To the Editor:
Genetic variability of HIV-1, due to its high mutation and recombination rates, complicates diagnosis, monitoring, and treatment. There are 3 phylogenetically distinct groups of HIV-1: M, O, and N; group M strains are further subdivided into subtypes designated as A–D, F–H, J, and K.1 Adding to the overall genetic complexity of HIV-1, increasing numbers of recombinant strains are being identified.1 Although it has been estimated that more than 95% of HIV-1 cases in North America are subtype B, this subtype represents <12% of infections worldwide.1 With increased immigration, travel, and military deployment, the distribution of nonsubtype B infections in North America is changing.2 A survey of HIV-1–infected patients in 11 states in the United States revealed that 5.1% harbored non-B and recombinant strains.3
HIV-1 diversity presents a challenge to accurate and reliable viral load measurement.4–7 The case presented herein illustrates the impact of HIV-1 genetic diversity on performance of viral load assays and the difficulty this presents for clinical management of patients.
A 25-year-old woman presented for care due to an upper respiratory infection. She had emigrated from Central America to the United States 8 months prior and was known to be HIV positive for 5 years. She acquired HIV infection from heterosexual contact and had never received antiretroviral therapy. The patient’s history included diagnoses of syphilis and gonorrhea in 1997 and pyelonephritis in 2004. She was asymptomatic until a few months before presentation when she started experiencing anorexia and recurrent episodes of diarrhea, resulting in significant weight loss. At presentation, her absolute CD4+ T-cell count was 76 cells per cubic millimeter and plasma viral load was <50 (1.7 log10) copies per milliliter based on the ultrasensitive format of COBAS AMPLICOR HIV-1 MONITOR Test v1.5 (Monitor v1.5; Roche Molecular Systems, Inc, Branchburg, NJ). The respiratory condition subsided with a short course of antibiotics. The patient was initiated on prophylaxis for Pneumocystis jeroveci. The CD4 cell count and viral load test were repeated, obtaining similar results. The patient was then referred to the University of Miami AIDS Clinical Research Unit. On examination, she was underweight with enlarged cervical lymph nodes.
Due to the low CD4 cell count (<100 cells/mm3) and a lack of detectable HIV-1 RNA in plasma by Monitor v1.5, surplus plasma from the same draw was tested in several additional viral load assays. A value of 90,840 (4.96 log10) copies per milliliter was reported by the VERSANT HIV–1 RNA 3.0 branched-chain assay (bDNA; Siemens Healthcare Diagnostics, Tarrytown, NY), whereas the Abbott RealTime HIV-1 assay (Abbott Molecular Inc, Des Plaines, IL) measured 251,189 (5.4 log10) copies per milliliter of HIV-1 RNA. Due to limited volume, the sample was diluted 1:10 in EDTA plasma before testing in the COBAS AmpliPrep/COBAS TaqMan HIV-1 Test (TaqMan HIV-1; Roche Molecular Systems, Inc.) at LabCorp (Research Triangle Park, NC); no HIV-1 RNA was detected. Subsequent testing of 4 serial 10-fold dilutions of virus cultured from the patient’s peripheral blood mononuclear cells corroborated the results obtained with plasma. Viral load values were 78,779 (4.9 log10), 9902 (4.0 log10), 759 (2.88 log10), and 70 (1.85 log10) copies per milliliter for RealTime HIV-1 and 29,938 (4.48 log10), 3657 (3.56 log10), 283 (2.45 log10), and <75 (1.88 log10) copies per milliliter using the bDNA assay. Thus, relative to the RealTime HIV-1 assay, bDNA consistently underquantified viral load by ~0.4 log10 copies per milliliter across the dilution series. The TaqMan HIV-1 test failed to detect all 4 members of the dilution series. Parallel testing of a control subtype B strain in the same matrix yielded the expected linear dilution profile in the TaqMan HIV-1 assay. To determine subtype, gag p24, pol IN, and env IDR were amplified and sequenced (GenBank accession numbers: FJ167345-FJ167347) as previously described.6,7 Phylogenetic analysis of sequences from all 3 regions clustered with high confidence to subtype F (subsubtype F1) reference sequences. Subsequent analyses showed that this strain is distinguishable from all known B/F circulating recombinant forms.
On a global scale, distribution of HIV-1 subtypes is changing, with non-B subtypes and recombinant forms becoming increasingly more common in countries where subtype B strains predominate.1,8 For example, subtype F, originally reported mainly in Central Africa, is becoming more prevalent in some South American countries. In Brazil and Argentina, subtype F represents up to 30% of HIV-1 cases, either as a pure subtype or a recombinant with subtype B.1,8 There is evidence that the prevalence of non-B subtypes in the United States is increasing and is higher than previously estimated.2,3 Due to the continual redistribution of subtypes and recombinant viruses, it has become imperative for clinicians to consider the potential for non-B infections in their patient populations. This is especially important if patients present with an undetectable viral load or if CD4+ T-cell count and viral load values are discordant.7 More insidious would be a situation where viral load is measured but significantly underestimated (>1 log10 copies/mL); this could compromise patient management and lead to severe clinical ramifications.
Nucleic acid assays designed to measure viral load require HIV-1 sequence-specific primers and/or probes. Natural polymorphisms occurring within primer and/or probe-binding sites can reduce the efficiency of hybridization resulting in failure to detect or under-quantification of the virus.6,7 In the present study, analysis of target regions for Monitor v1.5 and TaqMan HIV-1 tests (in gag p24) and for the RealTime HIV-1 (in pol IN) revealed that assay performance reflected the level of conservation within the respective primer and probe sites. For Monitor v1.5, 3 mismatches were present in the probe site and 4 mismatches were distributed across the reverse primer-binding site (Fig. 1). The probe site of TaqMan HIV-1 contained 2 internal mismatches and the reverse primer contained the same 4 mismatches observed for Monitor v1.5 (Fig. 1). In contrast, there were no mismatches in the RealTime HIV-1 primer sites and only 1 probe mismatch. The number and distribution of mismatches within the reverse primer and/or probe-binding sites of Monitor v1.5 and TaqMan HIV-1 likely provide the molecular basis for failure to detect this strain. The magnitude of underquantitation can be substantial.4–7 In this case, Monitor v1.5 and TaqMan HIV-1 tests underquantified this subtype F strain by >3 log10 copies per milliliter relative to RealTime HIV-1.
FIGURE 1.
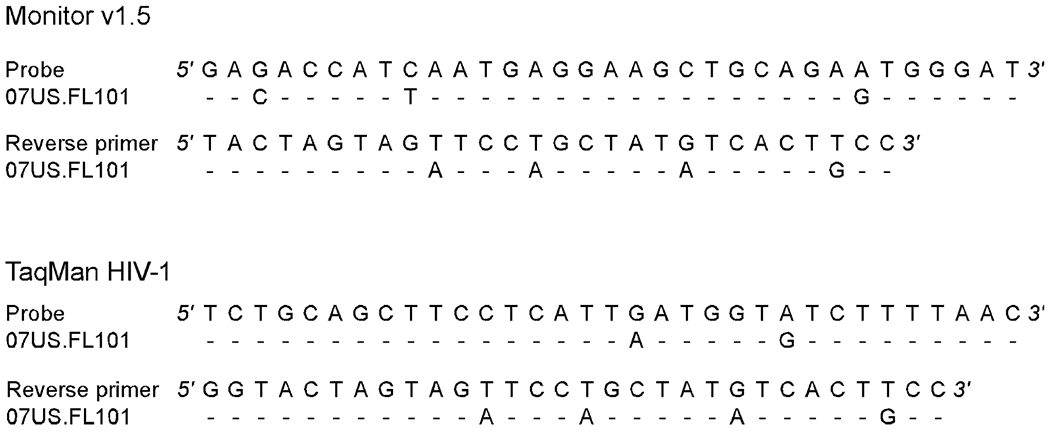
Primer and probe mismatch analysis. Alignment of the 07US.FL101 strain nucleotide sequence against Monitor v1.5 and TaqMan HIV-1 reverse primer and probe sequences.7,9 Dashes denote sequence identity.
Measurement of HIV-1 RNA is crucial for optimal clinical management of patients. Thus, underquantitation or failure to detect a particular HIV-1 strain can have severe consequences.10 It is important to recognize that there are substantial differences in performance between viral load assays and to utilize assays that provide group and subtype-independent performance. Optimal management of patients requires an understanding of the limitations of the assays utilized and a comprehensive analysis of clinical findings and laboratory data.
ACKNOWLEDGMENTS
We thank Dr. Richard Hodinka for assistance with bDNA testing and Priscilla Swanson for critical review of the article. Ilene Auerbach, PhD, of ACCESS Medical (Chicago, IL) assisted in writing this article.
REFERENCES
- 1.Hemelaar J, Gouws E, Ghys PD, et al. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20:W13–W23. [DOI] [PubMed] [Google Scholar]
- 2.Lin HH, Gaschen BK, Collie M, et al. Genetic characterization of diverse HIV-1 strains in an immigrant population living in New York City. J Acquir Immune Defic Syndr. 2006;41:399–404. [DOI] [PubMed] [Google Scholar]
- 3.Wheeler W, Mahle K, Bodnar U, et al. Antiretroviral drug resistance mutations and subtypes in drug naive persons newly diagnosed with HIV-1 infection, United States, March 2003 to October 2006. Paper presented at: 14th Conference on Retroviruses and Opportunistic Infections; February 25–28, 2007; Los Angeles, CA. [Google Scholar]
- 4.Damond F, Roquebert B, Bénard A, et al. Human immunodeficiency virus type 1 (HIV-1) plasma load discrepancies between the Roche COBAS AMPLICOR HIV-1 MONITOR Version 1.5 and the Roche COBAS AmpliPrep/COBAS TaqMan HIV-1 assays. J Clin Microbiol. 2007;45:3436–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gueudin M, Plantier JC, Lemée V, et al. Evaluation of the Roche Cobas TaqMan and Abbott RealTime extraction-quantification systems for HIV-1 subtypes. J Acquir Immune Defic Syndr. 2007;44:500–505. [DOI] [PubMed] [Google Scholar]
- 6.Swanson P, de Mendoza C, Joshi Y, et al. Impact of human immunodeficiency virus type 1 (HIV-1) genetic diversity on performance of four commercial viral load assays: LCx HIV RNA Quantitative, AMPLICOR HIV-1 MONITOR v1.5, VERSANT HIV-1 RNA 3.0, and NucliSens HIV-1 QT. J Clin Microbiol. 2005;43:3860–3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Truchsess I, Harris B, Schätzl HM, et al. The first B/G intersubtype recombinant form of human immunodeficiency virus type 1 (HIV-1) identified in Germany was undetected or underquantitated by some commercial viral load assays. J Med Virol. 2006;78:311–317. [DOI] [PubMed] [Google Scholar]
- 8.Teixeira SL, Bastos FI, Telles PR, et al. HIV-1 infection among injection and ex-injection drug users from Rio de Janeiro, Brazil: prevalence, estimated incidence and genetic diversity. J Clin Virol. 2004;31:221–226. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt M, Korn K, Nüabling CM, et al. First transmission of human immunodeficiency virus type 1 by a cellular blood product after mandatory nucleic acid screening in Germany. Transfusion. 2009;49:1836–1844. [DOI] [PubMed] [Google Scholar]
- 10.Geelen S, Lange J, Borleffs J, et al. Failure to detect a non-B HIV-1 subtype by the HIV-1 Amplicor Monitor test, version 1.5: a case of unexpected vertical transmission. AIDS. 2003; 17:781–782. [DOI] [PubMed] [Google Scholar]


