Abstract
Bench scale column studies were used to examine the partitioning of microorganisms between groundwater and a geologic medium and to examine the effect of hydrogeology (i.e., porous- versus fracture-flow) on organism partitioning. Replicated columns were constructed with intact basalt core segments that contained natural fractures and with the same basalt crushed into particles. The columns were perfused with groundwater, and upon reaching a steady state, the columns were sacrificed and the attached and unattached communities were analyzed by multiple approaches. The analyses included the total number of cells, the phylogenetic affiliation of the cells (i.e., the α, β, and γ subclasses of the class Proteobacteria and gram positives with high G+C DNA content) by fluorescent in situ hybridization (FISH), number and taxonomic affiliation by fatty acid methyl ester profiles of culturable heterotrophs, most-probable-number estimates of methanotrophs and phenol oxidizers, and whole-community sole carbon source utilization patterns from Biolog GN microplates. In the packed columns, about 99% of the total biomass (per cubic centimeter of porous medium) was attached to the geologic medium. Lack of equitable units precluded a comparison of attached and unattached biomasses in the fractured columns where the attached biomass was expressed per unit of surface area. Compositional differences in the attached and unattached communities were evidenced by (i) the recovery of Pseudomonas stutzeri, an Enterococcus sp., and Bacillus psychrophilus from the groundwater and not from the basalt, (ii) differences between community carbon source utilization patterns, and (iii) the relative abundances of different phylogenetic groups estimated by FISH in both column types. In the packed columns, attached communities were depleted of members of the α- and β-Proteobacteria subclasses in comparison to those in the corresponding groundwater. In the fractured columns, attached communities were enriched in gram-positive Bacteria and γ-Proteobacteria and depleted of β-Proteobacteria, in comparison to those in the corresponding groundwater. Segregation of populations and their activities, possibly modified by attachment to geologic media, may influence contaminant fate and transport in the subsurface and impact other in situ applications.
Although the presence of microbes in both geologic media and groundwaters is undisputed (44), little work has described the partitioning of populations and activities between communities that colonize geologic media and those that exist planktonically in groundwaters. Clearly, the relative mobility of the microorganisms (i.e., attached or unattached) would have a great influence on processes related to subsurface contaminant fate and transport (29, 57), the stability of geologic radioactive waste repositories (45), microbially enhanced oil recovery, and solution mining. For example, consider the strategies required to deliver a substrate or nutrient to an attached organism versus a planktonic organism that might be displaced by an injected volume of solution. A combined modeling and physical experimentation approach using a single bacterial population has indicated that organism partitioning influences the transport of dissolved substances through a porous medium (41). Strict modeling studies have shown that the distribution of degradative microorganisms attached to porous media can profoundly affect the fate of soluble organic contaminants in porous media (34). The effect of the distribution of attached microorganisms on a mobile contaminant can be further contrasted with that of a homogeneous distribution of unattached degrading organisms moving with a contaminant plume.
Studies of hydrocarbon biodegradation in porous media (12, 29) and of other activities in samples from subsurface (46), freshwater (52), and marine environments (55) have shown differences in activity between attached and unattached bacteria. Therefore, the type of sample (core or groundwater) used to characterize an aquifer and upon which to base a treatability study may greatly influence the predicted outcome of an in situ bioremediation attempt (1, 54). Furthermore, physiological studies of single bacterial populations have demonstrated substantial differences between cells in attached and unattached states with respect to traits such as cell size, reproductive rate, enzyme activity, and exopolymer production (39, 56). However, the direction of these alterations cannot be predicted and the causal factors responsible for these physiological changes remain unclear (56).
Recognizing that cell attachment is associated with variable physiological changes in microorganisms studied in pure culture, a simple description of the partitioning of mixed microbial populations and activities in aquifers would advance the assessment of in situ manipulation strategies and modeling of contaminant fate. While there is clearly exchange between the two compartments, it is probable that a dichotomy between attached and unattached organisms will be established under a stable set of conditions (28) and there will be a distribution of biodegradation rates (for a particular substance) between the two compartments.
Since the report of Harvey et al. (25), most researchers have generally concluded that attached bacteria dominate subsurface environments in biomass and activity and that planktonic cells are inactive subsets of the attached organisms or transients (1, 19, 26, 47). However, few studies have systematically compared the microbial communities in depth-paired core and groundwater samples from a single corehole. In sandy aquifers, Kolbel-Boelke et al. (32) and Godsy et al. (21) found more biomass attached to sediments than in the comparative groundwater while Bekins et al. (7) found a bimodal distribution of the relative abundances of planktonic bacteria (modes were 15 and 100% of the total cells) in a set of core and groundwater samples. All three studies reported differences in community composition between core and groundwater samples, with two studies reporting a predominance of methanogens in groundwater samples (7, 21). Therefore, the data on unconsolidated sedimentary aquifers indicate the potential importance of groundwater organisms at specific locations due to dominance of biomass or specific physiological groups.
The studies cited above were conducted in aquifers where groundwater flow is through porous media. However, many applications may occur in deeper, crystalline rock or consolidated sediments, where flow is confined to fractures. Bacteria associated with three samples of ashfall tuff were compared with those suspended in a sample of groundwater flowing from a fracture in the rock walls of tunnels at the Nevada Test Site (3). The researchers found about equal numbers of heterotrophic organisms in ashfall tuff and groundwater (per gram and per milliliter) but found that the identities of these groups of isolates were different. In a study of 16 paired core and groundwater samples from a single corehole in a fractured, quartz aquifer, the majority of organisms were planktonic and not attached to surfaces and there were qualitative differences in the types of organisms recovered from the two sample types (35). F. S. Colwell and R. M. Lehman (unpublished data) have observed little biomass and activity associated with cores compared to groundwater taken from the same depth in a single corehole in a basalt aquifer.
A review of the reports presented above suggests that hydrogeology, i.e., porous or fracture flow, may affect the partitioning of aquifer microbes. However, in deep, crystalline rock with low matrix porosity, where organisms are confined to fracture surfaces, the inability to aseptically collect fracture surfaces (while using drilling fluids) for microbiological analyses may have created artifactual comparisons. Furthermore, other variables that can affect the distribution of organisms, such as groundwater velocity and organic carbon concentrations, may be correlated with the hydrogeological regimen. To address the uncertainty regarding the relative importance of attached and unattached microorganisms in saturated subsurface environments and to examine the effect of hydrogeology (porous versus fracture flow) on organism partitioning, we utilized bench scale column studies. The use of laboratory columns allowed the control of groundwater velocity and carbon concentration. Replicated columns were constructed (i) with intact basalt core segments from the eastern Snake River Plain Aquifer that contained natural fractures and (ii) with the same basalt crushed into particles. The columns were perfused with groundwater and sacrificed upon reaching a steady state, and the attached and unattached communities were analyzed for microorganisms expected from previous studies of this aquifer and other subsurface environments, i.e., aerobic chemoheterotrophic bacteria (5, 19, 22, 24, 46). The results of these analyses were scaled with respect to units of volume and surface area to enable an equitable comparison between attached and unattached organisms.
MATERIALS AND METHODS
General experimental design.
The effects of the flow regimen (porous flow versus fracture flow) and the types of media sampled (rock versus groundwater) on the microbial communities were studied by comparing microbiological variables measured on the two sampling media in replicated columns packed with crushed basalt (n = 5) and constructed with fractured basalt core segments (n = 5). The microorganisms associated with the basalt were considered to be attached, and those associated with the groundwater were considered to be unattached. Differences between attached and unattached microbiological measurements within a single column type were tested for statistical significance at the P = 0.05 level by using a one-way analysis of variance (ANOVA). Comparisons between column types for either attached or unattached measurements were similarly performed. Values of measured variables are reported as the mean ±1 standard deviation of five independent replicates, unless otherwise stated.
Construction of flow units. (i) Fractured columns.
Several lengths of dense olivine-basalt core (8.3-cm diameter, freshly obtained by reverse-air rotary coring from the eastern Snake River Plain Aquifer, southeastern Idaho) from depths of 76 and 122 m in the saturated zone were identified that contained natural fractures oriented with the long axis of the core. The fractures were not induced by drilling and contained deposits of clay and calcite on their exposed surfaces. The two halves of these longitudinally fractured core pieces were paired as they occurred in situ and bound with tape. The core lengths were then cross-sectioned into 18-cm (nominal length) segments which were sterilized by autoclaving. No removal of surface deposits was attempted. Columns were constructed by coating the sides of the fractured core segments with epoxy (Hysol EPK 1C; Hysol, Seabrook, N.H.) and fitting the ends with concentric ring endplates cast with the same epoxy in Teflon molds with embedded stainless steel tubing connectors. Coating of the cores with epoxy was done in several thin layers while the core turned on a rotisserie to prevent sagging of the epoxy. The completed columns were refrigerated (4°C, several days) prior to use.
(ii) Packed columns.
Columns packed with crushed basalt were used to isolate the flow regimen as an experimental variable while holding the mineralogy constant. It is recognized that there are no natural analogue aquifers composed of particulate basalt. Nonfractured core segments adjacent to the fractured core segments and similar in terms of basalt density and degree of oxidation were selected. Basalt cores were sectioned by a hydraulic core splitter, and the resulting pieces were reduced in size with a crusher (Braun Chipmunk Crusher type VD; Bico, Burbank, Calif.) and a pulverizer (Bico Pulverizer type UD; Bico). The crushed basalt was sieved to collect particles retained by a sieve with a 1.6-mm mesh size but passed by a sieve with a 3.2-mm mesh size. The sized basalt particles were washed with water to remove fines; sterilized by autoclaving; slurried with filtered (0.2-μm pore size), deionized water; and packed into glass columns (17.8 by 8.1 cm) in 3-cm lifts. The columns were capped with plexiglass endplates tapped for nylon plumbing fittings. Between the endplates and the crushed basalt were a fabric mesh screen and a rubber gasket which sealed the columns upon tightening of a metal frame threaded through the endplates. All column components were either autoclaved or surface disinfected with dilute sodium hypochlorite solution. The columns were then enclosed in aluminum foil to exclude light. The completed columns were refrigerated (4°C, several days) prior to use.
Operation of columns.
The columns were clamped to a metal support lattice and operated in the upflow mode. Polyethylene tubing (1/8-in. diameter) and nylon and stainless steel fittings were used to connect the flow units to a peristaltic pump, source reservoir, and waste container. All tubing and fittings were newly purchased, flushed with dilute bleach solution, and thoroughly rinsed with filtered (0.2-μm pore size), deionized water. The columns were initially saturated with autoclaved, filtered (0.2-μm pore size), deionized water, air pockets were eliminated, and mass measurements were made to obtain porosities. After initial saturation, the column feed was changed to oligotrophic groundwater collected from the eastern Snake River Plain Aquifer. Groundwater in the eastern Snake River Plain Aquifer is described as a calcium-sodium-bicarbonate type that is slightly alkaline (ca. pH 8) and saturated with dissolved oxygen (60). The groundwater was pumped (following standard 3-well-volume purging) from a 213-m depth in an uncased well (USGS-103) located in a pristine location where the total organic carbon content is about 0.3 mg/liter (R. Bartholomay, U.S. Geological Survey, personal communication). Each set of columns, fracture flow or porous flow, had independent, multihead peristaltic pumps operating at different discharge rates to achieve the same flow velocity (1 m/day; similar to rates in the Snake River Plain Aquifer) and similar pore volume periods (ca. 5 h) in the columns. Following a time period of 14 days to allow achievement of a steady state between cell attachment and detachment (minimum time period defined in the pilot study described below), groundwater samples for microbiological analyses of unattached organisms were collected from the effluent tubing by using sterile, polypropylene centrifuge tubes. The columns were then sacrificed, and the attached biomass was obtained as described in the cell-harvesting section below.
Pilot study to determine steady state of microorganism exchange in columns.
One column of each type was prepared as described above and perfused with groundwater. Effluent samples of groundwater were collected at intervals of 1 day for a period of 9 days from the columns for direct cell counts and community level physiological profiling (CLPP) (methods described below). The time course of cell number in the effluent was plotted to determine when a steady state was achieved, i.e., when the number of cells in the effluent became constant. To determine if components of the total community were partitioning unequally over time despite a steady state with respect to the total cell number, principal-component plots of groundwater carbon source utilization patterns were constructed to determine if a stable community was established.
Cell harvesting from solids.
Once a sufficient volume of groundwater had been collected from the columns at the conclusion (14 days) of the replicated experiment, the flow of groundwater through the columns was terminated, the plumbing was disconnected, and the residual water was pushed from the columns with a gentle stream of filtered (0.2-μm pore size) nitrogen. Epoxied, fractured columns were split longitudinally along the axis of the natural fracture by using a disinfected hydraulic splitter. Fracture face-associated biomass was harvested by scrubbing the face with a disinfected toothbrush and repeatedly rinsing the removed material with sterile phosphate-buffered saline (PBS; 1.18 g of Na2HPO4, 0.223 g of NaHPO4 · H2O, 8.5 g of NaCl per liter, pH 7.3) into a sterile collection vessel. Biomass from both fracture faces of a given column was pooled, and the volume was normalized to 100 ml with PBS. Packed columns were extruded onto sterile aluminum foil, and 5-g subsamples of the particulate basalt were taken from the upper, middle, and lower portions of the column. The three 5-g samples for each column were pooled with 150 ml of sterile 0.1% (wt/vol) sodium pyrophosphate (pH 7.3) and blended in a high-speed blender. The supernatant containing suspensions of small particulate and colloidal basalt was used for all microbiological analyses except CLPP. The additional clarified supernatant required for CLPP was prepared by the extraction regimen described in the following section. Additional subsamples of crushed basalt were taken from the columns for gravimetric moisture determination.
Comparison of methods used to extract cells from basalt.
Preliminary experiments were performed to determine the most effective and time-efficient method for the removal of cells from particulate basalt and the creation of a clarified suspension of cells for CLPP. Based on previous studies, mechanical dispersion of particles by blending was selected (4, 6, 36). Three different extraction solutions were tested: PBS (pH 7.3), 0.1% sodium pyrophosphate (pH 7.3), and deionized water (>17.6 MΩ; no reliable pH value was obtained). Samples were blended in a high-speed blender (two 30-s bursts separated by a 30-s rest) and then decanted into 250-ml Erlenmeyer flasks, which were agitated at 150 rpm on a platform shaker for 24 h. Following dispersion and the 24-h period for cell desorption, three methods by which to separate extracted cells from abiotic particulates were tested: low-speed centrifugation (1,000 × g, 10 min), density gradient high-speed centrifugation (10,000 × g, 60 min) using Nycodenz (Sigma) as the density medium (4, 36), and gravity settling with particle flocculation using 0.25 g (per 100 ml of extractant) of an 8:5 MgCO3-CaCl2 · 2H2O mixture of salts (11). The experimental design was a three-by-three two-way ANOVA with extractant solution and separation method as the treatments (three levels each). The measured variates were the total number of cells enumerated by direct observation and community metabolic richness, as defined by the number of carbon sources oxidized by the supernatant in Biolog microplates (see methods below). The nine combinations of extractant solution and separation method were performed on three independent samples of basalt prepared as described below. The 27 individual trials were performed in a randomized sequence. Main effects of extractant and separation treatments on the total number of cells per gram of basalt extracted and metabolic richness of the extracts were tested with a two-way ANOVA, followed by multiple comparisons using Tukey's post-hoc test (61). The basalt was prepared by steeping 0.5 kg of crushed basalt for 3 days in 0.5 liter of concentrated groundwater on a platform shaker (100 rpm). The cells in the groundwater had been concentrated ca. 50 times by filtering (Gelman 0.2-μm [pore size] sterile, pleated capsule) and subsequently backflushing the cells from the filter with a smaller volume of filtered groundwater, which then contained ca. 107 cells/ml. Following steeping, the groundwater was drained from the basalt through cheesecloth and the basalt was divided into experimental aliquots, lyophilized, and stored at −80°C prior to thawing for use.
Microbiological analyses. (i) Direct cell counts.
Samples of groundwater and basalt extracts were fixed in 1% (final concentration) phosphate-buffered glutaraldehyde and refrigerated (4°C) prior to analysis (31). Aliquots of the fixed samples were filtered under vacuum onto 0.2-μm-pore-size black polycarbonate membrane filters with cellulose-acetate support filters, and the total number of cells was determined by direct counts of 4′6-diamidino-2-phenylindole (DAPI)-stained cells using epifluorescent illumination with a Zeiss no. 2 filter set (48). DAPI was applied at a staining concentration of 10 μg/ml for 30 min, followed by a filtered, deionized water rinse, drying of the filter, and mounting in immersion oil. A minimum of 5 fields and 200 cells were counted or 20 fields when 200 cells were not achieved.
(ii) Enumeration, isolation, and identification of culturable aerobic chemoheterotrophs.
The number of culturable aerobic heterotrophs was determined by standard spread plating onto R2A solid medium (49) amended with cycloheximide (50 mg/liter) after serial dilution of samples in PBS. Numbers of CFU were determined by counts performed 14 days after incubation at room temperature in the dark. Morphologically distinct isolates from each sample were identified by using colony size, color, consistency, edge, and elevation as parameters. These isolates were subsequently streaked until pure, and identification was performed by determining fatty acid methyl ester profiles with the MIDI system (MIDI, Inc., Newark, Del.) in accordance with the manufacturer's instructions.
(iii) Relative abundance of phylogenetic groups by FISH of whole cells.
Groundwater and basalt extract samples were preserved by freezing (−20°C) for fluorescent in situ hybridization (FISH) analysis using 16S rRNA-directed oligonucleotide probes to estimate the relative abundance of phylogenetic groups of Bacteria in the samples (2). Probes for Bacteria (2); the α, β, and γ subclasses of Proteobacteria (38); and gram-positive Bacteria with high G+C DNA content (50) were all linked with CY3 and purchased from Operon Technologies (Alameda, Calif.). Each probe was tested for specificity and cross-hybridization by using representative isolates (type strains obtained from the American Type Culture Collection) that have been commonly retrieved from subsurface environments (3, 8, 13, 62). Nonspecific binding of the olignucleotide probes was estimated by using a CY3-linked probe (NON338) that has a nucleotide sequence complementary to that of EUB338. The method of Glockner et al. (20) for concentrating cells on white polycarbonate filters and performing hybridization on filter fragments was used. The method was modified in that an unlaminated hydrophobic membrane filter (e.g., Millipore MITEX) was placed between the glass slide and the polycarbonate filter fragments, which kept the hybridization solution from draining through the polycarbonate filters. For each sample, two separate aliquots were sonicated (50 W at 45 kHz) for 10 min and filtered. For basalt extracts only, 0.05% Tween 80 was added prior to sonication. Following sonication, these aliquots were centrifuged (200 × g, 10 min) to deposit particulate matter. Also for basalt extracts only, prestaining with DAPI (10 μg/ml) after fixation suppressed nonspecific binding of the probes to colloidal basalt. Probes were grouped according to the required fixative, formamide percentage in the hybridization buffer, and NaCl concentration in the wash buffer so that probes EUB338, ALF1b, HGC1901, and NON338 were applied to an aliquot fixed with 50% ethanol and probes BET42A, GAM42A, and NON338 and no probe were applied to another aliquot fixed with 3.7% formaldehyde. Hybridizations were performed overnight at 46°C with 50 ng of probe in hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 8], 0.1% sodium dodecyl sulfate plus formamide [20% for EUB338, ALF1b, HGC1901, and NON338; 35% for BET42A, GAM42A, NON338, and no probe]) and followed by washing for 15 min at 48°C in buffer (20 mM Tris-HCl [pH 8], 0.1% sodium dodecyl sulfate, 5 mM EDTA plus NaCl [0.225 M for EUB338, ALF1b, HGC1901, and NON338; 0.080 M for BET42A, GAM42A, NON338, and no probe]). One hundred nanograms of competitive non-CY3-labeled GAM42A and BETA42A was used with CY3-labeled probes BET42A and GAM42A, respectively (38). Filter sections were dried and mounted on glass slides with Fluormount and viewed with a Zeiss no. 16 filter set. At least 500 cells or 20 fields were counted for each probe. Counts from NON338 probes were subtracted from all group probes, and the resulting values were expressed as the percentages of the total cells hybridizing to the EUB338 probe.
(iv) CLPP.
CLPP was performed by inoculating groundwater and basalt extracts into Biolog GN microplates, which test the respiration of 95 different sole carbon sources by the mixed communities (18). Microplates were incubated in a humidified atmosphere in the dark, and the A590 nm (i.e., production of reduced tetrazolium dye coupled to carbon source oxidation) in each of the microplate wells was recorded every 2 h for 1 week by using an automated microplate reader-stacker (Multiscan MCC 340 MKII; ICN Pharmaceuticals, Costa Mesa, Calif.). The appearance of a colored product (formazan), relative to a control microplate well containing no carbon source indicates utilization of the sole carbon source in that microplate well. Results were considered in two ways: (i) community metabolic richness, which is simply the sum of all positive carbon source utilization tests, and (ii) community carbon source utilization patterns based on the continuous absorbance readings for each of the 95 reactions. Community carbon source utilization patterns were analyzed by principal-components analysis (PCA), an ordination technique that reduces the dimensionality of the data set while maximizing the amount of the original variance retained in derived factors (51). Thus, the new factors or principal components reflect the most prominent intrinsic patterns in multivariate data sets. The first principal component contains the most variance present in the original data set, and each subsequent factor contains successively less of this original variance; the amount of variance associated with a factor is its eigenvalue. By using background-corrected microplate readings at equivalent average well color development (14, 15, 42), PCA was performed on the correlation matrix of the variables (R matrix) with no factor rotation. Each sample possesses a score on each of the new factors (factor score) which can be plotted in factor space (factor score plot). For each desired comparison, PCA was performed on the selected set of samples and the factor scores for principal components 1 and 2 were plotted to examine, in two dimensions, the characterization of samples by their community carbon source utilization patterns. The contributions of the original variables (sole carbon sources) to the discrimination among samples were expressed as factor loadings, which are simple correlation coefficients (when PCA is performed on the correlation matrix) relating the original variables to the derived factors.
(v) MPN estimates for methanotrophs and phenol oxidizers.
Enumeration of methanotrophs was performed by most-probable-number (MPN) analysis of sample dilutions in nitrate mineral salts liquid medium (9) modified by the addition of 1 mM (NH4)2SO4 to provide an alternative nitrogen source with complementary reduction of NaNO3 from 2 to 1 mM and the use of 16% methane in the headspace. For phenol oxidizers, the same mineral salts medium was used with the addition of 570 mg of phenol per liter and omission of the headspace methane. A six-level, five-vial MPN matrix for each sample and positive and negative controls was performed with 50-ml glass serum vials with static incubation. After 3 months of incubation, turbid vials were scored positive with confirmation of this presumptive evidence by direct observation of cells and analytical detection by gas chromatography of the disappearance of methane and phenol.
Selection and derivation of units used to express enumerations.
While some microbiological results are independent of mass once a threshold mass is reached (i.e., intensive properties), enumerations which are mass dependent (i.e., extensive properties) require appropriate units for equitable comparisons. Microbiological enumerations of groundwater, porous media, and fracture surfaces may be expressed in a variety of different units (Table 1). For comparison of attached and unattached bacteria in packed columns, the volume (cubic centimeters) of the porous medium was used. No equivalent units could be identified with which to compare enumerations of attached and unattached bacteria in fractured columns. Enumerations of unattached organisms from the two column types were compared in units of groundwater volume (milliliters) while comparison of attached organisms between the two column types required units of external surface area (square centimeters). External surface area (calculations described below) is distinguished from (i) total surface area, which is both internal and external and may be measured by gas adsorption techniques, and (ii) adjusted surface area, which is the total area minus the internal surface area inaccessible to an average bacterium, i.e., that internal surface area connected by pore throats 1 μm or less in diameter as estimated by pore throat dimension distributions derived from mercury porosimetry data.
TABLE 1.
Units for expression of enumerations of attached and unattached subsurface microorganisms
| Medium | Expression | Calculationa |
|---|---|---|
| Groundwater | Per ml or cm3 of groundwater | Default |
| Groundwater | Per cm3 of in situ porous medium | (Cells/ml) · n |
| Particulate solids | Per g dry weight of solids | (Cells/g [wet wt]) · (1 + %M) |
| Particulate solids | Per cm3 of in situ porous medium | (Cells/g [dry wt]) · (bulk density, g/cm3) |
| Particulate solids | Per cm3 of solids | (Cells/g [dry wt]) · (specific gravity, g/cm3) |
| Particulate solids | Per cm3 of equivalent pore volume | (Cells/g [dry wt]) · (g [dry wt]/ml of pore water) |
| Particulate solids | Per cm2 of external surface area | (Cells/g [dry wt]) · (g [dry wt]/SAext) |
| Particulate solids | Per cm2 of total surface area | (Cells/g [dry wt]) · (g [dry wt]/SAtot) |
| Particulate solids | Per cm2 of adjusted surface area | (Cells/g [dry wt]) · (g [dry wt]/SAadj) |
| Fracture face | Per cm2 of external surface area | (Cells/ml of extract) · (ml of extract/SAfract) |
Cells is a generic microbiological enumeration or other quantification, e.g., biomass; n is porosity; %M is percent moisture; SAext is the external surface area of particles; SAtot is the total surface area of particles; SAadj is the adjusted surface area of particles, which includes the external and internal surfaces accessible to bacteria; SAfract is the external surface area of fracture surfaces; ml of extract is the volume of fluid in which organisms removed from fracture faces are suspended.
The external surface area of fracture surfaces and particles was determined by micromorphometry using metallographically prepared specimens, an optical microscope, and image analysis software (NIH-Image; National Institutes of Health [http://rsb.info.nih.gov/nih-image]) to estimate the relative enhancement of linear distance due to topographical relief. For fracture face samples, rock sections containing the fracture face were cut, mounted, polished in both directions (parallel and perpendicular to the directions in which the original basalt flowed), and viewed at a magnification of ×500. The distance along the edge of the basalt fracture surface was determined and related to the shortest linear distance between the endpoints resulting in a linear distance enhancement factor (α1). The enhancement factor was applied to the gross geometric dimensions (length [l] and width [w]) of the fracture face to determine the external surface area. The total area of the number of fracture faces (ni) was calculated as follows: SAext = Σni(α1)2 (l × w). Estimates of variance for this linear enhancement factor were obtained within core sections by polishing to two depths, between core sections by analyzing two sections from a core segment, between core segments by analyzing two core segments taken from different locations in the total length of the core, and according to orientation (with respect to lava flow) within each section. Differences in the linear enhancement factor between sections within a core segment, between core segments, and between directions were tested by one-way ANOVA. Similar measurements were made of the perimeters of mounted particles of crushed basalt from packed columns by using the projected area perimeter of the particle (27) as a reference for calculating an enhancement factor (α2) and subsequently applying the enhancement factor to the formula for the surface area of a sphere. Thus, the external surface area of ni particle(s) was calculated as follows: SAext = πΣni(α2 · dPA)2. Estimates of variance due to individual particles and their size were obtained by performing the measurements on a subsample of 28 particles.
RESULTS
Pilot experiment to determine steady state.
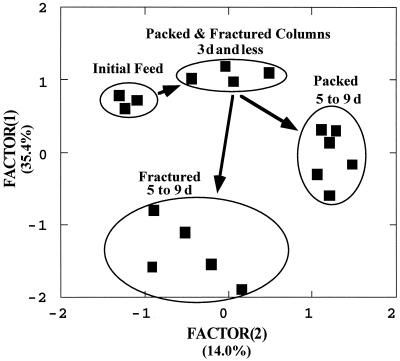
In the packed column, the number of cells in the effluent increased over time from initial values of ca. 105 cells/ml until about 4 days, when the numbers stabilized at ca. 106 cells/ml. In the fractured column, the number of cells in the effluent increased rapidly, leveling off at ca. 5 × 105 cells/ml within 1 or 2 days. A principal-components factor plot of the community sole carbon source utilization patterns of the effluent groundwaters showed a change from the initial feed water to similar patterns for both column types for the first 3 days and then divergence of the effluent from the two column types to separate steady states that existed from 5 to 9 days (Fig. 1). The average metabolic response obtained by averaging the responses of effluent samples to all of the carbon sources in the Biolog plate fell from an initial value of ca. 1.4 (A590) of the feed water over the first 5 days to stable values of 1.1 and 0.7 absorbance units for the packed and fractured columns, respectively. When these results are viewed collectively, it appears that a steady state of cell exchange between the column solid medium and groundwater was reached within 5 days of initial flooding of the columns and that appropriate sampling times for examination of steady-state partitioning would be scheduled subsequent to this period.
FIG. 1.
PCA factor plot of the community carbon source utilization patterns for effluent samples from both column types taken over a period of 9 days during a pilot study to determine the steady state of microorganism exchange within the columns. Replicate samples (n = 3) of the initial groundwater used for column feed are at the upper left part of the plot, while the effluent from fractured and packed columns for the first 3 days (d) is at top center of the plot. Effluent samples taken between 5 and 9 days after initial flooding of the columns show that the two column types arrived at independent steady states, as defined by the effluent community carbon source utilization patterns. n = 1 for the effluent sample for each type of column taken at each sampling date. Factors 1 and 2 accounted for 35.4 and 14.0% of the total variance in the data set, respectively.
Comparison of approaches to cell extraction from basalt.
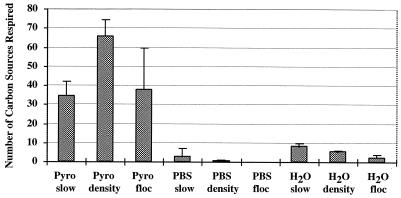
Results of the three-by-three ANOVA experiment to test the effect of the cell extraction protocol on crushed basalt did not show a main effect on the total number of cells extracted by either solution type (P = 0.127) or separation method (P = 0.704), although trends in the results indicated that sodium pyrophosphate generally resulted in higher cell yields (data not shown). In contrast, both solution type (P < 0.001) and separation method (P = 0.036) had a significant main effect on community metabolic richness (Fig. 2). There was an enhancement of cell extract metabolic richness by sodium pyrophosphate over both PBS and water (P < 0.001, Turkey's post-hoc test [61]). The density gradient method was significantly more effective than the slow-speed centrifugation method (P = 0.041) but not more effective than the flocculation method (P = 0.100). Based on these results, subsequent extractions were performed with sodium pyrophosphate. The flocculation method for particle separation was used since the density gradient method is considerably more labor intensive and did not provide clear improvement in extraction efficiency with sodium pyrophosphate.
FIG. 2.
Community metabolic richness (as measured by the number of sole carbon sources oxidized by a whole sample in Biolog GN microplates) of crushed basalt extracts under different conditions of extraction solution (first line of x-axis labels) and particle separation method (second line of x-axis labels). Pyro represents 0.1% sodium pyrophosphate; Pyro, PBS, and H2O refer to the extractant solutions described in Materials and Methods; slow, density, and floc refer to the particle separation methods described in Materials and Methods. The values shown are means with 1 standard deviation indicated; n = 3 for each treatment combination.
Derivation of units used to express microbiology.
The percent moisture of the wet crushed basalt averaged 33% ± 3% (n = 4). The porosity of the packed columns was 48.7% ± 0.7%, and the in situ bulk density of the columns was 1.61 ± 0.02 g/cm3. To calculate the external surface area of a fracture face, the linear enhancement factor (α1) of the length (l) and width (w) of the faces was found to be 2.20 ± 0.30. There was no statistically significant difference between factors calculated from different sections within a core segment, from different segments, or with respect to lava flow orientation. The total external surface (considering two fracture faces per column) averaged 1,297 ± 135 cm2 for the five fractured columns. Because extracts from both fracture faces were pooled in 100 ml of PBS, on average, there was 13 cm2 of external surface area/ml of extract. For particles, the projected area diameter (dPA) of the particles was 2.7 ± 0.6 mm (n = 28) and the linear enhancement factor (α2) was 1.47 ± 0.33 (n = 28). Based on counting of particles in multiple subsamples and regression analyses of these results, the average number of particles per gram of dry weight was 86 (95% confidence limits, 73 to 103); therefore, the external surface area of 1 g of dry, crushed basalt averaged 42.6 cm2.
Microbiology of groundwater compared to that of rock. (i) Extensive properties.
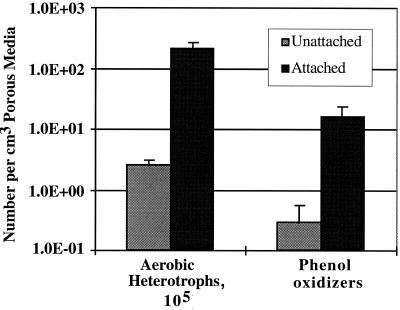
Estimates of total cell counts on basalt extracts were unreliable due to nonspecific binding of the DAPI stain by colloidal mineral material. Because total cell counts in the column effluent groundwater (data not shown) closely approximated the number of aerobic heterotrophs in the same samples, the numbers of heterotrophs were used as a proxy for the total number of cells, and, by extension, the total biomass. Considering the results obtained from the packed columns in terms of cubic centimeters of in situ porous medium, the number of attached heterotrophs was approximately 2 orders of magnitude greater than that of those that were unattached (Fig. 3). Similarly, the number of attached phenol oxidizers greatly exceeded that of those that were unattached. No methanotrophs were detected in any groundwater or rock extract samples.
FIG. 3.
Numbers of aerobic chemoheterotrophs (CFU) and phenol oxidizers, expressed per cubic centimeter of porous medium, in samples of effluent groundwater (Unattached) and crushed basalt extracts (Attached) from packed columns. The values shown are means with 1 standard deviation indicated (n = 5).
(ii) Intensive properties.
The number of aerobic, heterotrophic morphotypes recovered on R2A averaged eight for all sample types, including the feed groundwater (8.3 ± 1.6), and there was no significant difference between rock and water for either column type. Twenty morphologically distinct colonies were isolated from the sum of all samples and identified by fatty acid methyl ester profiles as follows (the MIDI database similarity coefficient is in parentheses following each identification): Hydrogenophaga pseudoflava (0.029), Paenibacillus macerans (0.048), Methylobacterium mesophilicum (0.301), Pseudomonas sp. (>0.230), Methylobacterium sp. (>0.343), Arthrobacter sp. (>0.326), Arthrobacter oxydans (0.373), Curtobacterium citreum (0.775), Pseudomonas stutzeri (0.801), Enterococcus sp. (>0.235), and Bacillus psychrophilus (0.147). Five isolates did not match any entry in the MIDI database, and four did not produce enough biomass for analysis. While most isolates were observed both attached and unattached (including gram-positive and gram-negative types), three isolates were only observed in the groundwater and not associated with the rock: P. stutzeri, an Enterococcus sp., and B. psychrophilus.
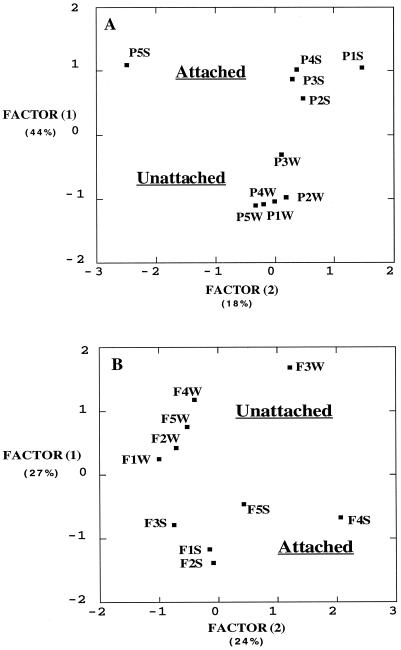
Community metabolic richness ranged from 78 to 92 sole carbon sources across all types of samples. Attached communities respired significantly less carbon sources than did their counterparts in both the packed (P < 0.001) and fractured (P = 0.04) columns. Principal-components plots of the community carbon source utilization patterns distinguished between attached and unattached communities in both column types (Fig. 4A and B). In the packed columns, attached communities demonstrated a preference for amino acids (median factor 1 loading for 20 amino acids was 0.763) while unattached communities tended to use more carbohydrates (median factor 1 loading for 28 carbohydrates was −0.463). However, a similar distribution of loadings was not observed in the fractured columns (amino acid median factor 1 loading, 0.273; carbohydrate median factor 1 loading, 0.167).
FIG. 4.
(A) PCA factor plot of community carbon source utilization patterns for effluent groundwater (Unattached) and crushed basalt extracts (Attached) from packed columns. Factors 1 and 2 accounted for 44 and 18% of the total variance in the data set, respectively. Individual observations are denoted with the letter P for packed column, followed by the number of the replicate column and either S for solid or W for water. (B) PCA factor plot of community carbon source utilization patterns for effluent groundwater (Unattached) and fracture face-associated biomass (Attached) from fractured core flood units. Factors 1 and 2 accounted for 27 and 24% of the total variance in the data set, respectively. Individual observations are denoted with the letter F for fractured core flood unit, followed by the number of the replicate column and either S for solid or W for water.
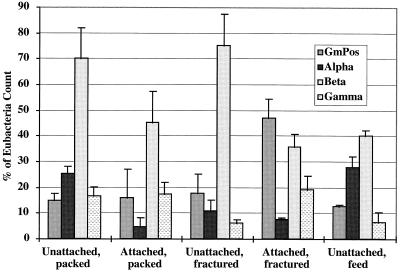
FISH enumerations indicated that the unattached communities in the effluents from both column types were dominated by β-Proteobacteria, compared to the respective attached communities, although the difference was only significant for the fractured columns (P = 0.014) (Fig. 5). The β-Proteobacteria seemed to be enriched in the laboratory column environment compared to the final column feed groundwater. The unattached communities were also slightly enriched in α-Proteobacteria compared to attached communities, although this difference is only significant (P = 0.008) in the case of the packed columns. For the fractured columns only, attached communities were significantly (P = 0.033) enriched in members of the gram-positive group with high G+C DNA content.
FIG. 5.
Phylogenetic affiliation of unattached (in groundwater) and attached (extracted from basalt) bacteria in fractured and packed columns by FISH using probes for gram positives (GmPos) with high G+C DNA content and α, β, and γ Proteobacteria. Counts are background corrected for nonspecific binding to NON338 and expressed as percentages of the total count hybridized to the Bacteria probe (EUB338) for each sample. Error bars indicate standard errors of independent replicate columns (except feed water—see below): unattached, packed, n = 4; attached, packed, n = 3; unattached, fractured, n = 4; attached, fractured, n = 5. Results obtained with the groundwater used as column feed, at the time of column sacrifice, are shown for comparison (four subsamples). The sum of percentages for a given sample type may not equal 100%, as averages of columns are presented and all counts were performed independently.
Microbiology of fractured core floods compared to that of packed columns. (i) Extensive properties.
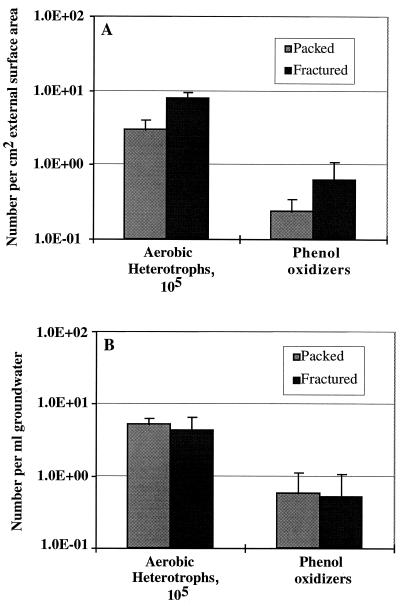
In terms of external surface area, the average number of heterotrophs (P < 0.001) and phenol oxidizers (P = 0.120) was higher on the fracture faces than on crushed basalt (Fig. 6A). If the organisms recovered from the crushed basalt include cells from internal portions of the particles, then the numbers of bacteria per unit of particle surface area were even lower than those of bacteria recovered from the fracture surfaces and the difference in phenol oxidizers could become significant. In contrast, there was no significant difference between the two column types in the numbers of heterotrophs and phenol oxidizers enumerated in the effluent groundwaters (Fig. 6B).
FIG. 6.
(A) Comparison of the numbers of aerobic chemoheterotrophs (CFU) and phenol oxidizers expressed per square centimeter of external surface area for crushed basalt extracts (Packed) and fracture faces (Fractured). The values shown are means with 1 standard deviation indicated (n = 5). (B) Comparison of the number of aerobic chemoheterotrophs (CFU) and phenol oxidizers expressed per milliliter for effluent groundwater from crushed-basalt columns (Packed) and that from fractured columns (Fractured). The values shown are means with 1 standard deviation indicated (n = 5).
(ii) Intensive properties.
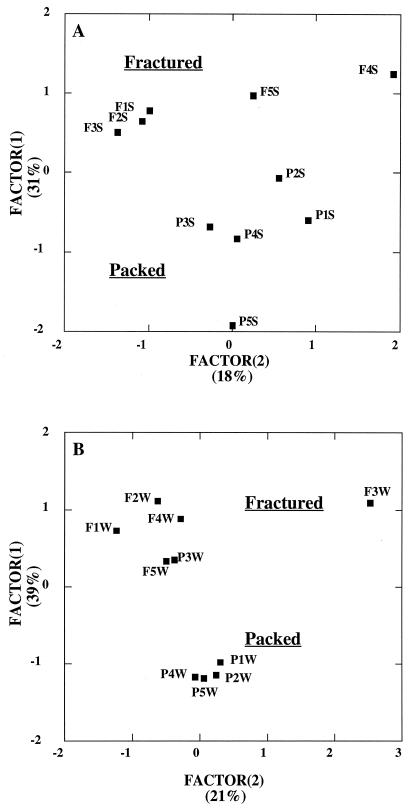
There was no difference between the two column types in the number of heterotrophic morphotypes (ca. 8) found in rock or water. Heterotrophic isolates recovered from groundwaters and rock from both column types were nearly identical, with one exception, C. citreum, which was observed in the crushed basalt and not on the fracture faces. There were no significant differences in the community metabolic richness when attached or unattached communities were compared between the two column types. Principal-components plots of the community carbon source utilization patterns distinguished extracts of the packed basalt from those removed from fracture surfaces (Fig. 7A). Similarly, groundwater communities from packed columns exhibited patterns differing from those of their counterparts from the fractured columns, with one exception (P3W) (Fig. 7B). Again, this segregation of communities was largely driven by differences in their respiration of carbohydrates and amino acids, which together comprised about 50% of the 96 substrates in the Biolog GN microplates. For the attached communities, median factor 1 loadings for carbohydrates and amino acids were 0.420 and −0.583, respectively, indicating stronger use of amino acids by communities attached to the crushed basalt. For unattached communities, median factor 1 loadings for carbohydrates and amino acids were −0.379 and 0.689, respectively, indicating stronger use of amino acids by planktonic communities in the fractured columns relative to the packed columns.
FIG. 7.
(A) PCA factor plot of community carbon source utilization patterns for crushed basalt extracts (Packed) and fracture faces (Fractured). Factors 1 and 2 accounted for 31 and 18% of the total variance in the data set, respectively. (B) PCA factor plot of community carbon source utilization patterns for effluent groundwater from crushed-basalt columns (Packed) and that from fractured columns (Fractured). Factors 1 and 2 accounted for 39 and 21% of the total variance in the data set, respectively.
Comparison of the FISH results of unattached communities between column types shows that there was a significant difference in the percentages of α-Proteobacteria (P = 0.034) and γ-Proteobacteria (P = 0.029) (Fig. 6). Considering the attached communities, a larger (borderline significance) percentage of gram-positive bacteria with high G+C DNA content was removed from the fracture faces than from the crushed basalt (P = 0.058). The unique phylogenetic profile of bacteria removed from the fracture faces was supported by visual observations of larger and more morphologically diverse cell forms in these samples compared to other samples.
DISCUSSION
Attached and unattached communities in porous-flow columns.
In the packed columns, about 99% of the total biomass (as estimated by numbers of aerobic heterotrophs using units of cubic centimeters of a porous medium) and 96% of the phenol oxidizers were found attached to the geologic medium. The general predominance of attached biomass in the packed columns was identical to that concluded from field sampling of unconsolidated, sedimentary aquifers (1, 19, 21, 25, 26, 32, 47). There were also qualitative differences in the communities as evidenced by (i) the recovery of several aerobic heterotrophs from the groundwater and not from the basalt (P. stutzeri, Enterococcus sp., and B. psychrophilus), (ii) the difference in community carbon source utilization patterns, and (iii) the relative abundances of α-Proteobacteria. Qualitative differences between attached and unattached communities have been commonly reported in unconsolidated, sedimentary aquifers (7, 21, 32), although these observations have depended strictly on culture-based assays. The corroborative results observed in our packed columns suggest a reasonable approximation of bacterial biomass partitioning under field conditions by our laboratory model.
Attached and unattached communities in fracture-flow columns.
In the fractured columns, no units allow equitable comparisons of biomass or other enumerations between attached and unattached components. Some volumetric units could be derived, but they would be dependent upon the size and distribution of fractures assumed. Differences were observed with high G+C DNA content in the community carbon source utilization patterns and relative abundances of gram positives with high G+C DNA content and β- and γ-Proteobacteria. Similar to the packed-column results, P. stutzeri, Enterococcus sp., and B. psychrophilus were recovered only from the groundwater and not from the fracture faces. In other crystalline rock aquifers, compositional differences between attached and unattached communities have been shown (3, 35), as well as differences in biomass based on the complete absence of measured organisms or activity associated with the rock (which avoids confoundment of unit comparisons) (F. S. Colwell and R. M. Lehman, unpublished data).
Effect of hydrogeology on community partitioning.
FISH results showed differences in the relative abundances of α- and β-Proteobacteria between the groundwaters from the two column types, and there was enrichment of gram-positive bacteria colonizing fracture faces versus the particulate basalt. Effects associated with hydrogeology were also seen for the number of aerobic heterotrophs and phenol oxidizers which were similar in the effluent groundwaters of the two columns but elevated in the attached communities of the fractured columns compared to those of the packed columns (Fig. 6A and B). Furthermore, differences between packed and fractured columns in the community carbon source utilization patterns of both waters and basalt extracts were observed (Fig. 7A and B). In this experiment, groundwater velocity was held constant between the two columns to eliminate large differences in shear forces which may affect community partitioning (37). No nutrient or carbon was added to the oligotrophic groundwater (<0.3 mg of organic carbon per liter) in either column type; however, it is possible that differential sorption of organic carbon occurred as crushing of the basalt exposed fresh faces that had not previously been in contact with groundwater. Differences in groundwater organic content have been reported to influence microbial partitioning between groundwater and geologic media (7, 25, 26). Differences in surface-associated organic carbon could have influenced cell attachment due to increased hydrophobic surface area or simple concentration of electron donors. Increased inorganic coatings, such as clay or calcite, on the fracture surface may have also altered the adsorption characteristics of the substrata.
Limitations and considerations in comparison of attached and unattached organisms.
Although the extension of the CLPP results to the whole community or to in situ activities is limited due to the assay's growth dependence and the small and selective percentage of the community assayed (16, 33, 53, 59), the technique is reliable in distinguishing communities based on this responsive fraction (14, 23, 33). The necessary assumption for extension to the larger community is that the assayed populations interact with the unassayed populations in the original community. It should be noted that the segment of the community assayed in the Biolog plate environment does not necessarily equate to the sum of culturable bacteria (16, 17, 30, 40, 58). In our study, the differences in utilization of broad classes of substrates (carbohydrates and amino acids) provided discrimination between carbon source utilization patterns of attached and unattached communities in the packed columns; however, there is little context in which to judge the ecological significance of this observation. Interpretation of variable loadings are limited to relative comparisons of the samples included in that particular analysis.
The definition of community composition by the identity of culturable organisms is also normally limited, but in our experiment, the total number of culturable aerobic heterotrophs approximated the numbers of total cells, allowing more general extension of these results. The equivalence of heterotroph enumerations and total cells (as measured in the effluent groundwaters) is the result of an enrichment effect of the laboratory column environment, as shown by the initial feed water, which contained 4.7 × 104 total cells/ml and 6.8 × 103 heterotrophic CFU/ml (data not shown), and the effluent groundwater from all of the columns, which contained about 5 × 105 total cells/ml (data not shown) (heterotrophic CFU/milliliter data are shown in Fig. 6B). Thus, total number of cells increased by an order of magnitude and the majority became culturable by our methods. This increase in culturability is similar to that seen when groundwater or a basalt core is incubated in minimal-salts medium with no added carbon in our laboratory (unpublished observations). Even though most of the cells were culturable, the number of morphotypes and identity of culturable heterotrophs minimally distinguished between attached and unattached communities; however, the FISH results allowed further discrimination by observation of in situ relative population abundances (based on phylogenetic groupings).
Due to the increase in cell culturability, the number of heterotrophs was used to approximate the total biomass that would have been represented by the total number of cells (absolute biomass obtainable with conversion factor) had the DAPI stain not bound so strongly to colloidal basalt particles. The unreliability of obtaining direct counts on DAPI-stained cells from basalt extracts may have confounded the determination of the effects of extraction approaches on total cell counts. This nonspecific binding of DAPI to basalt was used to advantage in the FISH analyses, as basalt extracts were DAPI stained prior to hybridization, reducing nonspecific binding of the labeled probe to colloidal material. Other methods of occupying nonspecific probe binding sites, including the use of an unlabeled NON338 probe and alternative fluors, were not as successful (data not shown). Further, the type of mountant used strongly interacted with the ability to visualize the intended fluor (CY3) and minimize the visualization of the fluor used to occupy nonspecific binding sites (DAPI). With our combination of fluors and basalt, Fluormount was found to outperform the other mountants tested (data not shown).
The most critical limitation in the comparison of attached and unattached communities is the quantitative extraction of cells from basalt. While we did examine the results of different extraction strategies, some cells were undoubtedly not successfully extracted from the crushed basalt. For the fractured columns, the method of mechanically removing the biomass from the fracture surface presented a different type of selectivity. Ideally, assays would be performed in situ on whole rock, although this approach precludes the use of some assays, such as CLPP, and presents great difficulty with others, such as the direct observation of stained cells. The use of multiple assays to describe attached and unattached communities strengthens comparative conclusions, as a single assay may be uniquely influenced by the extraction method. In dual-permeability, crystalline rock aquifers featuring a low-permeability matrix with highly conductive fractures, microorganisms should largely exist on the fracture faces (10). The ability to retrieve uncontaminated, intact fracture faces from the field for direct observation of cells would allow the most unbiased survey of attached organisms in fractured environments.
Consideration of units for comparison of attached and unattached organisms.
For properties that are largely mass independent once a threshold mass is reached (e.g., structural or metabolic diversity, relative population abundances), no consideration of the units used for comparison of attached and unattached organisms is necessary, but for density-dependent properties (e.g., any enumerations), appropriate units must be used for an equitable comparison (43, 47). Traditionally, planktonic organisms have been expressed per milliliter of water and sessile organisms have been expressed per gram of dry weight; these units are obviously not equivalent. The most satisfactory unit for sedimentary aquifers is probably the cubic centimeters of in situ porous media, as used previously by several authors (7, 21, 25, 54), which requires measurement of in situ bulk density. The use of equivalent pore volume for the expression of particle-associated biomass has proven useful for numerical modeling of contaminant transport in porous media (41), but for fractured environments, fracture distribution and widths would have to be assumed. For crystalline rock, where organisms reside on fracture surfaces, surface area seems most appropriate. The external surface area can be calculated by measuring a linear enhancement factor that accounts for the microtopography of the geologic medium, as was done in this study. For comparison of cell density in clastic deposits to that in fractured media, an expression for the external surface area of a particle based on an enhanced particle perimeter can be similarly described. Depending upon inclusion of the response of cells from the interior of the particles in the assay, the results may be scaled by including a measurement of internal surface area. The total surface area (largely internal) of porous particles is commonly measured by gas adsorption, an approach that may include pore space that is inaccessible to bacteria based on the dimensions of connecting pore throats. Therefore, internal surface area should be scaled to account for the internal surface area connected by pore throats through which a bacterium may pass. A consensus on the units used to express attached and unattached biomass would allow the use of a ratio or an enrichment factor that would simplify statistical comparisons of partitioned biomass under different environmental conditions.
Conclusions.
While it is clear that attachment can be associated with great changes in the metabolism of microorganisms (56) and has been shown to affect the biodegradation of organic contaminants (12, 29), there has been little consideration of the appropriate use of groundwater or solid geologic medium in feasibility studies or monitoring plans for in situ remediation. The current literature suggests the existence of qualitative differences between attached and unattached communities in sedimentary aquifers, while there is insufficient published data from crystalline aquifers to support a similar conclusion. The comparative importance of in situ activities of planktonic and sessile biomass and populations in porous- and fractured-medium settings remains unstudied, and conclusions may vary depending on whether activities are expressed per unit of biomass (i.e., specific activity). The relative mobility of a transforming population with respect to a target contaminant and the dependence of the rate of contaminant transformation on the organism's mobility may influence the outcome of in situ bioremediation attempts or the modeling of such processes.
ACKNOWLEDGMENTS
Funding for this project was provided by the Department of Energy Office of Environmental Management to the Idaho National Engineering and Environmental Laboratory operated by Bechtel BWXT, LLC, under contract DE-AC07-99ID13727.
The assistance of the following persons is acknowledged: Sandy Fox for performing the extraction experiment, Eric Robertson for construction and operation of the flow units, Joni Barnes and Louise Holmquist for troubleshooting FISH analyses, Jon Carmack for performance of the micromorphometry of basalt surfaces, and David Cummings for manuscript review.
REFERENCES
- 1.Alfreider A, Krossbacher M, Psenner R. Groundwater samples do not reflect bacterial densities and activity in subsurface systems. Water Res. 1997;31:832–840. [Google Scholar]
- 2.Amann R I, Krumholz L, Stahl D A. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amy P S, Haldeman D L, Ringelberg D, Hall D H, Russell C. Comparison of identification systems for classification of bacteria isolated from water and endolithic habitats within the deep subsurface. Appl Environ Microbiol. 1992;58:3367–3373. doi: 10.1128/aem.58.10.3367-3373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakken L R, Lindahl V. Recovery of bacterial cells from soil. In: Trevors J T, van Elsas J D, editors. Nucleic acids in the environment: methods and applications. Berlin, Germany: Springer-Verlag; 1995. pp. 9–27. [Google Scholar]
- 5.Balkwill D L. Numbers, diversity, and morphological characteristics of aerobic, chemoheterotrophic bacteria in deep subsurface sediments from a site in South Carolina. Geomicrobiology J. 1989;7:33–52. [Google Scholar]
- 6.Balkwill D L, Ghiorse W C. Characterization of subsurface bacteria associated with two shallow aquifers in Oklahoma. Appl Environ Microbiol. 1985;50:580–588. doi: 10.1128/aem.50.3.580-588.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bekins B A, Godsy E M, Warren E. Distribution of microbial physiologic types in an aquifer contaminated by crude oil. Microb Ecol. 1999;37:263–275. doi: 10.1007/s002489900149. [DOI] [PubMed] [Google Scholar]
- 8.Boivin-Jahns V, Ruimy R, Bianchi A, Daumas S, Christen R. Bacterial diversity in a deep-subsurface clay environment. Appl Environ Microbiol. 1996;62:3405–3412. doi: 10.1128/aem.62.9.3405-3412.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bowman J P, Jimenez L, Rosario I, Hazen T C, Sayler G S. Characterization of the methanotrophic bacterial community present in a trichloroethylene-contaminated subsurface groundwater site. Appl Environ Microbiol. 1993;59:2380–2387. doi: 10.1128/aem.59.8.2380-2387.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown D A, Kamineni D C, Sawicki J A, Beveridge T J. Minerals associated with biofilms occurring on exposed rock in a granitic underground research laboratory. Appl Environ Microbiol. 1994;60:3182–3191. doi: 10.1128/aem.60.9.3182-3191.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demezas D H, Bottomley P J. Autecology in rhizospheres and nodulating behavior of indigenous Rhizobium trifolii. Appl Environ Microbiol. 1986;52:1014–1019. doi: 10.1128/aem.52.5.1014-1019.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doong R-A, Chen T-F, Wu Y-W. Anaerobic dechlorination of carbon tetrachloride by free-living and attached bacteria under various electron-donor conditions. Appl Microbiol Biotechnol. 1997;47:317–323. [Google Scholar]
- 13.Fredrickson J K, Balkwill D L, Zachara J M, Li S W, Brockman F J, Simmons M A. Physiological diversity and distributions of heterotrophic bacteria in deep cretaceous sediments of the Atlantic coastal plain. Appl Environ Microbiol. 1991;57:402–411. doi: 10.1128/aem.57.2.402-411.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garland J L. Analysis and interpretation of community-level physiological profiles in microbial ecology. FEMS Microbiol Ecol. 1997;24:289–300. [Google Scholar]
- 15.Garland J L. Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol Biochem. 1996;28:213–221. [Google Scholar]
- 16.Garland J L, Cook K L, Loader C A, Hungate B A. The influence of microbial community structure and function of community-level physiological profiles. In: Insam H, Rangger A, editors. Microbial communities: functional versus structural approaches. Heidelberg, Germany: Springer; 1997. pp. 171–183. [Google Scholar]
- 17.Garland J L, Lehman R M. Dilution/extinction of community phenotypic characters to estimate relative structural diversity in mixed communities. FEMS Microbiol Ecol. 1999;30:333–343. doi: 10.1111/j.1574-6941.1999.tb00661.x. [DOI] [PubMed] [Google Scholar]
- 18.Garland J L, Mills A L. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-cource utilization. Appl Environ Microbiol. 1991;57:2351–2359. doi: 10.1128/aem.57.8.2351-2359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghiorse W C, Wilson J T. Microbial ecology of the terrestrial subsurface. Adv Appl Microbiol. 1988;33:107–173. doi: 10.1016/s0065-2164(08)70206-5. [DOI] [PubMed] [Google Scholar]
- 20.Glockner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trezesius K, Schleifer K-H. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;19:403–406. [Google Scholar]
- 21.Godsy E M, Goerlitz D F, Grbic Galic D. Methanogenic biodegradation of creosote contaminants in natural and simulated groundwater ecosystems. Ground Water. 1992;30:232–242. [Google Scholar]
- 22.Gounot A M. Microbial ecology of groundwaters. In: Gibert J, Danielopol D L, Stanford J A, editors. Groundwater ecology. San Diego, Calif: Academic Press; 1994. pp. 189–504. [Google Scholar]
- 23.Haack S K, Garchow H, Klug M J, Forney L J. Analysis of factors affecting the accuracy, reproducibility, and interpretation of microbial community carbon source utilization patterns. Appl Environ Microbiol. 1995;61:1458–1468. doi: 10.1128/aem.61.4.1458-1468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haldeman D L, Amy P D. Bacterial heterogeneity in deep subsurface tunnels at Rainier Mesa, Nevada Test Site. Microb Ecol. 1993;25:183–194. doi: 10.1007/BF00177194. [DOI] [PubMed] [Google Scholar]
- 25.Harvey R W, Smith R L, George L. Effect of organic contamination upon microbial distributions and heterotrophic uptake in a Cape Cod, Mass., aquifer. Appl Environ Microbiol. 1984;48:1197–1202. doi: 10.1128/aem.48.6.1197-1202.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hazen T C, Jimenez L, de Victoria G L, Fliermans C B. Comparison of bacteria from deep subsurface sediment and adjacent groundwater. Microb Ecol. 1991;22:293–304. doi: 10.1007/BF02540231. [DOI] [PubMed] [Google Scholar]
- 27.Hinds W C. Aerosol technology, properties, behavior and measurement of airborne particles. In co. New York, N.Y: John Wiley & Sons; 1982. [Google Scholar]
- 28.Hirsch P, Rades-Rohkohl E. Some special problems in the determination of viable counts of groundwater microorganisms. Microb Ecol. 1988;16:99–113. doi: 10.1007/BF02097408. [DOI] [PubMed] [Google Scholar]
- 29.Holm P E, Nielsen P H, Albrechtsen H-J, Christensen T H. Importance of unattached bacteria and bacteria attached to sediment in determining potentials for degradation of xenobiotic organic contaminants in an aerobic aquifer. Appl Environ Microbiol. 1992;58:3020–3026. doi: 10.1128/aem.58.9.3020-3026.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karthikeyan S, Wolfaardt G M, Korber D R, Caldwell D E. Functional and structural responses of a degradative microbial community to substrates with varying degrees of complexity in chemical structure. Microb Ecol. 1999;38:215–224. doi: 10.1007/s002489900171. [DOI] [PubMed] [Google Scholar]
- 31.Kepner R L, Pratt J R. Use of fluorochromes for direct enumeration of total bacteria in environmental samples: past and present. Microbiology. 1994;58:603–615. doi: 10.1128/mr.58.4.603-615.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolbel-Boelke J, Anders E-M, Nehrkorn A. Microbial communities in the saturated groundwater environment. II. Diversity of bacterial communities in a Pleistocene sand aquifer and their in vitro activities. Microb Ecol. 1988;16:31–48. doi: 10.1007/BF02097403. [DOI] [PubMed] [Google Scholar]
- 33.Konopka A, Oliver L, Turco R F., Jr The use of carbon substrate utilization patterns in environmental and ecological microbiology. Microb Ecol. 1998;35:103–115. doi: 10.1007/s002489900065. [DOI] [PubMed] [Google Scholar]
- 34.Laviolette R A, Watwood M E, Ginn T R, Stoner D L. Spatial disorder and degradation kinetics in intrinsic biodegradation schemes. J Phys Chem A. 1999;103:4480–4484. [Google Scholar]
- 35.Lehman R M, Roberto F F, Earley D, Bruhn D F, Brink S E, O'Connell S P, Delwiche M E, Colwell F S. Attached and unattached microbial communities in a 120-meter corehole in an acidic, crystalline rock aquifer. Appl Environ Microbiol. 2001;67:2095–2106. doi: 10.1128/AEM.67.5.2095-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindahl V, Bakken L R. Evaluation of methods for extraction of bacteria from soil. FEMS Microbiol Lett. 1995;16:135–142. [Google Scholar]
- 37.MacDonald T T, Kitanidis P K, McCarty P L, Roberts P V. Effects of shear detachment on biomass growth and in situ bioremediation. Ground Water. 1999;37:555–563. [Google Scholar]
- 38.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K-H. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 39.Marshall K C. Planktonic versus sessile life of prokaryotes. In: Balows A, Truper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications, second ed. New York, N.Y: Springer-Verlag; 1992. pp. 262–275. [Google Scholar]
- 40.Mills A L, Bouma J E. Strain and functional stability in gnotobiotic reactors. In: Insam H, Rangger A, editors. Microbial communities: functional versus structural approaches. Heidelberg, Germany: Springer; 1997. pp. 184–194. [Google Scholar]
- 41.Murphy E M, Ginn T R, Chilakapati A, Resch C T, Phillips J L, Wietsma T W, Spadoni C M. The influence of physical heterogeneity on microbial degradation and distribution in porous media. Water Resour Res. 1997;33:1087–1103. [Google Scholar]
- 42.O'Connell S, Lawson R D, Watwood M E, Lehman R M. BASIC program for reduction of data from community-level physiological profiling using Biolog microplates: rationale and critical interpretation of data. J Microbiol Methods. 2000;40:213–220. doi: 10.1016/s0167-7012(00)00128-7. [DOI] [PubMed] [Google Scholar]
- 43.Pedersen K. The deep subterranean biosphere. Earth-Sci Rev. 1993;34:243–260. [Google Scholar]
- 44.Pedersen K. Exploration of deep intraterrestrial microbial life: current perspectives. FEMS Microbiol Lett. 2000;185:9–16. doi: 10.1111/j.1574-6968.2000.tb09033.x. [DOI] [PubMed] [Google Scholar]
- 45.Pedersen K. Investigations of subterranean bacteria in deep crystalline bedrock and their importance for the disposal of nuclear waste. Can J Microbiol. 1996;42:382–391. [Google Scholar]
- 46.Pedersen K, Ekendahl S. Assimilation of CO2 and introduced organic compounds by bacterial communities in groundwater from southeastern Sweden deep crystalline bedrock. Microb Ecol. 1992;23:1–14. doi: 10.1007/BF00165903. [DOI] [PubMed] [Google Scholar]
- 47.Pedersen K, Ekendahl S. Distribution and activity of bacteria in deep granitic groundwaters of southeastern Sweden. Microb Ecol. 1990;20:37–52. doi: 10.1007/BF02543865. [DOI] [PubMed] [Google Scholar]
- 48.Porter K G, Feig Y S. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- 49.Reasoner D J, Geldreich E E. A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol. 1985;49:1–7. doi: 10.1128/aem.49.1.1-7.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roller C, Wagner M, Amann R, Ludwing W, Schleifer K. In situ probing of gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology. 1994;140:2849–2858. doi: 10.1099/00221287-140-10-2849. [DOI] [PubMed] [Google Scholar]
- 51.Rosswall T, Kvillner E. Principal-components and factor analysis for the description of microbial populations. Vol. 2. New York, N.Y: Plenum Press; 1978. pp. 1–48. [Google Scholar]
- 52.Simon M. Specific uptake rates of amino acids by attached and free-living bacteria in a mesotrophic lake. Appl Environ Microbiol. 1985;49:1254–1259. doi: 10.1128/aem.49.5.1254-1259.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smalla K, Wachtendorf U, Heuer H, Liu W-T, Forney L. Analysis of BIOLOG GN substrate utilization patterns by microbial communities. Appl Environ Microbiol. 1998;64:1220–1225. doi: 10.1128/aem.64.4.1220-1225.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas J M, Lee M D, Ward C H. Use of ground water in assessment of biodegradation potential in the subsurface. Environ Toxicol Chem. 1987;6:607–614. [Google Scholar]
- 55.Unanue M, Ayo B, Azua I, Barcina I, Iriberri J. Temporal variability of attached and free-living bacteria in coastal waters. Microb Ecol. 1992;23:27–39. doi: 10.1007/BF00165905. [DOI] [PubMed] [Google Scholar]
- 56.Van Loosdrecht M C M, Lyklema J, Norde W, Zehnder A J B. Influence of interfaces on microbial activity. Microbiol Rev. 1990;54:75–87. doi: 10.1128/mr.54.1.75-87.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Schie P M, Fletcher M. Adhesion of biodegradative anaerobic bacteria to solid surfaces. Appl Environ Microbiol. 1999;65:5082–5088. doi: 10.1128/aem.65.11.5082-5088.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verschuere L, Fievez V, Van Vooren L, Verstraete W. The contribution of individual populations to the Biolog pattern of model microbial communities. FEMS Microbiol Ecol. 1997;24:353–362. [Google Scholar]
- 59.Winding A, Hendrickson N B. Biolog assay for metabolic fingerprints of soil bacteria: incubation time and sensitivity. In: Insam H, Rangger A, editors. Microbial communities: functional versus structural approaches. Heidelberg, Germany: Springer; 1997. pp. 195–205. [Google Scholar]
- 60.Wood W W, Low W H. Aqueous geochemistry and diagenesis in the eastern Snake River Plain aquifer system, Idaho. Geol Soc Am Bull. 1986;97:1456–1466. [Google Scholar]
- 61.Zar J H. Biostatistical analysis, third ed. Upper Saddle River, N.J: Prentice Hall; 1996. [Google Scholar]
- 62.Zlatkin I V, Schneider M, Bruijn F J, Forney L J. Diversity among bacteria isolated from the deep subsurface. J Ind Microbiol. 1996;17:219–227. [Google Scholar]