Summary
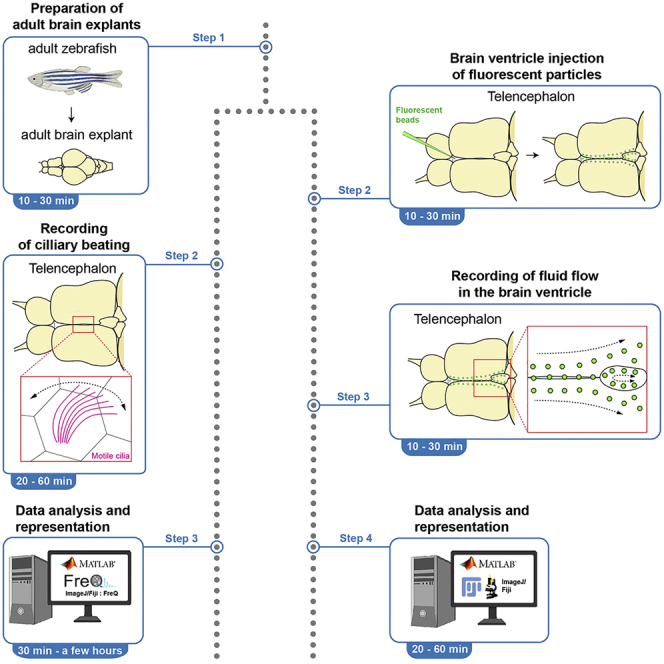
Motile cilia are hair-like structures that move and propel fluid, playing important roles in the physiology of organs. Here, we present a protocol to visualize and measure ciliary beating and cerebrospinal fluid (CSF) flow in the telencephalon of an adult zebrafish brain explant. We describe the preparation of brain explants, the recording of ciliary beating and CSF flow, and data analysis using ImageJ and MATLAB. These imaging and analysis techniques can be directly translated to other ciliated systems.
For complete details on the use and execution of this protocol, please refer to D’Gama et al. (2021).
Subject areas: Cell Biology, Microscopy, Model Organisms, Neuroscience
Graphical abstract
Highlights
-
•
Procedure to dissect an adult zebrafish brain
-
•
Approaches to record ciliary beating and fluid flow in the telencephalon
-
•
Data analysis tools for quantifying ciliary beating and fluid flow
-
•
Adaptable to other motile ciliated systems
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Motile cilia are hair-like structures that move and propel fluid, playing important roles in the physiology of organs. Here, we present a protocol to visualize and measure ciliary beating and cerebrospinal fluid (CSF) flow in the telencephalon of an adult zebrafish brain explant. We describe the preparation of brain explants, the recording of ciliary beating and CSF flow, and data analysis using ImageJ and MATLAB. These imaging and analysis techniques can be directly translated to other ciliated systems.
Before you begin
Before starting the experiment, experimenters should raise adult zebrafish (less than 1 year-old) with the desired genotype (either pigmented or pigment-less) and get approvals for animal experimentation. Prepare various materials and equipment such as surgery kits, solutions, microscopes, and image analysis software. Please see “key resources table” and “materials and equipment.”
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Instant Ocean Salt | https://www.webzoo.net/ | AS 1010006 |
| Sodium chloride (NaCl) | VWR | 27788.297 |
| D-(+)-Glucose | Sigma-Aldrich | G7021 |
| Potassium chloride (KCl) | Sigma-Aldrich | P5405 |
| Magnesium sulfate, heptahydrate (MgSO4·7H2O) | Sigma-Aldrich | M1880 |
| Potassium phosphate monobasic (KH2PO4) | Sigma-Aldrich | P5655 |
| Sodium hydrogen carbonate (NaHCO3) | VWR | 27780.360 |
| Calcium chloride, dihydrate (CaCl2·2H2O) | Sigma-Aldrich | C3881 |
| 70 kDa rhodamine B isothiocyanate-dextran (RITC-dextran) | Sigma-Aldrich | R9379 |
| SPHERO Fluorescent Yellow Particles 1% w/v, Φ = 1.33 μm | Spherotech | FP-0252-2 |
| Experimental models: Organisms/Strains | ||
| Wildtype adult zebrafish (male and female, less than 1 year-old) | n/a | n/a |
| Nacre (mitfab692) adult zebrafish (male and female, less than 1 year-old) | (Lister et al., 1999) | ZFIN:ZDB-ALT-010919-2 |
| Software and algorithms | ||
| ImageJ/Fiji | (Schindelin et al., 2012) | https://imagej.net/Fiji.html#Downloads |
| FreQ plugin for ImageJ/Fiji | This manuscript | https://github.com/hansenjn/FreQ/ |
| TrackMate plugin for ImageJ/Fiji | (Tinevez et al., 2017) | https://github.com/fiji/TrackMate/releases |
| MultiStackReg for ImageJ/Fiji | (Thevenaz et al., 1998) | http://bradbusse.net/downloads.html |
| MATLAB 2021a | MathWorks | https://se.mathworks.com/ |
| MATLAB script for Fast Fourier Transform analysis | MathWorks | https://ch.mathworks.com/help/matlab/ref/fft.html |
| CBF analysis using MATLAB | This manuscript | https://data.mendeley.com/datasets/p6sfdpjpj2/2 |
| Other | ||
| Pressure injector | Eppendorf | Femtojet 4i |
| Thin Wall Glass Capillaries (4 in, OD 1.0 mm, Filament) | World Precision Instruments | TW100F-4 |
| S78 Stage Micrometer (1 mm/0,01 mm, Reflected Light) | Graticules Optics | 02B00421 |
| Mineral oil | Sigma-Aldrich | M8410 |
| Petri dish | VWR | 391-0608 |
| FluoroDish | World Precision Instruments | FD3510-100 |
| Student Vannas Spring Scissor | Fine Science Tools | 91460-11 |
| Student Fine Scissor | Fine Science Tools | 91500-09 |
| Dumont #5 Forceps | Fine Science Tools | 11251-20 |
| Dumont #3c Forceps | Fine Science Tools | 11231-20 |
| Student Standard Pattern Forceps | Fine Science Tools | 91100-16 |
| Tungsten wire | Sigma-Aldrich | 267554-9.5G |
| Carbogen (95% O2/5% CO2) bottles | Linde Gas | Gasart 219 Carbogen |
| Plastic pipets | VWR | 612-1681 |
| Stereomicroscope | Olympus | SZX7 |
| Epifluorescent microscope | Olympus | BX51WI |
| 10× water immersion objective (NA 0.30) | Olympus | 1-U2M583 |
| 40× water immersion objective (NA 0.80) | Olympus | 1-U2M587 |
| Sutter Laser puller | Sutter Instrument | P-2000 |
| Manta camera (for ciliary beating analysis) | Allied Vision | Prosilica GT1930 |
| Ocular camera (Retiga ELECTRO CCD Camera) (for fluid flow analysis) | Q Imaging | 01-ELECTRO-M-14-C |
Materials and equipment
Artificial fish water (AFW)
| Reagent | Final concentration | Amount |
|---|---|---|
| Instant ocean salt | n/a | 0.2 g |
| reverse osmosis purified water | 1 L | 1 L |
| Total | 0.2 g/L | 1 L |
Note: Store AFW at room temperature (23°C–27°C) (maximal time for storage: 6 month). Before starting the experiment, AFW should be prechilled on ice. Volume of AFW can be modified upon needs of experimenters.
Artificial cerebrospinal fluid (aCSF)
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium Chloride (NaCl) | 131 mM | 7.247 g |
| D-(+)-Glucose | 10 mM | 3.964 g |
| Potassium chloride (KCl) | 2 mM | 0.149 g |
| Magnesium sulfate, heptahydrate (MgSO4·7H2O) | 2 mM | 0.394 g |
| Potassium dihydrogen phosphate (KH2PO4) | 1.25 mM | 0.170 g |
| Sodium hydrogen carbonate (NaHCO3) | 20 mM | 2.016 g |
| Calcium chloride, dihydrate (CaCl2·2H2O) | 2.5 mM | 0.294 g |
| reverse osmosis purified water | n/a | 1 L |
| Total | n/a | 1 L |
Note: We use the recipe described in (Turner et al., 1994). Store aCSF at 4°C (maximal time for storage: 3 month). Before starting the experiments, prepare two kinds of aCSF; prechilled aCSF and aCSF bubbled with carbogen (95% O2/5% CO2) at room temperature. Volume of aCSF can be modified upon needs of experimenters.
RITC-dextran mixture
| Reagent | Final concentration | Amount |
|---|---|---|
| 70 kDa rhodamine B isothiocyanate-dextran | 10 mg | |
| aCSF | n/a | 1 mL |
| Total | 10 mg/mL | 1 mL |
Note: Store the mixture at −20°C as aliquots (10 μL) (maximal time for storage: 2 years).
Mixture of fluorescent particles
| Reagent | Final concentration | Amount |
|---|---|---|
| SPHERO Fluorescent Yellow Particles 1% w/v, Φ = 1.33 μm | n/a | 2 μL |
| RITC-dextran mixture (10 mg/mL) | n/a | 10 μL |
| Total | n/a | 12 μL |
Note: Before starting the experiments, prepare the mixture fresh and keep it at room temperate in the dark before loading it into an injection needle. (Maximal time for storage: 8 h).
Preparation of the injection needles
The needles used for the injections are pulled with a puller machine (Sutter Instrument Co. Model P-2000) from thin-walled glass capillaries (1.00 mm; VWR), using the following settings: heat = 785, filament = 4, velocity = 40, delay = 220, pull = 70.
Note: Pulling condition can be adjusted to fit to pulling machines available in the laboratory.
Step-by-step method details
Preparation of the adult brain explant
Timing: 10–30 min
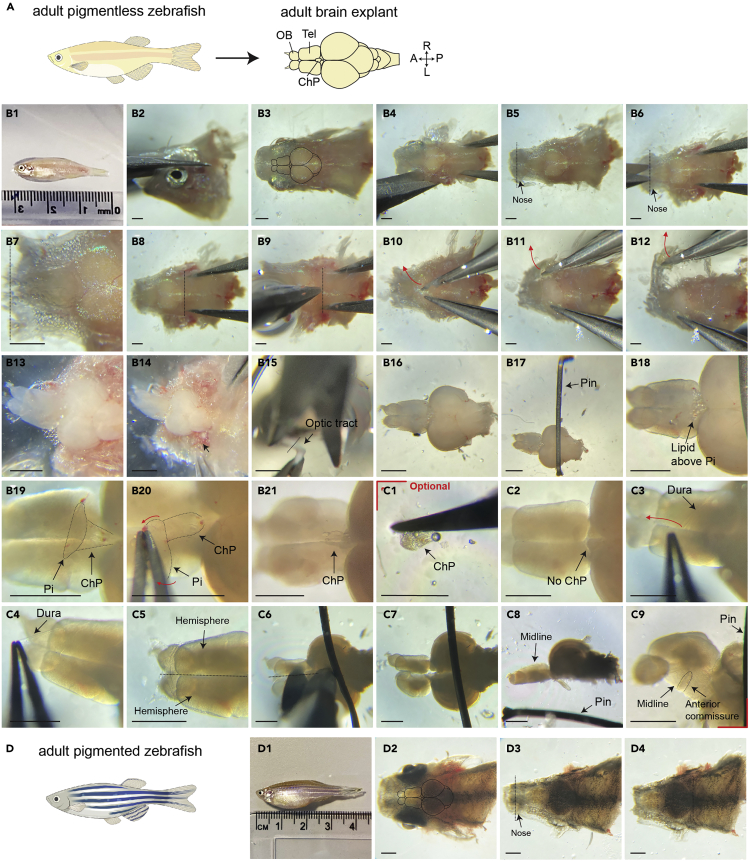
Dissecting intact brain explants (Figure 1A) is critical for the success of the experiment. Damaged adult brain explants can be a major cause of experimental failure. Experimenters should carefully prepare brain explants during the process described below.
-
1.Euthanize adult zebrafish with ice-cold AFW.
-
a.Gently move one adult zebrafish into ice-cold AFW.
-
b.Wait for a few minutes until there is no body and gill movements.
-
c.During the time, prepare a petri dish and fill it up with prechilled aCSF.
-
a.
Note: Hypothermic euthanasia by ice-cold water is not approved by the EU directive 2010/63/EU. Thus, this step could require an additional animal permit, depending on the legislation of each country. Alternatively, overdose of anesthetics could be used for euthanasia. Yet, we do not know if the use of anesthetics has an impact on fluid flow or ciliary beating.
-
2.Record information on the fish and prepare the sample.
-
a.Measure body length and log this information together with the gender (Figure 1B1).
-
b.Cut off the head posterior to the operculum, using scissors.
-
c.From here onwards, work as quickly as possible to avoid brain degradation interfering with the sample quality.
-
a.
-
3.Transfer the head into a petri dish filled with aCSF and proceed to the next steps using cold aCSF to maintain the brain explant alive.
-
a.The lifetime of a brain explant is very variable. However, we recommend the experimenters to finish all dissection steps within 30 min and imaging within 60 min.
-
a.
- 4.
CRITICAL: Do not damage the olfactory bulb. Loss of olfactory bulbs causes leakage or loss of fluid in the telencephalon, which will impair the measurement of fluid flow in the ventricle (troubleshooting Problem 1).
CRITICAL: If the nose part is not properly incised, olfactory bulb could be easily damaged when the dorsal skull is removed.
-
5.
Cut the dorsal skull nearby the cerebellum or brain stem using a small scissor (Figures 1B8 and 1B9).
-
6.Cautiously remove the dorsal part of skull by tearing both sides of the dorso-posterior skull, using fine forceps (Figures 1B10–1B12).
-
a.If there are still bones connected to the olfactory epithelium and olfactory bulb, carefully remove them with the forceps.
-
a.
CRITICAL: Be careful not to scratch the telencephalon, especially the dorsal part where the motile ciliated tela choroida is located. It will not be possible to image fluid flow in a telencephalon with a damaged tela choroida.
-
7.
Remove the ventral parts of the head gently using fine forceps (Figures 1B13 and 1B14). Then, cut optic tracts with a small scissor (Figure 1B15).
-
8.
Mount the brain explant on the small petri dish coated with Sylgard using a 10 mL plastic pipette (Figure 1B16) and fix it with a tungsten pin tightly (Figure 1B17) (Kermen et al., 2020a, 2020b).
Note: Be careful when fixing the explant with the tungsten pin since the pin could scratch the explant easily. Also, if the explant is not affixed firmly, the explant will move, which impairs data acquisition and analysis.
-
9.
Remove the lipid droplets above the pineal gland (Figures 1B18 and 1B19) and pull the pineal gland out very cautiously using fine forceps (Figures 1B20 and 1B21).
Note: The presence of the pineal gland hinders the recording of ciliary beating and fluid flow in the choroid plexus and part of the telencephalon located below the pineal gland. If the target areas are not obscured by the pineal gland, this step is not necessary.
CRITICAL: The pineal gland is located above the choroid plexus and they are connected to each other. Be extremely careful when detaching it. If the choroid plexus is pulled out together with the pineal gland, fluid flow measurement will be impossible since the choroid plexus is linked with the ventricular system. Loss of the choroid plexus may also impair ciliary beating in other areas due to leakage of CSF.
Optional: For imaging of ciliary beating at the telencephalic midline, first remove the choroid plexus (Figures 1C1 and 1C2). Then, take the dura off from the posterior to anterior part of telencephalon (Figures 1C3 and 1C4). Split the hemisphere of telencephalon using forceps or a small scissor (Figures 1C5–1C8) and mount the midline upside by fixing with a tungsten pin (Figure 1C9). Scratching brain areas is acceptable for recording ciliary beating if there is no direct injury at the surface of the midline.
Note: This dissection requires some practice but should be achievable by anyone following our recommendations.
Note: There is constant neurogenesis in the zebrafish brain, hence their brain keeps on growing during their life span. In our experience, it is easier to dissect bigger brain. Nevertheless, the same steps can be followed at any stages.
Figure 1.
Preparation of the adult brain explant
(A) Diagrams showing how the pigmentless adult brain explant is prepared.
(B and C) Detailed procedure for the dissection of the adult zebrafish brain. (C) Optional steps. Red marks indicate optional steps.
(D) Diagrams showing the key steps for the preparation of a brain explant from a pigmented zebrafish. The black lines delineate the border of the adult brain (B3, D2). The dotted straight lines label the cutting sites. Red arrows indicate pulling directions. Black arrows mark brain regions. Anterior is to the right in all panels. Abbreviation: Telencephalon (Tel), olfactory bulb (OB), choroid plexus (ChP), pineal gland (Pi), anterior (A), posterior (P), left (L) and right (R). Scale bar: 100 μm.
Recording of ciliary beating
Timing: 20–60 min
-
10.
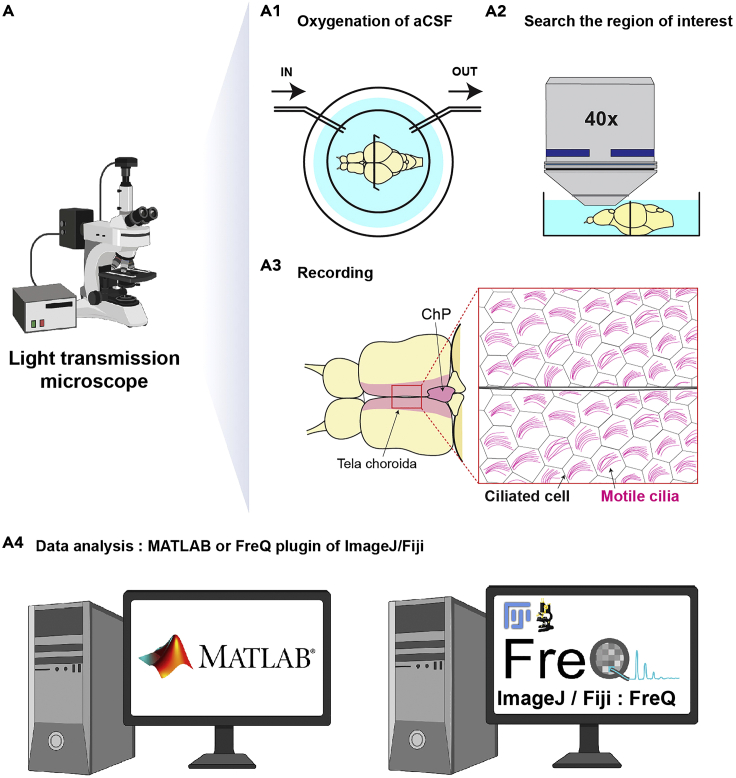
Mount the dish on the brightfield light transmission microscope and exchange to aCSF bubbled with carbogen (95% O2/5% CO2) at room temperature (Figure 2A1).
Note: Be cautious when exchanging cold aCSF to room temperature carbogenated aCSF via a perfusion/suction system. It may alter the location of the explant on the dish.
Note: It is important to exchange to oxygenated aCSF at room temperature for 30 s at least, because temperature could affect ciliary beating (Clary-Meinesz et al., 1992; Green et al., 1995) (troubleshooting Problem 2).
Note: We have a perfusion/suction system to exchange aCSF as shown in Figure 2A1. It is very important to perfuse the sample as much as possible to keep it healthy. We stop perfusion only when an externally applied fluid flow impacts the recording (e.g., motile cilia are exposed or measuring fluid flow).
Note: We do not use an incubator to maintain the temperature. However, we ensure that the temperature is consistent across animals.
-
11.Search the region of interest in the telencephalon (Figures 2A2 and 2A3) and then record the explant with 40× water immersion objective at circa 100 Hertz (Hz) frame rate for 30 s (Figures 2A2 and 2A3).
-
a.We record the explant using an upright Olympus microscope with a Manta camera and a custom designed software previously described (Reiten et al., 2017). The resolution with the 40× objective is 1 pixel = 0.314 μm.Note: Other microscope and cameras can be used for this recording as long as they have similar specifications in spatial and time resolution.
 CRITICAL: The frequency of acquisition needs to be at least 2–3 times the ciliary beating frequency. We usually record at 100 frames per second, which is circa 4–5 faster than the beating frequency and lead to good quality results.
CRITICAL: The frequency of acquisition needs to be at least 2–3 times the ciliary beating frequency. We usually record at 100 frames per second, which is circa 4–5 faster than the beating frequency and lead to good quality results. -
b.Multiple recordings are required to observe overall ciliary beating along the dorsal telencephalon. We recommend the experimenters to acquire recordings from multiple overlapping regions, and then collate the data after the image analysis.
-
c.The recorded files will be used in data analysis (Figure 2A4).
-
d.We recommend the following regions of interest to detect ciliary beating: the choroid plexus, the tela choroida (Figure 2A3) which is located in the superficial mid-dorsal telencephalon, and the ventral telencephalon on the midline surrounding the anterior commissure (Figure 1C9), as described in our prior study (D'Gama et al., 2021).
-
a.
Figure 2.
Measurement of ciliary beating in the adult telencephalon
(A) Diagrams presenting the overall procedure to measure ciliary beating in the brain explant using a light transmission microscope. (A1) Oxygenated artificial cerebrospinal fluid (aCSF) is delivered to the brain explant through a perfusion system (inlet is labeled as “IN” and outlet as “OUT”). (A2) A region of interest is identified using a 40× objective prior to the recording (A3). The pink areas indicate the regions where ciliary beating can be observed (the tela choroida and choroid plexus). The big red box on the right side indicates high magnification of the small red box in the left side. The clusters of magenta lines display motile cilia in the explant. (A4) The analysis can be performed using a MATLAB script or the FreQ plugin in ImageJ/Fiji.
Microinjection of fluorescent particles
Timing: 10–20 min
This step is only necessary for measuring cerebrospinal fluid (CSF) flow. Experimenters who only want to assess ciliary beating can skip this step.
-
12.
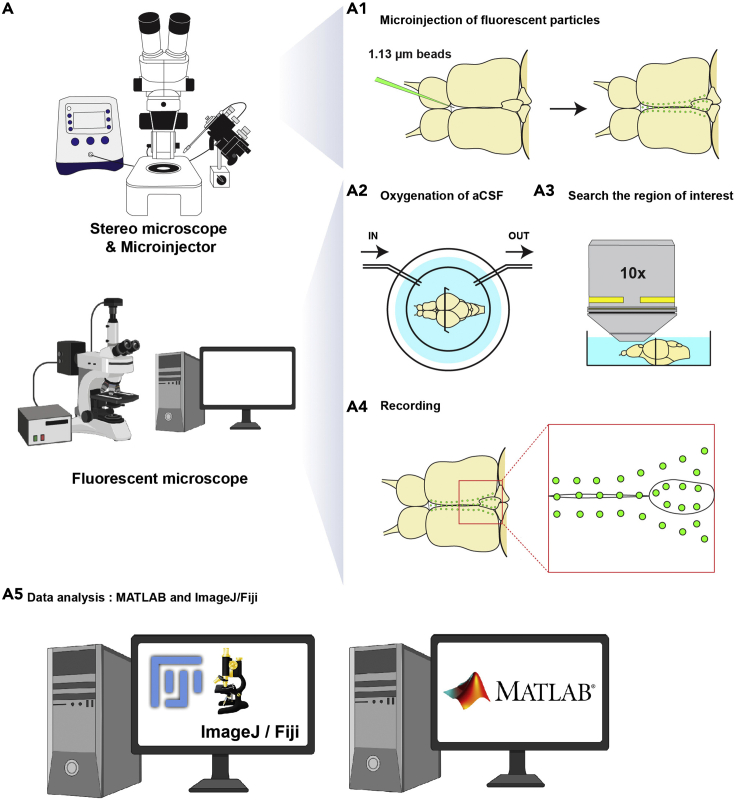
Load the mixture of fluorescent particles (SPHERO Fluorescent Yellow Particles in RITC-dextran) into the injection needle and mount on the needle holder connected with pressure injector (Femtojet 4i). Then, cut the needle tip open with fine forceps.
Note: Start loading the mixture of fluorescent particles after preparation of the brain explant as the mixture easily clogs the needle tip.
Note: Before loading the mixture in the needle, mix it by pipetting rather than using a vortex. In our experience, vortexing does not reduce the clogging of the needles. Also, it may reduce the quantity of the particles by attaching the particle to the tube wall.
Note: Cut the needle tip as little as possible since a large tip diameter could create a large hole in the tela choroida, promoting leakage of CSF.
-
13.Measure the size of a single droplet by injecting into mineral oil on a microscale.
-
a.The single droplet size can be 1–3 nL.
-
b.Try to ensure the reproducibility of your injection by using similar parameters on the pressure injector and needles with openings of similar size.
-
a.
-
14.
Carefully inject the fluorescent particles into the ventricle. Do not penetrate the brain parenchyma. We recommend injecting up to 15 nL in the middle cavity of the most anterior telencephalic area, posterior to the olfactory bulb (Figure 3A1), as this allows to carefully place the needle in the ventricle without touching the parenchyma.
Note: Slowly pull out the needle from the injected site to avoid leakage of CSF and fluorescent particles.
Note: Injection of over 15 nL could increase the ventricle size. This will alter CSF flow patterns in the telencephalon. The volume should be adapted to the size of the brain, e.g. if juvenile zebrafish is used for this analysis.
Note: The needle can be reused. In this case, do not let the needle tip dry. Keep it inside aCSF to prevent clogging of the tip.
Note: If the injection is successful, experimenters will be able to observe particles movements immediately using a light transmission stereomicroscope (Figure 3A1).
Figure 3.
Measurement of cerebrospinal fluid flow in the adult telencephalon
The illustration shows the overall procedure to record cerebrospinal fluid flow in the brain explant.
(A) Microinjection of fluorescent beads into the brain explant, using a stereo microscope and a microinjector.
(A1) Dorsal view of the brain explant. Before (left) and after (right) injection of fluorescent beads (1.13 μm diameter). Green colored needle indicates the injection needle containing the fluorescent beads. Green dots label the distributed fluorescent particles in the explant. (A2) Oxygenation of the brain explant with aCSF (dorsal view). (A3) A region of interest is identified using a 10× objective prior to the recording (A4). The red box in the right side indicates high magnification of the region of interest. Green dots mark the fluorescent beads in the brain. (A5) Data analysis using the TrackMate (Tinevez et al., 2017) plugin in ImageJ/Fiji or MATLAB script (this manuscript).
Recording of CSF flow
Timing: 10–30 min
-
15.
Mount the dish on a fluorescent microscope and exchange aCSF bubbled with carbogen (95% O2/5% CO2) at room temperature (Figure 3A2).
Note: Be cautious when exchanging cold aCSF to room temperature aCSF bubbled with carbogen. It often alters the location of the explant.
-
16.Find the region of interest in the telencephalon (Figure 3A3) and then record the brain explant with 10× water immersion objective at 50 frames per second (fps) for at least 1 min (Figure 3A4).
-
a.We record 2D timelapse of the explant using an upright epifluorescence microscope with an ocular camera and an Ocular software previously described (D'Gama et al., 2021). The resolution with the 10× objective is 1 pixel = 1.13 μm.Note: Other software and camera with similar specifications can be used for this recording.
-
b.The recorded files will be used in data analysis (Figure 3A5).Note: Record the CSF flow as soon as possible. The fluorescent particles tend to stick to the brain parenchyma and tela choroida over time, which will render the recording unusable.Note: Injecting into the brain parenchyma or an inappropriate volume of particles will impact the quality of the recording and of the analysis (Figure 9E). Hence, it is important that the experimenters optimize well their injections and their imaging parameters to ensure the best particle tracking.
-
a.
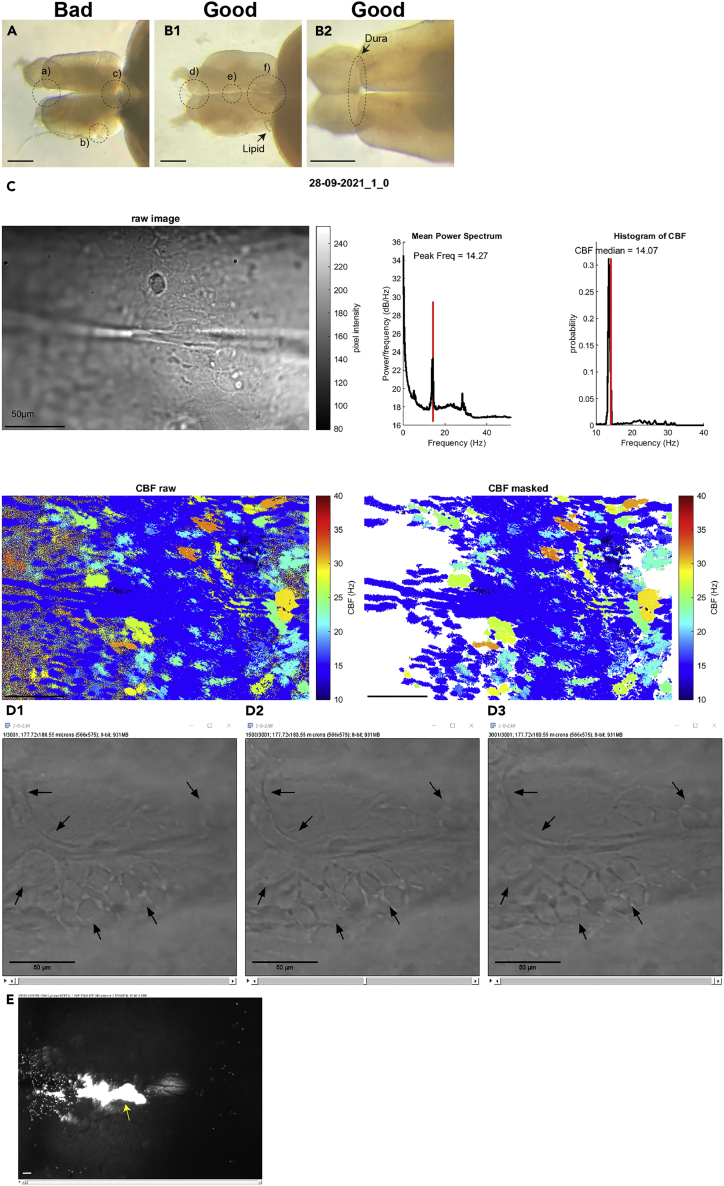
Figure 9.
Examples of good and bad brain explant and analysis
(A and B) Good (B) and bad (A) examples of the adult brain explant. Dotted circles are the areas that experimenters should be careful. Small letters mark each dotted circle that show bad (a)-c)) and good examples (d)-f)). Scale bar: 50 μm.
(C) The PNG output file of a MATLAB CBF analysis on a non-aligned recording file with some drift. Refer to the MATLAB CBF analysis from the aligned recording file (Figure 4D2). Note that there are lots of artefacts (appearing at the frequency of circa 10 Hz, blue in the ‘CBF raw’ and ‘CBF masked’ plots) visible on the mean power spectrum, CBF histogram and CBF heatmaps.
(D) Remaining drifts in an aligned recording file. Images of the first (D1), middle (D2) and last (D3) frame of the 30s recording file. Each black arrow indicates an identical pixel site on the image. Note the loss of sharpness of the image at the arrow locations reflecting z drift.
(E) Example of an unsuccessful fluorescent particle injection. The yellow arrow marks accumulated fluorescent particle in the brain parenchyma. Scale bar: 50 μm.
Data analysis of ciliary beating frequency using a MATLAB script
Timing: 30 min – several hours
To analyze ciliary beating frequency (CBF), we suggest two methods using a MATLAB script or an ImageJ/Fiji plugin (FreQ). The experimenters can choose their method of choice because both methods adapt a similar Fourier-based principle to analyze CBF (Figure 4A) (Haward et al., 2021; Reiten et al., 2017; Ringers et al., 2021). However, we recommend the experimenters to always use the same method for their analysis, since there are slight differences in their output results, as indicated in the limitations section and in Figure 10.
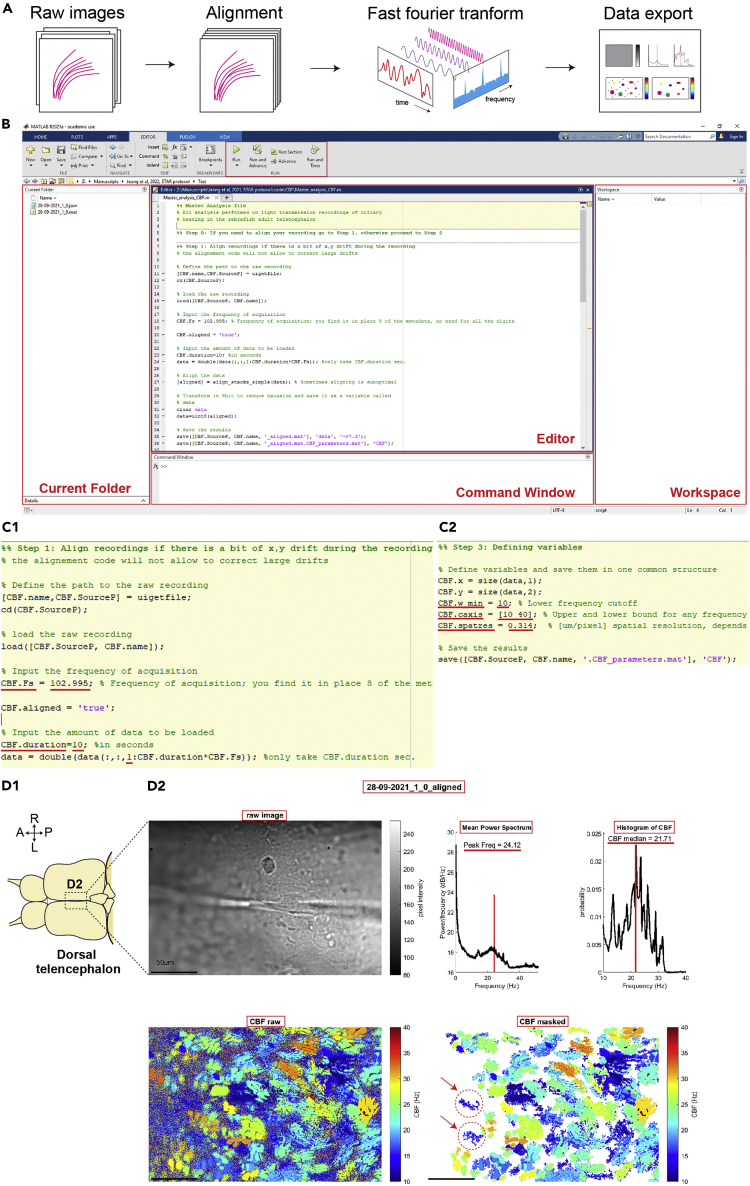
Figure 4.
Data analysis for ciliary beating images using a MATLAB script
(A) The diagrams show the principle underlying the analysis for ciliary beating using the provided MATLAB scripts.
(B) The interface of MATLAB.
(C) The partial screenshot of “step 1” (C1) and “step 3” (C2) in the MATLAB script. The numbers with red lines can be modified to reflect the experimental or analyzing conditions.
(D) Expected outcome of data analysis. (D1) Diagram of the brain explant (dorsal view). (D2) The PNG file output after analysis. The file contains the raw image, mean power spectrum, histogram of ciliary beating frequency (CBF) and color-coded raw/masked CBF. Red dotted circles with red arrows indicate difference between the results by the MATLAB script and FreQ plugin. Abbreviation: (D1) anterior (A), posterior (P), left (L) and right (R). (D2) Peak Frequency (Peak Freq) and Hertz (Hz). Scale bars are 50 μm.
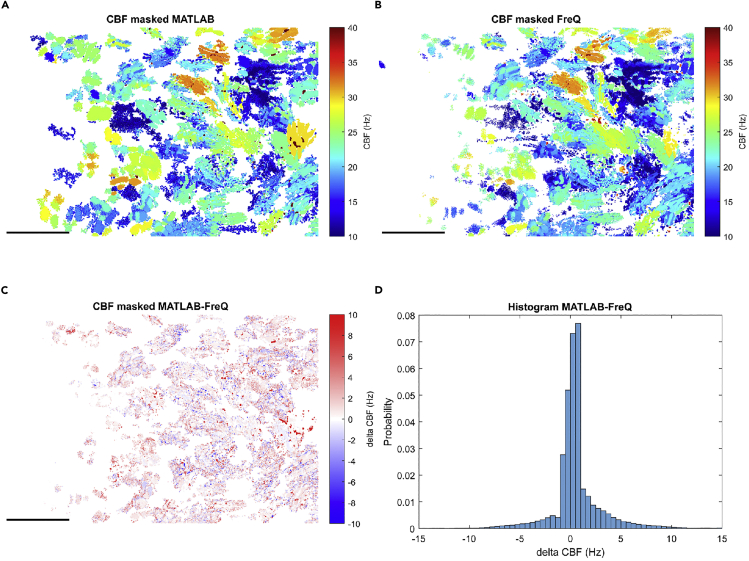
Figure 10.
Comparison of the data analysis for ciliary beating from the MATLAB script and FreQ plugin
(A and B) Color-coded masked CBF plots from the MATLAB script (A) and the FreQ plugin (B).
(C and D) The differences between the MATLAB and FreQ outputs are shown as a heat map of masked CBF (C) and histogram (D). Scale bars are 50 μm.
Before starting this analysis, install the software (MATLAB, 2021a or the latest version of ImageJ/Fiji with FreQ plugin). Here we describe detailed steps for users who are unfamiliar with MATLAB scripts or ImageJ/Fiji.
Be aware that the time of analysis will increase for larger image sizes and longer time of recording.
Since the analyses calculate the frequency of the intensity changes for each individual pixel of the recording, any drifts will lead to artifacts (Figure 9C). Hence it is crucial to make sure that there are no drifts in x and y axis prior to the analysis. If you observe drift in x,y, you may try to correct those with an alignment code prior to the CBF analysis described earlier (Reiten et al., 2017; Ringers et al., 2021). However, it is important that you check manually, frame by frame that, after correction, the sample is no longer drifting in the image. To do this you may pick 2–3 very obvious features in the image and check that these localize at the same pixel position in the image and do not change shape or rotate across all frames of the image. We provide in Figure 9D an example of a recording with a large drift that cannot be corrected with the alignment code.
Note: Drifts in z cannot be corrected. Hence it will not be possible to analyze recordings with z drift.
Note: We have included the raw and aligned datasets in Mendeley Data (https://data.mendeley.com/datasets/p6sfdpjpj2/2) to help the reader gain confidence in how to assess an aligned dataset
-
17.Download all provided MATLAB scripts (https://data.mendeley.com/datasets/p6sfdpjpj2/2) into one folder and set the Path to this folder.
-
a.To add the folders to the Path, right click the folders at the “Current Folder” tab and choose “Add to Path”- “Selected Folders and Subfolders”. This will change the gray lettered name of folder to black lettered the name of folder.
-
a.
Note: The MATLAB codes and raw recording files can be located in different folders.
Note: Adapt to the interface of MATLAB. (Figure 4B).
-
18.
Open the “Master_analysis_CBF” file using the MATLAB software.
Note: The file “Master analysis_CBF” contains detailed comments on how to proceed the steps. Following the comments will be helpful for the analysis.
-
19.To align the recording file, at the “step 1” of “Master_analysis_CBF” at the “Editor” tab, enter the value of the frequency of acquisition (Figure 4C1, red lines) as “CBF.Fs.”. Click the “Run Section” button. (Figure 4B) Then, select the raw recording file (.mat format) when the window “Select Files to Open” pop up.
-
a.To do “Run Section”, the section should be indicated with yellow color (Figure 4B).
-
b.This alignment step is optional, however, it will increase the quality of analysis if there are drifts. We recommend users to conduct this step and to visually inspect aligned files prior to further analysis to ensure that drifts are corrected. Note that the alignment codes will not be able to fix large drifts as shown in Figure 9D. Hence, we recommend experimenters to discard recordings with drifts that cannot be corrected.
-
c.Place the exact value of frequency for accurate analysis.
-
d.It will take longer time when the image size and length of recording are larger. The estimated end-time will be shown in the “Command Window” tab (Figure 4B). The time also changes depending on the computer specifications.
-
e.We usually analyze 10 s to measure ciliary beating frequency. However, it can be modified by the purpose of experimenters. Put the desired analysis time at “CBF.duration” and starting timepoint of analysis instead of “1” (Figure 4C1, redlines).
-
f.If there are already aligned files or users do not want to align the recording files, skip the “step 1” and proceed the “step 2” by clicking “Run Section.” Select the aligned or raw files after a new window “Select Files to Open” pop up. In case of using not aligned raw files, experimenters should enter extra information such as the frequency of acquisition and the duration of file for analysis when questions appear in the “Command Window.”
-
a.
-
20.Define variables like the cutoff and displaying range of frequency and spatial resolution of microscope at the “step 3”. Then, run the section (Figure 4C2).
-
a.We usually use 10 Hz for the lower cutoff frequency, 10–40 Hz for plotting frequency, based on our prior work (D'Gama et al., 2021). These values can be adapted based on the cilia beating properties of the tissue. Also, 0.314 μm/pixel is the spatial resolution of our microscope with a 40× objective. Experimenters can adjust the values for their purpose and specification of their microscope.
-
a.
-
21.
Perform the fast Fourier transform by running the “step 4” section. The analysis result will be saved as a PNG file and one structure array called CBF, containing all metadata and Fourier transform results.
Note: This step including the plotting and saving takes time. The time will change depending on the specification of the analyzing computer.
Note: The PNG file will show the raw image of the recording file, mean of power spectrum, histogram of CBF for the masked CSF heatmap, color-coded CBF heatmap and masked CBF heatmap (Figure 4D2).
Data analysis of ciliary beating frequency using the FreQ plugin
Timing: 30 min – several hours
To analyze CBF, experimenters could also use ImageJ/Fiji with the plugin FreQ. To use this plugin, users should install the plugin (https://github.com/hansenjn/FreQ/) and prepare the recording file as a TIF format. This approach provides another tool to analyze CBF. It could be more convenient for the users who are not familiar with handling MATLAB scripts or do not have access to MATLAB.
To correct small drifts, we suggest using an ImageJ plugin called “MultiStackReg” by Brad Busse prior to CBF analysis (http://bradbusse.net/downloads.html). The MultiStackReg plugin is an adaptation of another ImageJ plugin called “TurboReg” (http://bigwww.epfl.ch/thevenaz/turboreg/) (Thevenaz et al., 1998), which also needs to be installed.
Note: The MultiStackReg (= TurboReg) plugin uses the first frame in a time series as a reference and transforms the following frames to match the first frame. For the transformation, the user can select one of four different methods: Translation, Rigid Body, Scaled Rotation, and Affine. If you observe only drift in X or Y direction, a simple translation function suffices and should be selected, since translation does not change the scale in the images. Note, that the other transformation functions also allow to correct rotation of the sample or rotation around X and/or Y axis, as they can stretch and rotate the other frame images to match the first frame. This, however, may change relations of the distances between objects in the transformed frames and thus such transformed files should be only subjected to distance measures with caution. For a detailed description of the individual transformation methods please visit the TurboReg webpage: http://bigwww.epfl.ch/thevenaz/turboreg/.
Note: The FreQ plugin used in the following workflow cannot analyze images that have been modified in Fiji / ImageJ and not saved somewhere in your file system - if unsaved images are subjected to the analysis, an error message will appear. Make sure to save the images after registration before continuing with a FreQ analysis.
-
22.
Open ImageJ/Fiji after installation of the FreQ plugin.
Note: The format of the files should be TIF format to use this plugin.
Note: Experimenters can open recording files before starting the FreQ plugin.
-
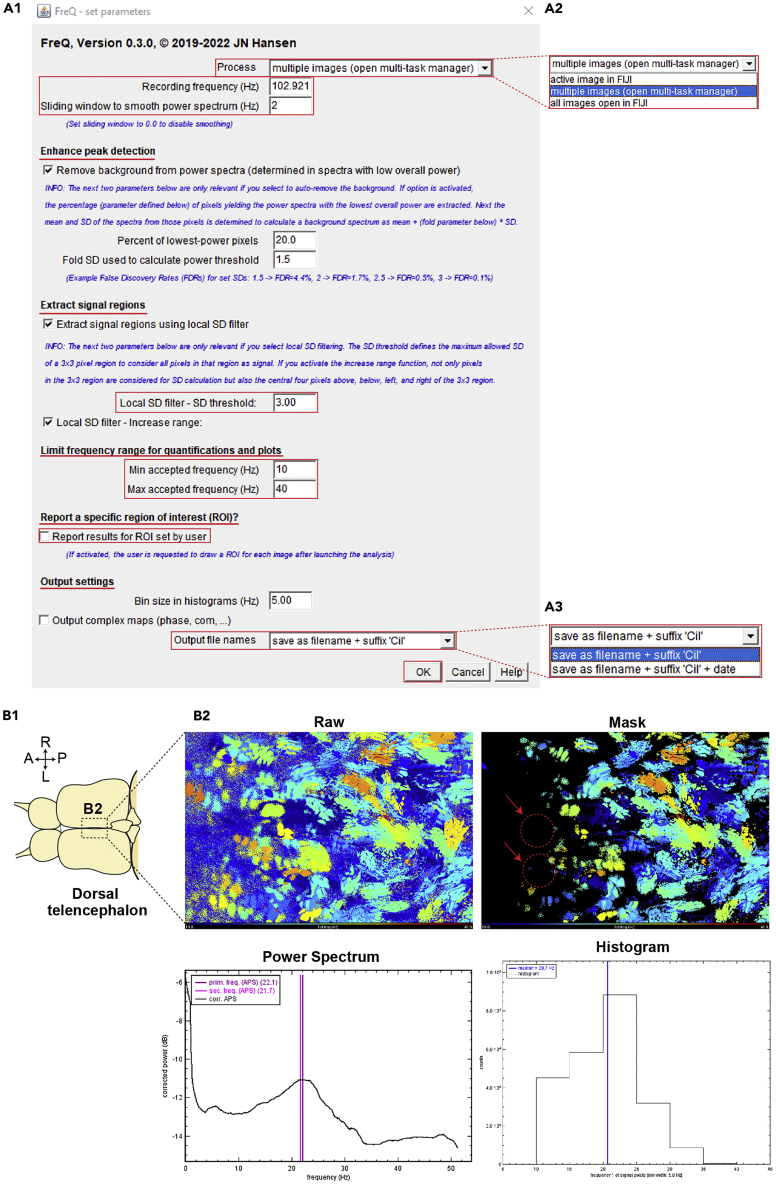
23.Start the FreQ plugin to set up parameters for analysis.
-
a.Click the taps, “Plugin”- “JNH”- “FreQ” to open it. Then, a new window will popup (Figure 5A1).
-
b.Enter values using the guidance described in the following steps.
-
a.
- 24.
-
25.
Enter the exact value of acquisition frequency in the “Recording frequency.”
-
26.In the “Sliding window to smooth power spectrum”, put the number to smooth power spectrum.
-
a.We used 2 for assessing CBF of the adult brain explant.
-
b.The value may be increased for recordings of samples with very instable beating frequencies to improve the detection of the average frequency or decreased for more sharp detection of the most predominant frequency instead of determining an averaged peak frequency.
-
a.
-
27.The section under “Enhance peak detection” provides an extra function that is not implemented in the MATLAB script above. We have experienced that this function can help to detect frequencies in unstable / noisy recordings (for examples, see Note below). Under such conditions, users may try the default settings for this section.
-
a.The default state of “Percent of lowest-power pixels” and “Fold SD” (standard deviation) used to calculate power threshold” are “20.0” and “1.5”, respectively.Note: These parameters may require optimization to your data set. Importantly, the percent of lowest-power pixels needs to be set in a way that the percentage does not exceed the percent of the image’s field-of-view where ciliary beating is visible. If the percentage is set too high, signals from ciliary beating in the image will influence background estimations. In turn, the background will be estimated too high. Thus, not all ciliary beating in the image will be quantified. This also means that this function should not be used if your whole image is showing beating cilia and if there are no image regions, where no ciliary beating is present. The SD for the power threshold also determines the background estimations and requires fine-tuning. If the SD is set too high, you will miss-out lower-powered frequencies in the results. If the SD and/or the percentage are set too low, you will still see a lot of noisy regions in the output maps.
-
b.“3.0” is usually used for the “Local SD filter” and corresponds to the value implemented in the MATLAB script.Note: Additional Examples: This function can help in analyzing recordings at extremely high temporal and spatial resolution (e.g. 3000 Hz, recordings where individual cilia are distinguishable in the image).In high-temporal-resolution recordings, it can occur that the whole sample shivers with an additional stable high frequency (e.g., 100 Hz and higher), which can be, e.g., caused by an electrical device placed on the microscope’s table or the microscope itself. This function can be used to suppress this stable, overlying frequency in the frequency peak detection.In high-spatial resolution recordings there maybe oscillations of background structures become apparent as an underlying oscillation at low amplitude (e.g., triggered by cilia remote from the focal plane). Such oscillations appear in the power spectrum at lower power but may distract the primary frequency peak detection and blur the whole spectrum. When using this function, the background frequency spectrum is determined from a region where no sharp cilia are present (low power region) and this spectrum is subtracted from all spectra so that detected peaks will reflect the obvious ciliary beating in the image.
-
a.
-
28.In the section of “Limit frequency range for quantification and plots”, enter the values of minimum and maximum frequency that experimenters desire to analyze. Then click “OK”. (Figure 5A1).
-
a.We commonly use the range from the minimum “10” to maximum “40” for our analysis. It can be set differently by users.
-
a.
Optional: If experimenters want to assess a specific region of interest (ROI), they can use it by activating “Report results for ROI set by user”. Then, for each analyzed image, FreQ will request the user to draw a ROI of interest and will output (in addition to all other results) results determined within that user-defined ROI only.
Optional: Users could select formats of output file names (Figures 5A1 and 5A3).
-
29.If the user had selected to process “multiple images (open multi-task-manager)” above, a new window will pop up that allows to make a list of files to be processed. Click the “select file individually” to find the recording files after a new window pop up.
-
a.Experimenters could select multiple files for the analysis.
-
a.
-
30.Press the button, “start processing”. Then, the process will start.
-
a.Longer / larger recording files will take more time to finish the analysis.
-
a.
-
31.After this analysis is finished, the analysis files will be saved automatically at the same folder that the recording files are located.
-
a.The list of output files is identical to the output of the MATLAB scripts and contains color-coded CBF heatmaps unmasked and masked as well as the average power spectrum and histogram of CBF (Figure 5B2).
-
a.
Figure 5.
Data analysis for ciliary beating using the FreQ plugin
(A) The screenshots show various setup conditions on the parameters of the analysis in the FreQ plugin.
(B) Expected outcome of the analysis. (B1) Diagram of the brain explant (dorsal view). (B2) The output PNG files after analysis. The files are color-coded raw/masked CBF, power spectrum of primary/secondary peak frequency, histogram of ciliary beating frequency (CBF). Red dotted circles with red arrows indicate difference between the results by the MATLAB script and FreQ plugin.
Data analysis of CSF flow
Timing: 10–30 min
To analyze the directionality of fluid flow in the adult brain explant, we use both ImageJ/Fiji and MATLAB script for the analysis. We describe the detailed steps below and provide a MATLAB script to examine the directionality of fluid flow.
-
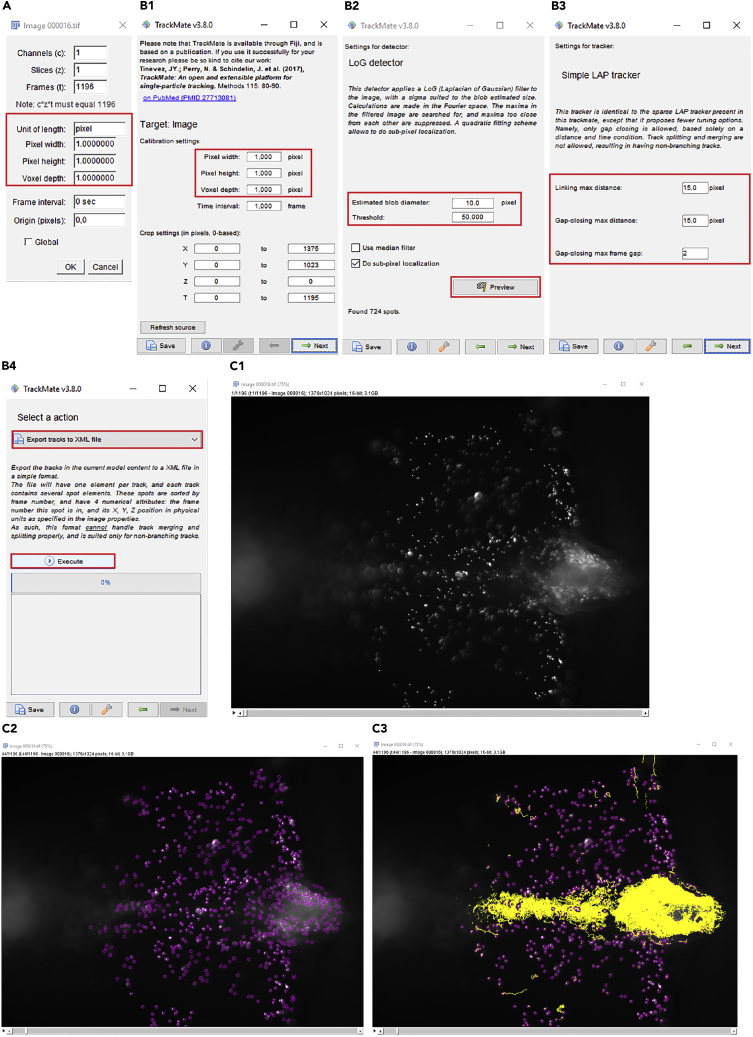
32.Open the recording file using ImageJ/Fiji program and convert the image scale to pixel (Figure 6A).
- a.
-
33.Open the “TrackMate” plugin (Tinevez et al., 2017) in the ImageJ/Fiji. Then, click the “Next” after checking that each value is written correctly (Figure 6B1).
-
a.To open “TrackMate” plugin, click the tables “Plugins”-“Tracking”-“TrackMate”.
-
b.All values should be “1.000” at the “calibration settings”. Whereas, “X” and “Y” of “Crop settings” indicate the pixel size of the image and “T” means the number of images, i.e., timepoints.
-
c.If the values do not match with recording images, try clicking “Refresh source” and then check the value again. If there is no change, start “TrackMate” plugin again after making sure that step 32 was done correctly.
-
a.
-
34.
Use the default detector, “LoG detector” and proceed “Next”. Set the “Estimated blob diameter: 10.0” and “Threshold: 50” (Figure 6B2). Then verify that fluorescent particles are detected by clicking “Preview” (Figures 6B2 and 6C2). Click “Next” to proceed.
Note: We use the 10 pixel of blob diameters and a lower threshold of 50 to detect the fluorescent particles. However, these values need to be modified based on the imaging conditions and particles used. Set these numbers to detect the fluorescent particles correctly (you can check which particles are detected using the “Preview” button).
Note: Detected fluorescent particles are labeled by purple circles (Figure 6C2).
Note: It will take time from a few second to a few minutes after clicking “Next”. During the step, TrackMate will provide various information including the number of found spots.
-
35.Proceed “Next” four times and select the default tracker “Simple LAP tracker.” Then, click “Next.” Use the default values of pixels for “Simple LAP tracker” and proceed “Next” (Figure 6B3).
-
a.Default values of “Simple LAP tracker”: Linking max distance: 15.0, Gap-closing max distance: 15.0 and Gap-closing max frame gap: 2.
-
a.
Note: These values should be adapted to your image settings.
-
36.
Yellow tracer lines by the particles will be shown at the image file (Figure 6C3). Proceed “Next.”
-
37.
Color-coded tracer lines will be shown at the image file. Proceed “Next” three times to see “Selection a action (Figure 6B4)”
-
38.Choose “Export tracks to XML file” at the “Selection a action.” Click “Execute” and then save the file in the desired folder.
-
a.The folder saving XML files should be in the added paths for running MATLAB codes.
-
b.The next steps on MATLAB will allow to plot the tracks. If you do not have access to MATLAB, you can use the inbuild plotting functions of TrackMate or other programing languages (e.g., R or Python) to plot the track of the particles.
-
a.
-
39.
Place all provided MATLAB codes and files in the folders that experimenters want to analyze.
Note: The MATLAB codes and raw recording files can be located in different folders.
-
40.
Open the “masterfile_PlotParticlesTracks” file using the MATLAB software and find the folders at the “Current Folder” tab. Then, add both folders containing the codes and recordings to the Path.
Note: To add the folders to the Path, right click the folders at the “Current Folder” tab and choose “Add to Path” > “Selected Folders and Subfolders”. This will change the gray lettered name of folder to black lettered the name of folder.
Note: The file “masterfile_PlotParticleTracks” contains detailed instructions on how to proceed with the data analysis. Following the comments will be helpful for the analysis.
-
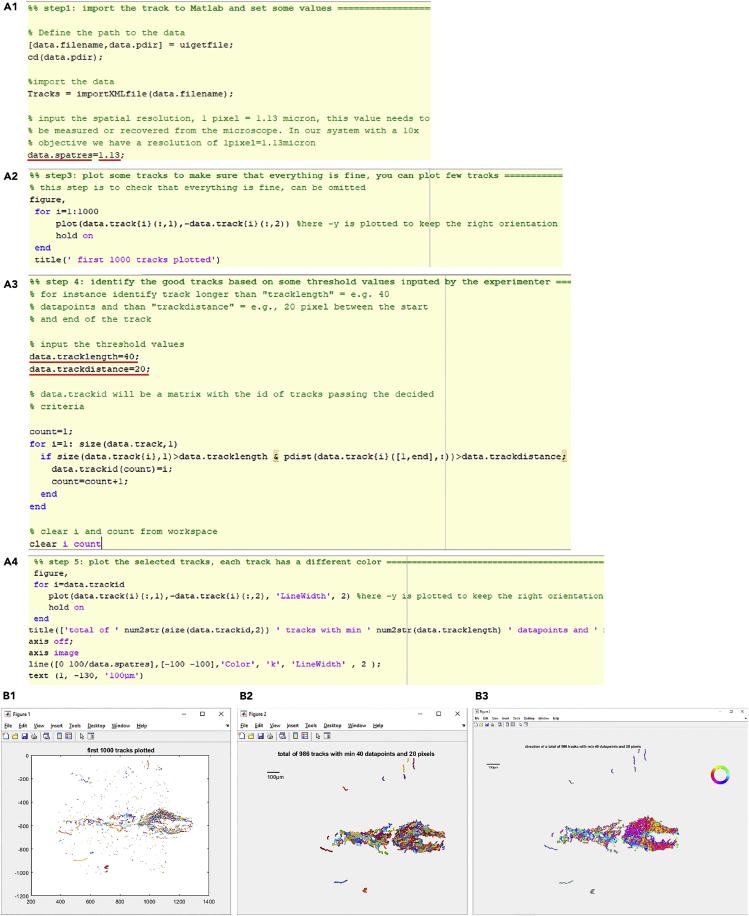
41.To import the track data, click “Run Section” at the “step1” of “masterfile_PlotParticleTracks” at the “Editor” tab (Figure 7A1).
-
a.Before click “Run Section”, put information of spatial resolution of data at the “data.spatres”. The resolution with our 10× objective is 1 pixel = 1.13 μm.
-
b.Confirm that the “data” and “Tracks” files appear in the “Workspace” as well as the defined path and data at the “Command Window” after running.
-
a.
-
42.
Run “step2” to recognize the traces of each particle in the image files and reorganize them in a new array “data.track”.
- 43.
-
44.Run “step4” to identify the good tracks based on some threshold values inputted by the experimenter (Figure 7A3).
-
a.The values of “tracklength” and “trackdistance” at the script can be modified by the experimenters to fit their imaging settings.
-
b.In our example, we have set a threshold of 40 datapoints of “tracklength” (corresponding to 0.8 s) and 20 pixels of “trackdistance” (corresponding to 22.6 μm). Hence, the code will identify tracks of the particles longer than 0.8 s and 22.6 μm.
-
a.
- 45.
-
46.Run “step6” to plot selected tracks color-coded based on the direction of the particle movement. It represents the angle of a particle from t=i to t=i+1.
-
a.This step will take time from a few seconds to minutes. The “Figure 3” will pop up after running (Figure 7B3). It will be automatically saved as PNG file format and the name will be the data file name combined with “tracks”.
-
a.
-
47.
Run “step7” to save the data.
Figure 6.
Data analysis for CSF flow using ImageJ/Fiji
(A and B) The screenshots show each step of the analysis using the TrackMate plugin in ImageJ/Fiji using our suggested parameters.
(C) The screenshots of recording files during the TrackMate analysis. Dorsal view of the adult brain explant with injected fluorescent particles. (C2) Particles detected by TrackMate are circled in magenta. (C3) Tracks detected by TrackMate are highlighted in yellow.
Figure 7.
Data plotting for CSF flow using a MATLAB script
(A) The screenshots show partial parts of the MATLAB script. The numbers with red lines can be modified based on the experimental or analyzing conditions.
(B) The figure files that are generated during analysis showing a subset of tracks (B1), all tracks that meet the experimenters’ thresholds with colors representing each track (B2) or the direction of movement (B3, direction is represented by the rose plot). Only the Figure 3 (B3) is saved.
Expected outcomes
This protocol provides the methods to measure ciliary beating and CSF flow in the telencephalon of an adult zebrafish brain explant. Combining light transmission microscope and fast Fourier transform-based analysis (D'Gama et al., 2021; Olstad et al., 2019; Reiten et al., 2017; Ringers et al., 2021) allows experimenters to examine the extent and frequency of ciliary beating. We also describe how to measure CSF flow upon injection of fluorescent particles into the brain ventricle and tracking via fluorescent imaging.
As described in our prior study (D'Gama et al., 2021), we observe ciliary beating in the choroid plexus, tela choroida (Figures 4D1 and 4D2) and ventral telencephalon on the midline surrounding the anterior commissure (data not shown). At adult stage, cells are mainly multiciliated (D'Gama et al., 2021). The CBF of the areas shows heterogenous values from 10 to 40 Hz (Figure 4D2), as described previously (D'Gama et al., 2021). Hence, we recommend the experimenters to take into account the CBF variability in control animals, prior to measure and report differences in treated or mutant conditions. We also provide here a comparison of the results obtained with our MATLAB codes and FreQ plugin (Figures 4D2, 5B2, and 10).
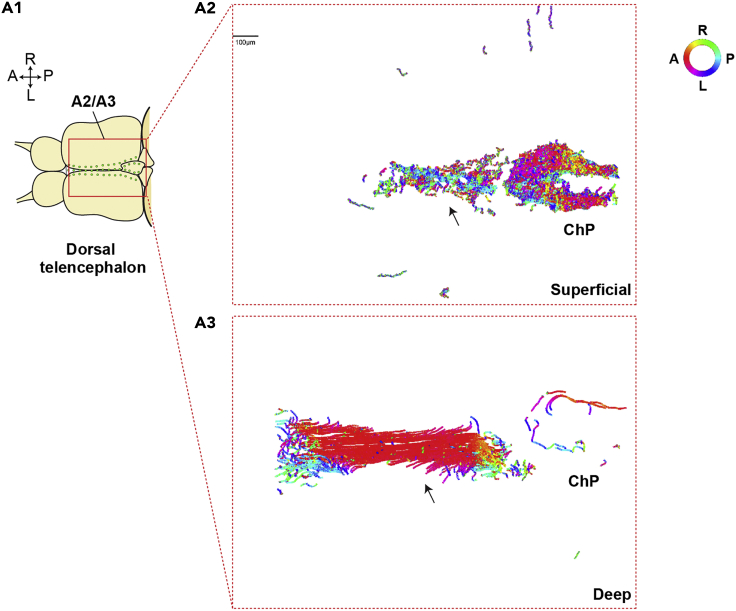
Using the methods described in this protocol, we report a directional CSF flow in the dorsal telencephalic ventricle. Note that CSF flows primarily from anterior to posterior in the superficial telencephalon (Figure 8A2) and posterior to anterior in the deeper area (Figure 8A3). It also flows rostrally anterior to the commissure and caudally posterior to the commissure in the ventral telencephalic area (data not shown) (D'Gama et al., 2021). In the choroid plexus, fluid moves throughout its cavity (Figures 8A2 and 8A3).
Figure 8.
Expected outcomes of CSF flow measurement
(A) Expected outcomes of CSF flow analysis. (A1) Diagram of the brain explant (dorsal view) injected with fluorescent particles. Color-coded particle tracks of superficial (A2) or deep (A3) parts of the dorsal telencephalic ventricle.
This protocol is also applicable to other ciliated tissues. For instance, we used identical analytical tools to measure ciliary beating of multiciliated cells in the zebrafish nose (Reiten et al., 2017; Ringers et al., 2021), mouse tracheal epithelial cultures (Haward et al., 2021), and acute slices of the murine lung (unpublished data). However, it may be difficult to apply this approach for the analysis of monociliated tissues because it is hard to detect ciliary beating of mono cilia by a light transmission microscope. The mono motile cilia can be detected in the zebrafish brain at 2 day-post-fertilization (dpf) stage (Olstad et al., 2019). We only performed the analysis using adult zebrafish over 2-month-old to observe robust ciliary beating and CSF flow. To analyze ciliary beating of monociliated cells, in the zebrafish embryonic system (Fame et al., 2016; Kramer-Zucker et al., 2005; Olstad et al., 2019; Thouvenin et al., 2020), we recommend the use of a fluorescent transgenic zebrafish line, such as e.g., Tg(b-actin2:arl13b-gfp (Borovina et al., 2010), combined with the MATLAB script provided here (Olstad et al., 2019). We would also recommend experimenters to use the SpermQ ImageJ/Fiji plugin (Hansen et al., 2018) for the analysis of individual motile cilia, as it provides additional parameters, including the tracking of the cilium over time. Primary cilia may also exhibit some movements over time, yet without a regular beating pattern and frequency. If experimenters would be interested to investigate the properties of primary cilia, we refer them to the CiliaQ ImageJ/Fiji plugin (Hansen et al., 2020, 2021b).
Limitations
We previously reported that the ventricular system of the telencephalon collapses in the adult brain explants as compared to the brain of a deeply anaesthetized animal (D'Gama et al., 2021). The reduced size of the ventricular cavity might affect ciliary beating and CSF flow and hence may be different in an in vivo situation. Nevertheless, ciliary beating is still maintained in brain explants. Thus, we assume that the directionality of fluid flow is similar between dissected brain explants and the in vivo ventricular system. Moreover, experiments using adult brain explants are ex vivo condition rather than in vivo. In the ex vivo condition, there are no body movement, heart beating and other factors which may affect CSF flow (Ringers et al., 2020). Thus, our ex vivo preparation only reflects cilia-driven CSF flow. There are currently technical limitations to observe ciliary beating and fluid flow in the adult zebrafish brain in vivo in a non-invasive manner because the skull and other tissues above the dorsal telencephalon hinder imaging cilia and fluid movements. If novel tools are developed to overcome these challenging issues, movements of cilia and fluid in live adult zebrafish could be assessed.
To assess CSF flow, we inject exogenous particles. Even though our protocol is well optimized to minimize the volume and hence the impact of injection of exogenous particles, it may still affect the ventricular system and CSF flow. It is crucial to limit the injected volume and avoid damage of the ventricles upon introduction of the needle, which could produce leakage of CSF and alter flow. We have also observed that our fluorescent beads tend to stick to the brain parenchyma over time and may therefore disturb particle movement. Different particles and coatings may reduce their adherence to brain tissues and provide better recordings. Nevertheless, there is no non-invasive method available currently to observe CSF flow without inserting exogenous substances.
In our protocol, we use single plane recording to detect ciliary beating and CSF flow. Because ciliary beating and CSF flow are three-dimensional (3D) movements, these 2-dimensional (2D) approaches are only approximation of the 3D movements. There are methods allowing fast imaging in 3D (Hansen et al., 2021a; Silva-Villalobos et al., 2014), but these require customized imaging setups and thus, unfortunately, are not broadly available to the community. We argue that by imaging in 2D with epifluorescence and transmission microscopes allows us to have a thicker view of the Z axis. This provides more information at the 3D level, compared to imaging single planes using confocal microscope. Future work could potentially compare 2D vs 3D level measurements, but this goes beyond the scope of this protocol.
Note that there are slight differences in the output of our MATLAB script and the FreQ plugin (Figures 4D2 and 5B2, indicated with red circles and Figure 10). Since we implemented an identical workflow in both tools (MATLAB and FreQ plugin), this deviation can only result from different implementation of the Fast Fourier Transform function in the package used by the FreQ plugin (edu.emory.mathcs.jtransforms.fft\DoubleFFT_1D.java & edu.emory.mathcs.utils\ConcurrencyUtils.java (Licenses: MPL 1.1/GPL 2.0/LGPL 2.1, Mozilla Public License Version 1.1, author: Piotr Wendykier)) and MATLAB. We observed that this difference impacts noise filtering, which generates slightly different color-coded CBF images. Since both tools are based on the same principles and produce very similar color-coded CBF images, this difference will not affect the result of CBF. Additionally, adjusting the settings for the plugin or MATLAB script (e.g., smoothing the power spectrum) may slightly change the absolute output values. We thus suggest experimenters to always use the same method and settings to compare their data.
Troubleshooting
Problem 1
Adult brain explants are damaged during the preparation steps (steps 1–9 in preparation of the adult brain explant).
Potential solution
We provide here few examples of damaged brains with segregated telencephalic hemispheres (Figure 9A, a), scratched dura or telencephalon (Figures 9A and 9B), a detached or damaged olfactory bulb and a removed choroid plexus (Figures 9A and 9C). There are tips to obtain good brain explants (Figures 9B1, d–f and 9B2) and avoid damages. First, cutting the olfactory nerve in the nose is important. Insufficient cutting of the nasal area will result in olfactory bulb damage and split of the telencephalic hemispheres. Be aware that cutting the nasal area too closely to the brain might also injure the olfactory bulbs. Second, when handling the explant with fine forceps or tungsten pins, experimenters should be very careful not to poke or scratch the telencephalic areas with the sharp tips of the tools. Third, the choroid plexus can be easily detached together with the pineal gland. Pulling and stretching the pineal gland slowly, slightly, and repetitively toward an anterior direction can help to remove it out from the explant without further damage.
Problem 2
Ciliary beating is not observed in the brain explant (steps 10 and 11 in recording of ciliary beating).
Potential solution
Ciliary beating of the brain explant will be affected by the environment and health of the preparation. Avoid cold or unoxygenated aCSF and perform experiments as soon as possible after the dissection. aCSF should be oxygenated and kept around room temperature throughout the recording. In addition, make sure that the brain explant is intact. Damages of the dura or brain parenchyma might also affect ciliary beating.
Problem 3
The injection needle is clogged and no particles are released upon injection (steps 12–14. In microinjection of fluorescent particles).
Potential solution
The mixture of fluorescent particles easily clogs the tip of the injection needle. Clean the tip before injection, according to the instructions of your injector. Keeping the injection needle in the air will enhance clogging. Thus, the tip should be immersed in aCSF between injections. Based on our experience, it is better to prepare new injection needle for each sample.
Problem 4
Fluorescent beads are not dispersed or cannot be observed in the brain ventricle after injection (steps 12–14. In microinjection of fluorescent particles).
Potential solution
Introducing the needle into the brain ventricle and pulling it out are critical for a successful injection of particles. We usually target the medio-rostral dura of the telencephalon (Figure 3A1) since the ventricle is wider and there is a better visibility to penetrate the tela without damaging the parenchyma. Pulling the tip out slowly is also important to prevent leakage of the particle and CSF in the ventricle. We recommend experimenters to do a single puncture of the dura. Repetitive punctures will damage the ciliated tela choroida.
Problem 5
The brain explant is moving during recording of ciliary beating or CSF flow (steps 10 and 11. in recording of ciliary beating and steps 15 and 16. recording of CSF flow).
Potential solution
Alignment of the recording images using the provided MATLAB codes or the MultiStackReg ImageJ plugin by Brad Busse (Thevenaz et al., 1998) is helpful to correct small drifts. For example, Figure 9C is the result from the pre-aligned recording from Figure 4D2. However, large drifting of the explant, in particular drift in depth, cannot be corrected using the code and plugin. Therefore, it is important to hold the brain explant tightly with the tungsten pins. It can also be effective to reduce the flow rate of oxygenated aCSF to avoid excessive movement.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Nathalie Jurisch-Yaksi (nathalie.jurisch-yaksi@ntnu.no).
Materials availability
There is no newly generated material in this paper.
Acknowledgments
This work was supported by RCN FRIPRO grants 314189 (N.J.Y.). Most diagrams were drawn by Adobe Illustrator and some images were created with BioRender. Work in the Wachten lab has been supported by grants from the Deutsche Forschungsgemeinschaft (DFG) – SFB 1454 – project number 432325352; TRR83 – project number 112927078; under Germany’s Excellence Strategy – EXC2151 – project number 390873048; SPP1926, SPP1726, FOR2743 as well as intramural funding from the University of Bonn.
Author contributions
Conceptualization, supervision, project administration, funding acquisition, visualization, manuscript writing, N.J.Y.; methodology, investigation, formal analysis, visualization, manuscript writing, I.J.; ImageJ-/Fiji-plugin FreQ, J.N.H. and D.W.; editing of the manuscript, all authors.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Inyoung Jeong, Email: inyoung.jung@ntnu.no.
Nathalie Jurisch-Yaksi, Email: nathalie.jurisch-yaksi@ntnu.no.
Data and code availability
The dataset included in this protocol is available on Mendeley data (https://data.mendeley.com/datasets/p6sfdpjpj2/2). MATLAB codes and ImageJ/Fiji plugin are deposited in GitHub (https://github.com/hansenjn/FreQ/; to allow long-term accessibility to all versions, the code for each release is also archived on Zenodo: https://dx.doi.org/10.5281/zenodo.5138071) and Mendeley data (https://data.mendeley.com/datasets/p6sfdpjpj2/2). Any additional information required to perform the protocol reported in this paper is available from the lead contact upon request.
References
- Borovina A., Superina S., Voskas D., Ciruna B. Vangl2 directs the posterior tilting and asymmetric localization of motile primary cilia. Nat. Cell Biol. 2010;12:407–412. doi: 10.1038/ncb2042. [DOI] [PubMed] [Google Scholar]
- Clary-Meinesz C.F., Cosson J., Huitorel P., Blaive B. Temperature effect on the ciliary beat frequency of human nasal and tracheal ciliated cells. Biol. Cell. 1992;76:335–338. doi: 10.1016/0248-4900(92)90436-5. [DOI] [PubMed] [Google Scholar]
- D'Gama P.P., Qiu T., Cosacak M.I., Rayamajhi D., Konac A., Hansen J.N., Ringers C., Acuña-Hinrichsen F., Hui S.P., Olstad E.W., et al. Diversity and function of motile ciliated cell types within ependymal lineages of the zebrafish brain. Cell Rep. 2021;37:109775. doi: 10.1016/j.celrep.2021.109775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fame R.M., Chang J.T., Hong A., Aponte-Santiago N.A., Sive H. Directional cerebrospinal fluid movement between brain ventricles in larval zebrafish. Fluids Barriers CNS. 2016;13:11. doi: 10.1186/s12987-016-0036-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green A., Smallman L.A., Logan A.C.M., Drake-Lee A.B. The effect of temperature on nasal ciliary beat frequency. Clin. Otolaryngol. Allied Sci. 1995;20:178–180. doi: 10.1111/j.1365-2273.1995.tb00040.x. [DOI] [PubMed] [Google Scholar]
- Hansen J.N., Gong A., Wachten D., Pascal R., Turpin A., Jikeli J.F., Kaupp U.B., Alvarez L. Multifocal imaging for precise, label-free tracking of fast biological processes in 3D. Nat. Commun. 2021;12:4574. doi: 10.1038/s41467-021-24768-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J.N., Kaiser F., Klausen C., Stüven B., Chong R., Bönigk W., Mick D.U., Möglich A., Jurisch-Yaksi N., Schmidt F.I., Wachten D. Nanobody-directed targeting of optogenetic tools to study signaling in the primary cilium. Elife. 2020;9:e57907. doi: 10.7554/eLife.57907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J.N., Rassmann S., Jikeli J.F., Wachten D. SpermQ(-)A simple analysis software to comprehensively study flagellar beating and sperm steering. Cells. 2018;8:10. doi: 10.3390/cells8010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J.N., Rassmann S., Stüven B., Jurisch-Yaksi N., Wachten D. CiliaQ: a simple, open-source software for automated quantification of ciliary morphology and fluorescence in 2D, 3D, and 4D images. Eur. Phys. J. E Soft Matter. 2021;44:18. doi: 10.1140/epje/s10189-021-00031-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haward F., Maslon M.M., Yeyati P.L., Bellora N., Hansen J.N., Aitken S., Lawson J., von Kriegsheim A., Wachten D., Mill P., et al. Nucleo-cytoplasmic shuttling of splicing factor SRSF1 is required for development and cilia function. Elife. 2021;10:e65104. doi: 10.7554/eLife.65104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermen F., Darnet L., Wiest C., Palumbo F., Bechert J., Uslu O., Yaksi E. Stimulus-specific behavioral responses of zebrafish to a large range of odors exhibit individual variability. BMC Biol. 2020;18:66. doi: 10.1186/s12915-020-00801-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermen F., Lal P., Faturos N.G., Yaksi E. Interhemispheric connections between olfactory bulbs improve odor detection. PLoS Biol. 2020;18:e3000701. doi: 10.1371/journal.pbio.3000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer-Zucker A.G., Olale F., Haycraft C.J., Yoder B.K., Schier A.F., Drummond I.A. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer's vesicle is required for normal organogenesis. Development. 2005;132:1907–1921. doi: 10.1242/dev.01772. [DOI] [PubMed] [Google Scholar]
- Lister J.A., Robertson C.P., Lepage T., Johnson S.L., Raible D.W. Nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development. 1999;126:3757–3767. doi: 10.1242/dev.126.17.3757. [DOI] [PubMed] [Google Scholar]
- Olstad E.W., Ringers C., Hansen J.N., Wens A., Brandt C., Wachten D., Yaksi E., Jurisch-Yaksi N. Ciliary beating compartmentalizes cerebrospinal fluid flow in the brain and regulates ventricular development. Curr. Biol. 2019;29:229–241.e6. doi: 10.1016/j.cub.2018.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiten I., Uslu F.E., Fore S., Pelgrims R., Ringers C., Diaz Verdugo C., Hoffman M., Lal P., Kawakami K., Pekkan K., et al. Motile-cilia-mediated flow improves sensitivity and temporal resolution of olfactory computations. Curr. Biol. 2017;27:166–174. doi: 10.1016/j.cub.2016.11.036. [DOI] [PubMed] [Google Scholar]
- Ringers C., Bialonski S., Hansen J.N., Ege M., Friedrich B.M., Jurisch-Yaksi N. Locally synchronized ciliary domains and tissue-scale cilia alignment underlie global metachronal wave patterns. bioRxiv. 2021 doi: 10.1101/2021.11.23.469646. Preprint at. [DOI] [Google Scholar]
- Ringers C., Olstad E.W., Jurisch-Yaksi N. The role of motile cilia in the development and physiology of the nervous system. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020;375:20190156. doi: 10.1098/rstb.2019.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Villalobos F., Pimentel J.A., Darszon A., Corkidi G., DArszon A. Imaging of the 3D dynamics of flagellar beating in human sperm. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2014;2014:190–193. doi: 10.1109/EMBC.2014.6943561. [DOI] [PubMed] [Google Scholar]
- Thevenaz P., Ruttimann U.E., Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 1998;7:27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- Thouvenin O., Keiser L., Cantaut-Belarif Y., Carbo-Tano M., Verweij F., Jurisch-Yaksi N., Bardet P.L., van Niel G., Gallaire F., Wyart C. Origin and role of the cerebrospinal fluid bidirectional flow in the central canal. Elife. 2020;9:e47699. doi: 10.7554/eLife.47699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez J.Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., Eliceiri K.W. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Turner R.W., Maler L., Deerinck T., Levinson S.R., Ellisman M.H. TTX-sensitive dendritic sodium channels underlie oscillatory discharge in a vertebrate sensory neuron. J. Neurosci. 1994;14:6453–6471. doi: 10.1523/jneurosci.14-11-06453.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset included in this protocol is available on Mendeley data (https://data.mendeley.com/datasets/p6sfdpjpj2/2). MATLAB codes and ImageJ/Fiji plugin are deposited in GitHub (https://github.com/hansenjn/FreQ/; to allow long-term accessibility to all versions, the code for each release is also archived on Zenodo: https://dx.doi.org/10.5281/zenodo.5138071) and Mendeley data (https://data.mendeley.com/datasets/p6sfdpjpj2/2). Any additional information required to perform the protocol reported in this paper is available from the lead contact upon request.