ABSTRACT.
A broad-spectrum antibiotic, typically amoxicillin, is included in many country guidelines as part of the management of uncomplicated severe acute malnutrition (SAM) in children without overt clinical symptoms of infection. Alternative antibiotics may be beneficial for children with SAM without increasing selection for beta-lactam resistance. We conducted a 1:1 randomized controlled trial of single dose azithromycin versus a 7-day course of amoxicillin for SAM. Children 6–59 months of age with uncomplicated SAM (mid-upper arm circumference < 11.5 cm and/or weight-for-height Z-score < −3) were enrolled in Boromo District, Burkina Faso, from June through October 2020. Rectal swabs were collected at baseline and 8 weeks after treatment and processed using DNA-Seq. We compared the resistome at the class level in children randomized to azithromycin compared with amoxicillin. We found no evidence of a difference in the distribution of genetic antibiotic resistance determinants to any antibiotic class 8 weeks after treatment. There was no difference in genetic macrolide resistance determinants (65% azithromycin, 65% placebo, odds ratio, OR, 1.00, 95% confidence interval, CI, 0.43–2.34) or beta-lactam resistance determinants (82% azithromycin, 83% amoxicillin, OR 0.94, 95% CI, 0.33–2.68) at 8 weeks. Although presence of genetic antibiotic resistance determinants to macrolides and beta-lactams was common, we found no evidence of a difference in the gut resistome 8 weeks after treatment. If there are earlier effects of antibiotics on selection for genetic antibiotic resistance determinants, the resistome may normalize by 8 weeks.
INTRODUCTION
The WHO recommends a broad-spectrum antibiotic as part of the care package for outpatient management of uncomplicated severe acute malnutrition (SAM).1 Children with uncomplicated SAM by definition do not have clinical signs of infection. However, malnutrition can suppress the immune system and mask clinical signs of infection, and infectious disease and mortality is common in children with SAM.2,3 Although many country guidelines recommend the use of amoxicillin for uncomplicated SAM, randomized controlled trials have found mixed evidence of its efficacy.4,5 Antibiotic use selects for antibiotic resistance,6,7 and unnecessary antibiotic use as part of uncomplicated SAM management could be contributing to rising antibiotic resistance in regions where SAM is common.
We conducted a randomized controlled trial evaluating azithromycin versus amoxicillin as part of outpatient SAM management.8,9 As part of the trial, we collected rectal swabs in a random subset of enrolled children for assessment of the gut resistome, which is defined as the collection of antibiotic resistance genes in a given environment.10 Here, we report resistome outcomes in children being managed for uncomplicated SAM receiving azithromycin compared with amoxicillin.
METHODS
Study setting and participants.
Participants were enrolled in six Centres de Santé et de Promotion Sociale (CSPS) in Boromo District, Burkina Faso. Participants were enrolled between June and October 2020 and the final follow-up visit was in December 2020. Participants were followed for 8 weeks as part of the trial. CSPS are government-run primary healthcare facilities that provide basic preventative and curative care and include an outpatient nutritional program to provide care for uncomplicated SAM. Study enrollment sites were rural. The study area experiences seasonal food insecurity from approximately August through October prior to the annual harvest.11 Participants were recruited via community-based screening based on mid-upper arm circumference (MUAC) or during weekly outpatient screening at the CSPS through the nutritional program. Children were eligible for inclusion in the trial if they were 6–59 months of age, had a MUAC < 11.5 cm, and/or weight-for-height Z-score (WHZ) < −3, had not been admitted to a nutritional program for SAM in the previous 14 days, had no antibiotic use in the previous 7 days, had no clinical complications requiring inpatient care or antibiotic treatment, had sufficient appetite according to a feeding test with ready-to-use therapeutic food (RUTF), had no reported allergies to macrolides or azalides, had no congenital abnormalities that would lead to growth faltering, and were planning to stay in the study area for the full 8-week study period.
Trial design.
Complete methods for the trial have been previously reported.9 The study was reviewed and approved by the Comité Institutionnel d’Ethique in Nouna, Burkina Faso, and the Institutional Review Board at the University of California, San Francisco. Written informed consent was obtained from the caregivers of all included participants. Eligible participants were randomized in a 1:1 fashion to a single, oral 20 mg/kg dose of azithromycin (Azithrin oral suspension 200 mg/5 mL, Strides Shashun Ltd., Bangalore, India) or a 7-day course of amoxicillin (80 mg/kg divided into two daily doses, Amoxicillin syrup, 250 mg/5 mL, Reyoung Pharmaceutical, Shandong, China) per national guidelines. Personnel at the enrollment facility administered and directly observed the first dose of amoxicillin and the dose of azithromycin. For participants in the amoxicillin arm, caregivers administered all remaining doses. Adherence to additional doses of amoxicillin was not monitored. All medications were prepared as pediatric oral suspension.
A random sample of approximately one-third of children enrolled in the parent trial was selected for inclusion in the resistance study, which involved collection of a rectal swab at baseline and 8 weeks from enrollment. At baseline, the study’s mobile data collection application randomly showed the interviewer if the child was selected for the swab study, and if so, the caregiver was offered participation. The same child was sampled at baseline and 8 weeks. Rectal samples were immediately placed in DNA/RNA Shield (Zymo Research, Irvine, CA) and transported to the laboratory in Nouna, Burkina Faso, where they were stored at −80°C until they were shipped to the United States for processing.
Laboratory methods.
Deoxyribonucleic acid was extracted using the ZymoBIOMICS DNA/RNA miniprep kit (Zymo Research, Irvine, CA) per manufacturer’s instructions. Double-stranded DNA was fragmented, size selected, and converted to Illumina libraries using the NEBNext Ultra II DNA Library Prep Kit (E7645) per manufacturer’s recommendations and amplified with 11 polymerase chain reaction (PCR) cycles. Samples were sequenced on the Illumina NovaSeq 6000 using 150-nucleotide paired-end sequencing. Human reads were removed as previously described.12 Nonhost read pairs were aligned to the MEGARes reference antimicrobial database using Burrows-Wheeler alignment with default settings.13 Only antimicrobial resistance determinants (ARDs) with a gene fraction of > 80% were identified as present in a sample to reduce false-positive ARD identification. Each ARD was classified at the class level, and a sample was determined to be resistant if even one ARD was detected.
Sample size.
The sample size was determined based on the trial’s primary aim as a pilot study to evaluate azithromycin versus amoxicillin for weight gain in children with uncomplicated SAM.9 A sample size of 50 children per arm (100 total) was chosen for the resistome outcome for feasibility and resource-related reasons. Assuming a prevalence of macrolide resistance of 65% in the amoxicillin arm, a sample size of 50 children per arm had approximately 80% power to detect a 30% absolute difference in the prevalence of genetic macrolide resistance determinants between arms.
Statistical methods.
We evaluated the prevalence of ARDs at the class level at 8 weeks in children receiving azithromycin compared with amoxicillin using unadjusted logistic regression models with a term for randomized treatment group as the sole predictor. We compared the normalized abundance of resistance reads to macrolides, beta-lactams, and other antibiotic classes in children receiving azithromycin compared with amoxicillin using linear regression models adjusted for the baseline normalized reads to each antibiotic class. P values were adjusted for the false discovery rate (FDR) to account for multiple comparisons. As a post-hoc analysis, we compared mean normalized reads to macrolide genetic resistance determinants between baseline and 8 weeks in the azithromycin arm and beta-lactam resistance determinants in the amoxicillin arm between baseline and 8 weeks using a paired t test to evaluate within-arm changes over time in reads. All analyses were conducted in R (The R Foundation for Statistical Computing, Vienna, Austria).
RESULTS
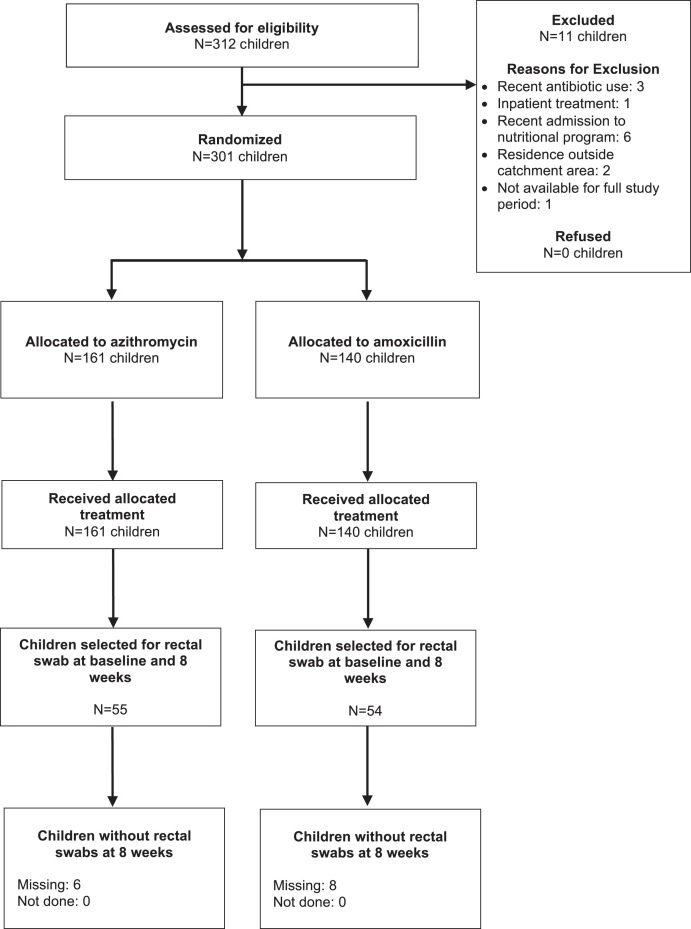
Of 301 children enrolled in the trial, 109 (36%) were randomly selected for inclusion in the resistance study, of whom 55 were in the azithromycin group and 54 were in the amoxicillin group (Figure 1). Among children enrolled in the resistance study, baseline characteristics were well balanced between arms (Table 1). Children were a median of 15 and 14 months and 58% and 52% were female in the azithromycin and amoxicillin arms, respectively. Baseline anthropometric measures were similar between the two arms. Children had a median MUAC of 11.3 cm and mean WHZ of −3.1 SD at baseline, which was similar to the full study population.8 At baseline, more than half of samples had genetic macrolide resistance determinants (azithromycin: 60%; amoxicillin: 54%) and the majority of samples had genetic beta-lactam resistance determinants (azithromycin: 89%, amoxicillin: 81%; Supplemental Table 1). Of children enrolled in the swab sub-study, six (11%) were lost to follow-up in the azithromycin arm and eight (15%) in the amoxicillin arm and did not have 8-week rectal swabs collected (Figure 1).
Figure 1.
Screening, enrollment, and follow-up of study participants.
Table 1.
Baseline characteristics by randomized treatment group among participants in the rectal swab study
| Azithromycin (N = 55) | Amoxicillin (N = 54) | |
|---|---|---|
| Age, months, median (IQR) | 15 (9–23) | 14 (10–18) |
| Female sex, N (%) | 32 (58%) | 28 (52%) |
| Breastfeeding, N (%) | 40 (73%) | 42 (78%) |
| Mother’s age, years, median (IQR) | 24 (21–30) | 26 (22–30) |
| MUAC, cm, median (IQR) | 11.3 (11.1–11.4) | 11.3 (11.2–11.4) |
| WHZ, mean (SD) | −3.1 (1.1) | −3.1 (0.9) |
| WAZ, mean (SD) | −3.4 (1.0) | −3.4 (1.0) |
| HAZ, mean (SD) | −2.3 (1.3) | −2.5 (1.5) |
HAZ = height-for-age Z-score; IQR = interquartile range; MUAC = mid-upper arm circumference; SD = standard deviation; WAZ = weight-for-age Z-score; WHZ = weight-for-height Z-score.
We found no evidence of a difference at 8-week posttreatment in the prevalence of ARDs in the gut in children randomized to azithromycin versus amoxicillin. Macrolide resistance determinants were present in 65% of samples in the azithromycin and amoxicillin groups (odds ratio [OR] 1.00, 95% confidence interval [CI] 0.43–2.34). Beta-lactam resistance determinants were present in 82% and 83% of the samples in the azithromycin and amoxicillin groups (OR 0.94, 95% CI 0.70–3.56). There was no statistically significant difference in genetic resistance determinants of any other antibiotic class by treatment group (Table 2).
Table 2.
Proportion of participants with genetic antibiotic resistance determinants by antibiotic class by treatment group at 8 weeks from enrollment
| Azithromycin (N = 49) | Amoxicillin (N = 46) | Odds ratio (95% CI) | P value* | |
|---|---|---|---|---|
| Macrolides | 32 (65%) | 30 (65%) | 1.00 (0.43–2.34) | 0.99 |
| Beta-lactams | 40 (82%) | 38 (83%) | 0.94 (0.33–2.68) | 0.99 |
| Fluoroquinolones | 29 (59%) | 22 (48%) | 1.58 (0.70–3.56) | 0.56 |
| Sulfonamides | 24 (49%) | 14 (30%) | 2.19 (0.95–5.09) | 0.52 |
| Trimethoprim | 38 (78%) | 26 (78%) | 0.96 (0.36–2.53) | 0.99 |
| Tetracyclines | 45 (92%) | 40 (87%) | 1.69 (0.44–6.41) | 0.77 |
| Rifampin | 19 (39%) | 15 (33%) | 1.31 (0.56–3.04) | 0.77 |
| Aminoglycosides | 31 (63%) | 23 (50%) | 1.72 (0.76–3.91) | 0.52 |
| Metronidazole | 24 (49%) | 29 (63%) | 0.56 (0.25–1.28) | 0.52 |
| Aminocoumarins | 10 (20%) | 4 (9%) | 2.69 (0.78–9.29) | 0.52 |
| Bacitracin | 19 (39%) | 12 (26%) | 1.79 (0.75–4.30) | 0.52 |
| Fosfomycins | 1 (2%) | 2 (4%) | 0.46 (0.04–5.23) | 0.77 |
| Elfamycins | 34 (69%) | 27 (59%) | 1.60 (0.69–3.71) | 0.56 |
| Lipopeptides | 2 (4%) | 1 (2%) | 1.91 (0.17–21.9) | 0.80 |
| Phenicol | 8 (16%) | 7 (15%) | 1.09 (0.36–3.28) | 0.99 |
| Glycopeptides | 0 | 0 | NA | NA |
| Cationic peptides | 30 (61%) | 22 (48%) | 1.72 (0.76–3.89) | 0.52 |
False discovery rate-adjusted P value.
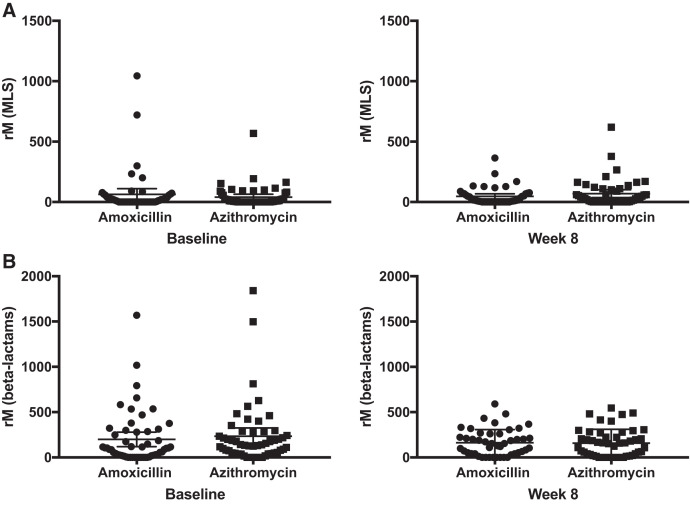
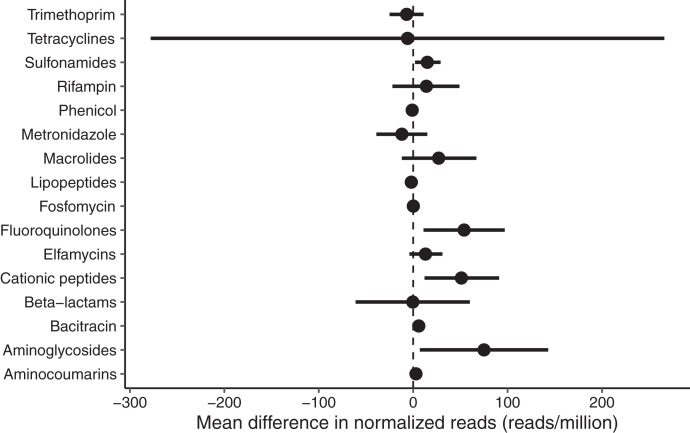
We found no significant difference in normalized read count of genetic azithromycin determinants to any antibiotic class after adjusting for multiple comparisons. Figure 2 shows the number of normalized ARD reads in reads per million sequencing reads for macrolides and beta-lactams at baseline and 8 weeks in the azithromycin and amoxicillin groups. In a linear regression model adjusted for baseline normalized ARD reads, we found no evidence of a difference in abundance of genetic macrolide (mean difference 27, 95% CI −12 to 67) or beta-lactam (mean difference −0.4, 95% CI −61 to 60) resistance determinants. There were no differences in abundance of normalized ARD reads to other antibiotic classes after adjustment for multiple comparisons (Figure 3). We found no evidence of a difference in abundance of normalized ARD reads before and after treatment to macrolide genetic resistance determinants in the azithromycin group (mean difference −29 for week 8 versus baseline, 95% CI −67 to 8, P = 0.15) or beta-lactam genetic resistance determinants in the amoxicillin group (mean difference 28 for week 8 versus baseline, 95% CI −63 to 118, P = 0.54).
Figure 2.
Normalized reads (rM, in reads/million) of genetic macrolide (MLS) resistance determinants (A) at baseline and 8 weeks and genetic beta-lactam resistance determinants (B) at baseline and 8 weeks.
Figure 3.
Mean difference in normalized reads (in reads/million) between children receiving azithromycin compared with amoxicillin. Differences above zero favor azithromycin; differences below zero favor amoxicillin.
DISCUSSION
We found no evidence of a difference in the resistome as defined by the prevalence of genetic ARDs in the gut of children with uncomplicated SAM who received azithromycin compared with amoxicillin 8 weeks after treatment. A broad-spectrum antibiotic, typically amoxicillin, is included in most country guidelines for the treatment of uncomplicated SAM in the absence of clinic symptoms of infection because acute malnutrition can suppress the immune system and mask symptoms of infection. Amoxicillin is one of the most commonly used antibiotics for childhood infection in Burkina Faso,14 and is routinely used in regions with a large burden of SAM for management of pneumonia and other common childhood infections.15,16 Furthermore, beta-lactam antibiotics belong to the WHO Access group of antibiotics due to their effectiveness against a wide range of organisms. Selection for beta-lactam resistance could reduce the efficacy of amoxicillin for critical treatment needs.
The present study evaluated the collection of genetic antibiotic resistance determinants in the gut. This approach does not provide information on phenotypic resistance in specific organisms, and thus does not necessarily imply functional resistance. High throughput sequencing has advantages in that it can identify all ARD genes. In contrast, phenotypic methods are organism-specific and PCR-based methods are limited in the number of genes screened. Genomic approaches as performed on the rectal samples in this study are typically species-independent and thus do not provide evidence about resistance in specific organisms, nor do they provide information about functional resistance.17 Genotypic resistance does not perfectly correlate with phenotypic resistance. However, as a clinical trial outcome (versus guidance for clinical antibiotic decision-making), evaluation of the resistome may provide more information efficiently on the overall impact of antibiotics on selection for resistance compared with microbiological methods.
Azithromycin has recently been shown to reduce all-cause childhood mortality when mass distributed to children aged 1–59 months in some settings with high childhood mortality in sub-Saharan Africa,18 with the strongest effects in young infants and those who were underweight at the time of treatment.19 Surveillance for antimicrobial resistance has shown that mass azithromycin distributions select for genotypic and phenotypic macrolide resistance, although the prevalence of resistance decreases once selection pressure is removed.7,20,21 Resistance selection with mass treatment may differ from individual-level treatment. In individually randomized trials of antibiotic treatment of children without SAM, azithromycin has generally been shown to lead to short-term selection for genetic macrolide resistance. For example, studies have found selection for genetic macrolide resistance determinants in the gut at 5 days after treatment and in the nasopharynx at 28 days but not 12 months after antibiotic exposure.12,22 The present study did not include a short-term timepoint. If azithromycin use selected for increased macrolide resistance relative to amoxicillin in the short term, this study could not detect it. Previous studies of mass treatment of azithromycin have suggested that co-selection for multiple antibiotic classes may occur with azithromycin, including an increase in genetic beta-lactam resistance determinants in children residing in communities receiving azithromycin compared with placebo.20 If bacteria have resistance to multiple antibiotic classes, use of one class of antibiotics could induce selection to resistant strains to other antibiotic classes.23 The lack of difference in resistome at 8 weeks may indicate that the gut resistome normalizes by 8 weeks or that co-selection occurred, masking any differences in resistance selection for specific antibiotic classes.
Children with acute malnutrition experience a large burden of infection, and previous studies have shown that infections in children with SAM are often resistant to first-line antibiotics.2,24 Relapse after recovery from SAM and default and readmission to SAM therapeutic feeding programs are not uncommon.25 These children may receive frequent antibiotic exposure if they are commonly given amoxicillin for SAM or other indications. Prevalence of genetic beta-lactam resistance determinants was high at baseline, although similar to that observed in another trial of antibiotics in a general population-based sample of children in Burkina Faso.12 This may be reflective of high background amoxicillin use among children in the study area.14,26 The degree to which selection for genetic antibiotic resistance determinants affects antibiotic efficacy is unknown.
This study has several limitations. Loss to follow-up was moderately high, with approximately 13% of children enrolled in the swab study lost by 8 weeks and not included in analyses. If loss to follow-up was differential with respect to randomized treatment assignment, this could lead to bias in estimates. The parent trial did not include a placebo arm. Because the national guidelines in Burkina Faso for management of uncomplicated SAM include a broad-spectrum antibiotic and the parent trial was a pilot trial for a full-scale efficacy trial,9 inclusion of a placebo arm in the trial was not considered ethical. Although we did not find evidence of a difference in antibiotic resistance determinants between antibiotic arms, how this would compare with no antibiotic treatment cannot be determined in this study. The amoxicillin arm required twice-daily dosing for 7 days by the child’s caregiver. We did not measure adherence to the amoxicillin regimen in this study. If caregivers missed a substantial number of doses, it is possible that results could have been biased toward the null. While the trial’s exclusion criteria excluded children who had received an antibiotic in the previous 7 days, it is possible that children could have recently (> 7 days) received an antibiotic that affected the composition of their resistome. We did not measure recent antibiotic exposure at baseline and therefore cannot comment on how prior antibiotic exposure affected the resistome. However, due to the randomized nature of the intervention, prior antibiotic exposure is not expected to bias by arm comparisons. Finally, data from this study arose from a pilot randomized controlled trial and the sample size was not large. As a result, confidence intervals are wide for some comparisons, indicating uncertainty.
We found no evidence of a difference in the gut resistome among children with uncomplicated SAM receiving azithromycin compared with amoxicillin. Although we hypothesized that children receiving azithromycin would have higher prevalence of macrolide resistance determinants and those receiving amoxicillin would have higher prevalence of beta-lactam resistance determinants, the prevalence of genetic antibiotic resistance determinants to those two classes was nearly identical between arms. The results of this study suggest that use of an alternative antibiotic as part of the management of uncomplicated SAM will not substantively affect the gut resistome in treated children. However, due to the use of genetic resistance determinants in the current study, we are unable to comment on whether the different antibiotic classes in the present study could have an impact on clinical outcomes for specific pathogens. Placebo-controlled trials are required to understand if antibiotics for uncomplicated SAM affect the resistome compared with no antibiotics to provide evidence of the cost-benefit balancing risk of selection for antibiotic resistance with efficacy of antibiotics for improving outcomes in SAM.
Supplemental Material
ACKNOWLEDGMENTS
We confirm that all ongoing and related trials for this drug/intervention are registered (#NCT03568643). This trial is registered at ClinicalTrials.gov (#NCT03568643, https://clinicaltrials.gov/ct2/show/NCT03568643).
Note: Supplemental table appears at www.ajtmh.org.
REFERENCES
- 1. World Health Organization , 2010. Guideline: Updates on the Management of Severe Acute Malnutrition in Infants and Children. Geneva, Switzerland: WHO. [PubMed] [Google Scholar]
- 2. Page A-L et al. 2013. Infections in children admitted with complicated severe acute malnutrition in Niger. PLOS One 8: e68699–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bourke CD Berkley JA Prendergast AJ , 2016. Immune dysfunction as a cause and consequence of malnutrition. Trends Immunol 37: 386–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Trehan I Goldbach HS LaGrone LN Meuli GJ Wang RJ Maleta KM Manary MJ , 2013. Antibiotics as part of the management of severe acute malnutrition. N Engl J Med 368: 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Isanaka S et al. 2016. Routine amoxicillin for uncomplicated severe acute malnutrition in children. N Engl J Med 374: 444–453. [DOI] [PubMed] [Google Scholar]
- 6. Lipsitch M Samore MH , 2002. Antimicrobial use and antimicrobial resistance: a population perspective. Emerg Infect Dis 8: 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. O’Brien K, Emerson P, Hooper PJ, Reingold AL, Dennis EG, Keenan JD, Lietman TM, Oldenburg CE, 2018. Antimicrobial resistance following mass azithromycin distribution for trachoma: a systematic review. Lancet Infect Dis 19: e14–e25. [DOI] [PubMed] [Google Scholar]
- 8. O’Brien KS et al. 2021. Comparing azithromycin to amoxicillin in the management of uncomplicated severe acute malnutrition in Burkina Faso: a pilot randomized trial. medRxiv. doi: 10.1101/2021.06.15.21258967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. O’Brien K Sié A Dah C Ourohire M Arzika AM Boudo V Lebas E Godwin WW Arnold BF Oldenburg CE , 2021. Azithromycin for uncomplicated severe acute malnutrition: study protocol for a pilot randomized controlled trial. Pilot Feasibility Stud 7: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Crofts TS Gasparrini AJ Dantas G , 2017. Next-generation approaches to understand and combat the antibiotic resistome. Nat Rev Microbiol 15: 422–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Belesova K Gasparrini A Sie A Sauerborn R Wilkinson P , 2018. Annual crop yield variation, child survival, and nutrition among subsistence farmers in Burkina Faso. Am J Epidemiol 187: 242–250. [DOI] [PubMed] [Google Scholar]
- 12. Oldenburg CE et al. 2019. Gut resistome after oral antibiotics in preschool children in Burkina Faso: a randomized controlled trial. Clin Infect Dis 70: 525–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lakin SM et al. 2016. MEGARes: an antimicrobial resistance database for high throughput sequencing. Nucleic Acids Res 45: D574–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sie A et al. 2019. Antibiotic prescriptions among children under age 5 in Nouna District, Burkina Faso. Am J Trop Med Hyg 100: 1121–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sié A et al. 2021. Indication for antibiotic prescription among children attending primary healthcare services in Rural Burkina Faso. Clin Infect Dis 1–4: 1288–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ginsburg AS Mvalo T Nkwopara E McCollum ED Ndamala CB Schmicker R Phiri A Lufesi N Izadnegahdar R May S , 2019. Placebo vs amoxicillin for nonsevere fast-breathing pneumonia in Malawian children aged 2 to 59 months: a double-blind, randomized clinical noninferiority trial. JAMA Pediatr 173: 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Belkum A Burnham CAD Rossen JWA Mallard F Rochas O Dunne WM , 2020. Innovative and rapid antimicrobial susceptibility testing systems. Nat Rev Microbiol 18: 299–311. [DOI] [PubMed] [Google Scholar]
- 18. Keenan JD et al. 2018. Azithromycin to reduce childhood mortality in sub-Saharan Africa. N Engl J Med 378: 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. O’Brien KS et al. 2020. Biannual azithromycin distribution and child mortality among malnourished children: a subgroup analysis of the MORDOR cluster-randomized trial in Niger. PLoS Med 17: e1003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Doan T et al. 2020. Macrolide and nonmacrolide resistance with mass azithromycin distribution. N Engl J Med 383: 1941–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haug S et al. 2010. The decline of pneumococcal resistance after cessation of mass antibiotic distributions for trachoma. Clin Infect Dis 51: 571–574. [DOI] [PubMed] [Google Scholar]
- 22. Bojang A et al. 2020. Impact of intrapartum oral azithromycin on the acquired macrolide resistome of infants’ nasopharynx: a randomized controlled trial. Clin Infect Dis 71: 3222–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lipsitch M , 2001. Measuring and interpreting associations between antibiotic use and penicillin resistance in Streptococcus pneumoniae. Clin Infect Dis 32: 1044–1054. [DOI] [PubMed] [Google Scholar]
- 24. Nalwanga D Musiime V Kizito S Kiggundu JB Batte A Musoke P Tumwine JK , 2020. Mortality among children under five years admitted for routine care of severe acute malnutrition: a prospective cohort study from Kampala, Uganda. BMC Pediatr 20: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stobaugh HC, Marie N, Mayberry A, Mcgrath M, Mark J, Lelijveld N, 2018. Relapse after severe acute malnutrition: a systematic literature review and secondary data analysis. 1–12. doi: 10.1111/mcn.12702. [DOI] [PMC free article] [PubMed]
- 26. Oldenburg CE et al. 2021. Distance to primary care facilities and healthcare utilization for preschool children in rural northwestern Burkina Faso: results from a surveillance cohort. BMC Health Serv Res 21: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.