Abstract
Sixteen percent of California ground squirrels (Spermophilus beecheyi) were found to be shedding an average of 53,875 Cryptosporidium parvum oocysts/g of feces. Male squirrels had a higher prevalence and higher intensity of shedding than did female squirrels. The majority of C. parvum isolates matched a bovine-murine genotype, with a few isolates resembling a porcine genotype. Higher intensities of shedding by males may enhance dissemination and genotypic mixing of this protozoa given males' proclivity to disperse to nonnatal colonies.
Rodents can achieve high population densities, which in combination with relatively high prevalences of fecal shedding of Cryptosporidium parvum oocysts (4, 7, 19, 22) can result in significant environmental loading rates for this parasite. California ground squirrels (Spermophilus beecheyi) live in grasslands, meadows, agricultural regions, and lower-elevation woodlands from central Washington to Baja California Norte in Mexico. In California, colonies of these rodents can achieve population densities from 8.4 to 92 adults/ha (1, 10, 14). Previous studies of gastrointestinal parasites in ground squirrels focused on Wyoming and Towsend's ground squirrel (Spermophilus elegans and Spermophilus towsendii), in which various species of helminths and coccidians (Eimeria sp.) were identified (15, 16, 18, 23). We conducted the following study to estimate the daily environmental loading rate of C. parvum for populations of California ground squirrels. Given the current interest in characterizing transmission cycles for genotypes of C. parvum (2, 3, 8, 11, 13, 24, 25), we also determined the distribution of C. parvum genotypes within this host population.
Sample collection.
Under a memorandum of understanding with the Wildlife Services Agency, U.S. Department of Agriculture Maricopa, Calif., California ground squirrels from throughout Kern County were dispatched by expert marksmen. Squirrels were collected from June 1998 through October 1998, sexed, and aged, and fecal samples were collected from the colon during necropsy (6, 20).
Enumeration of C. parvum.
Enumeration of oocysts was performed as previously described (12). Percent recovery of the immunofluorescent assay was determined by spiking two negative squirrel fecal samples with wild-type dairy calf C. parvum oocysts to a concentration of 104, 105, and 106 oocysts per g, with six replicates per squirrel per concentration, as previously described (12).
DNA extraction from C. parvum oocysts.
C. parvum positive fecal samples that had >30 oocysts per smear were purified as previously described (9, 21). For DNA extraction, a 2.0-ml aliquot of sample was centrifuged at 15,800 × g for 10 min. Supernatant was discarded, and oocysts were resuspended in 500 μl of 10% commercial bleach solution and incubated at 37°C for 10 min, which was followed by the addition of 1.0 ml of 10% sodium thiosulfate. The samples were centrifuged as before, and the oocysts were resuspended in a commercial sodium phosphate-buffered solution and transferred to a FastDNA Multimix tube (Bio 101, Vista, Calif.). DNA was extracted according to the manufacturer's recommendations (FastDNA; Bio 101) with minor modifications.
DNA amplification conditions.
Two procedures were used to determine the species and genotype of the isolates of Cryptosporidium. The first method was a nested PCR restriction fragment length polymorphism (RFLP) technique that targets the 18S small-subunit (SSU) rRNA gene locus of Cryptosporidium (24, 25). All procedures for the outer and inner (nested) PCR amplification have been previously described (24). The second PCR-RFLP genotyping method targeted the Cryptosporidium oocyst wall protein gene (COWP) (17). All procedures for the PCR amplification have been previously described (17). Negative and positive controls consisted of sterile H2O and a PCR-confirmed bovine C. parvum sample, respectively.
Restriction enzyme digest.
Ten-microliter aliquots of the various PCR products were electrophoresed in 2% agarose gel (Life Technologies, Gaithersburg, Md.) with a 100- to 1,500-bp DNA ladder (Promega Corporation, Madison, Wis.), with the size of the amplicons determined by a Bio-Rad Fluor-S MultiImager apparatus and Multi-Analyst, version 1.1, software (Bio-Rad, Hercules, Calif.). Ten microliters of nested PCR product from the SSU rRNA gene-based primers was digested with 10 U of VspI and SspI (Life Technologies and Boehringer Mannheim Corp. [Indianapolis, Ind.]) and 1 × buffer (Boehringer Mannheim Corp. and Life Technologies) (24). Amplicons obtained with the COWP gene-based primers were digested using 10 μl of PCR product mixed with 5 U of RsaI and 1 × buffer (Boehringer Mannheim Corp.). Digests from both PCR methods were incubated at 37°C for 1 h, and 10 μl was electrophoresed in 2% agarose gel (Life Technologies). Gels were stained and the sizes of restriction fragments were determined as before.
Estimation of daily fecal production and percent moisture of fecal pellets.
Naturally voided fecal pellets from 11 captive adult California ground squirrels were collected and weighed for a 24-h period. Squirrels had an ad libitum supply of water, oats, fresh weeds, and commercial rat chow. Fecal matter was then dried for 7 days at 70°C to determine percent moisture.
Statistical analyzes.
The mean number of oocysts per gram was compared across the various sex and age groups, using a square-root transformation of oocyst counts and either a Welch modified two-sample t test with unequal variances or a one-way analysis of variance (as explained in volume 1 of Guide to Statistics, p. 52–92 [Data Analysis Products Division, MathSoft, Seattle, Wash.], 1999). The prevalence of oocyst shedding was compared across these sex, age, and sex-by-age groupings using a likelihood ratio test (C. Mehta and N. Patel, StatXact 4.0: user manual [Cytel Software Corporation, Cambridge, Mass.], 1996).
Three hundred and nine California ground squirrels from 17 geographic locations were tested for C. parvum oocysts. Sixteen percent of squirrels were shedding C. parvum oocysts (Table 1). The mean concentration of C. parvum for positive squirrels was 53,875 oocysts/g of feces and for all 309 squirrels in the study was 8,543 oocysts/g of feces. Percentages of adults and juvenile squirrels shedding oocysts were not significantly different. Males were about 1.5 times more likely than females to be shedding oocysts (two-sided P = 0.10), with males also shedding higher concentrations of oocysts than females (two-sided P = 0.05) (Table 1). Male squirrels are more likely than females to disperse from natal colonies and emigrate to move to nonnatal colonies (1, 5, 6, 10). This tendency of males to disperse to other colonies can promote the dissemination of C. parvum to nonkin squirrel populations. Given the occurrence of active infection of C. parvum (12 to 22%) among the different age and sex classes of California ground squirrels during much of the season of natal dispersal (late summer through early winter), it is likely that mixed cryptosporidial infections can occur when infected males, with their 20% prevalence of active shedding of oocysts, emigrate to move into new colonies. Neither the prevalence nor oocyst concentration was significantly different between male juveniles, male adults, female juveniles, and female adults. These oocyst concentrations were adjusted for the percent recovery of the immunofluorescent assay (Table 2).
TABLE 1.
Intensity of shedding of C. parvum oocysts by California ground squirrels (S. beecheyi)
| Stratification | No. of positive squirrels/no. of sampled squirrels (%) | Mean oocysts/g (SD) of sample
|
|
|---|---|---|---|
| Positivea | Totalb | ||
| Age | |||
| Adults | 21/142 (15) | 44,025 (55,677) | 6,511 (26,186) |
| Juveniles | 28/167 (17) | 61,263 (131,024) | 10,272 (57,612) |
| Sex | |||
| Male | 24/119 (20)c | 69,101 (112,477) | 13,936d (56,931) |
| Female | 25/190 (13)c | 39,259 (97,297) | 5,166d (37,137) |
| Age and sex | |||
| Male juveniles | 12/55 (22) | 75,973 (148,857) | 16,576 (74,267) |
| Male adults | 12/64 (19) | 62,264 (64,745) | 11,675 (36,495) |
| Female juveniles | 16/112 (14) | 50,257 (119,791) | 7,180 (47,447) |
| Female adults | 9/78 (12) | 19,707 (28,665) | 2,274 (11,204) |
| Total | 49/309 (16) | 53,875 (104,988) | 8,543 (45,895) |
Arithmetic mean for the number of oocysts shed per gram of positive fecal samples from the specified population, adjusted for percent recovery of the direct immunofluorescent assay.
Arithmetic mean for the number of oocysts shed per gram of all fecal samples collected from the specified population, adjusted for percent recovery of the direct immunofluorescent assay.
Significantly different at the P = 0.05 level, with oocyst counts square-root transformed.
Significantly different at the P = 0.10 level.
TABLE 2.
Percent recovery for the direct immunofluorescent assaya
| Squirrel no. | Concn (oocysts/g) | Mean smear wtb (range) (g) | Mean no. (range) of oocysts detected | % Recoveryc (range) |
|---|---|---|---|---|
| 1 | 10,000 | 0.019 (0.015–0.022) | 18 (15–22) | 9 (7–13) |
| 100,000 | 0.017 (0.014–0.023) | 171 (156–182) | 10 (7–12) | |
| 1,000,000 | 0.016 (0.011–0.018) | 2,413 (2,135–2,654) | 15 (13–18) | |
| 2 | 10,000 | 0.018 (0.013–0.020) | 15 (11–18) | 9 (6–11) |
| 100,000 | 0.020 (0.015–0.022) | 246 (211–273) | 12 (10–15) | |
| 1,000,000 | 0.013 (0.011–0.016) | 2,449 (2,130–2,851) | 18 (17–20) |
Performed on two separate California ground squirrels for three different concentrations of C. parvum oocysts.
Mean of six replicate fecal samples per squirrel per concentration.
Percent recovery was calculated as n/(wc), where n is the number of oocysts counted in the smear, w is the weight in grams of the fecal smear, and c is the concentration of oocysts per gram of the spiked stool sample.
Based on these data, the daily environmental loading rate of squirrel C. parvum can be roughly calculated as follows: mean shedding intensity × mean daily fecal output × population density (or size). Mean shedding intensity was recalculated so that each sex and age class category was equally weighted, resulting in a mean intensity of 9,426 oocysts/g of feces. Fecal output by the 11 captive squirrels was 119.4 g (wet weight) and 56.7 g (dry weight). Total squirrel biomass was 7,624.8 g (range of 550 to 915 g per squirrel), therefore, daily fecal production (wet weight) was ∼2% of body weight. The average mass of California ground squirrels is 100 to 150 g for young pups, with adult females ranging from 450 to 650 g and adult males ranging from 650 to 1,000 g (6, 10). Population densities range from 8.4 to 92 adults per ha (1, 10, 14), resulting in biomass densities of 4,620 to 50,600 g/ha (based on a mean squirrel mass of 550 g). Therefore, the oocyst loading rate for ground squirrel populations was approximately 8.7 × 105 oocysts/ha/day for low density populations and 9.5 × 106 oocysts/ha/day for highly dense populations.
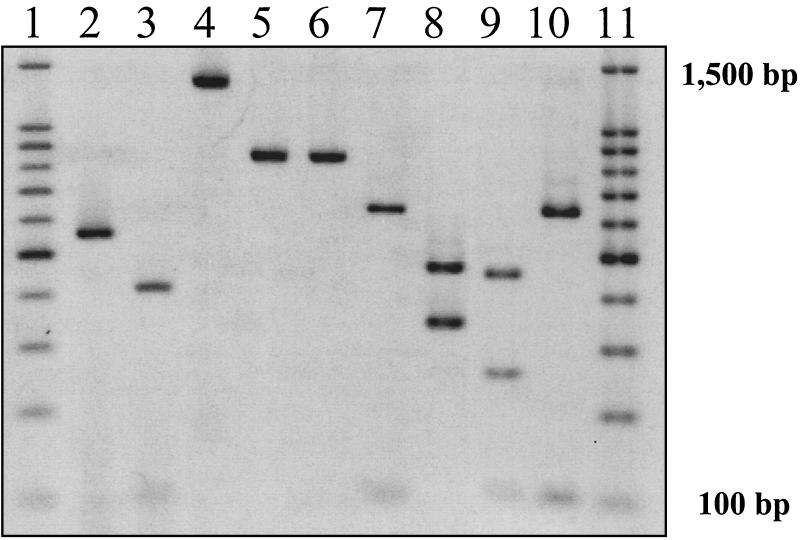
California ground squirrels in our population were infected with at least two genotypes of this protozoan. Specifically, isolates from nine squirrels were genotyped by the COWP method, resulting in an ∼550-bp amplicon and two visible fragments of 410 and 106 bp when digested with RsaI (Fig. 1). This C. parvum genotype is similar to isolates from both bovine and murine sources (17, 26). Using the nested PCR-RFLP technique for the SSU rRNA gene, the outer primer pair generated amplicons of 1,325 bp for 13 isolates (Fig. 1). The inner primer pair generated an amplicon of 831 bp for 11 of the 13 isolates (85%), and an amplicon of approximately 838 bp for 2 of the 13 isolates (15%). For the 11 isolates having the 831-bp internal amplicon, digestion with SspI resulted in visible fragments of 449, 254, and 119 bp. This nested PCR-RFLP pattern is consistent with previous isolations of C. parvum from bovine sources (24, 25). For the two isolates having the 838-bp internal amplicon, digestion with SspI resulted in visible fragments of 453 and 365 bp, a pattern consistent with previous isolates of C. parvum from porcine sources (24, 25). In contrast to the finding of two distinct genotypes from digestion with SspI, digestion with VspI of the 13 internal amplicons resulted in visible fragments of 628 and 102 to 104 bp, a pattern consistent with previous isolations of C. parvum from bovine sources, indicative of what is referred to as bovine B genotype (24, 25). Having two genotypes of C. parvum circulating within a single host population has also been observed for humans and cattle (2, 11, 13, 17, 24, 25). Whether mixed infections of C. parvum isolates from different squirrel colonies enhance the fitness of this protozoal parasite is unclear, but the occurrence of sexual multiplication (gametogony) by C. parvum suggests it is feasible that oocysts can be constructed from gamonts originating from different colonies and of different genotypes.
FIG. 1.
Confirmation of species and genotyping of C. parvum oocysts from California ground squirrels through one-step PCR-RFLP and nested PCR-RFLP analysis. Lane 1, 100- to 1,500-bp markers; lane 2, 550-bp amplicon from COWP gene primers; lane 3, 410 and 106-bp fragments from COWP RFLP analysis with RsaI; lane 4, 1,325-bp amplicon from SSU rRNA primary product; lane 5, 831-bp amplicon from SSU rRNA secondary product, genotype bovine B; lane 6, 838-bp amplicon from SSU rRNA secondary product, genotype porcine; lane 7, 628-bp and 102- to 104-bp fragments from porcine genotype with VspI; lane 8, 453- and 365-bp fragments from porcine genotype with SspI; lane 9, 449-, 254-, and 119-bp fragments from genotype bovine B with SspI; lane 10, 628-bp and 102- to 104-bp fragments from genotype bovine B with VspI; lane 11, 100- to 1,500-bp markers.
Acknowledgments
This work was supported in part by Section 15 of the Bureau of Land Management, Grazing Advisory Committee, Bakersfield, Calif.
We are especially grateful to Mark Jensen and Gary Simmons of the Wildlife Services Agency, USDA, for establishing a memorandum of understanding for this project.
REFERENCES
- 1.Boellstorff D E, Owings D H. Home range, population structure, and spatial organization of California ground squirrels. J Mammol. 1995;76:551–561. [Google Scholar]
- 2.Bonnin A, Fourmaux M N, Dubremetz J F, Nelson R G, Gobet P, Harly G, Buisson M, Puygautheir-Toubas D, Gabriel-Pospisil F, Naciri M, Camerlynck P. Genotyping human and bovine isolates of Cryptosporidium parvum by polymerase chain reaction-restriction fragment length polymorphism analysis of a repetitive DNA sequence. FEMS Microbiol Lett. 1996;137:207–211. doi: 10.1111/j.1574-6968.1996.tb08107.x. [DOI] [PubMed] [Google Scholar]
- 3.Carraway M, Tzipori S, Widmer G. Identification of genetic heterogeneity in the Cryptosporidium parvum ribosomal repeat. Appl Environ Microbiol. 1996;62:712–716. doi: 10.1128/aem.62.2.712-716.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chalmers R M, Sturdee A P, Bull S A, Miller A, Wright S E. The prevalence of Cryptosporidium parvum and C. muris in Mus domesticus, Apodemus sylvaticus and Clethrionomys glareolus in an agricultural system. Parasitol Res. 1997;83:478–482. doi: 10.1007/s004360050283. [DOI] [PubMed] [Google Scholar]
- 5.Holekamp K E. Dispersal in ground-dwelling sciurids. In: Murie J O, Michener G R, editors. The biology of ground-dwelling squirrels. Lincoln: University of Nebraska Press; 1984. pp. 297–320. [Google Scholar]
- 6.Holekamp K E, Nunes S. Seasonal variation in body weight, fat, and behavior of California ground squirrels (Spermophilus beecheyi) Can J Zool. 1989;67:1425–1433. [Google Scholar]
- 7.Mager A L, Standridge J, Kluender S M, Peterson L L. Source Water Protection Symposium: a focus on waterborne pathogens. San Francisco, Calif: American Water Works Association; 1998. Source and occurrence of pathogens in watersheds. [Google Scholar]
- 8.Morgan U M, Sargent K D, Deplazes P, Forbes D A, Spano F, Hertberg H, Elliot A, Thompson R C A. Molecular characterization of Cryptosporidium from various hosts. Parasitology. 1998;117:31–37. doi: 10.1017/s0031182098002765. [DOI] [PubMed] [Google Scholar]
- 9.Muller H M, Ranuci L, Pozio E, Crisanti A. A method for collecting large quantities of Cryptosporidium parvum. Parasitol Today. 1993;9:261–263. doi: 10.1016/0169-4758(93)90072-n. [DOI] [PubMed] [Google Scholar]
- 10.Owings D H, Borchert M, Virginia R. The behaviour of California ground squirrels. Anim Behav. 1977;25:221–230. [Google Scholar]
- 11.Peng M M, Xiao L, Freeman A R, Arrowood M J, Escalante A A, Weltman A C, Ong C S L, MacKenzie W R, Lal A A, Beard C B. Genetic polymorphism among Cryptosporidium isolates: evidence of two distinct human transmission cycles. Emerg Infect Dis. 1997;3:567–572. doi: 10.3201/eid0304.970423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pereira M D G C, Atwill E R, Jones T. Comparison of sensitivity of immunofluorescent microscopy to that of a combination of immunofluorescent microscopy and immunomagnetic separation for detection of Cryptosporidium parvum oocysts in adult bovine feces. Appl Environ Microbiol. 1999;65:3236–3239. doi: 10.1128/aem.65.7.3236-3239.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rochelle P A, Jutras E M, Atwill E R, DeLeon R, Stewart M H. Polymorphisms in the β-tubulin gene of Cryptosporidium parvum differentiate between isolates based on animal host but not geographic origin. J Parasitol. 1999;85:986–989. [PubMed] [Google Scholar]
- 14.Schitoskey F, Jr, Woodmansee S R. Energy requirements and diet of the California ground squirrel. J Wildl Manage. 1978;42:373–382. [Google Scholar]
- 15.Shults L M, Stanton N L. Helminth parasites of the Wyoming ground squirrel, Spermophilus elegans elegans Kennicott, 1863. Great Basin Nat. 1987;47:103–104. [Google Scholar]
- 16.Shults L M, Seville R S, Stanton N L, Menkens G E., Jr Eimeria sp. (Apicomplexa: eimeriidae) from Wyoming ground squirrels (Spermophilus elegans) and white tailed prairie dogs (Cynomys leucurus) in Wyoming. Great Basin Nat. 1990;50:327–331. [Google Scholar]
- 17.Spano F, Putignani L, McLauchling J, Casemore D P, Crisanti A. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrari and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol Lett. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- 18.Stanton N L, Shults L M, Parker M, Seville R S. Coccidian assemblages in the Wyoming ground squirrel, Spermophilus elegans elegans. J Parasitol. 1992;78:323–328. [PubMed] [Google Scholar]
- 19.Sturdee A P, Chalmers R M, Bull S A. Detection of Cryptosporidium oocysts in wild mammals of mainland Britain. Vet Parasitol. 1999;80:273–280. doi: 10.1016/s0304-4017(98)00226-x. [DOI] [PubMed] [Google Scholar]
- 20.Van Vuren D, Kuenzi A J, Loredo I, Leider A L, Morrison M L. Translocation as a nonlethal alternative for managing California ground squirrels. J Wildl Manage. 1997;61:351–359. [Google Scholar]
- 21.Weber R, Bryan T B, Juranek D D. Improved stool concentration procedure for detection of Cryptosporidium oocysts in fecal specimens. J Clin Microbiol. 1992;30:2869–2873. doi: 10.1128/jcm.30.11.2869-2873.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webster J P, MacDonald D W. Cryptosporidiosis reservoir in wild brown rats (Rattus norvegicus) in the UK. Epidemiol Infect. 1995;115:207–209. doi: 10.1017/s0950268800058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilber P G, Hanelt B, Van Horne B, Duszynski D W. Two new species and temporal changes in the prevalence of eimerians in a free-living population of Towsend's ground squirrels (Spermophilus towsendii) in Idaho. J Parasitol. 1994;80:251–259. [PubMed] [Google Scholar]
- 24.Xiao L, Escalante L, Yang C, Sulaiman I M, Escalante A A, Montali R J, Fayer R, Lal A A. Phylogenetic analysis of Cryptosporidium parasites based on the small subunit rRNA gene locus. Appl Environ Microbiol. 1999;65:1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao L, Morgan U M, Limor J, Escalante A, Arrowood M, Shulaw W, Thompson R C A, Fayer R, Lal A A. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl Environ Microbiol. 1999;65:3386–3391. doi: 10.1128/aem.65.8.3386-3391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao L, Limor J, Morgan U M, Sulaiman I M, Thomspon R C A, Lal A A. Sequence differences in the diagnostic target region of the oocyst wall protein gene of Cryptosporidium parasites. Appl Environ Microbiol. 2000;66:5499–5502. doi: 10.1128/aem.66.12.5499-5502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]