Abstract
A real-time PCR method was developed to quantitate viral DNA that includes duplex amplification, internal standardization, and two-color fluorescence detection without the need to generate an external standardization curve. Applied to human parvovirus B19 DNA, the linear range was from 102 to at least 5 × 106 copies per ml of sample. The coefficient of variation was 0.29 using a run control of 2,876 copies per ml. The method reduces the risk of false-negative results, yields high precision, and is applicable for other DNA targets.
Real-time PCR allows the monitoring of the generation of PCR products simultaneous with the amplification process; for example, by measuring the increase of fluorescence in the reaction vial caused by the cleavage of an appropriately labeled probe by the DNA polymerase used for amplification (6, 7). Numerous applications of real-time PCR have been published applying one-color fluorescence detection and quantitation using an external standardization curve generated by amplification of a known amount of standard, for instance, the quantitation of human papilloma virus, human herpes simplex virus, and hepatitis B virus DNA (1, 8, 9). External standardization, however, does not take into account the efficacy of the extraction procedure and the variability thereof. Thus, there is the chance of biased results and, in the worst case, a sample might be judged as negative, not due to true absence of target nucleic acids, but because of an unsuccessful extraction procedure. In order to overcome this shortcoming, an internally standardized real-time PCR method was developed. Quantitation of DNA of human parvovirus B19 was used as an example. The method consists of the coextraction of internal standards together with the nucleic acids of interest, followed by their simultaneous amplification, and the specific detection of the PCR products by appropriately labeled probes, including two-color fluorescence detection. This method offers the advantages of high precision and a reduced risk of false-negative results without the need to generate an external standardization curve.
First, the internal standards were constructed. Plasmid pTM/Parvo-wt is a derivative of pBluescript SK(−) (Stratagene; GenBank accession no. X52330) and harbors nucleotides 1324 to 1560 of human parvovirus B19 (GenBank accession no. M13178) cloned into the NotI and XhoI sites in forward direction. In addition, the sequence 5′-TCTAGA-3′ was inserted between nucleotides 1354 and 1355. Plasmids pTM/Parvo−23bp and pTM/Parvo+12bp were used as internal standards (further termed calibrators) and were derived from pTM/Parvo-wt by replacing the region corresponding to nucleotides 1355 to 1532 with the artificial sequences 5′-TAGCCTGGGA CGATTTTGCA TATAAGGGAA CAATTATAAA GCAGTCCTAA AGTTAATATG CTGTTAGAAG AAGAGGGGGT CAACATGAAG CGTAGTGAGT CATTAGCTAT GTTGACATTA GTCAAGTTGC TTACGAAAAG CAGATAGATT AGCTT-3′ and 5′-TAGCCTGGGA CGATTTTGCA TATAAGGGAA CCGGAAGATA GAGCGCGAAT TCCTATGAGA GGGCCTTAGA CAAGAAGATG TGTGACGATC TGGACGACAT AATGGTATAA TATAATGCTG TTAAGAAGAC GTTTCTAAGT ATGATATACA AGAAGTTCAA GTTGCTTACG AAAAGCAGAT AGATTAGCTT-3′, respectively. The plasmids were purified, quantitated by measuring the A260 units, and diluted to the appropriate concentration in a buffer consisting of 10 mM Tris-HCl (pH 8.0), 0.1 mM EDTA, and 10 μg of fish sperm DNA per ml. The source of human parvovirus B19 was a positive plasma sample, which was diluted in phosphate-buffered saline (PBS) supplemented with 5% human plasma. For each analysis, 4,500 and 1,500 copies of linearized pTM/Parvo−23bp and pTM/Parvo+12bp, respectively, were used. The extraction was done as described earlier (3), followed by real-time PCR using a 7700 SDS instrument plus Software version 1.6.3 from PE Biosystems. The PCR reaction was set up in a 50-μl volume containing a 15-μl sample extract, 1× TaqMan buffer A (PE Biosystems; 50 mM KCl, 10 mM Tris-HCl, 0.01 mM EDTA, 60 nM passive reference 1; pH 8.3), 5 mM MgCl2, a 200 nM concentration of each primer (PE Biosystems), a 100 nM concentration of each probe (PE Biosystems), 200 μM concentrations of each deoxynucleoside triphosphate, 9% glycerol (wt/vol), 0.05% gelatine (wt/vol), 0.01% Tween 20 (wt/vol), and 2.5 U of AmpliTaq Gold Polymerase (PE Biosystems). The specifications of primers and probes are given in Table 1. The amplification profile was a 10-min incubation at 95°C followed by 40 cycles, with each cycle consisting of denaturation at 95°C for 15 s and annealing-extension at 60°C for 1 min, applying a dual-color mode. The default settings were used for data processing, and the results were exported to a Microsoft Excel spreadsheet. The evaluation was done using the relation: NT/ml sample = 1/V × f × NC × 2(CtC − CtT), where NT and NC are the copies of target and calibrator, CtC and CtT are threshold cycle values of calibrator and target, f is the correction factor, and V is the volume of sample used per extraction.
TABLE 1.
Real-time PCR primers and probesa
| Primer or probe | Sequence (5′ to 3′) | Type of primer or probe | Tm (°C) |
|---|---|---|---|
| Parvo+1355.C | GGCAGCATGTGTTAAAGTGGAT | Target, forward | 58 |
| Parvo−1529.C | CCTGCTACATCATTAAATGGAAAGTT | Target, reverse | 58 |
| ART-FOR1.F | AGATAGCCTGGGACGATTTTG | Calibrator, forward | 57 |
| ART-REV1.C | GCTTAAGCTAATCTATCTGCTTTTCGTA | Calibrator, reverse | 58 |
| Ovarp-wt/T01 | ACCATGCCATATACTGGAACACTTTTAGCA | Target, probe | 65 |
| Ovarp-20/T01 | AACATAGCTAATGACTCACTACGCTTCATG | Calibrator, probe | 62 |
For each PCR primer and probe used, the name, the nucleic acid sequence, the type, and the theoretical melting temperature (Tm) are given. The B19-specific probe Ovarp-wt/T01 was labeled at the 5′ end with 6-FAM as the reporter dye. The probe Ovarp-20/T01 was specific for calibrator pTM/Parvo−23bp and was labeled at the 5′ end with VIC as the reporter dye. Both probes were linked with the quencher dye TAMRA at the 3′ end. Melting temperatures (Tm) given in degrees Celsius were calculated using Primer Express software version 1.0 (PE Biosystems).
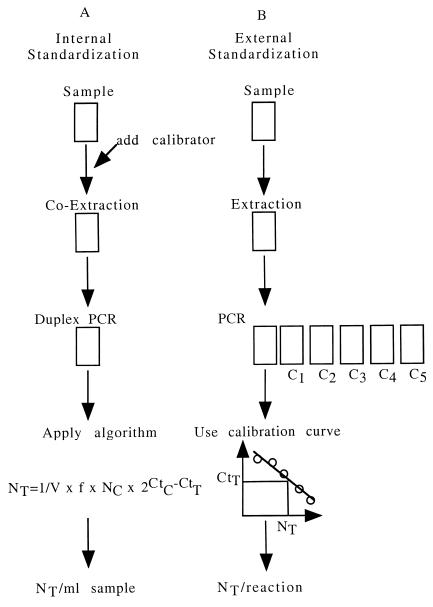
External standardization (Fig. 1B) is based on the amplification of the target nucleic acids in an extract or lysate and the determination of the so-called threshold cycle (Ct) value, which is defined as the cycle at which the fluorescence in a respective well exceeds the background fluorescence. The Ct value is used to calculate the copy numbers by applying a standardization curve generated by amplification of increasing amounts of standards, by determination of the Ct values, and by plotting these versus copy numbers of the standards used. The final result is given as copies per reaction. Using the novel approach of internal standardization (Fig 1A), a known amount of calibrator is coextracted with the target nucleic acids, amplified by real-time PCR applying two sets of primer pairs and appropriately labeled probes. Subsequently, the primary results of the analysis, the Ct values, are determined. Based on the known amount of calibrator and an experimentally determined correction factor f, the number of initial target molecules is calculated using the relation described above, and the final result is given in copies per milliliter of sample.
FIG. 1.
Principle of the assay. (A) Approach of internal standardization of real-time PCR. A known amount of calibrator is coextracted with the target nucleic acid, viral DNA of human parvovirus B19 concentrated by ultracentrifugation. Extraction is followed by the simultaneous amplification of target nucleic acids and calibrator, subsequent two-color fluorescence detection, and determination of the Ct values of the target and calibrator. The evaluation is done by applying the algorithm described in the text, and the final result is given in copies per milliliter of sample. (B) Approach of external standardization of real-time PCR. The approach of external standardization is done by amplification of the target nucleic acids in an extract or lysate of the sample, followed by determination of the Ct value. Using a calibration curve generated by amplification of increasing amounts of calibrator (C1 to C5), the final result is evaluated and given in copies per reaction. Note that the internal standardization, as opposed to the external standardization, does take into account the variation of the efficacy of the extraction procedure and further sample-related effects.
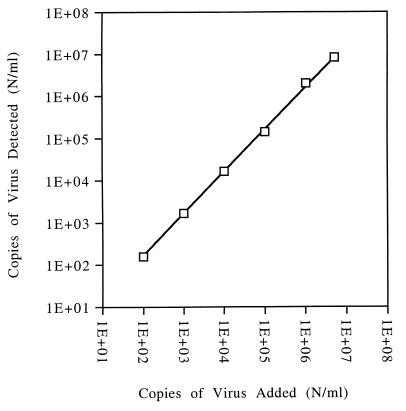
The accuracy of the assay was determined according to guidelines on validating analytical procedures (2). An amount of 4,044 copies of plasmid DNA of pTM/Parvo-wt was analyzed eight times, and a mean value of 5,431 was obtained. Therefore, in order to get an accurate result, an experimental value, obtained from either plasmid DNA or native viral DNA, had to be corrected by the ratio 4,044 divided by 5,431 or 0.7446. The assumption was made that plasmid and native viral DNA behave similarly. The linearity of the assay was determined by analyzing samples of an appropriate dilution series of a B19 DNA positive plasma sample. The results are plotted in Fig. 2, where the x axis shows the concentration of relative target nucleic acid, and the y axis shows the number of molecules detected. The data show that there is an excellent correlation from 102 to at least 5 × 106 copies per ml. The coefficient of correlation was 0.998. The intermediate precision is a measure of the degree of the variation of data obtained by a particular assay (2). To evaluate this parameter, 18 values of a run control, which were obtained by three different operators over a time period of 7 weeks, were analyzed. The statistical analysis of the final results is compiled in Table 2. The mean value of this run control was 2,876 copies per ml, and the standard deviation was 830, resulting in an absolute coefficient of variation of 0.29. The mean Ct values for the target and calibrator were 32.03 and 30.24, respectively. The standard deviations were 1.02 and 0.78, respectively. The selectivity of the assay was determined in order to detect any effect of a particular sample matrix on the results obtained. A sample consisting of 20 mM Tris-HCl (pH 7.0) plus 0.1% (wt/vol) bovine serum albumin was spiked with virus to a final concentration of 1,851 copies per ml and analyzed six times. The mean value obtained was 1,703 copies per ml and, therefore, the spike recovery was 92%. Since the observed decrease of 8% is statistically not significant, the conclusion is that no matrix effect exists.
FIG. 2.
Linearity of the assay. Aliquots of a serial dilution of human parvovirus B19, diluted in PBS supplemented with 5% human plasma, were analyzed. The x axis shows the concentration of the respective sample, and the y axis shows the result obtained. Both values are given in copies per milliliter of sample. The correlation factor was evaluated as 0.998 in the range from 102 to 5 × 106 copies/ml. The number of data per concentration was ≥3.
TABLE 2.
Intermediate precision of the assay obtained by repeated analyis of a run controla
| Value | Copies/ml | CtC | CtT |
|---|---|---|---|
| Mean | 2,876 | 30.24 | 32.03 |
| SEM | 196 | 0.18 | 0.24 |
| Median | 2,697 | 30.29 | 32.00 |
| SD | 830 | 0.78 | 1.02 |
| Minimum | 1,451 | 29.05 | 30.24 |
| Maximum | 4,675 | 31.64 | 34.07 |
| Range | 3,224 | 2.59 | 3.83 |
| Count | 18 | 18 | 18 |
Aliquots of a run control containing human parvovirus B19 at a concentration of 2,876 copies per ml of PBS supplemented with 5% human plasma were processed over a period of 7 weeks by three different operators. Column 2 shows the results given in copies per ml of sample. Columns 3 and 4 give the results of the Ct calibrator and target values.
An internally standardized real-time PCR method was used to quantitate nucleic acids, as exemplified by human parvovirus B19 DNA. Two species of calibrators were added, coextracted, and coamplified with the target sequences, whereby only the PCR products of one calibrator, those of pTM/Parvo−23bp, were detected by adding the appropriate probe. Thus, the PCR products of the second calibrator species, pTM/Parvo+12bp, were also generated but were not detected. Nevertheless, the approach of using two calibrators was pursued (3–5) because it offers the opportunity for making the assay even more precise once appropriate software and dyes are available. The software version used only allows simultaneous processing of two dyes, namely, FAM and VIC, whereby FAM was used to detect PCR products derived from the target sequences, i.e., B19 DNA, and VIC for PCR products of the calibrator pTM/Parvo−23bp. The sequences of the calibrators chosen for amplification are artificial and do not show any homology to other naturally occurring sequences. Thus, these calibrators may be used as universal calibrators in further assays, provided compatible amplification conditions for the target and calibrator can be found. The correction factor was introduced into the evaluation algorithm in order to increase the accuracy by correcting for minute differences in the amplification efficiencies of target nucleic acid and calibrator, as well as for probe-related parameters such as efficiency of synthesis, binding to PCR products, efficiency of cleavage by the DNA polymerase, and spectral characteristics of the fluorescent dyes coupled to the probe. The factor was obtained by measuring a known amount of plasmid DNA. Alternatively, an international standard for native viral B19 DNA could be used. The observed precision was good, especially considering that, opposed to external standardization, the variation in the efficacy of the extraction was included, since the calibrators were added at the beginning of the extraction procedure. Linearity was determined for the range from 102 to at least 5 × 106 copies per ml, with a correlation factor of 0.998. The feasible upper limit of the linear range was not fully determined and could very well be higher, further increasing the usefulness of the assay. In conclusion, the method described here to quantitate DNA by real-time PCR reduces the risk of false-negative results, yields high precision, and is applicable for a wide range of DNA targets.
Acknowledgments
We thank S. Bachler and S. Braun for expert technical assistance and E. Johnson-Froneberg for critical reading of the manuscript.
REFERENCES
- 1.Abe A, Inoue K, Tanaka T, Kato J, Kajiyama N, Kawaguchi R, Tanaka S, Yoshiba M, Kohara M. Quantitation of hepatitis B virus genomic DNA by real-time detection PCR. J Clin Microbiol. 1999;37:2899–2903. doi: 10.1128/jcm.37.9.2899-2903.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Food and Drug Administration. ICH guideline on validating analytical procedures, finalized #40,709. Washington, D.C.: Food and Drug Administration; 1995. [Google Scholar]
- 3.Gruber F, Falkner F G, Dorner F, Hammerle T. Precise quantitation of human parvovirus B19 DNA in biological samples by PCR. Biologicals. 1998;26:213–216. doi: 10.1006/biol.1998.0129. [DOI] [PubMed] [Google Scholar]
- 4.Hammerle T, Falkner F G, Dorner F. A sensitive PCR assay system for the quantitation of viral genome equivalents: hepatitis C virus (HCV) Arch Virol. 1996;141:2103–2114. doi: 10.1007/BF01718218. [DOI] [PubMed] [Google Scholar]
- 5.Hammerle T, Himmelspach M, Dorner F, Falkner F G. A sensitive PCR assay system for the quantitation of viral genome equivalents: human immunodeficiency virus type 1 (HIV-1) and hepatitis B virus (HBV) Arch Virol. 1997;142:1297–1306. doi: 10.1007/s007050050161. [DOI] [PubMed] [Google Scholar]
- 6.Heid C A, Stevens J, Livak K J, Williams P M. Real-time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 7.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′----3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Josefsson A, Livak K, Gyllensten U. Detection and quantitation of human papillomavirus by using the fluorescent 5′ exonuclease assay. J Clin Microbiol. 1999;37:490–496. doi: 10.1128/jcm.37.3.490-496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ryncarz A J, Goddard J, Wald A, Huang M L, Roizman B, Corey L. Development of a high-throughput quantitative assay for detecting herpes simplex virus DNA in clinical samples. J Clin Microbiol. 1999;37:1941–1947. doi: 10.1128/jcm.37.6.1941-1947.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]