ABSTRACT
Enterococcus faecalis is a common commensal bacterium in the gastrointestinal tract as well as a frequent nosocomial pathogen. The secreted metalloprotease gelatinase (GelE) is an important E. faecalis virulence factor that contributes to numerous cellular activities, such as autolysis, biofilm formation, and biofilm-associated antibiotic resistance. Expression of gelE has been extensively studied and is regulated by the Fsr quorum sensing system. Here, we identify two additional factors regulating gelatinase expression and activity in E. faecalis OG1RF. The Bph phosphatase is required for expression of gelE in an Fsr-dependent manner. Additionally, the membrane-anchored protein foldase PrsA is required for GelE activity, but not fsr or gelE gene expression. Disrupting prsA also leads to increased antibiotic sensitivity in biofilms independent of the loss of GelE activity. Together, our results expand the model for gelatinase production in E. faecalis, which has important implications for fundamental studies of GelE function in Enterococcus and also E. faecalis pathogenesis.
IMPORTANCE In Enterococcus faecalis, gelatinase (GelE) is a virulence factor that is also important for biofilm formation and interactions with other microbes as well as the host immune system. The long-standing model for GelE production is that the Fsr quorum sensing system positively regulates expression of gelE. Here, we update that model by identifying two additional factors that contribute to gelatinase production. The biofilm-associated Bph phosphatase regulates the expression of gelE through Fsr, and the peptidyl-prolyl isomerase PrsA is required for production of active GelE through an Fsr-independent mechanism. This provides important insight into how regulatory networks outside of the fsr locus coordinate expression of gelatinase.
KEYWORDS: biofilms, Enterococcus, gelatinase, quorum sensing
INTRODUCTION
Enterococcus faecalis is a commensal bacterium in the gastrointestinal tracts of hosts ranging from insects to humans (1). It is also a prevalent human pathogen that causes biofilm-associated infections, such as endocarditis, urinary tract infections, and infections at wounds and surgical sites (2, 3). A major virulence factor for E. faecalis is gelatinase (GelE), a secreted zinc metalloprotease that mediates chain length and autolysis as well as host intestinal epithelial permeability (4–6). GelE also processes numerous substrates, including the Candida albicans-inhibiting peptide EntV (EF1097, enterocin O16) (7, 8), the major E. faecalis autolysin AtlA (9), pheromone peptides that induce conjugative plasmid transfer in E. faecalis (5, 10), and a component of the enterohemorrhagic Escherichia coli type 3 secretion system (11). Expression of gelE is positively regulated by the well-studied Fsr quorum sensing system, which is encoded by the fsrABDC locus (12–14). Genotypic and phenotypic screens of E. faecalis clinical isolates have identified mutations that abrogate production of GelE, including a 23.9-kb deletion encompassing the fsr locus as well as truncations and IS256 insertions in fsrC (15–20).
Here, we identified two additional gene products that modulate gelatinase expression and activity in E. faecalis OG1RF. Using RNA sequencing (RNA-seq), we found that deletion of the gene encoding the biofilm-associated phosphatase Bph (Δbph) decreases expression of the fsr locus and thus gelE. Δbph also had decreased expression of an uncharacterized locus encoding multiple cell surface WxL domain proteins, which contribute to in vitro biofilm formation. Through separate experiments, we found that the peptidyl-prolyl isomerase (PPIase) PrsA is required for GelE activity in an Fsr-independent manner, and we hypothesize that PrsA is required for correct folding (and therefore activity) of secreted GelE. PrsA was previously linked to salt tolerance and virulence in E. faecalis (21). Additionally, PrsA homologs in other Gram-positive bacteria are required for activity of secreted proteins (22–24) and resistance to oxacillin and vancomycin (25, 26). We observed a similar pattern in OG1RF, although increased antibiotic sensitivity was detected in biofilms and colony growth on plates, but not liquid culture. These results describe two new regulators of the virulence factor gelatinase, highlight the global effects of disrupting bph and prsA in E. faecalis, and provide insight into biofilm-specific responses to antibiotics when secreted proteins are misfolded.
RESULTS
Deletion of bph decreases expression of gelE through the Fsr quorumsensing system.
We previously reported that E. faecalis Bph is a phosphatase required for biofilm formation, and we found widespread differences in protein expression in a Δbph mutant compared to the parental OG1RF strain (27). To determine whether these changes were due to differential gene expression, we used RNA-seq to compare transcripts present in planktonic cultures of OG1RF and Δbph after 2 and 4 h of growth. Using a significance cutoff of q < 0.05 and a log2 fold change cutoff of ±2, we identified 133 differentially expressed transcripts in Δbph at 2 h (42 upregulated, 91 downregulated) and 218 differentially expressed transcripts at 4 h (119 upregulated, 99 downregulated) (see Table S1 and Fig. S1A and B in the supplemental material). Differentially expressed genes included many involved in carbohydrate and nucleotide metabolism, membrane transport, peptidoglycan biosynthesis, ribosomal protein synthesis, protein secretion, and DNA replication and repair (Fig. 1C and D). Additionally, the ethanolamine utilization operon (OG1RF_11333-11351) plus genes involved in selenium and molybdenum metabolism and folate biosynthesis (OG1RF_11941-11962 [28] and OG1RF_11179-11189) were significantly upregulated at 4 h (see Table S1).
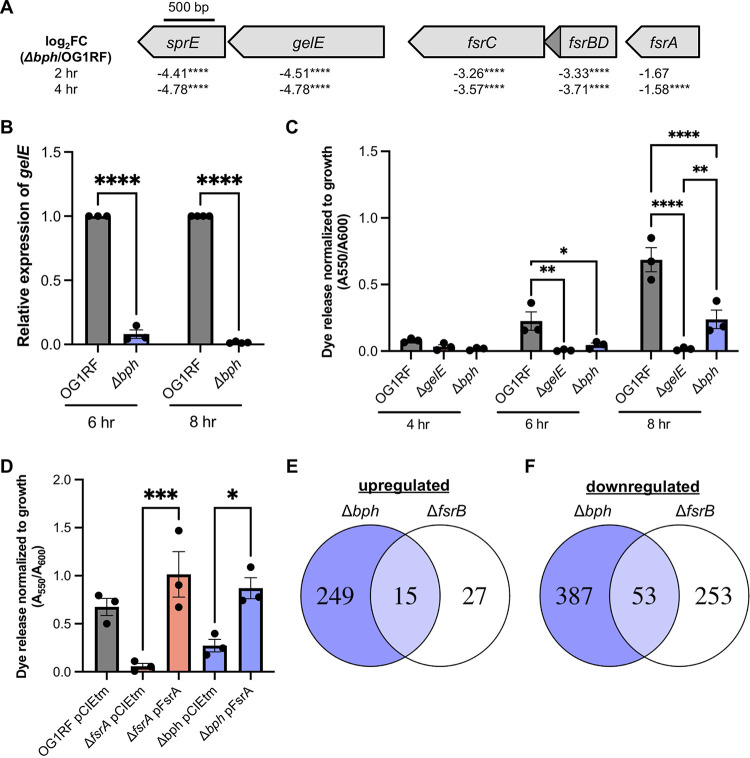
FIG 1.
Deletion of bph reduces gelE expression and GelE activity through reduced expression of the Fsr quorum sensing system. (A) Genetic organization of fsrABDC, gelE, and sprE, showing the direction the genes are encoded and log2 fold change (FC) values from RNA-seq at 2 and 4 h. ****, P < 0.0001. The fsrD open reading frame is shown as a dark gray triangle. (B) Relative levels of gelE transcripts in OG1RF and Δbph at 6 and 8 h as measured by qRT-PCR. Three biological replicates were tested at 6 h, and 4 biological replicates were tested at 8 h. ****, P < 0.0001 by two-way analysis of variance (ANOVA) with Sidak’s test for multiple comparisons. (C) Quantification of dye release (A550) from OG1RF, ΔgelE, and Δbph supernatants relative to growth (A600) at 4, 6, and 8 h. Three biological replicates were tested for each strain. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 by two-way ANOVA with Sidak’s test for multiple comparisons. (D) Rescue of gelatinase activity in Δbph by expression of fsrA from a pheromone-inducible plasmid. Three biological replicates were tested for all strains. *, P < 0.05; ***, P < 0.001 by one-way ANOVA with Sidak’s test for multiple comparisons. For panels B, C, and D, each data point represents an independent biological replicate, and error bars represent standard errors of the means. (E and F) Venn diagrams comparing upregulated and downregulated genes in Δbph and ΔfsrB mutants.
Surprisingly, expression of the Fsr quorum sensing system as well as the Fsr-regulated proteases gelE and sprE was significantly reduced in Δbph compared to OG1RF (Fig. 1A; see also Table S1 in the supplemental material), although we previously found that Δbph has a gelatinase (GelE)-positive phenotype on gelatin plates (27). Because fsr, gelE, and sprE expression increases with greater cell density, we used reverse transcription-quantitative PCR (qRT-PCR) to measure gelE expression in Δbph at 6 and 8 h. We measured an ~92% decrease in transcript levels relative to OG1RF at 6 h and an ~95% decrease at 8 h (Fig. 1B), suggesting that the Fsr system is downregulated throughout Δbph growth.
We next quantified GelE activity using dye release assays to resolve the incongruent expression and phenotype in the Δbph mutant. Gelatin plate-based assays can show cumulative GelE activity after overnight colony growth and may obscure subtle phenotypes. We isolated supernatants from planktonic OG1RF, ΔgelE, and Δbph cultures at 4, 6, and 8 h and mixed them with the protease substrate Azocoll. Proteases like GelE degrade the insoluble dye-protein complex and release soluble azo dyes, which can be quantified by measuring absorbance (29). Dye released by OG1RF supernatants increased over time, while no dye release was detected in the ΔgelE supernatants (Fig. 1C). Dye release from Δbph supernatants decreased 70 to 80% relative to that of OG1RF at each time point. Gelatinase activity in Δbph was significantly increased relative to that in ΔgelE at 8 h but not 4 or 6 h. This suggests that despite reduced gelE expression in Δbph, low levels of active GelE accumulate over time, resulting in the GelE-positive phenotype previously observed after overnight growth on plates (27).
Low levels of fsrA transcript in Δbph (see Table S1) could be caused by reduced gene expression or RNA degradation. We reasoned that if transcription of fsrA was reduced, then expressing fsrA from a plasmid could restore GelE activity. However, if fsrA transcripts were degraded in the Δbph background, then plasmid-borne fsrA would not restore GelE activity. We cloned fsrA into a pheromone-inducible backbone, induced expression in ΔfsrA and Δbph, and quantified GelE activity in the supernatant of planktonic cultures using Azocoll assays. In both strains, expression of fsrA from a plasmid significantly increased GelE activity relative to the empty vector control (Fig. 1D, 17.8-fold and 3.19-fold, respectively). Overall, we conclude that Bph regulates gelE expression through Fsr and that in the absence of bph, expression of both the Fsr quorum sensing system as well as genes outside the Fsr regulon are disrupted.
Comparison of differentially expressed genes in Δbph and ΔfsrB deletion mutants.
Next, we asked how much of the altered gene expression in the Δbph mutant could be linked to disruption of the Fsr quorum sensing system. We compared all differentially expressed genes identified using RNA-seq to published microarray data for a ΔfsrB mutant (30). In ΔfsrB, 15 of the 42 upregulated genes were shared with Δbph and 53 of the 306 downregulated genes were shared with Δbph (Fig. 1E and F). Of the 15 common upregulated genes, 8 are within the ethanolamine utilization operon (see Table S2). Shared downregulated genes included 5 genes predicted to encode an uncharacterized surface protein complex (OG1RF_10485-10489, OG1RF_10491). To determine whether reduced expression of OG1RF_10485-10491 contributes to the biofilm-deficient phenotype of Δbph (27), we obtained Tn mutants from an arrayed Tn library (31) and measured biofilm production in 96-well plates. OG1RF_10490::Tn had reduced biofilm relative to OG1RF at 6 h and 24 h, and OG1RF_10489::Tn and OG1RF_10492::Tn had reduced biofilm at 24 h (see Fig. S2). Interestingly, 4 genes in the enterococcal polysaccharide antigen (Epa) operon (OG1RF_11721 to OG1RF_11724) were also downregulated in both Δbph and ΔfsrB, although we did not previously observe changes associated with altered Epa synthesis in polysaccharides purified from Δbph (27, 32, 33).
The peptidyl-prolyl isomerase PrsA is required for GelE functionality independent of Fsr.
In a previous screen for Tn mutants with defects in biofilm formation, we found that a mutant with a Tn insertion in prsA (OG1RF_10423) had a GelE-negative phenotype on gelatin plates (34). PrsA is an extracellular parvulin-like PPIase (Fig. 2A) that is required for activity of secreted proteins and membrane proteins in numerous Gram-positive bacteria, including Bacillus anthracis (23), Bacillus subtilis (35), Listeria monocytogenes (24), Staphylococcus aureus (36), group A Streptococcus (37), and Streptococcus equi (38). In E. faecalis, PrsA is important for survival in high salt concentrations and virulence in Galleria mellonella (21), although no role in folding specific protein substrates has been described in Enterococcus. We confirmed the gelatinase-negative phenotype of the prsA Tn mutant (prsA::Tn) grown in both tryptic soy broth without added dextrose (TSB-D), or modified M9 medium supplemented with yeast extract and glucose (MM9-YEG) growth media, both of which were previously used when studying biofilm formation by prsA::Tn (Fig. 2B). Expression of prsA from a pheromone-inducible plasmid restored GelE activity on plates (Fig. 2B). We next measured GelE activity using dye release assays with supernatants from planktonic cells cultured for 6 h. Relative to OG1RF, dye release from prsA::Tn supernatants was reduced approximately 70% in both media, and dye release was restored in prsA::Tn by expression of prsA from a plasmid (Fig. 2C).
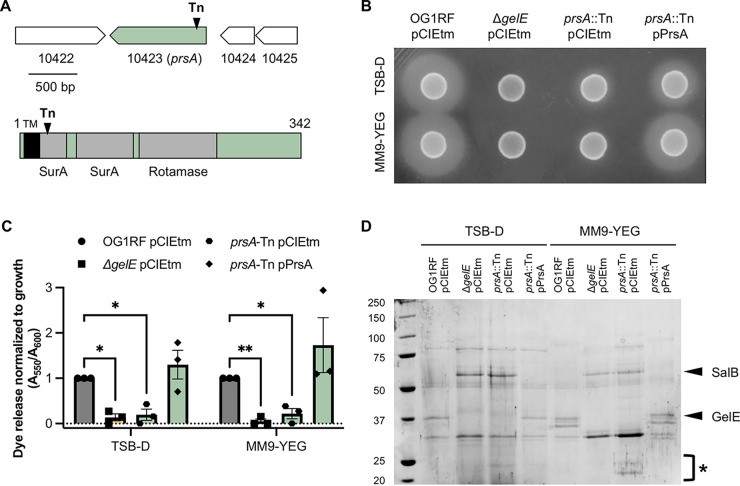
FIG 2.
PrsA is a predicted extracellular peptidyl-prolyl isomerase that is required for gelatinase functionality in OG1RF. (A) Cartoon showing the location and direction of prsA in the genome (top) and the PrsA protein features (bottom). The black triangle annotated with Tn indicates the location of the transposon insertion. A predicted transmembrane domain is shown as a black rectangle labeled with TM, and Pfam domains are shown as gray rectangles. TMHMM was used to predict transmembrane domains. (B) Gelatinase plate assay with OG1RF pCIEtm, ΔgelE pCIEtm and prsA::Tn pCIEtm, and prsA::Tn pPrsA. Hazy zones around colonies are indicative of gelatinase activity. Image is representative of three biological replicates. (C) Quantification of relative gelatinase activity compared to OG1RF in an Azocoll dye release assay. Three biological replicates were tested for each strain. *, P < 0.05; **, P < 0.01 by two-way ANOVA with Dunnett’s test for multiple comparisons. (D) SDS-PAGE evaluation of exoproteins using the same strains as in panels B and C. Prominent secreted proteins (SalB, GelE) are marked with arrows, and small protein bands in prsA::Tn are marked with *. Image is representative of three biological replicates.
Given that GelE processes secreted proteins (5, 39), a loss of GelE activity in the prsA mutant background could also affect other supernatant proteins. We used SDS-PAGE to evaluate supernatant proteins from planktonic cells grown in TSB-D and MM9-YEG and found that the prsA::Tn supernatant lacked the prominent GelE band (40) (Fig. 2D). The ΔgelE and prsA::Tn supernatants had similar protein banding patterns, and additional small protein bands were visible in the prsA::Tn samples (Fig. 2D, marked with *). Previous work showed the ΔgelE mutant had an increase in supernatant levels of the secreted protein SalB, which contributes to cell envelope integrity and cephalosporin resistance (39, 41, 42). Additionally, GelE processes recombinant SalB (42). We observed a band corresponding to SalB in the prsA::Tn supernatants (Fig. 2D). SalB levels are also increased in Δbph supernatants (27), and so decreased GelE levels in the Δbph and prsA::Tn supernatants likely lead to increased accumulation of SalB. Expression of prsA from a plasmid restored prsA::Tn supernatant proteins to that from parental OG1RF. We also observed minor differences in protein banding patterns between OG1RF and prsA::Tn in protoplast and cell wall samples (see Fig. S3 in the supplemental material). Together, these experiments demonstrate that disruption of prsA abrogates gelatinase production and alters the exoprotein profile of OG1RF.
Because gelE expression is reduced in Δbph, we also considered that GelE levels in prsA::Tn could be reduced due to changes in gene expression. We examined expression from fsrA, fsrB, and gelE promoters using lacZ promoter fusion constructs and found no significant differences in expression between prsA::Tn and OG1RF (Fig. 3A and B), suggesting that reduced GelE levels in prsA::Tn are not due to reduced transcription. We then asked whether the gelatinase-negative phenotypes of Δbph and prsA::Tn could be rescued by expression of gelE from a plasmid. We hypothesized that if the gelatinase-negative phenotype of prsA::Tn resulted from aberrant folding of GelE, then expression in trans would not rescue the phenotype in Azocoll assays. Nisin induction of pMSP3535::gelE in planktonic cultures significantly increased dye release from ΔgelE and Δbph supernatants (21.0-fold and 3.0-fold, respectively), but not from prsA::Tn (Fig. 3C). This demonstrates that the loss of GelE activity in prsA::Tn is not caused by changes in fsr or gelE gene expression.
FIG 3.
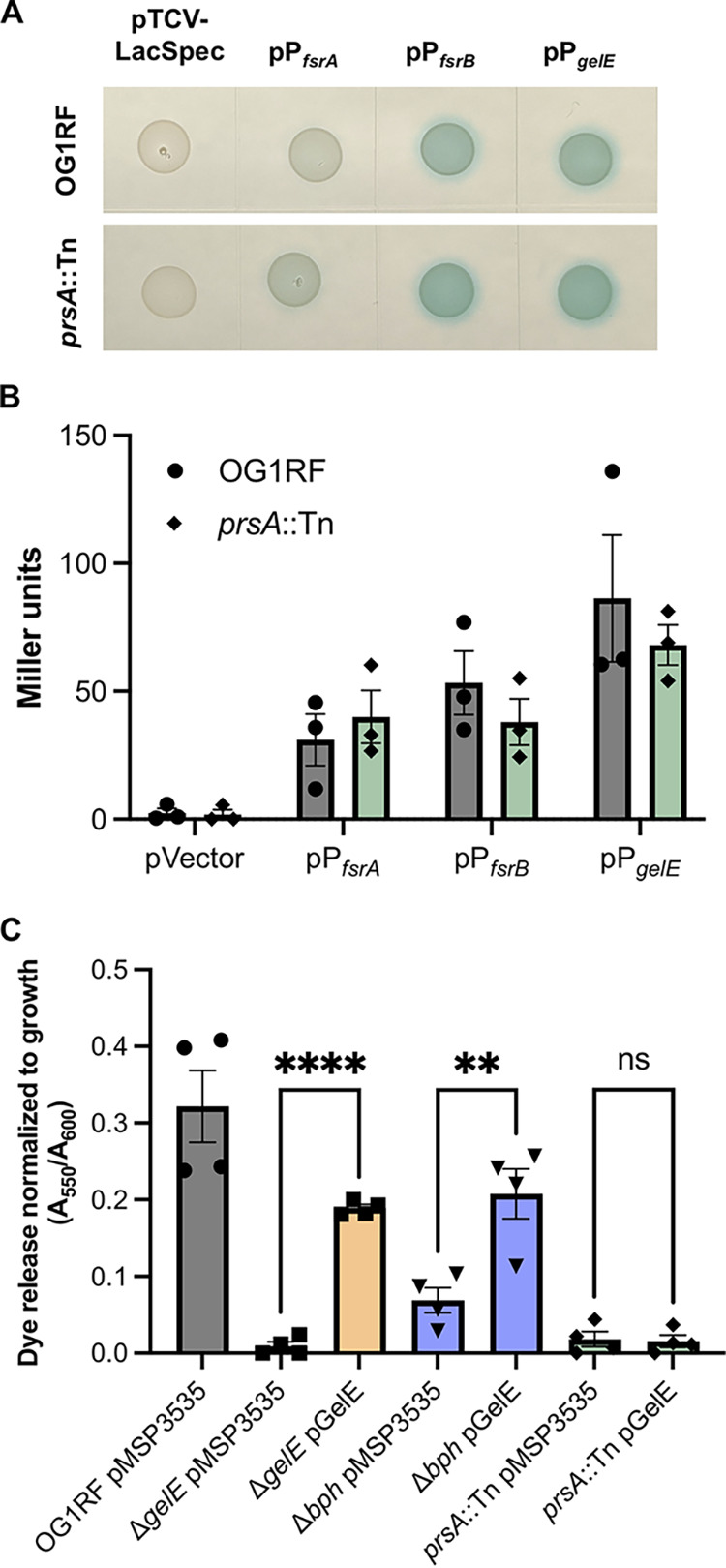
Loss of GelE in prsA::Tn is not due to reduced expression of the Fsr quorum sensing system. (A) Empty pTCV-LacSpec or derivatives carrying the promoter regions of fsrA, fsrB, or gelE were transformed into OG1RF and prsA::Tn and spotted on MM9-YEG agar plates supplemented with X-Gal. Images are representative of 3 independent experiments. (B) Promoter activities of the strains in panel A were quantified in β-galactosidase assays (n = 3). (C) Expression of gelE from a pheromone-inducible plasmid (pMSP3614) rescues gelatinase activity in ΔgelE and Δbph but not prsA::Tn (n = 4). In panels B and C, each data point represents a biological replicate. **, P < 0.01; ****, P < 0.0001 by one-way ANOVA with Sidak’s multiple-comparison test.
PrsA mediates biofilm-associated antibiotic resistance independently of GelE.
Deletion of prsA in S. aureus isolates leads to increased sensitivity to glycopeptides and oxacillin (25, 26), and disruption of prsA in B. subtilis results in destabilized penicillin-binding proteins (35). However, in E. faecalis V583, deletion of prsA did not alter susceptibility to antibiotics during growth on agar plates (21). Therefore, we wondered whether disrupting prsA in the OG1RF background would lead to altered susceptibility to cell wall-active antibiotics. We measured growth of OG1RF, ΔgelE, Δbph, and prsA::Tn in 2-fold dilution series of ampicillin, oxacillin, penicillin G, and vancomycin relative to untreated cultures.
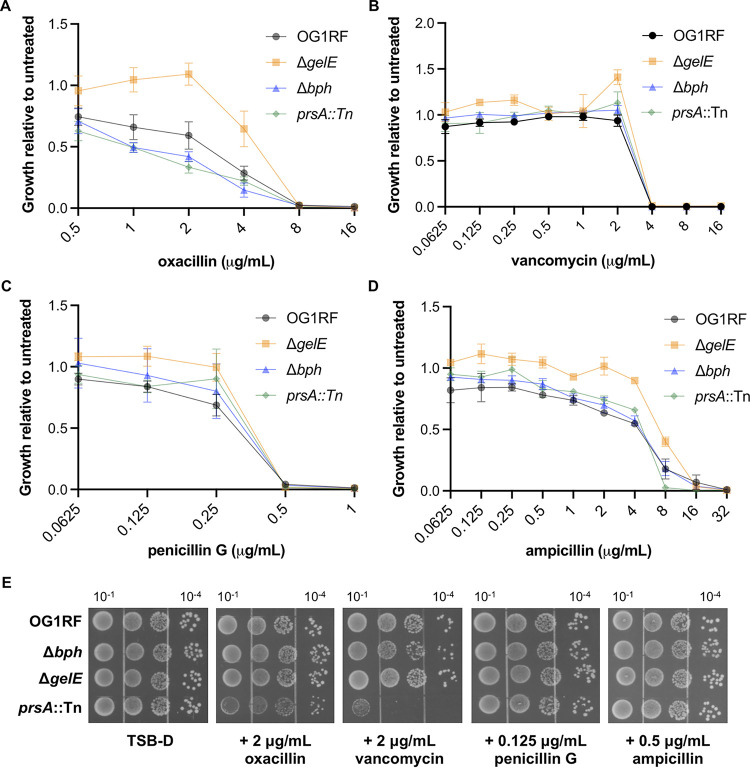
In liquid culture, no change in sensitivity was observed for prsA::Tn to oxacillin, penicillin G, or vancomycin (Fig. 4A to C). prsA::Tn was slightly more susceptible than the other strains to 8 μg/mL ampicillin (Fig. 4D). Interestingly, ΔgelE grew to a higher optical density at 600 nm (OD600) in subinhibitory antibiotic concentrations. Next, we spotted serial dilutions of each strain onto agar plates supplemented with subinhibitory concentrations of each antibiotic (empirically chosen based on the liquid growth assays). Surprisingly, growth of prsA::Tn but not the ΔgelE strain was strongly inhibited relative to that of OG1RF on plates containing 2 μg/mL oxacillin or vancomycin (Fig. 4E), even though no growth defect was observed in liquid culture for either antibiotic at this concentration. None of the mutant strains had growth defects on plates containing subinhibitory concentrations of ampicillin or penicillin G (Fig. 4E).
FIG 4.
Growth of OG1RF, ΔgelE, Δbph, and prsA::Tn in antibiotics that target the cell envelope. The indicated strains were grown in TSB-D in a 2-fold dilution series of oxacillin (A), vancomycin (B), penicillin G (C), and ampicillin (D). Growth at each concentration was calculated relative to untreated samples. Data points represent the average of 3 biological replicates, and error bars show standard errors of the means. (E) Overnight cultures were normalized to 107 CFU/mL and serially diluted. Dilutions (shown above plate images) were spotted onto TSB-D plates supplemented with the indicated subinhibitory concentrations of antibiotics. Gel images are representative of 3 biological replicates.
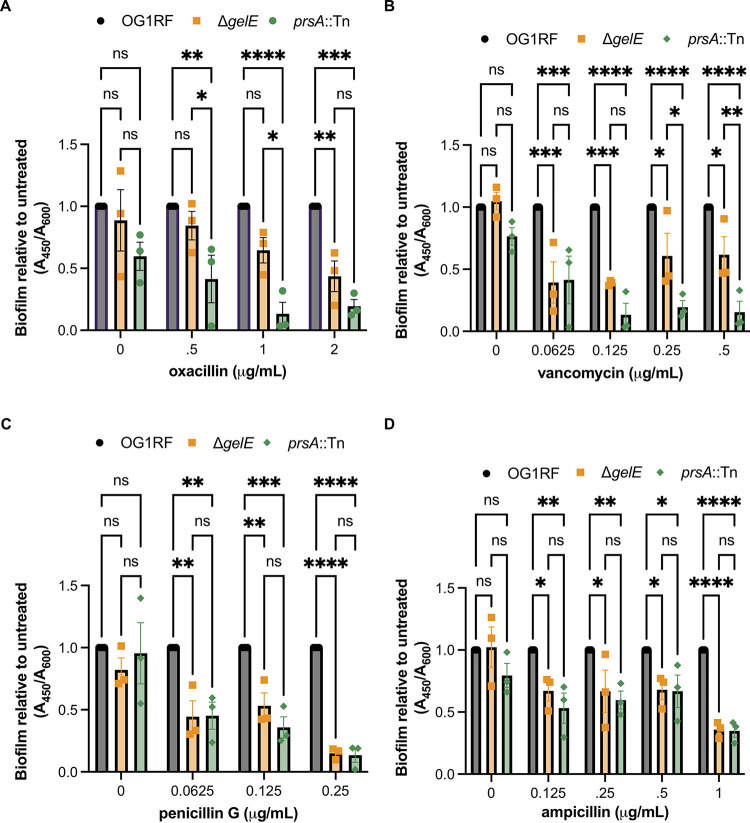
We previously reported that OG1RF mutants with Tn insertions in fsrA and gelE had decreased biofilm formation in the presence of subinhibitory concentrations of daptomycin, gentamicin, and linezolid (33). Therefore, we wondered whether reduced levels of GelE in our mutant strains would lead to altered biofilm production in subinhibitory concentrations of the cell wall-active antibiotics tested above. At 0.5 and 1 μg/mL oxacillin, prsA::Tn biofilm production was reduced relative to that of both OG1RF and ΔgelE (Fig. 5A). Similarly, prsA::Tn biofilm production was reduced relative to that of ΔgelE at 0.5 and 1 μg/mL vancomycin (Fig. 5B). This suggests that disruption of prsA could affect stability or folding of additional proteins besides GelE that are important for biofilm formation in the presence of these antibiotics. In subinhibitory concentrations of penicillin G and ampicillin, ΔgelE and prsA::Tn had reduced biofilm production relative to OG1RF, but there were no differences in biofilm quantity produced between the 2 mutant strains (Fig. 5C and D). Because the Δbph mutant has a severe biofilm defect even in the absence of antibiotics (27), it was difficult to interpret any antibiotic-associated changes in biofilm formation for this strain (see Fig. S4 in the supplemental matreial).
FIG 5.
prsA::Tn has reduced biofilm production independent of GelE in subinhibitory concentrations of oxacillin and vancomycin. The indicated strains were grown in a 2-fold dilution series of oxacillin (A), vancomycin (B), penicillin G (C), and ampicillin (D), and growth was measured as the A600. Biofilm material was stained with safranin and quantified as the A450. For each mutant, biofilm production was normalized to that of OG1RF. Data points represent independent biological replicates (n = 3), and error bars show standard errors of the means. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by two-way ANOVA with Tukey’s multiple comparison test.
Deletion of prsA increases efficiency of conjugative plasmid transfer.
GelE affects stability of aggregation substance proteins and pheromone peptides that mediate transfer of conjugative plasmids like pCF10, and plasmid transfer into strains lacking gelE is more efficient than into GelE-producing cells (5). We previously showed that the rate of transfer of plasmid pCF10 into Δbph was similar to that of parental OG1RF (27), suggesting that the reduction in GelE levels in this strain is not enough to affect conjugation. Here, we asked whether the prsA::Tn mutation affected plasmid transfer, since this Tn mutant does not produce active gelatinase. Donor cells (OG1Sp pCF10) and recipients (OG1RF, ΔgelE, or prsA::Tn) were mixed 1:1 in liquid culture, and transconjugants were quantified by plating on selective growth medium. As seen previously, transfer to ΔgelE recipients was increased ~2-fold relative to that for OG1RF. Consistent with the GelE-negative phenotype of prsA::Tn, transfer was also increased into these recipients (Fig. 6).
FIG 6.
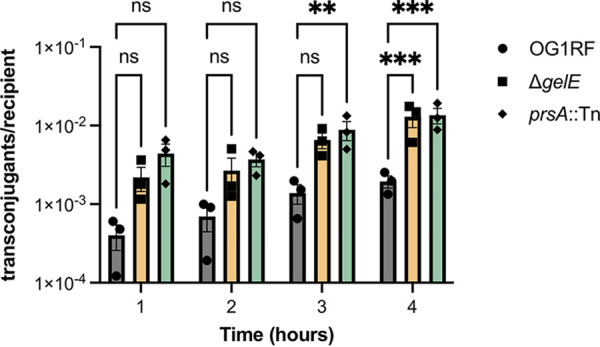
Disruption of prsA increases transfer of the conjugative plasmid pCF10. Donors (OG1Sp pCF10) and recipients (OG1RF, ΔgelE, or prsA::Tn) were mixed 1:1 and incubated at 37°C. Each data point represents a biological replicate (n = 3). **, P < 0.01; ***, P < 0.0001 by two-way ANOVA with Dunnett’s correction for multiple comparisons.
DISCUSSION
Gelatinase is an important virulence factor for E. faecalis, and regulation of gelE expression by the Fsr quorum sensing system has been extensively studied (12, 13). In 2004, Hancock and Perego proposed there might be another protein required for folding or processing of GelE, based on complications purifying active GelE overexpressed in E. coli (13). Here, we identified two additional proteins involved in gelatinase production and activity (Fig. 7). The biofilm-associated phosphatase Bph regulates gelE expression in an Fsr-dependent manner, as expression of the fsr locus is reduced in a Δbph mutant. The extracellular peptidyl isomerase PrsA acts upon GelE in an Fsr-independent mechanism, presumably by ensuring proper refolding of GelE as it is secreted from the cell. Interestingly, Δbph has a gelatinase-positive phenotype after overnight growth on gelatin plates (27), although here we showed a significant reduction in gelatinase activity at discrete time points during growth. This highlights the caution that must be taken when interpreting genetic screens that can obscure subtle phenotypes caused by reduced gene expression over a long period of time. Given that our findings update the longstanding model for gelatinase production, it is worth determining whether E. faecalis encodes other gene products that regulate gelatinase expression and activity. GelE and Fsr levels increased in an E. faecalis mutant lacking the ClpP protease, suggesting that additional factors beyond Fsr, Bph, and PrsA could regulate expression, activity, or stability of gelatinase (43).
FIG 7.
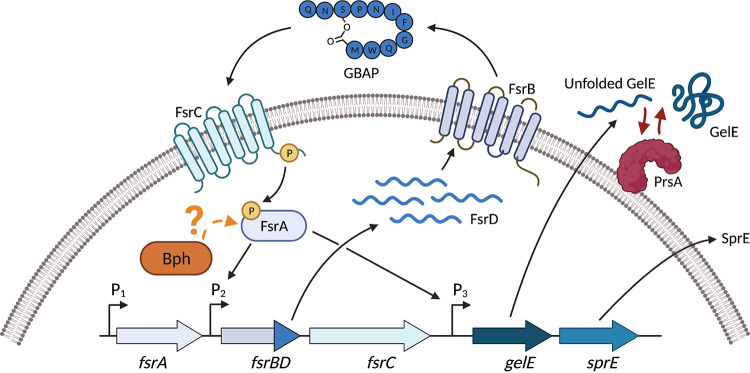
Updated model for gelatinase production in E. faecalis. The Fsr quorum sensing system positively regulates expression of the proteases GelE and SprE. Bph is required for expression of the fsr locus and thus regulation of GelE through an unknown mechanism (indicated by the question mark). The extracellular foldase PrsA is required for activity of secreted GelE, but not fsr or gelE gene expression. This figure was generated with Biorender.com.
Although our studies were done in vitro, the RNA-seq could provide physiological insight into the relevance of Bph in vivo. Multiple genes from the ethanolamine utilization (eut) operon were upregulated in both bph and fsrB mutants (30), and the loss of eut genes increased E. faecalis fitness during gastrointestinal tract (GIT) colonization (44). Therefore, overexpression of this operon may in turn decrease GIT fitness of Δbph. Additionally, genes that contribute to production of the enterococcal polysaccharide surface antigen (Epa) were downregulated, and Epa is required for GIT colonization and persistence (45–48). These results combined with our previous study on Bph activity demonstrate that loss of Bph has pleiotropic effects on quorum sensing, expression of virulence factors, and metabolism (27). However, one limitation of our study is that many E. faecalis genes are not assigned a K number or to a KEGG category, so there could be additional pathways that are differentially regulated in the bph deletion mutant that were not detected in our analysis.
An additional limitation of our study is that the target of Bph phosphatase activity is still unknown. Multiple transcription factors are differentially expressed in Δbph (see Table S1), so the overall differences in gene expression may represent the cumulative regulons of these proteins. Bph could potentially directly dephosphorylate transcription factors (like FsrA), but previous work did not identify differential protein phosphorylation using a gel-based approach (27). We do not think Bph acts directly on FsrA, as this would not explain the reduced expression of fsrA in the bph deletion mutant. Dephosphorylation of a small molecule, like nucleotides or second messengers, could cause global changes in gene expression. fsrBC, gelE, and sprE are also downregulated in a (p)ppGpp-null mutant (49), and recombinant Bph had phosphatase activity on (p)ppGpp in vitro (27). Therefore, additional work is required to disentangle the genetic networks disrupted in the Δbph mutant, identify the target of Bph phosphatase activity, and determine the importance of bph during colonization and infection.
This study also highlights differences in PrsA function across Gram-positive bacteria. In B. subtilis, prsA is essential and cannot be deleted in vitro without compensatory mutations or increased Mg2+ in growth medium (22, 35). Multiple PrsA proteins are produced by B. anthracis, L. monocytogenes, and group A Streptococcus (23, 24, 50). Although E. faecalis encodes multiple PPIases, it produces only one PrsA family protein, which is not essential for viability. In group A Streptococcus, PrsA is required for the proper folding of the secreted exotoxin SpeB (51). The prsA gene is encoded adjacent to speB, and they are cotranscribed. In E. faecalis, prsA is in a distal location relative to gelE, which is encoded adjacent to the Fsr quorum sensing system that regulates gelE expression. Additionally, trigger factor is also required for refolding of SpeB (52, 53), but Tig is not required for GelE activity in OG1RF (34).
Interestingly, neither Δbph nor prsA::Tn exactly phenocopied the ΔgelE mutant during growth in subinhibitory concentrations of vancomycin or β-lactam antibiotics, despite the reduced gelatinase levels in those strains. Multiple cell wall biosynthesis genes were downregulated in Δbph, which could affect sensitivity to antibiotics. We also observed protein bands in protoplast, cell wall, and supernatant fractions that were differentially expressed in the prsA::Tn mutant relative to OG1RF. This coupled with the biofilm-specific antibiotic sensitivity of prsA::Tn suggests that other proteins that sense or respond to antibiotics during biofilm growth could be misfolded in the absence of PrsA. As such, identifying additional targets of PrsA foldase activity (beyond GelE) could serve as a platform for better understanding biofilm-specific antibiotic resistance in E. faecalis. We also observed interesting differences in Δbph and prsA::Tn antibiotic sensitivity when we compared growth in liquid culture and on agar plates. We speculate that this is because growth on an agar plate could more closely resemble biofilm growth than planktonic growth, although this has yet to be evaluated for E. faecalis.
Given the importance of GelE in virulence, it is interesting to consider the potential interplay between bph, prsA, and gelatinase production during infections. In a rabbit subdermal abscess model, expression of bph and prsA increased after 2 h of infection, whereas bph and gelE were downregulated after 8 h (54). This suggests that regulation of gelatinase is critical for establishing an infection and in vivo survival. Additionally, numerous studies have documented a discrepancy between gelatinase genotype and phenotype in clinical isolates (16, 18, 55–58). However, these collections of isolates are traditionally screened for GelE activity using gelatin plates or tubes, which could obscure temporal changes in gelatinase production, as demonstrated in this study. A survey of environmental Enterococcus isolates found that GelE-positive E. faecalis strains lost the gelatinase phenotype with serial passaging even though gelE was still transcribed. (59). Therefore, additional analysis is needed to understand how Bph and PrsA control the dynamics of gelatinase production and how this affects the establishment and persistence of infections in humans.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used in this study are listed in Table S3 in the supplemental material. Bacteria were maintained as freezer stocks at −80°C in 25% glycerol. Strains were cultured in brain-heart infusion broth (BHI), tryptic soy broth without added dextrose (TSB-D), or modified M9 medium supplemented with yeast extract and glucose (MM9-YEG), as indicated. BHI and TSB-D were purchased from BD, and MM9-YEG was prepared as previously described (60). For solid media, agar was added to a final concentration of 1%. Unless indicated otherwise, overnight cultures were grown in the same medium used for experiments. Antibiotics were used at the following concentrations: erythromycin (10 μg/mL), fusidic acid (25 μg/mL), spectinomycin (50 μg/mL for E. coli, 250 to 1,000 μg/mL for E. faecalis), and tetracycline (5 μg/mL). When needed for induction of gene expression, cCF10 (Mimotopes) or nisin (Sigma) was added to cultures at 25 to 50 ng/mL or at 50 ng/mL, respectively.
Cloning.
Oligonucleotides used for cloning are listed in Table S3 in the supplemental material. Plasmids pML200 and pML201 are derivatives of pTCV-LacSpec (61) with the fsrA and fsrB promoters fused to lacZ, respectively. These plasmids were a generous gift from Lynn Hancock and Marta Perego. To obtain pML200, a PCR fragment encompassing ~1 kb upstream of the fsrA gene as well as a portion of the fsrA gene was digested with EcoRI and HincII and cloned into pTCV-LacSpec digested with EcoRI and SmaI. To obtain pML201, a PCR fragment encompassing ~500 bp that included a portion of the fsrA gene, the intergenic region, as well as the first 197 bp of the fsrB gene was digested with ApoI and HaeIII and cloned into pTCV-LacSpec digested with EcoRI and SmaI. Plasmid pDM7 is a derivative of pTCV-LacSpec plasmid with the gelE promoter fused to lacZ and was a generous gift from Dawn Manias. To create pheromone-inducible plasmids expressing fsrA and gelE, alleles were amplified from purified OG1RF genomic DNA using Pfu Ultra II polymerase (Agilent), digested with BamHI-HF and NheI-HF, then ligated to pCIEtm (27) treated with the same restriction enzymes. Restriction enzymes were purchased from New England Biolabs. Plasmid constructs were verified by Sanger sequencing at the University of Minnesota Genomics Center.
RNA-seq.
Overnight cultures were diluted to an OD600 of 0.05 in TSB-D. At 2 and 4 h, a volume equivalent to an OD600 of 1 was mixed with an equal volume of RNAprotect and incubated at room temperature for 10 min. Cells were pelleted and stored at −80°C. Pellets from 2 biological replicates were resuspended in 200 μL of buffer (10 mM Tris [pH 8.0], 1.0 mM EDTA) supplemented with lysozyme (30 mg/mL) and incubated at 37°C for 10 min. Total RNA was extracted with a Qiagen RNeasy kit following the manufacturer’s instructions and treated with Turbo DNase (Ambion/Thermo Scientific, Waltham, MA) following the manufacturer’s “rigorous treatment” protocol. Removal of contaminating genomic DNA was verified by conventional PCR using 16S primers (JD460s/JD461as [see Table S3 in the supplemental material]). RNA was submitted to the University of Minnesota Genomics Center for rRNA depletion (Illumina) and TruSeq stranded library preparation. Samples were pooled and sequenced (2 × 75-bp paired ends) on an Illumina NextSeq 550 in mid-output mode. Sequencing quality was evaluated using FastQC (62). Reads were trimmed with Trimmomatic (63) and imported into Rockhopper for analysis using standard settings (64, 65). Fold changes (log2) were calculated from Rockhopper expression values. Rockhopper-generated q values of ≤0.05 were considered statistically significant. For KEGG analysis, identifiers corresponding to OG1RF locus tags were obtained from KEGG and used as input for KEGG Mapper (66, 67).
qRT-PCR.
Overnight cultures were adjusted to an OD600 of 0.05 in fresh media and grown for 6 h, after which the equivalent of 1 mL cells at an OD600 of 1.0 was collected. Total RNA isolation, DNase treatment, and confirmation of genomic DNA removal were carried out as described above. An iScript Select cDNA synthesis kit (Bio-Rad) was used for cDNA synthesis using random hexamers with 50 μg/mL RNA in a 20-μL reaction mixture. Samples without reverse transcriptase were included to control for DNA contamination. cDNA was diluted 10-fold in sterile water, after which 2 μL was used per qRT-PCR mixture (iQ SYBR green Supermix, Bio-Rad). Total reaction volumes were 15 μL with 250 nM each primer (see Table S3). Reactions were run on an iCycler iQ5 (Bio-Rad), and the 2−ΔΔCT method (68) was used to analyze data and calculate the relative fold change normalized to relA, which is constitutively expressed in planktonic cultures for each strain.
Biofilm assays.
Cells were grown overnight and diluted 1:100 into fresh TSB-D. A 100-μL volume was added to a 96-well plate (Corning 3595) and incubated in a humidified chamber at 37°C for 6 or 24 h, as indicated. Overall growth was measured in a Biotek Synergy HT plate reader as the absorbance at 600 nm (A600). Planktonic cells were removed, and plates were washed gently three times with ultrapure water. Plates were dried overnight in a biosafety cabinet or on the lab bench. Adherent biofilm material was stained with 0.1% (wt/vol) safranin for 20 min at room temperature. Plates were washed and dried as described above. Biofilm material was measured in a Biotek Synergy HT plate reader as the A450. Media blanks were included on every plate, and the blank wells were also stained with safranin. Media blanks for A600 and A450 measurements were subtracted from all samples during analysis. Biofilm values are presented as safranin-stained material relative to overall cell growth (A450/A600) normalized to OG1RF. All assays had at least 3 biological replicates, each with technical duplicates.
Gelatinase and Azocoll assays.
Agar plate-based gelatinase assays were performed as described previously (34, 69). Azocoll assays using culture supernatants were modified from published protocols (29, 69). To prepare the substrate, 0.25 g Azocoll (azo dye-conjugated collagen; Sigma) was washed in 50 mL KPBS (potassium-phosphate buffered saline, pH 7.0) on a rotating platform shaker for 1 h at room temperature and then centrifuged (4,900 × g for 10 min). Azocoll was resuspended, and the wash was repeated. Following the second wash, the residue was resuspended in 50 mL KPBS containing 1 mM CaCl2. Washed Azocoll was stored at room temperature and used within 1 week. Strains were grown overnight in TSB-D supplemented with fusidic acid or tetracycline and were diluted to an OD600 of 0.05 in fresh medium with tetracycline and cCF10 as needed. Cultures were incubated statically at 37°C. At each time point, the OD600 was measured, and 1.5 mL of cells was pelleted in a tabletop centrifuge (6,080 × g for 2 min). A 500-μL aliquot of supernatant was mixed with 500 μL washed Azocoll in a 1.5-mL tube, and tubes were incubated at 37°C on a shaker for 24 h. Samples were centrifuged (16,000 × g for 5 min), and dye release was quantified in a Biotek Synergy HT plate reader as the A550. Data are expressed as dye release relative to growth (A550/OD600) from three biological replicates with technical replicates (3 per sample). Due to variations between batches of Azocoll, all replicates for a given experiment were done using Azocoll prepared from the same bottle.
β-Galactosidase activity assays.
Strains were grown overnight in BHI containing spectinomycin. For blue-white plate screening, 5 μL of each overnight culture was spotted on MM9-YEG plates containing spectinomycin and 200 μg/mL X-Gal (5-bromo-4- chloro-3-indolyl-β-d-galactopyranoside). Plates were incubated overnight at 37°C and photographed with a cell phone camera. To quantify β-galactosidase activity, overnight cultures were diluted to an OD600 of 0.05 in MM9-YEG and spectinomycin and incubated for 4 h at 37°C. The cells were pelleted (6,080 × g for 2 min) and stored at −80°C. β-Galactosidase expression was measured as previously described (70). Assays were performed in biological triplicates with technical duplicates.
SDS-PAGE.
Overnight cultures grown in either TSB-D or MM9-YEG (with tetracycline and cCF10, as needed) were diluted to an OD600 of 0.05 in fresh media. After 6 h of static incubation at 37°C, the equivalent of 1 mL of cells at an OD600 of 1.0 was pelleted in a tabletop centrifuge (6,080 × g) for 2 min. Pellets were resuspended in 100 μL TESL (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 25% sucrose, and 15 mg/mL lysozyme) and incubated at 37°C for 30 min. Fifteen microliters was mixed directly with 2× Laemmli sample buffer (Bio-Rad) for whole-cell lysates, and 85 μL was centrifuged (16,000 × g) for 1 min to separate the pellet (protoplast) from the supernatant (cell wall extract). Cell wall extracts were mixed with an equal volume of 2× Laemmli sample buffer. Protoplast samples were resuspended in 50 μL urea lysis buffer (8 M urea, 20 mM Tris–HCl [pH 7.5], 150 mM NaCl) and 50 μL 2× Laemmli sample buffer. Proteins in the culture supernatant were precipitated by mixing 1 volume of supernatant with 0.25 volumes of chilled 100% trichloroacetic acid on ice, pelleted in a tabletop centrifuge (16,000 × g) for 10 min, and washed twice with acetone. Pellets were dried and resuspended in 50 μL urea lysis buffer and 50 μL 2× Laemmli sample buffer. Samples were heated at 95°C before loading onto 10% Tris-glycine SDS-PAGE gels. Gels were run at 110 V, stained with Coomassie blue, destained, and imaged on a Bio-Rad Gel Doc EZ imager.
Antibiotic sensitivity assays.
Overnight cultures were grown in TSB-D and adjusted to 5 × 107 CFU/mL in fresh medium. Cells were diluted 1:50 into 96-well plates (Corning 3595) containing 100 μL of 2-fold serial dilutions of the indicated antibiotics prepared in TSB-D. Plates were incubated in a humidified chamber at 37°C for 24 h, after which OD600 measurements were taken in a BioTek Synergy H1 plate reader. Relative growth was quantified by dividing the OD600 at each antibiotic concentration by the OD600 of the untreated culture. The results shown are the mean OD600 values from three independent biological replicates with technical duplicates. To measure biofilm production in the presence of subinhibitory antibiotic concentrations, the 96-well plates used for antibiotic sensitivity assays were dried and stained with safranin as described above.
Statistical analysis.
Data analysis was performed using GraphPad Prism (version 9.2.0). Statistical tests and significance thresholds are described in the figure legends.
Data availability.
Files generated from RNA sequencing and data analysis have been deposited with NCBI GEO under accession number GSE198051.
ACKNOWLEDGMENTS
We thank Lynn Hancock, Marta Perego, and Dawn Manias for providing plasmid constructs. We acknowledge the Minnesota Supercomputing Institute (MSI) at the University of Minnesota (http://www.msi.umn.edu) and the University of Minnesota Genomics Center (https://genomics.umn.edu/) for providing resources that contributed to the results reported here. This work was supported by National Institutes of Health grants R35GM118079 and R01AI122742 to G.M.D., 1K99AI151080 to J.L.E.W., and the National Center for Advancing Translational Sciences grant UL1TR002494. E.B.R. was supported by the University of Minnesota Undergraduate Research Opportunities Program (UROP). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health’s National Center for Advancing Translational Sciences.
Footnotes
Supplemental material is available online only.
Contributor Information
Julia L. E. Willett, Email: jwillett@umn.edu.
Michael J. Federle, University of Illinois at Chicago
REFERENCES
- 1.Gilmore MS, Lebreton F, van Schaik W. 2013. Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr Opin Microbiol 16:10–16. 10.1016/j.mib.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ch'ng JH, Chong KKL, Lam LN, Wong JJ, Kline KA. 2019. Biofilm-associated infection by enterococci. Nat Rev Microbiol 17:82–94. 10.1038/s41579-018-0107-z. [DOI] [PubMed] [Google Scholar]
- 3.Fiore E, Van Tyne D, Gilmore MS. 2019. Pathogenicity of Enterococci. Microbiol Spectr 7. 10.1128/microbiolspec.GPP3-0053-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maharshak N, Huh EY, Paiboonrungruang C, Shanahan M, Thurlow L, Herzog J, Djukic Z, Orlando R, Pawlinski R, Ellermann M, Borst L, Patel S, Dotan I, Sartor RB, Carroll IM. 2015. Enterococcus faecalis gelatinase mediates intestinal permeability via protease-activated receptor 2. Infect Immun 83:2762–2770. 10.1128/IAI.00425-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waters CM, Antiporta MH, Murray BE, Dunny GM. 2003. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J Bacteriol 185:3613–3623. 10.1128/JB.185.12.3613-3623.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thurlow LR, Thomas VC, Narayanan S, Olson S, Fleming SD, Hancock LE. 2010. Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis. Infect Immun 78:4936–4943. 10.1128/IAI.01118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown AO, Graham CE, Cruz MR, Singh KV, Murray BE, Lorenz MC, Garsin DA. 2019. Antifungal activity of the Enterococcus faecalis peptide EntV requires protease cleavage and disulfide bond formation. mBio 10. 10.1128/mBio.01334-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dundar H, Brede DA, La Rosa SL, El-Gendy AO, Diep DB, Nes IF. 2015. The fsr quorum-sensing system and cognate gelatinase orchestrate the expression and processing of proprotein EF_1097 into the mature antimicrobial peptide enterocin O16. J Bacteriol 197:2112–2121. 10.1128/JB.02513-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stinemetz EK, Gao P, Pinkston KL, Montealegre MC, Murray BE, Harvey BR. 2017. Processing of the major autolysin of E. faecalis, AtlA, by the zinc-metalloprotease, GelE, impacts AtlA septal localization and cell separation. PLoS One 12:e0186706. 10.1371/journal.pone.0186706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mäkinen PL, Clewell DB, An F, Mäkinen KK. 1989. Purification and substrate specificity of a strongly hydrophobic extracellular metalloendopeptidase (“gelatinase”) from Streptococcus faecalis (strain 0G1-10). J Biol Chem 264:3325–3334. 10.1016/S0021-9258(18)94069-X. [DOI] [PubMed] [Google Scholar]
- 11.Cameron EA, Sperandio V, Dunny GM. 2019. Enterococcus faecalis enhances expression and activity of the enterohemorrhagic Escherichia coli type III secretion system. mBio 10. 10.1128/mBio.02547-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin X, Singh KV, Weinstock GM, Murray BE. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect Immun 68:2579–2586. 10.1128/IAI.68.5.2579-2586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock LE, Perego M. 2004. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J Bacteriol 186:5629–5639. 10.1128/JB.186.17.5629-5639.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakayama J, Chen S, Oyama N, Nishiguchi K, Azab EA, Tanaka E, Kariyama R, Sonomoto K. 2006. Revised model for Enterococcus faecalis fsr quorum-sensing system: the small open reading frame fsrD encodes the gelatinase biosynthesis-activating pheromone propeptide corresponding to staphylococcal agrd. J Bacteriol 188:8321–8326. 10.1128/JB.00865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopes MF, Simões AP, Tenreiro R, Marques JJ, Crespo MT. 2006. Activity and expression of a virulence factor, gelatinase, in dairy enterococci. Int J Food Microbiol 112:208–214. 10.1016/j.ijfoodmicro.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Hashem YA, Amin HM, Essam TM, Yassin AS, Aziz RK. 2017. Biofilm formation in enterococci: genotype-phenotype correlations and inhibition by vancomycin. Sci Rep 7:5733. 10.1038/s41598-017-05901-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maasjost J, Lüschow D, Kleine A, Hafez HM, Mühldorfer K. 2019. Presence of virulence genes in Enterococcus species isolated from meat turkeys in Germany does not correlate with chicken embryo lethality. Biomed Res Int 2019:1–10. 10.1155/2019/6147695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakayama J, Kariyama R, Kumon H. 2002. Description of a 23.9-kilobase chromosomal deletion containing a region encoding fsr genes which mainly determines the gelatinase-negative phenotype of clinical isolates of Enterococcus faecalis in urine. Appl Environ Microbiol 68:3152–3155. 10.1128/AEM.68.6.3152-3155.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez M, Calles-Enríquez M, del Rio B, Ladero V, Martín MC, Fernández M, Alvarez MA. 2015. IS256 abolishes gelatinase activity and biofilm formation in a mutant of the nosocomial pathogen Enterococcus faecalis V583. Can J Microbiol 61:517–519. 10.1139/cjm-2015-0090. [DOI] [PubMed] [Google Scholar]
- 20.Teixeira N, Santos S, Marujo P, Yokohata R, Iyer VS, Nakayama J, Hancock LE, Serror P, Silva Lopes M.dF. 2012. The incongruent gelatinase genotype and phenotype in Enterococcus faecalis are due to shutting off the ability to respond to the gelatinase biosynthesis-activating pheromone (GBAP) quorum-sensing signal. Microbiology (Reading) 158:519–528. 10.1099/mic.0.055574-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reffuveille F, Connil N, Sanguinetti M, Posteraro B, Chevalier S, Auffray Y, Rince A. 2012. Involvement of peptidylprolyl cis/trans isomerases in Enterococcus faecalis virulence. Infect Immun 80:1728–1735. 10.1128/IAI.06251-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kontinen VP, Sarvas M. 1993. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol Microbiol 8:727–737. 10.1111/j.1365-2958.1993.tb01616.x. [DOI] [PubMed] [Google Scholar]
- 23.Williams RC, Rees ML, Jacobs MF, Prágai Z, Thwaite JE, Baillie LWJ, Emmerson PT, Harwood CR. 2003. Production of Bacillus anthracis protective antigen is dependent on the extracellular chaperone, PrsA. J Biol Chem 278:18056–18062. 10.1074/jbc.M301244200. [DOI] [PubMed] [Google Scholar]
- 24.Alonzo F, Freitag NE. 2010. Listeria monocytogenes PrsA2 is required for virulence factor secretion and bacterial viability within the host cell cytosol. Infect Immun 78:4944–4957. 10.1128/IAI.00532-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jousselin A, Renzoni A, Andrey DO, Monod A, Lew DP, Kelley WL. 2012. The posttranslocational chaperone lipoprotein PrsA is involved in both glycopeptide and oxacillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 56:3629–3640. 10.1128/AAC.06264-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jousselin A, Manzano C, Biette A, Reed P, Pinho MG, Rosato AE, Kelley WL, Renzoni A. 2015. The Staphylococcus aureus chaperone PrsA is a new auxiliary factor of oxacillin resistance affecting penicillin-binding protein 2A. Antimicrob Agents Chemother 60:1656–1666. 10.1128/AAC.02333-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willett JL, Ji M, Dunny GM. 2019. Exploiting biofilm phenotypes for functional characterization of hypothetical genes in Enterococcus faecalis. NPJ Biofilms Microbiomes 5. 10.1038/s41522-019-0099-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Turanov AA, Hatfield DL, Gladyshev VN. 2008. In silico identification of genes involved in selenium metabolism: evidence for a third selenium utilization trait. BMC Genomics 9:251. 10.1186/1471-2164-9-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakayama J, Cao Y, Horii T, Sakuda S, Akkermans AD, de Vos WM, Nagasawa H. 2001. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol Microbiol 41:145–154. 10.1046/j.1365-2958.2001.02486.x. [DOI] [PubMed] [Google Scholar]
- 30.Bourgogne A, Hilsenbeck SG, Dunny GM, Murray BE. 2006. Comparison of OG1RF and an isogenic fsrB deletion mutant by transcriptional analysis: the Fsr system of Enterococcus faecalis is more than the activator of gelatinase and serine protease. J Bacteriol 188:2875–2884. 10.1128/JB.188.8.2875-2884.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dale JL, Beckman KB, Willett JLE, Nilson JL, Palani NP, Baller JA, Hauge A, Gohl DM, Erickson R, Manias DA, Sadowsky MJ, Dunny GM. 2018. Comprehensive functional analysis of the Enterococcus faecalis core genome using an ordered, sequence-defined collection of insertional mutations in strain OG1RF. mSystems 3. 10.1128/mSystems.00062-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korir ML, Dale JL, Dunny GM. 2019. Role of epaQ, a previously uncharacterized Enterococcus faecalis gene, in biofilm development and antimicrobial resistance. J Bacteriol 201. 10.1128/JB.00078-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dale JL, Cagnazzo J, Phan CQ, Barnes AM, Dunny GM. 2015. Multiple roles for Enterococcus faecalis glycosyltransferases in biofilm-associated antibiotic resistance, cell envelope integrity, and conjugative transfer. Antimicrob Agents Chemother 59:4094–4105. 10.1128/AAC.00344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willett JLE, Dale JL, Kwiatkowski LM, Powers JL, Korir ML, Kohli R, Barnes AMT, Dunny GM. 2021. Comparative biofilm assays using Enterococcus faecalis OG1RF identify new determinants of biofilm formation. mBio 12:e0101121. 10.1128/mBio.01011-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hyyryläinen H-L, Marciniak BC, Dahncke K, Pietiäinen M, Courtin P, Vitikainen M, Seppala R, Otto A, Becher D, Chapot-Chartier M-P, Kuipers OP, Kontinen VP. 2010. Penicillin-binding protein folding is dependent on the PrsA peptidyl-prolyl cis-trans isomerase in Bacillus subtilis. Mol Microbiol 77:108–127. 10.1111/j.1365-2958.2010.07188.x. [DOI] [PubMed] [Google Scholar]
- 36.Wiemels RE, Cech SM, Meyer NM, Burke CA, Weiss A, Parks AR, Shaw LN, Carroll RK. 2017. An intracellular peptidyl-prolyl cis/trans isomerase is required for folding and activity of the Staphylococcus aureus secreted virulence factor nuclease. J Bacteriol 199. 10.1128/JB.00453-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsen RJ, Sitkiewicz I, Ayeras AA, Gonulal VE, Cantu C, Beres SB, Green NM, Lei B, Humbird T, Greaver J, Chang E, Ragasa WP, Montgomery CA, Cartwright J, McGeer A, Low DE, Whitney AR, Cagle PT, Blasdel TL, DeLeo FR, Musser JM. 2010. Decreased necrotizing fasciitis capacity caused by a single nucleotide mutation that alters a multiple gene virulence axis. Proc Natl Acad Sci USA 107:888–893. 10.1073/pnas.0911811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikolo F, Zhang M, Harrington DJ, Robinson C, Waller AS, Sutcliffe IC, Black GW. 2015. Characterisation of SEQ0694 (PrsA/PrtM) of Streptococcus equi as a functional peptidyl-prolyl isomerase affecting multiple secreted protein substrates. Mol Biosyst 11:3279–3286. 10.1039/c5mb00543d. [DOI] [PubMed] [Google Scholar]
- 39.Shankar J, Walker RG, Wilkinson MC, Ward D, Horsburgh MJ. 2012. SalB inactivation modulates culture supernatant exoproteins and affects autolysis and viability in Enterococcus faecalis OG1RF. J Bacteriol 194:3569–3578. 10.1128/JB.00376-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Del Papa MF, Hancock LE, Thomas VC, Perego M. 2007. Full activation of Enterococcus faecalis gelatinase by a C-terminal proteolytic cleavage. J Bacteriol 189:8835–8843. 10.1128/JB.01311-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Djorić D, Kristich CJ. 2017. Extracellular SalB contributes to intrinsic cephalosporin resistance and cell envelope integrity in. J Bacteriol 199. 10.1128/JB.00392-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stinemetz EK. 2017. Evaluating the impact of post-translational modifications by the secreted zinc metalloprotease, GelE, on the major autolysin of E. faecalis, AtlA, and a stress-induced protein, SalB. The University of Texas MD Anderson Cancer Center UT Health Graduate School of Biomedical Sciences. [Google Scholar]
- 43.Zheng J, Wu Y, Lin Z, Wang G, Jiang S, Sun X, Tu H, Yu Z, Qu D. 2020. ClpP participates in stress tolerance, biofilm formation, antimicrobial tolerance, and virulence of Enterococcus faecalis. BMC Microbiol 20:30. 10.1186/s12866-020-1719-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaval KG, Singh KV, Cruz MR, DebRoy S, Winkler WC, Murray BE, Garsin DA. 2018. Loss of ethanolamine utilization in Enterococcus faecalis increases gastrointestinal tract colonization. mBio 9. 10.1128/mBio.00790-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rigottier-Gois L, Madec C, Navickas A, Matos RC, Akary-Lepage E, Mistou M-Y, Serror P. 2015. The surface rhamnopolysaccharide epa of Enterococcus faecalis is a key determinant of intestinal colonization. J Infect Dis 211:62–71. 10.1093/infdis/jiu402. [DOI] [PubMed] [Google Scholar]
- 46.Guerardel Y, Sadovskaya I, Maes E, Furlan S, Chapot-Chartier M-P, Mesnage S, Rigottier-Gois L, Serror P. 2020. Complete structure of the enterococcal polysaccharide antigen (EPA) of vancomycin-resistant Enterococcus faecalis V583 reveals that EPA decorations are teichoic acids covalently linked to a rhamnopolysaccharide Backbone. mBio 11. 10.1128/mBio.00277-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banla LI, Salzman NH, Kristich CJ. 2019. Colonization of the mammalian intestinal tract by enterococci. Curr Opin Microbiol 47:26–31. 10.1016/j.mib.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chatterjee A, Johnson CN, Luong P, Hullahalli K, McBride SW, Schubert AM, Palmer KL, Carlson PE, Duerkop BA. 2019. Bacteriophage resistance alters antibiotic-mediated intestinal expansion of enterococci. Infect Immun 87. 10.1128/IAI.00085-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colomer-Winter C, Gaca AO, Chuang-Smith ON, Lemos JA, Frank KL. 2018. Basal levels of (p)ppGpp differentially affect the pathogenesis of infective endocarditis in Enterococcus faecalis. Microbiology (Reading) 164:1254–1265. 10.1099/mic.0.000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Z-Y, Campeau A, Liu C-H, Gonzalez DJ, Yamaguchi M, Kawabata S, Lu C-H, Lai C-Y, Chiu H-C, Chang Y-C. 2021. Unique virulence role of post-translocational chaperone PrsA in shaping Streptococcus pyogenes secretome. Virulence 12:2633–2647. 10.1080/21505594.2021.1982501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma Y, Bryant AE, Salmi DB, Hayes-Schroer SM, McIndoo E, Aldape MJ, Stevens DL. 2006. Identification and characterization of bicistronic speB and prsA gene expression in the group A Streptococcus. J Bacteriol 188:7626–7634. 10.1128/JB.01059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lyon WR, Caparon MG. 2003. Trigger factor-mediated prolyl isomerization influences maturation of the Streptococcus pyogenes cysteine protease. J Bacteriol 185:3661–3667. 10.1128/JB.185.12.3661-3667.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lyon WR, Gibson CM, Caparon MG. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J 17:6263–6275. 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frank KL, Colomer-Winter C, Grindle SM, Lemos JA, Schlievert PM, Dunny GM. 2014. Transcriptome analysis of Enterococcus faecalis during mammalian infection shows cells undergo adaptation and exist in a stringent response state. PLoS One 9:e115839. 10.1371/journal.pone.0115839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roberts JC, Singh KV, Okhuysen PC, Murray BE. 2004. Molecular epidemiology of the fsr locus and of gelatinase production among different subsets of Enterococcus faecalis isolates. J Clin Microbiol 42:2317–2320. 10.1128/JCM.42.5.2317-2320.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strzelecki J, Hryniewicz W, Sadowy E. 2011. Gelatinase-associated phenotypes and genotypes among clinical isolates of Enterococcus faecalis in Poland. Pol J Microbiol 60:287–292. 10.33073/pjm-2011-041. [DOI] [PubMed] [Google Scholar]
- 57.Saffari F, Dalfardi MS, Mansouri S, Ahmadrajabi R. 2017. Survey for correlation between biofilm formation and virulence determinants in a collection of pathogenic and fecal Enterococcus faecalis isolates. Infect Chemother 49:176–183. 10.3947/ic.2017.49.3.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anderson AC, Jonas D, Huber I, Karygianni L, Wölber J, Hellwig E, Arweiler N, Vach K, Wittmer A, Al-Ahmad A. 2015. Enterococcus faecalis from food, clinical specimens, and oral sites: prevalence of virulence factors in association with biofilm formation. Front Microbiol 6:1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Macovei L, Ghosh A, Thomas VC, Hancock LE, Mahmood S, Zurek L. 2009. Enterococcus faecalis with the gelatinase phenotype regulated by the fsr operon and with biofilm-forming capacity are common in the agricultural environment. Environ Microbiol 11:1540–1547. 10.1111/j.1462-2920.2009.01881.x. [DOI] [PubMed] [Google Scholar]
- 60.Dunny GM, Clewell DB. 1975. Transmissible toxin (hemolysin) plasmid in Streptococcus faecalis and its mobilization of a noninfectious drug resistance plasmid. J Bacteriol 124:784–790. 10.1128/jb.124.2.784-790.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Del Papa MF, Perego M. 2008. Ethanolamine activates a sensor histidine kinase regulating its utilization in Enterococcus faecalis. J Bacteriol 190:7147–7156. 10.1128/JB.00952-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 63.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B. 2013. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 41:e140. 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tjaden B. 2015. De novo assembly of bacterial transcriptomes from RNA-seq data. Genome Biol 16:1. 10.1186/s13059-014-0572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kanehisa M, Sato Y. 2020. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci 29:28–35. 10.1002/pro.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 69.Zeng J, Teng F, Murray BE. 2005. Gelatinase is important for translocation of Enterococcus faecalis across polarized human enterocyte-like T84 cells. Infect Immun 73:1606–1612. 10.1128/IAI.73.3.1606-1612.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manias DA, Dunny GM. 2018. Expression of adhesive pili and the collagen-binding adhesin Ace is activated by ArgR family transcription factors in Enterococcus faecalis. J Bacteriol 200. 10.1128/JB.00269-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download jb.00129-22-s0001.xlsx, XLSX file, 0.7 MB (728.8KB, xlsx)
Table S2. Download jb.00129-22-s0002.xlsx, XLSX file, 0.06 MB (57.1KB, xlsx)
Fig. S1-S3; Table S3; Table S1 and S2 captions. Download jb.00129-22-s0003.pdf, PDF file, 2.7 MB (2.7MB, pdf)
Data Availability Statement
Files generated from RNA sequencing and data analysis have been deposited with NCBI GEO under accession number GSE198051.