Abstract
Localized disease heterogeneity on imaging extracted via radiomics approaches have recently been associated with disease prognosis and treatment response. Traditionally, radiomics analyses leverage texture operators to derive voxel- or region-wise feature values towards quantifying subtle variations in image appearance within a region-of-interest (ROI). With the goal of mining additional voxel-wise texture patterns from radiomic “expression maps”, we introduce a new RADIomic Spatial TexturAl descripTor (RADISTAT). This was driven by the hypothesis that quantifying spatial organization of texture patterns within an ROI could allow for better capturing interactions between different tissue classes present in a given region; thus enabling more accurate characterization of disease or response phenotypes. RADISTAT involves: (a) robustly identifying sub-compartments of low, intermediate, and high radiomic expression (i.e. heterogeneity) in a feature map and (b) quantifying spatial organization of sub-compartments via graph interactions. RADISTAT was evaluated in two clinically challenging problems: (1) discriminating nodal/distant metastasis from metastasis-free rectal cancer patients on post-chemoradiation T2w MRI, and (2) distinguishing tumor progression from pseudo-progression in glioblastoma multiforme using post-chemoradiation T1w MRI. Across over 800 experiments, RADISTAT yielded a consistent discriminatory signature for tumor progression (GBM) and disease metastasis (RCa); where its sub-compartments were associated with pathologic tissue types (fibrosis or tumor, determined via fusion of MRI and pathology). In a multi-institutional setting for both clinical problems, RADISTAT resulted in higher classifier performance (11% improvement in AUC, on average) compared to radiomic descriptors. Furthermore, combining RADISTAT with radiomic descriptors resulted in significantly improved performance compared to using radiomic descriptors alone.
Keywords: radiomics, graph organization, response characterization, rectal cancer, glioblastoma multiforme, tumor heterogeneity
I. Introduction
Radiomics, or the computerized extraction of subvisual appearance characteristics from radiographic images [1], has demonstrated great promise for capturing subtle differences between disease subtypes [2], as well as predicting patient outcomes [3] or treatment response [4], [5]. Radiomic features work by quantifying the texture or shape of the tumor and its surrounding environment, either by using region-wise [6], [7] or voxel-wise statistics [8], [9]. These features are then subsequently used as an input to machine learning classifiers to distinguish between disease or response phenotypes [5], [8], [10], [11].
One of the most popular radiomic operators are gray level co-occurrence matrix (GLCM) features [12], which capture image heterogeneity within a target region-of-interest (ROI) by quantifying higher-order derivatives in voxel-wise intensities within local neighborhoods. GLCM texture patterns on imaging have been linked to treatment response [5], [13] as well as tumor aggressiveness [14], [15]; and can be visualized as radiomic “expression maps”. However, a lesion ROI may comprise multiple pathologic tissue types, such as in rectal cancer (RCa) patients who develop a mixture of residual disease and benign treatment effects (fibrosis, ulceration) [16] in response to neoadjuvant chemoradiation therapy (CRT). This is illustrated in Figure 1 for two rectal cancer patients with different responses to CRT, whose ex vivo rectal pathology specimens (annotated for presence of residual tumor, fibrosis, and other treatment effects in different colors) show differences in organization and interactions between tissue types in Figures 1(c) and (f) [17]. Interestingly, the voxel-wise GLCM radiomic expression maps in Figures 1(b) and (e) on pre-operative in vivo MRIs for these patients also exhibit unique arrangements of red and blue sub-regions (red being over-expression and blue corresponding to under-expression of image heterogeneity), where these heterogeneity differences are likely driven by differences in the underlying pathologic sub-compartments [18].
Fig. 1.
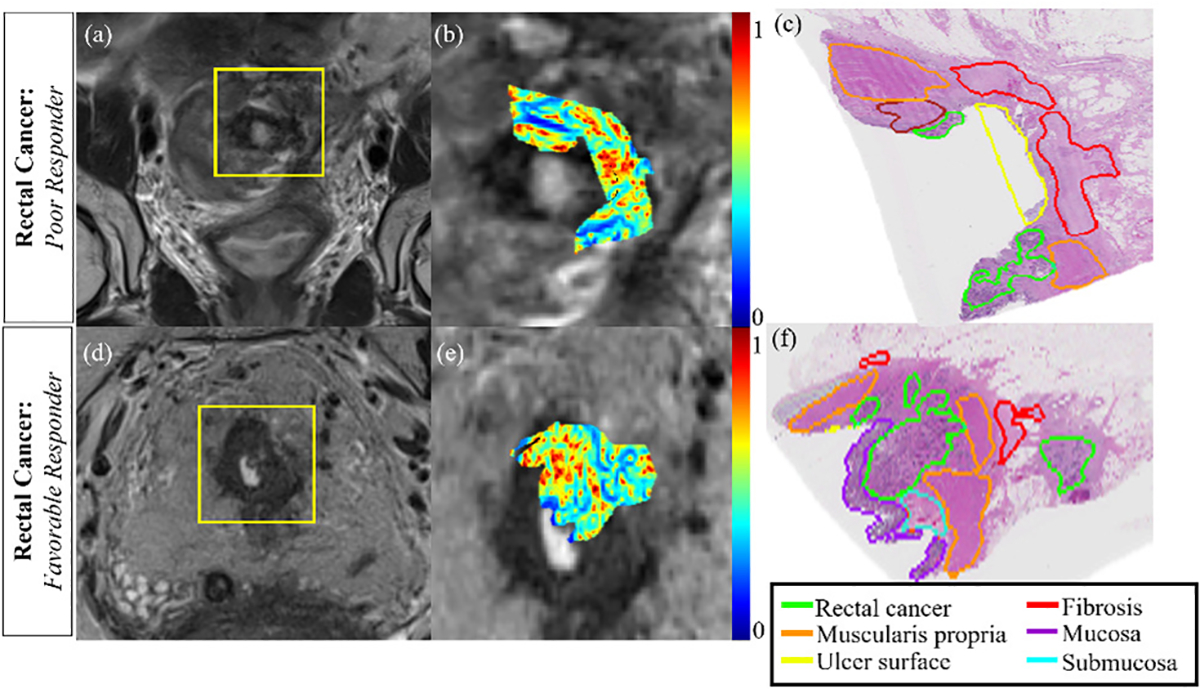
(a)(d) Pre-operative, post-CRT T2w MRIs for 2 rectal cancer patients (yellow box corresponds to rectal ROI). (b)(e) Radiomic expression maps corresponding to ROIs in (b), (e) which are in rough spatial correspondence to (c), (f) ex vivo pathology sections from post-CRT excised rectal cancer specimens from these patients. When comparing poor response (top row) to favorable response (bottom row), differential organization of tissue compartments can be observed within and around the tumor region (different pathologic tissue types delineated in different colors by an expert pathologist in (c), (f)). Note that regions of high (red) and low (blue) heterogeneity within radiomic expression maps in (b), (e) appear to be organized in a manner reflective of underlying pathologic tissue compartments in (c), (f).
However, traditional radiomic analysis would involve using region-wise statistics to simply categorize a lesion as “more heterogeneous” or “less heterogeneous”. This suggests an opportunity to exploit organizational differences within radiomic expression maps in order to capture a surrogate of pathologic tissue organization and interactions within diseased lesions on the radiographic scale.
II. Previous Work and Novel Contributions
There has been some related work with regards to quantitative characterization of imaging-based sub-regions within the tumor environment [19]–[23]. Typically, these sub-regions have been defined based on clustering PET/CT/MR intensities or contrast-enhancement parameters (via dynamic contrast enhanced MRI). Following this, volumetric [19], [21] or texture [20], [22], [23] features of these sub-regions have yielded relatively good performance in distinguishing patients based on survival or response to therapy in different cancers. In other words, these studies have primarily attempted to capture clustering-based composition of the tumor environment on imaging alone.
Gallego-Ortiz and Martel [24] utilized an Artificial Neural Network to quantify graph connectivity of intra-lesion subclusters defined using spatial and temporal enhancement in breast MRI. Incorporating these graph descriptors was found to improve the performance of the diagnostic classifier in identifying non-mass like breast lesions by 21%, compared to the original feature space. Similarly, Wu et al [25] quantified spatial interactions of intratumoral sub-regions (defined by clustering breast MR perfusion parameters) and used network analysis to predict response to chemotherapy while also interrogating related perfusion characteristics of breast tumor subtypes. While these studies have shown the potential for capturing spatial interactions between perfusion patterns on functional imaging, we are not aware of any studies that have specifically examined spatial organization differences in texture patterns between disease or response phenotypes.
In this work, we present a new RADIomic Spatial TexturAl descripTor (RADISTAT) to capture the relative spatial organization between compartments of radiomic over- or under-expression within a lesion ROI. This will be done by discretizing the radiomic heatmap into more stratified expression levels (or “sub-compartments”) and using graph-based constructs to analyze the relationships between these radiomic sub-compartments. Figure 2 depicts an overview of the steps involved in computing RADISTAT. The motivation behind RADISTAT originates from digital pathology, where graph-based features are commonly used for capturing the spatial arrangement of primitives (e.g. glands or nuclei) or tissue sub-compartments (e.g. stroma or epithelium); which have been shown to differ based on disease phenotypes and aggressiveness [26]. RADISTAT seeks to similarly characterize tissue heterogeneity on radiographic imaging by measuring the density and arrangement of radiomic expression sub-compartments, which may in turn be related to the underlying pathologic organization of tissue regions within the lesion environment. A preliminary implementation of 2D RADISTAT was previously presented in [27], and this work includes a more detailed examination of its performance in additional clinical settings, algorithmic parameter sensitivity, as well as evaluating the pathologic basis for RADISTAT. The specific novel contributions of this work are as follows:
RADISTAT represents one of the first efforts to capture the spatial organization of radiomic patterns within the lesion environment on structural imaging, using a unique superpixel clustering scheme. The latter approach incorporates both spatial and size constraints, rather than the conventional K equally sized clusters [28]. Additionally, clustering is performed on radiomic expression values, rather than the original imaging intensities.
RADISTAT uniquely re-quantizes the resulting superpixel organization by binning the resulting clusters into partitions of radiomic expression, enabling comparison of radiomic organization across datasets.
RADISTAT ultimately quantifies this organization of radiomic expression by measuring the relationships between distinct partitions via explicit graph construction and adjacency measures, and can be mined for an imaging-based surrogate of the spatial arrangement of underlying pathologic compartments.
RADISTAT will be uniquely evaluated for capturing spatial organization of radiomic expression in the context of two challenging clinical problems involving characterizing treatment response on post-treatment MR imaging: (a) discriminating metastastic from non-metastatic rectal cancers (RCa) after chemoradiation, and (b) distinguishing tumor progression from pseudo-progression (PsP) in chemoradiated glioblastoma multiforme (GBM) tumors. Both of these problems pose significant challenges for expert assessment due to significant tissue heterogeneity and overlapping appearance of tissue pathologies present after treatment (fibrosis, ulceration, edema confound residual disease in RCa [29], radiation necrosis in PsP appears similar to recurrent disease [30]). Capturing organizational differences in the post-treatment lesion environment in both these diseases offers a unique solution to the clinically relevant problem of accurately differentiating these confounding pathologies via radiographic imaging.
Fig. 2.
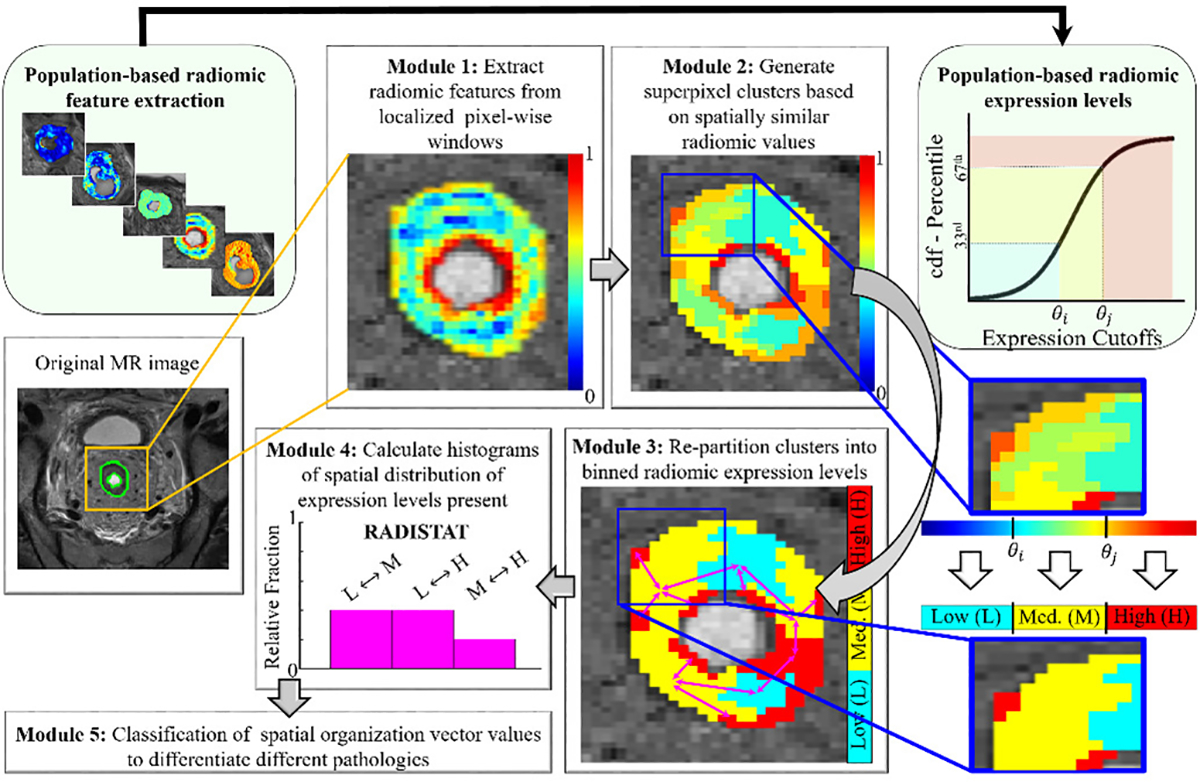
Overall workflow for extracting RADISTAT descriptor. Module 1 involves extracting radiomic features on a per-pixel basis within the region of interest (outlined in green), visualized here as a heatmap where red and blue correspond to high and low radiomic feature expression, respectively. In Module 2, superpixel-based clusters are computed from the radiomic expression map. The superpixel clusters are re-partitioned into a fixed number of binned expression levels in Module 3. The cutoff values for these binned radiomic expression levels is determined from population-based radiomic distributions (shown via connected light-green module blocks at the top). For ease of understanding, a representative re-partitioning of the radiomic map into 3 levels is shown alongside, e.g. low (L), medium (M), high (H)). Note that the number of levels in the RADISTAT partition map may be determined based on the application at hand. The final RADISTAT descriptor quantifies spatial adjacencies between pairwise combinations of expression levels (shown in Module 4). The RADISTAT descriptor can then be passed as a vector into a machine learning classifier for predicting different underlying pathologies (Module 5).
The rest of the paper is organized as follows. In Section III, algorithmic details for extracting the RADISTAT descriptor (both in 2D and 3D) are provided. The experimental design to demonstrate the utility of RADISTAT in the context of two problems, rectal tumors and brain lesions, is described in Section IV. Subsequently we present the results and discussion in Section V, followed by concluding remarks (Section VI).
III. Methodology
A. Notation
We denote a radiomic feature expression map , where C is a spatial grid of pixels c, in or . Every pixel, c ∈ C, is considered to be associated with a radiomic feature value f(c).
B. Superpixel Clustering of Radiomic Feature Expression Maps:
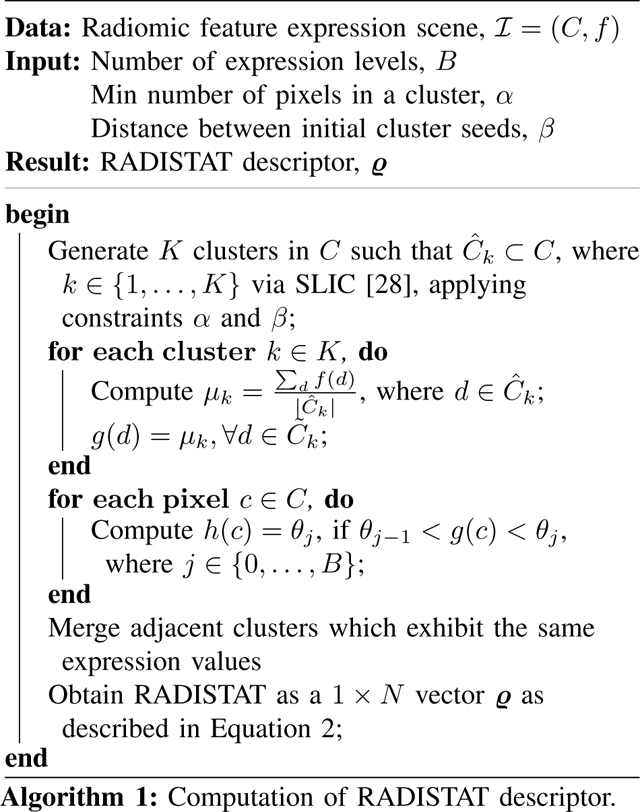
Superpixel clustering of is performed using a modified version of the simple linear iterative clustering (SLIC) algorithm [28], to generate K clusters, , k ∈ {1, …, K}. The goal is to robustly identify (in an unsupervised fashion) spatially contiguous regions in the radiomic expression map based on similar-valued pixels. In our SLIC implementation, K is implicitly defined based on 2 user-specified parameters: (1) the minimum number of pixels in a cluster (α), and (2) the distance between initial cluster seeds (β). Thus for each combination of α and β, different clusterings of will be obtained. Based on superpixel clustering, is quantized to obtain a cluster map , where for every , g(c) is the average radiomic feature value within cluster .
C. Re-partitioning of Superpixel Clusters into Expression Levels:
The desired number of expression levels across a cohort is set via a user-defined parameter B. Note that the number of superpixel clusters (K) resulting from the previous step can vary as a function of how many unique expression values there are in each (as well as α and β). The choice of B rebins the range of radiomic features to be consistent across a cohort and merges superpixel clusters accordingly.
Using the input parameter B, the range of all (across a cohort) is split into B equally spaced bins, yielding B + 1 thresholds θj, j ∈ {0, …, B}. These thresholds are used to re-quantize into an expression map, , where ∀c ∈ C, h(c) = θj, if θj−1 < g(c) < θj. As each can only have up to B unique values, any adjacent clusters which exhibit the same expression value are merged to yield M distinct partitions. A partition is defined as , where m ∈ {1, …, M}, and . For ease of notation, we also define the expression value of a partition as , if , h(c) = θj. Note that for ease of visualization, this step has been depicted in Figure 2 for B = 3. Each patient within a cohort may therefore have a different M (number of partitions) associated with it, but each distinct partition within each will only be assigned one of B expression values.
D. Constructing RADISTAT descriptor:
In this work, RADISTAT is based on quantifying the adjacency of each pairwise combination of B expression levels in , in order to capture the spatial interactions between them. Considering the case of low (L), medium (M), and high (H) expression (i.e. B = 3), there are 3 pairwise combinations: L-M, L-H, M-H. The adjacency of L-M is obtained by counting the number of times that has partitions with low and medium expression adjacent to each other (similarly for L-H and M-H). For this, an adjacency graph G = (V, E) is defined, where V = {vm}, m ∈ {1, …, M}, comprises the centroids of each of M partitions obtained in Step 2; and E = {emn}, m, n ∈ {1, …, M}, is a set of edges. An edge in E is defined when,
| (1) |
For every pair of expression levels θi and θj, i, j ∈ {1, …, B}, the spatial component is calculated as,
| (2) |
The resulting feature, RADISTAT, is a 1 × N vector ϱ = [ϱ1, …, ϱN], where is the total number of expression level pairs in . Complete methodological details for computing RADISTAT are summarized in Algorithm 1.
IV. Experimental Design
RADISTAT was evaluated within 2 clinical problems: (a) distinguishing which patients with RCa had metastatic disease on post-chemoradiation T2-weighted (T2w) MRI following neoadjuvant therapy, and (b) identifying tumor progression in glioblastoma multiforme (GBM) patients via Gadolinium-enhanced T1-weighted (T1w) MRI after chemoradiation.
A. Data description
1). Rectal cancer cohort:
A total of 73 RCa patient datasets were de-identified and retrospectively included in this institutional review board approved study (University Hospitals IRB #07-16-40, approval date: 12/03/2020) from two different institutions. All patients had been treated for a clinically staged cT2-T4 or cN+ locally advanced rectal carcinoma between August 2007 and January 2019 with standard-of-care neoadjuvant chemoradiation. For all included patients, a T2w MRI was available which had been acquired after treatment (and prior to surgery) together with pathology reports of surgically excised rectal specimens. Annotations of the rectal wall on post-chemoradiation T2w MRIs on sections suspected of containing residual tumor were obtained from expert radiologists for the two sites independently, which was considered the ROI for radiomic analysis. Patients were assigned to one of two treatment outcome groups based on pathologically confirmed metastatic disease (ypN+/ypM+ vs ypN0M0) after chemoradiation [31]; summarized in Table I.
TABLE I.
Summary of patient characteristics, imaging parameters, and outcome groups in discovery and validation cohorts for each clinical use case.
| Rectal Cancer (RCa) | Glioblastoma Multiforme (GBM) | |||
|---|---|---|---|---|
| Discovery (n=44) | Validation (n=29) | Discovery (n=56) | Validation (n=44) | |
|
| ||||
| Patient Characteristics | ||||
| Sex, men:women | 25:19 | 19:10 | 39:20 | 30:16 |
| Age, mean (range), years | 62.8 (30–83) | 58.2 (37–78) | 60.6 (26–74) | 55.6 (25–76) |
| Imaging Parameters | ||||
| MRI Protocol | T2w TSE | T2w TSE | Gd-T1w | Gd-T1w |
| In-plane resolution (mm) | 0.0256–0.97 | 0.313–0.898 | 1.000 | 1.000 |
| Slice thickness (mm) | 3.0–5.0 | 3.0–6.0 | 1.000 | 1.000 |
| Repetition time (msec) | 3253–12690 | 3400–13333 | 263–764 | 263–850 |
| Echo time (msec) | 67–110 | 84–166 | 5–20 | 5–20 |
| 3T | 44 | 1 | 0 | 0 |
| 1.5T | 0 | 28 | 56 | 44 |
| Pathologic groupings | ||||
| Mets: 13 | Mets: 13 | Prog: 36 | Prog: 32 | |
| Non-mets: 31 | Non-mets: 16 | PsP: 20 | PsP: 12 | |
2). Brain tumor cohort:
A total of 100 GBM patients were de-identified and retrospectively included in this institutional review board exempted study (University Hospitals IRB #STUDY20180757, exempt determination: 1/8/2019) from two different institutions. Patients had been identified by a review of all brain tumor patients who received chemoradiation treatment using Stupp protocol [32] at the respective institutions and who had an enhancing lesion within 3 months of treatment. All patients had a routine Gadolinium-enhanced T1w MRI after chemoradiation available together with histologic or imaging reports. To correct for known intensity differences, brain T1w MRI scans had undergone bias field correction, skull-stripping, and intensity standardization. Annotations of the enhancing lesion were obtained for every MRI slice with more than 5-mm of rim enhancement by an expert radiologist. Treatment outcomes were assigned based on labeling the enhancing lesion on follow-up scans as either pseudo-progression (PsP, a radiation-induced treatment effect) or tumor progression based on histologic analysis or follow-up imaging, as summarized in Table I.
B. Pre-processing and Radiomic feature map extraction
All imaging data and their respective annotations were resampled to 1 × 1 × 1 mm3 isotropic resolution in x-,y-, and z-directions. A total of 13 GLCM features [12] were extracted on a voxel-wise basis from each ROI, while considering intensity co-occurrences in all spatial directions within local 3 × 3 × 3 sliding-window neighborhoods. GLCM features were selected based on their wide usage in radiomic analyses, especially in the context of GBM [8] and RCa [4]. This resulted in 13 voxel-wise GLCM radiomic feature maps associated with each ROI, denoted . A single 2D section in the middle of the annotated ROI volume was identified and utilized for 2D analysis, while the entire volumetric ROI was used for 3D analysis.
C. Extraction of RADISTAT and parameter sensitivity analysis
Algorithm 1 was implemented in MATLAB (Mathworks, MA) to compute RADISTAT (ϱ); for each . The number of expression levels was set at B = 3 in all experiments, to correspond intuitively to low, medium, and high expression.
The RADISTAT partition map (Figure 2, Module 3) is dependent on superpixel parameters: (1) the minimum number of pixels in a cluster (α), and (2) the distance between initial cluster seeds (β). A range of parameters was considered for α ∈ {5, 10, 15, 20}, β ∈ {3, 5, 7, 9} in 2D, and α ∈ {10, 25, 50, 100}, β ∈ {3, 5, 7, 9} in 3D. This range of clustering hyperparameters was selected based on the size of ROIs being analyzed. These 16 possible α-β parameter combinations were evaluated for each of resulting in a total of 208 possible parameter variations of RADISTAT being evaluated in each of the 2D and 3D settings.
D. Extraction of radiomic descriptors for comparative assessment
As an alternative strategy, commonly utilized radiomic descriptors were extracted, including voxel-wise statistics [5], [33] and region-wise (“whole tumor”) features [6]. For each of the 13 voxel-wise GLCM features, 4 statistical descriptors (mean, variance, kurtosis, skewness) were computed from the radiomic distribution across the ROI (totaling 52 voxel-wise descriptors). Additionally, 22 region-wise GLCM descriptors were computed from pyRadiomics [6] while ensuring parameters aligned with those used in Section IV-B. Based on the number of datasets avaiable and to reduce the risk of overfitting, four top-ranked features were selected from 52 voxel-wise (denoted ς) as well as from 22 region-wise (denoted ξ) descriptors, based on a bootstrapped wilcoxon-based feature selection scheme [5].
E. Experimental evaluation
Both disease cohorts were segregated into discovery and validation sets on an institution-wise basis, as summarized in Table I. Classification performance for each of ϱ (RADISTAT for each , in 2D and 3D, separately), ς (best 4 region-wise), and ξ (best 4 voxel-wise) was evaluated via a binary linear discriminant analysis (LDA) classifier that was trained to distinguish between the 2 classes in each task (metastatic vs non-metastatic RCa, progression vs PsP in GBM). Additionally, the combination of RADISTAT with radiomic descriptors ([ς,ϱ] and [ξ,ϱ]) was also evaluated. An LDA classifier was specifically chosen to enable more fair comparisons between singleton and non-singleton descriptors by projecting each into a lower dimensional space [34]. In all experiments, the classifier was trained and optimized on the discovery cohort (using 50 runs of 3-fold cross-validation) and later evaluated in hold-out fashion on the validation cohort; with the area-under-the-receiver-operator-characteristic curve (AUC) used as a measure of performance.
To identify the optimal GLCM descriptor to use in conjunction with RADISTAT, the average cross-validated AUC for ϱ across all α − β parameters was computed for each . This was done separately for each classification task, in both 2D and 3D. The parameter sensitivity of this top-performing GLCM descriptor was further visualized via a surface plot of AUC against changes in α and β. Next, in the context of each clinical problem, the AUC achieved by the top-ranked ϱ was compared against corresponding AUCs achieved via (a) ς, (b) ξ, (c) [ς,ϱ], and (d) [ξ,ϱ]. A nonparametric Wilcoxon rank sum test was employed to assess statistical differences in AUC values in pairwise fashion, with the p-value adjusted via the Bonferroni correction.
To evaluate the pathologic basis of RADISTAT, a subset of 6 RCa patients were identified for whom digitized ex vivo pathology sections were available together with corresponding in vivo pre-operative, post-chemoradiation MRIs. Using a published co-registration technique [35], pathology sections were spatially aligned onto corresponding 2D T2w MRI sections which allowed for expert annotations of different tissue types (fibrosis, residual disease, ulceration) to be mapped onto MRI. This allowed for identifying which pathologic tissue types were present within different expression levels (low, medium, high), and thus the potential histopathologic tissue organization being reflected via RADISTAT.
V. Results and Discussion
A. Experiment 1: Evaluating RADISTAT for identifying metastatic disease in rectal cancers
Figure 3 illustrates representative 2D GLCM inertia feature heatmaps together with their corresponding RADISTAT partition maps, for rectal cancer patients with no metastatic disease (right half) and as well as patients who suffered metastatic disease (left half); after chemoradiation. Supplementary Figure S1 similarly depicts representative results in 3D for GLCM difference variance. RADISTAT partition maps and descriptors in both 2D (Figures 3(c)–(f)) and 3D (Supplementary Figures S1(c)–(f)) are seen to exhibit similar trends that are distinctly different between outcome groups. In patients with poor outcomes after chemoradiation, the rectal wall comprises 2 major expression levels (high and intermediate), while all 3 major expression levels appear in patients with favorable outcomes. Thus, the interactions between low, medium, and high expression in the RADISTAT map appear correspondingly skewed when comparing ϱ via histogram visualizations. Notably, these distinctive RADISTAT signatures were consistently associated with poor and favorable RCa outcomes across all α-β parameter combinations; illustrated by the outcome-wise median histograms for ϱ in Supplementary Figures S2 and S3. RADISTAT descriptors are also seen to maintain their distinctive signatures across both discovery (top row) and hold-out validation (bottom row).
Fig. 3.
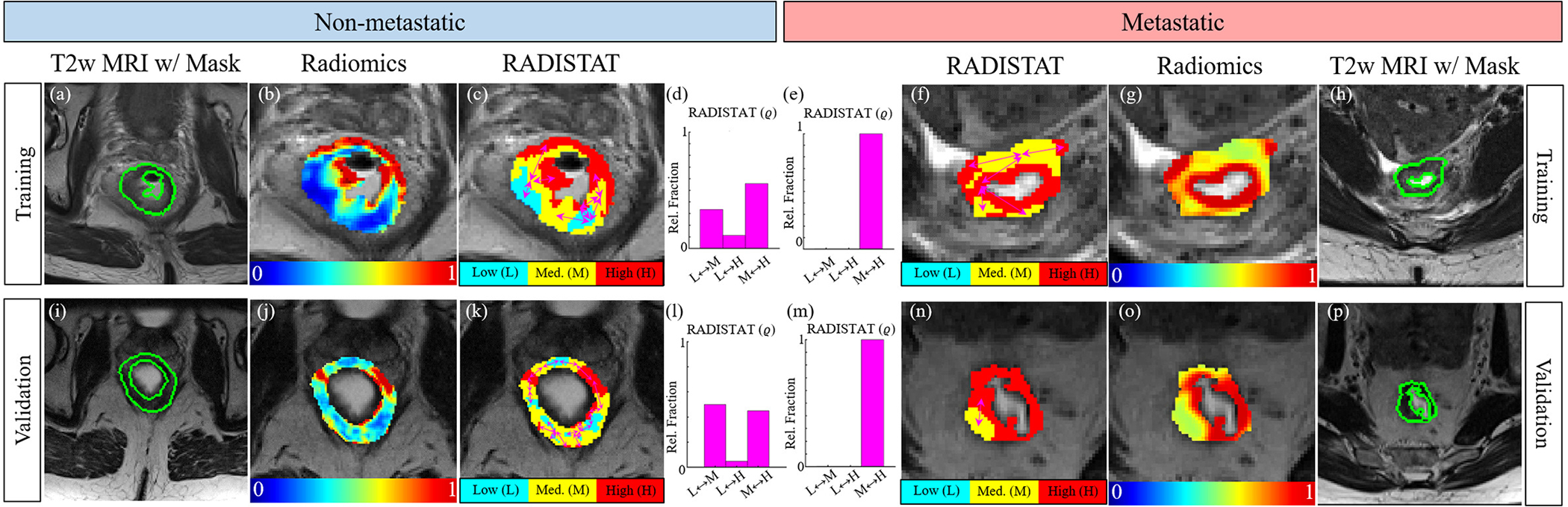
(a)(h),(i),(p) Expert-annotated rectal wall ROI delineated in green in representative patients suffering from metastasis (left half) as well as free of nodal or distant metastasis (right half) after chemoradiation. Note that the top row corresponds to discovery cohort while the bottom row corresponds to hold-out validation cohort (different institution). (b)(g),(j),(o) Heatmaps for GLCM inertia quantifying differential radiomic expression within the rectal wall on MRI; red corresponds to over-expression and blue to under-expression. (c)(f)(k),(n) RADISTAT partition maps accurately reflect distinct regions of radiomic over- and under-expression, which result in marked differences in ϱ (shown as histograms) between metastatic and non-metastatic RCa patients.
Based on the co-registered radiology-pathology cohort, Figure 4 shows that partitions with high radiomic expression (red regions) primarily overlap with pathologically mapped residual tumor annotations while low radiomic expression partitions (cyan regions) primarily appear within pathologically confirmed fibrotic tissue. This difference in radiomic expression levels was found to be significantly higher in residual tumor compared to fibrosis (p ≤ 0.001, Figure 4(f)). Thus, the 3-compartment organization associated with metastatic-free outcomes (Figure 3) may be the result of distinctive fibrotic stroma regions developing within the chemoradiated rectal wall, which appear as sub-regions of lower radiomic expression on rectal MRIs. Such morphologic alterations occur within the rectum as a result of chemoradiation-induced fibro-inflammatory changes that replace cancerous neoplastic glands, and have been associated with improved outcomes and longer disease-free survival [36]. Conversely, post-chemoradiated rectal tumors where fibrotic tissue regions may not be as distinctively present tend to be more biologically aggressive with a higher likelihood of metastasis [37]); reflected via a 2-compartment organization in RADISTAT. Our observations also resonate with recent radiomics studies of post-chemoradiation rectal MRIs [4], [13], which have observed more variation in GLCM inertia and other GLCM features in rectal cancer patients with favorable outcomes after therapy.
Fig. 4.
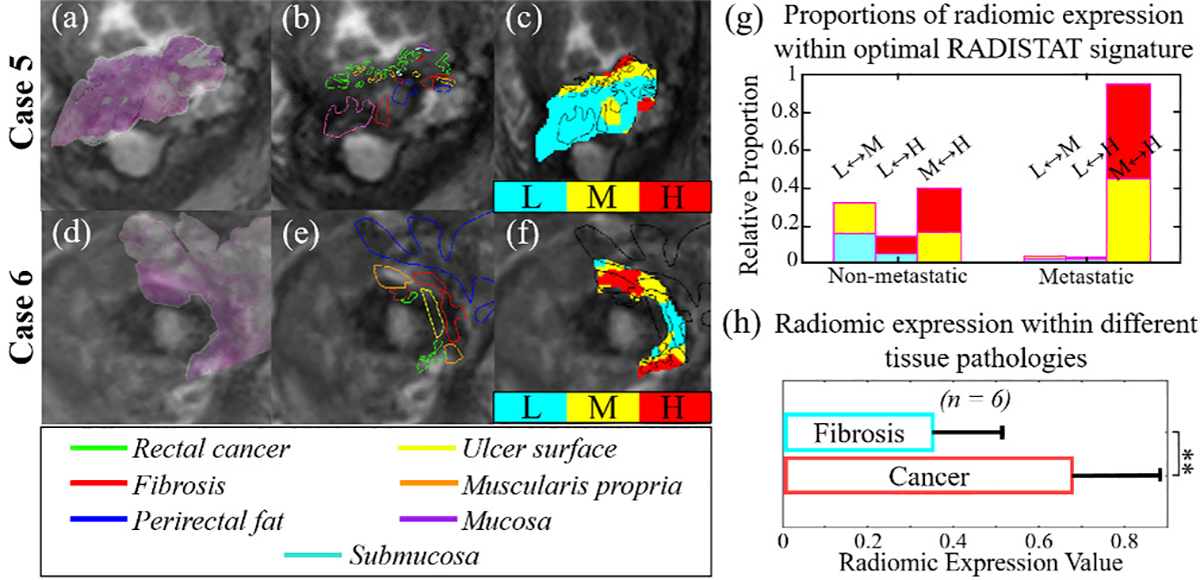
(a),(d) Fused image overlays of pre-operative in vivo T2w MRIs with corresponding ex vivo pathologic specimens for two representative patients, via a published co-registration approach [35]. (b),(e) Expert annotations of different tissue types (in different colors) spatially mapped onto the MRI sections. (c),(f) RADISTAT expression maps overlaid with pathologic annotations demonstrate overlap of different expression levels with different tissue types. (g) Histograms of optimal RADISTAT signature with colors denoting expression level pairs within each bar; note elevated interactions with over-expression (red) in metastatic disease compared to non-metastatic disease (more interactions with blue or under-expression). (h) Barplot shows significantly elevated radiomic expression within pathologically mapped residual tumor (red regions) compared to fibrosis (cyan regions), suggesting potential pathologic basis for interactions quantified via RADISTAT. **: p ≤ 0.001
Figure 5(a) depicts bar plots of RADISTAT classifier performance for each of the 13 GLCM features (classifier AUC averaged across all α-β combinations), with bars shaded differently for 2D and 3D analysis. Intuitively, RADISTAT performance is seen to vary depending on the GLCM feature it is being computed on. Features indicated via asterisks were selected for detailed analysis in Figure 3 and Table II. In Figures 5(b) and (c), RADISTAT is seen to perform relatively consistently in 62.5% (10/16) of the 16 hyperparameter combinations in 2D (within 1.5 times the standard deviation of the highest AUC, visualized as gray stripes on the surface plots), and in 43.75% (7/16) of hyperparameter combinations in 3D. RADISTAT performance appears to decline markedly for extreme parameter combinations (β ≪ α). This is likely because the initial seed spacing or the ROI being evaluated is too small to reach the minumum bumber of voxels to be considered a superpixel cluster; this could be resolved by limiting the parameter space to larger β and smaller α values.
Fig. 5.
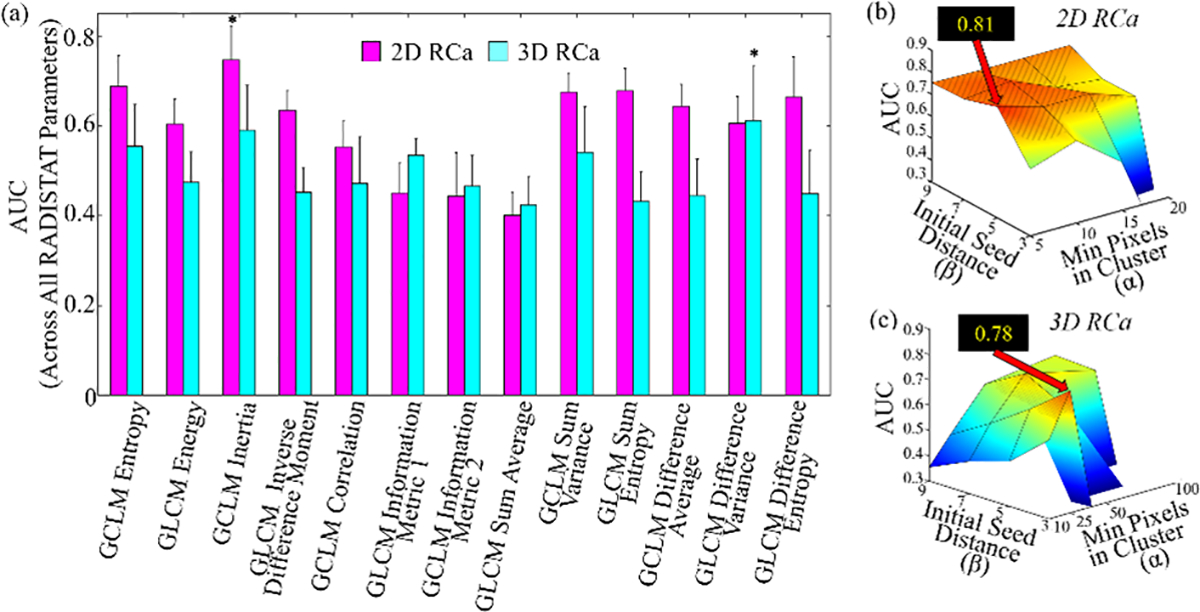
Bar plot summarizing mean AUCs achieved by RADISTAT in conjunction with each GLCM descriptor (, X-axis) averaged over all α-β combinations for the RCa classification task; for both 2D and 3D respectively. Error bars correspond to standard deviation in AUC over all α-β combinations. Asterisks denote the top-performing RADISTAT feature for each classification task. (b), (c) surface plots of AUC values of RADISTAT in 2D (top row) and 3D (bottom), for each classification task. Black call-out box in each subfigure corresponds to highest cross-validated AUC achieved in the discovery set, corresponding to different GLCM descriptors. Patterned gray lines indicate most stable performance of RADISTAT (within 1.5 times the standard deviation of the highest AUC).
TABLE II.
AUC values comparing performance of RADISTAT (ϱ) against region-wise and voxel-wise radiomic descriptors, as well as combinations thereof, via an LDA classifier. Discovery AUCs (across 50 runs of 3-fold cross validation) as well as hold-out validation AUCs are shown for each classification task, with the top-performing strategy bolded.
| Classification Task | Top 4 Voxel-wise GLCM ξ | Top 4 Region-wise GLCM ς | RADISTAT ϱ | Top 4 Voxel-wise GLCM + RADISTAT [ξ, ϱ] | Top 4 Region-wise GLCM + RADISTAT [ς, ϱ] | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| RCa: Metastatic vs Non-metastatic | 2D | Discovery (μ ± σ) | 0.52 ± 0.08 | 0.51 ± 0.08 | 0.81 ± 0.04*, ** | 0.77 ± 0.04* | 0.74 ± 0.05** |
| Validation | 0.49 | 0.51 | 0.72 | 0.70 | 0.57 | ||
|
|
|||||||
| 3D | Discovery (μ ± σ) | 0.62 ± 0.08 | 0.50 ± 0.08 | 0.78 ± 0.04*, ** | 0.74 ± 0.04* | 0.69 ± 0.07** | |
| Validation | 0.67 | 0.51 | 0.72 | 0.75 | 0.57 | ||
|
| |||||||
| GBM: Progression vs PsP | 2D | Training (μ ± σ) | 0.50 ± 0.06 | 0.52 ± 0.05 | 0.68 ± 0.03*, ** | 0.65 ± 0.04* | 0.67 ± 0.04** |
| Validation | 0.52 | 0.63 | 0.68 | 0.60 | 0.72 | ||
|
|
|||||||
| 3D | Training (μ ± σ) | 0.59 ± 0.06 | 0.49 ± 0.07 | 0.62 ± 0.06*, ** | 0.67 ± 0.04 * | 0.64 ± 0.06** | |
| Validation | 0.61 | 0.59 | 0.63 | 0.59 | 0.60 | ||
μ: mean AUC; σ: standard deviation of AUC (across all cross-validation runs)
: adjusted p ≤ 0.05 when comparing AUCs between a descriptor and ξ
: adjusted p ≤ 0.05 when comparing AUCs between a descriptor and ς
All p-values corrected for multiple comparisons using Bonferonni’s correction
When compared to radiomic descriptors, RADISTAT resulted in a markedly higher classification performance on the discovery set (ϱ AUCs = 0.78–0.81 vs ς or ξ AUCs = 0.50–0.72; Table II) for discriminating RCa patients with and without metastatic disease after chemoradiation. In hold-out validation, RADISTAT maintained this performance (AUCs = 0.72), while significantly outperforming radiomic descriptors (AUCs = 0.48–0.56). Interestingly, combining radiomic features with RADISTAT (i.e. [ς,ϱ] or [ξ,ϱ]) resulted in significantly higher AUCs compared to ς or ξ alone, and only marginally worse than RADISTAT alone. RADISTAT performance showed no difference between differently sized RCa lesions in 2D (AUCsmall = 0.82 ± 0.04 vs AUClarge = 0.83 ± 0.04, p > 0.05). In 3D, smaller RCa lesions were associated with higher RADISTAT performance (AUCsmall = 0.85 ± 0.03 vs AUClarge = 0.72 ± 0.10, p ≤ 0.05), although these trends were reversed in validation. One other work to-date has also demonstrated that radiomic feature heterogeneity in the rectal lesion is discriminant of lymph node metastasis alone [38] using region-wise multi-parametric radiomic features within the suspected tumor boundary and achieving AUCs of 0.79 in discovery and 0.78 in validation within a single institution. By contrast, RADISTAT demonstrates the presence of a distinctive spatial organization signature for metastatic disease within the entire rectal wall, while also achieving comparable AUCs in multi-institutional validation.
B. Experiment 2: Evaluating RADISTAT for distinguishing pseudo-progression from tumor progression in GBMs
Figure 7 depicts representative 2D GLCM energy feature maps together with their corresponding RADISTAT partition maps, for GBM patients with lesions exhibiting tumor progression (left half) and those exhibiting PsP (right half). RADISTAT partition maps and descriptors in Figures 7(c)–(f) and (k)–(n) capture distinctive spatial organizations of low, medium, and high expression clusters from the corresponding radiomic heatmaps. Lesions with PsP were seen to exhibit a markedly elevated interactions between medium and high (M-H) expression clusters, compared to tumor progression (elevated interactions of low and medium (L-M) expression clusters). These trends in ϱ were consistently associated with PsP and tumor progression across all α-β combinations as shown in Supplementary Figures S4 (for 2D) and S6 (for 3D), as well as between discovery and validation cohorts (top and bottom rows in Figure 7).
Fig. 7.
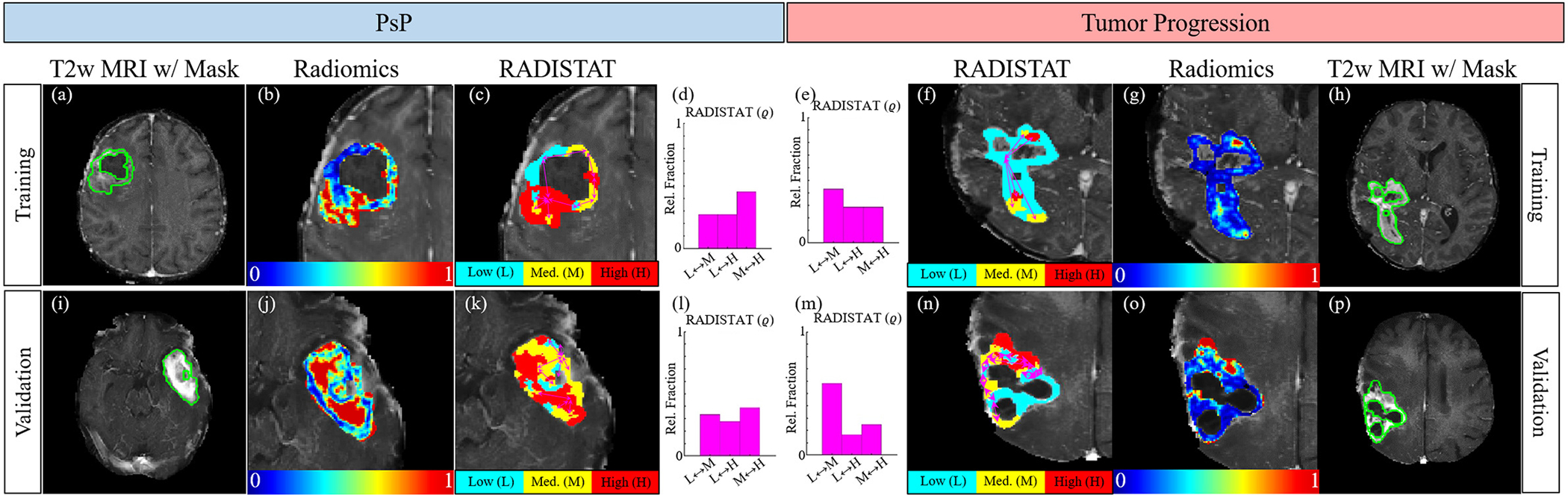
(a),(h) Radiologist annotations of tumor ROI in green for GBM patients from the training cohort with tumor progression (left half) and PsP (right half), after chemoradiation. (b),(g) 2D radiomic heatmaps for GLCM energy showing distinctive radiomic expression between the two groupings. (c),(f) Resulting RADISTAT partition maps reveal markedly different (d),(e) histograms for ϱ; reflecting underlying organizattional differences in regions with low (cyan), intermediate (yellow), and high (red) expression. These differential phenotypes are also maintained in (k)-(n) hold-out validation (different institution).
While all GBM tumors showed a 3-compartment organization via RADISTAT, cases with pseudo-progression were found to be associated with interactions between higher expression values (high and medium) within the lesion while tumor progression cases primarily exhibited interactions between lower radiomic expression values. As RADISTAT is designed to capture disease-specific heterogeneity on MRI, the elevated RADISTAT M-H component associated with pseudo-progression may arise from presence of radiation-induced necrosis; with the opposite trend observed in tumor progression cases. These organizational differences in texture patterns [39] may be driven by hemorrhagic changes associated with tumor recurrence compared to diffuse periventricular white matter changes associated with radiation necrosis [40].
Figure 6 depicts bar plots of RADISTAT classifier performance for each of the 13 GLCM features (classifier AUC averaged across all α-β combinations), with bars shaded differently for 2D and 3D analysis. Intuitively, RADISTAT performance is seen to vary depending on the GLCM feature it is being computed on. while features indicated via asterisks were selected for detailed analysis in Figure 7 and Table II. Parameter sensitivity of the top-performing RADISTAT descriptors (bars denoted by asterisks) can be further analyzed via surface plots in Figures 6(b) and (c). 2D GBM classification yielded very consistent performance via RADISTAT across all hyperparameter combinations (81.25%, or 13/16, Figure 6(b)). However, RADISTAT performance is relatively inconsistent across a majority of hyperparameter combinations for the 3D GBM experiments, likely due to clustering non-convergence (only 1 short green-yellow ridge in Figure 6(c)).
Fig. 6.
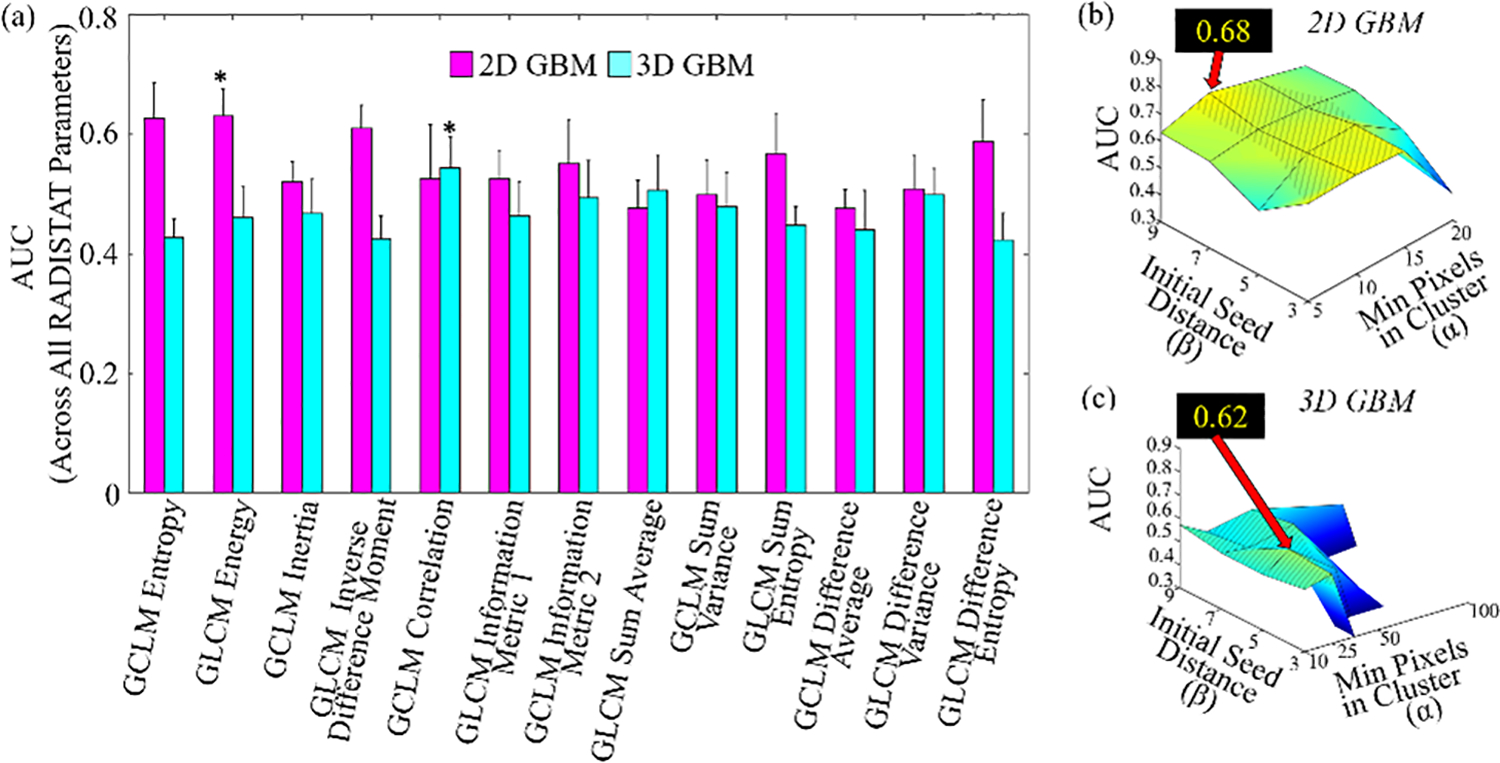
Bar plot summarizing mean AUCs achieved by RADISTAT in conjunction with each GLCM descriptor (, X-axis) averaged over all α-β combinations for the GBM classification task; for both 2D and 3D respectively. Error bars correspond to standard deviation in AUC over all α-β combinations. Asterisks denote the top-performing RADISTAT feature for each classification task. (b), (c) surface plots of AUC values of RADISTAT in 2D (top row) and 3D (bottom), for each classification task. Black call-out box in each subfigure corresponds to highest cross-validated AUC achieved in the discovery set, corresponding to different GLCM descriptors. Patterned gray lines indicate most stable performance of RADISTAT (within 1.5 times the standard deviation of the highest AUC).
In comparative analysis (summarized in Table II), RADISTAT resulted in marginally higher classification performance in both 2D and 3D (AUCs = 0.62–0.68) for distinguishing PsP from tumor progression than either of ς or ξ (AUCs=0.49–0.63); a trend that was maintained in hold-out validation (AUCs = 0.63–0.68) When considering [ς,ϱ] or [ξ,ϱ], both discovery and validation performance were significantly improved compared ς or ξ alone; as well as achieving the highest overall validation performance when combined with RADISTAT. RADISTAT performance showed no difference between differently sized GBM lesions in 2D (AUCsmall = 0.62 ± 0.05 vs AUClarge = 0.63 ± 0.04, p > 0.05). In 3D, smaller GBM lesions were associated with lower RADISTAT performance (AUCsmall = 0.37 ± 0.09 vs AUClarge = 0.52 ± 0.08, p ≤ 0.05), although these trends were reversed in validation. While the cohort used in our study is similar to that of Ismail et al [41], the lower classifier performance indicates the mixed ability of texture-based features in identifying pseudoprogression in GBM [42]. Our results are nevertheless comparable to several previous studies which have evaluated the ability of texture alone [39], [43], [44] in distinguishing pseudoprogression in GBM; albeit while utilizing a single MR sequence (GD T1w MRI) to define a spatial organization signature which was validated in a comparably sized multi-institutional cohort.
VI. Concluding Remarks
We have presented a new RADIomic Spatial TexturAl descripTor (RADISTAT) for quantifying the spatial arrangement of radiomic feature expression to distinguish disease and response phenotypes on imaging. We evaluated the discriminative ability of RADISTAT in the context of 2 challenging clinical problems in a multi-institutional setting: (a) discriminating metastastic from non-metastatic rectal cancers via post-chemoradiation T2w MRI, and (b) distinguishing tumor progression from pseudo-progression in chemoradiated glioblastoma multiforme on Gadolinium-enhanced T1w MRI. RADISTAT revealed distinctive spatial organization signatures to be associated with each disease condition, suggesting it may be able to accurately capture and exploit a surrogate of underlying interactions between tissue sub-compartments associated with these lesions.
RADISTAT was evaluated in conjunction with 13 different GLCM features popularly used in radiomic analysis [5], [8], [10], [11], in both 2D and 3D. The performance of RADISTAT was found to vary between GLCM features, suggesting that the differences in texture patterns captured by each feature results in differences in radiomic sub-compartments as well as in terms of organization and discriminability. However, mathematically related GLCM features yielded the highest AUC values for RADISTAT in both clinical problems (2D inertia for RCa, 2D energy for GBM, 3D difference variance for RCa, 3D correlation for GBM), indicating a common underlying organization phenotype in image texture patterns may exist for a specific disease. This is further confirmed by the similarity in the spatial organization phenotypes between 2D and 3D in both problems, suggesting similarities in the underlying tissue morphology and organization being captured via RADISTAT. While all of these top-ranked GLCM features are intended to capture intensity variability or dispersion in local pixel neighborhoods [12], both voxel- and region-wise radiomic descriptors of these distributions yielded subpar performance in all our experiments. In addition to the RADISTAT descriptor independently yielding relative high classifier performance in distinguishing between patient groups, combining RADISTAT with radiomic descriptors resulted in significantly improved AUCs comapred to using radiomic features alone. This suggests our approach may be capturing complementary new information related to spatial organization from radiomic texture feature maps compared to conventional radiomic descriptors, thus enabling more accurate disease sub-typing via MRI. The intuitive construction of the RADISTAT descriptor amay also be used to identify which aspects of the underlying disease heterogeneity are being captured; an intuition that may be “lost” when computing statistics from voxel-wise maps or region-wise features directly.
We do acknowledge some limitations to our study. While being multi-institutional, our individual cohorts for the 2 experiments may still be considered limited in size. In total, we evaluated over 170 patient datasets across 2 different tumor types, and performed over 800 experimental combinations to fully benchmark RADISTAT performance across feature types and parameter choices. Our comprehensive benchmarking of RADISTAT was primarily based on one class of radiomic features: GLCM-based feature maps, generated using a single window size. However, this class of features is also the most popularly employed in radiomic analysis, and we chose to focus on it based on demonstrated success in multiple disease classification problems including RCa and GBMs [5], [8], [10], [11]. Additional classes of radiomic and texture features could be explored in future work. We also opted to compute RADISTAT descriptors based on defining 3 “levels” for the radiomic expression map (intuitively corresponding to low (L), medium (M), and high (H) expression of texture patterns) as well as for a specific range of clustering hyperparameters (based on lesion size). These parameters may be specified differently based on the clinical question at hand, making it important to ensure these parameters appropriately tuned for a specific task as well as confirming that the superpixel clustering algorithm converges while still capturing informative sub-compartments within the ROI. In our 2D experiments, 5 ≤ α ≤ 15; 3 ≤ β ≤ 9 was found to yield the most stable results, while in 3D, 10 ≤ α ≤ 25; 3 ≤ β ≤ 9 was found to be most stable; across both classification tasks. Across the 4 major experiments in 2D and 3D, RADISTAT performed poorest in 3D when evaluated on the GBM cohort. The latter was mostly likely due to introduction of artifacts in the mask across the multiple post-processing steps applied to account for the significant imaging heterogeneity between institutions; which also negatively impacted voxel- and region-wise radiomic performance. Additionally, we had to restrict our 3D superpixel clustering algorithm to converge in a reasonable amount of time (< 24 hours per case), which meant that a few parameter combinations in the 3D GBM analysis were not evaluated by us. However, our parameter sensitivity analysis across 2D and 3D suggests robust performance for RADISTAT in a majority of the evaluations performed. While we did not specifically evaluate the sensitivity of RADISTAT to variations in ROI segmentation, previous studies of these cohorts [13], [41] have demonstrated excellent expert agreement in annotations as well as robustness of radiomic features to any minor annotation differences.
In the future, we plan to further examine the utility of RADISTAT across an expanded set of radiomic features as well as for other tumor and disease types, including to interrogate sex differences and race differences in tumor phenotypes [45]. Additionally, we plan to explore the value of performing superpixel partitioning and computing RADISTAT using the original signal intensity data rather than the radiomic features. This may be advantageous in a multi-parametric setting [46]. We will also seek to develop more advanced spatial correlation and graph construction approaches to combining radiology and pathology datasets for improved disease characterization. Open-source implementations of RADISTAT with sample data have been made available via GitHub (https://github.com/ccipd/RADISTAT-py).
Supplementary Material
Fig. S1. Representative 3D visualizations of radiomic and RADISTAT expression maps for RCa patients with metastatic disease (top row) and non-metastatic disease (bottom) after chemoradiation; from the discovery cohort. Visually apparent differences across the (a),(d) radiomic volume-rendered heatmaps for GLCM difference variance are captured via sub-regions of (b),(e) high, medium, and low radiomic expression via RADISTAT. Histograms reveal distinctive spatial phenotypes within the rectal wall that accurately discriminate between (c) metastatic and (f) non-metastatic rectal cancers after chemoradiation.
Fig. S2. Median histograms for 2D RADISTAT (based on GLCM inertia) per outcome group and for each α-β combination in RCa patients, indicating consistent trends between metastatic and non-metastatic disease.
Fig. S3. Median histograms for 3D RADISTAT (based on GLCM difference variance) per outcome group and for each α-β combination in RCa patients, with consistent trends between metastatic and non-metastatic disease.
Fig. S4. Median histograms for 2D RADISTAT (based on GLCM energy) per outcome group and for each α-β combination in GBM patients, with consistent trends between PsP and tumor progression.
Fig. S5. Median histograms for 3D RADISTAT (based on GLCM correlation) per outcome group and for each α-β combination in GBM patients, with consistent trends between PsP and tumor progression.
Fig. S6. Testing for statistically significant differences between RADISTAT components. Asterisks (*) indicate statistical significance using nonparametric ranksum test (p ≤ 0.05, corrected for multiple comparisons) between corresponding vertical pairs of spatial adjacency components between different outcomes. (a)-(b) RCa classification task. (c)-(d) GBM classification task.
VII. Acknowledgements
We would like to thank Kenneth Friedman, Joseph Willis, Sharon Stein, Conor P. Delaney, Rajmohan Paspulati, Justin T Brady, Katie Bingmer, Andrei Purysko, David Liska, and Matthew Kalady for access to anonymized rectal MRI scans and pathologic variables utilized in this study. Additionally, we thank Volodymyr Statsevych, R. Huang, and Manmeet Aahluwalia for making available anonymized brain tumor MRI scans and tumor outcomes that were utilized in our experiments.
Research reported in this publication was supported by the National Cancer Institute under 1U24CA199374-01, 1R01CA249992-01A1, 1R01CA202752-01A1, 1R01CA208236-01A1, 1R01CA216579-01A1, 1R01CA220581-01A1, 1R01CA257612-01A1, 1U01CA239055-01, 1U01CA248226-01, 1U54CA254566-01, 1F31CA216935-01A1, the National Heart, Lung and Blood Institute under 1R01HL151277-01A1, the National Institute of Biomedical Imaging and Bioengineering under 1R43EB028736-01, the National Center for Research Resources under 1C06RR12463-01, the VA Merit Review Award IBX004121A from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Service, the Office of the Assistant Secretary of Defense for Health Affairs through the Breast Cancer Research Program (W81XWH-19-1-0668), the Prostate Cancer Research Program (W81XWH-15-1-0558, W81XWH-20-1-0851), the Lung Cancer Research Program (W81XWH-18-1-0440, W81XWH-20-1-0595), the Peer Reviewed Cancer Research Program (W81XWH-16-1-0329, W81XWH-18-1-0404, W81XWH-21-1-0345), the Kidney Precision Medicine Project (KPMP) Glue Grant, the Ohio Third Frontier Technology Validation Fund, the Dana Foundation David Mahoney Neuroimaging Program, the V Foundation Translational Research Award, the Johnson & Johnson WiSTEM2D Award, the Clinical and Translational Science Collaborative of Cleveland (UL1TR0002548) from the National Center for Advancing Translational Sciences (NCATS) component of the National Institutes of Health and NIH roadmap for Medical Research, and the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, the Department of Defense, or the United States Government.
References
- [1].Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, Van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, et al. , Radiomics: extracting more information from medical images using advanced feature analysis, European journal of cancer 48 (4) (2012) 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kickingereder P, Neuberger U, Bonekamp D, Piechotta PL, Gotz M, Wick A, Sill M, Kratz A, Shinohara RT, Jones DT, Radbruch A, Muschelli J, Unterberg A, Debus J, Schlemmer H-P, Herold-Mende C, Pfister S, von Deimling A, Wick W, Capper D, Maier-Hein KH, Bendszus M, Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma, Neuro-Oncology 20 (2018) 848–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, et al. , Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach, Nature communications 5 (2014) 4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Horvat N, Veeraraghavan H, Khan M, Blazic I, Zheng J, Capanu M, Sala E, Garcia-Aguilar J, Gollub MJ, Petkovska I, Mr imaging of rectal cancer: radiomics analysis to assess treatment response after neoadjuvant therapy, Radiology 287 (3) (2018) 833–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Braman NM, Etesami M, Prasanna P, Dubchuk C, Gilmore H, Tiwari P, Plecha D, Madabhushi A, Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast dce-mri, Breast Cancer Research 19 (1) (2017) 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Griethuysen JJ, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RG, Fillon-Robin J-C, Pieper S, Aerts HJ, Computational radiomics system to decode the radiographic phenotype, Cancer Research 77 (21) (2017) e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJ, Dekker A, Fenstermacher D, et al. , Radiomics: the process and the challenges, Magnetic resonance imaging 30 (9) (2012) 1234–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Prasanna P, Patel J, Partovi S, Madabhushi A, Tiwari P, Radiomic features from the peritumoral brain parenchyma on treatment-naive multi-parametric mr imaging predict long versus short-term survival in glioblastoma multiforme: preliminary findings, European radiology 27 (10) (2017) 4188–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Thawani R, McLane M, Beig N, Ghose S, Prasanna P, Velcheti V, Madabhushi A, Radiomics and radiogenomics in lung cancer: a review for the clinician, Lung Cancer 115 (2018) 34–41. [DOI] [PubMed] [Google Scholar]
- [10].Soni N, Priya S, Bathla G, Texture analysis in cerebral gliomas: A review of the literature, American Journal of Neuroradiology 40 (6) (2019) 928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wibmer A, Hricak H, Gondo T, Matsumoto K, Veeraraghavan H, Fehr D, Zheng J, Goldman D, Moskowitz C, Fine SW, Reuter VE, Eastham J, Sala E, Vargas HA, Haralick texture analysis of prostate mri: utility for differentiating non-cancerous prostate from prostate cancer and differentiating prostate cancers with different gleason scores, European Radiology 25 (10) (2015) 2840–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Haralick RM, Shanmugam K, Dinstein IH, Textural features for image classification, IEEE Transactions on systems, man, and cybernetics (6) (1973) 610–621. [Google Scholar]
- [13].Alvarez-Jimenez C, Antunes JT, Talasila N, Bera K, Brady JT, Gollamudi J, Marerstein E, Kalady MF, Purysko A, Willis JE, Stein S, Friedman K, Paspulati R, Delaney CP, Romero E, Madabhushi A, Viswanath SE, Radiomic texture and shape descriptors of the rectal environment on post-chemoradiation t2-weighted mri are associated with pathologic tumor stage regression in rectal cancers: A retrospective, multi-institution study, Cancers 12 (8) (2020) e2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vallieres M, Kay-Rivest E, Perrin LJ, Liem X, Furstoss C, Aerts HJ, Khaouam N, Nguyen-Tan PF, Wang C-S, Sultanem K, Seuntjens J, El Naqa I, Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer, Scientific Reports 7 (10117). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Coroller TP, Agrawal V, Narayan V, Hou Y, Grossmann P, Lee SE, Mak RH, Aerts HJ, Radiomic phenotype features predict pathological response in non-small cell lung cancer, Radiotherapy and Oncology 119 (2016) 480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dworak O, Keilholz L, Hoffmann A, Pathological features of rectal cancer after preoperative radiochemotherapy, International journal of colorectal disease 12 (1) (1997) 19–23. [DOI] [PubMed] [Google Scholar]
- [17].Lindsay H, Näthke I, Rchanges in cell and tissue organization in cancer of the breast and colon, Current Opinions Cell Biology 0 (2014) 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Penzias G, Singanamalli A, Elliott R, Gollamudi J, Shih N, Feldman M, Stricker PD, Delprado W, Tiwari S, Böhm M, et al. , Identifying the morphologic basis for radiomic features in distinguishing different Gleason grades of prostate cancer on MRI: Preliminary findings, PloS one 13 (8) (2018) e0200730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Zhou M, Hall L, Goldgof D, Russo R, Balagurunathan Y, Gillies R, Gatenby R, Radiologically defined ecological dynamics and clinical outcomes in glioblastoma multiforme: preliminary results, Translational oncology 7 (1) (2014) 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhou M, Chaudhury B, Hall LO, Goldgof DB, Gillies RJ, Gatenby RA, Identifying spatial imaging biomarkers of glioblastoma multiforme for survival group prediction, Journal of Magnetic Resonance Imaging 46 (1) (2017) 115–123. [DOI] [PubMed] [Google Scholar]
- [21].Wu J, Gensheimer MF, Dong X, Rubin DL, Napel S, Diehn M, Loo BW Jr, Li R, Robust intratumor partitioning to identify high-risk subregions in lung cancer: a pilot study, International Journal of Radiation Oncology* Biology* Physics 95 (5) (2016) 1504–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wu J, Gong G, Cui Y, Li R, Intratumor partitioning and texture analysis of dynamic contrast-enhanced (dce)-mri identifies relevant tumor subregions to predict pathological response of breast cancer to neoadjuvant chemotherapy, Journal of Magnetic Resonance Imaging 44 (5) (2016) 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fan M, Cheng H, Zhang P, Gao X, Zhang J, Shao G, Li L, DCE-MRI texture analysis with tumor subregion partitioning for predicting Ki-67 status of estrogen receptor-positive breast cancers, Journal of Magnetic Resonance Imaging 48 (1) (2018) 237–247. [DOI] [PubMed] [Google Scholar]
- [24].Gallego-Ortiz C, Martel AL, A graph-based lesion characterization and deep embedding approach for improved computer-aided diagnosis of nonmass breast MRI lesions, Medical Image Analysis 51 (2019) 116–124. [DOI] [PubMed] [Google Scholar]
- [25].Wu J, Cao G, Sun X, Lee J, Rubin DL, Napel S, Kurian AW, Daniel BL, Li R, Intratumoral spatial heterogeneity at perfusion mr imaging predicts recurrence-free survival in locally advanced breast cancer treated with neoadjuvant chemotherapy, Radiology 288 (1) (2018) 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Corredor G, Wang X, Zhou Y, Lu C, Fu P, Syrigos K, Rimm DL, Yang M, Romero E, Schalper KA, Velcheti V, Madabhushi A, Spatial architecture and arrangement of tumor-infiltrating lymphocytes for predicting likelihood of recurrence in early-stage non-small cell lung cancer, Clinical Cancer Research 25 (2018) 1526–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Antunes J, Prasanna P, Madabhushi A, Tiwari P, Viswanath S, Radiomic spatial textural descriptor (radistat): Characterizing intra-tumoral heterogeneity for response and outcome prediction (2017) 468–476. [Google Scholar]
- [28].Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Süsstrunk S, Slic superpixels compared to state-of-the-art superpixel methods, IEEE transactions on pattern analysis and machine intelligence 34 (11) (2012) 2274–2282. [DOI] [PubMed] [Google Scholar]
- [29].Seo N, Kim H, Choo MS, Lim JS, Response assessment with mri after chemoradiotherapy in rectal cancer: Current evidences, Korean Journal of Radiology 20 (2019) 1003–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Parvez K, Parvez A, Zadeh G, The diagnosis and treatment of pseudoprogression, radiation necrosis and brain tumor recurrence, International Journal of Molecular Sciences 15 (2014) 11832–11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Weiser MR, Ajcc 8th edition: Colorectal cancer, Annals of Surgical Oncology 25 (6) (2018) 1454–1455. [DOI] [PubMed] [Google Scholar]
- [32].Stupp R, Mason WP, den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe JG, Denis and Cairncross, Eisenhauer E, Mirimanoff RO, Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma, New England Journal of Medicine 352 (2005) 987–996. [DOI] [PubMed] [Google Scholar]
- [33].Gillies RJ, Kinahan PE, Hricak H, Radiomics: images are more than pictures, they are data, Radiology 278 (2) (2015) 563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fisher R, The use of multiple measurements in taxonomic problems, Annals of Eugenics 7 (1936) 179–188. [Google Scholar]
- [35].Antunes J, Viswanath S, Brady JT, Crawshaw B, Ros P, Steele S, Delaney CP, Paspulati R, Willis J, Madabhushi A, Coregistration of preoperative mri with ex vivo mesorectal pathology specimens to spatially map post-treatment changes in rectal cancer onto in vivo imaging: preliminary findings, Academic radiology 25 (7) (2018) 833–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Shia J, Guillem JG, Moore HG, Tickoo SK, Qin J, Ruo L, Suriawinata A, Paty PB, Minsky BD, Weiser MR, et al. , Patterns of morphologic alteration in residual rectal carcinoma following preoperative chemoradiation and their association with long-term outcome, The American journal of surgical pathology 28 (2) (2004) 215–223. [DOI] [PubMed] [Google Scholar]
- [37].Hugen N, Van de Velde C, De Wilt J, Nagtegaal I, Metastatic pattern in colorectal cancer is strongly influenced by histological subtype, Annals of oncology 25 (3) (2014) 651–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhou X, Yi Y, Liu Z, Zhou Z, Lai B, Sun K, Li L, Huang L, Feng Y, Cao W, Tian J, Radiomics-based preoperative prediction of lymph node status following neoadjuvant therapy in locally advanced rectal cancer, Frontiers in Oncology 10 (2020) 604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tiwari P, Prasanna P, Wolansky L, Pinho M, Cohen M, Nayate A, Gupta A, Singh G, Hatanpaa K, Sloan A, et al. , Computer-extracted texture features to distinguish cerebral radionecrosis from recurrent brain tumors on multiparametric MRI: a feasibility study, American Journal of Neuroradiology 37 (12) (2016) 2231–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chan Y, Leung S, King A, Metreweli C, Late radiation injury to the temporal lobes: morphologic evaluation at mr imaging, Radiology 213 (1999) 800–1807. [DOI] [PubMed] [Google Scholar]
- [41].Ismail M, Hill V, Statsevych V, Huang R, Prasanna P, Correa R, Sigh G, Bera K, Beig N, Thawani R, Madabhushi A, Aahluwalia M, Tiwari P, Shape features of the lesion habitat to differentiate brain tumor progression from pseudo-progression on routine multi-parametric mri: A multi-site study, American Journal of Neuororadiology 39 (12) (2018) 2187–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Rowe LS, Butman JA, Mackey M, Shih JH, Cooley-Zgela T, Ning H, Gilbert MR, Smart DK, Camphausen K, Krauze AV, Differentiating pseudoprogression from true progression: analysis of radiographic, biologic, and clinical clues in gbm, Journal of Neurooncology 139 (5) (2018) 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Elshafeey N, Kotrotsou A, Hassan A, Elshafei N, Ahmed S, Abrol S, Agarwal A, Salek KE, Bergamaschi S, Acharya J, Moron FE, Law M, Fuller GN, Huse JT, Ziin PO, Colen RR, Multicenter study demonstrates radiomic features derived from magnetic resonance perfusion images identify pseudoprogression in glioblastoma, Nature Communications 10 (2019) 3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jang B-S, Jeon SH, Kim IH, Kim I. a., Prediction of pseudoprogression versus progression using machine learning algorithm in glioblastoma. sci rep, Scientific Reports 8 (12516). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Beig N, Singh S, Bera K, Prasanna P, Singh G, Chen J, Bamashmos A, Barnett A, Hunter K, Statsevych V, Hill V, Varadan V, Madabhushi A, Ahluwalia M, T. P, Sexually dimorphic radiogenomic models identify distinct imaging and biological pathways that are prognostic of overall survival in glioblastoma, Neuro-Onc 23 (2) (2021) 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Antunes J, Ofshteyn A, Wang E, Brady J, Willis J, Friedman K, Marderstein E, Kalady M, Stein S, Purysko A, Paspulati R, Gollamudi J, Madabhushi A, Viswanath S, Radiomic features of primary rectal cancers on baseline t2-weighted mri are associated with pathologic complete response to neoadjuvant chemoradiation: A multisite study, J Magn Reson Imaging 52 (5) (2020) 1531–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Representative 3D visualizations of radiomic and RADISTAT expression maps for RCa patients with metastatic disease (top row) and non-metastatic disease (bottom) after chemoradiation; from the discovery cohort. Visually apparent differences across the (a),(d) radiomic volume-rendered heatmaps for GLCM difference variance are captured via sub-regions of (b),(e) high, medium, and low radiomic expression via RADISTAT. Histograms reveal distinctive spatial phenotypes within the rectal wall that accurately discriminate between (c) metastatic and (f) non-metastatic rectal cancers after chemoradiation.
Fig. S2. Median histograms for 2D RADISTAT (based on GLCM inertia) per outcome group and for each α-β combination in RCa patients, indicating consistent trends between metastatic and non-metastatic disease.
Fig. S3. Median histograms for 3D RADISTAT (based on GLCM difference variance) per outcome group and for each α-β combination in RCa patients, with consistent trends between metastatic and non-metastatic disease.
Fig. S4. Median histograms for 2D RADISTAT (based on GLCM energy) per outcome group and for each α-β combination in GBM patients, with consistent trends between PsP and tumor progression.
Fig. S5. Median histograms for 3D RADISTAT (based on GLCM correlation) per outcome group and for each α-β combination in GBM patients, with consistent trends between PsP and tumor progression.
Fig. S6. Testing for statistically significant differences between RADISTAT components. Asterisks (*) indicate statistical significance using nonparametric ranksum test (p ≤ 0.05, corrected for multiple comparisons) between corresponding vertical pairs of spatial adjacency components between different outcomes. (a)-(b) RCa classification task. (c)-(d) GBM classification task.


