Abstract
Iron is a mineral essential for blood production and a variety of critical cellular functions. Altered iron metabolism has been increasingly observed in many diseases and disorders, but a comprehensive and mechanistic understanding of the cellular impact of impaired iron metabolism is still lacking. We examined the effects of iron overload or iron deficiency on cellular stress responses and autophagy which collectively regulate cell homeostasis and survival. Acute iron loading led to increased mitochondrial ROS (mtROS) production and damage, lipid peroxidation, impaired autophagic flux, and ferroptosis. Iron-induced mtROS overproduction is the mechanism of increased lipid peroxidation, impaired autophagy, and the induction of ferroptosis. Iron excess-induced ferroptosis was cell-type dependent and regulated by activating transcription factor 4 (ATF4). Upregulation of ATF4 mitigated iron-induced autophagic dysfunction and ferroptosis, whereas silencing of ATF4 expression impaired autophagy and resulted in increased mtROS production and ferroptosis. Employing autophagy-deficient hepatocytes and different autophagy inhibitors, we further showed that autophagic impairment sensitized cells to iron-induced ferroptosis. In contrast, iron deficiency activated the endoplasmic reticulum (ER) stress response, decreased autophagy, and induced apoptosis. Decreased autophagy associated with iron deficiency was due to ER stress, as reduction of ER stress by 4-phenylbutyric acid (4-PBA) improved autophagic flux. The mechanism of decreased autophagy in iron deficiency is a disruption in lysosomal biogenesis due to impaired posttranslational maturation of lysosomal membrane proteins. In conclusion, iron excess and iron deficiency cause different forms of cell stress and death in part through the common mechanism of impaired autophagic function.
Keywords: ATF4, ER stress, Ferroptosis, Lipid peroxidation, Mitochondria
Abbreviations: 4-PBA, 4-phenylbutyric acid; ALF, autophagosome-lysosome fusion; ATF4, activating transcription factor 4; Chlq, chloroquine; CHOP, C/EBP homologous protein; DFO, desferoxamine; ER, endoplasmic reticulum; FAC, ferric ammonium citrate; FeDex, iron dextran; Fer-1, ferrostatin-1; FTH1, ferritin heavy chain 1; Grp78, 78-kDa glucose-regulated protein; HAEC, human aortic endothelial cell; LAMP, lysosome-associated membrane protein; LC3B, microtubule-associated protein 1A/1B-light chain 3B; MT, MitoTEMPO; mtROS, mitochondrial ROS; NRF2, nuclear factor-erythroid factor 2-related factor 2; PERK, protein kinase RNA-like endoplasmic reticulum kinase; P-Hepa, primary mouse hepatocyte; PI, propidium iodide; TEM, transmission electron microscopy
1. Introduction
Iron is a vital micronutrient for cell metabolism and homeostasis [1]. Iron is utilized by various organelles to execute essential functions. Mitochondria are the primary site of intracellular iron utilization with most of the cellular iron utilized for the biogenesis of heme and iron-sulfur [Fe–S] clusters [2,3]. The [Fe–S] clusters are essential cofactors for proteins involved in tricarboxylic acid (TCA) cycle and electron transport in the mitochondria and for other cellular functions including DNA synthesis and protein translation in the endoplasmic reticulum (ER) [4,5]. In physiological settings, intracellular iron content is tightly controlled through the coordinated regulation of iron influx and efflux, and excess iron is stored in the ferritin complex [6,7]. In pathological conditions, iron transport and regulation may be abnormal leading to iron overload or iron deficiency that contributes to the pathogenesis and progression of many diseases including liver, metabolic, neurodegenerative, and cardiovascular diseases [8]. A comprehensive and mechanistic understanding of the precise effects of altered iron metabolism on cellular stress responses and cell homeostasis in these diseases remains lacking.
Excess iron causes oxidant stress that alters nucleic acids, lipids and proteins resulting in organelle damage and dysfunction. Iron-induced reactive oxygen species (ROS) production and mitochondrial dysfunction has been shown in cultured cells including neurons [9], cardiomyocytes [10], hepatocytes [11], and endothelial cells [12]. In vivo, a high-iron diet induces oxidant stress that results in mitochondrial DNA damage and dysfunction in rat liver [13]. Mitochondrial ROS (mtROS) can trigger the apoptotic pathway [14]. Recent studies have also suggested that mtROS overproduction mediates ferroptosis [[15], [16], [17], [18]], a necrotic death dependent on iron-catalyzed lipid peroxidation in the context of glutathione (GSH) deficiency [19]. Lysosomes, a site of ferritin-mediated iron storage and recycling [20], are also subject to regulation by altered iron metabolism. Excess iron deposition in lysosomes leads to lysosomal membrane permeabilization and release of cathepsins that induce cell death [21]. Lysosomes are a major component of the autophagy machinery that degrades damaged biomolecules and organelles, playing a critical role in cellular redox homeostasis, antioxidant defense, and cell survival [22,23]. Studies have shown that excess iron impairs autophagic flux by inhibiting autophagosome-lysosome fusion (ALF) and lysosomal acidification [24,25]. Despite the above depicted effects, it remains not understood how the ROS production, impaired mitochondrial and lysosomal integrity, and autophagic dysfunction integrate leading to iron-induced cell death. It is also unclear whether ER homeostasis is disrupted under the condition of iron overload playing a role in the integrative responses and cell death induction.
Iron deficiency can also be deleterious due to the indispensable role of iron in the biogenesis of [Fe–S] clusters for energy production and protein translation. Iron deficiency leads to swollen mitochondria and decreased mitochondrial respiration and ATP production in cultured cardiomyocytes [26]. We recently reported that iron deficiency decreases mitochondrial respiration in cultured human trophoblasts [27]. Mitochondrial damage was also observed in the liver of rats fed a low-iron diet [13], and in dopaminergic neurons with the loss-of-function of transferrin receptor 1 [28], an important importer of transferrin-bound iron. The effects of iron deficiency on ER homeostasis have been reported in cultured cardiomyocytes and neuroblastoma cells [26,29], but it is not known whether iron deficiency induces ER stress in vivo. Iron deficiency also modulates autophagy as iron chelation increases the number of LC3-II containing autophagosomes [30], but it is unclear whether this increase mediates improved autophagy or suggests impaired autophagic flux. Moreover, the precise mechanisms by which low iron induces ER stress response and altered autophagy have not been elucidated.
The current study employed a variety of cultured cell types and in vivo models to examine the effects of iron excess and iron deficiency on cellular stress responses, autophagy and cell survival. We demonstrate that with iron excess mitochondrial iron loading catalyzes overproduction of mtROS resulting in lipid peroxidation-dependent impairment of autophagic flux and induction of ferroptosis, which were prevented by upregulation of activating transcription factor 4 (ATF4). In contrast, iron deficiency induces ER stress and decreased autophagy through disruption of protein folding and maturation.
2. Materials and methods
2.1. Reagents
The materials and reagents used in the current study are listed in Supplementary Table 1.
2.2. Animal models
WT mice on a C57BL/6 background were purchased from The Jackson Laboratory. For all the studies mice were used at the age of 8–10 weeks. Iron dextran (FeDex) was injected intraperitoneally at the dose of 500 mg/kg body weight, and the livers were collected after 24 h. Mice with floxed ferroportin (Fpn) gene, Fpnf/f mice, were crossed with VillinCreERT2 mice to generate Fpnf/f;VillinCreERT2 mice [31]. Fpnf/f;VillinCreERT2 mice (2 weeks old), along with the control Fpnf/f mice, were treated with TAM (100 mg/kg/day) for two consecutive days to induce the intestinal epithelial cell (IEC)-specific knockout of Fpn (FpnΔIEC). At the age of 8 weeks, they were either sacrificed directly, or treated for 5 h with autophagy inhibitor chloroquine (Chlq, 60 mg/kg) or vehicle for autophagic flux analysis. Atg5f/f mice were crossbred with AlbuminCreERT2 (AlbCreERT2) mice to generate Atg5f/f;AlbCreERT2 mice for TAM-inducible knockout of Atg5 gene specifically in the hepatocytes [32]. All animal experiments were approved by the Institutional Animal Care and Use Committee of Emory University and in accordance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals.
2.3. Cell culture
Cell lines, and their culture media and supplements are listed in Supplementary Table 1. Primary mouse hepatocytes (P-Hepa) were isolated by liver perfusion as previously described [33]. Briefly, mouse liver was pre-perfused with Leffert's buffer (HEPES 10 mM, KCl 3 mM, NaCl 130 mM, NaH2PO4 1 mM, glucose 10 mM, pH 7.4) containing EGTA, followed by perfusion with Liberase that consists of collagenase I and II. Dissociated hepatocytes were purified on a Percoll gradient. Cell viability is determined by Trypan blue staining, and only isolations that have a viability of >90% were used. Hepatocytes were plated in collagen-coated 12-well plates at the density of 2 × 105 cells per well for 3 h in William's E medium supplemented with 5% FBS, 1x P/S, 10 mM HEPES, 1x insulin and 40 ng/ml Dexamethasone. Hepatocytes were then cultured overnight in fresh medium containing 5% FBS, 1x P/S and 10 mM HEPES prior to any treatments. Atg5f/f;AlbCreERT2 hepatocytes were treated with 4-hydroxyltamoxifen (4-OHT) for 24 h to induce the knockout of ATG5 in vitro. All cells were cultured in a 5% CO2 humidified incubator at 37 °C. Cells for live cell imaging analysis were seeded in 12-well plates.
2.4. Cell transfection and infection
293T cells were transfected with shRNA that targets ATF4, pLKO.1/shATF4 (Sigma-Aldrich, TRCN0000013573), or scrambled shRNA by using Lipofectamine 2000 following the manufacturer's instruction. Stable expression of shATF4 or shCont was selected with 3 μg/ml puromycin for two passages. shATF4 and shCont cells were transiently transfected with pMRX-IP-GFP-LC3-RFP reporter (Addgene, #84573) for the analysis of autophagic flux. Lentiviral particles carrying V5 tagged human ATF4 (DNASU, HsCD00434003) or the backbone (Addgene, #25890) were prepared as previously described [34]. HepG2 cells were infected with lentivirus in the presence of 5 μg/ml polybrene, and stable expression of ATF4 was selected for two passages with 3 μg/ml blasticidin. Selection antibiotics were excluded from cells that were seeded for treatment.
2.5. Quantitative RT-PCR analysis
Mouse livers were homogenized, and total RNA was extracted with the RNeasy Mini Kit. Three μg of total RNA was used for first strand cDNA synthesis using SuperScript III First-Strand Synthesis System according to the manufacturer's instruction. Quantitative PCR was performed with SsoAdvanced Universal SYBR Green Mastermix SYBR on QuantStudio 3 real-time PCR system. PCR primer sequences are as follows: LC3-forward: 5′-CAT GAG CGA GTT GGT CAA GA -3′ and LC3-reverse: 5′- TTG ACT CAG AAG CCG AAG GT -3’; p62-forward: 5′- GAA GAA GCA GGA TGG AGC AC -3′ and p62-reverse: GCT TTC GTT GGA AAA ATG GA -3’; and Rpl13a-forward: 5′- ATG ACA AGA AAA AGC GGA TG -3′ and Rpl13a-reverse 5′-CTT TTC TGC CTG TTT CCG TA -3’.
2.6. Western blot analysis
Cultured cells were lysed in lysis buffer supplemented with protease and phosphatase inhibitors. Liver tissues were homogenized in the same lysis buffer. The crude lysates were sonicated for 2 × 15 s and spun at 14,000 g for 15 min. Protein concentration of the supernatant was determined by Bicinchoninic Acid Protein Assay. Protein lysates were heated at 37 °C for 30 min in 1 × Laemmli buffer, separated on SDS-PAGE gel, and transferred to nitrocellulose membrane for immunoblotting with the corresponding antibodies. Densitometric analysis was performed by using ImageJ software.
2.7. Cell death assay
Necrotic death was detected by staining cultured cells with propidium iodide (PI, 2 μg/ml) in the culture medium. Following 30 min incubation, cells were washed with PBS prior to fluorescent imaging by an Olympus IX83 system with a HAMAMATSU camera. The numbers of PI+ cells and total numbers of cells per field were counted, from which the percentage of PI+ cell death was calculated. Please note that many P-Hepa have two nuclei and show two red puncta from PI staining if they have undergone necrosis or ferroptosis.
2.8. Confocal immunofluorescence
Immunofluorescence staining was performed, as previously described [35]. In brief, cryosections of the liver, and cultured cells, were fixed with 4% paraformaldehyde, permeated with PBS containing 0.2% Triton X-100 for 10 min. Specimens were then blocked with 5% goat serum before incubation with primary antibodies for 1 h at room temperature. After 3 washes with PBS, specimens were incubated for 30 min with Alexa Fluor–conjugated secondary antibodies, phalloidin and DAPI, washed, and then mounted with ProLong Diamond Antifade mounting medium for visualization under a Leica SP8 fluorescence microscope. Fluorescence intensity of the obtained images was quantified using ImageJ software.
2.9. Detection of mitochondrial ROS, Fe2+, and mitochondrial membrane potential (MMP)
Cells were treated with ferric ammonium citrate (FAC), a commonly used form of iron [36]. Cells were then incubated with 5 μM Mito-FerroGreen and/or 5 μM MitoSOX in HBSS for the detection of Fe2+ and mtROS, respectively. MMP was determined by incubating FAC-treated and untreated cells with 2 μM JC-1 fluorescent probe, which yields green fluorescence at low concentrations while lights up in red when it accumulates in the mitochondria at high concentrations. Thirty minutes after incubation, cells were washed two times with HBSS buffer and subject to live imaging. Fluorescence intensity was quantified using ImageJ software.
2.10. Lipid peroxidation assay
Cultured cells were treated with FAC for 24 h, and then incubated for 30 min with 2 μM BODIPY® 581/591 C11 (Image-iT Lipid Peroxidation Kit). After washing with PBS, cells were imaged at the emission of ∼590 nm (red) and ∼510 nm (green). A shift from red to green fluorescence signal suggests lipid peroxidation.
2.11. Transmission electron microscopy (TEM)
Monolayers of P-Hepa and HepG2 cells were fixed overnight in Karnovsky's fixative (2.5% glutaraldehyde and 4% paraformaldehyde) in 0.1 M cacodylate buffer. After washing with 0.1 M cacodylate buffer, cells were subjected to post fixation with 1% osmium tetroxide. Fixed cells were then dehydrated through an ascending ethanol series, followed by infiltration with Eponate 12 Epoxy resin (Ted Pella, Inc.). Specimens were then polymerized and sectioned at 80–90 nm using a diamond knife (Diatome, Hatfield, PA) and a Leica Ultracut S ultramicrotome (Leica Microsystems Inc., Buffalo Grove, IL). Sections were mounted onto 200 mesh copper grids, and stained in 5% Uranyl Acetate and Reynold's Lead Citrate. The sections were imaged with a JEOL JEM-1400 transmission electron microscope (Tokyo, Japan) using a Gatan US1000 CCD camera (Gatan, Inc., Pleasanton, CA).
2.12. Autophagic flux analysis
Autophagic flux was analyzed by using Bafilomycin A1 (BafA1) or Chlq as an inhibitor of autophagy and by utilizing GFP-LC3-RFP reporter [37]. Cells were treated with 0.5 μM BafA1 or 100 μM Chlq or vehicle for 4 h, and the expression of LC3B-II and p62 was determined by Western blotting and quantified by densitometric analysis. The GFP-LC3-RFP reporter was transiently expressed in 293 T cells. Forty-eight hours after the treatments, fluorescent GFP-LC3 and RFP-LC3 was directly imaged by the Olympus IX83 system. The same exposure was applied in imaging the treated and untreated cells.
2.13. Statistical analysis
Statistical significance was determined by one-way analysis of variance. Data are presented as mean ± SE. A value of P < 0.05 was considered significant.
3. Results
3.1. Excess iron induces oxidative responses and changes in autophagy but not ER stress proteins
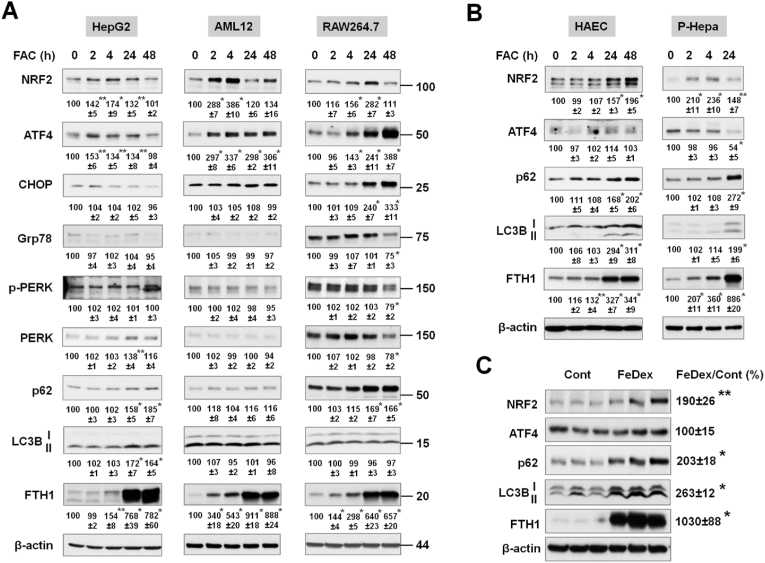
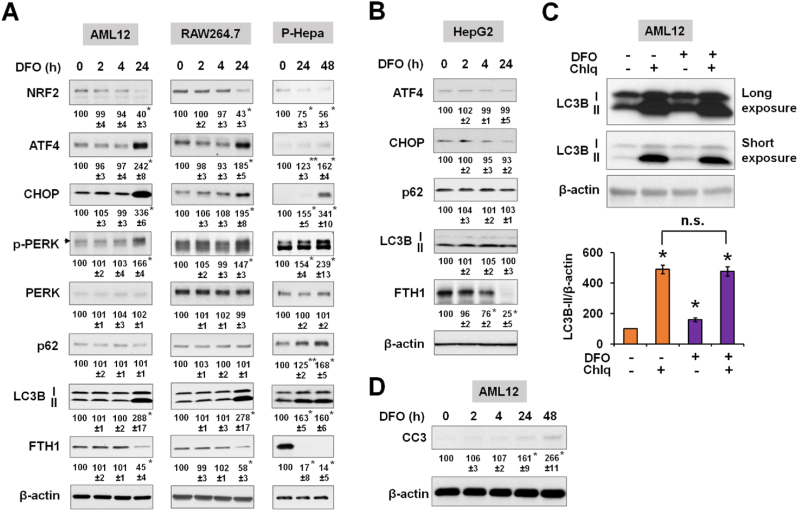
As cells respond differently to altered iron content, we determined the effects of iron excess in multiple cell types including the human hepatoma cell line HepG2, a nontransformed mouse hepatocyte cell line AML12, and the mouse macrophage cell line RAW264.7, as well as the primary cells including P-Hepa and human aortic endothelial cells (HAECs) (Fig. 1). These cell types were selected because hepatocytes and macrophages play critical roles in iron storage and recycling and vascular endothelial cells are one of the main targets of iron overload-mediated injury. Intracellular iron overload was achieved by treating cells with FAC for various time points to determine acute (2 h and 4 h) and chronic (24 h and 48 h) effects. As a measure of the effects of excess iron on cellular stress pathways and autophagy, protein expression were determined by immunoblotting for the oxidative stress marker nuclear factor-erythroid factor 2-related factor 2 (NRF2), integrated stress response marker ATF4, ER stress markers C/EBP homologous protein (CHOP), 78-kDa glucose-regulated protein (Grp78) and phosphorylated protein kinase RNA-like endoplasmic reticulum kinase (P-PERK), and the autophagy factors microtubule-associated protein 1A/1B-light chain 3B (LC3B) and p62. The status of intracellular iron loading was determined by immunoblotting ferritin heavy chain 1 (FTH1) [38].
Fig. 1.
Excess iron induces differential expression of stress and autophagy proteins. (A) Human hepatoma HepG2 cells, mouse AML12 hepatocytes, and mouse macrophage-like RAW264.7 cells, and (B) primary human aortic endothelial cells (HAECs) and primary mouse hepatocytes (P-Hepa) were treated with 100 μM FAC for the indicated times. The expression of stress proteins (NRF2, ATF4, CHOP, Grp78, and phospho-PERK), autophagy markers (p62 and LC3B), and ferritin heavy chain 1 (FTH1, a maker of iron loading) was determined by Western blotting. Numbers below the blots represent mean ± SE (n = 3 independent experiments), with the untreated control set at 100. For LC3B protein, the active isoform LC3B-II was quantified. (C) C57BL/6 mice were injected (i.p.) with iron dextran (FeDex, 500 mg/kg/b.w.) or PBS as a control. After 24 h, livers were harvested for the analysis of the expression of stress and autophagy proteins. Data are mean ± SE (n = 6 mice of two independent experiments). *, P < 0.01; and **, P < 0.05 compared to the control.
All of the cell lines examined showed increases in NRF2 expression in response to FAC treatment. Induction of NRF2 was more acute and robust in AML12 cells than HepG2 and RAW264.7 cells (Fig. 1A). ATF4 induction was much greater in AML12 and RAW264.7 cells than in HepG2 cells (Fig. 1A). None of the ER stress markers including CHOP, Grp78 and P-PERK was induced in HepG2 and AML12 cells, with only increased CHOP expression in RAW264.7 cells, overall suggesting the lack of ER stress induction by excess iron (Fig. 1A). The expression of LC3B-II, the active isoform, and p62 was increased in HepG2 cells (Fig. 1A). In contrast, LC3B expression was not significantly altered in AML12 and RAW264.7 cells (Fig. 1A). The cell context-dependent regulation of autophagy was not due to the magnitude of iron loading based on the induction of FTH1 (Fig. 1A), although FTH1 was strongly induced at an earlier time point (2 h) in AML12 cells. The effects of excess iron on the expression of NRF2, ATF4 and autophagy factors were further examined in primary cultured cells, HAECs and P-Hepa. Both cell types exhibited increased expression of NRF2, p62 and LC3B-II, but not of ATF4 (Fig. 1B). The effects of iron loading on the autophagic markers LC3B-II and p62 were more robust in primary cells as compared to cell lines. The in vivo effects of acute iron loading on the expression of oxidative and autophagy markers were determined in the liver, the primary site of excess iron deposition, by treating WT mice with FeDex. FeDex injection drastically increased the expression of FTH1 (Fig. 1C). Likewise, the expression of NRF2, LC3B (particularly LC3B-II) and p62 were significantly increased, but ATF4 expression was unchanged (Fig. 1C). Altogether, our in vitro and in vivo results demonstrate that acute iron overload induces an oxidative response and altered expression of autophagic proteins but does not cause an ER stress response. Our findings also suggest a potential role of ATF4 in regulating autophagy.
3.2. Iron excess induces an impairment of autophagic flux that potentiates ferroptosis
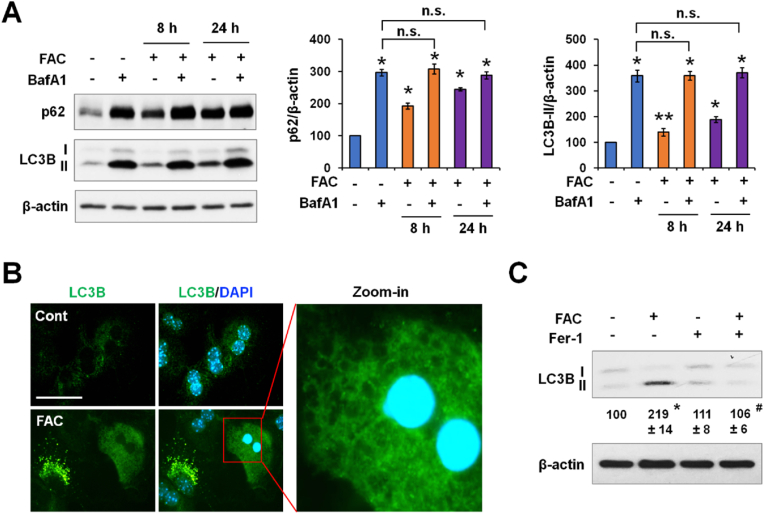
Increased LC3B-II expression can indicate stimulation of autophagy or impaired autophagic flux. Recent studies have suggested that iron excess decreases autophagic flux by inhibiting ALF in cultured neurons and myoblasts [24,39]. We determined whether increased LC3B-II expression in P-Hepa, HepG2 cells and HAECs was a result of decreased flux by using autophagy inhibitor BafA1 or Chlq [40]. FAC induced a significant increase in p62 and LC3B-II expression in P-Hepa at both the 8 h and 24 h time points (Fig. 2A). Compared to BafA1 alone, FAC/BafA1 cotreatment did not increase p62 or LC3B-II level further at the examined times (Fig. 2A). Similar findings were seen in HepG2 cells (Fig. S1A) and HAECs (Fig. S1B) by using Chlq. These results indicate decreased autophagic flux in hepatocytes and endothelial cells by iron excess. Consistently, FAC treatment induced a profound accumulation of LC3B-positive puncta in P-Hepa (Fig. 2B). Some of the iron-treated P-Hepa displayed strong but diffuse LC3B staining (arrowhead) throughout the cytoplasm as well as nuclear shrinkage. At high magnification, these cells contained large vacuoles and had a complete loss of integrity (Fig. 2B), denoting cell death. Iron-loaded P-Hepa did not show cell surface staining for Annexin V, a marker of apoptosis (data not shown). Instead, FAC-treated P-Hepa were stained for PI in the nuclei, which was abolished by pretreatment with Ferrostatin-1 (Fer-1, an inhibitor of ferroptosis), but not with Z-VAD-FMK (an inhibitor of apoptosis) or Necrostatin-1 (an inhibitor of necroptosis) (Fig. S2). Likewise, FAC-induced lipid peroxidation was mitigated by Fer-1, but not by Z-VAD-FMK or Necrostatin-1 (Fig. S3). FAC-induced PI staining was also observed in HepG2 cells and HAECs (Fig. S4), but not in AML12 and RAW264.7 cells where autophagy was unaltered. Importantly, we show that iron-induced accumulation of LC3B-II was abrogated by Fer-1 (Fig. 2C). These results demonstrate that excess iron induces lipid peroxidation-dependent ferroptosis, and that scavenging lipid peroxides rescues iron-induced inhibition of autophagic flux.
Fig. 2.
Iron excess induces lipid peroxidation-dependent impairment of autophagy. (A) P-Hepa were treated with 100 μM FAC for 8 h or 24 h in the presence or not of 0.5 μM Bafilomycin A1 (BafA1) during the last 4 h. Changes in LC3B-II expression were quantified relative to the untreated control (set at 100) after standardized to β-actin. Data are presented as mean ± SE (n = 3 independent experiments). *, P < 0.01; **, P < 0.05 compared to the control; and n. s., not significant. (B) Representative images of LC3B (green) and nuclei staining (DAPI, blue) in P-Hepa treated with or not FAC for 24 h. Bar, 50 μm. (C) P-Hepa were pretreated with or not 5 μM ferrostatin-1 (Fer-1) prior to treatment with 100 μM FAC. The relative changes in LC3B-II expression were calculated relative to that of the control (set at 100). Data are mean ± SE (n = 3). *, P < 0.01 compared to the control; and #, P < 0.01 compared to FAC alone. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
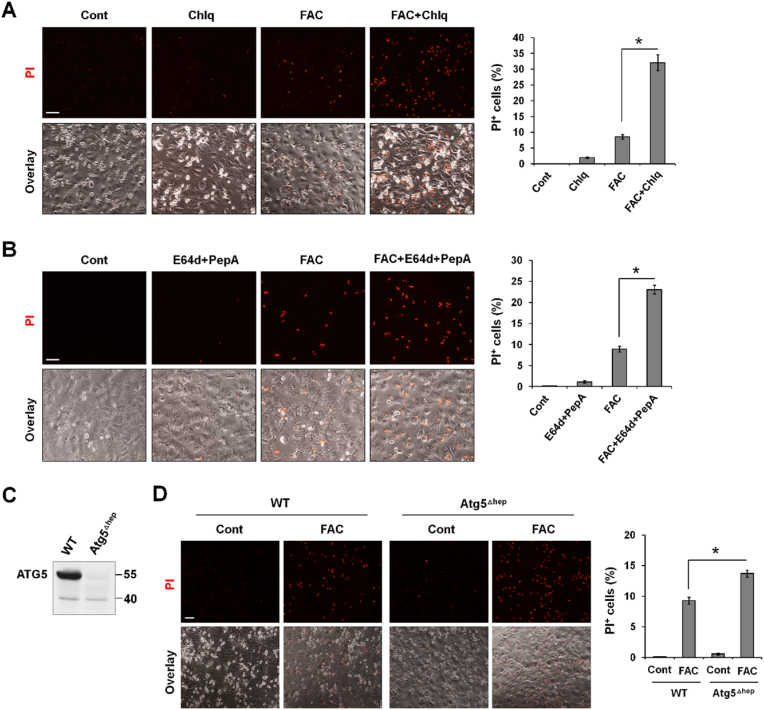
Given that excess iron only induced ferroptosis in cells that exhibited autophagic dysfunction, and the critical role that autophagy plays in the removal of oxidized molecules and damaged organelles [41], we wondered whether impaired autophagy contributes to iron excess-induced ferroptosis by using P-Hepa cell model. Compared to FAC alone, FAC/Chlq cotreatment resulted in an approximately 3.7-fold increase in the percentage of PI+ cells (Fig. 3A). E64d and pepstatin A, which suppress lysosomal proteases, are also commonly autophagy inhibitors [42]. Likewise, compared to FAC alone, FAC and E64d/pepstatin A cotreatment led to a 2.6-fold increase in the percentage of PI-positive cells (Fig. 3B). We did not utilize BafA1 in determining iron-induced cell death because it interferes with intracellular iron mobilization by elevating pH in endosomes and lysosomes [43]. The promoting effects of autophagic dysfunction on iron-induced ferroptosis was further explored in ATG5-deficient P-Hepa [32]. Atg5 was deleted in cultured P-Hepa (ATG5Δhep) by treatment with 4-OHT (Fig. 3C). Consistent with the above scenarios wherein autophagic flux is impaired, autophagy deficiency also potentiated FAC-induced ferroptosis as indicated by the increased rate of PI-positive cell death in ATG5Δhep cells relative to control cells (Fig. 3D). These findings demonstrates that autophagic dysfunction potentiates ferroptosis, at least in the context of iron excess.
Fig. 3.
Impaired autophagy potentiates iron-induced ferroptosis. (A) P-Hepa were treated with 50 μM FAC and/or 50 μM Chlq for 8 h, and (B) P-Hepa were treated with 50 μM FAC and/or 30 μM E64d/30 μM pepstatin A (PepA) for 16 h. Ferroptosis was detected by staining with PI, and the percentage of PI + cells over the total numbers of cells per field was quantified. Data are shown as mean ± SE (n = 15 fields of three independent experiments). *, P < 0.01. (C) Cultured P-Hepa from Atg5f/f;AlbCreERT2 mice were treated with 4-hydroxytamoxifen (4-OHT, Atg5Δhep) or vehicle as a control (WT), and the expression of ATG5 was examined by Western blotting. (D) Atg5Δhep and WT cells were treated with or not 50 μM FAC for 24 h, followed by staining with PI. The percentage of PI+ cells was quantified. Data are mean ± SE (n = 9 fields of three independent experiments). *, P < 0.01. Bars, 100 μm.
3.3. Iron-induced mtROS production underlies lipid peroxidation-dependent ferroptosis
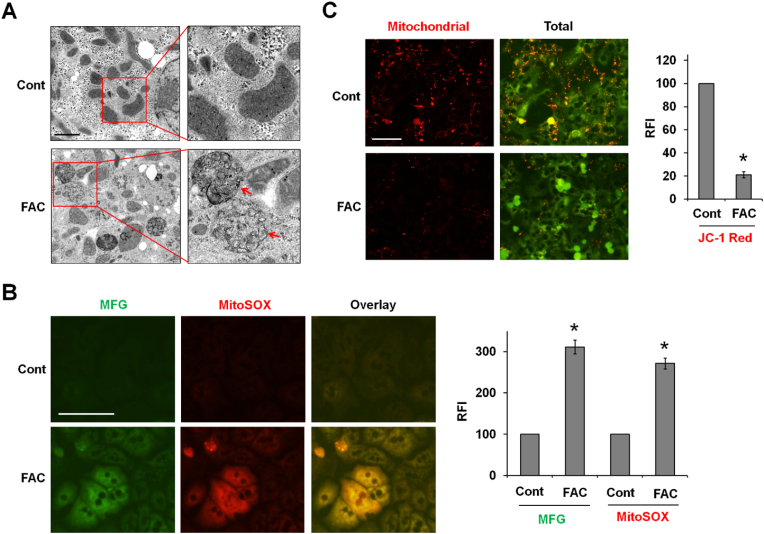
Iron catalyzed ROS damages organelles. TEM imaging showed ruptured mitochondria with a loss of crista (arrows), an important feature of ferroptosis, in FAC-treated P-Hepa (Fig. 4A). We speculated that iron is loaded in the mitochondria catalyzing the production of mtROS mediating mitochondrial damage. To address this possibility, Fe2+, the redox active form of iron, was stained with Mito-FerroGreen, and mtROS was detected by MitoSOX. We observed a strong overlay of mitochondrial Fe2+ and ROS following 6-h FAC treatment (Fig. 4B). Consistently, FAC also induced strong increases in total Fe2+ and ROS as determined by RhoNox-1 and H2DCFDA, respectively (Fig. S5). By using fluorescent probe JC-1, we further showed that FAC treatment led to a reduction in MMP as indicated by the markedly decreased intensity of the red fluorescence (Fig. 4C). These findings suggest that acute iron loading in mitochondria causes mtROS overproduction and loss of MMP that results in mitochondrial damage.
Fig. 4.
Iron is loaded in the mitochondria inducing ROS production, loss of MMP and mitochondrial damage. (A) P-Hepa were treated for 24 h with 100 μM FAC, and organellular structural changes were examined by transmission electron microscope (TEM). Arrow, ruptured mitochondria. Bar, 2 μm. (B) P-Hepa were treated with 100 μM FAC for 6 h, and mitochondrial Fe2+ and mtROS were detected by 5 μM Mito-FerroGreen (MFG) and 5 μM MitoSOX, respectively. Relative fluorescence intensity (RFI) was quantified with ImageJ on fields with similar cell density between the control and FAC-treated groups. Data are mean ± SE (n = 9 fields of three independent experiments), with the control set at 100. *, P < 0.01 compared to the control. Bar, 100 μm. (C) P-Hepa, treated for 24 h with 100 μM FAC, were stained with JC-1 probe that yields red fluorescence when accumulated in the mitochondria. Green fluorescence denotes JC-1 of non-mitochondrial location at low concentrations. RFI of the red fluorescence was quantified, and data are presented as mean ± SE (n = 9 fields of three independent experiments). *, P < 0.01 compared to the control. Bar, 100 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
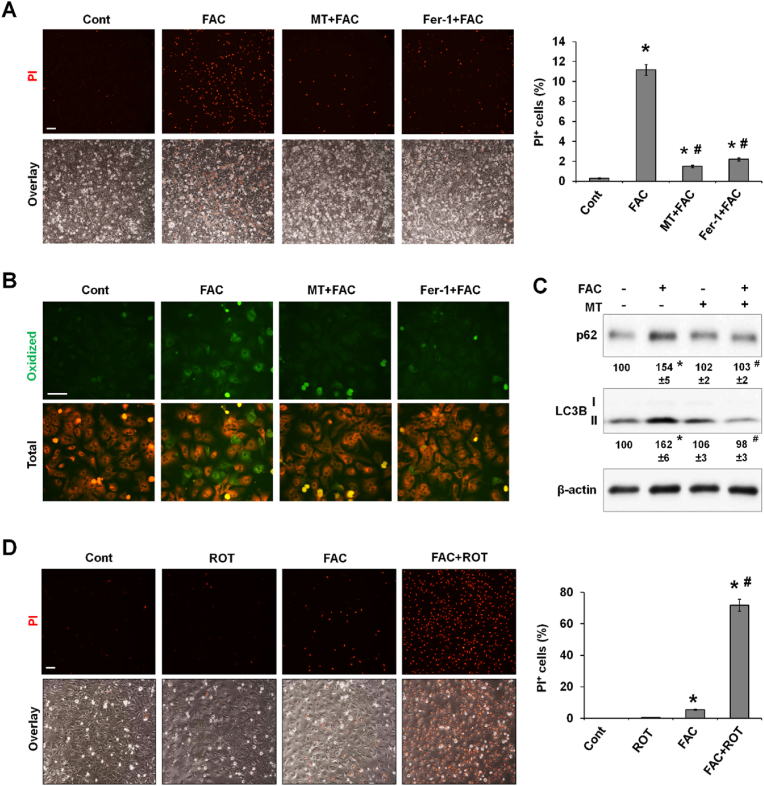
The above findings suggested that overproduced mtROS might be responsible for FAC-induced ferroptosis. We examined whether the commonly used mitochondria-targeted ROS scavenger MitoTEMPO (MT) rescues iron-induced ferroptosis. Importantly, FAC-induced ferroptosis was abolished by both MT and Fer-1 in P-Hepa (Fig. 5A). By staining with the fluorescence probe BODIPY 581/591 which turns green when oxidized, we demonstrated that MT blocked iron-mediated lipid peroxidation as effectively as the ferroptosis inhibitor Fer-1 (Fig. 5B). MT also prevented iron-induced accumulation of LC3B-II and p62 (Fig. 5C), indicating improved autophagic flux. These data suggest that mtROS overproduction increases lipid peroxidation as the underlying mechanism of iron-induced ferroptosis.
Fig. 5.
Overproduction of mtROS is indispensable for iron-induced lipid peroxidation, autophagic dysfunction, and ferroptosis. (A) P-Hepa were treated with 100 μM FAC for 24 h in the presence or not of MitoTEMPO (MT, 10 μM) and Fer-1 (5 μM). Ferroptosis was examined by staining with PI. The percentage of PI + cells over the total numbers of cells per field was quantified and is presented as mean ± SE (n = 9 fields of three independent experiments). (B) P-Hepa were treated with 100 μM FAC for 24 h in the presence or not of MT and Fer-1. Lipid peroxidation was detected using fluorescent probe BODIPY 581/591 C11, with oxidized lipid in green fluorescence. (C) The expression of autophagy markers, p62 and LC3B, was determined in P-Hepa treated for 24 h by 100 μM FAC with or not MT pretreatment. Data shown are mean ± SE (n = 3 independent experiments). (D) P-Hepa were treated with 300 nM rotenone (ROT) and/or 20 μM FAC for 6 h. Ferroptosis was examined by staining with PI. The percentage of PI+ cells was quantified and is shown as mean ± SE (n = 9 fields of three independent experiments). *, P < 0.01 compared to the control; #, P < 0.01 compared to FAC treatment. Bars, 100 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Rotenone, a selective inhibitor of mitochondrial complex I, induces mtROS production and causes apoptosis at high concentrations [44]. We asked whether an elevation in mtROS induced by low dose rotenone in combination with a non-toxic FAC concentration sensitizes hepatocytes to iron-dependent ferroptosis. Rotenone alone did not induce PI positivity, and FAC at 20 μM only led to a small number of PI positive cells (Fig. 5D). In contrast, massive PI staining resulted when P-Hepa were treated by FAC and rotenone together, suggesting that in the presence of low levels of elevated mtROS production even a small increase in mitochondrial iron can trigger ferroptosis.
3.4. Induction of ATF4 protects autophagy and cell survival from iron toxicity
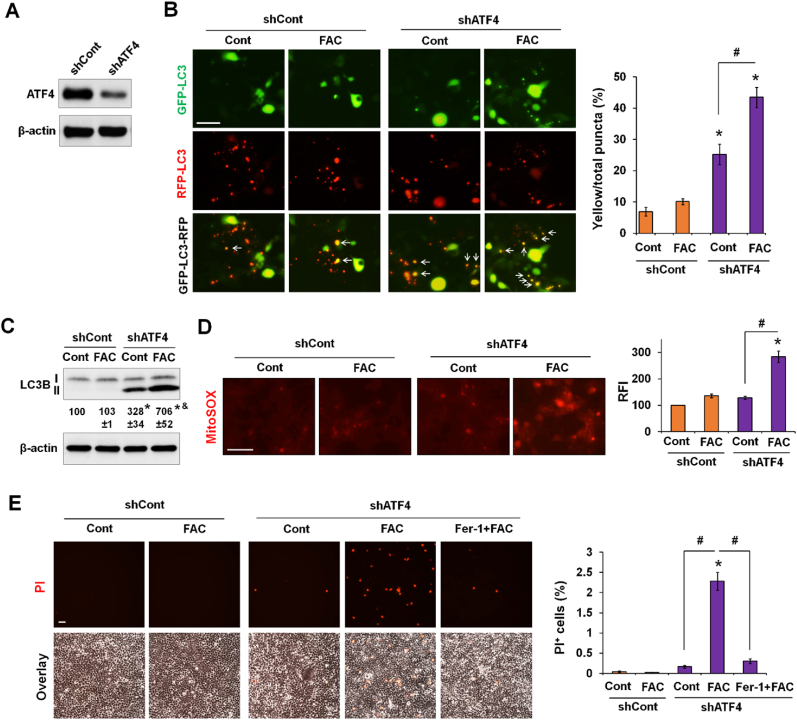
Our earlier findings suggested that ATF4 maintains autophagy and cell survival against iron toxicity. To test the potential role of ATF4, we adopted 293 T cells, a commonly used highly transfectable model cell line. Of note, 293 T cells are like RAW264.7 and AML12 cells by showing upregulated ATF4 and unchanged LC3B expression in response to iron excess (Fig. S6). Thus, the utilization of 293 T cells helps ascertain whether silencing ATF4 leads to a loss of protection of autophagy and cell survival against iron toxicity. ATF4 Knockdown of ATF4 was achieved by transfecting 293 T cells with a shATF4 plasmid construct, with scrambled shRNA transduction as a control (Fig. 6A). 293 T/shATF4 and shCont cells were then transfected with GFP-LC3-RFP, an autophagic flux reporter in which GFP-LC3, but not RFP-LC3, is sensitive to degradation in the acidic environment of the autolysosome [37]. An increase in the percentage of yellow puncta relative to the total number demonstrates impaired autophagic flux. In control cells, only 7% of the total puncta were yellow under control conditions, and treatment with FAC did not induce a significant change (Fig. 6B). In contrast, ATF4 knockdown decreased autophagic flux in control condition as indicated by the 25% yellow puncta, which was significantly increased to 43% by FAC treatment, suggesting a further decrease in autophagic flux (Fig. 6B). Consistently, compared to the control cells, ATF4-deficient cells elicited an accumulation of LC3B-II, which was potentiated by FAC (Fig. 6C). Moreover, ATF4 knockdown led to a marked increase in FAC-induced mtROS production (Fig. 6D). Compared to the control cells, shATF4 cells were sensitized to FAC-induced cell death that was abrogated by ferroptosis inhibitor Fer-1 (Fig. 6E). Earlier data showed that HepG2 cells did not manifest upregulation of ATF4 but increased LC3B-II accumulation and ferroptosis in response to iron excess. Herein, we further showed that expressing an exogenous V5-ATF4 in HepG2 cells impeded FAC-induced accumulation of LC3B-II and p62 (Fig. S7A) and iron-induced cell death (Fig. S7B). Collectively, our findings in 293 T and HepG2 cells indicate a role of ATF4 in sustaining autophagic function and preventing iron-induced mtROS-mediated ferroptosis.
Fig. 6.
ATF4 deficiency leads to iron-dependent autophagic dysfunction, mtROS production and ferroptosis. (A) 293 T cells with stable knockdown of ATF4 (shATF4) or transduction with scrambled shRNA (shCont). (B) 293 T/shATF4 and shCont cells transiently expressing GFP-LC3-RFP were treated with or not 100 μM FAC for 24 h. Representative images are shown (left). The numbers of yellow puncta were quantified, and the percentages of yellow (white arrow) vs the total numbers of puncta are presented as mean ± SE (n = 15 fields of three independent experiments). (C) LC3B-II expression in 293 T/shATF4 and shCont cells that were treated with or not FAC for 24 h. Data shown are mean ± SE (n = 3 independent experiments). (D-E) 293 T/shATF4 and shCont cells were treated with or not 100 μM FAC or 5 μM Fer-1. After 24 h, cells were then stained with MitoSOX or PI. Relative fluorescence intensity (RFI) of MitoSOX staining per field was analyzed by ImageJ, and the percentage of PI + cells over the total numbers of cells per field was quantified. Data are mean ± SE (n = 9 fields of three independent experiments). *, P < 0.01 compared to untreated shCont cells; &, P < 0.01 relative to untreated shATF4 cells; and #, P < 0.01. Bars, 50 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.5. Iron deficiency induces ER stress and decreased autophagy
To compare the effects of increased and decreased iron, we then determined the impact of iron deficiency on cellular stress responses and autophagy in the different cell species. Iron deficiency was induced by desferoxamine (DFO), and again FTH1 expression was measured to indicate intracellular iron stores. We showed earlier that AML12 and RAW264.7 cells were insensitive to iron overload in terms of changes in autophagy and cell survival. Conversely, both cell lines were sensitive to iron deficiency by displaying an ER stress response as indicated by increased expression of phospho-PERK, ATF4 and CHOP, as well as increased LC3B expression (Fig. 7A). These effects were also seen in P-Hepa, but not in HepG2 cells (Fig. 7B). Intriguingly, the expression of oxidative stress marker NRF2 was decreased in prolonged iron deficiency (Fig. 7A). We continued to assess how autophagic flux is regulated by iron deficiency by using the autophagy inhibitor Chlq in AML12 cells. Again, DFO induced a significant increase in LC3B-II expression, but DFO/Chlq co-treatment did not increase LC3B-II further compared to Chlq alone (Fig. 7C), suggesting decreased autophagic flux in iron deficiency. Moreover, prolonged iron deficiency led to apoptosis as indicated by the increased expression of cleaved caspase-3 (CC3) in DFO-treated AML12 cells (Fig. 7D).
Fig. 7.
Iron deficiency induces ER stress response and attenuates autophagic flux in in vitro. (A) AML12, RAW264.7 and P-Hepa were treated with 100 μM DFO for the indicated times. The expression of stress proteins (NRF2, ATF4, CHOP, and phospho-PERK), autophagy markers (p62 and LC3B), and FTH1 was determined by Western blotting. Data are mean ± SE (n = 3 independent experiments), with the untreated control set at 100. For LC3B protein, the active isoform LC3B-II was quantified. Arrowhead denotes the upper bands are phospho-PERK (p-PERK). (B) Protein expression of ER stress and autophagy factors in DFO-treated HepG2 cells. (C) AML12 cells were treated with 100 μM DFO for 48 h in the presence or not of 100 μM chloroquine (Chlq) during the last 4 h. Changes in LC3B-II expression were quantified relative to untreated control (set at 100). Data are mean ± SE (n = 3 independent experiments). (D) The expression of cleaved caspase-3 (CC3) in DFO-treated AML12 cells. Data are mean ± SE (n = 3). *, P < 0.01; and **, P < 0.05 compared to the untreated control. n. s., not significant.
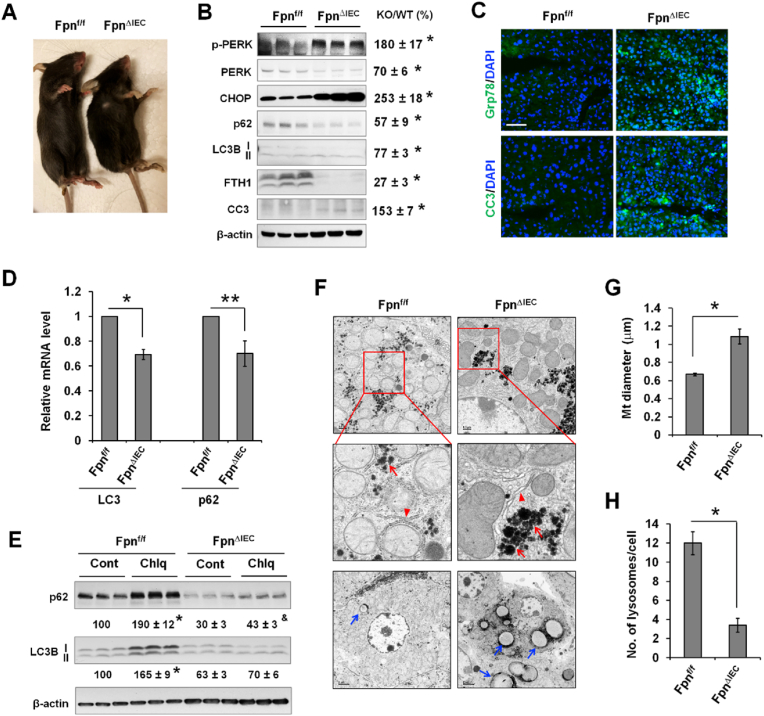
The in vivo effects of iron deficiency on the ER stress response and autophagy in the liver were then determined by using intestinal epithelial-specific knockout of iron exporter ferroportin (Fpn), FpnΔIEC mice. Consistent with previous reports [31,45], at 6 weeks after the knockout of Fpn, FpnΔIEC mice showed growth retardation and became visibly anemic (toes in gray color) (Fig. 8A) and iron deficient as indicated by the markedly decreased expression of FTH1 (Fig. 8B). Compared to control Fpnf/f mice, FpnΔIEC mice exhibited significantly elevated expression of phospho-PERK and CHOP (Fig. 8B), indicating increased ER stress. The expression of CC3 was also increased in iron-deficient livers (Fig. 8B). Consistently, fluorescent staining revealed increased expression of Grp78 and CC3 in the liver of FpnΔIEC mice (Fig. 8C). Unlike DFO-induced acute iron deficiency in cultured cells, FpnΔIEC liver with prolonged iron deficiency exhibited a significant decrease in the protein expression of p62 and LC3B (Fig. 8B). ER stress potentially induces transcriptional reprogramming that leads to transcriptional suppression of many genes [46,47]. Indeed, LC3 and p62 transcripts were significantly decreased in FpnΔIEC liver (Fig. 8D), accounting for the reduction in LC3B and p62 proteins. Moreover, Chlq significantly increased LC3B-II expression by 65% in the control liver, but did not induce any significant change in FpnΔIEC liver (Fig. 8E). Likewise, Chlq increased p62 expression by 90% in the control liver, but only led to a 43% increase in FpnΔIEC liver. The decreased accumulation of LC3B-II and p62 in response to Chlq suggests decreased autophagic flux in iron-deficient livers.
Fig. 8.
Iron deficiency induces ER stress, and decreased autophagy through attenuation of lysosome biogenesis in vivo in the liver. (A) Gross appearance of iron deficient FpnΔIEC mice and the control Fpnf/f mice at 6 weeks after the injection of tamoxifen. Note smaller size and pale toes of FpnΔIEC mice. (B) The expression of ER stress proteins (phospho-PERK and CHOP), autophagy markers (p62 and LC3B), and apoptotic marker (cleaved caspase-3, CC3) was determined by Western blotting. The percentage changes were calculated after standardized to β-actin and are presented as KO/WT. Data are mean ± SE (n = 6 mice of two independent experiments). *, P < 0.01. (C) Fluorescent staining of Grp78 and CC3 in the liver of FpnΔIEC and Fpnf/f mice. Bar, 50 μm. (D) qPCR analysis of the mRNA expression of LC3 and p62 in the liver of FpnΔIEC and Fpnf/f mice using ribosomal protein Rpl13a as an internal control. Data are mean ± SE (n = 3 mice per group). *, P < 0.01, and **, P < 0.05. (E) FpnΔIEC and control Fpnf/f mice were given Chlq (60 mg/kg). After 5 h, the livers were collected and the expression of LC3B and p62 was determined by Western blotting. The percentage changes in LC3B-II and p62 expression were calculated relative to untreated Fpnf/f mice (set at 100). Results are presented as mean ± SE (n = 3 mice per group). *, P < 0.01 compared to untreated Fpnf/f mice; and&, P < 0.05 compared to untreated FpnΔIEC mice. (F) TEM analysis of the intracellular structures of the hepatocytes of FpnΔIEC and Fpnf/f mice. Red arrowhead, ER; red arrow, glycogen granules; and blue arrow, lipid droplets. (G) Quantified diameter of the mitochondria in hepatocytes of FpnΔIEC and Fpnf/f mice (n = 100 mitochondria, 10 mitochondria per hepatocyte of two mice). *, P < 0.01. (H) Quantified number of lysosomes per hepatocyte of FpnΔIEC and Fpnf/f mice (n = 20 hepatocytes of two mice). *, P < 0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Iron deficiency-induced ER stress was recapitulated by TEM analysis where slightly dilated ER was seen in FpnΔIEC hepatocytes (Fig. 8F). We have also seen increased accumulation of glycogen (red arrow) and lipid droplets (blue arrow) in FpnΔIEC hepatocytes (Fig. 8F). Consistent with previous reports [26,48], mitochondria were significantly enlarged in iron-deficient FpnΔIEC hepatocytes (Fig. 8G, Fig. S8), Strikingly, compared to wild-type hepatocytes, FpnΔIEC hepatocytes contained significantly fewer numbers of lysosomes (Fig. 8H), which at least in part accounting for the decrease in autophagic flux.
Collectively, in vitro and in vivo data both show iron deficiency induces ER stress and decreased autophagic flux, which are correlated. Prolonged iron deficiency in mouse liver leads to impaired lysosomal biogenesis that is at least partially responsible for decreased autophagy.
3.6. Decreased autophagic flux by iron deficiency is dependent on the induction of ER stress
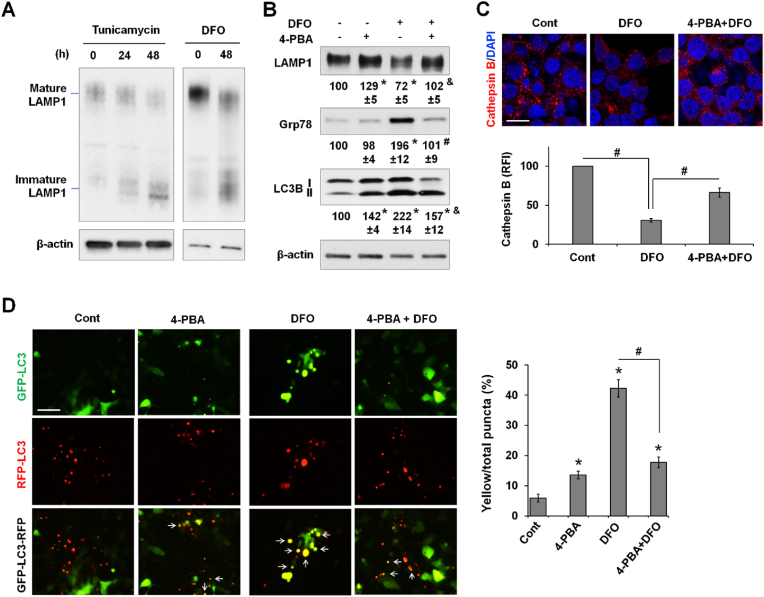
Our earlier findings in cultured cell and in the liver indicated a positive correlation between the induction of ER stress and autophagic dysfunction under a low-iron state. This relationship was also seen in 293 T cells (Fig. S4). We sought to determine how disturbed ER homeostasis associated with iron deficiency leads to decreased autophagic flux. LAMP1 is a glycosylated protein that is integral to lysosome biogenesis and autophagy [49]. We observed that DFO and tunicamycin, an inducer of ER stress by inhibiting N-glycosylation of proteins, both reduced the maturation of LAMP1 (Fig. 9A). Alleviation of ER stress by 4-phenylbutyric acid (4-PBA), a chemical chaperone that assists protein folding and inhibits ER stress, suppressed DFO-induced Grp78 expression and improved LAMP1 maturation (Fig. 9B). Importantly, 4-PBA attenuated DFO-induced accumulation of LC3B-II.
Fig. 9.
Decreased autophagy in iron deficiency is associated with ER stress. (A) The expression of mature and immature forms of LAMP1 in 293 T cells treated by tunicamycin (2 μM), an ER stress inducer, or DFO (100 μM). (B) The expression of mature LAMP1, Grp78, and LC3B in 293 T cells treated for 48 h by DFO (100 μM) and/or 4-PBA (10 mM), an inhibitor of ER stress. The percentage changes were relative to the untreated condition (set at 100). Data are presented as mean ± SE (n = 3 independent experiments). *, P < 0.01 compared to the untreated condition; #, P < 0.01, and &, P < 0.05 compared to DFO treatment alone. (C) Cathepsin B staining in 293 T cells treated by DFO in the presence or not of 4-PBA. Relative fluorescence intensity (RFI) of cathepsin B staining was quantified and is shown as mean ± SE (n = 9 fields of three independent staining). Bar, 10 μm. (D) 293 T cells expressing GFP-LC3-RFP reporter were treated by DFO and/or 4-PBA for 48 h. Representative fluorescent images are shown (left). The percentages of yellow (white arrow) vs the total numbers of puncta were calculated and are shown as mean ± SE (n = 15 fields of three independent experiments). *, P < 0.01 compared to the untreated control; and #, P < 0.01. Bar, 50 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We speculated that defects in the maturation of lysosomal proteins impairs lysosomal biogenesis. Cathepsin B, a lysosomal protease [50], was stained to examine potential changes in the number of lysosomes. LysoTracker was not used because it labels lysosomes as well as late endosomes [51]. DFO treatment resulted in a marked decrease in the number of cathepsin B-positive puncta, which was partially rescued by co-treatment with 4-PBA (Fig. 9C). By using autophagic flux reporter GFP-LC3-RFP, we further showed that DFO induced a marked increase in the percentage of yellow puncta, which was significantly attenuated by cotreatment with 4-PBA (Fig. 9D), suggesting that restoration of protein maturation and inhibition of ER stress improve autophagic flux. Of note, compared to the untreated control, 4-PBA by itself also led to a slight but significant increase in the percentage of yellow puncta. Altogether our results indicate that low iron availability disturbs ER homeostasis and protein maturation, at least in part accounting for impaired lysosome biogenesis and decreased autophagy.
4. Discussion
Disturbed iron homeostasis leading to iron deficiency or iron overload contributes to various pathological conditions. Iron deficiency leads to anemia, impaired lipid metabolism and lack of energy [52]. In recent years, iron overload has received much attention because of its detrimental role in the process of liver diseases, metabolic disorders, neurodegenerative disorders, and cardiovascular diseases [8]. An in-depth understanding of the molecular events resulting from impaired iron metabolism may allow the identification of novel therapeutic targets for these disorders. In the present study, we examined the deleterious effects of iron excess and iron deficiency on cellular homeostasis focusing on the regulation of the oxidative and ER stress responses, autophagy, and cell survival. We demonstrated that iron excess increased mtROS production and lipid peroxidation leading to impaired autophagic flux and ferroptosis (Fig. 10). Iron-induced lipid peroxidation was dependent on the overproduction of mtROS, which was attenuated by the upregulated ATF4. Elevation of ATF4 expression, and treatment with scavengers for mtROS and lipid peroxides, all mitigated iron-induced ferroptosis and the impairment in autophagy. Moreover, the impairment in autophagy potentiated iron-dependent ferroptosis. In contrast, prolonged iron deficiency led to impaired protein folding and maturation and ER stress that decrease autophagic flux contributing to the induction of apoptosis. The decrease in autophagic flux resulted, at least in part, from decreased lysosome biogenesis due to impaired lysosomal protein maturation.
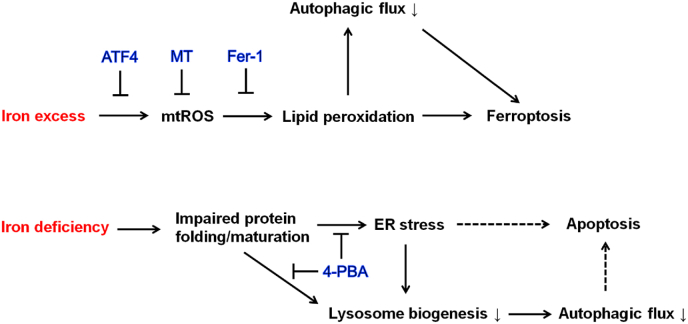
Fig. 10.
A schematic model of the impacts of iron excess and iron deficiency on cellular stress responses, autophagy and cell survival. On the one hand, iron excess induces the production of mtROS that causes lipid peroxidation-dependent induction of ferroptosis and inhibition of autophagic flux. Decreased autophagy further potentiates iron excess-induced ferroptosis. Upregulation of ATF4, and treatment with mtROS scavenger MT or lipid peroxide scavenger Fer-1 impede lipid peroxidation and the impairment of autophagic flux, preventing the induction of ferroptosis. On the other hand, iron deficiency impairs protein folding and maturation resulting in ER stress and decreased lysosome biogenesis that together leads to decreased autophagic flux. Improving protein maturation and alleviation of ER stress by 4-PBA rescues lysosome biogenesis and autophagic flux. It is speculated that ER stress and impaired autophagic flux both contribute to the induction of apoptosis.
Iron has the potential to induce cellular oxidant damage because it catalyzes ROS production. The main source of ROS is mitochondria [53]. ROS play important physiological roles including the regulation of cell proliferation and differentiation, adaptation to hypoxia, and regulation of autophagy and immunity. Overproduction of ROS, however, induces organelle damage, cell death and proinflammatory responses underlying the pathogenesis and progression of many diseases including liver diseases, metabolic diseases, neurodegenerative disorders, and cardiovascular diseases. We observed that in iron excess iron loading in the mitochondria was increased by staining with a mitochondrial Fe2+-specific probe. The intensity of mitochondrial Fe2+ staining correlated well with mtROS levels, suggesting a crucial role for iron in mtROS production. Our TEM analysis suggested that mitochondria are the primary target of damage by robust iron loading due to overwhelming production of mtROS. The increased production of mtROS resulting from iron loading likely results from the redox active Fe2+-mediated Fenton reaction and mtROS generation from increased mitochondrial respiration. The role of mitochondria in ferroptosis remains controversial. An earlier study showed that cancer cells with deleted mitochondrial DNA are not different from their parental cells in ferroptosis sensitivity to erastin, an inhibitor of xCT transporter and GSH synthesis [19]. Recent work, however, demonstrated that mitochondria participate in erastin-mediated ferroptosis [15]. Our study identified an indispensable role of mitochondria in iron excess-mediated lipid peroxidation and ferroptosis in hepatocytes. This finding is consistent with recent reports that mitochondria-targeted antioxidant MT rescued mitochondrial damage and ferroptosis in doxorubicin-induced cardiomyopathy [16,18]. We have not determined the level of mitochondrial iron required to generate sufficient mtROS to mediate ferroptosis. The amount of iron required to cause ferroptotic death likely varies depending on the environmental setting. Although 20 μM FAC was insufficient to induce ferroptosis in unstressed hepatocytes, it caused massive ferroptosis in hepatocytes pretreated with rotenone, a mitochondrial complex I inhibitor. These findings indicate that in conditions where pathological level of mtROS exists the threshold of iron content required to trigger ferroptosis is significantly lower because of the amplifying role of iron in ROS generation.
Consistent with previous reports [24,39], we have also shown that acute iron loading causes decreased autophagic flux. Importantly, our findings further demonstrate that overproduced mtROS and increased lipid peroxidation account for iron-induced impairment of autophagy as MT and Fer-1 both ablated LC3B-II accumulation. This finding is supported by a previous report that lipid peroxidation product 4-hydroxynonenal (4-HNE) at high concentrations impairs autophagic flux in primary neurons [54]. Autophagic dysfunction leads to the accumulation of oxidized molecules, damaged organelles and ROS [41]. We have recently shown that deficiency in autophagy promotes inflammatory cytokines-induced necrosis of hepatocytes [32]. By using ATG5-deficient hepatocytes and different autophagy inhibitors, we demonstrated in this study that cells with impaired autophagy are sensitized to iron excess-induced ferroptosis. Previous studies have suggested that autophagy is required for erastin-induced ferroptosis in that autophagic machinery is required for the release of labile iron through ferritinophagy [55,56]. Our findings suggest that, at least in the scenario when iron excess is achieved from an external source, impaired autophagic flux potentially promotes ferroptosis. The present findings and published work together suggest a balanced autophagic function is important to prevent cells from developing ferroptosis in physiological and diseased conditions.
Our current findings identified a novel role of ATF4 in preventing iron toxicity by sustaining mitochondrial homeostasis. ATF4 is a master regulator of integrated stress responses including oxidative stress, ER stress, amino acid deprivation, and viral infection [57]. Recent studies demonstrated that ATF4 is activated upon mitochondrial stress to suppress ATP-dependent respiration and mediate mitochondrial repairment [58]. ATF4 is involved in amino acid metabolism by regulating the expression of amino acid transporter [59], of which xCT mediates cystine uptake for GSH synthesis and is a target of erastin-induced ferroptosis [19,60]. Our finding of the anti-ferroptotic role of ATF4 in the context of iron excess is supported by a previous report that knockdown of ATF4 rendered glioma cells susceptible to erastin-induced ferroptosis [61]. ATF4 was also found to promote oxidative death in primary cultured neurons [62], suggesting a context dependency of ATF4 in cell survival regulation. Our current study further identified a potentially important role of ATF4 in sustaining autophagic function under basal and stressed (e.g., iron overload) conditions. The dual protective effects of ATF4 on autophagy and cell survival are consistent with our findings that impaired autophagy potentiates iron excess-dependent ferroptosis.
Unlike iron excess, which induced oxidative stress and mitochondrial damage, iron deficiency induced the ER stress response. Perturbed ER homeostasis has previously been reported in low iron states, but the exact mechanism has not been elucidated. We showed that iron deficiency-associated ER stress was rescued by treatment with 4-PBA, a pharmacological drug that interacts with hydrophobic domains of misfolded proteins and prevents their aggregation [63]. Our data suggest that iron deficiency induces the ER stress response through impaired protein folding and maturation at the site of ER. Indeed, maturation of LAMP1, primarily N-linked glycosylation, was impeded in iron-deficient 293 T cells. Impaired LAMP1/2 maturation under ER stress was previously observed in trophoblasts [64]. Impaired protein maturation likely resulted from decreased biogenesis of [Fe–S] clusters, which is a co-factor for many proteins to execute fundamental functions in various organelles including the ER. Increased loading of fatty acids is known as a strong risk factor for UPR induction [65]. Given that iron deficiency leads to lipid accumulation as observed in the current study and in previous reports [66], it is likely that lipid overload also contributes to low-iron induced ER stress.
An earlier study suggested that iron chelation activates autophagy because of increased expression of LC3B-II [30]. However, autophagy inhibitors were not included to ascertain the underlying cause of LC3B-II accumulation. By using autophagy inhibitor Chlq and flux reporter GFP-LC3-RFP, we demonstrated that iron deficiency leads to decreased autophagic flux. Decreases in lysosome biogenesis in prolonged iron deficiency accounts at least in part for the decreased autophagy. Mechanistically, it is likely that low in iron availability causes impaired function of enzymes that catalyze posttranslational protein maturation, leading to unfolded protein response (UPR) or ER stress. Thus, LAMP1/2 which are integral for the biogenesis of lysosomes may be dysfunctional because they require N-glycosylation as part of the maturation process. Indeed, treatment by 4-PBA partially rescued DFO-induced impairment of LAMP1 maturation and decreases in the number of lysosomes and autophagic flux. The role of ER stress in the regulation of autophagy has previously been reported in different contexts from stimulation to inhibition of autophagy [67]. Ganley et al. showed that induction of ER stress by thapsigargin, a potent inhibitor of sarco/endoplasmic reticulum Ca2+-ATPases, inhibits ALF by blocking the recruitment of Rab 7 to autophagosomes [68]. Lipid overload-induced ER stress also inhibits autophagy by disrupting ALF [69]. It is worth to note that a potential role of transcription factor EB (TFEB), which regulates lysosome biogenesis at the transcription level [70], in lysosomal biogenesis impairment associated with iron deficiency may be determined in future studies.
In conclusion, the current study demonstrates that acute iron overload induces an mtROS-dependent increase of lipid peroxidation leading to impaired autophagic flux and ferroptosis, whereas upregulation of ATF4 protects mitochondria and autophagy from iron toxicity, thus blocking iron excess-induced ferroptosis. Conversely, we have also highlighted that intracellular iron deficiency induces ER stress and decreased autophagy. Downregulated autophagy associated with iron deficiency is at least partially due to reduced lysosomal biogenesis in ER stress condition. Our study provides a detailed molecular understanding for how disrupted cellular iron homeostasis affects cellular function and cell survival.
Author contributions
Conception and design: EN, MJC, PH; Acquisition of data: YW, MW, YL, HT, SB, PH; Analysis and interpretation of data: MW, SB, PH; Writing, review and revision of the manuscript: SS, EN, MJC, PH.
Declaration of competing interest
The authors declare no conflict of interests.
Acknowledgements
This study was supported by National Institute of Health grant R01DK125647 (to P.H.), R01DK044234 (to M.J.C. and S.S.), and VA Research and Development Merit Review Award BX000136-08 (to S.S.). Robert P. Apkarian Integrated Electron Microscopy Core is supported by the Emory University School of Medicine, the Emory College of Arts and Sciences, and the Georgia Clinical & Translational Science Alliance. We would like to thank Dr. Adam Gracz for sharing the Olympus IX83 live imaging microscope.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102407.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Cairo G., Bernuzzi F., Recalcati S. A precious metal: iron, an essential nutrient for all cells. Genes Nutr. 2006;1:25–39. doi: 10.1007/BF02829934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Read A.D., Bentley R.E., Archer S.L., Dunham-Snary K.J. Mitochondrial iron-sulfur clusters: structure, function, and an emerging role in vascular biology. Redox Biol. 2021;47 doi: 10.1016/j.redox.2021.102164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muckenthaler M.U., Rivella S., Hentze M.W., Galy B. A red carpet for iron metabolism. Cell. 2017;168:344–361. doi: 10.1016/j.cell.2016.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pain D., Dancis A. Roles of Fe-S proteins: from cofactor synthesis to iron homeostasis to protein synthesis. Curr. Opin. Genet. Dev. 2016;38:45–51. doi: 10.1016/j.gde.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rouault T.A. Biogenesis of iron-sulfur clusters in mammalian cells: new insights and relevance to human disease. Dis Model Mech. 2012;5:155–164. doi: 10.1242/dmm.009019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson G.J., Frazer D.M. Current understanding of iron homeostasis. Am. J. Clin. Nutr. 2017;106:1559S. doi: 10.3945/ajcn.117.155804. 66S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganz T. Systemic iron homeostasis. Physiol. Rev. 2013;93:1721–1741. doi: 10.1152/physrev.00008.2013. [DOI] [PubMed] [Google Scholar]
- 8.Dev S., Babitt J.L. Overview of iron metabolism in health and disease. Hemodial. Int. 2017;21(Suppl 1):S6–S20. doi: 10.1111/hdi.12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang H., Chen J., Lu H., Zhou M., Chai Z., Hu Y. Iron-induced generation of mitochondrial ROS depends on AMPK activity. Biometals. 2017;30:623–628. doi: 10.1007/s10534-017-0023-0. [DOI] [PubMed] [Google Scholar]
- 10.Sumneang N., Siri-Angkul N., Kumfu S., Chattipakorn S.C., Chattipakorn N. The effects of iron overload on mitochondrial function, mitochondrial dynamics, and ferroptosis in cardiomyocytes. Arch. Biochem. Biophys. 2020;680 doi: 10.1016/j.abb.2019.108241. [DOI] [PubMed] [Google Scholar]
- 11.Uchiyama A., Kim J.S., Kon K., Jaeschke H., Ikejima K., Watanabe S., et al. Translocation of iron from lysosomes into mitochondria is a key event during oxidative stress-induced hepatocellular injury. Hepatology. 2008;48:1644–1654. doi: 10.1002/hep.22498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He H., Qiao Y., Zhou Q., Wang Z., Chen X., Liu D., et al. Iron overload damages the endothelial mitochondria via the ROS/ADMA/DDAHII/eNOS/NO pathway. Oxid. Med. Cell. Longev. 2019;2019 doi: 10.1155/2019/2340392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walter P.B., Knutson M.D., Paler-Martinez A., Lee S., Xu Y., Viteri F.E., et al. Iron deficiency and iron excess damage mitochondria and mitochondrial DNA in rats. Proc. Natl. Acad. Sci. U. S. A. 2002;99:2264–2269. doi: 10.1073/pnas.261708798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Redza-Dutordoir M., Averill-Bates D.A. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta. 2016;1863:2977–2992. doi: 10.1016/j.bbamcr.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Gao M., Yi J., Zhu J., Minikes A.M., Monian P., Thompson C.B., et al. Role of mitochondria in ferroptosis. Mol Cell. 2019;73:354–363 e3. doi: 10.1016/j.molcel.2018.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tadokoro T., Ikeda M., Ide T., Deguchi H., Ikeda S., Okabe K., et al. Mitochondria-dependent ferroptosis plays a pivotal role in doxorubicin cardiotoxicity. JCI Insight. 2020;5 doi: 10.1172/jci.insight.132747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao C., Liu X., Zhang Y., Lei G., Yan Y., Lee H., et al. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature. 2021;593:586–590. doi: 10.1038/s41586-021-03539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang X., Wang H., Han D., Xie E., Yang X., Wei J., et al. Ferroptosis as a target for protection against cardiomyopathy. Proc. Natl. Acad. Sci. U. S. A. 2019;116:2672–2680. doi: 10.1073/pnas.1821022116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mancias J.D., Wang X., Gygi S.P., Harper J.W., Kimmelman A.C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014;509:105–109. doi: 10.1038/nature13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Y., Epstein D.L., Liton P.B. Intralysosomal iron induces lysosomal membrane permeabilization and cathepsin D-mediated cell death in trabecular meshwork cells exposed to oxidative stress. Invest. Ophthalmol. Vis. Sci. 2010;51:6483–6495. doi: 10.1167/iovs.10-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X., Yu L., Xu H. Lysosome calcium in ROS regulation of autophagy. Autophagy. 2016;12:1954–1955. doi: 10.1080/15548627.2016.1212787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navarro-Yepes J., Burns M., Anandhan A., Khalimonchuk O., del Razo L.M., Quintanilla-Vega B., et al. Oxidative stress, redox signaling, and autophagy: cell death versus survival. Antioxidants Redox Signal. 2014;21:66–85. doi: 10.1089/ars.2014.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jahng J.W.S., Alsaadi R.M., Palanivel R., Song E., Hipolito V.E.B., Sung H.K., et al. Iron overload inhibits late stage autophagic flux leading to insulin resistance. EMBO Rep. 2019;20 doi: 10.15252/embr.201947911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kao J.K., Wang S.C., Ho L.W., Huang S.W., Chang S.H., Yang R.C., et al. Chronic iron overload results in impaired bacterial killing of THP-1 derived macrophage through the inhibition of lysosomal acidification. PLoS One. 2016;11 doi: 10.1371/journal.pone.0156713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoes M.F., Grote Beverborg N., Kijlstra J.D., Kuipers J., Swinkels D.W., Giepmans B.N.G., et al. Iron deficiency impairs contractility of human cardiomyocytes through decreased mitochondrial function. Eur. J. Heart Fail. 2018;20:910–919. doi: 10.1002/ejhf.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sangkhae V., Fisher A.L., Wong S., Koenig M.D., Tussing-Humphreys L., Chu A., et al. Effects of maternal iron status on placental and fetal iron homeostasis. J. Clin. Invest. 2020;130:625–640. doi: 10.1172/JCI127341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matak P., Matak A., Moustafa S., Aryal D.K., Benner E.J., Wetsel W., et al. Disrupted iron homeostasis causes dopaminergic neurodegeneration in mice. Proc. Natl. Acad. Sci. U. S. A. 2016;113:3428–3435. doi: 10.1073/pnas.1519473113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seo Y.A., Li Y., Wessling-Resnick M. Iron depletion increases manganese uptake and potentiates apoptosis through ER stress. Neurotoxicology. 2013;38:67–73. doi: 10.1016/j.neuro.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pullarkat V., Meng Z., Donohue C., Yamamoto V.N., Tomassetti S., Bhatia R., et al. Iron chelators induce autophagic cell death in multiple myeloma cells. Leuk. Res. 2014;38:988–996. doi: 10.1016/j.leukres.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Hanudel M.R., Czaya B., Wong S., Rappaport M., Namjoshi S., Chua K., et al. Enteral ferric citrate absorption is dependent on the iron transport protein ferroportin. Kidney Int. 2021;101:711–719. doi: 10.1016/j.kint.2021.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen Y., Malik S.A., Amir M., Kumar P., Cingolani F., Wen J., et al. Decreased hepatocyte autophagy leads to synergistic IL-1 beta and TNF mouse liver injury and inflammation. Hepatology. 2020;72:595–608. doi: 10.1002/hep.31209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen Y., Cingolani F., Malik S.A., Wen J., Liu Y., Czaja M.J. Sex-specific regulation of interferon-gamma cytotoxicity in mouse liver by autophagy. Hepatology. 2021;74:2745–2758. doi: 10.1002/hep.32010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao L., Bartnikas T., Chu X., Klein J., Yun C., Srinivasan S., et al. Hyperglycemia promotes microvillus membrane expression of DMT1 in intestinal epithelial cells in a PKCalpha-dependent manner. Faseb. J. 2019;33:3549–3561. doi: 10.1096/fj.201801855R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He P., Zhao L., Zhu L., Weinman E.J., De Giorgio R., Koval M., et al. Restoration of Na+/H+ exchanger NHE3-containing macrocomplexes ameliorates diabetes-associated fluid loss. J. Clin. Invest. 2015;125:3519–3531. doi: 10.1172/JCI79552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fisher A.L., Srole D.N., Palaskas N.J., Meriwether D., Reddy S.T., Ganz T., et al. Iron loading induces cholesterol synthesis and sensitizes endothelial cells to TNFalpha-mediated apoptosis. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.101156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ni H.M., Bockus A., Wozniak A.L., Jones K., Weinman S., Yin X.M., et al. Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy. 2011;7:188–204. doi: 10.4161/auto.7.2.14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hentze M.W., Caughman S.W., Rouault T.A., Barriocanal J.G., Dancis A., Harford J.B., et al. Identification of the iron-responsive element for the translational regulation of human ferritin mRNA. Science. 1987;238:1570–1573. doi: 10.1126/science.3685996. [DOI] [PubMed] [Google Scholar]
- 39.Fernandez B., Fdez E., Gomez-Suaga P., Gil F., Molina-Villalba I., Ferrer I., et al. Iron overload causes endolysosomal deficits modulated by NAADP-regulated 2-pore channels and RAB7A. Autophagy. 2016;12:1487–1506. doi: 10.1080/15548627.2016.1190072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klionsky D.J., Abdel-Aziz A.K., Abdelfatah S., Abdellatif M., Abdoli A., Abel S., et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)(1) Autophagy. 2021;17:1–382. doi: 10.1080/15548627.2020.1797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee J., Giordano S., Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem. J. 2012;441:523–540. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M., Khambu B., Zhang H., Kang J.H., Chen X., Chen D., et al. Suppression of lysosome function induces autophagy via a feedback down-regulation of MTOR complex 1 (MTORC1) activity. J. Biol. Chem. 2013;288:35769–35780. doi: 10.1074/jbc.M113.511212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yambire K.F., Rostosky C., Watanabe T., Pacheu-Grau D., Torres-Odio S., Sanchez-Guerrero A., et al. Impaired lysosomal acidification triggers iron deficiency and inflammation in vivo. Elife. 2019;8 doi: 10.7554/eLife.51031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li N., Ragheb K., Lawler G., Sturgis J., Rajwa B., Melendez J.A., et al. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J. Biol. Chem. 2003;278:8516–8525. doi: 10.1074/jbc.M210432200. [DOI] [PubMed] [Google Scholar]
- 45.Schwartz A.J., Converso-Baran K., Michele D.E., Shah Y.M. A genetic mouse model of severe iron deficiency anemia reveals tissue-specific transcriptional stress responses and cardiac remodeling. J. Biol. Chem. 2019;294:14991–15002. doi: 10.1074/jbc.RA119.009578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arensdorf A.M., Rutkowski D.T. Endoplasmic reticulum stress impairs IL-4/IL-13 signaling through C/EBPbeta-mediated transcriptional suppression. J. Cell Sci. 2013;126:4026–4036. doi: 10.1242/jcs.130757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chikka M.R., McCabe D.D., Tyra H.M., Rutkowski D.T. C/EBP homologous protein (CHOP) contributes to suppression of metabolic genes during endoplasmic reticulum stress in the liver. J. Biol. Chem. 2013;288:4405–4415. doi: 10.1074/jbc.M112.432344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu W., Barrientos T., Mao L., Rockman H.A., Sauve A.A., Andrews N.C. Lethal cardiomyopathy in mice lacking transferrin receptor in the heart. Cell Rep. 2015;13:533–545. doi: 10.1016/j.celrep.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eskelinen E.L. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol. Aspect. Med. 2006;27:495–502. doi: 10.1016/j.mam.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 50.Turk V., Turk B., Turk D. Lysosomal cysteine proteases: facts and opportunities. EMBO J. 2001;20:4629–4633. doi: 10.1093/emboj/20.17.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mundy D.I., Li W.P., Luby-Phelps K., Anderson R.G. Caveolin targeting to late endosome/lysosomal membranes is induced by perturbations of lysosomal pH and cholesterol content. Mol. Biol. Cell. 2012;23:864–880. doi: 10.1091/mbc.E11-07-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lal A. Iron in health and disease: an update. Indian J. Pediatr. 2020;87:58–65. doi: 10.1007/s12098-019-03054-8. [DOI] [PubMed] [Google Scholar]
- 53.Chen Q., Vazquez E.J., Moghaddas S., Hoppel C.L., Lesnefsky E.J. Production of reactive oxygen species by mitochondria: central role of complex III. J. Biol. Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- 54.Dodson M., Wani W.Y., Redmann M., Benavides G.A., Johnson M.S., Ouyang X., et al. Regulation of autophagy, mitochondrial dynamics, and cellular bioenergetics by 4-hydroxynonenal in primary neurons. Autophagy. 2017;13:1828–1840. doi: 10.1080/15548627.2017.1356948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hou W., Xie Y., Song X., Sun X., Lotze M.T., Zeh H.J., 3rd, et al. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016;12:1425–1428. doi: 10.1080/15548627.2016.1187366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao M., Monian P., Pan Q., Zhang W., Xiang J., Jiang X. Ferroptosis is an autophagic cell death process. Cell Res. 2016;26:1021–1032. doi: 10.1038/cr.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pakos-Zebrucka K., Koryga I., Mnich K., Ljujic M., Samali A., Gorman A.M. The integrated stress response. EMBO Rep. 2016;17:1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quiros P.M., Prado M.A., Zamboni N., D'Amico D., Williams R.W., Finley D., et al. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 2017;216:2027–2045. doi: 10.1083/jcb.201702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 60.Lim J.K.M., Delaidelli A., Minaker S.W., Zhang H.F., Colovic M., Yang H., et al. Cystine/glutamate antiporter xCT (SLC7A11) facilitates oncogenic RAS transformation by preserving intracellular redox balance. Proc. Natl. Acad. Sci. U. S. A. 2019;116:9433–9442. doi: 10.1073/pnas.1821323116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen D., Fan Z., Rauh M., Buchfelder M., Eyupoglu I.Y., Savaskan N. ATF4 promotes angiogenesis and neuronal cell death and confers ferroptosis in a xCT-dependent manner. Oncogene. 2017;36:5593–5608. doi: 10.1038/onc.2017.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lange P.S., Chavez J.C., Pinto J.T., Coppola G., Sun C.W., Townes T.M., et al. ATF4 is an oxidative stress-inducible, prodeath transcription factor in neurons in vitro and in vivo. J. Exp. Med. 2008;205:1227–1242. doi: 10.1084/jem.20071460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kolb P.S., Ayaub E.A., Zhou W., Yum V., Dickhout J.G., Ask K. The therapeutic effects of 4-phenylbutyric acid in maintaining proteostasis. Int. J. Biochem. Cell Biol. 2015;61:45–52. doi: 10.1016/j.biocel.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 64.Nakashima A., Cheng S.B., Kusabiraki T., Motomura K., Aoki A., Ushijima A., et al. Endoplasmic reticulum stress disrupts lysosomal homeostasis and induces blockade of autophagic flux in human trophoblasts. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-47607-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karaskov E., Scott C., Zhang L., Teodoro T., Ravazzola M., Volchuk A. Chronic palmitate but not oleate exposure induces endoplasmic reticulum stress, which may contribute to INS-1 pancreatic beta-cell apoptosis. Endocrinology. 2006;147:3398–3407. doi: 10.1210/en.2005-1494. [DOI] [PubMed] [Google Scholar]
- 66.Crooks D.R., Maio N., Lane A.N., Jarnik M., Higashi R.M., Haller R.G., et al. Acute loss of iron-sulfur clusters results in metabolic reprogramming and generation of lipid droplets in mammalian cells. J. Biol. Chem. 2018;293:8297–8311. doi: 10.1074/jbc.RA118.001885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rashid H.O., Yadav R.K., Kim H.R., Chae H.J. ER stress: autophagy induction, inhibition and selection. Autophagy. 2015;11:1956–1977. doi: 10.1080/15548627.2015.1091141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ganley I.G., Wong P.M., Gammoh N., Jiang X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell. 2011;42:731–743. doi: 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miyagawa K., Oe S., Honma Y., Izumi H., Baba R., Harada M. Lipid-induced endoplasmic reticulum stress impairs selective autophagy at the step of autophagosome-lysosome fusion in hepatocytes. Am. J. Pathol. 2016;186:1861–1873. doi: 10.1016/j.ajpath.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 70.Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.