Abstract
Since 2020, developed countries have rapidly shared both publicly and academically relevant wastewater surveillance information. Data on SARS-CoV-2 circulation is pivotal for guiding public health policies and improving the COVID-19 pandemic response. Conversely, low- and middle-income countries, such as Latin America and the Caribbean, showed timid activities in the Wastewater-Based Epidemiology (WBE) context. In these countries, isolated groups perform viral wastewater monitoring, and the data are unevenly shared or accessible to health agencies and the scientific community. This manuscript aims to highlight the relevance of a multiparty effort involving research, public health, and governmental agencies to support usage of WBE methodology to its full potential during the COVID-19 pandemic as part of a joint One Health surveillance approach. Thus, in this study, we explored the results obtained from wastewater surveillance in different regions of Brazil as a part of the COVID-19 Wastewater Monitoring Network ANA (National Water Agency), MCTI (Ministry of Science, Technology, and Innovations) and MS (Ministry of Health). Over the epidemiological weeks of 2021 and early 2022, viral RNA concentrations in wastewater followed epidemiological trends and variations. The highest viral loads in wastewater samples were detected during the second Brazilian wave of COVID-19. Corroborating international reports, our experience demonstrated usefulness of the WBE approach in viral surveillance. Wastewater surveillance allows hotspot identification, and therefore, early public health interventions. In addition, this methodology allows tracking of asymptomatic and oligosymptomatic individuals, who are generally underreported, especially in emerging countries with limited clinical testing capacity. Therefore, WBE undoubtedly contributes to improving public health responses in the context of this pandemic, as well as other sanitary emergencies.
Keywords: COVID-19, Wastewater-based epidemiology, Wastewater surveillance, SARS-CoV-2, Sewage, Brazil
Graphical Abstract
1. Introduction
High mortality rates were reported worldwide during the COVID-19 pandemic. By February 2022, it was estimated that SARS-CoV-2 was responsible for approximately 380.322.000 infected persons and 5.681.000 deaths [1]. To date, more than 59.357.000 cases of COVID-19 have been reported since the pandemic began [2]. Most COVID-19-related discussions in the region have focused on Brazil [3], [4], which was one of the global epicenters of the disease [5], [6], [7].
On February 26, 2020, the first case was confirmed in São Paulo, the largest city in Latin America [8], [9]. In March, additional surveillance, restriction, and non-pharmacological measures were implemented by the Ministry of Health [10] in an attempt to mitigate the effects of the pandemic. However, these initial efforts were insufficient. In May 2022, the total number of COVID-19 deaths in the country exceeded 663.000 and more than 30.448.000 cases were confirmed [1].
In Brazil, the Brazilian Health Regulatory Agency (ANVISA) initially approved the AstraZeneca and Sinovac vaccines for emergency use on January 17, 2021 and both have been manufactured in Biomanguinhos, Fiocruz and Butantan Institute, respectively. Approximately 70% of the entire population has already been vaccinated with the second/unique doses since 2021 January 18, 2021, when São Paulo state initiated vaccination of the elderly and health professionals [11]. Vaccination may have been essential for reducing the number of hospitalizations and deaths during the second wave of the disease in the country. However, emergence of the Omicron variant (B.1.1.529) in the country has caused a huge increase in COVID-19 cases, especially in the first weeks of 2022 [12]. On January 28, 2022, approximately 270,000 new cases of COVID-19 were reported in 24 h, which was the highest since the beginning of the pandemic [1].
Studies performed by Chen et al. [13], Parasa et al. [14], and Zheng et al. [15] have demonstrated that SARS-CoV-2 RNA can be found in feces during viral infection. The presence of viral RNA in stool and other human excreta allows viral detection in domestic wastewater. Thus, wastewater surveillance can be a low-cost strategy of quick response and can be integrated with other public health interventions to serve as an additional tool in decision-making [16], [17], [18], [19], [20]. The wastewater monitoring approach is referred as Wastewater-Based Epidemiology (WBE) and it has been used to track numerous infectious diseases [21], [22], [23], [24], [25], [26]. This strategy supports the concept of health promotion through basic sanitation and is absolutely aligned with the UN Sustainable Development Goals, Good Health and Well-being, and Sustainable Cities and Communities concepts as part of a global agenda to promote equity and health.
Collecting and analyzing data on the occurrence and quantification of SARS-CoV-2 in wastewater can provide the basis for an early warning system (EWS) to track viral circulation in any context. The EWS can be especially useful for low- and middle-income countries, where financial resources such as viral and antibody testing capabilities, hospital infrastructure, qualified staff, and personal protective equipment (PPE) may be limited. Effective EWS can also be used to identify COVID-19 hotspots and guide action and resource allocation, which includes testing and tracking strategies [24], [27], [28], [29], [30].
Countries such as Finland, Hungary, Luxembourg, the Netherlands, Spain, and Turkey have already included WBE as a nationwide strategy to monitor SARS-CoV-2 spread, whereas the United States, Canada, Australia, France, Switzerland, and the United Kingdom have established regional monitoring strategies [31]. Likewise, South Africa, an emerging country such as Brazil, has recently created a wastewater surveillance panel (https://www.samrc.ac.za/wbe/) which presents weekly results for more than 45 monitoring points in different cities.
To date, more than 58 countries have detected SARS-CoV-2 RNA in wastewater, and 276 universities worldwide are conducting research on this topic. In Latin America, there are ongoing studies carried out by universities and research institutions in Mexico, Colombia, Ecuador, Argentina, and Chile [32], [33], [34], [35], [36], [37]. In Brazil, the first initiatives were carried out independently in the states of Minas Gerais (MG), Rio de Janeiro (RJ), and Rio Grande do Sul (RS) [38]. Currently, other regions of the country using the WBE approach, but through a cooperative work (monitoring network) include: the cities of Belo Horizonte–Minas Gerais (MG), Curitiba–Paraná (PR), Fortaleza–Ceará (CE), Recife–Pernambuco (PE), Rio de Janeiro–Rio de Janeiro (RJ), and the Federal District (DF). Another joint initiative is the COVID-19 Wastewater Monitoring Network ANA/MCTI/MS, which is promoted by the National Water and Basic Sanitation Agency, the Ministry of Science, Technology and Innovation, and the Ministry of Health, which monitors the ABC region in São Paulo and the cities of Foz do Iguaçu–Paraná (PR), Goiânia–Goiás (GO), and the Federal District (DF).
This manuscript highlights the relevance of a multiparty effort involving research, public health, and governmental agencies to support usage of WBE to its full potential during the COVID-19 pandemic as part of a joint One Health surveillance approach. The results and experiences of the COVID-19 Wastewater Monitoring Network ANA/MCTI/MS (ABC Region, Foz do Iguaçu, Goiânia, and Federal District) are presented.
2. Material and methods
2.1. Sampling sites and sample collection
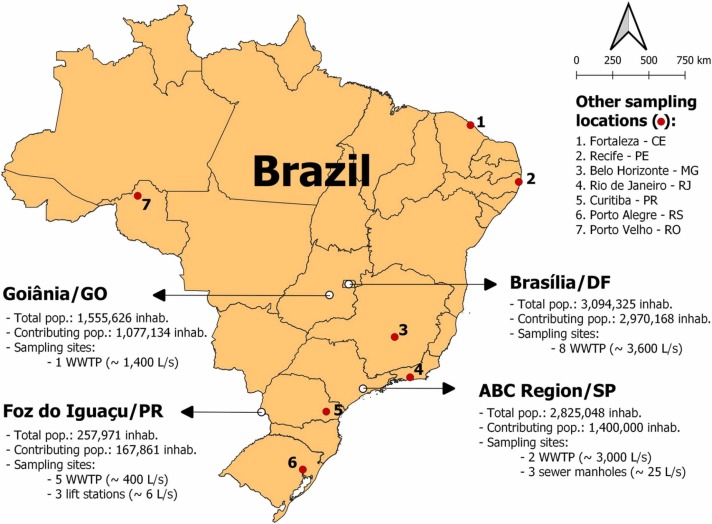
Sewage samples were collected from epidemiological week (EW) 01/2021 (January 3, 2021 to January 9, 2021) to EW 03/2022 (January 16, 2022 to January 22, 2022) at the entrance of wastewater treatment plants (WWTP), and in other locations of the sewer system, such as lift stations and sewer manholes. Fig. 1 summarizes the municipalities and regions where WBE initiatives were carried out in Brazil. This includes those monitored by the ANA/MCTI/MS COVID-19 Wastewater Monitoring Network (ABC Region, Foz do Iguaçu, Goiânia, and Federal District) and other Brazilian initiatives (red dots).
Fig. 1.
Wastewater surveillance sites in Brazil, according to geographical distribution, population, and type of sampling sites.
As estimated by the Brazilian Institute of Geography and Statistics (IBGE) for 2021, the ABC region of São Paulo, Federal District, and Goiânia are highly populated areas (from 1.555.626 to 3.094.325 inhabitants) and have heterogeneous socioeconomic characteristics. In contrast, Foz do Iguaçu, located in southern Brazil, is a less populated city with 257.971 inhabitants [39].
In the WWTPs, 24 h composite sampling of 1000 mL (proportional to the hourly flow rate) was performed using a refrigerated automatic sampler (storage temperature of 4 °C). In the sewer manholes and lift stations, a 4 h semi-composite sampling of 1000 mL (proportional to time) was carried out using the same automatic sampler. Semi-composite sampling was performed within a time interval (in this case, 4 h) lower than the standard of 24 h. Although it may not accurately represent the daily variation in sewage, it is an alternative for sampling sites which are difficult to access, such as sewer manholes, where it is not possible to maintain an automatic sampler for long periods [17], [40]. The 1000 mL samples were divided into two aliquots of 40 mL and then stored at 4 °C for a maximum of 36 h. The remaining sample (920 mL) was used for physicochemical and RNA-sequencing analyses (not presented in this manuscript).
2.2. Viral RNA detection and quantification
Methods for detecting the virus in wastewater samples from the ABC Region, Foz do Iguaçu, Goiânia, and Federal District are presented below:
Viral particles were concentrated using a precipitation method [17], [41]. Briefly, 40 mL of sample was centrifuged (8000 x g/120 min/4 °C) using polyethylene glycol 8000 (PEG 8000) and sodium chloride (NaCl). The pellet formed after centrifugation was resuspended in 0.4 mL of 1x PBS (pH 7.2). For sample cleaning, acidic phenol (1 mL) was added to the resuspended pellet and centrifuged (12,000 x g/10 min/4 ºC) (Claro et al., 2021). The liquid was then transferred to a microtube containing lysis buffer (0.3 mL). RNA extraction was performed using the PureLink Viral RNA/DNA Mini Kit (Thermo Fisher Scientific) according to the protocol of the manufacturer. The concentration of RNA was measured using Nanodrop Lite (Thermo Fisher Scientific) to assess quality of the genetic material (nucleic acid) extraction process.
To detect and quantify SARS-CoV-2 RNA, reaction mixtures were prepared using the SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase (Thermo Fischer Scientific, Waltham, MA, USA) for the targeted nucleocapsid (N1 and N2) genomic regions [42]. The sequences and concentrations of primers and probes used (Thermo Fisher Scientific) are listed in Table S1. Components of the reaction contained 10 μL 2x reaction mix (0.4 mM of each dNTP, 3.2 mM MgSO4), 1.5 μL probe and primer mix (FAM-labelled probe, forward and reverse primers), 0.35 μL SuperScript III RT/Platinum Taq mix (Mg++ and dNTP are not used), 3.15 μL nuclease free-water, and 5 μL RNA template to a final volume of 20 μL. RT-qPCR was performed on a CFXOpus 96 thermal cycler (Bio-Rad, Hercules, CA, USA). The thermal cycling conditions for RT-qPCR assays were as follows: initial incubation at 50 °C for 30 min and initial denaturation at 95 °C for 3 min, followed by 45 cycles of denaturation at 95 °C for 3 s and primer annealing and extension reaction at 55 °C for 30 s (acquiring fluorescence in the green filter).
RT-qPCR assays for SARS-CoV-2 were performed in duplicates. A 10-fold dilution series of standard RNAs was prepared (2019-nCoV_N_Positive Control Cat. PC67102, Norgen) to obtain standard curves.
The calibration curves for N1 (y = −3.217x + 41.431) and N2 (y = −3.118x + 41.919) showed linear dynamics, as shown in Fig. S1. The values of efficiency and R² for N1 and N2 were 104.6% and 0.990%, and 109.3% and 0.998, respectively. The limit of detection (LOD) was 3 and 4 genome copies for N1 and N2, respectively. Table S2 presents the calibration curve parameters considering N1 and N2. Samples with Ct values of < 40 were considered positive for SARS-CoV-2 [19], [41].
Following the protocols described by Rajal et al. [43] and Boxus et al. [44], bovine respiratory syncytial virus (Inforce™ 3) (Zoetis, US) RT-qPCR reactions were performed to evaluate the recovery of concentration methods. A normalization step was performed according to previous studies by our research group [17], [45]. Further, the wastewater samples were spiked with plasmids (pET28a) to determine whether the nucleic acid could be recovered from wastewater. After validating the concentration method, a commercial vaccine containing bovine respiratory syncytial virus (BRSV) was used to verify the efficiency of viral recovery. This virus was chosen because of its structural similarity with SARS-CoV-2; it is an RNA enveloped virus that loses its envelope similar to SARS-CoV-2 due to the presence of surfactants when eliminated in the wastewater system. However, viral RNA can be detected in this matrix because it is protected by a protein (nucleoprotein N). Recoveries ranged from 20% to 65%.
An inhibition test was performed to verify the influence of inhibitors on RT-qPCR performance. The RNA samples were processed, concentrated, and diluted 10x and no significant difference was detected. Thus, it was decided to use concentrated samples in all analyses.
2.3. Prevalence estimation and statistical analysis
Prevalence estimation of SARS-CoV-2 infected individuals for each location were based on the viral load quantified in the wastewater samples according to the following equations [17], [25], [46], [47]:
| (1) |
| (2) |
where CRNA = SARS-CoV-2 RNA concentration measured in wastewater samples (genome copies/L), F is the wastewater volumetric flow rate (L/day), α is the fecal load (g/person/day), and β is the SARS-CoV-2 shedding rate of an infected individual (genome copies/g). In Eq. (1), the Monte Carlo statistical model was incorporated because the α and β data presented in the literature have a wide range of variation. Hence, we implemented a model with 10.000 random samples for α and β products (viral load excreted by an infected individual through feces). The parameter values/ranges and their respective statistical distributions are shown in Table S3.
Additionally, viral RNA concentration data from sewage samples were compared with the number of reported clinical cases accumulated over 7 days. Spearman's correlation was used with a significance level of 95% (p < 0.05). Confirmed COVID-19 cases and other epidemiological statistics, such as the respective epidemiological weeks, were accessed from the official Brazilian COVID-19 database [48].
Statistical analysis was performed using Origin Pro 8.6, and the Monte Carlo simulation was implemented in Microsoft Excel.
3. Results and discussion
The WBE approach for SARS-CoV-2 has been used in different Brazilian cities to identify hotspots and provide data for the early warning of new outbreaks (EWS). Thus, monitoring wastewater allows local and effective health interventions, such as enhancing clinical testing, tracking asymptomatic and oligosymptomatic individuals, and tailoring additional preventive strategies. Different cities in Brazil have been monitored since the beginning of the pandemic in March 2020. In the ABC region, wastewater monitoring began in June 2020 as part of the COVID-19 Wastewater Monitoring Network (ANA/MCTI/MS). WBE started between February and April 2021 in the other studied locations such as Foz do Iguaçu, Goiânia, and the Federal District. All data were regularly shared with the Ministry of Health and local health secretariats to complement epidemiological surveillance and strengthen the COVID-19 pandemic response. In this context, weekly reports have been published by government authorities [49], [50]. Wastewater data from Foz do Iguaçu were also computed by the municipal health secretariat dashboard, which is available for public consultation [51].
In this study, we mainly discuss wastewater data from the ABC Region, Foz do Iguaçu, Goiânia, and the Federal District, throughout 2021 and early 2022. Results of wastewater surveillance, COVID-19 confirmed cases (7-day moving average/100k inhabitants), and prevalence estimates for each city studied are presented in Fig. 2.
Fig. 2.
SARS-CoV-2 viral load assessed in wastewater from ABC Region (a), Foz do Iguaçu (b), Goiânia (c) and Federal District (d) in 2021 and 2022, according to clinical data and predicted prevalence. TNP: test not performed; ND: not detected.
The ABC region, Federal District, and Goiânia are highly populated areas/municipalities with approximately 2.8, 3.1, and 1.5 million inhabitants, respectively. In contrast, Foz do Iguaçu, which is located in the southern region of Brazil, is a less populated city with approximately 260.000 inhabitants [39]. As shown in Fig. 2, there were differences in the patterns of moving averages of the new cases. These distinct states were able to adopt local policies and guidelines concerning social distancing and other pharmacological preventive measures, which could also partially explain the regional epidemiological patterns of viral circulation. Finally, the Brazilian population is diverse and has characteristic regional habits. Despite these factors, a more detailed investigation is necessary for a complete understanding of the observed behaviors, which will certainly be the object of future work.
Furthermore, there are different socioeconomic contexts in each assessed location, especially the most populous ones (ABC Region, Federal District, and Goiânia). Investigating the correlation of wastewater viral loads with the social vulnerability index of each evaluated sub-basin is encouraged in future studies.
In contrast, the fluctuations observed in the wastewater data (viral load) and prevalence estimates can also be verified in the reported clinical data (7-day moving average/100k inhabitants) for all the evaluated localities. In Brazil, an upsurge in new cases and hospitalizations was observed in March 2021. This second wave of COVID-19 cases was more intense than the first one observed in 2020 [52]. Unsurprisingly, the highest viral loads in the wastewater samples were detected in these months, with values ranging from 106–108 genome copies/L.
In addition to the peak of new cases observed in March 2021, another peak was observed in June 2021 in the ABC region and in the city of Foz do Iguaçu, and in August 2021 in the city of Goiânia and the Federal District. It is noteworthy that Goiânia and the Federal District are in the central part of Brazil, approximately 130 miles apart, with near latitudes. Similarly, Foz do Iguaçu and the ABC region are also at close latitudes, although they are at a greater distance from each other.
After the second peak of the disease (between June and August 2021), there was a significant drop in the number of new clinical cases and hospitalizations. In the ABC region, the 7-day moving average of notifications after September 2021 was less than 10 new cases per 100,000 inhabitants. The same positive mark was reached in October 2021 in the city of Goiânia and in November 2021 in the city of Foz do Iguaçu and the Federal District. This scenario may have been favored by the vaccination campaign against COVID-19 which was well accepted by Brazilian citizens [53]. As of February 2022, 352.047.311 vaccine doses had been administered [1], [11]. Approximately 70% of the entire population is vaccinated with second/unique doses [11].
As shown in Fig. 2, high concentrations of SARS-CoV-2 RNA titers were measured in wastewater samples, even when the 7-day moving average of new COVID-19 cases dropped sharply. Asymptomatic and unreported cases may have contributed to this effect. These cases are not tracked by classical epidemiological surveillance, but by wastewater surveillance. The number of underreported cases may have increased with improvement in vaccination, as the vaccine reduces clinical severity but does not completely prevent infection [54]. Reducing disease severity tends to increase underreporting, which makes it difficult to determine the effectiveness of vaccination strategies [55].
Emergence of the highly mutated Omicron variant (B.1.1.529) caused an increase in COVID-19 infections in different countries worldwide [56]. More than 38 countries across all six WHO regions have been affected by the new variant since it was first detected in South Africa in November 2021 [57]. Many recent studies have shown that this variant is highly contagious and resistant to currently used vaccines, but also appears to cause milder disease [58].
In Brazil, the Omicron variant was the cause of the third wave of COVID-19. There was a significant increase in number of infections in the first epidemiological week across the country [59]. Fig. 2 shows this new peak of clinical cases in epidemiological weeks 1, 2, and 3 of 2022, especially in the Federal District and cities of Foz do Iguaçu and Goiânia. As shown in Fig. 2, an increase in viral loads in the same week can also be observed. Viral load levels in the cities of Foz do Iguaçu and Goiânia were as high as those measured during the second wave of COVID-19, when many hospitalizations and deaths were observed worldwide.
The correlation between the concentration of SARS-CoV-2 RNA fragments in sewage and the number of new cases accumulated over 7 days was tested. Spearman's correlation coefficient was used, and the results obtained for these four locations are listed in Table 1.
Table 1.
Spearman correlation coefficients for each evaluated locality.
| City or Region | ABC Region | Foz do Iguaçu | Goiânia | Federal District |
|---|---|---|---|---|
| Spearman’s Rho | 0.41 | 0.57 | 0.63 | 0.61 |
| p-value | 0.003 | 0.00003 | 0.0001 | 0.002 |
A positive and significant correlation between the viral load in sewage and clinical cases was verified, with a significance level of 95% (p < 0.05). The values of Spearman's rho correlation coefficient ranged between 0.41 and 0.61.
To date, few studies have demonstrated a strong correlation between these variables. Correlation coefficients between 0.18 and 0.87 were found in the literature [19], [60], [61], [62], [63]. Medema et al. [19] observed correlation coefficients of up to 0.79 for Dutch cities in the early stages of the COVID-19 pandemic. This was the first peer-reviewed paper on SARS-CoV-2 Wastewater Surveillance. Amereh et al. [60] also found a strong correlation between incidence of COVID-19 and the viral load in sewage (R² = 0.80, p < 0.001) by evaluating seven WWTPs in Tehran, Iran from September 2020 to April 2021. Kitamura et al. [63] observed a higher correlation between variables when considering the number of new COVID 19 cases based on the onset date rather than on the reported date.
The modelled prevalence of infection based on the wastewater results for each sampling location is presented in Table 2. This table also shows the observed prevalence (average, minimum, and maximum) for the evaluated period, which was calculated from the number of confirmed clinical cases reported by the health authorities.
Table 2.
Modelled and reported prevalence of infection for each sampling location during monitoring months.
| City/Region | Months | Modelled prevalence of infection (%) median (90% CI) |
Reported prevalence of infection (%) mean (minimum – maximum) |
|---|---|---|---|
| ABC Region | Jan 2021 | 0.1 (0.05 – 0.9) | 0.02 (0.004 – 0.03) |
| Feb | 3.0 (1.1 – 18.7) | 0.02 (0.003 – 0.03) | |
| Mar | 6.6 (2.4 – 41.9) | 0.03 (0.006 – 0.05) | |
| Apr | 0.5 (0.2 – 3.4) | 0.02 (0.006 – 0.04) | |
| May | 0.2 (0.1 – 1.3) | 0.02 (0.004 – 0.04) | |
| June | 0.4 (0.1 – 2.3) | 0.02 (0.005 – 0.07) | |
| July | 0.002 (0.0007 – 0.01) | 0.02 (0.006 – 0.04) | |
| Aug | 0.4 (0.1 – 2.4) | 0.01 (0.003 – 0.02) | |
| Sept | 0.4 (0.2 – 2.6) | 0.01 (0.0001 – 0.03) | |
| Oct | 0.1 (0.05 – 0.8) | 0.002 (0.0002 – 0.01) | |
| Nov | 0.02 (0.006 – 0.1) | 0.003 (0.00004 – 0.01) | |
| Dec | 0.01 (0.005 – 0.1) | 0.001 (0.00004 – 0.004) | |
| Jan 2022 | 0.6 (0.2 – 4.0) | 0.004 (0.0001 – 0.02) | |
| Foz do Iguaçu | Feb 2021 | 2.6 (0.9 – 16.1) | 0.05 (0.01 – 0.2) |
| Mar | 8.2 (3.0 – 51.9) | 0.07 (0.02 – 0.2) | |
| Apr | 0.2 (0.1 – 1.3) | 0.03 (0.006 – 0.05) | |
| May | 0.3 (0.1 – 2.2) | 0.04 (0.008 – 0.07) | |
| June | 0.1 (0.04 – 0.6) | 0.04 (0.01 – 0.1) | |
| July | 0.002(0.001 – 0.01) | 0.02 (0.004 – 0.03) | |
| Aug | 0.2 (0.1 – 1.5) | 0.02 (0.004 – 0.04) | |
| Sept | 0.1 (0.02 – 0.3) | 0.01 (0.003 – 0.03) | |
| Oct | 0.2 (0.1 – 1.5) | 0.01 (0.0008 – 0.03) | |
| Nov | 0.1 (0.03 – 0.4) | 0.006 (0.002 – 0.02) | |
| Dec | 0.1 (0.03 – 0.6) | 0.007 (0.002 – 0.02) | |
| Jan 2022 | 0.7 (0.2 – 4.2) | 0.3 (0.03 – 0.7) | |
| Goiânia | May 2021 | 0.5 (0.2 – 2.9) | 0.03 (0.01 – 0.05) |
| June | 0.2 (0.1 – 1.1) | 0.03 (0.01 – 0.05) | |
| July | 0.1 (0.03 – 0.5) | 0.04 (0.01 – 0.07) | |
| Aug | 0.1 (0.03 – 0.5) | 0.04 (0.006 – 0.06) | |
| Sept | 0.01 (0.003 – 0.05) | 0.02 (0.005 – 0.03) | |
| Oct | 0.003 (0.001 – 0.02) | 0.007 (0.001 – 0.02) | |
| Nov | 0.005 (0.002 – 0.03) | 0.007 (0.0005 – 0.02) | |
| Dec | 0.003 (0.001 – 0.02) | 0.004 (0.0001 – 0.02) | |
| Jan 2022 | 0.04 (0.02 – 0.3) | 0.04 (0.0005 – 0.1) | |
| Federal District | Apr 2021 | 0.5 (0.2 – 3.3) | 0.03 (0.02 – 0.05) |
| May | 0.1 (0.04 – 0.7) | 0.02 (0.01 – 0.03) | |
| June | 0.1 (0.04 – 0.7) | 0.02 (0.01 – 0.04) | |
| July | 0.1 (0.03 – 0.4) | 0.02 (0.01 – 0.04) | |
| Aug | 0.1 (0.05 – 0.8) | 0.02 (0.01 – 0.04) | |
| Sept 2022 | 0.2 (0.1 – 1.2) | 0.02 (0.01 – 0.04) |
As shown in Table 2, wastewater prevalence estimates were generally significantly higher (approximately ten times) than those reported in clinical cases. In some cases, the predicted values were 100 times higher than those observed in the months of February and March (peak of the second wave of COVID-19) for the ABC Region and Foz do Iguaçu. A higher viral load (CRNA × F, Eq. (1) numerator) tends to increase the variability of prevalence estimates and, consequently, the imprecision of results. In contrast, the data from Goiânia showed greater agreement between the predicted and observed values, especially when considering confidence intervals.
Our findings corroborate those of previous studies. In Massachusetts (USA), Wu et al. [41] estimated prevalence values between 0.1% and 5.0%, which are much higher than those observed by clinical surveillance (approximately 0.026%). Likewise, in another study performed by our research group (COVID-19 Wastewater Monitoring Network ANA/MCTI/MS), predicted values for data collected between June 9, 2020, and April 7, 2021 were approximately 10 times higher than the reported values [17].
As recommended by the CDC [42], prevalence estimates should not be used to support decision-making because there are still many uncertainties in the calculations. For example, the viral load excreted by an infected individual through feces (Eq. 1 denominator) can vary by up to 5 logs [25], [64]. The values of the SARS-CoV-2 shedding rate (β) used to calculate the number of infected in this study were those reported by Kitajima et al. [65], which ranged between 5.79 and 8.11 log10 genome copies.g feces-1. However, according to some authors, this range of variation may even be greater. For instance, Wölfel et al. [64] observed a shedding rate between 2.56 and 7.67 log10 genome copies.g feces-1 in infected individuals one week after symptom onset. The fecal load (α), which is another parameter of Eq. (1), also shows great variation. According to Rose et al. [66], the daily fecal mass (or fecal load) produced by individuals from low-income countries usually ranges from 75.0 to 520.0 g per person (with an average value of 243.0 ± 130.2 g.person-1. d-1). Therefore, significant variability is expected in the predicted data, even when the Monte Carlo probabilistic model is used.
Using the same prevalence estimation model, Ahmed et al. [46] found that there are opportunities for refinement as more data becomes available. The authors used a sensitivity analysis to verify that the model was highly dependent on values of the SARS-CoV-2 shedding rate (β). The number of infected individuals and prevalence estimates showed a high correlation (> 0.9) with SARS-CoV-2 RNA titers in stool samples. However, the model used did not include the percentage of infected individuals shedding viral RNA in their stool, despite there being great variability in this data. Some studies have indicated that approximately 60–70% of infected individuals excrete virus fragments through feces [15], [67]. There also seems to be a geographic variation for this parameter, as studies carried out in China found a cohort of 28% [68], while in Germany, a value of 88% was obtained for the same [64].
Another limitation of the model is that it considers the shedding of viral RNA only through feces, and recent studies have shown other important pathways of contribution, such as sputum [69]. As SARS-CoV-2 is an infectious respiratory virus, it is mainly detected in respiratory tract samples (70–100%). According to Li et al. [69], release of sputum into sewers can increase the concentrations of SARS-CoV-2 RNA fragments in sewage samples by up to 70 times. However, this source has not been considered in previous studies on prevalence modeling.
Other factors to consider are: (i) an infected individual may shed viral RNA in the feces for 22 days (interquartile range from 17 to 31 days) [15]; (ii) sanitary wastewater is a complex matrix with different chemical and biological compounds that can promote SARS-CoV-2 RNA degradation [70], [71], especially in hot-climate regions such as Latin American countries; (iii) there are numerous uncertainties and limitations associated with sampling and sample preservation procedures, in addition to the analytical procedures for concentration, detection, and quantification of the virus in environmental samples [72], [73], [74], [75]; and (iv) the WBE approach is population-based and includes asymptomatic cases, whereas sampling is limited to symptomatic cases in classic epidemiological surveillance [16], [72].
Although it is not yet possible to make an absolute comparison between predicted prevalence and observed/reported prevalence, wastewater data can be used to preview trends, identify hotspots, and complement clinical surveillance data. As it is a population approach, WBE makes it possible to evaluate large areas based on a few samples. In addition to reducing surveillance costs, it enables the tracking of asymptomatic and oligosymptomatic individuals who are usually not detected by clinical surveillance [30], [76].
In this respect, all Brazilian cities and regions mentioned throughout the present work strongly advocate the use of WBE as a complementary tool for epidemiological surveillance. Strengthening holistic health surveillance through the integration of epidemiological, environmental, and genomic approaches will be one of the major legacies of the COVID-19 pandemic.
There are still many challenges for the effective implementation of WBE as a public health policy in emerging countries, starting with sampling strategies, as their sanitation systems are very precarious [34]. On average, the percentage of wastewater collected was very low, at approximately 54.1% [77]. However, Brazil is a contrasting country, including areas with adequate sanitation systems and others that are not even served by potable water systems. Data from the latest national survey on water supply and sewage revealed that in the southeast region, most municipalities (95.9%) had access to sewage services, whereas in the other Brazilian regions, this percentage was below 50%: 49.0%, 40.9%, 38.1%, and 13.8% in the northeastern, southern, midwestern, and northern regions, respectively [78]. Significant variability is observed in health indicators even in states and cities with proper sanitation, particularly in areas of socioeconomic vulnerability with irregular occupations [79]. Faced with this challenging scenario, how can the virus be monitored in environmental samples? There are several challenges and opportunities to this end. Particularly, this question must be addressed in some Latin American, Caribbean, African, and Asian countries, where surface water contaminated by the discharge of human excreta can be an alternative source for viral monitoring. Some studies have shown that the viral RNA concentrations in polluted rivers are similar to those found in wastewater from developed countries [34], [38].
In this manuscript, we present some local and ongoing efforts conducted in Brazil. Despite the multiple challenges imposed by a long and winding road along the COVID-19 pandemic, public health authorities and scientists remain absolutely committed to continue presenting knowledge and innovation for public health to face COVID-19 and other sanitary emergencies.
The use of the WBE approach should be encouraged, even in emerging countries, not only for COVID-19 surveillance but also as a tool to detect other emerging and re-emerging diseases of international concern. Evidently, there are many challenges, especially related to the representativeness of samples in regions that do not have adequate sanitation conditions, which should be explored in future studies.
4. Conclusions
Wastewater-based epidemiology is an important tool that has emerged to help monitor infectious diseases. Currently, it is widely used in monitoring COVID-19, providing essential information on the magnitude of spread of the disease while considering symptomatic, oligosymptomatic, and asymptomatic cases. Generally, the results of sewage monitoring anticipate clinical events, thereby making it possible to plan actions and make decisions.
This manuscript presents some Brazilian wastewater surveillance experiences, including the monitoring of SARS-CoV-2 in the ABC region, Foz do Iguaçu, Goiânia, and the Federal District, throughout 2021 and early 2022. In general, the fluctuations observed in the wastewater data (viral load) can also be verified from the reported clinical data (7-day moving average). The peaks of new cases in the second and third waves of COVID-19 coincided with the peaks of viral RNA concentration in the wastewater samples. A positive and significant correlation between the viral load in wastewater and clinical cases was verified, with a significance level of 95% (p < 0.05). The values of Spearman's rho correlation coefficient ranged between 0.41 and 0.61. However, the wastewater prevalence estimates were significantly higher (about ten times) than those reported in clinical cases. Therefore, an absolute comparison between the predicted and observed prevalence is not recommended. However, wastewater data can be used to preview trends, identify hotspots, and complement clinical surveillance data. In this way, the municipalities mentioned in this work have used sewage monitoring data through their health secretariats.
We expect to have learned from the COVID-19 crisis, and use these lessons to address or even prevent similar future events. This difficult moment has encouraged the use of wastewater surveillance in different Brazilian municipalities, at least at the research level. Thus, the implementation and improvement of the WBE approach is a legacy of the COVID-19 pandemic.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to acknowledge financial support from the following Brazilian institutions: the Brazilian National Council of Scientific and Technological Development (CNPq) in partnership with the Ministry of Science, Technology and Innovations (MCTI) and Ministry of Health (MoH), Secretariat of Science, Technology, Innovation and Strategic Inputs – Decit/SCTIE 07/2020 (Research to cope with COVID-19, its consequences, and other severe acute respiratory syndromes – Processes 402432/2020-7 and 401832/2020-1). The authors are also grateful to the Federal District Research Foundation (Process 00092-00000366/2020-59) and for the combined support of the INCT Sustainable Sewage Treatment Plants and the National Water and Sanitation Agency (TED 07/2020/ANA: Creation of the COVID-Wastewater Network: Expansion of Covid Wastewater Monitoring to other regions/cities in Brazil, Process 02501.005256/2020-40). This study was also supported by the Labor Prosecution Service, CNPq (grant number:402694/2020-1), and FAPEG (grant number:202010267000273). Itaipu Binational and Itaipu Technological Park for financial support (grant number:4500049462-NIT.FP4.005) and Basic Sanitation Company of Paraná (SANEPAR) and Basic Sanitation Company of the State of São Paulo (SABESP).
CRediT authorship contribution statement
Rodrigo de Freitas Bueno: Conceptualization, Project administration; Resources, Supervision, Writing – original draft. Ieda Carolina Mantovani Claro: Laboratory analysis and data processing. Matheus Ribeiro Augusto: Writing – original draft. Adriana Feliciano Alves Duran: Methodology, Data curation. Lívia de Moraes Bomediano Camillo: Data curation, Methodological review. Aline Diniz Cabral: Data curation, Methodological review. Fernando Fabriz Sodré: Conceptualization, Supervision; Writing – original draft. Cristina Celia Silveira Brandão: Supervision, Writing – original draft. Carla Simone Vizzotto: Methodology, Data curation, Methodological review. Rafaella Silveira: Methodology; Data curation. Geovana de Melo Mendes: Data curation, Methodological review. Andrea Fernandes Arruda: Methodology, Data curation. Núbia Natália de Brito: Data curation, Methodological review. Bruna Aparecida Souza Machado: Data curation, Methodological review. Gabriela Rodrigues Mendes Duarte: Conceptualization, Supervision, Writing – original draft. Maria de Lourdes Aguiar-Oliveira: Writing – review & editing.
Editor: Yang Liu
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jece.2022.108298.
Appendix A. Supplementary material
Supplementary material.
.
Supplementary material.
.
Data Availability
No data was used for the research described in the article.
References
- 1.W.H.O., Coronavirus (COVID-19) Dashboard, World Heal. Organ. (2022). 〈https://covid19.who.int/〉.
- 2.Reuters, C.O.V.I.D.–19 Tracker: Latin America and the Caribbean, Reuters Graph. (2022). 〈https://graphics.reuters.com/world-coronavirus-tracker-and-maps/regions/latin-america-and-the-caribbean/〉.
- 3.Monteiro de Oliveira M., Fuller T.L., Brasil P., Gabaglia C.R., Nielsen-Saines K. Controlling the COVID-19 pandemic in Brazil: a challenge of continental proportions. Nat. Med. 2020;26:1505–1506. doi: 10.1038/s41591-020-1071-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kupek E. How many more? Under‐reporting of the COVID‐19 deaths in Brazil in 2020. Trop. Med. Int. Heal. 2021;26:1019–1028. doi: 10.1111/tmi.13628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor L. Covid-19: Brazil’s hospitals close to collapse as cases reach record high. BMJ. 2021;372:n800. doi: 10.1136/bmj.n800. [DOI] [PubMed] [Google Scholar]
- 6.The Lancet COVID-19 in Brazil: “so what? Lancet. 2020;395:1461. doi: 10.1016/S0140-6736(20)31095-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marson F.A.L., Ortega M.M. COVID-19 in Brazil. Pulmonology. 2020;26:241–244. doi: 10.1016/j.pulmoe.2020.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Oliveira W.K., Duarte E., de França G.V.A., Garcia L.P. Como o Brasil pode deter a COVID-19. Epidemiol. Serviços Saúde. 2020;29 doi: 10.5123/S1679-49742020000200023. [DOI] [PubMed] [Google Scholar]
- 9.Brasil, Boletim Epidemiológico N. 02 - Infecção Humana pelo Novo Coronavírus (2019-nCoV) (In portuguese), 2020.
- 10.Brasil, Boletim Epidemiológico N. 05 – Ampliação da Vigilância, Medidas Não Farmacológicas e Descentralização do Diagnóstico Laboratorial (In portuguese), 2020.
- 11.Brasil, Vacinômetro (In portuguese), Ministério Da Saúde. (2021). 〈https://www.gov.br/saude/pt-br/vacinacao〉.
- 12.Taylor L. Covid-19: Brazil sees omicron cases soar but data blackout obscures true impact. BMJ. 2022:o133. doi: 10.1136/bmj.o133. [DOI] [PubMed] [Google Scholar]
- 13.Chen L., Lou J., Bai Y., Wang M. COVID-19 disease with positive fecal and negative pharyngeal and sputum viral tests. Am. J. Gastroenterol. 2020;115:790. doi: 10.14309/ajg.0000000000000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parasa S., Desai M., Thoguluva Chandrasekar V., Patel H.K., Kennedy K.F., Roesch T., Spadaccini M., Colombo M., Gabbiadini R., Artifon E.L.A., Repici A., Sharma P. Prevalence of gastrointestinal symptoms and fecal viral shedding in patients with coronavirus disease 2019: a systematic review and meta-analysis. JAMA Netw. Open. 2020;3:1–14. doi: 10.1001/jamanetworkopen.2020.11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng S., Fan J., Yu F., Feng B., Lou B., Zou Q., Xie G., Lin S., Wang R., Yang X., Chen W., Wang Q., Zhang D., Liu Y., Gong R., Ma Z., Lu S., Xiao Y., Gu Y., Zhang J., Yao H., Xu K., Lu X., Wei G., Zhou J., Fang Q., Cai H., Qiu Y., Sheng J., Chen Y., Liang T. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., Edson J., Nguyen T.M.H., O’Brien J.W., Simpson S.L., Sherman P., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claro I.C.M., Cabral A.D., Augusto M.R., Duran A.F.A., Graciosa M.C.P., Fonseca F.L.A., Speranca M.A., Bueno R.F. Long-term monitoring of SARS-COV-2 RNA in wastewater in Brazil: a more responsive and economical approach. Water Res. 2021;203 doi: 10.1016/j.watres.2021.117534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prado T., Fumian T.M., Mannarino C.F., Resende P.C., Motta F.C., Eppinghaus A.L.F., Chagas do Vale V.H., Braz R.M.S., da J., de Andrade S.R., Maranhão A.G., Miagostovich M.P. Wastewater-based epidemiology as a useful tool to track SARS-CoV-2 and support public health policies at municipal level in Brazil. Water Res. 2021;191 doi: 10.1016/j.watres.2021.116810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 20.Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J., Buttiglieri G., Calabro V., Carducci A., Castiglioni S., Cetecioglu Gurol Z., Chakraborty S., Costa F., Curcio S., De Los Reyes F.L., Delgado Vela J., Farkas K., Fernandez-Casi X., Gerba C., Gerrity D., Girones R., Gonzalez R., Haramoto E., Harris A., Holden P.A., Islam M.T., Jones D.L., Kasprzyk-Hordern B., Kitajima M., Kotlarz N., Kumar M., Kuroda K., La Rosa G., Malpei F., Mautus M., McLellan S.L., Medema G., Meschke J.S., Mueller J., Newton R.J., Nilsson D., Noble R.T., Van Nuijs A., Peccia J., Perkins T.A., Pickering A.J., Rose J., Sanchez G., Smith A., Stadler L., Stauber C., Thomas K., Van Der Voorn T., Wigginton K., Zhu K., Bibby K. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54:7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- 21.La Rosa G., Della Libera S., Iaconelli M., Ciccaglione A.R., Bruni R., Taffon S., Equestre M., Alfonsi V., Rizzo C., Tosti M.E., Chironna M., Romanò L., Zanetti A.R., Muscillo M. Surveillance of hepatitis A virus in urban sewages and comparison with cases notified in the course of an outbreak, Italy 2013. BMC Infect. Dis. 2014;14:419. doi: 10.1186/1471-2334-14-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prevost B., Lucas F.S., Goncalves A., Richard F., Moulin L., Wurtzer S. Large scale survey of enteric viruses in river and waste water underlines the health status of the local population. Environ. Int. 2015;79:42–50. doi: 10.1016/j.envint.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 23.WHO, Polio Surveillance: Tracking Progress towards Eradication Worldwide, 2013–2014, (2015). https://www.who.int/wer/2015/wer9021/en/. [PubMed]
- 24.Daughton C.G. Wastewater surveillance for population-wide Covid-19: the present and future. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saththasivam J., El-Malah S.S., Gomez T.A., Jabbar K.A., Remanan R., Krishnankutty A.K., Ogunbiyi O., Rasool K., Ashhab S., Rashkeev S., Bensaad M., Ahmed A.A., Mohamoud Y.A., Malek J.A., Abu Raddad L.J., Jeremijenko A., Abu Halaweh H.A., Lawler J., Mahmoud K.A. COVID-19 (SARS-CoV-2) outbreak monitoring using wastewater-based epidemiology in Qatar. Sci. Total Environ. 2021;774 doi: 10.1016/j.scitotenv.2021.145608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sodré F.F., Brandão C.C.S., Vizzotto C.S., Maldaner A.O. Epidemiologia do esgoto como estratégia para monitoramento comunitário, mapeamento de focos emergentes e elaboração de sistemas de alerta rápido para CoVID-19. Quim. Nova. 2020;43:515–519. doi: 10.21577/0100-4042.20170545. [DOI] [Google Scholar]
- 27.Bhattacharya P., Kumar M., Islam M.T., Haque R., Chakraborty S., Ahmad A., Niazi N.K., Cetecioglu Z., Nilsson D., Ijumulana J., van der Voorn T., Jakariya M., Hossain M., Ahmed F., Rahman M., Akter N., Johnston D., Ahmed K.M. Prevalence of SARS-CoV-2 in communities through wastewater surveillance—a potential approach for estimation of disease burden. Curr. Pollut. Rep. 2021;7:160–166. doi: 10.1007/s40726-021-00178-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Y., Oishi W., Maruo C., Saito M., Chen R., Kitajima M., Sano D. Early warning of COVID-19 via wastewater-based epidemiology: potential and bottlenecks. Sci. Total Environ. 2021;767 doi: 10.1016/j.scitotenv.2021.145124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar M., Joshi M., Shah A.V., Srivastava V., Dave S. Wastewater surveillance-based city zonation for effective COVID-19 pandemic preparedness powered by early warning: a perspectives of temporal variations in SARS-CoV-2-RNA in Ahmedabad, India. Sci. Total Environ. 2021;792 doi: 10.1016/j.scitotenv.2021.148367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naughton C.C., Roman F.A., Grace A., Alvarado F., Tariqi A.Q., Deeming M.A., Bibby K., Bivins A., Rose J.B., Medema G., Ahmed W., Katsivelis P., Allan V., Sinclair R., Zhang Y., Kinyua M.N. Show us the data: global COVID-19 wastewater monitoring efforts, Equity, and Gaps. MedRxiv. 2021 doi: 10.1101/2021.03.14.21253564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrios M.E., Díaz S.M., Torres C., Costamagna D.M., Blanco Fernández M.D., Mbayed V.A. Dynamics of SARS-CoV-2 in wastewater in three districts of the Buenos Aires metropolitan region, Argentina, throughout nine months of surveillance: a pilot study. Sci. Total Environ. 2021;800 doi: 10.1016/j.scitotenv.2021.149578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giraud-Billoud M., Cuervo P., Altamirano J.C., Pizarro M., Aranibar J.N., Catapano A., Cuello H., Masachessi G., Vega I.A. Monitoring of SARS-CoV-2 RNA in wastewater as an epidemiological surveillance tool in Mendoza, Argentina. Sci. Total Environ. 2021;796 doi: 10.1016/j.scitotenv.2021.148887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guerrero-Latorre L., Ballesteros I., Villacrés-Granda I., Granda M.G., Freire-Paspuel B., Ríos-Touma B. SARS-CoV-2 in river water: implications in low sanitation countries. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosiles-González G., Carrillo-Jovel V.H., Alzate-Gaviria L., Betancourt W.Q., Gerba C.P., Moreno-Valenzuela O.A., Tapia-Tussell R., Hernández-Zepeda C. Environmental surveillance of SARS-CoV-2 RNA in Wastewater and Groundwater in Quintana Roo, Mexico. Food Environ. Virol. 2021 doi: 10.1007/s12560-021-09492-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coronado Y., Navarro R., Mosqueda C., Valenzuela V., Pérez J.P., González-Mendoza V., de la Torre M., Rocha J. SARS-CoV-2 in wastewater from Mexico City used for irrigation in the Mezquital Valley: quantification and modeling of geographic dispersion. Environ. Manag. 2021;68:580–590. doi: 10.1007/s00267-021-01516-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carrillo-Reyes J., Barragán-Trinidad M., Buitrón G. Surveillance of SARS-CoV-2 in sewage and wastewater treatment plants in Mexico. J. Water Process Eng. 2021;40 doi: 10.1016/j.jwpe.2020.101815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de M., Aguiar-Oliveira L., Campos A., Matos A.R., Rigotto C., Sotero-Martins A., Teixeira P.F.P., Siqueira M.M. Wastewater-based epidemiology (WBE) and viral detection in polluted surface water: a valuable tool for COVID-19 surveillance—a brief review. Int. J. Environ. Res. Public Health. 2020;17:9251. doi: 10.3390/ijerph17249251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.IBGE, Panorama do Brasil (In portuguese), IBGE Cid. (2021). 〈https://cidades.ibge.gov.br/brasil/panorama〉.
- 40.Augusto M.R., Claro I.C.M., Siqueira A.K., Sousa G.S., Caldereiro C.R., Duran A.F.A., de Miranda T.B., de L., Bomediano Camillo M., Cabral A.D., de Freitas Bueno R. Sampling strategies for wastewater surveillance: evaluating the variability of SARS-COV-2 RNA concentration in composite and grab samples. J. Environ. Chem. Eng. 2022;10 doi: 10.1016/j.jece.2022.107478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu F., Zhang J., Xiao A., Gu X., Lee W.L., Armas F., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N., Duvallet C., Poyet M., Moniz K., Washburne A.D., Erickson T.B., Chai P.R., Thompson J., Alm E.J. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. MSystems. 2020;5:1–9. doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.C.D.C., Wastewater Surveillance Data Reporting and Analytics, Centers Dis. Control Prev. (2021). 〈https://www.cdc.gov/healthywater/surveillance/wastewater-surveillance/data-reporting-analytics.html〉.
- 43.Rajal V.B., McSwain B., Thompson D., Leutenegger C., Kildare B., Wuertz S. Validation of hollow fiber ultrafiltration and real-time PCR using bacteriophage PP7 as surrogate for the quantification of viruses from water samples. Water Res. 2007;41:1411–1422. doi: 10.1016/j.watres.2006.12.034. [DOI] [PubMed] [Google Scholar]
- 44.Boxus M., Letellier C., Kerkhofs P. Real time RT-PCR for the detection and quantitation of bovine respiratory syncytial virus. J. Virol. Methods. 2005;125:125–130. doi: 10.1016/j.jviromet.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 45.Cabral A.D., Claro I.C.M., Augusto M.R., Friolani V.N., Bezerra C.D.E., Graciosa M.C.P., Fonseca F.L.A., Speranca M.A., de R., Bueno F. Padronização de método de concentração e extração de ácidos nucleicos em amostras de esgoto sanitário: uma ferramenta de baixo custo para ser utilizada na vigilância epidemiológica de SARS-CoV-2. Eng. Sanit. Ambient. 2021;26:1043–1049. doi: 10.1590/s1413-415220200370. [DOI] [Google Scholar]
- 46.Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hemalatha M., Kiran U., Kuncha S.K., Kopperi H., Gokulan C.G., Mohan S.V., Mishra R.K. Surveillance of SARS-CoV-2 spread using wastewater-based epidemiology: Comprehensive study. Sci. Total Environ. 2021;768 doi: 10.1016/j.scitotenv.2020.144704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brasil, Notificações de Síndrome Gripal (In portuguese), OpenDataSUS. (2020). 〈https://opendatasus.saude.gov.br/dataset/notificacoes-de-sindrome-gripal〉.
- 49.MCTI, Rede Vírus MCTI (In portuguese), Ministério Da Ciência Tecnol. e Inovação. (2021). 〈http://redevirus.mcti.gov.br/novidades/aguas-residuais/〉.
- 50.A.N.A., Monitoramento COVID Esgotos. Boletins, notas e apresentações sobre o projeto (in portuguese), Agência Nac. Águas e Saneam. Básico. (2021). 〈https://www.gov.br/ana/pt-br/assuntos/acontece-na-ana/monitoramento-covid-esgotos〉.
- 51.SMS, Vigilância epidemiológica, painel coronavírus - Foz do Iguaçu (In portuguese), Secr. Munic. Da Saúde. (2021). 〈https://datastudio.google.com/u/0/reporting/9c4e18e4–68c3–425c-a656–20fe415adeea/page/2CmaB〉.
- 52.Souza F.S.H., Hojo-Souza N.S., da Silva C.M., Guidoni D.L. Second wave of COVID-19 in Brazil: younger at higher risk. Eur. J. Epidemiol. 2021;36:441–443. doi: 10.1007/s10654-021-00750-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Victora C.G., Castro M.C., Gurzenda S., Medeiros A.C., França G.V.A., Barros A.J.D. Estimating the early impact of vaccination against COVID-19 on deaths among elderly people in Brazil: Analyses of routinely-collected data on vaccine coverage and mortality. EClinicalMedicine. 2021;38 doi: 10.1016/j.eclinm.2021.101036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sah P., Fitzpatrick M.C., Zimmer C.F., Abdollahi E., Juden-Kelly L., Moghadas S.M., Singer B.H., Galvani A.P. Asymptomatic SARS-CoV-2 infection: a systematic review and meta-analysis. Proc. Natl. Acad. Sci. 2021;118 doi: 10.1073/pnas.2109229118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albani V., Loria J., Massad E., Zubelli J. COVID-19 underreporting and its impact on vaccination strategies. BMC Infect. Dis. 2021;21:1–13. doi: 10.1186/s12879-021-06780-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khan N.A., Al-Thani H., El-Menyar A. The emergence of new SARS-CoV-2 variant (Omicron) and increasing calls for COVID-19 vaccine boosters-the debate continues. Travel Med. Infect. Dis. 2022;45 doi: 10.1016/j.tmaid.2021.102246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poudel S., Ishak A., Perez-Fernandez J., Garcia E., León-Figueroa D.A., Romaní L., Bonilla-Aldana D.K., Rodriguez-Morales A.J. Highly mutated SARS-CoV-2 Omicron variant sparks significant concern among global experts – what is known so far. Travel Med. Infect. Dis. 2022;45 doi: 10.1016/j.tmaid.2021.102234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwon D. Omicron’s molecular structure could help explain its global takeover. Nature. 2022 doi: 10.1038/d41586-022-00292-3. [DOI] [PubMed] [Google Scholar]
- 59.Fujita D.M., dos Santos Soares G., Sartori G.P., Henrique da Silva Nali L. COVID-19 and Influenza coinfection: the rise of Ômicron and H3N2 in Brazil – 2022. Travel Med. Infect. Dis. 2022;46 doi: 10.1016/j.tmaid.2022.102262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amereh F., Jahangiri-rad M., Mohseni-Bandpei A., Mohebbi S.R., Asadzadeh-Aghdaei H., Dabiri H., Eslami A., Roostaei K., Aali R., Hamian P., Rafiee M. Association of SARS-CoV-2 presence in sewage with public adherence to precautionary measures and reported COVID-19 prevalence in Tehran. Sci. Total Environ. 2022;812 doi: 10.1016/j.scitotenv.2021.152597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robotto A., Lembo D., Quaglino P., Brizio E., Polato D., Civra A., Cusato J., Di Perri G. Wastewater-based SARS-CoV-2 environmental monitoring for Piedmont, Italy. Environ. Res. 2021;203 doi: 10.1016/j.envres.2021.111901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nemudryi A., Nemudraia A., Wiegand T., Surya K., Buyukyoruk M., Cicha C., Vanderwood K.K., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Rep. Med. 2020;1 doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kitamura K., Sadamasu K., Muramatsu M., Yoshida H. Efficient detection of SARS-CoV-2 RNA in the solid fraction of wastewater. Sci. Total Environ. 2021;763 doi: 10.1016/j.scitotenv.2020.144587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 65.Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rose C., Parker A., Jefferson B., Cartmell E. The characterization of feces and urine: a review of the literature to inform advanced treatment technology. Crit. Rev. Environ. Sci. Technol. 2015;45:1827–1879. doi: 10.1080/10643389.2014.1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saguti F., Magnil E., Enache L., Churqui M.P., Johansson A., Lumley D., Davidsson F., Dotevall L., Mattsson A., Trybala E., Lagging M., Lindh M., Gisslén M., Brezicka T., Nyström K., Norder H. Surveillance of wastewater revealed peaks of SARS-CoV-2 preceding those of hospitalized patients with COVID-19. Water Res. 2021;189 doi: 10.1016/j.watres.2020.116620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang W., Du R.-H., Li B., Zheng X.-S., Yang X.-L., Hu B., Wang Y.-Y., Xiao G.-F., Yan B., Shi Z.-L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li X., Kulandaivelu J., Guo Y., Zhang S., Shi J., O’Brien J., Arora S., Kumar M., Sherchan S.P., Honda R., Jackson G., Luby S.P., Jiang G. SARS-CoV-2 shedding sources in wastewater and implications for wastewater-based epidemiology. J. Hazard. Mater. 2022;432 doi: 10.1016/j.jhazmat.2022.128667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in Water and Wastewater. Environ. Sci. Technol. Lett. 2020;7:937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li X., Zhang S., Shi J., Luby S.P., Jiang G. Uncertainties in estimating SARS-CoV-2 prevalence by wastewater-based epidemiology. Chem. Eng. J. 2021;415 doi: 10.1016/j.cej.2021.129039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bivins A., Kaya D., Bibby K., Simpson S.L., Bustin S.A., Shanks O.C., Ahmed W. Variability in RT-qPCR assay parameters indicates unreliable SARS-CoV-2 RNA quantification for wastewater surveillance. Water Res. 2021 doi: 10.1016/j.watres.2021.117516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hamouda M., Mustafa F., Maraqa M., Rizvi T., Aly Hassan A. Wastewater surveillance for SARS-CoV-2: lessons learnt from recent studies to define future applications. Sci. Total Environ. 2021;759 doi: 10.1016/j.scitotenv.2020.143493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Street R., Malema S., Mahlangeni N., Mathee A. Wastewater surveillance for Covid-19: an African perspective. Sci. Total Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van der Voorn T., van den Berg C., Bhattacharya P., Quist J. Never waste a crisis: drawing first lessons from the COVID-19 pandemic to tackle the water crisis. ACS EST Water. 2021;1:8–10. doi: 10.1021/acsestwater.0c00041. [DOI] [Google Scholar]
- 77.SNIS, Sistema Nacional de Informações sobre Saneamento: 25o Diagnóstico dos Serviços de Água e Esgotos - 2019., Ministério do Desenvolvimento Regional/Secretaria Nacional de Saneamento, Brasília, DF, 2020.
- 78.IBGE, Pesquisa nacional de saneamento básico Coordenação de População e Indicadores Sociais. Rio De. Jan. 2017:2020. [Google Scholar]
- 79.Silva M.E.D. Interfaces entre Desenvolvimento, Meio Ambiente e Sustentabilidade. Atena Ed. 2021 doi: 10.22533/at.ed.601211103. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary material.
Data Availability Statement
No data was used for the research described in the article.