Abstract
Hepatocellular carcinoma (HCC) is characterized by its high degrees of both inter- and intratumoral heterogeneity. Its complex tumor microenvironment is also crucial in promoting tumor progression. Recent advances in single-cell RNA sequencing provide an important highway to characterize the underlying pathogenesis and heterogeneity of HCC in an unprecedented degree of resolution. This review discusses the up-to-date discoveries from the latest studies of HCC with respect to the strength of single-cell RNA sequencing. We discuss its use in the dissection of the landscape of the intricate HCC ecosystem and highlight the major features at cellular levels, including the malignant cells, different immune cell types, and the various cell-cell interactions, which are crucial for developing effective immunotherapies. Finally, its translational applications will be discussed. Altogether, these explorations may give us some hints at the tumor growth and progression and drug resistance and recurrence, particularly in this era of personalized medicine.
Keywords: Hepatocellular Carcinoma, Immune Tumor Microenvironment, Single-Cell RNA-Sequencing
Abbreviations used in this paper: CNV, copy number variation; CSC, cancer stem cell; DC, dendritic cell; FACS, fluorescence-activated cell sorting; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; ILC, innate lymphoid cell; ITH, intratumoral heterogeneity; MAIT, mucosal-associated invariant T cell; NK, natural killer; scRNA-seq, single-cell RNA-sequencing; snRNA-seq, single nucleus RNA-sequencing; TAM, tumor-associated macrophage; TCGA, The Cancer Genome Atlas; TF, transcription factor; TME, tumor microenvironment; Treg, regulatory T cell
Summary.
Single-cell RNA-sequencing (scRNA-seq) provides a cutting-edge method to better understand the heterogeneity of the hepatocellular carcinoma microenvironment by capturing the whole transcription expression of thousands of various individual cells. This review summarizes and discusses the latest achievements on hepatocellular carcinoma via scRNA-seq. Insights from scRNA-seq analysis would help advance prospective studies for personalized medicine and targeted therapy.
Hepatocellular carcinoma (HCC) is the major form of primary liver cancer and constitutes >85% of the cases.1 Despite continuing efforts in the investigation of its pathogenesis, the current understanding remains far from adequate, largely owing to its extremely heterogeneous composition. Regarding the cancer cells, liver cancer stem cells (CSCs) constitute subtle but substantial fractions and are believed to exert adverse effects leading to refractory disease and metastasis. Many lines of evidence have indicated the complex involvement of tumor microenvironment (TME), particularly the immune cells, as well as their crosstalk in HCC.2,3 The immune cells in the TME play a pivotal role in tumor immunosuppression. In this review, we (1) give an overview of the single-cell RNA sequencing (scRNA-seq) platforms and procedures; (2) summarize scRNA-seq use and the latest insights from such studies in HCC, ranging from the HCC cell population and the intricate immune ecosystem in the TME to cell-cell interaction; and (3) discuss scRNA-seq translational applications.
scRNA-seq Platforms and Procedures
scRNA-seq vs bulk-cell RNA-seq
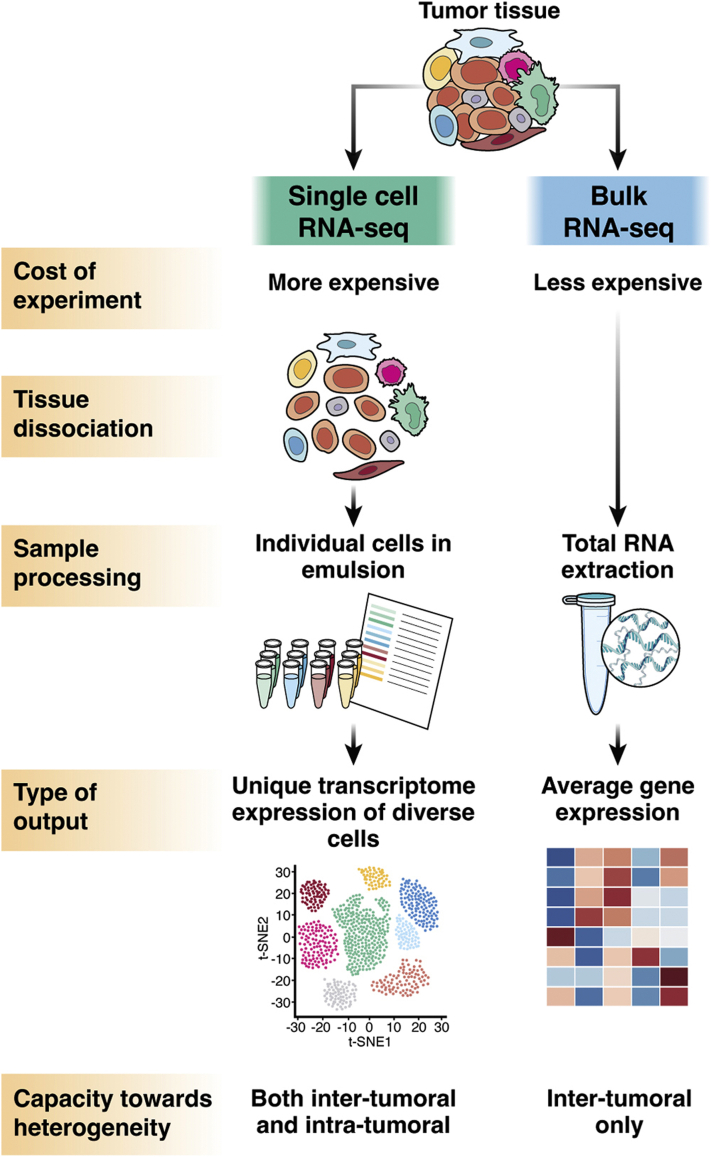
Nowadays, RNA-seq is a widely adopted profiling strategy to interrogate the whole transcriptome of tens of thousands of genes. Nevertheless, the traditional “bulk-cell” RNA-seq approach suffers from mixing and adding up data from multiple cells, which inevitably masks the signals from individual cells, particularly the less abundant cell types. This is especially problematic for studying HCC, which is highly heterogeneous and composed of a rich content of malignant cells, immune cells, and stromal cells in the TME. Despite the fact that the cellular deconvolution algorithms provide computational estimation on cell type composition based on bulk-cell data,4,5 they still have fundamental limitations (eg, use of inappropriate reference or lack of suitable reference). In contrast, scRNA-seq overcomes the major limitations of bulk-cell approach (Figure 1). It allows us to differentiate cell types and analyze the heterogeneous characteristics, including the biological functions of different cell subpopulations and their possible interactions, and helps better understanding of the cellular and molecular regulatory mechanisms within the tumor ecosystem.
Figure 1.
Comparison of scRNA-seq and bulk RNA-seq.
scRNA-seq Platforms
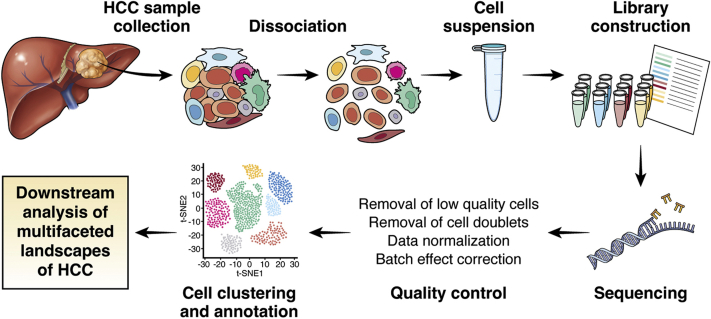
scRNA-seq begins with single-cell capture, followed by conversion of RNA into complementary DNA, amplification, and library preparation (Figure 2). Regarding single-cell capture, there are different ways ranging from manual to microfluidic handling.6 Importantly, they differ in terms of their handling capacity and the number of genes that they can typically detect. There are currently 2 most frequently used platforms, utilizing plate-based (eg, Smart-seq2)7 and droplet-based (eg, 10X Genomics Chromium platforms [10X Genomics, Pleasanton, CA]) single-cell capture. In general, the plate-based platform sequences full-length transcripts and detects more genes per cell. This is particularly useful in targeting low-abundance transcripts and identifying alternative splicing events. Also, it is less limited by cell size. However, the number of cells analyzed is lower. On the other hand, the droplet-based platform has far greater throughput of experiment and can capture cells in the scale of thousands to even tens of thousands of cells. It is therefore particularly powerful to investigate the cellular landscapes and detect rare cell types or subclones of cells. However, it comes with a cost of providing sequencing information of only short intervals at either the 5′ or 3′ end of a gene, and there are also more frequent occurrences of dropout events for low-expression genes.8 Besides, its utility is sometimes limited by the fluidic tube diameter inside the system that poses an upper limit to the size of the cells that can be studied. The cell suspension must be ensured to contain single cells free of doublets and clumps beforehand, and this will rely very much on the efficiency of the tissue digestion process, the removal of undigested cell clumps, and the single-cell enrichment process such as using fluorescence-activated cell sorting (FACS). Overall, because cell viability of different subpopulations may be differently affected by the previously mentioned processes (eg, immune cells have higher viability, while hepatocytes are very fragile), investigators must keep in mind whether some cell types will be over- or underrepresented when analyzing the final single-cell sequencing dataset.9
Figure 2.
Experimental workflow of scRNA-seq on clinical HCC tissue samples using microfluidics platform.
Single-Cell Capture
To obtain single-cell suspension, the tissues under study are usually dissociated by physical homogenization and enzymatic digestion at 37°C for certain duration of time to help release the single cells from the tissues. Any undigested tissue debris is filtered away by cell strainer of certain pore sizes (eg, 100 μm). To further remove clumps of cells, stepwise filtration using cell strainer of gradually decreasing pore sizes is recommended. On the other hand, for peripheral blood mononuclear cells, they can be easily harvested by collecting the buffy coat layer upon the use of Ficoll isolation protocol. Viability of cells and number of live cells available can then be determined by trypan blue staining. Depending on the cell types of the target populations to be investigated, FACS or magnetic-activated cell sorting can be used to enrich the particular cell type population according to the different cell surface markers recognized by the respective antibodies conjugated to the appropriate fluorophore or magnetic beads, respectively (eg, CD45 positivity for immune cells, CD45 negativity for nonimmune cells). FACS provides added advantage of eliminating doublets by proper gating of forward or side scatter signals, and dead cells by cell viability dye. Furthermore, our experience shows that subpopulations of different cell types will have their viability affected differently by the previous sample preparation processes. For instance, myeloid cells in peripheral blood mononuclear cells usually exist as single cells with high viability, thus sparing the need for FACS into single cells. On the other hand, cells in cancer tissues are subjected to the dissociation process and FACS, which inevitably creates some stress and poses adverse effects on their viability. Besides, whether cell type enrichment processes, such as FACS or magnetic-activated cell sorting for particular cell type populations, are included before scRNA-seq, may affect the distribution of different cell types, such as tumor-infiltrating myeloid cells and lymphoid cells, HCC cells and other nonmalignant stromal cells, in the samples under examination. After collecting sufficient cells of the enriched cell types, the cell suspension with single cells can be subjected to single-cell capture process.
Use of scRNA-seq to Analyze Different Cell Types in the Tumor
Previous Reports Using scRNA-seq on HCC
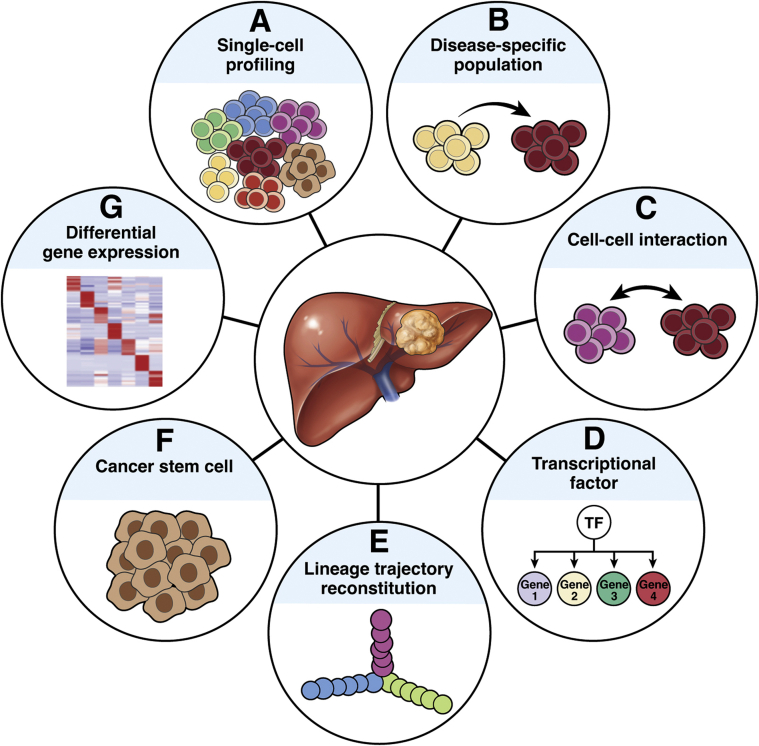
To date, the scRNA-seq technique has been increasing adopted in HCC investigation. The previous reports are summarized in Table 1.2,3,10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 The samples in these reports consisted mostly of cells from primary tumors, while circulating tumor cells were studied in a few reports.15,16 In brief, scRNA-seq has been reported to allow delineation of cell type abundance, cell-cell interactions, transition in cellular status, clonal evolution, heterogeneity landscape, and lineage hierarchy in terms of marker gene expression, mutation and inferred copy number variation (CNV) status, and gene expression profiling at single-cell resolution (Figure 3).
Table 1.
A Summary of the Studies on Human HCC Using the scRNA-seq Technique
| Reference | Samples | Platform | Cells | Patients | Cell Types | Data Accession Number |
|---|---|---|---|---|---|---|
| 2 | HCC | 10X | 43,645 | 8 | All cell types | SRP318499 |
| 3 | HCC, adjacent tissues | MIRALCS | 16,498 | 18 | All cell types | CNP0000650 |
| 10 | HCC, cell lines | Smart-seq | 118 | 1 | HCC cells, HuH1 cells, HuH7 cells | n/a |
| 11 | HCC (HBV + nonviral), adjacent tissues, PBMCs | CyTOF | n/a | 23 | Immune lineages | n/a |
| 12 | HCC PDTX | Fluidigm C1 | 139 | n/a | CSC clusters | n/a |
| 13 | HCC | 10X | 38,553 | 2 | All cell types | E-MTAB-5905; GSE112271; E-MTAB-5899; E-MTAB-8127; E-MTAB-5878 |
| 14 | HCC, nontumor liver tissues | 10X | 41,698 | 7 | Immune cell lineages | CRA002308 |
| 15 | HCC | Smart-seq2 | 113 | 10 | CTCs | EGAS00001005204 |
| 16 | HCC whole blood | Smart-seq2 | 38 | 6 | CTCs | n/a |
| 17 | HCC; normal human hepatocytes | CEL-Seq2/Smart-seq2 | 938 cells (420 HCC cells), 200 healthy hepatocytes | 2 | HCC cells (nonimmune cells) | SRP165160; SRP275756 |
| 18 | HCC | 10X | 5753 | 1 | All cell types | n/a |
| 19 | HCC PDTX | BD Rhapsody | 10,602 | n/a | All cell types | GSE175716 |
| 20 | iPSCs, hepatoblasts, hepatic organoids | Fluidigm C1 | 424 | n/a | iPSC, hepatoblast, hepatic organoid | GSE139382 |
| 21 | HCC and paired normal liver | 10X | 5782 (HCC cells); 11,394 (normal liver cells) | 4 | All cell types | EGAS00001005194 |
| 22 | HCC and NT | Smart-seq2 | 405 | 6 | All cell types | GSE154906 |
| 23 | Mouse HCC cells | 10X | 27,327 | n/a | All cell types | GSE157561 |
| 24 | HCC and iCCA biopsies | 10X | 56,721 | 44 | All cell types | GSE151530 |
| 25 | HCC | 10X | 17,432,600 | 39 | Immune cell lineages | CRA001276 |
| 26 | HCC, NT, PBMC | Smart-seq2 | 5063 | 6 | T cells | EGAS00001002072; GSE98638 |
| 27 | HCC, adjacent tissues, hepatic lymph nodes, ascitic fluid, PBMC | 10X; Smart-seq2 | 66,187 (10X Genomics); 11,134 (Smart-Seq2) | 16 | CD45+ immune cells | HRA000069; EGAS00001003 |
| 28 | HCC, iCCA | 10X | 5082 | 19 | All cell types | GSE125449 |
10X, 10X Genomics Chromium platform; CSC, cancer stem cell; CTC, circulating tumor cell; CyTOF, cytometry by time of flight; iCCA, intrahepatic cholangiocarcinoma; iPSC, induced pluripotent stem cell; MIRALCS, microwell full-length mRNA amplification and library construction system; n/a, not available; NT, nontumorous liver; PBMC, peripheral blood mononuclear cell; PDTX, patient-derived tumor xenograft.
Figure 3.
Applications of scRNA-seq technique on HCC. An overview of the current main research capabilities for scRNA-seq data is shown. A wide variety of aspects can be reached through scientific analyses, including (A) depicting the single-cell profiles of HCC, (B) distinguishing the disease-specific population, (C) interpreting cell-cell communication across intricate tumor microenvironment, (D) identifying the key transcriptional factors and their regulatory networks, (E) reconstructing the lineage trajectory, (F) capturing the cancer stem cell population and exploring its potential applications in treatment, and (G) deciphering the differential expression genes on particular cell types or subsets.
HCC Cancer Cells
The vast majority of cells within a tumor consists of hepatocytes that have undergone malignant transformation. While the malignant cells usually display a patient-specific clustering pattern,28 somatic mutations analysis revealed the intratumoral heterogeneity of malignant cells across different regions of a tumor.13 To define malignant cells in HCC for computational analysis is important. To this end, high expression of epithelial marker genes has been used to assign their epithelial origin.28 In addition, CNV analysis is also a conventional method to distinguish malignant cells from other nonmalignant cells, as malignant cells have more frequent copy number aberrations,29 with a major CNV group enriched in the tumor cells in individual HCC cases.2
The most striking feature of most cancer types is tumor heterogeneity, both intertumoral and intratumoral heterogeneity. To address the intratumoral heterogeneity (ITH) as found across multiple regions within the same tumor tissue. In a previous study, differential gene expression was used to capture the gene expression landscapes of different clones within the tumors, and the resulting differential expression signatures were computed to derive a score to predict the prognosis of patients.13 The resulting ITH signatures were found to correlate with early tumor recurrence and unfavorable prognostic biomarker α-fetoprotein. Further scRNA-seq on 21,143 and 17,410 cells from 3 and 4 tumoral regions, respectively, from 2 HCC patients’ tumor samples revealed both transcriptomic and regulatory ITH. Clusters distinct to specific regions were found. Upon further single-cell regulatory network inference and clustering analysis, a poorly differentiated region was found to show a distinct gene regulatory network activation pattern corresponding to pluripotency signaling, while another region was enriched with activation pattern of the ETS transcription factor (TF) family. This study hence uncovers the heterogeneity in transcriptome and TF activation network across multiple regions of HCC tumors by examining the differentially expressed genes and concerned gene regulatory networks across cell clusters in the scRNA-seq.
Another study investigated the HCC and corresponding nontumorous livers from 6 HCC patients using Smart-seq2 method22 and analyzed the differentially expressed genes among the heterogeneous subclones in HCC tissues. Upon co-network and functional annotation analyses on the scRNA-seq data based on the gene ontology and KEGG, different clusters were found to have different enriched genes corresponding to various functional abnormalities, including epithelial-to-mesenchymal transition, Wnt signaling, PI3K-Akt signaling, and various metabolic-related pathways. Pseudotime analysis was performed to construct the trajectory of various HCC subclones, and the shifting trend in the expression profile of TFs suggests the paths of the subpopulations toward metabolic disorders. Intriguingly, by comparing the HCC clusters and the hepatocyte cluster, one of the TFs examined, MLXIPL, was found to be elevated along pseudotime, and MLXIPL overexpression at protein level was associated with poor prognosis in a bigger cohort of HCC patients.
In another study,17 by modified CEL-Seq2 platform, 938 single cells (including 420 HCC cells) of 2 HCC patients with chronic hepatitis B virus (HBV) infection were analyzed. The complementary scRNA-seq platform Smart-Seq2 was also applied to intensely cover the full-length RNA-transcripts. By mapping the fused HBV-host transcripts in the scRNA-seq data, the HBV integration sites were investigated at the single-cell level. Interestingly, although patients shared similar serum HBV DNA levels, HBV RNA was found to have different expression levels in the tumor cells. The scRNA-seq analysis allowed the investigation of virus heterogeneity within HCC tumors. It also demonstrated a positive correlation between HBV-RNA levels and the tumor differentiation markers as well as some HBV-related host factors, like liver-specific transcription factor HLF that enhances HBV replication, among the individual HCC cells. Additionally, the clustering of the analyzed HCC cells according to HBV integration sites allowed the mapping of the clonal origin of the cells.
Besides scRNA-seq on clinical HCC tissue samples, the transcriptomes of HCC cells in mouse HCC and other HCC models have also been studied. For instance, scRNA-seq was performed on patient-derived HCC xenografts to study sorafenib resistance in HCC.19 By comparing the sorafenib-resistant and sorafenib-sensitive groups, a special cluster was identified in the resistant group. Pseudotime analysis also showed different trajectory differentiation processes between the sorafenib-resistant and sorafenib-sensitive groups. SCENIC (single-cell regulatory network inference and clustering) analysis revealed that the hypoxic HIF pathway regulatory genes, JUN, FOS, and JUND, are particularly more highly expressed in a cell cluster specific to the resistant group. In a hepatocyte-specific, Shp2 knockout, myc-driven spontaneous HCC mouse model,23 the scRNA-seq data derived from the HCC tumor cells that were subjected to zonation-based clustering revealed heterogeneity in myc expression. Moreover, it was shown that the myc+ tumors were derived from the rare Shp2-positive cells. On the other hand, scRNA-seq was carried out in induced pluripotent stem cells (iPSCs), the induced day 9 hepatoblasts, day 21 human hepatic organoids, and primary human hepatocytes.20 The data were compared with those of primary human adult and fetal hepatocytes from the Gene Expression Omnibus database to study the global transcriptome changes to identify key pathways for early liver development. The study identified 2 clusters in the hepatoblasts, including one that resembled iPSCs and another one that resembled hepatocytes and hepatic organoids, suggesting a possible transition of the cell populations from iPSC state to differentiated hepatocytes and hepatic organoids. On the other hand, the transcriptome of hepatic organoid cells was more closely related to adult hepatocytes than to fetal hepatocytes.
HCC CSCs
Evidence suggests the existence of CSCs in HCC, and they could partly be responsible for the tumor heterogeneity, treatment resistance, tumor relapse, and metastasis.10 CSCs are a group of cancer cells possessing self-renewal capacity.30 The hepatic CSCs are heterogeneous in terms of phenotype, function, and transcription at single-cell level.10 Different CSC subpopulations have distinct surface markers, and they are independently associated with HCC prognosis. Interestingly, a significant correlation has also been found among hypoxia-related tumor diversity, cancer stemness, and TME polarization.10 This initial work suggests the biodiversity of CSC subpopulations in HCC,28 which was subsequently exemplified in another study.24 Related research by Ho et al12 used patient-derived HCC xenografts in a mouse model as proof of concept and identified 2 distinct major cell populations according to epithelial cell adhesion molecule expression. More importantly, the results indicate the characteristic stemness-related features of CD24+/CD44+/EPCAM+ cell subclone in HCC.12
Intricate Immune Ecosystem
Cancer cells, immune cells, and stromal cells are the main constituents of HCC. Cancer cells acquire some unique molecular characteristics to enable their effective evasion from the immune surveillance during tumor initiation and sustain tumor development at subsequent stages.31 On the other hand, the anti-cancer immune system may acquire transformation, upon the action of specific key genes or signaling pathways, to become tolerant or even supportive of cancer development.32 To this end, the immune cells are regarded as the major contributors to tumor immunosuppression, resistance to anti-tumor therapies, and tumor clearance. Studies have revealed that the HCC immune ecosystem involves the lymphocyte-derived (T cells, B cells, plasma cells, and natural killer [NK] cells) and myeloid-derived (dendritic cells [DCs] and tumor-associated macrophages [TAMs]) components.3,14,28 Among all the immune cell types in HCCs, T cells constitute the highest proportion (36%), followed by NK cells (29%) and macrophages (25%), while DCs are the least abundant (1%).14 Moreover, although the HCC cancer cells display significant degrees of intra- and intertumoral heterogeneity, immune cell types are shared among different patients3 but may vary in their abundance. In general, there is a trend of transition of the TME toward immunosuppressive and exhaustive status,2,28 which could be further exacerbated in HBV-associated HCC11 to elicit various cancer-promoting properties of the TME.2,11
Lymphocytes in the TME
In HCC tumors, tumor-infiltrating T cells have attracted a great deal of attention due to their frequent interaction with the tumor cells. They can mediate unique cytotoxic immune response that provides anti-tumor capacity under endogenous condition as well as upon the action of immunotherapy. Tumor-infiltrating T cells are further classified into CD8+ T, CD4+ T, regulatory T (Treg) cells, mucosal-associated invariant T (MAIT) cells, and so forth. Based on the analysis of T cell receptor sequences, evidence has demonstrated that there is concomitant clonal expansion of T cells upon their tumor infiltration, resulting in their different subtypes that possess specific molecular and functional properties.26 Pseudotime trajectory and RNA velocity analyses are commonly used to reconstruct the transformation process of proliferative T cells into its exhausted states.27 Regarding CD8+ T cells, recent studies have shown tumor-infiltrating CD8+ T cells possess exhausted characteristics as demonstrated by expression of PD-1 and TIGIT, impairing their endogenous anti-tumor function.2,26 HCCs at advanced stages have more clonally expanded and exhausted CD8+ T cells, as compared with HCCs at early stage.14,26 In relapsed HCC, reduced T cell proliferation, together with increased CD8+ fraction and diversity, could be observed.3 It is also worth mentioning that, in early relapse and primary HCCs, a study uncovered the accumulation of CD8+ T cells in the early relapse HCCs, and these cells had an innate-like, low cytotoxic, and low clonal expansion phenotype. Importantly, unlike the usual exhaustive T cell status identified in primary HCC, early relapse cases had fewer activated T cells that were not exhausted, and evidence also hinted at the important role of memory CD8+ T cells during tumor recurrence.3 Besides, the etiological differences also play an important role. There was more enrichment of CD8+ resident memory T cells in viral-related HCCs than non–viral-related ones, demonstrating the stimulatory effects exerted by the virus.11 Apart from CD8+ T cells, the functions of other types of T cells are also important in carcinogenesis. For instance, decreased CD4+ T cells were found in early-relapse HCC as compared with primary HCC, and this may reflect the impaired anti-tumor system.3 Moreover, CD4/CD8 double-positive T cells were found in leading-edge regions, with concurrent expression of PD-1/HLA-DR/ICOS/CD45RO, and displayed an elevated level of interferon gamma, tumor necrosis factor α, and PD-1 upon stimulation. This implicates the functional consequence of their unique distribution in the tumor.25 In contrast, a high proportion of Treg cells with massive clonal expansion has been observed in HCC microenvironment in comparison with healthy control cells, and high expression of PD-1 in Treg cells may also indicate their immunosuppressive feature by suppressing the cytotoxicity mediated by effector CD8+ T cells.2,26 Nevertheless, one controversy is the origin of these clonally accumulated Treg cells. Whether they proliferate and migrate from peripheral lymph organs or proliferate locally within TME cannot be fully distinguished.26 In addition, although MAITs are unique innate-like T cells resisting against bacterial and viral infections,33 their cell fraction was markedly reduced in HCC, and this could be validated by The Cancer Genome Atlas (TCGA) cohort as indicated by the extremely low expression of MAIT marker.26 Apart from that, a recent study discovered the differential expression of tumor-dependent cytokine gradients between HCC tumor and nontumor samples, which mediated tumor innate lymphoid cell (ILC) composition toward activated tumor ILC2s and ILC1s. Tumors with high ILC2-to-ILC1 ratio correlated with high expression of interleukin-33 that promoted ILC2 generation, which was associated with better survival. It gave us a hint that the regulation of tumor-dependent cytokine gradients may be useful for the plasticity change of tumor ILC, and further application on the clinical anti-tumor therapy.34 All these findings reflect the impaired anti-tumor immunity and the immunosuppression within the TME.
There is no doubt that T cells play pivotal defense function against tumor cells, but the function of B cells could not be ignored. B cells mainly participate in the humoral immunity. Studies have reported an increase of total B cells in HCC as compared with healthy control, adjacent nontumorous tissue, or even peripheral blood samples, albeit their overall abundance is lower than T cells.3 More intriguingly, there was a high percentage of plasma cells within HCC, which may accelerate the progression of hepatocarcinogenesis, both in humans and mice.29,35 The inverse correlation between the proportion of plasma cells in patients and their survival provides a hint for the prognosis of HCC.29 The pseudotime cell trajectory analysis revealed continuous change in humoral immunity during the development of HCC, with concomitant and increasing immune responses. Taken together, the B cells or the plasma cells should be considered in the study of HCC development.29
During the process of tumor invasion, NK cells also play vital roles in immune response and immune checkpoint therapies.36, 37, 38 Zhang et al27 revealed the presence of circulating NK cells and liver-resident NK cells in HCC samples via scRNA-seq. Another study reported the patient-specific feature of NK cells, which is different from the traditional immune cell types and reflect the varied genetic origin or adaptability source of NK cells.14 Similarly, NK cells displayed exhausted status during hepatocarcinogenesis.2 Collectively, these outcomes demonstrate how tumor cells take advantage of immune cells in supporting various aspects of HCC development.
Myeloid cells in the TME
Myeloid cells originate from myeloid progenitors, and they ultimately differentiate into monocytes of macrophages, DCs, and mature granulocytes. In HCC TME, they are abundant and are often associated with poor prognosis. They participate in tumor initiation, progression, angiogenesis, metastasis, and therapeutic resistance processes.39 Particular attention has been paid to TAMs and DCs by scRNA-seq.
In HCC, there exists plasticity regarding TAMs in response to the ever-changing TME or the interaction with the cancer cells. Sun et al3 described an increase in the proportion of TAMs in HCC when compared with the paired nontumorous livers, indicating the potential inflammatory signatures produced by the TME.27 Besides, there is possible ambiguity and difficulty for the distinction of different states of TAM via scRNA-seq, despite studies having shown the coexistence of M1 and M2 by diffusion map with unique gene features.27 Nonetheless, there remain some methods that can help to distinguish the potential polarity of TAMs, such as judging from the expression of core genes for each polarization program. In general, the TAMs in HCC are more toward immunosuppressive state, and this implies less M1 and more M2 TAMs in the TME to support tumor invasion and metastasis.2,14 Therefore, the properties of TAMs can also be used to predict the progress of HCC. Indeed, advanced HCCs possess more M2-like TAMs with stronger lipid metabolism characteristics, indicating that tumor progression is potentially supported by the M2 macrophage through enhanced lipid metabolism.14
DCs are capable of tumor-antigen recognition, antigen presentation, and activating tumor-specific T cell response.40 Unfortunately, there are very few studies focusing on DCs in HCC. A previous study revealed the variable fractions of DCs among patients that ranged from 0.1% to 20% of the total immune fraction.3 Zhang et al27 found one unique subset of DCs characterized by the high expression of LAMP3 gene, and this specific subset could also be detected in lung cancer and breast cancer via scRNA-seq.41,42 It was subsequently revealed that LAMP3+ DCs migrated from tumors to hepatic lymph nodes, suggesting the persistent inflammatory response during tumorigenesis.27
Cell-Cell Interactions
The cells within the TME are not separate individual entities. They constantly undergo a wide range of cell-cell interactions supporting various aspects of HCC development. Notably, the tumor communities have been poorly characterized. How the various heterogeneous cancer cell subtypes, immune cells, and stromal cells cooperate with each other and among them is still elusive.10 The advances of scRNA-seq technology have enabled systematic and comprehensive studies of cell-cell interactions in HCC. Regarding cell-cell interactions analysis, common approaches utilize the respective gene expression levels of curated ligand-receptor pairs to evaluate crosstalk between cell types.43 Existing efforts provide valuable resources documenting precise components of the interacting ligand-receptor complexes. More importantly, there are also statistical frameworks that allow the evaluation of cell-cell interactions using scRNA-seq data. For instance, CellPhoneDB44,45 is a popular tool to perform cell-cell interaction analysis. It consists of a comprehensive repository of ligands, receptors, and their interactions. The definition of ligand-receptor pairs is critical to the subsequent statistical inference because prediction on cell-cell interactions is solely restricted to the documented interacting partners. Regarding the protein interacting relationship, it can be exerted by secretory or membrane proteins. Because individual tools are based on different database of ligand-receptor pairs, the apparent difference between their findings may partly be attributable to this variability.
Interactions between cancer cells and immune subsets
The well-known communication between cancer cells and immune cells is via the immune checkpoint PD-1-PD-L1 axis,46 which illustrates immune escape mechanism and provides guideline for the immune checkpoint therapeutics through targeting the cell-cell interactions. However, investigations on the cellular interaction between malignant cells and immune cells are insufficient in HCC samples with the scRNA-seq method. There have been few studies reporting on the interaction between immune and malignant cells. In a recent study by Ho et al,2 they examined the expression of both co-stimulatory and co-inhibitory immune checkpoint molecules in tumor-infiltrating immune cells and the complementary antigen-presenting cells. They identified a profound co-inhibitory signal via the TIGIT-NECTIN2 axis, with NECTIN2 being the most prominently expressed gene in the PVR gene family. Moreover, such TIGIT-NECTIN2 interaction could be exemplified by significant association between TIGIT and NECTIN2 expression in patients’ HCC tumors, and overexpression of NECTIN2 in HCCs as compared with the corresponding nontumorous livers, suggesting its function as a putative tumor evasion strategy. Using a co-culturing system together with an anti-Nectin2–neutralizing antibody or knockout of Nectin2 in mouse HCC cells, they provided experimental evidence supporting that Nectin2 could suppress T cell proliferation. Further in vivo observation was obtained using hydrodynamic tail-vein injection and orthotopic injection mouse models for knockout and knockdown of Nectin2, respectively. Tumor shrinkage and restoration of T cell tumor infiltration were achieved upon suppression of Nectin2, highlighting the roles of Nectin2 in reducing T cell activity and tumor infiltration in HCC.
Interaction within immune subsets
The communications among different immune cell types also elicit supportive mechanism to facilitate HCC development. As a main cellular component in the tumor niche, the different immune cell types and their connection within have attracted much attention recently. Intense research has revealed that the accumulation of M2 macrophages in HCC promotes the production of tumor-promoting plasma cells, which then could inhibit the effect of T cells, indicating the negative regulatory relationship between the plasma cells and T cells.29 There is suggestive evidence using mouse models that, under the stimulation by activated CD4+ T cells, TAM-mediated cell-cell contact is necessary for B cell survival and maturation.35 In contrast, depletion of B cells could suppress the generation of TAMs and stimulate the anti-tumor response by T cells to inhibit tumor growth. CXCL10 production by TAMs could bind to the CXC chemokine receptor 3 on B cells, which markedly increased the proportion of plasma cells by triggering the preferential accumulation of the latter in the invading edge of HCC.35 Besides, another study observed the potential inverse correlation of T cells and TAMs in HCC samples, which implies the function of TAMs in suppressing T cell tumor infiltration.2 Beyond that, some researchers explored the correlation between DCs and other immune cells through paired ligand-receptor analysis. They discovered that the LAMP3+ DCs could regulate diverse subtypes of lymphocytes by expressing multiple immune-related ligands (eg, PD-L1 and PD-L2). Besides, LAMP3+ DCs could interact with NK cells, particularly via NECTIN2-CD226 and NECTIN2-TIGIT on circulating NK and liver-resident NK cells, respectively.27
Translational Implications and Clinical Implementation and their Barriers
Translational Implications and Clinical Implementation
scRNA-seq has been adopted to detect and profile circulating HCC tumor cells, which can be enriched upon CD45-negative selection.15,16,47 For clinical diagnosis and prognosis of HCCs, scRNA-seq has been used to identify marker genes related to the molecular regulatory mechanisms to correlate with the survival of HCC patients.48 scRNA-seq has helped identify specific proliferating tumor cell clusters and the corresponding gene signature associated with poorer prognosis in HCC.18 The identification of transcriptome signatures associated with therapeutic response or treatment resistance can improve treatment guidance and efficacy monitoring.49 For instance, when combined with CIBERSORT analysis on bulk-seq data, scRNA-seq has been used in the construction of risk model that was validated in both the TCGA and International Cancer Genome Consortium cohorts and showed good predictive power in prognosis as well as immunotherapy efficacy.50 These can help reveal the immune cell composition in patients insensitive to immunotherapy to allow elucidation of the underlying mechanisms for such resistance to immune checkpoint inhibitors.49 The use of gene signatures to discriminate the TCGA cohort into distinct subtypes inspires future endeavor to refine HCC stratification for better molecular understanding and targeted treatments for precision medicine. Furthermore, by combining scRNA-seq and plasma RNA analysis, cell type–specific RNA transcripts can be identified to determine the origin of plasma RNA. This may help future development of novel noninvasive RNAs.21
Barriers to Clinical Implementation
However, there are barriers to the clinical implementation of scRNA-seq. First, scRNA-seq data interpretation can be affected by small changes in gene expression due to the sensitive nature of the technology.48 Some artificial changes may be introduced during the sample processing (eg, the tissue dissociation protocol, storage conditions of the RNA or complementary DNA samples).49 It is also much influenced by the cell yield and quality as well as by the cellular state of the transcriptome at the time of single-cell capture. For instance, it was reported that not all eukaryotic cells carried out transcription at the same rate, as transcription occurs in pulses, making the results much determined at the instant of single-cell capture. This may lead to bias in the final results interpretation.49 Second, with regard to the logistic aspects, lack of standardization in HCC sample collection and processing across different medical centers presents an issue.48 As a result, batch effects arise from sample processing procedures and the use of different scRNA-seq platforms and analytical methods. Also, there is a lack of guidelines to define the quality control standards, the removal of technical artifacts and data interpretation.51 Many parameters can be manually adjusted during the sequencing and analytical processes. These include the types of sequencing libraries (3′ end enriched, full transcript, or targeted-seq, etc.), sequencing depth, mapping techniques, imputations, normalization processes, and differential expression definition.9 Other uncontrollable parameters include the input cell number and quality as well as the single-cell capture efficiency of the cell isolation platform chosen.9 The combinations of these varying parameters will lead to many possible ways in the interpretation of the data. Therefore, the lack of standardized analysis pipelines reduces the reproducibility and interpretability of the ultimate analytical results across centres.9 Third, the high cost of instruments and reagents required also creates hurdles to the general implementation of scRNA-seq in routine clinical practice in HCC management.29 Sometimes, the amount of sample materials is limited (eg, biopsy tissues), and this may affect the representativeness of the various cell types in the scRNA-seq data.51
Conclusions and Future Directions
The revolution on sequencing technology has led to unprecedented comprehension of HCC at the molecular level. Notably, scRNA-seq has revealed the changes that render each individual cell type unique. Dramatic improvements in scRNA-seq in recent years have helped the understanding of the cellular behavior collectively and mutual regulatory mechanisms within the HCC ecosystem.29 As summarized in this review, different research groups have studied the HCC microenvironment using scRNA-seq techniques, laying important foundations for future research. For instance, the landscape of HCC milieu, with a main focus on the immune ecosystem, in different tumor stages (eg, primary HCC vs relapsed HCC)3 and the cellular communications within and across multiple cell types have been unveiled. On the other hand, the heterogeneity, origin, and plasticity of liver CSCs, which consist of a unique subset of cells with stemness features and trigger therapy resistance and tumor recurrence, have also been uncovered.52 In fact, there are interesting studies describing single-cell investigations related to hepatic stellate cells, which are critical to the pathogenesis of liver fibrosis53,54 and intrahepatic cholangiocarcinoma.55, 56, 57 Together with the aforementioned investigations in HCC, they represent the major parts in the hepatocarcinogenesis spectrum of primary liver cancer.
Nevertheless, there are methodological and technological challenges in scRNA-seq that remain to be solved. scRNA-seq is sensitive to the quality of the input sample, tissue dissociation, and sample preparation procedures, which can have adverse effects leading to bias on the analysis outcome. Single-nucleus RNA-seq (snRNA-seq) isolates individual nuclei from specimen. It reduces bias introduced during experimental procedures and is more compatible with frozen archival samples. Different variants of snRNA-seq have been applied in studying various human tissue types.58, 59, 60, 61 In a recent study by Andrews et al,62 the transcriptomic landscape of the human liver was studied by matched scRNA-seq and snRNA-seq. The use of snRNA-seq revealed the presence of rare subtypes of liver mesenchymal cells, while lymphocytes were only distinguishable by scRNA-seq. Findings suggest the importance of using both technologies in obtaining a more complete cellular map of the liver. On the other hand, CITE-seq (cellular indexing of transcriptomes and epitopes by sequencing) is a newly reported method that utilizes oligonucleotide-labelled antibodies to enable simultaneous measurement of surface protein expression and intracellular transcriptome at single-cell readout.63 This technology has been subsequently extended to perform cell hashing64 and ECCITE-seq (expanded CRISPR-compatible cellular indexing of transcriptomes and epitopes by sequencing).65 Taken together, emerging single-cell sequencing strategies allow more comprehensive and in-depth investigation of liver cancer, pinpointing different cellular components at unprecedented resolution. As the scRNA-seq results may not represent the overall state of the tumor due to the limited loading quantity of samples, multiple samples may have to be taken from different parts of the tumor with inevitable increase in sequencing cost. Stereoscopic structure of a cell is another factor that is worth considering. Although the current scRNA-seq technique makes it possible for the exploration of cellular elements of the TME (eg, immune cells, cancer-associated fibroblasts), the already dissociated tumor does not allow illustration of the intercellular interactions in a 3-dimensional manner. To cope with this, the emergence of spatial transcriptomics raises hopes for deeper investigation of TME. This will be pivotal to the development of novel treatment options including immune therapies and newer molecular targeted drugs. With the combo of atezolizumab and bevacizumab that is now the first-line systemic drug treatment for advanced HCC and with further successful clinical trials of immune checkpoint inhibitors, there will be more to come in the near future.66
Last, the barriers to the clinical implementation of scRNA-seq in HCC management need to be addressed. There should be more clinical trials for large multicenter cohorts on the use of scRNA-seq for every aspect of HCC management to explore its clinical utility.9,49 Studies are much needed on how every step and parameter in the scRNA-seq and analytical processes may affect the final interpretation of the scRNA-seq data for generating clinically relevant conclusion and decision.51
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding The study was supported by the National Natural Science Foundation of China (81872222), Health and Medical Research Fund (03142836 and 07182546), Hong Kong Research Grants Council General Research Fund (17100021 and 17117019), Hong Kong Research Grants Council Theme-based Research Scheme (T12-704/16-R), Innovation and Technology Commission grant for State Key Laboratory of Liver Research, University Development Fund of The University of Hong Kong, and Loke Yew Endowed Professorship award. Irene Oi-Lin Ng is Loke Yew Professor in Pathology.
References
- 1.El-Serag H.B., Rudolph K.L. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Ho D.W.H., Tsui Y.M., Chan L.K., Sze K.M.F., Zhang X., Cheu J.W.S., Chiu Y.T., Lee J.M.F., Chan A.C.Y., Cheung E.T.Y., Yau D.T.W., Chia N.H., Lo I.L.O., Sham P.C., Cheung T.T., Wong C.C.L., Ng I.O.L. Single-cell RNA sequencing shows the immunosuppressive landscape and tumor heterogeneity of HBV-associated hepatocellular carcinoma. Nat Commun. 2021;12:3684. doi: 10.1038/s41467-021-24010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun Y., Wu L., Zhong Y., Zhou K., Hou Y., Wang Z., Zhang Z., Xie J., Wang C., Chen D., Huang Y., Wei X., Shi Y., Zhao Z., Li Y., Guo Z., Yu Q., Xu L., Volpe G., Qiu S., Zhou J., Ward C., Sun H., Yin Y., Xu X., Wang X., Esteban M.A., Yang H., Wang J., Dean M., Zhang Y., Liu S., Yang X., Fan J. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell. 2021;184:404–421.e16. doi: 10.1016/j.cell.2020.11.041. [DOI] [PubMed] [Google Scholar]
- 4.Avila Cobos F., Alquicira-Hernandez J., Powell J.E., Mestdagh P., De Preter K. Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat Commun. 2020;11:5650. doi: 10.1038/s41467-020-19015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin H., Liu Z. A benchmark for RNA-seq deconvolution analysis under dynamic testing environments. Genome Biol. 2021;22:102. doi: 10.1186/s13059-021-02290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Svensson V., Vento-Tormo R., Teichmann S.A. Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc. 2018;13:599–604. doi: 10.1038/nprot.2017.149. [DOI] [PubMed] [Google Scholar]
- 7.Picelli S., Bjorklund A.K., Faridani O.R., Sagasser S., Winberg G., Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- 8.Wang X., He Y., Zhang Q., Ren X., Zhang Z. Direct comparative analyses of 10X Genomics Chromium and Smart-seq2. Genomics Proteomics Bioinformatics. 2021;19:253–266. doi: 10.1016/j.gpb.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuksin M., Morel D., Aglave M., Danlos F.X., Marabelle A., Zinovyev A., Gautheret D., Verlingue L. Applications of single-cell and bulk RNA sequencing in onco-immunology. Eur J Cancer. 2021;149:193–210. doi: 10.1016/j.ejca.2021.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Zheng H., Pomyen Y., Hernandez M.O., Li C., Livak F., Tang W., Dang H., Greten T.F., Davis J.L., Zhao Y., Mehta M., Levin Y., Shetty J., Tran B., Budhu A., Wang X.W. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology. 2018;68:127–140. doi: 10.1002/hep.29778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim C.J., Lee Y.H., Pan L., Lai L.Y., Chua C., Wasser M., Lim T.K.H., Yeong J., Toh H.C., Lee S.Y., Chan C.Y., Goh B.K.P., Chung A., Heikenwalder M., Ng I.O.L., Chow P., Albani S., Chew V. Multidimensional analyses reveal distinct immune microenvironment in hepatitis B virus-related hepatocellular carcinoma. Gut. 2019;68:916–927. doi: 10.1136/gutjnl-2018-316510. [DOI] [PubMed] [Google Scholar]
- 12.Ho D.W.H., Tsui Y.M., Sze K.M.F., Chan L.K., Cheung T.T., Lee E., Sham P.C., Tsui S.K.W., Lee T.K.W., Ng I.O.L. Single-cell transcriptomics reveals the landscape of intra-tumoral heterogeneity and sternness-related subpopulations in liver cancer. Cancer Lett. 2019;459:176–185. doi: 10.1016/j.canlet.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Losic B., Craig A.J., Villacorta-Martin C., Martins S.N., Akers N., Chen X.T., Ahsen M.E., von Felden J., Labgaa I., D'Avola D., Allette K., Lira S.A., Furtado G.C., Garcia-Lezana T., Restrepo P., Stueck A., Ward S.C., Fiel M.I., Hiotis S.P., Gunasekaran G., Sia D., Schadt E.E., Sebra R., Schwartz M., Llovet J.M., Thung S., Stolovitzky G., Villanueva A. Intratumoral heterogeneity and clonal evolution in liver cancer. Nat Commun. 2020;11:291. doi: 10.1038/s41467-019-14050-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song G.H., Shi Y., Zhang M.Y., Goswami S., Afridi S., Meng L., Ma J.Q., Chen Y., Lin Y.P., Zhang J., Liu Y.M., Jin Z.J., Yang S.X., Rao D.N., Zhang S., Ke A.W., Wang X.Y., Cao Y., Zhou J., Fan J., Zhang X.M., Xi R.B., Gao Q. Global immune characterization of HBV/HCV-related hepatocellular carcinoma identifies macrophage and T-cell subsets associated with disease progression. Cell Discov. 2020;6:90. doi: 10.1038/s41421-020-00214-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Y.F., Wu L., Liu S.P., Jiang M.M., Hu B., Zhou K.Q., Guo W., Xu Y., Zhong Y., Zhou X.R., Zhang Z.F., Liu G., Liu S., Shi Y.H., Ji Y., Du M., Li N.N., Li G.B., Zhao Z.K., Huang X.Y., Xu L.Q., Yu Q.C., Peng D.H., Qiu S.J., Sun H.C., Dean M., Wang X.D., Chung W.Y., Dennison A.R., Zhou J., Hou Y., Fan J., Yang X.R. Dissecting spatial heterogeneity and the immune-evasion mechanism of CTCs by single-cell RNA-seq in hepatocellular carcinoma. Nat Commun. 2021;12:4091. doi: 10.1038/s41467-021-24386-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen V.L., Huang Q., Harouaka R., Du Y., Lok A.S., Parikh N.D., Garmire L.X., Wicha M.S. A dual-filtration system for single-cell sequencing of circulating tumor cells and clusters in HCC. Hepatol Commun. 2022 Jan 23 doi: 10.1002/hep4.1900. [E-pub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Juhling F., Saviano A., Ponsolles C., Heydmann L., Crouchet E., Durand S.C., El Saghire H., Felli E., Lindner V., Pessaux P., Pochet N., Schuster C., Verrier E.R., Baumert T.F. Hepatitis B virus compartmentalization and single-cell differentiation in hepatocellular carcinoma. Life Sci Alliance. 2021;4 doi: 10.26508/lsa.202101036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang J., Chen W., Ye J., Ni C., Zhai W. Single-cell transcriptomics analysis reveals intratumoral heterogeneity and identifies a gene signature associated with prognosis of hepatocellular carcinoma. Biosci Rep. 2022;42 doi: 10.1042/BSR20212560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan X., Wu Y., Zhang S., Liu Z., Fan Q., Fang S., Qiao S., Sun F., Liang C. Activation of FcRn mediates a primary resistance response to sorafenib in hepatocellular carcinoma by single-cell RNA sequencing. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.709343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan Y., Chen X., Wu M., Zhu W., Arslan A., Takeda S., Nguyen M.H., Majeti R., Thomas D., Zheng M., Peltz G. The phosphatidylethanolamine biosynthesis pathway provides a new target for cancer chemotherapy. J Hepatol. 2020;72:746–760. doi: 10.1016/j.jhep.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vong J.S.L., Ji L., Heung M.M.S., Cheng S.H., Wong J., Lai P.B.S., Wong V.W.S., Chan S.L., Chan H.L.Y., Jiang P., Chan K.C.A., Chiu R.W.K., Lo Y.M.D. Single cell and plasma RNA sequencing for RNA liquid biopsy for hepatocellular carcinoma. Clin Chem. 2021;67:1492–1502. doi: 10.1093/clinchem/hvab116. [DOI] [PubMed] [Google Scholar]
- 22.Dong X., Wang F., Liu C., Ling J., Jia X., Shen F., Yang N., Zhu S., Zhong L., Li Q. Single-cell analysis reveals the intra-tumor heterogeneity and identifies MLXIPL as a biomarker in the cellular trajectory of hepatocellular carcinoma. Cell Death Discov. 2021;7:14. doi: 10.1038/s41420-021-00403-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen W.S., Liang Y., Zong M., Liu J.J., Kaneko K., Hanley K.L., Zhang K., Feng G.S. Single-cell transcriptomics reveals opposing roles of Shp2 in Myc-driven liver tumor cells and microenvironment. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.109974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma L., Wang L., Khatib S.A., Chang C.W., Heinrich S., Dominguez D.A., Forgues M., Candia J., Hernandez M.O., Kelly M., Zhao Y., Tran B., Hernandez J.M., Davis J.L., Kleiner D.E., Wood B.J., Greten T.F., Wang X.W. Single-cell atlas of tumor cell evolution in response to therapy in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. J Hepatol. 2021;75:1397–1408. doi: 10.1016/j.jhep.2021.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng B., Wang D., Qiu X., Luo G., Wu T., Yang S., Li Z., Zhu Y., Wang S., Wu R., Sui C., Gu Z., Shen S., Jeong S., Wu X., Gu J., Wang H., Chen L. Trajectory and functional analysis of PD-1(high) CD4(+)CD8(+) T cells in hepatocellular carcinoma by single-cell cytometry and transcriptome sequencing. Adv Sci (Weinh) 2020;7 doi: 10.1002/advs.202000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng C., Zheng L., Yoo J.K., Guo H., Zhang Y., Guo X., Kang B., Hu R., Huang J.Y., Zhang Q., Liu Z., Dong M., Hu X., Ouyang W., Peng J., Zhang Z. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–1356.e16. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q., He Y., Luo N., Patel S.J., Han Y., Gao R., Modak M., Carotta S., Haslinger C., Kind D., Peet G.W., Zhong G., Lu S., Zhu W., Mao Y., Xiao M., Bergmann M., Hu X., Kerkar S.P., Vogt A.B., Pflanz S., Liu K., Peng J., Ren X., Zhang Z. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019;179:829–845.e20. doi: 10.1016/j.cell.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Ma L., Hernandez M.O., Zhao Y., Mehta M., Tran B., Kelly M., Rae Z., Hernandez J.M., Davis J.L., Martin S.P., Kleiner D.E., Hewitt S.M., Ylaya K., Wood B.J., Greten T.F., Wang X.W. Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer Cell. 2019;36:418–430.e6. doi: 10.1016/j.ccell.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang S., Liu Z., Wu D., Chen L., Xie L. Single-cell RNA-seq analysis reveals microenvironmental infiltration of plasma cells and hepatocytic prognostic markers in HCC with cirrhosis. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.596318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Najafi M., Farhood B., Mortezaee K. Cancer stem cells (CSCs) in cancer progression and therapy. J Cell Physiol. 2019;234:8381–8395. doi: 10.1002/jcp.27740. [DOI] [PubMed] [Google Scholar]
- 31.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez H., Hagerling C., Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 2018;32:1267–1284. doi: 10.1101/gad.314617.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toubal A., Nel I., Lotersztajn S., Lehuen A. Mucosal-associated invariant T cells and disease. Nat Rev Immunol. 2019;19:643–657. doi: 10.1038/s41577-019-0191-y. [DOI] [PubMed] [Google Scholar]
- 34.Heinrich B., Gertz E.M., Schaffer A.A., Craig A., Ruf B., Subramanyam V., McVey J.C., Diggs L.P., Heinrich S., Rosato U., Ma C., Yan C., Hu Y., Zhao Y., Shen T.W., Kapoor V., Telford W., Kleiner D.E., Stovroff M.K., Dhani H.S., Kang J., Fishbein T., Wang X.W., Ruppin E., Kroemer A., Greten T.F., Korangy F. The tumour microenvironment shapes innate lymphoid cells in patients with hepatocellular carcinoma. Gut. 2022;71:1161–1175. doi: 10.1136/gutjnl-2021-325288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei Y., Lao X.M., Xiao X., Wang X.Y., Wu Z.J., Zeng Q.H., Wu C.Y., Wu R.Q., Chen Z.X., Zheng L.M., Li B., Kuang D.M. Plasma cell polarization to the immunoglobulin G phenotype in hepatocellular carcinomas involves epigenetic alterations and promotes hepatoma progression in mice. Gastroenterology. 2019;156:1890–1904.e16. doi: 10.1053/j.gastro.2019.01.250. [DOI] [PubMed] [Google Scholar]
- 36.Bottcher J.P., Bonavita E., Chakravarty P., Blees H., Cabeza-Cabrerizo M., Sammicheli S., Rogers N.C., Sahai E., Zelenay S., Sousa C.R.E. NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell. 2018;172:1022–1037.e14. doi: 10.1016/j.cell.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smyth M.J., Hayakawa Y., Takeda K., Yagita H. New aspects of natural-killer-cell surveillance and therapy of cancer. Nat Rev Cancer. 2002;2:850–861. doi: 10.1038/nrc928. [DOI] [PubMed] [Google Scholar]
- 38.Barry K.C., Hsu J., Broz M.L., Cueto F.J., Binnewies M., Combes A.J., Nelson A.E., Loo K., Kumar R., Rosenblum M.D., Alvarado M.D., Wolf D.M., Bogunovic D., Bhardwaj N., Daud A.I., Ha P.K., Ryan W.R., Pollack J.L., Samad B., Asthana S., Chan V., Krummel M.F. A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat Med. 2018;24:1178–1191. doi: 10.1038/s41591-018-0085-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wan S., Kuo N., Kryczek I., Zou W., Welling T.H. Myeloid cells in hepatocellular carcinoma. Hepatology. 2015;62:1304–1312. doi: 10.1002/hep.27867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song S.S., Yuan P.F., Wu H.X., Chen J.Y., Fu J.J., Li P.P., Lu J.T., Wei W. Dendritic cells with an increased PD-L1 by TGF-beta induce T cell anergy for the cytotoxicity of hepatocellular carcinoma cells. Int Immunopharmacol. 2014;20:117–123. doi: 10.1016/j.intimp.2014.02.027. [DOI] [PubMed] [Google Scholar]
- 41.Wu S.Z., Al-Eryani G., Roden D.L., Junankar S., Harvey K., Andersson A., Thennavan A., Wang C., Torpy J.R., Bartonicek N., Wang T., Larsson L., Kaczorowski D., Weisenfeld N.I., Uytingco C.R., Chew J.G., Bent Z.W., Chan C.L., Gnanasambandapillai V., Dutertre C.A., Gluch L., Hui M.N., Beith J., Parker A., Robbins E., Segara D., Cooper C., Mak C., Chan B., Warrier S., Ginhoux F., Millar E., Powell J.E., Williams S.R., Liu X.S., O'Toole S., Lim E., Lundeberg J., Perou C.M., Swarbrick A. A single-cell and spatially resolved atlas of human breast cancers. Nat Genet. 2021;53:1334–1347. doi: 10.1038/s41588-021-00911-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leader A.M., Grout J.A., Maier B.B., Nabet B.Y., Park M.D., Tabachnikova A., Chang C., Walker L., Lansky A., Le Berichel J., Troncoso L., Malissen N., Davila M., Martin J.C., Magri G., Tuballes K., Zhao Z., Petralia F., Samstein R., D'Amore N.R., Thurston G., Kamphorst A.O., Wolf A., Flores R., Wang P., Muller S., Mellman I., Beasley M.B., Salmon H., Rahman A.H., Marron T.U., Kenigsberg E., Merad M. Single-cell analysis of human non-small cell lung cancer lesions refines tumor classification and patient stratification. Cancer Cell. 2021;39:1594–1609.e12. doi: 10.1016/j.ccell.2021.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Armingol E., Officer A., Harismendy O., Lewis N.E. Deciphering cell-cell interactions and communication from gene expression. Nat Rev Genet. 2021;22:71–88. doi: 10.1038/s41576-020-00292-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vento-Tormo R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., Park J.E., Stephenson E., Polanski K., Goncalves A., Gardner L., Holmqvist S., Henriksson J., Zou A., Sharkey A.M., Millar B., Innes B., Wood L., Wilbrey-Clark A., Payne R.P., Ivarsson M.A., Lisgo S., Filby A., Rowitch D.H., Bulmer J.N., Wright G.J., Stubbington M.J.T., Haniffa M., Moffett A., Teichmann S.A. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 46.Zitvogel L., Kroemer G. Targeting PD-1/PD-L1 interactions for cancer immunotherapy. Oncoimmunology. 2012;1:1223–1225. doi: 10.4161/onci.21335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang K., Wang X., Meng C., He L., Sang X., Zheng Y., Xu H. The application of single-cell sequencing technology in the diagnosis and treatment of hepatocellular carcinoma. Ann Transl Med. 2019;7:790. doi: 10.21037/atm.2019.11.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aliya S., Lee H., Alhammadi M., Umapathi R., Huh Y.S. An overview on single-cell technology for hepatocellular carcinoma diagnosis. Int J Mol Sci. 2022;23:1402. doi: 10.3390/ijms23031402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lei Y., Tang R., Xu J., Wang W., Zhang B., Liu J., Yu X., Shi S. Applications of single-cell sequencing in cancer research: progress and perspectives. J Hematol Oncol. 2021;14:91. doi: 10.1186/s13045-021-01105-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu J., Chen Y., Zhang X., Guo J., Xu K., Li L. A novel prognostic model based on single-cell RNA sequencing data for hepatocellular carcinoma. Cancer Cell Int. 2022;22:38. doi: 10.1186/s12935-022-02469-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun G., Li Z., Rong D., Zhang H., Shi X., Yang W., Zheng W., Sun G., Wu F., Cao H., Tang W., Sun Y. Single-cell RNA sequencing in cancer: applications, advances, and emerging challenges. Mol Ther Oncolytics. 2021;21:183–206. doi: 10.1016/j.omto.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee T.K., Guan X.Y., Ma S. Cancer stem cells in hepatocellular carcinoma - from origin to clinical implications. Nat Rev Gastroenterol Hepatol. 2022;19:26–44. doi: 10.1038/s41575-021-00508-3. [DOI] [PubMed] [Google Scholar]
- 53.Krenkel O., Hundertmark J., Ritz T.P., Weiskirchen R., Tacke F. Single cell RNA sequencing identifies subsets of hepatic stellate cells and myofibroblasts in liver fibrosis. Cells. 2019;8:503. doi: 10.3390/cells8050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Payen V.L., Lavergne A., Alevra Sarika N., Colonval M., Karim L., Deckers M., Najimi M., Coppieters W., Charloteaux B., Sokal E.M., El Taghdouini A. Single-cell RNA sequencing of human liver reveals hepatic stellate cell heterogeneity. JHEP Rep. 2021;3 doi: 10.1016/j.jhepr.2021.100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Affo S., Nair A., Brundu F., Ravichandra A., Bhattacharjee S., Matsuda M., Chin L., Filliol A., Wen W., Song X., Decker A., Worley J., Caviglia J.M., Yu L., Yin D., Saito Y., Savage T., Wells R.G., Mack M., Zender L., Arpaia N., Remotti H.E., Rabadan R., Sims P., Leblond A.L., Weber A., Riener M.O., Stockwell B.R., Gaublomme J., Llovet J.M., Kalluri R., Michalopoulos G.K., Seki E., Sia D., Chen X., Califano A., Schwabe R.F. Promotion of cholangiocarcinoma growth by diverse cancer-associated fibroblast subpopulations. Cancer Cell. 2021;39:883. doi: 10.1016/j.ccell.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang W., He H., Wang T., Su N., Zhang F., Jiang K., Zhu J., Zhang C., Niu K., Wang L., Yuan X., Liu N., Li L., Wei W., Hu J. Single-cell transcriptomic analysis reveals a hepatic stellate cell-activation roadmap and myofibroblast origin during liver fibrosis in mice. Hepatology. 2021;74:2774–2790. doi: 10.1002/hep.31987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang M., Yang H., Wan L., Wang Z., Wang H., Ge C., Liu Y., Hao Y., Zhang D., Shi G., Gong Y., Ni Y., Wang C., Zhang Y., Xi J., Wang S., Shi L., Zhang L., Yue W., Pei X., Liu B., Yan X. Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J Hepatol. 2020;73:1118–1130. doi: 10.1016/j.jhep.2020.05.039. [DOI] [PubMed] [Google Scholar]
- 58.Wu H., Kirita Y., Donnelly E.L., Humphreys B.D. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol. 2019;30:23–32. doi: 10.1681/ASN.2018090912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bakken T.E., Hodge R.D., Miller J.A., Yao Z., Nguyen T.N., Aevermann B., Barkan E., Bertagnolli D., Casper T., Dee N., Garren E., Goldy J., Graybuck L.T., Kroll M., Lasken R.S., Lathia K., Parry S., Rimorin C., Scheuermann R.H., Schork N.J., Shehata S.I., Tieu M., Phillips J.W., Bernard A., Smith K.A., Zeng H., Lein E.S., Tasic B. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS One. 2018;13 doi: 10.1371/journal.pone.0209648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Habib N., Avraham-Davidi I., Basu A., Burks T., Shekhar K., Hofree M., Choudhury S.R., Aguet F., Gelfand E., Ardlie K., Weitz D.A., Rozenblatt-Rosen O., Zhang F., Regev A. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods. 2017;14:955–958. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cavalli M., Diamanti K., Pan G., Spalinskas R., Kumar C., Deshmukh A.S., Mann M., Sahlen P., Komorowski J., Wadelius C. A multi-omics approach to liver diseases: integration of single nuclei transcriptomics with proteomics and HiCap bulk data in human liver. OMICS. 2020;24:180–194. doi: 10.1089/omi.2019.0215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andrews T.S., Atif J., Liu J.C., Perciani C.T., Ma X.Z., Thoeni C., Slyper M., Eraslan G., Segerstolpe A., Manuel J., Chung S., Winter E., Cirlan I., Khuu N., Fischer S., Rozenblatt-Rosen O., Regev A., McGilvray I.D., Bader G.D., MacParland S.A. Single-cell, single-nucleus, and spatial RNA sequencing of the human liver identifies cholangiocyte and mesenchymal heterogeneity. Hepatol Commun. 2022;6:821–840. doi: 10.1002/hep4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stoeckius M., Zheng S., Houck-Loomis B., Hao S., Yeung B.Z., Mauck W.M., 3rd, Smibert P., Satija R. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 2018;19:224. doi: 10.1186/s13059-018-1603-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mimitou E.P., Cheng A., Montalbano A., Hao S., Stoeckius M., Legut M., Roush T., Herrera A., Papalexi E., Ouyang Z., Satija R., Sanjana N.E., Koralov S.B., Smibert P. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat Methods. 2019;16:409–412. doi: 10.1038/s41592-019-0392-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Inarrairaegui M., Melero I., Sangro B. Immunotherapy of hepatocellular carcinoma: facts and hopes. Clin Cancer Res. 2018;24:1518–1524. doi: 10.1158/1078-0432.CCR-17-0289. [DOI] [PubMed] [Google Scholar]