THE SULFUR-OXIDIZING PROKARYOTES
Biological oxidation of hydrogen sulfide to sulfate is one of the major reactions of the global sulfur cycle. Reduced inorganic sulfur compounds (referred to below as sulfur) are exclusively oxidized by prokaryotes, and sulfate is the major oxidation product. Sulfur oxidation in members of the Eukarya is mediated by lithoautotrophic bacterial endosymbionts (44).
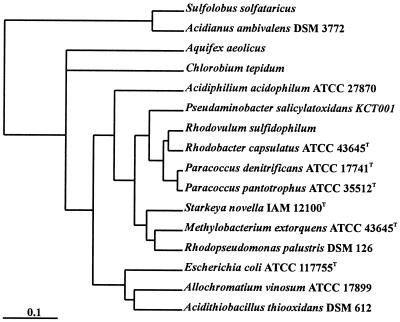
The sulfur-oxidizing prokaryotes are phylogenetically diverse (Fig. 1). In the domain Archaea aerobic sulfur oxidation is restricted to members of the order Sulfolobales (21, 58), and in the domain Bacteria sulfur is oxidized by aerobic lithotrophs or by anaerobic phototrophs. The nonphototrophic obligate anaerobe Wolinella succinogenes oxidizes hydrogen sulfide to polysulfide during fumarate respiration (41). The ecology, physiology, and biochemistry of sulfur-oxidizing bacteria have been reviewed previously. The neutrophilic chemolithotrophic bacteria have been reviewed by Kelly et al. (31, 32, 35) and Takakuwa (63), the acidophilic sulfur-oxidizing bacteria have been reviewed by Harrison (23) and Pronk et al. (46), and the molecular genetics of Acidithiobacillus ferrooxidans has been reviewed by Rawlings and Kusano (49). The sulfur metabolism of phototrophic bacteria has been reviewed by Brune (9, 10) and Trüper and Fischer (65). The physiology and genetics of both phototrophic and lithotrophic sulfur-oxidizing prokaryotes have been discussed recently (18).
FIG. 1.
Phylogenetic tree based on 16S rRNA gene sequence analysis of P. pantotrophus and different sulfur-oxidizing bacteria. Bar = 1 inferred nucleotide change per 100 nucleotides.
Prokaryotes oxidize hydrogen sulfide, sulfur, sulfite, thiosulfate, and various polythionates under alkaline (57), neutral, or acidic conditions (23). Aerobic sulfur-oxidizing prokaryotes belong to genera like Acidianus (18), Acidithiobacillus (36), Aquaspirillum (19), Aquifex (25), Bacillus (2), Beggiatoa (60), Methylobacterium (15, 34), Paracoccus, Pseudomonas (19), Starkeya (33), Sulfolobus (58), Thermithiobacillus (36), Thiobacillus, and Xanthobacter (19) and are mainly mesophilic. Phototrophic anaerobic sulfur-oxidizing bacteria are mainly neutrophilic and mesophilic (10, 65) and belong to genera like Allochromatium (formerly Chromatium [26]), Chlorobium, Rhodobacter, Rhodopseudomonas, Rhodovulum, and Thiocapsa (9). Lithoautotrophic growth in the dark has been described for Thiocapsa roseopersicina, Allochromatium vinosum, and other purple sulfur bacteria, as well as for purple nonsulfur bacteria like Rhodovulum sulfidophilum (formerly Rhodobacter sulfidophilus) (24), Rhodocyclus genatinosus, and Rhodopseudomonas acidophila (39, 56). This capacity may be based on related biochemical mechanisms of sulfur oxidation in lithotrophic and phototrophic bacteria.
Autotrophic bacteria fix carbon dioxide either via the reductive pentose phosphate cycle or via the reductive tricarboxylic acid cycle. Methylotrophic bacteria fix formaldehyde either via the ribulose monophosphate route or via the serine pathway (16, 61). Reductant released from sulfur oxidation is used in lithotrophic bacteria for aerobic respiration and carbon dioxide reduction, while in anaerobic phototrophic bacteria reductant is used mainly for carbon dioxide fixation. Sulfur oxidation by methylotrophic bacteria has been observed upon growth with methylated sulfur compounds, such as dimethyl sulfide (34). Few reports have focused on the sulfur-oxidizing potential of these organisms (15, 16, 61, 62) due to the major interest in their methylotrophic characteristics.
Two groups of sulfur-oxidizing lithotrophic bacteria have been distinguished previously; members of one group are able to utilize polythionates, and members of the other group are not able to do this (18, 35). On the basis of physiological and biochemical data, at least two major pathways have been proposed for different sulfur-oxidizing bacteria: (i) the sulfur oxidation pathway and (ii) the S4 intermediate pathway involving polythionates (35). Here we discuss the sox gene cluster of Paracoccus pantotrophus and the biochemistry and functions of the encoded proteins. On the basis of our analysis, together with available genomic data, we concluded that different metabolic reactions merge into a common mechanism in lithotrophic and phototrophic sulfur-oxidizing bacteria.
THE SULFUR-OXIDIZING SYSTEM OF P. PANTOTROPHUS
sox gene cluster.
P. pantotrophus (48) is a gram-negative, neutrophilic, facultatively lithoautotrophic bacterium that is able to grow with thiosulfate or with molecular hydrogen as an energy source and with a large variety of carbon sources. The gene cluster of P. pantotrophus coding for sulfur-oxidizing ability (Sox) comprises at least two transcriptional units with 15 genes. Seven genes, soxXYZABCD, code for proteins essential for sulfur oxidation in vitro. These genes and soxFGH are induced by thiosulfate (11, 20, 42, 51a, 69, 70). Four open reading frames (ORFs), ORF1, ORF2, and shxVW, are located upstream of soxX (Fig. 2), and the shxVW ORFs are cotranscribed (F. Bardischewsky and C. G. Friedrich, submitted for publication).
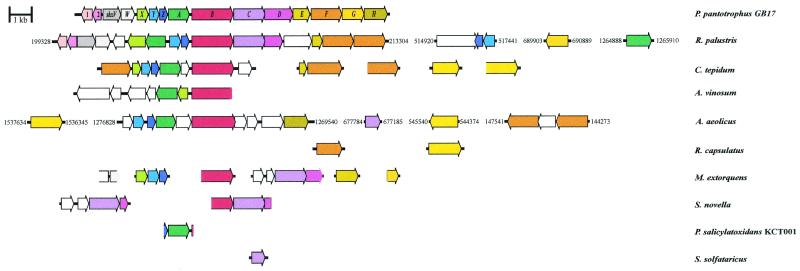
FIG. 2.
Schematic map of the sox gene cluster of P. pantotrophus and identified or putative sox genes of other sulfur-oxidizing bacteria. ORFs predicting homologous proteins are indicated by the same color. ORFs that encode proteins not homologous to Sox proteins of P. pantotrophus are white. The numbers indicate the positions of putative sox genes in the complete genomes. Accession numbers of the ORFs are given in Table 1.
The Sox proteins of P. pantotrophus are located in the periplasm, as shown by selective extraction of periplasmic proteins (20), and signal peptides have been predicted for all of the proteins except SoxZ (Table 1). The Sox proteins are transported to the periplasm either via the Sec system or via the Tat system. Twin arginine motifs in the signal peptides are diagnostic for protein transport via the Tat system (6, 8, 68) and are predicted for SoxY, SoxB, SoxC, SoxF, SoxG, and possibly SoxH (Table 1). The SoxY and SoxZ proteins do not contain a prosthetic group, redox center, or metal (20), and SoxZ is likely to be cotransported through the cytoplasmic membrane by SoxY in hitchhiker fashion, as previously shown for the heterodimeric hydrogenases of Escherichia coli (51) and Ralstonia eutropha (7).
TABLE 1.
sox genes of P. pantotrophus and genes thought to be involved in sulfur oxidation in other thiobacteria
| Organism | Gene | No. of amino acids | % Identity to P. pantotrophusa | Signal peptide with twin arginine motif | Putative function | Reference(s) | Accession no. |
|---|---|---|---|---|---|---|---|
| P. pantotrophus | orf1 | 149 | −c | Transcriptional regulator | 20 | X79242PDSOXf | |
| R. palustris | orf685 | 124 | 47.1 (104) | −c | RRPA00685d | ||
| P. pantotrophus | orf2 | 142 | − | Thioredoxin | This study | ||
| R. palustris | orf708 | 142 | 41.4 (116) | − | RRPA00708d | ||
| P. pantotrophus | shxV | 245 | − | Reductant transporter | 20 | X79242PDSOXf | |
| R. palustris | orf618 | 247 | 46.4 (237) | − | RRPA00618d | ||
| P. pantotrophus | shxW | 186 | − | Thioredoxin | 20 | X79242PDSOXf | |
| R. palustris | orf651 | 197 | 42.0 (176) | − | RRPA00651d | ||
| M. extorquens | orf239b | 119b | 47.6 (105) | + (TRRQFLA) | RMQ00239d | ||
| P. pantotrophus | soxX | 157 | − | Cytochrome c | 20 | X79242PDSOXf | |
| R. palustris | orf625 | 247 | 47.5 (40) | − | RRPA00625d | ||
| C. tepidum | orf1709 | 118 | 29.7 (37) | − | RCL01709d | ||
| A. vinosum | soxX | 135 | 32.5 (80) | − | 13 | ||
| M. extorquens | orf2551b,g | 158bg | 47.2 (125) | − | RMQ02551dg | ||
| P. pantotrophus | soxY | 140 | + (SRREALW) | Sulfur covalently binding protein | 20 | X79242PDSOXf | |
| R. palustris | orf660 | 154 | 44.5 (146) | + (SRREALA) | RRPA00660d | ||
| R. palustris | orf2261 | 156 | 34.9 (140) | ? (DRRQMLI) | RRPA02261d | ||
| C. tepidum | orf1710g | 153g | 37.5 (120) | + (SRRDFCR) | RCL01710dg | ||
| A. aeolicus | aq1810 | 150 | 30.1 (136) | + (TRRDFLK) | 14 | AE000757e | |
| M. extorquens | orf2552 | 152 | 56.7 (60) | + (NRRQALA) | RMQ02552d | ||
| P. pantotrophus | soxZ | 109 | −c | Sulfur compound chelating protein | 20 | X79242PDSOXf | |
| R. palustris | orf674 | 109 | 54.1 (109) | −c | RRPA00674d | ||
| R. palustris | orf2280 | 111 | 36.1 (108) | −c | RRPA02280d | ||
| C. tepidum | orf1711 | 101 | 37.0 (81) | −c | RCL01711d | ||
| A. aeolicus | aq1809 | 110 | 36.4 (110) | −c | 14 | AE000757e | |
| M. extorquens | orf2553 | 109 | 49.5 (107) | −c | RMQ02553d | ||
| P. salicylatoxidans | soxZb | 33b | 62.5 (32) | ? | 43 | ||
| P. pantotrophus | soxA | 290 | − | Cytochrome c (diheme) | 20 | X79242PDSOXf | |
| R. palustris | orf637 | 273 | 28.5 (144) | − | Cytochrome c (monoheme) | RRPA00637d | |
| R. palustris | orf2737 | 341 | 22.5 (120) | −c | Cytochrome c (diheme) | RRPA02737d | |
| C. tepidum | orf1712 | 286 | 29.5 (176) | − | Cytochrome c (monoheme) | RCL01712d | |
| A. vinosum | soxA | 281 | 30.0 (207) | − | Cytochrome c (monoheme) | 13 | |
| A. aeolicus | aq1807 | 267 | 29.2 (178) | − | Cytochrome c (diheme) | 14 | AE000757e |
| P. salicylatoxidans | soxA | 286 | 46.0 (276) | − | Cytochrome c (diheme) | 43 | AJ404005 |
| P. pantotrophus | soxB | 564 | + (TRREFIQ) | Sulfate thiol esterase | 20, 69 | X79242PDSOXf | |
| R. palustris | orf525 | 565 | 59.8 (565) | + (SRREFLQ) | RPA00525d | ||
| C. tepidum | orf1714g | 584g | 47.9 (376) | + (SRREFLR) | RCL01714dg | ||
| A. vinosum | soxB | 522b | 49.9 (519) | + (SRREFVR) | 13 | ||
| A. aeolicus | orf724 | 592 | 40.5 (551) | + (TRRDFLE) | Quinol-oxidase polypeptide 1 | 14 | AE000757e |
| M. extorquens | orf2550b | 374b | 65.7 (370) | ? | RMQ02550d | ||
| S. novella | soxBb | 309b | 62.3 (308) | ? | AF139113e | ||
| P. salicylatoxidans | soxBb | 7b | 85.7 (7) | + (SRREF..) | 43 | ||
| P. pantotrophus | soxC | 430 | + (SRRAFLR) | Sulfur dehydrogenase | 70 | X79242PDSOXf | |
| R. palustris | orf545 | 431 | 61.3 (408) | + (NRRRFLG) | RRPA00545d | ||
| M. extorquens | orf3760b,g | 403bg | 45.6 (410) | + (NRRAFLR) | RMQ03760dg | ||
| A. aeolicus | aq979 | 200 | 25.2 (119) | −c | 14 | AE000716c | |
| S. novella | soxC | 427 | 61.2 (428) | + (DRRRFLN) | AF139113c | ||
| S. novella | sorA | 405 | 26.5 (385) | + (NRRQILK) | Sulfite-cytochrome c oxidoreductase subunit A | 28 | AF154565e |
| S. solfataricus | 209 | 37.0 (100) | + (NRRDFLK) | J. van der Oost | |||
| P. pantotrophus | soxD | 384 | − | Cytochrome c | 70 | X79242PDSOXf | |
| R. palustris | orf644 | 251 | 52.7 (201) | − | RRPA00644d | ||
| M. extorquens | orf3759b,g | 190bg | 47.2 (176) | − | RMQ03789d, g | ||
| S. novella | soxDb | 84b | 55.2 (67) | − | AF139113e | ||
| S. novella | sorB | 108 | 60.0 (10) | − | c-type heme subunit of sorA | 28 | AF154565e |
| P. pantotrophus | soxE | 236 | − | Cytochrome c | 70 | X79242PDSOXf | |
| R. palustris | orf701 | 139 | 49.4 (77) | − | RRPA00701d | ||
| M. extorquens | orf3227 | 305 | 43.0 (107) | − | RMQ03227d | ||
| P. pantotrophus | soxF | 420 | + (TRRSLIA) | Flavoprotein | 51a, 70 | X79242PDSOXf | |
| R. palustris | orf554 | 421 | 57.1 (422) | + (NRRDVFK) | RRPA00554d | ||
| R. palustris | orf566 | 430 | 48.1 (420) | + (SRRGMIR) | RPA00566d | ||
| C. tepidum | orf1707g | 430g | 40.2 (420) | + (SRRTFNR) | RCL01707d, g | ||
| C. tepidum | orf258 | 458 | 46.3 (382) | + (SRRDFNK) | RCL00258d | ||
| A. aeolicus | dhsU | 436 | 30.5 (410) | + (NRRDVFK) | Flavocytochrome c sulfide dehydrogenase | 14 | AE000679e |
| A. aeolicus | fccB | 417 | 30.1 (379) | + (DRRNLLL) | Sulfide dehydrogenase, flavoprotein subunit | 14 | AE000679e |
| R. capsulatus | orf443 | 340 | 26.7 (318) | + (TRRHLAL) | RCL00443d | ||
| P. pantotrophus | soxG | 303 | + (SRRHFLA) | Thiol esterase | 51a | X79242PDSOXf | |
| M. extorquens | orf2512b | 145b | 32.7 (113) | ? | RMQ02512d | ||
| P. pantotrophus | soxH | 317 | ? (MRRLILC) | Thiol esterase | 51a | X79242PDSOXf | |
| A. aeolicus | cphA1 | 326 | 27.2 (268) | − | beta-Lactamase precursor | 14 | AE000757e |
| R. palustris | orf708 | 142 | − | Thiol-disulfide isomerase and thioredoxins | RRPA00708d | ||
| A. aeolicus | rhdA2 | 293 | − | Thiosulfate sulfur transferase | 14 | AE000757c | |
| A. aeolicus | aq1806 | 206 | − | 14 | AE000757c | ||
| A. aeolicus | trxA2 | 135 | − | Thioredoxin | 14 | AE000757c | |
| C. tepidum | orf717b | 444b | ? | Sulfide-quinone oxidoreductase | RCL00717d | ||
| C. tepidum | orf540 | 390 | − | Sulfide-quinone oxidoreductase | RCL00540d | ||
| A. aeolicus | sqr | 430 | − | Sulfide-quinone reductase | 14 | AE000777e | |
| R. capsulatus | sqr | 427 | − | Sulfide-quinone reductase | 53, 54 | X97478f |
The numbers in parentheses indicate the sizes of the amino acid overlaps.
The ORF is incomplete.
There is no signal peptide.
IGwit database accession number.
GenBank database accession number.
EMBL data library accession number.
Based on our analysis, we concluded that some sequences contain errors which affect the lengths of the ORFs, as follows: in orf1710 of C. tepidum the predicted start is 72 nucleotides upstream of the designated start; in orf1714 of C. tepidum the predicted start is 96 nucleotides downstream of the designated start; in orf3760 of M. extorquens the predicted start is at nucleotide 1838 due to a frameshift aound position 1426 upstream of the designated start at nucleotide 1384; in orf3759 of M. extorquens the predicted start is at nucleotide 639 due to a frameshift around position 505 downstream of the designated start at nucleotide 518; and in orf1707 of C. tepidum the predicted start is 90 nucleotides upstream of the designated start.
ORF1 predicts a transcriptional regulator of the ArsR family, and ORF2 predicts a periplasmic thioredoxin. Both of these ORFs are oriented divergent to the sox gene cluster (Fig. 2). shxV predicts a protein with six transmembrane channel-forming helices and a conserved cysteine at helices 1 and 4. ShxV is structurally related to CcdA of P. pantotrophus, which is involved in cytochrome c biogenesis. CcdA mediates transport of reductant from the cytoplasm to the periplasm for rereduction of the periplasmic apoprotein prior to heme addition. Disruption of ccdA results in a complete deficiency of c-type cytochromes, which in turn affects metabolic reactions involving c-type cytochromes, including sulfur oxidation (4). Disruption of shxV by the Ω-kanamycin interposon disables mutant GBΩV so that it cannot grow lithotrophically with thiosulfate and with molecular hydrogen. Cytochrome c formation is not affected in GBΩV, demonstrating that ShxV has a function distinct from that of CcdA. Addition of cysteine to the medium restores growth of strain GBΩV with hydrogen but not growth of this strain with thiosulfate. However, the thiosulfate-oxidizing activity of mixotrophically grown GBΩV is increased about 20-fold by 0.3 mM cystine and 2-fold by 0.3 mM cysteine, suggesting that there is a periplasmic oxidation reaction. shxW predicts a periplasmic thioredoxin that is unusual with respect to the predicted distribution of β-strands and α-helices, which very likely specify its role either in a redox reaction or for transport of reductant (Bardischewsky and Friedrich, submitted).
The soxEFGH genes are located downstream of soxD (Fig. 2). These genes are coexpressed with the sox structural genes, and thiosulfate-induced formation of SoxFGH has been demonstrated. However, a function could not be determined for these proteins since an in-frame deletion in soxF and complementation did not reveal obvious phenotypes of the mutants (51a).
Biochemistry of the Sox system of P. pantotrophus.
Seven sox structural genes code for four proteins. SoxXA, SoxYZ, SoxB, and SoxCD are required for sulfur-dependent cytochrome c reduction. SoxXA is a heterodimeric c-type cytochrome; SoxX (molecular mass, 14,834 Da) is a monoheme subunit, and SoxA (molecular mass including the heme moieties, 30,452 Da) is a diheme subunit. SoxYZ is composed of SoxY and SoxZ, which have predicted molecular masses of 10,977 and 11,718 Da, respectively. Neither SoxY nor SoxZ contains a cofactor or metal. The molecular mass of SoxY as determined by electrospray mass spectrometry (11,094 Da) differs from the predicted molecular mass of the mature protein by 117 Da (20). Thiosulfate covalently bound to SoxY accounts for 112 Da of the difference, and the higher molecular mass suggests that such an adduct is formed. In native gradient polyacrylamide gel electrophoresis gels SoxYZ appears as a heterodimer and as a 50-kDa α2β2 heterotetramer, and two forms of the heterodimer that have molecular masses of 29 and 31 kDa are observed (20).
The monomeric SoxB protein contains two manganese atoms (70), and the predicted molecular mass (58,611 Da) is identical to the molecular mass determined by electrospray mass spectrometry. Previous differences between the predicted and determined molecular masses (20) were resolved by analysis of the almost complete primary sequence and the N-terminal 32-pyroglutamate, which also identified the cleavage site of the signal peptidase (G. Grelle, unpublished data).
SoxCD is an α2β2 heterotetramer (molecular mass, 190,000 Da); SoxC is the molybdenum cofactor-containing subunit (molecular mass, 43,897 Da), and SoxD is the diheme c-type cytochrome (molecular mass including the two heme moieties, 38,815 Da) (47).
soxE predicts a diheme c-type cytochrome (molecular mass of the mature protein, 23,616 Da), and it is thought that SoxE is associated with SoxF (70). soxF encodes a 42,832-Da monomeric flavoprotein that includes a flavin moiety, as determined by electrospray mass spectrometry (51a). The primary sequence of SoxF is very similar to the primary sequence of FccB (70), the flavoprotein of flavocytochrome c-sulfide dehydrogenase of the phototrophic bacterium A. vinosum (71). SoxF does not exhibit sulfide dehydrogenase activity and appears not to be associated with a cytochrome (51a). soxG codes for a 29,657-Da polypeptide with two zinc binding motifs. The homodimeric SoxG molecule contains 0.90 atom of zinc per subunit. soxH codes for a 32,317-Da protein with a zinc binding motif and another metal binding motif. By using homodimeric SoxH it has been determined that there is 0.29 atom of zinc and 0.20 atom of copper per subunit (51a).
Catalytic properties of the Sox system of P. pantotrophus.
The Sox system reconstituted from SoxXA, SoxYZ, SoxB, and SoxCD mediates thiosulfate-, sulfite-, sulfur-, and hydrogen sulfide-dependent cytochrome c reduction, and each of the proteins alone is catalytically inactive (20, 51a). Thiosulfate is oxidized by the system according to equation 1, and sulfite is oxidized according to equation 2. Sulfite is also oxidized without SoxCD. This result excludes a function as a sulfite dehydrogenase and suggests a novel function for this molybdoenzyme:
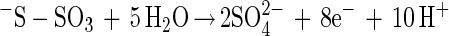 |
1 |
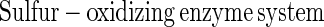 |
 |
2 |
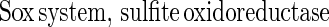 |
Without SoxCD 2 mol of electrons is produced per mol of thiosulfate, and addition of SoxCD increases the yield to eight electrons (20). Since no free intermediate of sulfur oxidation is observed, we suggest that SoxCD acts as a dehydrogenase at a protein-bound sulfur atom, and this protein is designated sulfur dehydrogenase. This suggestion is in accordance with the phenotype of a mutant with an in-frame deletion in soxC that is not able to grow lithotrophically with thiosulfate but is still able to oxidize thiosulfate, although at a low rate (51a, 70).
SOX SYSTEMS OF OTHER SULFUR-OXIDIZING BACTERIA
Oxidation of hydrogen sulfide to sulfur.
Phototrophic bacteria like Rhodobacter capsulatus, Pelodictyon luteolum, and some Chlorobium species anaerobically oxidize hydrogen sulfide to sulfur in the light. In vitro, hydrogen sulfide is oxidized to sulfur by some phototrophic and chemotrophic bacteria via a flavocytochrome c-sulfide dehydrogenase (equation 3) (22, 66) and a sulfide-quinone reductase (equation 4) (53). Flavocytochrome c of A. vinosum is not required for phototrophic growth with hydrogen sulfide, as shown by disruption of the cytochrome subunit fccA, since no obvious phenotype is observed for a fccA::Ω mutant (50).
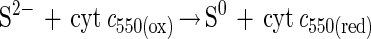 |
3 |
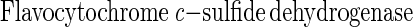 |
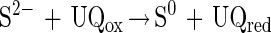 |
4 |
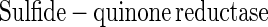 |
Therefore, it has been suggested that sulfide-quinone reductase is essential for growth of A. vinosum with hydrogen sulfide (50). Sulfide-quinone reductase has been shown to be essential for phototrophic growth of R. capsulatus with hydrogen sulfide (53). Hydrogen sulfide is the only sulfur substrate that R. capsulatus utilizes for phototrophic growth which is oxidized to sulfur, while A. vinosum oxidizes hydrogen sulfide to sulfate (18). Thus, the enzyme systems that oxidize hydrogen sulfide to sulfur and to sulfate are probably different.
SoxF of P. pantotrophus is not required for oxidation of reduced inorganic sulfur compounds in vitro (20). Moreover, a mutant with an in-frame deletion in soxF grows with thiosulfate, just as the wild-type does (51a). Two flavoprotein homologues have been observed in different genomes of sulfur-oxidizing bacteria (Fig. 2). Since the functions of these homologues are unknown, it is not known if they can complement each other.
Sulfide-quinone reductase has been characterized from Chlorobium (55), R. capsulatus (54), and the cyanobacterium Oscillatoria limnetica (3). Sulfide-quinone reductase activity is also present in P. pantotrophus (52), but it does not account for lithotrophic hydrogen sulfide oxidation since an insertional mutant with a mutation in the soxB gene is not able to oxidize hydrogen sulfide and thiosulfate (11).
Oxidation of sulfur to sulfate.
The first thiosulfate-oxidizing enzyme system analyzed in detail (equation 1) was that of Paracoccus versutus (formerly Thiobacillus versutus [30]). This system (reviewed in reference 35) is composed of enzyme A (molecular mass, 16,000 Da), enzyme B (60,000 Da), hexameric cytochrome c551 (260,000 Da) with 43,000-Da subunits, which is intimately associated with sulfite dehydrogenase (44,000 Da), and homodimeric cytochrome c552.5 (35). An important study has demonstrated that enzyme A binds thiosulfate to sulfite as a competitive inhibitor (40). In spite of differences in the subunit compositions of enzyme A, cytochrome c552.5, and cytochrome c551-sulfite dehydrogenase compared to SoxYZ, SoxXA, and SoxCD, respectively, the similar partial primary sequences of P. versutus Sox proteins (20, 69) suggest that the system is similar to that of P. pantotrophus with respect to structure and function (18):
 |
5 |
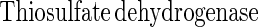 |
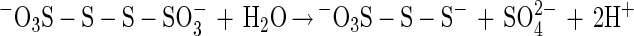 |
6 |
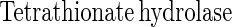 |
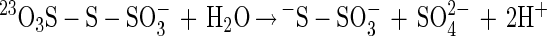 |
7 |
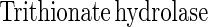 |
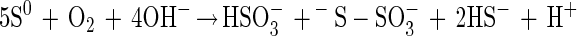 |
8 |
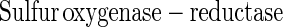 |
From other sulfur-oxidizing bacteria different enzymes involved in oxidation or hydrolysis of inorganic sulfur compounds have been characterized (equations 5 to 7). The reactions, however, do not involve oxidation of sulfur to sulfate. From Acidianus brierleyi a sulfur oxygenase was described (17), and from Acidianus ambivalens a sulfur oxygenase-reductase was described (37, 38). The two proteins probably function identically and produce sulfite, hydrogen sulfide, and thiosulfate from sulfur and molecular oxygen (equation 8). No link to energy metabolism has been reported (38), and the significance of a sulfur oxygenase-reductase in energy metabolism is not clear. This enzyme has not been detected in Bacteria.
Sulfite oxidoreductase oxidizes sulfite to sulfate (equation 2) and has been characterized from Starkeya novella and some members of the Eukarya (28, 64). Sulfite oxidoreductase is present in S. novella at high specific activity, and its function clearly differs from that of SoxCD. Thiosulfate dehydrogenase oxidizes thiosulfate to tetrathionate (equation 5) and is present in various litho- or phototrophic sulfur-oxidizing bacteria (9, 31). Trithionate hydrolase yields thiosulfate and sulfate. In general, polythionate hydrolysis yields polysulfide sulfate esters [O3S-S-(S-)x]2−, and these esters decompose spontaneously to sulfur and thiosulfate (59).
ANALYSIS OF GENOME SEQUENCES
Proteins homologous to the Sox proteins of P. pantotrophus were found in members of the domain Bacteria but not in members of the domain Archaea. From the complete genome of Sulfolobus solfataricus only a periplasmic sulfite dehydrogenase was predicted (J. van der Oost, personal communication). Proteins homologous to the Sox proteins of P. pantotrophus were detected in members of the domain Bacteria, such as the thermophile Aquifex aeolicus (25), the moderately thermophilic green bacterium Chlorobium tepidum (67), and the purple bacterium Rhodopseudomonas palustris (65). For the facultative methylotroph Methylobacterium extorquens (1, 45) essential Sox proteins were deduced from the partial genome sequence (Table 1). Partial sox gene clusters with the same order of genes as P. pantotrophus were detected in S. novella and Pseudaminobacter salicylatoxidans KCT001 (Fig. 2), both of which grow with tetrathionate (29, 43). It was predicted that these proteins are located in the periplasm and are transported to the periplasm from the cytoplasm via either the Tat system or the Sec system (6, 18, 68).
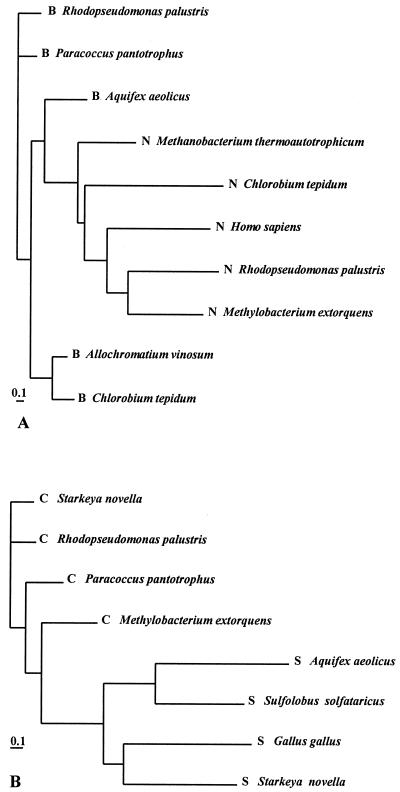
Analysis of genome data allows workers to differentiate the function of SoxB and SoxC from the function of their homologues. SoxB proteins exhibit significant identities to 5′-nucleotidases but are distinct from these molecules on a protein phylogenetic tree (Fig. 3A). In view of the differences and since phosphate is not involved in sulfur oxidation but is isosteric to sulfate, we suggest that SoxB proteins function as sulfate thiohydrolases but not as phosphatases.
FIG. 3.
Phylogenetic relationships of SoxB and SoxC homologous proteins of different bacteria. (A) Proteins that were identified as SoxB homologues (B) and 5′-nucleotidases (N). (B) SoxC (C) and sulfite oxidoreductase (S) homologous proteins The bar indicates the estimated distance of accepted point mutations per 100 amino acids.
It has been suggested that SoxC proteins act as dehydrogenases with the protein-bound sulfur atom, while their homologues, sulfite oxidoreductases (like SorA), oxidize free sulfite to sulfate. Both types of enzymes are molybdenum hydroxylases, and the suggested different functions of SorA and SoxC coincide with the separate positions of the molecules on the protein phylogenetic tree (Fig. 3B).
sox genes of phototrophic sulfur-oxidizing bacteria.
The sox gene cluster of R. palustris is similar to that of P. pantotrophus and comprises 16 genes equivalent to ORF1, ORF2, shxVW, and soxXAYZBCDEF1F2; soxF1 and soxF2 predict flavoproteins. Two hypothetical genes are in the cluster, while a soxG homologue is located at some distance. A second set of soxA and soxYZ genes and a gene predicting a sulfatase are located outside the sox gene cluster (Fig. 2). The soxA gene in the sox cluster, RRPA00637, predicts a monoheme, and the soxA gene outside the sox gene cluster, RRPA02737, predicts a diheme cytochrome (Fig. 2; Table 1).
In a partial genome sequence of C. tepidum a sox gene cluster comprising eight genes equivalent to soxF1XYZAB followed by a gene predicting a thioredoxin was detected. A hypothetical gene separated soxB and soxA. Outside the cluster were genes predicting two flavoproteins, one SoxE homologue, and two sulfide-quinone reductases. Genes equivalent to shxV, shxW, and soxCD essential for lithotrophic growth of P. pantotrophus with thiosulfate have not been detected in the incomplete genome sequence of C. tepidum (Fig. 2).
In A. vinosum a partial sox gene cluster that included soxB and soxXA was identified by using PCR technology (13). These genes may be essential for sulfur oxidation in this organism since inactivation of fccA encoding the cytochrome subunit of flavocytochrome c-sulfide dehydrogenase and inactivation of the aprBA locus encoding adenosine-5′-phosphosulfate reductase did not affect hydrogen sulfide oxidation (50) or sulfite oxidation (12).
R. capsulatus oxidizes hydrogen sulfide to sulfur, and this activity is linked to sulfide-quinone reductase. Disruption of sqrA results in an inability to grow phototrophically with hydrogen sulfide. Thus, this system is clearly different from that described for other sulfur-oxidizing bacteria which oxidize inorganic reduced sulfur compounds to sulfate. From the available genome sequence data for R. capsulatus no Sox protein homologue except a flavoprotein has been detected.
sox genes of lithotrophic sulfur-oxidizing bacteria.
A. aeolicus is an obligately aerobic chemolithotrophic bacterium. This organism requires molecular hydrogen for lithoautotrophic growth and does not grow with thiosulfate alone. However, thiosulfate increases the cellular yield, indicating that it is used for energy conservation (25). The sox gene cluster of A. aeolicus comprises 10 genes. A gene predicting a thioredoxin is followed by genes equivalent to soxYZAB and soxH. soxB and soxH are separated by three ORFs; one of these ORFs predicts a thiosulfate sulfur transferase (rhodanese), while two ORFs have unknown functions. The sulfite dehydrogenase homologue-encoding gene is separated from the sox gene cluster of A. aeolicus. Also, two genes predicting homologues of the flavoprotein SoxF of P. pantotrophus and two putative sqr genes are separated from the gene cluster (Fig. 2). The incomplete genomic sequence of M. extorquens predicts that there are 10 sox genes, 5 of which are incomplete, and these 10 genes are equivalent to shxV′W′, soxXYZ, soxB′, soxCD′, soxE, and soxG′. This finding suggests that M. extorquens is a sulfur-oxidizing bacterium. In fact, this organism utilizes thiosulfate, which increases the cellular yield during mixotrophic growth, and sulfur dehydrogenase (SoxCD) antigens are detected in cells grown with thiosulfate but not in cells grown without thiosulfate (J. Fischer and C. G. Friedrich, unpublished data).
In the tetrathionate-oxidizing organism S. novella, soxC predicting sulfur dehydrogenase has been identified together with truncated soxB′ and soxD′ genes. The gene order soxB′CD′ is identical to that in P. pantotrophus and may indicate that the gene clusters are similar. The sorAB genes encode sulfite oxidoreductase and may be separated from soxB′CD′ (Fig. 2; Table 1). The proposal that there are two thiosulfate-oxidizing systems in S. novella was based on the presence of two sulfite dehydrogenase homologues, SorAB and SoxCD (29). The proposed different functions of SoxCD of S. novella and SorAB bring into question whether there are two sulfur oxidation pathways and whether SorAB is the only representative of the second pathway. Moreover, induced S. novella cells oxidize sulfite at a high rate, which P. pantotrophus is unable to do, and the latter organism lacks sulfite oxidoreductase activity (5, 11). In light of the genomic data, a second pathway for thiosulfate oxidation appears to be unlikely, and confirmation of this hypothesis will require inactivation of either SorAB or SoxCD in S. novella.
In the tetrathionate-oxidizing organism P. salicylatoxidans KCT001 a soxZ′AB′ gene order is observed (Fig. 2). The predicted diheme c-type cytochrome SoxA is highly homologous to SoxA of P. pantotrophus and the corresponding cytochromes of other phototrophic or lithotrophic sulfur-oxidizing bacteria (Table 1). A partial soxZ gene is located upstream of soxA, and downstream a truncated ORF predicts a partial leader peptide of SoxB. The order of these genes is identical to the order in P. pantotrophus (Fig. 2). Disruption of soxA of P. salicylatoxidans KCT001 results in an inability to grow with and to oxidize thiosulfate and tetrathiothionate (43), demonstrating that SoxA is essential for sulfur oxidation and that polythionates and thiosulfate may be oxidized to sulfate by the same system.
Common Sox proteins of different sulfur-oxidizing bacteria.
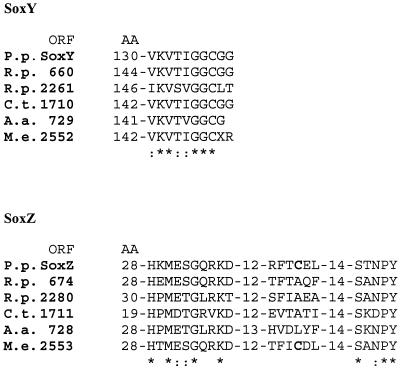
The different Sox systems are located exclusively in the periplasm. According to the available data, the sox genes appear to be clustered, and homologues of SoxA, SoxB, SoxY, and SoxZ are present in phototrophic, lithotrophic, and methylotrophic bacteria which oxidize reduced sulfur compounds to sulfate (in M. extorquens a SoxA homologue is missing from the incomplete genome sequence). The Sox proteins share the novel motif of SoxY and conserved regions of SoxZ and SoxB. The novel motif (V/I)KV(T/S)(V/I)GGC is located at the C terminus of SoxY (Fig. 4), and the cysteine residue is predicted to bind to the sulfur atom that is oxidized to sulfate. This prediction is based on the difference between the determined molecular mass of SoxY and the molecular mass predicted from the nucleotide sequence and determined from the amino acid sequence (20).
FIG. 4.
Predicted sulfur binding motifs of SoxY proteins and conserved signatures of SoxZ proteins of different sulfur-oxidizing bacteria. P. p., P. pantotrophus; R. p., R. palustris; C. t., C. tepidum; A. a., A. aeolicus; M. e., M. extorquens; AA, amino acid sequence.
SoxZ proteins contain two signatures, HXM(E/D)(T/S) GXK(D/T) and SX(N/D)PY (Fig. 4). The cysteine residue present in SoxZ of P. pantotrophus and M. extorquens appears not to be required for linkage of the proteins for cotransport through the cytoplasmic membrane since periplasmic SoxZ proteins from other sources lack this cysteine. Sulfur oxidation in both organisms is strictly aerobic, while sulfur oxidation in phototrophic bacteria is anaerobic and sulfur oxidation in A. aeolicus is microaerophilic. Some phototrophic bacteria grow lithotrophically in the dark under microaerophilic conditions with reduced sulfur compounds (27, 39). Therefore, the cysteine residue of SoxZ may play a role in coordination of the Sox proteins or in aerobic sulfur oxidation. Some SoxA homologues are either monoheme or diheme cytochromes. The difference may also have significance for aerobic or anaerobic sulfur metabolism in phototrophic sulfur bacteria or for differences in the initial reaction of sulfur oxidation.
PROPOSAL OF THE MECHANISM OF SULFUR OXIDATION
Functions of the Sox proteins.
The reconstituted Sox system of P. pantotrophus performs sulfite-, thiosulfate-, sulfur-, and hydrogen sulfide-dependent cytochrome c reduction (20, 51a). Oxidation of sulfite by SoxXA, SoxYZ, and SoxB is consistent with the finding that SoxCD does not function as a sulfite dehydrogenase but functions as a sulfur dehydrogenase (20, 51a). The c-type cytochrome SoxXA may (i) act as a specific electron mediator, (ii) fuse SoxY with the sulfur substrate in an oxidative reaction (equation 9 [see below]), or (iii) fuse SoxY and SoxZ in acrobic lithotrophs, thus acting as a heme enzyme. The catalytic properties of the Sox proteins of P. pantotrophus and the cysteine motif of SoxY suggest a mechanism for sulfur oxidation. The cysteine residue of SoxY is exposed at the C terminus (Fig. 4). Thus, its sulfur atom is located at the tip of the protein and is able to swing to either SoxXA, SoxCD, or SoxB.
Proposed reactions and intermediates.
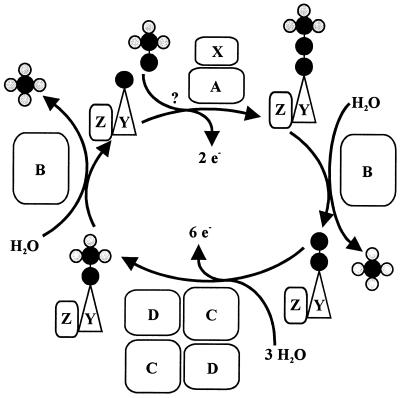
The N-terminal amino acid sequence of SoxZ is highly homologous to that of enzyme A of P. versutus (20), suggesting that the function of SoxYZ is identical to the function of enzyme A. Enzyme A of P. versutus binds thiosulfate and sulfite. It has been suggested that binding of sulfite occurs at a protein disulfide group (40). By analogy, complexes of the two substrates may be similarly formed by SoxYZ of P. pantotrophus. Each subunit contains a single cysteine whose thiols are able to form a disulfide bond. SoxZ molecules of phototrophic bacteria lack a cysteine residue, and SoxXA may initiate oxidation of thiosulfate to form SoxY-thiocysteine-S-sulfate (equation 9). SoxB would hydrolyze sulfate from the thiocysteine-S-sulfate residue to give S-thiocysteine (equation 10). SoxCD could then oxidize the outer sulfur atom to SoxY-cysteine-S-sulfate (equations 11 to 13). Finally, sulfate may again be hydrolyzed and removed by SoxB to regenerate the cysteine residue of SoxY (equation 14). The sequence of reactions is summarized in Fig. 5.
FIG. 5.
Model of the sequence of the sulfur oxidation reactions of P. pantotrophus.
Sulfite oxidation does not require SoxCD. Sulfite may also be added to SoxY to form SoxY-cysteine-S-sulfate (equation 15), which would be subsequently hydrolyzed by SoxB, yielding sulfate (equation 14). Hydrogen sulfide may be initially oxidized by SoxXA to form an S-thiocysteine residue of SoxY (equation 16) with further oxidation (equations 11 to 13) and hydrolysis (equation 14), and sulfur may be initially oxidized similarly, as proposed for hydrogen sulfide.
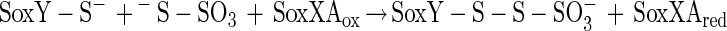 |
9 |
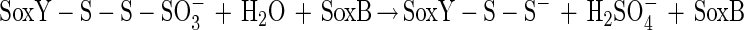 |
10 |
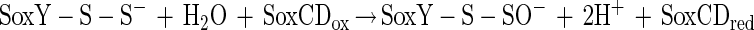 |
11 |
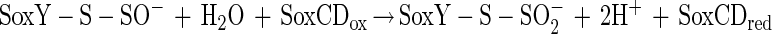 |
12 |
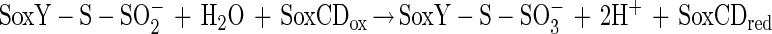 |
13 |
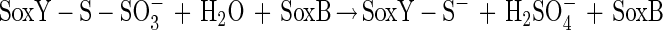 |
14 |
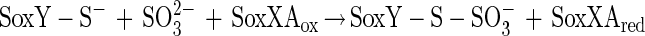 |
15 |
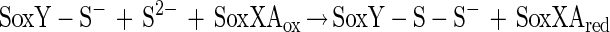 |
16 |
Tetrathionate is hydrolyzed by tetrathionate hydrolase to sulfate and thioperoxymonosulfate ([O3S-S-S]2−). Its spontaneous decomposition to sulfur and thiosulfate (59) enables the same Sox system to oxidize both products. SoxYZAB proteins appear to be crucial for sulfur oxidation and are present in all sulfur-oxidizing bacteria, as deduced from the available data (Fig. 2). In the hyperthermophile A. aeolicus a SoxC homologue has not been detected, while SorA appears to be present. Also, ShxVW homologues are missing from the genome of A. aeolicus. In this hyperthermophile initiation of sulfur oxidation may occur as it does in other bacteria but may proceed differently, as proposed here.
CONCLUSION
The sulfur-oxidizing enzyme system of P. pantotrophus is able to oxidize different reduced inorganic sulfur compounds. It is proposed that the sulfur atom oxidized binds covalently to the cysteine residue of the SoxY protein to form S-thiocysteine. The outer sulfur atom is oxidized by the sulfur dehydrogenase SoxCD, and sulfate is hydrolyzed by the sulfatase SoxB. From available genome sequence data for sulfur-oxidizing bacteria evidence has emerged that similar proteins are present in anaerobic phototrophic and aerobic lithotrophic bacteria but not in the archaeon S. solfataricus. Thus, oxidation of sulfur to sulfate may be mediated by very similar systems in bacteria. However, differences may involve the mechanism of linkage of the sulfur atom to be oxidized with SoxY during aerobic and anaerobic sulfur metabolism in lithotrophic and phototrophic bacteria. Also, the systems may differ with respect to the specificities of the actual sulfur substrates.
ACKNOWLEDGMENTS
We thank Christiane Dahl for making available the nucleotide sequence-predicting sox genes of A. vinosum prior to publication, John van der Oost for performing the BLAST search of the S. solfataricus genome for Sox protein homologues of P. pantotrophus, and Erko Stackebrandt for providing the 16S ribosomal DNA sequence of P. salicylatoxidans KCT001 prior to publication.
This study was supported by grant Fr318/6-3 from the Deutsche Forschungsgemeinschaft and the Ministerium für Wissenschaft und Forschung des Landes Nordrhein-Westfalen.
REFERENCES
- 1.Anthony C. The biochemistry of the methylotrophs. New York, N.Y: Academic Press; 1982. [Google Scholar]
- 2.Aragno M. Aerobic chemolithoautotrophic bacteria. In: Kristjansson J K, editor. Thermophilic bacteria. Boca Raton, Fla: CRC Press; 1991. pp. 7–103. [Google Scholar]
- 3.Arieli B, Shahak Y, Taglicht D, Hauska D, Padan E. Purification and characterization of sulfide-quinone reductase, a novel enzyme driving anoxygenic photosynthesis in Oscillatoria limnetica. J Biol Chem. 1994;269:5705–5711. [PubMed] [Google Scholar]
- 4.Bardischewsky F, Friedrich C G. Identification of ccdA in Paracoccus pantotrophus GB17: disruption of ccdA causes complete deficiency in c-type cytochromes. J Bacteriol. 2001;183:257–263. doi: 10.1128/JB.183.1.257-263.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beffa T, Fischer C, Aragno M. Growth and respiratory oxidation of reduced sulfur compounds by intact cells of Thiobacillus novellus (type strain) grown on thiosulfate. Curr Microbiol. 1993;26:323–326. [Google Scholar]
- 6.Berks B C. A common export pathway for proteins binding complex redox cofactors. Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 7.Bernhard M, Friedrich B, Siddiqui A. Ralstonia eutropha is blocked in Tat-mediated protein export. J Bacteriol. 2000;182:581–588. doi: 10.1128/jb.182.3.581-588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogsch E G, Sargent F, Stanley N R, Berks B C, Robinson C, Palmer T. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J Biol Chem. 1998;273:18003–18006. doi: 10.1074/jbc.273.29.18003. [DOI] [PubMed] [Google Scholar]
- 9.Brune D C. Sulfur oxidation by phototrophic bacteria. Biochim Biophys Acta. 1989;975:189–221. doi: 10.1016/s0005-2728(89)80251-8. [DOI] [PubMed] [Google Scholar]
- 10.Brune D C. Sulfur compounds as photosynthetic electron donors. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer; 1995. pp. 847–870. [Google Scholar]
- 11.Chandra T S, Friedrich C G. Tn5-induced mutations affecting sulfur-oxidizing ability (Sox) of Thiosphaera pantotropha. J Bacteriol. 1986;166:446–452. doi: 10.1128/jb.166.2.446-452.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahl C. Insertional gene inactivation in a phototrophic sulphur bacterium: APS-reductase-deficient mutants of Chromatium vinosum. Microbiology. 1996;142:3363–3372. doi: 10.1099/13500872-142-12-3363. [DOI] [PubMed] [Google Scholar]
- 13.Dahl C, Kuever J, Kräling M. Genes encoding a thiosulfate-oxidizing multienzyme complex in phototrophic and chemotrophic sulfur bacteria. Biospektrum. 2001;7:98. [Google Scholar]
- 14.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olson G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 15.de Zwart J M M, Nelisse P N, Kuenen J G. Isolation and characterization of Methylophaga sulfidovorans, sp. nov.: an obligately methylotrophic, aerobic, dimethyl sulfide oxidizing bacterium from a microbial mat. FEMS Microbiol Ecol. 1996;20:261–270. [Google Scholar]
- 16.de Zwart J M M, Sluis J M R, Kuenen J G. Competition for dimethyl sulfide and hydrogen sulfide by Methylophaga sulfidovorans and Thiobacillus thioparus T5 in continuous cultures. Appl Environ Microbiol. 1997;63:3318–3322. doi: 10.1128/aem.63.8.3318-3322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emmel T, Sand W, König W A, Bock E. Evidence for the existance of a sulphur oxygenase in Sulfolobus brierleyi. J Gen Microbiol. 1986;132:3415–3420. [Google Scholar]
- 18.Friedrich C G. Physiology and genetics of sulfur-oxidizing bacteria. Adv Microb Physiol. 1998;39:235–289. doi: 10.1016/s0065-2911(08)60018-1. [DOI] [PubMed] [Google Scholar]
- 19.Friedrich C G, Mitrenga G. Oxidation of thiosulfate by Paracoccus denitrificans and other hydrogen bacteria. FEMS Microbiol Lett. 1981;10:209–212. [Google Scholar]
- 20.Friedrich C G, Quentmeier A, Bardischewsky F, Rother D, Kraft R, Kostka S, Prinz H. Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J Bacteriol. 2000;182:4677–4687. doi: 10.1128/jb.182.17.4677-4687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuchs T, Huber H, Burggraf S, Stetter K O. 16S rDNA-based phylogeny of the archaeal order Sulfolobales and reclassification of Desulfurolobus ambivalens as Acidianus ambivalens comb. nov. Syst Appl Microbiol. 1996;19:56–60. [Google Scholar]
- 22.Fukumori Y P, Yamanaka I. Flavocytochrome c of Chromatium vinosum. Some enzymatic properties and subunit structure. J Biochem (Tokyo) 1979;85:1405–1414. doi: 10.1093/oxfordjournals.jbchem.a132467. [DOI] [PubMed] [Google Scholar]
- 23.Harrison A P., Jr The acidophilic thiobacilli and other acidophilic bacteria that share their habitat. Annu Rev Microbiol. 1984;38:265–292. doi: 10.1146/annurev.mi.38.100184.001405. [DOI] [PubMed] [Google Scholar]
- 24.Hiraishi A, Umeda Y. Intragenic structure of the genus Rhodobacter: transfer of Rhodobacter sulfidophilus and related marine species to the genus Rhodovulum gen. nov. Int J Syst Bacteriol. 1994;44:15–23. [Google Scholar]
- 25.Huber R, Stetter K O. Embryonic ELS. No. 785998. Houndmills, England: Macmillan; 1999. Aquificales; pp. 1–7. [Google Scholar]
- 26.Imhoff J F, Süling J, Petri R. Phylogenetic relationship and taxonomic reclassification of Chromatium species and related purple sulfur bacteria. Int J Syst Bacteriol. 1998;48:1029–1043. [Google Scholar]
- 27.Kämpf C, Pfennig N. Chemoautotrophic growth of Thiocystis violacea, Chromatium gracile and C. vinosum in the dark at various O2-concentrations. J Basic Microbiol. 1986;26:517–531. [Google Scholar]
- 28.Kappler U, Bennett B, Rethmeier J, Schwarz G, McEwan A G, Dahl C. Sulfite:cytochrome c oxidoreductase from Thiobacillus novellus—purification, characterization and molecular biology of a heterodimeric member of the sulfite oxidase family. J Biol Chem. 2000;275:13202–13212. doi: 10.1074/jbc.275.18.13202. [DOI] [PubMed] [Google Scholar]
- 29.Kappler U, Friedrich C G, Trüper H G, Dahl C. Evidence for two pathways of thiosulfate oxidation in Starkeya novella (formerly Thiobacillus novellus) Arch Microbiol. 2001;175:102–111. doi: 10.1007/s002030000241. [DOI] [PubMed] [Google Scholar]
- 30.Katayama Y, Hiraishi A, Kuraishi H. Paracoccus thiocyanatus sp. nov., a new species of thiocyanate-utilizing facultative chemolithotroph, and transfer of Thiobacillus versutus to the genus Paracoccus as Paracoccus versutus comb. nov. with emendation of the genus. Microbiology. 1995;141:1469–1477. doi: 10.1099/13500872-141-6-1469. [DOI] [PubMed] [Google Scholar]
- 31.Kelly D P. Biochemistry of the chemolithotrophic oxidation of inorganic sulfur. Phil Trans R Soc Lond B Biol Sci. 1982;298:499–528. doi: 10.1098/rstb.1982.0094. [DOI] [PubMed] [Google Scholar]
- 32.Kelly D P. Physiology and biochemistry of unicellular sulfur bacteria. In: Schlegel H G, Bowien B, editors. Autotrophic bacteria. Berlin, Germany: Springer; 1989. pp. 193–217. [Google Scholar]
- 33.Kelly D P, McDonald I R, Wood A P. Proposal for the reclassification of Thiobacillus novellus as Starkeya novella gen. nov., comb. nov., in the α-subclass of the Proteobacteria. Int J Syst Evol Microbiol. 2000;50:1797–1802. doi: 10.1099/00207713-50-5-1797. [DOI] [PubMed] [Google Scholar]
- 34.Kelly D P, Smith N A. Organic sulfur compounds in the environment. Adv Microb Ecol. 1990;11:345–385. [Google Scholar]
- 35.Kelly D P, Shergill J K, Lu W-P, Wood A P. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Leeuwenhoek. 1997;71:95–107. doi: 10.1023/a:1000135707181. [DOI] [PubMed] [Google Scholar]
- 36.Kelly D P, Wood A P. Reclassification of some species of Thiobacillus to the newly designated genera Acidithiobacillus gen. nov., Halobacillus gen. nov. and Thermithiobacillus gen. nov. Int J Syst Evol Microbiol. 2000;50:511–516. doi: 10.1099/00207713-50-2-511. [DOI] [PubMed] [Google Scholar]
- 37.Kletzin A. Coupled enzymatic production of sulfite, thiosulfite, and hydrogen sulfide from sulfur: purification and properties of a sulfur-oxygenase/reductase from the facultatively anaerobic archaebacterium Desulfurolobus ambivalens. J Bacteriol. 1989;171:1638–1643. doi: 10.1128/jb.171.3.1638-1643.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kletzin A. Molecular characterization of the sor gene, which encodes the sulfur-oxygenase/reductase of the thermophilic archaeum Desulfurolobus ambivalens. J Bacteriol. 1992;174:5854–5859. doi: 10.1128/jb.174.18.5854-5859.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kondratieva E N. Chemolithotrophy of phototrophic bacteria. In: Schlegel H G, Bowien B, editors. Autotrophic bacteria. Berlin, Germany: Springer; 1989. pp. 283–287. [Google Scholar]
- 40.Lu W-P, Swoboda B E P, Kelly D P. Properties of the thiosulfate-oxidizing multi-enzyme system from Thiobacillus versutus. Biochim Biophys Acta. 1985;828:116–122. [Google Scholar]
- 41.Macy J M, Schröder I, Thauer R K, Kröger A. Growth of Wolinella succinogenes on H2S plus fumarate and on formate plus sulfur as energy sources. Arch Microbiol. 1986;144:147–150. [Google Scholar]
- 42.Mittenhuber G, Sonomoto K, Egert M, Friedrich C G. Identification of the DNA region responsible for sulfur-oxidizing ability of Thiosphaera pantotropha. J Bacteriol. 1991;173:7340–7344. doi: 10.1128/jb.173.22.7340-7344.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukhopadhyaya P N, Deb C, Lahiri C, Roy P. A soxA gene, encoding a diheme cytochrome c, and a sox locus, essential for sulfur oxidation in a new sulfur lithotrophic bacterium. J Bacteriol. 2000;182:4278–4287. doi: 10.1128/jb.182.15.4278-4287.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson D C, Fisher C R. Chemoautotrophic and methanotrophic endosymbiotic bacteria at deep-sea vents and seeps. In: Karl D M, editor. The microbiology of deep-sea hydrothermal vents. Boca Raton, Fla: CRC Press; 1995. pp. 125–167. [Google Scholar]
- 45.Peel D, Quayle J R. Microbial growth on C1-compounds: isolation and characterization of Pseudomonas AM1. Biochem J. 1961;81:465–469. doi: 10.1042/bj0810465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pronk J T, Meulenberg R, Hazeu W, Bos P, Kuenen J G. Oxidation of reduced inorganic sulphur compounds by acidophilic thiobacilli. FEMS Microbiol Rev. 1990;75:293–306. [Google Scholar]
- 47.Quentmeier A, Kraft R, Kostka S, Klockenkämper R, Friedrich C G. Characterization of a new type of sulfite dehydrogenase from Paracoccus pantotrophus GB17. Arch Microbiol. 2000;173:117–125. doi: 10.1007/s002039900118. [DOI] [PubMed] [Google Scholar]
- 48.Ralney F A, Kelly D P, Stackebrandt E, Burghardt J, Hiraishi A, Katayama Y, Wood A P. A re-evaluation of the taxonomy of Paracoccus denitrificans and a proposal for the combination Paracoccus pantotrophus comb. nov. Int J Syst Bacteriol. 1999;49:645–651. doi: 10.1099/00207713-49-2-645. [DOI] [PubMed] [Google Scholar]
- 49.Rawlings D E, Kusano T. Molecular genetics of Thiobacillus ferrooxidans. Microbiol Rev. 1994;58:39–55. doi: 10.1128/mr.58.1.39-55.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reinartz M, Tschäpe J, Brüser T, Trüper H G, Dahl C. Sulfide oxidation in the phototrophic sulfur bacterium Chromatium vinosum. Arch Microbiol. 1998;170:59–68. doi: 10.1007/s002030050615. [DOI] [PubMed] [Google Scholar]
- 51.Rodrigue A, Chanal A, Beck K, Müller M, Wu L-F. Cotranslocation of a periplasmic enzyme complex by a hitchhiker mechanism through the bacterial Tat pathway. J Biol Chem. 1999;274:13223–13228. doi: 10.1074/jbc.274.19.13223. [DOI] [PubMed] [Google Scholar]
- 51a.Rother, D., H.-J. Henrich, A. Quentmeier, F. Bardischewsky, and C. G. Friedrich. Novel genes of the sox gene cluster, mutagenesis of the flavoprotein SoxF, and evidence for a general sulfur-oxidizing system in Paracoccus pantotrophus GB17. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 52.Schütz M, Klughammer C, Griesbeck C, Quentmeier A, Friedrich C G, Hauska G. Sulfide-quinone reductase activity in membranes of the chemotrophic bacterium Paracoccus denitrificans GB17. Arch Microbiol. 1998;170:353–360. [Google Scholar]
- 53.Schütz M, Maldener I, Griesbeck C, Hauska G. Sulfide-quinone reductase from Rhodobacter capsulatus: requirement for growth, periplasmic location, and extension of gene sequence analysis. J Bacteriol. 1999;181:6516–6523. doi: 10.1128/jb.181.20.6516-6523.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schütz M, Shahak I, Padan E, Hauska D. Sulfide-quinone reductase from Rhodobacter capsulatus: purification, cloning and expression. J Biol Chem. 1997;272:9890–9894. doi: 10.1074/jbc.272.15.9890. [DOI] [PubMed] [Google Scholar]
- 55.Shahak Y, Arieli B, Padan E, Hauska G. Sulfide-quinone reductase (SQR) activity in Chlorobium. FEBS Lett. 1992;299:127–130. doi: 10.1016/0014-5793(92)80230-e. [DOI] [PubMed] [Google Scholar]
- 56.Siefert E, Pfennig N. Chemoautotrophic growth of Rhodopseudomonas species with hydrogen and chemotrophic utilization of methanol and formate. Arch Microbiol. 1979;122:177–182. [Google Scholar]
- 57.Sorokin D Y, Lysenko A M, Mityushina L L, Tourova T P, Jones B E, Rainey F A, Robertson L A, Kuenen J G. Thioalkalimicrobium aerophilum gen. nov., sp. nov., and Thioalkalimicrobium sibericum sp. nov., and Thioalkalivibrio versutus gen. nov., sp. nov., Thioalkalivibrio nitratis sp. nov., and Thioalkalivibrio denitrificans sp. nov., novel obligately alkaliphilic and obligately chemolithoautotrophic sulfur-oxidizing bacteria from soda lakes. Int J Syst Evol Microbiol. 2001;51:565–580. doi: 10.1099/00207713-51-2-565. [DOI] [PubMed] [Google Scholar]
- 58.Stetter K O, Fiala G, Huber G, Huber H, Segerer A. Hyperthermophilic microorganisms. FEMS Microbiol Rev. 1990;75:117–124. [Google Scholar]
- 59.Steudel R, Holdt G, Göbel T, Hazeu W. Chromatographic separation of higher polythionates SnO62− (n=3…22) and their detection in cultures of Thiobacillus ferrooxidans: molecular composition of bacterial sulfur excretions. Angewandte Chemie. 1987;26:151–153. [Google Scholar]
- 60.Strohl W R. Genus I. Beggiatoa. 1989. pp. 2091–2097. [Google Scholar]
- 61.Suylen G M H, Kuenen J G. Chemostat enrichment and isolation of Hyphomicrobium EG, a dimethylsulfide oxidizing methylotroph, and reevaluation of Thiobacillus MS1. Antonie Leeuwenhoek J Microbiol. 1986;52:281–293. doi: 10.1007/BF00428640. [DOI] [PubMed] [Google Scholar]
- 62.Suylen G M H, Sefess G C, Kuenen J G. Chemolithotrophic potential of a Hyphomicrobium species capable of growth on methylated sulfur compounds. Arch Microbiol. 1986;146:192–198. [Google Scholar]
- 63.Takakuwa S. Biochemical aspects of microbial oxidation of inorganic sulfur compounds. In: Oae S, Okuyama T, editors. Organic sulfur chemistry. Biochemical; 1992. pp. 1–43. aspects. CRC Press, Boca Raton, Fla. [Google Scholar]
- 64.Toghrol F, Southerland W M. Purification of Thiobacillus novellus sulfite oxidase. Evidence for the presence of heme and molybdenum. J Biol Chem. 1983;258:6762–6766. [PubMed] [Google Scholar]
- 65.Trüper H-G, Fischer U. Anaerobic oxidation of sulfur compounds as electron donors for bacterial photosynthesis. Phil Trans R Soc Lond. 1982;298:529–542. [Google Scholar]
- 66.Visser J M, de Jong G A H, Robertson L A, Kuenen J G. A novel membrane-bound flavocytochrome c sulfide dehydrogenase from the colorless sulfur bacterium Thiobacillus sp. W5. Arch Microbiol. 1997;167:295–301. doi: 10.1007/s002030050447. [DOI] [PubMed] [Google Scholar]
- 67.Wahlund T M, Woese C R, Castenholz R W, Madigan M T. A thermophilic green sulfur bacterium from New Zealand hot springs, Chlorobium tepidum sp. nov. Arch Microbiol. 1991;156:81–90. [Google Scholar]
- 68.Weiner J H, Bilous P T, Shaw G M, Lubitz S P, Frost L, Thomas G H, Cole J A, Turner R J. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 69.Wodara C, Kostka S, Egert M, Kelly D P, Friedrich C G. Identification and sequence analysis of the soxB gene essential for sulfur oxidation of Paracoccus denitrificans GB17. J Bacteriol. 1994;176:6188–6191. doi: 10.1128/jb.176.20.6188-6191.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wodara C, Bardischewsky F, Friedrich C G. Cloning and characterization of sulfite dehydrogenase, two c-type cytochromes, and a flavoprotein of Paracoccus denitrificans GB17: essential role of sulfite dehydrogenase in lithotrophic sulfur oxidation. J Bacteriol. 1997;179:5014–5023. doi: 10.1128/jb.179.16.5014-5023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yamanaka T, Fukumori Y, Okunuki K. Preparation of subunits of flavocytochromes c derived from Chlorobium limicola f. thiosulfatophilum and Chromatium vinosum. Anal Biochem. 1979;95:209–213. doi: 10.1016/0003-2697(79)90207-0. [DOI] [PubMed] [Google Scholar]