Abstract
Sleep deprivation (SD) is very common in modern society and regarded as a potential causal mechanism of several clinical disorders. Previous neuroimaging studies have explored the neural mechanisms of SD using magnetic resonance imaging (MRI) from static (comparing two MRI sessions [one after SD and one after resting wakefulness]) and dynamic (using repeated MRI during one night of SD) perspectives. Recent SD researches have focused on the dynamic functional brain organization during the resting‐state scan. Our present study adopted a novel metric (temporal variability), which has been successfully applied to many clinical diseases, to examine the dynamic functional connectivity after SD in 55 normal young subjects. We found that sleep‐deprived subjects showed increased regional‐level temporal variability in large‐scale brain regions, and decreased regional‐level temporal variability in several thalamus subregions. After SD, participants exhibited enhanced intra‐network temporal variability in the default mode network (DMN) and increased inter‐network temporal variability in numerous subnetwork pairs. Furthermore, we found that the inter‐network temporal variability between visual network and DMN was negative related with the slowest 10% respond speed (β = −.42, p = 5.57 × 10−4) of the psychomotor vigilance test after SD following the stepwise regression analysis. In conclusion, our findings suggested that sleep‐deprived subjects showed abnormal dynamic brain functional configuration, which provides new insights into the neural underpinnings of SD and contributes to our understanding of the pathophysiology of clinical disorders.
Keywords: dynamic functional connectivity, psychomotor vigilance test, resting‐state functional magnetic resonance imaging, sleep deprivation, temporal variability
Our present study adopted a novel metric (temporal variability), which has been successfully applied to many clinical diseases, to examine the dynamic functional connectivity after SD. Regional‐level and network‐level temporal variabilities were significantly increased after SD and correlated with the psychomotor vigilance test performance. These findings suggested that sleep‐deprived subjects showed abnormal dynamic brain functional configuration, which provides new insights into the neural underpinnings of SD and contributes to our understanding of the pathophysiology of clinical disorders.
1. INTRODUCTION
Sleep deprivation (SD) has been a common problem in modern society. Previous studies have reported that 35% of adults in the United States sleep less than 7 h during a typical 24‐h day, and this phenomenon is more prominent among US middle school students (Bandyopadhyay & Sigua, 2019) and older adults (Mander et al., 2017). It has been well‐documented that insufficient sleep is detrimental to human health and causes cognitive dysfunctions, even after one night of SD (Hudson et al., 2020; Itani et al., 2017; Porras‐Segovia et al., 2019; Tobaldini et al., 2017; Van Craenenbroeck, 2019). Prominently, attention lapses, impaired working memory, hindering decision making, impaired emotional processing are associated with one single night of SD (Cassé‐Perrot et al., 2016; Cousins & Fernández, 2019; Tempesta et al., 2018). Furthermore, recent studies have demonstrated that SD is one of the reasons for the pathogenic mechanism of various neurological disorders such as Alzheimer's disease and Parkinson's disease (Bishir et al., 2020). Patients with psychiatric disorders suffer from sleep deficit during the early stage of these diseases, and several researches have revealed that this deficit may be a potential causal mechanism of psychosis (Waite et al., 2020). Thus, elucidating the underlying mechanisms of SD is an important goal in basic and clinical neuroscience.
A growing body of neuroimaging researches have explored the neural underpinnings of SD by using magnetic resonance imaging (MRI; Chee & Zhou, 2019; Krause et al., 2017; Li et al., 2021; Zhang et al., 2021). Recently, our group has carried out a series of studies on this research area. By comparing SD session with resting wakefulness (RW) session, we have reported poorer performance of inhibitory control and deficits in the cerebral activation of inhibitory control after SD (Zhao, Zhang, Fei, et al., 2019). We further have found that one night of SD could induce alterations in brain structure (Sun, Zhao, et al., 2020), and the structural MRI data in RW session could linearly predict the alterations of response inhibition after SD (Zhao et al., 2018). Within resting‐state functional MRI (rsfMRI) data, our group have reported the disrupted resting state functional organization in hippocampal subregions using the masked independent component analysis (Zhao, Zhang, Zhu, et al., 2019), and increased interhemispheric resting‐state functional connectivity (FC) which may reflect the compensatory mechanisms after SD (Zhu et al., 2016). Using diffusion tensor imaging, we have examined the relationship between the inter‐individual difference in cognitive performance with white matter characteristics after SD (Zhu et al., 2017). These studies used a typically experimental paradigm that run two MRI scans (one after SD and one after RW). However, this type of researches depicted the modulated brain after SD, rather than the modulation processes during SD. For this issue, our group increases the number of MRI scan during the whole time of SD, and explored the dynamic changes of brain response to sustained attention task and working memory task during one night of SD (Zhu et al., 2018; Y. Zhu et al., 2019). Above mentioned studies have revealed the neural mechanisms of SD from static and dynamic perspectives, which give us a deeper understanding of SD.
Besides the dynamic analysis using repeated fMRI, recent SD researches have investigated the effects of SD on the dynamic functional brain organization on the order of seconds to minutes over the resting state scan based on a sliding‐window method. Li et al. and Xu et al. (C. Li et al., 2020; Xu et al., 2018) have shown altered dwell time and transit between FC states after SD. Long et al. (Long et al., 2021) have examined the age‐related changes in FC variability of the thalamus after partial SD, and reported reduced FC variability between the left thalamus and the left superior parietal cortex in young adults after 3 h of sleep restriction. However, the FC states analysis measured the patterns of whole brain dynamic FC (Allen et al., 2014; Bolton et al., 2020), rather than the dynamic FC architecture of a specific region. The FC variability analysis quantifies the fluctuations of FC between two brain regions (Kucyi & Davis, 2014; Liu et al., 2018), lack of global information. Recently, the temporal variability of functional architecture associated with a specific region, a novel measure of functional brain dynamics at the mesoscale introduced by Zhang et al. (2016), is different from the above two methods which measures the interregional property of dynamic FC (Hu et al., 2018). This approach allows the coupling analysis between the temporal variability of a region and its neural activities, and can localize regions showing significant variability changes between groups and regions showing significant variability correlated with behavior, thus helping to revealing the underlying neuroimaging mechanisms of brain disorders (Sun et al., 2019; Zhang et al., 2016). It has been successfully applied to many diseases including schizophrenia (Deng et al., 2019; Dong et al., 2019; Yue et al., 2018), major depressive disorder (Hou et al., 2018), stroke (Hu et al., 2018), Parkinson's disease (H. Zhu et al., 2019), and Alzheimer's disease (Gu et al., 2020). These researches suggested that the temporal variability might provide new insights into the neural underpinnings of brain disorders. Considering that SD may be a potential causal factor of these disorders (Reeve et al., 2015; Waite et al., 2020), and that experimentally controlled SD is a valuable experimental medicine model of schizophrenia (Ettinger & Kumari, 2015; Kumari & Ettinger, 2020), performing research on temporal variability after SD may promote our understanding of the pathophysiology of clinical disorders. However, the effects of SD on the temporal variability have not been depicted.
In this present study, we investigated the effects of one night of SD on the temporal variability in 55 healthy young subjects. Firstly, we measured the regional‐level temporal variability of FC architecture after RW and SD, using the method introduced by Zhang et al. (2016), and compared the differences of the temporal variability between RW and SD states. Similarly, the intra‐network and inter‐network temporal variability of FC architecture was analyzed using the method introduced by (Sun et al., 2019). Finally, we explored the correlations between the temporal variability and vigilant attention using stepwise regression analysis.
2. MATERIALS AND METHODS
2.1. Subjects
Sixty‐eight righted‐handed healthy young subjects were recruited from Xidian University in this study. The recruitment criteria were similar to those in our previous researches (Sun, Zhao, et al., 2020; Zhao, Zhang, Fei, et al., 2019; Zhao, Zhang, Zhu, et al., 2019; Zhao et al., 2018). All participants had normal sleep schedules of 7–9 h per night between 10:00 p.m. and 8:00 a.m. They had no history of smoking, self‐reported medical, psychiatric, neurological or sleep disorders, and were free of any abused alcohol or drugs. Furthermore, all of them did not present an extreme morning or extreme evening type, assessed by the Morningness‐Eveningness questionnaire (Horne & Ostberg, 1976) which was the most used and cited questionnaire. Subjects with the total score ranging from 16 to 30 were determined to be the extreme evening type. Subjects were divided into the extreme morning type with the total score ranging from 70 to 86.
We performed the pre‐experiment for the first subject. However, this subject was removed from this study owing to incomplete data. Two subjects opted out of this study during the SD session. Three subjects were discarded due to the abnormality in brain structure. Furthermore, one subject was abandoned by reason that the MRI scanner was broken during the SD session scan. Another six participants were excluded who showed abnormal head motion (see Section 2.5.1 for details). Therefore, 55 subjects were included in the final analysis in total with the mean age of 20.58 ± 1.83 years (range 18–23 years; 29 males, 26 females).
All participants declared that they did not smoke or consume any stimulants, medications, alcohol or caffeine (coffee, tea, cola, etc.) for at least 24 h before the formal experiment. All subjects provided written informed consent prior to participation and were compensated for their time. All research procedures were conducted in accordance with the Declaration of Helsinki and approved by the institutional research ethics committee of the Xijing Hospital of the Fourth Military Medical University.
2.2. Experimental procedure
The experimental procedure in the present study was similar to those that in our previous researches (Zhao et al., 2018; Zhao, Zhang, Zhu, et al., 2019). All subjects were scheduled for three visits to the laboratory. For the first visit, subjects underwent the screening process, and were informed of the experimental procedures and given instructions about the psychomotor vigilance test (PVT). Subjects were also requested to keep a sleep diary throughout the experiment to verify subjects' compliance to a regular sleep schedule. After 1 week, subjects who met the inclusion criteria performed the second visit. During this visit, subjects underwent the RW session or SD session. During the third visit, subjects underwent the other session. For the RW session, subjects slept as usual with at least 7 h of sleep and reported to the laboratory at 7:30 a.m. After performing the PVT task, subjects underwent the MR scanning at 8:00 a.m. For the SD session, subjects waked up regularly according to their sleep schedules (at 8:00 a.m. at the latest) after 7–9 h of sleeping and were required no naps or vigorous physical activities in the daytime. Subjects reported to the laboratory before 10:00 p.m., and were not allowed to sleep from 10:00 p.m. to 8:00 a.m. In order to prevent subjects from falling asleep in this period, they were monitored by experimenters in the laboratory, and they could do some nonstrenuous activities such as reading and watching videos in this period. After finishing the PVT task, subjects were administered the MR scanning at 8:00 a.m. The second and third visits were administered in a randomized, cross‐over fashion with at least 1 week apart to minimize possible residual effects of SD on cognition (Van Dongen et al., 2003). During this week, subjects continued their usual daily activities, but were not permitted to perform shiftwork or stay up all night. They were also not permitted to do vigorous physical activities 1 day before scanning.
2.3. Psychomotor vigilance test
We used PVT to measure vigilant attention, which is the cognitive domain most severely impaired by SD (Lim & Dinges, 2008; Yang et al., 2018). The procedure of this task in the present study was similar to our previous research (Sun, Zhao, et al., 2020). Firstly, a red fixation cross appeared in the center of a black background on the screen and remained for 2 s. Then, the red fixation cross disappeared, and the black background screen was presented for a random duration of 2–10 s. After that, a red target circle was displayed and participants were instructed to press a button as quickly as possible with their right index finger. They were required to press the button within 30 s. If the participant responded, the red target circle disappeared and the real‐time reaction time (RT) was displayed on the screen to provide feedback regarding their performance. The feedback was presented 1 s after the response. If the participant did not respond, the displayed real‐time RT was 0 ms. The whole process of this task lasted for 7 min.
The primary behavioral measurements of interest in the PVT were (1) the number of trails; (2) lapse number (the lapse was defined as the trail with RT >500 ms); (3) the mean RT of all trials; (4) the RT of the fastest 10% trials (10% fast RT); (5) the reciprocal of RT of the slowest 10% trials (10% slow 1/RT); (6) the standard deviation of RT (sd_RT); and (7) coefficient of variation of RT (cv_RT).
2.4. MRI data acquisition
MRI data were acquired from a 3T GE MR750 scanner at Department of Radiology, Xijing Hospital, The Air Force Medical University, Xi'an, China. A standard 8‐channel head coil was used together with a restraining foam pad to minimize head motion and diminish scanner noise. The 3D T1‐weighted structural images were obtained during the second visit with the following scanning parameters: repetition time (TR) = 8.2 ms; echo time (TE) = 3.18 ms; field of view (FOV) = 256 × 256 mm2; matrix = 512 × 512; in‐plane resolution = 0.5 × 0.5 mm2; slice thickness = 1 mm; 196 sagittal slices; flip angle = 9°. The rsfMRI data were collected within 7 min using the following parameters: TR = 2 s; TE = 30 ms; FOV = 240 × 240 mm2; matrix = 64 × 64; in‐plane resolution = 3.75 × 3.75 mm2; slice thickness = 3.5 mm; 45 axial slices; flip angle = 90°, and 210 volumes were acquired for each subject.
In order to avoid sleeping during the rsfMRI scanning, subjects were required to relax, stay still, remain awake with their eyes open and focus on a fixation cross. The fixation cross was projected onto a screen positioned at the head of the magnet bore and viewed with a mirror attached to the head coil. After the rsfMRI scanning, subjects were asked whether they were awake during the scanning and all the subjects confirmed that they were awake.
2.5. Data analysis
2.5.1. Preprocessing
Preprocessing was performed using the Statistical Parametric Mapping (SPM12, www.fil.ion.ucl.ac.uk/spm/software/spm12/) and Functional Connectivity Toolbox (CONN version 17f, https://www.nitrc.org/projects/conn). The first five volumes of the rsfMRI data for each session were discarded for signal stabilization, and for participants to become accustomed to the scanning noise. Then, the remaining 205 volumes of the rsfMRI were preprocessed with the CONN's default preprocessing pipeline which included 7 steps. (1) Functional realignment and unwarp. The remaining rsfMRI data was realigned to the middle volume of each session using the SPM12 realign and unwarp procedure (Andersson et al., 2001) to perform the head motion correction. In order to control the influence of the head motion on the results, subjects with translation of more than 2 mm or rotation of more than 2° over the course of each session were excluded from the final analyzed group. Six participants were discarded with higher head motion. (2) The realigned functional data were centered to (0, 0, 0) coordinates. (3) Slice‐timing correction. According to the sequential nature of the fMRI acquisition protocol, the slice order of the rsfMRI was temporal misaligned, which was time‐shifted using the SPM12 slice‐timing correction procedure (Henson et al., 1999). (4) Functional outlier detection. Functional scans were subjected to artifact and motion outlier identification for scrubbing using the Artifact Detection Toolbox (ART; https://www.nitrc.org/projects/artifact_detect/) according to intermediate settings (97th percentiles in normative sample). Timepoints were flagged as outliers with global blood oxygenation level dependent (BOLD) signal changes above 5 standard deviation and framewise displacement above 0.9 mm. (5) Functional direct segmentation and normalization. The slice‐timing corrected rsfMRI data were segmented into gray matter, white matter, and cerebrospinal fluid (CSF), and normalized into the standard Montreal Neurological Institute (MNI) space adopting default Tissue Probability Maps using SPM12 unified segmentation and normalization procedure (Ashburner & Friston, 2005), with a voxel size of 2 × 2 × 2 mm3. (6) The 3D T1‐weighted structural images were center to (0, 0, 0) coordinates. (7) Structural segmentation and normalization. The structural data were segmented into gray matter, white matter, and CSF, and normalized into the standard MNI space with a voxel size of 2 × 2 × 2 mm3, using the similar procedure that applied to the functional data. (8) Functional smoothing. In order to increase the BOLD signal‐to‐noise ratio, functional data were spatially smoothed using a Gaussian kernel with 8 mm full width at half maximum.
Finally, the denoising pipeline was applied to remove potential confounders from the BOLD signal using an anatomical component‐based noise correction procedure (aCompCor; Behzadi et al., 2007). Confounders included 10 noise components from white matter and CSF (five components each) with the principal component analysis, 12 motion‐related parameters (3 translation and 3 rotation parameters plus their associated first‐order derivatives), and scrubbing parameters obtained from the functional outlier detection. Then, the temporal band‐pass filtering was implemented with a frequency window of 0.008–0.09 Hz to focus on slow‐frequency fluctuations and minimize the influence of physiological, head‐motion and other noise sources. The linear detrending was also performed to remove linear trends within each functional session.
2.5.2. Temporal variability of regional‐level FC architecture
Figure 1 illustrated the analysis pipeline for computing the tree types of temporal variabilities. The temporal variability of the FC architecture associated with a brain region was evaluated based on the previously described method (Zhang et al., 2016). For the regional‐level temporal variability, data processing was divided into five major steps. (1) The whole brain was segmented into N ROIs according to a parcellation brain atlas. (2) The mean time course of each ROI was extracted by averaging the time series of all voxels in the corresponding ROI. (3) Averaged time series of all ROIs were segmented into M nonoverlapping windows with an equal length of L. (4) Within the ith window, a N × N FC matrix (denoted as F i , i = 1, 2, 3, …, M) was generated by computing pairwise Pearson's correlation among N ROIs using the averaged windowed time series. The kth row (or kth column) in F i , that is, F i (k,:), is a N‐dimensional vector and characterizes the FC profile of the kth ROI at the ith time window. We denoted it as F i,k for convenience. (5) The regional‐level temporal variability of the kth ROI () was computed by comparing the FC profile of region k at different windows based on the following formulation:
In this equation, denotes the Pearson's correlation coefficient between the FC architecture of the kth ROI at the ith time window and the jth time window. is the averaged correlation coefficient of the FC architecture of the kth ROI between any two different time windows, which measures the averaged similarity of FC profile associated with brain region k across different time windows. A deduction from 1 indicates temporal variability of FC architecture of the region k.
FIGURE 1.
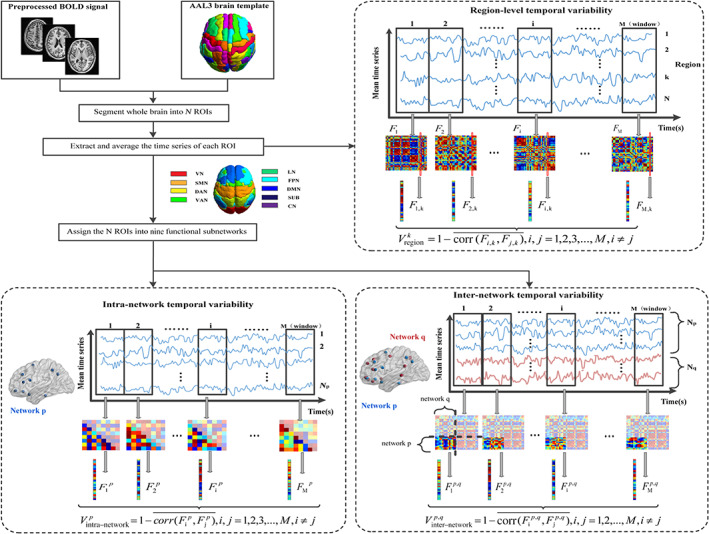
The analysis pipeline for computing the tree types of temporal variabilities. First, the whole brain was segmented into N ROIs according to a parcellation brain atlas such as the AAL3 template. Second, the mean time course of each ROI was extracted from the preprocessed BOLD signal. Third, the mean time series of all ROIs were segmented into M nonoverlapping windows. Forth, within the ith window, a N × N FC matrix F i was generated. We extracted the kth column in F i and denoted it as F i,k . Then, we calculated the regional‐level temporal variability of the kth ROI (). For the network‐level temporal variability, the N ROIs were assigned into nine functional subnetworks. For the network p, N p ROIs were assigned into this network. Within the ith window, a N p *(N p − 1)/2 FC matrix was obtained and reshaped as a 1D vector (). Similar with the regional‐level temporal variability, we computed the intra‐network temporal variability of network p (). However, for the inter‐network variability () between network p with N p ROIs and network q with N q ROIs, a N p *N q FC matrix between these two networks in the ith window was calculated and reshaped as a 1D vector (denoted as ). Finally, we calculated according to the corresponding formula
However, several considerations of the specific analysis processes needed to be explained. Firstly, for the parcellation brain atlas, we choose the automated anatomical labeling atlas 3 (AAL3) template to parcellate the whole brain into 166 ROIs. However, the locus coeruleus, ventral tegmental area, raphe nuclei, substantia nigra, red nucleus, and reuniens nucleus were excluded which should be used with caution for their smaller voxels (Rolls et al., 2020). Therefore, the whole brain was segmented into 152 ROIs (i.e., N = 152). This atlas was used because it has named brain areas that can be related to neurology, and because it subdivided well a large brain area into smaller sections with more specific functions (Rolls et al., 2021). Secondly, to reduce the influence of the starting point of the window for the BOLD signal, we segmented the time series L − 1 times for a given window length L, and the segmented time series started from the sth point (s = 1, 2, …, L − 1). with different starting points s (s = 1, 2, …, L − 1) was calculated, and the obtained L − 1 were averaged as the temporal variability of the kth ROI with the window length L. Thirdly, to avoid the arbitrary choice of the window length, with different window length L (L = 10, 11, 12, …, 30 volumes, equal to 20, 22, 24, …, 60 s) was also calculated. This range was determined based on several previous studies on temporal variability (Dong et al., 2019; Long et al., 2020; Sun et al., 2019; Sun, Zhang, et al., 2020), and we chose their union. The corresponding window number M was the rounded down 205/L. We took the averaged over different window length L as the final temporal variability of the kth ROI.
2.5.3. Temporal variability of network‐level FC architecture
In addition to calculating the temporal variability of the regional‐level FC profile, the temporal variability of FC at the network level was further characterized following recently published procedures (Dong et al., 2019; Sun et al., 2019). There were two major types of network‐level temporal variability: intra‐network variability, which characterizes the changes of FC within subnetwork over time, and inter‐network variability. For this analysis, three major steps should be performed. (1) The N ROIs were assigned into nine functional subnetworks, which were consisted of the subcortical network (SUB), cerebellum (CN) and the seven networks defined by Yeo et al. (2014) including visual network (VN), sensorimotor network (SMN), dorsal attention network (DAN), ventral attention network (VAN), limbic network (LN), frontal‐parietal network (FPN), and default mode network (DMN). (2) For the pth network, N p ROIs were assigned into this network. The number of ROIs of each network satisfied the following formula:
N p *(N p − 1)/2 FCs within this network in the ith window were obtained and reshaped as a 1D vector (denoted as , i = 1, 2, 3, …, M; p = 1, 2, 3, …, 9). Similar with the regional‐level temporal variability, the intra‐network temporal variability of network p ( in short) was defined according to the following equation:
(3) In order to measure the inter‐network variability () between network p with N p ROIs and network q with N q ROIs, N p *N q FCs between these two networks in the ith window were calculated and reshaped as a 1D vector (denoted as , i = 1, 2, 3, …, M; p = 1, 2, 3, …, 9; q = 1, 2, 3, …, 9; p q). Then, was defined as:
Furthermore, in order to improve the robustness of the result, we computed and with different starting points s (s = 1, 2, …, L − 1) and multiple window length L (L = 10, 11, 12, …, 30 volumes) using the similar procedure for the regional‐level temporal variability, and took an average as the final intra‐network variability and inter‐network variability.
2.6. Validation analysis
In order to verify the consistency of the results, we also measured these three variables with the Shen‐268 functional atlas which is an FC‐based brain parcellation generated with a rsfMRI dataset from 79 healthy adults based on the groupwise clustering of voxel‐wise FCs (Shen et al., 2013). This atlas is more suitable for rsfMRI studies because it provides highly homogeneous and functionally coherent brain parcellations, compared with other anatomical information‐based parcellations (Arslan et al., 2018), and it has been widely used in many functional brain studies (Beaty et al., 2018; Bertolero et al., 2015; Rosenberg et al., 2016; Wen et al., 2020).
2.7. Statistical analysis
First, the seven primary behavioral measurements of PVT were compared between RW and SD sessions using paired t‐test with false discovery rate (FDR) correction for multiple comparisons (p <.05). Secondly, in order to explore the effects of SD on temporal variability, we compared the above mentioned three types of temporal variabilities (regional‐level, intra‐network, inter‐network) between RW and SD states using paired t‐test with FDR correction (p <.05). To improve the robustness of the results, we extracted the overlapped results showing significant changes in regional‐level and network‐level temporal variability after SD using the AAL3 template and Shen‐268 functional atlas.
Thirdly, we performed stepwise regression analysis to explore the relationship between temporal variability and behavioral measures of PVT. In statistics, stepwise regression is a method of fitting regression models in which the choice of predictive variables is carried out by an automatic procedure. This method can screen and eliminate the variables which are marginal important and cause multicollinearity, and identify the important variates. Therefore, in consideration of the highly correlation between the seven performance measures, we input these performance measures into the regression model as independent variables, with the temporal variability as the dependent variable, and picked out the independent variables that have significant effect on the dependent variable. This analysis was implemented in the overlapped regions showing significant changes of regional‐level temporal variability (n = 37), one network showing significant changes of intra‐network temporal variability, and the common subnetwork pairs showing significant changes of inter‐network temporal variability (n = 15) after SD using the above two atlases. Moreover, this analysis was accomplished for the SD state and the changes between SD and RW (the differences in temporal variability as the dependent variable and the differences in performance measures as independent variables) with the AAL3 atlas. Then, the FDR correction for multiple comparisons was performed with p <.05 (37 * 2 repeated comparisons for the regional‐level temporal variability, 2 repeated comparisons for the intra‐network temporal variability, and 15*2 repeated comparisons for the inter‐network temporal variability) using MATLAB 2019b. Furthermore, considering the dimensional differences among temporal variability and seven performance measures, independent variables and dependent variable were normalized before stepwise regression analysis.
3. RESULTS
3.1. Behavioral results
Poorer performance of PVT was observed after SD with less number trails (p = .04, p(FDR) = .04), slower mean RT (p = 1.99 × 10−6, p(FDR) = 3.49 × 10−6), more lapse (p = 5.71 × 10−9, p(FDR) = 2.00 × 10−8), increased 10% fast RT (p = 0.0032, p(FDR) = .0037), decreased 10% slow 1/RT (p = 2.33 × 10−11, p(FDR) = 1.63 × 10−10), greater sd_RT (p = 8.34 × 10−6, p(FDR) = 1.17 × 10−5), increased cv_RT (p = 3.78 × 10−7, p(FDR) = 8.81 × 10−7, Table 1).
TABLE 1.
Behavioral changes after SD
| Behavioral measurements | RW | SD | t value | p value | p(FDR) value |
|---|---|---|---|---|---|
| Number trails | 55.38 ± 2.46 | 54.45 ± 2.81 | −2.10 | .04 | 0.04 |
| Lapse | 1.73 ± 2.51 | 7.66 ± 7.22 | 6.91 | 5.71 × 10−9 | 2.00 × 10−8 |
| Mean RT (ms) | 341.74 ± 41.15 | 475.90 ± 209.27 | 5.33 | 1.99 × 10−6 | 3.49 × 10−6 |
| 10% fast RT (ms) | 274.76 ± 25.59 | 286.54 ± 35.02 | 3.09 | .0032 | 0.0037 |
| 10% slow 1/RT(s) | 2.22 ± 0.38 | 1.50 ± 0.74 | −8.39 | 2.33 × 10−11 | 1.63 × 10−10 |
| sd_RT | 68.71 ± 52.32 | 450.69 ± 586.60 | 4.93 | 8.34 × 10−6 | 1.17 × 10−5 |
| cv_RT | 19.74 ± 11.92 | 72.92 ± 67.92 | 5.78 | 3.78 × 10−7 | 8.81 × 10−7 |
Note: Data are presented as mean ± standard deviation.
Abbreviations: cv_RT, the coefficient of variation of RT; sd_RT, the standard deviation of RT; SD, sleep deprivation. RT, reaction time; RW, rested wakefulness.
3.2. Temporal variability of regional‐level FC architecture
For the RW session, subjects showed low regional‐level temporal variability among a large of brain regions (Figure 2a), especially in VN (including the bilateral cuneus, bilateral calcarine, bilateral lingual gyrus, and bilateral fusiform gyrus), DMN (including the bilateral lateral orbital gyrus (OFClat), bilateral medial of superior frontal gyrus (Frontal_Sup_Medial), bilateral medial orbital of superior frontal gyrus (Frontal_Med_Orb), and left inferior frontal gyrus pars orbitalis (Frontal_Inf_Orb)), and VAN (including the bilateral insula, bilateral supramarginal gyrus (SMG), and bilateral middle cingulate & paracingulate gyri (MCC)) with the AAL3 atlas. High regional‐level temporal variability was observed in SUB (including thalamus, putamen and nucleus accumbens (N_Acc)), CN, and bilateral anterior cingulate cortex, supracallosal (ACC_sup) (Figure 2a). These results suggested that the functional architecture of regions in VN, DMN and VAN was highly correlated across different time windows, and the dynamic FC time series between SUB, CN, ACC_sup, and other regions were asynchronous.
FIGURE 2.
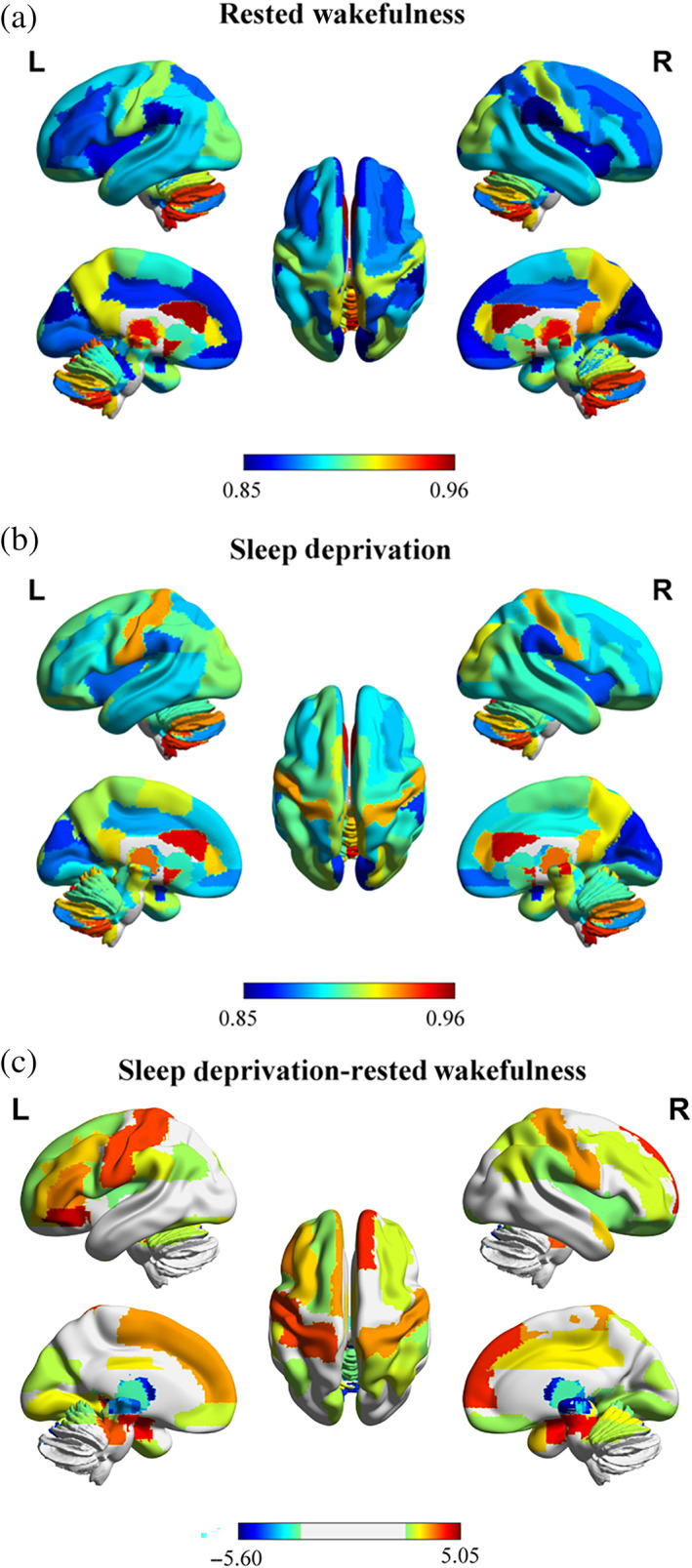
Whole‐brain regional‐level temporal variability topography on AAL3 template after rested wakefulness and sleep deprivation. (a) After rested wakefulness (RW). The color scale represents regional‐level temporal variability. (b) After sleep deprivation (SD). The color scale represents regional‐level temporal variability. (c) Significant changes between RW and SD with FDR correction for multiple comparisons (p <.05). The color scale represents t value. The positive t values mean SD > RW; the negative t values mean SD < RW. R, right. L, left. These figures were constructed using the BrainNet Viewer (http://www.nitrc.org/projects/bnv/; Xia et al., 2013)
For the SD session, low regional‐level temporal variability covered bilateral cuneus, bilateral calcarine, bilateral amygdala, bilateral intralaminar thalamus and bilateral lateral posterior thalamus (Figure 2b). Subjects showed high regional‐level temporal variability in bilateral ACC_sup, bilateral N_Acc, left ventral posterolateral thalamus, bilateral mediodorsal medial magnocellular thalamus, and cerebellum (Figure 2b) with the AAL3 atlas.
Compared with the RW session, subjects exhibited significant increased regional‐level temporal variability in several brain regions after SD with FDR correction (p <.05). The involved regions included the bilateral OFClat, left Frontal_Inf_Orb, bilateral Frontal_Sup_Medial, bilateral hippocampus (HIP), bilateral supplementary motor area (SMA), bilateral postcentral gyrus, bilateral lobule III of cerebellar hemisphere (Cerebellum_3), and so on (Table 2 and Figure 2C). Furthermore, significant reduced regional‐level temporal variability was found in thalamus subregions and vermis after SD with FDR correction (p <.05).
TABLE 2.
Significant changes of regional‐level temporal variability after SD with AAL3 atlas
| Brain region | Hemisphere | RW | SD | t value | p value | p(FDR) value |
|---|---|---|---|---|---|---|
| Superior frontal gyrus, dorsolateral | L | 0.89 ± 0.04 | 0.90 ± 0.03 | 2.53 | .0144 | .0384 |
| Middle frontal gyrus | L | 0.87 ± 0.04 | 0.89 ± 0.03 | 3.29 | .0018 | .0082 |
| R | 0.87 ± 0.03 | 0.89 ± 0.03 | 2.86 | .0061 | .0220 | |
| Inferior frontal gyrus, opercular part | L | 0.88 ± 0.04 | 0.89 ± 0.04 | 2.54 | .0140 | .0384 |
| Inferior frontal gyrus, triangular part | L | 0.87 ± 0.04 | 0.89 ± 0.03 | 3.68 | .0005 | .0030 |
| IFG pars orbitalis | L | 0.88 ± 0.03 | 0.90 ± 0.03 | 4.66 | 1.87 × 10−5 | .0004 |
| Rolandic operculum | L | 0.87 ± 0.04 | 0.89 ± 0.03 | 2.41 | .0194 | .0492 |
| Supplementary motor area | L | 0.89 ± 0.03 | 0.91 ± 0.03 | 3.57 | .0008 | .0038 |
| Superior frontal gyrus, medial | L | 0.87 ± 0.04 | 0.89 ± 0.03 | 3.54 | .0008 | .0040 |
| R | 0.86 ± 0.04 | 0.89 ± 0.04 | 4.20 | .0001 | .0008 | |
| Superior frontal gyrus, medial orbital | L | 0.86 ± 0.04 | 0.88 ± 0.03 | 2.82 | .0067 | .0236 |
| Gyrus rectus | R | 0.87 ± 0.04 | 0.88 ± 0.03 | 2.58 | .0127 | .0358 |
| Anterior orbital gyrus | R | 0.89 ± 0.04 | 0.90 ± 0.03 | 2.51 | .0152 | .0398 |
| Lateral orbital gyrus | L | 0.88 ± 0.04 | 0.90 ± 0.03 | 5.04 | 5.53 × 10−6 | .0002 |
| Insula | R | 0.86 ± 0.04 | 0.87 ± 0.03 | 2.44 | .0182 | .0470 |
| Middle cingulate and paracingulate gyri | R | 0.87 ± 0.04 | 0.89 ± 0.03 | 3.13 | .0028 | .0120 |
| Hippocampus | L | 0.89 ± 0.03 | 0.92 ± 0.02 | 4.41 | 5.01 × 10−5 | .0005 |
| R | 0.89 ± 0.03 | 0.92 ± 0.03 | 4.42 | 4.85 × 10−5 | .0005 | |
| Cuneus | L | 0.85 ± 0.04 | 0.87 ± 0.03 | 2.70 | .0092 | .0285 |
| Lingual gyrus | L | 0.87 ± 0.04 | 0.89 ± 0.04 | 3.03 | .0037 | .0152 |
| R | 0.88 ± 0.04 | 0.90 ± 0.04 | 2.62 | .0115 | .0339 | |
| Fusiform gyrus | L | 0.88 ± 0.03 | 0.90 ± 0.04 | 2.73 | .0086 | .0273 |
| Postcentral gyrus | L | 0.91 ± 0.03 | 0.93 ± 0.02 | 4.04 | .0002 | .0012 |
| R | 0.91 ± 0.02 | 0.93 ± 0.02 | 3.60 | .0007 | .0037 | |
| Superior parietal gyrus | R | 0.87 ± 0.03 | 0.89 ± 0.03 | 2.60 | .0071 | .0239 |
| Inferior parietal gyrus, excluding supramarginal, and angular gyri | R | 0.87 ± 0.04 | 0.89 ± 0.04 | 2.81 | .0068 | .0236 |
| Supramarginal gyrus | L | 0.86 ± 0.04 | 0.88 ± 0.04 | 2.95 | .0047 | .0187 |
| R | 0.85 ± 0.04 | 0.87 ± 0.04 | 2.53 | .0144 | .0384 | |
| Angular gyrus | L | 0.89 ± 0.03 | 0.90 ± 0.03 | 2.61 | .0118 | .0339 |
| R | 0.89 ± 0.03 | 0.90 ± 0.03 | 2.91 | .0053 | .0200 | |
| Temporal pole: superior temporal gyrus | R | 0.89 ± 0.04 | 0.91 ± 0.03 | 3.27 | .0019 | .0084 |
| Lobule III of cerebellar hemisphere | L | 0.86 ± 0.03 | 0.88 ± 0.04 | 4.44 | 4.45 × 10−5 | .0005 |
| R | 0.87 ± 0.04 | 0.89 ± 0.04 | 3.94 | .0002 | .0015 | |
| Lobule IV, V of vermis | 0.94 ± 0.02 | 0.92 ± 0.03 | −2.65 | .0105 | .0320 | |
| Lobule VII of vermis | 0.92 ± 0.03 | 0.90 ± 0.04 | −4.55 | 3.10 × 10−5 | .0004 | |
| Lobule VIII of vermis | 0.92 ± 0.03 | 0.90 ± 0.04 | −4.56 | 2.96 × 10−5 | .0004 | |
| Lobule IX of vermis | 0.94 ± 0.02 | 0.93 ± 0.02 | −2.87 | .0059 | .0220 | |
| Lobule X of vermis | 0.94 ± 0.02 | 0.92 ± 0.04 | −4.30 | 7.16 × 10−5 | .0006 | |
| Thalamus, anteroventral nucleus | L | 0.91 ± 0.03 | 0.89 ± 0.03 | −4.71 | 1.80 × 10−5 | .0004 |
| R | 0.92 ± 0.03 | 0.89 ± 0.04 | −5.49 | 1.10 × 10−6 | 7.08 × 10−5 | |
| Lateral posterior | L | 0.90 ± 0.04 | 0.86 ± 0.04 | −5.60 | 7.51 × 10−7 | 7.08 × 10−5 |
| R | 0.89 ± 0.03 | 0.87 ± 0.04 | −4.30 | 7.14 × 10−5 | .0006 | |
| Ventral anterior | L | 0.91 ± 0.03 | 0.88 ± 0.04 | −3.60 | .0007 | .0037 |
| R | 0.91 ± 0.03 | 0.89 ± 0.04 | −3.43 | .0012 | .0056 | |
| Ventral lateral | L | 0.92 ± 0.03 | 0.89 ± 0.04 | −5.03 | 5.74 × 10−6 | .0002 |
| R | 0.92 ± 0.03 | 0.89 ± 0.04 | −4.34 | 6.35 × 10−5 | .0006 | |
| Ventral posterolateral | R | 0.94 ± 0.03 | 0.92 ± 0.03 | −3.88 | .0003 | .0017 |
| Intralaminar | L | 0.89 ± 0.03 | 0.87 ± 0.04 | −3.87 | .0003 | .0017 |
| R | 0.89 ± 0.03 | 0.86 ± 0.04 | −4.36 | 5.97 × 10−5 | .0006 | |
| Mediodorsal medial magnocellular | L | 0.95 ± 0.02 | 0.93 ± 0.03 | −2.92 | .0051 | .0197 |
| Mediodorsal lateral parvocellular | L | 0.94 ± 0.03 | 0.92 ± 0.03 | −2.74 | .0084 | .0273 |
| R | 0.94 ± 0.02 | 0.92 ± 0.03 | −2.61 | .0118 | .0339 | |
| Lateral geniculate | L | 0.92 ± 0.03 | 0.91 ± 0.03 | −3.26 | .0021 | .0093 |
| R | 0.92 ± 0.02 | 0.90 ± 0.04 | −4.14 | .0001 | .0009 | |
| Medial Geniculate | L | 0.91 ± 0.03 | 0.88 ± 0.04 | −4.56 | 3.02 × 10−5 | .0004 |
| R | 0.91 ± 0.03 | 0.88 ± 0.04 | −4.64 | 2.23 × 10−5 | .0004 | |
| Pulvinar anterior | L | 0.91 ± 0.03 | 0.88 ± 0.04 | −5.43 | 1.40 × 10−6 | 7.08 × 10−5 |
| R | 0.92 ± 0.03 | 0.88 ± 0.04 | −4.83 | 1.19 × 10−5 | .0003 | |
| Pulvinar medial | R | 0.92 ± 0.03 | 0.90 ± 0.05 | −3.98 | .0002 | .0014 |
| Pulvinar inferior | R | 0.87 ± 0.03 | 0.88 ± 0.04 | 2.73 | .0085 | .0273 |
Note: Data are presented as mean ± standard deviation.
Abbreviations: L, left; R, right; RW, rested wakefulness; SD, sleep deprivation.
3.3. Temporal variability of network‐level FC architecture
As shown in Figure 3 and Table S1, sleep‐deprived subjects showed increased intra‐network temporal variability in DMN and SMN, while decreased intra‐network temporal variability was found in SUB and CN after FDR corrected (p <.05) when using the AAL3 atlas.
FIGURE 3.
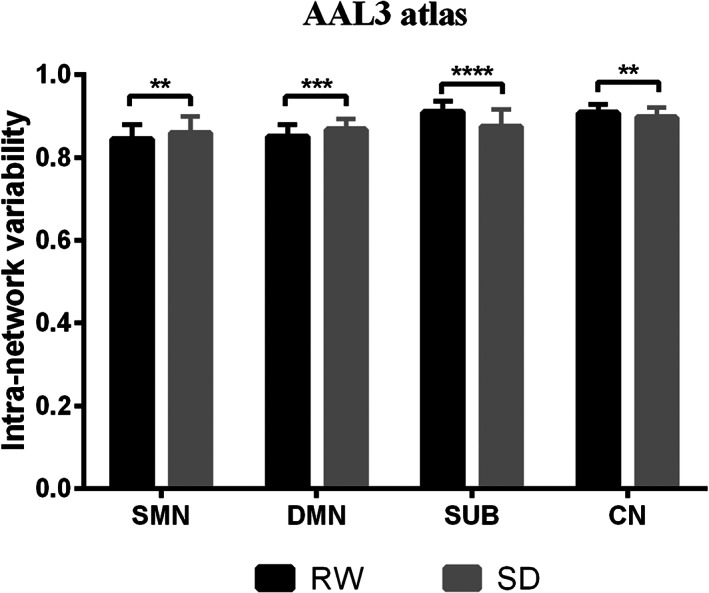
Significant changes in intra‐network temporal variability between RW and SD with the AAL3 atlas. Subjects showed increased intra‐network temporal variability in DMN and SMN and decreased in SUB and CN after SD with false discovery rate (FDR) correction (p <.05). CN, cerebellum; DMN, default mode network; RW, rested wakefulness; SD, sleep deprivation; SMN, sensorimotor network; SUB, subcortical network. ****p <.0001. ***p <.001. **p <.01
Among all the 36 inter‐network temporal variability (9 × 8/2), we found 20 significant increased inter‐network temporal variability and one decreased inter‐network temporal variability (SUB‐CN) after SD with FDR correction (p <.05) using the AAL3 atlas (Table 3 and Figure 4).
TABLE 3.
The significant changes of inter‐network temporal variability between RW and SD with AAL3 atlas
| Paired network | RW | SD | t value | p value | p(FDR) value |
|---|---|---|---|---|---|
| VN‐SMN | 0.90 ± 0.03 | 0.92 ± 0.03 | 4.13 | .0001 | .0015 |
| VN‐DMN | 0.92 ± 0.02 | 0.94 ± 0.02 | 3.32 | .0016 | .0065 |
| SMN‐DAN | 0.89 ± 0.04 | 0.91 ± 0.05 | 2.79 | .0073 | .0164 |
| SMN‐VAN | 0.82 ± 0.06 | 0.86 ± 0.06 | 3.58 | .0007 | .0038 |
| SMN‐FPN | 0.91 ± 0.02 | 0.93 ± 0.03 | 3.45 | .0011 | .0049 |
| SMN‐DMN | 0.90 ± 0.03 | 0.91 ± 0.02 | 3.24 | .0021 | .0074 |
| SMN‐CN | 0.94 ± 0.02 | 0.95 ± 0.02 | 3.05 | .0035 | .0098 |
| DAN‐LN | 0.93 ± 0.04 | 0.95 ± 0.04 | 2.37 | .0215 | .0408 |
| DAN‐FPN | 0.84 ± 0.06 | 0.86 ± 0.05 | 3.20 | .0023 | .0075 |
| DAN‐DMN | 0.87 ± 0.05 | 0.90 ± 0.04 | 3.00 | .0041 | .0106 |
| DAN‐CN | 0.92 ± 0.04 | 0.94 ± 0.04 | 2.27 | .0272 | .0489 |
| VAN‐LN | 0.90 ± 0.04 | 0.93 ± 0.04 | 4.68 | 1.98 × 10−5 | .0007 |
| VAN‐FPN | 0.86 ± 0.05 | 0.89 ± 0.05 | 3.60 | .0007 | .0038 |
| VAN‐DMN | 0.85 ± 0.04 | 0.87 ± 0.04 | 3.05 | .0036 | .0098 |
| VAN‐CN | 0.93 ± 0.03 | 0.95 ± 0.03 | 2.65 | .0105 | .0223 |
| LN‐FPN | 0.91 ± 0.03 | 0.92 ± 0.03 | 2.88 | .0057 | .0138 |
| LN‐DMN | 0.89 ± 0.02 | 0.91 ± 0.02 | 3.79 | .0004 | .0027 |
| FPN‐DMN | 0.88 ± 0.03 | 0.90 ± 0.03 | 4.10 | .0001 | .0015 |
| FPN‐CN | 0.94 ± 0.02 | 0.95 ± 0.02 | 2.39 | .0205 | .0408 |
| DMN‐CN | 0.94 ± 0.01 | 0.95 ± 0.02 | 2.25 | .0285 | .0489 |
| SUB‐CN | 0.95 ± 0.01 | 0.94 ± 0.02 | −4.05 | .0002 | .0015 |
Note: Data are presented as mean ± standard deviation.
Abbreviations: CN, cerebellum; DAN, dorsal attention network; DMN, default mode network; FPN, frontal‐parietal network; LN, limbic network; RW, rested wakefulness; SD, sleep deprivation; SMN, sensorimotor network; SUB, subcortical network; VAN, ventral attention network; VN, visual network.
FIGURE 4.
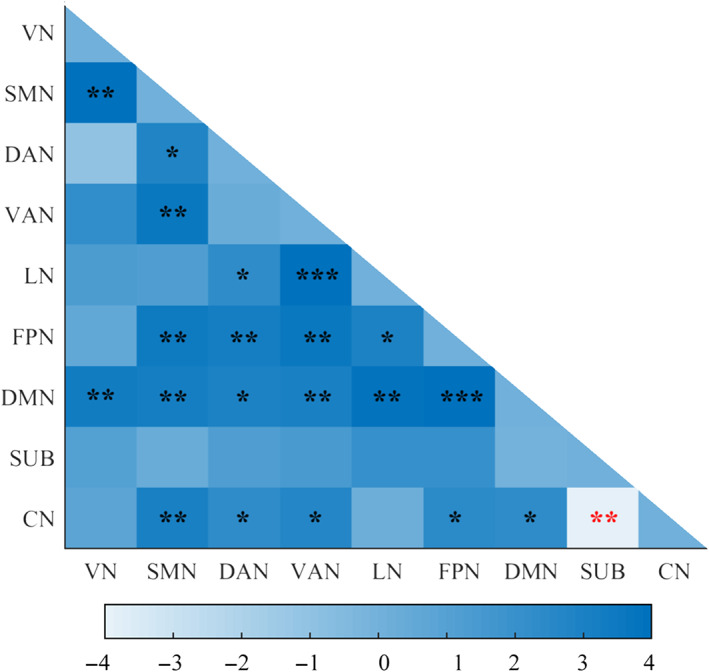
Significant changes in inter‐network temporal variability between RW and SD using the AAL3 atlas after FDR correction (p <.05). The color scale represents t value. The positive t values mean SD > RW; the negative t values mean SD < RW. Black asterisks (*) indicate the subnetwork pairs showing significantly increased inter‐network temporal variability after SD compared with RW. Red asterisks indicate the subnetwork pairs showing significantly decreased inter‐network temporal variability after SD compared with RW. ****p <.0001. ***p <.001. **p <.01. *p <.05. CN, cerebellum; DAN, dorsal attention network; DMN, default mode network; LN, limbic network; FDR, false discovery rate; FPN, frontal‐parietal network; SD, sleep deprivation; SMN, sensorimotor network; SUB, subcortical network; VAN, ventral attention network; VN, visual network; RW, rested wakefulness
3.4. Validation results
For the regional‐level temporal variability, subjects showed significant increased regional‐level temporal variability in a wide range of brain regions which spanned VN, SMN, DAN, VAN, LN, FPN, DMN and CN after SD with FDR correction (p <.05, Figure S1) using the Shen‐268 functional atlas. We also found significant reduced regional‐level temporal variability in thalamus and cerebellum.
In order to improve the robustness of the results, we extracted the overlapped brain regions showing significant changes in regional‐level temporal variability after SD using the two brain region templates. We found that compared with RW session, the regional‐level temporal variability was significant changed after SD under both the AAL3 and Shen‐268 atlas in VN (including the left cuneus, bilateral lingual gyrus and left fusiform gyrus), SMN (including the left Rolandic operculum, bilateral postcentral gyrus and left SMA), the right superior parietal gyrus, VAN (including the right insula, right MCC, and bilateral SMG), LN (including the right gyrus rectus, right HIP, and right temporal pole: superior temporal gyrus), FPN (including the bilateral middle frontal gyrus, left inferior frontal gyrus, opercular part, left inferior frontal gyrus, triangular part, left anterior orbital gyrus, and right inferior parietal gyrus), and DMN (including the left superior frontal gyrus, dorsolateral, left Frontal_Inf_Orb, bilateral Frontal_Sup_Medial, left Frontal_Med_Orb, left OFClat, and bilateral angular gyrus). All of these brain regions exhibited increased regional‐level temporal variability after SD. Meanwhile, we also found that several thalamus subregions showed reduced regional‐level temporal variability after SD using both the two brain templates (Table S2 and Figure 5).
FIGURE 5.
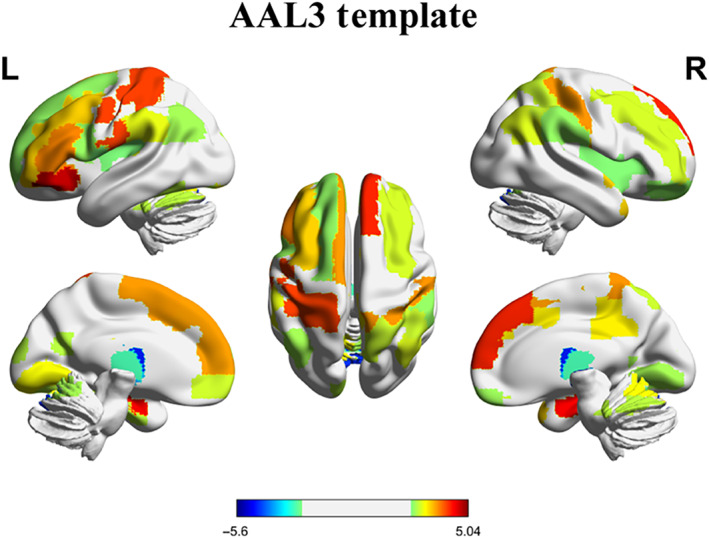
The overlapped brain regions showing significant changes of regional‐level temporal variability using the AAL3 template and Shen‐268 functional atlas after SD. The color scale represents t value. The positive t values mean SD > RW; the negative t values mean SD < RW. R, right. L, left. RW, rested wakefulness; SD, sleep deprivation. This figure was constructed using the BrainNet Viewer (http://www.nitrc.org/projects/bnv/; Xia et al., 2013)
For the intra‐network temporal variability, higher intra‐network temporal variability in DAN, FPN and DMN was observed after SD after FDR correction (p <.05) using the Shen‐268 functional atlas. No network showed significant decreased intra‐network temporal variability after SD (Figure S2 and Table S1). However, the intra‐network temporal variabilities in DMN in AAL3 atlas and in Shen‐268 functional atlas had significant difference between RW and SD, and showed the similar change trend.
For the inter‐network temporal variability, we found 19 significant increased inter‐network temporal variability and 4 decreased inter‐network temporal variability (between LN, VN, VAN, DMN, and SUB) after SD with FDR correction (p <.05) in the Shen‐268 functional atlas (Table S3 and Figure S3). However, there were 15 inter‐network temporal variability which showed significant changes after SD using the two atlases (Figure 6): VN‐SMN, VN‐DMN, SMN‐DAN, SMN‐DMN, DAN‐LN, DAN‐FPN, DAN‐DMN, DAN‐CN, VAN‐FPN, VAN‐DMN, VAN‐CN, LN‐FPN, FPN‐DMN, FPN‐CN, and DMN‐CN. All of these subnetwork pairs showed increased inter‐network temporal variability after SD.
FIGURE 6.
The common subnetwork pairs showing significant changes of inter‐network temporal variability after SD using the AAL3 atlas and Shen‐268 functional atlas. All of these subnetwork pairs showed increased inter‐network temporal variability after SD. CN, cerebellum; DAN, dorsal attention network; DMN, default mode network; FPN, frontal‐parietal network; LN, limbic network; SD, sleep deprivation; SMN, sensorimotor network; VAN, ventral attention network; VN, visual network. These figures were constructed using the BrainNet Viewer (http://www.nitrc.org/projects/bnv/; Xia et al., 2013)
3.5. Correlations between temporal variability and clinical variables
For SD state, we found that only the 10% slow 1/RT could be an important predictor of the inter‐network temporal variability between VN and DMN (β = −.42, p = 5.57 × 10−4) following the stepwise regression analysis (Figure 7). These results suggest that the 10% slow 1/RT have significant negative influence on the VN‐DMN inter‐network temporal variability. However, no significant relationship between the differences of temporal variability and the differences in PVT measurements.
FIGURE 7.
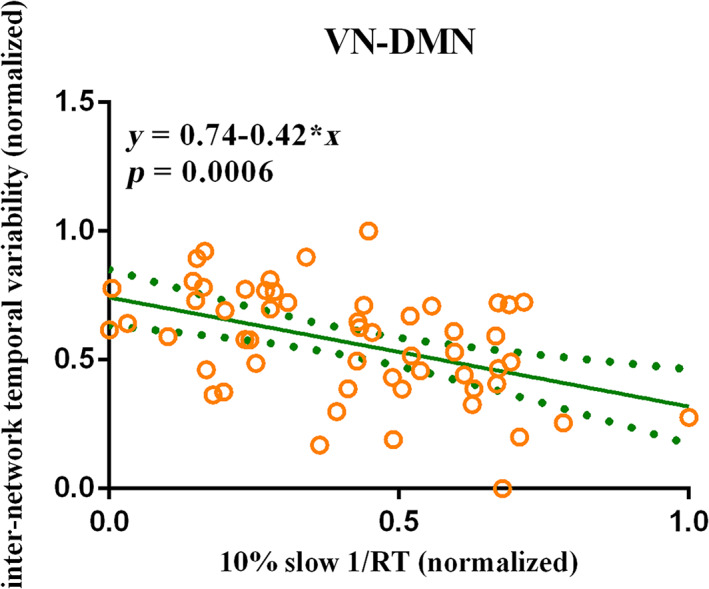
The temporal variability is correlated with the performance of PVT in SD state. The 10% slow 1/RT showed significant negative correlation with the inter‐network temporal variability between VN and DMN in SD state after FDR correlation with the AAL3 atlas (β = −.42, p = 5.57 × 10−4) following the stepwise regression analysis. Considering the dimensional differences among temporal variability and performance measures, the 10% slow 1/RT and the inter‐network temporal variability between VN and DMN were normalized. The green solid line indicated the linear regression of the correlation. The green dotted line indicated the error bars. DMN, default mode network; FDR, false discovery rate; PVT, psychomotor vigilance test; RT, reaction time; SD, sleep deprivation; VN, visual network
4. DISCUSSION
In the present study, we adopted a novel metric (temporal variability) to explore the resting state dynamic FC alterations after SD. Our main findings were as follows. (1) At the regional‐level, compared with RW session, sleep‐deprived subjects showed widespread significant alterations including increased temporal variability in several brain regions such as visual cortices, primary sensorimotor cortices, FPN, DMN, and so on, as well as decreased temporal variability was found in several thalamus subregions after SD (Figure 5 and Table S2). (2) For the intra‐network temporal variability, increased intra‐network temporal variability in DMN were observed after SD. (3) Subjects exhibited increased inter‐network temporal variability in 15 subnetwork pairs such as VN‐DMN, FPN‐DMN, and so on after SD (Figures 4 and 6). (4) The inter‐network temporal variability between VN and DMN showed significant negative relationship with the 10% slow 1/RT of PVT in SD state with the AAL3 atlas (Figure 7). These results suggest that sleep‐deprived subjects showed widespread functional network abnormalities in term of temporal variability, and the abnormal temporal variability also correlated with behavior performance and thus offering new insights in understanding SD.
For the RW state, we found that very low regional‐level temporal variability was showed in regions of VN, DMN, and VAN, such as cuneus, calcarine, fusiform gyrus, medial frontal gyrus and insula, which is consistent with the findings of Zhang et al. in healthy control subjects (Zhang et al., 2016; Figure 2a and Figure S1a). However, high regional‐level temporal variability was showed in regions of SUB and CN, which was not found by Zhang et al. These different findings may result from the different brain templates. Zhang et al. calculate the regional‐level temporal variability in the 90‐regions of AAL template which exclude CN and take the thalamus as a whole ROI. We measured the regional‐level temporal variability in 152 regions of AAL3 template and 268 regions of Shen atlas, which contain the CN and divide the thalamus into several subregions.
After SD, increased regional‐level temporal variability was found in large‐scale regions of functional network (VN, SMN, VAN, LN, FPN, and DMN), which indicated that the dynamical FC series between these regions and all other regions remained independent. Previous neuroimaging studies have shown increased regional spontaneous fluctuations in visual cortex and sensorimotor cortex after SD using the amplitude of low‐frequency fluctuations (ALFFs) (Cai et al., 2021), percent amplitude of fluctuation (PerAF) (Zeng et al., 2020) and regional homogeneity (ReHo) (Dai et al., 2012). Qi et al. (2021) have explored the effect of SD on FC in insula, which is a core hub of VAN. They find increased FC between the insula and prefrontal cortex and ACC, and reduced FC between the insula and temporal, parietal, and occipital regions after SD. For the LN, Li et al. have found that SD reduces the FC between the hippocampus and SMA, SFG, and temporal regions, and increases the FC with thalamus, using a ROI‐to‐voxel analysis (Chengyang et al., 2017). Furthermore, our previous study has investigated the effect of SD on the FC of hippocampus subregions using the masked ICA, and found differential effects of SD on the FC in specific hippocampal regions (Zhao, Zhang, Zhu, et al., 2019). Previous studies have reported that cognitive decline after SD is associated with imbalances in functional brain networks such as DMN, FPN, and DAN (Wirsich et al., 2018). Abnormal FC within FPN and DMN after SD have been showed in several rsfMRI studies (Dai et al., 2020; Yeo et al., 2015). Jointly with our results, these findings suggested that brain regions showed abnormal functional integration after SD.
An interesting finding was the significant decreased regional‐level temporal variability in several thalamus subregions after SD, which suggested that the enhanced synchronization of FC time series between thalamus and all other regions during the scanning time. Thalamus is a vital region which seems to integrate neural activity from widespread neocortical inputs and outputs (Postuma & Dagher, 2006). The vast majority of studies show that thalamus plays a key central role in the sleep–wake pathway and involved in a variety of brain cognitive functions (Chee et al., 2008, 2010; Gent et al., 2018; Krause et al., 2017). Thalamus has strong reciprocal connections with the cerebral cortex which suggests that thalamus modulates and facilitates communication in all areas of the cerebral cortex (Shao et al., 2013). The higher synchronization in thalamus may contribute to maintaining cognitive performance when arousal is low after SD. This result is consistent with previous studies that found increased thalamic activation during working memory task and attention tasks (Ma et al., 2015), increased ALFF in the thalamus (Cai et al., 2021), and increased voxel‐mirrored homotopic connectivity (Zhu et al., 2016) after SD. These results might represent a compensatory mechanism to maintain cognitive performance after SD.
For the intra‐network temporal variability, we found inconsistent results between AAL3 atlas and Shen‐268 template. Increased intra‐network temporal variability in DMN and SMN and decreased intra‐network temporal variability in SUB and CN were showed with the AAL3 atlas after SD. However, for the Shen‐268 functional atlas, higher intra‐network temporal variability in DAN, FPN, and DMN was observed after SD. These inconsistent findings may result from the different brain partitions. AAL3 atlas is an update on the AAL which is widely used in neuroimaging research. AAL3 atlas subdivide several brain which are of interest in many neuroimaging investigations into small regions, such as ACC and thalamus (Rolls et al., 2020). The Shen‐268 functional atlas is an FC‐based brain parcellation using graph‐theory‐based approaches (Shen et al., 2013), which segments the whole brain into more small regions. These two partitions have given rise to different number and scope of brain regions in each network when we assign the N ROIs into nine functional subnetworks. For example, if one region in AAL3 distributes over two networks, we assign this region into the networks in which the region has more voxels. Therefore, the discrepant assigned networks may cause different intra‐network temporal variability, and then result in the inconsistent findings.
For the inter‐network temporal variability, we found that 15 inter‐network temporal variability was significant changed after SD using both the two templates, and that all of them were increased after SD. These results implied an unstable FC architecture and more dynamic interactions between subnetworks. Previous studies have reported the abnormal FC patterns among cortical networks after SD, including the stronger FC between DAN and DMN, and decreased FC between FPN and DMN (Dai et al., 2020; Yeo et al., 2015). Furthermore, recent studies have investigated the alterations of time‐varying fluctuations of FC after SD in global level using a sliding‐window correlations approach, and found that the mean dwell time and fraction of two brain connectivity states were significantly altered after SD (C. Li et al., 2020; Xu et al., 2018). Together with our observations, these results suggested widespread aberrant dynamic brain network reconfigurations after SD.
Taken together, we found that the regional‐level temporal variability in several brain regions of DMN, the intra‐network temporal variability in DMN and the inter‐network temporal variability between DMN and other subnetworks such as VN, SMN, DAN, VAN, FPN, and CN were increased after SD. Furthermore, the inter‐network temporal variability between VN and DMN was significant negative related with the 10% slow 1/RT of PVT in SD state with the AAL3 atlas using the stepwise regression analysis. These results suggested that subjects who showed poor behavior performance after SD, would exhibit greater asynchrony of the dynamical FC time series between VN and DMN. Previous studies have reported hypoconnectivity within DMN (De Havas et al., 2012) and between the right precuneus (a key node of DMN) and the right middle frontal gyrus after SD (B. Li et al., 2020). Moreover, the decreased precuneus FC is correlated with the alertness decline after SD. Jointly with our present observations, these results suggest that SD disrupts resting DMN function. DMN is a very special brain network because it is often more activated in the resting state than in the task state, is the neural basis of the self and is related to maintaining the awake state. Therefore, the increased temporal variability in DMN and the relationship with PVT may indicate that DMN is a hub network in the neural foundations of SD‐related cognitive deficit.
Considering that SD may be a potential causal factor of several neurological disorders and psychiatric disorders (Bishir et al., 2020; Waite et al., 2020), we then discussed the relationship between the significant changes of brain functional variability in SD and clinical diseases. Dong et al. (2019) have examined the alterations of region‐level and network‐level temporal variability in schizophrenia, and reported increased regional‐level temporal variability in thalamus and regions of VN, SMN, DAN, and VAN, decreased in regions of FPN and DMN; increased intra‐network temporal variability in VN, SMN and thalamus, decreased in FPN and DMN; increased inter‐network temporal variability (VN‐thalamus, SMN‐VAN, and SMN‐thalamus) and decreased between FPN and DMN compared to healthy subjects. Zhu, et al. (H. Zhu et al., 2019) have performed the similar analysis in Parkinson's disease, and found increased regional‐level temporal variability in putamen and regions of VN, SMN and CN; increased intra‐network temporal variability in VN, salience network and SUB; increased inter‐network temporal variability in several subnetwork pairs associated with SMN, VN, SUB, and CN compared to normal people. Furthermore, Gu et al. (2020) have reported decreased regional‐level temporal variability in several regions in VN, SMN, control network, and DMN in Alzheimer's disease. Combining these findings with our results, we found aberrant temporal variability in VN and SMN both after SD and in the above‐mentioned diseases, which indicated that the brain function in VN and SMN might be more vulnerable. We also found abnormal temporal variability in thalamus, FPN and DMN both after SD and in schizophrenia. These results were partially in agreement with the viewpoint that experimentally controlled SD was a valuable experimental medicine model of schizophrenia (Ettinger & Kumari, 2015; Kumari & Ettinger, 2020). However, the alteration of temporal variability in thalamus, FPN and DMN in sleep‐deprived subjects and schizophrenia patients was in the reverse direction. Although recent studies have found that SD‐induced cognitive impairments are similar to those in schizophrenia patients and asserted that SD is a useful schizophrenia model (Kumari & Ettinger, 2020), the reverse alterations indicated that the similarity between the neural mechanisms of SD and schizophrenia should be further studied.
The present study has several limitations that should be considered. Firstly, one night of SD can cause substantial deterioration of multiple types of cognitive performance. We only examined the relationship between temporal variability metrics and PVT performance. Further researches should investigate variability metrics and their association with other cognitive performance to establish the relationship between dynamic analysis and behavioral performance. Secondly, compared the results of AAL3 atlas with Shen‐268 functional, we did not entirely replicate the findings from one atlas with the other which were also found in H. Zhu et al. (2019). These findings suggested that the temporal variability might be partly dependent on brain template. Further researches should study the stability of the three temporal variability measures using test retest data and more brain templates. Thirdly, we did not objectively monitor the real‐time sleep state during resting‐state scans. In further studies, electroencephalography (EEG) monitoring is necessary to exclude the interference of microsleeps. Forth, in this study, we did not straightforwardly explore the relationship between SD and clinical diseases. Further studies should explain this relationship by vertical comparison that long‐term follow up sleep‐deprived subjects. Furthermore, we monitored subjects' sleep before scanning using sleep diary. Further researches should use actigraphy or EEG to verify subjects' compliance to a regular sleep schedule.
5. CONCLUSIONS
In conclusion, the present study explored the effect of SD on dynamic FC from temporal variability perspective. We identified increased regional‐level temporal variability in large‐scale regions of functional network (VN, SMN, VAN, LN, FPN, and DMN), and decreased regional‐level temporal variability in several thalamus subregions after SD. Increased intra‐network temporal variability in DMN was observed after SD. Several subnetwork pairs also showed increased inter‐network temporal variability after SD. Furthermore, the inter‐network temporal variability between DMN and VN were negative correlated with the performance of PVT. These findings suggested that participants showed widespread abnormal dynamic FC configuration which provide new insights into the neural underpinnings of SD and advance our understanding of the pathophysiology of clinical disorders.
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
Supporting information
Figure S1 Whole‐brain regional‐level temporal variability topography on Shen‐264 functional template after rested wakefulness and sleep deprivation. (a) After rested wakefulness (RW). The color scale represents regional‐level temporal variability. (b) After sleep deprivation (SD). The color scale represents regional‐level temporal variability. (c) Significant changes between RW and SD with false discovery rate (FDR) correction for multiple comparisons (p <.05). The color scale represents t value. The positive t values mean SD > RW; the negative t values mean SD < RW. R, right. L, left. These figures were constructed using the BrainNet Viewer (http://www.nitrc.org/projects/bnv/; Xia et al., 2013).
Figure S2 Significant changes in intra‐network temporal variability between RW and SD with the Shen‐268 functional atlas. Higher intra‐network temporal variability in DAN, FPN, and DMN was observed after SD after false discovery rate (FDR) correction (p <.05). No network showed significant changed intra‐network variability after SD. DMN, default mode network; DAN, dorsal attention network; FPN, frontal‐parietal network; RW, rested wakefulness; SD, sleep deprivation. ****p <.0001. ***p <.001.
Figure S3 Significant changes in inter‐network temporal variability between RW and SD using the Shen‐268 functional atlas after FDR correction (p <.05). The color scale represents t value. The positive t values mean SD > RW; the negative t values mean SD < RW. Black asterisks (*) indicate the subnetwork pairs showing significantly increased inter‐network temporal variability after SD compared with RW. Red asterisks indicate the subnetwork pairs showing significantly decreased inter‐network temporal variability after SD compared with RW. ****p <.0001. ***p <.001. **p <.01. *p <.05. CN, cerebellum; DAN, dorsal attention network; DMN, default mode network; FDR, false discovery rate; LN, limbic network; FPN, frontal‐parietal network; SD, sleep deprivation; SMN, sensorimotor network; SUB, subcortical network; RW, rested wakefulness; VAN, ventral attention network; VN, visual network.
Table S1 The significant changes of intra‐network temporal variability between RW and SD.
Table S2 The overlapped brain regions which showed significant changes in regional‐level temporal variability after SD using the AAL3 template and Shen‐268 functional atlas.
Table S3 The significant changes of inter‐network temporal variability between RW and SD with SHEN‐264 atlas.
ACKNOWLEDGMENT
We thank the subjects whose participation enabled this work.
Sun, J. , Zhao, R. , He, Z. , Chang, M. , Wang, F. , Wei, W. , Zhang, X. , Zhu, Y. , Xi, Y. , Yang, X. , & Qin, W. (2022). Abnormal dynamic functional connectivity after sleep deprivation from temporal variability perspective. Human Brain Mapping, 43(12), 3824–3839. 10.1002/hbm.25886
Jinbo Sun and Rui Zhao authors contributed equally to this work.
Funding information Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine, Grant/Award Number: ZYYCXTD‐C‐202004; National Science Foundation of China, Grant/Award Numbers: 81801772, 81901827; Natural Science basic Research Program of Shaanxi Province, Grant/Award Numbers: 2020JQ‐836, 2020JQ‐837, 2021JQ‐211, 2022JM‐146; the Fundamental Research Funds for the Central Universities, Grant/Award Number: XJS201209; the National Key RD Program of China, Grant/Award Number: 2021YFF0306500; the PhD start‐up fund of Xi'an Polytechnic University, Grant/Award Number: BS201914
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- Allen, E. A. , Damaraju, E. , Plis, S. M. , Erhardt, E. B. , Eichele, T. , & Calhoun, V. D. (2014). Tracking whole‐brain connectivity dynamics in the resting state. Cerebral Cortex, 24, 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, J. L. R. , Hutton, C. , Ashburner, J. , Turner, R. , & Friston, K. (2001). Modeling geometric deformations in EPI time series. NeuroImage, 13, 903–919. [DOI] [PubMed] [Google Scholar]
- Arslan, S. , Ktena, S. I. , Makropoulos, A. , Robinson, E. C. , Rueckert, D. , & Parisot, S. (2018). Human brain mapping: A systematic comparison of parcellation methods for the human cerebral cortex. NeuroImage, 170, 5–30. [DOI] [PubMed] [Google Scholar]
- Ashburner, J. , & Friston, K. J. (2005). Unified segmentation. NeuroImage, 26, 839–851. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay, A. , & Sigua, N. L. (2019). What is sleep deprivation? American Journal of Respiratory Critical Care Medicine, 199, P11–P12. [DOI] [PubMed] [Google Scholar]
- Beaty, R. E. , Kenett, Y. N. , Christensen, A. P. , Rosenberg, M. D. , Benedek, M. , Chen, Q. , Fink, A. , Qiu, J. , Kwapil, T. R. , Kane, M. J. , & Silvia, P. J. (2018). Robust prediction of individual creative ability from brain functional connectivity. Proceedings of the National Academy of Sciences of the United States of America, 115, 1087–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behzadi, Y. , Restom, K. , Liau, J. , & Liu, T. T. (2007). A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. NeuroImage, 37, 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolero, M. A. , Yeo, B. T. , & D'Esposito, M. (2015). The modular and integrative functional architecture of the human brain. Proceedings of the National Academy of Sciences of the United States of America, 112, E6798–E6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishir, M. , Bhat, A. , Essa, M. M. , Ekpo, O. , Ihunwo, A. O. , Veeraraghavan, V. P. , Mohan, S. K. , Mahalakshmi, A. M. , Ray, B. , Tuladhar, S. , Chang, S. , Chidambaram, S. B. , Sakharkar, M. K. , Guillemin, G. J. , Qoronfleh, M. W. , & Ojcius, D. M. (2020). Sleep deprivation and neurological disorders. BioMed Research International, 2020, 5764017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton, T. A. W. , Morgenroth, E. , Preti, M. G. , & Van De Ville, D. (2020). Tapping into multi‐faceted human behavior and psychopathology using fMRI brain dynamics. Trends in Neurosciences, 43, 667–680. [DOI] [PubMed] [Google Scholar]
- Cai, Y. , Mai, Z. , Li, M. , Zhou, X. , & Ma, N. (2021). Altered frontal connectivity after sleep deprivation predicts sustained attentional impairment: A resting‐state functional magnetic resonance imaging study. Journal of Sleep Research, 30, e13329. [DOI] [PubMed] [Google Scholar]
- Cassé‐Perrot, C. , Lanteaume, L. , Deguil, J. , Bordet, R. , Auffret, A. , Otten, L. , Blin, O. , Bartrés‐Faz, D. , & Micallef, J. (2016). Neurobehavioral and cognitive changes induced by sleep deprivation in healthy volunteers. CNS & Neurological Disorders ‐ Drug Targets, 15, 777–801. [DOI] [PubMed] [Google Scholar]
- Chee, M. W. , Tan, J. C. , & Parimal, S. (2010). Sleep deprivation and its effects on object‐selective attention. Neuroimage, 49, 1903–1910. [DOI] [PubMed] [Google Scholar]
- Chee, M. W. , Tan, J. C. , Zheng, H. , Parimal, S. , Weissman, D. H. , Zagorodnov, V. , & Dinges, D. F. (2008). Lapsing during sleep deprivation is associated with distributed changes in brain activation. The Journal of Neuroscience, 28, 5519–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chee, M. W. L. , & Zhou, J. (2019). Functional connectivity and the sleep‐deprived brain. Progress in Brain Research, 246, 159–176. [DOI] [PubMed] [Google Scholar]
- Chengyang, L. , Daqing, H. , Jianlin, Q. , Haisheng, C. , Qingqing, M. , Jin, W. , Jiajia, L. , Enmao, Y. , Yongcong, S. , & Xi, Z. (2017). Short‐term memory deficits correlate with hippocampal‐thalamic functional connectivity alterations following acute sleep restriction. Brain Imaging Behavior, 11, 954–963. [DOI] [PubMed] [Google Scholar]
- Cousins, J. N. , & Fernández, G. (2019). The impact of sleep deprivation on declarative memory. Progress in Brain Research, 246, 27–53. [DOI] [PubMed] [Google Scholar]
- Dai, C. , Zhang, Y. , Cai, X. , Peng, Z. , Zhang, L. , Shao, Y. , & Wang, C. (2020). Effects of sleep deprivation on working memory: Change in functional connectivity between the dorsal attention, default mode, and fronto‐parietal networks. Frontiers in Human Neuroscience, 14, 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, X. J. , Gong, H. H. , Wang, Y. X. , Zhou, F. Q. , Min, Y. J. , Zhao, F. , Wang, S. Y. , Liu, B. X. , & Xiao, X. Z. (2012). Gender differences in brain regional homogeneity of healthy subjects after normal sleep and after sleep deprivation: A resting‐state fMRI study. Sleep Medicine, 13, 720–727. [DOI] [PubMed] [Google Scholar]
- De Havas, J. A. , Parimal, S. , Soon, C. S. , & Chee, M. W. (2012). Sleep deprivation reduces default mode network connectivity and anti‐correlation during rest and task performance. NeuroImage, 59, 1745–1751. [DOI] [PubMed] [Google Scholar]
- Deng, Y. , Liu, K. , Cheng, D. , Zhang, J. , Chen, H. , Chen, B. , Li, Y. , Wang, W. , Kong, Y. , & Wen, G. (2019). Ventral and dorsal visual pathways exhibit abnormalities of static and dynamic connectivities, respectively, in patients with schizophrenia. Schizophrenia Research, 206, 103–110. [DOI] [PubMed] [Google Scholar]
- Dong, D. , Duan, M. , Wang, Y. , Zhang, X. , Jia, X. , Li, Y. , Xin, F. , Yao, D. , & Luo, C. (2019). Reconfiguration of dynamic functional connectivity in sensory and perceptual system in schizophrenia. Cerebral Cortex, 29, 3577–3589. [DOI] [PubMed] [Google Scholar]
- Ettinger, U. , & Kumari, V. (2015). Lancet Psychiatry. Effects of sleep deprivation on inhibitory biomarkers of schizophrenia: Implications for drug development., 2, 1028–1035. [DOI] [PubMed] [Google Scholar]
- Gent, T. C. , Bassetti, C. , & Adamantidis, A. R. (2018). Sleep‐wake control and the thalamus. Current Opinion in Neurobiology, 52, 188–197. [DOI] [PubMed] [Google Scholar]
- Gu, Y. , Lin, Y. , Huang, L. , Ma, J. , Zhang, J. , Xiao, Y. , & Dai, Z. (2020). Abnormal dynamic functional connectivity in Alzheimer's disease. CNS Neuroscience & Therapeutics, 26, 962–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson, R. N. A. , Buechel, C. , Josephs, O. , & Friston, K. J. (1999). The slice‐timing problem in event‐related fMRI. NeuroImage, 9, 125. [Google Scholar]
- Horne, J. A. , & Ostberg, O. (1976). A self‐assessment questionnaire to determine morningness‐eveningness in human circadian rhythms. International Journal of Chronobiology, 4, 97–110. [PubMed] [Google Scholar]
- Hou, Z. , Kong, Y. , He, X. , Yin, Y. , Zhang, Y. , & Yuan, Y. (2018). Increased temporal variability of striatum region facilitating the early antidepressant response in patients with major depressive disorder. Progress in Neuro‐Psychopharmacology & Biological Psychiatry, 85, 39–45. [DOI] [PubMed] [Google Scholar]
- Hu, J. , Du, J. , Xu, Q. , Yang, F. , Zeng, F. , Weng, Y. , Dai, X. J. , Qi, R. , Liu, X. , Lu, G. , & Zhang, Z. (2018). Dynamic network analysis reveals altered temporal variability in brain regions after stroke: A longitudinal resting‐state fMRI study. Neural Plasticity, 2018, 9394156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, A. N. , Van Dongen, H. P. A. , & Honn, K. A. (2020). Sleep deprivation, vigilant attention, and brain function: A review. Neuropsychopharmacology, 45, 21–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itani, O. , Jike, M. , Watanabe, N. , & Kaneita, Y. (2017). Short sleep duration and health outcomes: A systematic review, meta‐analysis, and meta‐regression. Sleep Medicine, 32, 246–256. [DOI] [PubMed] [Google Scholar]
- Krause, A. J. , Simon, E. B. , Mander, B. A. , Greer, S. M. , Saletin, J. M. , Goldstein‐Piekarski, A. N. , & Walker, M. P. (2017). The sleep‐deprived human brain. Nature Reviews Neuroscience, 18, 404–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucyi, A. , & Davis, K. D. (2014). Dynamic functional connectivity of the default mode network tracks daydreaming. NeuroImage, 100, 471–480. [DOI] [PubMed] [Google Scholar]
- Kumari, V. , & Ettinger, U. (2020). Controlled sleep deprivation as an experimental medicine model of schizophrenia: An update. Schizophrenia Research, 221, 4–11. [DOI] [PubMed] [Google Scholar]
- Li, B. , Zhang, L. , Zhang, Y. , Chen, Y. , Peng, J. , Shao, Y. , & Zhang, X. (2020). Decreased functional connectivity between the right Precuneus and middle frontal gyrus is related to attentional decline following acute sleep deprivation. Frontiers in Neuroscience, 14, 530257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. Z. , Cao, Y. , Zhang, Y. , Chen, Y. , Gao, Y. H. , Peng, J. X. , Shao, Y. C. , & Zhang, X. (2021). Relation of decreased functional connectivity between left thalamus and left inferior frontal gyrus to emotion changes following acute sleep deprivation. Frontiers in Neurology, 12, 642411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C. , Fronczek‐Poncelet, J. , Lange, D. , Hennecke, E. , Kroll, T. , Matusch, A. , Aeschbach, D. , Bauer, A. , Elmenhorst, E. M. , & Elmenhorst, D. (2020). Impact of acute sleep deprivation on dynamic functional connectivity states. Human Brain Mapping, 41, 994–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, J. , & Dinges, D. F. (2008). Sleep deprivation and vigilant attention. Annals of the New York Academy of Sciences, 1129, 305–322. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Liao, X. , Xia, M. , & He, Y. (2018). Chronnectome fingerprinting: Identifying individuals and predicting higher cognitive functions using dynamic brain connectivity patterns. Human Brain Mapping, 39, 902–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, Y. , Liu, Z. , Chan, C. K. Y. , Wu, G. , Xue, Z. , Pan, Y. , Chen, X. , Huang, X. , Li, D. , & Pu, W. (2020). Altered temporal variability of local and large‐scale resting‐state brain functional connectivity patterns in schizophrenia and bipolar disorder. Frontiers in Psychiatry, 11, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, Z. , Zhao, J. , Chen, D. , & Lei, X. (2021). Age‐related abnormalities of thalamic shape and dynamic functional connectivity after three hours of sleep restriction. PeerJ, 9, e10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, N. , Dinges, D. F. , Basner, M. , & Rao, H. (2015). How acute total sleep loss affects the attending brain: A meta‐analysis of neuroimaging studies. Sleep, 38, 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mander, B. A. , Winer, J. R. , & Walker, M. P. (2017). Sleep and Human Aging. Neuron, 94, 19–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porras‐Segovia, A. , Pérez‐Rodríguez, M. M. , López‐Esteban, P. , Courtet, P. , Barrigón, M. M. L. , López‐Castromán, J. , Cervilla, J. A. , & Baca‐García, E. (2019). Contribution of sleep deprivation to suicidal behaviour: A systematic review. Sleep Medicine Reviews, 44, 37–47. [DOI] [PubMed] [Google Scholar]
- Postuma, R. B. , & Dagher, A. (2006). Basal ganglia functional connectivity based on a meta‐analysis of 126 positron emission tomography and functional magnetic resonance imaging publications. Cerebral Cortex, 16, 1508–1521. [DOI] [PubMed] [Google Scholar]
- Qi, J. , Li, B. Z. , Zhang, Y. , Pan, B. , Gao, Y. H. , Zhan, H. , Liu, Y. , Shao, Y. C. , & Zhang, X. (2021). Altered insula‐prefrontal functional connectivity correlates to decreased vigilant attention after total sleep deprivation. Sleep Medicine, 84, 187–194. [DOI] [PubMed] [Google Scholar]
- Reeve, S. , Sheaves, B. , & Freeman, D. (2015). The role of sleep dysfunction in the occurrence of delusions and hallucinations: A systematic review. Clinical Psychology Review, 42, 96–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls, E. T. , Cheng, W. , & Feng, J. (2021). Brain dynamics: The temporal variability of connectivity, and differences in schizophrenia and ADHD. Translational Psychiatry, 11, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls, E. T. , Huang, C. C. , Lin, C. P. , Feng, J. , & Joliot, M. (2020). Automated anatomical labelling atlas 3. NeuroImage, 206, 116189. [DOI] [PubMed] [Google Scholar]
- Rosenberg, M. D. , Finn, E. S. , Scheinost, D. , Papademetris, X. , Shen, X. , Constable, R. T. , & Chun, M. M. (2016). A neuromarker of sustained attention from whole‐brain functional connectivity. Nature Neuroscience, 19, 165–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, Y. , Wang, L. , Ye, E. , Jin, X. , Ni, W. , Yang, Y. , Wen, B. , Hu, D. , & Yang, Z. (2013). Decreased thalamocortical functional connectivity after 36 hours of total sleep deprivation: Evidence from resting state FMRI. PLoS One, 8, e78830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, X. , Tokoglu, F. , Papademetris, X. , & Constable, R. T. (2013). Groupwise whole‐brain parcellation from resting‐state fMRI data for network node identification. NeuroImage, 82, 403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J. , Liu, Z. , Rolls, E. T. , Chen, Q. , Yao, Y. , Yang, W. , Wei, D. , Zhang, Q. , Zhang, J. , Feng, J. , & Qiu, J. (2019). Verbal creativity correlates with the temporal variability of brain networks during the resting state. Cerebral Cortex, 29, 1047–1058. [DOI] [PubMed] [Google Scholar]
- Sun, J. , Zhang, Q. , Li, Y. , Meng, J. , Chen, Q. , Yang, W. , Wei, D. , & Qiu, J. (2020). Plasticity of the resting‐state brain: Static and dynamic functional connectivity change induced by divergent thinking training. Brain Imaging and Behavior, 14, 1498–1506. [DOI] [PubMed] [Google Scholar]
- Sun, J. , Zhao, R. , Yang, X. , Deng, H. , Zhu, Y. , Chen, Y. , Yuan, K. , Xi, Y. , Yin, H. , & Qin, W. (2020). Alteration of brain gray matter density after 24 h of sleep deprivation in healthy adults. Frontiers in Neuroscience, 14, 754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempesta, D. , Socci, V. , De Gennaro, L. , & Ferrara, M. (2018). Sleep and emotional processing. Sleep Medicine Reviews, 40, 183–195. [DOI] [PubMed] [Google Scholar]
- Tobaldini, E. , Costantino, G. , Solbiati, M. , Cogliati, C. , Kara, T. , Nobili, L. , & Montano, N. (2017). Sleep, sleep deprivation, autonomic nervous system and cardiovascular diseases. Neuroscience Biobehavioral Reviews, 74, 321–329. [DOI] [PubMed] [Google Scholar]
- Van Craenenbroeck, E. M. (2019). Sleep deprivation and increased cardiovascular risk: A wake‐up call! European Journal of Preventive Cardiology, 28, 187–188. [DOI] [PubMed] [Google Scholar]
- Van Dongen, H. P. A. , Greg, M. , Mullington, J. M. , & Dinges, D. F. (2003). The cumulative cost of additional wakefulness: Dose‐response effects on neurobehavioral functions and sleep physiology from chronic sleep restriction and Total sleep deprivation. Sleep:2, 26, 117–126. [DOI] [PubMed] [Google Scholar]
- Waite, F. , Sheaves, B. , Isham, L. , Reeve, S. , & Freeman, D. (2020). Sleep and schizophrenia: From epiphenomenon to treatable causal target. Schizophrenia Research, 221, 44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, X. , Wang, R. , Yin, W. , Lin, W. , Zhang, H. , & Shen, D. (2020). Development of dynamic functional architecture during early infancy. Cerebral Cortex, 30, 5626–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirsich, J. , Rey, M. , Guye, M. , Bénar, C. , Lanteaume, L. , Ridley, B. , Confort‐Gouny, S. , Cassé‐Perrot, C. , Soulier, E. , Viout, P. , Rouby, F. , Lefebvre, M.‐N. , Audebert, C. , Truillet, R. , Jouve, E. , Payoux, P. , Bartrés‐Faz, D. , Bordet, R. , Richardson, J. C. , … Pharmacog Consortium . (2018). Brain networks are independently modulated by donepezil, sleep, and sleep deprivation. Brain Topography, 31, 380–391. [DOI] [PubMed] [Google Scholar]
- Xia, M. , Wang, J. , & He, Y. (2013). BrainNet viewer: A network visualization tool for human brain connectomics. PLoS One, 8, e68910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, H. , Shen, H. , Wang, L. , Zhong, Q. , Lei, Y. , Yang, L. , Zeng, L. L. , Zhou, Z. , Hu, D. , & Yang, Z. (2018). Impact of 36 h of total sleep deprivation on resting‐state dynamic functional connectivity. Brain Research, 1688, 22–32. [DOI] [PubMed] [Google Scholar]
- Yang, F. N. , Xu, S. , Chai, Y. , Basner, M. , Dinges, D. F. , & Rao, H. (2018). Sleep deprivation enhances inter‐stimulus interval effect on vigilant attention performance. Sleep, 41, zsy189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo, B. T. , Krienen, F. M. , Chee, M. W. , & Buckner, R. L. (2014). Estimates of segregation and overlap of functional connectivity networks in the human cerebral cortex. NeuroImage, 88, 212–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo, B. T. , Tandi, J. , & Chee, M. W. (2015). Functional connectivity during rested wakefulness predicts vulnerability to sleep deprivation. NeuroImage, 111, 147–158. [DOI] [PubMed] [Google Scholar]
- Yue, J. L. , Li, P. , Shi, L. , Lin, X. , Sun, H. Q. , & Lu, L. (2018). Enhanced temporal variability of amygdala‐frontal functional connectivity in patients with schizophrenia. NeuroImage Clinical, 18, 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, B. , Zhou, J. , Li, Z. , Zhang, H. , Li, Z. , & Yu, P. (2020). Altered percent amplitude of fluctuation in healthy subjects after 36 h sleep deprivation. Frontiers in Neurology, 11, 565025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Cheng, W. , Liu, Z. , Zhang, K. , Lei, X. , Yao, Y. , Becker, B. , Liu, Y. , Kendrick, K. M. , Lu, G. , & Feng, J. (2016). Neural, electrophysiological and anatomical basis of brain‐network variability and its characteristic changes in mental disorders. Brain, 139, 2307–2321. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Dai, C. , Shao, Y. , Peng, J. , Yang, Y. , & Hou, Y. (2021). Decreased functional connectivity in the reward network and its relationship with negative emotional experience after total sleep deprivation. Frontiers in Neurology, 12, 641810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, R. , Zhang, X. , Fei, N. , Zhu, Y. , Sun, J. , Liu, P. , Yang, X. , & Qin, W. (2019). Decreased cortical and subcortical response to inhibition control after sleep deprivation. Brain Imaging Behavior, 13, 638–650. [DOI] [PubMed] [Google Scholar]
- Zhao, R. , Zhang, X. , Zhu, Y. , Fei, N. , Sun, J. , Liu, P. , Yang, X. , & Qin, W. (2018). Prediction of the effect of sleep deprivation on response inhibition via machine learning on structural magnetic resonance imaging data. Frontiers in Human Neuroscience, 12, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, R. , Zhang, X. , Zhu, Y. , Fei, N. , Sun, J. , Liu, P. , Yang, X. , & Qin, W. (2019). Disrupted resting‐state functional connectivity in hippocampal subregions after sleep deprivation. Neuroscience, 398, 37–54. [DOI] [PubMed] [Google Scholar]
- Zhu, H. , Huang, J. , Deng, L. , He, N. , Cheng, L. , Shu, P. , Yan, F. , Tong, S. , Sun, J. , & Ling, H. (2019). Abnormal dynamic functional connectivity associated with subcortical networks in Parkinson's disease: A temporal variability perspective. Frontiers in Neuroscience, 13, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y. , Feng, Z. , Xu, J. , Fu, C. , Sun, J. , Yang, X. , Shi, D. , & Qin, W. (2016). Increased interhemispheric resting‐state functional connectivity after sleep deprivation: A resting‐state fMRI study. Brain Imaging Behavior, 10, 911–919. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. , Wang, L. , Xi, Y. , Dai, T. , Fei, N. , Liu, L. , Xu, Z. , Yang, X. , Fu, C. , Sun, J. , Xu, J. , Shi, D. , Tian, J. , Yin, H. , & Qin, W. (2017). White matter microstructural properties are related to inter‐individual differences in cognitive instability after sleep deprivation. Neuroscience, 365, 206–216. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. , Xi, Y. , Fei, N. , Liu, Y. , Zhang, X. , Liu, L. , Xu, Z. , Sun, J. , Yang, X. , Yin, H. , Tian, J. , & Qin, W. (2018). Dynamics of cerebral responses to sustained attention performance during one night of sleep deprivation. Journal of Sleep Research, 27, 184–196. [DOI] [PubMed] [Google Scholar]
- Zhu, Y. , Xi, Y. , Sun, J. , Guo, F. , Xu, Y. , Fei, N. , Zhang, X. , Yang, X. , Yin, H. , & Qin, W. (2019). Neural correlates of dynamic changes in working memory performance during one night of sleep deprivation. Human Brain Mapping, 40, 3265–3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Whole‐brain regional‐level temporal variability topography on Shen‐264 functional template after rested wakefulness and sleep deprivation. (a) After rested wakefulness (RW). The color scale represents regional‐level temporal variability. (b) After sleep deprivation (SD). The color scale represents regional‐level temporal variability. (c) Significant changes between RW and SD with false discovery rate (FDR) correction for multiple comparisons (p <.05). The color scale represents t value. The positive t values mean SD > RW; the negative t values mean SD < RW. R, right. L, left. These figures were constructed using the BrainNet Viewer (http://www.nitrc.org/projects/bnv/; Xia et al., 2013).
Figure S2 Significant changes in intra‐network temporal variability between RW and SD with the Shen‐268 functional atlas. Higher intra‐network temporal variability in DAN, FPN, and DMN was observed after SD after false discovery rate (FDR) correction (p <.05). No network showed significant changed intra‐network variability after SD. DMN, default mode network; DAN, dorsal attention network; FPN, frontal‐parietal network; RW, rested wakefulness; SD, sleep deprivation. ****p <.0001. ***p <.001.
Figure S3 Significant changes in inter‐network temporal variability between RW and SD using the Shen‐268 functional atlas after FDR correction (p <.05). The color scale represents t value. The positive t values mean SD > RW; the negative t values mean SD < RW. Black asterisks (*) indicate the subnetwork pairs showing significantly increased inter‐network temporal variability after SD compared with RW. Red asterisks indicate the subnetwork pairs showing significantly decreased inter‐network temporal variability after SD compared with RW. ****p <.0001. ***p <.001. **p <.01. *p <.05. CN, cerebellum; DAN, dorsal attention network; DMN, default mode network; FDR, false discovery rate; LN, limbic network; FPN, frontal‐parietal network; SD, sleep deprivation; SMN, sensorimotor network; SUB, subcortical network; RW, rested wakefulness; VAN, ventral attention network; VN, visual network.
Table S1 The significant changes of intra‐network temporal variability between RW and SD.
Table S2 The overlapped brain regions which showed significant changes in regional‐level temporal variability after SD using the AAL3 template and Shen‐268 functional atlas.
Table S3 The significant changes of inter‐network temporal variability between RW and SD with SHEN‐264 atlas.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


