Abstract
Discovery of the CRISPR-Cas (clustered regularly interspaced short palindromic repeat, CRISPR-associated) system a decade ago has opened new possibilities in the field of precision medicine. CRISPR-Cas was initially identified in bacteria and archaea to play a protective role against foreign genetic elements during viral infections. The application of this technique for the correction of different mutations found in the Duchenne muscular dystrophy (DMD) gene led to the development of several potential therapeutic approaches for DMD patients. The mutations responsible for Duchenne muscular dystrophy mainly include exon deletions (70% of patients) and point mutations (about 30% of patients). The CRISPR-Cas 9 technology is becoming increasingly precise and is acquiring diverse functions through novel innovations such as base editing and prime editing. However, questions remain about its translation to the clinic. Current research addressing off-target editing, efficient muscle-specific delivery, immune response to nucleases, and vector challenges may eventually lead to the clinical use of the CRISPR-Cas9 technology. In this review, we present recent CRISPR-Cas9 strategies to restore dystrophin expression in vitro and in animal models of DMD.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13311-022-01197-9.
Keywords: CRISPR-Cas, Gene therapy, DMD gene, Dystrophin, Duchenne muscular dystrophy
CRISPR-Cas9 Origin and Translation to Health Sciences
While sequencing the iap gene responsible for alkaline phosphate isoenzyme conversion in Escherichia coli, an unusual structure flanking at the 3′end of the iap gene was found. The structure was made of five highly homologous repeated sequences of 29 nucleotides separated by a sequence of 32 nucleotides called a spacer sequence [1]. These sequences have subsequently been characterized as CRISPR-Cas (clustered regularly interspaced short palindromic repeat-CRISPR-associated) genes present among Archaea and Bacteria [2]. With time, it was shown that the CRISPR-Cas system confers resistance against viral infection in prokaryotes through an adaptive immunity [3]. It has been found that the memory of previous viral infections was stored as spacer sequences [4]. During the infection, Archaea and Bacteria are capable of integrating a DNA fragment of the invasive agent between the repetitive sequences using the CRISPR system, thus creating a new spacer sequence [4, 5]. These spacers were subsequently used to detect and destroy the same viral invaders following different mechanism mediated by CRISPR-associated activities [6]. This process follows three important steps. The first step represents the acquisition phase where the spacer sequences are integrated between conserved repetitive sequences to form the CRISPR array. The second step is known as the expression and maturation phase where the CRISPR array is transcribed to form the CRISPR RNAs (crRNAs), which are unique sequences complementary to the foreign DNA sequences. In the last step, which is the interference phase, the crRNAs serve as guides to form a stable complex with the CRISPR-associated proteins to target and cleave the invasive DNA elements [7]. The CRISPR-Cas system is divided into two classes. Class I is comprised of multimeric crRNA effectors and includes CRISPR-Cas type I, III, and IV. Class II is comprised of monomeric crRNA effectors and includes CRISPR-Cas type II, V, and VI. These six CRISPR types are also divided in at least 33 subtypes (16 subtypes for class I and 17 subtypes for class II), which differ from the Cas gene repository, the Cas operon organization, and the CRISPR array features [8, 9]. A deep understanding of this system resulted in the identification of their involvement in cellular pathways other than prokaryotic immunity. These included their implication in DNA repair, gene regulation, and genome evolution [10]. The growing knowledge of the CRISPR system has led to the use of crRNA-based programmable tools with different CRISPR-associated proteins like Cas9 for diagnostic and therapeutic purposes [11–13]. The CRISPR-Cas system is increasingly gaining interest in various fields of precision medicine [14, 15].
The Use of CRISPR Mechanism for Genome Editing
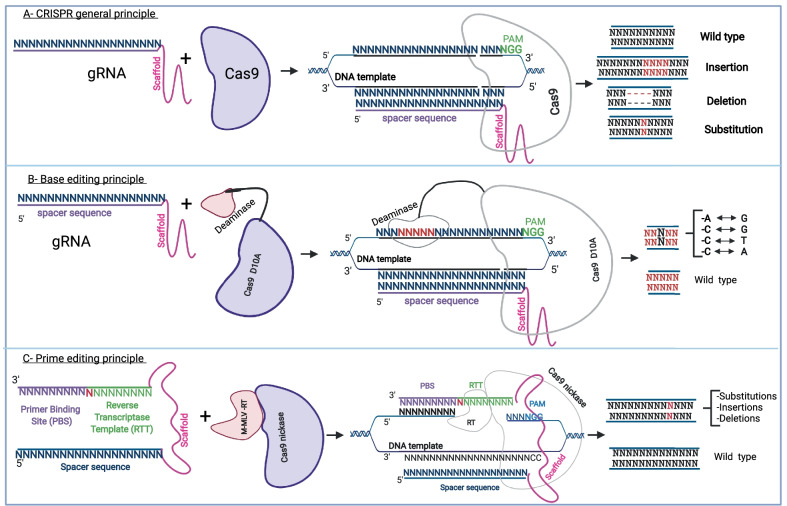
CRISPR-associated protein 9 (Cas9) belongs to the type II-A CRISPR-Cas system and is mostly used for gene modification. To this end, Cas9 forms a complex with a tracrRNA-crRNA duplex and the target DNA sequence [10, 16]. For genome editing, the tracrRNA-crRNA duplex was combined into a single guide RNA (sgRNA) made of a chemically synthesized spacer sequence (a sequence of 20 nucleotides) linked with a constant sequence known as the scaffold. The Cas9 possess two nuclease domains known as the RuvC, which cleaves the DNA strand non-complementary to the spacer, and the HNH, which cleaves the DNA strand complementary to the spacer. The RuvC domain is made of three discontinuous sequences (RuvC-I, RuvC-II, and RuvC-III) and an alpha helicoidal loop found between the first and the second sequence. The HNH domain is inserted between the second and the third sequence. The C-terminal domain of Cas9 binds with the protospacer adjacent motif (PAM) sequence and stabilizes the interaction of the sgRNA with the target protospacer sequence [17–19]. The PAM sequence is a conserved motif of 2 to 5 nucleotides, which is NGG for the Streptococcus Pyogenes Cas9 (SpCas9) protein. The PAM is immediately adjacent to the protospacer sequence [20]. In prokaryotes, the PAM sequence is required to distinguish foreign DNA and is required for the nuclease activity of the Cas enzyme [10]. In eukaryotic genome editing practices, the PAM sequence is also required for the efficient nuclease activity of Cas9. The detection of the PAM sequence initiates the ATP-independent separation of the double-stranded DNA from that point and allows the formation of the sgRNA-target DNA sequence duplex [17, 21, 22]. It is important to note that the sgRNA is previously complexified as a stable ribonucleoprotein complex with the Cas9 nuclease. The nuclease activity of Cas9 immediately follows the formation of the PAM-ribonucleoprotein (Cas9 and sgRNA) and its binding with the target DNA sequence [17, 21]. The major CRISPR mechanisms for genome editing are described in Fig. 1.
Fig. 1.
General CRISPR mechanism for genome modification. A The basic principle of CRISPR Cas9 gene modification which uses a guide RNA and a Cas9 to mediate the nuclease activity that could result in insertions, deletions, or INDELs. B The basic mechanism for base editing, which uses a sgRNA (spacer sequence + scaffold) and a Cas9 nickase fused with a cytosine or an adenine deaminase. Depending on the deaminase used, after binding with a specific DNA template, a cytosine is modified into a thymine or an adenosine into a guanine. C shows the two important components of prime editing: a PE2 protein and a pegRNA. The pegRNA is an extended sgRNA including in addition to the spacer sequence and the constant sequence (scaffold), a reverse transcriptase template (RTT), and a primer binding site (PBS). PE2 is a Cas9 nickase fused with an engineered reverse transcriptase enzyme from M-MLV. After the binding of the PE2 protein and pegRNA with a specific DNA target, the possible modification outcomes are an insertion, a deletion, or a substitution of a few nucleotides
CRISPR-Cas9 Application in Duchenne Muscular Dystrophy
The Disease
Duchenne muscular dystrophy (DMD) is a lethal X-linked recessive disease characterized by progressive muscle wasting resulting from mutations in the DMD gene coding for the dystrophin protein [23]. These mutations lead to an absence of the dystrophin protein under the sarcolemma of patients suffering from DMD (Fig. 2A). The dystrophin protein is essential for maintaining the integrity of the muscle fibers as well as the stability of their membrane. In the absence of dystrophin, the muscle fibers are damaged during contractions [24]. At an early age (1 to 3 years old), DMD patients start showing noticeable symptoms like delayed walking and frequent falls, and experience difficulties in running and climbing stairs. At that age range, children suffering from DMD show bulkier muscles around the calf, pelvis, and thigh compared to their peers, indicative of a pseudo muscle hypertrophy. Muscle weakening and loss of ambulation start by the age of 8 and progress continuously from there. DMD patients generally die between 18 and 30 years of age as a result of cardiorespiratory complications [25, 26]. The prevalence of the disease is estimated at 19.8 per 100,000 live male births, 7.1 cases per 100,000 living males, and 2.8 cases per 100,000 individuals globally [27]. The prevalence of the disease is estimated at 2.2 per 100,000 males in the UK, 10.9 per 100,000 males in France, 1.9 per 100,000 males in USA, and 6.1 per 100,000 males in Canada [28].
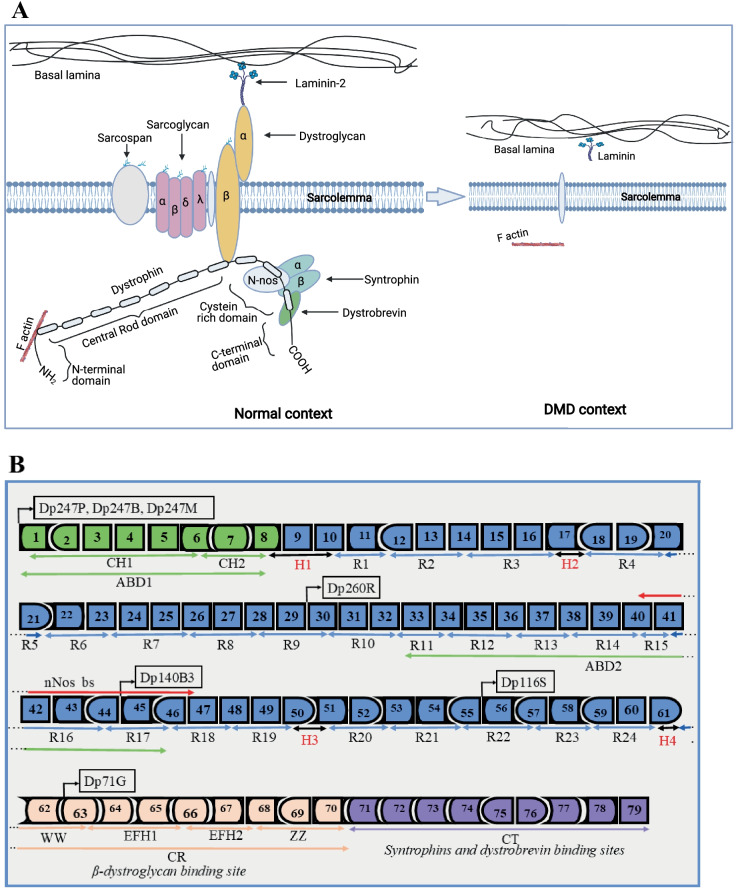
Fig. 2.
Structure of dystrophin complex and dystrophin gene and regulator elements. A The approximate structure of the dystrophin complex in the normal context and in a DMD context. In the normal context, dystrophin is present under the sarcolemma and is properly attached to other components to form the dystrophin complex. In the DMD context, there is an absence of the dystrophin complex which results in muscle fiber degradation during muscle contraction. B The dystrophin gene (exons 1 to 79) with different functional domains. The N-terminal actin binding domain (ABD1 in green) with different dystrophin promoters (Dp427P, Dp427B, Dp427M, Dp260R, Dp140B3, Dp116S, and Dp71G, respectively, for cerebellar Purkinje cells, brain, muscle, retina, brain, Schwann cells, and general) and calponin-homology motifs (CH1 and CH2). The central rod domain (blue color) with spectrin-like repeats R1 to R24, the second actin-binding domain (ABD2), the nNos-binding site (nNos bs), and the spreading of proline rich hinge regions (H1, H2, H3, and H4 in red). The cysteine-rich domain contains the WW domain, EF hands, and ZZ domain. The C terminal domain serves as binding site for syntrophins and dystrobrevin glycoprotein
Molecular Characteristics of the Disease
The DMD gene represents 0.1% of the human genome and is thus considered one of the largest in the human genome. It is composed of 79 exons representing approximately 2.2 mega-bases, which encode four important segments of the dystrophin protein [29]. The first segment is the actin-binding amino terminal domain (ABD1) which binds dystrophin through the subsarcolemmal actin network to the contractile apparatus in skeletal muscle cells [30]. The second segment is the dystrophin central rod domain. It is composed of 24 spectrin-like repeats which interact with the microtubules, membrane phospholipids, and a variety of other proteins [29, 31]. The third segment is the cysteine-rich domain which is required for the maintenance of the dystrophin under the sarcolemma [32]. The last segment is the carboxy terminal domain which provides the binding sites for systrophins and dystrobrevin and is thus involved in protein–protein interaction [29]. These four segments interact with other components of the cell cytoskeleton to form the so-called dystrophin complex which is important for the membrane stability (Fig. 2A, B) [33]. Dystrophin protein has different isoforms whose expressions are associated with different polyadenylation signal sites, messenger RNA splice events, and muscle-specific promoters [34, 35]. The Dp427m isoform is the most implicated in DMD [35]. Mutations in the DMD gene leading to DMD include exonic and intronic duplications accounting for 10 to 15% of DMD mutations, small insertions and deletions (3%), point mutations (nonsense and missense mutations, splice site mutations, and mid intronic mutations, 26%), and single or multi-exon deletions (60 to 70%) [29, 36]. The regions spanning exons 2 to 10 and exons 45 to 55 have been characterized as mutation hotspots [37, 38]. Some exons in these regions are described as the most mutated. These include exon 51 (14% total DMD mutation and 21% exon deletions), exon 53 (10% total DMD mutation and 15% exon deletions), exon 45 (9% total DMD mutation and 13% exon deletions), and exons 44 and 43 (both 7% total DMD mutation and 11% exon deletions) [36].
CRISPR-Cas9 Approaches Requiring Double-Strand Breaks
The nuclease variants for CRISPR used in this approach are designed to induce a double-strand break in a specific DNA sequence. Using CRISPR-Cas9 to induce cuts in the DNA, it is possible to restore dystrophin expression in the cells of DMD patients. This occurs by inducing the deletion of complete exons, the skipping of an exon following a splice modification, the restoration of the normal reading frame by inducing micro-insertions or micro-deletions (INDELs), or the formation of a hybrid exon (Fig. 3).
Fig. 3.
Different CRISPR approaches to mediate the dystrophin restoration in DMD patients. A The outcome of DMD exon 52 deletion and the two primary methods to restore dystrophin expression: (1) splice sites modifications: the donor site (gt) may be modified to ga or the acceptor site (ag) may be modified to gg. (2) The insertion in exon 51 of one nucleotide (A in red) to reframe exon 53 while conserving the same first amino acid codon for exon 53. B represents a strategy where a double-strand break can be made to generate an INDEL (micro-insertion or micro-deletion) in the exon preceding a stop codon. This stop codon was induced by a frameshift mutation which is compensated for by the engineered INDEL to restore the normal reading frame for the next exon. C The strategy where two single guide RNA can be used to completely delete one or several exons to restore the normal reading. D The strategy leading to the formation of a hybrid exon by inducing double-strand breaks. This hybrid exon not only restores the normal reading frame but also produces a dystrophin protein, which has a normal structure thus including a normal spectrin-like repeat structure at the junction site of the two exons. E The point mutation strategy where the abnormal nucleotide (G) in the figure forming a stop codon (TAG) is directly modified into another nucleotide (T in the figure) thus removing the stop codon
Deletion of Complete Exons
Many studies have demonstrated that pairs of specific sgRNAs may be used to induce cuts in introns, thus restoring the normal reading frame for DMD genes with a deletion or a duplication of one or several exons [39–41] (Fig. 3A, C). One or several exons have to be deleted to restore the normal reading frame. Xiang et al. [42] demonstrated that it is possible to use SpCas9 guided by a pair of sgRNAs to restore the ORF in a DMD gene which contained a frameshift mutation. The authors used patient fibroblasts carrying a deletion in exon 45 which was responsible for the frameshift. The exon 45 deletion is a common deletion in DMD patients, representing approximately 9% of total DMD mutations and 13% of exon deletions [36]. Exon 44 was deleted in some fibroblasts which were transdifferentiated into myoblasts and subsequently induced to fuse, forming myotubes expressing dystrophin. The repair outcome was estimated at 60% [42]. This approach has also been successfully used by many authors [43–53].
Formation of a Hybrid Exon
Iyombe et al. [54] and Duchêne et al. [55] demonstrated that by using the CRISPR technology to induce cuts in exons preceding and following a DMD patient deletion, it was possible to create hybrid exons either 50–54 or 47–58 resulting in the expression of a dystrophin protein with a normal structure, i.e., a normal spectrin-like repeat at the junction between the two exons (Fig. 3D). As the DMD gene section spanning exons 47–58 is one of the most important mutation hotspots [38], restoring the normal ORF by cutting within exons 48 and 57 represents a potential treatment for about 1/3 DMD cases. Duchêne et al. [55] used the Streptococcus aureus Cas9 (SaCas9) because its small size permitted it to be packaged with the two sgRNAs into a single adeno-associated virus (AAV). In vitro experiments were done with myoblasts from DMD patients carrying the deletion exons 51–53, exons 49–50, exons 51–56, and exons 50–52. In vivo experiments were done in the humanized dystrophic del52hDMD/mdx mouse model [56] using an AAV9 vector. The DNA correction by non-homologous end joining (NHEJ) following double-strand breaks generated by SaCas9 resulted in 83 to 86% of normal nucleotide sequences indicating the potential of in vivo therapeutic application. The hybrid exon 47–58 coded for a dystrophin protein with a hybrid spectrin-like repeat (SLR18/SLR23) with a normal structure. Dystrophin expression was restored not only in most skeletal muscle fibers of del52hDMD/mdx mouse but also in the cardiomyocytes.
Exon Skipping Following a Splice Site Modification
In this strategy, researchers explored the possibility of modifying either the splice acceptor site (5′NAG) or the splice donor site (5′NGT) of the frameshifted DMD gene caused by different exon deletions (Fig. 3A). The splice site modifications induced the skipping of one or many exons during the messenger RNA maturation process where introns are eliminated and exons are bound together to form a mature messenger RNA. This mature RNA is then be translated into the dystrophin protein in the case of DMD [57]. Many studies attempted to use this approach to restore the dystrophin expression for frameshift deletions [58–61]. Generally, sgRNAs are designed to induce a double-strand break in at least one of the splice sites to generate INDELs which modify the nucleotides of the splice sites through NHEJ, all the while maintaining the exon attached to the proximal or adjacent intron during the splice events. Min et al. [62] used this approach to successfully skip exons 43 and 45 in induced pluripotent stem cells (iPSCs) from a DMD patient carrying the exon 44 deletion. The exon 44 deletion disrupted the ORF of the DMD gene and accounts for about 7% of total DMD mutations and 11% of exon deletions [36]. These authors used sgRNAs to target the splice junction of exon 43 to mediate its skipping during the splicing process resulting in the in-frame junction of exon 42 to exon 45. They also designed a sgRNA to target the 5′ boundary site of exon 45 to reframe the DMD gene by linking exon 43 to exon 46. Dystrophin expression was detected by western blot and immunostaining in cardiomyocytes obtained from the differentiation of patient’s iPSCs. Intraperitoneal injection of 5 × 1013 AAV9/kg in mdx mice carrying the exon 44 deletion resulted in the expression of dystrophin in 93% of myofibers in tibialis anterior (TA), triceps, diaphragm, and cardiomyocytes followed by a significant improvement in mouse muscle function [62]. Long et al. [46] used this approach to correct the deletion of exons 48 to 50, an intronic mutation (intron 47), and for the duplication of exons 55 to 59 in iPSCs from DMD patients. Zhang et al. [63] also used this strategy for dystrophin restoration in a mouse model carrying the exon 44 deletion.
INDEL-Derived Reframing
INDELs refer to a variety of nucleotide micro-insertions or micro-deletions in a DNA fragment or in human genome that results from a double-strand DNA break [64]. Generating INDELs in the DMD gene harboring frameshift mutations can reframe the DMD gene to permit the expression of the dystrophin protein (Figs. 1A and 3B). Koo et al. [65] achieved NHEJ-mediated repair using Campylobacter jejuni Cas9 (CjCas9) nuclease and a sgRNA in the DMD knockout mouse harboring a frameshift mutation in exon 23 suggesting that in vivo DMD frameshift correction using CjCas9 has great potential as a therapeutic approach for DMD. These authors injected 5 × 1011 vg/kg of AAV serotype 2 or 9 encoding a CjCas9 nuclease and one sgRNA targeting a sequence of exon 23 in the TA of the mdx mouse with a stop codon in exon 23. Results at 8 weeks post-injection revealed 39 ± 4% dystrophin positive fibers in mice with one base pair insertion (cytidine nucleotide) and 28 ± 6% of mice carrying 14 base pairs deletion. INDEL levels were estimated at 8 ± 0.7% and 3 ± 0.6% respectively. Treated mice harboring the 1 bp insertion or 14 bp deletion showed an increase in muscle-specific maximal force. Min et al. [62] also demonstrated that the insertion of a single adenine nucleotide using a pair of sgRNA at the 5′ boundary of exon 45 through INDELs led to the reframing of exon 45 followed by increased dystrophin protein expression. The integration of the inverted terminal repeat was reported at 0.2% and 1.1% at the genomic level and mRNA level respectively. Many other studies successfully used this approach for dystrophin restoration in vitro and in animal models of DMD [47, 60, 63, 66–68].
Consequences of This Approach
DNA double-stranded breaks are considered one of the most dangerous types of DNA damage because of their high mutagenic potential and their propensity for losing genetic information [69]. Experiments using Cas9 to induce double-stranded breaks and lead the dystrophin expression are regularly associated with important INDEL levels. Imprecision in DNA double-strand repair leads to random mutations such as INDELs or point mutations [69, 70].
CRISPR Cas9 Approaches Mediating Single-Strand Break
In the human genome, DNA single-strand breaks represent the most common type of DNA damage with more than 10 000 single-strand breaks per cell each day [71]. To overcome the DNA double-strand break-related damages and imprecisions when attempting precise genome modifications, Cas proteins have been developed to specifically break only one strand of DNA [18, 19]. The wild-type Cas9 nuclease contains two nuclease domains: HNH and RuvC. These domains allow the nuclease to cleave the non-PAM and PAM strands of the DNA respectively. Specific modifications at different catalytic residues have made it possible to inhibit one of the nuclease domains, thus promoting DNA single-strand breaks. These modifications include the D10A substitution that inactivates the RuvC domain as well as the N863A and H840A substitutions that each inactivate the HNH nuclease domain [18, 19]. Modified Cas9 protein with nickase properties are used in this approach for precise modifications of the DMD gene.
Base Editing
Initially developed by Komor et al. [72], cytosine base editing (CBE) is a system that induces the chemical modification of a cytidine into an uridine without inducing a double-strand break. The uridine is subsequently replaced by a thymine during DNA duplication. CBE is programmed through a sgRNA and is composed of a Cas9 nickase D10A fused with a cytidine deaminase and the uracil DNA glycosylase inhibitor (UGI). The UGI acts to favor the insertion of the desired modification by inhibiting the effect of uracil DNA glycosylase during the base excision repair mechanism. The deaminase has an editing window of about 5 nucleotides approximately 15 nucleotides upstream of the PAM. CBE mediates the direct conversion of a cytosine into a thymine (C-to-T) in the strand not binding with the sgRNA (Fig. 1B).
Adenine base editing (ABE) was subsequently developed. It uses an SpCas9 nickase fused with an adenosine deaminase to induce the chemical modification of an adenosine into an inosine, which is a nucleotide equivalent to guanine. Thus, there is an A-to-G modification [73]. The initial cytosine and adenine base editing were previously only able to mediate the conversion of bases within the same class, meaning a purine could be exchanged for a purine (A-to-G or G-to-A) and a pyrimidine for a pyrimidine (C-to-T or T-to-C). Kurt et al. [74] recently developed the CRISPR C-to-G or G-to-C transversion base editors for a targeted DNA modification with reduced INDELs. Another group [75] also developed the C-to-A modification with high editing efficiency and tested it in 30 endogenous sites. Base editors are constantly being improved to reduce their potential for off target events [76–78]. As documented below, base editing has been used to develop different gene therapy approaches for the eventual treatment of DMD.
Base Editing Mediating Exon Skipping
Chemello et al. [79] used the ABEmax adenine base editor to target the splice acceptor site of exon 50 and the splice donor site of exon 52 in the DMD gene of a mouse model of DMD carrying an exon 51 deletion. The exon 51 deletion in DMD gene leads to a frameshift resulting in the formation of a premature stop codon in exon 52. The exon 51 deletion is one of the most frequent DMD exon deletions representing about 14% of total DMD mutations and 21% of exon deletions [36]. By modifying the splice sites, these authors intended to skip exon 50 or exon 52, leading to the restoration of the normal reading frame and the expression of dystrophin. The base editor was split and packaged into two AAV9 (dual-AAV9) because of the packaging limitation size of a single AAV [80]. The intramuscular injection of 5 × 1010 viral genomes (vg) per leg for each AAV9 in the tibialis anterior of the mouse led to an edition of 35 ± 1.5% after 3 weeks with bystander editing (other adenine present in the editing windows) of 6.7 ± 0.9% and 10.7 ± 1.2%. The dystrophin protein was detected by western blot. Substantial reduction in fibrosis, necrotic myofibers, and regenerating fibers with a significant reduction in centralized nuclei were observed after histological analysis. Experiments were also performed with human iPSC-derived cardiomyocytes. Dystrophin was again detected by western blot and the targeted editing percentage was estimated at 87 ± 4.1% with bystander editing of 29.3 ± 4.3% and 5% [79]. Importantly, these observed bystander edits are inconsequential for the final dystrophin transcript as they are present in the exon that will be skipped. This approach was also used in other studies to demonstrate base editing’s ability to restore dystrophin expression [81, 82]
Base Editing Mediating Nonsense Mutation Corrections
Xu et al. [83] recently used a modified adenine base editor (iABE-NGA) to mediate the correction of the nonsense mutation in the mdx4cv mouse model of DMD leading to dystrophin expression. The mdx4cv mouse is characterized by the presence of a premature stop codon in the exon 53 of the DMD gene leading to the absence of dystrophin expression under the sarcolemma [84]. A systemic delivery of the editor was done through a single dose injection of 1 × 1014 vg/kg in the vein of the mouse tail using a dual-AAV9 encoding for the two parts (N and C terminal half) of the iABE-NGA under the MHCK7 promotor. A group of treated mice showed after the quantification of the entire heart sections that 41.9 ± 10.5% of cardiomyocytes became dystrophin positive 5 weeks after the treatment compared to control mice that were dystrophin negative. Dystrophin-positive fibers were also observed in the gastrocnemius (10% in average) and diaphragm (5% in average). Western blot confirmed the dystrophin expression in these different muscles with different expression levels consistent with the immunostaining experiments. A 10-month impact study of the initial single-dose treatment showed 95.9 ± 1.6% dystrophin-positive fibers in mouse hearts. The cDNA Sanger sequencing from cells isolated from the heart, 5 weeks after the treatment, showed 32.6 ± 2% editing of the adenine (A) into a guanine (G). The mice kept for 10 months showed 84.6 ± 2.6% A-to-G modification. These results were accompanied with histopathology improvement and important anti-AAV9 capsid and anti-iABE-NGA titers detected in mouse serum. The transcriptomic analysis from the mdx4cv mouse heart samples indicated 34 to 185 RNA editing events with 22 shared among the three involved mice. Some of the affected genes including Myom2, RyR2, and Myh7 were indicated by authors to play important roles in the heart [83]. Such events were also reported in other studies [76, 85]. The base editing inducing the non-sense mutation correction was also experimented in other studies [86, 87].
Prime Editing
Prime editing is a novel gene editing approach which can convert any nucleotide into any other nucleotide and introduce small insertions or deletions in a DNA sequence [88]. Prime editing uses a Streptococcus pyogene Cas9 nickase H840A fused with an engineered Moloney murine leukemia virus reverse transcriptase through a flexible linker. The complex is guided to a specific DNA fragment by a modified sgRNA known as the prime editing guide RNA (pegRNA). This pegRNA contains the scaffold sequence and the spacer sequence (a sequence of 20 nucleotides) of the sgRNA. The pegRNA also includes a reverse transcriptase template (RTT) sequence (a sequence of 10 to 20 nucleotides containing the nucleotide modification(s) to be made) and a primer binding site (PBS) sequence (a sequence of 7 to 17 nucleotides) serving as primer to initiate the synthesis by the reverse transcriptase of a new DNA fragment containing the desired modification (Fig. 1C). Prime editing has been rapidly optimized to improve the editing efficiency by modifying the nuclear localisation signals [89], the annealing temperature [90], and the cellular determinants [91]. It has also been possible to delete large DNA fragments ranging from 1 to 10 kb in the genome with an editing efficacy of up to 30% [92, 93].
Prime Editing Mediating Insertions in DMD Gene
The prime editing technology can insert one or several nucleotides in the DMD gene to correct frame shifts and restore dystrophin expression. Chemello et al. [79] successfully used this approach to insert 2 nucleotides at the position +1 from the nicking site in exon 52. This was performed in cardiomyocytes derived from iPSC belonging to a DMD patient with an exon 51 deletion. The deletion of exon 51 in the DMD gene led to a frameshift mutation with the formation of a premature stop codon in exon 52. The adenine and cytosine (AC) dinucleotide introduced in the antisense strand of exon 52 reached up to 54% editing using the PE3 approach, which uses an additional sgRNA to nick at the position +52 from the first nick site induced by the pegRNA. This modification resulted in dystrophin expression confirmed by immunohistochemistry and western blot analyses. It also permitted the normalization of contractile abnormalities of human DMD cardiomyocytes.
Prime Editing Mediating Point Mutations of DMD Gene
This approach uses a specific nucleotide modification to change a stop codon into an amino acid codon leading to the expression of the full-length dystrophin. Rousseau et al. [94] successfully demonstrated the ability of prime editing to mediate the specific point mutations in many exons of the DMD gene. These authors designed and used different pegRNAs varying in PBS and RTT lengths to reproduce the c.883C > T mutation in exon 9 and c.4996 C > T mutation in exon 35 of the DMD gene of HEK 293 T cells. The editing percentage ranged from 6 to 10% and 4 to 8% respectively. Three consecutive treatments increased the mutation rate to an average of 14% and 16% respectively.
Perspectives and Challenges
Using CRISPR gene editing to create potential treatments for Duchenne muscular dystrophy is evolving rapidly. There are many published experimental strategies to modify the DMD gene and mediate the dystrophin expression and follow-up of histological and physiological benefits. Many of the proposed approaches start by screening several sgRNAs to find the one that can mediate the highest activity of the Cas9 nuclease. Sometimes, it is easy to choose the best sgRNA among the many possibilities due to the presence of many possible PAMs. However, in other cases, there is only one possible PAM and thus only one sgRNA. In other cases, there are no available PAMs close enough to the target sequence. Walton et al. [95] developed various PAM-relaxed Cas9 variants to overcome the PAM compatibility limitation for CRISPR Cas9 genome editing [96]. These variants were rapidly adapted for prime editing experiments [97]. However, to date, these variants have not yet been used for the modification of the DMD gene. Delivery approaches involving viral and nonviral methods [89, 98–100] for in vivo CRISPR experiments to spread the editors in various muscles and the heart remain challenging for CRISPR DMD gene modifications. Recently, new AAV serotypes have been developed to overcome tissue-specific promotor limitations in the gene therapy of DMD [101, 102]. The MyoAAV 2A serotype was shown to be 250 times more efficient than AAV9 [101] and can be easily adapted for the delivery of genome-specific modifiers like base or prime editors. Whatever CRISPR technology is used to mediate high-frequency modifications, important questions remain regarding possible immune responses to Cas9 [83, 103], transcriptomic adverse events and other off-target effects [83, 104, 105] are not totally understood and controlled. As yet, there are no CRISPR-Cas9 experimental treatments for DMD that have been translated to a clinical trial. However, this is not too far from becoming a reality since similar experiments for other hereditary and nonhereditary diseases are already undergoing phase I/II clinical trials. As of 22/10/2021, there were 35 CRISPR-Cas9 studies (among 51 CRISPR studies) in the US clinical trial database.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by grants from the Canadian Institute of Health Research (CIHR), the Foundation for Cell and Gene Therapy (Jesse’s Journey), and the TheCell network of the Fond de Recherche du Québec en Santé (FRQS).
Required Author Forms
Disclosure forms provided by the authors are available with the online version of this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ishino Y, Shinagawa H, Makino K, et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jansen R, van Embden JDA, Gaastra W, et al. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 3.Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 4.Bolotin A, Quinquis B, Sorokin A, et al. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 5.Pourcel C, Salvignol G, Vergnaud GY. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 6.Makarova KS, Aravind L, Wolf YI, et al. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hille F, Richter H, Wong SP, et al. The biology of CRISPR-Cas: backward and forward. Cell. 2018;172:1239–1259. doi: 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 8.Makarova KS, Wolf YI, Iranzo J, et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020;18:67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang W, Marraffini LA. CRISPR-Cas: new tools for genetic manipulations from bacterial immunity systems. Annu Rev Microbiol. 2015;69:209–228. doi: 10.1146/annurev-micro-091014-104441. [DOI] [PubMed] [Google Scholar]
- 10.Hille F, Charpentier E. CRISPR-Cas: biology, mechanisms and relevance. Philos Trans R Soc B. 2016;371:20150496. doi: 10.1098/rstb.2015.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bikard D, Euler CW, Jiang W, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Citorik RJ, Mimee M, Lu TK. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol. 2014;32:1141–1145. doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomaa AA, Klumpe HE, Luo ML, et al. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. MBio. 2014;5:e00928–e1013. doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang B. CRISPR/Cas gene therapy. J Cell Physiol. 2021;236:2459–2481. doi: 10.1002/jcp.30064. [DOI] [PubMed] [Google Scholar]
- 15.Ledford H. Landmark CRISPR trial shows promise against deadly disease. Nature [Internet]. 2021 [cited 2021 Oct 23]; Available from: https://www.nature.com/articles/d41586-021-01776-4. [DOI] [PubMed]
- 16.Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013;10:726–737. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jinek M, Jiang F, Taylor DW, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimasu H, Ran FA, Hsu PD, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jinek M, Chylinski K, Fonfara I, et al. A Programmable dual-RNA–guided DNA Endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Li J, Zhao H, et al. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell. 2015;163:840–853. doi: 10.1016/j.cell.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 21.Ishida K, Gee P, Hotta A. Minimizing off-target mutagenesis risks caused by programmable nucleases. Int J Mol Sci. 2015;16:24751–24771. doi: 10.3390/ijms161024751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sinkunas T, Gasiunas G, Fremaux C, et al. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nowak KJ, Davies KE. Duchenne muscular dystrophy and dystrophin: pathogenesis and opportunities for treatment. EMBO Rep. 2004;5:872–876. doi: 10.1038/sj.embor.7400221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blake DJ, Weir A, Newey SE, et al. Function and genetics of dystrophin and dystrophin-related proteins in muscle. Physiol Rev. 2002;82:291–329. doi: 10.1152/physrev.00028.2001. [DOI] [PubMed] [Google Scholar]
- 25.Ciafaloni E, Kumar A, Liu K, et al. Age at onset of first signs or symptoms predicts age at loss of ambulation in Duchenne and Becker muscular dystrophy: data from the MD STARnet. J Pediatr Rehabil Med. 2016;9:5–11. doi: 10.3233/PRM-160361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duchenne muscular dystrophy | Genetic and Rare Diseases Information Center (GARD) – an NCATS Program [Online]. Available from: https://rarediseases.info.nih.gov/diseases/6291/duchenne-muscular-dystrophy. Accessed Feb 25, 2021.
- 27.Crisafulli S, Sultana J, Fontana A, et al. Global epidemiology of Duchenne muscular dystrophy: an updated systematic review and meta-analysis. Orphanet J Rare Dis. 2020;15:141. doi: 10.1186/s13023-020-01430-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryder S, Leadley RM, Armstrong N, et al. The burden, epidemiology, costs and treatment for Duchenne muscular dystrophy: an evidence review. Orphanet J Rare Dis. 2017;12:79. doi: 10.1186/s13023-017-0631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Q, McNally EM. The Dystrophin Complex: structure, function and implications for therapy. Compr Physiol. 2015;5:1223–1239. doi: 10.1002/cphy.c140048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fealey ME, Horn B, Coffman C, et al. Dynamics of dystrophin’s actin-binding domain. Biophys J. 2018;115:445–454. doi: 10.1016/j.bpj.2018.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mias-Lucquin D, Morais RDS, Chéron A, et al. How the central domain of dystrophin acts to bridge F-actin to sarcolemmal lipids. J Struct Biol. 2020;209:107411. doi: 10.1016/j.jsb.2019.107411. [DOI] [PubMed] [Google Scholar]
- 32.Bies RD, Caskey CT, Fenwick R. An intact cysteine-rich domain is required for dystrophin function. J Clin Invest. 1992;90:666–672. doi: 10.1172/JCI115909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Omairi S, Hau K-L, Collins-Hooper H, et al. Regulation of the dystrophin-associated glycoprotein complex composition by the metabolic properties of muscle fibres. Sci Rep. 2019;9:2770. doi: 10.1038/s41598-019-39532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doorenweerd N, Mahfouz A, van Putten M, et al. Timing and localization of human dystrophin isoform expression provide insights into the cognitive phenotype of Duchenne muscular dystrophy. Sci Rep. 2017;7:12575. doi: 10.1038/s41598-017-12981-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dystrophin isoforms and their expression [Online]. Available from: https://www.dmd.nl/isoforms.html. Accessed May 29, 2021.
- 36.Bladen CL, Salgado D, Monges S, et al. The TREAT-NMD DMD Global Database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum Mutat. 2015;36:395–402. doi: 10.1002/humu.22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liechti-Gallati S, Koenig M, Kunkel LM, et al. Molecular deletion patterns in Duchenne and Becker type muscular dystrophy. Hum Genet. 1989;81:343–348. doi: 10.1007/BF00283688. [DOI] [PubMed] [Google Scholar]
- 38.Neri M, Rossi R, Trabanelli C, et al. The genetic landscape of dystrophin mutations in Italy: a nationwide study. Front Genet. 2020;11:131. doi: 10.3389/fgene.2020.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dara M, Razban V, Mazloomrezaei M, et al. Dystrophin gene editing by CRISPR/Cas9 system in human skeletal muscle cell line (HSkMC) Iran J Basic Med Sci. 2021;24:1153–1158. doi: 10.22038/IJBMS.2021.54711.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dara M, Razban V, Talebzadeh M, et al. Using CRISPR/Cas9 system to knock out exon 48 in DMD gene. Avicenna J Med Biotechnol. 2021;13:54–57. doi: 10.18502/ajmb.v13i2.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young CS, Hicks MR, Ermolova NV, et al. A Single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell Stem Cell. 2016;18:533–540. doi: 10.1016/j.stem.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiang X, Zhao X, Pan X, et al. Efficient correction of Duchenne muscular dystrophy mutations by SpCas9 and dual gRNAs. Mol Ther - Nucleic Acids. 2021;24:403–415. doi: 10.1016/j.omtn.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu L, Park KH, Zhao L, et al. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol Ther. 2016;24:564–569. doi: 10.1038/mt.2015.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Refaey ME, Xu L, Gao Y, et al. In vivo genome editing restores dystrophin expression and cardiac function in dystrophic mice. Circ Res. 2017;121:923–929. doi: 10.1161/CIRCRESAHA.117.310996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bengtsson NE, Hall JK, Odom GL, et al. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat Commun. 2017;8:14454. doi: 10.1038/ncomms14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Long C, Amoasii L, Mireault AA, et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ousterout DG, Kabadi AM, Thakore PI, et al. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li HL, Fujimoto N, Sasakawa N, et al. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Rep. 2014;4:143–154. doi: 10.1016/j.stemcr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tabebordbar M, Zhu K, Cheng JKW, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nelson CE, Hakim CH, Ousterout DG, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hakim CH, Wasala NB, Nelson CE, et al. AAV CRISPR editing rescues cardiac and muscle function for 18 months in dystrophic mice. JCI Insight. 2018;3:e124297. doi: 10.1172/jci.insight.124297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nelson CE, Wu Y, Gemberling MP, et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat Med. 2019;25:427–432. doi: 10.1038/s41591-019-0344-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu L, Lau YS, Gao Y, et al. Life-Long AAV-mediated CRISPR genome editing in dystrophic heart improves cardiomyopathy without causing serious lesions in mdx mice. Mol Ther. 2019;27:1407–1414. doi: 10.1016/j.ymthe.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iyombe-Engembe J-P, Ouellet DL, Barbeau X, et al. Efficient restoration of the dystrophin gene reading frame and protein structure in DMD myoblasts using the CINDEL method. Mol Ther Nucleic Acids. 2016;5:e283. doi: 10.1038/mtna.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duchêne BL, Cherif K, Iyombe-Engembe J-P, et al. CRISPR-induced deletion with SaCas9 restores dystrophin expression in dystrophic models in vitro and in vivo. Mol Ther. 2018;26:2604–2616. doi: 10.1016/j.ymthe.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Veltrop M, van Vliet L, Hulsker M, et al. A dystrophic Duchenne mouse model for testing human antisense oligonucleotides. PLoS ONE. 2018;13:e0193289. doi: 10.1371/journal.pone.0193289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gehring NH, Roignant J-Y. Anything but Ordinary – Emerging splicing mechanisms in eukaryotic gene regulation. Trends Genet. 2021;37:355–372. doi: 10.1016/j.tig.2020.10.008. [DOI] [PubMed] [Google Scholar]
- 58.Long C, Li H, Tiburcy M, et al. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci Adv. 2018;4:eaap9004. doi: 10.1126/sciadv.aap9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ifuku M, Iwabuchi KA, Tanaka M, et al. Restoration of dystrophin protein expression by exon skipping utilizing CRISPR-Cas9 in myoblasts derived from DMD patient iPS cells. Methods Mol Biol. 2018;1828:191–217. doi: 10.1007/978-1-4939-8651-4_12. [DOI] [PubMed] [Google Scholar]
- 60.Amoasii L, Hildyard JCW, Li H, et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science. 2018;362:86–91. doi: 10.1126/science.aau1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maggio I, Stefanucci L, Janssen JM, et al. Selection-free gene repair after adenoviral vector transduction of designer nucleases: rescue of dystrophin synthesis in DMD muscle cell populations. Nucleic Acids Res. 2016;44:1449–1470. doi: 10.1093/nar/gkv1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Min Y-L, Li H, Rodriguez-Caycedo C, et al. CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Sci Adv. 2019;5:eaav4324. doi: 10.1126/sciadv.aav4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, Li H, Min Y-L, et al. Enhanced CRISPR-Cas9 correction of Duchenne muscular dystrophy in mice by a self-complementary AAV delivery system. Sci Adv. 2020;6:eaay6812. doi: 10.1126/sciadv.aay6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mullaney JM, Mills RE, Pittard WS, et al. Small insertions and deletions (INDELs) in human genomes. Hum Mol Genet. 2010;19:R131–R136. doi: 10.1093/hmg/ddq400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koo T, Lu-Nguyen NB, Malerba A, et al. Functional rescue of dystrophin deficiency in mice caused by frameshift mutations using Campylobacter jejuni Cas9. Mol Ther. 2018;26:1529–1538. doi: 10.1016/j.ymthe.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y, Long C, Li H, et al. CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci Adv. 2017;3:e1602814. doi: 10.1126/sciadv.1602814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, Nishiyama T, Li H, et al. A consolidated AAV system for single-cut CRISPR correction of a common Duchenne muscular dystrophy mutation. Mol Ther - Methods Clin Dev. 2021;22:122–132. doi: 10.1016/j.omtm.2021.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amoasii L, Long C, Li H, et al. Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci Transl Med. 2017;9:eaan8081. doi: 10.1126/scitranslmed.aan8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vítor AC, Huertas P, Legube G, et al. Studying DNA double-strand break repair: an ever-growing toolbox. Front Mol Biosci. 2020;7:24. doi: 10.3389/fmolb.2020.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bennett EP, Petersen BL, Johansen IE, et al. INDEL detection, the ‘Achilles heel’ of precise genome editing: a survey of methods for accurate profiling of gene editing induced INDELs. Nucleic Acids Res. 2020;48:11958–11981. doi: 10.1093/nar/gkaa975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hossain MA, Lin Y, Yan S. Single-strand break end resection in genome integrity: mechanism and regulation by APE2. Int J Mol Sci. 2018;19:2389. doi: 10.3390/ijms19082389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Komor AC, Kim YB, Packer MS, et al. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kurt IC, Zhou R, Iyer S, et al. CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat Biotechnol. 2021;39:41–46. doi: 10.1038/s41587-020-0609-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao D, Li J, Li S, et al. Glycosylase base editors enable C-to-A and C-to-G base changes. Nat Biotechnol. 2021;39:35–40. doi: 10.1038/s41587-020-0592-2. [DOI] [PubMed] [Google Scholar]
- 76.Grünewald J, Zhou R, Garcia SP, et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature. 2019;569:433–437. doi: 10.1038/s41586-019-1161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thuronyi BW, Koblan LW, Levy JM, et al. Continuous evolution of base editors with expanded target compatibility and improved activity. Nat Biotechnol. 2019;37:1070–1079. doi: 10.1038/s41587-019-0193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu Y, Leete TC, Born DA, et al. Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity. Nat Commun. 2020;11:2052. doi: 10.1038/s41467-020-15887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chemello F, Chai AC, Li H, et al. Precise correction of Duchenne muscular dystrophy exon deletion mutations by base and prime editing. Sci Adv. 2021;7:eabg4910. doi: 10.1126/sciadv.abg4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu Z, Yang H, Colosi P. Effect of genome size on AAV vector packaging. Mol Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yuan J, Ma Y, Huang T, et al. Genetic modulation of RNA splicing with a CRISPR-guided cytidine deaminase. Mol Cell. 2018;72:380–394.e7. doi: 10.1016/j.molcel.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 82.Winter J, Luu A, Gapinske M, et al. Targeted exon skipping with AAV-mediated split adenine base editors. Cell Discov. 2019;5:1–12. doi: 10.1038/s41421-019-0109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu L, Zhang C, Li H, et al. Efficient precise in vivo base editing in adult dystrophic mice. Nat Commun. 2021;12:3719. doi: 10.1038/s41467-021-23996-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chapman VM, Miller DR, Armstrong D, et al. Recovery of induced mutations for X chromosome-linked muscular dystrophy in mice. PNAS. 1989;86:1292–1296. doi: 10.1073/pnas.86.4.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jin S, Zong Y, Gao Q, et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science. 2019;364:292–295. doi: 10.1126/science.aaw7166. [DOI] [PubMed] [Google Scholar]
- 86.Ryu S-M, Koo T, Kim K, et al. Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat Biotechnol. 2018;36:536–539. doi: 10.1038/nbt.4148. [DOI] [PubMed] [Google Scholar]
- 87.Kim K, Ryu S-M, Kim S-T, et al. Highly efficient RNA-guided base editing in mouse embryos. Nat Biotechnol. 2017;35:435–437. doi: 10.1038/nbt.3816. [DOI] [PubMed] [Google Scholar]
- 88.Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu P, Liang S-Q, Zheng C, et al. Improved prime editors enable pathogenic allele correction and cancer modelling in adult mice. Nat Commun. 2021;12:2121. doi: 10.1038/s41467-021-22295-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin Q, Jin S, Zong Y, et al. High-efficiency prime editing with optimized, paired pegRNAs in plants. Nat Biotechnol. 2021;39:923–927. doi: 10.1038/s41587-021-00868-w. [DOI] [PubMed] [Google Scholar]
- 91.Chen PJ, Hussmann JA, Yan J, et al. Enhanced prime editing systems by manipulating cellular determinants of editing outcomes. Cell. 2021;184:5635–5652.e29. doi: 10.1016/j.cell.2021.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Choi J, Chen W, Suiter CC, et al. Precise genomic deletions using paired prime editing. Nat Biotechnol. 2021;1–9. [DOI] [PMC free article] [PubMed]
- 93.Jiang T, Zhang X-O, Weng Z, et al. Deletion and replacement of long genomic sequences using prime editing. Nat Biotechnol. 2021;1–8. [DOI] [PMC free article] [PubMed]
- 94.Rousseau J, Mbakam CH, Guyon A, et al. Specific mutations in genes responsible for Alzheimer and for Duchenne muscular dystrophy introduced by base editing and PRIME editing. bioRxiv. 2020.
- 95.Walton RT, Christie KA, Whittaker MN, et al. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368:290–296. doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hu JH, Miller SM, Geurts MH, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kweon J, Yoon J-K, Jang A-H, et al. Engineered prime editors with PAM flexibility. Mol Ther. 2021;29:2001–2007. doi: 10.1016/j.ymthe.2021.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.O’Keeffe Ahern J, Lara-Sáez I, Zhou D, et al. Non-viral delivery of CRISPR–Cas9 complexes for targeted gene editing via a polymer delivery system. Gene Ther. 2021;1–14. [DOI] [PMC free article] [PubMed]
- 99.Li J, Røise JJ, He M, et al. Non-viral strategies for delivering genome editing enzymes. Adv Drug Deliv Rev. 2021;168:99–117. doi: 10.1016/j.addr.2020.09.004. [DOI] [PubMed] [Google Scholar]
- 100.Chuang Y-F, Phipps AJ, Lin F-L, et al. Approach for in vivo delivery of CRISPR/Cas system: a recent update and future prospect. Cell Mol Life Sci. 2021;78:2683–2708. doi: 10.1007/s00018-020-03725-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tabebordbar M, Lagerborg KA, Stanton A, et al. Directed evolution of a family of AAV capsid variants enabling potent muscle-directed gene delivery across species. Cell. 2021;184:4919–4938.e22. doi: 10.1016/j.cell.2021.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mendell JR, Sahenk Z, Lehman K, et al. Assessment of systemic delivery of rAAVrh74.MHCK7.micro-dystrophin in children with Duchenne muscular dystrophy: a nonrandomized controlled trial. JAMA Neurol. 2020;77:1122. doi: 10.1001/jamaneurol.2020.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Crudele JM, Chamberlain JS. Cas9 immunity creates challenges for CRISPR gene editing therapies. Nat Commun. 2018;9:3497. doi: 10.1038/s41467-018-05843-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang L, Xue W, Zhang H, et al. Eliminating base-editor-induced genome-wide and transcriptome-wide off-target mutations. Nat Cell Biol. 2021;23:552–563. doi: 10.1038/s41556-021-00671-4. [DOI] [PubMed] [Google Scholar]
- 105.Dastidar S, Majumdar D, Tipanee J, et al. Comprehensive transcriptome-wide analysis of spliceopathy correction of myotonic dystrophy using CRISPR-Cas9 in iPSCs-derived cardiomyocytes. Mol Ther. 2022;30:75–91. doi: 10.1016/j.ymthe.2021.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.