ABSTRACT
Fluoroquinolones are potentially effective against Elizabethkingia anophelis. We investigated the MIC, mutant prevention concentration (MPC), and target gene mutations of fluoroquinolones in E. anophelis. Eighty-five E. anophelis isolates were collected from five hospitals in Taiwan. The MIC and MPC of ciprofloxacin and levofloxacin were examined for all E. anophelis except 17 isolates, in which ciprofloxacin MPC could not be determined due to drug precipitation caused by overly high drug concentration. Mutations in the quinolone resistance-determining regions of DNA gyrase (GyrA and GyrB) and topoisomerase IV (ParC and ParE) in the clinical isolates and fluoroquinolone-selected mutants were examined. Overall, 23.5% and 71.8% of the isolates tested were susceptible to ciprofloxacin and levofloxacin, respectively. The MPC50 of ciprofloxacin was 128 mg/L, and the MPC50 of levofloxacin was 51.2 mg/L. The MPC50/MIC50 ratio for ciprofloxacin was 64, whereas that for levofloxacin was 25.6. The coefficient of determination between the MPC and MIC for ciprofloxacin and levofloxacin was 0.72 and 0.56, respectively, in the linear regression analysis. Preexisting mutations in GyrA (S83I, S83R, and D87Y) were identified in 18 clinical isolates, all of which were resistant to both ciprofloxacin and levofloxacin. Additional amino acid substitutions in GyrA were identified in all ciprofloxacin- and levofloxacin-selected mutants. Furthermore, GyrB alterations (D431N or D431H) were found in nine levofloxacin-treated isolates. Given that maintaining the serum concentrations of fluoroquinolones above MPCs is impossible under presently recommended doses, the selection of mutant E. anophelis strains seems inevitable.
KEYWORDS: Elizabethkingia anophelis, mutant prevention concentration, ciprofloxacin, levofloxacin, quinolone-resistance determining region
INTRODUCTION
Elizabethkingia spp. are Gram-negative, aerobic, and glucose-nonfermenting bacteria that colonize and survive in soils, water, and hospital environments (1–3). Elizabethkingia anophelis has recently been identified as responsible for lethal infections in humans, particularly in immunocompromised patients. Patients with E. anophelis infection have a mortality rate of 24% to 60% (4–10). This pathogen is usually nonsusceptible to most penicillins, cephalosporins, carbapenems, β-lactam-β-lactamase inhibitor combinations, and aminoglycosides, but it demonstrates variable susceptibilities to fluoroquinolones (4–10). Several mechanisms are involved in the antimicrobial resistance of E. anophelis, such as the production of β-lactamases, carbapenemase, and aminoglycoside acetyltransferase, target gene mutations, and overexpression of efflux systems, etc. (4–10). However, it is not clear whether there is cross-resistance between fluoroquinolones and the remaining antibiotic classes for the drug resistance in E. anophelis.
In the face of the drug-resistant microbes, the concept of the mutant prevention concentration (MPC), the minimum antibiotic concentration inhibiting the growth of the least-susceptible mutant subpopulation, was proposed (11). If the antimicrobial concentrations fall between the MPC and MIC—that is, in the so-called mutant selection window—resistant mutants can be selected (12, 13). Maintaining the concentration of antimicrobial agents above the MPC has been suggested to minimize the selection of mutant strains (12, 13).
The mechanisms for fluoroquinolone resistance include (i) target-mediated resistance: mutations in the quinolone resistance-determining regions (QRDRs) of genes encoding DNA gyrase (i.e., gyrA and gyrB) and DNA topoisomerase IV (i.e., parC and parE); (ii) plasmid-mediated resistance: extrachromosomal genetic elements that encode proteins to disturb quinolone−enzyme interactions, alter drug metabolism, or increase quinolone efflux; and (iii) chromosome-mediated resistance: underexpression of porins or the overexpression of cellular efflux pumps to reduce cellular drug concentration (14). Gene mutations in QRDRs have been established as the main mechanism of fluoroquinolone resistance in E. anophelis (9, 15). To prevent E. anophelis from developing fluoroquinolone resistance, understanding the MPC of fluoroquinolones in this pathogen is essential. However, the fluoroquinolone MPC has never been determined in E. anophelis. In the present study, we investigated the MPCs of levofloxacin and ciprofloxacin against E. anophelis isolates collected from five hospitals in Taiwan. Furthermore, nucleotide alterations in the QRDRs of the clinical isolates and the selected resistant subpopulations were investigated.
RESULTS
Antimicrobial susceptibility.
The MICs of ciprofloxacin and levofloxacin ranged from 0.5 to >512 mg/L and from 0.5 to 128 mg/L, respectively (Table 1). Overall, 71.8% of the 85 isolates were susceptible to levofloxacin, but only 23.5% were susceptible to ciprofloxacin.
TABLE 1.
Antimicrobial susceptibility testing of 85 Elizabethkingia anophelis isolates
| Antimicrobial agent | MIC (mg/L) |
Interpretation of susceptibility, n (%) |
|||||
|---|---|---|---|---|---|---|---|
| Lowest | Highest | MIC50c | MIC90d | Susceptible | Intermediate | Resistant | |
| Ciprofloxacina | 0.5 | >512 | 2 | 512 | 20 (23.5) | 26 (30.6) | 39 (45.9) |
| Levofloxacinb | 0.5 | 128 | 2 | 32 | 61 (71.8) | 3 (3.5) | 21 (24.7) |
Susceptible MIC, ≤1 mg/L; intermediate MIC, 2 mg/L; resistant MIC, ≥4 mg/L.
Susceptible MIC, ≤2 mg/L; intermediate MIC, 4 mg/L; resistant MIC, ≥8 mg/L.
MIC at which 50% of the isolates tested are inhibited.
MIC at which 90% of the isolates tested are inhibited.
MPC determination.
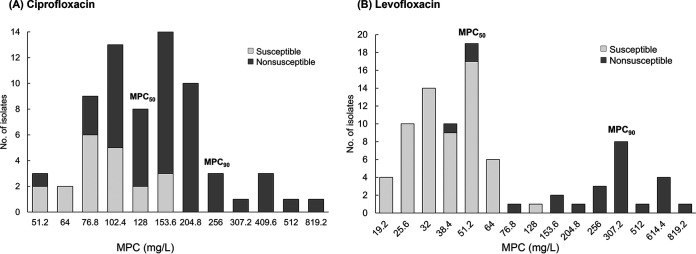
All isolates (n = 85) were tested for levofloxacin provisional MPC (MPCpr) and MPC. However, 17 isolates exhibited ciprofloxacin MPCpr >1,024 mg/L, and their MPC could not be determined due to drug precipitation caused by overly high drug concentration. In total, 68 isolates were tested to determine the ciprofloxacin MPCpr and MPC. Figure 1 presents the ciprofloxacin MPCs of 68 isolates and levofloxacin MPCs of 85 isolates. The MPC50 of ciprofloxacin was 128 mg/L and that of levofloxacin was 51.2 mg/L. When only ciprofloxacin-susceptible (MIC ≤1 mg/L) or levofloxacin-susceptible isolates (MIC ≤2 mg/L) were examined, the MPC of ciprofloxacin was 51.2–153.6 mg/L, and the MPC of levofloxacin was 19.2–128 mg/L.
FIG 1.
Mutant prevention concentration (MPC) of fluoroquinolones in Elizabethkingia anophelis. (A) MPC of ciprofloxacin in 68 isolates. (B) MPC of levofloxacin in 85 isolates.
Relationship between MIC and MPC.
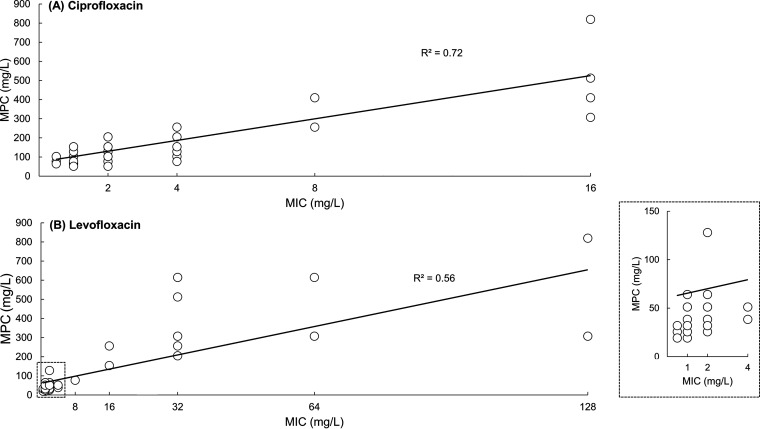
Overall, the MPC/MIC ratios of ciprofloxacin were higher than were those of levofloxacin (Table 2). The MPC/MIC ratios of ciprofloxacin ranged from 16 to ≥128, and those of levofloxacin ranged from <16 to <128. The MPC50/MIC50 ratio for ciprofloxacin and levofloxacin was 64 and 25.6, respectively. The MPC90/MIC90 ratio for ciprofloxacin was 153.6, whereas that for levofloxacin was 51.2. When only the susceptible portions of isolates (ciprofloxacin MIC ≤1 mg/L or levofloxacin MIC ≤2 mg/L) were included, the MPC/MIC ranges of ciprofloxacin and levofloxacin were 51.2 to 204.8 and 12.8 to 64, respectively. The coefficient of determination (R2) between the MPC and MIC for ciprofloxacin was 0.72 and that for levofloxacin was 0.56 (Fig. 2).
TABLE 2.
MPC/MIC ratios of fluoroquinolones in E. anophelis isolates
| Antimicrobial agent | No. (%) of isolates having ratios of MPC/MIC |
MPC50/MIC50a | MPC90/MIC90b | ||||
|---|---|---|---|---|---|---|---|
| <16 | 16 to <32 | 32 to <64 | 64 to <128 | ≥128 | |||
| Ciprofloxacin (n = 68) | 0 | 5 (7.4) | 27 (39.7) | 26 (38.2) | 10 (14.7) | 64 | 153.6 |
| Levofloxacin (n = 85) | 20 (23.5) | 34 (40) | 25 (29.4) | 6 (7.1) | 0 | 25.6 | 51.2 |
The 50th percentile of mutant prevention concentration (MPC)/MIC ratio among the isolates tested.
bThe 90th percentile of MPC/MIC ratio among the isolates tested.
FIG 2.
Correlation between the MIC and MPC of fluoroquinolones in E. anophelis. (A) Relationship determined for ciprofloxacin in 68 isolates. (B) Relationship determined for levofloxacin in 85 isolates. The inset of the square represents the isolates which are more susceptible to levofloxacin.
QRDR mutations.
Preexisting mutations in the QRDR of GyrA were identified in 18 clinical isolates (Table 3). All 18 isolates were resistant to both ciprofloxacin and levofloxacin. The amino acid alterations included S83I (AGC→ATC; n = 15), S83R (AGC→AGA; n = 2), and D87Y (GAT→TAT; n = 1). A mutation in ParC (P134T; CCG→ACG) was detected in an isolate with MICs for ciprofloxacin and levofloxacin of 4 and 2 mg/L, respectively.
TABLE 3.
Preexisting mutations in the quinolone-resistance determining regions of 85 E. anophelis isolates
| Site of mutation | No. of isolate | Ciprofloxacin MIC (mg/L) | Levofloxacin MIC (mg/L) |
|---|---|---|---|
| No mutation (n = 66) | 5 | 0.5 | 0.5 |
| 1 | 1 | 0.5 | |
| 14 | 1 | 1 | |
| 15 | 2 | 1 | |
| 11 | 2 | 2 | |
| 13 | 4 | 2 | |
| 1 | 4 | 4 | |
| 1 | 8 | 4 | |
| 1 | 8 | 8 | |
| 1 | 8 | 16 | |
| 1 | 16 | 2 | |
| 1 | 16 | 4 | |
| 1 | 16 | 16 | |
| S83I in GyrA (n = 15) | 4 | 256 | 32 |
| 2 | 512 | 32 | |
| 1 | 512 | 64 | |
| 2 | >512 | 32 | |
| 3 | >512 | 64 | |
| 3 | >512 | 128 | |
| S83R in GyrA (n = 2) | 1 | 128 | 16 |
| 1 | 256 | 32 | |
| D87Y in GyrA (n = 1) | 1 | 16 | 16 |
| P134T in ParC (n = 1) | 1 | 4 | 2 |
Additional QRDR changes in GyrA were identified in all 68 ciprofloxacin-selected mutants (Table 4). By contrast, no isolate was found to have induced nucleotide substitutions in GyrB, ParC, or ParE. The most common ciprofloxacin-induced mutation was S83I in GyrA, followed by S83R in GyrA. Four isolates (5.9%) had double mutations in GyrA, specifically S83R/D87Y (MPC = 153.6 mg/L), S83R/S84P (MPC = 204.8 mg/L), S83R/D87H (MPC = 307.2 mg/L), and S83I/D87Y (MPC = 512 mg/L). One isolate had triple substitutions in GyrA (H78Y/S83I/D87Y; MPC = 204.8 mg/L). The detailed information on the QRDR mutations is shown in Table S2 in the supplemental material.
TABLE 4.
Preexisting and induced mutations after ciprofloxacin treatment in the QRDRs of E. anophelis isolatesa
| Ciprofloxacin MPC (mg/L) | Preexisting QRDR change in the original collection |
QRDR change in the selected mutant |
||
|---|---|---|---|---|
| GyrA | ParC | GyrA | ||
| 51.2 (n = 3) | S83I (3) | |||
| 64 (n = 2) | S83I (2) | |||
| 76.8 (n = 9) | S83I (7), S83R (2) | |||
| 102.4 (n = 13) | S83I (12), S83I/R (1)b | |||
| 128 (n = 8) | S83I (7), A119K (1) | |||
| 153.6 (n = 14) | P134T (1) | S83I (13), S83R/D87Y (1) | ||
| 204.8 (n = 10) | S83I (7), S83I/R (1),b S83R/S84P (1), H78Y/S83I/D87Y (1) | |||
| 256 (n = 3) | S83I (3) | |||
| 307.2 (n = 1) | S83R/D87H (1) | |||
| 409.6 (n = 3) | S83I (3) | |||
| 512 (n = 1) | D87Y (1) | S83I/D87Y (1) | ||
| 819.2 (n = 1) | S83I (1) | |||
| N/A (n = 17)c | S83I (15), S83R (2) | N/Ac | ||
Parentheses represent the number of isolates with quinolone-resistance determining region (QRDR) mutations.
Nucleotide mutation detected in different colonies of the same isolate.
MPCs and induced QRDR changes could not be examined in 17 isolates due to drug precipitation caused by overly high drug concentrations.
As for levofloxacin-selected mutants (Table 5 and Table S2), QRDR changes in GyrA were detected in all 85 isolates: 71 isolates acquiring additional mutations and 14 having only preexisting mutations. The most common additional mutation was G81D (n = 63), followed by S83I (n = 15). QRDR mutations in GyrB (D431N or D431H) were found in nine isolates, all of which harbored preexisting S83I or S83R mutations. No additional mutations were detected in ParC and ParE in the levofloxacin-selected mutants. Double mutations in GyrA were identified in 16 isolates (18.8%), including G81D/S83I (n = 11; MPC = 19.2–64 mg/L), S83R/D87Y (n = 3; MPC = 153.6–256 mg/L), D82N/S83I (n = 1; MPC = 38.4 mg/L), and S83I/D87N (n = 1; MPC = 307.2 mg/L). No isolates had triple mutations in GyrA.
TABLE 5.
Preexisting and induced mutations after levofloxacin treatment in the QRDRs of E. anophelis isolatesa
| Levofloxacin MPC (mg/L) | Preexisting QRDR change in the original collection |
QRDR change in the selected mutant |
||
|---|---|---|---|---|
| GyrA | ParC | GyrA | GyrB | |
| 19.2 (n = 4) | G81D (3), G81D/S83I (1) | |||
| 25.6 (n = 10) | G81D (8), G81D/S83I (2) | |||
| 32 (n = 14) | G81D (10), G81D/S83I (4) | |||
| 38.4 (n = 10) | G81D (8), G81D/S83I (1), D82N/S83I (1) | |||
| 51.2 (n = 19) | G81D (16), G81D/S83I (2), S83I (1) | |||
| 64 (n = 6) | P134T (1) | G81D (5), G81D/S83I (1) | ||
| 76.8 (n = 1) | S83I (1) | |||
| 128 (n = 1) | G81D (1) | |||
| 153.6 (n = 2) | S83R (1) | S83I (1), S83R/D87Y (1) | ||
| 204.8 (n = 1) | S83R (1) | S83R/D87Y (1) | D431N (1) | |
| 256 (n = 3) | S83I (1), D87Y (1) | G81D (1), S83I (1), S83R/D87Y (1) | ||
| 307.2 (n = 8) | S83I (8) | S83I (7), S83I/D87N (1) | D431H (5) | |
| 512 (n = 1) | S83I (1) | S83I (1) | D431H (1) | |
| 614.4 (n = 4) | S83I (4) | S83I (4) | D431H (1) | |
| 819.2 (n = 1) | S83I (1) | S83I (1) | D431H (1) | |
Parentheses represent the number of isolates with QRDR mutations.
DISCUSSION
Many studies have demonstrated that ciprofloxacin and levofloxacin were potentially effective against E. anophelis (4–10). With the extensive use of antibiotics, resistance to fluoroquinolones has emerged as a global public health concern. To prevent the widespread emergence of antimicrobial resistance, optimizing strategies for the therapeutic administration of antibiotics is imperative.
Given that it reflects the minimum concentration of the least drug-susceptible mutant subpopulation, the MPC can be applied to the formulation of antimutant dosing strategies (16). Maintaining drug concentrations above the MPC can prevent bacteria from acquiring antimicrobial resistance under selective pressure. However, the determination of MPC is time-consuming and labor-intensive and thus cannot be routinely performed in clinical practice. Sanders (17) indicated that fluoroquinolone concentrations for stepwise mutant selection ranged from 4-fold to 8-fold MIC and suggested that fluoroquinolones with more than 8-fold MIC were least likely to be selected for resistance. Ho wever, some studies showed a poor relationship between the MPC and MIC for fluoroquinolones in Escherichia coli and Pseudomonas aeruginosa (18, 19). Herein, the MPC of ciprofloxacin against E. anophelis was 16 to ≥128 times the MIC, and the MPC of levofloxacin was <16 to <128 times the MIC. The MPC and MIC of fluoroquinolones against E. anophelis was not very strongly correlated, particularly for levofloxacin (R2 = 0.56). The MIC corresponding to individual E. anophelis isolates was not highly predictive of the MPC.
The MPCs of ciprofloxacin and levofloxacin in the fluoroquinolone-susceptible isolates exceeded 51.2 and 19.2 mg/L, respectively. Previous studies indicated that the peak serum concentration of ciprofloxacin following a 400-mg intravenous injection and that of levofloxacin following a 750-mg intravenous injection was only approximately 6.7 and 8.1 mg/L, respectively (20, 21). Although these peak serum concentrations are above the MIC50 of ciprofloxacin and levofloxacin for E. anophelis, they are substantially lower than the MPC50. Thus, under presently recommended standard doses of ciprofloxacin and levofloxacin, efforts to prevent mutation emergence in the treatment of E. anophelis infection would not be effective. Combination therapy may be necessary to minimize the selection of mutants when using fluoroquinolones to treat patients with E. anophelis infection (22).
Herein, 21.2% of the clinical isolates harbored amino acid substitutions of S83I, S83R, or D87Y in GyrA, and all of those isolates exhibited resistance to ciprofloxacin and levofloxacin. Except for D87Y, mutations of S83I or S83R in GyrA are commonly associated with fluoroquinolone resistance (23, 24). For the selected mutants, our study found that the most common additional mutation following ciprofloxacin and levofloxacin treatment was S83I and G81D in GyrA, respectively. GyrB mutations were detected in nine isolates following levofloxacin therapy. Neither ParC nor ParE mutations were identified with ciprofloxacin- or levofloxacin-treated E. anophelis. Drug resistance to fluoroquinolones in a stepwise manner has been observed in many bacteria. Mutations in gyrA are often recovered first, followed by additional mutations in parC when treated with higher concentration of fluoroquinolones (25, 26). However, our study identified nine isolates with second-step gyrB mutations in addition to their first-step gyrA mutations after high-level levofloxacin treatment. The second-step amino acid alterations in the ParC of E. anophelis was not observed in our study. The reason for this result remains unclear.
Conclusions.
The findings from this study suggest that MPCs of fluoroquinolones in E. anophelis should be determined by experiments owing to not very strong correlation between the MIC and MPC. Due to unattainably high serum drug levels over MPC, the potential for resistance against these fluoroquinolones in the treatment of E. anophelis infection seems undeniable. Including another class of antibiotic as part of an antimicrobial strategy may be a reasonable approach for limiting the selection of fluoroquinolone-resistant mutants in treating this emerging infection.
MATERIALS AND METHODS
Ethics approval.
This study was conducted in accordance with the tenets of the Declaration of Helsinki and the national standards of Taiwan, and the study protocol received institutional review board approval (approval no. EMRP-109-007). The need for informed consent was waived because the retrospective analysis of clinical isolates posed no more than a minimal risk of harm to the patients from which they were collected.
Study setting and bacterial strains.
Between 2016 and 2019, 85 clinical isolates of Elizabethkingia spp. were collected from five hospitals in Taiwan, including 28 isolates from E-Da Hospital, 26 from Kaohsiung Medical University Hospital, 15 from National Cheng Kung University Hospital, 9 from E-Da Cancer Hospital, and 7 from Taichung Veterans General Hospital. These isolates were routinely collected from patients at each hospital according to clinical requirements and were then stored as glycerol stocks at −80°C until use. The precise species of the isolates was reidentified using 16S rRNA gene sequencing as described in our previous study (9).
MIC determination.
Ciprofloxacin and levofloxacin were obtained from Sigma-Aldrich (St. Louis, MO, USA). Their MICs against E. anophelis isolates were examined using 96-well broth microdilution panels in accordance with the manufacturer’s instructions (Thermo Fisher Scientific, Oakwood Village, OH, USA). The breakpoints for susceptibility testing were appraised under the interpretive standards for “other non-Enterobacteriaceae” from Clinical & Laboratory Standards Institute guidelines (27). An MIC of ≤1 mg/L indicated susceptibility to ciprofloxacin, whereas MICs of 2 and ≥4 mg/L corresponded to intermediate sensitivity and resistance, respectively. MICs of ≤2, 4, and ≥8 mg/L indicated susceptibility, intermediate sensitivity, and resistance to levofloxacin, respectively.
MPC determination.
MPC determination was conducted as described previously (12, 28). In brief, the frozen bacteria were thawed and subcultured on Muller-Hinton (MH) agar plates (Becton, Dickinson and Company, Sparks, MD, USA) and incubated overnight in 5% CO2 at 37°C. Bacteria were then placed in 25 mL of cation-adjusted MH broth (CAMHB; Becton, Dickinson and Company) and incubated overnight at 37°C on a shaker operating at 180 rpm. The turbidity of the culture medium was adjusted to OD 1.8 (approximately 1 × 1010 CFU/mL). Aliquots of 1 mL were concentrated by centrifugation at 8,000 × g for 10 min at 10°C. The supernatants were discarded, and the pellets were resuspended in 200 μL of CAMHB. Next, 200-μL aliquots containing at least 1010 CFU/mL were spread on MH agar plates with a panel of geometric sequences of different concentrations of antibiotics (1×, 2×, 4×, …512 × MIC) and then incubated in 5% CO2 for 72 h at 37°C. The plates were screened visually for growth. The lowest antibiotic concentration that inhibited bacterial growth was recorded as the MPCpr (12). To refined the MPC of each isolate, the aforementioned procedures were repeated, and the bacteria were inoculated on agar plates with corresponding antibiotic concentrations of 20% linear reductions (i.e., 80% and 60%) of the MPCpr values (28). The lowest concentration inhibiting bacterial growth was designated as the MPC. For each isolate, the MPC/MIC ratio was calculated. The MPC50/MIC50 and MPC90/MIC90 ratios represent the 50th and 90th percentiles of the MPC/MIC ratios among the isolates tested, respectively.
QRDR mutation identification.
Bacteria colonies that survived on each MH agar plate with the highest concentration of fluoroquinolones below the MPC were regarded as potentially resistant mutants. Nucleotide sequences were examined for the mutations in QRDRs of gyrA, gyrB, parC, and parE in three randomly selected colonies of the originally collected isolates and potentially fluoroquinolone-resistant strains. Total DNA from was prepared using a Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA). The PCR amplicons were sequenced using an Applied Biosystems 3730xl DNA Analyzer. The primers for amplification and sequencing of QRDRs are listed in Table S1 in the supplemental material. The conditions used for QRDR amplification were an initial extended denaturation step of 94°C for 5 min, followed by 30 cycles of 30 s at 94°C, 30 s at 55°C, 1 min at 72°C, and a final 5 min at 72°C.
Statistical analysis.
Linear regression analysis was performed to determine the relationships between the MICs and MPCs of ciprofloxacin and levofloxacin by using IBM SPSS Statistics for Windows, version 24 (IBM Corp., Armonk, NY, USA). The coefficient of determination, R2, between the MIC and MPCs was calculated.
ACKNOWLEDGMENTS
This work was supported by grants EDAHP109046 from E-Da Hospital and MOST 109-2314-B-214-006-MY2 from the Ministry of Science and Technology, Taiwan.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Nicholson AC, Gulvik CA, Whitney AM, Humrighouse BW, Graziano J, Emery B, Bell M, Loparev V, Juieng P, Gartin J, Bizet C, Clermont D, Criscuolo A, Brisse S, McQuiston JR. 2018. Revisiting the taxonomy of the genus Elizabethkingia using whole-genome sequencing, optical mapping, and MALDI-TOF, along with proposal of three novel Elizabethkingia species: Elizabethkingia bruuniana sp. nov., Elizabethkingia ursingii sp. nov., and Elizabethkingia occulta sp. nov. Antonie Van Leeuwenhoek 111:55–72. doi: 10.1007/s10482-017-0926-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore LSP, Owens DS, Jepson A, Turton JF, Ashworth S, Donaldson H, Holmes AH. 2016. Waterborne Elizabethkingia meningoseptica in adult critical care. Emerg Infect Dis 22:9–17. doi: 10.3201/eid2201.150139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yung CF, Maiwald M, Loo LH, Soong HY, Tan CB, Lim PK, Li L, Tan NW, Chong CY, Tee N, Thoon KC, Chan YH. 2016. Elizabethkingia anophelis and association with tap water and handwashing, Singapore. Emerg Infect Dis 24:1730–1733. doi: 10.3201/eid2409.171843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin JN, Lai CH, Yang CH, Huang YH. 2019. Elizabethkingia infections in humans: from genomics to clinics. Microorganisms 7:295. doi: 10.3390/microorganisms7090295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lau SK, Chow WN, Foo CH, Curreem SO, Lo GC, Teng JL, Chen JH, Ng RH, Wu AK, Cheung IY, Chau SK, Lung DC, Lee RA, Tse CW, Fung KS, Que TL, Woo PC. 2016. Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci Rep 6:26045. doi: 10.1038/srep26045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perrin A, Larsonneur E, Nicholson AC, Edwards DJ, Gundlach KM, Whitney AM, Gulvik CA, Bell ME, Rendueles O, Cury J, Hugon P, Clermont D, Enouf V, Loparev V, Juieng P, Monson T, Warshauer D, Elbadawi LI, Walters MS, Crist MB, Noble-Wang J, Borlaug G, Rocha EPC, Criscuolo A, Touchon M, Davis JP, Holt KE, McQuiston JR, Brisse S. 2017. Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat Commun 8:15483. doi: 10.1038/ncomms15483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Navon L, Clegg WJ, Morgan J, Austin C, McQuiston JR, Blaney DD, Walters MS, Moulton-Meissner H, Nicholson A. 2016. Notes from the field: investigation of Elizabethkingia anophelis cluster–Illinois, 2014–2016. MMWR Morb Mortal Wkly Rep 65:1380–1381. doi: 10.15585/mmwr.mm6548a6. [DOI] [PubMed] [Google Scholar]
- 8.Chew KL, Cheng B, Lin RT, Teo JW. 2018. Elizabethkingia anophelis is the dominant Elizabethkingia species found in blood cultures in Singapore. J Clin Microbiol 56:e01445-17. doi: 10.1128/JCM.01445-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin JN, Lai CH, Yang CH, Huang YH, Lin HH. 2018. Clinical manifestations, molecular characteristics, antimicrobial susceptibility patterns and contributions of target gene mutation to fluoroquinolone resistance in Elizabethkingia anophelis. J Antimicrob Chemother 73:2497–2502. doi: 10.1093/jac/dky197. [DOI] [PubMed] [Google Scholar]
- 10.Breurec S, Criscuolo A, Diancourt L, Rendueles O, Vandenbogaert M, Passet V, Caro V, Rocha EPC, Touchon M, Brisse S. 2016. Genomic epidemiology and global diversity of the emerging bacterial pathogen Elizabethkingia anophelis. Sci Rep 6:30379. doi: 10.1038/srep30379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong Y, Zhao X, Domagala J, Drlica K. 1999. Effect of fluoroquinolone concentration on selection of resistant mutants of Mycobacterium bovis BCG and Staphylococcus aureus. Antimicrob Agents Chemother 43:1756–1758. doi: 10.1128/AAC.43.7.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blondeau JM, Zhao X, Hansen G, Drlica K. 2001. Mutant prevention concentrations of fluoroquinolones for clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother 45:433–438. doi: 10.1128/AAC.45.2.433-438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith HJ, Walters M, Hisanaga T, Zhanel GG, Hoban DJ. 2004. Mutant prevention concentrations for single-step fluoroquinolone-resistant mutants of wild-type, efflux-positive, or ParC or GyrA mutation-containing Streptococcus pneumoniae isolates. Antimicrob Agents Chemother 48:3954–3958. doi: 10.1128/AAC.48.10.3954-3958.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aldred KJ, Kerns RJ, Osheroff N. 2014. Mechanism of quinolone action and resistance. Biochemistry 53:1565–1574. doi: 10.1021/bi5000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jian MJ, Cheng YH, Chung HY, Cheng YH, Yang HY, Hsu CS, Perng CL, Shang HS. 2019. Fluoroquinolone resistance in carbapenem-resistant Elizabethkingia anophelis: phenotypic and genotypic characteristics of clinical isolates with topoisomerase mutations and comparative genomic analysis. J Antimicrob Chemother 74:1503–1510. doi: 10.1093/jac/dkz045. [DOI] [PubMed] [Google Scholar]
- 16.Zhao X, Drlica K. 2001. Restricting the selection of antibiotic-resistant mutants: a general strategy derived from fluoroquinolone studies. Clin Infect Dis 33(Suppl 3):S147–156. doi: 10.1086/321841. [DOI] [PubMed] [Google Scholar]
- 17.Sanders CC. 2001. Mechanisms responsible for cross-resistance and dichotomous resistance among the quinolones. Clin Infect Dis 32(Suppl 1):S1–8. doi: 10.1086/319369. [DOI] [PubMed] [Google Scholar]
- 18.Marcusson LL, Olofsson SK, Komp Lindgren P, Cars O, Hughes D. 2005. Mutant prevention concentrations of ciprofloxacin for urinary tract infection isolates of Escherichia coli. J Antimicrob Chemother 55:938–943. doi: 10.1093/jac/dki136. [DOI] [PubMed] [Google Scholar]
- 19.Hansen GT, Zhao X, Drlica K, Blondeau JM. 2006. Mutant prevention concentration for ciprofloxacin and levofloxacin with Pseudomonas aeruginosa. Int J Antimicrob Agents 27:120–124. doi: 10.1016/j.ijantimicag.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Catchpole C, Andrews JM, Woodcock J, Wise R. 1994. The comparative pharmacokinetics and tissue penetration of single-dose ciprofloxacin 400 mg iv and 750 mg po. J Antimicrob Chemother 33:103–110. doi: 10.1093/jac/33.1.103. [DOI] [PubMed] [Google Scholar]
- 21.Chow AT, Fowler C, Williams RR, Morgan N, Kaminski S, Natarajan J. 2001. Safety and pharmacokinetics of multiple 750-milligram doses of intravenous levofloxacin in healthy volunteers. Antimicrob Agents Chemother 45:2122–2125. doi: 10.1128/AAC.45.7.2122-2125.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drlica K, Zhao X. 2007. Mutant selection window hypothesis updated. Clin Infect Dis 44:681–688. doi: 10.1086/511642. [DOI] [PubMed] [Google Scholar]
- 23.Weigel LM, Anderson GJ, Tenover FC. 2002. DNA gyrase and topoisomerase IV mutations associated with fluoroquinolone resistance in Proteus mirabilis. Antimicrob Agents Chemother 46:2582–2587. doi: 10.1128/AAC.46.8.2582-2587.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beka L, Fullmer MS, Colston SM, Nelson MC, Talagrand-Reboul E, Walker P, Ford B, Whitaker IS, Lamy B, Gogarten JP, Graf J. 2018. Low-level antimicrobials in the medicinal leech select for resistant pathogens that spread to patients. mBio 9:e01328-18. doi: 10.1128/mBio.01328-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan XS, Fisher LM. 1997. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob Agents Chemother 41:471–474. doi: 10.1128/AAC.41.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan XS, Fisher LM. 1998. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob Agents Chemother 42:2810–2816. doi: 10.1128/AAC.42.11.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clinical and Laboratory Standards Institute. 2020. Performance standards for antimicrobial susceptibility testing: twenty-sixth informational supplement M100-S30. CLSI, Wayne, PA. [Google Scholar]
- 28.Sun C, Hao J, Dou M, Gong Y. 2015. Mutant prevention concentrations of levofloxacin, pazufloxacin and ciprofloxacin for A. baumannii and mutations in gyrA and parC genes. J Antibiot (Tokyo) 68:313–317. doi: 10.1038/ja.2014.150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2. Download aac.00301-22-s0001.pdf, PDF file, 0.1 MB (98.3KB, pdf)