Abstract
Emergence of SARS-CoV-2 variants and waning of vaccine/infection-induced immunity pose threats to curbing the COVID-19 pandemic. Effective, safe, and convenient booster vaccines are in need. We hypothesized that a variant-modified mucosal booster vaccine might induce local immunity to prevent SARS-CoV-2 infection at the port of entry. The beta-variant is one of the hardest to cross-neutralize. Herein, we assessed the protective efficacy of an intranasal booster composed of beta variant-spike protein S1 with IL-15 and TLR agonists in previously immunized macaques. The macaques were first vaccinated with Wuhan strain S1 with the same adjuvant. A total of 1 year later, negligibly detectable SARS-CoV-2-specific antibody remained. Nevertheless, the booster induced vigorous humoral immunity including serum- and bronchoalveolar lavage (BAL)-IgG, secretory nasal- and BAL-IgA, and neutralizing antibody against the original strain and/or beta variant. Beta-variant S1-specific CD4+ and CD8+ T cell responses were also elicited in PBMC and BAL. Following SARS-CoV-2 beta variant challenge, the vaccinated group demonstrated significant protection against viral replication in the upper and lower respiratory tracts, with almost full protection in the nasal cavity. The fact that one intranasal beta-variant booster administrated 1 year after the first vaccination provoked protective immunity against beta variant infections may inform future SARS-CoV-2 booster design and administration timing.
Keywords: SARS-CoV-2, beta variant, booster vaccine, adjuvanted subunit vaccine, intranasal mucosal vaccine
Significance Statement.
Emergence of SARS-CoV-2 variants and waning of vaccine-induced immunity pose threats to curbing the COVID-19 pandemic. Effective, safe, and convenient booster vaccines are in need. Here, we tested the immunogenicity and protective efficacy of a SARS-CoV-2 subunit booster using beta-variant S1 as antigen 1 year after the first vaccination. We found that administrated intranasally, the 1-year booster induced both systemic immunity and mucosal IgA responses against both the original strain and the beta variant. Most importantly, in addition to protection against lung infection, significant protection was observed in the nasal cavity after beta-variant viral challenge, suggesting the advantage of mucosal vaccination with beta-variant antigen. This study informs future SARS-CoV-2 booster design and administration timing.
Introduction
Emergence of novel SARS-CoV-2 variants of concern (VOC) threatens the efforts to curb the COVID-19 pandemic. Some variants demonstrated significantly reduced neutralization sensitivity to sera from convalescent and vaccinated individuals. A recent study assessed the cross-reactive neutralizing responses to different variants including B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.429 (Epsilon), B.1.526 (Iota), and B.1.617.2 (Delta) in mRNA-1273-vaccinated individuals, and found that the beta variant had the lowest antibody recognition (1). To date, the beta variant seems to be one of the most resistant variants to convalescent and vaccinated sera (1, 2). This variant was first detected in South Africa in October 2020 from samples collected at Eastern Cape Province in early August (3, 4). Since then, it quickly spread within South Africa and to the other parts of the world. By December 2020, it spread all over the world, accounted for 87% of viruses sequenced in South Africa, and became the dominant strain in Zambia (4, 5). Multiple mutations were found in this variant with K417N, E484K, and N501Y as key substitutions (6). It had 5-fold enhanced affinity to ACE2 compared to the original virus (7), and from several—to up to 10-fold (2, 8) reduction in neutralization ability in convalescent and vaccinated individuals. A total of two studies have shown that the beta variant can partially or completely escape three classes of therapeutically relevant antibodies and convalescent sera (9, 10).
Meanwhile, waning immunity after vaccination has led to a gradual decline of vaccine efficacy against SARS-CoV-2 infections (11–15). Recently, more SARS-CoV-2 breakthrough infections in vaccinated individuals, and resurgence of SARS-CoV-2 cases have been observed. Starting December 2021, the Omicron (B.1.1.529) variant, which shares the key mutations K417N, E484K/A, and N501Y with the beta variant, became the predominantly circulating variant (16). In addition to enhanced transmissibility, Omicron, similar to the beta variant, exhibited substantial immune escape (17, 18). Based on the previous experience with other coronaviruses and the current situation, extra boosters have been authorized. For the general population, it is anticipated that a booster, ideally targeting circulating viral variants, will be needed, when the immunity induced by the original vaccine cannot provide adequate protection against the circulating viral variants (19). Since a large number of individuals have been vaccinated with the vaccines comprised of antigens from the SARS-CoV-2 original Wuhan strain, data on immunogenicity and protective efficacy of a variant booster to vaccinees, who have previously received the original vaccines, would be urgently needed (20, 21). Recent studies have shown that intranasal administration of different platforms of SARS-CoV-2 vaccines induce protective immunity in preclinical animal models (22–26).
Herein, we performed a proof-of-concept study to test the immunogenicity and efficacy of an adjuvanted SARS-CoV-2 beta variant subunit booster in rhesus macaques that were vaccinated with the same vaccine platform except that the spike protein S1 was from the original Wuhan strain. We found that 1 year after the first vaccination, almost no detectable immunity was present in these macaques. However, an intranasal booster with the adjuvanted beta variant S1 subunit vaccine induced vigorous humoral and cellular immunity against both the original and beta variant antigens. Most importantly, secretory IgA responses against S1 from both the original Wuhan strain and the beta variant were detected in the nasal cavity, which was consistent with the almost full protection we observed against the beta variant in the nasal cavity after viral challenge. Whether this mucosal vaccine can protect against viral transmission, and whether the mucosal IgA response is responsible for the protection in the nasal cavity merits further investigation. Importantly, our data showed that the 1-year intranasal booster with beta variant S1 protein reinvigorated SARS-CoV-2-specific immune responses and led to significant protection against beta variant challenge. This study may provide important information regarding the timing of booster immunizations and the type of antigens included in the booster, and the value, and design of intranasal mucosal vaccines.
Results
Robust systemic and mucosal humoral responses against S1 from the original Wuhan strain and beta variant were elicited after intranasal variant booster
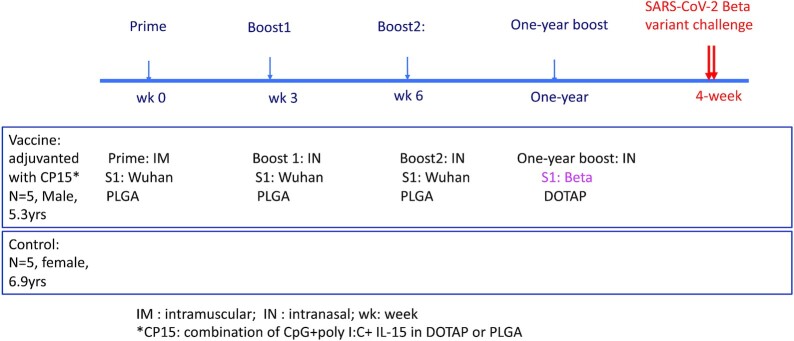
In this study, we took advantage of five Indian rhesus macaques that had been vaccinated 1 year earlier with the S1 protein from the original Wuhan strain (Table S1, Supplementary Material). The vaccine included 100 µg of S1 and CP15 adjuvant, which was composed of IL-15 and TLR agonists (CpG and Poly I: C) incorporated in PLGA nanoparticles as used in our previous study (22). The macaques were first primed with the vaccine intramuscularly (IM) at week 0, and then boosted with the same vaccine intranasally (IN) at weeks 3 and 6 (Figure 1). A total of 100 µg of S1 per dose was used based on our previous HIV and SARS-CoV-2 vaccine studies (22, 27). S1 with the sequence of the original Wuhan strain was used in the first three vaccinations. When evaluating the S1-specific IgG antibody responses, we found that this vaccine regimen induced a moderate level of humoral immune responses in serum and bronchoalveolar lavage (BAL) fluid (Figure 2a). Compared to the half-maximal binding titer of 25,209 induced by IM-primed and -boosted alum-adjuvanted subunit vaccine (22), the peak median serum half-maximal binding titer was only 945 (Figure 2a). Moreover, the vaccine-induced immunity also waned with time. After 1 year, the IgG responses in the vaccinated animals were comparable to those of the naïve controls (Figure 2a).
Fig. 1.
Schematic diagram of vaccination and viral challenge.
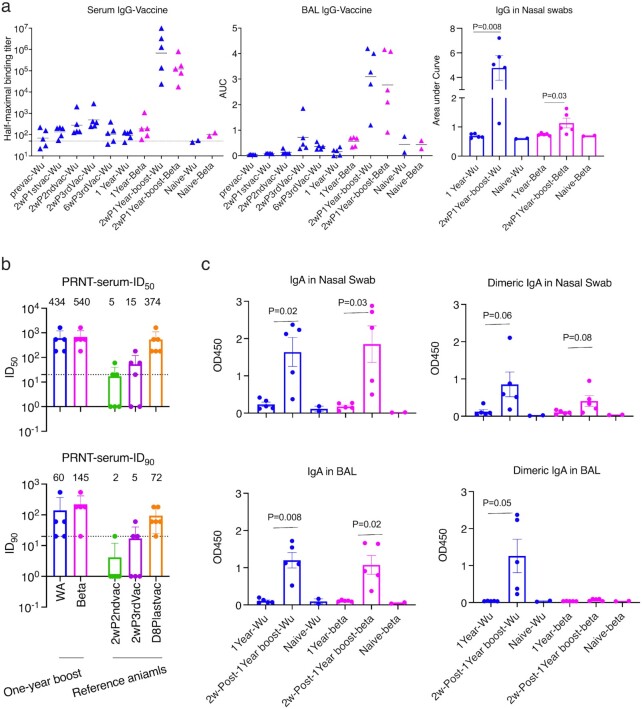
Fig. 2.
Humoral immune responses against SARS-CoV-2 spike protein 1 (S1) in vaccinated macaques. (a) The kinetics of S1-specific binding IgG titers in serum, BAL, and nasal swabs. Bars indicate geometric means of half-maximal binding titers and means of AUC. (b) PRNT titers in the serum samples of the vaccinated animals at 2-week after 1-year boost. PRNT data against Washington strain from six reference macaques were shown at the right. These macaques were primed IM with S1 adjuvanted with alum and boosted IN three times with S1 + CP15. Geometric mean ± geometric SD are shown. (c) S1-specific IgA and dimeric IgA responses in nasal swabs and BAL samples. Paired t tests were used to compare the humoral responses after the booster. WA: WA1/2020 D614G SARS-CoV-2 strain; Wu: Wuhan original strain; and Beta: B.1.351 variant. The dashed lines indicate the detection limits. Data are shown as mean ± SEM. Blue color indicates the S1 protein or the virus from Wuhan or WA strain, and magenta color indicates from beta variant.
We then gave the animals one intranasal booster with S1 from the beta variant adjuvanted with CP15 in 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) nanoparticles. After the booster, significant anamnestic responses were elicited. The Log (half-maximal binding titer) of serum IgG titer reached 5.83 for the original Wuhan strain, and 5.08 for the beta variant, compared to the highest IgG titer of 2.77 logs at 2 weeks post the third vaccination 1 year earlier (Figure 2a). The booster also led to the induction of a substantial increase of binding IgG responses against both the original Wuhan strain and the beta variant S1 in BAL and nasal swabs (NS; Figure 2a).
High titers of live virus neutralization antibody (Nab) responses against both Washtington WA1/2020 D614G SARS-CoV-2 (WA) strain and the beta variant were detected in the serum. The geometric mean titers (GMT) of Nab were 434 and 540 for ID50, and 60 and 145 for ID90 for the WA strain and the beta variant, respectively (Figure 2b). Given the fact that the beta variant has been one of the most difficult strains to neutralize so far (1), boosting with beta variant S1 might account for this improvement and suggest the potential benefit of switching antigens from the original WA strain to a variant. It is noteworthy that boosting with the variant S1 still induced a strong anamnestic response against the original priming Wuhan S1.
IgA and dimeric IgA responses in BAL and NS were also examined, as IgA, especially dimeric IgA, displays high binding avidity to pathogens, and thus is more potent at preventing mucosal pathogen infections (28, 29). Right before the 1-year booster, no S1 (original or beta variant)-specific IgA, or dimeric IgA responses were detected, and the antibody titers were comparable to the basal levels of naïve animals (Figure 2c). Consistent with IgG and neutralization responses, the 1-year booster enhanced IgA responses in NS and BAL samples with similar antibody titers against S1 from the original strain and the beta variant (Figure 2c). However, dimeric IgA responses against beta variant were not induced in BAL samples, whereas increased dimeric IgA responses were observed in BAL against the Wuhan strain and in NS against both strains (Figure 2c). Given the high variability in mucosal sample collection, we normalized the mucosal humoral responses to total IgG and total IgA and found that the normalization did not change the conclusions (Figure S1, Supplementary Material).
Since Omicron has become the predominant variant, we tested the binding titers against Omicron after the 1-year booster. No change or no more than a 3.3-fold decrease in titer for Omicron was observed compared to original strain in the serum and mucosal samples (Figure S2, Supplementary Material).
Overall, our results showed that the 1-year booster induced robust S1-specific antibody responses in serum and BAL, including potent neutralizing antibody (Nab) responses in peripheral blood. Most importantly, mucosal IgA responses were induced in NS and BAL that were comparable against both the original priming Wuhan strain and the beta variant, except dimeric IgA responses against beta variant in BAL.
Variant S1-specific cellular responses were induced after the 1-year booster
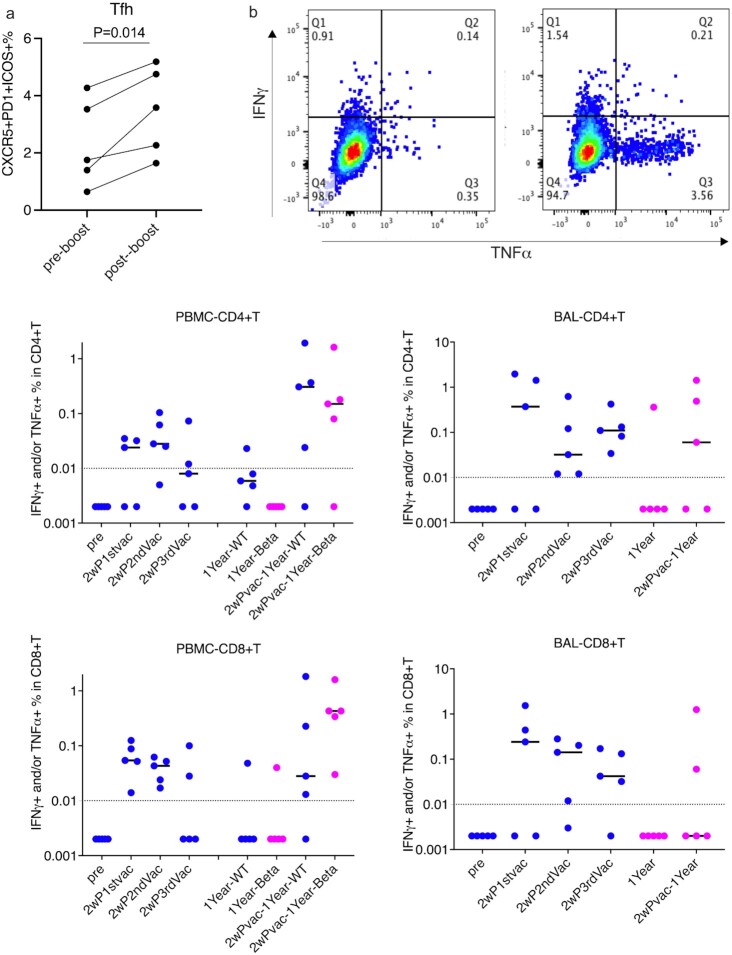
As circulating T follicular helper cells (Tfh) play a significant role in orchestrating humoral responses (30, 31), we measured the Tfh frequencies in the peripheral blood mononuclear cells (PBMC)s. The 1-year booster significantly increased the frequencies of CXCR5+PD1+ ICOS+ CD4+ Tfh in the PBMCs (Figure 3a). The vaccine-induced S1-specific T cell responses in PBMC and BAL samples of the vaccinated animals were evaluated by intracellular cytokine staining. S1-specific type 1 helper T cell responses (Th1) and CD8+ T cell that secrete tumor necrosis factor (TNF)-α, and/or interferon (IFN)-γ were induced after the first vaccination (Figure 3b). Though the responses were persistent in most of the vaccinated animals, no further enhancement of the responses was observed after the second and third vaccinations. We did not detect a significant number of IL-2-producing cells. For CD8+ T cell responses, especially the responses in PBMC, we observed a declining trend with each vaccination (less so in BAL). In any case, the responses waned to under the detection limit in most of the animals after 1 year. After the administration of the 1-year beta-variant booster, the S1-specific CD8+ T cell responses were successfully recalled in all five PBMC samples and CD4+ responses in four out of five (Figure 3b). Even though the route of the 1-year booster was intranasal, S1-specific CD4+ T cells were induced only in 3 BAL samples, and CD8+ T cells in only two. One possibility could be the migration of antigen-specific T cell to the nasal cavity.
Fig. 3.
T cell responses against SARS-COV-2 spike protein 1 (S1) in PBMC and BAL samples of the vaccinated macaques. (a) The frequencies of CXCR5+PD1+ ICOS+ CD4+ Tfh in the PBMCs before and after 1-year booster. (b) The frequencies of IFNγ and/or TFNα-producing CD4+ and CD8+ T cells were stained and measured after stimulation with S1 either from Wuhan strain or from beta variant for 18 h in PBMC and BAL samples. The upper plots showed a representative cytokine gating of CD4+T cells in medium-only control (left) and S1-stimulated (right) BAL sample from the same vaccinated animal. Dashed lines indicate the detection limits. Bars indicate medians. Blue color indicates the S1 protein from Wuhan strain, and magenta color indicates from beta variant.
As the frequencies of antigen-specific T cell responses were low, we further assessed the kinetics of total Th1 and Th2 subsets after stimulation with Phorbol 12-myristic 13-acetate (PMA) and ionomycin. There were no significant alterations after the first three vaccinations in the prior year (Figure S1, Supplementary Material). However, the 1-year boost resulted in a slight increase of Th1 responses in PBMC while the Th2 responses did not change (Figure S3, Supplementary Material).
Vaccinated animals demonstrated significant protection in BAL, and NS against SARS-CoV-2 beta variant replication
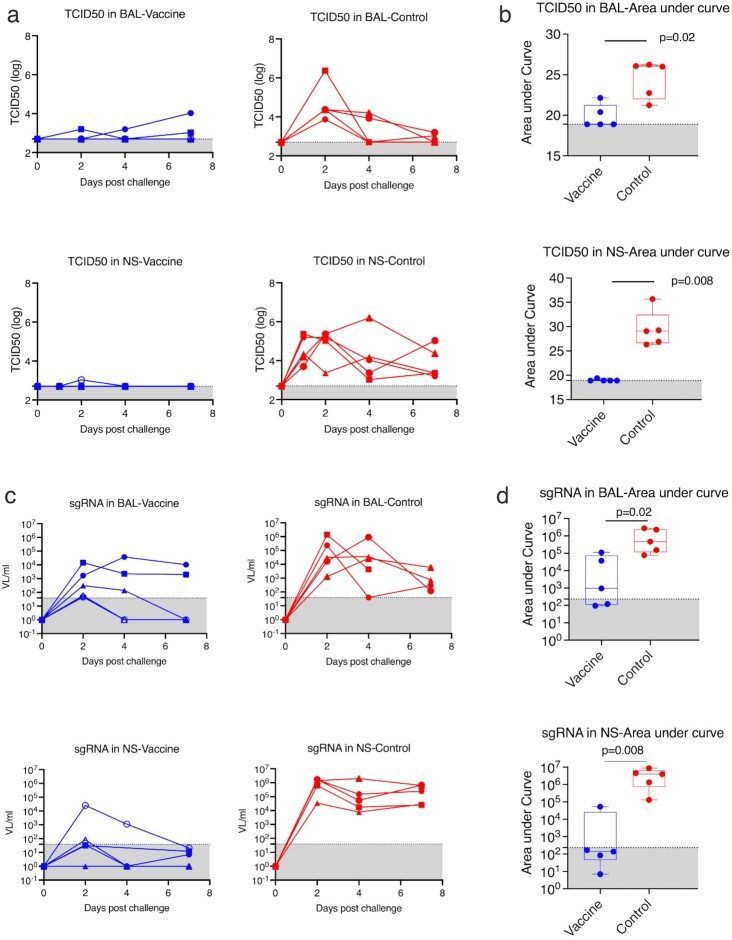
To test the protective efficacy against SARS-CoV-2 beta variant, five vaccinated and five naïve macaques were challenged with 1.0 × 10^5 TCID50 SARS-CoV-2 beta variant (isolate beta variant B.1.351, in-house generated stock from BEI Resources, NR-54974) through intranasal (1 mL) and intratracheal (1 mL) routes 4 weeks after the last vaccination. Viral tissue culture infectious dose 50 titers (TCID50) were measured in the collected NS and lung BAL samples. Replicating viruses were detected in both NS and BAL samples of all five naïve animals, indicating that the viral inoculation was successfully delivered and propagated in the upper and lower airways (Figure 4a). It is worth mentioning that the inoculation of SARS-CoV-2 beta variant led to prolonged detection of replicating virus in the nasal turbinate of the naïve animals. High levels of viral replication were present in all five naïve animals at day 7 postvirus challenge. In contrast, the vaccinated animals demonstrated almost full protection in NS: only one animal showed a small blip at day 2 postviral challenge, while four other vaccinated animals were free of replicating virus during the 7-days postchallenge period (Figure 4a). The vaccinated group showed significant reduction of viral replication in both nasal turbinate and lungs compared to naïve controls, based on the area under curves (AUC) over all time points (Figure 4b). Since vaccine-induced antibody was present in the mucosal samples and had the potential to interfere with the TCID50 assay (although TCID50 would still reflect viable (non-neutralized) virus in those fluids), we measured the subgenomic viral RNA (sg RNA) using real-time PCR. With this more sensitive method, more data-points turned positive; however, the overall conclusions were still valid (Figure 4c and d). A future transmission study is needed to test whether this mucosal booster can prevent forward transmission to other animals.
Fig. 4.
Viral burden in the NS and BAL samples after SARS-CoV-2 beta variant intranasal and intratracheal challenges. (a) TCID50 titer of the viral burdens in NS and BAL samples of individual animals (n = 5 in the vaccine group and n = 5 in the control group). (b) AUC over time after challenge was calculated for each animal, representing total viral burdens. The total viral burdens were compared between vaccine and control groups in NS and BAL. (c) SARS-CoV-2 sgRNA in mucosal samples of individual animals. (d) AUC of sgRNA for vaccinated and naïve groups. Dashed lines indicate the detection limits. Gray areas show the undetectable areas. Box and whiskers with min to max were shown in the graph.
Histopathology in the lungs after viral infection suggested protection in the lungs of vaccinated animals
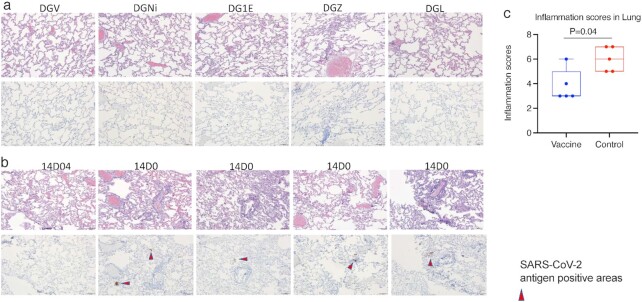
As reported in the previous study (22), the mucosal vaccine is safe. The vaccinations were well-tolerated. Throughout the whole course of this study, we did not observe any adverse effects in the vaccinated animals. When the animals were necropsied on day 7, sections of lung were evaluated immunohistochemically for SARS-CoV-2 virus antigen and histologically for the presence of SARS-CoV-2-associated inflammation. None of the five vaccinated animals demonstrated immunoreactivity to viral antigens, while virus antigens were detected in the lung sections of the four out of five animals in the control group (Figure 5a and b). Predominantly perivascular to interstitial inflammation was observed in the control group. An inflammation score was given to each animal blindly by a certified pathologist based on the evaluation of lung infiltration collected at the time of necropsy at day 7 post-SARS-CoV-2 challenges (Table S2, Supplementary Material). As beta variant led to persistent viral replication in the lungs of both naïve and vaccinated animals, the inflammation scores even in the vaccinated animals were not zero at day 7 postinfection, which is consistent with the viral load data (Figure 4a). However, the inflammation score was significantly more severe in the control group than in the vaccinated group (Figure 5c).
Fig. 5.
Histopathology in the lungs at day 7 post-SARS-CoV-2 challenge. H&E and immunohistochemistry to detect SARS-COV-2 antigens were performed in the vaccinated (a) and naïve (b) animals. The upper rows of (a) and (b) were H&E staining, while the lower rows of (a) and (b) were immunohistochemistry of SARS-CoV-2 detection. All images 10x (scale bar = 100 um). (c) inflammation scores in the lung were compared between the vaccinated and naïve groups. Mann–Whitney test was used for comparison. Box and whiskers with min to max were shown in the graph.
Discussion
An additional booster vaccine is needed to curb the resurgence of SARS-CoV-2 cases. We demonstrated here that the 1-year beta variant mucosal booster given IN elicited high quality immune responses and mediated protection against subsequent SARS-CoV-2 beta variant viral challenge in rhesus macaques. Notably, the protection in the upper respiratory tract seemed to be better than that in the lower respiratory tract, which is different from most of the systemic vaccines (32–35). The protection against viral replication in the nasal cavity is especially encouraging, indicating its potential to prevent viral spread and transmission. During SARS-CoV-2 infection, the nasal mucosa is usually the first site of viral replication, so the local immunity induced by vaccination might be able to abort viral replication here before it disseminates systemically and may also prevent spread to other individuals. Indeed, we found that high titers of mucosal IgA responses against both original and variant spike proteins were induced in the nasal mucosa, which might account for the efficient clearing of the virus in situ. These findings show the promise of a nasal mucosal vaccine as a booster rather than another systemic (IM) vaccine dose.
Waning immunity over time after vaccination/infection is contributing significantly to the resurgence of SARS-CoV-2 cases (14, 15, 36). Though the immune correlates of protection have not been fully established, Nab responses are believed to be one of the major protective mechanisms (37–39). To evaluate the durability, one study found that the half-life of Nab was biphasic, with a rapid initial decline over 61 days, and then a more gradual tapering after the first 2 months out to 104 days (40), while the other study found that Nab exhibited a biphasic decay with an extended half-life of > 200 days (41). Though prolonged humoral and cellular immunity up to 10 months or 1 year has been reported in SARS-CoV-2-convalescent individuals (42–44), the durability of the protective immunity against SARS-CoV-2 infection remains unknown.
The emergence of SARS-CoV-2 VOC might partially account for the reported decreased vaccine effectiveness after 6 months (12, 16, 45). These variants either have high infectious potency or evade the immunity induced by SARS-CoV-2 infection or vaccination. The beta variant was one of the variants that has substantial immune evasive capacity (1, 17, 18). In this study, we have switched the S1 from original Wuhan strain to that of the beta variant, which led to successful elicitation of systemic and mucosal immune responses against both the original strain and the beta variant, and most importantly mediated protection against subsequent SARS-CoV-2 beta variant challenge. Incorporating S1 from the beta variant into the booster vaccine might account for the observed robust protection. Interestingly, we also observed the discrepancy in dimeric IgA between the original and beta strains. This suggested that the antibody repertoires in the nasal cavity and lung were different. One possibility could be that the booster-induced dimeric IgA antibody responses in the nasal cavity had better cross-reactivity to the beta variant than those in the lung. This could be achieved through induction of non-RBD antibody repertoire, which was less affected by the variant. Another possibility was the de nova induction of beta-specific dimeric IgA responses in the nasal cavity, but not in the lung. In line with this, the IgG responses in the nasal cavity were different from those in the BAL and serum. It has been shown by another study that serum IgG and BAL IgG correlated strongly with each other, suggesting BAL IgG derives from serum (46). Nasal IgG, on the other hand, could be induced at the mucosal site. The antibody repertoire in the nasal cavity, where the vaccines were directly instilled, was more imprinted by the first several original strain vaccine doses (i.e. “original antigenic sin”).
One caveat of this study was the unbalanced sex and age distribution of the vaccinated (all male) and control (all female) animals, which is due to the difficulty in finding the matched control animals during the pandemic. As the females tend to have innately slightly higher responses than males, considering the sex imbalance in control (all female) and vaccinated (all male) groups, the difference in inflammation scores in the lungs at the time of necropsy may not be purely due to the vaccine. On the other hand, more importantly, the greater protective response in the vaccinees would not likely be a sex difference because that would be expected to have the opposite effect (47). The vaccinated animals were 5.3 years (64 months) old when they were challenged by SARS-CoV-2 virus, while the control animal were 6.9 years old (83 months). Though there were 1.6 years (19 months) of age difference between the two groups, both groups were considered young adult macaques (5 to < 20 years) (48, 49).
A dramatic increase in antibody titers after the 1-year booster was observed (more than 3 log of increase compared to the highest titers 1 year before for serum IgG titers). This is consistent with what we have found in a previous study, where the booster at 4 months induced much higher quality SARS-CoV-2 specific immune responses than the booster at 3 weeks did (22). This could be due to the DOTAP nanoparticles with beta S1 incorporated in the 1-year booster rather than the PLGA nanoparticles with Wuhan S1 used before. Another possibility is that the longer interval between the booster and the previous vaccinations enhances the immune responses. Similar phenomena were reported in AstraZeneca (AZ) and inactivated vaccine trials, as well as in the standard hepatitis B viral vaccine regimen. In the AZ trial, a longer prime-boost interval (> 12 weeks) led to higher vaccine efficacy compared to shorter interval (< 6 weeks) (50). In an inactivated vaccine trial, 6 or more months between the second and third vaccinations also induced a remarkable increase in antibody levels compared to a 4-week interval (51). Thus, these studies should be taken into consideration when deciding the timing of an additional booster.
The CP15 adjuvanted vaccine described here was not very effective as a prime vaccine. It did not induce robust immune responses compared to an alum adjuvanted vaccine (22). A total of 1 year after the first vaccination, no virus-specific humoral or cellular immunity was detected. Nevertheless, the 1-year booster elicited high quality immune responses, and mediated protection against subsequent beta variant challenge, which suggested that the vaccinations in the prior year generated persistent SARS-CoV-2 specific immune memory. The specific immune memory may include antigen-specific long-lived B memory cells in bone memory and/or innate cell-mediated trained immunity (52), which warrants future investigation. Though the humoral and cellular immune responses waned to undetectable levels after 1 year, the immune memory persisted, which facilitated the later recall responses, when boosted. Moreover, our data suggest that a weaker variant-modified booster vaccine might be sufficient to induce protective immunity in previously vaccinated hosts. These findings may help guide future prime-boosting regimens for COVID-19.
Materials and Methods
Animals. A total of 10 Indian-origin adult male rhesus macaques (Macaca mulatta), 3- to 8-y-old, were enrolled in the study. The animals tested seronegative for cercopithecine herpesvirus 1, SIV, simian type-D retrovirus, simian T lymphotropic virus type 1, and SARS-CoV-2 prior to study assignment.
Vaccine design and inoculation. A total of five previously primed male macaques were included in the vaccine group. The average age of these vaccinated macaques was 64 months (5.3 years) old. Since during the COVID-19 pandemic, we were not able to obtain matched males, we had to include five female macaques in the SARS-CoV-2-naive control group. The average age of the control macaques was 83 months (6.9 years) old. If anything, female macaques would be expected to make stronger immune responses than males, so the sex difference would not account for any greater immune response in the vaccinated male animals (47). The five naïve control animals had been exposed to HIV envelope protein/glycopeptide vaccination more than 1 year before, but had not been infected or challenged. The five macaques in the vaccine group were primed at Week 0 (administrated IM) and boosted at Week 3 (administered IN) and Week 6 (administered IN) with SARS-CoV-2 S1 protein (WA strain) with alum or CP15 adjuvant in PLGA nanoparticles. The CP15 adjuvant was composed of 200 µg per dose of D-type CpG oligodeoxynucleotide, 1 mg per dose of Poly I: C (InvivoGen), and 200 µg per dose of recombinant human IL-15 (Sino Biological). A total of 1 year later, a boost was given to the remaining five animals with S1 protein from the beta variant adjuvanted with CP15. 100 µg of recombinant SARS-CoV-2 (2019-nCoV) spike S1 protein (Cat: 40591-V08H and 40591-V08H10, Sino Biological, endotoxin level: < 0.001 U/µg) was used per dose. S1 protein and CP15 were formulated in nanoparticles in PLGA (Alchem Laboratories) for the first two doses and the last (1-year) boost was in DOTAP (100 µL per dose; Roche). For immunization, the CP15 adjuvanted vaccine was given either IM in 1 mL of volume, or IN in a volume of 50 µL per nostril, while the animals were anesthetized. After vaccination, blood, NS, and BAL fluid samples were collected at the times noted and analyzed.
NS and BAL sample collection. Nasal secretions were collected and stored at −80°C after either using cotton-tipped swabs and then in 1 mL of PBS buffer containing 0.1% BSA, 0.01% thimerosal, and 750 Kallikrein inhibitor units of aprotinin (27) for prechallenge stage, or using Copan flocked swabs and in virus transport medium for postchallenge stage. BAL samples were collected as described before (22). Briefly, while the animals were under anesthesia, up to 10 mL/kg of sterile saline were instilled into and sucked out of the lungs. Large pieces were removed by passing through a 100-µm cell strainer (prechallenge). The BAL fluid was collected after centrifugation and stored at −20°C for analysis. The BAL cells were washed with R10 medium (RPMI-1640 with 10% fetal bovine serum) before subsequent treatment or cryopreservation.
ELISA assay to detect S1-specific antibody responses. The BAL samples were concentrated using Amicon Ultra centrifugal filter units (10 kDa cutoff, Millipore Sigma), and the total IgG and IgA were determined using the Rhesus Monkey IgG-UNLB (Southern Biotech), and the Monkey IgA ELISA development kit (HRP; MabTech), respectively, following the manufacturer's protocol as described before (22). NS samples were put into 1 mL of 1XPBS buffer containing 0.1% BSA, 0.01% thimerosal, and 750 Kallikrein inhibitor units of aprotinin (Sigma) and stored at −80°C. NS were thawed, and the recovered solution was passed through a 5-µm PVDF microcentrifugal filter unit (Millipore, Billerica, MA). The buffer flow-through was collected and stored at −20° until analysis. ELISA assays were run as described before (22), and the detail protocol was in the supplemental material. Total IgG and IgA in Bal and NS have been measured for normalization as previously described (22).
Plaque reduction neutralization test (PRNT). The PRNT was performed in duplicate as described before (22). Vero E6 cells (ATCC, cat. no. CRL-1586), and 30 pfu challenge titers of SARS-CoV-2 virus USA-WA1/2020 strain or Vero TMPRSS2 cells (obtained from Dr. Adrian Creanga and Barney Graham, VRC, NIAID, Bethesda, MD) and same titer of the beta variant (B.1.351, SRA strain) was used to test the PRNT titers against the WA or beta variant of SARS-CoV-2 (53). Serum samples of 3-fold serial dilution starting from 1:20, and up to final dilution of 1:4,860 were incubated with 30 pfu of SARS-CoV-2 virus for 1 h at 37 °C. The serial dilutions of virus–serum mixtures were then added onto Vero E6 cell monolayers in cell culture medium with 1% agarose for 1 h at 37 °C with 5% CO2. The plates were fixed and stained after 3 days of culture. Specifically, after fixation, the methanol is discarded, and the monolayers stained with 250 µL per well of 0.2% crystal violet (20% MeOH and 80% dH2O) for 30 min at room temperature. The plates are finally washed once with PBS or dH2O and let dry for ∼15 min. The plaques in each well are recorded and the IC50 and IC90 titers are calculated based on the average number of plaques detected in the virus control wells. A control (rabbit) reference serum with established titer (5,400 IC50) is included in each assay set-up to serve as an internal positive control. For optimal assay performance, the IC50 value of the positive control should test within the nearest dilution above and below the expected value, i.e. between 1,800 and 16,200. ID50 and ID90 were calculated as the highest serum dilution resulting in 50% and 90% reduction of plaques, respectively.
Intracellular cytokine staining assay. SARS-CoV-2-specific T cells were measured from BAL and PBMC samples by flow cytometric intracellular cytokine analysis, as previously described (22, 54, 55), and the detailed protocol is in the supplemental material. The antigen-specific T cell responses were reported as the frequencies of cytokine-positive cells in the samples stimulated with S1 overlapping peptide pools (PepTivator SARS-CoV-2 Prot_S1, and PepTivator SARS-CoV-2 Prot_S B.1.351 Mutation Pool/WT reference Pool, from Milteny Biotech Inc.) minus those in the medium-only control from the same animal at each time-point. If the medium control had a higher frequency of cytokine-positive cells than that of the S1 protein-stimulated sample in the matched animal, an arbitrary number of “0.001” was assigned to each cytokine as a negative on the log scale.
SARS-CoV-2 beta variant viral challenge. A total of 4 weeks after the 1-year boost, five vaccinated and five control animals were challenged with 1 × 10^5 TCID50 SARS-CoV-2 virus beta variant (seed stock obtained from BEI Resources; NR-54974, B.1.351, SRA strain). The challenge stock was grown in Calu-3 cells and was deep sequenced, which confirmed the expected sequence identity with no mutations in the Spike protein greater than > 2.5% frequency and no mutations elsewhere in the virus at > 13% frequency. The same beta variant stock was used in the earlier macaque challenge study at the same facility (32). To make sure that the virus was delivered to both upper and lower airway simultaneously, the diluted virus was given IN and intratracheally, each route with 1 mL (0.5 mL for each nostril). NS and BAL fluid samples were collected after challenge to measure the viral load.
TCID50 assays to measure viral loads. Vero TMPRSS2 cells (obtained from the Vaccine Research Center-NIAID) were plated at 25,000 cells/well in DMEM + 10% FBS + Gentamicin and the cultures were incubated at 37°C, 5.0% CO2. Cells should be 80% to 100% confluent the following day. Medium was aspirated and replaced with 180 µL of DMEM + 2% FBS + gentamicin. A total of 20 µL of sample was added to top row in quadruplicate and mixed using a P200 pipettor five times. Using the pipettor, 20 µL was transferred to the next row, and repeated down the plate (columns A–H) representing 10-fold dilutions. The tips were disposed for each row and repeated until the last row. Positive (virus stock of known infectious titer in the assay) and negative (medium only) control wells were included in each assay set-up. The plates were incubated at 37°C, 5.0% CO2 for 4 days. The cell monolayers were visually inspected for cytopathic effect (CPE). Noninfected wells will have a clear confluent cell layer while infected cells will have cell rounding. The presence of CPE was marked on the lab form as a + and absence of CPE as −. The TCID50 value was calculated using the Read–Muench formula. For optimal assay performance, the TCID50 value of the positive control should test within 2-fold of the expected value.
Subgenomic viral assay. SARS-CoV-2 RNA levels were monitored by reverse transcription PCR by Duke Human Vaccine Institute-Immunology and Virology Quality Assessment Center using Qiagen QIAsymphony DSP Virus/Pathogen Midi Kit (96)/QIAgility/Applied Biosystems QuantStudio 3 Real-Time PCR System as previously described (22). The limit of quantification (LOQ) for this assay is approximately 31 RNA cp/mL (1.49 log10) with 800 µL of sample.
Histopathology and immunohistochemistry of lung sections. A total of 7 days after SARS-CoV-2 viral challenge all the animals were necropsied and the lung tissue specimens were collected, fixed, processed, and embedded in paraffin blocks and sectioned at a thickness of 5 µm as described in the previous study (22). Briefly, hematoxylin and eosin (H&E) sections were examined under light microscopy and scored by a board-certified veterinary pathologist, who was blind to the groups. A rabbit polyclonal SARS-CoV-2 antibody (GeneTex) was used immunohistochemically to stain for the presence of SARS-CoV-2 virus antigen. An Olympus BX51 brightfield microscope was used, and representative photomicrographs were captured using an Olympus DP73 camera.
Statistical analysis. Prism version 8 (Graph Pad) was used for statistical analyses. AUC values were calculated for viral load, and Mann–Whitney and paired t tests were used for group comparisons as shown in the figures. A P-value less than 0.05 was considered significant, and all statistical tests were 2-tailed.
Study approval. Vaccination was performed at the National Institutes of Health NCI Animal Facility, Bethesda, MD, an American Association for the Accreditation of Laboratory Animal Care (AAALAC)-accredited facility with PHS Approved Animal Welfare Assurance (Assurance ID A4149-01). Animal Protocol No. VB-037 was approved by the NCI Animal Care and Use Committee (ACUC) to conduct the study. A total of 2 weeks before viral challenge, all 10 animals were moved to a qualified BSL3 facility at BIOQUAL, Inc.. The SARS-CoV-2 viral challenge study was approved and performed under BIOQUAL's IACUC approved Protocol No. 20–107.
Supplementary Material
ACKNOWLEDGEMENTS
The NIH nonhuman primate reagent resource provided mouse antirhesus J chain antibody. The staff of the Laboratory Animal Sciences Program, Frederick National Laboratory for Cancer Research, provided technical support and animal care. The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-53780, and Isolate beta variant B.1.351 and NR-54974. We thank Shelby O'Connor for sequencing of the beta variant challenge stock, Adrian Creanga and Barney Graham (VRC, NIAID, Bethesda, MD) for the Vero TMPRSS2 cells. We would like to thank Logan Eiermann, Naomi Williamson, and Nicole De Naeyer from the Duke Human Vaccine Institute for the sgRNA measurements. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Notes
Competing Interest: The authors declare no competing interest.
Contributor Information
Yongjun Sui, Vaccine Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
Jianping Li, Vaccine Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
Hanne Andersen, BIOQUAL Inc., Rockville, MD 20850, USA.
Roushu Zhang, Department of Chemistry and Biochemistry, University of Maryland, College Park, MD 20742, USA.
Sunaina K Prabhu, Department of Chemistry and Biochemistry, University of Maryland, College Park, MD 20742, USA.
Tanya Hoang, Vaccine Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
David Venzon, Biostatistics and Data Management Section, Center of for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
Anthony Cook, BIOQUAL Inc., Rockville, MD 20850, USA.
Renita Brown, BIOQUAL Inc., Rockville, MD 20850, USA.
Elyse Teow, BIOQUAL Inc., Rockville, MD 20850, USA.
Jason Velasco, BIOQUAL Inc., Rockville, MD 20850, USA.
Laurent Pessaint, BIOQUAL Inc., Rockville, MD 20850, USA.
Ian N Moore, Infectious Disease Pathogenesis Section, National Institute of Allergy and Infectious Diseases, Rockville, MD 20852, USA.
Laurel Lagenaur, Vaccine Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
Jim Talton, Alchem Laboratories, Alachua, FL 32615, USA.
Matthew W Breed, Laboratory Animal Sciences Program, Frederick National Laboratory for Cancer Research, Rockville, MD 20850, USA.
Josh Kramer, Laboratory Animal Sciences Program, Frederick National Laboratory for Cancer Research, Rockville, MD 20850, USA.
Kevin W Bock, Infectious Disease Pathogenesis Section, National Institute of Allergy and Infectious Diseases, Rockville, MD 20852, USA.
Mahnaz Minai, Infectious Disease Pathogenesis Section, National Institute of Allergy and Infectious Diseases, Rockville, MD 20852, USA.
Bianca M Nagata, Infectious Disease Pathogenesis Section, National Institute of Allergy and Infectious Diseases, Rockville, MD 20852, USA.
Hyoyoung Choo-Wosoba, Biostatistics and Data Management Section, Center of for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
Mark G Lewis, BIOQUAL Inc., Rockville, MD 20850, USA.
Lai-Xi Wang, Department of Chemistry and Biochemistry, University of Maryland, College Park, MD 20742, USA.
Jay A Berzofsky, Vaccine Branch, Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD 20892, USA.
Funding
This work was supported by funding from the Intramural Research Program, National Institutes of Health, National Cancer Institute, Center for Cancer Research funding Z01 BC-011941 to J.A.B.
Authors' Contributions
Y.S. and J.A.B. designed and interpreted the project. Y.S. and J.L. processed the samples and ran the cellular assays. J.L., T.H., R.Z., S.P., and Y.S. performed the antibody assays. L.P. performed the PRNT assays. J.T. and Y.S. prepared the PLGA nanoparticle, and other vaccines. I.M., K.B., M.M., and B.M.N. performed pathology. H.A., A.C., R.B., E.T., J.V., M.B., and J.K. led the animal studies. Y.S., J.A.B., H.A., L.L., M.L., and L.W. participated in the study design and interpreted the experiments. D.V., H.C., and Y.S. performed the statistical analyses. Y.S. and J.A.B. wrote the manuscript with input from all the coauthors.
Data Availability
All data is included in the manuscript and/or supporting information.
References
- 1. Pegu A, et al. . 2021. Durability of mRNA-1273 vaccine-induced antibodies against SARS-CoV-2 variants. Science. 373:1372–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang P, et al. . 2021. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 593:130–135. [DOI] [PubMed] [Google Scholar]
- 3. Tegally H, et al. . 2021. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 592:438–443. [DOI] [PubMed] [Google Scholar]
- 4. Abdool Karim SS, de Oliveira T. 2021. New SARS-CoV-2 variants - clinical, public health, and vaccine implications. N Engl J Med. 384:1866–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mwenda M, et al. . 2021. Detection of B.1.351 SARS-CoV-2 variant strain - Zambia, December 2020. Morb Mortal Wkly Rep. 70:280–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Singh J, et al. . 2021. Structure-function analyses of new SARS-CoV-2 variants B.1.1.7, B.1.351 and B.1.1.28.1: clinical, diagnostic, therapeutic and public health implications. Viruses. 13:439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ramanathan M, Ferguson ID, Miao W, Khavari PA. 2021. SARS-CoV-2 B.1.1.7 and B.1.351 spike variants bind human ACE2 with increased affinity. Lancet Infect Dis. 21:1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Edara VV, et al. . 2021. Infection- and vaccine-induced antibody binding and neutralization of the B.1.351 SARS-CoV-2 variant. Cell Host Microbe. 29:516–521.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wibmer CK, et al. . 2021. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 27:622–625. [DOI] [PubMed] [Google Scholar]
- 10. Weisblum Y, et al. . 2020. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 9:e61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goldberg Y, et al. . 2021. Waning immunity after the BNT162b2 vaccine in Israel. N Engl J Med. 385:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thomas SJ, et al. . 2021. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine through 6 months. N Engl J Med. 385:1761–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Canaday DH, et al. . 2021. Significant reduction in vaccine-induced antibody levels and neutralization activity among healthcare workers and nursing home residents 6 months following coronavirus disease 2019 BNT162b2 mRNA vaccination. Clin Infect Dis. 73:35174389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Levin EG, et al. . 2021. Waning immune humoral response to BNT162b2 Covid-19 vaccine over 6 months. N Engl J Med. 385:e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tartof SY, et al. . 2021. Effectiveness of mRNA BNT162b2 COVID-19 vaccine up to 6 months in a large integrated health system in the USA: a retrospective cohort study. Lancet North Am Ed. 398:1407–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Viana R, et al. . 2022. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. 603:679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dejnirattisai W, et al. . 2022. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 185:467–484.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wilks SH, et al. . 2022. Mapping SARS-CoV-2 antigenic relationships and serological responses. bioRxiv. DOI: 10.1101/2022.01.28.477987. [DOI] [PubMed] [Google Scholar]
- 19. Krause PR, et al. . 2021. Considerations in boosting COVID-19 vaccine immune responses. Lancet North Am Ed. 398:1377–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu K, et al. . 2021. Variant SARS-CoV-2 mRNA vaccines confer broad neutralization as primary or booster series in mice. Vaccine. 39:7394–7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Choi A, et al. . 2021. Safety and immunogenicity of SARS-CoV-2 variant mRNA vaccine boosters in healthy adults: an interim analysis. Nat Med. 27:2025–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sui Y, et al. . 2021. Protection against SARS-CoV-2 infection by a mucosal vaccine in rhesus macaques. JCI Insight. 6:e148494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hassan AO, et al. . 2020. A sngle-dose intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell. 183:169–184.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu S, et al. . 2020. A single dose of an adenovirus-vectored vaccine provides protection against SARS-CoV-2 challenge. Nat Commun. 11:4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ku MW, et al. . 2021. Intranasal vaccination with a lentiviral vector protects against SARS-CoV-2 in preclinical animal models. Cell Host Microbe. 29, 236–249.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feng L, et al. . 2020. An adenovirus-vectored COVID-19 vaccine confers protection from SARS-COV-2 challenge in rhesus macaques. Nat Commun. 11:4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sui Y, et al. . 2019. Mucosal vaccine efficacy against intrarectal SHIV is independent of anti-Env antibody response. J Clin Invest. 129:1314–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Johansen FE, Kaetzel CS. 2011. Regulation of the polymeric immunoglobulin receptor and IgA transport: new advances in environmental factors that stimulate pIgR expression and its role in mucosal immunity. Mucosal Immunol. 4:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cerutti A, Chen K, Chorny A. 2011. Immunoglobulin responses at the mucosal interface. Annu Rev Immunol. 29:273–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schmitt N, Bentebibel SE, Ueno H. 2014. Phenotype and functions of memory Tfh cells in human blood. Trends Immunol. 35:436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iwamoto Y, Ueno H. 2022. Circulating T follicular helper subsets in human blood. Methods Mol Biol. 2380:29–39. [DOI] [PubMed] [Google Scholar]
- 32. Yu J, et al. . 2021. Protective efficacy of Ad26.COV2.S against SARS-CoV-2 B.1.351 in macaques. Nature. 596:423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Corbett KS, et al. . 2020. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N Engl J Med. 383:1544–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mercado NB, et al. . 2020. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 586:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Doremalen N, et al. . 2020. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 586:578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Keehner J, et al. . 2021. Resurgence of SARS-CoV-2 infection in a highly vaccinated health system workforce. N Engl J Med. 385:1330–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yu J, et al. . 2020. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 369:806–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sui Y, Bekele Y, Berzofsky JA. 2021. Potential SARS-CoV-2 immune correlates of protection in infection and vaccine immunization. Pathogens. 10:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Corbett KS, et al. . 2021. Immune correlates of protection by mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. Science. 373:eabj0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu X, et al. . 2021. Dynamics of neutralizing antibody responses to SARS-CoV-2 in patients with COVID-19: an observational study. Signal Transduct Target Ther. 6:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cohen KW, et al. . 2021. Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell Rep Med. 2:100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alsayb MA, et al. . 2021. Prolonged humoral and cellular immunity in COVID-19-recovered patients. Saudi J Biol Sci. 28:4010–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dan JM, et al. . 2021. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 371:eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Feng C, et al. . 2021. Protective humoral and cellular immune responses to SARS-CoV-2 persist up to 1 year after recovery. Nat Commun. 12:4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Levine-Tiefenbrun M, et al. . 2021. Viral loads of Delta-variant SARS-CoV-2 breakthrough infections after vaccination and booster with BNT162b2. Nat Med. 27:2108–2110. [DOI] [PubMed] [Google Scholar]
- 46. Fahy RJ, Diaz PT, Hart J, Wewers MD. 2001. BAL and serum IgG levels in healthy asymptomatic HIV-infected patients. Chest. 119:196–203. [DOI] [PubMed] [Google Scholar]
- 47. Musich T, et al. . 2020. A prime/boost vaccine regimen alters the rectal microbiome and impacts immune responses and viremia control post-simian immunodeficiency virus infection in male and female rhesus macaques. J Virol. 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Didier ES, Sugimoto C, Bowers LC, Khan IA, Kuroda MJ. 2012. Immune correlates of aging in outdoor-housed captive rhesus macaques (Macaca mulatta). Immun Ageing. 9:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Simmons HA. 2016. Age-associated pathology in rhesus macaques (Macaca mulatta). Vet Pathol. 53:399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Voysey M, et al. . 2021. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet North Am Ed. 397:881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zeng G, et al. . 2022. Immunogenicity and safety of a third dose of CoronaVac, and immune persistence of a two-dose schedule, in healthy adults: interim results from two single-centre, double-blind, randomised, placebo-controlled phase 2 clinical trials. Lancet Infect Dis. 22:483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chumakov K, et al. . 2021. Old vaccines for new infections: exploiting innate immunity to control COVID-19 and prevent future pandemics. Proc Natl Acad Sci. 118:e2101718118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Perera RA, et al. . 2020. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Euro Surveill. 25:2000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lamoreaux L, Roederer M, Koup R. 2006. Intracellular cytokine optimization and standard operating procedure. Nat Protoc. 1:1507–1516. [DOI] [PubMed] [Google Scholar]
- 55. Sui Y, et al. . 2010. Innate and adaptive immune correlates of vaccine and adjuvant-induced control of mucosal transmission of SIV in macaques. Proc Natl Acad Sci. 107:9843–9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is included in the manuscript and/or supporting information.