Abstract
Prevalence of Salmonella enterica on a Danish pig farm presenting recurrent infections was investigated. A comparison of the pulsed-field gel electrophoresis patterns of fecal isolates from piggeries, waste slurry, and agricultural soil amended with Salmonella-contaminated animal waste (slurry) and subclinical isolates from the same farm (collected in 1996 and later) showed identical patterns, indicating long-term persistence of the Salmonella enterica serovar Typhimurium DT12 clone in the herd environment. Furthermore, when Salmonella-contaminated slurry was disposed of on the agricultural soil (a common waste disposal practice), the pathogen was isolated up to 14 days after the spread, indicating potentially high risks of transmission of the pathogen in the environment, animals, and humans.
The distribution and prevalence of Salmonella enterica in food production animal herds are challenges for safe food production (8, 19). Human food-borne disease outbreaks that are associated with pig products not only are a public health concern but also have economic importance worldwide and in Denmark, where the annual production is about 23 million pigs (2). A wide variety of phage types and genotypes of S. enterica serovar Typhimurium have previously been identified in Danish pig production (7, 15, 17), and the most frequently isolated phage type of serovar Typhimurium in 1993, 1994, and 1998 was definitive phage type 12 (DT12) (4, 5). Previous investigations have shown that this phage type mainly has been spread clonally by trading of animals (3, 8), while Mutalib et al. (13) have reported the isolation of identical phage types of S. enterica serovar Enteritidis in clinical and environmental samples from poultry, indicating clonal survival. Although the same genotype of multiresistant serovar Typhimurium DT104 has repeatedly been isolated from several herds for up to 16 months (4), it is not clear whether persistence was due to chronic subclinical infection in pigs or to the persistence of Salmonella in the herd environment. Furthermore, the survival of Salmonella in agricultural soil amended with contaminated slurry (animal waste) used as a fertilizer poses a potential risk for the transmission of infection (9, 11) and may also contribute to the “reinoculation” of the pathogen in the herd environment. Thus, the present investigation focused on the persistence of the serovar Typhimurium DT12 clone in pig herds as well as the length of its survival in agricultural soil amended with Salmonella-contaminated slurry.
The Danish pig farm selected for this study is part of ongoing, larger animal husbandry and environment investigations being carried out on local pig farms by the Danish Veterinary Laboratory (17). The aim of these investigations has been to look for the presence or persistence of Salmonella in asymptomatic animals, their feed and feeding troughs, and the piggery environment, as this farm has had a history of recurrent clinical and asymptomatic Salmonella infections. The waste from piggeries consisted of slurry collected and stored in large slurry tanks located on the farm premises, and this is disposed of by being spread on agricultural fields once a year. Porcine fecal samples were collected from different sites (but as far as possible from the same area) in the piggeries, animal feed (including feeding troughs), and slurry. While the herd was monitored at least 10 samples in all were collected each time at regular quarterly intervals from 1998 to 2000. After screening of all samples using the conventional culture methods mentioned below (5), the results were recorded as Salmonella positive or negative and not as percent positive, due to the apparent complexity of the environmental samples. We were aware that there could be a continuous “replenishing” of Salmonella organisms as a result of fecal shedding by asymptomatic (carrier) animals. However, the purpose of these (environmental) investigations was to see how long the pathogen persisted or could be isolated from the herd environment and the animal waste. As a result, serological tests on animals were not performed or planned in these studies.
Furthermore, investigations were carried out to determine the risks associated with the spreading of Salmonella-contaminated slurry on agricultural soil by monitoring the length of persistence and survival of Salmonella in the treated soil. Ten samples each were collected from untreated soil (agricultural soil on which no slurry had been spread for at least a year) and treated soil (agricultural soil on which slurry had been spread during the past year or annually). The soil samples were taken by removing an upper 10-cm layer from the surface. On day 0, i.e., the day on which slurry was spread, the 10 agricultural soil samples were collected to ascertain the absence of Salmonella in soil prior to the spreading of Salmonella-contaminated slurry. Similarly, the slurry was also tested for the presence of Salmonella cells. In the case of treated soil, the samples were collected the same day shortly after the slurry had been spread, and later samplings were collected from soil that had been ploughed to mix and distribute the fertilizer. The treated soil was monitored for the presence of Salmonella by sampling on a weekly basis. These soil samples (untreated and treated soil) were collected from the same area; i.e., 10 samples were collected on each occasion from predetermined sites that were 1 m apart.
As a general practice after collection of samples from the animals, their environment and feed were tested for the presence of Salmonella by preculture in enriched, buffered peptone water and the use of selective media (5). The isolates characterized in the present investigation are listed in Table 1. The strains were identified as serovar Typhimurium according to the Kauffman-White typing scheme (16). Phage typing was performed according to the scheme described by Callow (6) and modified by Anderson et al. (1).
TABLE 1.
Salmonella serovar Typhimurium isolated from piggeries, slurry, and agricultural soil samples of the investigated Danish pig farm with recurring Salmonella infections
| Isolate(s) | Source | Sampling date |
|---|---|---|
| 9622953 | Clinical sample | 1 October 1996 |
| 9720733 | Clinical sample | 24 February 1997 |
| 9724024 | Clinical sample | 29 October 1997 |
| S 55 | Slurry | 29 June 1998 |
| S 62–S 65 | Slurry, piggery | 17 July 1998 |
| S 75–S 77 | Slurry | 11 February 2000, 10 March 2000 |
| S 78–S 85 | Piggery | 10 March 2000 |
| S 86 | Slurry | 11 April 2000 |
| S 87–S 103 | Treated soila | 11 April 2000, 13 April 2000 |
| S 105 | Slurry | 17 April 2000 |
| S 106–S 116 | Treated soil | 17 April 2000, 25 April 2000 |
| S 118–S 123 | Piggery | 12 May 2000 |
Agricultural soil amended with Salmonella-contaminated slurry.
All isolates were typed using pulsed-field gel electrophoresis (PFGE) to investigate the relationships among different isolates. Preparation of total DNA and the experimental setup were as described in reference 10 except that plugs had a final agarose concentration of 0.7% and 0.1 mg of proteinase K per ml was used for proteolysis. Restriction enzyme digestion was for 4 h, using 20 U of BlnI enzyme in 50 μl of restriction buffer (Amersham Life Science, Buckinghamshire, England), after which the plugs were preincubated in the appropriate buffer at 37°C for 30 min. Electrophoresis was performed in a CHEF-DR III electrophoresis system (Bio-Rad Laboratories) at 14°C using 0.5× Tris-borate-EDTA running buffer and 1.0% SeaKem GTG agarose gel (FMC Bioproducts, Rockland, Maine).
Electrophoresis conditions were as follows: for phase one, the initial switch time was 17.0 s and the final switch time was 26.0 s for 17.5 h; for phase two, the initial switch time was 50.0 s and the final switch time was 60.0 s for 5 h. PFGE was performed using 7.0 V/cm and a 120° angle. The molecular size marker used was the Lambda Ladder PFG marker (New England Biolabs). For visualization of the DNA, the gel was stained in ethidium bromide (2 μg/ml) for 10 min, destained in water for 20 min, and photographed with Polaroid film on UV transilluminator.
During the monitoring period of June 1998 to May 2000, serovar Typhimurium was isolated from 16 different samples collected from various environmental sites of the piggeries (including pig feces and bedding) and from 8 of the samples collected from slurry tanks (Table 1). Thus, not all samples were positive on each sampling occasion, and only the dates of positive Salmonella isolations are shown in Table 1. Investigations of seasonal variations in the isolation of Salmonella were not planned, and such variations were not clearly evident in this study due to small sample size. None of the samples from animal feed and feeding troughs was positive for Salmonella during the entire monitoring period.
Waste slurry sampled immediately before being spread on agricultural soil also tested positive for Salmonella, while all 10 samples of untreated soil were found to be negative for the presence of Salmonella. Investigations were continued to monitor the treated soil on a weekly sampling regimen (10 soil samples each time) for the survival of Salmonella in soil amended with contaminated slurry, until the treated soil sampled was negative on day 21. Of the 30 soil samples collected on three occasions, 26 were positive for Salmonella. Nine of the 10 soil samples were positive immediately after the spreading (day 0), while 7, 5, and 5 were positive 2, 6, and 14 days after the spreading, respectively (Table 1).
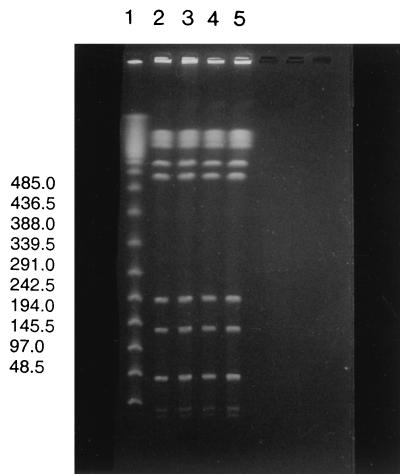
All serovar Typhimurium isolates were of phage type DT12. All isolates had identical PFGE patterns with restriction enzyme BlnI (PFGE patterns of selected isolates are shown in Fig. 1). Thus, the serovar Typhimurium isolates from the farmhouse environment, piggeries, and agricultural soil amended with Salmonella-contaminated slurry were indistinguishable by PFGE using this restriction enzyme. Similar results were obtained when the restriction enzyme XbaI was used separately and when the isolates were compared with isolates from asymptomatic animals on the same farm from which porcine fecal samples were collected in October 1996, February 1997, and again in October 1997 (data not shown). The results obtained from these investigations indicate that the clone has apparently persisted and survived in the farmhouse environment or in pigs with asymptomatic infection that shed these bacteria, explaining possible recrudescence of infection in the herd.
FIG. 1.
PFGE typing of selected Salmonella serovar Typhimurium strains isolated from piggeries, slurry, and soil amended with Salmonella-contaminated slurry (treated soil). Lane 1 contains molecular size markers (kilobases) (Lambda Ladder PFG marker; New England Biolabs). Lanes 2 to 5 contain DNA from selected Salmonella serovar Typhimurium isolates from piggeries, slurry, and agricultural soil amended with Salmonella-contaminated slurry (i.e., treated soil), as follows: lane 2, isolate S 83—piggery sample (tested on 10 March 2000); lane 3, isolate S 86—slurry sample (tested before being spread on agriculture soil on 11 March 2000); lane 4, isolate S 88—treated soil (sampled on 11 March 2000); lane 5, isolate S 114—treated soil sampled 14 days after spreading of slurry. All serovar Typhimurium DNA samples shown were digested with the restriction enzyme BlnI.
The similarity in PFGE patterns among clinical isolates and fecal isolates from piggeries, slurry, and agricultural soil amended with Salmonella-contaminated slurry indicates that the clone either survived in or was never eradicated from the herd environment. Furthermore, it is also possible that the clone has been continuously reintroduced into the herd environment by asymptomatic animals as a result of fecal shedding. PFGE analysis using one or two enzymes, although crude, is an effective method for determining that the isolates are essentially clonal, although we are aware that fine mapping using more enzymes would likely reveal differences between isolates. However, according to Tenover et al. (18), bacterial isolates that are indistinguishable by PFGE are unlikely to demonstrate substantial differences by other typing techniques. This corresponds well with reports by other groups who also used PFGE to identify epidemiological and clonal relationships within Salmonella (14, 15). In the present study, isolates from asymptomatic animals and those taken from environmental samples had identical phage types and were indistinguishable by PFGE. This finding correlates well with the findings of Mutalib et al. (13) that the same serovar Enteritidis phage types were found in samples from clinically diseased animals and in the poultry environment. Those authors also identified isolates in rodents and birds with the same phage type as those found in the environment.
Our investigations corroborate previous studies showing that a single clone of serovar Typhimurium can persist on farm premises for a long time (12, 17, 20). Furthermore, the isolation of Salmonella from piggeries and their environment, manure, or slurry over the entire period of monitoring brought to light the limitations of the current methods of isolation with regard to their sensitivity and specificity. Compared to molecular methods such as Salmonella-specific PCR, the conventional culture method (preculture in enriched, buffered peptone water followed by the use of selective media) (5) proved to be the most reliable and sensitive method for the isolation of viable Salmonella cells from complex environmental samples (S. Baloda, unpublished data). Limitations of the methods for detecting viable cells are due to the complexity of the samples, the low number of pathogens spread over a large surface area (soil and slurry), or the probable presence of the viable but nonculturable forms of Salmonella due to environmental stress conditions. Thus, negative results obtained for individual soil samples or after 14 days must be seen in the light of different parameters before they are considered to be true negative, and hence potential risks associated with the spreading of Salmonella-contaminated slurry on agricultural soil must be taken into account.
The persistence of serovar Typhimurium strains in piggeries and fields should give rise to further control measures related to the handling of manure and slurry and the disinfection of pig production facilities. It is thus important to point out that despite the limitations of the methods of isolation and detection encountered for the environmental samples mentioned above, the isolation of viable Salmonella cells from agricultural soil under natural environmental conditions even after 14 days of the spread of contaminated slurry is a great risk factor. This is important, since our laboratory investigations using terrestrial microcosms under controlled conditions indicate that serovar Typhimurium clones DT104 and DT12 can survive up to 299 days (S. Baloda, unpublished data). The survival of the pathogen in contaminated soil can facilitate the spread of pathogens (infection) via grazing farm animals, birds, cats, dogs, rodents, and even humans. As a result, effective waste management practices should be devised in view of the long-term survival potential of this zoonotic pathogen in soil amended with Salmonella-contaminated slurry.
Acknowledgments
This project is financed by a grant from the Danish Ministry of Food, Agriculture and Fisheries (MIL-97).
Cooperation extended by the respective farmers is gratefully acknowledged. We thank Kristian Møller for critical comments and Eva Pedersen for help with PFGE analysis.
REFERENCES
- 1.Anderson E S, Ward L R, de Saxe M J, de Sa J D H. Bacteriophage-typing designations of Salmonella Typhimurium. J Hyg Camb. 1977;78:297–300. doi: 10.1017/s0022172400056187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. Annual report on zoonoses in Denmark, 1998. Copenhagen, Denmark: Danish Veterinary Laboratory; 1998. .. [Google Scholar]
- 3.Bager F, Baggesen D L. CNEVA Salmonella and salmonellosis. Ploufragan/Saint-Brieuc, France: World Veterinary Poultry Association; 1992. Epidemiological typing of porcine Salmonella isolates; pp. 184–190. [Google Scholar]
- 4.Baggesen D L, Christensen J, Nielsen A C, Svenmark B, Nielsen B. Characterisation of Salmonella enterica isolated from swine herds in a cross-sectional study of the Danish swine production. Washington, D.C.: ISECSP; 1999. [Google Scholar]
- 5.Baggesen D L, Wegener H C, Bager F, Stege H, Christensen J. Herd prevalence of Salmonella enterica infections in Danish slaughter pigs determined by microbiological testing. Prev Vet Med. 1996;26:201–213. [Google Scholar]
- 6.Callow B R. A new phage-typing scheme for Salmonella typhimurium. J Hyg Camb. 1959;57:346–359. doi: 10.1017/s0022172400020209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen J, Baggesen D L, Nielsen A C, Nielsen B. Prevalence of Salmonella enterica in pigs before start of Danish Salmonella Control Program (1993/1994) and four years later (1998) Washington, D.C.: ISECP; 1999. pp. 333–335. [Google Scholar]
- 8.Davies R H, Wray C. Persistence of Salmonella enteritidis in poultry units and poultry food. Br Poult Sci. 1996;37:589–596. doi: 10.1080/00071669608417889. [DOI] [PubMed] [Google Scholar]
- 9.Forshell L P, Ekesbo I. Survival of salmonellas in composted and not composted solid animal manure. Zentbl Vetmed Reihe B. 1993;40:654–658. doi: 10.1111/j.1439-0450.1993.tb00188.x. [DOI] [PubMed] [Google Scholar]
- 10.Gautom R K. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J Clin Microbiol. 1997;35:2977–2980. doi: 10.1128/jcm.35.11.2977-2980.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinonen-Tanski H, Niskanen E M, Salmela P, Lanki E. Salmonella in animal slurry can be destroyed by aeration at low temperatures. J Appl Microbiol. 1998;85:277–281. doi: 10.1046/j.1365-2672.1998.00487.x. [DOI] [PubMed] [Google Scholar]
- 12.McLaren I M, Wray C. Epidemiology of Salmonella typhimurium infection in calves: persistence of salmonellae on calf units. Vet Rec. 1991;129:461–462. doi: 10.1136/vr.129.21.461. [DOI] [PubMed] [Google Scholar]
- 13.Mutalib A, McDonough P, Shin S, Patten V, Lein D. Salmonella Enteritidis in commercial layer farms in New York State: environmental survey results and significance of available monitoring tests. J Vet Diagn Investig. 1992;4:416–418. doi: 10.1177/104063879200400408. [DOI] [PubMed] [Google Scholar]
- 14.Olsen J E, Skov M N, Threlfall E J, Brown D J. Clonal lines of Salmonella enterica serotype Enteritidis documented by IS200-, ribo-, pulsed-field gel electrophoresis and RFLP typing. J Med Microbiol. 1994;40:15–22. doi: 10.1099/00222615-40-1-15. [DOI] [PubMed] [Google Scholar]
- 15.On S L, Baggesen D L. Determination of clonal relationships of Salmonella Typhimurium by numerical analysis of macrorestriction profiles. J Appl Microbiol. 1997;83:699–706. doi: 10.1046/j.1365-2672.1997.00295.x. [DOI] [PubMed] [Google Scholar]
- 16.Popoff M Y, Minor L L. Antigenic formulas of the Salmonella serovar. WHO Collaboration Centre for Reference and Research on Salmonella. Paris, France: Institut Pasteur; 1997. [Google Scholar]
- 17.Sandvang D, Jensen L B, Baggesen D L, Baloda S B. Persistence of Salmonella enterica serotype Typhimurium clone in Danish pig production units and farmhouse environment studied by pulsed field gel electrophoresis (PFGE) FEMS Microbiol Lett. 2000;187:21–25. doi: 10.1111/j.1574-6968.2000.tb09130.x. [DOI] [PubMed] [Google Scholar]
- 18.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Threlfall J E, Rowe B, Ward L R. A comparison of multiple drug resistance in salmonellas from humans and food animals in England and Wales, 1981 and 1990. Epidemiol Infect. 1993;111:189–197. doi: 10.1017/s0950268800056892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Twiddy N, Hopper D W, Wray C, McLaren I. Persistence of S. typhimurium in calf rearing premises. Vet Rec. 1988;122:399. doi: 10.1136/vr.122.16.399-b. [DOI] [PubMed] [Google Scholar]