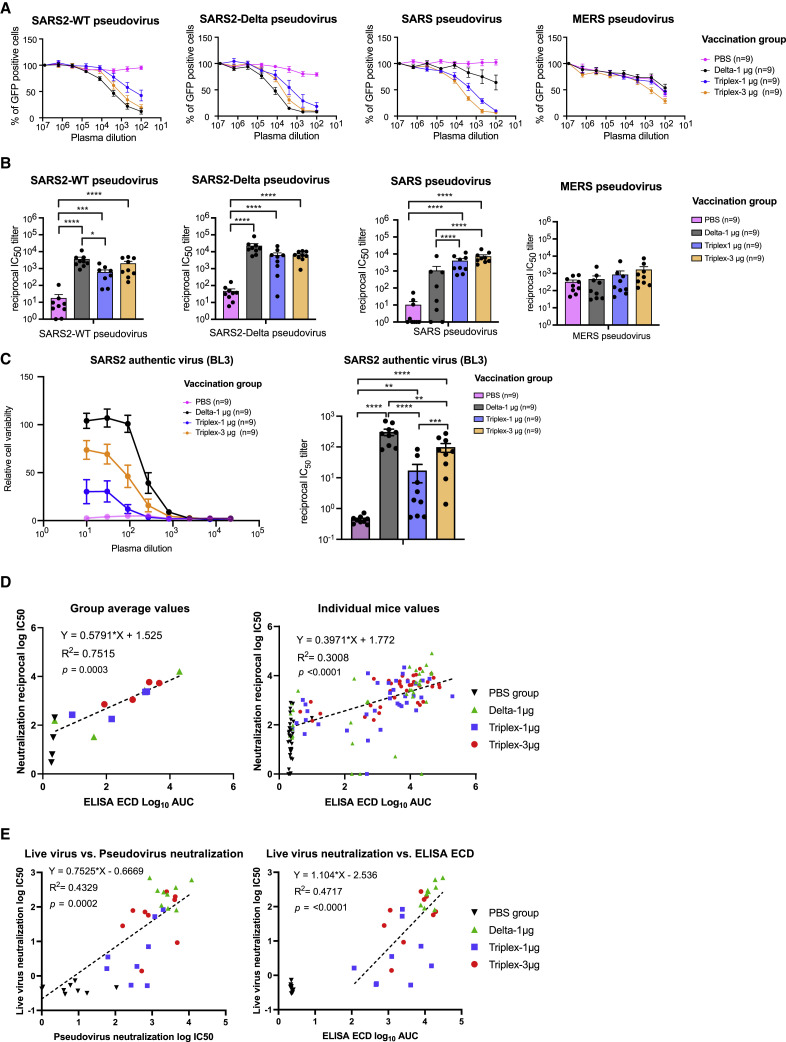
We went on to examine the neutralizing antibody response in the pseudovirus assay. All three Delta and Triplex-CoV LNP-mRNA vaccines induced marked increase in neutralizing antibodies against SARS2 WT/WA-1, Delta, and SARS pseudoviruses (Figures 2A and 2B), which mimicked the overall titer landscape of binding antibodies in ELISA. All three LNP-mRNA groups (1 μg Delta, 1 μg and 3 μg Triplex-CoV) elicited potent neutralization activity against SARS2 Delta in the plasma of the vaccinated animals (Figures 2A and 2B). In addition, both Triplex-CoV LNP-mRNA groups (1 μg and 3 μg Triplex-CoV) elicited potent neutralization activity against SARS in the plasma samples of the vaccinated animals (Figures 2A and 2B). Despite the lack of SARS mRNA, the Delta-alone group of LNP-mRNA also elicited a substantial level of anti-SARS neutralization antibody response in a fraction of animals (4/9 above background) with high variation, although significantly lower than those of the Triplex-CoV groups (Figures 2A and 2B), again potentially due to the similarity between the two species (Figure S1A). The significantly higher antibody titer against SARS and higher robustness highlighted superior SARS protection efficacy of Triplex-CoV vaccine over Delta vaccine alone against SARS. Moderate neutralization activity against MERS was observed at this dosing scheme, with the PBS group showing a relatively high background level of neutralization (Figures 2A and 2B). Similar to ELISA, the neutralization activities are at similarly high levels between the 1 μg and 3 μg Triplex-CoV groups for SARS and SARS2 spikes (Figures 2A and 2B), while there is a trend of dose-dependent increase albeit statistically insignificant. We validated the neutralization against authentic virus of SARS-CoV-2 in a Biosafety Level 3 (BL3) setting, where the plasma samples from all three LNP-mRNA groups (1 μg Delta, 1 μg and 3 μg Triplex-CoV) showed significant neutralization activity (Figure 2C). Similar to the observations in the BL2 pseudovirus assay, the Triplex vaccination at both doses showed a lower level of neutralization against SARS2 as compared with Delta vaccination alone (Figure 2C). Overall, antibody binding against ECD (ELISA) significantly correlated with neutralization activities for all groups or all mice among spikes and pseudoviruses tested (Figure 2D). Consistent with various previous reports (Bewley et al., 2021; Doria-Rose et al., 2021; Liu et al., 2020; Nie et al., 2020; Riepler et al., 2020), neutralization activity against authentic virus (BL3) significantly correlated with neutralization activities against pseudovirus (BL2), and correlated with binding antibody titers by ELISA (Figure 2E).
Figure 2.
Neutralizing antibody responses induced by Triplex LNP-mRNA vaccination against SARS-CoV-2 Delta, SARS-CoV, and MERS-CoV in vivo
(A) Neutralization titration curves of plasma from mice treated with PBS, Delta, and Triplex-CoV LNP-mRNA against WT and Delta SARS-CoV-2, SARS-CoV, and MERS-CoV pseudoviruses. The percentage of GFP-positive cells reflected the infection rate of host cells by pseudovirus and was plotted against the dilution factors of mouse plasma to quantify neutralizing antibody titers.
(B) Neutralizing antibody titers in the form of reciprocal IC50 derived from fitting the titration curves with a logistic regression model. Each dot represents data from one mouse, and each group contains nine mice (n = 9, one independent experiment).
(C) Neutralization assay using authentic virus in BL3 setting. Neutralization curves and titer quantification dot plots (n = 9).
(D) Correlation of neutralization log10 IC50 versus antibody titers against ECD of same coronavirus spike, by individual mouse, or by averaged group (n = 9 mice × 4 antigens).
(E) Correlation between BL3 authentic virus neutralization and BL2 pseudovirus neutralization, and between BL3 authentic virus neutralization and ELISA, by individual mouse (n = 9 mice × 1 antigen).
Statistical information is provided in STAR Methods. See also Figure S1.
Immune responses to duplex coronavirus LNP-mRNA vaccination centering on MERS
As the levels of Triplex-CoV-induced MERS binding and neutralizing antibodies were relatively lower than those of SARS and SARS2 in the Triplex-CoV groups (Figures 1 and 2), we sought to test single and duplex vaccination schemes for MERS LNP-mRNA (Figure 3A). A proline mutation engineered prefusion MERS-CoV spike antigen has been previously generated and purified (Pallesen et al., 2017), which provided the basis for a MERS mRNA vaccine in clinical development (Corbett et al., 2020). To test how the MERS HexaPro spike LNP-mRNA can work in combination with SARS2 Delta or SARS LNP-mRNA as duplex vaccines, we designed a duplex vaccine experiment for MERS LNP-mRNA. In the single vaccine scheme, we used the MERS LNP-mRNA alone (MERS Singlet). In the duplex vaccine schemes, we mixed the MERS plus SARS, or MERS plus SARS2 Delta mRNAs, for the formulation of LNP-mRNAs (MERS Duplexes).
Figure 3.
In vivo antibody responses induced by Duplex LNP-mRNA vaccination against MERS-CoV, in combination with SARS-CoV-2 Delta or SARS-CoV
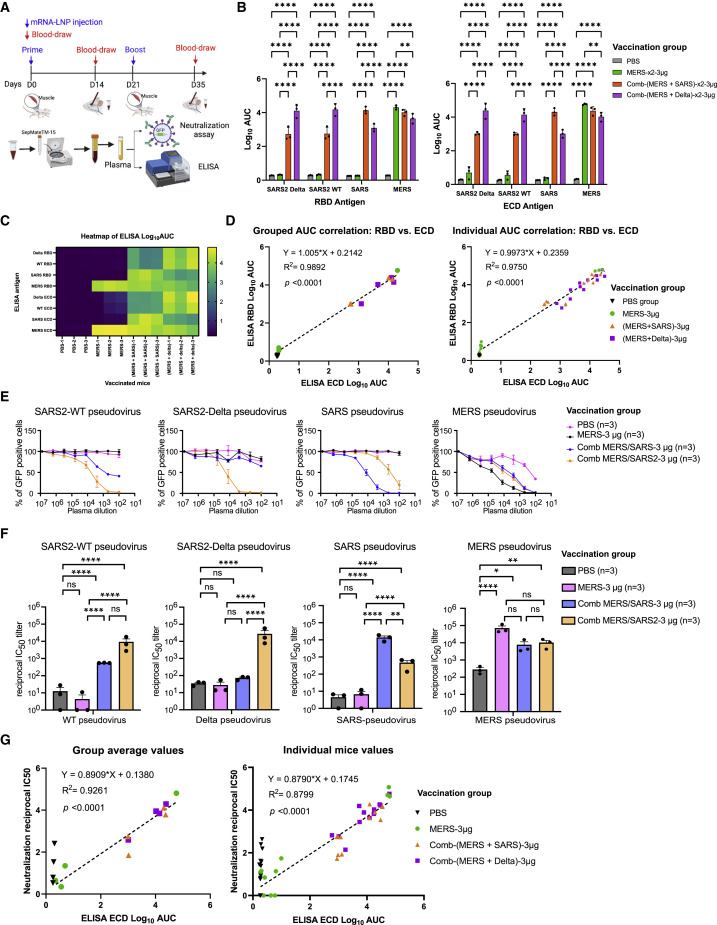
(A) Schematics of vaccination schedule of the MERS Singlet and Duplex combo LNP-mRNA formulations, as well as downstream assays to evaluate the antibody responses and other immunological profiles. Two Duplexes were evaluated, (MERS + SARS) or (MERS + SARS2 Delta).
(B) Dot-box plots summarizing binding antibody titers of plasma from mice administered with PBS or different LNP-mRNAs (n = 3 mice, one independent experiment) against RBD or ECD of SARS-CoV-2 WT/WA-1 and Delta variant, as well as SARS and MERS spikes.
(C) Heatmap of antibody titers of individual mice (one column represents one mouse, n = 3) against eight spike antigens in ELISA (one row represents one antigen).
(D) Correlation of antibody titers against RBD (y value) and ECD (x value) of same coronavirus spike, by individual mouse, or by averaged group (n = 3 × 4 antigens).
(E) Neutralization titration curves of plasma from mice treated with PBS control, or LNP-mRNA formulations with MERS alone or in Duplexes (MERS + SARS) or (MERS + SARS2 Delta); all tested against WT/WA-1 and Delta SARS-CoV-2, SARS-CoV, and MERS-CoV pseudoviruses. The percentage of GFP-positive cells reflected the infection rate of host cells by pseudovirus and was plotted against the dilution factors of mouse plasma to quantify neutralizing antibody titers (n = 3).
(F) Neutralizing antibody titers in the form of reciprocal IC50 derived from fitting the titration curves with a logistic regression model. Each dot represents data from one mouse, and each group contains three mice (n = 3).
(G) Correlation of neutralization IC50 versus antibody titers against ECD of same coronavirus spike, by individual mouse, or by averaged group (n = 3 × 4 antigens).
Statistical information is provided in STAR Methods. See also Figures S5 and S6.
Mice vaccinated with 3 μg MERS LNP-mRNA Singlet elicited high titers of MERS binding antibodies with little or no cross-reactivity to WT, Delta, or SARS spikes (Figures 3B, 3C, and S1E), suggesting that the MERS LNP-mRNA, when used alone at high dose, has sufficient immunogenicity. Combined with an equal mass of MERS LNP-mRNA, Delta, or SARS LNP-mRNA, the two MERS Duplexes also exhibited strong binding antibody titers against cognate antigens (Delta and SARS spike, respectively, plus MERS spike) (Figures 3B, 3C, and S1E). Meanwhile they also showed cross-reactive response to counterpart spike (e.g., MERS + SARS2 Delta against SARS spike, or vice versa) at a lower level than the cognate response (Figures 3B, 3C, and S1E). Similar to the Triplex experiment, the ELISA ECD activity highly correlated with RBD (Figure 3D).
We again tested the neutralization activities using the same pseudovirus assays. Mice vaccinated with 3 μg MERS LNP-mRNA Singlet elicited potent MERS neutralizing antibody response with little or no cross-reactivity to WT, Delta, or SARS spikes (Figures 3E and 3F), suggesting that the antibodies induced by MERS vaccination alone does not cross-react with SARS or SARS2. Both MERS Duplexes also exhibited strong neutralization activities against MERS, as well as cognate species (MERS + SARS2 Delta for SARS2; and MERS + SARS for SARS) (Figures 3E and 3F). Interestingly, although the MERS + SARS LNP-mRNA elicited binding antibodies that cross-reacted with both WT and Delta spike antigens (Figure 3B), the induced cross-reactive antibodies only significantly neutralized the WT but not Delta pseudovirus (Figure 3F). Consistent with prior Triplex-CoV experiment, the ELISA ECD panel correlated well with RBD panel results (Figure 3G) and tend to have higher titers than the RBD panel. Overall, such neutralization activities also significantly correlated with antibody binding against ECD (ELISA) for all groups or all mice among the spike antigens and pseudoviruses tested (Figure 3G).
Single-cell immune repertoire mapping of multiplexed LNP-mRNA vaccinated animals
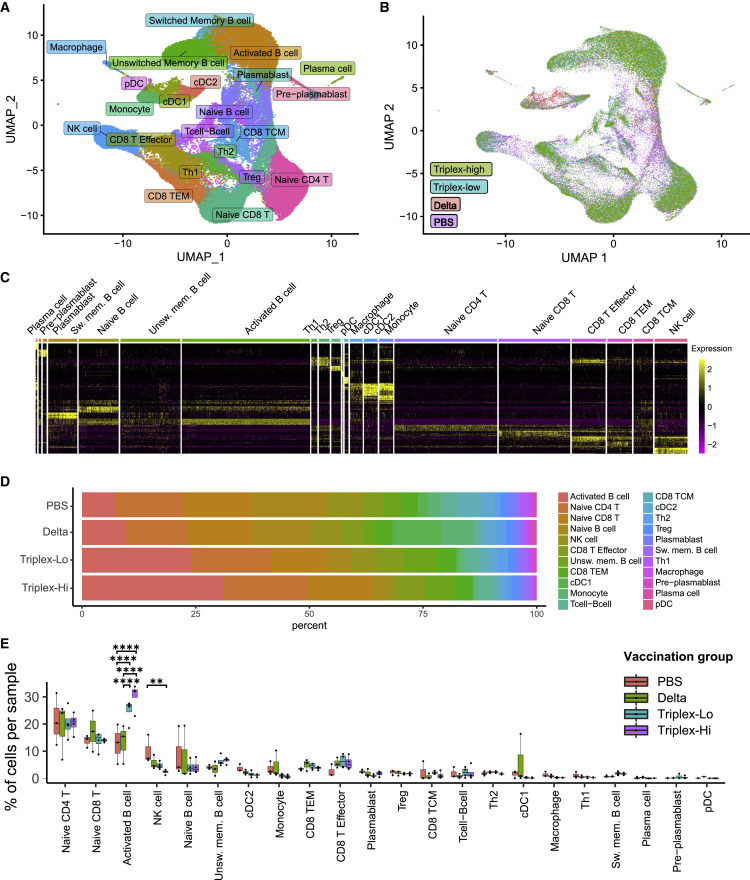
To gain insights into the global composition and transcriptional landscape of the immune cells, we performed scRNA-seq and single-cell gene expression (scGEX) for immune transcriptomics on the PBMC samples of Delta and Triplex LNP-mRNA vaccinated animals. The use of PBMC samples allows collection of immune cell samples without sacrificing mice so that it is possible to monitor live animals’ antibody response over time. As visualized in an overall uniform manifold approximation and projection (UMAP), from a total of 12 animals from four vaccination groups (Delta 1 μg and Triplex-CoV 1 μg and 3 μg dose groups, plus a placebo control group [PBS]), we sequenced the transcriptomes of a total of 91,526 single cells (Data S1), which were visualized in reduced dimensional space by UMAP and clustered to identify cell population structure (Figures 4A and 4B). Using the expression of a set of canonical cell-type-specific markers, we identified 21 cell clusters as distinct immune cell populations (Figures 4A and S2A–S2D). In this dataset, the identified cell clusters include various subsets of B lymphocytes (naive B cell, activated B cell, unswitched memory B cell, switched memory B cell, pre-plasmablast, plasmablast, and plasma cell); T lymphocytes of various subsets (naive CD8 T cell, CD8 T effector, CD8 central memory T cell, CD8 effector memory T cell, naive CD4 T cell, T helper 1 [Th1] type CD4 T cell, Th2 type CD4 T cell, regulatory T cell [Treg]); dendritic cells (DCs) of various subsets (pDC, cDC1, cDC2); and other immune cells (natural killer [NK] cells, macrophages, and monocytes) (Figure 4A). These immune cell populations have distinct gene expression signatures that clearly defined each population against others (Figure 4C): for example, distinct expression (in terms of both mean expression level and percentage in cluster) of Cd19 + H2-Aa + Ighd + Fcer2a + Cd27 − defines activated B cells; Cd9 + Sdc1 + Cd19 − Pax5 -lo defines plasma cells (Figures 4C, S2A, and S2B). Similarly, for T cell subset examples, Cd3d + Cd4 + Tbx21 + Gzmb + marks Th1 CD4 T cells; Cd3d + Cd4 + Foxp3 + Il2ra + marks Tregs; Cd3d + Cd8b1 + Ccr7 + Cd44 − Tcf7 + defines naive CD8 T cells; Cd3d + Cd8b1 + Tcf7 − Cd44 + Ccr7 − defines CD8 effector T cells; Itgam + Itgax + Cd24a − Sirpa + defines cDC2 cells; Itgam − Itgax + Bst2 + Siglech + defines pDC; Ncr1 defines NK cells; and Itgam + Csf1r + Cd14 + defines monocytes (Figures 4C, S2A, and S2B).
Figure 4.
Single-cell transcriptomics of animals vaccinated by multiplexed LNP-mRNA vaccine against SARS-CoV-2, SARS-CoV, and MERS-CoV in mice
(A) UMAP visualization of all 91,526 cells pooled across samples and conditions. All identified clusters are shown with cell identities assigned, based on the expression of cell-type-specific markers.
(B) UMAP visualization, colored by vaccination groups: PBS, Delta, MixCoV-lo (i.e., Triplex 1 μg), and MixCoV-hi (i.e., Triplex 3 μg). n = 3 mice, one independent experiment.
(C) Heatmap showing the population clusters with distinct expression patterns. Rows represent the scaled expression of the top ten genes that were differentially expressed in each cluster, relative to all other cells, based on Wilcoxon rank sum analysis.
(D) Stacked bar plot depicting the proportion of different immune populations for each vaccination group.
(E) Dot-whisker plot of immune cell proportions by cell type for each vaccination group: PBS, Delta, MixCoV-lo, and MixCoV-hi; n = 3 mice each group.
Statistical information is provided in STAR Methods. See also Figures S2–S4.
We then quantified the fractions of each cell type in each sample to reveal a full picture of immune cell compositions in all vaccination groups profiled (Figures 4D and 4E). With these quantitative fractions, we then compared the systemic immune cell compositions between placebo and vaccinated animals (Figure 4D). While most of the clusters did not show significant difference at a gross cell population level, three populations (activated B cells, unswitched memory B cells, and NK cells) showed significant differences between groups (Figures 4D and 4E). Interestingly, Triplex-CoV/MixCoV at both high and low doses of vaccination showed a significantly increased level of activated B cell populations compared with both PBS and Delta groups (Figures 4D and 4E). Both activated and memory B cell populations have been previously implicated for their important roles in SARS-CoV-2 immunity (Newell et al., 2021; Quast and Tarlinton, 2021; Sokal et al., 2021).
Transcriptomic signatures of B and T cell populations of triplex LNP-mRNA vaccinated animals
To examine the transcriptomic changes in the immune cell subpopulations upon vaccination, we performed differential expression analysis in the matched subpopulations between PBS and the several LNP-mRNA groups. We focused on the major adaptive immune cell populations, i.e., the pan-activated B cell population (including all identified activated B cell subsets, merged as “B cell”), pan-activated CD4 T cell population (all identified activated CD4 T cell subsets, “CD4 T cell”), and pan-activated CD8 T cell population (all identified activated CD8 T cell subsets, “CD8 T cell”). Vaccination caused substantial transcriptome changes in the host animals’ B cells, CD4 T cells, and CD8 T cells, as evidenced by the differential gene expression from vaccinated (Delta, Triplex-CoV/MixCoV low- and high-dose groups) compared with the PBS group (Data S1; Figures S3 and S4). To gain a broad, unbiased view of these transcriptomic changes, we performed a series of gene set and pathway analyses. These analyses revealed a number of altered pathways in the vaccinated animals’ B cells, CD4 T cells, and CD8 T cells compared with the PBS group (Data S1 and Figure S3A). Because the altered pathways are diverse, we also performed clustering analysis to uncover the key signal by grouping them into “supra-pathways” where multiple gene sets of similar function were altered. This network analysis of enriched pathways of differentially expressed genes highlighted the most significantly enriched member pathways (as meta-pathway) for the main adaptive immune cell types (B and T cells) for the three vaccination groups (Figure S3B).
To further distinguish the directions, we also created ridge density plots, showing the expression log fold change meta-pathway genes between different vaccination groups in different cell types (Figure S4B). Consistent with the prior observations, the differentially expressed pathways in B cells include leukocyte/lymphocyte-mediated immunity in all three vaccination groups compared with PBS (Data S1; Figures S3B and S4B). A top enriched pathway of the differentially expressed genes in B cells is B cell activation, where all three vaccines induced a higher expression of these genes (Figures S4B and S4C). In CD4 and CD8 T cells, common gene sets are observed, including immune system processes, immune cell differentiation, and T cell activation, consistent with the expected induction from vaccination (Figures S3B, S4B, and S4C). Interestingly, in T cells in the differentially expressed genes in all three vaccines, besides regulation of T cell activation, leukocyte proliferation, leukocyte differentiation, defense response to virus, and immune responses, basic fundamental pathways are also enriched, especially those involved in core cellular and metabolic functions such as apoptosis, translation, ubiquitin ligase activity, oxidative phosphorylation, mitochondria electron transport, and respiratory chain activities (Figures S4B, S4C, and S3B), consistent with the expectation that T cells are metabolically active upon vaccination. The Triplex vaccination induced strong B cell activation pathway clusters in B cells as well as immune cell differentiation and metabolic activity gene sets in T cells (Figures S3B, S4B, and S4C). These data reveal the broad gene expression signatures at the pathway and cluster levels across the main adaptive immune cells (B and T cells) for the three vaccination groups studied. The transcriptomic signatures are largely coherent with the literature stating that these pathways are important for immunity against coronavirus infection and host defense (Stephenson et al., 2021; Zhang et al., 2020) as well as vaccine-induced immune responses (Arunachalam et al., 2021; Cao et al., 2021). These data revealed meta-pathway-level gene expression changes in the B and T cells’ transcriptomes of the animals receiving multiplexed vaccination.
Direct comparison of sequential versus simultaneous vaccination schedules for LNP-mRNA vaccination against three species
As observed above, Triplex LNP-mRNA vaccination is associated with reduction of antibody responses (Figures 1, 2, and 3), we hypothesized that splitting such vaccination into separate doses may be a strategy to mitigate this loss of effectiveness. We therefore sought to perform a sequential vaccination schedule and test it in parallel with simultaneous vaccination with mRNAs in mixture (Figure 5A). In the “Sequential” vaccination schedule, vaccinations of SARS-CoV-2 Delta, MERS-CoV, and SARS-CoV were given in sequence separated by 3 weeks, each with 1 μg of LNP-mRNA prime and 1 μg of LNP-mRNA boost 3 weeks apart. In the “Mixture” vaccination schedule, vaccinations of SARS-CoV-2 Delta, MERS-CoV, and SARS-CoV were given simultaneously, each at 1 μg o LNP-mRNA (3 μg total) for both prime and boost. To generate comparable data, we started the first dose on the same day (day 0) and harvested the blood sample on the same day (day 119, i.e., 4 months post first dose) for both Sequential and Mixture schedules (Figure 5A).
Figure 5.
Direct comparison of sequential versus mixture vaccination schedules against SARS-CoV-2 Delta, MERS-CoV, and SARS-CoV
(A) Schematics of sequential versus mixture vaccination schedules and sampling. In the Sequential vaccination schedule, vaccinations of SARS-CoV-2 Delta, MERS-CoV, and SARS-CoV were given in sequence separated by 3 weeks, each with 1 μg of LNP-mRNA prime and 1 μg LNP-mRNA boost 3 weeks apart. In the Mixture vaccination schedule, vaccinations of SARS-CoV-2 Delta, MERS-CoV, and SARS-CoV were given simultaneously, each at 1 μg of LNP-mRNA (3 μg total) for both prime and boost. The first dose and the blood sample harvest were done on the same day for both sequential and mixture schedules for comparison.
(B) Dot-box plots summarizing binding antibody titers of plasma from mice administered with PBS, Sequential, or Mixture LNP-mRNA vaccinations (n = 4 mice, one independent experiment) against RBD or ECD of SARS-CoV-2 WT/WA-1 and Delta variant, as well as SARS and MERS spikes.
(C) Neutralization titration curves of plasma from mice treated with PBS, Sequential, or Mixture LNP-mRNA vaccinations (n = 4 each, one independent experiment); all tested against WT/WA-1 and Delta SARS-CoV-2, SARS-CoV, and MERS-CoV pseudoviruses. The percentage of GFP-positive cells reflected the infection rate of host cells by pseudovirus and was plotted against the dilution factors of mouse plasma to quantify neutralizing antibody titers.
(D) Neutralizing antibody titers in the form of reciprocal IC50 derived from fitting the titration curves with a logistic regression model. Each dot represents data from one mouse, and each group contains three mice (n = 4).
(E) Blocking ELISA antibody titers of plasma from different vaccination groups against Delta (left), SARS (middle), and MERS (right) ECDs in the presence of competing reagents including PBS (negative control), Delta, SARS, or MERS ECDs. Statistical significance was analyzed between groups of different blockers (n = 4, one independent experiment).
Statistical information is provided in STAR Methods. See also Figures S6 and S7.
We measured the antibody titers from plasma samples of both Sequential and Mixture LNP-mRNA vaccinated animals (Figures 5B, S5, and S6). While all vaccinated animals showed certain antibody responses across all antigens tested (SARS2 WT/WA1, SARS2 Delta, SARS, MERS; both ECD and RBD), the Sequential vaccination group showed significantly higher antibody responses than the Mixture group across all conditions, i.e., across all antigens from these three species (Figure 5B). Similar to the results above, the ELISA ECD activity highly correlated with that of RBD (Figure S6B). We tested the neutralization activities using the same pseudovirus assays (Figures 5C and 5D). Again, mice in the Sequential vaccination schedule showed significantly higher neutralization activities than those in the Mixture vaccination group and across all three species (Figures 5C and 5D). It is notable that similarly to the previous experiment (Figure 2), the MERS neutralization activity was almost completely lost at this time point in the Mixture vaccination group, yet the Sequential vaccination group retained significant activity above background (Figure 5D). Overall, such neutralization activities significantly correlated with ECD ELISA for all groups or all mice among the spike antigens and pseudoviruses tested (Figure S6C). These data suggested that, for LNP-mRNA vaccination against three coronavirus species under the conditions tested, vaccination in sequence can elicit more potent antibody responses than vaccination simultaneously in mixture.
To comprehensively evaluate the cognate and cross-reactive antibody response induced by the Sequential and Mixture LNP-mRNA vaccination, we conducted a blocking ELISA whereby soluble spike antigens or competing agents partially block the plasma antibody response to the homologous or heterologous spike antigen coated on the ELISA plates. The antibody response of Sequential and Triplex samples at matched time points (day 35 in Figure 1 and day 119 in Figure 5) in the presence and absence of competing agents were directly compared in blocking ELISA. In the absence of competing agents (PBS control), the Triplex’s antibody titers against all three spikes (Delta, SARS, and MERS ECDs) significantly declined over time (day 35 versus day 119 in Figures 5E and S7). Sequential samples on day 35 before exposure to SARS and MERS antigens only displayed low or moderate activity against MERS and SARS ECDs. At 2 weeks after final immunization, Sequential samples (day 119) showed a universal trend of higher antibody titers than Triplex samples (day 35). Under equal-mass condition of antigen mRNA in both Sequential and Triplex vaccination, antibody response to SARS was greater than that of Delta, the antibody titer of which was higher than MERS, indicative of distinct immunogenicity of spike antigens from different coronavirus species.
Compared with heterologous blockers, homologous blockers (same spike as ELISA antigen) unequivocally led to greater titer reduction, which ranged from a 30% to 70% decrease and represents the maximum achievable blocking effect under current conditions (Figures 5E and S7). Significant titer reductions by heterologous blockers were associated with cross-species antibodies and were observed in Sequential vaccination (day 119) response to Delta ECD by SARS blocker and Sequential/Delta vaccination (day 35) response to SARS ECD by MERS blocker (comparison bracket colored red in Figure 5E). Most heterologous blockers mediated very limited antibody titer reduction, suggesting that cross-coronavirus species antibodies, if they exist, only account for a small population of Sequential or Triplex vaccine-induced antibodies. The fact that no heterologous blocker induced significant titer changes in the Triplex group suggests that simultaneous exposure to all three coronavirus spike antigens mainly elicits species-specific antibodies, not cross-species antibodies. In most cases, Sequential (day 35) or Delta vaccination showed stronger cross-reactivity or heterologous blocking effect than other vaccination schemes (Figure S7), except for Sequential vaccination (day 119) response to Delta ECD by SARS blocker. It is worth noting that despite the absence of Delta antigen stimulation in Sequential vaccination since day 21, subsequent SARS and MERS antigen immunizations further elevated the antibody titer against Delta. The significant blocking effect of SARS ECD on Sequential vaccination (day 119) response to Delta ECD revealed that the Delta titer increase by heterologous boosters was mainly mediated by SARS antigen and not MERS antigen. Interestingly, the Sequential vaccination (day 119 versus day 35) lost its strong heterologous blocking effect in SARS and MERS ELISA panels (Figure S7), suggesting that the SARS and MERS antigens predominantly elicited cognate species-specific antibodies, eclipsing the cross-species antibodies observed in Delta vaccination.
Discussion
Protective vaccines are the keys to control the ongoing and potential future coronavirus pandemics. Coronavirus is a group of viral species that can constantly evolve to become highly contagious and pathogenic to humans. Pathogenic coronaviruses have emerged multiple times and infected human populations, several of which (SARS-CoV, MERS-CoV, and SARS-CoV-2) have caused severe diseases and fatalities (Cui et al., 2019; Dong et al., 2020; Zhu et al., 2020). Several existing less pathogenic coronavirus species (e.g., NL63, 2293, OC43, and HKU1) have been reported to have evolved hundreds to tens of thousands of years ago, and might have evolved to be coexisting with humans without causing severe symptoms (Ye et al., 2020). Therefore, it is critical to have vaccines against multiple coronavirus species, ideally as pan-coronavirus vaccines, not only to help fight the current pandemic but also to prevent the re-emergence of the previously existing pathogenic species, as well as the constantly evolving and lurking coronavirus diseases as probable future outbreaks. Equally importantly, there remains a long-standing need to gain the fundamental understanding of the immune response and the immunological landscape of joint host responses in the context of multiplex coronavirus vaccine.
Various prior efforts led to the development of SARS and MERS vaccine candidates, although at earlier stages of development (Bosaeed et al., 2021; Folegatti et al., 2020; Li et al., 2020; Pallesen et al., 2017; Su et al., 2021). The COVID-19 pandemic urged an international effort for rapid development of vaccines against SARS-CoV-2 (Tregoning et al., 2021), leading to multiple successful candidates including the highly efficacious mRNA vaccines (Baden et al., 2021; Polack et al., 2020). However, all of these vaccines target a single species and may not offer sufficient protection against other pathogenic species. A small number of “pan-coronavirus” vaccine candidates have been recently generated and tested in animal models, using protein antigen nanocage or mRNA encoding chimeric spike, with the focus on SARS-CoV and SARS-CoV-2 and several other non-pathogenic viruses (Cohen et al., 2021; Martinez et al., 2021). Multiplex LNP-mRNA vaccine against the more lethal species such as SARS-CoV and MERS-CoV also need to be rigorously tested.
However, to our knowledge no study has tested the multiplexing of mRNA vaccines against three major pathogenic coronavirus species (MERS/SARS/SARS2) in the triplex setting or in sequence. Our study generated a full-length MERS LNP-mRNA vaccine construct and tested it alone, in combination with SARS-CoV vaccine, SARS-CoV-2 vaccine, and in triplex. Our study directly generated mRNA vaccine candidates tested in several LNP-mRNA combinations against MERS-CoV, SARS-CoV, and SARS-CoV-2, and profiled the immune responses at the single-cell level. Neither of the studies above provides comparison of different vaccine schedules, while our study directly compares sequential versus mixture vaccination schemes.
When simultaneously administered with same dose of SARS, SARS2 Delta, and MERS LNP-mRNA in the Triplex formulation, mice generated MERS binding and neutralizing titers severalfold lower than those of SARS and SARS2, which showed similar titer levels. Several factors might account for the lower titer of MERS. The immunogenicity of MERS spike LNP-mRNA was lower than those of SARS and SARS2 Delta spikes at the same dose in the Triplex vaccine. The simultaneous vaccination with multiplexing may also have a negative impact on MERS LNP-mRNA as it did for Delta LNP-mRNA as discussed above, perhaps due to immunodominance (Angeletti and Yewdell, 2018), although never tested in a multiplexed LNP-mRNA vaccination setting before. It is also possible that coexpression of distinct spikes from different coronavirus species would reduce the surface density of homotrimer spikes, thereby hindering antigen recognition and antibody production. Interestingly MERS LNP-mRNA works better in a duplex setting than in a triplex setting, where its antigen is composed of 50% in duplex as opposed to 33% in triplex. The immune response of triplex and duplex LNP-mRNA vaccines reported here has implications for future multiplex coronavirus vaccine development.
Our study reported the antibody responses of triplex and duplex LNP-mRNA vaccines based on MERS spike in combination with SARS and/or SARS2 Delta spikes. The level of cross-reactivity of induced antibodies was in concordance with the sequence identity between vaccine antigen and binding antigen tested in ELISA and pseudovirus assays. Different from prior studies, the antiviral spectrum we tested here covers three highly pathogenic coronavirus species in the Betacoronavirus genus, and goes beyond the group 2b coronavirus category (Sarbecoviruses), as it includes MERS in the Merbecovirus subgenus. Our data showed that because of low sequence similarity, the vaccine based on Sarbecovirus (SARS and SARS2) provides little or no protection against MERS, the most fatal coronavirus to date with a 35% mortality rate. To broaden the vaccine’s anti-coronavirus spectrum, we designed and tested the triplex and duplex LNP-mRNA vaccine including SARS, SARS2, and MERS.
In order to obtain sufficient and broad protection of neutralizing antibodies in multiplex vaccine against these coronavirus species, the relative composition or the scheme of vaccination need to be carefully considered in the future. In addition to the multiplexing approach we showed in this study, there are other ways of inducing protective antibodies against SARS2 Delta, SARS, and MERS. The production and manufacturing procedures of multiplexed LNP-mRNA formulations, such as mixing, normalization, and encapsulation, may benefit from further optimization and testing in the future. Alternatively, the three spike LNP-mRNAs can be given sequentially to avoid negative interactions between spike antigens seen in triplex vaccine. In fact, this is one of the clinical precautions whereby individuals are advised to take the COVID-19 mRNA vaccine at least 2 weeks away from taking other vaccines. Consistent with this notion, our data with direct comparisons in animal vaccination experiments suggested that giving the mRNA vaccine shots in sequence may benefit from higher antibody titers over a long period of time rather than giving mRNAs simultaneously in mixture. We directly compared antibody titers 14 days after the final dose of sequential vaccination and triplex vaccination (Figure S5, Delta-sequential versus MixCoV-2weeks). MixCoV group antibody titers against SARS1 and SARS2 variants were comparable with the Delta-sequential group, while MERS immunity tended to be lower than with sequential vaccination. This is potentially due to competition in antigens, immunodominance, and other reasons. Three months after the final dose of triplex vaccine, the titers of triplex vaccination declined by ∼10-fold and were significantly lower than those of sequential vaccination, which maintained high antibody titers against all three coronavirus pathogens at day 119, partially due to continuous vaccine boosting over time.
Because of the waning immunity of coronavirus vaccines (Bergwerk et al., 2021; Goldberg et al., 2021), the general public is recommended to receive a booster shot of COVID-19 vaccine. Thus, vaccination in sequence may be beneficial regardless. These observations and considerations may be informative for LNP-mRNA vaccination against multiple coronavirus species. Although SARS-CoV and MERS case numbers are small, a pandemic or endemic can arise from a handful of cases, or even one infection, if not contained properly. Given that there are diverse coronavirus species with several of them being pathogenic and many of them being potentially pathogenic in future human exposures, multiplexed vaccination against these species will be critical. If the efficacy is comparable with that of monovalent COVID vaccine, multivalent or pan-COVID vaccines will be naturally more appealing to the general population as they have more potential to curb future coronavirus pandemic or endemic outbreaks. A recent commentary summarized the pipelines developing pan-COVID vaccines from multiple non-profit organizations and vaccine producers (Dolgin, 2022), which highlighted the importance and demand of this type of vaccine. Future design of pan-coronavirus vaccines may need to seek a balance between protection breadth and depth by choosing the correct number of spike antigens across coronavirus lineages.
We performed head-to-head sequential vaccination in comparison with simultaneous vaccination in mixture, showing that sequential vaccination showed higher antibody responses at the endpoint. These observations and considerations may be informative for LNP-mRNA vaccination against multiple coronavirus species. Because of the waning immunity of coronavirus vaccines (Goldberg et al., 2021; Levin et al., 2021), the general public, especially the immunocompromised, is recommended to receive booster shot(s) for COVID-19 vaccine. Thus, vaccination in sequence may be beneficial regardless. The direct comparison between simultaneous and sequential vaccination offers insights into optimization of vaccination schedules to provide broad and potent protective antibody immunity against three major pathogenic coronavirus species. Given that there are diverse coronavirus species with several of them being pathogenic and many of them being potentially pathogenic in future human exposures, multiplexed vaccination against two or more species will be critical. Future design of pan-coronavirus vaccines may need to seek a balance between protection breadth and depth by choosing the correct number of spike antigens across coronavirus lineages. In summary, this study provided LNP-mRNA vaccine constructs designed to target SARS-CoV, SARS-CoV-2 Delta, and MERS-CoV, as well as direct in vivo animal testing and single-cell immune profiling results of multiplexed combinations as well as comparative vaccination schedules.
Limitations of the study
Vaccine evaluations were exclusively performed in mice to allow a multiarm study with sufficient sample size. Non-human primate (NHP) represents a model closer to human biology. An NHP study would take a step closer to translate findings from bench to bedside. The goal of this study is to design a vaccine schedule targeting three circulating pathogenic coronaviruses, while the newly emerging coronavirus or SARS-CoV-2 variants are not included in the study. Developing vaccine candidates to protect against newly emerging coronavirus or SARS-CoV-2 variants would be a critical direction. In addition, this study lacks a comparator vaccine, such as irrelevant mRNA vaccine or protein-based COVID vaccine, as a control, which could show non-specific background from irrelevant mRNA or differences in vaccine types. However, owing to the nature of the differences in the vaccine scheme, there is no way of conducting perfect comparisons. Our comparison is one of the reasonable ways of comparing the immune responses exactly at the same time post vaccination. Our data, limited with its models, nevertheless provide a message that the overall immunity against the three pathogenic coronaviruses is superior as a sequential vaccination scheme versus simultaneous (i.e., spacing it out is better than taking all at once).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse secondary antibody | Fisher Scientific | Cat#31439; RRID: AB_1957654 |
| PE - anti-human FC antibody | Biolegend | Cat#410708;RRID:AB_2565786 |
| Bacterial and virus strains | ||
| SARS-CoV-2 Delta pseudovirus | This study | Sidi Chen Lab |
| SARS-CoV pseudovirus | This study | Sidi Chen Lab |
| MERS-CoV pseudovirus | This study | Sidi Chen Lab |
| SARS-CoV-2 Authentic virus (WA01) | This study | Wilen Lab |
| Chemicals, peptides, and recombinant proteins | ||
| DPBS | Kline | Cat#14190144 |
| TWEEN-20 | Sigma-Aldrich | Cat# P1379 |
| Fetal Bovine Serum | Sigma Aldrich | Cat#F4135-500ML |
| DMEM | Kline | Cat#11995065 |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | Cat#15140122 |
| ACK Lysing Buffer | Lonza | Cat#BP10-548E |
| ACE2–Fc chimera | Genescript | Cat#Z03484 |
| 50TS microplate washer | Fisher Scientific | Cat#BT50TS16 |
| 100 μm cell strainer | Corning | Cat#352360 |
| 40 μm cell strainer | Corning | Cat#352340 |
| Gibson Assembly Master Mix - 50 rxn | NEB | Cat#E2611L |
| HiscribeTM T7 ARCA mRNA Kit (with tailing) | NEB | Cat#E2060S |
| Phusion Flash High-Fidelity PCR Master Mix | ThermoFisher | Cat#F548L |
| E-Gel™ Low Range Quantitative DNA Ladder | ThermoFisher | Cat#12373031 |
| QIAquick Gel Extraction Kit | Qiagen | Cat#28706 |
| EndoFree® Plasmid Maxi Kit | Qiagen | Cat#12362 |
| Quant-it™ RiboGreen™ RNA Assay Kit | ThermoFisher | Cat#R11490 |
| Tetramethylbenzidine substrate | Biolegend | Cat#421101 |
| Glow-discharged formvar/carbon-coated copper grid | Electron Microscopy Sciences | FCF400-Cu-50 |
| 2% (w/v) uranyl formate | Electron Microscopy Sciences | Cat#22450 |
| Library Construction Kit, 16 rxns | 10X Genomics | Cat#1000190 |
| Live/Dead aqua fixable stain | Thermofisher | Cat#L34976 |
| GenVoy-ILM T Cell Kit for mRNA with Spark Cartridges | Precision Nanosystems | Cat#1000683 |
| GenVoy-ILM | Precision Nanosystems | Cat#NWW0042 |
| BSA | Fisher Scientific | BP1600-100 |
| Bovine Serum Albumin | Sigma Aldrich | Cat#A9418-100G |
| EDTA | Kline | Cat#AB00502-01000 |
| BbSl | Kline | Cat#R3539L |
| Polyethylenimine (PEI) | POLYSCIENCES INC | Cat#24765-1 |
| Macron™ 2796-05 Phosphoric Acid, 85% | Avantor | Cat#MK-2796-05 |
| Polyethylenimine HCl MAX, Linear, Mw 40,000 (PEI MAX 40000) | POLYSCIENCES INC | Cat#24765-1 |
| Tris-Cl pH 7.5 | Boston Bioproducts | Cat#IBB-594 |
| N1-Methylpseudouridine-5′-Triphosphate - (N-1081) | TriLink (NC) | Cat#N-1081-1 |
| Sucrose | Thomas | Cat#C987K85 (EA/1) |
| Tetramethylbenzidine | Biolegend | Cat#421101 |
| SARS-CoV-2 (2019-nCoV) Spike S1+S2 ECD-His Recombinant Protein | SINO | Cat#40589-V08B1 |
| SARS-CoV-2 (2019-nCoV) Spike RBD | Quote UQ7100 | Cat#40592-V08B |
| SARS-CoV-2 B.1.617.2 Spike RBD(L452R,T478K) | SINO | Cat#40592-V08H90 |
| SARS-CoV-2 B.1.617.2 Spike S1+S2 (ECD, His Tag) | SINO | Cat#40589-V08B16 |
| SARS-CoV Spike ECD | SINO | Cat#40634-V08B |
| SARS-CoV Spike RBD (ECD, His Tag) | Fisher | Cat#50-196-4017 |
| MERS-CoV Spike RBD | Fisher | Cat#50-201-9463 |
| MERS-CoV Spike ECD | SINO | Cat#40069-V08B |
| Chromium Next GEM Single Cell 5ʹ Kit v2, 16 rxns PN-1000263 | 10X Genomics | Cat#PN-1000263 |
| Chromium Next GEM Chip K Single Cell Kit, 16 rxns PN-1000287 | 10X Genomics | Cat#PN-1000287 |
| Dual Index Kit TT Set A, 96 rxns PN-1000215 | 10X Genomics | Cat#PN-1000215 |
| SPRIselect - 60 mL | Beckman Coulter | Cat#B23318 |
| SepMate™-15 (IVD) | STEMCELL | Cat#85415 |
| Lymphoprep™ | STEMCELL | Cat#07851 |
| Deposited data | ||
| Single cell RNA-seq data of Vaccinated animals | This study | GEO: GSE207141 |
| Flow cytometry data. | This study | Mendeley Data: https://doi.org/10.17632/nvrdxn35tb.1 |
| Code used for data analysis | This study | Zenodo: https://zenodo.org/badge/latestdoi/512286275 |
| Experimental models: Cell lines | ||
| HEK293FT | ThermoFisher | Catalog Number: R70007 |
| HKE293T-hACE2 | Schmidt et al., 2020 | Gift from Dr Bieniasz’ lab |
| Huh-7 | CLS | Cat#300156 |
| Experimental models: Organisms/strains | ||
| C57BL/6Ncr | Charles River | strain #556 |
| Oligonucleotides | ||
| pVP31gB1 | IDT | GACACCACAGATGCTGTGAGGGACCCACAGACCTTGGAGATTCTGG ACATCACACCATGTTCCTTTGGAGGAGTGTCTGTGATTACACCTGGC ACCAACACCAGCAACCAGGTGGCTGTGCTCTACCAGGATGTGAACT GTACTGAGGTGCCTGTGGCTATCCATGCTGACCAACTTACACCAAC CTGGAGGGTCTACAGCACAGGCAGCAATGTGTTCCAGACCAGGGC TGGCTGTCTGATTGGAGCAGAGCATGTGAACAACTCCTATGAGTGT GACATCCCAATTGGAGCAGGCATCTGTGCCTCCTACCAGACCCAG ACCAACAGCCCAGGCTCTGCATCTTCTGTGGCAAGCCAGAGCATC ATTGCCTACACAATGAGTCTGGGAGCAGAGAACTCTGTGGCTTAC AGCAACAACAGCATTGCCATCCCAACCAACTTCACCATCTCTGTGA CCACAGAGATTCTGCCTGTGAGTATGACCAAGACCTCTGTGGACT GTACAATGTATATCTGTGGAGACAGCAC |
| pVp31gB2 | IDT | ATGTATATCTGTGGAGACAGCACAGAGTGTAGCAACCTGCTGCTC CAATATGGCTCCTTCTGTACCCAACTTAACAGGGCTCTGACAGGC ATTGCTGTGGAACAGGACAAGAACACCCAGGAGGTGTTTGCCCA GGTGAAGCAGATTTACAAGACACCTCCAATCAAGGACTTTGGAGG CTTCAACTTCAGCCAGATTCTGCCTGACCCAAGCAAGCCAAGCAA GAGGTCCCCTATTGAGGACCTGCTGTTCAACAAGGTGACCCTGG CTGATGCTGGCTTCATCAAGCAATATGGAGACTGTCTGGGAGAC ATTGCTGCCAGGGACCTGATTTGTGCCCAGAAGTTCAATGGACT GACAGTGCTGCCTCCACTGCTGACAGATGAGATGATTGCCCAAT ACACCTCTGCCCTGCTGGCTGGCACCATCACCTCTGGCTGGAC CTTTGGAGCAGGACCAGCCCTCCAAATCCCATTTCCAATGCAGA TGGCTTACAGGTTCAATGGCATTGGAGTGACCCAGAATGTGCTC TATGAGAACCAGAAACTGATTGCCAACCAGTTCAACTCTGCCATT GGCAAGATTCAGGACTCCCTGTCCAGCACACCATCTGCCCTGG GCAAACTCCAAGATGTGGTGAACCAGAATGCCCAGGCTCTGAA CACCCTGGTGAAGCAACTTTCCAGCAACTTTGGAGCCATCTCCT CTGTGCTGAATGACATCCTGAGCAGACTGGACCCACCAGAGGC TGAGGTCCAGATTGACAGACTGATTAC |
| pVP31bF1 | IDT | CAGAGAGAACCCGCCACCATgTTTGTGTTCCTGGTGCTGCTG CCAC |
| pVP31bR1 | IDT | TGCGTGCATGCAGTACCAGCTCGAGTCAGGTGTAGTGCAGTTTC ACTCC |
| pVP33bgB1 | IDT | CAGAGAGAACCCGCCACCATgTTCATCTTCCTGCTGTTCCTGACCC TGACCTCTGGCTCTGACCTGGACAGGTGTACCACCTTTGATGATGT CCAGGCTCCAAACTACACCCAACACACCTCCAGTATGAGGGGAGT CTACTACCCTGATGAGATTTTCAGGTCTGACACCCTCTACCTGACC CAGGACCTGTTCCTGCCATTCTACAGCAATGTGACAGGCTTCCAC ACCATCAACCACACCTTTGACAACCCTGTGATTCCATTCAAGGATG GCATCTACTTTGCTGCCACAGAGAAGAGCAATGTGGTGAGGGGCT GGGTGTTTGGCAGCACAATGAACAACAAGAGCCAGTCTGTGATTA TCATCAACAACAGCACCAATGTGGTGATTAGGGCTTGTAACTTTGA ACTGTGTGACAACCCATTCTTTGCTGTGAGCAAGCCTATGGGCAC CCAGACCCACACAATGATTTTTGACAATGCCTTCAACTGTACCTTT GAATACATCTCTGATGCCTTCTCCCTGGATGTGTCTGAGAAGTCTG GCAACTTCAAACACCTGAGGGAGTTTGTGTTCAAGAACAAGGATG GCTTCCTCTATGTCTACAAGGGCTACCAACCAATTGATGTGGTGA GGGACCTGCCATCTGGCTTCAACACCCTGAAACCAATCTTCAAAC TGCCACTGGGCATCAACATCACCAACTTCAGGGCTATCCTGACAG CCTTCAGCCCTGCCCAGGACACCTGGGGCACCTCTGCTGCTGCC TACTTTGTGGGCTACCTGAAACCAACCACCTTTATGCTGAAATATG ATGAGAATGGCACCATCACAGATGCTGTGGACTGTAGCCAGAAC CCACTGGCTGAACTGAAATGTTCTGTGAAGTCCTTTGAGATTGAC AAGGGCATCTACCAGACCAGCAACTTCAGGGTGGTGCCATCTGG AGATGTGGTGAGGTTTCCAAACATCACCAACCTGTGTCCATTTGG AGAG |
| pVP33bgB2 | IDT | ACCTGTGTCCATTTGGAGAGGTGTTCAATGCCACCAAGTTTCCAT CTGTCTATGCCTGGGAGAGGAAGAAGATTAGCAACTGTGTGGCT GACTACTCTGTGCTCTACAACAGCACCTTCTTCAGCACCTTCAAG TGTTATGGAGTGTCTGCCACCAAACTGAATGACCTGTGTTTCAGC AATGTCTATGCTGACTCCTTTGTGGTGAAGGGAGATGATGTGAGA CAGATTGCCCCTGGACAAACAGGAGTGATTGCTGACTACAACTA CAAACTGCCTGATGACTTTATGGGCTGTGTGCTGGCTTGGAACA CCAGGAACATTGATGCCACCAGCACAGGCAACTACAACTACAAA TACAGATACCTGAGACATGGCAAACTGAGACCATTTGAGAGGGA CATCAGCAATGTGCCATTCAGCCCTGATGGCAAGCCATGTACTC CTCCTGCCCTGAACTGTTACTGGCCACTGAATGACTATGGCTTC TACACCACCACAGGCATTGGCTACCAACCATACAGGGTGGTGG TGCTGTCCTTTGAACTGCTGAATGCCCCTGCCACAGTGTGTGGA CCAAAACTGAGCACAGACCTGATTAAGAACCAGTGTGTGAACTT CAACTTCAATGGACTGACAGGCACAGGAGTGCTGACACCATCC AGCAAGAGGTTCCAACCATTCCAACAGTTTGGCAGGGATGTGT CTGACTTCACAGACTCTGTGAGGGACCCAAAGACCTCTGAGAT TCTGGACATCAGCCCATGTTCCTTTGGAGGAGTGTCTGTGATTA CACCTGGCACCAATGCCTCCTCTGAGGTGGCTGTGCTCTACCA GGATGTGAACTGTACTGATGTGAGCACAGCCATCCATGCTGAC CAACTTACACCTGCCTGGAGGATTTACAGCACAGGCAACAATG TGTTCCAGACCCAGGCTGGCTGTCTGATTGGAGCAGAGCATGT GGACACCTCCTATGAGTGTGACATCCCAATTGGAGCAGGCATC TGTGCCTCCTACCACACAGTGTCCCTGC |
| pVP33bgB3 | IDT | ACCACACAGTGTCCCTGCTGAGGAGCACCAGCCAGAAGAGCAT TGTGGCTTACACAATGAGTCTGGGAGCAGACTCCAGCATTGCC TACAGCAACAACACCATTGCCATCCCAACCAACTTCAGCATCA GCATCACCACAGAGGTGATGCCTGTGAGTATGGCTAAGACCTC TGTGGACTGTAATATGTATATCTGTGGAGACAGCACAGAGTGTG CCAACCTGCTGCTCCAATATGGCTCCTTCTGTACCCAACTTAAC AGGGCTCTGTCTGGCATTGCTGCTGAACAGGACAGGAACACCA GGGAGGTGTTTGCCCAGGTGAAGCAGATGTATAAGACACCAAC CCTGAAATACTTTGGAGGCTTCAACTTCAGCCAGATTCTGCCTG ACCCACTGAAACCAACCAAGAGGTCCCCAATTGAGGACCTGCT GTTCAACAAGGTGACCCTGGCTGATGCTGGCTTTATGAAGCAAT ATGGAGAGTGTCTGGGAGACATCAATGCCAGGGACCTGATTTG TGCCCAGAAGTTCAATGGACTGACAGTGCTGCCTCCACTGCTG ACAGATGATATGATTGCTGCCTACACAGCAGCCCTGGTGTCTG GCACAGCCACAGCAGGCTGGACCTTTGGAGCAGGACCAGCC CTCCAAATCCCATTTCCAATGCAGATGGCTTACAGGTTCAATG GCATTGGAGTGACCCAGAATGTGCTCTATGAGAACCAGAAGC AGATTGCCAACCAGTTCAACAAGGCTATCAGCCAGATTCAGG AGTCCCTGACCACCACCCCAACAGCCCTGGGCAAACTCCAA GATGTGGTGAACCAGAATGCCCAGGCTCTGAACACCCTGGTG AAGCAACTTTCCAGCAACTTTGGAGCCATCTCCTCTGTGCTG AATGACATCCTGAGCAGACTGGACCCACCAGAGGCTGAGGT CCAGATTGACAGACTG |
| pVP33bgB4 | IDT | GGTCCAGATTGACAGACTGATTACAGGCAGACTCCAATCCCTC CAAACCTATGTGACCCAACAACTTATCAGGGCTGCTGAGATTA GGGCATCTGCCAACCTGGCTGCCACCAAGATGAGTGAGTGTG TGCTGGGACAAAGCAAGAGGGTGGACTTCTGTGGCAAGGGCT ACCACCTGATGAGTTTTCCACAGGCTGCCCCTCATGGAGTGGT GTTCCTGCATGTGACCTATGTGCCAAGCCAGGAGAGGAACTTC ACCACAGCCCCTGCCATCTGCCATGAGGGCAAGGCTTACTTTC CAAGGGAGGGAGTGTTTGTGTTCAATGGCACCTCCTGGTTCAT CACCCAGAGGAACTTCTTCAGCCCACAGATTATCACCACAGAC AACACCTTTGTGTCTGGCAACTGTGATGTGGTGATTGGCATCAT CAACAACACAGTCTATGACCCACTCCAACCTGAACTGGACTCC TTCAAGGAGGAACTGGACAAATACTTCAAGAACCACACCAGCC CTGATGTGGACCTGGGAGACATCTCTGGCATCAATGCCTCTGT GGTGAACATCCAGAAGGAGATTGACAGACTGAATGAGGTGGC TAAGAACCTGAATGAGTCCCTGATTGACCTCCAAGAACTGGGC AAATATGAACAATACATCAAGTGGCCATGGTATGTGTGGCTGG GCTTCATTGCTGGACTGATTGCCATTGTGATGGTGACCATCCT GCTGTGTTGTATGACCTCCTGTTGTTCCTGTCTGAAAGGAGCC TGTTCCTGTGGCTCCTGTTGTAAGTTTGATGAGGATGACTCTG AACCTGTGCTGAAAGGAGTGAAACTGCACTACACCTAACTCG AGCTGGTACTGCATGCACGCAA |
| pVP34cgB1 | IDT | CAGAGAGAACCCGCCACCATgATTCACTCTGTGTTCCTGCTGATGTT CCTGCTGACACCAACAGAGTCCTATGTGGATGTGGGACCTGACTCT GTGAAGTCTGCCTGTATTGAGGTGGACATCCAACAGACCTTCTTTG ACAAGACCTGGCCAAGACCAATTGATGTGAGCAAGGCTGATGGCA TCATCTACCCACAGGGCAGGACCTACAGCAACATCACCATCACCTA CCAGGGACTGTTTCCATACCAGGGAGATCATGGAGATATGTATGTC TACTCTGCTGGTCATGCCACAGGCACCACACCACAGAAACTGTTTG TGGCTAACTACAGCCAGGATGTGAAGCAGTTTGCCAATGGCTTTGT GGTGAGGATTGGAGCAGCAGCCAACAGCACAGGCACAGTGATTAT CAGCCCAAGCACCTCTGCCACCATCAGGAAGATTTACCCTGCCTTT ATGCTGGGCTCCTCTGTGGGCAACTTCTCTGATGGCAAGATGGGC AGGTTCTTCAACCACACCCTGGTGCTGCTGCCTGATGGCTGTGGC ACCCTGCTGAGGGCTTTCTACTGTATCTTGGAACCAAGGTCTGGCA ACCACTGTCCTGCTGGCAACTCCTACACCTCCTTTGCCACCTACCA CACACCTGCCACAGACTGTTCTGATGGCAACTACAACAGGAATGC CTCCCTGAACTCCTTCAAGGAATACTTCAACCTGAGGAACTGTACC TTTATGTACACCTACAACATCACAGAGGATGAGATTTTGGAGTGGTT TGGCATCACCCAGACAGCCCAGGGAGTGCATCTGTTCTCGAGCAG ATATGTGGACCTCTATGGAGGCAATATGTTCCAGTTTGCCACCCTG CCTGTCTATGACACCATCAAATACTACAGCATCATCCCACACAGCA TCAGGAGCATCCAGTCTGACAGGAAGGCTTGGGCTGCCTTCTATG TCTACAAACTCCAACCACTGACCTTCCTGCTGGACTTCTCTGTGGA TGGCTACATCAGGAGGGCTATTGACTGTGGCTTCAATGACCTGAG CCAACTTCACTGTTCCTATGAGTC |
| pVP34cgB2 | IDT | CCTGTTTGGCTCTGTGGCTTGTGAACACATCTCCAGCACAATGAGT CAATACAGCAGGAGCACCAGGAGTATGCTGAAAAGGAGGGACAG CACATATGGACCACTCCAAACACCTGTGGGCTGTGTGCTGGGACT GGTGAACTCCTCCCTGTTTGTGGAGGACTGTAAACTGCCACTGGG ACAATCCCTGTGTGCCCTGCCTGACACACCAAGCACCCTGACAC CAAGGTCTGTGAGGTCTGTGCCTGGAGAGATGAGACTGGCAAGC ATTGCCTTCAACCACCCAATCCAGGTGGACCAACTTAACTCCTCC TACTTCAAACTGAGCATCCCAACCAACTTCTCCTTTGGAGTGACC CAGGAATACATCCAGACCACCATCCAGAAGGTGACAGTGGACTG TAAGCAATATGTGTGTAATGGCTTCCAGAAGTGTGAACAACTTCT GAGGGAATATGGACAATTCTGTAGCAAGATAAACCAGGCTCTTC ATGGAGCCAACCTGAGACAGGATGACTCTGTGAGGAACCTGTT TGCCTCTGTGAAGTCCAGCCAGTCCAGCCCAATCATCCCTGGC TTTGGAGGAGACTTCAACCTGACCCTGTTGGAACCGGTGAGCA TCAGCACAGGCAGCAGGTCTGCCAGGTCTCCAATTGAGGACCT GCTGTTTGACAAGGTGACCATTGCTGACCCTGGCTATATGCAG GGCTATGATGACTGTATGCAACAGGGACCTGCCTCTGCCAGG GACCTGATTTGTGCCCAATATGTGGCTGGCTACAAGGTGCTG CCTCCACTGATGGATGTGAATATGGAGGCTGCCTACACCTCC TCCCTGCTGGGCAGCATTGCTGGAGTGGGCTGGACTGCAGG ACTGTCCCCATTTGCTGCCATCCCATTTCCACAGAGCATCTTC TACAGACTGAATGGAGTGGGCATCACCCAACAGGTGCTGTCT GAGAACCAGAAACTGATTGCCAACAAGTTCAACCAGGCTCTG |
| pVP34cgB3 | IDT | AACAAGTTCAACCAGGCTCTGGGAGCTATGCAGACAGGCTTCA CCACCACCCCAGAGGCTTTCCAGAAGGTCCAGGATGCTGTGA ACAACAATGCCCAGGCTCTGAGCAAACTGGCATCTGAACTGAG CAACACCTTTGGAGCCATCTCTGCTAGCATTGGAGACATCATC CAGAGACTGGATCCACCAGAACAGGATGCCCAGATTGACAGA CTGATAAATGGCAGACTGACCACCCTGAATGCCTTTGTGGCTC AACAACTTGTGAGGTCTGAGTCTGCTGCCCTGTCTGCCCAACT TGCCAAGGACAAGGTGAATGAGTGTGTGAAGGCTCAAAGCAA GAGGTCTGGCTTCTGTGGACAAGGCACCCACATTGTGTCCTTT GTGGTGAATGCCCCAAATGGACTCTACTTTATGCATGTGGGCT ACTACCCAAGCAACCACATTGAGGTGGTGTCTGCCTATGGACT GTGTGATGCTGCCAACCCAACCAACTGTATTGCCCCTGTGAAT GGCTACTTCATCAAGACCAACAACACCAGGATTGTGGATGAGT GGTCCTACACAGGCTCCTCCTTCTATGCCCCTGAACCAATCAC CTCCCTGAACACCAAATATGTGGCTCCACAGGTGACCTACCAG AACATCAGCACCAACCTGCCTCCTCCACTGCTGGGCAACAGC ACAGGCATTGACTTCCAGGATGAACTGGATGAGTTCTTCAAGA ATGTGAGCACCAGCATCCCAAACTTTGGCTCCCTGACCCAGA TAAACACCACCCTGCTGGACCTGACCTATGAGATGCTGTCCC TCCAACAGGTGGTGAAGGCTCTGAATGAGTCCTACATTGACC TGAAAGAACTGGGCAACTACACCTACTACAACAAGTGGCCAT GGTACATCTGGCTGGGCTTCATCGCTGGCCTGGTGGCCCTG GCGCTGTGCGTGTTCTTCATCCTGTGCTGCACCGGCTGCGG CACCAACTGCATGGGCAAGCTGAAGTGCAACAGGTGCTGCG ACAGGTACGAGGAGTACGACCTGGAGCCCCACAAGGTGCAC GTACATTAACTCGAGCTGGTACTGCATGCACGCA |
| pVP39gB1 | IDT | AGAGAGAACCCGCCACCATgTTTGTGTTCCTGGTGCTGCTGCC ACTGGTGTCCAGCCAGTGTGTGAACCTGAGGACCAGGACCCA ACTTCCTCCTGCCTACACCAACTCCTTCACCAGGGGAGTCTAC TACCCTGACAAGGTGTTCAGGTCCTCTGTGCTGCACAGCACCC AGGACCTGTTCCTGCCATTCTTCAGCAATGTGACCTGGTTCCAT GCCATCCATGTGTCTGGCACCAATGGCACCAAGAGGTTTGACA ACCCTGTGCTGCCATTCAATGATGGAGTCTACTTTGCCAGCACA GAGAAGAGCAACATCATCAGGGGCTGGATTTTTGGCACCACCC TGGACAGCAAGACCCAGTCCCTGCTGATTGTGAACAATGCCAC CAATGTGGTGATTAAGGTGTGTGAGTTCCAGTTCTGTAATGACC CATTCCTGGACGTCTACTACCACAAGAACAACAAGTCCTGGATG GAGTCTGGCGTCTACTCCTCTGCCAACAACTGTACCTTTGAATA TGTGAGCCAACCATTCCTGATGGACTTGGAGGGCAAGCAGGGC AACTTCAAGAACCTGAGGGAGTTTGTGTTCAAGAACATTGATGG CTACTTCAAGATTTACAGCAAACACACACCAATCAACCTGGTGA GGGACCTGCCACAGGGCTTCTCTGCCTTGGAACCACTGGTGG ACCTGCCAATTGGCATCAACATCACCAGGTTCCAGACCCTGCT GGCTCTGCACAGGTCCTACCTGACACCTGGAGACTCCTCCTCT GGCTGGACAGCAGGAGCAGCAGCCTACTATGTGGGCTACCTC CAACCAAGGACCTTCCTGCTGAAATACAATGAGAATGGCACCA TCACAGATGCTGTGGACTGTGCCCTGGACCCACTGTCTGAGAC CAAGTGTACCCTGAAATCCTTCACAGTGGAGAAGGGCATCTAC CAGACCAGCAACTTCAGGGTCCAAC |
| pVP39gB2 | IDT | CCAGCAACTTCAGGGTCCAACCAACAGAGAGCATTGTGAGGTT TCCAAACATCACCAACCTGTGTCCATTTGGAGAGGTGTTCAAT GCCACCAGGTTTGCCTCTGTCTATGCCTGGAACAGGAAGAGG ATTAGCAACTGTGTGGCTGACTACTCTGTGCTCTACAACTCTG CCTCCTTCAGCACCTTCAAGTGTTATGGAGTGAGCCCAACCAA ACTGAATGACCTGTGTTTCACCAATGTCTATGCTGACTCCTTTG TGATTAGGGGAGATGAGGTGAGACAGATTGCCCCTGGACAAA CAGGCAAGATTGCTGACTACAACTACAAACTGCCTGATGACTT CACAGGCTGTGTGATTGCCTGGAACAGCAACAACCTGGACAG CAAGGTGGGAGGCAACTACAACTACAGGTACAGACTGTTCAG GAAGAGCAACCTGAAACCATTTGAGAGGGACATCAGCACAGA GATTTACCAGGCTGGCAGCAAGCCATGTAATGGAGTGGAGG GCTTCAACTGTTACTTTCCACTCCAATCCTATGGCTTCCAACC AACCAATGGAGTGGGCTACCAACCATACAGGGTGGTGGTGC TGTCCTTTGAACTGCTCCATGCCCCTGCCACAGTGTGTGGAC CAAAGAAGAGCACCAACCTGGTGAAGAACAAGTGTGTGAAC TTCAACTTCAATGGACTGACAGGCACAGGAGTGCTGACAGA GAGCAACAAGAAGTTCCTGCCATTCCAACAGTTTGGCAGGG ACATTGCTGACACCACAGATGCTGTGAGGGACCCACAGACC TTGGAGATTCTGGACATCACACCATGTTCCTTTGGAGGAGTG TCTGTGATTACACCTGGCACCAACACCAGCAACCAGGTGGC TGTGCTCTACCAGGGCGTGAACTGTACTGAGGTGCCTGTGG CTATCCATGCTGACCAACTTACACCAACCTGGAGGGTCTACA GCACAGGAAGCAACGTGTTCCAGACCAGGG |
| pVP39gB3 | IDT | GCAACGTGTTCCAGACCAGGGCTGGCTGTCTGATTGGAGCA GAGCATGTGAACAACTCCTATGAGTGTGACATCCCAATTGGA GCAGGCATCTGTGCCTCCTACCAGACCCAGACCAACAGCAG GGGCTCTGCATCTTCTGTGGCAAGCCAGAGCATCATTGCCTA CACAATGAGTCTGGGAGCAGAGAACTCTGTGGCTTACAGCA ACAACAGCATTGCCATCCCAACCAACTTCACCATCTCTGTGA CCACAGAGATTCTGCCTGTGAGTATGACCAAGACCTCTGTG GACTGTACAATGTATATCTGTGGAGACAGCACAGAGTGTAG CAACCTGCTGCTCCAATATGGCTCCTTCTGTACCCAACTTAA CAGGGCTCTGACAGGCATTGCTGTGGAACAGGACAAGAACA CCCAGGAGGTGTTTGCCCAGGTGAAGCAGATTTACAAGACA CCTCCAATCAAGGACTTTGGAGGCTTCAACTTCAGCCAGATT CTGCCTGACCCAAGCAAGCCAAGCAAGAGGTCCCCTATTGA GGACCTGCTGTTCAACAAGGTGACCCTGGCTGATGCTGGCT TCATCAAGCAATATGGAGACTGTCTGGGAGACATTGCCGCCA GGGACCTGATTTGTGCCCAGAAGTTCAATGGACTGACAGTG CTGCCTCCACTGCTGACAGATGAGATGATTGCCCAATACAC CTCTGCCCTGCTGGCTGGCACCATCACCTCTGGCTGGACC TTTGGAGCAGGACCAGCCCTCCAAATCCCATTTCCAATGCA GATGGCTTACAGGTTCAATGGCATTGGAGTGACCCAGAATG TGCTCTATGAGAACCAGAAACTGATTGCCAACCAGTTCAAC TCTGCCATTGGCAAGATTCAGGACTCCCTGTCCAGCACAC CATCTGCCCTGGGCAAACTCCAAAACGTGGTGAACCAGAA TGCCCAGGCTCTGAACACCCTGGTGAAGCAACTTTCCAGC AACTTTGGAGCCAT |
| pVP39gB4 | IDT | CCAGCAACTTTGGAGCCATCTCCTCTGTGCTGAATGACATCC TGAGCAGACTGGACCCACCAGAGGCTGAGGTCCAGATTGAC AGACTGATTACAGGCAGACTCCAATCCCTCCAAACCTATGTG ACCCAACAACTTATCAGGGCTGCTGAGATTAGGGCATCTGCC AACCTGGCTGCCACCAAGATGAGTGAGTGTGTGCTGGGACA AAGCAAGAGGGTGGACTTCTGTGGCAAGGGCTACCACCTGA TGAGTTTTCCACAGTCTGCCCCTCATGGAGTGGTGTTCCTGC ATGTGACCTATGTGCCTGCCCAGGAGAAGAACTTCACCACAG CCCCTGCCATCTGCCATGATGGCAAGGCTCACTTTCCAAGGG AGGGAGTGTTTGTGAGCAATGGCACCCACTGGTTTGTGACCC AGAGGAACTTCTATGAACCACAGATTATCACCACAGACAACA CCTTTGTGTCTGGCAACTGTGATGTGGTGATTGGCATTGTGAA CAACACAGTCTATGACCCACTCCAACCTGAACTGGACTCCTTC AAGGAGGAACTGGACAAATACTTCAAGAACCACACCAGCCCT GATGTGGACCTGGGAGACATCTCTGGCATCAATGCCTCTGTG GTGAACATCCAGAAGGAGATTGACAGACTGAATGAGGTGGCT AAGAACCTGAATGAGTCCCTGATTGACCTCCAAGAACTGGGC AAATATGAACAATACATCAAGTGGCCATGGTACATCTGGCTGG GCTTCATTGCTGGACTGATTGCCATTGTGATGGTGACCATAAT GCTGTGTTGTATGACCTCCTGTTGTTCCTGTCTGAAAGGCTGT TGTTCCTGTGGCTCCTGTTGTAAGTTTGATGAGGATGACTCTG AACCTGTGCTGAAAGGAGTGAAACTGCACTACACCTGACTCG AGCTGGTACTGCATGCACGCA |
| pVP35gB1 | IDT | cactatagggagacccaagctggctagccaccATgTTCATCTTCCTGCTGTT CCTGACCCTGACCTCTGGCTCTGACCTGGACAGGTGTACCACC TTTGATGATGTCCAGGCTCCAAACTACACCCAACACACCTCCAG TATGAGGGGAGTCTACTACCCTGATGAGATTTTCAGGTCTGACA CCCTCTACCTGACCCAGGACCTGTTCCTGCCATTCTACAGCAAT GTGACAGGCTTCCACACCATCAACCACACCTTTGACAACCCTG TGATTCCATTCAAGGATGGCATCTACTTTGCTGCCACAGAGAAG AGCAATGTGGTGAGGGGCTGGGTGTTTGGCAGCACAATGAACA ACAAGAGCCAGTCTGTGATTATCATCAACAACAGCACCAATGTG GTGATTAGGGCTTGTAACTTTGAACTGTGTGACAACCCATTCTTT GCTGTGAGCAAGCCTATGGGCACCCAGACCCACACAATGATTT TTGACAATGCCTTCAACTGTACCTTTGAATACATCTCTGATGCCT TCTCCCTGGATGTGTCTGAGAAGTCTGGCAACTTCAAACACCTG AGGGAGTTTGTGTTCAAGAACAAGGATGGCTTCCTCTATGTCTA CAAGGGCTACCAACCAATTGATGTGGTGAGGGACCTGCCATCT GGCTTCAACACCCTGAAACCAATCTTCAAACTGCCACTGGGCA TCAACATCACCAACTTCAGGGCTATCCTGACAGCCTTCAGCCC TGCCCAGGACACCTGGGGCACCTCTGCTGCTGCCTACTTTGT GGGCTACCTGAAACCAACCACCTTTATGCTGAAATATGATGAG AATGGCACCATCACAGATGCTGTGGACTGTAGCCAGAACCCA CTGGCTGAACTGAAATGTTCTGTGAAGTCCTTTGAGATTGACA AGGGCATCTACCAGACCAGCAACTTCAGGGTGGTGCCATCTG GAGATGTGGTGAGGTTTCCAAACATCACCAACCTGTGTCCATT TGGAGAGGTG |
| pVP35gB2 | IDT | TGTCCATTTGGAGAGGTGTTCAATGCCACCAAGTTTCCATCTGT CTATGCCTGGGAGAGGAAGAAGATTAGCAACTGTGTGGCTGA CTACTCTGTGCTCTACAACAGCACCTTCTTCAGCACCTTCAAG TGTTATGGAGTGTCTGCCACCAAACTGAATGACCTGTGTTTCA GCAATGTCTATGCTGACTCCTTTGTGGTGAAGGGAGATGATGT GAGACAGATTGCCCCTGGACAAACAGGAGTGATTGCTGACTA CAACTACAAACTGCCTGATGACTTTATGGGCTGTGTGCTGGCT TGGAACACCAGGAACATTGATGCCACCAGCACAGGCAACTAC AACTACAAATACAGATACCTGAGACATGGCAAACTGAGACCAT TTGAGAGGGACATCAGCAATGTGCCATTCAGCCCTGATGGCA AGCCATGTACTCCTCCTGCCCTGAACTGTTACTGGCCACTGAA TGACTATGGCTTCTACACCACCACAGGCATTGGCTACCAACCA TACAGGGTGGTGGTGCTGTCCTTTGAACTGCTGAATGCCCCTG CCACAGTGTGTGGACCAAAACTGAGCACAGACCTGATTAAGAA CCAGTGTGTGAACTTCAACTTCAATGGACTGACAGGCACAGGA GTGCTGACACCATCCAGCAAGAGGTTCCAACCATTCCAACAGT TTGGCAGGGATGTGTCTGACTTCACAGACTCTGTGAGGGACCC AAAGACCTCTGAGATTCTGGACATCAGCCCATGTTCCTTTGGAG GAGTGTCTGTGATTACACCTGGCACCAATGCCTCCTCTGAGGT GGCTGTGCTCTACCAGGATGTGAACTGTACTGATGTGAGCACA GCCATCCATGCTGACCAACTTACACCTGCCTGGAGGATTTACA GCACAGGCAACAATGTGTTCCAGACCCAGGCTGGCTGTCTGAT TGGAGCAGAGCATGTGGACACCTCCTATGAGTGTGACATCCCA ATTGGAGCAGG |
| pVP35gB3 | IDT | GACATCCCAATTGGAGCAGGCATCTGTGCCTCCTACCACACAGT GTCCCTGCTGAGGAGCACCAGCCAGAAGAGCATTGTGGCTTAC ACAATGAGTCTGGGAGCAGACTCCAGCATTGCCTACAGCAACAA CACCATTGCCATCCCAACCAACTTCAGCATCAGCATCACCACAG AGGTGATGCCTGTGAGTATGGCTAAGACCTCTGTGGACTGTAAT ATGTATATCTGTGGAGACAGCACAGAGTGTGCCAACCTGCTGCT CCAATATGGCTCCTTCTGTACCCAACTTAACAGGGCTCTGTCTG GCATTGCTGCTGAACAGGACAGGAACACCAGGGAGGTGTTTGC CCAGGTGAAGCAGATGTATAAGACACCAACCCTGAAATACTTTG GAGGCTTCAACTTCAGCCAGATTCTGCCTGACCCACTGAAACC AACCAAGAGGTCCTTCATTGAGGACCTGCTGTTCAACAAGGTG ACCCTGGCTGATGCTGGCTTTATGAAGCAATATGGAGAGTGTC TGGGAGACATCAATGCCAGGGACCTGATTTGTGCCCAGAAGTT CAATGGACTGACAGTGCTGCCTCCACTGCTGACAGATGATATG ATTGCTGCCTACACAGCAGCCCTGGTGTCTGGCACAGCCACA GCAGGCTGGACCTTTGGAGCAGGAGCAGCCCTCCAAATCCCA TTTGCTATGCAGATGGCTTACAGGTTCAATGGCATTGGAGTGA CCCAGAATGTGCTCTATGAGAACCAGAAGCAGATTGCCAACCA GTTCAACAAGGCTATCAGCCAGATTCAGGAGTCCCTGACCACC ACCAGCACAGCCCTGGGCAAACTCCAAGATGTGGTGAACCAG AATGCCCAGGCTCTGAACACCCTGGTGAAGCAACTTTCCAGCA ACTTTGGAGCCATCTCCTCTGTGCTGAATGACATCCTGAGCAGA CTGGACAAGGTGGAGGCTGAGGTCCAGATTGACAGACTGATTA CAGGC |
| pVP35gB4 | IDT | TTGACAGACTGATTACAGGCAGACTCCAATCCCTCCAAACCTAT GTGACCCAACAACTTATCAGGGCTGCTGAGATTAGGGCATCTGC CAACCTGGCTGCCACCAAGATGAGTGAGTGTGTGCTGGGACAA AGCAAGAGGGTGGACTTCTGTGGCAAGGGCTACCACCTGATGA GTTTTCCACAGGCTGCCCCTCATGGAGTGGTGTTCCTGCATGTG ACCTATGTGCCAAGCCAGGAGAGGAACTTCACCACAGCCCCTG CCATCTGCCATGAGGGCAAGGCTTACTTTCCAAGGGAGGGAGT GTTTGTGTTCAATGGCACCTCCTGGTTCATCACCCAGAGGAACT TCTTCAGCCCACAGATTATCACCACAGACAACACCTTTGTGTCT GGCAACTGTGATGTGGTGATTGGCATCATCAACAACACAGTCTA TGACCCACTCCAACCTGAACTGGACTCCTTCAAGGAGGAACTG GACAAATACTTCAAGAACCACACCAGCCCTGATGTGGACCTGG GAGACATCTCTGGCATCAATGCCTCTGTGGTGAACATCCAGAA GGAGATTGACAGACTGAATGAGGTGGCTAAGAACCTGAATGAG TCCCTGATTGACCTCCAAGAACTGGGCAAATATGAACAATACAT CAAGTGGCCATGGTATGTGTGGCTGGGCTTCATTGCTGGACTG ATTGCCATTGTGATGGTGACCATCCTGCTGTGTTGTATGACCTC CTGTTGTTCCTGTCTGAAAGGAGCCTGTTCCTGTGGCTCCTGTT GTAAGTTTGATGAGGATGACTCTGAACCTGTGCTGAAAGGAGT GAAACTGCACTACACCTAAggatccgagctcggtaccaagcttaagtttaaa ccgctgatcagcctcgactg |
| pVP35F1 | IDT | tatagggagacccaagctggctagccaccATgTTCATCTTCCTGCTGTTC CTGA |
| pVP37R | IDT | aagcttggtaccgagctcggatccTTAACAACAGGAGCCACAGGAACAG |
| pVP36gB1 | IDT | cactatagggagacccaagctggctagccaccATgATTCACTCTGTGTTCC TGCTGATGTTCCTGCTGACACCAACAGAGTCCTATGTGGATGT GGGACCTGACTCTGTGAAGTCTGCCTGTATTGAGGTGGACAT CCAACAGACCTTCTTTGACAAGACCTGGCCAAGACCAATTGAT GTGAGCAAGGCTGATGGCATCATCTACCCACAGGGCAGGAC CTACAGCAACATCACCATCACCTACCAGGGACTGTTTCCATA CCAGGGAGATCATGGAGATATGTATGTCTACTCTGCTGGTCA TGCCACAGGCACCACACCACAGAAACTGTTTGTGGCTAACTA CAGCCAGGATGTGAAGCAGTTTGCCAATGGCTTTGTGGTGAG GATTGGAGCAGCAGCCAACAGCACAGGCACAGTGATTATCA GCCCAAGCACCTCTGCCACCATCAGGAAGATTTACCCTGCCT TTATGCTGGGCTCCTCTGTGGGCAACTTCTCTGATGGCAAGA TGGGCAGGTTCTTCAACCACACCCTGGTGCTGCTGCCTGAT GGCTGTGGCACCCTGCTGAGGGCTTTCTACTGTATCTTGGAA CCAAGGTCTGGCAACCACTGTCCTGCTGGCAACTCCTACAC CTCCTTTGCCACCTACCACACACCTGCCACAGACTGTTCTGA TGGCAACTACAACAGGAATGCCTCCCTGAACTCCTTCAAGGA ATACTTCAACCTGAGGAACTGTACCTTTATGTACACCTACAAC ATCACAGAGGATGAGATTTTGGAGTGGTTTGGCATCACCCAG ACAGCCCAGGGAGTGCATCTGTTCTCGAGCAGATATGTGGA CCTCTATGGAGGCAATATGTTCCAGTTTGCCACCCTGCCTGT CTATGACACCATCAAATACTACAGCATCATCCCACACAGCAT CAGGAGCATCCAGTCTGACAGGAAGGCTTGGGCTGCCTTCT ATGTCTACAAACTCCAACCACTGACCTT |
| pVP36gB2 | IDT | ACTCCAACCACTGACCTTCCTGCTGGACTTCTCTGTGGATGG CTACATCAGGAGGGCTATTGACTGTGGCTTCAATGACCTGAG CCAACTTCACTGTTCCTATGAGTCCTTTGATGTGGAGTCTGGA GTCTACTCTGTGTCCTCCTTTGAGGCTAAGCCATCTGGCTCT GTGGTGGAACAGGCTGAGGGAGTGGAGTGTGACTTCAGCCC ACTGCTGTCTGGCACACCTCCACAGGTCTACAACTTCAAGAG ACTGGTGTTCACCAACTGTAACTACAACCTGACCAAACTGCT GTCCCTGTTCTCTGTGAATGACTTCACTTGTAGCCAGATTAGC CCTGCTGCCATTGCCAGCAACTGTTACTCCTCCCTGATTCTG GACTACTTCTCCTACCCACTGAGTATGAAGTCTGACCTGTCT GTGTCCTCTGCTGGACCAATCAGCCAGTTCAACTACAAGCAG TCCTTCAGCAACCCAACTTGTCTGATTCTGGCTACAGTGCCA CACAACCTGACCACCATCACCAAGCCACTGAAATACTCCTAC ATCAACAAGTGTAGCAGACTGCTGTCTGATGACAGGACAGAG GTGCCACAACTAGTGAATGCCAACCAATACAGCCCATGTGTG AGCATTGTGCCAAGCACAGTGTGGGAGGATGGAGACTACTA CAGGAAGCAACTTAGCCCATTGGAGGGAGGAGGCTGGCTGG TGGCATCTGGCAGCACAGTGGCTATGACAGAACAACTCCAAA TGGGCTTTGGCATCACAGTCCAATATGGCACAGACACCAACT CTGTGTGTCCAAAATTGGAGTTTGCCAATGACACCAAGATTGC CAGCCAACTTGGCAACTGTGTGGAATACTCCCTCTATGGAGT GTCTGGCAGGGGAGTGTTCCAGAACTGTACTGCTGTGGGAG TGAGACAACAGAGGTTTGTCTATGATGCCTACCAGAACCTGG TGGGCTACTACTCTGATGATGGCAACTACTACTGT |
| pVP36gB3 | IDT | GATGATGGCAACTACTACTGTCTGAGGGCTTGTGTGTCTGTGC CTGTGTCTGTGATTTATGACAAGGAGACCAAGACCCATGCCAC CCTGTTTGGCTCTGTGGCTTGTGAACACATCTCCAGCACAATG AGTCAATACAGCAGGAGCACCAGGAGTATGCTGAAAAGGAGG GACAGCACATATGGACCACTCCAAACACCTGTGGGCTGTGTG CTGGGACTGGTGAACTCCTCCCTGTTTGTGGAGGACTGTAAA CTGCCACTGGGACAATCCCTGTGTGCCCTGCCTGACACACCA AGCACCCTGACACCAAGGTCTGTGAGGTCTGTGCCTGGAGAG ATGAGACTGGCAAGCATTGCCTTCAACCACCCAATCCAGGTG GACCAACTTAACTCCTCCTACTTCAAACTGAGCATCCCAACCA ACTTCTCCTTTGGAGTGACCCAGGAATACATCCAGACCACCAT CCAGAAGGTGACAGTGGACTGTAAGCAATATGTGTGTAATGG CTTCCAGAAGTGTGAACAACTTCTGAGGGAATATGGACAATTC TGTAGCAAGATAAACCAGGCTCTTCATGGAGCCAACCTGAGA CAGGATGACTCTGTGAGGAACCTGTTTGCCTCTGTGAAGTCC AGCCAGTCCAGCCCAATCATCCCTGGCTTTGGAGGAGACTT CAACCTGACCCTGTTGGAACCGGTGAGCATCAGCACAGGCA GCAGGTCTGCCAGGTCTGCCATTGAGGACCTGCTGTTTGACA AGGTGACCATTGCTGACCCTGGCTATATGCAGGGCTATGATG ACTGTATGCAACAGGGACCTGCCTCTGCCAGGGACCTGATTT GTGCCCAATATGTGGCTGGCTACAAGGTGCTGCCTCCACTGA TGGATGTGAATATGGAGGCTGCCTACACCTCCTCCCTGCTGG GCAGCATTGCTGGAGTGGGCTGGACTGCAGGACTGTCCTCC TTTGCTGCCATCCCATTTGCCCAG |
| pVP36gB4 | IDT | TGCCATCCCATTTGCCCAGAGCATCTTCTACAGACTGAATGGA GTGGGCATCACCCAACAGGTGCTGTCTGAGAACCAGAAACTG ATTGCCAACAAGTTCAACCAGGCTCTGGGAGCTATGCAGACA GGCTTCACCACCACCAATGAGGCTTTCCAGAAGGTCCAGGAT GCTGTGAACAACAATGCCCAGGCTCTGAGCAAACTGGCATCT GAACTGAGCAACACCTTTGGAGCCATCTCTGCTAGCATTGGA GACATCATCCAGAGACTGGATGTGTTGGAACAGGATGCCCAG ATTGACAGACTGATAAATGGCAGACTGACCACCCTGAATGCC TTTGTGGCTCAACAACTTGTGAGGTCTGAGTCTGCTGCCCTG TCTGCCCAACTTGCCAAGGACAAGGTGAATGAGTGTGTGAAG GCTCAAAGCAAGAGGTCTGGCTTCTGTGGACAAGGCACCCA CATTGTGTCCTTTGTGGTGAATGCCCCAAATGGACTCTACTTT ATGCATGTGGGCTACTACCCAAGCAACCACATTGAGGTGGTG TCTGCCTATGGACTGTGTGATGCTGCCAACCCAACCAACTGT ATTGCCCCTGTGAATGGCTACTTCATCAAGACCAACAACACC AGGATTGTGGATGAGTGGTCCTACACAGGCTCCTCCTTCTAT GCCCCTGAACCAATCACCTCCCTGAACACCAAATATGTGGCT CCACAGGTGACCTACCAGAACATCAGCACCAACCTGCCTCC TCCACTGCTGGGCAACAGCACAGGCATTGACTTCCAGGATG AACTGGATGAGTTCTTCAAGAATGTGAGCACCAGCATCCCAA ACTTTGGCTCCCTGACCCAGATAAACACCACCCTGCTGGAC CTGACCTATGAGATGCTGTCCCTCCAACAGGTGGTGAAGGC TCTGAATGAGTCCTACATTGACCTGAAAGAACTGGGCAACTA CACCTACTACAACAAGTGGCCATGGTACATCTGGCTGGGCT TCATCGCTGGCCTGGTGGCCCTGGCGCTGTGCGTGTTCTTC ATCCTGTGCTGCACCGGCTGCGGCACCAACTGCATGGGCA AGCTGAAGTGCAACAGGTGCTGCGACAGGTACGAGGAGTA CGACCTGGAGCCCCACAAGGTGCACGTACATTAAggatccga gctcggtaccaagcttaagtttaaaccgctgatcagcctcgactg |
| pVP36F | IDT | tatagggagacccaagctggctagccaccATgATTCACTCTGTGTTCCT GCTGA |
| pVP38R | IDT | agcttggtaccgagctcggatccTTAGCAGCACCTGTTGCACTTCAG CTTG |
| del19R1 | IDT | cttaagcttggtaccgagctcggatccTCAACAACAGGAGCCACAGGA ACAAC |
| pVP30F1 | IDT | AGACTGGACAAGGTGGAGGCTGAGGTCCAGATTGACAGACTGA TTACAG |
| pVP40gB1 | IDT | tagggagacccaagctggctagccaccATGTTTGTGTTCCTGGTGCTGCT GCCACTGGTGTCCAGCCAGTGTGTGAACCTGAGGACCAGGAC CCAACTTCCTCCTGCCTACACCAACTCCTTCACCAGGGGAGTC TACTACCCTGACAAGGTGTTCAGGTCCTCTGTGCTGCACAGCA CCCAGGACCTGTTCCTGCCATTCTTCAGCAATGTGACCTGGTT CCATGCCATCCATGTGTCTGGCACCAATGGCACCAAGAGGTT TGACAACCCTGTGCTGCCATTCAATGATGGAGTCTACTTTGCC AGCACAGAGAAGAGCAACATCATCAGGGGCTGGATTTTTGGC ACCACCCTGGACAGCAAGACCCAGTCCCTGCTGATTGTGAAC AATGCCACCAATGTGGTGATTAAGGTGTGTGAGTTCCAGTTCT GTAATGACCCATTCCTGGACGTCTACTACCACAAGAACAACAA GTCCTGGATGGAGTCTGGCGTCTACTCCTCTGCCAACAACTG TACCTTTGAATATGTGAGCCAACCATTCCTGATGGACTTGGAG GGCAAGCAGGGCAACTTCAAGAACCTGAGGGAGTTTGTGTTC AAGAACATTGATGGCTACTTCAAGATTTACAGCAAACACACAC CAATCAACCTGGTGAGGGACCTGCCACAGGGCTTCTCTGCCT TGGAACCACTGGTGGACCTGCCAATTGGCATCAACATCACCA GGTTCCAGACCCTGCTGGCTCTGCACAGGTCCTACCTGACAC CTGGAGACTCCTCCTCTGGCTGGACAGCAGGAGCAGCAGCC TACTATGTGGGCTACCTCCAACCAAGGACCTTCCTGCTGAAA TACAATGAGAATGGCACCATCACAGATGCTGTGGACTGTGCC CTGGACCCACTGTCTGAGACCAAGTGTACCCTGAAATCCTTC ACAGTGGAGAAGGGCATCTAC |
| pVP40gB2 | IDT | AGTGGAGAAGGGCATCTACCAGACCAGCAACTTCAGGGTCC AACCAACAGAGAGCATTGTGAGGTTTCCAAACATCACAAACC TGTGTCCATTTGGAGAGGTGTTCAATGCCACCAGGTTTGCCT CTGTCTATGCCTGGAACAGGAAGAGGATTAGCAACTGTGTG GCTGACTACTCTGTGCTCTACAACTCTGCCTCCTTCAGCAC CTTCAAGTGTTATGGAGTGAGCCCAACCAAACTGAATGACC TGTGTTTCACCAATGTCTATGCTGACTCCTTTGTGATTAGGG GAGATGAGGTGAGACAGATTGCCCCTGGACAAACAGGCAA GATTGCTGACTACAACTACAAACTGCCTGATGACTTCACAG GCTGTGTGATTGCCTGGAACAGCAACAACCTGGACAGCAA GGTGGGAGGCAACTACAACTACAGGTACAGACTGTTCAGG AAGAGCAACCTGAAACCATTTGAGAGGGACATCAGCACAG AGATTTACCAGGCTGGCAGCAAGCCATGTAATGGAGTGGA GGGCTTCAACTGTTACTTTCCACTCCAATCCTATGGCTTCC AACCAACCAATGGAGTGGGCTACCAACCATACAGGGTGGT GGTGCTGTCCTTTGAACTGCTCCATGCCCCTGCCACAGTG TGTGGACCAAAGAAGAGCACAAACCTGGTGAAGAACAAGT GTGTGAACTTCAACTTCAATGGACTGACAGGCACAGGAGT GCTGACAGAGAGCAACAAGAAGTTCCTGCCATTCCAACAG TTTGGCAGGGACATTGCTGACACCACAGATGCTGTGAGGG ACCCACAGACCTTGGAGATTCTGGACATCACACCATGTTC CTTTGGAGGAGTGTCTGTGATTACACCTGGCACCAACACC AGCAACCAGGTGGCTGTGCTCTACCAGGGCGTGAACTGT ACTGAGGTGCCTGTGGCTATCCATGCTGACCAACTTACC CCAACCTG |
| pVP40gB3 | IDT | GACCAACTTACCCCAACCTGGAGGGTCTACAGCACAGGC AGCAACGTGTTCCAGACCAGGGCTGGCTGTCTGATTGGA GCAGAGCATGTGAACAACTCCTATGAGTGTGACATCCCA ATTGGAGCAGGCATCTGTGCCTCCTACCAGACCCAGACC AACAGCAGGAGGAGGGCAAGGTCTGTGGCAAGCCAGAG CATCATTGCCTACACAATGAGTCTGGGAGCAGAGAACTC TGTGGCTTACAGCAACAACAGCATTGCCATCCCAACCAAC TTCACCATCTCTGTGACCACAGAGATTCTGCCTGTGAGTA TGACCAAGACCTCTGTGGACTGTACAATGTATATCTGTGG AGACAGCACAGAGTGTAGCAACCTGCTGCTCCAATATGG CTCCTTCTGTACCCAACTTAACAGGGCTCTGACAGGCATT GCTGTGGAACAGGACAAGAACACCCAGGAGGTGTTCGCC CAAGTGAAGCAGATTTACAAGACACCTCCAATCAAGGACT TTGGAGGCTTCAACTTCAGCCAGATTCTGCCTGACCCAAG CAAGCCAAGCAAGAGGTCCTTCATTGAGGACCTGCTGTTC AACAAGGTGACCCTGGCTGATGCTGGCTTCATCAAGCAAT ATGGAGACTGTCTGGGAGACATTGCCGCCAGGGACCTGA TTTGTGCCCAGAAGTTCAATGGACTGACAGTGCTGCCTCC ACTGCTGACAGATGAGATGATTGCCCAATACACCTCTGCC CTGCTGGCTGGCACCATCACCTCTGGCTGGACCTTTGGA GCAGGAGCAGCCCTCCAAATCCCATTTGCTATGCAGATG GCTTACAGGTTCAATGGCATTGGAGTGACCCAGAATGTG CTCTATGAGAACCAGAAACTGATTGCCAACCAGTTCAACT CTGCCATTGGCAAGATTCAGGACTCCCTGTCCAGCACAG CCTCTGCCCTGGGCAAGCTCCAAAACGTGGTGAACCAG AATGCCCAGGCTCTGAACACCCTGGTGAAGCAACTTTCC AGCAACTTTGGAGCCATCTCCTCTGTGCTGAATGACATC CTGAGCAGACTGGACAAGGTGGAGGCTGAGGTCCAGAT TGACAG |
| Recombinant DNA | ||
| pcDNA3.1 | Addgene | Cat# V790-20 |
| pHIVNLGagPol | Schmidt et al., 2020 | Gift from Dr Bieniasz’ lab |
| pCCNanoLuc2AEGFP | Schmidt et al., 2020 | Gift from Dr Bieniasz’ lab |
| pCCNanoLuc2AEGFP plasmid | Schmidt et al. | Gift from Dr Bieniasz’ lab |
| pVP39 (SARS-CoV-2 B.1.61.72 variant (6P)) | This study | Sidi Chen Lab |
| pVP33b (SARS-CoV (6P)) | This study | Sidi Chen Lab |
| pVP34c (MERS-CoV (6P)) | This study | Sidi Chen Lab |
| pVP31b (WT spike (6P) | This study | Sidi Chen Lab |
| SARS-CoV-2 Delta plasmid | This study | Sidi Chen Lab |
| SARS-CoV plasmid | This study | Sidi Chen Lab |
| MERS-CoV plasmid | This study | Sidi Chen Lab |
| Software and algorithms | ||
| FlowJo software 9.9.6 | FlowJo, LLC | https://www.flowjo.com |
| GraphPad Prism 8.0 | GraphPad Software Inc | https://www.graphpad.com/scientific-software/prism/ |
| Pymol | Schrödinger | http://www.pymol.org/ |
| Cell Ranger v3.1.0 | 10X Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/installation |
| Loupe V(D)J Browser | 10X Genomics | https://support.10xgenomics.com/single-cell-vdj/software/visualization/latest/installation |
| Trimmomatic | Bolger et al., Bioinformatics, 2014 | https://github.com/timflutre/trimmomatic |
| mixcr | Bolotin et al., Nat Methods, 2015 | https://github.com/milaboratory/mixcr |
| R | R project | https://www.r-project.org |
| Seurat R package | Satija et al., 2015 | https://satijalab.org/seurat/index.html |
| plyr R package | Wickham (2011). Journal of Statistical Software | http://www.jstatsoft.org/v40/i01/ |
| dplyr R package | Wickham et al. (2022). dplyr: A Grammar of Data Manipulation. R package version 1.0.9 | https://CRAN.R-project.org/package=dplyr |
| patchwork R package | Pedersen (2020). patchwork: The Composer of Plots. R package version 1.1.1 | https://CRAN.R-project.org/package=patchwork |
| ggplot2 R package | Wickham. (2016). ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York | https://ggplot2.tidyverse.org |
| ggrepel R package | Slowikowski (2021). ggrepel: Automatically Position Non-Overlapping Text Labels with 'ggplot2'. R package version 0.9.1 | https://CRAN.R-project.org/package=ggrepel |
| limma R package | Ritchie et al. (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 43(7), e47 | https://git.bioconductor.org/packages/limma |
| edgeR R package | Robinson et al., Bioinformatics 2010; McCarthy et al., Nucleic Acid Research 2012 |
https://git.bioconductor.org/packages/edgeR |
| stringr R package | Wickham (2019). stringr: Simple, Consistent Wrappers for Common String Operations. R package version 1.4.0 | https://CRAN.R-project.org/package=stringr |
| ggridges R package | Wilke (2021). ggridges: Ridgeline Plots in 'ggplot2'. R package version 0.5.3 | https://CRAN.R-project.org/package=ggridges |
| igraph R package | Csardi, G., & Nepusz, T. (2006). The Igraph Software Package for Complex Network Research. InterJournal 2006, Complex Systems, 1695. | https://igraph.org |
| network R package | Butts (2008). network: a Package for Managing Relational Data in R. Journal of Statistical Software, 24 (2) | https://CRAN.R-project.org/package=network |
| sna R package | Carter T. Butts (2020). sna: Tools for Social Network Analysis. R package version 2.6 | https://CRAN.R-project.org/package=sna |
| Immunarch R package | ImmunoMind Team, Zenodo, 2019 | https://github.com/immunomind/immunarch |
| Circlize R package | Gu et al., Bioinformatics, 2014 | https://cran.r-project.org/package=circlize |
| Pheatmap R package | Kolde (2019) | https://cran.r-project.org/package=pheatmap |
| Future R package | Bengtsson (2021) | https://cran.r-project.org/package=future |
| SeuratWrappers R package | Satija et al. (2020) | https://github.com/satijalab/seurat-wrappers |
| glmGamPoi R package | Ahlmann-Eltze and Huber, Bioinformatics, 2021 | https://github.com/const-ae/glmGamPoi |
| Other | ||
| SARS-CoV-2 Delta-LNP-mRNA vaccine candidate | This study | Sidi Chen Lab |
| SARS-CoV-LNP-mRNA vaccine candidate | This study | Sidi Chen Lab |
| MERS-CoV-LNP-mRNA vaccine candidate | This study | Sidi Chen Lab |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sidi Chen (sidi.chen@yale.edu).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact. Certain materials such as vaccine candidates will be shared with a completed Materials Transfer Agreement.
Experimental model and subject details
Institutional approval
This study has received institutional regulatory approval. All recombinant DNA (rDNA) and biosafety work were performed under the guidelines of Yale Environment, Health and Safety (EHS) Committee with approved protocols (Chen-15-45, 18-45, 20-18, 20-26). All animal work was performed under the guidelines of Yale University Institutional Animal Care and Use Committee (IACUC) with approved protocols (Chen-2018-20068; Chen-2020-20358; Chen 2021-20068).
Animals
M. musculus (mice), 6–8 weeks old females of C57BL/6Ncr were purchased from Charles River. M. musculus (mice) used for immunogenicity study. Animals were housed in individually ventilated cages in a dedicated vivarium with clean food, water, and bedding. Animals are housed with a maximum of 5 mice per cage, at regular ambient room temperature (65–75°F, or 18–23°C), 40–60% humidity, and a 14 h:10 h light cycle. All experiments utilize randomized littermate controls.
Cell line
HEK293T (ThermoFisher), Huh-7 and 293T-hACE2 (Dr Bieniasz’ lab) cell lines were cultured in complete growth medium, Dulbecco’s modified Eagle’s medium (DMEM; ThermoFisher) supplemented with 10% Fetal bovine serum (FBS, Hyclone), 1% penicillin-streptomycin (Gibco) (D10 media for short). Cells were typically passaged every 1–2 days at a split ratio of 1:2 or 1:4 when the confluency reached about 80%.
Mouse immunization
6–8 weeks old female C57BL/6Ncr (B6) mice were purchased from Charles River and used for vaccine immunogenicity study. Animals were housed in individually ventilated cages in a dedicated vivarium with clean food, water, and bedding. A maximum of 5 mice was allowed in each cage, at regular ambient room temperature (65–75°F, or 18–23°C), 40–60% humidity, and a 14 h:10 h day/night cycle. All experiments utilize randomized littermate controls. A standard two-dose schedule given 21 days apart was adopted (Polack et al., 2020), unless otherwise noted. Three sets of immunization experiments were performed: Triplex dosage testing, MERS Duplex testing and Schedule comparison testing.
For the Triplex dosage testing experiment, 1 μg Delta LNP-mRNA, 1 μg or 3 μg Triplex-CoV LNP-mRNA (equal mass mixture of Delta, MERS and SARS mRNA) were diluted to the same volume with 1X PBS and inoculated into mice intramuscularly during prime and boost.
For the MERS Duplex testing experiment, 3 μg MERS LNP-mRNA, 3 μg equal-mass mRNA mixture of MERS+SARS or MERS+Delta spikes at same concentration were inoculated into mice intramuscularly during prime and boost.
For the Schedule comparison testing experiment, 1 μg Delta, MERS and SARS LNP-mRNA were sequentially inoculated into mice during prime and boost.
Control mice received 50 μL PBS at prime and boost at the same matched time points in all experiments.
Method details
Coronavirus spike sequence alignment
The spike sequence used to produce the LNP-mRNA vaccines were aligned using Clustal Omega (Goujon et al., 2010) and visualized in Jalview (Waterhouse et al., 2009).
Plasmid construction
The spike cDNA of SARS-CoV (Genbank accession AAP13567.1) and MERS-CoV (Genbank accession AFS88936.1) were purchased from Sino Biological (Cat # VG40150-G-N and VG40069-G-N, respectively). cDNA of SARS-CoV-2 B.1.617.2 (Delta variant) (Liu et al., 2021) were synthesized as gBlocks (IDT). The spike sequences were cloned by Gibson Assembly (NEB) into pcDNA3.1 plasmid for the mRNA transcription and pseudovirus assay. The plasmids for the pseudotyped virus assay including pHIVNLGagPol and pCCNanoLuc2AEGFP are gifts from Dr. Bieniasz’ lab (Schmidt et al., 2020). The C-terminal 19 (for SARS-CoV and SARS-CoV-2) or 16 (for MERS-CoV) amino acids were deleted in the spike sequence for the pseudovirus assay. To improve expression and retain prefusion conformation, six prolines (HexaPro variant, 6P) (Wrapp et al., 2020) were introduced to the SARS-CoV-2, SARS-CoV and MERS-CoV spike sequence at the homologous sites in the mRNA transcription plasmids. The furin site of SARS-CoV-2 spike (RRAR) were replaced with a GSAS short stretch to keep S1 and S2 subunits connected in the spike.
In vitro mRNA transcription and vaccine formulation
Codon-optimized mRNA encoding HexaPro spikes of SARS-CoV-2 WT, Delta, SARS-CoV and MERS-CoV were synthesized in vitro using an Hiscribe™ T7 ARCA mRNA Kit (with tailing) (NEB, Cat # E2060S), with 50% replacement of uridine by N1-methyl-pseudouridine. A linearized DNA template containing the spike open reading frame flanked by 5′ untranslated region (UTR), 3′ UTR and 3′-end polyA tail was used as for mRNA transcription. The linearization of DNA templates was achieved by digesting circular plasmids with BbsI restriction enzyme, followed by gel purification.
The mRNA was synthesized and purified by following the manufacturer’s instructions and kept frozen at −80°C until further use. In brief, the synthesized mRNA was purified by spin column-based method using Monarch RNA cleanup kit (NEB, Cat No. T2040L). The mRNA was encapsulated in lipid nanoparticles using the NanoAssemblr® Ignite™ machine (Precision Nanosystems). For the MixCoV vaccine, equal mass of SARS, MERS and Delta spike mRNA were mixed before encapsulated by lipid nanoparticles. All procedures are following the guidance of manufacturers. In brief, GenVoy ILM lipid mixture was mixed with transcribed mRNA in the low pH formulation buffer 1 on Ignite instrument at a molar ratio of 6:1 (LNP: mRNA), similar to previously described (Corbett et al., 2020; Hassett et al., 2019). The GenVoy ILM contains 50% PNI ionizable lipids, 10% DSPC, 37.5% cholesterol and 2.5% PNI stabilizer. The LNP encapsulated mRNA was buffer exchanged to PBS using 30kDa Amicon filter (MilliporeSigma™ UFC901024). Sucrose was added as a cryoprotectant. The particle size of mRNA-LNP was determined by DLS machine (DynaPro NanoStar, Wyatt, WDPN-06) and TEM described below. The encapsulation rate and mRNA concentration were measured by Quant-iT™ RiboGreen™ RNA Assay (ThermoFisher).
In vitro mRNA expression and receptor binding validation of translated spikes
HEK293T cells were electroporated with mRNA encoding SARS, MERS or Delta spikes using Neon™ Transfection System 10 μL Kit following the standard protocol provided by manufacturer. After 12 h, the cells were collected and resuspended. To detect surface-protein expression, the cells were stained with ACE2–Fc chimera (Genscript, Z03484) or DPP4-Fc (Sino Biological, 10688-H01H) in MACS buffer (D-PBS with 2 mM EDTA and 0.5% BSA) for 30 min on ice. Thereafter, cells were washed twice and incubated with PE–anti-human FC antibody (Biolegend, 410708) in MACS buffer for 30 min on ice. Data acquisition was performed on BD FACSAria II Cell Sorter (BD). Analysis was performed using FlowJo software.
Negative-stain TEM
5 μL of the sample was deposited on a glow-discharged formvar/carbon-coated copper grid (Electron Microscopy Sciences, catalog number FCF400-Cu-50), incubated for 1 min and blotted away. The grid was washed briefly with 2% (w/v) uranyl formate (Electron Microscopy Sciences, catalog number 22450) and stained for 1 min with the same uranyl formate buffer. Images were acquired using a JEOL JEM-1400 Plus microscope with an acceleration voltage of 80 kV and a bottom-mount 4k × 3k charge-coupled device camera (Advanced Microscopy Technologies, AMT).
Sample collection, plasma and PBMCs isolation
At the defined time points, usually two weeks post the last dose of boost unless otherwise noted (e.g., day 35, or day 119, as noted in the schematics), blood was retro-orbitally collected from mice. The PBMCs and plasma were isolated from blood via SepMate-15 (StemCell Technologies). 200 μL blood was immediately diluted with 800 ul PBS with 2% FBS. The diluted blood was then added to SepMate-15 tubes with 5mL Lymphoprep (StemCell Technologies). 1200 x g centrifugation for 20 minutes was applied to isolate RBCs, PBMCs and plasma. 200ul diluted plasma was collected from the surface layer. Then the solution at the top layer containing PBMCs was poured to a new tube. PBMCs were washed once with PBS + 2% FBS before being used in downstream analysis. The separated plasma was used in ELISA and neutralization assay. PBMCs were collected for single cell profiling using a 10xGenomics platform.
ELISA
The 384-well ELISA plates were coated with 3 μg/mL of antigens overnight at 4 degree. The antigen panel used in the ELISA assay includes SARS-CoV-2 spike S1+S2 ECD and RBD of 2019-nCoV WT (Sino Biological, ECD 40589-V08B1 and RBD 40592-V08B), Delta variant B.1.617.2 (SINO, ECD 40589-V08B16 and RBD 40592-V08H90), SARS-CoV (ECD Sino Biological 40634-V08B and RBD Fisher 50-196-4017) and MERS-CoV (ECD Sino Biological and RBD Fisher 50-201-9463). Plates were washed with PBS plus 0.5% Tween 20 (PBST) three times using the 50TS microplate washer (Fisher Scientific, NC0611021) and blocked with 0.5% BSA in PBST at room temperature for one hour. Plasma was serially diluted twofold or fourfold starting at a 1:500 dilution. Samples were added to the coated plates and incubate at room temperature for one hour, followed by washes with PBST five times. Anti-mouse secondary antibody (Fisher, Cat# A-10677) was diluted to 1:2500 in blocking buffer and incubated at room temperature for one hour. Plates were washed five times and developed with tetramethylbenzidine substrate (Biolegend, 421101). The reaction was stopped with 1 M phosphoric acid, and OD at 450 nm was determined by multimode microplate reader (PerkinElmer EnVision 2105). The binding response (OD450) were plotted against the dilution factor in log10 scale to display the dilution-dependent response. The area under curve of the dilution-dependent response (Log10 AUC) was calculated to evaluate the potency of the serum antibody binding to spike antigens.
Blocking ELISA
0.6 μg/mL ECDs of Delta (Sino 40589-V08B16), MERS (40069-V08B) and SARS (Sino 40634-V08B) were coated to 384-well plate at 4 degree overnight. Low-density antigen was coated in blocking ELISA to ensure the blocking effect can be observed. The coated plate was then washed with PBST (0.5% Tween-20) three times and blocked with 2% BSA in PBST for 1 hour at room temperature. Equal volume of blocking agents at 5 μg/mL was mixed with serially diluted plasma and incubated at room temperature for 30 min before added to the plate. The blocking agents include PBS as negative control, Delta ECD, SARS ECD or MERS ECD. The conditions used in blocking ELISA was based on the optimized competition ELISA conditions in our previous study(Fang et al., 2022). After 1 hour incubation with the plasma and blocking agents, the plate was washed with PBST 5 times and incubated with anti-mouse secondary antibody (Fisher, Cat# A-10677) for 1 hour. Then the plate was washed five times with PBST, developed with tetramethylbenzidine substrate and fixed with 1M phosphoric acid. The OD450 was quantified by multimode microplate reader (PerkinElmer EnVision 2105). The normalized blocking effect was calculated by normalizing the AUC reduction by blocking reagents with AUC difference between plasma samples of PBS and vaccination groups.
Pseudovirus neutralization assay
HIV-1 based SARS-CoV-2 WT, B.1.617.2 (delta) variant, SARS and MERS pseudotyped virions were generated using corresponding spike sequences, and applied in neutralization assays. The pseudotyped virus was packaged using a coronavirus spike plasmid, a reporter vector and a HIV-1 structural protein expression plasmid. The reporter vector, pCCNanoLuc2AEGFP, and HIV-1 structural/regulatory proteins (pHIVNLGagPol) expression plasmid were from Bieniasz lab. The spike plasmid for SARS-CoV-2 WT pseudovirus truncated 19 C-terminal amino acids of S protein (SARS-CoV-2-Δ19) and was from Bieniasz lab. Spike plasmids expressing C-terminally truncated SARS-CoV-2 B.1.617.2 variant S protein (Delta variant-Δ19), SARS-CoV S protein (SARS-CoV-Δ19) and MERS S protein (MERS-CoV-Δ16) were generated based on the pSARS-CoV-2-Δ19. Briefly, 293T cells were seeded in 150 mm plates, and transfected with 21 μg pHIVNLGagPol, 21 μg pCCNanoLuc2AEGFP, and 7.5 μg of corresponding spike plasmids, in the presence of 198 μL PEI. At 48 h after transfection, the 20-mL supernatant was harvested and filtered through a 0.45-μm filter, and concentrated before aliquoted and frozen in −80°C.
The SARS-CoV and SARS-CoV-2 pseudovirus neutralization assays were performed on 293T-hACE2 cell, while the MERS-CoV neutralization assay was performed on Huh-7 cells. One day before infection, 293T-hACE2 cells were plated in a 96 well plate with 0.01 × 106 cells per well. In the next day, plasma collected from PBS or LNP-mRNA immunized mice were 5-fold serially diluted with complete growth medium starting from 1:100. 55 μL aliquots of diluted plasma were mixed with the same volume of SARS-CoV-2 WT, Delta variant, SARS or MERS pseudovirus. The mixture was incubated for 1 hr in the 37°C incubator, supplied with 5% CO2. Then 100 μL of mixtures were added into 96-well plates with 293T-hACE2 or Huh-7 cells. Plates were incubated at 37°C for 48 hr. Then host cells were collected and the percent of GFP-positive cells were analyzed with Attune NxT Acoustic Focusing Cytometer (ThermoFisher). The 50% inhibitory concentration (IC50) was calculated with a four-parameter logistic regression using GraphPad Prism (GraphPad Software Inc.). If the curve of individual mouse fails to produce positive fit (i.e. negative titer), suggestive of no neutralization activity, the value was converted to zero.
Authentic virus neutralization assay
Mouse plasma samples were serially diluted, then incubated with SARS-CoV-2 isolate USA-WA1/2020 for 1 h at 37°C. Vero-E6 overexpressing ACE2/TMPRSS2 was added to the plasma/virus mixture such that the final MOI was 1. Cell viability was measured at 72 hpi using CellTiter Glo.
Correlation analysis
Correlation analysis of ELISA, pseudovirus neutralization and authentic virus neutralization data were performed using the respective data collected. Linear regression model was used to evaluate the correlations between ELISA RBD and ECD AUCs, pseudovirus neutralization and authentic virus neutralization log10 IC50. Model fitting and statistical analysis were performed in Graphpad Prism9.1.2. Correlations of data points from either individual mouse, or group average of different vaccination groups, were analyzed separately. The vaccination-group ELISA AUC or neutralization log10 IC50 were calculated from the average of individual value in each group. Due to assay-dependent PBS background level, only non-PBS data points were included in the correlation analysis.
Single cell RNA-seq
PBMCs were collected from mRNA-LNP vaccinated and control mice were collected as described above for mouse immunization and sample collection, and normalized to 1000 cells/μL. Standard volumes of cell suspension were loaded to achieve targeted cell recovery to 10000 cells. The samples were subjected to 14 cycles of cDNA amplification. Following this, gene expression (GEX) libraries were prepared according to the manufacturer’s protocol (10x Genomics). All libraries were sequenced using a NovaSeq 6000 (Illumina) with 2∗150 read length.
Single cell data analysis for immune repertoire profiling and transcriptomic signatures
Both standard established pipelines and custom scripts were used for processing and analyzing single cell GEX data. Illumina sequencing data were processed using the Cellranger v6.0.1 (10x Genomics) pipeline, aligning reads to the mm10 reference transcriptome and aggregating all samples. Cellranger outputs were then preprocessed using a modified Seurat v4.0.5 workflow with the R statistical programming language(Satija et al., 2015). Briefly, individual sample data sets were filtered for quality cells (200–2000 RNA features and <5% mitochondrial RNA), log-normalized, scaled, and quality features were selected to calculate low-dimensional “anchors” (reciprocal-PCR dimensional reduction, k = 20, anchors = 2000), which were used to integrate the different sample data sets(Stuart et al., 2019). Integrated single-cell data were scaled, centered, clustered by shared nearest neighbors graph (k = 20, first 12 PCA dimensions, chosen by the elbow plot method) with modularity optimization (Louvain algorithm with multilevel refinement, empirically chosen resolution = 0.31). Clustered cells were visualized in low-dimensional space by uniform manifold approximation and projection (UMAP; first 12 PCA dimensions) (McInnes et al., 2018), and clusters were labeled as immune cell types via canonical marker expression, based on scaled-mean expression and expression detection rate for the cluster. Immune cell subtypes were identified for B cells, plasma cells, activated CD4 T cells, and mononuclear myeloid cells by sub-setting the cells of each group, rescaling with mt-RNA % as a covariate, centering, UMAP dimensional reduction as before (first 14, 11, 16, and 10 PCA dimensions for B cells, plasma cells, activated CD4 T cells, and myeloid cells, respectively), and clustering was performed as previously described (empirically chosen modularity resolution = 0.20, 0.10, 0.25, and 0.10 for B cells, plasma cells, activated CD4 T cells, and myeloid cells, respectively), but canonical marker genes were used as features. To show that the cell type populations displayed distinct transcriptional profiles, markers were identified for each cluster vs all other cells using Wilcoxon rank sum testing of scaled data (SeuratWrappers::RunPrestoAll R function), while down-sampling to 5000 cells per cluster. The top 10 mean log fold change genes were selected from each cell type to visualize by heatmap with hierarchical clustering.
Differential expression was performed using the edgeR analysis pipeline and quasi-likelihood (QL) F tests (Lun et al., 2016; Soneson and Robinson, 2018). Specifically, raw single-cell expression data were filtered to include genes with >5% detection rate across all cells, genes were TMM-normalized, fitted to a QL negative binomial generalized linear model using trended dispersion estimates with cell detection rate and treatment as covariates, and empirical Bayes QL F tests were performed with treatment as the coefficient equal to zero under the null hypothesis (Soneson and Robinson, 2018).
Pathway enrichment analyses were performed for differentially expressed genes (DEG; absolute log2(x+1) expression fold-change > 0.5, FDR-adjusted p value (q) < 0.01) using the gost function of the gProfiler2 R package (Kolberg et al., 2020; Raudvere et al., 2019) with biological process gene ontologies (GO) for mus musculus, an adjusted p value-ordered gene list, and known genes as the domain for the statistics. In addition, the analysis p values were adjusted for multiple testing using the gProfiler gSCS method. Results were filtered to include GO terms ≤600 genes in size that intersected >2 DEG, an absolute activation score (mean log2(x+1) expression fold change of GO term DEGs) > 0.5, and an adjusted p < 0.01. Network analyses were performed by (1) creating network graphs with filtered pathway results as nodes and GO term similarity coefficients as edges (coefficients = 50% jaccard +50% overlap scores; edge similarity threshold = 0.375), (2) finding graph clusters via the Leiden algorithm using the modularity method with similarity coefficients as weights (resolution = 0.5, iterations = 1000), and (3) labeling clusters by their most significant GO term (meta-pathway). Meta-pathway genes were visualized by heatmap, using log-normalized, scaled expression for GO term genes that were differentially expressed in vaccination groups compared to the PBS control. Custom R scripts were used for generating various plots.
Schematic illustrations
Schematic illustrations were created with Affinity Designer or BioRender.
Quantification and statistical analysis
Standard statistics
Standard statistical methods were applied to non-high-throughput experimental data. The statistical methods are described in figure legends and/or supplementary Excel tables. In the dot-box plots of all figures, each dot represents data from one mouse. Data are shown as mean ± s.e.m. plus individual data points in plots. The statistical significance was labeled as follows: n.s., not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. Statistical differences were assessed by two-way ANOVA with Tukey’s correction for multiple testing. Non-significant comparisons are not shown, unless otherwise noted as n.s., not significant. Prism (GraphPad Software) and RStudio were used for these analyses. Additional information can be found in the supplemental excel tables.
Replication, randomization, blinding and reagent validations
Replicate experiments have been performed for all key data shown in this study.
Biological or technical replicate samples were randomized where appropriate. In animal experiments, mice were randomized by littermates.
Experiments were not blinded
NGS data processing were blinded using metadata. Subsequent analyses were not blinded.
Commercial antibodies were validated by the vendors, and re-validated in house as appropriate. Custom antibodies were validated by specific antibody - antigen interaction assays, such as ELISA. Isotype controls were used for antibody validations.
Cell lines were authenticated by original vendors, and re-validated in lab as appropriate.
All cell lines tested negative for mycoplasma.
Acknowledgments
We thank various members from the Chen and Wilen labs for discussions and support. We thank staff from various Yale core facilities (Keck, YCGA, HPC, YARC, CBDS, and others) for technical support. We thank Drs. Tsemperouli, Karatekin, Lin, Wang, Castaldi, and others for providing equipment and related support. We thank various support from the Department of Genetics, Institutes of Systems Biology and Cancer Biology, Dean’s Office of Yale School of Medicine, and the Office of Vice Provost for Research. This work is supported by DoD PRMRP IIAR (W81XWH-21-1-0019) and discretionary funds to S.C., as well as Burroughs Wellcome Fund and NIH grants (K08AI128043, R01AI148467) to C.B.W. The transmission electron microscopy core is supported by NIH grant GM132114. The sequencing core YCGA is supported by NIH grant 1S10OD018521.
Author contributions
S.C. conceived the study and designed it with L.P., Z.F., P.A.R., J.J.P., X.Z., and M.B.D. L.P. and Z.F. performed most experiments. P.A.R. performed most next-generation sequencing data analyses. A.M. performed BL3 neutralization. Q.L. assisted with experiments. B.Z. assisted with data analysis. S.C., C.B.W., and H.Z. secured funding and supervised the work. L.P., Z.F., P.A.R., and S.C. prepared the manuscript with input from all authors.
Declaration of interests
Yale University has filed a patent application related to the data described herein (inventors: S.C., L.P., Z.F., J.J.P., X.Z., M.B.D.) and has committed to rapidly executable nonexclusive royalty-free licenses to intellectual property rights for the purpose of making and distributing products to prevent, diagnose, and treat COVID-19 infection during the pandemic and for a short period thereafter. S.C. is a scientific Founder of EvolveImmune Tx and Cellinfinity Bio, unrelated to this study.
Published: August 2, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111160.
Supplemental information
Data and code availability
All data generated or analyzed during this study are included in this article, supplemental information, and source data files. Specifically, source data and statistics for non-high-throughput experiments are provided in a supplementary table excel file. Additional Supplemental Items are available from Mendeley Data: https://doi.org/10.17632/nvrdxn35tb. Processed data and statistics for NGS experiments are provided in Data S1. The raw NGS data have been deposited to the Gene Expression Omnibus (GEO) and are publicly available (GEO: GSE207141). The original codes of data analysis are available in Github (Zenodo: https://doi.org/10.5281/zenodo.6814583, https://zenodo.org/badge/latestdoi/512286275).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abdelrahman Z., Li M., Wang X. Comparative review of SARS-CoV-2, SARS-CoV, MERS-CoV, and Influenza A respiratory viruses. Front. Immunol. 2020;11:552909. doi: 10.3389/fimmu.2020.552909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alluwaimi A.M., Alshubaith I.H., Al-Ali A.M., Abohelaika S. The coronaviruses of animals and birds: their Zoonosis, vaccines, and models for SARS-CoV and SARS-CoV2. Front. Vet. Sci. 2020;7:582287. doi: 10.3389/fvets.2020.582287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeletti D., Yewdell J.W. Understanding and manipulating viral immunity: antibody immunodominance enters center stage. Trends Immunol. 2018;39:549–561. doi: 10.1016/j.it.2018.04.008. [DOI] [PubMed] [Google Scholar]
- Arunachalam P.S., Scott M.K.D., Hagan T., Li C., Feng Y., Wimmers F., Grigoryan L., Trisal M., Edara V.V., Lai L., et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature. 2021;596:410–416. doi: 10.1038/s41586-021-03791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi S., Hook L.M., Pardi N., Wang F., Myles A., Cancro M.P., Cohen G.H., Weissman D., Friedman H.M. Nucleoside-modified mRNA encoding HSV-2 glycoproteins C, D, and E prevents clinical and subclinical genital herpes. Sci. Immunol. 2019;4:eaaw7083. doi: 10.1126/sciimmunol.aaw7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. Overseas. Ed. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson H. A Unifying Framework for Parallel and Distributed Processing in R using Futures. The R Journal. 2021 doi: 10.32614/RJ-2021-048. [DOI] [Google Scholar]
- Bergwerk M., Gonen T., Lustig Y., Amit S., Lipsitch M., Cohen C., Mandelboim M., Levin E.G., Rubin C., Indenbaum V., et al. Covid-19 breakthrough infections in vaccinated Health Care workers. N. Engl. J. Med. 2021;385:1474–1484. doi: 10.1056/NEJMoa2109072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley K.R., Coombes N.S., Gagnon L., McInroy L., Baker N., Shaik I., St-Jean J.R., St-Amant N., Buttigieg K.R., Humphries H.E., et al. Quantification of SARS-CoV-2 neutralizing antibody by wild-type plaque reduction neutralization, microneutralization and pseudotyped virus neutralization assays. Nat. Protoc. 2021;16:3114–3140. doi: 10.1038/s41596-021-00536-y. [DOI] [PubMed] [Google Scholar]
- Bosaeed M., Balkhy H.H., Almaziad S., Aljami H.A., Alhatmi H., Alanazi H., Alahmadi M., Jawhary A., Alenazi M.W., Almasoud A., et al. Safety and immunogenicity of ChAdOx1 MERS vaccine candidate in healthy Middle Eastern adults (MERS002): an open-label, non-randomised, dose-escalation, phase 1b trial. Lancet. Microbe. 2021;3:e11–e20. doi: 10.1016/S2666-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butts C.T. network: a Package for Managing Relational Data in R. Journal of Statistical Software. 2008;24(2) https://www.jstatsoft.org/v24/i02/paper [Google Scholar]
- Cao Q., Wu S., Xiao C., Chen S., Chi X., Cui X., Tang H., Su W., Zheng Y., Zhong J., et al. Integrated single-cell analysis revealed immune dynamics during Ad5-nCoV immunization. Cell Discov. 2021;7:64. doi: 10.1038/s41421-021-00300-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen A.A., Gnanapragasam P.N.P., Lee Y.E., Hoffman P.R., Ou S., Kakutani L.M., Keeffe J.R., Wu H.J., Howarth M., West A.P., et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science. 2021;371:735–741. doi: 10.1126/science.abf6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn B.A., Cirillo P.M., Murphy C.C., Krigbaum N.Y., Wallace A.W. SARS-CoV-2 vaccine protection and deaths among US veterans during 2021. Science. 2021;375:331–336. doi: 10.1126/science.abm0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin E. Pan-coronavirus vaccine pipeline takes form. Nat. Rev. Drug Discov. 2022;21:324–326. doi: 10.1038/d41573-022-00074-6. [DOI] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose N., Suthar M.S., Makowski M., O'Connell S., McDermott A.B., Flach B., Ledgerwood J.E., Mascola J.R., Graham B.S., Lin B.C., et al. Antibody persistence through 6 Months after the second dose of mRNA-1273 vaccine for Covid-19. N. Engl. J. Med. 2021;384:2259–2261. doi: 10.1056/NEJMc2103916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z., Peng L., Filler R., Suzuki K., McNamara A., Lin Q., Renauer P.A., Yang L., Menasche B., Sanchez A., et al. Omicron-specific mRNA vaccination alone and as a heterologous booster against SARS-CoV-2. Nat. Commun. 2022;13:3250. doi: 10.1038/s41467-022-30878-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folegatti P.M., Bittaye M., Flaxman A., Lopez F.R., Bellamy D., Kupke A., Mair C., Makinson R., Sheridan J., Rohde C., et al. Safety and immunogenicity of a candidate Middle East respiratory syndrome coronavirus viral-vectored vaccine: a dose-escalation, open-label, non-randomised, uncontrolled, phase 1 trial. Lancet Infect. Dis. 2020;20:816–826. doi: 10.1016/S1473-3099(20)30160-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyn A.W., Ramos da Silva J., Rosado V.C., Bliss C.M., Pine M., Mui B.L., Tam Y.K., Madden T.D., de Souza Ferreira L.C., Weissman D., et al. A multi-targeting, nucleoside-modified mRNA Influenza virus vaccine provides broad protection in mice. Mol. Ther. 2020;28:1569–1584. doi: 10.1016/j.ymthe.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg Y., Mandel M., Bar-On Y.M., Bodenheimer O., Freedman L., Haas E.J., Milo R., Alroy-Preis S., Ash N., Huppert A. Waning immunity after the BNT162b2 vaccine in Israel. N. Engl. J. Med. 2021;385:e85. doi: 10.1056/NEJMoa2114228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38:W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., et al. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S., Yuzhakov O., Woods A., Deterling J., Hassett K., Shaw C.A., Ciaramella G. Multi-antigenic human cytomegalovirus mRNA vaccines that elicit potent humoral and cell-mediated immunity. Vaccine. 2018;36:1689–1699. doi: 10.1016/j.vaccine.2018.01.029. [DOI] [PubMed] [Google Scholar]
- Kolberg L., Kerimov N., Peterson H., Alasoo K. Co-expression analysis reveals interpretable gene modules controlled by trans-acting genetic variants. Elife. 2020;9:e58705. doi: 10.7554/eLife.58705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. pheatmap: Pretty Heatmaps. R package version 1.0.12. 2019 https://CRAN.R-project.org/package=pheatmap. [Google Scholar]
- Latif A.A., Mukaratirwa S. Zoonotic origins and animal hosts of coronaviruses causing human disease pandemics: a review. Onderstepoort J. Vet. Res. 2020;87:e1–e9. doi: 10.4102/ojvr.v87i1.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin E.G., Lustig Y., Cohen C., Fluss R., Indenbaum V., Amit S., Doolman R., Asraf K., Mendelson E., Ziv A., et al. Waning immune humoral response to BNT162b2 Covid-19 vaccine over 6 months. N. Engl. J. Med. 2021;385:e84. doi: 10.1056/NEJMoa2114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.D., Chi W.Y., Su J.H., Ferrall L., Hung C.F., Wu T.C. Coronavirus vaccine development: from SARS and MERS to COVID-19. J. Biomed. Sci. 2020;27:104. doi: 10.1186/s12929-020-00695-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Liu Y., Xia H., Zou J., Weaver S.C., Swanson K.A., Cai H., Cutler M., Cooper D., Muik A., et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature. 2021;596:273–275. doi: 10.1038/s41586-021-03693-y. [DOI] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Lun A.T.L., Chen Y., Smyth G.K. It's DE-licious: a recipe for differential expression analyses of RNA-seq experiments using quasi-likelihood methods in edgeR. Methods Mol. Biol. 2016;1418:391–416. doi: 10.1007/978-1-4939-3578-9_19. [DOI] [PubMed] [Google Scholar]
- Martinez D.R., Schäfer A., Leist S.R., De la Cruz G., West A., Atochina-Vasserman E.N., Lindesmith L.C., Pardi N., Parks R., Barr M., et al. Chimeric spike mRNA vaccines protect against Sarbecovirus challenge in mice. Science. 2021;373:991–998. doi: 10.1126/science.abi4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes L., Healy J., Saul N., Großberger L. UMAP: uniform manifold approximation and projection. J. Open Source Softw. 2018;3:861. [Google Scholar]
- Newell K.L., Clemmer D.C., Cox J.B., Kayode Y.I., Zoccoli-Rodriguez V., Taylor H.E., Endy T.P., Wilmore J.R., Winslow G.M. Switched and unswitched memory B cells detected during SARS-CoV-2 convalescence correlate with limited symptom duration. PLoS One. 2021;16:e0244855. doi: 10.1371/journal.pone.0244855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., et al. Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nat. Protoc. 2020;15:3699–3715. doi: 10.1038/s41596-020-0394-5. [DOI] [PubMed] [Google Scholar]
- Pallesen J., Wang N., Corbett K.S., Wrapp D., Kirchdoerfer R.N., Turner H.L., Cottrell C.A., Becker M.M., Wang L., Shi W., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. USA. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen T.L. patchwork: The Composer of Plots. R package version 1.1.1. https://CRAN.R-project.org/package=patchwork
- Peiris J.S.M., Yuen K.Y., Osterhaus A.D.M.E., Stöhr K. The severe acute respiratory syndrome. N. Engl. J. Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast I., Tarlinton D. B cell memory: understanding COVID-19. Immunity. 2021;54:205–210. doi: 10.1016/j.immuni.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riepler L., Rossler A., Falch A., Volland A., Borena W., von Laer D., Kimpel J. Comparison of four SARS-CoV-2 neutralization assays. Vaccines (Basel) 2020;9:13. doi: 10.3390/vaccines9010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R., Butler A., Hoffman P., Stuart T. SeuratWrappers: Community-Provided Methods and Extensions for the Seurat Object. R package Version 0.3.0. 2020 [Google Scholar]
- Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F., Weisblum Y., Muecksch F., Hoffmann H.H., Michailidis E., Lorenzi J.C.C., Mendoza P., Rutkowska M., Bednarski E., Gaebler C., et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J. Exp. Med. 2020;217:e20201181. doi: 10.1084/jem.20201181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slowikowski K. ggrepel: Automatically Position Non-Overlapping Text Labels with ‘ggplot2’. R package version 0.9.1. https://CRAN.R-project.org/package=ggrepel
- Sokal A., Chappert P., Barba-Spaeth G., Roeser A., Fourati S., Azzaoui I., Vandenberghe A., Fernandez I., Meola A., Bouvier-Alias M., et al. Maturation and persistence of the anti-SARS-CoV-2 memory B cell response. Cell. 2021;184:1201–1213.e14. doi: 10.1016/j.cell.2021.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C., Robinson M.D. Bias, robustness and scalability in single-cell differential expression analysis. Nat. Methods. 2018;15:255–261. doi: 10.1038/nmeth.4612. [DOI] [PubMed] [Google Scholar]
- Stephenson E., Reynolds G., Botting R.A., Calero-Nieto F.J., Morgan M.D., Tuong Z.K., Bach K., Sungnak W., Worlock K.B., Yoshida M., et al. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 2021;27:904–916. doi: 10.1038/s41591-021-01329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Du L., Jiang S. Learning from the past: development of safe and effective COVID-19 vaccines. Nat. Rev. Microbiol. 2021;19:211–219. doi: 10.1038/s41579-020-00462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tregoning J.S., Flight K.E., Higham S.L., Wang Z., Pierce B.F. Progress of the COVID-19 vaccine effort: viruses, vaccines and variants versus efficacy, effectiveness and escape. Nat. Rev. Immunol. 2021;21:626–636. doi: 10.1038/s41577-021-00592-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V'Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. stringr: Simple, Consistent Wrappers for Common String Operations. R package version 1.4.0. https://CRAN.R-project.org/package=stringr
- Wickham H., François R., Henry L., Müller K. dplyr: A Grammar of Data Manipulation. R package version 1.0.9. https://CRAN.R-project.org/package=dplyr
- Wilke C.O. ggridges: Ridgeline Plots in ‘ggplot2’. R package version 0.5.3. https://CRAN.R-project.org/package=ggridges
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z.W., Yuan S., Yuen K.S., Fung S.Y., Chan C.P., Jin D.Y. Zoonotic origins of human coronaviruses. Int. J. Biol. Sci. 2020;16:1686–1697. doi: 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang J.Y., Wang X.M., Xing X., Xu Z., Zhang C., Song J.W., Fan X., Xia P., Fu J.L., Wang S.Y., et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat. Immunol. 2020;21:1107–1118. doi: 10.1038/s41590-020-0762-x. [DOI] [PubMed] [Google Scholar]
- Zhu Z., Lian X., Su X., Wu W., Marraro G.A., Zeng Y. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir. Res. 2020;21:224. doi: 10.1186/s12931-020-01479-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article, supplemental information, and source data files. Specifically, source data and statistics for non-high-throughput experiments are provided in a supplementary table excel file. Additional Supplemental Items are available from Mendeley Data: https://doi.org/10.17632/nvrdxn35tb. Processed data and statistics for NGS experiments are provided in Data S1. The raw NGS data have been deposited to the Gene Expression Omnibus (GEO) and are publicly available (GEO: GSE207141). The original codes of data analysis are available in Github (Zenodo: https://doi.org/10.5281/zenodo.6814583, https://zenodo.org/badge/latestdoi/512286275).
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.