Abstract
Anti-SARS-CoV-2 antibodies are crucial for protection from future COVID-19 infections, limiting disease severity, and control of viral transmission. While patients with the most common type of hematologic malignancy, B cell lymphoma, often develop insufficient antibody responses to messenger RNA (mRNA) vaccines, vaccine-induced T cells would have the potential to ‘rescue’ protective immunity in patients with B cell lymphoma. Here we report the case of a patient with B cell lymphoma with profound B cell depletion after initial chemoimmunotherapy who received a total of six doses of a COVID-19 mRNA vaccine. The patient developed vaccine-induced anti-SARS-CoV-2 antibodies only after the fifth and sixth doses of the vaccine once his B cells had started to recover. Remarkably, even in the context of severe treatment-induced suppression of the humoral immune system, the patient was able to mount virus-specific CD4+ and CD8+ responses that were much stronger than what would be expected in healthy subjects after two to three doses of a COVID-19 mRNA vaccine and which were even able to target the Omicron ‘immune escape’ variant of the SARS-CoV-2 virus. These findings not only have important implications for anti-COVID-19 vaccination strategies but also for future antitumor vaccines in patients with cancer with profound treatment-induced immunosuppression.
Keywords: Vaccination, COVID-19, IMMUNOLOGY, T-Lymphocytes, Hematologic Neoplasms
COVID-19 is caused by SARS-CoV-2, which contains the spike (S) and nucleocapsid (N) proteins.1 2 The S protein has S1 and S2 domains and the virus uses the receptor-binding domain (RBD) within S1 to bind to ACE-2 receptor3 and enter normal cells such as the pneumocytes in the lungs.1 4 Unfortunately, patients with hematologic malignancies and COVID-19 show dramatically increased mortality rate,5 6 which correlates with the intensity of prior antilymphoma treatments.5–7
Disease-induced or vaccine-induced anti-SARS-CoV-2 antibodies are crucial for protection from future COVID-19 infections, limiting disease severity, and control of viral transmission.8 9 Unfortunately, patients with the most common type of hematologic malignancy, namely B cell lymphoma, often develop insufficient antibody responses to messenger RNA (mRNA) vaccines due to the immunosuppression caused by their anti-B cell treatments.10 In addition to antibody responses, antiviral T cells have been shown to improve survival in patients with COVID-19,11 including patients with hematologic cancers,12 and vaccine-induced T cells have the potential to ‘rescue’ protective immunity in patients with B cell lymphoma. However, it is not entirely clear whether patients with B cell lymphoma are capable of mounting a vaccine-induced T cell response in the framework of treatment-induced immunosuppression and whether such T cells would be able to recognize and target immune escape variants such as Omicron.
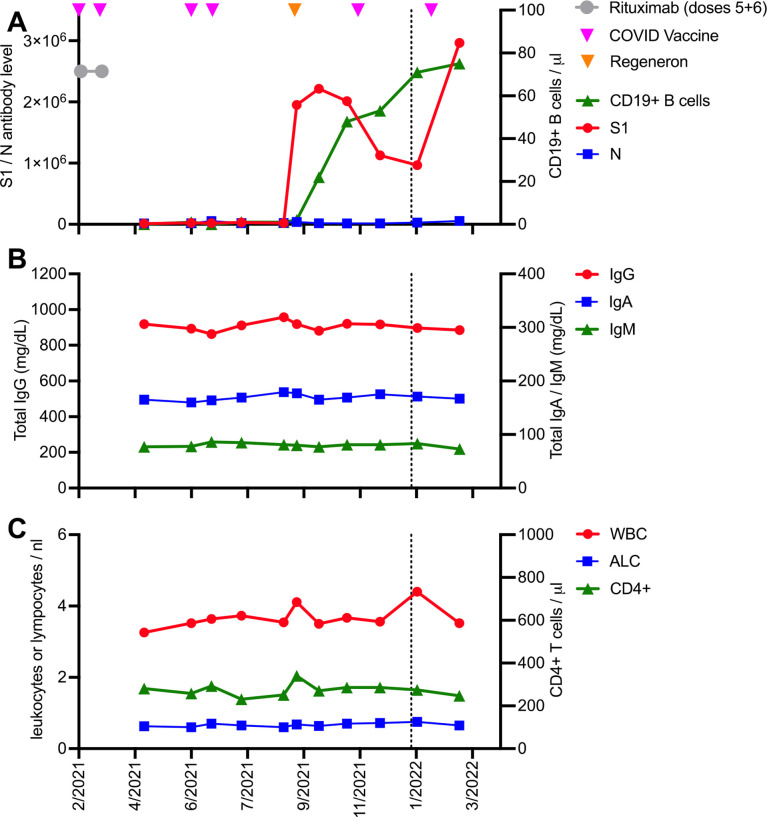
In this study we performed a comprehensive monitoring of anti-SARS-CoV-2 antibody and T cell immunity in a patient with B cell lymphoma with profound immunosuppression receiving multiple doses of a COVID-19 mRNA vaccine (For methods used please see online supplemental methods and online supplemental tables 1–3). The patient is a man in his early 70s with diffuse large B cell lymphoma involving the left cervical chain (stage 1) who received four cycles of R-CHOP (rituximab/cyclophosphamide/doxorubicin/vincristine/prednisone) followed by two cycles of rituximab alone. He achieved complete remission which was sustained. While the patient was under treatment with the final two doses of rituximab (figure 1A and online supplemental figure 1), he simultaneously received the first two doses of the BNT162b2 COVID-19 mRNA vaccine (online supplemental figure 1). At that time, he did not have any B cells in his peripheral blood (figure 1A), and accordingly he did not develop antibodies against the S protein of SARS-CoV-2 (figure 1A). Two more doses of the same vaccine did not lead to the development of endogenous antiviral antibodies, and as a consequence he received Regeneron’s antibody cocktail REGN-COV2 off-label as an alternative prophylactic measure (figure 1A). Shortly thereafter, anti-S1 antibodies became detectable, presumably due to the exogenous antibodies persisting in his blood (figure 1A). In October 2021 the patient’s B cell counts finally started to recover from anti-CD20 treatment (figure 1A). Off-label he received a fifth dose of the COVID-19 vaccine, with normal B cells detectable but still low, which led to a stabilization of total anti-S antibody levels (figure 1A) without any additional doses of the REGN-COV2 antibody cocktail, presumably representing early signs of an initial endogenous humoral immune response to the fifth dose of the vaccine. A sixth dose of the same mRNA COVID-19 vaccine given after normalization of B cell numbers led to a substantial increase in anti-S1 antibody levels. We performed a comprehensive analysis of vaccine-induced T cell and B cell responses between administration of the fifth and sixth doses of the vaccine (figure 1A).
jitc-2022-004953supp002.pdf (102.7KB, pdf)
jitc-2022-004953supp003.pdf (45.4KB, pdf)
jitc-2022-004953supp001.pdf (3.5MB, pdf)
Figure 1.
Time course of immune parameters including anti-SARS-CoV-2 antibodies in a patient with lymphoma receiving multiple COVID-19 vaccinations. (A) Absolute numbers of peripheral blood B cells and levels of antibodies directed against SARS-CoV proteins S1 and N after one dose of Regeneron’s antibody cocktail REGN-COV2 and multiple doses of the BNT162b2 COVID-19 vaccine, respectively. (B) Absolute serum concentrations of IgG, IgA, and IgM immunoglobulins over time. (C) Absolute white blood cell count (WBC), absolute lymphocyte count (ALC), and number of peripheral blood CD4+ T cells over time. The dotted line indicates the timepoint when comprehensive immunomonitoring was performed.
When we asked whether the lack of a vaccine-induced anti-SARS-CoV-2 antibody response to the first four doses was due to an unspecific and global treatment-induced and/or disease-induced immunosuppression, we found that the patient indeed showed lower levels of total IgG, IgM, and IgA immunoglobulins compared with a group of healthy controls (online supplemental figure 2A). However, there was no decline in immunoglobulins over time (figure 1B), and even more importantly the patient with B cell-depleted lymphoma maintained normal levels of IgG antibodies against recall antigens such as influenza A, tetanus toxoid, and Epstein-Barr virus even after six cycles of anti-B cell lymphoma treatment (online supplemental figure 2B).
Taking a closer look at the anti-SARS-CoV-2 immunity our patient had developed after five doses of the vaccine, we detected, in agreement with the routine laboratory assay (figure 1A), IgG antibodies directed against the S1, RBD (online supplemental figure 2C), and S2 proteins, with no detectable anti-N antibodies (online supplemental figure 2E). The antibody titers were lower compared with those of seven healthy controls at 4 weeks after the second dose of a COVID-19 mRNA vaccine (online supplemental figure 2C); however, they still led to an almost 100% antiviral neutralizing activity (online supplemental figure 2D). Consistent with our longitudinal analyses of peripheral B cell numbers in this patient who initially showed therapy-induced B cell depletion (figure 1A), we were able to detect a significant number of CD19+/CD20+ B cells (online supplemental figure 2F), including CD19+ B cells secreting IgG antibodies against the S protein (online supplemental figure 2G), after five doses of the vaccine. Unfortunately, the comparably low antibody titers in our patient were further reduced when binding to the Omicron variant instead of the ancestral S1 and RBD proteins (online supplemental figure 2C). The reduced binding resulted in a dramatically diminished anti-Omicron neutralizing activity of the polyclonal antibodies (online supplemental figure 2D).
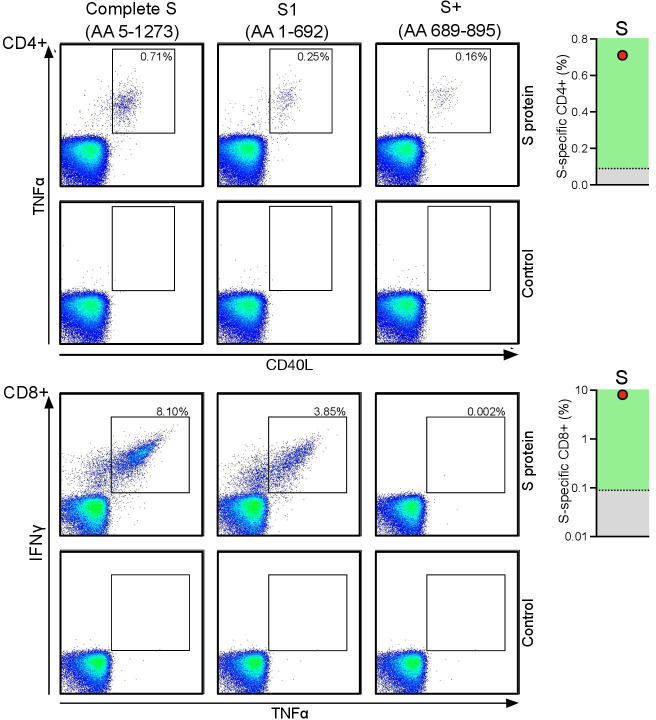
We next asked whether our patient evidenced anti-SARS-CoV-2 T cells despite an only very slowly developing humoral antiviral immune component, which was still suboptimal after the fifth dose of the vaccine. We found that the patient actually showed vaccine-induced CD4+ and CD8+ T cell responses in their blood targeting the S protein of the SARS-CoV-2 virus that were much stronger than what we had observed in a group of healthy individuals who had received two doses of the mRNA vaccine (figure 2). Unfortunately, we were not able to measure T cell responses in our patient after only two doses of the vaccine; however, after five doses, the number of SARS-CoV-2-specific CD8+ T cells was almost 100 times higher than what we had observed in healthy vaccinated individuals after the two initial doses (figure 2). For both CD4+ and CD8+ T cells targeting the S protein, most of the immunodominant epitopes of the anti-SARS-CoV-2 CD4+ T cells were within the S1 component of the fusion protein (figure 2).
Figure 2.
Vaccine-induced SARS-CoV-2-specific T cells in a patient with B cell lymphoma after multiple doses of a COVID-19 mRNA vaccine. After the patient had received five doses of the COVID-19 mRNA vaccine, T cells specific for the S protein of the SARS-CoV-2 were identified ex vivo after short-term stimulation of the total peripheral blood mononuclear cells (PBMC) using libraries of overlapping peptides covering the complete sequence of the protein. Intracellular staining of cytokines followed by flow cytometry served as the read-out assay. SARS-CoV-2-specific CD4+ T cells (upper panel) were defined as tumor necrosis factor (TNF) α/CD40L (CD154) double-positive CD3+CD4+ T cells, and SARS-CoV-2-specific CD8+ T cells (lower panel) were defined as interferon (IFN) γ/TNFα double-positive CD3+CD8+ T cells. The number of vaccine-induced CD4+ and CD8+ T cells specific for the complete sequence of the S fusion protein was compared with the number of T cells from the same individual recognizing the N-terminal S1 protein or the N-terminal portion (amino acids 689–895) of the S2 protein (‘S+’). Background levels were typically <0.01% of all CD4+ or CD8+ T cells. Plots on the right show the patient’s S-specific CD4+ and CD8+ T cells (red dot) in relation to the median number (dotted line) of the same T cells from a group of six healthy control subjects, where the results were available from 4 weeks after the second dose of a COVID-19 mRNA vaccine. mRNA, messenger RNA.
It has previously been shown that SARS-CoV-2 vaccination is capable of inducing T cells with the potential to cross-recognize the Omicron variant13 14; however, to the best of our knowledge, the same phenomenon has not been demonstrated in patients with B cell lymphoma. When we examined cross-recognition by our patient’s T cells, we found that, while there was a certain decrease in T cell reactivity when exposed to the Omicron variant of the S protein, most of the patient’s polyclonal vaccine-induced CD4+ and CD8+ T cells also recognized this immune escape variant of the ancestral SARS-CoV-2 virus (online supplemental figure 3). While this certainly cannot serve as evidence of a protective function of the vaccine-induced T cells, it has indeed previously been shown that postinfection and vaccine-induced anti-SARS-CoV-2 T cells can play a protective role,12 15–17 and our patient never developed COVID-19 despite multiple known close exposures to family members with a proven infection.
We have shown here that a patient with B cell lymphoma whose B cells were initially depleted following antilymphoma chemoimmunotherapy was not able to mount an antibody response to four doses of a COVID-19 mRNA vaccine. The patient only developed vaccine-induced anti-SARS-CoV-2 antibodies once his B cells had started to recover. Importantly, even in the context of severe treatment-induced suppression of the humoral immune system, the patient was able to mount SARS-CoV-2-specific CD4+ and CD8+ responses that were much stronger than what would be expected in healthy subjects after two to three rounds of a COVID-19 mRNA vaccine and which were even able to target the Omicron ‘immune escape’ variant18 of the virus. These findings not only have important implications for anti-COVID-19 vaccination strategies but also for future antitumor vaccines in patients with cancer with profound treatment-induced immunosuppression.
Acknowledgments
The authors recognize Hong Zhou for technical assistance.
Footnotes
DA and RJK contributed equally.
Contributors: DA designed the study, performed the experiments, analyzed the data, made the figures, and wrote the manuscript. RJK collected patient data and samples, analyzed the data, and wrote the manuscript. DO, TI, and EG processed patient samples and performed the experiments. XF and PDB performed the experiments and wrote the manuscript. NMH, TL, SD, JC, and APR analyzed the data and wrote the manuscript.
Funding: This study was funded by two grants from the Kahlert Foundation (to DA) and was supported in part by the Intramural Research Program of the National Cancer Institute, National Institute of Allergy and Infectious Diseases, and National Institute of Dental and Craniofacial Research.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Ethics statements
Patient consent for publication
Obtained.
Ethics approval
This study involves human participants and was approved by the IRB of the University of Maryland. Participants gave informed consent to participate in the study before taking part.
References
- 1. Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579:270–3. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020;382:727–33. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 2020;11:1620. 10.1038/s41467-020-15562-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ge X-Y, Li J-L, Yang X-L, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013;503:535–8. 10.1038/nature12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee LYW, Cazier J-B, Starkey T, et al. COVID-19 prevalence and mortality in patients with cancer and the effect of primary tumour subtype and patient demographics: a prospective cohort study. Lancet Oncol 2020;21:1309–16. 10.1016/S1470-2045(20)30442-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vijenthira A, Gong IY, Fox TA, et al. Outcomes of patients with hematologic malignancies and COVID-19: a systematic review and meta-analysis of 3377 patients. Blood 2020;136:2881–92. 10.1182/blood.2020008824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Busca A, et al. COVID-19 and CAR-T cells: current challenges and future directions-a report from the EPICOVIDEHA survey by EHA-IDWP. Blood Adv 2022. 10.1182/bloodadvances.2021005616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bergwerk M, Gonen T, Lustig Y, et al. Covid-19 breakthrough infections in vaccinated health care workers. N Engl J Med 2021;385:1474–84. 10.1056/NEJMoa2109072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lucas C, Klein J, Sundaram ME, et al. Delayed production of neutralizing antibodies correlates with fatal COVID-19. Nat Med 2021;27:1178–86. 10.1038/s41591-021-01355-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Herishanu Y, Avivi I, Aharon A, et al. Efficacy of the BNT162b2 mRNA COVID-19 vaccine in patients with chronic lymphocytic leukemia. Blood 2021;137:3165–73. 10.1182/blood.2021011568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sattler A, Angermair S, Stockmann H, et al. SARS-CoV-2-specific T cell responses and correlations with COVID-19 patient predisposition. J Clin Invest 2020;130:6477–89. 10.1172/JCI140965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bange EM, Han NA, Wileyto P, et al. CD8+ T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat Med 2021;27:1280–9. 10.1038/s41591-021-01386-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tarke A, Coelho CH, Zhang Z, et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from alpha to omicron. Cell 2022;185:847–59. 10.1016/j.cell.2022.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu J, Chandrashekar A, Sellers D, et al. Vaccines elicit highly conserved cellular immunity to SARS-CoV-2 omicron. Nature 2022;603:493–6. 10.1038/s41586-022-04465-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kundu R, Narean JS, Wang L, et al. Cross-Reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts. Nat Commun 2022;13:80. 10.1038/s41467-021-27674-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McMahan K, Yu J, Mercado NB, et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 2021;590:630–4. 10.1038/s41586-020-03041-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guo L, Wang G, Wang Y, et al. SARS-CoV-2-specific antibody and T-cell responses 1 year after infection in people recovered from COVID-19: a longitudinal cohort study. Lancet Microbe 2022;3:e348–56. 10.1016/S2666-5247(22)00036-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu L, Iketani S, Guo Y, et al. Striking antibody evasion manifested by the omicron variant of SARS-CoV-2. Nature 2022;602:676–81. 10.1038/s41586-021-04388-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-004953supp002.pdf (102.7KB, pdf)
jitc-2022-004953supp003.pdf (45.4KB, pdf)
jitc-2022-004953supp001.pdf (3.5MB, pdf)