Abstract
3D bioprinting is an emerging additive manufacturing technique to fabricate constructs for human disease modeling. However, current cell-laden bioinks lack sufficient biocompatibility, printability, and structural stability needed to translate this technology to preclinical and clinical trials. Here, a new class of nanoengineered hydrogel-based cell-laden bioinks is introduced, that can be printed into 3D, anatomically accurate, multicellular blood vessels to recapitulate both the physical and chemical microenvironments of native human vasculature. A remarkably unique characteristic of this bioink is that regardless of cell density, it demonstrates a high printability and ability to protect encapsulated cells against high shear forces in the bioprinting process. 3D bioprinted cells maintain a healthy phenotype and remain viable for nearly one-month post-fabrication. Leveraging these properties, the nanoengineered bioink is printed into 3D cylindrical blood vessels, consisting of living co-culture of endothelial cells and vascular smooth muscle cells, providing the opportunity to model vascular function and pathophysiology. Upon cytokine stimulation and blood perfusion, this 3D bioprinted vessel is able to recapitulate thromboinflammatory responses observed only in advanced in vitro preclinical models or in vivo. Therefore, this 3D bioprinted vessel provides a potential tool to understand vascular disease pathophysiology and assess therapeutics, toxins, or other chemicals.
Keywords: cell-laden bioink, disease models, regenerative medicine, 3D bioprinting, vascular tissue
1. Introduction
3D bioprinting, a subclass of fused deposition modeling or extrusion printing, is capable of producing heterogeneous, tissue-shaped constructs in a layer-by-layer fashion with embedded cells, demonstrating enormous promise to recapitulate the native, multi-cellular vascular anatomy.[1–3] Some of the earlier pioneering work demonstrated that 3D bioprinting can be used to engineer small vessels[4,5] that can be a perfused via multiple approaches[6–8] and can be used for various biomedical applications including scaffold engraftment and 3D tissue models for evaluating drugs efficacy.[9] In past few years, a range of hydrogel bioinks[10] are introduced to design these complex 3D bioprinted vascular structures, however, there is currently a limitation in available bioinks (cell-laden hydrogel ink) that are able to mimic the vascular composition and architectures of native tissues. This is mostly attributed to the lack of a bioink with high printability, modular mechanical properties, and ability to localize and deposit a high-density of living cells into complex 3D architectures. Specifically, hydrogels for direct extrusion are not mechanically resilient for high-fidelity 3D bioprinting.[10,11] For example, uncrosslinked hydrogels (precursor solutions) are often too fluidic for high-fidelity extrusion of anatomically accurate structures, while crosslinked hydrogels are too brittle to yield through a needle without fracturing. In addition, current bioinks are not able to maintain construct integrity and fidelity while sustaining a high cell viability and phenotypic maintenance. Taken together, limitations of current bioinks restrict the potential to fabricate anatomically accurate, multilayered tissues with the innate ability to replicate human pathophysiological function.
Here, we address past limitations by introducing a nanoengineered bioink that is bioprinted into 3D models of blood vessels. This approach offers improved spatio-temporal resolution on a macro-structure and tissue-level micro-structure, achieving what is currently not possible with currently available bioinks.[12] Specifically, we demonstrated that endothelial cells (EC) and vascular smooth muscle cells (VSMC) can be co-cultured into a cylindrical vessel utilizing 3D bioprinting technology (Figure 1). To achieve this, we developed a high viscosity colloidal bioink, termed nanoengineered extracellular matrix (nECM), composed of gelatin methacryloyl (GelMA), poly(ethylene glycol) diacrylate (PEGDA), and two dimensions (2D) nanosilicates (nSi, also known as nanoclay[13]). This GelMA-PEGDA-nSi composite allows for cellular interactions and tunable mechanical properties (PEGDA molecular weight and concentration) while still maintaining a stabilized network for extrusion (nanosilicates reinforcement). Specifically, this optimized nECM bioink is reinforced by clay–clay and polymer–clay interactions, permitting extrusion of high-fidelity scaffolds that can be subsequently crosslinked via UV light. The designed nECM bioink enables for the fabrication of free-standing vessels of varying lumen sizes to accurately reflect upon diverse human vessel geometries.[14–16] Subsequently, we show that this nECM bioink can be used to engineer a 3D vascular model and predict human thromboinflammation upon cytokine insult. This data lays the foundation for this 3D model to be applied in preclinical settings of vascular medicine, including drug discovery, and analysis of chemicals and toxins.
Figure 1.
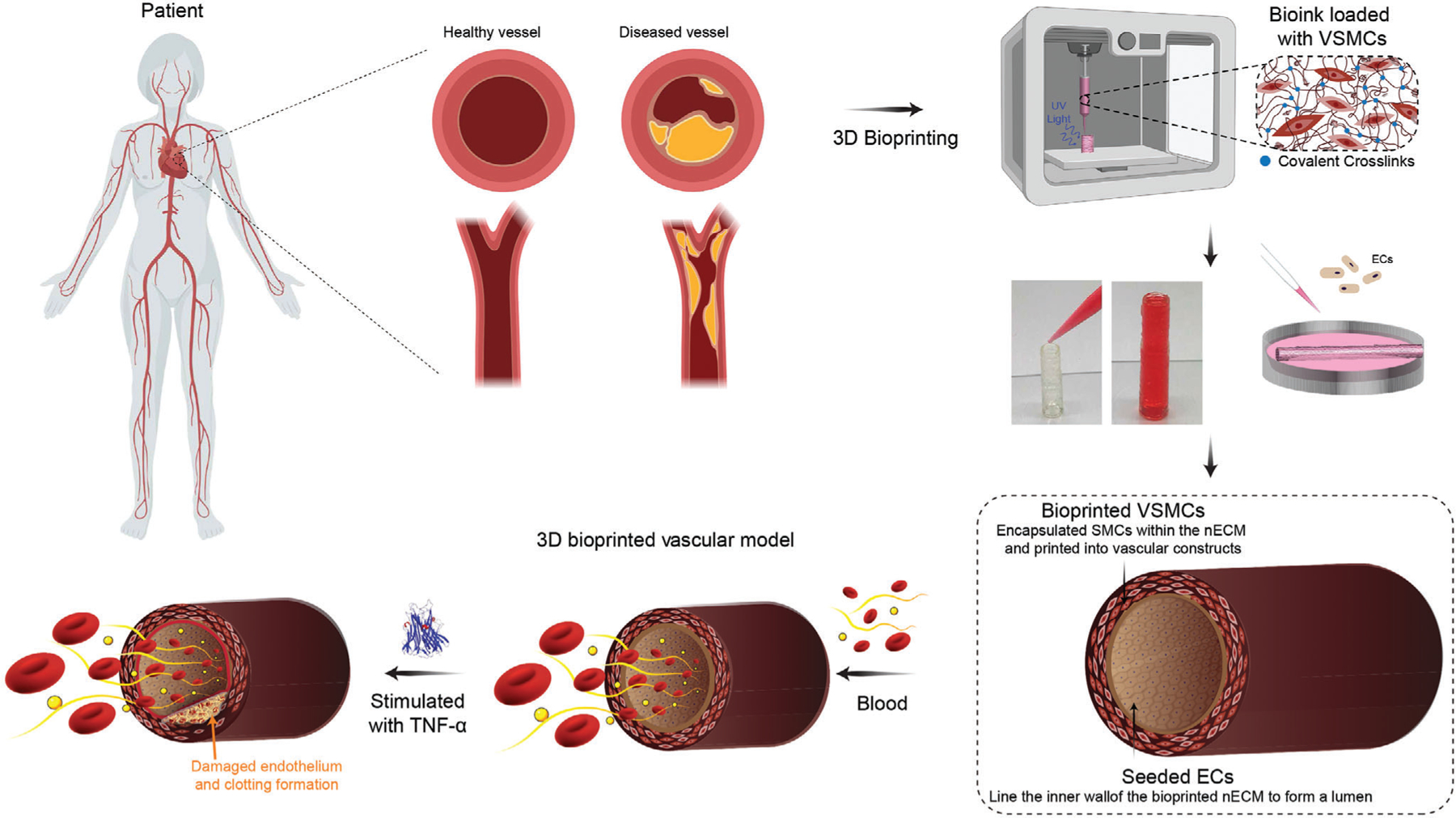
Fabrication of 3D vascular model. 3D bioprinting can be used to replicate structure and pathophysiological function of a healthy and diseased blood vessel. nECM bioinks are developed for 3D bioprinting of vascular constructs by first printing VSMCs and then subsequently seeding with ECs to form a confluent monolayer. This fabrication technique permits the construction of a 3D bioprinted vascular model and can recapitulate responses of vascular activation.
Results and Discussion
2.1. Designing Bioink with High Printability and Print Fidelity
Bioinks for 3D bioprinting must flow through a needle (printability) and maintain a deposited shape (print fidelity) to fabricate constructs with complex geometries. Shear-thinning biomaterials, also referred as pseudoplastic or thixotropic, demonstrate high viscosities at low-shear rates that linearly decreases with increasing shear rates.[17–19] This behavior is vital for 3D bio-printing, as it depicts the ability of a bioink to be extruded through a printing gauge (low-viscosity at high shear), but maintain its deposited shape on the printer bed (high-viscosity at low-shear). The incorporation of nanosilicates to polymeric precursor solutions, such as GelMA-PEGDA, serves as a rheological modifier due to its ability to form a physically crosslinked shear-thinning network (Figures S1 and S2, Supporting Information). Nanosilicates (Na+0.7[(Si8Mg55Li0.3)O20(OH)4]−0.7) are a 2D, trioctahedral, smectite clay nanoparticles (20–50 nm diameter; 1–2 nm thickness).[20–23] These nanosilicateshave a negative surface charge on face due to the presence of oxygen and a positive edge charge due to cleaved Mg2+ and Li2+ in aqueous environments.[20–23] This anisotropic charge distribution facilitates interactions between the edge of one nanosilicates with the face of another, forming a “house-of-cards” structure.[24,25] Aside from particle-particle interactions, nanosilicates-polymer interactions also occur through hydrogen bonding, ionic interactions, and electrostatic interactions.[21,22,26] For example, charged groups present on polymer backbones (GelMA) may interact with the opposite surface/edge charge of the nanosilicates via electrostatic interactions.[27,28] Additionally, the hydrogen from the ethylene carbons of PEGDA chains have also been shown to absorb and desorb onto the nanosilicates surface.[20,29–32] Thus, the unique interactions between nanosilicates, GelMA-nanosilicates, and PEGDA-nanosilicates allows this nano-colloidal network to exhibit high printability for 3D bioprinting applications (Figure S3, Supporting Information).
During 3D bioprinting, a bioink must first shear-thin (transition from a high viscous gel to a low viscosity fluid), and subsequently rapidly recover its internal structure to retain the integrity of printed structure. The rate of structural recovery is essential to limit lateral spreading or pooling onto the printer bed. To assess for recoverability, we have designed a peak-hold analysis to monitor the change in viscosity of bioink as it transitions through different stages in the printing process (Figure 2A and Figure S4, Supporting Information). To do this, constants defined from the Power–Law model were applied as an input within the Rabinowitsch equation (Equation (1)), where Q is the volumetric flow rate, R is the radius the material is confined to (i.e., extruder, needle gauge, printer bed), and n is the flow behavior index (defined in Equation (2)). Bioinks were first exposed to a low-shear rate (3.02 s−1) to quantify the preliminary or pre-shear viscosity, resembling the shear within the extruder. After one minute, samples were exposed to high-shear rates (23,000 s−1) for 10 s, mimicking those in the needle during extrusion, followed by a low-shear rate (0.01 s−1) for 2 min, representing the shear experienced on the printer-bed after deposition. The results support the Newtonian nature of GelMA and PEGDA, limiting their use for 3D printing. However, the shear-thinning behavior exhibited upon the addition of nanosilicates to GelMA-PEGDA composite permits for rapid rebuilding of the internal structure of bioink, permitting for shape retention of the extrudate on the printing bed.
Figure 2.
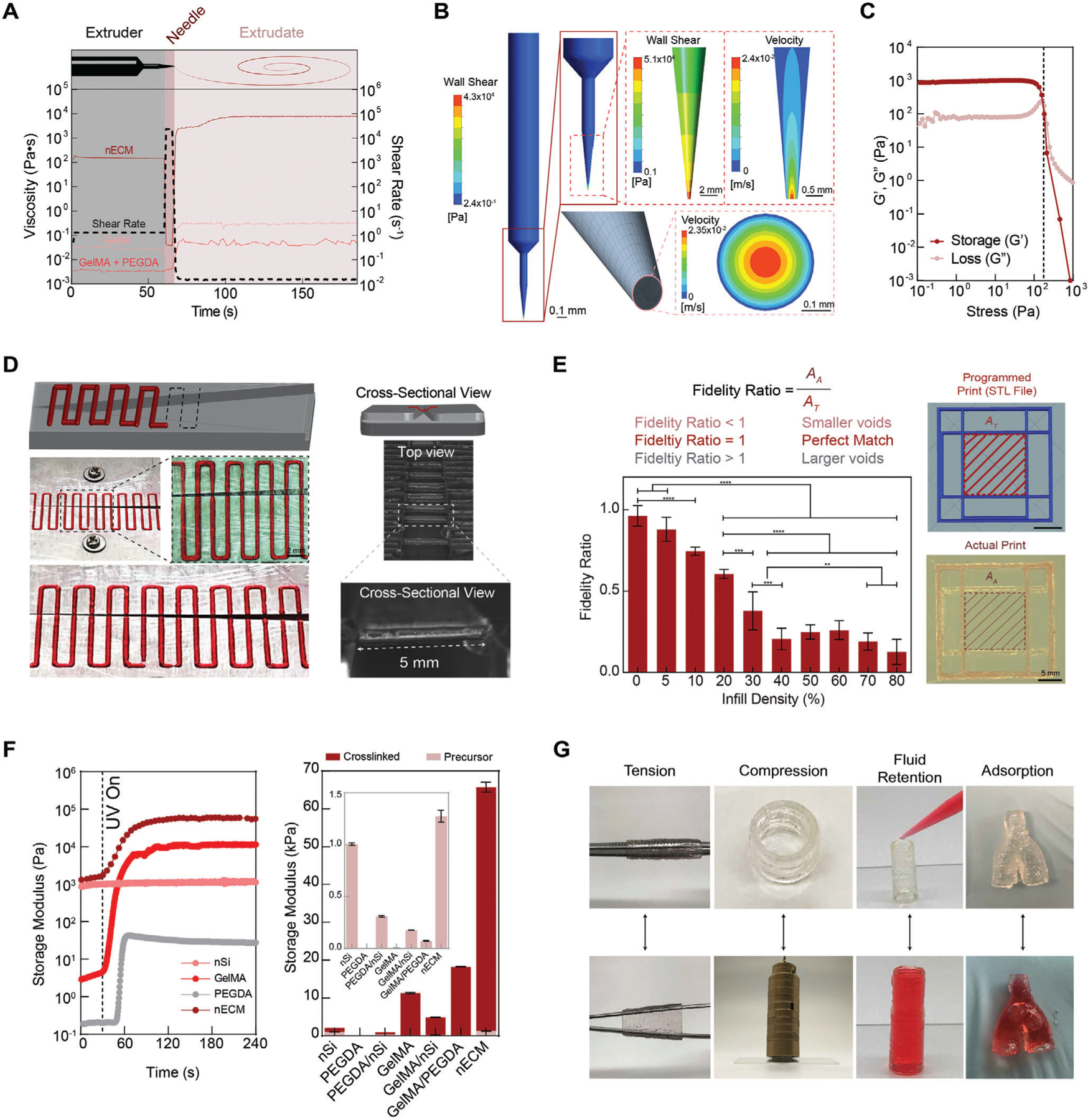
Rheological and biophysical characterization of nECM bioink. A) Utilizing the flow-behavior index and consistency index, shear-rates are defined for specific locations within the printing process, represented by the dotted line corresponding to the right y-axis. When applied to GelMA and GelMA/PEGDA, the viscosity (left y-axis) remains below 1 Pa·s. However, when applied to the nECM, the bioink remains viscous when in the extruder, while rapid decreases when flowing through the needle gauge, permitting for extrusion. Post-extrusion, the extrudate rapidly recovers a high viscosity, permitting for shape-retention when deposited onto the printing bed. B) Rheological modeling of the nECM behavior throughout the printing process demonstrates a plug like flow, with the wall shear stress highest at the walls. C) A stress sweep validates the rheological modeling presented in B. D) The nECM is able to bridge a gap, up to a length of 5 mm, without any supporting structures, demonstrating the bioinks ability to support itself. E) Fidelity ratio is dependent on the infill density of printed lattice networks. Vascular assemblies, containing no infill density due to their hollow nature, present a fidelity ratio ≈1 (ideal match) and thus supports enhanced reliability of the optimized nECM formulation to construct programmed scaffolds. F) Time-sweep with exposure to UV light after 30 s demonstrates the addition of nanosilicates does not impede on covalent crosslinking of polymer precursor solutions post-printing and significantly increases the storage of the storage modulus compared to the bioinks constituents. G) Manipulations of printed structures after curing via tension, compression, fluid retention, and adsorption validate the mechanical integrity of the printed nECM constructs. Multiple replicates of samples are performed for rigor (n = 5) and statistics analysis was performed using one-way ANOVA with posthoc Turkey analysis (* = p < 0.05, ** = p < 0.05, *** = p < 0.005, and **** = p < 0.0001).
| (1) |
Taken cohesively, shear-thinning and recoverable properties exhibited by the nECM bioink is hypothesized to shield encapsulated cells from damaging shear stresses during extrusion.[33–35] Utilizing the Power–Law model constants, ANSYS modeling was used to predict the wall shear stress within different regions of the extruder (barrel, connector, and needle gauge) (Figure 2B). The velocity profile at the printing gauge exhibits a plug-like flow, demonstrating material extrusion first from the center of the gauge and subsequently outward. It is hypothesized that this velocity profile provides protection to encapsulated cells, shielding them from damaging shear stresses. To validate these results, stress-sweeps were performed to calculate the stress required to cause yielding of the material through the extruder (Figure 2C). The results obtained confirm that the ANSYS model recapitulates the stresses exposed to the nECM during the printing process. Specifically, the stresses exhibited in the ANSYS model (4208 Pa) exceeded those measured in the stress sweeps (178 Pa), demonstrating at these regions of the extruder, the material yields through the gauge and is deposited. In addition, the stress-sweeps performed demonstrates the addition of nanosilicates increases the yield stress needed for extrusion (Figure S4, Supporting Information). For example, the Newtonian nature of the GelMA, PEGDA, and GelMA/PEGDA solutions causes negligible yield stresses, limiting control over the extruded volume. However, solutions with nanosilicates exhibit higher yield stresses, thus providing more control over the volume being deposited and therefore improving print fidelity of bioink.
The flow behavior described above demonstrates the ability of the bioink to be extruded, however to fabricate intricate geometries, such as the anatomy of a blood vessel, bioinks must be able to support deposition of subsequent layers without compromising the shapes. Thus, bioinks must be able to bridge over gaps without support material present. When printing GelMA, PEGDA, and GelMA/PEGDA across a gap of varying lengths, the material pools along the surface, demonstrating the lack of any shape retention. However, upon the addition of nanosilicates, the nECM bioink was able to bridge across a gap up to 5 mm in distance, illustrating the potential of the nECM to print complex networks (Figure 2D).
Although self-support is essential to produce anatomically accurate vasculature, bridging needs to be taken into consideration along with line fidelity and infill densities. Currently, print fidelity is most commonly characterized through straight-lines, comparing the measured thickness of the line with the needle gauge diameter and printing parameters.[36–38] However, physiological structures do not exists in straight geometries, consequently multiple parameters, such as bridging, line deposition, and infill densities should be holistically characterized together. This can be accounted for through fidelity ratio quantification, defined as the ratio of the programmed or theoretical void area (AT) to the actual void area of the print (AA) (see method section in Supporting Information for details). The results show high print fidelity ratio (AA/AT >0.8) for low infill density, which is utilized to print hollow geometries, such as vascular models (Figure 2E). With an increased infill density, the print fidelity ratio decreases. This is due to the compounding deviations observed as the infill density is increased. Explicitly, when comparing the printed model to the actual printed construct, deviations observed increase as more volume is being extruded. Thus, all deviations are reflected in the print fidelity ratio.
Post-printing, the nECM bioink is cross-linked via exposure to UV light, locking the structure into the deposited shape through covalent bonds between the acrylate groups on the PEGDA and GelMA. To verify the nanosilicates do not impede on this bonding and that printing stresses do not disrupt the polymer’s chemical structures, a time sweep was performed, measuring the storage modulus upon UV-exposure after 30 s (Figure 2F). All materials containing GelMA, PEGDA, or their combination exhibited an increase in the storage modulus after exposure to UV light. This increase corresponds to a rise in the material’s elastic response, therefore supporting the occurrence of crosslinking. However, as expected, nanosilicates independently did not manifest any change in the storage modulus, translating to no crosslinking. Thus, nanosilicates can be extruded and printed into complex shapes, but cannot be fixed into a stable construct that can be manipulated. However, when combined in the nECM, nanosilicates provide necessary rheological properties to deposit bioinks into complex geometries, while subsequent crosslinking of GelMA and PEGDA provide mechanical stability under physiological conditions (Figure 2G). Overall, this nECM bioink permits for direct extrusion of a mechanically resilient, shear-thinning, and recoverable bioinks for the deposition of high-fidelity constructs.
2.2. Bioprinted nECM Conserves Phenotype of Vascular Endothelial Cells and Smooth Muscle Cells
Introducing cells within the bioink can alter interactions between bioink components, affecting both the rheological properties and extrusion process (Figure 3A). To ensure that the encapsulation of VSMCs within the nECM bioink does not affect printing parameters and construct fidelity, different densities of VSMCs (0.5–10 million mL−1) were incorporated into the nECM bioink and the rheological characteristics assessed (Figure 3B). Interestingly, as the cell density was increased, the consistence index (K) decreased correspondingly. This is hypothesized to be due to the disruption of polymer–polymer and polymer–nanosilicates interactions due to the presence of cells. However, even though a decrease in K was observed, all densities maintained a shear-thinning, thixotropic behavior (n < 1) as well as recoverable characteristics, thus all densities can be used for 3D bioprinting applications. Utilizing a cell density of 2.5 million VSMCs per milliliter, cells maintained a high viability (>85%) post-extrusion on days 1, 3, and 7 (Figure 3C and Figure S5D, Supporting Information). A true validation of VSMCs density to in vivo conditions may be difficult since there is very sparse data regarding VSMCs density in vivo, which is heterogeneous across vessels and varies based on dynamic cues and microenvironmental factors. Therefore, we evaluated if a broad range of VSMCs encapsulation densities (1–10 millions mL−1) and selected a cell density of 2.5 million cells mL−1 for developing our vascular model.
Figure 3.
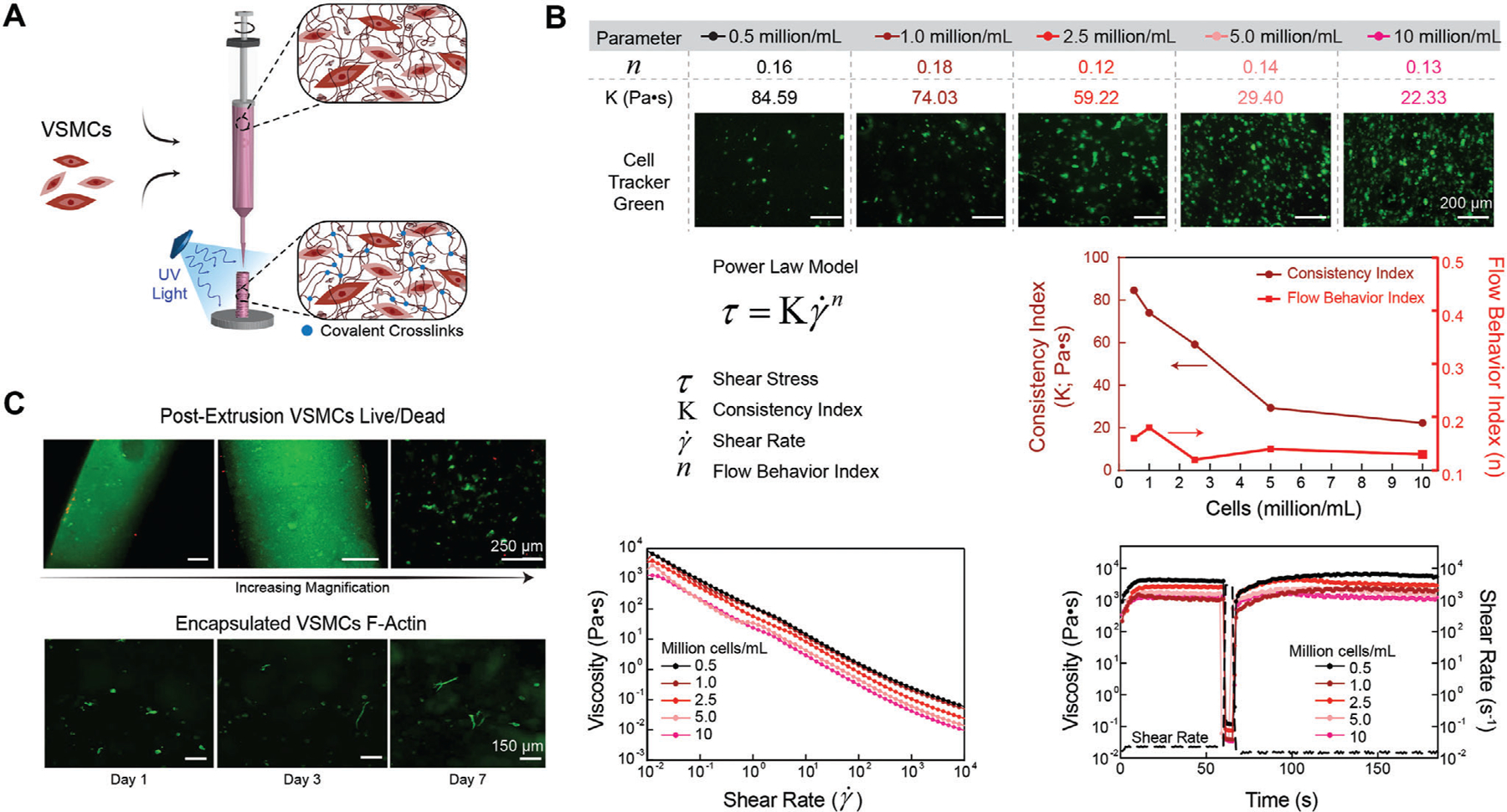
3D Bioprinting using nECM bioink. A) Schematic illustrating VSMC encapsulation and extrusion for 3D bioprinting. B) All cell densities encapsulated within the nECM maintained a high low-shear viscosity, shear thinning profile (flow behavior index < 1), and recoverability optimal for 3D bioprinting. However, as the encapsulation density was increased, the consistency index decreased in a logarithmic manner. C) Directly after extrusion, VSMCs maintained a high cell viability, illustrating an increase in magnification from left to right (top row). After crosslinking the nECM, encapsulated VSMCs demonstrate an increase in spreading and elongation between days 1, 3, and 7 (bottom row).
Aside from cells affecting material interactions, cells are sensitive and can be affected by a multitude of environmental factors, such as material constituents and shear forces within the 3D bioprinting processes that can have a major impact on cellular metabolic activity, viability, and phenotypic maintenance. To assess for these variables, ECs and VSMCs were monitored for 30 days, demonstrating long-term maintenance of metabolic activity, independent of the fabrication technique being used (i.e., seeding, encapsulating, printing) (Figure 4A). We observed an increase in metabolic activity, demonstrating vascular cell pro-liferation when interacting with the nECM. This was further confirmed through live cell analysis of the VSMCs on days 1, 7, and 30 (Figure 4B). In addition, cell viability and phenotypic maintenance (ECs: vascular endothelial-cadherin, VE-cadherin; VSMCs: α-smooth muscle actin, α-SMA) was assessed on days 1, 3, and 7 with immunohistochemistry techniques (Figure 4C,D). Both cell types demonstrated a high viability (> 80%) as well as an increase in proliferation over time. Phenotypic maintenance was also validated using quantitative real-time polymerase chain reaction (qRT-PCR), demonstrating no significant difference in cell-specific markers (ECs: VE-cadherin, vWF, and eNOS; VSMCs: ACTA2, CNN1, and TAGLN) between nECM bioink and tissue culture polystyrene (TCPS) controls. In addition, no calcification was observed when VSMCs are cultured within the nECM for 30 days. Therefore, the nECM bioink is able to maintain construct integrity and deposition fidelity upon cell encapsulation, independent of the cell density being used, as well as support improved biocompatibility (i.e., cellular viability and phenotypic maintenance).
Figure 4.
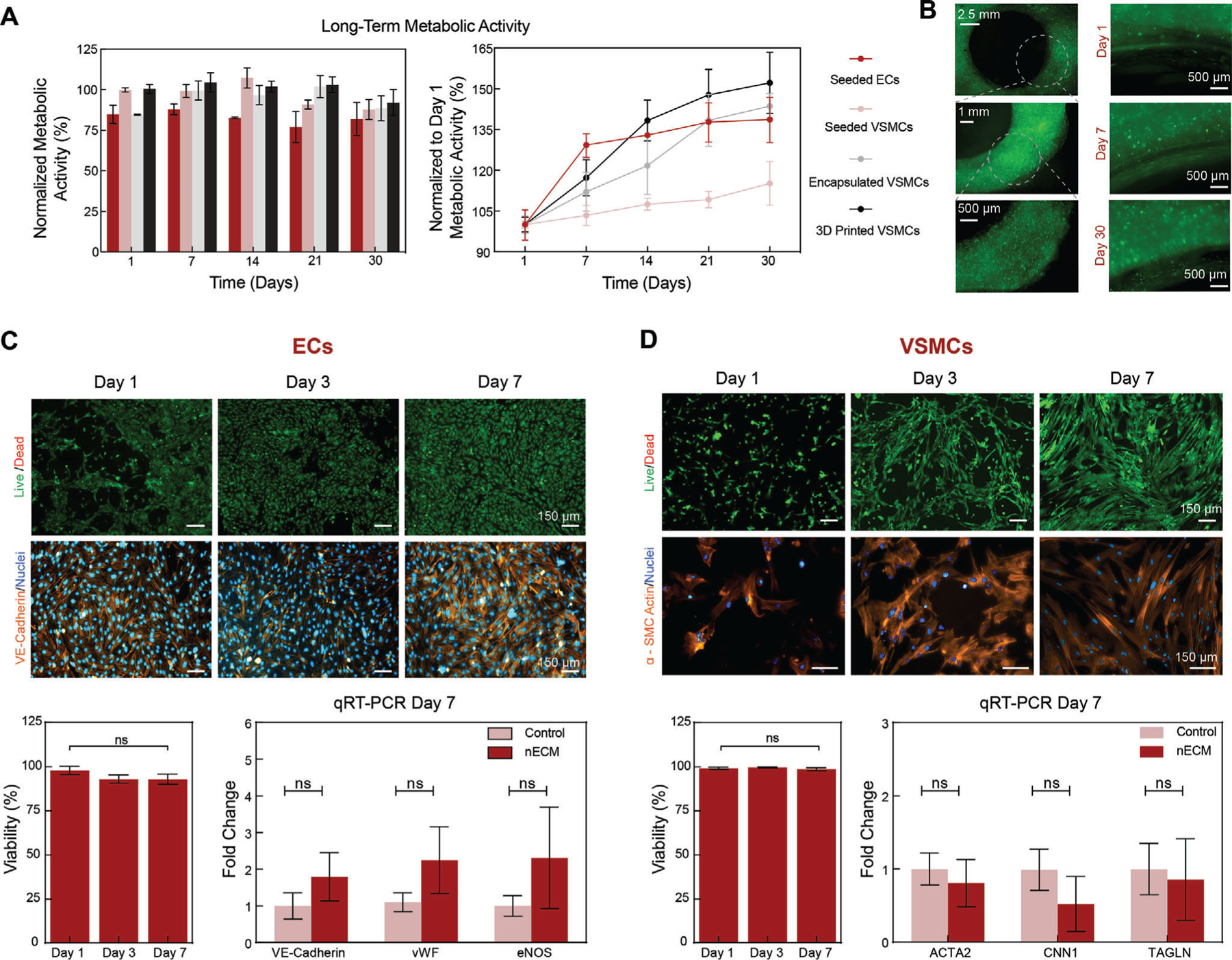
Metabolic activity, viability, and phenotypic maintenance of vascular cells. A) Long-term metabolic activity was maintained up to 30 days when normalized to a TCPS control. Normalizing to Day 1, demonstrated an increase in metabolic activity independent of the fabrication technique being used, correlating to cellular proliferation. B) Cells imaged via cell tracker show increase in proliferation between days 1 and 30. C) ECs and D) VSMCs demonstrate a high viability (> 80%) and increase in cell number from Day 1 to Day 7. In addition, both (C) ECs and (D) SMCs maintain a healthy phenotype as illustrated in immunohistochemistry staining on days 1, 3, and 7 as well as verified with qRT-PCR on day 7, showing no significant difference when compared to TCPS controls. Multiple replicates of samples are performed for rigor (n = 5) and statistics analysis was performed using one-way ANOVA with posthoc Turkey analysis (ns = not significant).
2.3. 3D Bioprinting Anatomically Sized Vascular Structures
Currently, 3D printed vascular models are limited to sacrificial printing techniques to fabricate micron-sized, hollow channels in bulk-casted hydrogels.[3,8,39,40] In a recent approach, small-diameter blood vessel grafts containing both functional endothelial and muscular cell layers are fabricated.[41] Although this method accurately recapitulates small blood vessels (i.e., arteriole), it may not be always sufficient to study macro-sized vessels (i.e., arteries, aorta). In addition, the sacrificial printing approach requires high cell densities and yields a cross-sectional channel geometry that may not be cylindrical. Importantly, these models do not effectively support the co-culture and crosstalk of endothelial and muscle cells, and include perfusion capability. Thus, there is a need to introduce models that recapitulate the anatomical macro-structure as well as the tissue- and cell-level micro-structure (i.e., cellular composition and physiological communication) to monitor, assess, quantify, and eventually treat vascular diseases.[42]
To address these limitations, we utilize the nECM bioink printing platform for the direct fabrication of free-standing vessels of varying diameters, including macro-scale vessels to model larger geometries, such as the carotid artery or the aorta. Utilizing our nECM bioink, VSMCs are extruded into cylindrical constructs, localizing VSMCs within the nECM bioink to form a living medial layer. Next, ECs were seeded on the extruded geometry, forming a confluent monolayer that recapitulates the vascular lumen (Figure 5A). Both cells were combined and localization of seeded ECs and VSMCs embedded within the nECM were verified to establish a co-culture model (Figure 5A). These results show the versatility of 3D bioprinting to obtain multi-cellular structure consisting of ECs on inside surface, VSMCs within vessel wall and fibroblast can be seed on the outside surface of the 3D printed structure.
Figure 5.
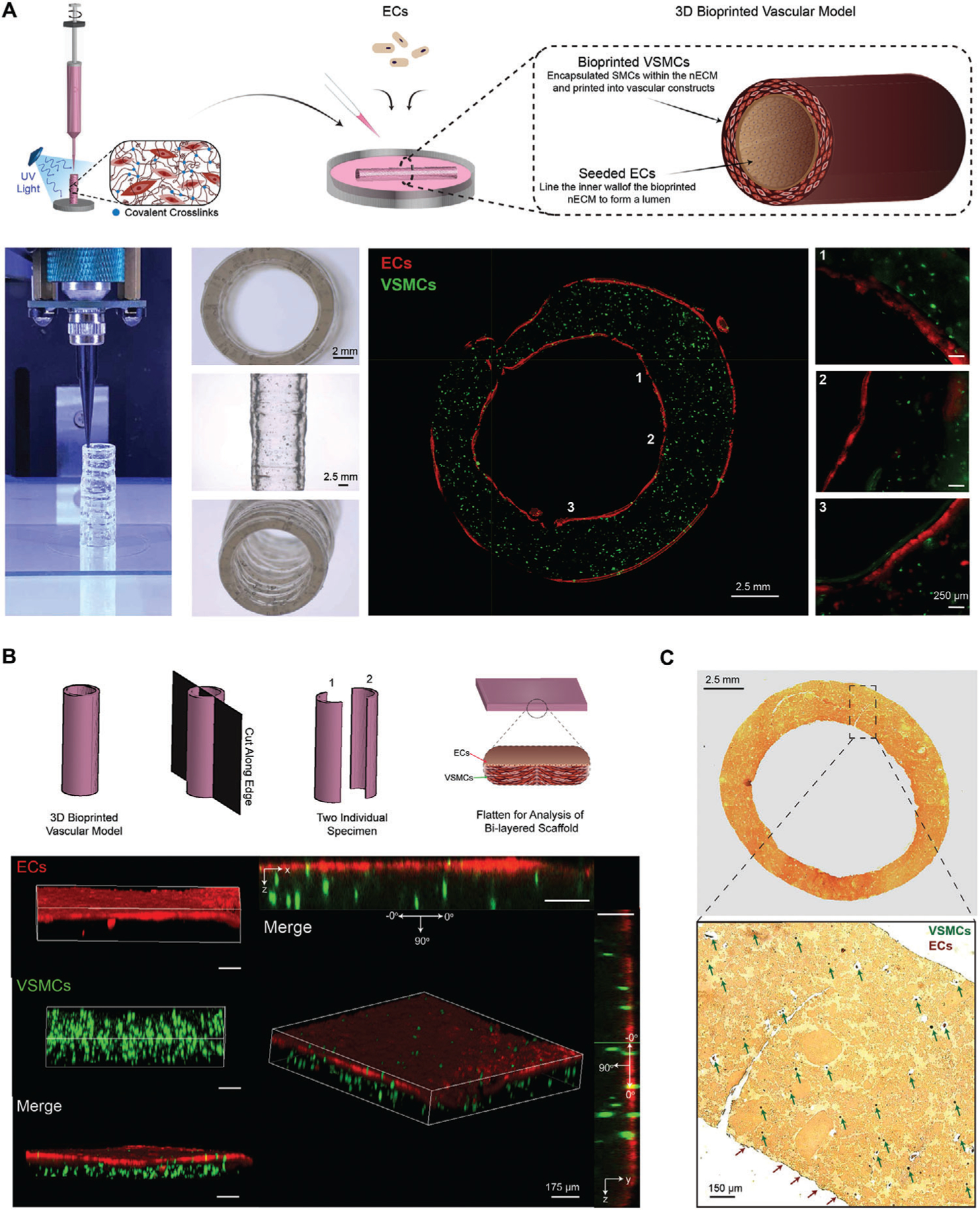
3D bioprinted vascular model. A) Depiction of the method used to fabricate the 3D bioprinted vascular model with EC and VSMC co-culture. Staining with different cell tracker, EC-VSMC co-culture is depicted with spatial-temporal control to recapitulate the structure of native human vasculature. B) Schematic illustrating the En Face characterization technique, including cutting the model into two independent samples for cell localization verification. Staining with cell tracker, ECs are shown to be localized on the top surface (x,y-plane) and VSMCs localized in 3D (z-plane). C) VVG histology staining, illustrating elastin and nuclei as black, collagen as red, and cytoplasmic elements as yellow. VSMC nuclei are distributed throughout the 3D printed construct (some shown with the green arrows), whereas EC nuclei are located on the surface of the printed construct (red arrows).
En Face is a technique used to characterize and image harvested vascular conduits in vivo.[43,44] Specifically, conduits are cut into two independent specimen that are laid flat between two glass slides for further analysis (Figure 5B). Employing this technique to our 3D bioprinted vascular model, we observed that VSMCs (green) are evenly distributed throughout the nECM in 3D (z-plane), while ECs (red) make a confluent monolayer on the nECM, localized on the x,y-plane (Figure 5B). In addition, quantifying 3D bioprinted VSMC directionality utilizing the En Face technique, VSMCs demonstrated a directionality between 60 and 90°, reflecting an arrangement perpendicular to the seeded endothelium (Figure S6, Supporting Information). We hypothesize this alignment to be caused by the unique interaction between shear stress, strain, and deposition path exhibited on nECM encapsulated VSMC during the bioprinting process. This alignment could be influenced by introduction of fluid shear forces, changing polymer molecular weight, printing speed as well as nozzle size.
In addition to illustrating EC-VSMC localization, Verhoeff van Gieson (VVG) histological analysis was performed (Figure 5C). This stain is utilized to visualize blood vessel microstructural organization, where elastin and nuclei are stained black, collagen stained red, and cytoplasmic materials stained yellow.[45–48] Given GelMA is a derivative of collagen and the main component of the nECM bioink, the printed construct demonstrates a significant amount of red-stained material. VSMC nuclei, some identified by green arrows, are shown to be distributed throughout the nECM bioink construct in 3D. Furthermore, EC nuclei, as identified by the red arrows, are localized to the 2D surface of the printed construct. Taken collectively, this data illustrates the ability of our nECM bioink to fabricate constructs with living EC and VSMC co-culture, providing a tool to create anatomically accurate cellular architectures innate to human blood vessels.
2.4. 3D Bioprinted Blood Vessel Mimics Thromboinflammatory Outcomes
Next, we set out to determine if this 3D bioprinted vessel is able to recapitulate the geometry and obstruction observed in native human vessels. To achieve this, utilizing optical coherence tomography (OCT), we imaged a left anterior descending artery and compared it to a fabricated 3D bioprinted vessel with no endothelium present (Figure 6A). After clotting overnight, no significant difference in percent obstruction was observed between the human artery and our 3D bioprinted model.
Figure 6.
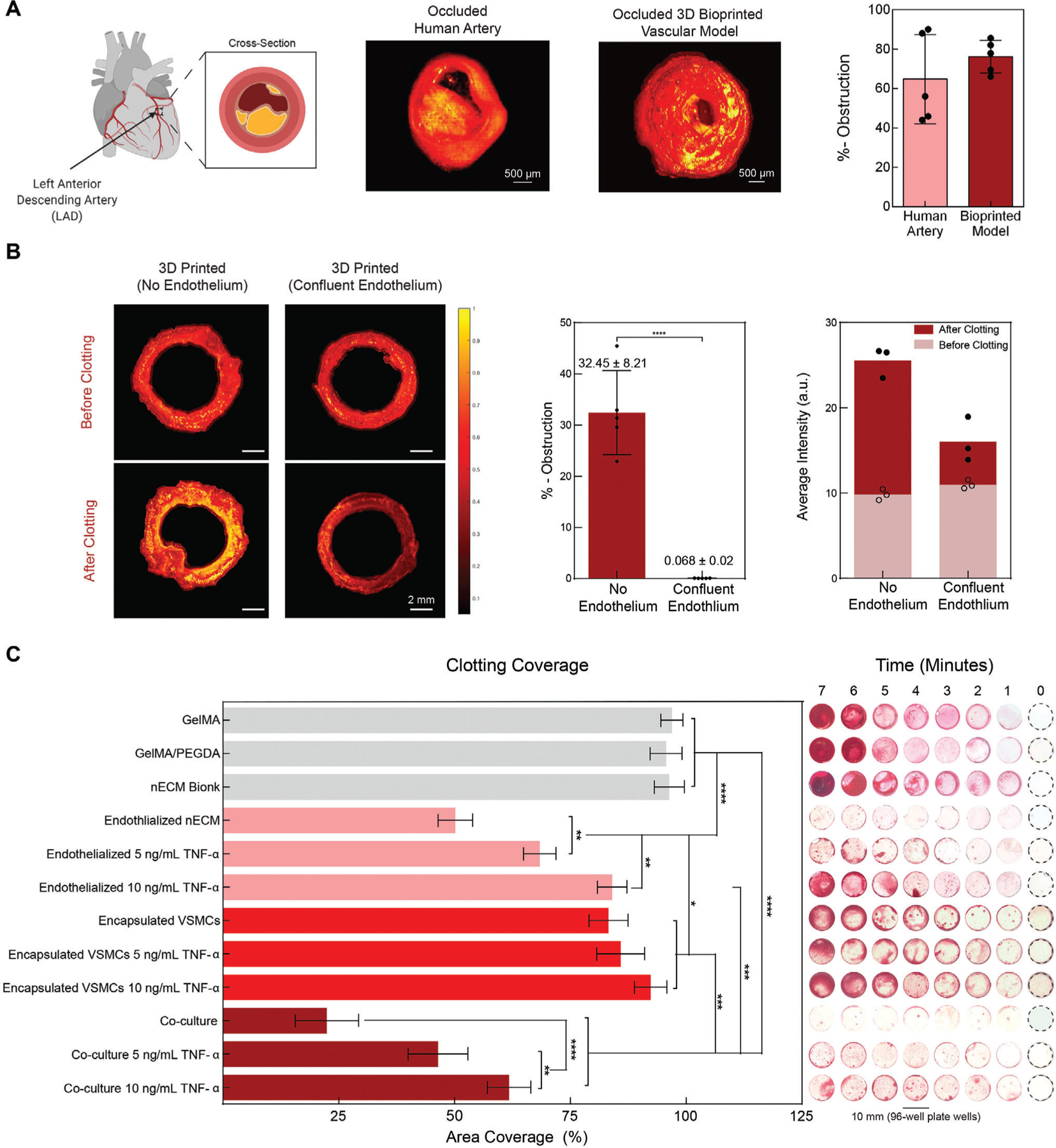
Functional validation of 3D bioprinted vascular model. A) OCT is used to determine obstruction of the lumen following blood perfusion, demonstrating the formation of diseased model. Comparing a clotted human left anterior descending artery to the occluded bioprinted vascular model (no endothelium), similar geometries are achieved and no significant difference in obstruction is observed after 12 h of clotting. B) OCT of a 3D printed (no endothelium) and 3D printed (confluent endothelium) was performed in pre- and post-stenosed models. When no endothelium was present, a significant amount of clotting is depicted, increasing the percent obstruction as compared to when a confluent endothelium is present. Comparing the average intensities between groups demonstrated a significant increase, further supporting clotting formation. C) To exemplify dose-dependent sensitivity, static clotting experiments were performed. With no lumen formation (GelMA, GelMA+PEGDA, nECM, encapsulated VSMCs, Encapsulated VSMCs with 5 ng mL−1 TNF-α, and encapsulated VSMCs with 10 ng mL−1 TNF-α), there was a significant increase in area coverage after 7 min. However upon the addition of ECs, the area coverage significantly decreases and demonstrates a dose-dependent effect on percent area coverage with TNF-α stimulation. Multiple replicates of samples were performed for rigor (n = 5) and one-way ANOVA with posthoc Turkey analysis (* = p < 0.05, ** = p < 0.05, *** = p < 0.005, and **** = p < 0.0001) was used for statistics analysis.
Furthermore, we also wanted to assess whether the fabricated model is able to accurately mimic vascular thromboinflammatory responses upon activating the endothelium through cytokine-induced injury. To do this, we stimulated 3D bioprinted vessels with TNF-α and hypothesized that, like in vivo or in other physiomimetic in vitro models,[49,50] we would observe disruption in barrier function and blood clotting in a dose-dependent manner (Figure 6B). Utilizing OCT, untreated bioprinted vessels perfused with blood, revealed no obstruction of the lumen, mimicking a healthy vessel (0.07±0.02% obstruction, Figure 6B). On the contrary, when no endothelium was formed and blood was perfused through only the nECM (red), significant clotting (yellow) was observed with OCT, demonstrating 32±8% obstruction. Given the difference in refractive index between the nECM and clotted blood, clot formation can further be validated by the average intensity differences presented before and after clotting. Specifically, before clotting, no significant difference was reflected in the average intensity between the two groups. However, after clotting, a significant difference in average intensity was observed and directly correlated to the presence of blood. Explicitly, higher intensities are presented with no endothelium (25±2 a.u) and lower intensities with a confluent endothelium (16±3 a.u), when compared to the baseline average intensity of the nECM itself (10±1 a.u).
Subsequently, additional experimentation revealed that after 7 min of blood exposure, high clotting coverage was observed for GelMA (97±2%), GelMA/PEGDA (96±3.5%), and the nECM (96±3%) (Figure 6C). However, the presence of a confluent EC layer significant reduced clotting coverage (50±4%). When ECs cultured on the nECM were treated with 5 and 10 ng mL−1 of TNF-α for 18 h, a dose-dependent trend was observed. Explicitly, ECs treated with 5 ng mL−1 exhibited regions of disrupted barrier function, causing an increase in clotting and therefore percent coverage (68±3.5%). When the ECs were treated with a higher dose of TNF-a (10 ng mL−1), the barrier function is further compromised compared to the 5 ng mL−1 dosage. Correspondingly, this leads to further clotting as measured through percent coverage (84±3%). Next, clotting coverage of encapsulated VSMCs in the absence of lumen formation demonstrated was evaluated. A high clotting coverage was observed in encapsulated VSMCs after 7 min of blood exposure, irrespective of the TNF-α dosage delivered. (VSMCs: 83 ± 4%; VSMCs with 5 ng mL−1 TNF-α: 86 ± 5%; VSMCs with 10 ng mL−1 of TNF-α: 92 ± 4%) This might be attributed to the absence of a lumen, correlating to results observed on GelMA, GelMA + PEGDA, and nECM bioink controls. Nonetheless, upon the incorporation of an endothelium on encapsulated VSMCs, a significantly altered clotting phenomena was observed. Specifically, a significant reduction in clotting coverage was observed upon co-culturing ECs and VSMCs (22 ± 7%) compared to relative individual cultures, suggesting cellular cross-talk and communication within the 3D printed model. It is hypothesized that EC-VSMC communication modifies EC expression and potentially alters cellular phenotype.[51–53] For example, it has been shown that in the presence of VSMCs, ECs adhere slower and present a less thrombotic phenotype.[53–56] Yet, EC-VSMC co-culture still preserved a dose-dependent response to TNF-α stimulation, as illustrated in the clotting coverage after 7 min. Inclusively, this demonstrates the ability of this 3D bioprinted vascular model to recapitulate responses of vascular activation and blood perfusion, which has currently only been demonstrated in animal models or microfluidic organ-on-chips, but not in 3D printed vessel of a large cylindrical anatomy.
To further assess for EC-VSMC communication, we compared a stimulated 3D bioprinted model (10 ng mL−1 TNF-α) to a control, healthy EC-VSMC co-culture model, specifically assessing for EC barrier disruption and permeability to the underlying nECM (Figure 7A,B). EC monoculture demonstrated exposure of the underlying nECM bioink, as depicted by white arrows in Figure 6B. The exposure to underlying nECM and EC permeability is further increased upon TNF-α stimulation. This observation supports the hypothesis that stimulation disrupts the EC integrity and increases exposure of the underlying ECM, leading to the increased clotting coverage previously observed. However, upon VSMC-EC co-culture within the vascular model, ECs form a confluent monolayer with tight VE-cadherin junction on the nECM, improving upon EC integrity and decreasing nECM exposure. Upon stimulating the co-culture model with TNF-α, VE-cadherin junctions decrease, however the confluent EC monolayer is conserved and continues to inhibit EC permeability or exposure of the underlying nECM to blood. Furthermore, ECs without stimulation in both mono- and co-culture systems did not demonstrate any form of. However, upon exposure to 10 ng mL−1 of TNF-α, an increase in cellular alignment was observed along the 30°–60° direction. This observed alignment within this direction is hypothesized to occur as a survival mechanism by ECs to recover from an inflamed response. Specifically, conservation EC junctions and directionality promotes the release of growth factors, encouraging healing of the damaged monolayer and promoting re-endothelialization.[53,54,56]
Figure 7.
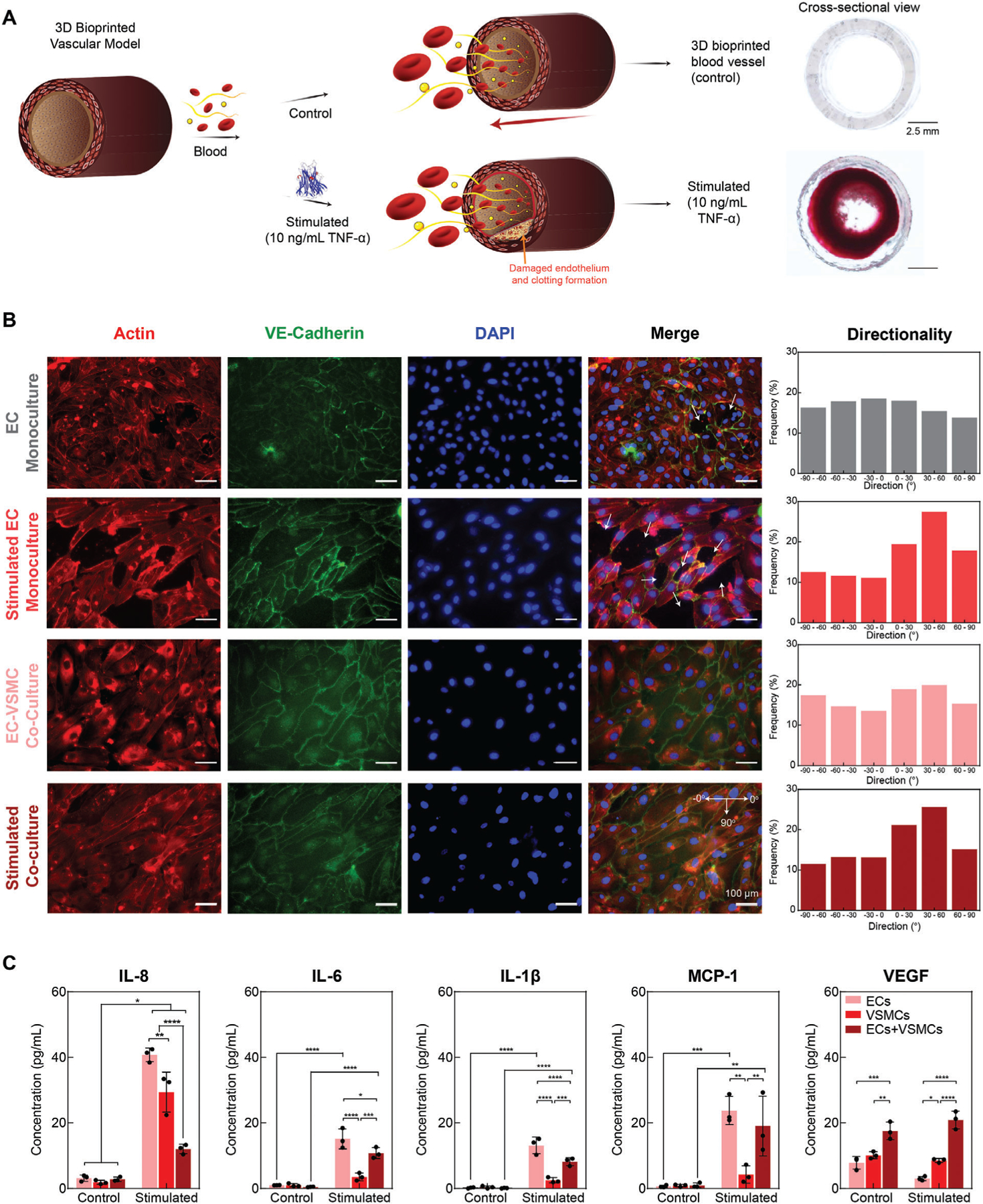
Vascular cell communication. A) Schematic demonstrating clotting formation upon stimulation with TNF-α. TNF-α disrupts EC barrier function, initiating clotting. B) EC monoculture demonstrates exposure of the underlying matrix (white arrows), yet the area of exposed nECM bioink is increased upon TNF-α stimulation. Upon co-culture, a healthy, confluent monolayer is formed with VE-Cadherin junctions connecting the ECs. Upon stimulation, VE-Cadherin is decreased, however a confluent monolayer of ECs is still present. Upon stimulation, ECs tend to align, demonstrating an increased alignment along 30 –60 direction. C) Co-Culturing of ECs and VSMCs modified EC expression, resulting in a reduction of inflammatory markers IL-8, IL-6, IL-1β, and MCP-1 as well as increases VEGF expression, motivating a more atheroprotective phenotype. Multiple replicates of samples were performed for rigor (n = 5) and one-way ANOVA with posthoc Turkey analysis (* = p < 0.05, ** = p < 0.05, *** = p < 0.005, and **** = p < 0.0001) was used for statistics analysis.
In addition, EC mono-culture demonstrated an increase in inflammatory markers, specifically interleukin-8 (IL-8), interleukin-6 (IL-6), interleukin-1β (IL-1β), and monocyte chemoattractant protein-1 (MCP-1) (Figure 7C). 3D bioprinted VSMCs cultured independently significantly decreased the inflammatory response in comparison to EC monoculture. This decrease in inflammatory markers was also observed in co-culturing of VSMCs and ECs together, supporting previous results of VSMCs promoting a less thrombotic EC phenotype.[53–55] Furthermore, the presence of VSMCs also significantly increased the expression of vascular endothelial growth factor (VEGF). This increase is hypothesized to promote re-endothelialization of the exposed surface.[57]
One limitation of this work is the lack of flow within the model as well as achieving mechanical properties within the physiological range to that of a native vessel (MPa range). Another limitation of this work is the inability to isolate the inside of the 3D bioprinted hollow channel, leading to an inefficiency in cell seeding and causing ECs to adhere to all surfaces, inside and out. In future, a bioreactor could be designed, aiming to isolate the inner channel from the outside surface of the printed construct. This would improve upon cellular localization and consent for the inclusion of fibroblasts to fabricate tri-layered vascular tissues. In addition, we hope to modulate polymeric and printing properties in the future to further our understanding of 3D bioprinting on VSMCs alignment, in health and diseases conditions. By gaining a cohesive understanding of these unique interactions, we can fabricate tissue models that more accurately recapitulate native tissue architectures. Another unique advantage of this system is to sequester therapeutic proteins within the bioprinting structure for prolong duration due to presence of nanosilicates. This aspect is useful to mimic physiological features of health or disease conditions.
3. Conclusion
In summary, we report a new class of nanoengineered hydrogel-based cell-laden bioinks that can be printed into 3D, anatomically accurate, multicellular blood vessels to recapitulate both the physical and chemical microenvironments native to human vasculature. A remarkably unique characteristic of this bioink is that regardless of cell density, it demonstrates a high printability and ability to protect encapsulated cells against high shear forces in the bioprinting process. 3D Bioprinted cells maintain a healthy phenotype and remain viable for nearly one-month post-fabrication. Leveraging these properties, the nanoengineered bioink is printed into 3D cylindrical blood vessels, consisting of living co-culture of ECs and VSMC, providing the opportunity to model vascular function and pathophysiology. Upon cytokine stimulation and blood perfusion, this 3D bioprinted vessel is able to recapitulate thromboinflammatory responses observed only in advanced in vitro preclinical models or in vivo. Therefore, this 3D bioprinted vessel provides a potential tool to understand vascular disease pathophysiology and assess therapeutics, toxins, or other chemicals in preclinical trials.
4. Experimental Section
Materials and Synthesis:
Nanosilicates (nSi) (Laponite XLG), acquired from BYK Additive and Instruments, was stored under vacuum to prevent particle interaction with water. PEGDA, with a molecular weight of 10 kilodaltons, was purchased from Polysciences Incorporated. Porcine gelatin (Type A, 300 Bloom), methacrylic anhydride, and Irgacure 2959 (2-Hydroxy-4′-(2-hydroxyethoxy)-2-methylpropiophenone) was purchased from Sigma Aldrich. GelMA (80% methacrylated) was synthetized using procedures previously reported.[32] In short, 10 g of porcine gelatin was dissolved into 100 mL of 1× phosphate buffered saline (PBS). The solution was heated on a stir plate for 1 h at 65 °C to obtain a transparent, homogeneous solution. Following dissolution, 8 mL of methacrylic anhydride was slowly added dropwise over several minutes and allowed to react for 3 h. To stop the reaction, 400 mL of 1× PBS, heated to 40 °C was added to the system and held stable for 15 min. The synthesized solution was then allowed to dialyze for 7 days, changing the deionized water twice daily, followed by sterile filtering (0.2 μm cellulose membrane) and lyophilization.
Bioink Fabrication:
All bioink fabrication was performed in a biosafety cabinet after ethanol and UV sterilization. All beakers and stir-bars used were sprayed with 70% ethanol and left in the BSC overnight under UV light. Desired volumes of GelMA and PEGDA were dissolved into heated deionized water (40 °C) under constant mixing on a stir plate. Once a transparent solution was obtained (≈5 min), 0.03% w/v Irgacure 2959 was added and stirred for an additional 3 min. Nanosilicates was then slowly incorporated into the hydrogel precursor solution and mixed until a change in viscosity was observed (≈5 min). Bioink formulations were stored overnight at 40 °C before use. Subsequent crosslinking of bioink formulations was obtained via exposure to UV light at 7 mW cm−2 for 60 s. UV duration was determined through the assessment of cellular spreading of encapsulated VSMCs after 1 and 7 days of culture after exposure to 7 mW cm−2 of UV light for 20, 30, 60, or 120 s (Figure S5A, Supporting Information). When performing this optimization, inhomogeneity (non-crosslinked regions) were observed in printed constructs after 20 or 40 s or UV exposure. However, longer exposure times (60 or 120 s) exhibited a homogenous, elastic structure. Furthermore, improved cellular spreading was observed in constructs exposed to 60 s of UV light compared to the rounded morphology depicted from samples after 120 s of UV exposure. Thus, 60 s of UV exposure at 7 mW cm−2 is used.
Custom-Made 3D Bioprinter:
The 3D bioprinter was designed and developed by modifying a commercial ANET A8 3D printer kit to utilize screw extrusion. The thermoplastic extruder assembly was replaced with a 3D printed screw extruder assembly, which held a stepper motor, guide rail, and a modified clay extruder. Firmware changes were made to accommodate the new extruder motor as needed. All prints used a 400 μm interior diameter tapered extruder tip. Printed shapes were created using Solid-works and exported as STL files. STL files were processed in Slic3r Prusa Edition to convert them into G-code printer instructions. Repetier Host was used to control the 3D printer.
Physiological Stability:
Equilibrium water uptake was calculated by soaking crosslinked hydrogels in water and media for 24 h in the incubator (37 °C; 5% CO2; n = 4). The mass of the hydrogel as prepared (T = 0 h) was compared to the mass of the hydrogel after swelling (T = 24 h) and recorded as Qo (Equation (2)). Water uptake was collected for combinations of GelMA (5, 7.5, and 10 wt%) and PEG (10k)DA (0, 1, 2.5, and 5 wt%) as well as GelMA, PEG(10k)DA, and altering amounts of nanosilicates (7.5 wt% Gelma + 2.5 wt% PEG(10k)DA + 1–5 wt% nanosilicates ranging in increments of 1 wt%).
| (2) |
Uniaxial Compression:
An ADMET MTEST Quattro eXpert 7600 Single Column Testing System equipped with a 25 lb load cell was used for all uniaxial compression testing. Utilizing a biopsy punch, hydrogels were casted into 2 mm thick sheets and stamped into 7 mm diameter samples (n = 4). Each sample’s diameter and height were measured with a digital caliper and variations of measurements were incorporated into modulus calculations. Unconstrained samples were compressed 20% of the measured height and returned to starting position at a strain rate of 1 mm min−1. Raw data for cyclic compression was processed using Excel to calculate the compressive modulus as the slope of the linear region of the data (0 to 20% strain). Compression data was collected for combinations of GelMA (5, 7.5, and 10 wt%) and PEG(10k)DA (0, 1, 2.5, and 5 wt%) as well as GelMA, PEG(10k)DA, and varying amounts of nanosilicates (7.5 wt% Gelma + 2.5 wt% PEG(10k)DA + 1–5 wt% nanosilicates ranging in increments of 1 wt%). In addition, compression was also performed on a combination of GelMA (7.5 wt%), PEGDA (2.5 wt%), and nanosilicates (4 wt%) utilizing various molecular weights of PEGDA (700 Da, 3.4 kDa, 8 kDa, and 10 kDa).
Rheological Characterization:
Rheological experimentation was performed using a stress-controlled Discovery Hybrid Rheometer 2 (DHR-2, TA Instruments) with a 40 mm diameter, parallel plate geometry attached. The geometry was equilibrated to 37 °C and a gap height of 0.25 mm in combination with a solvent trap for all experiments unless otherwise noted. Rotational shear-rate sweeps were performed between 10−2 and 104 s−1 to determine the power-law region and shear behavior. Power-law parameter’s n (flow behavior index) and K (consistency index) was calculated utilizing the TRIOS software (TA instruments) and verified using excel. Rotational time sweeps were executed at three consecutive shear rates (s−1): 3.02 (60 s), 23,000 (5 s), and 0.01 (120 s) to characterize the shear recoverability of precursor solutions. Oscillatory stress sweeps were performed between 10−1 and 103 Pa with a frequency of 1 Hz. Both storage and loss moduli were monitored in order to measure the material’s yield stress. UV crosslinking kinetics were characterized utilizing an oscillatory time sweep with a stress of 1 Pa and frequency of 1 Hz. After 30 s, samples were exposed to UV light (7 mW cm−2) and the storage modulus observed.
Numerical Modeling:
A 3D model of the extruder was fabricated utilizing Solidworks 2018 and imported into ANSYS Spaceclaim (ANSYS 19.2) in order to initiate the computational simulation. Upon importing, a 3D mesh was rendered with 3D linear elements. In whole, the model was formatted as a steady laminar problem. Specifically, material properties were inputted such that the viscosity profile of the ink followed the power law model (Equation (3)). The inlet boundary condition was specified as 4.97 × 10−6 m s−1, based off of the volumetric flow rate in which the bioink is printed. All walls within the system were assumed to have no-slip boundary conditions. As the ink was extruded onto the printing bed, the outlet boundary condition was established as 0 Pa (gauge pressure). The problem was then solved for continuity, mass balance, momentum balance, and energy balance to model the bioink flow patterns through the 3D extruder.
| (3) |
3D Print Fidelity:
All printed shapes were designed in Solidworks, exported into STL files, loaded and modified in Slic3r, and exported into G-code instructions for the printer. All printed constructs were programmed to have a layer height of 200 μm and a speed of 0.20 mm s−1. When necessary, 4 μL/mL of food coloring was added to aid in visualization of printed constructs. All precursors were stored and printed at 37 °C utilizing a 400 μm diameter gauge. To characterize the nECM’s bridging ability, a device was fabricated to assess the bioinks ability to deposit over a gap and support itself without any underlying materials. Two metal plates were angled and screwed into a stationary bottom plate, permitting for a varied gap along the x-axis. A rectangular shape of varying infill densities (5 and 10% infill to ensure visualization of gap distance) was printed over the varied distances. In addition, to characterize the fidelity ratio, 2 cm × 2 cm squares were printed at various infill densities (n = 3; 0, 5, 10, 20, 30, 40, 50, 60, 70, and 80%) programmed in the Slic3r software. Infill areas were calculated in ImageJ, utilizing a raster technique to produce as an overall average per sample. The print fidelity ratio was quantified as the ratio of the programmed or theoretical void area (AT) to the actual void area of the print (AA) (Equation (4)).
| (4) |
If the fidelity ratio is less than one, voids are smaller than the theoretical area. This is common for hydrogels due to variables such as over-extrusion, pooling, extrudate swell, and/or low print velocities. On the contrary, if the fidelity ratio is greater than 1, the area printed is larger than the theoretical, programmed void, translating to conditions such as air bubbles in the extruder, clogging, drag, and/or fast print velocity. Ideally, the printing ratio should be equivalent to one, meaning the printed void is equivalent to the theoretical void. The results obtained support the hypothesis that as the infill density is increased, errors are compounding, leading to a logarithmic decease in the fidelity ratio (Figure 2E). Due to the nature of this work directed at modeling vasculature, the geometries constructed consist of hollow channels with no infill density. Thus, the printing parameters defined yield an optimal fidelity ratio (≈1) for the scaffolds being constructed. Lastly, all hollow vascular constructs, which presented an optimal fidelity ratio, was printed with two perimeters (wall thickness ≈1 mm) to provide structural integrity for the large vascular prints.
In Vitro Cell Studies:
To evaluate the nECMs ability to maintain vascular cell viability, phenotype, and metabolic activity, human umbilical vein ECs (Lonza CC-2517) and human umbilical artery smooth muscle cells (VSMCs; Lonza CC-2579) were seeded on 4 mm diameter samples and used between 5 and 15 passages and a density of 500,000 cells per gel. ECs were cultured in EGM-2 medium supplemented with bullet kits (Lonza CC-3156 and CC-4176) and VSMCs in SmGM-2 medium supplemented with bullet kits (Lonza CC-3181 and CC-4149) in the incubator (37 °C; 5% CO2; n = 4 per time point and per assay) for 1, 3, and 7 days. Viability was analyzed through a live–dead assay. Specifically, cells were stained with a solution of ethidium homodimer (4 μL/mL) and calcein AM (2 μL/mL) in PBS for 30 min, and rinsed with PBS. To assure viable cells were maintaining their proper phenotype on the nECM, cell-specific antibodies were utilized for staining, explicitly vascular endothelial-cadherin (VE-cadherin monoclonal antibody; Invitrogen 36–1900) for ECs and α-smooth muscle actin (α-SMA monoclonal antibody; Invitrogen MA1–26 017) for VSMCs. In detail, hydrogels were rinsed with PBS and fixed with 4% paraformaldehyde at each time point. When staining, cells were permeabilized using 0.1% Triton ×−100 in a PBS-bovine serum albumin solution to block non-specific binding. Once permeabilized, gels were rinsed with PBS, exposed to 1:100 ratio of primary antibody (Rabbit anti-VE cadherin polyclonal antibody, ECs; rabbit anti-α SMA polyclonal antibody), washed with PBS, incubated in the same buffer with a secondary antibody (goat anti-rabbit conjugated with Alexa Fluor), and rinsed again with PBS. After antibody-staining, gels were stained with a 1:2000 Hoechst dye solution to stain for cell nuclei. All imaging was conducted on a Zeiss Axio Observer Z1 Microscope. All images were randomly taken on the surface of 4 replicates, processed in ImageJ, and one representative image presented. In addition to immunohistochemical staining, real-time quantitative reverse transcription PCR (RTq-PCR) was performed (n ≥ 3). To begin with, cells were seeded on TCPS controls or on bioink samples in non-adherent 6-well plates. After 7 days of culture, cells were trypsinized (3 min at 37 °C) to remove cells from the gels and pelleted by centrifugation. RNA was isolated and collected via a Zymo Research Quick-RNA Miniprep Extraction Kit following the manufacturer’s protocol. Initial quality of nucleic material was evaluated using a NanoDrop One/One Microvolume UV–Vis spectrophotometer (ThermoFisher Scientific), assessing the 260/280 absorbance ratios (≈2.0). Following RNA isolation, cDNA was synthesized via SuperScript III Reverse Transcriptase (Thermo Fisher), following manufacturer’s protocol. All primers (GAPDH; EC: vascular endothelial cadherin—VE-cadherin, von Willebrand Factor—vWF, and endothelial nitric oxide synthase 3—eNOS; VSMCs: α-smooth muscle actin—ACTA2, calponin 1—CNN1, and transgelin—TAGLN) were purchased from Bio-Rad. SYBR Green reagent was then used for amplification quantification using an Applied Biosystems QuantStudio 12K Flex. Expression and fold-change values were calculated from fluorescence and normalization to GAPDH values as well as to TCPS controls utilizing Excel [log2(TCPS controls/nECM gels)]. Metabolic activity was assessed utilizing an AlamarBlue assay on seeded ECs, seeded VSMCs, encapsulated VSMCs, and printed VSMCs on or within the nECM to ensure metabolic activity of cells was maintained (Bio-Rad). AlamarBlue assays were performed on days 1, 7, 14, 21, and 30. All seeded samples were normalized to TCPS controls, while encapsulated VSMCs samples were normalized to seeded VSMCs on the nECM and printed VSMCs samples were normalized to encapsulated VSMCs.
Bioprinted Vascular Model:
All 3D Bioprinting was conducted inside of a biosafety cabinet. Specifically, all parts of the printer (i.e., extruder, printer body, needle gauge) was sprayed to ethanol and put under UV sterilization over night to ensure a sterile environment for 3D bioprinting of VSMCs. To visualize the distribution of encapsulated VSMCs within the nECM bioink, cells were incubated with 5 μm CellTracker Green (Invitrogen) in 1× PBS for 30 min prior to passaging, utilizing the manufacturer’s protocol. Cells were then passaged, distributed through careful stirring into the nECM bioink, and printed into 5 mm tall hollow cylinders. After printing, constructs were crosslinked with UV light, placed into wells plates with the scaffold lying on its side. After 24 h of VSMC encapsulation, one T75-flask of ECs was exposed to 5 μm CellTracker Red (Invitrogen) in 1× PBS for 30 min prior to passaging. ECs were then passaged, seeded on the bottom half of the printed inner channel, and cultured in 50:50 SmGM-2 to EGM-2 media for 6 h. The 50:50 EC:VSMC media ratio utilized was determined through assessment of cellular metabolic activity on days 1, 3, and 7, supporting the proliferation of ECs while the maintenance of VSMCs (Figure S5B, Supporting Information). After 6 h, constructs were rotated 180° and EC seeding repeated (one T75-flask of ECs stained with CellTracker Red seeded within the hollow channel). VSMCs and ECs were cultured for 3 days and images were randomly taken of 5 replicates utilizing Zeiss LSM 780 NLO Multiphoton Microscope. Utilizing the same culture technique, samples were prepared for histological analysis. After 3 days of EC-VSMC co-culture, samples were washed with PBS and fixed using 10% neutral buffered formalin in the fridge overnight. Once fixed, samples were put into 70% ethanol and ramped to higher concentrations of ethanol for sample dehydration and paraffin wax embedding. Once infiltrated with wax, a microtome was used to section the 3D printed construct into 5 μm thick samples for VVG staining. Following VVG staining, images were randomly taken of 3 replicates utilizing a Zeiss Axio Observer Z1 Microscope. All images were processed in ImageJ, presenting one representative image.
Validation of 3D Bioprinted Vascular Model:
To validate the use of the nECM 3D bioprinted vascular model, clotting experiments were performed and analyzed with clinically relevant techniques. First, nECM and nECM constituents were prepared, crosslinked, punched into 6 mm diameter biopsies, and placed into a 96 well plate (n = 5). Samples were then exposed to coagulation-activated blood (10 μL of 0.1 m calcium chloride (CaCl2) and 100 μL of bovine blood containing sodium citrate) at 37°C for a pre-defined duration of time (varying between 1 and 7 min; increments of 1 min). Fresh bovine blood was obtained from Texas A&M College of Veterinary Medicine (College Station) and the experiments were performed within 72 h. After the specific duration of time, any uncoagulated supernatant was removed and the sample washed three times with 1× PBS, leaving any clotted blood. Samples were then imaged with a Zeiss SteRIO Discovery v 8 stereomicroscope and the images quantified in ImageJ. This process was repeated on endothelialized nECM constructs cultured for 7 days as well as endothelialized constructs with damaged barrier function. Samples containing ECs were exposed to human tumor necrosis factor alpha (TNF-α, Sigma-T6674; 5 and 10 ng mL−1 in EGM-2 medium) for 18 h in the EC-VSMC after the 7 days of culture, disrupting barrier function. After TNF-α treatment, samples were washed two times with 1× PBS and exposed to coagulation-activated blood, as described above. Utilizing clinical techniques, 3D printed nECM constructs with and without lumen formation were fabricated (n = 3). After 7 days of EC culture, for those with lumen formation, samples were washed with 1× PBS and exposed to coagulation-activated blood at 37°C for 7 min. After 7 min, samples were carefully washed three times with 1× PBS and imaged with a benchtop OCT system. Image reconstruction and 3D rendering were performed using MATLAB and percent obstruction calculated by measuring the pre-stenosed and post-stenosed inner diameters. Lastly, cytokine markers were assessed by collecting media effluents after 7 days of the respective culture (EC monoculture, stimulated EC monoculture, VSMC monoculture, stimulated VSMC monoculture, EC-VSMC co-culture, and stimulated EC-VSMC co-culture). Effluents were analyzed for specific cytokines using MILLIPLEX MAP Human cytokine/chemokine magnetic bead panel kit containing the beads tagged with specific antibodies against the target cytokines and performed using the manufacturer’s protocol.
Statistical Analysis:
All results are expressed as the mean ± deviation (n = 4–5). Statistical analysis of all quantitative data was performed via one-way ANOVA with posthoc Turkey analysis using GraphPad Prism (v 6.01). Statistical significance is demonstrated as * = p < 0.05, ** = p < 0.05, *** = p < 0.005, and **** = p < 0.0001.
Supplementary Material
Acknowledgements
The authors acknowledge Dr. Rola Barhoumi Mouneimne of the Texas A&M Veterinary Medicine and Biomedical Sciences Image Analysis Lab for her assistance with multiphoton imaging of printed constructs. The authors would also like to acknowledge the Texas A&M University Veterinary Medicine and Biomedical Sciences CVM Histology Lab for their assistance performing histology. Some of the schematics are draw using Biorender.com. K.G. acknowledges financial support from Texas A&M University Graduate Diversity fellowship. The authors would like to acknowledge financial support from Texas A&M Engineering Experiment Station (TEES), the National Institute of Biomedical Imaging and Bioengineering (NIBIB) (DP2 EB026265 to A.K.G. and R21 EB025945 to A.J), the National Science Foundation (CBET 1705852 and 1944322), and the Office of the President at Texas A&M University for X-Grant and T3 (to both A.K.G. and A.J.). J.P.C. acknowledges support from the National Heart Lung and Blood Institute, 1R01HL148338, 1R01HL133254, HL133254, and 1R01HL157790.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Karli A. Gold, Biomedical Engineering, College of Engineering, Texas A&M University, College Station, TX 77843, USA
Biswajit Saha, Biomedical Engineering, College of Engineering, Texas A&M University, College Station, TX 77843, USA.
Navaneeth Krishna Rajeeva Pandian, Biomedical Engineering, College of Engineering, Texas A&M University, College Station, TX 77843, USA.
Brandon K. Walther, Biomedical Engineering, College of Engineering, Texas A&M University, College Station, TX 77843, USA; Department of Cardiovascular Sciences, Houston Methodist Research Institute, Houston Methodist Hospital, Houston, TX 77030, USA
Jorge A. Palma, Biomedical Engineering, College of Engineering, Texas A&M University, College Station, TX 77843, USA
Javier Jo, Electrical and Computer Engineering, College of Engineering, The University of Oklahoma, Norman, OK 73019, USA.
John P. Cooke, Department of Cardiovascular Sciences, Houston Methodist Research Institute, Houston Methodist Hospital, Houston, TX 77030, USA
Abhishek Jain, Biomedical Engineering, College of Engineering, Texas A&M University, College Station, TX 77843, USA; Department of Cardiovascular Sciences, Houston Methodist Research Institute, Houston Methodist Hospital, Houston, TX 77030, USA; Medical Physiology, College of Medicine, Texas A&M Health Science Center, Bryan, TX 77807, USA.
Akhilesh K. Gaharwar, Biomedical Engineering, College of Engineering, Texas A&M University, College Station, TX 77843, USA; Interdisciplinary Program in Genetics, Texas A&M University, College Station, TX 77843, USA; Material Science and Engineering, College of Engineering, Texas A&M University, College Station, TX 77843, USA; Center for Remote Health Technologies and Systems, Texas A&M University, College Station, TX 77843, USA
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- [1].Murphy SV, Atala A, Nat. Biotechnol 2014, 32, 773. [DOI] [PubMed] [Google Scholar]
- [2].Kang H-W, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A, Nat. Biotechnol 2016, 34, 312. [DOI] [PubMed] [Google Scholar]
- [3].Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA, Proc. Natl. Acad. Sci. U. S. A 2016, 113, 3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Norotte C, Marga FS, Niklason LE, Forgacs G, Biomaterials 2009, 30, 5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Marga F, Jakab K, Khatiwala C, Shepherd B, Dorfman S, Hubbard B, Colbert S, Forgacs G, Biofabrication 2012, 4, 022001. [DOI] [PubMed] [Google Scholar]
- [6].Kolesky DB, Truby RL, Gladman AS, Busbee TA, Homan KA, Lewis JA, Adv. Mater 2014, 26, 3124. [DOI] [PubMed] [Google Scholar]
- [7].Grigoryan B, Paulsen SJ, Corbett DC, Sazer DW, Fortin CL, Zaita AJ, Greenfield PT, Calafat NJ, Gounley JP, Ta AH, Science 2019, 364, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Grigoryan B, Paulsen SJ, Corbett DC, Sazer DW, Fortin CL, Zaita J, Greenfield PT, Calafat NJ, Gounley JP, Ta AH, Johansson F, Randles A, Rosenkrantz JE, Louis-Rosenberg JD, Galie P, Stevens KR, Miller JS, Science 2019, 364, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Pradhan S, Banda OA, Farino CJ, Sperduto JL, Keller KA, Taitano R, Slater JH, Adv. Healthcare Mater 2020, 9, 1901255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chimene D, Kaunas R, Gaharwar AK, Adv. Mater 2020, 32, 1902026. [DOI] [PubMed] [Google Scholar]
- [11].Chimene D, Lennox KK, Kaunas RR, Gaharwar AK, Ann. Biomed. Eng 2016, 44, 2090. [DOI] [PubMed] [Google Scholar]
- [12].Moroni L, Burdick JA, Highley C, Lee SJ, Morimoto Y, Takeuchi S, Yoo JJ, Nat. Rev. Mater 2018, 3, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Gaharwar AK, Cross LM, Peak CW, Gold K, Carrow JK, Brokesh A, Singh KA, Adv. Mater 2019, 31, 1900332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hansen F, Mangell P, Sonesson B, Länne T, Ultrasound Med. Biol 1995, 21, 1. [DOI] [PubMed] [Google Scholar]
- [15].Konig G, McAllister TN, Dusserre N, Garrido SA, Iyican C, Marini A, Fiorillo A, Avila H, Wystrychowski W, Zagalski K, Maruszewski M, Jones AL, Cierpka L, de la Fuente LM, L’Heureux N, Biomaterials 2009, 30, 1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Schöneberg J, De Lorenzi F, Theek B, Blaeser A, Rommel D, Kuehne AJ, Kießling F, Fischer H, Sci. Rep 2018, 8, 10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Escudier MP, Gouldson IW, Jones DM, Exp. Fluids 1995, 18, 225. [Google Scholar]
- [18].Pilavtepe M, Recktenwald SM, Schuhmann R, Emmerich K, Willenbacher N, J. Rheol 2018, 62, 593. [Google Scholar]
- [19].Sears NA, Wilems TS, Gold KA, Lan Z, Cereceres SN, Dhavalikar PS, Foudazi R, Cosgriff-Hernandez EM, Adv. Mater. Technol 2019, 4, 1800343. [Google Scholar]
- [20].Mourchid A, Delville A, Lambard J, LeColier E, Levitz P, Langmuir 1995, 11, 1942. [Google Scholar]
- [21].Ruzicka B, Zaccarelli E, Soft Matter 2011, 7, 1268. [Google Scholar]
- [22].Thompson DW, Butterworth JT, J. Colloid Interface Sci 1992, 151, 236. [Google Scholar]
- [23].Mongondry P, Tassin JF, Nicolai T, J. Colloid Interface Sci 2005, 283, 397. [DOI] [PubMed] [Google Scholar]
- [24].Wang Q, Mynar JL, Yoshida M, Lee E, Lee M, Okuro K, Kinbara K, Aida T, Nature 2010, 463, 339. [DOI] [PubMed] [Google Scholar]
- [25].Laponite Performance Additives, Technical Information B-RI 21, BYK Additives & Instruments, Germany, 2014. [Google Scholar]
- [26].Ruzicka B, Zulian L, Ruocco G, Philos. Mag 2007, 87, 449. [Google Scholar]
- [27].Pawar N, Bohidar HB, J. Chem. Phys 2009, 131, 045103. [DOI] [PubMed] [Google Scholar]
- [28].Gaharwar AK, Kishore V, Rivera C, Bullock W, Wu CJ, Akkus O, Schmidt G, Macromol. Biosci 2012, 12, 779. [DOI] [PubMed] [Google Scholar]
- [29].Suman K, Joshi YM, Langmuir 2018, 34, 13079. [DOI] [PubMed] [Google Scholar]
- [30].Nelson A, Cosgrove T, Langmuir 2004, 20, 10382. [DOI] [PubMed] [Google Scholar]
- [31].Lorthioir C, Khalil M, Wintgens V, Amiel C, Langmuir 2012, 28, 7859. [DOI] [PubMed] [Google Scholar]
- [32].Chimene D, Peak CW, Gentry JL, Carrow JK, Cross LM, Mondragon E, Cardoso GB, Kaunas R, Gaharwar AK, ACS Appl. Mater. Interfaces 2018, 10, 9957. [DOI] [PubMed] [Google Scholar]
- [33].Peak CW, Stein J, Gold KA, Gaharwar AK, Langmuir 2018, 34, 917. [DOI] [PubMed] [Google Scholar]
- [34].Marquardt LM, Heilshorn SC, Curr. Stem Cell Rep 2016, 2, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Howell DW, Peak CW, Bayless KJ, Gaharwar AK, Adv. Biosyst 2018, 2, 1800092. [Google Scholar]
- [36].Sears NA, Dhavalikar PS, Cosgriff-Hernandez EM, Macromol. Rapid Commun 2016, 37, 1369. [DOI] [PubMed] [Google Scholar]
- [37].Kyle S, Jessop ZM, Al-Sabah A, Whitaker IS, Adv. Healthcare Mater 2017, 6, 1700264. [DOI] [PubMed] [Google Scholar]
- [38].Wilson SA, Cross LM, Peak CW, Gaharwar AK, ACS Appl. Mater. Interfaces 2017, 9, 43449. [DOI] [PubMed] [Google Scholar]
- [39].Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen D-HT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS, Nat. Mater 2012, 11, 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Skylar-Scott MA, Uzel SGM, Nam LL, Ahrens JH, Truby RL, Damaraju S, Lewis JA, Sci. Adv 2019, 5, eaaw2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gao G, Kim H, Kim BS, Kong JS, Lee JY, Park BW, Chae S, Kim J, Ban K, Jang J, Park H-J, Cho D-W, Appl. Phys. Rev 2019, 6, 041402. [Google Scholar]
- [42].Gold K, Gaharwar AK, Jain A, Biomaterials 2019, 196, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ko KA, Fujiwara K, Krishnan S, Abe JI, J. Visualized Exp 2017, 123, e55460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jelev L, Surchev L, J. Anat 2008, 212, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wang R, Raykin J, Li H, Gleason RL Jr., Brewster LP, Biomech. Model. Mechanobiol 2014, 13, 1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ong CS, Fukunishi T, Liu RH, Nelson K, Zhang H, Wieczorek E, Palmieri M, Ueyama Y, Ferris E, Geist G, Youngblood B, Johnson J, Hibino N, Tissue Eng., Part C 2017, 728, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Huang AH, Balestrini JL, Udelsman BV, Zhou KC, Zhao L, Ferruzzi J, Starcher BC, Levene MJ, Humphrey JD, Niklason LE, Tissue Eng., Part C 2016, 22, 524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kumar N, Kumar P, Badagabettu SN, Prasad K, Kudva R, Vasudevarao RC, Plast. Surg. Int 2014, 2014, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jain A, van der Meer AD, Papa A-L, Barrile R, Lai A, Schlechter BL, Otieno MA, Louden CS, Hamilton GA, Michelson AD, Frelinger AL, Ingber DE, Biomed. Microdevices 2016, 18, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Mathur T, Singh KA, Pandian NKR, Tsai SH, Hein TW, Gaharwar AK, Flanagan JM, Jain A, Lab Chip 2019, 19, 2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wang YH, Yan ZQ, Shen BR, Zhang L, Zhang P, Jiang ZL, Eur. J. Cell Biol 2009, 88, 701. [DOI] [PubMed] [Google Scholar]
- [52].Xu S, He Y, Vokurkova M, Touyz RM, Hypertension 2009, 54, 427. [DOI] [PubMed] [Google Scholar]
- [53].Wallace CS, Truskey GA, Am. J. Physiol.: Heart Circ. Physiol 2010, 299, H338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wallace CS, Strike SA, Truskey GA, Am. J. Physiol.: Heart Circ. Physiol 2007, 293, H1978. [DOI] [PubMed] [Google Scholar]
- [55].Imberti B, Seliktar D, Nerem RM, Remuzzi A, Endothelium 2002, 9, 11. [DOI] [PubMed] [Google Scholar]
- [56].Balcells M, Martorell J, Olive C, Santacana M, Chitalia V, Cardoso AA, Edelman ER, Circulation 2010, 121, 2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Dulak J, Józkowicz A, Dembinska-Kiec A, Guevara I, Zdzienicka A, Zmudzinska-Grochot D, Florek I, Wójtowicz A, Szuba A, Cooke JP, Arterioscler., Thromb., Vasc. Biol 2000, 20, 659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


