Abstract
For diagnosis of citrus bacterial canker by PCR, an internal standard is employed to ensure the quality of the DNA extraction and that proper requisites exist for the amplification reaction. The ratio of PCR products from the internal standard and bacterial target is used to estimate the initial bacterial concentration in citrus tissues with lesions.
Citrus bacterial canker (CBC), caused by Xanthomonas axonopodis pv. citri, is a serious disease of most citrus species and cultivars in many citrus-producing areas worldwide (1). In Florida, CBC is currently under eradication because this disease threatens to reduce fruit quality, to cause premature leaf and fruit abscission, and to restrict foreign and domestic markets by regulatory measures that prevent disease spread (1, 13). When CBC-like symptoms are detected in a new location, the disease is confirmed by the isolation of the putative X. axonopodis pv. citri population from the lesion and inoculation of leaves of a grapefruit seedling to reproduce the symptoms. Although CBC causes characteristic leaf and fruit symptoms, rapid, specific, and reliable methods for pathogen identification are critical for disease diagnosis in quarantine programs.
PCR is a principal tool for plant disease diagnosis (9). However, routine application of PCR for the detection of pathogens in plant tissue is restricted by the presence of plant factors that inhibit the amplification of nucleic acids (16). This condition can result in a false-negative diagnosis when the pathogen is actually present. Another limitation of the traditional PCR methods is their inability to quantify the initial amount of target sequence present in the tissue. Use of competitive PCR avoids these two obstacles through simultaneous coamplification of an internal standard with the specific target sequence in one reaction (17).
Internal standard construction.
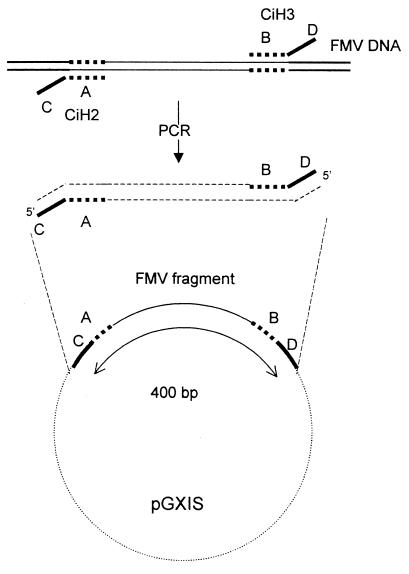
An internal standard plasmid was constructed by following an approach similar to that described for Agrobacterium tumefaciens (11). Primers CiH2 and CiH3 (Fig. 1) containing 5′ termini identical to those of primers designed to amplify a fragment inside a plasmid in X. axonopodis pv. citri (7) and 3′ termini homologous to a sequence from Figwort mosaic virus (FMV) were used for PCR using FMV DNA as a target (Fig. 2). PCR was carried out with a 50-μl volume containing 1× Taq buffer, 3 mM MgCl2, 0.1 μM each primer, 0.2 mM each deoxynucleoside triphosphate, and 1 U of Taq polymerase (Promega, Madison, Wis.). The amplified product of 400 bp was cloned into the pGEM-T vector (Promega) to create plasmid pGXIS, and competent Escherichia coli (strain JM109) cells were transformed. Adjusted concentrations of purified pGXIS were added and used as internal standards for PCRs. To optimize the concentration of plasmid pGXIS in the reactions, without creating an appreciable decrease in the sensitivity of the PCR assay for detection of the bacterium, a titration of the internal standard was performed. An extract from an asymptomatic leaf or fruit from a mature grapefruit tree was amended with a suspension of X. axonopodis pv. citri to achieve bacterial concentrations ranging from 0 to 108 CFU/ml. DNA was extracted using a protocol previously described (2, 10) with an improvement of the DNA precipitation step by the use of Pellet Paint coprecipitant (Novagen, Darmstadt, Germany) (14). Amplifications were carried out with 25-μl volumes containing 1× Taq buffer, 3 mM MgCl2, 0.2 μM concentrations of primers 2 and 3 (7), 0.2 mM each deoxynucleoside triphosphate, and 1 U of Taq polymerase (Promega) with a profile of 93°C (30 s), 58°C (30 s), and 72°C (45 s) for 40 cycles plus an initial step of 94°C for 5 min and a final step of 72°C for 10 min. PCR products were visualized in agarose gels. Reactions were performed in the presence of pGXIS concentrations of 0.015, 0.15, 1.5, and 15 pg/μl. Serial dilutions of the plant extracts amended with X. axonopodis pv. citri were also plated on kasugamycin-cephalexin-Bravo-amended medium (4). After 72 h, colonies that appeared were counted to check initial concentrations of X. axonopodis pv. citri in the suspensions.
FIG. 1.
Sequences 2 and 3 are primers designed to amplify a fragment of the X. axonopodis pv. citri plasmid (7). CiH2 and CiH3 are the primers designed for internal standard construction. Fragments underlined are those corresponding to FMV DNA.
FIG. 2.
pGXIS construction. A and B are sequences in FMV DNA that were incorporated into the 3′ termini of primers CiH2 and CiH3; C and D are the sequences from primers 2 and 3 that specifically amplify a fragment in the X. axonopodis pv. citri plasmid (7) and are placed at the 5′ termini in primers CiH2 and CiH3. CiH2 (C plus A) and CiH3 (D plus B) were used to amplify the fragments from FMV DNA. The PCR product (400 bp) was cloned into the pGEM-T vector to result in pGXIS. In quantitative PCR, primers 2 (C) and 3 (D) were used for amplifications.
Amplification from the internal standard resulted in a 400-bp DNA fragment, whereas amplification from target X. axonopodis pv. citri yielded a 222-bp fragment. The two fragments were easily differentiated in agarose gels. pGXIS at a concentration of 0.015 pg/μl provided a sensitivity for detection of X. axonopodis pv. citri in plant material comparable to that obtained without the addition of this plasmid in the PCR. At 0.015 pg of pGXIS per μl, X. axonopodis pv. citri was detected when the initial concentration was 102 CFU/ml, whereas at 0.15 and 1.5 pg of pGXIS per μl, detection was possible only when the initial concentration was above 104 and 105 CFU/ml, respectively. At concentrations of bacteria greater than 109 CFU/ml, only the 222-bp fragment from the target sequence in X. axonopodis pv. citri was amplified, whereas at concentrations of bacteria below 102 CFU/ml, only the 400-bp fragment from the internal standard was detected. The sensitivity of the PCR with pGXIS is similar to that previously described for this bacterium using the same primers before the addition of an internal standard (7) and is adequate for detection of endophytic and epiphytic populations of X. axonopodis pv. citri from diseased plants (3, 5, 6, 12). Moreover, this PCR assay is sufficiently sensitive to detect bacteria in initial ages of infection before visible symptoms develop or to detect epiphytic populations in asymptomatic trees (1, 15). Although the sensitivity (102 CFU/ml) of our quantitative method is slightly lower than that described by other authors using nested PCR (8), our method does not require a second round of PCR. A one-step PCR minimizes false-positive results and facilitates the analysis for quantification purposes.
Quantification of PCR products and constructions of calibration curves.
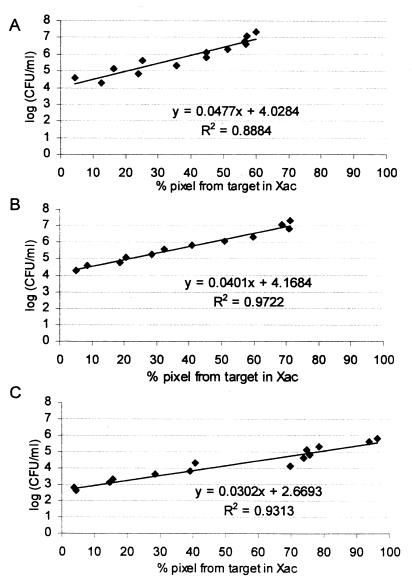
Images from visualized PCR products were recorded by a video camera and were processed with an Alphaimager (Alpha Innotech Corporation, San Leandro, Calif.). Their densities were quantified as average pixel densities. Peaks from an internal standard and target sequences were compared, and their density ratios were used to estimate initial bacterial concentrations relative to the calibration curves. These calibrations were established by comparing the densities of PCR products obtained from serial dilutions of known bacterial concentrations in plant material and known concentrations of pGXIS. The density ratio was defined as the ratio of amplified product generated from the target sequence (222 bp) in X. axonopodis pv. citri to the product of the internal standard (400 bp). This ratio was calculated for each concentration of pGXIS (0.015, 0.15, and 1.5 pg/μl) employed in the PCR. Linear-regression equations were derived for each concentration of pGXIS to relate the initial bacterial concentration (log CFU per milliliter) to the density ratio. For each concentration of pGXIS, at least 12 measurements were taken from at least two different DNA extractions and PCRs (Fig. 3).
FIG. 3.
Linear regressions for calibration of PCR products generated from the target sequence in serial dilutions of X. axonopodis pv. citri (Xac) in grapefruit leaves and from pGXIS at concentrations of 1.5 (A), 0.15 (B), and 0.015 (C) pg/μl. Each point represents a measure of the percentage of the pixel density from the PCR product obtained from the target sequence (222 bp) relative to the total pixel density generated from the target and pGXIS (400 bp) PCR products. Also shown are equations that resulted from a linear-regression analysis.
Based on the regression curve derived from an internal standard at 0.015 pg/μl, bacterial DNA concentration was related to bacterial populations ranging from 103 to 106 CFU/ml. At 0.15 pg of pGXIS per μl, quantification of X. axonopodis pv. citri was possible from 104 to 108 CFU/ml, and at 1.5 pg of pGXIS per μl, quantification was possible from 104 to almost 109 CFU/ml. Bacteria in extracts from fruit gave results similar to those obtained from leaf extracts (data not shown).
Quantification of X. axonopodis pv. citri in naturally infected samples.
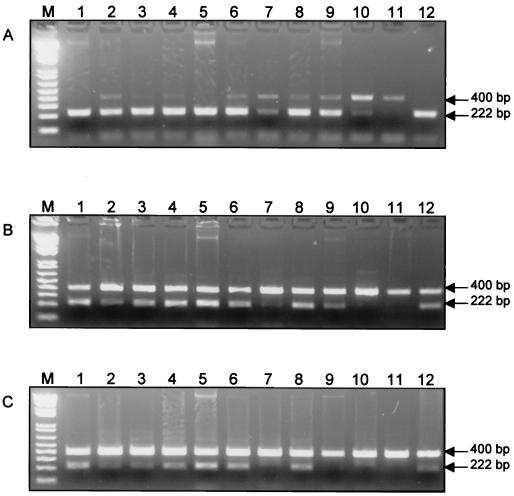
DNA extraction from 17 samples of lesions suspected to be caused by X. axonopodis pv. citri in grapefruit, Mexican lime, lemon, Persian lime, and sweet orange tree leaves and grapefruit and lemon fruits (Table 1) were examined in a competitive PCR utilizing pGXIS at concentrations of 0.015, 0.15, and 1.5 pg/μl. Coamplification of pGXIS and DNA from X. axonopodis pv. citri was obtained from lesions of CBCs on naturally infected leaves and fruits of different citrus species using three concentrations of the internal standard (Fig. 4). All samples contained bacterial concentrations ranging from 104 to 106 CFU/ml, and results required the use of only one pGXIS concentration and one standard curve (Table 1). In many cases, averages and standard deviations of bacterial concentrations were derived from two or three pGXIS standard curves. In only one of the positive samples (XI00-00080) was the quantitative-PCR method not sensitive enough to accurately quantify a low bacterial population. Although the target sequence for X. axonopodis pv. citri was detected in this sample, the 222-bp band at the lowest pGXIS concentration was too faint to accurately estimate the population. In another sample (XI00-00075), the conditions for PCR were sufficient and the band from pGXIS was obtained but bacterial detection was not possible because the concentration of bacteria was below the detection limit of 102 CFU/ml.
TABLE 1.
Quantification of X. axonopodis pv. citri in lesions from naturally infected plants by PCR using pGXIS as the competitive element
| Sample | Speciesa | Sample type | % of pixels that were from the target sequenceb with a pGXIS concn (pg/μl) of:
|
Avg from different pGXIS concns (log10 CFU/ml)c | ||
|---|---|---|---|---|---|---|
| 1.5 | 0.15 | 0.015 | ||||
| S1a | Grapefruit | Leaf | 34.2 | 46.4 | 100 | 5.8 ± 0.2 |
| S1b | Grapefruit | Leaf | 0 | 13.6 | 89.1 | 5.0 ± 0.3 |
| 5W | Lemon | Fruit | 10.9 | 33.3 | 100 | 4.9 ± 0.6 |
| 5B | Lemon | Fruit | 22.1 | 45.2 | 96.7 | 5.5 ± 0.4 |
| S2W | Grapefruit | Leaf | 39.1 | 49.9 | 100 | 6.0 ± 0.2 |
| S3B | Grapefruit | Leaf | 28.1 | 41.4 | 90.8 | 5.5 ± 0.2 |
| 8W | Grapefruit | Leaf | 0 | 0 | 51.1 | 4.3 |
| 7B | Lemon | Leaf | 25.6 | 36.7 | 93.0 | 5.4 ± 0.2 |
| X00-00048 | Persian lime | Leaf | 0 | 12.8 | 85.8 | 5.0 ± 0.3 |
| XI00-00080 | Grapefruit | Fruit | 0 | 0 | 0 | <2.7 |
| XI00-00075 | Grapefruit | Fruit | 0 | 0 | 0 | <2.7 |
| X00-12906 | Mexican lime | Leaf | 11.3 | 33.0 | 100 | 4.9 ± 0.6 |
| XI00-143a | Sweet orange | Leaf | 0 | 0 | 63.6 | 4.6 |
| XI00-143b | Sweet orange | Leaf | 0 | 0 | 70.1 | 4.8 |
| X00-13042 | Mexican | Leaf | 38.4 | 52.5 | 100 | 6.1 ± 0.2 |
| Lime | ||||||
| X00-13043 | Grapefruit | Leaf | 34.5 | 37.1 | 100 | 5.6 ± 0.1 |
| X00-13044 | Lemon | Leaf | 0 | 13.4 | 100 | 4.7 |
Grapefruit, Citrus paradisi (Macf.); lemon, C. limon (L) Burm. f.; Persian lime, C. latifolia Tan.; Mexican lime, C. aurantifolia (Christm.) Swing; sweet orange, C. sinensis (L.) Osbeck.
Percentage of the pixels from PCR product obtained from the target sequence (222 bp) relative to the total number of pixels generated from target and pGXIS (400-bp) PCR products using three concentrations of pGXIS in PCRs.
Arithmetic mean and standard deviation obtained with the three bacterial concentrations calculated from the equation that resulted from each pGXIS concentration.
FIG. 4.
PCR products generated after coamplification from the target sequences in X. axonopodis pv. citri (222 bp) and pGXIS (400 bp) at different concentrations: 0.015 pg/μl (A), 0.15 pg/μl (B), and 1.5 pg/μl (C). Lane M, 1-kb Plus DNA ladder (Life Technologies); lanes 1 (S1a), 2 (S1b), 5 (S2W), 6 (S3B), and 7 (8W), lesions on leaves of a grapefruit tree; lanes 3 (5W) and 4 (5B), lesions on fruits of a lemon tree; lane 8 (7B), lesions on a leaf of a lemon tree; lane 9 (X00-00048), lesions on a leaf of a Persian lime tree; lanes 10 (XI00-00080) and 11 (XI00-00075), lesions on a fruit of a grapefruit tree; and lane 12 (X00-13044), lesion on a leaf of a Mexican lime tree.
The addition of the internal control in PCRs confirms the CBC diagnosis by ensuring the attainment of at least one PCR product, allows the quantification of bacterial concentration in plant samples, and thereby aids in the study of the pathogen in quarantine situations.
Acknowledgments
We thank X. Sun and M. Peacock from the Florida Department of Agriculture and Consumer Services and the Division of Plant Industry and T. Riley from the USDA for kindly providing some of the naturally infected samples analyzed. We also thank W. O. Dawson and his team for their cooperation, especially S. Gowda for providing FMV DNA, and M. A. Ayllón, L. W. Timmer, and K. R. Chung for critically reading the manuscript. We are also grateful to Diana Drouillard for her assistance.
J. Cubero is a recipient of a postdoctoral fellowship from the Spanish Ministerio de Educación y Ciencia. Research is funded by USDA-APHIS (99-8100-0560-CA) and the Florida Citrus Production Research Advisory Council (981-29).
Footnotes
Florida Agricultural Experiment Station Journal Series no. R-07912.
REFERENCES
- 1.Civerolo E. Bacterial canker disease of citrus. J Rio Grande Vall Hortic Soc. 1984;37:127–145. [Google Scholar]
- 2.Cubero J, Martínez M C, Llop P, López M M. A simple and efficient PCR method for the detection of Agrobacterium tumefaciens in plant tumours. J Appl Microbiol. 1999;86:591–602. doi: 10.1046/j.1365-2672.1999.00700.x. [DOI] [PubMed] [Google Scholar]
- 3.Egel D S, Graham J H, Riley T D. Population dynamics of strains of Xanthomonas campestris differing in aggressiveness on Swingle citrumelo and grapefruit. Phytopathology. 1991;81:666–671. [Google Scholar]
- 4.Graham J H, Gottwald T R. Variation in aggressiveness of Xanthomonas campestris pv. citrumelo associated with citrus bacterial spot in Florida citrus nurseries. Phytopathology. 1990;80:190–196. [Google Scholar]
- 5.Graham J H, McGuire R G, Miller J W. Survival of Xanthomonas campestris pv. citri in citrus plant debris and soil in Florida and Argentina. Plant Dis. 1987;71:1094–1098. [Google Scholar]
- 6.Graham J H, Gottwald T R, Fardelmann D. Cultivar-specific interactions for strains of Xanthomonas campestris from Florida that cause citrus canker and citrus bacterial spot. Plant Dis. 1990;74:753–756. [Google Scholar]
- 7.Hartung J S, Daniel J F, Pruvost O P. Detection of Xanthomonas campestris pv. citri by the polymerase chain reaction. Appl Environ Microbiol. 1993;59:1143–1148. doi: 10.1128/aem.59.4.1143-1148.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartung J S, Pruvost O P, Villemot I, Alvarez A. Rapid and sensitive colorimetric detection of Xanthomonas axonopodis pv. citri by immunocapture and nested-polymerase chain reaction. Phytopathology. 1996;86:95–101. [Google Scholar]
- 9.Henson J, French R. The polymerase chain reaction and plant disease diagnosis. Annu Rev Phytopathol. 1993;31:81–109. doi: 10.1146/annurev.py.31.090193.000501. [DOI] [PubMed] [Google Scholar]
- 10.Llop P, Caruso P, Cubero J, Morente C, López M M. A. simple extraction procedure for efficient routine detection of pathogenic bacteria in plant material by polymerase chain reaction. J Microbiol Methods. 1999;37:23–31. doi: 10.1016/s0167-7012(99)00033-0. [DOI] [PubMed] [Google Scholar]
- 11.Sachadyn P, Kur J. The construction and use of a PCR internal control. Mol Cell Probes. 1998;12:259–262. doi: 10.1006/mcpr.1998.0170. [DOI] [PubMed] [Google Scholar]
- 12.Stall R E, Miller J W, Marco G M, De Echenique B I C. Population dynamics of Xanthomonas citri causing cancrosis of citrus in Argentina. Proc Fla State Hortic Soc. 1980;93:10–14. [Google Scholar]
- 13.Stall R E, Seymour C P. Canker, a threat to the Gulf Coast states. Plant Dis. 1983;67:581–585. [Google Scholar]
- 14.Taggart E M, Bryngton C L, Hillyard D R, Robinson J E, Carrol K C. Enhancement of the AMPLICOR Enterovirus PCR test with a coprecipitant. J Clin Microbiol. 1999;36:3408–3409. doi: 10.1128/jcm.36.11.3408-3409.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Timmer L W, Zitlko S E, Gottwald T R. Population dynamics of Xanthomonas campestris pv. citri on symptomatic and asymptomatic citrus leaves under various environmental conditions. Proc Int Soc Citricult. 1996;1:448–451. [Google Scholar]
- 16.Wilson I G. Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol. 1997;63:3741–3751. doi: 10.1128/aem.63.10.3741-3751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmerman K, Mannhalter J W. Technical aspects of quantitative competitive PCR. BioTechniques. 1996;21:268–279. doi: 10.2144/96212rv01. [DOI] [PubMed] [Google Scholar]