Abstract
Chromatin is the context for all DNA-based molecular processes taking place in the cell nucleus. The initial chromatin structure at the site of the DNA damage determines both, lesion generation and subsequent activation of the DNA damage response (DDR) pathway. In turn, proceeding DDR changes the chromatin at the damaged site and across large fractions of the genome. Ubiquitination, besides phosphorylation and methylation, was characterized as an important chromatin post-translational modification (PTM) occurring at the DNA damage site and persisting during the duration of the DDR. Ubiquitination appears to function as a highly versatile “signal-response” network involving several types of players performing various functions. Here we discuss how ubiquitin modifiers fine-tune the DNA damage recognition and response and how the interaction with other chromatin modifications ensures cell survival.
Keywords: chromatin, DNA damage response, DNA repair, double-strand breaks, ubiquitination
Introduction
DNA damage is the major threat to genome stability, especially in the form of a double-strand break (DSB). Various exogenous and endogenous damaging agents can cause DSBs. Unrepaired DSBs are one of the most harmful damage types, causing cell cycle arrest, apoptosis, or malignant transformation (Bartkova et al., 2005; Gorgoulis et al., 2005; Negrini et al., 2010). To cope with the threat, DNA damage response (DDR) and repair systems have evolved. There are two major DSB repair pathways: non-homologous end joining (NHEJ) and homologous recombination (HR). The principal difference between them, which is also reflected in the fidelity of repair, is whether or not DSB ends are resected, i.e. endure 5′–3′ degradation of one strand at each side of the break. The NHEJ, or mammalian classical NHEJ (cNHEJ), is a very fast repair pathway that employs ligation of ends with minimum resection and sequence homology. In contrast, the multistep HR is driven by broken ends resection and homology search with the single-strand DNA using an intact copy of the damaged locus as a template for repair. The NHEJ is functional throughout the cell cycle while the HR is a preferable choice in the S-G2 stages of the cell cycle when a sister chromatid is available for template-guided repair. Naturally, DNA damage and the DDR both happen in the context of chromatin. Chromatin, comprising DNA together with associated histone and non-histone proteins, determines the structure and function of the genome. At the primary level, chromatin is constituted by approximately 147 base pairs of DNA wrapped twice around eight core histones (H2A, H2B, H3, H4) forming nucleosomes (Luger et al., 1997). Nucleosome arrays compact and fold further into high-order structures creating complex multilevel 3D chromatin architecture. With each new level of folding, more and more factors are involved in the decision process that regulates the chromatin structure. In addition, DNA and histones can be chemically modified. Histone post-translational modifications (PTM), such as phosphorylation, methylation, acetylation, ubiquitination as well as histone variants modulate histone dynamics and DNA association. Together with the linker histone H1 and many non-histone proteins these factors directly define the physical properties of the chromatin fiber including secondary structure, accessibility, phase separation properties, and mobility, hence ultimately affecting damage induction, recognition, and processing.
Chromatin Remodeling and Modifications Associated With the DNA Double Strand Break Response
In an undamaged state, chromatin exists in a variety of functionally and topologically different domains, which can change dynamically. Based on the degree of compaction, originally based upon contrast staining with basic dyes (Heitz, 1928) chromatin is classified into more compacted, usually defined as transcriptionally inert heterochromatin and open, transcriptionally active, gene-rich euchromatin. Each of them is characterized by unique epigenetic and structural properties. Chromatin conformation reflects the role of the underlying sequences in the genome: centromeric and pericentromeric regions of chromosomes are organized into constitutive heterochromatin, which is believed to stay repressed among all cell types (Saksouk et al., 2015). On the other hand, facultative heterochromatin contains genes, which are silent in the corresponding cell type-dependent on the developmental state. In response to various stimuli, facultative heterochromatin is remodeled into euchromatin and vice versa, which ensures that the correct genes are expressed.
The pre-existing conformation of chromatin is a factor that affects DNA damage induction. Additional factors include the cell cycle stage (Ward and Chen, 2001), transcriptional activity (Falk et al., 2008; Herrera-Moyano et al., 2014; Prendergast et al., 2020; Bayona-Feliu and Aguilera, 2021), DNA secondary structure such as G-quadruplexes (Kumari et al., 2019), as well as the type of DNA damage and its source (e.g. LET (linear energy transfer) of ionizing radiation (Löbrich et al., 1996; Sutherland et al., 2001; Radulescu et al., 2004). When exposed to ionizing irradiation, the more compacted heterochromatin environment has been reported to physically shield DNA from damage and thus decrease the frequency and severity of the lesions formed (Elia and Bradley, 1992; Yoshikawa et al., 2008; Takata et al., 2013; Cannan et al., 2014; Brambilla et al., 2020). The number of water molecules available for radiolysis and subsequent DNA damage generation in condensed regions is considered to be lower than in decondensed ones. Along with this line, tightly packed nucleosomes in heterochromatin should show decreased accessibility for damage sensors and repair proteins, and consequently, decreased repair efficiency. Yet, this hypothesis of the impervious heterochromatin was questioned (Caridi et al., 2017). Chromatin compaction affects lesion induction, but compacted regions are not as irresponsive as was suggested before (Jakob et al., 2011; Chagin et al., 2019). The complex relationship between the initial chromatin structure and the DDR was summarized in two models. The “access-repair-restore” model came first and depicted chromatin as an obstacle for the repair machinery (Smerdon, 1991). Later this model was revised according to the evidence that chromatin actively participates in the DDR and provides the necessary context for repair. The updated model was named “prime-repair-restore”, where the prime step comprises both the contribution of the initial chromatin landscape and its remodeling to facilitate damage signaling (Soria et al., 2012). At the prime step, the PTMs on histone and non-histone proteins, corresponding to the pre-existing eu- or heterochromatin epigenetic landscape, change to the ones of the repair-associated epigenetic landscape, thus, promoting the formation of a 3D repair domain. Interaction of proteins and successful crosstalk between damage signaling and repair mechanism at the sites of DSBs is achieved by a highly controlled network of PTMs. The latter changes protein stability, charge, activity, structure, or interaction with other players of the pathway. Among the various PTMs of proteins at the sites of DNA lesions are phosphorylation, acetylation, methylation, poly(ADP-ribosyl)ation, ubiquitination, and sumoylation (Kouzarides, 2007). The de novo establishment of the PTMs is catalyzed by writer enzymes, with the removal of the PTMs being done by eraser enzymes and the transduction of the signal downstream in the cascade being carried out by reader proteins. The chromatin modifications and their role are, therefore, highly dynamic and tightly regulated at multiple levels.
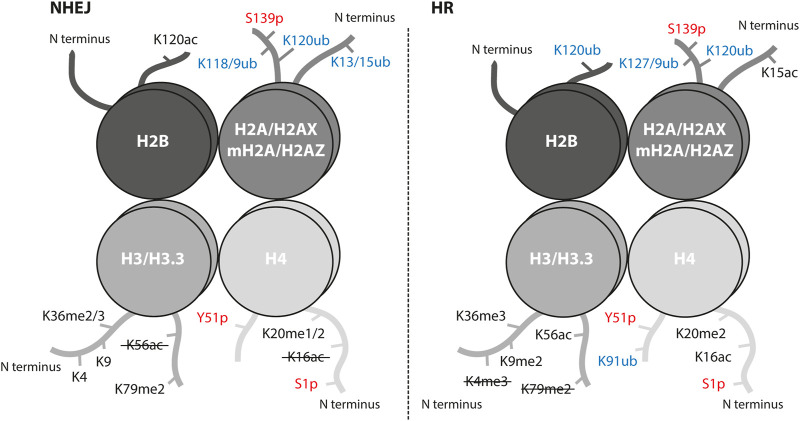
To maintain genome stability, two major DSB repair pathways (non-homologous end joining (NHEJ) and homologous recombination (HR)) have evolved. Both pathways are characterized by a different chromatin landscape (Figure 1).
FIGURE 1.
Histone post-translational modifications associated with NHEJ and HR repair pathways. Interaction of proteins and successful crosstalk between damage signaling and repair mechanism at the sites of DSBs is achieved by a highly controlled network of PTMs. Histone PTMs play a role in DSB repair pathway choice and serve as binding sites for repair proteins. Ubiquitination marks in blue, phosphorylation marks in red; ub, ubiquitination; p, phosphorylation; ac, acetylation; me, methylation. The crossed-out mark means it is removed in the repair pathway.
The key event in DSB repair is the recognition of DSB by the MRN complex (MRE11-RAD50-NBS1) and phosphorylation of serine 139 (termed γH2AX) on the histone variant H2AX by the PI3 kinase-related protein kinases ATM, ATR and DNA-PKcs (Rogakou et al., 1998; Sedelnikova et al., 2002). This signaling step is common for all DSBs independent of the repair pathway choice. The γH2AX-mediated downstream cascade involves such reader and writer enzymes as MDC1, RNF8, and RNF168 (Kolas et al., 2007; Doil et al., 2009). The RNF8-RNF168 ubiquitination cascade recruits main regulators of the repair pathway choice, 53BP1 for NHEJ and BRCA1 for HR. The RNF8-RNF168 axis promotes two chromatin modification marks, which 53BP1 binds in a bivalent mode: the H2AK15ub mark and the H4K20me2 mark (Acs et al., 2011; Mallette et al., 2012; Fradet-Turcotte et al., 2013). Interestingly, the H2AK15 site is a target for both ubiquitination and acetylation and is crucial for repair pathway decisions (Figure 1). When HR is preferred, i.e. in S/G2 phases, the TIP60/NuA4 acetyltransferase complex acetylates H2AK15 and directly blocks ubiquitination which impairs the binding of 53BP1 (Jacquet et al., 2016). The histone acetyltransferase (HAT) TIP60 is also responsible for acetylation of the H4K16 site which physically inhibits 53BP1 binding to H4K20me2 and, thus, promotes HR (Figure 1) (Tang et al., 2013). On the other hand, the removal of H4K16ac from chromatin by the deacetylases HDAC1 and HDAC2 promotes NHEJ (Miller et al., 2010). The removal or absence of methylation at H4K20 (H4K20me0) was reported to guide the pathway choice toward HR by opening the binding site for BRCA1-BARD1, the main HR player (Nakamura et al., 2019). As H4K20me0 is abundant in post-replicative cells, these data provide evidence for how HR can be promoted once the sister chromatid is available as a template. The BRCA1-BARD1 ubiquitinates H2AK127/129 that is read by the ubiquitin reader SMARCAD1 to start a cascade of chromatin remodeling and blocks 53BP1 from binding on damaged chromatin (Densham et al., 2016). There are several chromatin modifications at the DSB sites that mediate the pathway choice in cooperation with the transcription activity of damaged loci, e.g. H3K36me3, H2BK120ac, H2AK118/119ub (Shanbhag et al., 2010; Clouaire et al., 2018). Usually, these modifications are associated with the NHEJ repair pathway but some were shown to be involved in both and additionally regulated by other epigenetic marks or factors (Figure 1).
There are many more chromatin modifications that cooperate at the site of DNA damage to fine-tune the damage signaling, control chromatin remodeling and ensure a successful repair (Figure 1; Table 1). Combined, these data reveal the importance and complexity of chromatin remodeling in DDR.
TABLE 1.
Histone modifications in the DDR.
Local and Global Chromatin Changes Mediated by Ubiquitination
Ubiquitination is one of the major chromatin modifications in unperturbed chromatin as well at the sites of DSB in particular. It was discovered as a process of covalent protein modification (Goldknopf et al., 1977) when small chemical modifications like phosphorylation and acetylation were already reported to affect protein properties and function. In the 1980s ubiquitination was shown to be part of the protein degradation pathway via the 26S proteasome and for a long time, the proteasomal role of ubiquitin was the only known (Hershko and Ciechanover, 1998). In the last decades, however, an ever-growing number of studies described a non-proteolytical or non-classical role of ubiquitination in intracellular signaling, membrane trafficking, DNA repair, and cell cycle (Dwane et al., 2017; Liao et al., 2022). Through multiple large-scale quantitative proteomic screens, it was confirmed that there are more than 1000 players in the ubiquitin system and more than 10,000 known individual ubiquitination sites in human proteins, which could mean that every cellular protein is ubiquitinated at some point in its existence (Kim et al., 2011; Wagner et al., 2011; Clague et al., 2015). Despite the intense research on ubiquitination, which led to the Nobel Prize in Chemistry for Aaron Ciechanover, Avram Hershko, and Irwin Rose in 2004, there is still a lot unknown about its players and their regulation.
Ubiquitin-dependent Signaling Network
Ubiquitin (Ub) is a highly conserved 76 amino acids protein, its name due to its massive abundance in all eukaryotic cell types (Goldstein et al., 1975). Ubiquitin is the main building block in ubiquitination, which is a process of covalent attachment of ubiquitin to a target protein. There are four genes encoding ubiquitin in the human genome: UBB, UBC, UBA52, and RPS27A (Kimura and Tanaka, 2010). Translation of these genes does not directly produce free active ubiquitin but requires cleavage of precursors by deubiquitinases (DUB). In addition, DUBs can reverse ubiquitination by cleaving the isopeptide bond between ubiquitin and its substrate protein. In humans there are nearly 90 DUB genes, which can be classified into seven classes (Clague et al., 2019).
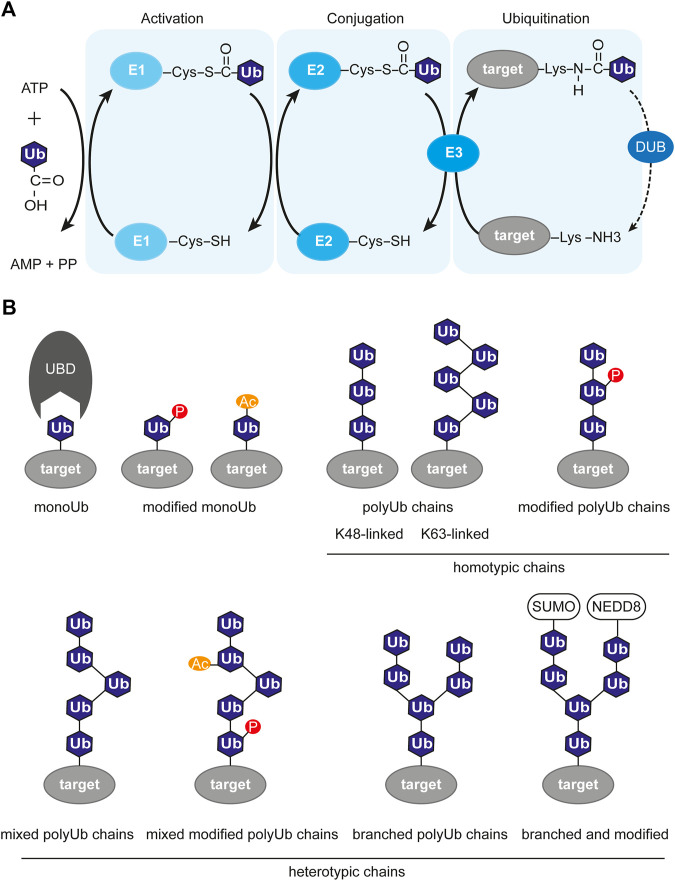
The enzymatic cascade of ubiquitination includes three steps and three types of enzymes (Pickart, 2004) (Figure 2A). At first, the free ubiquitin moiety is activated by the E1 enzyme in an ATP-dependent manner by building a thioester bond between the C-terminal Gly carboxyl group of Ub and the Cys in the active site of E1. Then, the activated Ub is delivered to the Cys residue of the E2 conjugating enzyme via E1-E2 thioester transfer. The third step is the substrate-specific transfer of the ubiquitin chain to the target molecule. This is achieved by the substrate-specific E3 ligase, which recognizes the E2-Ub complex and catalyzes the formation of an isopeptide bond between the C-terminal carboxyl group of a Ub moiety and an ε-NH2 group on a lysine on the target protein. There is recent evidence that ubiquitination could also take place on other residues like cysteine, serine, and threonine (McClellan et al., 2019; Mabbitt et al., 2020). There are two ubiquitin E1 genes, around 40 E2, around 90 DUBs, and around 663 E3 ubiquitin ligases genes in the human genome representing around 5% of the total number of genes (Li et al., 2008; George et al., 2018). Interestingly, the number of E3 ubiquitin ligases in the human genome is larger than the number of kinases (518 genes) (Li et al., 2008). The difference in number between the three classes of ubiquitination enzymes evolutionary originates from the ability of one E2 enzyme to cooperate with several different E3 ligases depending on the context, the so-called combinatorial effect. Although there are some E2 proteins able to mediate the direct transfer of Ub to the target, in most cases the E3 ligase is the enzyme that promotes selectivity and specificity of the ubiquitination.
FIGURE 2.
Ubiquitin-dependent signaling system. (A) Enzymatic cascade of ubiquitination involving three enzymes. The first step is activation of ubiquitin and covalent attachment to the E1 enzyme. The second step is conjugation, where ubiquitin is transferred to the E2 enzyme. The last step is the transfer of (poly)-ubiquitination to it’s target molecule mediated by a E3 ligase. The ubiquitination process is opposed by deubiquitination, which is the removal of the ubiquitin mark from the target protein by deubiquitinase (DUB). (B) Variety of ubiquitin chains, with or without modifications. P, phosphorylation; Ac, acetylation; SUMO, SUMOylation; NEDD8, NEDDylation; Ub, ubiquitin; UBD, ubiquitin-binding domain; target, target protein harboring ubiquitin modification.
The complexity of the ubiquitin system is also expanded by the fact that ubiquitin itself contains seven lysine residues available for modification: K6, K11, K27, K29, K33, K48, and K63. Multiple ubiquitination events result in the formation of elongated polyubiquitin chains with various structures depending on which lysine was modified (Figure 2B). Apart from the seven internal lysines, there is the eighth site for ubiquitination, which is the methionine at position 1. This results in the formation of non-canonical “linear” chains (Ikeda et al., 2011; Iwai et al., 2014). The topology of the chains defines their function. The K48-linked chains are predominant in cells and usually target their substrate for degradation (Finley, 2009), while K63- and K6-linked chains were shown to regulate DDR (Elia et al., 2015; Takahashi et al., 2018). The wide variety of all the ubiquitination types, like monoubiquitination, multi-monoubiquitination, homotypic polyubiquitination (through the same lysine residue), and heterotypic polyubiquitination (mixed, branched) was named the “ubiquitination code” (Komander and Rape, 2012). The ubiquitin code is steadily expanding with recent findings that ubiquitin itself can be further modified post-translationally by, e.g., phosphorylation at eleven potential sites (Thr7, Thr12, Thr14, Ser20, Ser57, Tyr59, Thr66 (Peng et al., 2003; Lundby et al., 2012; Swaney et al., 2013)), acetylation (Ohtake et al., 2015) and SUMOylation (Hendriks et al., 2014). Those additional Ub modifications may alter its recognition by E3 ligases or Ub-binding proteins. The readers for these modifications are mostly unknown.
The ubiquitination network not only includes writers but also erasers and readers for its function. Some of the proteins might have more than one function and each function is activated by a different signal. While the E3 ligases (RING, HECT, and RBR families) write the ubiquitination and DUBs erase it, the ubiquitination readers are the proteins containing ubiquitin-binding domains (UBD). There are at least 20 types of UBDs present on a wide range of signaling proteins (Dikic et al., 2009). Interestingly, certain types of UBDs are present in proteins enriched at the sites of DSB, namely UIM (ubiquitin-interacting motif), MIU (motif interacting with ubiquitin), and UBZ (ubiquitin-binding zinc-finger) domains (Table 2). The affinity of a single UBD towards ubiquitin marks is low, and, therefore, various mechanisms have evolved to increase the sensitivity and specificity of recognition. Some Ub readers contain multiple UBDs, e.g., the BRCA1-interacting protein RAP80 contains two UIM domains recognizing K63-linked polyubiquitin chains (Sato et al., 2009). Another way for UBD-containing proteins to ensure correct context-specific reading of ubiquitination is to cooperate. Such cooperation was reported for RNF168, RNF169, and RAD18 (Panier et al., 2012), containing bipartite modules composed of UBD and LR motifs (LRM) sequences. Therefore the specific binding is achieved by dual binding of both the ubiquitin binding domain as well as the target specific binding domain. Novel mechanisms of Ub reading and novel UBDs are yet to be identified.
TABLE 2.
Ubiquitin-binding domains.
Taken together, all the variations of the ubiquitin code, alternative Ub modifications, complex networks of regulating proteins, and a large number of putative ubiquitin ligases with still unknown functions constitute a complex, highly dynamic, and largely unexplored ubiquitin-dependent signaling system. Due to its remarkable versatility and exceptional ability to fine-tune a signal transfer, ubiquitination is ideally suited for the regulation of dynamic and complex cellular processes like DSB signaling and repair.
Local Chromatin Changes: Damage Signaling
The first ubiquitin modifier identified in the DNA damage response was the DNA repair gene RAD6, an E2 enzyme in S. cerevisiae (Jentsch et al., 1987). It was also in the DDR where the first evidence of a non-proteolytic function of K63-linked ubiquitination was observed (Spence et al., 1995). Since then evidence has accumulated confirming an essential role of the ubiquitination system in the cellular response to DNA damage.
The first DDR-associated ubiquitination event takes place already within minutes after DNA break induction, and it is the monoubiquitination of histone H2AX at lysine 119/120 by the canonical Polycomb Repressive Complex 1 (PRC1) complex including BMI1-RING1 proteins (Wu C.-Y. et al., 2011; Pan et al., 2011). Additionally, H2A gets ubiquitinated by PRC1 at the same residues and it is believed that this modification induces transcriptional repression, in turn facilitating repair (Chou et al., 2010; Ismail et al., 2010). H2AXK119/120ub modification happens in a DSB-dependent manner and possibly even earlier than the γH2AX formation. It was reported that the H2AXK119/120ub maintains the γH2AX foci stability by directly recruiting ATM to breaks, thus playing a significant role in initiating DNA damage signaling. Additionally, ATM is activated by the MRN complex in the absence of H2AX ubiquitination (Uziel et al., 2003). Since γH2AX is the apical modification event in the reactions cascade of damage response, H2AXK119/120 monoubiquitination subsequently indirectly affects the downstream repair factors. The phosphorylated H2AX is recognized by MDC1 which, when phosphorylated by ATM, tethers both L3MBTL2 and RNF8 to the vicinity of the DNA lesion (Stucki et al., 2005; Nowsheen et al., 2018). At the sites of DSB, the E3 ligase RNF8 performs various types of ubiquitination on different targets: firstly, it polyubiquitinates L3MBTL2 via K63-linkage formation, and secondly, histone H2A (Mailand et al., 2007; Stewart et al., 2009; Panier et al., 2012). RNF8-mediated ubiquitination attracts another E3 ligase RNF168 whose activity, in turn, is essential for chromatin rearrangements and recruitment of 53BP1 and BRCA1 via K63- and K27-linked ubiquitination (Gatti et al., 2015). Attracted by RNF8-mediated ubiquitination, RNF168 primes monoubiquitination of H2A at lysine 13 and 15 and with the help of RNF8 builds K63-linked chains (Doil et al., 2009; Mattiroli et al., 2012). RNF168 binds modified H2A itself and further accumulates at the DSBs, thereby spreading the ubiquitination. Interestingly, RNF168 activity is mediated not only by recognizing upstream ubiquitination but also by binding an acidic patch on the nucleosome (Leung et al., 2014; Mattiroli et al., 2014). This fact once again confirms how precisely and multitiered the ubiquitination is regulated to ensure the specificity of the signal.
A recent finding revealed another functional layer in the RNF8/RNF168 cascade. It was reported that RNF8 E3 ligase and UBC13 E2 conjugating enzyme mediate the K63-linked ubiquitination of H1 linker histones and, thus, provide a binding platform for RNF168 (Thorslund et al., 2015). This step is required for the further accumulation of ubiquitination at sites of DSBs. The RNF8/RNF168-mediated ubiquitination initiates the recruitment of other ubiquitin modifiers to the vicinity of DNA damage, e.g., HERC2, RAD18, BRCA1, and RNF169 (Huang et al., 2009; Bekker-Jensen et al., 2010; Krais et al., 2021).
Ubiquitination takes the form of a highly regulated dynamic network with multiple interconnected levels of regulation. One layer is the removal of Ub modifications by DUBs. The ubiquitination of H2A at lysine 13/15 is erased by USP51 and USP3 enzymes (Nicassio et al., 2007; Lancini et al., 2014; Sharma et al., 2014; Wang et al., 2016). Other Ub marks catalyzed by the RNF8/RNF168 tandem are removed by USP11 and USP44 DUBs (Mosbech et al., 2013; Yu et al., 2016). Another way to regulate the ubiquitination at DSBs is by controlling the protein levels of E3 ligases. So TRIP12 and UBR5 ubiquitin modifiers negatively control the RNF168 levels by ubiquitin-mediated degradation and, therefore, restrain ubiquitination spreading around damaged chromatin (Gudjonsson et al., 2012), while the RNF8 turnover was reported to be maintained by the VCP/p97/Ataxin complex in a proteasome-dependent manner (Singh et al., 2019). VCP/p97 is an ATP dependent segregase that extracts ubiquitinated proteins from chromatin and shuffles them to the proteasome. Remarkably, the negative regulation of ubiquitination at DSBs can be achieved not only by active removal of the mark or its writers but by changing the message of the signal as well. Accumulation of the K48- and K6-linked chains at the lesion targets proteins to proteasomal degradation, thus controlling the subsequent recruitment of repair factors (Acs et al., 2011; Meerang et al., 2011; Ali et al., 2018). Finally, there is evidence that there might be a non-catalytic competition present at the damage sites. The paralog of RNF168, RNF169 competes for RNF168-generated ubiquitination products with other proteins and, by that, it affects RNF8/RNF168 kinetics and removes 53BP1 from damaged chromatin (Chen et al., 2012; Poulsen et al., 2012; Kitevski-LeBlanc et al., 2017; An et al., 2018).
The RNF8/RNF168 is one of the multiple ubiquitination axes, which take place in DDR. The monoubiquitination of histone H2B at lysine 120 mediated by the RNF20/RNF40 E3 ligases happens in response to DNA damage. The modified H2BK120 attracts the chromatin remodeler SMARCA5 thereby promoting chromatin relaxation and facilitating the recruitment of repair factors (Moyal et al., 2011; Nakamura et al., 2011; So et al., 2019). The removal of the H2BK120ub mark from the chromatin by USP22 and the SAGA complex was reported to regulate the early stage of damage response, however, a lot about this axis is still to be identified.
There is another ubiquitination axis identified that promotes RAP80/BRCA1-A, RAD18, and 53BP1 recruitment to DSBs. Remarkably, it is based on the crosstalk between K63- and K11-polyubiquitin signaling mediated by one reader and eraser protein, Cezanne (Wu et al., 2019). Downstream of RNF8/RNF168, the DUB Cezanne in complex with Cezanne2 binds to K63-linked ubiquitin chains at the sites of damage and removes K11-linked ubiquitination from mixed polyUb chains as well. As the RAP80/BRCA1-A are not able to bind mixed K63-/K11-polyubiquitin chains, the erasure of K11 facilitates its binding.
There are other ubiquitination cascades at the sites of the DSBs but many of them are largely understudied. Not much is known about histones H3 and H4 ubiquitination in the DDR. The CUL4-DDB1-ROC1 complex was shown to ubiquitinate mammalian H3 and H4 histones in vivo and in vitro in response to UV irradiation (Wang et al., 2006). The homolog of this complex in yeast, however, appeared as a hit in a genetic screen for players of homologous recombination (Moss et al., 2010). Interestingly, the ubiquitination targets of this E3 ligase complex in two different DDR pathways are different as well: while in the nucleotide excision repair (NER) pathway the histones get ubiquitinated, in response to DSBs the CUL4-DDB1-ROC1 complex ubiquitinates the Spd1 protein. This shows that one ubiquitin writer can modify histone and non-histone targets and, therefore, serve various functions in different repair pathways depending on the stimulus.
Another E3 ligase was reported to ubiquitinate histone H4 in response to the DNA damage. BBAP monoubiquitinates H4K91 and protects cells exposed to hydroxyurea and doxorubicin (Yan et al., 2009). Although the levels of γH2AX and MDC1 were not affected in the BBAP knockdown cells, the lack of the H4K91ub mark significantly delayed 53BP1 recruitment to the DSBs. Subsequent investigation of the chromatin marks H4K20me2 and H2AK15 necessary for 53BP1 to bind damaged chromatin showed that their levels were decreased upon the BBAP knockdown. Based on these data, the authors suggested that the H4K91ub is somehow required prior for the H4K20me1/2 and other PTMs to take place at the damage sites, however, the exact mechanism is unknown.
Local Chromatin Changes: DNA Repair Pathway Choice
Ubiquitination is a fundamental part of DNA damage signaling. DSB repair pathway choice is also modulated by various ubiquitin modifiers. As mentioned above, there are two main DSB repair pathways, NHEJ and HR. The principal difference is reflected in the fidelity of repair, whether or not DSB ends are resected, that is 5′–3′ degradation of one strand at each side of the break. The NHEJ, or mammalian canonical NHEJ (cNHEJ), is a very fast repair pathway that employs end ligation with minimum resection and sequence homology. In contrast, the multi-step HR is driven by homology search and broken ends resection, where single-strand DNA uses an intact copy of the damaged locus as a template for repair and, therefore, requires significant homology between sequences. As a result, this difference leads to the concept of HR being the pathway that provides the most accurate repair of DSB, while NHEJ might serve as a source of point mutations and small deletions.
The correct choice of repair pathway at every DSB is of utmost importance for cell survival. Both pathways co-exist in a cell at the same time and partially compete for the DSBs (Beucher et al., 2009). Multiple factors affect the repair pathway decision in a spatio-temporal manner. One of the approaches suggested structuring our knowledge about all these factors and their coordination with each other in a way we could predict the pathway choice in a so-called “decision tree” (Scully et al., 2019). The first factor which affects the pathway choice is open DNA ends themselves. Depending on the source of damage and initial chromatin state, the composition of the break may significantly vary, which in turn affects the binding affinity of damage sensor proteins, e.g. Ku70/80 shows weak binding affinity to long ssDNA (Mimori and Hardin, 1986). The Ku70/80 binding to the break initiates NHEJ, while the displacement of the Ku70/80 heterodimer from the broken DNA ends is a necessary step to initiate resection and turn the repair in direction of HR (Langerak et al., 2011). The Ku70/80 removal from the ends can be achieved by ubiquitin-mediated degradation via K48-linked chains added to Ku70 (Postow et al., 2008; Ismail et al., 2015), followed by VCP/p97 mediated extraction from chromatin. Another important factor in the pathway choice is the cell cycle stage (Hustedt and Durocher, 2016). The NHEJ is functional throughout the cell cycle while the HR is a preferable choice in the S-G2 stages when a sister chromatid is available for template-guided repair, therefore the cell cycle stage is directly related to the resection (Mao et al., 2008). The cell cycle status can be directly transmitted from the cell cycle-dependent kinases (CDK) to the repair machinery, e.g. through phosphorylation of resection players CtIP and EXO1 (Tomimatsu et al., 2014; Anand et al., 2016). In G1 cells instead, CtIP gets ubiquitinated and targeted for degradation by the anaphase-promoting complex APC/C thus blocking resection (Lafranchi et al., 2014). Recently another way for the APC/C complex to regulate pathway choice in a ubiquitin-dependent manner was identified. The authors showed that in G1 cells BRCA1 recruitment to DSB is obstructed due to the USP1-mediated removal of K63-linked polyubiquitin chains from histones. In contrast, in S-G2 phases, USP1 is ubiquitinated by the APC/C complex and degraded. This process requires Chk1 activation as well (Ha et al., 2017). Aside from USP1, there were more DUB enzymes identified lately that modulate pathway choice by facilitating retention of resection proteins CtIP and EXO1 at the sites of damage, namely USP4 and UCHL5 (Liu et al., 2015; Wijnhoven et al., 2015).
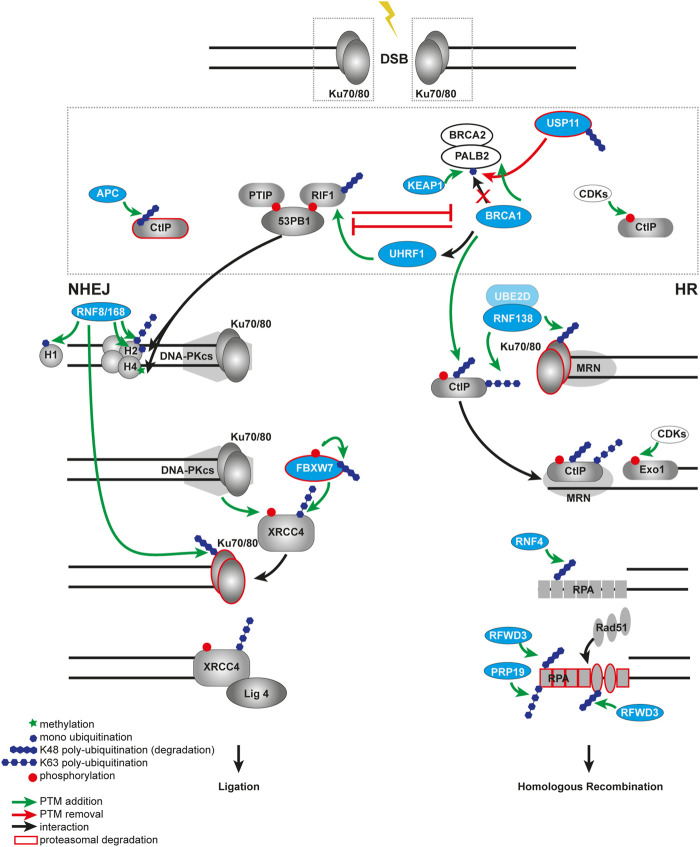
Another ubiquitin modifier that affects repair pathway choice is the E3 ligase RNF138 (Figure 3). Interestingly, RNF138 promotes HR by ubiquitination of two substrates. First, RNF138 ubiquitinates CtIP and causes its retention at DSBs, second, RNF138 ubiquitinates the Ku80 subunit of the Ku70/80 heterodimer which results in Ku70/80 displacement (Ismail et al., 2015; Schmidt et al., 2015). The ubiquitination of Ku80 happens in the S phase only and, thus, there must be an additional cell cycle-dependent regulation mechanism.
FIGURE 3.
Ubiquitination in DSB repair pathway choice. Open DNA ends are rapidly recognized by the Ku70/80 heterodimer. The choice between NHEJ and HR at the DSB is controlled by the competition between 53BP1 and BRCA1. In a NHEJ-permissive environment, 53BP1 builds a complex via ATM in a phosphorylation-dependent manner with its binding partners RIF1 and PTIP. The 53BP1-PTIP-RIF1 complex prevents recruitment of BRCA1 to the break and, therefore, antagonizes HR. The E3 ubiquitin ligase KEAP1 ubiquitinates PALB2 at the BRCA1 binding site and prevents assembling of the BRCA1-BRCA2-PALB2 complex, thus, promoting NHEJ. This process is balanced by the DUB USP11 and controlled by the cell cycle. BRCA1 antagonizes NHEJ by impairing retention of RIF1 at DSB sites in a CtIP- or UHRF1-dependent manner. Ubiquitin-dependent degradation of CtIP by the APC/C in NHEJ prevents resection process, in contrast to HR, where CtIP is not degraded but phosphorylated by the CDKs. The RNF8-RNF168-mediated ubiquitination on chromatin provides a binding platform for both 53BP1 and BRCA1. Later RNF8 also ubiquitinates Ku70/80 to facilitate its proteasomal degradation and removal from the broken DNA ends. In a HR-permissive environment the E3 ubiquitin ligase RNF138 ubiquitinates Ku to remove it from DSB much earlier compared to NHEJ, allowing end-resection factors to gain access to the deprotected ends. RNF138 also ubiquitinates CtIP, enhancing its retention on damaged chromatin. At the later steps of NHEJ the ligation of broken ends promoted by FBXW7 that recognizes phosphorylated XRRC4 and subsequently ubiquitinates it. In contrast during the later steps of HR, RPA SUMOylation-guided ubiquitination by RNF4 promotes RPA turnover and RAD51 recruitment. Multi-site ubiquitination of RPA by the ubiquitin ligases RFWD3 and PRP19 also facilitates HR.
The RNF138-mediated dual ubiquitination of two different targets in one process is an important example of the versatility of the ubiquitin-dependent network in DDR. Another example is when one target can be ubiquitinated by multiple different writers, e.g. the Ku70/80 can be removed from DSB ends by various ubiquitin E3 ligases. The RNF8 and SCF-FBXL12 ligases promote ubiquitination and subsequent removal of Ku70/80 as well (Feng and Chen, 2012; Postow and Funabiki, 2013).
At the sites of DSBs, 53BP1 and BRCA1 proteins antagonize each other in repair pathway choice. It was shown that both of them are recruited to the break via the RNF8/RNF168 ubiquitination cascade. We discussed earlier that 53BP1 uses bivalent recognition of both RNF8/RNF168-mediated H2AK15ub ubiquitination and H4K20me2 methylation to bind to damaged chromatin. In undamaged cells, the methylation mark is hidden from 53BP1 interaction by KDM4A (JMJD2A) and L3MBTL1, but at DSB sites both of them are targeted to proteasomal degradation (Acs et al., 2011; Mallette et al., 2012; Fradet-Turcotte et al., 2013). Once bound to chromatin in G1, 53BP1 promotes NHEJ through the recruitment of its interaction factors telomere-associated protein RIF1, PTIP, Shieldin and CST complex and REV7 and suppression of BRCA1 (Chapman et al., 2013; Di Virgilio et al., 2013; Wang et al., 2014; Xu et al., 2015; Mirman et al., 2018; Noordermeer et al., 2018). Moreover, a new ubiquitin-dependent mechanism for suppression of HR in G1 cells was identified (Orthwein et al., 2015). The ubiquitination of PALB2, one of the interacting partners for BRCA1, by KEAP1 E3 ligase complex blocks its interaction with BRCA1 and thus HR in G1 cells (Figure 3) (Orthwein et al., 2015). The DUB USP11 that removes ubiquitin from PALB2, promoting BRCA1–PALB2–BRCA2 complex formation in S/G2 cells, opposes this process (Schoenfeld et al., 2004; Orthwein et al., 2015). Interestingly, recent study described the role of USP11 in HR in centromeric regions in G1 in the absence of the sister chromatid and taking advantage of centromeric transcripts forming DNA-RNA hybrids. The DUB USP11 was shown to promote the recruitment of the RAD51-BRCA1-BRCA2 complex to centromeres by interaction with the centromeric histone CENP-A and HJURP (Yilmaz et al., 2021). In contrast, the HR-favoring conditions are characterized by ubiquitination of histone H2A at K125/127/129 by BRCA1-BARD1 (Kalb et al., 2014). The H2AK125/127/129ub in response to DSBs supports SMARCAD1 recruitment, which in turn facilitates repositioning of 53BP1 to the periphery of the repair focus, and promotes resection (Densham et al., 2016). Later the same group also identified the DUB USP48 for negative regulation of BRCA1-mediated resection (Uckelmann et al., 2018). The very recent studies further expanded our knowledge about the BRCA1-BARD1 repair mechanism in post-replicative chromatin (Becker et al., 2021; Hu et al., 2021). With help of cryo-electron microscopy, the authors revealed that BRCA1-BARD1 specifically binds to nucleosomal histones, DNA, and H2AK13/K15 monoubiquitination at the DSB sites and subsequently promotes ubiquitination at the C- terminus of H2A (Hu et al., 2021). Via multivalent interactions BARD1 mediates recruitment of BRCA1 to ubiquitinated chromatin, but other ubiquitin modifiers control the BARD1 protein itself. Newly identified E3 ligase RNF19A ubiquitinates BARD1 to prevent its retention and homologous recombination in the NHEJ-favored chromatin context (Zhu et al., 2021). Even more mechanisms of how BRCA1 antagonizes 53BP1 were identified. Several studies reported that BRCA1 impairs the retention of 53BP1 interacting partner RIF1 at the damaged chromatin in the S phase in either a CtIP-dependent manner (Escribano-Díaz et al., 2013) or via UHRF1 E3 ligase recruitment (Zhang H. et al., 2016). Interestingly, CtIP ubiquitination by BRCA1 does not result in its degradation but instead strengthens its role in the G2/M checkpoint. BRCA1 recruitment to DSBs was shown to be mediated by deubiquitination events as well (Li et al., 2017). The DUB enzyme USP13 deubiquitinates RAP80 which facilitates its interaction with BRCA1 and subsequent recognition of the K63-ubiquitinated chromatin (histones) in the vicinity of DSBs.
Ubiquitination affects DNA repair pathway choice at all steps of DDR. For example, during RPA-coated ssDNA formation and homology search, the E3 ligases PRP19 and RFWD3 control the stability of RPA and RAD51 nucleofilaments, thus, promoting HR (Figure 3). At the sites of DNA damage, RFWD3 polyubiquitinates both RPA and RAD51 in a phosphorylation-dependent manner, which requires the help of ATM and ATR (Dubois et al., 2017; Inano et al., 2017). In this case, VCP/p97 mediates eviction of RPA and RAD51 polyubiquitinated by RFWD3 from early DSB repair sites for proteosomal degradation. The E3 ligase PRP19 was reported to polyubiquitinate RPA via K63 linkages and promote ATRIP accumulation and subsequent ATR activation (Maréchal et al., 2014). Together these ubiquitin modifiers support late-HR progression in time and space.
Despite the seeming abundance of our knowledge about ubiquitination-guided repair pathway choice, a lot of questions remain unanswered: are there novel and yet undiscovered ubiquitin modifiers in DDR and what steps do they regulate; how do ubiquitin modifiers interact with DSB signaling network; how does ubiquitin code modulate DDR signaling etc. There is no doubt that a lot of new ubiquitin modifiers and regulation mechanisms are to be identified in the future.
The ubiquitination events are omnipresent throughout all steps of DDR from damage recognition to successful repair of the lesion. Ubiquitination signaling orchestrates DDR not only locally at the site of broken ends but also participates in the formation of a repair domain at a larger scale.
Global Chromatin Changes
Chromatin domains affect DSB formation and get affected by it at multiple hierarchically organized structural levels. On higher folding levels newly built breaks initiate the formation of repair domains, microscopically visible as foci, in the case of radiation known as radiation-induced foci (IRIF). The foci can be visualized by immunofluorescent labeling of various damage signaling and repair proteins but the most important for chromatin structure study is the labeling of γH2AX. The phosphorylation of H2AX spreads far beyond the actual break location over megabase-pair distance (Rogakou et al., 1999). It is generally believed that every single focus at the conventional microscopy level represents one break and the large spreading of phosphorylation was an intriguing fact back then. There is a lot of evidence that chromatin architecture controls the formation of the γH2AX-decorated repair domain. The super-resolution light microscopy showed that CTCF protein, which restrains cohesin-mediated loop extrusion and therefore essentially shapes the 3D organization of large chromatin domains, is juxtaposed to γH2AX foci. Moreover, the depletion of CTCF heavily impaired DDR signaling and repair efficiency in cells subjected to radiation (Natale et al., 2017). Other studies utilizing endonuclease AsiSI for targeted DSB generation in mammalian cells showed NIPBL-, MRN-, and ATM-mediated accumulation of cohesin at the DSB site (Arnould et al., 2021). With help of chromosome conformation capture mapping and chromatin immunoprecipitation (ChIP) assays, the authors revealed a surprising discrepancy between γH2AX spreading and accumulation of its kinase ATM around the break that suggests that H2AX phosphorylation is not just a linear, ATM-dependent process. Their study highlights the one-sided loop extrusion on either side of the break as a prime mechanism of γH2AX domain formation. Also, chromatin structural domains (TAD, topologically associated domain) are a template for the repair domain (Arnould et al., 2021). Thus, DDR utilizes a large-scale chromatin structure remodeling for genome maintenance.
Interestingly, similar to γH2AX a cohesin-dependent arrangement on chromatin was reported for other repair proteins like 53BP1, ubiquitination events, and H1 histone eviction (Clouaire et al., 2018; Ochs et al., 2019). Remarkably, ubiquitination of damaged chromatin follows γH2AX spreading almost precisely (Clouaire et al., 2018), not being restricted to the vicinity of the break. It is evident now that various histone/non-histone ubiquitin modifications and ubiquitin modifiers contribute as well to the formation of the 3D repair-permissive domain to facilitate the repair and, in the end, to maintain chromatin topology at sites of DNA breakage.
The chromatin domains vary in their properties. Pre-existing heterochromatin allows the entrance of some macromolecules and chromatin remodelers, e.g. due to the formation of a phase-separated domain with selective permeability (Gibson et al., 2019; Wang et al., 2019). Liquid-liquid phase separation (LLPS) as a force driving and maintaining chromatin compartmentalization is promoted by the chromatin scaffold, histone modifications, and their multiple reader proteins (Snead and Gladfelter, 2019). A newly formed lesion starts a cascade of changes in the chromatin context, which leads to its reorganization at a large scale to form a 3D domain outlining a repair-prone environment. In recent years, a growing number of publications highlighted the role of phase separation in DNA damage response. All levels, initiation, spatial organization/clustering of damage, maintenance of damage domain 3D architecture, and repair are affected (Kilic et al., 2019; Pessina et al., 2019; Singatulina et al., 2019; Oshidari et al., 2020; Frattini et al., 2021; Ghodke et al., 2021; Levone et al., 2021; Miné-Hattab et al., 2021). Remarkably, PTMs, namely ubiquitination and its crosstalk play a significant role in it. For example, it was proposed that 53BP1-seeded phase separation requires both interaction between 53BP1 and H4K20me2, as well as 53BP1 interaction with RNF168-mediated, DNA damage-induced H2AK15 ubiquitin marks (Kilic et al., 2019). When the 53BP1 LLPS is affected, the downstream DNA repair is impaired. Therefore modifiers like RNF169, TRIP12, and UBR5 that antagonize the RNF168-mediated ubiquitination spreading, indirectly control phase separation at the sites of the DSBs. Another seeding factor for the phase separation in DDR is poly(ADP-ribosyl)ation (PARylation), which is mediated by PARP1 activation. It was shown before that PAR chains can synergize with various intrinsically disordered proteins, e.g. FUS, to initiate LLPS, however, a recent study described a DUB enzyme to bind PAR chains in a damage-dependent manner (Kim et al., 2021). The USP39 deubiquitinase is recruited to the DSBs by the poly(ADP-ribosyl)ation with help of its tripartite RG domain, thus inducing liquid demixing and promoting NHEJ. The authors also suggested that the USP39 function is required for HR, however, it is not related to PAR or phase separation. Additionally, the USP39 activity is an important example of PAR-ubiquitination crosstalk in the DDR.
The ubiquitination and other PTMs on histones and non-histone proteins not only change the chemical properties of the target proteins but could also control the abundance of other molecules like RNA in the phase-separated domain. However, our understanding of how ubiquitination contributes to the phase separation in damaged chromatin is only starting to form, and more studies are needed to uncover this network.
It is exciting to think about how phase separation integrates with active molecular processes at the DNA damage sites. Although some repair proteins like 53BP1 are observed to initiate the phase separation, upstream γH2AX and MDC1 accumulation do not seem to be involved in it (Kilic et al., 2019). This fact hints that the formation of the repair domain is a product of multiple different processes, and their interplay is yet to be investigated.
Chromatin responds dynamically to the damage. Upon DNA damage, mammalian and yeast chromatin domains go through decompaction and expansion, which in turn changes the mobility and accessibility of the damaged locus. It is well documented now that the mobility of damaged locus increases, reflecting exploration of the nuclear space by DSB end during “homology search” (Hauer and Gasser, 2017; Herbert et al., 2017; Miné-Hattab et al., 2017). However, undamaged loci in damaged cells were observed to elevate their mobility as well but to a lesser extent, which points out that changes in chromatin mobility are a general feature of the whole-genome response to DSBs (Chiolo et al., 2011; Miné-Hattab et al., 2017; Caridi et al., 2018; Smith et al., 2019). Moreover, the DSBs in Drosophila and mouse cells were observed to relocalize to subnuclear domains facilitating repair: from pericentromeric heterochromatin regions to the nuclear periphery in flies and from the core to the outer layer of chromocenters in mice (Chiolo et al., 2011; Jakob et al., 2011; Tsouroula et al., 2016). Tsouroula et al. and Natale et al. additionally showed that damage-induced global expansion in centromeric and pericentromeric domains was not accompanied by the removal of silent chromatin marks and was not connected to the relocalization happening in mammals (Tsouroula et al., 2016; Natale et al., 2017). This finding supported the previously stated integral function of localized recompaction in checkpoint signaling but not in repair (Burgess et al., 2014). This suggests that chromatin compaction may regulate the timing of different steps of the DDR. In contrast to pericentromeric DSBs, centromeric DSBs were not shown to relocalize. Instead, they trigger new deposition of centromeric histone variant CENP-A, which allows the specific recruitment of DUB USP11 to damaged centromeres to facilitate RAD51 engagement and initiation of HR (Yilmaz et al., 2021).
The transient chromatin decompaction occurs at the sites of DSBs independently of the repair pathway. Such decompaction and accompanying it increased mobility correlated to significant depletion of nucleosomes on DNA with proteolytic degradation of 30–40% of core histones in yeast (Hauer et al., 2017). A similar effect was shown when core histone reduction was achieved by deletion of genes coding for histones H3 and H4 (Liang et al., 2012). Recent work of the Gasser group further explored the chromatin remodeling effect mediated by histone depletion and revealed the paramount role of ubiquitin ligases and chromatin ubiquitination in the mobility of the damaged site in yeast (Cheblal et al., 2020; Challa et al., 2021). They showed that five ubiquitin-conjugating factors are recruited to the damage sites in a checkpoint- and INO80C-dependent manner which results in depletion of core and H1 histones up to 20–40%, chromatin decompaction, and enhanced DNA locus mobility (Challa et al., 2021). Remarkably, a homolog of one of them was proposed to cause chromatin decompaction in mammalian cells as well. The RNF20/40 ligases (Bre1 in yeast) promote the formation of the H2BK120ub mark in damaged cells, which facilitates chromatin relaxation and repair proteins recruitment by promoting nucleosome disassembly (Chernikova et al., 2010; Moyal et al., 2011; Nakamura et al., 2011; Zheng et al., 2018; So et al., 2019; Liu et al., 2021). Interestingly, the monoubiquitination of histone H2B itself directly impairs the compaction of higher-order chromatin structures and results in open conformation (Fierz et al., 2011). The mechanism is different from the acetylation-mediated decompaction and in contrast to it, monoubiquitination spreads over larger distances. Lastly, the H2BK120ub was shown to crosstalk with other chromatin modifications at the damage sites, namely H4K20 and H4K9 methylation, which could lead to a pleiotropic chromatin reorganization.
Speaking of chromatin mobility in DDR, we cannot ignore the clustering of DNA damage. The clustering is thought to be happening to DSBs which require resection of broken ends or concentration of several damaged sites into the large domain to facilitate repair (Aymard et al., 2017; Schrank et al., 2018; Schrank and Gautier, 2019). It was recently identified that actin filament nucleators are the driving force for the DSB clustering and are a factor in the repair (Schrank et al., 2018). At last, the clustering can also protect DSBs from improper repair, where one of the examples is coordination between end resection degree and amount of damage inflicted on cells.
Crosstalk Between Ubiquitination and Other Chromatin Modifications in DDR
Crosstalk between various PTMs on histone and non-histone proteins lies at the basis of DNA damage signaling and repair. A single PTM affects the properties and function of its target, but only coordination of phosphorylation, acetylation, methylation, poly(ADP-ribosyl)ation, ubiquitination, and their readers ensure the spatiotemporal recruitment of repair proteins and chromatin remodeling at the sites of damage. There are several classifications of crosstalk proposed, e.g. based on how modifications are hierarchically organized in a cascade (Dantuma and van Attikum, 2016). According to it, the PTMs can be serial, when one modification sequentially promotes other modifications; parallel, when one reader containing multiple binding domains recognizes multiple independent modifications; and combinatorial, when two or more modifications are equally required for one reader to induce a signal. Above, we described some of the examples of the ubiquitin-dependent regulatory cascades, but the ubiquitination crosstalk with other PTMs constitutes an integrated signaling network in DDR.
Phosphorylation and Ubiquitination
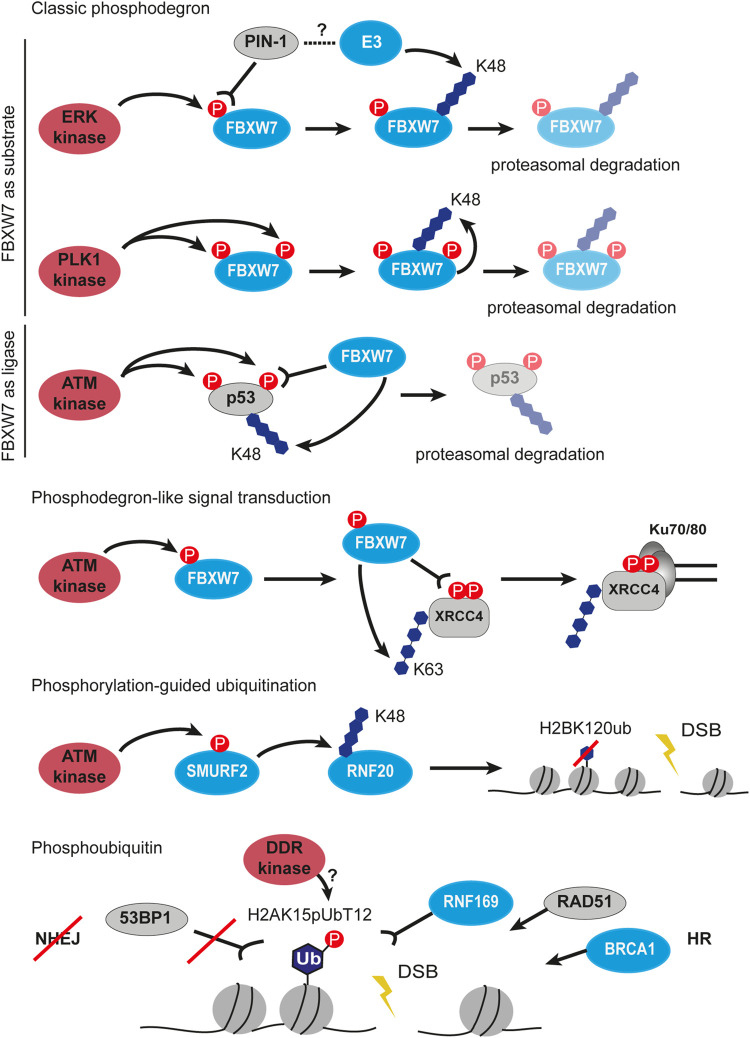
Phosphorylation is among the most abundant and most studied modifications at the sites of DNA damage. Crosstalk between phosphorylation and ubiquitination is essential for the correct execution of DDR. One of the examples of this crosstalk is phosphorylation-guided proteasomal degradation with help of phosphodegrons. Phosphodegrons are one or several residues on a target protein that can be phosphorylated by a kinase and subsequently bound by an E3 ubiquitin ligase to initiate proteasomal degradation via K48-linked ubiquitin chains (Ang and Wade Harper, 2005). At the sites of DSB, the NHEJ E3 ligase FBXW7 stability is controlled by phosphorylation and the creation of the phosphodegron signal (Figure 4). The extracellular signal-regulated kinase (ERK) directly interacts with and phosphorylates FBXW7 at Thr205, which results in the degradation of FBXW7 in a PIN-1-dependent manner by an unknown E3 ligase (Ji et al., 2015). When FBXW7 is phosphorylated at Ser58 and Thr284 by kinase PLK1, it autoubiquitinates itself which also leads to its degradation (Xiao et al., 2016). Remarkably, FBXW7 can not only be a substrate for simultaneous phosphorylation and ubiquitination but also acts as the degradation-inducing E3 ligase for other substrates, e.g. p53 (Figure 4) (Cui et al., 2020). In response to etoposide-induced DNA damage, ATM phosphorylates p53 on Ser33 and Ser37, which facilitates the FBXW7 binding and subsequent p53 degradation. The knockdown/knockout or chemical inhibition of FBXW7 increased p53 protein half-life upon DNA damage in an MDM2-independent manner, therefore sensitizing cells to radiation and etoposide treatment. Thus, the phosphorylation and ubiquitination crosstalk is finely tuned to ensure the protection of genome stability. Importantly, the “phosphodegron” signal does not necessarily result in degradation (Figure 4). Phosphorylated by ATM at Ser26, FBXW7 is recruited to the sites of DSB, where it recognizes another phosphorylated protein XRCC4. Upon phosphorylation at Ser325/326 by DNA-PKcs, XRCC4 is bound and ubiquitinated by FBXW7 via K63-chains at lysine 296 (Figure 4). The K63-linked polyubiquitination of XRCC4 enhances its association with the Ku70/80 complex to facilitate NHEJ repair (Zhang Q. et al., 2016). Recently, a systematic analysis of MARKs-phosphorylated degron motifs recognized by FBXW7 identified new targets among chromatin proteins (Singh et al., 2022). The exact mechanism remains elusive, however, it provides a look at the large regulation network managed by phosphorylation and ubiquitination.
FIGURE 4.
Crosstalk between phosphorylation and ubiquitination in DSB signaling and repair. First three panels describe the classic phosphodegron situations highlighting FBXW7 as substrate to phosphorylation and as phosphorylation reading ubiquitin ligase. Next, phosphodegron-like signal transduction by FBXW7 is shown, which results in ubiquitination and stabilization of XRCC4 at Ku70/80 covered DSB. The next panel depicts phosphorylation-guided ubiquitination involving sequential signal transfer by two ubiquitin ligases SMURF2 and RNF20. The last example of the crosstalk between phosphorylation and ubiquitination is the phosphorylation of the ubiquitin moiety. The H2AK15pUbT12 is recognized by the HR factors such as RNF169, RAD51 and BRCA1, but prevents binding of 53BP1 at DSB. P, phosphorylation, Ub, ubiquitin.
Phosphodegron cascades are possible due to the proteins containing both phosphorylation and ubiquitination sites. A comprehensive statistical analysis of the sequence and spatial distributions of PTMs revealed that among all the PTM pairs studied, phosphorylation and ubiquitination sites have a smaller distance between each other on the same proteins compared to random distance distribution (Korkuć and Walther, 2017). This close proximity of PTM sites is encoded in the protein sequence and evolutionary conserved. The close vicinity allows regulatory interference between ubiquitination and phosphorylation by steric interaction/hindrance. For example, ubiquitination sites enrichment near the domain activation loop and in the glycine-rich region of kinases were reported to serve for reversible inhibition (Swaney et al., 2013). The same proteome-wide study identified 466 yeast proteins with 2100 phosphorylation sites co-occurring with 2189 ubiquitination sites using mass spectrometry-based methods. Interestingly, greater conservation of phosphorylation on ubiquitinated proteins was observed than the other way around, which highlights the existence of crosstalk directionality where phosphorylation tends to precede ubiquitination (Swaney et al., 2013).
One of the central phosphorylation events in DDR is γH2AX formation mediated by ATM, ATR, and DNA-PKcs. Above we have already discussed in detail the γH2AX-dependent chromatin-associated RNF8/RNF168 ubiquitination axis (Kolas et al., 2007; Doil et al., 2009). Yet ATM phosphorylates various substrates in various damage response contexts and subsequently promotes various multifaceted ubiquitination events. Together with the CHK2 kinase, ATM phosphorylates CABIN1 which acts as a negative regulator of p53 activity. The phosphorylated CABIN1 is recognized and targeted to the ubiquitin-dependent proteasomal degradation mediated by the CRL4-DDB2 ubiquitin ligase complex, thus activating p53 in response to the genotoxic stress (Choi et al., 2013). There are more negative regulators of p53 at the damage sites, HDMX (human ortholog of Mdmx) is one of them as well. It was shown that in response to the lesion ATM phosphorylates HDMX at Ser395 and promotes its degradation by the E3 ligase HDM2 (human ortholog of Mdm2) (Pereg et al., 2005). The precise regulation of the ATM-mediated p53 activation and stabilization remains elusive due to the abundance of the phosphorylation-ubiquitination circuits. It was recently reported that the E3 ligase Mdm2 can be phosphorylated by ATM as well which leads to its autoubiquitination and subsequent degradation (Magnussen et al., 2020). More evidence is required to fill the gap in our knowledge about the organization of separate cascades and their crosstalk within the DDR network.
Apart from the p53 regulation, ATM was observed to phosphorylate Ser384 on the E3 ligase SMURF2, an interaction partner of RNF20 (Figure 4) (Tang et al., 2020). In turn, phosphoSMURF2 binds and ubiquitinates RNF20 promoting its proteasomal degradation. The SMURF2-mediated RNF20 ubiquitination results in a decrease of H2BK120 ubiquitination, thereby promoting chromatin compaction and protecting cells from DNA damage. However, in the absence of phosphorylation or SMURF2 knockout, the repair was shown to proceed faster than in wild-type cells as measured by γH2AX disappearance, probably, due to excessive relaxation of chromatin and facilitated recruitment of repair proteins. Despite faster DSB repair, the mouse embryonic fibroblasts with the mutant SMURF2 showed higher sensitivity to etoposide treatment which means that timely chromatin remodeling is of utmost importance for DDR (Tang et al., 2020).
Phosphoubiquitin (pUb) was shown for the first time to play a role in DNA damage signaling and repair only recently (Figure 4). Walser and colleagues described that the ubiquitin moiety phosphorylated at Thr12 (pUbT12) is used for the formation of a new chromatin mark H2AK15pUbT12 (Walser et al., 2020). Authors reported that the H2AK15pUbT12 levels increase in response to the radiation- and etoposide-induced DSBs and return back to the basal state after DNA repair. The H2AK15pUbT12 is differently recognized by ubiquitin readers in DDR: the phosphorylation of H2AK15ub abolishes 53BP1 binding while being still bound by RNF169. Thus, chromatin modified by pUbT12 is inaccessible to 53BP1 but permissive to HR proteins like RNF169, RAD51, and the BRCA1/BARD1 complex, which makes the H2AK15pUbT12 an HR-specific chromatin modification. Interestingly, the pUbT12 prevents the removal of the H2AK15pUbT12 mark by DUB USP51 from chromatin which results in the retention of ubiquitination and formation of the HR-prone environment. The DDR kinase responsible for this phosphorylation on ubiquitin is unknown as well as its mechanism, although the authors revealed that it depends on preceding ubiquitination events mediated by RNF168 (Walser et al., 2020). Taken together, the phosphoubiquitin modifications represent a new type of DDR signaling.
Poly(ADP-Ribosyl)ation and Ubiquitination
Poly(ADP-ribose) (PAR) is a PTM that consists of at least two or more ADP-ribose molecules covalently linked by glycosidic ribose–ribose bonds. Formation of the poly(ADP-ribose) chains or PARylation can occur at different amino acid residues, including aspartate, glutamate, and lysine residues, mediated by the family of the poly(ADP-ribose) polymerases (PARPs) (Leung, 2014). At the sites of DNA damage, the most abundant PARP is poly(ADP-ribose) polymerase 1 (PARP1) whose recruitment results in rapid and reversible PARylation at histone, non-histone proteins, and PARP1 itself (Tallis et al., 2014).
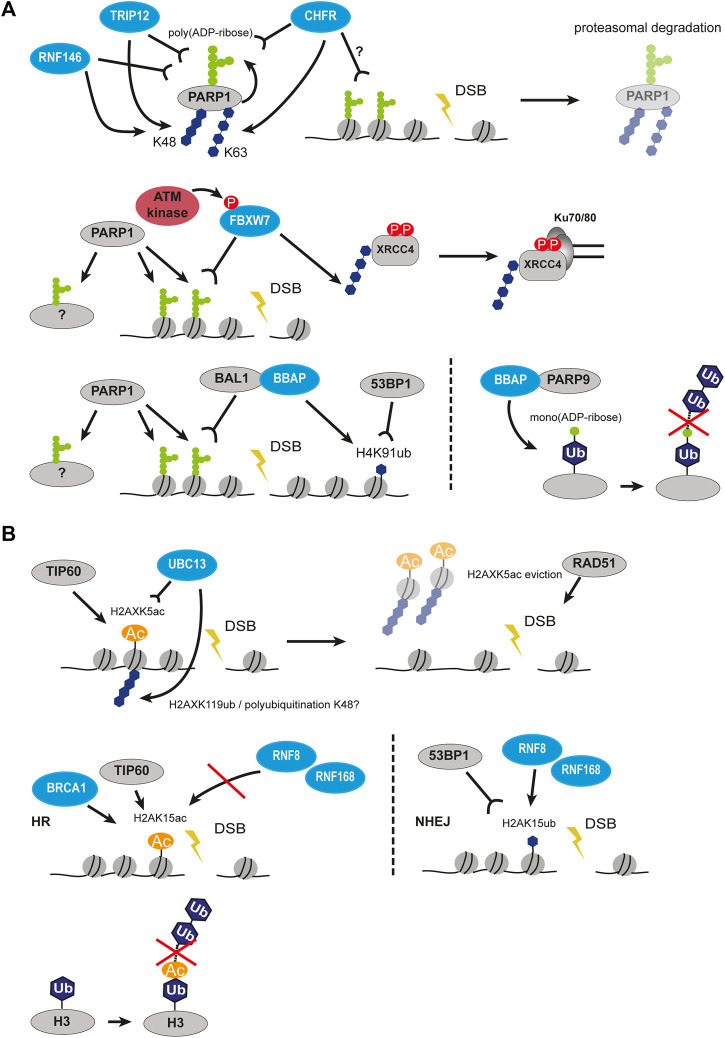
Poly(ADP-ribosyl)ation has extensive crosstalk with other PTM types in DDR, including ubiquitination. CHFR is one of the E3 ubiquitin ligases recruited to DSBs by direct interaction between its zinc finger domain and PAR chains (Figure 5A) (Oberoi et al., 2010). Interestingly, CHFR shares structural similarities with RNF8, with the exception that RNF8 is recruited to lesions in a phosphorylation-dependent manner. At the sites of DSBs, CHFR ubiquitinates auto-PARylated PARP1 with the K48- and K63-linked ubiquitin chains and promotes PARP1 degradation (Liu et al., 2013). It is thought that CHFR can bind and ubiquitinate PARylated histones as well, however, direct evidence is missing. Ubiquitination of PARP1 by CHFR leads to extraction of PARP1 from chromatin and spatiotemporal restriction of PAR spreading, thus controlling the early stage of damage response in a negative feedback loop. CHFR, therefore, belongs to the early-response ubiquitin modifiers in DDR. The E3 ligase TRIP12 can recognize the PAR chains on PARP1 and promote its removal from chromatin as well (Gatti et al., 2020). Another E3 ubiquitin ligase promoting proteasomal degradation of repair proteins and PARP1 in a PAR-dependent manner is RNF146 (Figure 5A) (Kang et al., 2011). Interestingly, RNF146 requires PARylation not only for interaction with its targets but also for ubiquitin ligase activity (DaRosa et al., 2015). The RNF146 RING domain responsible for ubiquitination requires non-covalent binding to PAR to undergo conformational change and switch from an inactive to an active state. Therefore, PARylation serves both as an upstream signal to ubiquitination and as a physical activator of ubiquitin modifiers.
FIGURE 5.
Crosstalk between PARylation and ubiquitination, as well as acetylation and ubiquitination in DSB signaling and repair. (A) Crosstalk between PARylation and ubiquitination. PARP1 auto-PARylates itself followed by recognition of PAR chains by several ubiquitin ligases such as CHFR, RNF146, TRIP12 and subsequent polyubiquitination. This results in proteasomal degradation of PARP1. PARP1-mediated modification of histones at DSBs can be recognized by FBXW7, which polyubiquitinates XRRC4 and leads to its interaction with Ku70/80. On the other hand, PARP1-mediated modification of histones can also be recognized by BAL1-BBAP which leads to ubiquitination of H4K91 creating a binding site for 53BP1. Interaction of BBAP with PARP9 leads to mono(ADP-ribosyl)ation of ubiquitin moiety preventing elongation of ubiquitin chain. (B) Crosstalk between acetylation and ubiquitination. TIP60 acetylates H2AXK5 creating a recognition site for UBC13, which then polyubiquitinates H2AXK119 resulting in eviction of H2AX. H2AK15 can be acetylated or ubiquitinated. When acetylated by TIP60, it is recognized by BRCA1 but prevents the action of RNF8/RNF168, thus promoting HR. When ubiquitinated by RNF8/RNF168, it is recognized by 53BP1 promoting NHEJ. Acetylation of the ubiquitin moiety similarly to mono(ADP-ribosyl)ation prevents elongation of ubiquitin chain. P, phosphorylation, Ac, acetylation, Ub, ubiquitin.
The master regulator of DNA repair BRCA1 was reported to recruit to the sites of DSBs in a PAR-dependent manner as well (Li et al., 2013; Li and Yu, 2013; Hu et al., 2014). It requires the BRCT domain of the BRCA1 interaction partner BARD1 to bind PAR and initiate the homologous recombination pathway. This example of the poly(ADP-ribosyl)ation and ubiquitination crosstalk and its importance for cell survival provides a strategy for cancer therapies.
The F-box containing ubiquitin modifiers were observed to have crosstalk with poly(ADP-ribosyl)ation as well. The mammalian FBXL10-RNF68-RING1B ubiquitin ligase complex (FRRUC) is known as part of the non-canonical PRC1.1 which monoubiquitinates H2A histones on K119 to initiate transcriptional repression in undamaged cells (Wang et al., 2004; Wu et al., 2013). The FRRUC complex is rapidly recruited to the sites of DNA damage in a PARP1- and TIMELESS-dependent manner, however, the exact mechanism is still unknown (Rona et al., 2018). When at the DSBs, the FRRUC non-canonical PRC1 complex facilitates recruitment of the canonical PRC1 complexes containing the E3 ligases BMI1 and MEL18 and mediates H2AK119 ubiquitination. The FRRUC complex was shown to be required for the transcriptional repression, the H2A/H2A.Z histone exchange, and HR damage signaling. Interestingly, depletion of FRRUC impaired the K63-linked ubiquitination by the RNF8/RNF168 axis, but not γH2AX or MDC1 foci formation (Rona et al., 2018). Another F-box E3 ligase that binds PAR chains at double-strand breaks was already mentioned in the previous chapter. FBXW7 is a factor in the NHEJ pathway and is recruited to the sites of DSBs in the ATM-mediated manner. How exactly was not clear until it was discovered that the WD40 domain of FBXW7 binds to PARP1-produced PAR chains (Zhang et al., 2019). Thus, the rapid recruitment of FBXW7 to PAR on damaged chromatin promotes ATM-dependent phosphorylation and retention at damage sites, XRCC4 ubiquitination, and activation of NHEJ. Which PARylated targets are exactly recognized by the FBXW7 reader domain is yet to be discovered, since PAR chains occur not only on histones but on PARP1, PARP2, and repair proteins as well. It is important to mention, that due to almost immediate poly(ADP-ribosyl)ation at the DSB sites, its readers are recruited rapidly too.
The BBAP E3 ligase (also known as DTX3L) was shown to selectively ubiquitinate histone H4 and indirectly promote 53BP1 recruitment to DSBs (Yan et al., 2009). Together with its partner, the macrodomain-containing protein BAL1, BBAP forms a complex that specifically binds PARP1-mediated PAR chains (Figure 5A). Interestingly, it is the BAL1 protein that recognizes PAR and BBAP is tethered to the sites of DNA damage via its interaction with BAL1 (Yan et al., 2013). Only together with BAL1 BBAP ligase is able to recruit to DSB and initiate the early ubiquitination wave, which ensures correct damage signaling and repair. The study also revealed that the PARP-BAL1-BBAP ubiquitination axis is functionally independent and nonredundant from the ATM-MDC1-RNF8 one, and they both significantly impair repair both at early and late timepoints (Yan et al., 2013). Strikingly, 53BP1, RAP80, and BRCA1 recruitment via PARP1-mediated BAL1-BBAP ubiquitination and via γH2AX-dependent RNF8-induced ubiquitination were shown to be separate mechanisms as well, which hints that the recruitment of the major repair proteins is a multi-regulated process. The BBAP ligase can form a complex with other partners apart from BAL1. Another study showed that BBAP can interact with PARP9, a mono(ADP-ribosyl)transferase, which was reported as being enzymatically inactive (Figure 5A) (Yang et al., 2017). When heterodimerized with BBAP, the BBAP-PARP9 heterodimer complex mediates reversible NAD + -dependent mono(ADP-ribosyl)ation of the ubiquitin moiety on Gly76 which is used for conjugation to substrates. As a result, the ADP-ribosylated Ub can no longer be used for ubiquitination and that restricts the E3 ligase activity of BBAP. It was observed that the BBAP-PARP9 complex is recruited to sites of DNA damage. The function of BBAP in NHEJ is regulated by the NAD + concentration and PAR chains. This, in turn, modulates ubiquitin mono(ADP-ribosyl)ation by PARP9. The authors suggested that under normal conditions the effect of ubiquitin mono(ADP-ribosyl)ation is not necessary to inhibit ubiquitin-conjugation, but rather to suppress the formation of polyubiquitin chains by restricting the reaction to a single round of ubiquitin transfers (Yang et al., 2017).
BBAP E3 ligase belongs to the family of the DELTEX ubiquitin ligases (DTX1 to DTX4 and DTX3L), which contain the RING domain and the conserved Deltex C-terminal (DTC) domain. A recent comprehensive study on the interactome and ubiquitination targets of the DTX2 E3 ligase found a large share of the DDR proteins among them (Ahmed et al., 2020). The DTC domain was reported as a new PAR-binding domain involved in PAR-guided ubiquitination by DTX2. This process is organized similarly to the phosphodegrons but in this case, DTX2 ubiquitinates proteins modified by PARP1/PARP2.
As PAR-chains and intrinsically disordered proteins are involved in the formation of phase-separated domains, there might be more ubiquitin modifiers to modulate this process at the sites of DNA damage.
Acetylation and Ubiquitination
The acetyltransferase TIP60 has several effects on ubiquitination in DDR. The TRRAP-TIP60 complex together with the ATPase p400 acetylates histones H4, H2AX, and ATM and promotes chromatin relaxation at DSBs (Sun et al., 2005; Murr et al., 2006; Ikura et al., 2007; Xu et al., 2010). This facilitates the binding of RNF8/RNF168 and subsequent ubiquitination in the vicinity of breaks. Activating transcription factor 2 ATF2 and E3 ligase complex Cul3/Roc1 in turn control the stability of TIP60 and, therefore, activity of ATM, which requires to be acetylated upon DNA damage (Bhoumik et al., 2008). The depletion of either component of the TRRAP-TIP60 complex impaired both acetylation and ubiquitination at chromatin and in turn hindered recruitment of 53BP1 and BRCA1. In response to DNA damage, TIP60 acetylates H2AX histone on K5 independently of its phosphorylation (Figure 5B). In turn, acetylated H2AX is required for the UBC13-mediated polyubiquitination of H2AX on K119 (Ikura et al., 2007). This acetylation-guided ubiquitination cascade promotes H2AX release from chromatin, which further relaxes the structure and facilitates RAD51 recruitment. Interestingly, the acetylation of the H2A N-terminal region in pre-existing chromatin was found to directly impair subsequent RNF20-RNF40-dependent ubiquitination of H2BK120 (Wojcik et al., 2018). This can bring novel insight into how acetylation regulates downstream ubiquitination in DDR.
The competition between acetylation and ubiquitination for the same residues was shown to regulate pathway choice and chromatin structure at the damage sites. H2AK15 acetylated by TIP60 blocks the ubiquitination and therefore inhibits 53BP1 recruitment promoting HR (Figure 5B) (Jacquet et al., 2016). Another site that can be both exclusively acetylated and ubiquitinated is H2BK120. The levels of acetylation were reported to increase in response to DSBs and the levels of ubiquitinated H2BK120 decreased (Clouaire et al., 2018). As both modifications were shown to alter the higher-order compaction state of the chromatin fiber (Fierz et al., 2011), it is clear that the acetylation-ubiquitination switch is an important part of chromatin remodeling at the sites of the breaks.
The ubiquitin modifiers can be tethered to DSBs by binding acetylated histone marks. The bromodomain-containing E3 ligase TRIM66 was observed to bind unmethylated H3R2-H3K4 and H3K56ac in a combinational manner at the damage sites (Chen et al., 2019). In wild-type cells, H3K56ac is rapidly deacetylated by SIRT6, HDAC1, and HDAC2 at the very early stage of DDR, but the knockdown of TRIM66 causes retention of this mark and impairs repair (Tjeertes et al., 2009; Miller et al., 2010; Battu et al., 2011; Toiber et al., 2013). TRIM66 was found to localize to DSBs in an H3K56ac-dependent manner and subsequently recruit SIRT6 to induce deacetylation, however, it is unknown if this regulatory circuit involves ubiquitination activity of TRIM66.
Moreover, the ubiquitin moiety can be acetylated on either K6 or K48 which does not affect the monoubiquitination but inhibits the formation of K11-, K48-, and K63-linked polyubiquitin chains (Ohtake et al., 2015). Interestingly, one of the targets getting modified with acetylated ubiquitin is histone H3 (Figure 5B). Another study analyzed the effect of acetylated ubiquitin on the catalytic activity of the E1 enzyme UBA1. It was observed that acetylation on all seven internal lysines can impair the conformational change required for the E1-E2 transfer and Ub-conjugation to the E2 enzyme, thus resulting in impaired target ubiquitination (Lacoursiere and Shaw, 2021). The role of acetylated ubiquitin in DDR signaling is yet to be investigated.
SUMOylation and Ubiquitination
The addition of SUMO (small ubiquitin-related modifier) is also involved in DDR. There are three functionally redundant SUMO proteins encoded in mammalian cells, SUMO1, SUMO2, and SUMO3. However very similar to ubiquitination, SUMOylation uses its own enzymatic system consisting of an E1 activating enzyme (SAE1/SAE2), an E2 ligase (UBC9 also known as UBE2I), and various E3 ligases (Celen and Sahin, 2020). Ubiquitination has tight crosstalk with SUMOylation, the cascades are various in their directionality and organization. SUMO proteins recruit to IRIF 4 h after damage (Galanty et al., 2009). The recruitment occurs in a PIAS1- and PIAS4-dependent manner, two E3 ligases of the SUMO system. It was found that the RNF8/RNF168-mediated ubiquitination on chromatin is required for the SUMO signaling at DSBs and the SUMOylation is functionally divided into 53BP1-SUMO1 and BRCA1-SUMO2/3 pathways (Figure 6A). So far SUMOylation was reported for multiple repair proteins, including MDC1, 53BP1, BRCA1, RPA, CtIP, RNF8, and HERC2 (Galanty et al., 2009, 2012; Morris et al., 2009; Danielsen et al., 2012; Luo et al., 2012; Yin et al., 2012; Locke et al., 2021). Many DDR ubiquitin modifiers require being SUMOylated for their E3 ligase activity, e.g. BRCA1/BARD1 heterodimer. Interestingly, SUMO signaling not only relies on RNF8 and K63-linked ubiquitination on chromatin but also participates in ubiquitination stability and spreading (Danielsen et al., 2012).
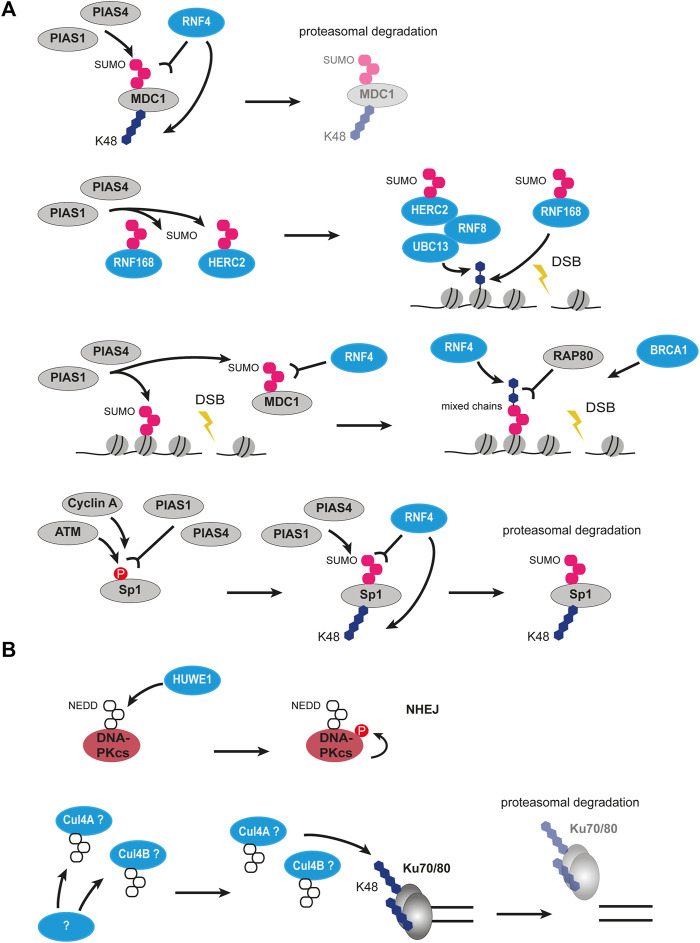
FIGURE 6.
Crosstalk between SUMOylation and ubiquitination, as well as NEDDylation and ubiquitination in DSB signaling and repair. (A) Crosstalk between SUMOylation and ubiquitination. MDC1 can be SUMOylated by PIAS1 and PIAS4 and then recognized by RNF4 leading to its polyubiquitination and degradation. PIAS1 and PIAS4 can also SUMOylate RNF168 and HERC2 leading to their activation. Activated HERC2 interacts with RNF8 and UBC13 and ubiquitinates histones together with activated RNF168. PIAS1 and PIAS4-mediated SUMOylation of MDC1 and histones recruits RNF4, which builds mixed SUMO-ubiquitin chains recognized by RAP80. Phosphorylated Sp1 is recognized PIAS1 and PIAS4 leading to its SUMOylation. SUMOylated Sp1 is recognized by RNF4 and gets polyubiquitinated and degraded. (B) Crosstalk between NEDDylation and ubiquitination. HUWE1 ubiquitin ligase can NEDDylate DNA-PKcs leading to its autophosphorylation and promoting NHEJ. NEDDylation of potentially cullins leads to polyubiquitination of Ku70/80 and their degradation. The enzyme performing NEDDylation is unknown. P, phosphorylation; SUMO, SUMOylation; NEDD8, NEDDylation; U, ubiquitin.
One of the essential SUMO cascades at the sites of DNA damage is mediated by the SUMO-targeted ubiquitin ligase (STUbL) RNF4. RNF4 is recruited to DSBs in a PIAS1-, PIAS4-dependent manner and requires its SUMO interaction motifs (Galanty et al., 2012). When bound to SUMOylated proteins like MDC1, RNF4 ubiquitinates them and targets them for proteasomal degradation thus ensuring the correct timing of DDR steps (Figure 6A). The lack of RNF4 results in severe DNA repair defects and constant phosphorylation of H2AX histone as a signal of persistent damage (Luo et al., 2012; Yin et al., 2012). Additionally, deSUMOylation by the deSUMOylase SENP2 provides a further pathway specific switch. It was shown that the extraction of MDC1 after RNF4 mediated ubiquitination and simultaneous SUMOylation is blocked by the SENP2 mediated deSUMOylation and thus promotes NHEJ (Garvin et al., 2019). Aside from repair proteins removal, RNF4 was proposed to facilitate ubiquitination signaling at the DSBs. The RAP80 protein is an important mediator of BRCA1 recruitment to damaged chromatin and requires both SUMO and ubiquitin modifications (Guzzo et al., 2012; Hu et al., 2012). RNF4 is thought to produce the hybrid SUMO-ubiquitin chains, which tether RAP80 and subsequently BRCA1 promoting HR (Figure 6A). The crucial role of SUMOylation and ubiquitination crosstalk in DNA repair pathway choice is getting more evidence with time. A recent study identified Sp1 as a target for SUMO-dependent RNF4 ubiquitination (Swift and Azizkhan-Clifford, 2022). The cascade involves DNA damage-induced phosphorylation of Sp1 by ATM and Cyclin A/CDK2 upon entry in the S phase, which is necessary for subsequent SUMOylation of Sp1 on K16 (Figure 6A). The serial events of phosphorylation and SUMOylation on Sp1 are recognized by RNF4 which ubiquitinates Sp1 and initiates its degradation. As Sp1 affects the localization of 53BP1, its degradation results in the removal of both Sp1 and 53BP1 from DSBs. The search for targets of RNF4 is ongoing and the number of players identified in SUMOylation-ubiquitination crosstalk is growing (Kumar et al., 2017).
Neddylation and Ubiquitination
NEDD8 (neural precursor cell expressed developmentally downregulated 8) is another ubiquitin-like protein that shares the highest similarity to ubiquitin, it is ∼60% identical and ∼80% homologous to ubiquitin (Jentsch and Pyrowolakis, 2000). However, as SUMOylation, the neddylation has its own enzymatic system in tight coordination with ubiquitination, and some ubiquitin ligases were shown to be able to perform neddylation due to the high similarity between NEDD8 and ubiquitin. The modification with NEDD8 was identified as the second most abundant ubiquitin-like modification following SUMO that accumulates at DSBs (Ma et al., 2013). At the DSBs, this NEDD8 accumulation was observed to be mediated by the E3 ubiquitin ligase RNF111 on the histone H4 N-terminus. Interestingly, H4 polyneddylation can be recognized by the MIU domain of RNF168, thus providing another regulating node for the RNF8/RNF168 ubiquitination axis (Ma et al., 2013).
Neddylation-guided ubiquitination at the sites of DNA damage regulates repair machinery and repair pathway choice as well. Ku70/80 was reported to be ubiquitinated on K195, K265, and K481 in Ku80 and K114 in Ku70 in a neddylation-dependent manner by a yet unknown factor (Figure 6B) (Brown et al., 2015). The ubiquitination of Ku70/80 causes its disassembly from the broken ends and therefore can promote resection-dependent repair. Consistently with the fact that cullins are the predominant targets of neddylation, the authors observed CUL4A and CUL4B as neddylation-dependent interacting partners of Ku70/80.
The dynamic regulation of neddylation and deneddylation at the DSBs was confirmed to guide repair pathway choice by controlling the length of CtIP-resected DNA (Jimeno et al., 2015). The authors reported that the RNF111/UBE2M-dependent neddylation inhibits resection and promotes NHEJ, while the deneddylation is required to initiate HR. At the sites of DNA damage, CtIP and its partner BRCA1 constitutively interact with neddylated proteins, therefore the balance between neddylation and its removal is crucial for the correct repair choice. Interestingly, the neddylation level on damaged chromatin regulates not only NHEJ/HR choice but also the fine balance between various variants of HR (Jimeno et al., 2015).
As a further example, DNA-PKcs was shown to be polyneddylated at its kinase domain by the E3 ubiquitin ligase HUWE1 (Figure 6B) (Guo et al., 2020). This modification is necessary for the autophosphorylation of DNA-PKcs on Ser2056 and efficient NHEJ.
There are more ubiquitin-like modifications, which have been described recently such as UFMylation, FATylation and ISGylation but little is known about their roles in DSB signaling and repair.
Ubiquitination, DNA Damage and Disease
Due to the abundance and versatility of the ubiquitin network, the dysregulation of any of its components can potentially lead to pathogenesis. Ubiquitination defects were reported to cause cancer, neurodegenerative diseases, immune pathologies, and muscle atrophy-related diseases (Popovic et al., 2014; Zheng et al., 2016). Since the first successful clinical application of proteasomal inhibitors (Hideshima et al., 2001), the search for a druggable ubiquitinome never stopped (Table 3).
TABLE 3.
Inhibitors of ubiquitin modifiers.
| Target | Name | References |
|---|---|---|
| VCP | NMS-873 | Magnaghi et al. (2013) |
| p97 | CB-5083 | Kilgas et al. (2021) |
| MDM2 | Nutlin | Vassilev et al. (2004) |
| MDM2 | APG-115 | Rasco et al. (2019a) |
| MDM2 | CGM097 | Holzer et al. (2015) |
| E3 ligases | PROTACs | Bondeson et al. (2015) |
| TRIM24 | dTRIM24 | Gechijian et al. (2018) |
| Cullin-RING ligases | MLN4924 | Zhao et al. (2014); Tong et al. (2017) |
| KEAP1 | Omaveloxolone | Lynch et al. (2019) |
| Proteasome components | PS-341 | Teicher et al. (1999) |
| Proteasome components | Marizomib | Potts et al. (2011) |
| Proteasome components | MLN9708 | Kupperman et al. (2010) |
| Proteasome components | NPI-0052 | Chauhan et al. (2005) |
| Proteasome components | CEP-18770 | Piva et al. (2008) |
| E3 ligases | CC-122 | Rasco et al. (2019b) |
| E3 ligases | CC-220 | Bjorklund et al. (2020) |
| SMURF1 | HS-152 | Tian et al. (2019) |
| DUBs | VLX1570 | Wang et al. (2015) |
The ubiquitination defects in DNA damage and repair are mostly related to genome instability. A well-known example are germline mutations in the BRCA1 gene, often in the RING domain region, that predispose to breast and ovarian cancer (Maxwell and Domchek, 2012). BRCA1 recruitment is mediated by the RNF8/RNF168 ubiquitination signaling and its dysregulation leads to severe consequences. Mutations inactivating the RNF168 gene were shown to cause the RIDDLE syndrome, a rare disease characterized by radiosensitivity, immunodeficiency, dysmorphic features, pulmonary failure, and learning disabilities (Stewart et al., 2009, 2007). The RNF168 can harbor various mutations and therefore result in different symptoms. A nonsense mutation leading to the loss of both ubiquitin-binding domains MIU1 and MIU2 was described to cause ataxia, growth retardation, microcephaly, immunodeficiency, and radiosensitivity (Devgan et al., 2011). Interestingly, the disease phenotype was accompanied by persistent chronic inflammation from unrepaired DNA damage that caused the idiopathic pulmonary fibrosis. RNF168 mutated in MIU1 and 2 was not able to retain 53BP1 and BRCA1 at the sites of the damage, thus impairing the RNF8-RNF168-HERC2-BRCA1 chromatin ubiquitin ligase cascade in DDR (Devgan et al., 2011). The paramount role of the ubiquitin ligases in maintaining genome stability is highlighted in the Fanconi anemia disease. Fanconi anemia is a rare disorder that results in bone marrow failure, cancer predisposition, and genomic instability. Fanconi anemia patients suffer chromosome fragility and hypersensitivity to drugs that induce DNA interstrand crosslinks. Often patients develop solid tumors such as squamous cell carcinomas in their 20s (Pollard and Gatti, 2009). There are more than 20 genes identified in the Fanconi anemia (FA) pathway that assemble in multiple complexes in the FA–BRCA DNA-damage response network (Wang, 2007; Fang et al., 2020). Eight of them (FANCA, B, C, E, F, G, L, and M) form the so-called FA core complex together with FANCA-associated polypeptides FAAP100 and FAAP24, a nuclear E3 ubiquitin ligase complex that monoubiquitinates the FANCD2/FANCI heterodimer (FA-ID complex) in response to damage. This results in the stabilization of the FA-ID complex on DNA and subsequent interaction with BRCA2, PALB2, RAD51, and BRIP1 to promote homologous recombination (Garcia-Higuera et al., 2001; Alcón et al., 2020). Most FA cases harbor mutations leading to a ubiquitination defect (Tan and Deans, 2017). The FA-ID ubiquitination is required for prevention of bone marrow failure, and FA patients with FANCD2 mutations were reported to have an early onset of bone marrow failure (Boisvert and Howlett, 2014; Tan et al., 2020). Depletion of any FA proteins responsible for the ubiquitination sensitizes cisplatin-resistant human lung adenocarcinoma cells to cisplatin treatment (Chen et al., 2016). Mutations in the FA genes deregulate the DDR and the predisposition to the BRCA1-and BRCA2-dependent cancers (Kais et al., 2016).
Various combinations of mutations in genes coding for different ubiquitin modifiers result in various pathological phenotypes. The genetic deletion of RNF8 and CHFR sensitizes mice to ionizing radiation and results in the development of T-cell lymphoma, emphasizing the importance of the combined action of these phospho- and PAR-targeted ubiquitin ligases in the DDR (Wu J. et al., 2011). The RNF8 knockout in mice causes growth retardation, sensitivity to ionizing radiation, impaired spermatogenesis, and defective immunoglobulin class-switch-recombination, similar as RNF168 and 53BP1 knockouts (Manis et al., 2004; Li et al., 2010; Santos et al., 2010). Interestingly, similar effects as for RNF8 and RNF168 knock out were observed for RNF4 deficiency, which links SUMOylation to ubiquitination cascades in DDR (Vyas et al., 2013).
The susceptibility of the ubiquitination cascades involved in DDR are not only the source of genetic instability in normal cells but can be also used to sensitize cancer cells to ionizing radiation or chemotherapy. A well-known example is PARP inhibition, which leads to impaired homologous recombination in cells with BRCA1 and BRCA2 mutations (Bryant et al., 2005). The VCP/p97 complex mediates the turnover of K48-linked ubiquitin-labeled proteins at the sites of DSBs thus facilitating timely repair (Meerang et al., 2011). The VCP/p97 inhibitor NMS-873 was shown to induce unfolded protein response, autophagy, and cell death (Magnaghi et al., 2013). Additionally, VCP/p97 inhibition by the specific small-molecule inhibitor CB-5083 leads to cell death after IR due to the excessive MRN-mediated resection (Kilgas et al., 2021). Many tumors overexpress MDM2 (human analog HDM2), the E3 ubiquitin ligase that ubiquitinates p53 and negatively controls its levels thus promoting cell survival (Ladanyi et al., 1993). Therefore, the inhibitors of MDM2 are promising anti-cancer drugs. One of the most potent MDM2 small-molecule inhibitors identified in high-throughput screening is the Nutlin family, analogs of cis-imidazoline (Vassilev et al., 2004). Currently two MDM2 inhibitors are in clinical trials: APG-115 is in Phase I, and CGM097 in Phase II (Holzer et al., 2015; Rasco et al., 2019a; Rasco et al., 2019b). Inhibiting MDM2 activity, the inhibitors restore the anti-cancer activity of p53 and lead to apoptosis. The MLN4924 inhibitor blocks Cullins NEDDylation and inactivates Cullin-RING ligases which, in turn, triggers cell cycle arrest, apoptosis, senescence, and autophagy in many cancer cells (Zhao et al., 2014; Tong et al., 2017). In addition, it was reported that the accumulation of substrates of the Cullin-RING ligases (p21, p27, Wee1), trigger DNA damage, and induce cell cycle arrest at the G2/M stage (Tong et al., 2017).
The development of new small-molecule inhibitors targeting ubiquitin modifiers in the DDR pathways for the treatment of cancer and other diseases is underway and requires the expansion of our knowledge about the ubiquitination network.
Conclusion
Although plenty of novel ubiquitin modifiers controlling DDR pathways were discovered in recent years, there is still missing knowledge. Due to the immense complexity of the ubiquitin-dependent signaling system, DNA damage signaling and repair are precisely spatiotemporally regulated in the chromatin context. Recent studies unveiled novel functions of the ubiquitin cascades at the DSBs, such as phase separation, chromatin mobility, DSB clustering, and homology search. An additional layer of regulation, the phosphorylation of ubiquitin, was recently reported for its role in DDR. In the coming years, many novel ubiquitin modifiers and ubiquitination cascades are expected to be discovered and more emphasis will be placed on the crosstalk between ubiquitination and other PTMs taking place at the damage sites. At last, the knowledge we gain about the ubiquitination system in DDR would give us a potent tool to develop therapeutic approaches for cancer and other diseases.
Acknowledgments
We apologize to those researchers whose work could not be cited. We would like to thank all past and present members of the lab for their support and passion about epigenetics.
Author Contributions
KK, AR, and MC substantially contributed to the design of the review and interpreting the relevant literature, KK wrote the manuscript, and AR and MC revised it critically.
Funding
Our laboratory is supported by grants from the German Research Foundation (DFG), the Federal Ministry of Education and Research (BMBF), and the Hessian Ministry of Higher Education, Research, Science and the Arts (HMWK).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
- ATM
ataxia-telangiectasia mutated
- ATR
ATM and RAD3-related
- CDK
cell cycle-dependent kinase
- CFS
common fragile sites
- cNHEJ
canonical NHEJ
- DDR
DNA damage response
- DNA-PKcs
DNA-dependent protein kinase catalytic subunit
- DSB
double-strand break
- DTC
Deltex C-terminal domain
- DUB
deubiquitinase
- ERK
extracellular signal-regulated kinase
- FA
Fanconi anemia
- FRRUC
FBXL10-RNF68-BMI1 ubiquitin ligase complex
- HDAC
histone deacetylase
- HR
homologous recombination
- IR
ionizing radiation
- IRIF
irradiation-induced focus
- LET
linear energy transfer
- LLPS
liquid-liquid phase separation
- MIU
motif interacting with ubiquitin
- NEDD8
neural precursor cell expressed developmentally downregulated 8
- NER
nucleotide excision repair
- NHEJ
non-homologous end joining
- PAR
poly(ADP-ribose)
- PARP
poly (ADP-ribose) polymerase
- PI3
phosphoinositide 3
- PI3KK
phosphoinositide 3 kinase-related protein kinase
- polyUb
polyubiquitin
- PTM
post-translational modification
- pUb
phosphoubiquitin
- RIDDLE
Radiosensitivity, ImmunoDeficiency Dysmorphic features and LEarning difficulties syndrom
- ssDNA
single-strand DNA
- STUbL
SUMO-targeted ubiquitin ligase
- SUMO
small ubiquitin-like modifier
- TAD
topologically associated domain
- TSA
Trichostatin A
- Ub
ubiquitin
- UBD
ubiquitin-binding domain
- UBZ
ubiquitin-binding zinc-finger
- UIM
ubiquitin-interacting motif
References
- Acs K., Luijsterburg M. S., Ackermann L., Salomons F. A., Hoppe T., Dantuma N. P. (2011). The AAA-ATPase VCP/p97 Promotes 53BP1 Recruitment by Removing L3MBTL1 from DNA Double-Strand Breaks. Nat. Struct. Mol. Biol. 18, 1345–1350. 10.1038/nsmb.2188 [DOI] [PubMed] [Google Scholar]
- Ahmed S. F., Buetow L., Gabrielsen M., Lilla S., Chatrin C., Sibbet G. J., et al. (2020). DELTEX2 C-Terminal Domain Recognizes and Recruits ADP-Ribosylated Proteins for Ubiquitination. Sci. Adv. 6. 10.1126/sciadv.abc0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagoz M., Katsuki Y., Ogiwara H., Ogi T., Shibata A., Kakarougkas A., et al. (2015). SETDB1, HP1 and SUV39 Promote Repositioning of 53BP1 to Extend Resection during Homologous Recombination in G2 Cells. Nucleic Acids Res. 43, 7931–7944. 10.1093/nar/gkv722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcón P., Shakeel S., Chen Z. A., Rappsilber J., Patel K. J., Passmore L. A. (2020). FANCD2-FANCI Is a Clamp Stabilized on DNA by Monoubiquitination of FANCD2 during DNA Repair. Nat. Struct. Mol. Biol. 27, 240–248. 10.1038/s41594-020-0380-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M. A. M., Strickfaden H., Lee B. L., Spyracopoulos L., Hendzel M. J. (2018). RYBP Is a K63-Ubiquitin-Chain-Binding Protein that Inhibits Homologous Recombination Repair. Cell Rep. 22, 383–395. 10.1016/j.celrep.2017.12.047 [DOI] [PubMed] [Google Scholar]
- Amendola P. G., Zaghet N., Ramalho J. J., Vilstrup Johansen J., Boxem M., Salcini A. E. (2017). JMJD-5/KDM8 Regulates H3K36me2 and Is Required for Late Steps of Homologous Recombination and Genome Integrity. PLoS Genet. 13, e1006632. 10.1371/journal.pgen.1006632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An L., Dong C., Li J., Chen J., Yuan J., Huang J., et al. (2018). RNF169 Limits 53BP1 Deposition at DSBs to Stimulate Single-Strand Annealing Repair. Proc. Natl. Acad. Sci. U.S.A. 115, E8286–E8295. 10.1073/pnas.1804823115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand R., Ranjha L., Cannavo E., Cejka P. (2016). Phosphorylated CtIP Functions as a Co-factor of the MRE11-RAD50-NBS1 Endonuclease in DNA End Resection. Mol. Cell 64, 940–950. 10.1016/j.molcel.2016.10.017 [DOI] [PubMed] [Google Scholar]
- Andersen P. L., Zhou H., Pastushok L., Moraes T., McKenna S., Ziola B., et al. (2005). Distinct Regulation of Ubc13 Functions by the Two Ubiquitin-Conjugating Enzyme Variants Mms2 and Uev1A. J. Cell Biol. 170, 745–755. 10.1083/jcb.200502113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang X. L., Wade Harper J. (2005). SCF-Mediated Protein Degradation and Cell Cycle Control. Oncogene 24, 2860–2870. 10.1038/sj.onc.1208614 [DOI] [PubMed] [Google Scholar]
- Arnould C., Rocher V., Finoux A.-L., Clouaire T., Li K., Zhou F., et al. (2021). Loop Extrusion as a Mechanism for Formation of DNA Damage Repair Foci. Nature 590, 660–665. 10.1038/s41586-021-03193-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymard F., Aguirrebengoa M., Guillou E., Javierre B. M., Bugler B., Arnould C., et al. (2017). Genome-wide Mapping of Long-Range Contacts Unveils Clustering of DNA Double-Strand Breaks at Damaged Active Genes. Nat. Struct. Mol. Biol. 24, 353–361. 10.1038/nsmb.3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymard F., Bugler B., Schmidt C. K., Guillou E., Caron P., Briois S., et al. (2014). Transcriptionally Active Chromatin Recruits Homologous Recombination at DNA Double-Strand Breaks. Nat. Struct. Mol. Biol. 21, 366–374. 10.1038/nsmb.2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayrapetov M. K., Gursoy-Yuzugullu O., Xu C., Xu Y., Price B. D. (2014). DNA Double-Strand Breaks Promote Methylation of Histone H3 on Lysine 9 and Transient Formation of Repressive Chromatin. Proc. Natl. Acad. Sci. U.S.A. 111, 9169–9174. 10.1073/pnas.1403565111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmik A. A., Sonawane S. K., Chinnathambi S. (2021). The Extracellular HDAC6 ZnF UBP Domain Modulates the Actin Network and Post-translational Modifications of Tau. Cell Commun. Signal. 19, 49. 10.1186/s12964-021-00736-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J., Hořejší Z., Koed K., Krämer A., Tort F., Zieger K., et al. (2005). DNA Damage Response as a Candidate Anti-cancer Barrier in Early Human Tumorigenesis. Nature 434, 864–870. 10.1038/nature03482 [DOI] [PubMed] [Google Scholar]
- Battu A., Ray A., Wani A. A. (2011). ASF1A and ATM Regulate H3K56-Mediated Cell-Cycle Checkpoint Recovery in Response to UV Irradiation. Nucleic Acids Res. 39, 7931–7945. 10.1093/nar/gkr523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayona-Feliu A., Aguilera A. (2021). The Role of Chromatin at Transcription-Replication Conflicts as a Genome Safeguard. Biochem. Soc. Trans. 49, 2727–2736. 10.1042/BST20210691 [DOI] [PubMed] [Google Scholar]
- Becker J. R., Clifford G., Bonnet C., Groth A., Wilson M. D., Chapman J. R. (2021). BARD1 Reads H2A Lysine 15 Ubiquitination to Direct Homologous Recombination. Nature 596, 433–437. 10.1038/s41586-021-03776-w [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S., Danielsen J. R., Fugger K., Gromova I., Nerstedt A., Lukas C., et al. (2010). HERC2 Coordinates Ubiquitin-dependent Assembly of DNA Repair Factors on Damaged Chromosomes. Nat. Cell Biol. 12, 80–86. 10.1038/ncb2008 [DOI] [PubMed] [Google Scholar]
- Beucher A., Birraux J., Tchouandong L., Barton O., Shibata A., Conrad S., et al. (2009). ATM and Artemis Promote Homologous Recombination of Radiation-Induced DNA Double-Strand Breaks in G2. EMBO J. 28, 3413–3427. 10.1038/emboj.2009.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoumik A., Singha N., O'Connell M. J., Ronai Z. e. A. (2008). Regulation of TIP60 by ATF2 Modulates ATM Activation. J. Biol. Chem. 283, 17605–17614. 10.1074/jbc.M802030200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund C. C., Kang J., Amatangelo M., Polonskaia A., Katz M., Chiu H., et al. (2020). Iberdomide (CC-220) Is a Potent Cereblon E3 Ligase Modulator with Antitumor and Immunostimulatory Activities in Lenalidomide- and Pomalidomide-Resistant Multiple Myeloma Cells with Dysregulated CRBN. Leukemia 34, 1197–1201. 10.1038/s41375-019-0620-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert R. A., Howlett N. G. (2014). The Fanconi Anemia ID2 Complex: Dueling Saxes at the Crossroads. Cell Cycle 13, 2999–3015. 10.4161/15384101.2014.956475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomar M. G., D'Souza S., Bienko M., Dikic I., Walker G. C., Zhou P. (2010). Unconventional Ubiquitin Recognition by the Ubiquitin-Binding Motif within the Y Family DNA Polymerases ι and Rev1. Mol. Cell 37, 408–417. 10.1016/j.molcel.2009.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondeson D. P., Mares A., Smith I. E. D., Ko E., Campos S., Miah A. H., et al. (2015). Catalytic In Vivo Protein Knockdown by Small-Molecule PROTACs. Nat. Chem. Biol. 11, 611–617. 10.1038/nchembio.1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan M. V., Lee J., Ward I. M., Kim J.-E., Thompson J. R., Chen J., et al. (2006). Structural Basis for the Methylation State-specific Recognition of Histone H4-K20 by 53BP1 and Crb2 in DNA Repair. Cell 127, 1361–1373. 10.1016/j.cell.2006.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla F., Garcia-Manteiga J. M., Monteleone E., Hoelzen L., Zocchi A., Agresti A., et al. (2020). Nucleosomes Effectively Shield DNA from Radiation Damage in Living Cells. Nucleic Acids Res. 48, 8993–9006. 10.1093/nar/gkaa613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. S., Lukashchuk N., Sczaniecka-Clift M., Britton S., le Sage C., Calsou P., et al. (2015). Neddylation Promotes Ubiquitylation and Release of Ku from DNA-Damage Sites. Cell Rep. 11, 704–714. 10.1016/j.celrep.2015.03.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant H. E., Schultz N., Thomas H. D., Parker K. M., Flower D., Lopez E., et al. (2005). Specific Killing of BRCA2-Deficient Tumours with Inhibitors of poly(ADP-Ribose) Polymerase. Nature 434, 913–917. 10.1038/nature03443 [DOI] [PubMed] [Google Scholar]
- Burgess R. C., Burman B., Kruhlak M. J., Misteli T. (2014). Activation of DNA Damage Response Signaling by Condensed Chromatin. Cell Rep. 9, 1703–1717. 10.1016/j.celrep.2014.10.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S., Ismail I. H., Young L. C., Poirier G. G., Hendzel M. J. (2013). Polycomb Repressive Complex 2 Contributes to DNA Double-Strand Break Repair. Cell Cycle 12, 2675–2683. 10.4161/cc.25795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannan W. J., Tsang B. P., Wallace S. S., Pederson D. S. (2014). Nucleosomes Suppress the Formation of Double-Strand DNA Breaks during Attempted Base Excision Repair of Clustered Oxidative Damages. J. Biol. Chem. 289, 19881–19893. 10.1074/jbc.M114.571588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caridi C. P., D’Agostino C., Ryu T., Zapotoczny G., Delabaere L., Li X., et al. (2018). Nuclear F-Actin and Myosins Drive Relocalization of Heterochromatic Breaks. Nature 559, 54–60. 10.1038/s41586-018-0242-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caridi P. C., Delabaere L., Zapotoczny G., Chiolo I. (2017). And yet, it Moves: Nuclear and Chromatin Dynamics of a Heterochromatic Double-Strand Break. Phil. Trans. R. Soc. B 372, 20160291. 10.1098/rstb.2016.0291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho S., Vítor A. C., Sridhara S. C., Martins F. B., Raposo A. C., Desterro J. M., et al. (2014). SETD2 Is Required for DNA Double-Strand Break Repair and Activation of the P53-Mediated Checkpoint. eLife 3, e02482. 10.7554/eLife.02482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celen A. B., Sahin U. (2020). Sumoylation on its 25th Anniversary: Mechanisms, Pathology, and Emerging Concepts. FEBS J. 287, 3110–3140. 10.1111/febs.15319 [DOI] [PubMed] [Google Scholar]
- Chagin V. O., Reinhart B., Becker A., Mortusewicz O., Jost K. L., Rapp A., et al. (2019). Processive DNA Synthesis Is Associated with Localized Decompaction of Constitutive Heterochromatin at the Sites of DNA Replication and Repair. Nucleus 10, 231–253. 10.1080/19491034.2019.1688932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challa K., Schmid C. D., Kitagawa S., Cheblal A., Iesmantavicius V., Seeber A., et al. (2021). Damage-induced Chromatome Dynamics Link Ubiquitin Ligase and Proteasome Recruitment to Histone Loss and Efficient DNA Repair. Mol. Cell 81, 811–829. 10.1016/j.molcel.2020.12.021 [DOI] [PubMed] [Google Scholar]
- Chapman J. R., Barral P., Vannier J.-B., Borel V., Steger M., Tomas-Loba A., et al. (2013). RIF1 Is Essential for 53BP1-dependent Nonhomologous End Joining and Suppression of DNA Double-Strand Break Resection. Mol. Cell 49, 858–871. 10.1016/j.molcel.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan D., Catley L., Li G., Podar K., Hideshima T., Velankar M., et al. (2005). A Novel Orally Active Proteasome Inhibitor Induces Apoptosis in Multiple Myeloma Cells with Mechanisms Distinct from Bortezomib. Cancer Cell 8, 407–419. 10.1016/j.ccr.2005.10.013 [DOI] [PubMed] [Google Scholar]
- Cheblal A., Challa K., Seeber A., Shimada K., Yoshida H., Ferreira H. C., et al. (2020). DNA Damage-Induced Nucleosome Depletion Enhances Homology Search Independently of Local Break Movement. Mol. Cell 80, 311–326. 10.1016/j.molcel.2020.09.002 [DOI] [PubMed] [Google Scholar]
- Chen J., Feng W., Jiang J., Deng Y., Huen M. S. Y. (2012). Ring Finger Protein RNF169 Antagonizes the Ubiquitin-dependent Signaling Cascade at Sites of DNA Damage. J. Biol. Chem. 287, 27715–27722. 10.1074/jbc.M112.373530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Wang Z., Guo X., Li F., Wei Q., Chen X., et al. (2019). TRIM66 Reads Unmodified H3R2K4 and H3K56ac to Respond to DNA Damage in Embryonic Stem Cells. Nat. Commun. 10, 4273. 10.1038/s41467-019-12126-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Li J., Chen Y.-C., Qian H., Chen Y.-J., Su J.-Y., et al. (2016). The Functional Status of DNA Repair Pathways Determines the Sensitization Effect to Cisplatin in Non-small Cell Lung Cancer Cells. Cell Oncol. 39, 511–522. 10.1007/s13402-016-0291-7 [DOI] [PubMed] [Google Scholar]
- Chernikova S. B., Dorth J. A., Razorenova O. V., Game J. C., Brown J. M. (2010). Deficiency in Bre1 Impairs Homologous Recombination Repair and Cell Cycle Checkpoint Response to Radiation Damage in Mammalian Cells. Radiat. Res. 174, 558–565. 10.1667/RR2184.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung W. L., Turner F. B., Krishnamoorthy T., Wolner B., Ahn S.-H., Foley M., et al. (2005). Phosphorylation of Histone H4 Serine 1 during DNA Damage Requires Casein Kinase II in S. cerevisiae . Curr. Biol. 15, 656–660. 10.1016/j.cub.2005.02.049 [DOI] [PubMed] [Google Scholar]
- Chiolo I., Minoda A., Colmenares S. U., Polyzos A., Costes S. V., Karpen G. H. (2011). Double-strand Breaks in Heterochromatin Move outside of a Dynamic HP1a Domain to Complete Recombinational Repair. Cell 144, 732–744. 10.1016/j.cell.2011.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.-Y., Jang H., Roe J.-S., Kim S.-T., Cho E.-J., Youn H.-D. (2013). Phosphorylation and Ubiquitination-dependent Degradation of CABIN1 Releases P53 for Transactivation upon Genotoxic Stress. Nucleic Acids Res. 41, 2180–2190. 10.1093/nar/gks1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou D. M., Adamson B., Dephoure N. E., Tan X., Nottke A. C., Hurov K. E., et al. (2010). A Chromatin Localization Screen Reveals Poly (ADP Ribose)-Regulated Recruitment of the Repressive Polycomb and NuRD Complexes to Sites of DNA Damage. Proc. Natl. Acad. Sci. U.S.A. 107, 18475–18480. 10.1073/pnas.1012946107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury D., Keogh M.-C., Ishii H., Peterson C. L., Buratowski S., Lieberman J. (2005). γ-H2AX Dephosphorylation by Protein Phosphatase 2A Facilitates DNA Double-Strand Break Repair. Mol. Cell 20, 801–809. 10.1016/j.molcel.2005.10.003 [DOI] [PubMed] [Google Scholar]
- Chowdhury D., Xu X., Zhong X., Ahmed F., Zhong J., Liao J., et al. (2008). A PP4-Phosphatase Complex Dephosphorylates γ-H2AX Generated during DNA Replication. Mol. Cell 31, 33–46. 10.1016/j.molcel.2008.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague M. J., Heride C., Urbé S. (2015). The Demographics of the Ubiquitin System. Trends Cell Biol. 25, 417–426. 10.1016/j.tcb.2015.03.002 [DOI] [PubMed] [Google Scholar]
- Clague M. J., Urbé S., Komander D. (2019). Breaking the Chains: Deubiquitylating Enzyme Specificity Begets Function. Nat. Rev. Mol. Cell Biol. 20, 338–352. 10.1038/s41580-019-0099-1 [DOI] [PubMed] [Google Scholar]
- Clouaire T., Rocher V., Lashgari A., Arnould C., Aguirrebengoa M., Biernacka A., et al. (2018). Comprehensive Mapping of Histone Modifications at DNA Double-Strand Breaks Deciphers Repair Pathway Chromatin Signatures. Mol. Cell 72, 250–262. 10.1016/j.molcel.2018.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P. J., Ju B. G., Telese F., Wang X., Glass C. K., Rosenfeld M. G. (2009). Tyrosine Dephosphorylation of H2AX Modulates Apoptosis and Survival Decisions. Nature 458, 591–596. 10.1038/nature07849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. Potts B., X. Albitar M., C. Anderson K., Baritaki S., Berkers C., Bonavida B., et al. (2011). Marizomib, a Proteasome Inhibitor for All Seasons: Preclinical Profile and a Framework for Clinical Trials. Ccdt 11, 254–284. 10.2174/156800911794519716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui D., Xiong X., Shu J., Dai X., Sun Y., Zhao Y. (2020). FBXW7 Confers Radiation Survival by Targeting P53 for Degradation. Cell Rep. 30, 497–509. 10.1016/j.celrep.2019.12.032 [DOI] [PubMed] [Google Scholar]
- Danielsen J. R., Povlsen L. K., Villumsen B. H., Streicher W., Nilsson J., Wikström M., et al. (2012). DNA Damage-Inducible SUMOylation of HERC2 Promotes RNF8 Binding via a Novel SUMO-Binding Zinc Finger. J. Cell Biol. 197, 179–187. 10.1083/jcb.201106152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma N. P., Attikum H. (2016). Spatiotemporal Regulation of Posttranslational Modifications in the DNA Damage Response. EMBO J. 35, 6–23. 10.15252/embj.201592595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daou S., Hammond-Martel I., Mashtalir N., Barbour H., Gagnon J., Iannantuono N. G., et al. (2015). The BAP1/ASXL2 Histone H2A Deubiquitinase Complex Regulates Cell Proliferation and Is Disrupted in Cancer. J. Biol. Chem. 290, 28643–28663. 10.1074/jbc.M115.661553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaRosa P. A., Wang Z., Jiang X., Pruneda J. N., Cong F., Klevit R. E., et al. (2015). Allosteric Activation of the RNF146 Ubiquitin Ligase by a poly(ADP-Ribosyl)ation Signal. Nature 517, 223–226. 10.1038/nature13826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C., Lucia M. S., Hansen K. C., Tyler J. K. (2009). CBP/p300-mediated Acetylation of Histone H3 on Lysine 56. Nature 459, 113–117. 10.1038/nature07861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A., Mondal P., Dalui S., Das C., Roy S. (2021). Molecular Characterization of Substrate‐induced Ubiquitin Transfer by UBR7‐PHD Finger, a Newly Identified Histone H2BK120 Ubiquitin Ligase. FEBS J. 289, 1842–1857. 10.1111/febs.16262 [DOI] [PubMed] [Google Scholar]
- Delgado-Díaz M. R., Martín Y., Berg A., Freire R., Smits V. A. J. (2014). Dub3 Controls DNA Damage Signalling by Direct Deubiquitination of H2AX. Mol. Oncol. 8, 884–893. 10.1016/j.molonc.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Densham R. M., Garvin A. J., Stone H. R., Strachan J., Baldock R. A., Daza-Martin M., et al. (2016). Human BRCA1-BARD1 Ubiquitin Ligase Activity Counteracts Chromatin Barriers to DNA Resection. Nat. Struct. Mol. Biol. 23, 647–655. 10.1038/nsmb.3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devgan S. S., Sanal O., Doil C., Nakamura K., Nahas S. A., Pettijohn K., et al. (2011). Homozygous Deficiency of Ubiquitin-Ligase Ring-Finger Protein RNF168 Mimics the Radiosensitivity Syndrome of Ataxia-Telangiectasia. Cell Death Differ. 18, 1500–1506. 10.1038/cdd.2011.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Virgilio M., Callen E., Yamane A., Zhang W., Jankovic M., Gitlin A. D., et al. (2013). Rif1 Prevents Resection of DNA Breaks and Promotes Immunoglobulin Class Switching. Science 339, 711–715. 10.1126/science.1230624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I., Wakatsuki S., Walters K. J. (2009). Ubiquitin-binding Domains - from Structures to Functions. Nat. Rev. Mol. Cell Biol. 10, 659–671. 10.1038/nrm2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D. H., Pepperkok R., et al. (2009). RNF168 Binds and Amplifies Ubiquitin Conjugates on Damaged Chromosomes to Allow Accumulation of Repair Proteins. Cell 136, 435–446. 10.1016/j.cell.2008.12.041 [DOI] [PubMed] [Google Scholar]
- Douglas P., Zhong J., Ye R., Moorhead G. B. G., Xu X., Lees-Miller S. P. (2010). Protein Phosphatase 6 Interacts with the DNA-dependent Protein Kinase Catalytic Subunit and Dephosphorylates γ-H2AX. Mol. Cell. Biol. 30, 1368–1381. 10.1128/MCB.00741-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan M.-R., Smerdon M. J. (2014). Histone H3 Lysine 14 (H3K14) Acetylation Facilitates DNA Repair in a Positioned Nucleosome by Stabilizing the Binding of the Chromatin Remodeler RSC (Remodels Structure of Chromatin). J. Biol. Chem. 289, 8353–8363. 10.1074/jbc.M113.540732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois J.-C., Yates M., Gaudreau-Lapierre A., Clément G., Cappadocia L., Gaudreau L., et al. (2017). A Phosphorylation-And-Ubiquitylation Circuitry Driving ATR Activation and Homologous Recombination. Nucleic Acids Res. 45, 8859–8872. 10.1093/nar/gkx571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwane L., Gallagher W. M., Ní Chonghaile T., O'Connor D. P. (2017). The Emerging Role of Non-traditional Ubiquitination in Oncogenic Pathways. J. Biol. Chem. 292, 3543–3551. 10.1074/jbc.R116.755694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ea C.-K., Deng L., Xia Z.-P., Pineda G., Chen Z. J. (2006). Activation of IKK by TNFα Requires Site-specific Ubiquitination of RIP1 and Polyubiquitin Binding by NEMO. Mol. Cell 22, 245–257. 10.1016/j.molcel.2006.03.026 [DOI] [PubMed] [Google Scholar]
- Elia A. E. H., Boardman A. P., Wang D. C., Huttlin E. L., Everley R. A., Dephoure N., et al. (2015). Quantitative Proteomic Atlas of Ubiquitination and Acetylation in the DNA Damage Response. Mol. Cell 59, 867–881. 10.1016/j.molcel.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia M. C., Bradley M. O. (1992). Influence of Chromatin Structure on the Induction of DNA Double Strand Breaks by Ionizing Radiation. Cancer Res. 52, 1580–1586. [PubMed] [Google Scholar]
- Escribano-Díaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J. T. F., Tkáč J., et al. (2013). A Cell Cycle-dependent Regulatory Circuit Composed of 53BP1-RIF1 and BRCA1-CtIP Controls DNA Repair Pathway Choice. Mol. Cell 49, 872–883. 10.1016/j.molcel.2013.01.001 [DOI] [PubMed] [Google Scholar]
- Falk M., Lukášová E., Kozubek S. (2008). Chromatin Structure Influences the Sensitivity of DNA to γ-radiation. Biochimica Biophysica Acta (BBA) - Mol. Cell Res. 1783, 2398–2414. 10.1016/j.bbamcr.2008.07.010 [DOI] [PubMed] [Google Scholar]
- Fang C.-B., Wu H.-T., Zhang M.-L., Liu J., Zhang G.-J. (2020). Fanconi Anemia Pathway: Mechanisms of Breast Cancer Predisposition Development and Potential Therapeutic Targets. Front. Cell Dev. Biol. 8, 160. 10.3389/fcell.2020.00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher D., Wellinger R. J. (2010). Methylated H3K4, a Transcription-Associated Histone Modification, Is Involved in the DNA Damage Response Pathway. PLoS Genet. 6, e1001082. 10.1371/journal.pgen.1001082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Chen J. (2012). The E3 Ligase RNF8 Regulates KU80 Removal and NHEJ Repair. Nat. Struct. Mol. Biol. 19, 201–206. 10.1038/nsmb.2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo O., Allis C. D., Nussenzweig A. (2004). Phosphorylation of Histone H2B at DNA Double-Strand Breaks. J. Exp. Med. 199, 1671–1677. 10.1084/jem.20032247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierz B., Chatterjee C., McGinty R. K., Bar-Dagan M., Raleigh D. P., Muir T. W. (2011). Histone H2B Ubiquitylation Disrupts Local and Higher-Order Chromatin Compaction. Nat. Chem. Biol. 7, 113–119. 10.1038/nchembio.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D. (2009). Recognition and Processing of Ubiquitin-Protein Conjugates by the Proteasome. Annu. Rev. Biochem. 78, 477–513. 10.1146/annurev.biochem.78.081507.101607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fnu S., Williamson E. A., De Haro L. P., Brenneman M., Wray J., Shaheen M., et al. (2011). Methylation of Histone H3 Lysine 36 Enhances DNA Repair by Nonhomologous End-Joining. Proc. Natl. Acad. Sci. U.S.A. 108, 540–545. 10.1073/pnas.1013571108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradet-Turcotte A., Canny M. D., Escribano-Díaz C., Orthwein A., Leung C. C. Y., Huang H., et al. (2013). 53BP1 Is a Reader of the DNA-Damage-Induced H2A Lys 15 Ubiquitin Mark. Nature 499, 50–54. 10.1038/nature12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frattini C., Promonet A., Alghoul E., Vidal-Eychenie S., Lamarque M., Blanchard M.-P., et al. (2021). TopBP1 Assembles Nuclear Condensates to Switch on ATR Signaling. Mol. Cell 81, 1231–1245. 10.1016/j.molcel.2020.12.049 [DOI] [PubMed] [Google Scholar]
- Fu Q.-S., Zhou C.-J., Gao H.-C., Jiang Y.-J., Zhou Z.-R., Hong J., et al. (2009). Structural Basis for Ubiquitin Recognition by a Novel Domain from Human Phospholipase A2-Activating Protein. J. Biol. Chem. 284, 19043–19052. 10.1074/jbc.M109.009126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y., Belotserkovskaya R., Coates J., Jackson S. P. (2012). RNF4, a SUMO-Targeted Ubiquitin E3 Ligase, Promotes DNA Double-Strand Break Repair. Genes Dev. 26, 1179–1195. 10.1101/gad.188284.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galanty Y., Belotserkovskaya R., Coates J., Polo S., Miller K. M., Jackson S. P. (2009). Mammalian SUMO E3-Ligases PIAS1 and PIAS4 Promote Responses to DNA Double-Strand Breaks. Nature 462, 935–939. 10.1038/nature08657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M. S., Timmers C., Hejna J., et al. (2001). Interaction of the Fanconi Anemia Proteins and BRCA1 in a Common Pathway. Mol. Cell 7, 249–262. 10.1016/s1097-2765(01)00173-3 [DOI] [PubMed] [Google Scholar]
- Garvin A. J., Walker A. K., Densham R. M., Chauhan A. S., Stone H. R., Mackay H. L., et al. (2019). The deSUMOylase SENP2 Coordinates Homologous Recombination and Nonhomologous End Joining by Independent Mechanisms. Genes Dev. 33, 333–347. 10.1101/gad.321125.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M., Imhof R., Huang Q., Baudis M., Altmeyer M. (2020). The Ubiquitin Ligase TRIP12 Limits PARP1 Trapping and Constrains PARP Inhibitor Efficiency. Cell Rep. 32, 107985. 10.1016/j.celrep.2020.107985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti M., Pinato S., Maiolica A., Rocchio F., Prato M. G., Aebersold R., et al. (2015). RNF168 Promotes Noncanonical K27 Ubiquitination to Signal DNA Damage. Cell Rep. 10, 226–238. 10.1016/j.celrep.2014.12.021 [DOI] [PubMed] [Google Scholar]
- Ge C., Che L., Du C. (2015). The UBC Domain Is Required for BRUCE to Promote BRIT1/MCPH1 Function in DSB Signaling and Repair Post Formation of BRUCE-USP8-BRIT1 Complex. PLoS ONE 10, e0144957. 10.1371/journal.pone.0144957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechijian L. N., Buckley D. L., Lawlor M. A., Reyes J. M., Paulk J., Ott C. J., et al. (2018). Functional TRIM24 Degrader via Conjugation of Ineffectual Bromodomain and VHL Ligands. Nat. Chem. Biol. 14, 405–412. 10.1038/s41589-018-0010-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- George A. J., Hoffiz Y. C., Charles A. J., Zhu Y., Mabb A. M. (2018). A Comprehensive Atlas of E3 Ubiquitin Ligase Mutations in Neurological Disorders. Front. Genet. 9, 29. 10.3389/fgene.2018.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodke I., Remisova M., Furst A., Kilic S., Reina-San-Martin B., Poetsch A. R., et al. (2021). AHNAK Controls 53BP1-Mediated P53 Response by Restraining 53BP1 Oligomerization and Phase Separation. Mol. Cell 81, 2596–2610. 10.1016/j.molcel.2021.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson B. A., Doolittle L. K., Schneider M. W. G., Jensen L. E., Gamarra N., Henry L., et al. (2019). Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 179, 470–484. 10.1016/j.cell.2019.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldknopf I. L., French M. F., Musso R., Busch H. (1977). Presence of Protein A24 in Rat Liver Nucleosomes. Proc. Natl. Acad. Sci. U.S.A. 74, 5492–5495. 10.1073/pnas.74.12.5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein G., Scheid M., Hammerling U., Schlesinger D. H., Niall H. D., Boyse E. A. (1975). Isolation of a Polypeptide that Has Lymphocyte-Differentiating Properties and Is Probably Represented Universally in Living Cells. Proc. Natl. Acad. Sci. U.S.A. 72, 11–15. 10.1073/pnas.72.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F., Chiu L.-Y., Cox B., Aymard F., Clouaire T., Leung J. W., et al. (2015). Screen Identifies Bromodomain Protein ZMYND8 in Chromatin Recognition of Transcription-Associated DNA Damage that Promotes Homologous Recombination. Genes Dev. 29, 197–211. 10.1101/gad.252189.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F., Clouaire T., Aguirrebengoa M., Legube G., Miller K. M. (2017). Histone Demethylase KDM5A Regulates the ZMYND8-NuRD Chromatin Remodeler to Promote DNA Repair. J. Cell Biol. 216, 1959–1974. 10.1083/jcb.201611135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis V. G., Vassiliou L.-V. F., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., et al. (2005). Activation of the DNA Damage Checkpoint and Genomic Instability in Human Precancerous Lesions. Nature 434, 907–913. 10.1038/nature03485 [DOI] [PubMed] [Google Scholar]
- Gudjonsson T., Altmeyer M., Savic V., Toledo L., Dinant C., Grøfte M., et al. (2012). TRIP12 and UBR5 Suppress Spreading of Chromatin Ubiquitylation at Damaged Chromosomes. Cell 150, 697–709. 10.1016/j.cell.2012.06.039 [DOI] [PubMed] [Google Scholar]
- Guo Z., Wang S., Xie Y., Han Y., Hu S., Guan H., et al. (2020). HUWE1-dependent DNA-PKcs Neddylation Modulates its Autophosphorylation in DNA Damage Response. Cell Death Dis. 11, 400. 10.1038/s41419-020-2611-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzo C. M., Berndsen C. E., Zhu J., Gupta V., Datta A., Greenberg R. A., et al. (2012). RNF4-dependent Hybrid SUMO-Ubiquitin Chains Are Signals for RAP80 and Thereby Mediate the Recruitment of BRCA1 to Sites of DNA Damage. Sci. Signal. 5, ra88. 10.1126/scisignal.2003485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha K., Ma C., Lin H., Tang L., Lian Z., Zhao F., et al. (2017). The Anaphase Promoting Complex Impacts Repair Choice by Protecting Ubiquitin Signalling at DNA Damage Sites. Nat. Commun. 8, 15751. 10.1038/ncomms15751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauer M. H., Gasser S. M. (2017). Chromatin and Nucleosome Dynamics in DNA Damage and Repair. Genes Dev. 31, 2204–2221. 10.1101/gad.307702.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauer M. H., Seeber A., Singh V., Thierry R., Sack R., Amitai A., et al. (2017). Histone Degradation in Response to DNA Damage Enhances Chromatin Dynamics and Recombination Rates. Nat. Struct. Mol. Biol. 24, 99–107. 10.1038/nsmb.3347 [DOI] [PubMed] [Google Scholar]
- Heitz E. (1928). Das Heterochromatin der Moose. Jahrb. Wiss. Bot. 69, 762–818. Gebrüder Borntraeger. [Google Scholar]
- Hendriks I. A., D'Souza R. C. J., Yang B., Verlaan-de Vries M., Mann M., Vertegaal A. C. O. (2014). Uncovering Global SUMOylation Signaling Networks in a Site-specific Manner. Nat. Struct. Mol. Biol. 21, 927–936. 10.1038/nsmb.2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry K. W., Wyce A., Lo W.-S., Duggan L. J., Emre N. C. T., Kao C.-F., et al. (2003). Transcriptional Activation via Sequential Histone H2B Ubiquitylation and Deubiquitylation, Mediated by SAGA-Associated Ubp8. Genes Dev. 17, 2648–2663. 10.1101/gad.1144003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert S., Brion A., Arbona J. M., Lelek M., Veillet A., Lelandais B., et al. (2017). Chromatin Stiffening Underlies Enhanced Locus Mobility after DNA Damage in Budding Yeast. EMBO J. 36, 2595–2608. 10.15252/embj.201695842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Moyano E., Mergui X., García-Rubio M. L., Barroso S., Aguilera A. (2014). The Yeast and Human FACT Chromatin-Reorganizing Complexes Solve R-Loop-Mediated Transcription-Replication Conflicts. Genes Dev. 28, 735–748. 10.1101/gad.234070.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. (1998). The Ubiquitin System. Annu. Rev. Biochem. 67, 425–479. 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- Hideshima T., Richardson P., Chauhan D., Palombella V. J., Elliott P. J., Adams J., et al. (2001). The Proteasome Inhibitor PS-341 Inhibits Growth, Induces Apoptosis, and Overcomes Drug Resistance in Human Multiple Myeloma Cells. Cancer Res. 61, 3071–3076. [PubMed] [Google Scholar]
- Hodson C., Cole A. R., Lewis L. P. C., Miles J. A., Purkiss A., Walden H. (2011). Structural Analysis of Human FANCL, the E3 Ligase in the Fanconi Anemia Pathway. J. Biol. Chem. 286, 32628–32637. 10.1074/jbc.M111.244632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer P., Masuya K., Furet P., Kallen J., Valat-Stachyra T., Ferretti S., et al. (2015). Discovery of a Dihydroisoquinolinone Derivative (NVP-Cgm097): A Highly Potent and Selective MDM2 Inhibitor Undergoing Phase 1 Clinical Trials in P53wt Tumors. J. Med. Chem. 58, 6348–6358. 10.1021/acs.jmedchem.5b00810 [DOI] [PubMed] [Google Scholar]
- Horn V., Uckelmann M., Zhang H., Eerland J., Aarsman I., le Paige U. B., et al. (2019). Structural Basis of Specific H2A K13/K15 Ubiquitination by RNF168. Nat. Commun. 10, 1751. 10.1038/s41467-019-09756-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M. B., Shifat R., Johnson D. G., Bedford M. T., Gabrusiewicz K. R., Cortes-Santiago N., et al. (2016). TIE2-mediated Tyrosine Phosphorylation of H4 Regulates DNA Damage Response by Recruiting ABL1. Sci. Adv. 2, e1501290. 10.1126/sciadv.1501290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q., Botuyan M. V., Zhao D., Cui G., Mer E., Mer G. (2021). Mechanisms of BRCA1-BARD1 Nucleosome Recognition and Ubiquitylation. Nature 596, 438–443. 10.1038/s41586-021-03716-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Paul A., Wang B. (2012). Rap80 Protein Recruitment to DNA Double-Strand Breaks Requires Binding to Both Small Ubiquitin-like Modifier (SUMO) and Ubiquitin Conjugates. J. Biol. Chem. 287, 25510–25519. 10.1074/jbc.M112.374116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Petit S. A., Ficarro S. B., Toomire K. J., Xie A., Lim E., et al. (2014). PARP1-driven Poly-ADP-Ribosylation Regulates BRCA1 Function in Homologous Recombination-Mediated DNA Repair. Cancer Discov. 4, 1430–1447. 10.1158/2159-8290.CD-13-0891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Huen M. S. Y., Kim H., Leung C. C. Y., Glover J. N. M., Yu X., et al. (2009). RAD18 Transmits DNA Damage Signalling to Elicit Homologous Recombination Repair. Nat. Cell Biol. 11, 592–603. 10.1038/ncb1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Teng L., Li L., Liu T., Li L., Chen D., et al. (2004). ZNF216 Is an A20-like and IκB Kinase γ-Interacting Inhibitor of NFκB Activation. J. Biol. Chem. 279, 16847–16853. 10.1074/jbc.M309491200 [DOI] [PubMed] [Google Scholar]
- Huang Q., Qin D., Pei D., Vermeulen M., Zhang X. (2022). UBE2O and USP7 Co‐regulate RECQL4 Ubiquitinylation and Homologous Recombination‐mediated DNA Repair. FASEB J. 36, e22112. 10.1096/fj.202100974RRR [DOI] [PubMed] [Google Scholar]
- Husnjak K., Elsasser S., Zhang N., Chen X., Randles L., Shi Y., et al. (2008). Proteasome Subunit Rpn13 Is a Novel Ubiquitin Receptor. Nature 453, 481–488. 10.1038/nature06926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hustedt N., Durocher D. (2016). The Control of DNA Repair by the Cell Cycle. Nat. Cell Biol. 19, 1–9. 10.1038/ncb3452 [DOI] [PubMed] [Google Scholar]
- Huyen Y., Zgheib O., DiTullio Jr R. A., Gorgoulis V. G., Zacharatos P., Petty T. J., et al. (2004). Methylated Lysine 79 of Histone H3 Targets 53BP1 to DNA Double-Strand Breaks. Nature 432, 406–411. 10.1038/nature03114 [DOI] [PubMed] [Google Scholar]
- Ikeda F., Deribe Y. L., Skånland S. S., Stieglitz B., Grabbe C., Franz-Wachtel M., et al. (2011). SHARPIN Forms a Linear Ubiquitin Ligase Complex Regulating NF-Κb Activity and Apoptosis. Nature 471, 637–641. 10.1038/nature09814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T., Tashiro S., Kakino A., Shima H., Jacob N., Amunugama R., et al. (2007). DNA Damage-dependent Acetylation and Ubiquitination of H2AX Enhances Chromatin Dynamics. Mol. Cell. Biol. 27, 7028–7040. 10.1128/MCB.00579-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inano S., Sato K., Katsuki Y., Kobayashi W., Tanaka H., Nakajima K., et al. (2017). RFWD3-Mediated Ubiquitination Promotes Timely Removal of Both RPA and RAD51 from DNA Damage Sites to Facilitate Homologous Recombination. Mol. Cell 66, 622–634. 10.1016/j.molcel.2017.04.022 [DOI] [PubMed] [Google Scholar]
- Ismail I. H., Andrin C., McDonald D., Hendzel M. J. (2010). BMI1-mediated Histone Ubiquitylation Promotes DNA Double-Strand Break Repair. J. Cell Biol. 191, 45–60. 10.1083/jcb.201003034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail I. H., Gagné J.-P., Genois M.-M., Strickfaden H., McDonald D., Xu Z., et al. (2015). The RNF138 E3 Ligase Displaces Ku to Promote DNA End Resection and Regulate DNA Repair Pathway Choice. Nat. Cell Biol. 17, 1446–1457. 10.1038/ncb3259 [DOI] [PubMed] [Google Scholar]
- Iwai K., Fujita H., Sasaki Y. (2014). Linear Ubiquitin Chains: NF-Κb Signalling, Cell Death and beyond. Nat. Rev. Mol. Cell Biol. 15, 503–508. 10.1038/nrm3836 [DOI] [PubMed] [Google Scholar]
- Jacquet K., Fradet-Turcotte A., Avvakumov N., Lambert J.-P., Roques C., Pandita R. K., et al. (2016). The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation. Mol. Cell 62, 409–421. 10.1016/j.molcel.2016.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob B., Splinter J., Conrad S., Voss K.-O., Zink D., Durante M., et al. (2011). DNA Double-Strand Breaks in Heterochromatin Elicit Fast Repair Protein Recruitment, Histone H2AX Phosphorylation and Relocation to Euchromatin. Nucleic Acids Res. 39, 6489–6499. 10.1093/nar/gkr230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., McGrath J. P., Varshavsky A. (1987). The Yeast DNA Repair Gene RAD6 Encodes a Ubiquitin-Conjugating Enzyme. Nature 329, 131–134. 10.1038/329131a0 [DOI] [PubMed] [Google Scholar]
- Jentsch S., Pyrowolakis G. (2000). Ubiquitin and its Kin: How Close Are the Family Ties? Trends Cell Biol. 10, 335–342. 10.1016/s0962-8924(00)01785-2 [DOI] [PubMed] [Google Scholar]
- Ji S., Qin Y., Shi S., Liu X., Hu H., Zhou H., et al. (2015). ERK Kinase Phosphorylates and Destabilizes the Tumor Suppressor FBW7 in Pancreatic Cancer. Cell Res. 25, 561–573. 10.1038/cr.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Xu Y., Price B. D. (2010). Acetylation of H2AX on Lysine 36 Plays a Key Role in the DNA Double-Strand Break Repair Pathway. FEBS Lett. 584, 2926–2930. 10.1016/j.febslet.2010.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimeno S., Fernández-Ávila M. J., Cruz-García A., Cepeda-García C., Gómez-Cabello D., Huertas P. (2015). Neddylation Inhibits CtIP-Mediated Resection and Regulates DNA Double Strand Break Repair Pathway Choice. Nucleic Acids Res. 43, 987–999. 10.1093/nar/gku1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kais Z., Rondinelli B., Holmes A., O’Leary C., Kozono D., D’Andrea A. D., et al. (2016). FANCD2 Maintains Fork Stability in BRCA1/2-Deficient Tumors and Promotes Alternative End-Joining DNA Repair. Cell Rep. 15, 2488–2499. 10.1016/j.celrep.2016.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb R., Mallery D. L., Larkin C., Huang J. T. J., Hiom K. (2014). BRCA1 Is a Histone-h2a-specific Ubiquitin Ligase. Cell Rep. 8, 999–1005. 10.1016/j.celrep.2014.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalocsay M., Hiller N. J., Jentsch S. (2009). Chromosome-wide Rad51 Spreading and SUMO-H2A.Z-dependent Chromosome Fixation in Response to a Persistent DNA Double-Strand Break. Mol. Cell 33, 335–343. 10.1016/j.molcel.2009.01.016 [DOI] [PubMed] [Google Scholar]
- Kang H. C., Lee Y.-I., Shin J.-H., Andrabi S. A., Chi Z., Gagné J.-P., et al. (2011). Iduna Is a poly(ADP-Ribose) (PAR)-dependent E3 Ubiquitin Ligase that Regulates DNA Damage. Proc. Natl. Acad. Sci. U.S.A. 108, 14103–14108. 10.1073/pnas.1108799108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkhanis V., Wang L., Tae S., Hu Y.-J., Imbalzano A. N., Sif S. (2012). Protein Arginine Methyltransferase 7 Regulates Cellular Response to DNA Damage by Methylating Promoter Histones H2A and H4 of the Polymerase δ Catalytic Subunit Gene, POLD1. J. Biol. Chem. 287, 29801–29814. 10.1074/jbc.M112.378281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury-Haddad H., Guttmann-Raviv N., Ipenberg I., Huggins D., Jeyasekharan A. D., Ayoub N. (2014). PARP1-dependent Recruitment of KDM4D Histone Demethylase to DNA Damage Sites Promotes Double-Strand Break Repair. Proc. Natl. Acad. Sci. U.S.A. 111, E728–E737. 10.1073/pnas.1317585111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana S., Kruhlak M. J., Kim J., Tran A. D., Liu J., Nyswaner K., et al. (2014). A Macrohistone Variant Links Dynamic Chromatin Compaction to BRCA1-dependent Genome Maintenance. Cell Rep. 8, 1049–1062. 10.1016/j.celrep.2014.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilgas S., Singh A. N., Paillas S., Then C.-K., Torrecilla I., Nicholson J., et al. (2021). p97/VCP Inhibition Causes Excessive MRE11-dependent DNA End Resection Promoting Cell Killing after Ionizing Radiation. Cell Rep. 35, 109153. 10.1016/j.celrep.2021.109153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic S., Lezaja A., Gatti M., Bianco E., Michelena J., Imhof R., et al. (2019). Phase Separation of 53 BP 1 Determines Liquid‐like Behavior of DNA Repair Compartments. EMBO J. 38, e101379. 10.15252/embj.2018101379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. J., Lee S. Y., Gong F., Battenhouse A. M., Boutz D. R., Bashyal A., et al. (2019). Systematic Bromodomain Protein Screens Identify Homologous Recombination and R-Loop Suppression Pathways Involved in Genome Integrity. Genes Dev. 33, 1751–1774. 10.1101/gad.331231.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. J., Lee S. Y., Hwang Y., Kim S., Chung J. M., Park S., et al. (2021). USP39 Promotes Non-homologous End-Joining Repair by poly(ADP-Ribose)-Induced Liquid Demixing. Nucleic Acids Res. 49, 11083–11102. 10.1093/nar/gkab892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., et al. (2011). Systematic and Quantitative Assessment of the Ubiquitin-Modified Proteome. Mol. Cell 44, 325–340. 10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y., Tanaka K. (2010). Regulatory Mechanisms Involved in the Control of Ubiquitin Homeostasis. J. Biochem. 147, 793–798. 10.1093/jb/mvq044 [DOI] [PubMed] [Google Scholar]
- Kitevski-LeBlanc J., Fradet-Turcotte A., Kukic P., Wilson M. D., Portella G., Yuwen T., et al. (2017). The RNF168 Paralog RNF169 Defines a New Class of Ubiquitylated Histone Reader Involved in the Response to DNA Damage. eLife 6. 10.7554/eLife.23872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas N. K., Chapman J. R., Nakada S., Ylanko J., Chahwan R., Sweeney F. D., et al. (2007). Orchestration of the DNA-Damage Response by the RNF8 Ubiquitin Ligase. Science 318, 1637–1640. 10.1126/science.1150034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D., Rape M. (2012). The Ubiquitin Code. Annu. Rev. Biochem. 81, 203–229. 10.1146/annurev-biochem-060310-170328 [DOI] [PubMed] [Google Scholar]
- Korkuć P., Walther D. (2017). Towards Understanding the Crosstalk between Protein Post-translational Modifications: Homo- and Heterotypic PTM Pair Distances on Protein Surfaces Are Not Random. Proteins 85, 78–92. 10.1002/prot.25200 [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin Modifications and Their Function. Cell 128, 693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Krais J. J., Wang Y., Patel P., Basu J., Bernhardy A. J., Johnson N. (2021). RNF168-mediated Localization of BARD1 Recruits the BRCA1-PALB2 Complex to DNA Damage. Nat. Commun. 12, 5016. 10.1038/s41467-021-25346-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., González-Prieto R., Xiao Z., Verlaan-de Vries M., Vertegaal A. C. O. (2017). The STUbL RNF4 Regulates Protein Group SUMOylation by Targeting the SUMO Conjugation Machinery. Nat. Commun. 8, 1809. 10.1038/s41467-017-01900-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari N., Vartak S. V., Dahal S., Kumari S., Desai S. S., Gopalakrishnan V., et al. (2019). G-Quadruplex Structures Contribute to Differential Radiosensitivity of the Human Genome. iScience 21, 288–307. 10.1016/j.isci.2019.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupperman E., Lee E. C., Cao Y., Bannerman B., Fitzgerald M., Berger A., et al. (2010). Evaluation of the Proteasome Inhibitor MLN9708 in Preclinical Models of Human Cancer. Cancer Res. 70, 1970–1980. 10.1158/0008-5472.CAN-09-2766 [DOI] [PubMed] [Google Scholar]
- Lacoursiere R. E., Shaw G. S. (2021). Acetylated Ubiquitin Modulates the Catalytic Activity of the E1 Enzyme Uba1. Biochemistry 60, 1276–1285. 10.1021/acs.biochem.1c00145 [DOI] [PubMed] [Google Scholar]
- Ladanyi M., Cha C., Lewis R., Jhanwar S. C., Huvos A. G., Healey J. H. (1993). MDM2 Gene Amplification in Metastatic Osteosarcoma. Cancer Res. 53, 16–18. [PubMed] [Google Scholar]
- Lafranchi L., Boer H. R., Vries E. G., Ong S. E., Sartori A. A., Vugt M. A. (2014). APC/CCdh1controls CtIPstability during the Cell Cycle and in Response toDNAdamage. EMBO J. 33, 2860–2879. 10.15252/embj.201489017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancini C., van den Berk P. C. M., Vissers J. H. A., Gargiulo G., Song J.-Y., Hulsman D., et al. (2014). Tight Regulation of Ubiquitin-Mediated DNA Damage Response by USP3 Preserves the Functional Integrity of Hematopoietic Stem Cells. J. Exp. Med. 211, 1759–1777. 10.1084/jem.20131436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak P., Mejia-Ramirez E., Limbo O., Russell P. (2011). Release of Ku and MRN from DNA Ends by Mre11 Nuclease Activity and Ctp1 Is Required for Homologous Recombination Repair of Double-Strand Breaks. PLoS Genet. 7, e1002271. 10.1371/journal.pgen.1002271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H., Kang B.-H., Jang H., Kim T. W., Choi J., Kwak S., et al. (2015). AKT Phosphorylates H3-Threonine 45 to Facilitate Termination of Gene Transcription in Response to DNA Damage. Nucleic Acids Res. 43, 4505–4516. 10.1093/nar/gkv176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A. K. L. (2014). Poly(ADP-ribose): an Organizer of Cellular Architecture. J. Cell Biol. 205, 613–619. 10.1083/jcb.201402114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J. W., Agarwal P., Canny M. D., Gong F., Robison A. D., Finkelstein I. J., et al. (2014). Nucleosome Acidic Patch Promotes RNF168- and RING1B/BMI1-dependent H2AX and H2A Ubiquitination and DNA Damage Signaling. PLoS Genet. 10, e1004178. 10.1371/journal.pgen.1004178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levone B. R., Lenzken S. C., Antonaci M., Maiser A., Rapp A., Conte F., et al. (2021). FUS-Dependent Liquid-Liquid Phase Separation Is Important for DNA Repair Initiation. J. Cell Biol. 220. 10.1083/jcb.202008030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Halaby M.-J., Hakem A., Cardoso R., El Ghamrasni S., Harding S., et al. (2010). Rnf8 Deficiency Impairs Class Switch Recombination, Spermatogenesis, and Genomic Integrity and Predisposes for Cancer. J. Exp. Med. 207, 983–997. 10.1084/jem.20092437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Lu L.-Y., Yang C.-Y., Wang S., Yu X. (2013). The FHA and BRCT Domains Recognize ADP-Ribosylation during DNA Damage Response. Genes Dev. 27, 1752–1768. 10.1101/gad.226357.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Yu X. (2013). Function of BRCA1 in the DNA Damage Response Is Mediated by ADP-Ribosylation. Cancer Cell 23, 693–704. 10.1016/j.ccr.2013.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Bengtson M. H., Ulbrich A., Matsuda A., Reddy V. A., Orth A., et al. (2008). Genome-Wide and Functional Annotation of Human E3 Ubiquitin Ligases Identifies MULAN, a Mitochondrial E3 that Regulates the Organelle's Dynamics and Signaling. PLoS ONE 3, e1487. 10.1371/journal.pone.0001487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Liu L., Yang S., Song N., Zhou X., Gao J., et al. (2014). Histone Demethylase KDM5B Is a Key Regulator of Genome Stability. Proc. Natl. Acad. Sci. U.S.A. 111, 7096–7101. 10.1073/pnas.1324036111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Luo K., Yin Y., Wu C., Deng M., Li L., et al. (2017). USP13 Regulates the RAP80-BRCA1 Complex Dependent DNA Damage Response. Nat. Commun. 8, 15752. 10.1038/ncomms15752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D., Burkhart S. L., Singh R. K., Kabbaj M.-H. M., Gunjan A. (2012). Histone Dosage Regulates DNA Damage Sensitivity in a Checkpoint-independent Manner by the Homologous Recombination Pathway. Nucleic Acids Res. 40, 9604–9620. 10.1093/nar/gks722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Sumara I., Pangou E. (2022). Non-proteolytic Ubiquitylation in Cellular Signaling and Human Disease. Commun. Biol. 5, 114. 10.1038/s42003-022-03060-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Wu J., Paudyal S. C., You Z., Yu X. (2013). CHFR Is Important for the First Wave of Ubiquitination at DNA Damage Sites. Nucleic Acids Res. 41, 1698–1710. 10.1093/nar/gks1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Yan J., Wang X., Chen J., Wang X., Dong Y., et al. (2021). RPA-Mediated Recruitment of Bre1 Couples Histone H2B Ubiquitination to DNA Replication and Repair. Proc. Natl. Acad. Sci. U.S.A. 118. 10.1073/pnas.2017497118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhang H., Wang X., Tian Q., Hu Z., Peng C., et al. (2015). The Deubiquitylating Enzyme USP4 Cooperates with CtIP in DNA Double-Strand Break End Resection. Cell Rep. 13, 93–107. 10.1016/j.celrep.2015.08.056 [DOI] [PubMed] [Google Scholar]
- Lobrich, P. K. Cooper and B. Rydber M., Cooper P. K., Rydberg B. (1996). Non-random Distribution of DNA Double-Strand Breaks Induced by Particle Irradiation. Int. J. Radiat. Biol. 70, 493–503. 10.1080/095530096144680 [DOI] [PubMed] [Google Scholar]
- Locke A. J., Hossain L., McCrostie G., Ronato D. A., Fitieh A., Rafique T. A., et al. (2021). SUMOylation Mediates CtIP's Functions in DNA End Resection and Replication Fork Protection. Nucleic Acids Res. 49, 928–953. 10.1093/nar/gkaa1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997). Crystal Structure of the Nucleosome Core Particle at 2.8 Å Resolution. Nature 389, 251–260. 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- Lundby A., Secher A., Lage K., Nordsborg N. B., Dmytriyev A., Lundby C., et al. (2012). Quantitative Maps of Protein Phosphorylation Sites across 14 Different Rat Organs and Tissues. Nat. Commun. 3, 876. 10.1038/ncomms1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Zhang H., Wang L., Yuan J., Lou Z. (2012). Sumoylation of MDC1 Is Important for Proper DNA Damage Response. EMBO J. 31, 3008–3019. 10.1038/emboj.2012.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch D. R., Farmer J., Hauser L., Blair I. A., Wang Q. Q., Mesaros C., et al. (2019). Safety, Pharmacodynamics, and Potential Benefit of Omaveloxolone in Friedreich Ataxia. Ann. Clin. Transl. Neurol. 6, 15–26. 10.1002/acn3.660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Chen Y., Zhang F., Yang C.-Y., Wang S., Yu X. (2013). RNF111-dependent Neddylation Activates DNA Damage-Induced Ubiquitination. Mol. Cell 49, 897–907. 10.1016/j.molcel.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabbitt P. D., Loreto A., Déry M.-A., Fletcher A. J., Stanley M., Pao K.-C., et al. (2020). Structural Basis for RING-Cys-Relay E3 Ligase Activity and its Role in Axon Integrity. Nat. Chem. Biol. 16, 1227–1236. 10.1038/s41589-020-0598-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macůrek L., Lindqvist A., Voets O., Kool J., Vos H. R., Medema R. H. (2010). Wip1 Phosphatase Is Associated with Chromatin and Dephosphorylates γH2AX to Promote Checkpoint Inhibition. Oncogene 29, 2281–2291. 10.1038/onc.2009.501 [DOI] [PubMed] [Google Scholar]
- Magnaghi P., D'Alessio R., Valsasina B., Avanzi N., Rizzi S., Asa D., et al. (2013). Covalent and Allosteric Inhibitors of the ATPase VCP/p97 Induce Cancer Cell Death. Nat. Chem. Biol. 9, 548–556. 10.1038/nchembio.1313 [DOI] [PubMed] [Google Scholar]
- Magnussen H. M., Ahmed S. F., Sibbet G. J., Hristova V. A., Nomura K., Hock A. K., et al. (2020). Structural Basis for DNA Damage-Induced Phosphoregulation of MDM2 RING Domain. Nat. Commun. 11, 2094. 10.1038/s41467-020-15783-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., et al. (2007). RNF8 Ubiquitylates Histones at DNA Double-Strand Breaks and Promotes Assembly of Repair Proteins. Cell 131, 887–900. 10.1016/j.cell.2007.09.040 [DOI] [PubMed] [Google Scholar]
- Mallette F. A., Mattiroli F., Cui G., Young L. C., Hendzel M. J., Mer G., et al. (2012). RNF8- and RNF168-dependent Degradation of KDM4A/JMJD2A Triggers 53BP1 Recruitment to DNA Damage Sites. EMBO J. 31, 1865–1878. 10.1038/emboj.2012.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis J. P., Morales J. C., Xia Z., Kutok J. L., Alt F. W., Carpenter P. B. (2004). 53BP1 Links DNA Damage-Response Pathways to Immunoglobulin Heavy Chain Class-Switch Recombination. Nat. Immunol. 5, 481–487. 10.1038/ni1067 [DOI] [PubMed] [Google Scholar]
- Mao Z., Bozzella M., Seluanov A., Gorbunova V. (2008). DNA Repair by Nonhomologous End Joining and Homologous Recombination during Cell Cycle in Human Cells. Cell Cycle 7, 2902–2906. 10.4161/cc.7.18.6679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A., Li J.-M., Ji X. Y., Wu C.-S., Yazinski S. A., Nguyen H. D., et al. (2014). PRP19 Transforms into a Sensor of RPA-ssDNA after DNA Damage and Drives ATR Activation via a Ubiquitin-Mediated Circuitry. Mol. Cell 53, 235–246. 10.1016/j.molcel.2013.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H., Hawke D., Kobayashi R., Verreault A. (2005). A Role for Cell-Cycle-Regulated Histone H3 Lysine 56 Acetylation in the DNA Damage Response. Nature 436, 294–298. 10.1038/nature03714 [DOI] [PubMed] [Google Scholar]
- Mattiroli F., Uckelmann M., Sahtoe D. D., van Dijk W. J., Sixma T. K. (2014). The Nucleosome Acidic Patch Plays a Critical Role in RNF168-dependent Ubiquitination of Histone H2A. Nat. Commun. 5, 3291. 10.1038/ncomms4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiroli F., Vissers J. H. A., van Dijk W. J., Ikpa P., Citterio E., Vermeulen W., et al. (2012). RNF168 Ubiquitinates K13-15 on H2A/H2AX to Drive DNA Damage Signaling. Cell 150, 1182–1195. 10.1016/j.cell.2012.08.005 [DOI] [PubMed] [Google Scholar]
- Maxwell K. N., Domchek S. M. (2012). Cancer Treatment According to BRCA1 and BRCA2 Mutations. Nat. Rev. Clin. Oncol. 9, 520–528. 10.1038/nrclinonc.2012.123 [DOI] [PubMed] [Google Scholar]
- McClellan A. J., Laugesen S. H., Ellgaard L. (2019). Cellular Functions and Molecular Mechanisms of Non-lysine Ubiquitination. Open Biol. 9, 190147. 10.1098/rsob.190147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerang M., Ritz D., Paliwal S., Garajova Z., Bosshard M., Mailand N., et al. (2011). The Ubiquitin-Selective Segregase VCP/p97 Orchestrates the Response to DNA Double-Strand Breaks. Nat. Cell Biol. 13, 1376–1382. 10.1038/ncb2367 [DOI] [PubMed] [Google Scholar]
- Meyer H. H., Wang Y., Warren G. (2002). Direct Binding of Ubiquitin Conjugates by the Mammalian P97 Adaptor Complexes, P47 and Ufd1-Npl4. EMBO J. 21, 5645–5652. 10.1093/emboj/cdf579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millan-Zambrano G., Santos-Rosa H., Puddu F., Robson S. C., Jackson S. P., Kouzarides T. (2018). Phosphorylation of Histone H4T80 Triggers DNA Damage Checkpoint Recovery. Mol. Cell 72, 625–635. 10.1016/j.molcel.2018.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. M., Tjeertes J. V., Coates J., Legube G., Polo S. E., Britton S., et al. (2010). Human HDAC1 and HDAC2 Function in the DNA-Damage Response to Promote DNA Nonhomologous End-Joining. Nat. Struct. Mol. Biol. 17, 1144–1151. 10.1038/nsmb.1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori T., Hardin J. A. (1986). Mechanism of Interaction between Ku Protein and DNA. J. Biol. Chem. 261, 10375–10379. 10.1016/S0021-9258(18)67534-9 [DOI] [PubMed] [Google Scholar]
- Miné-Hattab J., Heltberg M., Villemeur M., Guedj C., Mora T., Walczak A. M., et al. (2021). Single Molecule Microscopy Reveals Key Physical Features of Repair Foci in Living Cells. eLife 10. 10.7554/eLife.60577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miné-Hattab J., Recamier V., Izeddin I., Rothstein R., Darzacq X. (2017). Multi-scale Tracking Reveals Scale-dependent Chromatin Dynamics after DNA Damage. MBoC 28, 3323–3332. 10.1091/mbc.E17-05-0317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirman Z., Lottersberger F., Takai H., Kibe T., Gong Y., Takai K., et al. (2018). 53BP1-RIF1-shieldin Counteracts DSB Resection through CST- and Polα-dependent Fill-In. Nature 560, 112–116. 10.1038/s41586-018-0324-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. R., Boutell C., Keppler M., Densham R., Weekes D., Alamshah A., et al. (2009). The SUMO Modification Pathway Is Involved in the BRCA1 Response to Genotoxic Stress. Nature 462, 886–890. 10.1038/nature08593 [DOI] [PubMed] [Google Scholar]
- Mosammaparast N., Kim H., Laurent B., Zhao Y., Lim H. J., Majid M. C., et al. (2013). The Histone Demethylase LSD1/KDM1A Promotes the DNA Damage Response. J. Cell Biol. 203, 457–470. 10.1083/jcb.201302092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosbech A., Lukas C., Bekker-Jensen S., Mailand N. (2013). The Deubiquitylating Enzyme USP44 Counteracts the DNA Double-Strand Break Response Mediated by the RNF8 and RNF168 Ubiquitin Ligases. J. Biol. Chem. 288, 16579–16587. 10.1074/jbc.M113.459917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss J., Tinline-Purvis H., Walker C. A., Folkes L. K., Stratford M. R., Hayles J., et al. (2010). Break-induced ATR and Ddb1-Cul4Cdt2 Ubiquitin Ligase-dependent Nucleotide Synthesis Promotes Homologous Recombination Repair in Fission Yeast. Genes Dev. 24, 2705–2716. 10.1101/gad.1970810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyal L., Lerenthal Y., Gana-Weisz M., Mass G., So S., Wang S.-Y., et al. (2011). Requirement of ATM-dependent Monoubiquitylation of Histone H2B for Timely Repair of DNA Double-Strand Breaks. Mol. Cell 41, 529–542. 10.1016/j.molcel.2011.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R., Loizou J. I., Yang Y.-G., Cuenin C., Li H., Wang Z.-Q., et al. (2006). Histone Acetylation by Trrap-Tip60 Modulates Loading of Repair Proteins and Repair of DNA Double-Strand Breaks. Nat. Cell Biol. 8, 91–99. 10.1038/ncb1343 [DOI] [PubMed] [Google Scholar]
- Nakada S., Tai I., Panier S., Al-Hakim A., Iemura S.-I., Juang Y.-C., et al. (2010). Non-canonical Inhibition of DNA Damage-dependent Ubiquitination by OTUB1. Nature 466, 941–946. 10.1038/nature09297 [DOI] [PubMed] [Google Scholar]
- Nakamura K., Kato A., Kobayashi J., Yanagihara H., Sakamoto S., Oliveira D. V. N. P., et al. (2011). Regulation of Homologous Recombination by RNF20-dependent H2B Ubiquitination. Mol. Cell 41, 515–528. 10.1016/j.molcel.2011.02.002 [DOI] [PubMed] [Google Scholar]
- Nakamura K., Saredi G., Becker J. R., Foster B. M., Nguyen N. V., Beyer T. E., et al. (2019). H4K20me0 Recognition by BRCA1-BARD1 Directs Homologous Recombination to Sister Chromatids. Nat. Cell Biol. 21, 311–318. 10.1038/s41556-019-0282-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natale F., Rapp A., Yu W., Maiser A., Harz H., Scholl A., et al. (2017). Identification of the Elementary Structural Units of the DNA Damage Response. Nat. Commun. 8, 15760. 10.1038/ncomms15760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini S., Gorgoulis V. G., Halazonetis T. D. (2010). Genomic Instability - an Evolving Hallmark of Cancer. Nat. Rev. Mol. Cell Biol. 11, 220–228. 10.1038/nrm2858 [DOI] [PubMed] [Google Scholar]
- Nicassio F., Corrado N., Vissers J. H. A., Areces L. B., Bergink S., Marteijn J. A., et al. (2007). Human USP3 Is a Chromatin Modifier Required for S Phase Progression and Genome Stability. Curr. Biol. 17, 1972–1977. 10.1016/j.cub.2007.10.034 [DOI] [PubMed] [Google Scholar]
- Niu X., Chen W., Bi T., Lu M., Qin Z., Xiao W. (2019). Rev1 Plays Central Roles in Mammalian DNA‐damage Tolerance in Response to UV Irradiation. FEBS J. 286, 2711–2725. 10.1111/febs.14840 [DOI] [PubMed] [Google Scholar]
- Noordermeer S. M., Adam S., Setiaputra D., Barazas M., Pettitt S. J., Ling A. K., et al. (2018). The Shieldin Complex Mediates 53BP1-dependent DNA Repair. Nature 560, 117–121. 10.1038/s41586-018-0340-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowsheen S., Aziz K., Aziz A., Deng M., Qin B., Luo K., et al. (2018). L3MBTL2 Orchestrates Ubiquitin Signalling by Dictating the Sequential Recruitment of RNF8 and RNF168 after DNA Damage. Nat. Cell Biol. 20, 455–464. 10.1038/s41556-018-0071-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor H. F., Lyon N., Leung J. W., Agarwal P., Swaim C. D., Miller K. M., et al. (2015). Ubiquitin‐Activated Interaction Traps ( UBAIT S) Identify E3 Ligase Binding Partners. EMBO Rep. 16, 1699–1712. 10.15252/embr.201540620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Hagan H. M., Mohammad H. P., Baylin S. B. (2008). Double Strand Breaks Can Initiate Gene Silencing and SIRT1-dependent Onset of DNA Methylation in an Exogenous Promoter CpG Island. PLoS Genet. 4, e1000155. 10.1371/journal.pgen.1000155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberoi J., Richards M. W., Crumpler S., Brown N., Blagg J., Bayliss R. (2010). Structural Basis of poly(ADP-Ribose) Recognition by the Multizinc Binding Domain of Checkpoint with Forkhead-Associated and RING Domains (CHFR). J. Biol. Chem. 285, 39348–39358. 10.1074/jbc.M110.159855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs F., Karemore G., Miron E., Brown J., Sedlackova H., Rask M.-B., et al. (2019). Stabilization of Chromatin Topology Safeguards Genome Integrity. Nature 574, 571–574. 10.1038/s41586-019-1659-4 [DOI] [PubMed] [Google Scholar]
- Ohkuni K., Gliford L., Au W.-C., Suva E., Kaiser P., Basrai M. A. (2022). Cdc48Ufd1/Npl4 Segregase Removes Mislocalized Centromeric Histone H3 Variant CENP-A from Non-centromeric Chromatin. Nucleic Acids Res. 50, 3276–3291. 10.1093/nar/gkac135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake F., Saeki Y., Sakamoto K., Ohtake K., Nishikawa H., Tsuchiya H., et al. (2015). Ubiquitin Acetylation Inhibits Polyubiquitin Chain Elongation. EMBO Rep. 16, 192–201. 10.15252/embr.201439152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orthwein A., Noordermeer S. M., Wilson M. D., Landry S., Enchev R. I., Sherker A., et al. (2015). A Mechanism for the Suppression of Homologous Recombination in G1 Cells. Nature 528, 422–426. 10.1038/nature16142 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Oshidari R., Huang R., Medghalchi M., Tse E. Y. W., Ashgriz N., Lee H. O., et al. (2020). DNA Repair by Rad52 Liquid Droplets. Nat. Commun. 11, 695. 10.1038/s41467-020-14546-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai C.-C., Deegan R. S., Subramanian L., Gal C., Sarkar S., Blaikley E. J., et al. (2014). A Histone H3K36 Chromatin Switch Coordinates DNA Double-Strand Break Repair Pathway Choice. Nat. Commun. 5, 4091. 10.1038/ncomms5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M.-R., Peng G., Hung W.-C., Lin S.-Y. (2011). Monoubiquitination of H2AX Protein Regulates DNA Damage Response Signaling. J. Biol. Chem. 286, 28599–28607. 10.1074/jbc.M111.256297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S., Ichijima Y., Fradet-Turcotte A., Leung C. C. Y., Kaustov L., Arrowsmith C. H., et al. (2012). Tandem Protein Interaction Modules Organize the Ubiquitin-dependent Response to DNA Double-Strand Breaks. Mol. Cell 47, 383–395. 10.1016/j.molcel.2012.05.045 [DOI] [PubMed] [Google Scholar]
- Pei H., Zhang L., Luo K., Qin Y., Chesi M., Fei F., et al. (2011). MMSET Regulates Histone H4K20 Methylation and 53BP1 Accumulation at DNA Damage Sites. Nature 470, 124–128. 10.1038/nature09658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., et al. (2003). A Proteomics Approach to Understanding Protein Ubiquitination. Nat. Biotechnol. 21, 921–926. 10.1038/nbt849 [DOI] [PubMed] [Google Scholar]
- Pereg Y., Shkedy D., de Graaf P., Meulmeester E., Edelson-Averbukh M., Salek M., et al. (2005). Phosphorylation of Hdmx Mediates its Hdm2- and ATM-dependent Degradation in Response to DNA Damage. Proc. Natl. Acad. Sci. U.S.A. 102, 5056–5061. 10.1073/pnas.0408595102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessina F., Giavazzi F., Yin Y., Gioia U., Vitelli V., Galbiati A., et al. (2019). Functional Transcription Promoters at DNA Double-Strand Breaks Mediate RNA-Driven Phase Separation of Damage-Response Factors. Nat. Cell Biol. 21, 1286–1299. 10.1038/s41556-019-0392-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister S. X., Ahrabi S., Zalmas L.-P., Sarkar S., Aymard F., Bachrati C. Z., et al. (2014). SETD2-dependent Histone H3K36 Trimethylation Is Required for Homologous Recombination Repair and Genome Stability. Cell Rep. 7, 2006–2018. 10.1016/j.celrep.2014.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C. M. (2004). Back to the Future with Ubiquitin. Cell 116, 181–190. 10.1016/s0092-8674(03)01074-2 [DOI] [PubMed] [Google Scholar]
- Pinato S., Gatti M., Scandiuzzi C., Confalonieri S., Penengo L. (2011). UMI, a Novel RNF168 Ubiquitin Binding Domain Involved in the DNA Damage Signaling Pathway. Mol. Cell. Biol. 31, 118–126. 10.1128/MCB.00818-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinato S., Scandiuzzi C., Arnaudo N., Citterio E., Gaudino G., Penengo L. (2009). RNF168, a New RING Finger, MIU-Containing Protein that Modifies Chromatin by Ubiquitination of Histones H2A and H2AX. BMC Mol. Biol. 10, 55. 10.1186/1471-2199-10-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piva R., Ruggeri B., Williams M., Costa G., Tamagno I., Ferrero D., et al. (2008). CEP-18770: A Novel, Orally Active Proteasome Inhibitor with a Tumor-Selective Pharmacologic Profile Competitive with Bortezomib. Blood 111, 2765–2775. 10.1182/blood-2007-07-100651 [DOI] [PubMed] [Google Scholar]
- Pollard J. M., Gatti R. A. (2009). Clinical Radiation Sensitivity with DNA Repair Disorders: an Overview. Int. J. Radiat. Oncology*Biology*Physics 74, 1323–1331. 10.1016/j.ijrobp.2009.02.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic D., Vucic D., Dikic I. (2014). Ubiquitination in Disease Pathogenesis and Treatment. Nat. Med. 20, 1242–1253. 10.1038/nm.3739 [DOI] [PubMed] [Google Scholar]
- Postow L., Funabiki H. (2013). An SCF Complex Containing Fbxl12 Mediates DNA Damage-Induced Ku80 Ubiquitylation. Cell Cycle 12, 587–595. 10.4161/cc.23408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L., Ghenoiu C., Woo E. M., Krutchinsky A. N., Chait B. T., Funabiki H. (2008). Ku80 Removal from DNA through Double Strand Break-Induced Ubiquitylation. J. Cell Biol. 182, 467–479. 10.1083/jcb.200802146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen M., Lukas C., Lukas J., Bekker-Jensen S., Mailand N. (2012). Human RNF169 Is a Negative Regulator of the Ubiquitin-dependent Response to DNA Double-Strand Breaks. J. Cell Biol. 197, 189–199. 10.1083/jcb.201109100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prag G., Watson H., Kim Y. C., Beach B. M., Ghirlando R., Hummer G., et al. (2007). The Vps27/Hse1 Complex Is a GAT Domain-Based Scaffold for Ubiquitin-dependent Sorting. Dev. Cell 12, 973–986. 10.1016/j.devcel.2007.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendergast L., McClurg U. L., Hristova R., Berlinguer-Palmini R., Greener S., Veitch K., et al. (2020). Resolution of R-Loops by INO80 Promotes DNA Replication and Maintains Cancer Cell Proliferation and Viability. Nat. Commun. 11, 4534. 10.1038/s41467-020-18306-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J., Bunker R. D., Schenk A. D., Cavadini S., Gill M. E., Abdulrahman W., et al. (2019). Structural Basis of BRCC36 Function in DNA Repair and Immune Regulation. Mol. Cell 75, 483–497. 10.1016/j.molcel.2019.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulescu I., Elmroth K., Stenerlöw B. (2004). Chromatin Organization Contributes to Non-randomly Distributed Double-Strand Breaks after Exposure to High-LET Radiation. Radiat. Res. 161, 1–8. 10.1667/rr3094 [DOI] [PubMed] [Google Scholar]
- Ramachandran S., Haddad D., Li C., Le M. X., Ling A. K., So C. C., et al. (2016). The SAGA Deubiquitination Module Promotes DNA Repair and Class Switch Recombination through ATM and DNAPK-Mediated γH2AX Formation. Cell Rep. 15, 1554–1565. 10.1016/j.celrep.2016.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasco D. W., Lakhani N. J., Li Y., Men L., Wang H., Ji J., et al. (2019a). A Phase I Study of a Novel MDM2 Antagonist APG-115 in Patients with Advanced Solid Tumors. JCO 37, 3126. 10.1200/JCO.2019.37.15_suppl.3126 [DOI] [Google Scholar]
- Rasco D. W., Papadopoulos K. P., Pourdehnad M., Gandhi A. K., Hagner P. R., Li Y., et al. (2019b). A First-In-Human Study of Novel Cereblon Modulator Avadomide (CC-122) in Advanced Malignancies. Clin. Cancer Res. 25, 90–98. 10.1158/1078-0432.CCR-18-1203 [DOI] [PubMed] [Google Scholar]
- Rego M. A., Kolling F. W., Vuono E. A., Mauro M., Howlett N. G. (2012). Regulation of the Fanconi Anemia Pathway by a CUE Ubiquitin-Binding Domain in the FANCD2 Protein. Blood 120, 2109–2117. 10.1182/blood-2012-02-410472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu F. E., Horton J. R., Mullally J. E., Heroux A., Cheng X., Wilkinson K. D. (2006). The Ubiquitin Binding Domain ZnF UBP Recognizes the C-Terminal Diglycine Motif of Unanchored Ubiquitin. Cell 124, 1197–1208. 10.1016/j.cell.2006.02.038 [DOI] [PubMed] [Google Scholar]
- Rizzo A. A., Salerno P. E., Bezsonova I., Korzhnev D. M. (2014). NMR Structure of the Human Rad18 Zinc Finger in Complex with Ubiquitin Defines a Class of UBZ Domains in Proteins Linked to the DNA Damage Response. Biochemistry 53, 5895–5906. 10.1021/bi500823h [DOI] [PubMed] [Google Scholar]
- Rogakou E. P., Boon C., Redon C., Bonner W. M. (1999). Megabase Chromatin Domains Involved in DNA Double-Strand Breaks In Vivo . J. Cell Biol. 146, 905–916. 10.1083/jcb.146.5.905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. (1998). DNA Double-Stranded Breaks Induce Histone H2AX Phosphorylation on Serine 139. J. Biol. Chem. 273, 5858–5868. 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Rona G., Roberti D., Yin Y., Pagan J. K., Homer H., Sassani E., et al. (2018). PARP1-dependent Recruitment of the FBXL10-RNF68-RNF2 Ubiquitin Ligase to Sites of DNA Damage Controls H2A.Z Loading. Z loadingeLife 7. 10.7554/eLife.38771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksouk N., Simboeck E., Déjardin J. (2015). Constitutive Heterochromatin Formation and Transcription in Mammals. Epigenetics Chromatin 8, 3. 10.1186/1756-8935-8-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos M. A., Huen M. S. Y., Jankovic M., Chen H.-T., López-Contreras A. J., Klein I. A., et al. (2010). Class Switching and Meiotic Defects in Mice Lacking the E3 Ubiquitin Ligase RNF8. J. Exp. Med. 207, 973–981. 10.1084/jem.20092308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Fujita H., Yoshikawa A., Yamashita M., Yamagata A., Kaiser S. E., et al. (2011). Specific Recognition of Linear Ubiquitin Chains by the Npl4 Zinc Finger (NZF) Domain of the HOIL-1L Subunit of the Linear Ubiquitin Chain Assembly Complex. Proc. Natl. Acad. Sci. U.S.A. 108, 20520–20525. 10.1073/pnas.1109088108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Yoshikawa A., Mimura H., Yamashita M., Yamagata A., Fukai S. (2009). Structural Basis for Specific Recognition of Lys 63-linked Polyubiquitin Chains by Tandem UIMs of RAP80. EMBO J. 28, 2461–2468. 10.1038/emboj.2009.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt C. K., Galanty Y., Sczaniecka-Clift M., Coates J., Jhujh S., Demir M., et al. (2015). Systematic E2 Screening Reveals a UBE2D-Rnf138-CtIP axis Promoting DNA Repair. Nat. Cell Biol. 17, 1458–1470. 10.1038/ncb3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld A. R., Apgar S., Dolios G., Wang R., Aaronson S. A. (2004). BRCA2 Is Ubiquitinated In Vivo and Interacts with USP11, a Deubiquitinating Enzyme that Exhibits Prosurvival Function in the Cellular Response to DNA Damage. Mol. Cell. Biol. 24, 7444–7455. 10.1128/MCB.24.17.7444-7455.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrank B., Gautier J. (2019). Assembling Nuclear Domains: Lessons from DNA Repair. J. Cell Biol. 218, 2444–2455. 10.1083/jcb.201904202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrank B. R., Aparicio T., Li Y., Chang W., Chait B. T., Gundersen G. G., et al. (2018). Nuclear ARP2/3 Drives DNA Break Clustering for Homology-Directed Repair. Nature 559, 61–66. 10.1038/s41586-018-0237-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully R., Panday A., Elango R., Willis N. A. (2019). DNA Double-Strand Break Repair-Pathway Choice in Somatic Mammalian Cells. Nat. Rev. Mol. Cell Biol. 20, 698–714. 10.1038/s41580-019-0152-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedelnikova O. A., Rogakou E. P., Panyutin I. G., Bonner W. M. (2002). Quantitative Detection of125IdU-Induced DNA Double-Strand Breaks with γ-H2AX Antibody. Radiat. Res. 158, 486–492. 10.1667/0033-7587(2002)158[0486:qdoiid]2.0.co;2 [DOI] [PubMed] [Google Scholar]
- Shanbhag N. M., Rafalska-Metcalf I. U., Balane-Bolivar C., Janicki S. M., Greenberg R. A. (2010). ATM-Dependent Chromatin Changes Silence Transcription in Cis to DNA Double-Strand Breaks. Cell 141, 970–981. 10.1016/j.cell.2010.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N., Zhu Q., Wani G., He J., Wang Q.-E., Wani A. A. (2014). USP3 Counteracts RNF168 via Deubiquitinating H2A and γH2AX at Lysine 13 and 15. Cell Cycle 13, 106–114. 10.4161/cc.26814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singatulina A. S., Hamon L., Sukhanova M. V., Desforges B., Joshi V., Bouhss A., et al. (2019). PARP-1 Activation Directs FUS to DNA Damage Sites to Form PARG-Reversible Compartments Enriched in Damaged DNA. Cell Rep. 27, 1809–1821. 10.1016/j.celrep.2019.04.031 [DOI] [PubMed] [Google Scholar]
- Singh A. N., Oehler J., Torrecilla I., Kilgas S., Li S., Vaz B., et al. (2019). The P97-Ataxin 3 Complex Regulates Homeostasis of the DNA Damage Response E3 Ubiquitin Ligase RNF 8. EMBO J. 38, e102361. 10.15252/embj.2019102361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Zeke A., Reményi A. (2022). Systematic Discovery of FBXW7-Binding Phosphodegrons Highlights Mitogen-Activated Protein Kinases as Important Regulators of Intracellular Protein Levels. Ijms 23, 3320. 10.3390/ijms23063320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagsvold T., Aasland R., Hirano S., Bache K. G., Raiborg C., Trambaiolo D., et al. (2005). Eap45 in Mammalian ESCRT-II Binds Ubiquitin via a Phosphoinositide-Interacting GLUE Domain. J. Biol. Chem. 280, 19600–19606. 10.1074/jbc.M501510200 [DOI] [PubMed] [Google Scholar]
- Slupianek A., Jozwiakowski S. K., Gurdek E., Skorski T. (2009). BCR/ABL Kinase Interacts with and Phosphorylates the RAD51 Paralog, RAD51B. Leukemia 23, 2308–2310. 10.1038/leu.2009.164 [DOI] [PubMed] [Google Scholar]
- Smerdon M. J. (1991). DNA Repair and the Role of Chromatin Structure. Curr. Opin. Cell Biol. 3, 422–428. 10.1016/0955-0674(91)90069-b [DOI] [PubMed] [Google Scholar]
- Smith M. J., Bryant E. E., Joseph F. J., Rothstein R. (2019). DNA Damage Triggers Increased Mobility of Chromosomes in G1-phase Cells. MBoC 30, 2620–2625. 10.1091/mbc.E19-08-0469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snead W. T., Gladfelter A. S. (2019). The Control Centers of Biomolecular Phase Separation: How Membrane Surfaces, Ptms, and Active Processes Regulate Condensation. Mol. Cell 76, 295–305. 10.1016/j.molcel.2019.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So C. C., Ramachandran S., Martin A. (2019). E3 Ubiquitin Ligases RNF20 and RNF40 Are Required for Double-Stranded Break (DSB) Repair: Evidence for Monoubiquitination of Histone H2B Lysine 120 as a Novel Axis of DSB Signaling and Repair. Mol. Cell. Biol. 39. 10.1128/MCB.00488-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone K., Piao L., Nakakido M., Ueda K., Jenuwein T., Nakamura Y., et al. (2014). Critical Role of Lysine 134 Methylation on Histone H2AX for γ-H2AX Production and DNA Repair. Nat. Commun. 5, 5691. 10.1038/ncomms6691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G., Polo S. E., Almouzni G. (2012). Prime, Repair, Restore: the Active Role of Chromatin in the DNA Damage Response. Mol. Cell 46, 722–734. 10.1016/j.molcel.2012.06.002 [DOI] [PubMed] [Google Scholar]
- Spence J., Sadis S., Haas A. L., Finley D. (1995). A Ubiquitin Mutant with Specific Defects in DNA Repair and Multiubiquitination. Mol. Cell. Biol. 15, 1265–1273. 10.1128/MCB.15.3.1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G. S., Panier S., Townsend K., Al-Hakim A. K., Kolas N. K., Miller E. S., et al. (2009). The RIDDLE Syndrome Protein Mediates a Ubiquitin-dependent Signaling Cascade at Sites of DNA Damage. Cell 136, 420–434. 10.1016/j.cell.2008.12.042 [DOI] [PubMed] [Google Scholar]
- Stewart G. S., Stankovic T., Byrd P. J., Wechsler T., Miller E. S., Huissoon A., et al. (2007). RIDDLE Immunodeficiency Syndrome Is Linked to Defects in 53BP1-Mediated DNA Damage Signaling. Proc. Natl. Acad. Sci. U.S.A. 104, 16910–16915. 10.1073/pnas.0708408104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M., Clapperton J. A., Mohammad D., Yaffe M. B., Smerdon S. J., Jackson S. P. (2005). MDC1 Directly Binds Phosphorylated Histone H2AX to Regulate Cellular Responses to DNA Double-Strand Breaks. Cell 123, 1213–1226. 10.1016/j.cell.2005.09.038 [DOI] [PubMed] [Google Scholar]
- Sun Y., Jiang X., Chen S., Fernandes N., Price B. D. (2005). A Role for the Tip60 Histone Acetyltransferase in the Acetylation and Activation of ATM. Proc. Natl. Acad. Sci. U.S.A. 102, 13182–13187. 10.1073/pnas.0504211102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Jiang X., Xu Y., Ayrapetov M. K., Moreau L. A., Whetstine J. R., et al. (2009). Histone H3 Methylation Links DNA Damage Detection to Activation of the Tumour Suppressor Tip60. Nat. Cell Biol. 11, 1376–1382. 10.1038/ncb1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland B. M., Bennett P. V., Schenk H., Sidorkina O., Laval J., Trunk J., et al. (2001). Clustered DNA Damages Induced by High and Low LET Radiation, Including Heavy Ions. Phys. Med. 17 (Suppl. 1), 202–204. [PubMed] [Google Scholar]
- Swaney D. L., Beltrao P., Starita L., Guo A., Rush J., Fields S., et al. (2013). Global Analysis of Phosphorylation and Ubiquitylation Cross-Talk in Protein Degradation. Nat. Methods 10, 676–682. 10.1038/nmeth.2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift M. L., Azizkhan-Clifford J. (2022). DNA Damage-Induced Sumoylation of Sp1 Induces its Interaction with RNF4 and Degradation in S Phase to Remove 53BP1 from DSBs and Permit HR. DNA Repair 111, 103289. 10.1016/j.dnarep.2022.103289 [DOI] [PubMed] [Google Scholar]
- Takahashi T. S., Hirade Y., Toma A., Sato Y., Yamagata A., Goto-Ito S., et al. (2018). Structural Insights into Two Distinct Binding Modules for Lys63-Linked Polyubiquitin Chains in RNF168. Nat. Commun. 9, 170. 10.1038/s41467-017-02345-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata H., Hanafusa T., Mori T., Shimura M., Iida Y., Ishikawa K., et al. (2013). Chromatin Compaction Protects Genomic DNA from Radiation Damage. PLoS ONE 8, e75622. 10.1371/journal.pone.0075622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallis M., Morra R., Barkauskaite E., Ahel I. (2014). Poly(ADP-ribosyl)ation in Regulation of Chromatin Structure and the DNA Damage Response. Chromosoma 123, 79–90. 10.1007/s00412-013-0442-9 [DOI] [PubMed] [Google Scholar]
- Tan W., Deans A. J. (2017). A Defined Role for Multiple Fanconi Anemia Gene Products in DNA-Damage-Associated Ubiquitination. Exp. Hematol. 50, 27–32. 10.1016/j.exphem.2017.03.001 [DOI] [PubMed] [Google Scholar]
- Tan W., van Twest S., Leis A., Bythell-Douglas R., Murphy V. J., Sharp M., et al. (2020). Monoubiquitination by the Human Fanconi Anemia Core Complex Clamps FANCI:FANCD2 on DNA in Filamentous Arrays. eLife 9. 10.7554/eLife.54128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Cho N. W., Cui G., Manion E. M., Shanbhag N. M., Botuyan M. V., et al. (2013). Acetylation Limits 53BP1 Association with Damaged Chromatin to Promote Homologous Recombination. Nat. Struct. Mol. Biol. 20, 317–325. 10.1038/nsmb.2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L.-Y., Thomas A., Zhou M., Zhang Y. E. (2020). Phosphorylation of SMURF2 by ATM Exerts a Negative Feedback Control of DNA Damage Response. J. Biol. Chem. 295, 18485–18493. 10.1074/jbc.RA120.014179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teicher B. A., Ara G., Herbst R., Palombella V. J., Adams J. (1999). The Proteasome Inhibitor PS-341 in Cancer Therapy. Clin. Cancer Res. 5, 2638–2645. [PubMed] [Google Scholar]
- Thorslund T., Ripplinger A., Hoffmann S., Wild T., Uckelmann M., Villumsen B., et al. (2015). Histone H1 Couples Initiation and Amplification of Ubiquitin Signalling after DNA Damage. Nature 527, 389–393. 10.1038/nature15401 [DOI] [PubMed] [Google Scholar]
- Tian M., Zeng T., Liu M., Han S., Lin H., Lin Q., et al. (2019). A Cell-Based High-Throughput Screening Method Based on a Ubiquitin-Reference Technique for Identifying Modulators of E3 Ligases. J. Biol. Chem. 294, 2880–5770. 10.1074/jbc.RA118.003822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjeertes J. V., Miller K. M., Jackson S. P. (2009). Screen for DNA-Damage-Responsive Histone Modifications Identifies H3K9Ac and H3K56Ac in Human Cells. EMBO J. 28, 1878–1889. 10.1038/emboj.2009.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toiber D., Erdel F., Bouazoune K., Silberman D. M., Zhong L., Mulligan P., et al. (2013). SIRT6 Recruits SNF2H to DNA Break Sites, Preventing Genomic Instability through Chromatin Remodeling. Mol. Cell 51, 454–468. 10.1016/j.molcel.2013.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toma A., Takahashi T. S., Sato Y., Yamagata A., Goto-Ito S., Nakada S., et al. (2015). Structural Basis for Ubiquitin Recognition by Ubiquitin-Binding Zinc Finger of FAAP20. PLoS ONE 10, e0120887. 10.1371/journal.pone.0120887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomimatsu N., Mukherjee B., Catherine Hardebeck M., Ilcheva M., Vanessa Camacho C., Louise Harris J., et al. (2014). Phosphorylation of EXO1 by CDKs 1 and 2 Regulates DNA End Resection and Repair Pathway Choice. Nat. Commun. 5, 3561. 10.1038/ncomms4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S., Si Y., Yu H., Zhang L., Xie P., Jiang W. (2017). MLN4924 (Pevonedistat), a Protein Neddylation Inhibitor, Suppresses Proliferation and Migration of Human Clear Cell Renal Cell Carcinoma. Sci. Rep. 7, 5599. 10.1038/s41598-017-06098-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsouroula K., Furst A., Rogier M., Heyer V., Maglott-Roth A., Ferrand A., et al. (2016). Temporal and Spatial Uncoupling of DNA Double Strand Break Repair Pathways within Mammalian Heterochromatin. Mol. Cell 63, 293–305. 10.1016/j.molcel.2016.06.002 [DOI] [PubMed] [Google Scholar]
- Typas D., Luijsterburg M. S., Wiegant W. W., Diakatou M., Helfricht A., Thijssen P. E., et al. (2015). The De-ubiquitylating Enzymes USP26 and USP37 Regulate Homologous Recombination by Counteracting RAP80. Nucleic Acids Res. 43, 6919–6933. 10.1093/nar/gkv613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uckelmann M., Densham R. M., Baas R., Winterwerp H. H. K., Fish A., Sixma T. K., et al. (2018). USP48 Restrains Resection by Site-specific Cleavage of the BRCA1 Ubiquitin Mark from H2A. Nat. Commun. 9, 229. 10.1038/s41467-017-02653-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel T., Lerenthal Y., Moyal L., Andegeko Y., Mittelman L., Shiloh Y. (2003). Requirement of the MRN Complex for ATM Activation by DNA Damage. EMBO J. 22, 5612–5621. 10.1093/emboj/cdg541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L. T., Vu B. T., Graves B., Carvajal D., Podlaski F., Filipovic Z., et al. (2004). In Vivo activation of the P53 Pathway by Small-Molecule Antagonists of MDM2. Science 303, 844–848. 10.1126/science.1092472 [DOI] [PubMed] [Google Scholar]
- Vazquez B. N., Thackray J. K., Simonet N. G., Kane‐Goldsmith N., Martinez‐Redondo P., Nguyen T., et al. (2016). SIRT 7 Promotes Genome Integrity and Modulates Non‐homologous End Joining DNA Repair. EMBO J. 35, 1488–1503. 10.15252/embj.201593499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas R., Kumar R., Clermont F., Helfricht A., Kalev P., Sotiropoulou P., et al. (2013). RNF4 Is Required for DNA Double-Strand Break Repair In Vivo . Cell Death Differ. 20, 490–502. 10.1038/cdd.2012.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., et al. (2011). A Proteome-wide, Quantitative Survey of In Vivo Ubiquitylation Sites Reveals Widespread Regulatory Roles. Mol. Cell. Proteomics 10, M111013284. 10.1074/mcp.M111.013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeman T. P., Wang Q., Feng J., Wang X.-F. (2012). Bat3 Facilitates H3K79 Dimethylation by DOT1L and Promotes DNA Damage-Induced 53BP1 Foci at G1/G2 Cell-Cycle Phases. EMBO J. 31, 2169–2181. 10.1038/emboj.2012.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser F., Mulder M. P. C., Bragantini B., Burger S., Gubser T., Gatti M., et al. (2020). Ubiquitin Phosphorylation at Thr12 Modulates the DNA Damage Response. Mol. Cell 80, 423–436. 10.1016/j.molcel.2020.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., et al. (2004). Role of Histone H2A Ubiquitination in Polycomb Silencing. Nature 431, 873–878. 10.1038/nature02985 [DOI] [PubMed] [Google Scholar]
- Wang H., Zhai L., Xu J., Joo H.-Y., Jackson S., Erdjument-Bromage H., et al. (2006). Histone H3 and H4 Ubiquitylation by the CUL4-DDB-ROC1 Ubiquitin Ligase Facilitates Cellular Response to DNA Damage. Mol. Cell 22, 383–394. 10.1016/j.molcel.2006.03.035 [DOI] [PubMed] [Google Scholar]
- Wang J., Aroumougame A., Lobrich M., Li Y., Chen D., Chen J., et al. (2014). PTIP Associates with Artemis to Dictate DNA Repair Pathway Choice. Genes Dev. 28, 2693–2698. 10.1101/gad.252478.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Gao Y., Zheng X., Liu C., Dong S., Li R., et al. (2019). Histone Modifications Regulate Chromatin Compartmentalization by Contributing to a Phase Separation Mechanism. Mol. Cell 76, 646–659. 10.1016/j.molcel.2019.08.019 [DOI] [PubMed] [Google Scholar]
- Wang Q., Goh A. M., Howley P. M., Walters K. J. (2003). Ubiquitin Recognition by the DNA Repair Protein hHR23a. Biochemistry 42, 13529–13535. 10.1021/bi035391j [DOI] [PubMed] [Google Scholar]
- Wang T., Liu N. S., Seet L.-F., Hong W. (2010). The Emerging Role of VHS Domain-Containing Tom1, Tom1L1 and Tom1L2 in Membrane Trafficking. Traffic 11, 1119–1128. 10.1111/j.1600-0854.2010.01098.x [DOI] [PubMed] [Google Scholar]
- Wang W. (2007). Emergence of a DNA-Damage Response Network Consisting of Fanconi Anaemia and BRCA Proteins. Nat. Rev. Genet. 8, 735–748. 10.1038/nrg2159 [DOI] [PubMed] [Google Scholar]
- Wang X., D'Arcy P., Caulfield T. R., Paulus A., Chitta K., Mohanty C., et al. (2015). Synthesis and Evaluation of Derivatives of the Proteasome Deubiquitinase Inhibitor B-AP15. Chem. Biol. Drug Des. 86, 1036–1048. 10.1111/cbdd.12571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhang H., Liu J., Cheruiyot A., Lee J.-H., Ordog T., et al. (2016). USP51 Deubiquitylates H2AK13,15ub and Regulates DNA Damage Response. Genes Dev. 30, 946–959. 10.1101/gad.271841.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward I. M., Chen J. (2001). Histone H2AX Is Phosphorylated in an ATR-dependent Manner in Response to Replicational Stress. J. Biol. Chem. 276, 47759–47762. 10.1074/jbc.C100569200 [DOI] [PubMed] [Google Scholar]
- Wijnhoven P., Konietzny R., Blackford A. N., Travers J., Kessler B. M., Nishi R., et al. (2015). USP4 Auto-Deubiquitylation Promotes Homologous Recombination. Mol. Cell 60, 362–373. 10.1016/j.molcel.2015.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik F., Dann G. P., Beh L. Y., Debelouchina G. T., Hofmann R., Muir T. W. (2018). Functional Crosstalk between Histone H2B Ubiquitylation and H2A Modifications and Variants. Nat. Commun. 9, 1394. 10.1038/s41467-018-03895-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-Y., Kang H.-Y., Yang W.-L., Wu J., Jeong Y. S., Wang J., et al. (2011a). Critical Role of Monoubiquitination of Histone H2AX Protein in Histone H2AX Phosphorylation and DNA Damage Response. J. Biol. Chem. 286, 30806–30815. 10.1074/jbc.M111.257469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Chen Y., Lu L.-Y., Wu Y., Paulsen M. T., Ljungman M., et al. (2011b). Chfr and RNF8 Synergistically Regulate ATM Activation. Nat. Struct. Mol. Biol. 18, 761–768. 10.1038/nsmb.2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Johansen J. V., Helin K. (2013). Fbxl10/Kdm2b Recruits Polycomb Repressive Complex 1 to CpG Islands and Regulates H2A Ubiquitylation. Mol. Cell 49, 1134–1146. 10.1016/j.molcel.2013.01.016 [DOI] [PubMed] [Google Scholar]
- Wu X., Liu S., Sagum C., Chen J., Singh R., Chaturvedi A., et al. (2019). Crosstalk between Lys63- and Lys11-Polyubiquitin Signaling at DNA Damage Sites Is Driven by Cezanne. Genes Dev. 33, 1702–1717. 10.1101/gad.332395.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A., Li H., Shechter D., Ahn S. H., Fabrizio L. A., Erdjument-Bromage H., et al. (2009). WSTF Regulates the H2A.X DNA Damage Response via a Novel Tyrosine Kinase Activity. Nature 457, 57–62. 10.1038/nature07668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao D., Yue M., Su H., Ren P., Jiang J., Li F., et al. (2016). Polo-Like Kinase-1 Regulates Myc Stabilization and Activates a Feedforward Circuit Promoting Tumor Cell Survival. Mol. Cell 64, 493–506. 10.1016/j.molcel.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Xu G., Chapman J. R., Brandsma I., Yuan J., Mistrik M., Bouwman P., et al. (2015). REV7 Counteracts DNA Double-Strand Break Resection and Affects PARP Inhibition. Nature 521, 541–544. 10.1038/nature14328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Sun Y., Jiang X., Ayrapetov M. K., Moskwa P., Yang S., et al. (2010). The P400 ATPase Regulates Nucleosome Stability and Chromatin Ubiquitination during DNA Repair. J. Cell Biol. 191, 31–43. 10.1083/jcb.201001160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Kim Y.-S., Yang X.-P., Li L.-P., Liao G., Xia F., et al. (2007). The Ubiquitin-Interacting Motif-Containing Protein RAP80 Interacts with BRCA1 and Functions in DNA Damage Repair Response. Cancer Res. 67, 6647–6656. 10.1158/0008-5472.CAN-07-0924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., Dutt S., Xu R., Graves K., Juszczynski P., Manis J. P., et al. (2009). BBAP Monoubiquitylates Histone H4 at Lysine 91 and Selectively Modulates the DNA Damage Response. Mol. Cell 36, 110–120. 10.1016/j.molcel.2009.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q., Xu R., Zhu L., Cheng X., Wang Z., Manis J., et al. (2013). BAL1 and its Partner E3 Ligase, BBAP, Link Poly(ADP-Ribose) Activation, Ubiquitylation, and Double-Strand DNA Repair Independent of ATM, MDC1, and RNF8. Mol. Cell. Biol. 33, 845–857. 10.1128/MCB.00990-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.-S., Jividen K., Spencer A., Dworak N., Ni L., Oostdyk L. T., et al. (2017). Ubiquitin Modification by the E3 Ligase/ADP-Ribosyltransferase Dtx3L/Parp9. Mol. Cell 66, 503–516. e5. 10.1016/j.molcel.2017.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K., Moldovan G.-L., D'Andrea A. D. (2010). RAD18-dependent Recruitment of SNM1A to DNA Repair Complexes by a Ubiquitin-Binding Zinc Finger. J. Biol. Chem. 285, 19085–19091. 10.1074/jbc.M109.100032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Ding Y., Zhou C., Wen Y., Zhang N. (2019). Structural and Functional Studies of USP20 ZnF‐UBP Domain by NMR. Protein Sci. 28, 1606–1619. 10.1002/pro.3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz D., Furst A., Meaburn K., Lezaja A., Wen Y., Altmeyer M., et al. (2021). Activation of Homologous Recombination in G1 Preserves Centromeric Integrity. Nature 600, 748–753. 10.1038/s41586-021-04200-z [DOI] [PubMed] [Google Scholar]
- Yin Y., Seifert A., Chua J. S., Maure J.-F., Golebiowski F., Hay R. T. (2012). SUMO-Targeted Ubiquitin E3 Ligase RNF4 Is Required for the Response of Human Cells to DNA Damage. Genes Dev. 26, 1196–1208. 10.1101/gad.189274.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa Y., Mori T., Magome N., Hibino K., Yoshikawa K. (2008). DNA Compaction Plays a Key Role in Radioprotection against Double-Strand Breaks as Revealed by Single-Molecule Observation. Chem. Phys. Lett. 456, 80–83. 10.1016/j.cplett.2008.03.009 [DOI] [Google Scholar]
- Young L. C., McDonald D. W., Hendzel M. J. (2013). Kdm4b Histone Demethylase Is a DNA Damage Response Protein and Confers a Survival Advantage Following γ-Irradiation. J. Biol. Chem. 288, 21376–21388. 10.1074/jbc.M113.491514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., Liu K., Mao Z., Luo J., Gu W., Zhao W. (2016). USP11 Is a Negative Regulator to γH2AX Ubiquitylation by RNF8/RNF168. J. Biol. Chem. 291, 959–967. 10.1074/jbc.M114.624478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Liu H., Chen Y., Yang X., Wang P., Liu T., et al. (2016a). A Cell Cycle-dependent BRCA1-UHRF1 Cascade Regulates DNA Double-Strand Break Repair Pathway Choice. Nat. Commun. 7, 10201. 10.1038/ncomms10201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Karnak D., Tan M., Lawrence T. S., Morgan M. A., Sun Y. (2016b). FBXW7 Facilitates Nonhomologous End-Joining via K63-Linked Polyubiquitylation of XRCC4. Mol. Cell 61, 419–433. 10.1016/j.molcel.2015.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Mady A. S. A., Ma Y., Ryan C., Lawrence T. S., Nikolovska-Coleska Z., et al. (2019). The WD40 Domain of FBXW7 Is a poly(ADP-Ribose)-Binding Domain that Mediates the Early DNA Damage Response. Nucleic Acids Res. 47, 4039–4053. 10.1093/nar/gkz058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Morgan M. A., Sun Y. (2014). Targeting Neddylation Pathways to Inactivate Cullin-RING Ligases for Anticancer Therapy. Antioxidants Redox Signal. 21, 2383–2400. 10.1089/ars.2013.5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Huang T., Zhang L., Zhou Y., Luo H., Xu H., et al. (2016). Dysregulation of Ubiquitin-Proteasome System in Neurodegenerative Diseases. Front. Aging Neurosci. 8, 303. 10.3389/fnagi.2016.00303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S., Li D., Lu Z., Liu G., Wang M., Xing P., et al. (2018). Bre1-dependent H2B Ubiquitination Promotes Homologous Recombination by Stimulating Histone Eviction at DNA Breaks. Nucleic Acids Res. 46, 11326–11339. 10.1093/nar/gky918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Huang J., Huang H., Li H., Yi P., Kloeber J. A., et al. (2021). RNF19A-mediated Ubiquitination of BARD1 Prevents BRCA1/BARD1-dependent Homologous Recombination. Nat. Commun. 12, 6653. 10.1038/s41467-021-27048-3 [DOI] [PMC free article] [PubMed] [Google Scholar]