Highlights
-
•
CBX7 was down-regulated, while twist and EphA2 were up-regulated in BLBC.
-
•
EphA2 or twist silencing inhibited BLBC cell proliferation and metastasis.
-
•
Twist bond to EphA2 and increased the expression of EphA2.
-
•
CBX7 blocked the binding of twist to EphA2 and inhibited EphA2 expression.
-
•
CBX7 regulated BLBC growth and metastasis via Twist/EphA2 axis.
Keywords: Basal-like breast cancer, CBX7, EphA2, Metastasis, Twist1
Abbreviations: BLBC, Basal-like breast cancer; EMT, epithelial-mesenchymal transition; CBX7, Chromobox protein homolog 7; CCK-8, Cell Counting Kit-8; IHC, Immunohistochemistry
Abstract
Background
Basal-like breast cancer (BLBC) is an important subtype of breast cancer. Twist1 is a key transcription factor in BLBC metastasis, which serves a key role in tumorigenesis. The potential mechanism of Twist1 in BLBC remains to be elucidated. Here, we explored the role and molecular mechanism of Twist1 in BLBC.
Methods
The levels of CBX7, Twist1 and EphA2 in BLBC tissues and cells were determined by Western blot. ChIP and dual-luciferase reporter assays confirmed the interaction between CBX7, Twist1 and EphA2 promoter. The cellular functions were analyzed by CCK-8, colony formation, wound healing and Transwell assays. Expression of EMT related proteins was analyzed by Western blot. IHC measured the expression of CBX7, Twist1 and EphA2 in tumor tissues.
Results
CBX7 was down-regulated in BLBC tissues and cells, whereas Twist1 and EphA2 were up-regulated. Twist1 silencing inhibited the cell migration, invasion and cancer metastasis of BLBC through targeting EphA2 and regulating EphA2 expression. Additionally, CBX7 blocked the binding of Twist1 to EphA2 promoter and inhibited EphA2 expression and suppressed BLBC growth and metastasis via Twist1/EphA2 axis.
Conclusion
CBX7 suppresses BLBC growth and metastasis through Twist1/EphA2 pathway. Our study may provide evidence and new therapeutic targets for the comprehensive treatment of BLBC.
Introduction
Breast cancer is one of the most common malignant tumors and the leading cause of cancer death among women worldwide [1]. According to the different gene expression profiles, breast cancer is divided into four subtypes: Tubular A (Luminal A), Tubular B (Luminal B), ErbB2 and basal-like [2]. Basal-like breast cancer (BLBC) expresses low levels of estrogen receptor, progesterone receptor and epidermal growth factor receptor 2, and highly expresses several basal markers such as cytokeratins 5/6 and cytokeratin14 [3]. BLBC is prone to early metastasis and spread to the brain and lungs [4]. High invasiveness and metastasis of BLBC contribute to its poor clinical outcome and prognosis [5]. Studies showed that BLBC cells are more prone to epithelial-mesenchymal transition (EMT), thereby triggering tumor cell metastasis more easily [6]. Therefore, probing the molecular mechanism of BLBC metastasis has a very important role in the identification of new therapeutic targets.
Twist1 is a member of the basic helix-loop-helix protein family and is a transcription factor with a helix-loop-helix domain [7]. Studies have found that Twist1 was highly expressed in many malignant tumors such as breast cancer, and it was proved to be related to tumor metastasis and prognosis [8]. Subsequent studies confirmed that Twist1, which acted as transcription factors, can regulate cell migration and invasion of EMT target genes [9]. For example, Twist1 can bind to E-box, and can also bind to the Slug promoter region to regulate the transcription of the genes, thereby inhibiting E-cadherin and promoting the progress of EMT [10,11]. As a transcription factor, Twist1 usually binds to the promoter regions of a variety of target genes and initiates the transcription process. Our previous data have shown that Twist1 can bind to the promoter of the receptor tyrosine kinase ROR1 to up-regulate ROR1 transcription and promote breast cancer metastasis [12]. However, the regulatory mechanism of Twist1 in breast cancer is still not fully characterized.
EphA2 belongs to the Eph kinase family, which is the largest family of receptor tyrosine kinases [13]. When EphA2 binds with its ligand Ephrin A1, they regulate cell-cell interactions and many diseases, including breast cancer [14]. EphA2 is significantly highly expressed in BLBC, and its high expression often leads to poor prognosis [15]. In our previous research, we screened out a series of potential Twist1 target genes through gene chip analysis of cell lines overexpressing Twist1, and EphA2 was suddenly listed. We verified that EphA2 was significantly increased in T-47D cells overexpressing Twist1. Thus, we speculated that Twist1 could regulate EphA2 to promote the metastasis of BLBC.
Chromobox protein homolog 7 (CBX7), an important member from the Chromobox family, participates in the formation of polycomb repressive complex and is found to be crucial for regulating breast cancer progression. For example, CBX7 expression was strongly correlated with breast tumor subtype aggressiveness and the proliferation markers [16]. In addition, previous studies found that CBX7 bound to E-box and hindered Twist1′s transcription of the miR-199a promoter [17]. According to the TCGA database analysis, CBX7 is low expressed in BLBC. Therefore, we speculate that CBX7 hinders Twist1′s transcriptional regulation of EphA2, and then down-regulates EphA2 to affect the EMT and metastasis of BLBC.
In this study, we identified that Twist1 as a novel factor for the transcriptional activation of EphA2 in BLBC. CBX7 blocked the binding of Twist1 to EphA2 promoter and suppressed BLBC growth and metastasis through Twist1/EphA2 axis. Our results provide potential therapeutic agent for the treatment of BLBC.
Materials and methods
Clinical specimens
50 cases of basal-like breast cancer tissues and the adjacent tissues were obtained from surgically resected samples of BLBC patients from March 2018 to May 2019 in Xiangya Third Hospital of Central South University. Patients did not receive any radiotherapy or chemotherapy previously. Written informed consent was provided by patients. Samples were collected and stored in liquid nitrogen. Our research was approved by Clinical Research Ethics Committee of Xiangya Third Hospital of Central South University (No.2019-S144).
Cell culture
Human basal cell-like breast cancer cells (MCF-7, T47D, SK-BR-3, Hs578T, MDA-MB-231) and normal breast epithelial cells (MCF-10A) were provided by the American Type Culture Collection (ATCC, Manassas, VA, USA). These cells were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) containing 10% FBS (Gibco, Grand Island, NY, USA) and 1% Penicillin-Streptomycin (Invitrogen) at 37 °C in 5% CO2.
Cell transfection
The short hairpin RNA (shRNA) targeting EphA2 (shEphA2–1, shEphA2–2), Twist1 (shTwist1–1, shTwist1–2) and CBX7 (shCBX7) were purchased from Sigma (St Louis, MO, USA). The open reading frames of Twist1 or CBX7 genes were cloned into the pLenti6.3/V5-puro vector by gene cloning. Constructs were cotransfected with packaging plasmids into HEK-293T cells. Viral fluids were collected after 24 h. For stable cell generation, the BLBC cell lines were infected and selected with puromycin.
Western blot
Cells were lysed by RIPA to extract the total protein. The protein concentration was determined using the BCA method. Then 10% SDS-PAGE electrophoresis was performed and the membrane was transferred to the PVDF membrane (Millipore, Boston, MA, USA), followed by blocking with 5% skim milk for 1 h. The membranes were incubated with the primary antibody at 4 °C overnight: anti-CBX7 (ab21873, 1:1000, Abcam, Cambridge, MA, USA), anti-Twist1 (#69,366, 1:1000, Cell Signaling Technology, Danvers, MA, USA), anti-EphA2 (34–7400, 1:1000, Thermo Fisher Scientific, Waltham, MA, USA), anti-E-cadherin (ab133597, 1:1000, Abcam), anti-N-cadherin (ab76011, 1:1000, Abcam), anti-Vimentin (#5741, 1:1000, Cell Signaling Technology), anti-Slug-1 (ab27568, 1:1000, Abcam). The next day, the secondary antibody (#7074, 1:1000, Cell Signaling Technology) was incubated for 1 h. Finally, the X-ray film was developed.
Cell counting kit-8 (CCK-8) assay
Cells were seeded on 96-well plates at a density of 3000 cells/well. Cell viability was tested at 0, 24, 48 and 72 h after incubation. 100 μL CCK-8 reagent (Beyotime, Shanghai, China) was added into each well for 2 h at 37 °C. The optical density value was determined at 450 nm by a microplate reader (Bio-Rad Laboratories Inc, Hercules, CA, USA).
Colony formation assay
Cells were cultured on 6-well plates (500 per well) and incubated at 37 °C with 5% CO2 for 2 weeks. Colonies were fixed with 4% paraformaldehyde for 30 min and then stained using 0.5% crystal violet solution (Sigma). Finally, cell colonies were observed under a microscope (Olympus, Tokyo, Japan).
Wound healing assay
For detecting cell migration ability, cells were incubated on six-well plates. When the cell density reached 80% confluence, three scratches were made across the bottom of the plates in each well with a 200 μL pipette tip. The cells were then cultured in serum-free medium with proliferation inhibitor of ERK (PD98059, 10 μM, Cell Signaling Technology). After 24 h, the scratch areas were photographed.
Transwell assay
Firstly, the inner side of the filter membrane was coated with Matrigel or without Matrigel at 37 °C for 2 h. Hs578T and MDA-MB-231 cells were pretreated with proliferation inhibitor of ERK (PD98059, 10 μM). After the matrigel was solidified, Hs578T and MDA-MB-231 cells were resuspended with 200 μL serum-free DMEM medium, and then inoculated into the upper chamber of Transwell chamber. Then, DMEM medium containing 20% serum was added into the lower chamber. After cultured at 37 °C and 5% CO2 for 12 h, the invaded cells were fixed in 4% formaldehyde, stained with 0.1% crystal violet, and observed under a microscope (Olympus).
Method for predicting E-box
First, the human EphA2 promoter sequence (−2000–1) was found according to UCSC (http://genome-asia.ucsc.edu/), and then the sequence of the E-box (−689 to −683 of the EphA2 promoter region) was found according to the palindrome sequence CACGTG of the E-box.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed in Hs578T and MDA-MB-231 cells with a ChIP Assay Kit (Cell Signaling Technology). Cells were crosslinked with 1% formaldehyde for 10 min at room temperature. After that, cells were lysed using lysis buffer with protease inhibitors. Aliquots of lysates were used for each immunoprecipitation reaction with an equal amount of anti-Twist1 antibody (#69,366, Cell Signaling Technology), anti-CBX7 antibody (ab91431, Abcam) or IgG (#2729, Cell Signaling Technology) overnight at 4 °C with Protein A/G magnetic beads (Thermo Fisher Scientific). One-fourth of the chromatin solution was used for total input. Subsequently, the immunoprecipitates were reverse cross-linked. The precipitated DNA was analyzed using RT-qPCR with the specific primers of EphA2 promoter. Primer sequence of Twist1 binding to EphA2 promoter: Forward: 5′-TCCCGTTTGTTTCACTCACA-3′, 20 nt; Reverse: 5′-CTGGCATGGGTTGATAGGTT-3′, 20 nt.
Primer sequence of CBX7 binding to EphA2 promoter (E-box): Forward: 5′-TCAAAACCTATCAACCCATGC-3′, 21 nt; Reverse: 5′- AGAAACAACATGCCCAAAGG-3′, 20 nt.
Dual-luciferase reporter assay
The potential binding sequences of Twist1 on the EphA2 promoter or CBX7 (E-box) on EphA2 promoter were mutated using the QuikChange™ Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). EphA2 promoter region (−831 to −822) containing the wild-type (WT-) or mutant (Mut-) Twist1 binding site (WT-Twist1, AGGCAGGTGT; Mut-Twist1, TCCGTCCACA), and EphA2 promoter region (−689 to −683) containing WT-E-box (CACGTG) or Mut-E-box (TATGAT) were cloned into pGL3-basic vector (Promega, Madison, WI, USA). Cells were seeded in 24-well plates until growth to 80% confluence. The EphA2 promoter constructs and Twist1 and/or CBX7 were co-transfected into the indicated cells using Lipofectamine 3000 reagent following the manufacturer's protocol. The luciferase activity was conducted after 48 h of transfection with Dual luciferase reporter system (Promega).
Immunofluorescence
Cells were plated on a petri dish. After attachment, cells were fixed with 4% paraformaldehyde (Sigma) for 15 min. Next, cells were permeabilized with 0.5% Triton X-100 solution (Sigma) for 5 min. Goat serum blocking solution was used for blocking for 30 min. Cells were probed using anti-E-cadherin (ab133597, 1:1000, Abcam), anti-N-cadherin (ab76011, 1:1000, Abcam) at 4 °C overnight. The second antibody was labeled with fluorescent dye (A0516, A0562, 1:200, Beyotime) and incubated in the dark for 1 h. DAPI (1:5000, Beyotime) staining was performed for 5 min to stain the nuclei. Finally, slides were mounted and the protein localization and expression were observed under a laser confocal microscope (Olympus).
Immunohistochemistry (IHC)
Paraffin embedded tissues were sectioned, deparaffinized and rehydrated. Antigen retrieval was conducted with 10 mM citrate buffer (pH 6.0) in microwave. Following incubation with 5% normal goat serum for 30 min, slices were incubated at 4 °C overnight with primary antibodies against CBX7 (ab21873, 1:200, Abcam), Twist1 (#69,366, 1:200, Cell Signaling Technology), EphA2 (34–7400, 1:500, Thermo Fisher Scientific). After incubation by HRP-labeled secondary antibody (#7074, 1:1000, Cell Signaling Technology) for 1 h, slides were visualized with a light microscope (Olympus).
Nude mice xenograft
Five-week-old female BABL/c nude mice (n = 8/group) were obtained from Shanghai laboratory Animal Center of Chinese Academy of Sciences (Shanghai, China). For tumorigenesis experiment, mice were subcutaneously injected with 2 × 106 stably transfected cells in 150 μL PBS. Tail vein injection (150 μL, 2 × 106 cells) was conducted to verify metastasis. Tumor weights were measured and tumor volume = (Width2 ˟ Length)/2. After 45 days, the mice were sacrificed and lung tissues were taken out. The metastatic nodules in lungs were counted. HE staining was used to detect the tumor metastases in lung. The mice xenograft tumor experiments were permitted by the Animal Ethics Committee of Xiangya Third Hospital of Central South University (No.2019-S144).
Statistical analysis
All the data were expressed as means ± SD. The significance of difference between any two samples was analyzed using Student's t-test with GraphPad Prism 7 (Graphpad, La Jolla, CA, USA). ANOVA evaluated differences among multiple-group comparison. The relationship between Twist1 and CBX7, EphA2 in BLBC tissues was tested by using Pearson analysis. The association between CBX7, Twist1, EphA2 expression and clinicopathological characteristics of BLBC patients was analyzed with chi-square test. At least three independent experiments were performed. A P value <0.05 was considered as statistically significant.
Results
CBX7 was down-regulated, while Twist1 and EphA2 were increased in BLBC tissues and cells
We firstly assayed CBX7, Twist1 and EphA2 expression by IHC in BLBC tissues and normal adjacent tissues. Compared with adjacent normal tissues, CBX7 was lowly expressed, while Twist1 and EphA2 were highly expressed in BLBC tissues (Fig. 1A). Moreover, Western blot revealed that the protein levels of CBX7, Twist1 and EphA2 had similar trends to the above results (Fig. 1B). Chi-square test showed that EphA2 expression was not associated with the age and gender, but was related to BLBC grade, stage and lymph node metastasis. High expression of EphA2 was related to the advanced type of BLBC and lymph node metastasis (Table 1). Moreover, low expression of CBX7 or high expression of Twist1 and EphA2 was related to the advanced grade, invasion and lymph node metastasis of BLBC (Fig. 1C). In addition, Pearson analysis showed that CBX7 was negatively correlated with EphA2, while Twist1 and EphA2 were positively correlated in BLBC tissues (Fig. 1D). Furthermore, compared with breast epithelial cells, CBX7 was down-regulated, while Twist1 and EphA2 were up-regulated in breast cancer cells. Hs578T and MDA-MB-231 were the most obvious cell lines (Fig. 1E). These results suggest that CBX7, Twist1 and EphA2 may regulate BLBC progression.
Fig. 1.
CBX7 was down-regulated, while Twist1 and EphA2 were increased in BLBC. (A) IHC examined CBX7, Twist1 and EphA2 expression in BLBC tissues and adjacent normal breast tissues. Scar bar = 50 µm. (B) The levels of CBX7, Twist1 and EphA2 in BLBC tissues (n = 10) and adjacent normal breast tissues (n = 10) were determined by Western blot. (C) Chi-square test was used to detect the correlation between CBX7, Twist1, EphA2 expression and clinicopathological characteristics of BLBC patients. (D) Pearson analysis showed the correlation between the expression of CBX7 and EphA2, Twist1 and EphA2 in BLBC tissues. (E) The levels of CBX7, Twist1 and EphA2 in five breast cancer cell lines and breast epithelial cells were determined by Western blot. *P<0.05, **P<0.01, ***P<0.001.
Table 1.
The correlation between EphA2 expression and clinicopathological characteristics of BLBC patients.
| EphA2 expression | |||||
| Variable | (n = 50) | Low | High | P value | |
| Age | |||||
| <60years | 19 | 9 | 10 | 0.4929 | |
| ≥60 years | 26 | 15 | 11 | ||
| Gender | |||||
| Male | 22 | 8 | 14 | 0.1806 | |
| Female | 27 | 15 | 12 | ||
| Grade | |||||
| Low | 9 | 3 | 6 | 0.0440* | |
| High | 41 | 29 | 10 | ||
| stage | |||||
| I+II | 11 | 4 | 7 | 0.0041** | |
| III+IV | 42 | 35 | 7 | ||
| Lyph node status | |||||
| N0 | 40 | 30 | 10 | 0.02638* | |
| N1, N2 | 10 | 2 | 5 | ||
* P<0.05, **P<0.01.
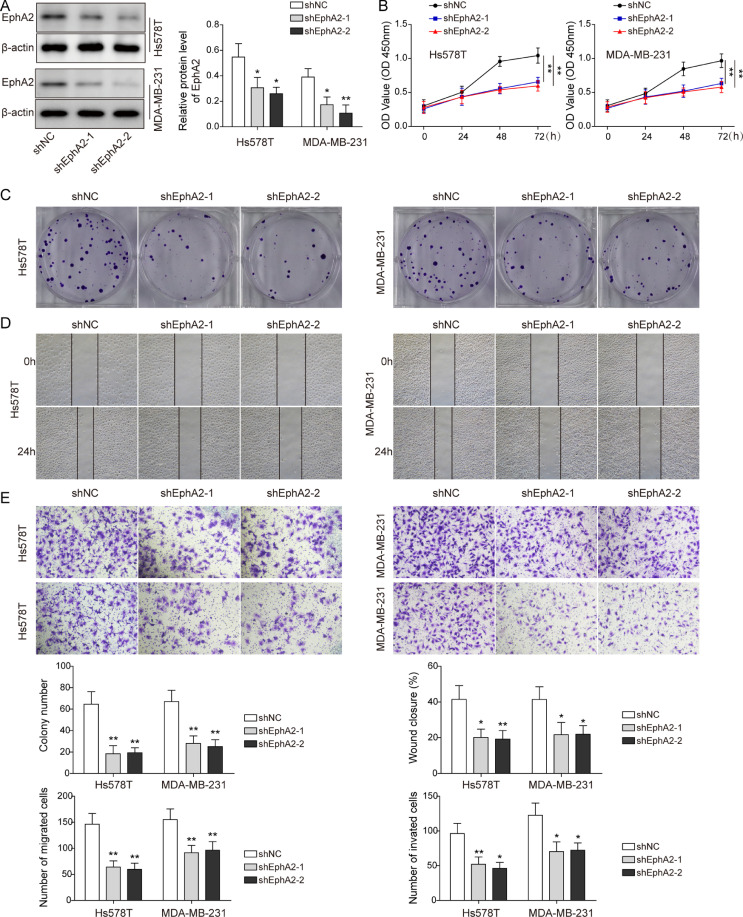
EphA2 knockdown inhibited the proliferation and metastasis of BLBC
To investigate the role of EphA2 in BLBC, cancer cells were transfected with shEphA2. Compared to control cells, EphA2 was obviously reduced in cells with EphA2 silencing (Fig. 2A). Cancer cell viability, proliferation and migration were greatly suppressed with EphA2 silencing (Fig. 2B-D). Additionally, knockdown of EphA2 impaired cell invasion ability (Fig. 2E). Besides, EphA2 downregulation increased the fluorescence intensity of E-cadherin, but decreased N-cadherin intensity (Supplemental Figure 1A). Moreover, knockdown of EphA2 increased the expression of E-cadherin, but decreased the expression of N-cadherin, Slug-1 and Vimentin (Supplemental Figure 1B). To further explore the role of EphA2 in BLBC growth and metastasis in vivo, cells with shEphA2 were injected into nude mice. EphA2 silencing reduced tumor volume and weight compared with control group (Supplemental Figure 1C). The lung lesions and metastatic nodules were significantly reduced with EphA2 silencing (Supplemental Figure 1D). Collectively, these data suggest that EphA2 knockdown suppresses proliferation and metastasis of BLBC.
Fig. 2.
EphA2 knockdown inhibited the proliferation and metastasis of BLBC. Cells were transfected with shEphA2 or shNC. (A) Western blot measured EphA2 protein level. (B-D) CCK-8, colony formation, wound healing experiments detected cell viability, proliferation and migration. (E) Transwell assay tested cell migration and invasion. Data are the means ± SD for three independent experiments. *P<0.05, **P<0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Knockdown of Twist1 suppressed the proliferation and metastasis of BLBC
Next, cancer cells were transfected with shTwist1. Knockdown of Twist1 significantly decreased the expression level of Twist1 (Fig. 3A). Besides, with Twist1 silencing, cell growth, migration and invasion abilities were all impaired (Fig. 3B-E). Twist1 knockdown upregulated the E-cadherin fluorescence intensity, but downregulated N-cadherin intensity (Supplemental Figure 2A). Additionally, Twist1 silencing increased E-cadherin level. However, the levels of N-cadherin, Slug-1 and Vimentin were decreased (Supplemental Figure 2B). For in vivo experiments, we found that with Twist1 knockdown, the tumor became smaller, lost weight and reduced volume (Supplemental Figure 2C). The lung lesions and metastatic nodules were greatly reduced with Twist1 silencing (Supplemental Figure 2D). Thus, Twist1 knockdown inhibits proliferation and metastasis of BLBC.
Fig. 3.
Knockdown of Twist1 suppressed the proliferation and metastasis of BLBC. Cancer cells were transfected with shTwist1. (A) Western blot measured Twist1 protein level. (B-D) CCK-8, colony formation, wound healing experiments detected cell viability, proliferation and migration. (E) Transwell assay tested cell migration and invasion. Data are the means ± SD for three independent experiments. *P<0.05, **P<0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
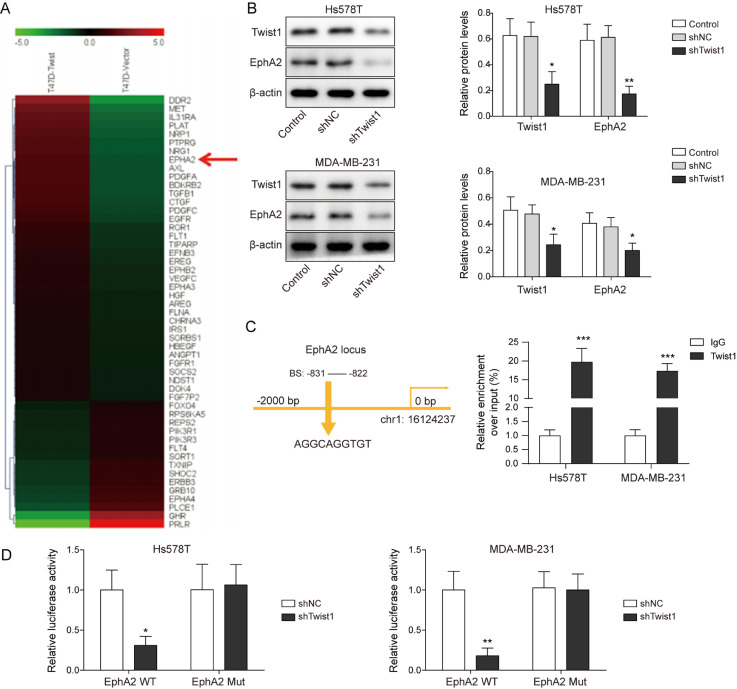
Twist1 bound to EphA2 promoter and increased the expression of EphA2
By analyzing the gene chip results of the Twist1 overexpression cell line, we found that EphA2 was also highly expressed in Twist1-overexpression T-47D cells (Fig. 4A). Cancer cells were then transfected with shTwist1. Decreased levels of Twist1 and EphA2 were observed in cells with shTwist1 compared to control group (Fig. 4B). Additionally, ChIP assay indicated that the binding of Twist1 to EphA2 promoter was decreased with Twist1 knockdown (Fig. 4C). Dual-luciferase reporter assay revealed that with Twist1 silencing, the binding of Twist1 to wild EphA2 promoter was reduced, but the binding to mutant EphA2 promoter remained unchanged (Fig. 4D). These data demonstrate that Twist1 binds to EphA2 promoter and increases the expression of EphA2.
Fig. 4.
Twist1 bound to EphA2 promoter and increased the expression of EphA2. (A) EphA2 expression increased in gene chip. Hs578T and MDA-MB-231 cells were transfected with shTwist1 or negative controls. (B) Levels of Twist1 and EphA2 were detected by Western blot. (C) ChIP assay detected Twist1 binding to EphA2 promoter. (D) Dual luciferase reporter assay detected the binding of Twist1 to EphA2 promoter mutant and wild site. *P<0.05, **P<0.01.
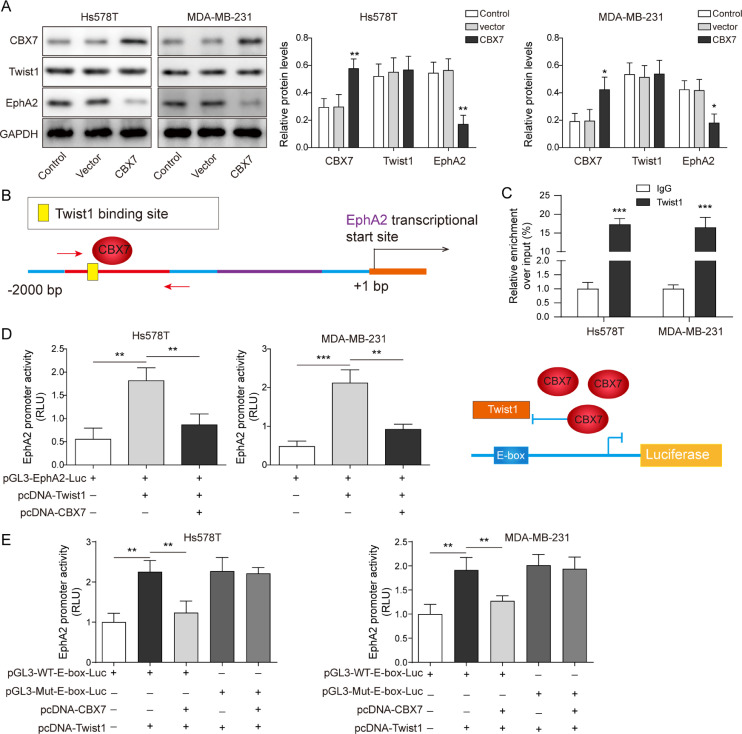
CBX7 blocked the binding of Twist1 to EphA2 promoter and transcriptionally inhibited the expression of EphA 2
We further explored the regulatory relationship between CBX7 and Twist1 in BLBC development. Cancer cells were overexpressed with CBX7. Overexpression of CBX7 increased CBX7 expression, but decreased EphA2 expression (Fig. 5A). We predicted that CBX7 could target E-box and prevent Twist1 from binding to EphA2 promoter, thereby inhibiting Twist1′s transcriptional regulation of EphA2 (Fig. 5B). To test this hypothesis, ChIP assay indicated that the CBX7 binding to the E-box on EphA2 promoter was increased with CBX7 overexpression (Fig. 5C). Dual-luciferase reporter assay revealed that with CBX7 overexpression, the binding of CBX7 to WT-Twist1 increased, which hindered the transcriptional regulation of EphA2 by Twist1 (Fig. 5D). We further constructed pGL3-WT-E-box-Luc and pGL3-Mut-E-box-Luc for dual-luciferase experiments. The results showed that in pGL3-Mut-E-box-Luc group, CBX7 did not bind to the E-box, resulting in increased binding of Twist1 to EphA2 promoter and increased luciferase activity (Fig. 5E). These results suggest that CBX7 blocks the binding of Twist1 to EphA2 promoter and transcriptionally inhibits the expression of EphA2.
Fig. 5.
CBX7 blocked the binding of Twist1 to EphA2 promoter and transcriptionally inhibited the expression of EphA2. Hs578T and MDA-MB-231 cells were overexpressed with CBX7. (A) The levels of CBX7, Twist1 and EphA2 were detected by Western blot. (B) The schematic diagram showed the regulatory relationship between CBX7 and Twist1 on the EphA2 promoter. (C) ChIP assay detected CBX7 binding to the E-box (CACGTG) on EphA2 promoter. (D) Dual luciferase reporter assay detected the binding of CBX7 and Twist1 to EphA2 promoter. (E) We further constructed pGL3-WT-E-box-Luc and pGL3-Mut-E-box-Luc. Dual luciferase experiment s confirmed that E-Box sequence was indispensable for CBX7 and Twist1 competition. Data are the means ± SD for three independent experiments. *P<0.05, **P<0.01, ***P<0.001.
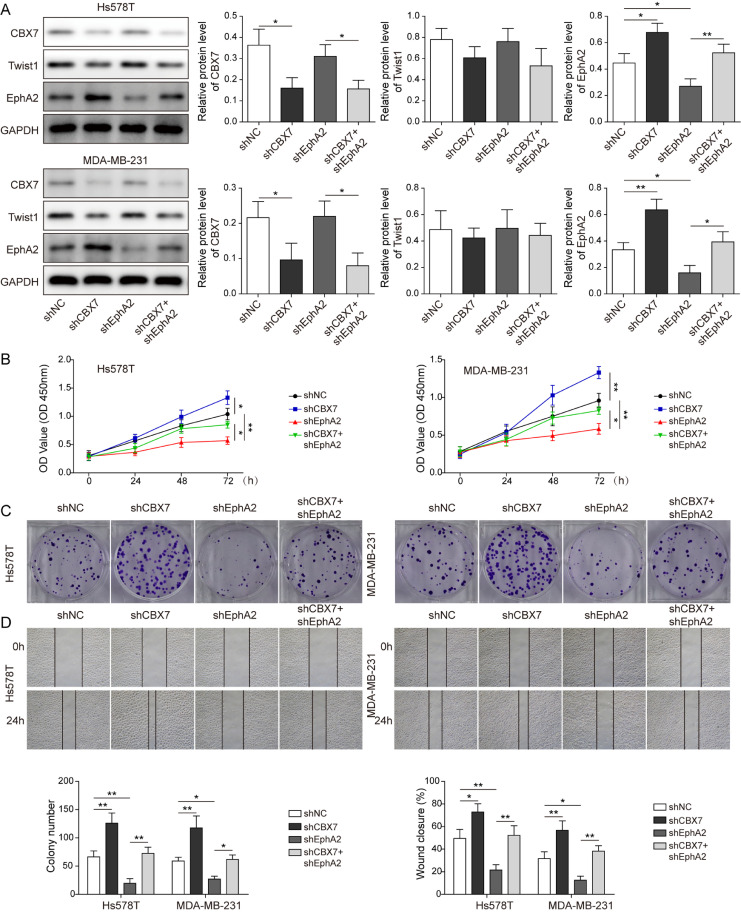
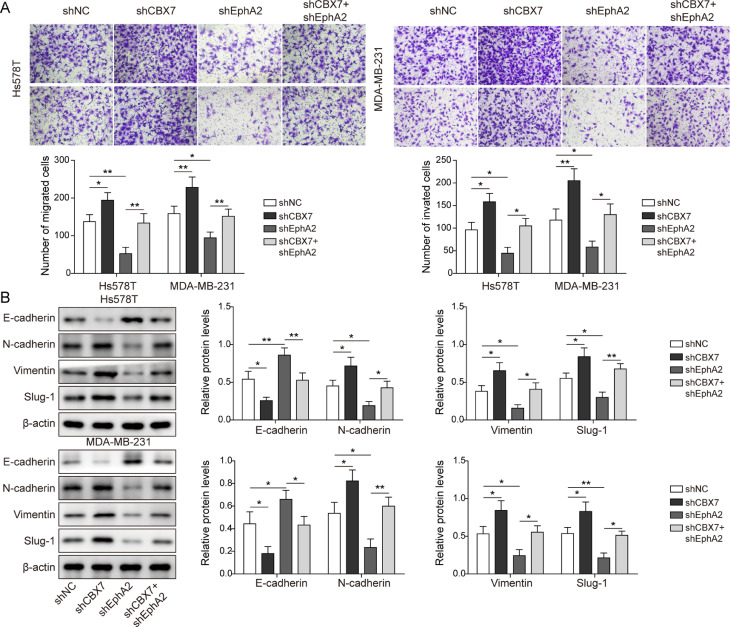
CBX7 regulated the proliferation and metastasis of BLBC via twist1/ EphA 2 axis
To better understand the correlation between CBX7, Twist1 and EphA2 in BLBC progression, shCBX7 and shEphA2 were transfected into cancer cells. EphA2 was decreased when EphA2 was knocked down. CBX7 knockdown reduced CBX7 expression, while the expression of EphA2 increased in cells with shCBX7 (Fig. 6A). Besides, overexpression of CBX7 increased the expression of CBX7 (Supplemental Figure 4A). Additionally, cell viability, proliferation, migration and invasion abilities were greatly impaired in cell with EphA2 silencing compared to control group. On the other hand, knockdown of CBX7 enhanced cell viability, proliferation, migration and invasion, which partially overturned the effect of EphA2 silencing (Figs. 6B-D and 7A). Meanwhile, CBX7 overexpression inhibited cell viability, proliferation, migration and invasion abilities compared to the control cells (Supplemental Figure 4B-E). Moreover, knockdown of EphA2 upregulated E-cadherin expression, while downregulated N-cadherin, Slug-1 and Vimentin levels. However, knockdown of CBX7 partially eliminated the effect of EphA2 silencing on the above proteins (Fig. 7B). Furthermore, the stable Hs578T and MDA-MB-231 cells transfected with shCBX7 or shEphA2 were injected into nude mice. Knockdown of EphA2 inhibited the tumor growth compared to the control groups, but cotransfection with shCBX7 overturned this effect (Supplemental Figure 3A and B). Similarly, IHC analysis showed that EphA2 expression decreased with EphA2 silencing. CBX7 knockdown reduced CBX7 expression, while the expression of EphA2 increased with CBX7 silencing (Supplemental Figure 3C). In addition, the tumor metastasis of EphA2 knockdown mice in lung was reduced obviously, while the tumor metastasis was aggravated with CBX7 silencing (Supplemental Figure 3D). Meanwhile, CBX7 overexpression inhibited the tumor growth and metastasis compared to the control group (Supplemental Figure 5). Collectively, these results indicate that CBX7 regulates the proliferation and metastasis of BLBC via Twist1/EphA2 axis.
Fig. 6.
CBX7 regulated the proliferation of BLBC via Twist1/EphA2 axis. Cells were transfected with shCBX7 or shEphA2. (A) Western blot determined CBX7, Twist1 and EphA2 protein levels. (B-D) CCK-8, colony formation, wound healing experiments tested cell viability, proliferation and migration. Data are the means ± SD for three independent experiments. *P<0.05, **P<0.01, ***P<0.001.
Fig. 7.
CBX7 regulated the metastasis and EMT of BLBC via Twist1/EphA2 axis. Cells were transfected with shCBX7 or shEphA2. (A) Transwell assay tested cell migration and invasion. (B) Western blot analyzed EMT related proteins. Data are the means ± SD for three independent experiments. *P<0.05, **P<0.01, ***P<0.001.
Discussion
BLBC is a distinct breast cancer subtype and has strong invasiveness and metastatic potential with poor prognosis [18]. Therefore, understanding of molecular mechanisms of BLBC metastasis can be of great importance [19]. Here, we identified that BLBC expressed high levels of Twist1 and EphA2, but low level of CBX7. In addition, we revealed a suppressive role of CBX7 in modulating the metastasis of BLBC through Twist1/EphA2 axis.
There is evidence that Twist1 is a key factor that endows cells with malignant traits during BLBC metastasis. Shi et al. reported that inhibition of Twist1-BRD4 association suppressed BLBC cell invasion [20,21]. Similar to previous studies, we found that Twist1 was up-regulated in BLBC tissues and cells. Meanwhile, high expression of Twist1 appeared to be associated with the malignancy grade of BLBC. Besides, repression of Twist1 inhibited the proliferation, EMT and metastasis of BLBC, suggesting that Twist1 modulates BLBC progression.
We next verified for the first time that EphA2 was a downstream regulator of Twist1 in BLBC development. It has been shown that aberrant EphA2 level is involved in modulating the initiation and development of BLBC. For instance, EphA2 expression was enriched in BLBC, and loss of EphA2 function suppressed tumor growth [22]. Consistent with these findings, our analysis of BLBC data proved that EphA2 was greatly increased in BLBC. EphA2 expression was related to BLBC grade, stage and lymph node metastasis. Additionally, Twist1 was positively correlated with EphA2 expression in BLBC tissues. Furthermore, functional assays indicated that EphA2 silencing suppressed the proliferation, EMT and metastasis of BLBC. Similarly, EphA2/Lyn/Twist1 mechanotransduction pathway drove EMT, invasion, and metastasis in breast cancer [23]. EphA2 and Twist1 were highly expressed in the head and neck squamous cell carcinoma patients [24].
We further explored the mechanism of Twist1 and EphA2 on BLBC and revealed that Twist1 directly targeted EphA2 and induced EphA2 level. Importantly, CBX7 blocked the binding of Twist1 to EphA2 and transcriptionally reduced the expression of EphA2. Literature has revealed that loss of CBX7 contributes to the development of BLBC. For example, CBX7 was associated with tamoxifen sensitivity, as well as chemosensitivity in breast tumors [25]. Notably, CBX7 prevented Twist1 from accessing the promoters of its target genes by directly targeting the promoters [17]. Here, we demonstrated that CBX7 was lowly expressed in BLBC and was negatively correlated with EphA2. Additionally, CBX7 regulated the proliferation and metastasis of BLBC via Twist1/EphA2 axis. We also adopted a xenograft model to investigate the effects of CBX7, Twist1 and EphA2 on BLBC in vivo and provided the first evidence that CBX7 suppressed the tumor growth and lung metastasis of BLBC xenografts through Twist1 and EphA2.
In conclusion, aberrant expression levels of CBX7, Twist1 and EphA2 were observed in BLBC tissues and cells. At the mechanistic level, CBX7 suppressed BLBC growth and metastasis through blocking the binding of Twist1 to EphA2 promoter. Thus, our study may provide innovative therapeutic targets for the diagnosis and prevention of BLBC distant metastasis. In addition, the cause of CBX7 downregulation in BLBC may due to the crosstalk between lncRNA/miRNA and CBX7 during BLBC progression, which remains elusive and requires further investigation.
Funding
This work was supported by Youth Fund Project of National Natural Science Foundation of China (81903027), and Youth Fund Project of Natural Science Foundation of Hunan Province (2021JJ40914).
Ethical approval
Written informed consent was provided by patients. Our research was approved by Clinical Research Ethics Committee of Xiangya Third Hospital of Central South University (No.2019-S144). The mice xenograft tumor experiments were permitted by the Animal Ethics Committee of Xiangya Third Hospital of Central South University (No.2019-S144).
Consent for publication
The informed consent was obtained from study participants.
Availability of data and material
All data generated or analyzed during this study are included in this article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
CRediT authorship contribution statement
Tao Dai: Conceptualization, Formal analysis, Methodology, Writing – original draft. Yiqi Liu: Investigation, Supervision, Validation. Renxian Cao: Data curation, Resources, Visualization. Jingying Cao: Funding acquisition, Project administration, Software, Writing – review & editing.
Conflict of Interest
The authors declare that there is no conflict of interest.
Acknowledgements
We would like to give our sincere gratitude to the reviewers for their constructive comments.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101468.
Appendix. Supplementary materials
References
- 1.Harbeck N., Gnant M. Breast cancer. Lancet. 2017;389(10074):1134–1150. doi: 10.1016/S0140-6736(16)31891-8. [DOI] [PubMed] [Google Scholar]
- 2.Jang M.H., Kim H.J., Kim E.J., Chung Y.R., Park S.Y. Expression of epithelial-mesenchymal transition-related markers in triple-negative breast cancer: ZEB1 as a potential biomarker for poor clinical outcome. Hum. Pathol. 2015;46(9):1267–1274. doi: 10.1016/j.humpath.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Milioli H.H., Tishchenko I., Riveros C., Berretta R., Moscato P. Basal-like breast cancer: molecular profiles, clinical features and survival outcomes. BMC Med. Genomics. 2017;10(1):19. doi: 10.1186/s12920-017-0250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itou J., Tsukihara H., Nukatsuka M., Toi M., Takechi T. 5-Chloro-2,4-dihydroxypyridine, CDHP, prevents lung metastasis of basal-like breast cancer cells by reducing nascent adhesion formation. Cancer Med. 2018;7(2):463–470. doi: 10.1002/cam4.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuo H.D., Wu Yao W. The role and the potential regulatory pathways of high expression of forkhead box C1 in promoting tumor growth and metastasis of basal-like breast cancer. J. BUON. 2016;21(4):818–825. [PubMed] [Google Scholar]
- 6.Zhang J., Lin X., Wu L., Huang J.J., Jiang W.Q., Kipps T.J., et al. Aurora B induces epithelial-mesenchymal transition by stabilizing Snail1 to promote basal-like breast cancer metastasis. Oncogene. 2020;39(12):2550–2567. doi: 10.1038/s41388-020-1165-z. [DOI] [PubMed] [Google Scholar]
- 7.Zhao Z., Rahman M.A., Chen Z.G., Shin D.M. Multiple biological functions of Twist1 in various cancers. Oncotarget. 2017;8(12):20380–20393. doi: 10.18632/oncotarget.14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y., Qin L., Sun T., Wu H., He T., Yang Z., et al. Twist1 promotes breast cancer invasion and metastasis by silencing Foxa1 expression. Oncogene. 2017;36(8):1157–1166. doi: 10.1038/onc.2016.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J., Yuan W., Wu L., Tang Q., Xia Q., Ji J., et al. PDGF-D promotes cell growth, aggressiveness, angiogenesis and EMT transformation of colorectal cancer by activation of Notch1/Twist1 pathway. Oncotarget. 2017;8(6):9961–9973. doi: 10.18632/oncotarget.14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onder T.T., Gupta P.B., Mani S.A., Yang J., Lander E.S., Weinberg R.A. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68(10):3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- 11.Casas E., Kim J., Bendesky A., Ohno-Machado L., Wolfe C.J., Yang J. Snail2 is an essential mediator of Twist1-induced epithelial mesenchymal transition and metastasis. Cancer Res. 2011;71(1):245–254. doi: 10.1158/0008-5472.CAN-10-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao J., Wang X., Dai T., Wu Y., Zhang M., Cao R., et al. Twist promotes tumor metastasis in basal-like breast cancer by transcriptionally upregulating ROR1. Theranostics. 2018;8(10):2739–2751. doi: 10.7150/thno.21477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.London M., Gallo E. The EphA2 and cancer connection: potential for immune-based interventions. Mol. Biol. Rep. 2020;47(10):8037–8048. doi: 10.1007/s11033-020-05767-y. [DOI] [PubMed] [Google Scholar]
- 14.Zhou L., Lu X., Zhang B., Shi Y., Li Z. EphA2 as a new target for breast cancer and its potential clinical application. Int. J. Clin. Exp. Pathol. 2021;14(4):484–492. [PMC free article] [PubMed] [Google Scholar]
- 15.Liang Z., Wang X., Dong K., Li X., Qin C., Zhou H. Expression Pattern and Prognostic Value of EPHA/EFNA in Breast Cancer by Bioinformatics Analysis: revealing Its Importance in Chemotherapy. Biomed. Res. Int. 2021;2021 doi: 10.1155/2021/5575704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iqbal M.A., Siddiqui S., Ur Rehman A., Siddiqui F.A., Singh P., Kumar B., et al. Multiomics integrative analysis reveals antagonistic roles of CBX2 and CBX7 in metabolic reprogramming of breast cancer. Mol. Oncol. 2021;15(5):1450–1465. doi: 10.1002/1878-0261.12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J., Alvero A.B., Nuti S., Tedja R., Roberts C.M., Pitruzzello M., et al. CBX7 binds the E-box to inhibit TWIST-1 function and inhibit tumorigenicity and metastatic potential. Oncogene. 2020;39(20):3965–3979. doi: 10.1038/s41388-020-1269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei W., Zhang X., Sun S., Xia B., Liang X., Cui Y., et al. Assessment of basal-like breast cancer by circulating tumor DNA analysis. Oncol. Lett. 2018;15(5):7389–7396. doi: 10.3892/ol.2018.8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J.T., Tan C.C., Wu X.R., He R., Zhang X., Wang Q.S., et al. FOXF2 deficiency accelerates the visceral metastasis of basal-like breast cancer by unrestrictedly increasing TGF-β and miR-182-5p. Cell Death Differ. 2020;27(10):2973–2987. doi: 10.1038/s41418-020-0555-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi J., Wang Y., Zeng L., Wu Y., Deng J., Zhang Q., et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25(2):210–225. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi J., Cao J., Zhou B.P. Twist-BRD4 complex: potential drug target for basal-like breast cancer. Current Pharmac. Des. 2015;21(10):1256–1261. doi: 10.2174/1381612821666141211153853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song W., Hwang Y., Youngblood V.M., Cook R.S., Balko J.M., Chen J., et al. Targeting EphA2 impairs cell cycle progression and growth of basal-like/triple-negative breast cancers. Oncogene. 2017;36(40):5620–5630. doi: 10.1038/onc.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fattet L., Jung H.Y., Matsumoto M.W., Aubol B.E., Kumar A., Adams J.A., et al. Matrix rigidity controls epithelial-mesenchymal plasticity and tumor metastasis via a mechanoresponsive EPHA2/LYN complex. Dev. Cell. 2020;54(3):302–316. doi: 10.1016/j.devcel.2020.05.031. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W., Lin P., Sun B., Zhang S., Cai W., Han C., et al. Epithelial-mesenchymal transition regulated by EphA2 contributes to vasculogenic mimicry formation of head and neck squamous cell carcinoma. Biomed. Res. Int. 2014;2014 doi: 10.1155/2014/803914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang Y.K., Lin H.Y., Chen C.F., Zeng D. Prognostic values of distinct CBX family members in breast cancer. Oncotarget. 2017;8(54):92375–92387. doi: 10.18632/oncotarget.21325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.