Fungi are amazing organisms, being able to use almost any surface (e.g., bathroom tile, skin, or leaves) for growth. Unfortunately, they also are proficient at colonizing and using plants, humans, and animals as substrates. During the last two decades, the incidence of human fungal infections, especially involving immunocompromised patients, has dramatically increased (27, 41, 57). This is in part due to the tremendous advances in medicine that permit the saving of patients with neoplastic and immunocompromising diseases who would otherwise not have survived. It is ironic that many of these patients succumb to fungal infections for which there are few or no drugs available for treatment. Encouragingly, naturally occurring antifungal proteins and peptides, as well as synthetic derivatives, have the potential to be very interesting clinical leads.
Fungi are an extremely diverse group of organisms, with about 250,000 species widely distributed in essentially every ecosystem. Muller and Loeffler (124) estimate that the weight of fungi on Earth exceeds that of humans; Armillaria bulbosa, a tree root pathogen, is reported to be among the largest and oldest organisms on the planet (162). Humans and other animals are exposed to fungi from the moment of birth. Fortunately, only 200 or so species are pathogenic to mammals, although many nonpathogenic fungi cause allergy symptoms (3). The majority of fungal exposures and infections are self-limiting in intact animal hosts (76). However, in patients with compromised immune systems, infections even by fungal organisms with low virulence can be life threatening; for example, systemic fungal infections of leukemia patients account for 50% of fatalities (101, 141). Nosocomial bloodstream infections have a similar fatality rate (107).
Plants are also exposed to a large number of pathogenic fungi; although they do not have an immune system, plants have evolved a variety of potent defense mechanisms, including the synthesis of low-molecular-weight compounds, proteins, and peptides that have antifungal activity (16, 18, 47, 80, 104, 127, 151, 159, 177). Similarly, bacteria, insects, mollusks, fungi, and mammals synthesize a number of proteins and peptides that are antifungal (13, 19, 20, 30, 49, 51–54, 58, 68, 69, 79, 83, 109–111, 122, 126, 128, 153, 188, 189, 192). These proteins appear to be involved in either constitutive or induced resistance to fungal attack. It is a testament to the efficacy of these defenses that plants and animals, including humans, do so well against pathogenic fungi.
There are hundreds of antifungal peptides and proteins known, with more being discovered almost daily. This brief review will focus on proteins with molecular masses of greater than ∼5 kDa, about 50 amino acids in length; this choice is somewhat arbitrary, for there is not consensus concerning where a peptide ends and a protein begins. Even eliminating the small proteins (peptides), the list of antifungal proteins is large and daunting. The reader is directed to a review concerning antifungal peptides and proteins of less than 5 kDa (30). Given the diverse and varied types of proteins, this review will be an overview of the primary classes of antifungal proteins. Thirteen classes of antifungal proteins will be described: PR-1 proteins, (1,3)β-glucanases, chitinases, chitin-binding proteins, thaumatin-like (TL) proteins, defensins, cyclophilin-like protein, glycine/histidine-rich proteins, ribosome-inactivating proteins (RIPs), lipid-transfer proteins (LTPs), killer proteins (killer toxins), protease inhibitors, and other proteins. These proteins have been named primarily on the basis of either their mechanism of action, (e.g., glucanases), their structure (e.g., cysteine rich), or their similarity to a known “type” protein. To confuse the nomenclature further is the fact that several proteins can be and have been classified into more than one group. It is unfortunate that a standard nomenclature based on structure or some other unifying property(ies) of these proteins has not been proposed or adopted.
FUNGAL CELL WALL STRUCTURE
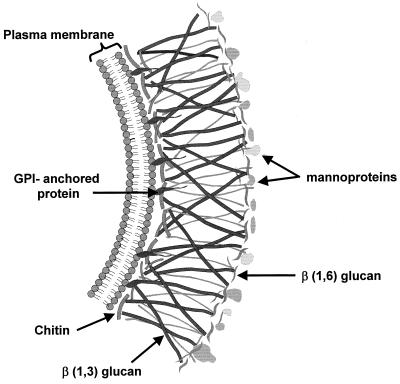
The fungal cell wall protects the organism against a hostile environment and relays signals for invasion and infection of a likely plant, animal, or human host. The cell wall of fungi and yeasts is synthesized at each hyphal apex in a complex assembly sequence (45, 105, 149). For example, the walls of Neurospora crassa and Candida albicans are composed of chitin, (1–3)β-d-glucan, (1,6)β-glucans, lipids, and peptides embedded in a protein matrix. The fungal wall affords a clear and discernible difference between fungi and their plant and animal hosts, providing an experimental target for antifungal antibiotics. A schematic of a typical fungal cell wall is shown in Fig. 1. It is important to note that fungi have significant internal turgor pressure so that even slight perturbation of the cell wall results in fungal cell lysis (54, 73, 118–120).
FIG. 1.
Schematic of fungal cell wall. GPI, glycophosphatidylinositol.
Several classes of antifungal proteins involve inhibition of the synthesis of the fungal cell wall or disrupt cell wall structure and/or function; others perturb fungal membrane structure, resulting in fungal cell lysis. The assays for antifungal activity include microtiter broth assays, agar diffusion assays, broth microdilution assays (43), and in planta assays (the determination of resistance of transgenic plants overexpressing a protein of interest). Most of the antifungal proteins described below are quite potent, with MICs in the micromolar or microgram-per-milliliter range, equivalent to MICs of many of the currently used agricultural and pharmaceutical antifungal compounds.
ANTIFUNGAL PROTEINS
PR proteins.
Plants when exposed to pathogens such as fungi and viruses produce low-molecular-weight antimicrobial compounds called phytoalexins, antimicrobial peptides, and small proteins (e.g., thionins [11, 40], defensins [14], hevein-like proteins, and knottin-like peptides [154]) and up-regulate a number of antimicrobial proteins. These plant proteins, called pathogenesis-related (PR) proteins, have been classically divided into five groups, PR-1, -2, -3, -4, and -5, based on serological and amino acid sequence analyses (180). Recently, another 6 groups of proteins have been suggested for inclusion as PR proteins, bringing the total to 11 groups. The reader is directed to a number of reviews concerning PR proteins, their regulation, and possible roles in plant defense (80, 163, 180, 194).
Each of the five classical groups of PR proteins has two subclasses: a basic subclass found in the plant cell vacuole and an acidic subclass usually found in the extracellular space (reference 80 and references therein). Each group has members with antifungal activity, and cognates of most groups have been found in a diversity of other organisms. The mechanisms of antifungal action of only the PR-2 and PR-3 groups of proteins have been clearly identified.
(i) PR-1 proteins.
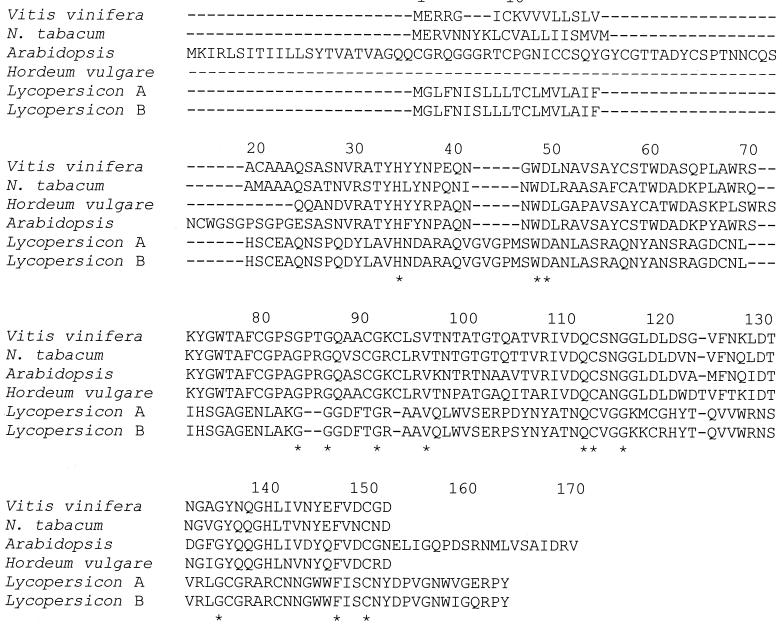
PR-1 proteins are accumulated to high levels after pathogen infection and are antifungal both in planta (transgenic plants overexpressing tobacco PR-1) and in vitro (129, 165). PR-1 proteins have been found in rice, wheat, maize, tobacco, Arabidopsis thaliana barley, and many other plants (1, 15, 117, 125, 145); an alignment of seven PR-1 proteins is shown in Fig. 2. Note that although these proteins are from diverse sources, they are remarkably similar (at least 35% identity). PR-1 proteins have antifungal activity at the micromolar level against a number of plant pathogenic fungi, including Uromyces fabae, Phytophthora infestans, and Erysiphe graminis (129). PR-1 proteins have molecular masses of ∼15 to 17 kDa and have homology to the superfamily of cysteine-rich proteins. Although the precise mechanism of antifungal activity is not understood for plant PR-1 proteins, a PR-1-like protein, helothermine, from the Mexican banded lizard interacted with membrane-channel proteins of target cells, inhibiting the release of Ca2+ (123). Whether antifungal plant PR-1 proteins act by this mechanism is not known but is suspected.
FIG. 2.
Amino acid sequence alignment of representative PR-1 proteins from Arabidopsis thaliana (accession no. P33154), Brassica napus (rape, accession no. T08154), Solanum tuberosum (potato, accession no. CAB58263), Lycopersicon esculentum (tomato, accession no. CAA04881), Sambucus nigra (elder, accession no. Q41359), Nicotiana tabacum (tobacco, accession no. S10205), Hordeum vulgare (barley, accession no. Q05968), and Zea mays (maize, accession no. T02054). Alignments were performed with the ClustalW program (http://clustalw.genome.ad.jp/); ∗ indicates amino acid identity.
(ii) PR-2 proteins (β-glucanses).
PR-2 proteins have (1,3)β-endoglucanase activity in vitro and have been grouped into three classes on the basis of amino acid sequence analysis (8, 25, 95, 103, 113, 131, 140). Class I glucanases are basic proteins of ∼ 33 kDa and are found in the plant vacuole. Classes II and III include acidic, extracellular proteins of about 36 kDa. The major structural difference between class I proteins and the other two classes is that class I proteins are synthesized as preproproteins that are processed prior to being enzymatically active. PR-2 proteins have been found in a wide variety of plants, including tobacco, A. thaliana, peas, grains, and fruits (25, 77, 146); the proteins are active in vitro at micromolar levels (∼50 μg/ml) against a wide number of fungi, including human and plant pathogens (e.g., Rhizoctonia solani, C. albicans, and Aspergillus fumigatus). The antifungal activity of PR-2 proteins has been convincingly demonstrated by a number of in vitro enzyme and whole-cell assays (163) as well as in planta using transgenic plants overexpressing a PR-2 protein (71).
The antifungal activity of plant (1,3)β-glucanases is thought to occur by PR-2 proteins hydrolyzing the structural (1,3)β-glucan present in the fungal cell wall, particularly at the hyphal apex of filamentous molds where glucan is most exposed, resulting in a cell wall that is weak. This weakened cell wall results in cell lysis and cell death.
(iii) PR-3 proteins (chitinases).
A number of enzymatic assays have shown PR-3 proteins to have in vitro chitinase activity. Most PR-3 proteins have molecular masses of between 26 and 43 kDa (131, 187). Chitinases (both plant PR-3 chitinases and chitinases from other sources) have been divided into five groups. class I chitinases contain an N-terminal cysteine-rich domain of ∼40 amino acids (also known as the wheat germ agglutinin domain), a chitin-binding hevein-like domain, a highly conserved central portion, and a hinge region; most class I proteins have molecular masses of ∼32 kDa. Class II proteins are similar in amino acid sequence to class I proteins, but they lack the N-terminal cysteine-rich domain and have molecular masses of 27 to 28 kDa. Class IV proteins resemble class I chitinases but are significantly smaller due to four major deletions. Class III proteins do not share amino acid sequence homology to any other class and have molecular masses of ∼28 to 30 kDa. Class V chitinases show sequence similarities to bacterial exochitinases and have molecular masses of 41 to 43 kDa. In addition to chitinases, a chitosanase (chitosan is deacetylated chitin) from Streptomyces strain N174 with antifungal activity has been isolated (119), and its X-ray structure has been determined.
Chitinases have been isolated from fungi (74, 112), plants (tobacco [114], cucumber, beans [198], peas, grains [63], and many others [37, 96, 112, 121, 150, 193]), and bacteria (22) and have potent antifungal activity against a wide variety of human and plant pathogens, including Trichoderma reesei, Alternaria solani, A. radicina, Fusarium oxysporum, R. solani, Guignardia bidwellii, Botrytis cinerea, and Coprinus comatus. By analogy with β-glucanases, the mode of action of PR-3 proteins is relatively straightforward: PR-3 proteins are endochitinases that cleave cell wall chitin polymers in situ, resulting in a weakened cell wall and rendering fungal cells osmotically sensitive. Not surprisingly, PR-2 (β-glucanases) and PR-3 (chitinases) proteins act synergistically in inhibiting fungal growth, both in vitro and in planta (71).
(iv) PR-4 (chitin-binding) proteins.
PR-4 proteins are chitin-binding proteins, have molecular masses of 13-14.5 kDa, and have been classified into two groups (42, 56, 143, 179). Class I proteins have amino acid sequence similarities to hevein (a chitin-binding polypeptide [42, 85, 179]) and belong to the superfamily of chitin-binding lectins. Class II proteins lack the chitin-binding domain. PR-4 proteins have been isolated from potato, tobacco, barley, tomato, and many other plants (42, 56, 85, 143, 179); an alignment of six PR-4 proteins is shown in Fig. 3. Note that the PR-4 proteins from the diverse sources share common sequences. Both classes of proteins have potent antifungal activity against a wide variety of human and plant pathogens (e.g., Trichoderma harzianum, Fusarium culmorum, F. graminearum, and B. cinerea). The antifungal activity of class I proteins is likely the result of protein binding to nascent fungal cell wall β-chitin. By mechanisms not understood, this results in disrupted cell polarity, with concomitant inhibition of growth (13). The mechanism of action of class II proteins (which lack the chitin-binding hevein domain but are antifungal nonetheless) is not understood.
FIG. 3.
Amino acid sequence alignment of selected PR-4 proteins from Vitis vinifera (accession no. AAC33732), Nicotiana tabacum (common tobacco, accession no. S23799), Arabidopsis thaliana (accession no. P43082), Hordeum vulgare (barley-barwin, accession no. A43474), and Lycopersicon esculentum (tomato, accession no. P04284 and Q04108). Alignments were performed with the ClustalW program; ∗ indicates amino acid identity.
Chitin-binding proteins and peptides that have antifungal activity but are not PR proteins have been isolated from a number of sources, including bacteria (13), plants, insects, and crustaceans (19, 29, 61, 76, 83, 131, 136). These non-PR-4 chitin-binding proteins include the tachystatins (75, 135) (horseshoe crab, 6.8 to 7.4 kDa), the penaeidins (31–33) (penaeid shrimp, 5.5 to 6.6 kDa), antifungal protein 1 (AFP1) (13) (Streptomyces tendae, 9.8 kDa), and others. Fungi inhibited by these proteins include plant and human pathogens, e.g., Paecilomyces variotii, Aspergillus spp., F. oxysporum, N. crassa, B. cinerea, and Alternaria brassicola. It is likely that these proteins act by a mechanism similar to that of the class I PR-4 proteins, namely, binding to cell wall chitin and disrupting cell polarity, thus leading to inhibition of fungal growth.
(v) PR-5 (TL) proteins.
PR-5 proteins share significant amino acid homology to thaumatin (a sweet-tasting [to humans] protein from the South African ketemfe berry bush) and are known as TL proteins. TL proteins have been isolated from A. thaliana (59, 60), corn (62, 148), soybeans, rice, wheat, tobacco (81), tomato (161), pumpkin (21), beans (196), barley (55), flax (12), and many other plants (122, 182, 184). The majority of PR-5 proteins have molecular masses of ∼22 kDa and are stabilized by eight disulfide bonds. This highly stabilized structure allows PR-5 proteins to be very resistant to protease degradation (148). The X-ray structures have been determined for two PR-5 proteins and thaumatin (82, 134).
Although the precise mechanism of action of PR-5 proteins is not completely understood, there are a number of interesting observations that may eventually lead to a unified hypothesis for how these proteins function to kill fungi (24, 66, 147, 158, 186). First, several TL proteins cause cell permeability changes in fungal cells with a cell wall but have no or little effect on protoplasts (148). For example, zeamatin (a TL protein from corn) caused very rapid cell lysis of N. crassa, even at 4°C; lysis occurred primarily at subapical regions (148). Second, a number of PR-5 proteins bind (1,3)β-glucan and have detectable in vitro (1,3)β-glucanase activity (47, 176). Third, zeamatin inhibits insect α-amylase and mammalian trypsin activities in vitro (152a). Fourth, osmotin, a TL protein from tobacco, causes perturbations in the regulation of fungal cell wall assembly (200, 201). Fifth, zeamatin and nikkomycin act in synergy, reducing the amount of zeamatin required for cell killing up to 1,000-fold (148). These disparate observations are difficult to assimilate into one mechanism of action. Regardless of the precise mode of action of TL proteins, they are fungicidal against a wide number of plant and human pathogens in vitro. Importantly, one protein, zeamatin, has shown efficacy in a murine vaginal model of C. albicans infection (D. A. Stevens et al., submitted for publication). It may be that certain PR-5 proteins can be developed into human therapeutics.
Defensins.
Defensins are a diverse group of low-molecular-mass cysteine-rich proteins found in mammals, fungi (89), insects (91), and plants (14, 16). The insect and mammalian defensins are quite small (3 to 5 kDa) and form voltage-dependent ion channels in plasma membranes (92, 93, 171). Thionins are also small (3 to 5 kDa) cysteine-rich peptides that are toxic to fungi (171). Neither the mammalian defensins, insect defensins, nor thionins will be described in this review, for they are generally smaller than 5 kDa.
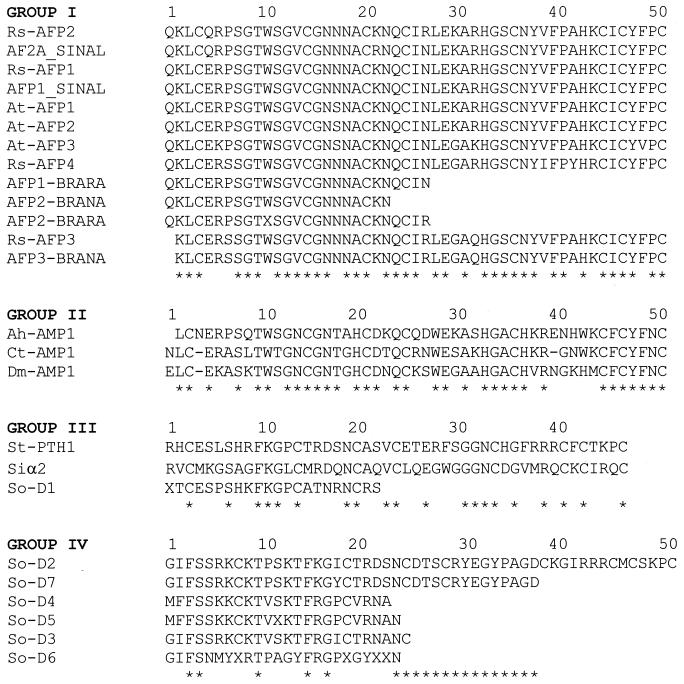
Plant and fungal defensins are cysteine-rich proteins ranging from 45 to 54 amino acids, are positively charged, and in most cases contain four disulfide bonds that stabilize each protein in solution (4, 5, 38, 49, 87, 88, 106, 110, 135, 155, 168, 169, 170, 181). In addition, most defensins are highly oligomeric (many subunits of 4 to 5 kDa) in situ (168, 169). Defensins are classified into four groups. Group I defensins cause morphological changes in susceptible fungi and are known as morphogenic defensins, group II proteins inhibit fungal growth but do not cause morphological changes (nonmorphogenic group), group III are inactive against test fungi but inhibit α-amylases in vitro, and group IV are unique in terms of antifungal specificity and structure (155). An amino acid alignment of a number of plant and fungal defensins is shown in Fig. 4. Note the high degree of similarity within each group. In addition, the positions of the cysteine residues are conserved in group I, II, and III proteins. No other significant homology exists between groups.
FIG. 4.
Amino acid sequence alignment of selected group I to IV defensins. Rs-AFP 1 to 4 are from Raphanus sativas (radish, accession no. P30225, P30230, 024332, and 024331), At-AFP1 to 3 are from Arabidopsis thaliana (thale cress, accession no. P30224, 080995, and 080994), AFP2-BRANA (rape, accession no. P30226) and AFP3-BRANA (rape, accession no. Q39313) are from Brassica napus, AFP1-BRARA (accession no. P30227) and AFP2-BRARA (accession no. P30228) are from B. rapa, AFP1_SINAL (white mustard, accession no. P30231) and AF2A_SINAL (white mustard, accession no. P30232) are from Sinapis alba, Ct-AMP1 (accession no. S66219) is from Clitoria ternate, Ah-AMP1 (common horse chestnut, accession no. S66218) is from Aesculus hippocastanum, Dm-AMP1 (bedding dahlia seeds, accession no. AAB34972) is from Dahlia merckii, St-PTH1 (potato tubers, accession no. AAB 31351) is from Solanum tuberosum, Siα2 (sorghum, accession no. P21924) is from Sorghum bicolor, and So-D1- to -7 (spinach) are from Spinacia oleracea. Alignments were done with the ClustalW program;∗ indicates amino acid identity.
In contrast to mammalian and insect defensins, plant defensins do not form channels either in artificial bilayers or in artificial liposomes (38), and they do not show significant hyphal permeabilization activity (171). In studies using N. crassa, Theviseen and coworkers have shown that treatment of hyphae with the defensins from radish (Rs-AFP2) or dahlia (DM-AMP1) caused K+ efflux and Ca2+ uptake through binding to specific cell membrane receptors (171–174). Although not tested with other fungi, it is likely that fungal inhibition occurs through this mechanism, i.e., ion efflux. Defensins are broadly active, inhibiting a large number of human and plant fungal pathogens, including B. cinerea, Alternaria brassicola, F. culmorum, F. oxysporum, F. solani, and C. albicans at micromolar levels.
Cyclophilin-like protein.
Cyclophilins are a highly conserved group of proteins that are the intracellular receptors for cyclosporin; they have been found in a wide variety of organisms, including bacteria, plants, animals, and fungi (137). Recently an 18-kDa protein was isolated from mung bean (Phaseolus mungo) with activity against R. solani, F. oxysporum, B. cinerea, and Coprinus comatus (199). This protein, called mungin, showed significant homology to cyclophilins and inhibited α- and β-glucosidases in vitro. However, the antifungal mechanism of action of mungin is not known.
Glycine/histidine-rich proteins.
Insects synthesize a number of glycine/histidine-rich antifungal proteins and polypeptides, including those from Holotrichia diomphalia larvae (holotrichin, 84 amino acids [97]), Sarcophaga peregrina (flesh fly, AFP, 67 amino acids [68]), and Tenebrio molitor (tenecin, 49 amino acids [28, 96, 98, 99]). An alignment of these proteins is shown in Fig. 5. Note that they are extremely rich in glycine and histidine, which comprise as much as 80% of the amino acids. Importantly, fungi inhibited included C. albicans, the most common human pathogen (e.g., the 50% inhibitory concentration of tenecin is ∼8 μg/ml [28]). The mechanism of action of these proteins is not understood.
FIG. 5.
Amino acid sequence alignment of selected glycine/histidine-rich proteins. Tenecin 3 (yellow mealworm, accession no. AAA97579.1) is from Tenebrio molitor, AFP (flesh fly, accession no. BAA02954.1) is from Sarcophaga peregrina, and holotricin 3 (accession no. BAA02889) is from Holotrichia diomphalia. Alignments were done with the ClustalW program, ∗ indicates amino acid identity.
RIPs.
RIPs are RNA N-glycosidases that depurinate rRNA, resulting in the arrest of protein synthesis due to ribosome damage (7, 39, 65, 94, 144, 167). Plant RIPs inhibit mammalian, bacterial, fungal, and plant protein syntheses in vitro and in vivo (67). As an aside, how plants protect themselves from the action of their own RIPs is a subject of very interesting research. RIPs have been classified into three groups. Type 1 RIPs are single-chain N-glycosidases with molecular masses of 11 to 30 kDa. Type 2 RIPs contain two chains, a cell-binding lectin (B chain) and an N-glycosidase (A chain), with molecular masses of ∼60 kDa (202); type 2 RIPs include toxic members such as ricin and nontoxic members such as ebulin 1 (44) and nigrin b. Type 3 RIPs consist of four chains organized as two dimers of type 2 RIPs. RIPs have been isolated from a number of plants (Mirabilis expansa [183], Pisum sativum [90, 197] Momordica charantia [100], Ricinus communis [6], Viscum album, and many others [35, 50, 102, 138, 178, 185, 190, 195]) as well as from fungi, e.g., Aspergillus giganteus (α-sarcin [51, 188]) Unfortunately, the antifungal activities of only a few of the many RIPs have been described.
RIPs have intrinsic antifungal activity due to their ability to inactive fungal ribosomes in vitro and, presumably, in situ. Recent studies with a type 2 RIP showed that the cell-binding B chain (lectin) binds to fungal cells, forming a channel allowing the N-glycosidase A-chain entry into cells, resulting in RNA damage (191, 202). Precisely how type I RIPs which do not have a cell-binding chain inhibit fungi, i.e., how are they internalized, is not known. Both type I and type 2 RIPs show broad activity against a number of plant and human pathogenic fungi as well as toxicity against mammalian cells (some type 2 RIPs are highly toxic to animals, likely because of the presence of the cell-binding B chain) (132, 142, 146).
LTPs.
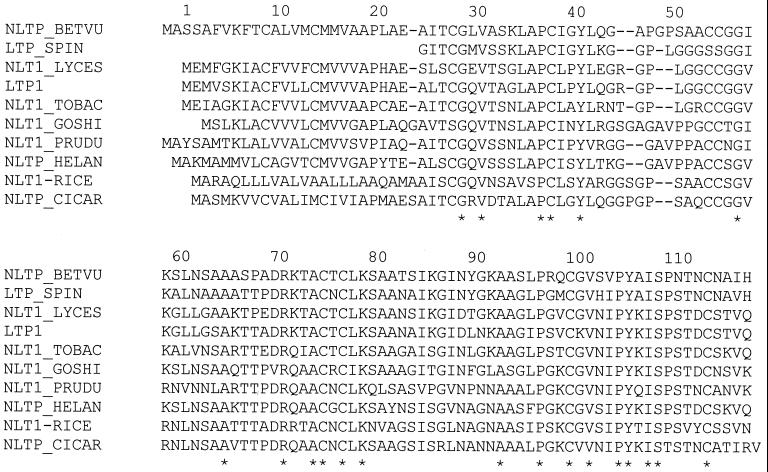
LTPs have the ability to transfer phospholipids between membranes. LTPs are small proteins (∼8.7 kDa) of ∼90 amino acids stabilized by four disulfide bonds with a central tunnel-like hydrophobic cavity. They have been isolated from a number of sources, including mammals, plants, fungi, and bacteria (17, 26, 115, 116, 130, 154, 166, 175), and may play several in vivo roles, including lipid exchange between cytoplasmic organelles and, importantly, defense against pathogens (48). An alignment of a number of LTPs is shown in Fig. 6. Note that although the proteins are from diverse sources, they have striking homologies (between 37 to 90% identity). LTPs are active in vitro against a number of bacteria and fungi; however, the mechanism of action is not known. It may be that these proteins insert themselves into the fungal cell membrane, and the central hydrophobic cavity forms a pore, allowing efflux of intracellular ions and thus leading to fungal cell death. How this is related to their lipid transfer function is not clear.
FIG. 6.
Amino acid sequence alignment of selected LTPs. NLTP_BETVU (beet, accession no. Q43748) is from Beta vulgaris, LTP_SPIN (common spinach, accession no. S00060) is from Spinacia oleracea, NLT1_LYCES (tomato, accession no. P93224) is from Lycopersicon esculentum, LTP1 (tomato, accession no. AAB07486) is from L. pennellii, NLT1_TOBAC (common tobacco, accession Q42952) is from Nicotiana tabacum, NLT1_GOSHI (upland cotton, accession no. 42762) is from Gossypium hirsutum, NLT1_PRUDU (almond, accession no. Q43017) is from Prunus dulcis, NLTP_HELAN (common sunflower, accession no. Q39950) is from Helianthus annuus, NLT1-RICE (rice, accession no. T02038) is from Oryza sativa, and NLTP_CICAR (chickpea, accession no. 023758) is from Cicer arietinum. Alignments were done with the ClustalW program; ∗ indicates amino acid identity.
Killer proteins (killer toxins).
A number of yeasts secrete proteins that are lethal to sensitive fungal cells. These proteins, called killer proteins or killer toxins, are encoded either by double-stranded RNA, linear double-stranded plasmid DNA, or nuclear genes (2, 23, 70, 108, 153). Fungal cells secreting a killer toxin are resistant to their own toxin but are sensitive to other toxins. Saccharomyces cerevisiae, Ustilago maydis, Hanseniaspora uvarum, Zygosaccharomyces bailii, Phaffia rhodozyma, Kluveromyces lactis, and several Pichia species secrete a number of killer proteins (reviewed in reference 108). Over 20 individual killer toxins have been identified, varying in molecular mass from 10.7 to 156.5 kDa (58, 84). The killer toxins have broad, potent antifungal activity against a number of human and plant pathogens (including Pneumocystis carinii [157])—MICs vary from 20 μg/ml to far less. Although they have varied mechanisms of action, the first step of killer protein activity involves binding of the protein to specific cell surface receptors. Once bound, killer proteins are internalized and can disrupt cell wall synthesis, DNA synthesis, and K+ channel activity, inhibit (1,3)β-glucan synthesis, or arrest the cell cycle (2, 36, 78, 79, 164). Any one of these effects leads to inhibition of fungal growth and to fungal cell death.
Protease inhibitors.
Protein inhibitors of serine (e.g., trypsin and chymotrypsin) and cysteine proteases have emerged as a class of antifungal proteins that have potent activity against plant and animal pathogens. Cysteine protease inhibitors have been isolated from a number of plants and form a fourth group of cystatins, the phytocystatins (10, 72, 86, 139). The phytocystatins are single polypeptides of 10 to 12 kDa and share common structural motifs. Although phytocystatins are active against plant pathogens such as F. solani (MIC of 20 μg/disk in an disk agar diffusion assay) and Trichoderma reesei (250 ng/disk) (72), the mechanism of antifungal activity is not understood.
Serine protease inhibitors that have antifungal activity also have the interesting property of inhibiting α-amylase activity from insects but not from bacterial or mammalian sources (152a). These proteins are bifunctional, inhibiting enzymes as well as inhibiting insect and fungal growth. Blanco-Labra and Iturbe-Chinas identified a bifunctional α-amlyase/trypsin inhibitor from corn (10); later it was found that this protein was identical to zeamatin (147, 148). We have recently confirmed that at high trypsin/zeamatin and α-amylase/zeamatin molar ratios, zeamatin inhibits trypsin and insect α-amylase activities in vitro (152a). Other bifunctional proteins from ragi (Eleusine coracana), wheat, and barley have been isolated and characterized (9, 46, 133, 152, 160). Only a few of these proteins have been tested for in vitro antifungal activity, with zeamatin being the most extensively characterized. The mechanism of antifungal activity of these proteins is not fully understood.
Other proteins.
New proteins that have antifungal activity but do not neatly fall into any of the above classes are being discovered at a rapid pace. Only a few can be mentioned here. Viridin, a novel protein isolated from the culture medium of Trichoderma viride, has a molecular mass of 65 kDa and is active against sensitive fungi at 6 μM (52, 53). Snakin-1 isolated from potato has a molecular mass of 6.9 kDa and is active at 10 μM (156). A 30-kDa protein with very potent antifungal activity (50 ng/disk in an agar diffusion assay) was isolated from Engelmann's daisy (Engelmannia pinnatifida); this protein showed 35 to 50% identity to self-incompatibility glycoproteins, not previously known to be antifungal (64). The mechanism of action of none of these proteins is known.
CONCLUSIONS
Antifungal proteins and polypeptides have been isolated from diverse groups of organisms, including plants, fungi, bacteria, insects, and animals (both vertebrates and invertebrates). The mechanisms of action of these proteins are as varied as their sources and include fungal cell wall polymer degradation, membrane channel and pore formation, damage to cellular ribosomes, inhibition of DNA synthesis, and inhibition of the cell cycle. The mode of action of many proteins remains unknown and is the subject of active research. The range of fungi inhibited by antifungal proteins is extremely broad, with plant pathogens and humans pathogens being sensitive at micromolar levels; in some cases, even more potent inhibition was found.
The genes encoding many antifungal proteins are currently being used by agribusiness to create genetically modified plants that have increased fungal resistance in the field. Whether these transgenic plants and the crops derived from them gain acceptance in the marketplace remains to be seen. Equally important, antifungal proteins and peptides are being tested for use as pharmaceutical agents for the treatment of human and animal fungal diseases. This is particularly exciting since the modes of action of these proteins are vastly different from the currently used therapeutics, resistance to which is becoming a clinical problem. There are a number of antifungal proteins in various stages of preclinical development, and the results of these experiments and of the subsequent human clinical trials are eagerly anticipated.
ACKNOWLEDGMENTS
This review could not have been completed without the much needed assistance of Samatha Renault, Rebecca Schimoler-O'Rourke, Shelly Wilson, and Tamara Kay Miller.
This work was supported by institutional funds from MycoLogics, Inc.
REFERENCES
- 1.Agrawal G K, Jwa N S, Rakwal R. A novel rice (Oryza sativa L.) acidic PR1 gene highly responsive to cut, phytohormones, and protein phosphatase inhibitors. Biochem Biophys Res Commun. 2000;274:157–165. doi: 10.1006/bbrc.2000.3114. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed A, Sesti F, Ilan N, Shih T M, Sturley S L, Goldstein S A. A molecular target for viral killer toxin: TOK1 potassium channels. Cell. 1999;99:283–291. doi: 10.1016/s0092-8674(00)81659-1. [DOI] [PubMed] [Google Scholar]
- 3.Ainsworth G, editor. The fungi, an advance treatise. New York, N.Y: Academic Press; 1973. [Google Scholar]
- 4.Almeida M S, Cabral K M, Zingali R B, Kurtenbach E. Characterization of two novel defense peptides from pea (Pisum sativum) seeds. Arch Biochem Biophys. 2000;378:278–286. doi: 10.1006/abbi.2000.1824. [DOI] [PubMed] [Google Scholar]
- 5.Alves A L, De Samblanx G W, Terras R R, Cammue B P, Broekaert W F. Expression of functional Raphanus sativusantifungal protein in yeast. FEBS Lett. 1994;348:228–232. doi: 10.1016/0014-5793(94)00597-4. [DOI] [PubMed] [Google Scholar]
- 6.Arias F J, Rojo M A, Ferreras J M, Iglesias R, Munoz R, Soriano F, Mendez E, Barbieri L, Girbes T. Isolation and characterization of two new N-glycosidase type-1 ribosome-inactivating proteins, unrelated in amino-acid sequence, from Petrocoptisspecies. Planta. 1994;194:487–491. doi: 10.1007/BF00714460. [DOI] [PubMed] [Google Scholar]
- 7.Barbieri L, Batelli M G, Stirpe F. Ribosome-inactivating proteins from plants. Biochim Biophys Acta. 1993;1154:237–282. doi: 10.1016/0304-4157(93)90002-6. [DOI] [PubMed] [Google Scholar]
- 8.Beffa R, Meins F., Jr Pathogenesis-related function of plant β-1,3-glucanases investigated by antisense transformation—a review. Gene. 1996;179:97–103. doi: 10.1016/s0378-1119(96)00421-0. [DOI] [PubMed] [Google Scholar]
- 9.Behnke C A, Yee V C, Trong I L, Pedersen L C, Stenkamp R E, Kim S S, Reeck G R, Teller D C. Structural determinants of the bifunctional corn Hageman factor inhibitor: x-ray structure at 1.95 Åresolution. Biochemistry. 1998;37:15277–15288. doi: 10.1021/bi9812266. [DOI] [PubMed] [Google Scholar]
- 10.Blanco-Labra A, Iturbe-Chinas F A. Purification and characterization of an α-amylase inhibitor from maise (Zea maize) J Food Biochem. 1980;5:1–17. [Google Scholar]
- 11.Bloch C, Jr, Patel S U, Baud F, Zvelebil M J, Carr M D, Sadler P J, Thornton J M. 1H NMR structure of an antifungal γ-thionin protein SIα1: similarity to scorpion toxins. Proteins. 1998;32:334–349. doi: 10.1002/(sici)1097-0134(19980815)32:3<334::aid-prot9>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 12.Borgmeyer J R, Smith C E, Huynh Q K. Isolation and characterization of a 25 kDa antifungal protein from flax seeds. Biochem Biophys Res Commun. 1992;187:480–487. doi: 10.1016/s0006-291x(05)81519-0. [DOI] [PubMed] [Google Scholar]
- 13.Bormann C, Baier D, Horr I, Raps C, Berger J, Jung G, Schwartz H. Characterization of a novel, antifungal, chitin-binding protein Streptomyces tendaeTu901 that interferes with growth polarity. J Bacteriol. 1999;181:7421–7429. doi: 10.1128/jb.181.24.7421-7429.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broekaert W F, Terras F R, Cammue B P, Osborn R W. Plant defensins: novel antimicrobial peptides as components of host defense system. Plant Physiol. 1995;108:1353–1358. doi: 10.1104/pp.108.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bryngelsson T, Sommer-Knudsen J, Gregersen P L, Collinge D B, Ek B, Thordal-Christensen H. Purification, characterization, and molecular cloning of basic PR-1-type pathogenesis-related proteins from barley. Mol Plant Microbe Interact. 1994;7:267–275. doi: 10.1094/mpmi-7-0267. [DOI] [PubMed] [Google Scholar]
- 16.Bull J, Mauch F, Hertig C, Regmann G, Dudler R. Sequence and expression of a wheat gene that encodes a novel protein associated with pathogen defense. Mol Plant Microbe Interact. 1992;5:516–519. doi: 10.1094/mpmi-5-516. [DOI] [PubMed] [Google Scholar]
- 17.Cammue B P, Thevissen K, Hendriks M, Eggermont K, Goderis I J, Proost P, Van Damme J, Osborn R W, Guerbette F, Kader J C. A potent antimicrobial protein from onion seeds showing sequence homology to plant lipid transfer proteins. Plant Physiol. 1995;109:445–455. doi: 10.1104/pp.109.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caruso C, Caporale C, Chilosi G, Vacca F, Bertini L, Magro P, Poerio E, Buonocore V. Structural and antifungal properties of a pathogenesis-related protein from wheat kernel. J Protein Chem. 1996;15:35–44. doi: 10.1007/BF01886809. [DOI] [PubMed] [Google Scholar]
- 19.Chae K S, Lee I H, Choi C S, Kim H R. Purification and characterization of chitin-binding proteins from the hemolymph of sweet potato hornworm, Agrius convolvuli. Comp Biochem Physiol B. 1999;124:475–481. doi: 10.1016/s0305-0491(99)00143-1. [DOI] [PubMed] [Google Scholar]
- 20.Charlet M, Chernysh S, Philippe H, Hetru C, Hoffmann J A, Bulet P. Innate immunity. Isolation of several cysteine-rich antimicrobial peptides from the blood of a mollusk, Mytilus edulis. J Biol Chem. 1996;271:21808–21813. doi: 10.1074/jbc.271.36.21808. [DOI] [PubMed] [Google Scholar]
- 21.Cheong N E, Choi Y O, Kim W Y, Bae I S, Cho M J, Hwang I, Kim J W, Lee S Y. Purification and characterization of an antifungal PR-5 protein from pumpkin leaves. Mol Cells. 1997;7:214–219. [PubMed] [Google Scholar]
- 22.Chernin L S, De la Fuente L, Sobolev V, Haran S, Vorgias C E, Oppenheim A B, Chet I. Molecular cloning, structural analysis, and expression in Escherichia coli of a chitinase gene from Enterobacter agglomerans. Appl Environ Microbiol. 1997;63:834–839. doi: 10.1128/aem.63.3.834-839.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clausen M, Krauter R, Schachermayr G, Potrykus I, Sautter C. Antifungal activity of a virally encoded gene in transgenic wheat. Nat Biotechnol. 1999;18:446–449. doi: 10.1038/74521. [DOI] [PubMed] [Google Scholar]
- 24.Coca M A, Damsz B, Yun D J, Hasegawa P M, Bressan R A, Narasimhan M L. Heterotrimeric G-proteins of a filamentous fungus regulate cell wall composition and susceptibility to a plant PR-5 protein. Plant J. 2000;22:61–69. doi: 10.1046/j.1365-313x.2000.00718.x. [DOI] [PubMed] [Google Scholar]
- 25.Cote F, Cutt J R, Asselin A, Klessig D F. Pathogenesis-related acidic β-1,3-glucanase genes of tobacco are regulated by both stress and developmental signals. Mol Plant Microbe Interact. 1991;4:173–181. doi: 10.1094/mpmi-4-173. [DOI] [PubMed] [Google Scholar]
- 26.Coutos-Thevenot P, Jounenne T, Maes O, Guerbette F, Grosbois M, Le Caer J P, Boulay M, Deloire A, Kader J C, Guern J. Four 9-kDa proteins excreted by somatic embryos of grapevine are isoforms of lipid-transfer proteins. Eur J Biochem. 1993;217:885–889. doi: 10.1111/j.1432-1033.1993.tb18317.x. [DOI] [PubMed] [Google Scholar]
- 27.Cox G, Perfect J. Fungal infections. Curr Opin Infect Dis. 1993;6:422–426. [Google Scholar]
- 28.Dae-Hee K, Lee Y T, Lee Y J, Chung J H, Lee B L, Choi B S, Younghoon L. Bacterial expression of tenecin 3, and insect antifungal protein isolated from Tenebrio molitor, and its efficient purification. Mol Cells. 1998;8:786–789. [PubMed] [Google Scholar]
- 29.De Bolle M F, David K M, Rees S B, Vanderleyden J, Cammue B P, Broekaert W F. Cloning and characterization of a cDNA encoding an antimicrobial chitin-binding protein from amaranth, Amaranthus caudatus. Plant Mol Biol. 1993;22:1187–1190. doi: 10.1007/BF00028991. [DOI] [PubMed] [Google Scholar]
- 30.De Lucca A J, Walsh T J. Antifungal peptides: novel therapeutic compounds against emerging pathogens. Antimicrob Agents Chemother. 1999;43:1–11. doi: 10.1128/aac.43.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Destoumieux D, Bulet P, Loew D, Van Dorsselaer A, Rodriguez J, Bachere E. Penaeidins, a new family of antimicrobial peptides isolated from the shrimp Penaeus vannamei (Decapoda) J Biol Chem. 1997;272:28398–28406. doi: 10.1074/jbc.272.45.28398. [DOI] [PubMed] [Google Scholar]
- 32.Destoumieux D, Munoz M, Bulet P, Bachere E. Penaeidins, a family of antimicrobial peptides from penaeid shrimp (Crustacea, Decapoda) Cell Mol Life Sci. 2000;57:1260–1271. doi: 10.1007/PL00000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Destoumieux D, Munoz M, Cosseau C, Rodriguez J, Bulet P, Comps M, Bachere E. Penaeidins, antimicrobial peptides with chitin-binding activity, are produced and stored in shrimp granulocytes and released after microbial challenge. J Cell Sci. 2000;113:461–469. doi: 10.1242/jcs.113.3.461. [DOI] [PubMed] [Google Scholar]
- 34.Does M P, Houterman P M, Dekker H L, Cornelissen B J. Processing, targeting, and antifungal activity of stinging nettle agglutinin in transgenic tobacco. Plant Physiol. 1999;120:421–432. doi: 10.1104/pp.120.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong T X, Ng T B, Yeung H W, Wong R N. Isolation and characterization of a novel ribosome-inactivating protein, β-kirilowin, from the seeds of Trichosanthes kirilowii. Biochem Biophys Res Commun. 1994;199:387–393. doi: 10.1006/bbrc.1994.1241. [DOI] [PubMed] [Google Scholar]
- 36.Eisfeld K, Riffer F, Mentges J, Schmitt M J. Endocytotic uptake and retrograde transport of a virally encoded killer toxin in yeast. Mol Microbiol. 2000;37:926–940. doi: 10.1046/j.1365-2958.2000.02063.x. [DOI] [PubMed] [Google Scholar]
- 37.Esake M, Teramoto T. cDNA cloning, gene expression and secretion of chitinase in winged bean. Plant Cell Physiol. 1998;39:349–356. doi: 10.1093/oxfordjournals.pcp.a029376. [DOI] [PubMed] [Google Scholar]
- 38.Fant F, Vranken W, Broekaert W, Borremans F. Determination of the three-dimensional solution structure of Raphanus sativus antifungal protein 1 by 1H NMR. J Mol Biol. 1998;279:257–270. doi: 10.1006/jmbi.1998.1767. [DOI] [PubMed] [Google Scholar]
- 39.Ferreras J M, Iglesias R, Barbieri L, Alegre C, Bolognesi A, Rojo M A, Carbajales M L, Escarmis C, Girbes T. Effects and molecular action of ribosome-inactivating proteins on ribosomes from Streptomyces lividans. Biochim Biophys Acta. 1995;1243:85–93. [PubMed] [Google Scholar]
- 40.Florach D E, Stiekema W J. Thionins: properties, possible biological roles and mechanisms of action. Plant Mol Biol. 1994;26:25–37. doi: 10.1007/BF00039517. [DOI] [PubMed] [Google Scholar]
- 41.Fox J L. Fungal infection rates are increasing. ASM News. 1993;59:515–518. [Google Scholar]
- 42.Friedrich L, Moyer M, Ward E, Ryals J. Pathogenesis-related protein 4 is structurally homologous to the carboxy-terminal domains of hevein, Win-1 and Win-2. Mol Gen Genet. 1991;230:113–119. doi: 10.1007/BF00290658. [DOI] [PubMed] [Google Scholar]
- 43.Galgiani J, Bartlett M, Ghannoum M, Espinel-Ingroff A, Lancaster A, Odds F. Reference method for broth dilution antifungal susceptibility testing of yeasts: tentative standard. NCCLS document M27-T. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1995. [Google Scholar]
- 44.Girbes T, Citores L, Iglesias R, Ferreras J M, Munoz R, Rojo M A, Arias F J, Garcia J R, Mendez E, Calonge M. Ebulin 1, a nontoxic novel type 2 ribosome-inactivating protein from Sambucus ebulusL. leaves. J Biol Chem. 1993;268:18195–18199. [PubMed] [Google Scholar]
- 45.Gooday G, Gow N. Enzymology of tip growth in fungi. In: Health I B, editor. Tip growth in plant and fungal cells. San Diego, Calif: Academic Press; 1990. pp. 31–58. [Google Scholar]
- 46.Gourinath S, Alam N, Srinivasan A, Betzel C, Singh T P. Structure of the bifunctional inhibitor of trypsin and α-amylase from ragi seeds at 2.2 Åresolution. Acta Crystallogr D. 2000;56:287–293. doi: 10.1107/s0907444999016601. [DOI] [PubMed] [Google Scholar]
- 47.Grenier J, Potvin C, Asselin A. Barley pathogenesis-related proteins with fungal cell wall lytic activity inhibit the growth of yeasts. Plant Physiol. 1993;103:1277–1283. doi: 10.1104/pp.103.4.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guerbette F, Grosbois M, Jolliot-Croquin A, Kader J C, Zachowski A. Lipid-transfer proteins from plants: structure and binding properties. Mol Cell Biochem. 1999;192:157–161. [PubMed] [Google Scholar]
- 49.Gun Lee D, Shin S Y, Maeng C Y, Jin Z Z, Kim K L, Hahm K S. Isolation and characterization of a novel antifungal peptide from Aspergillus niger. Biochem Biophys Res Commun. 1999;263:646–651. doi: 10.1006/bbrc.1999.1428. [DOI] [PubMed] [Google Scholar]
- 50.Guo B Z, Cleveland T E, Brown R L, Widstrom N W, Lynch R E, Russin J S. Distribution of antifungal proteins in maize kernel tissues using immunochemistry. J Food Prot. 1999;62:295–299. doi: 10.4315/0362-028x-62.3.295. [DOI] [PubMed] [Google Scholar]
- 51.Hao J J, Xu Y Z, Geng C D, Liu W Y, Wang Ed, Gong Z Z, Ulbrich N. Purification of α-sarcin and an antifungal protein from Aspergillus giganteusby blue sepharose CL-6B affinity chromatography. Protein Expr Purif. 1998;14:295–301. doi: 10.1006/prep.1998.0957. [DOI] [PubMed] [Google Scholar]
- 52.Hao J J, Geng C, Xie W, Gong Z, Liu W Y, Wang E. Isolation and characterization of viridin, a new 65 kDa antifungal protein from the mould Trichoderma viride. Biol Chem. 1999;380:1243–1245. doi: 10.1515/BC.1999.158. [DOI] [PubMed] [Google Scholar]
- 53.Hao J J, Ye J Q, Yang Q, Gong Z Z, Liu W Y, Wang E. A silent antifungal protein (AFP)-like gene lacking two introns in the mould Trichoderma viride. Biochim Biophys Acta. 2000;1475:119–124. doi: 10.1016/s0304-4165(00)00047-7. [DOI] [PubMed] [Google Scholar]
- 54.Harold F, Caldwell J. Tips and currents: electrobiology of Aprical growth. In: Health I, editor. Tip growth in plant and fungal cells. San Diego, Calif: Academic Press; 1990. pp. 59–90. [Google Scholar]
- 55.Hejgaard J, Jacobsen S, Svendsen I. Two antifungal thaumatin-like proteins from barley grain. FEBS Lett. 1991;291:127–131. doi: 10.1016/0014-5793(91)81119-s. [DOI] [PubMed] [Google Scholar]
- 56.Hejgaard J, Jacobsen S, Bjorn S E, Kragh K M. Antifungal activity of chitin-binding PR-4 type proteins from barley grain and stressed leaf. FEBS Lett. 1992;307:389–392. doi: 10.1016/0014-5793(92)80720-2. [DOI] [PubMed] [Google Scholar]
- 57.Herbrecht R. The changing epidemiology of fungal infections: are the lipid-based forms of amphotericin B an advance? Eur J Haematol. 1996;56:12–17. doi: 10.1111/j.1600-0609.1996.tb01347.x. [DOI] [PubMed] [Google Scholar]
- 58.Hiraga K, Hayashi S, Kitazawa M, Oda K. Isolation and some properties of a novel killer toxin-like protein produced by Streptomycessp. F-287. Biosci Biotechnol Biochem. 1999;63:1037–1044. doi: 10.1271/bbb.63.1037. [DOI] [PubMed] [Google Scholar]
- 59.Hu X, Reddy A S. Nucleotide sequence of a cDNA clone encoding a thaumatin-like protein from Arabidopsis. Plant Physiol. 1995;107:305–306. doi: 10.1104/pp.107.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hu X, Reddy A S. Cloning and expression of a PR5-like protein from Arabidopsis: inhibition of fungal growth by bacterially expressed protein. Plant Mol Biol. 1997;34:949–959. doi: 10.1023/a:1005893119263. [DOI] [PubMed] [Google Scholar]
- 61.Huang X, Xie W, Gong Z. Characteristics and antifungal activity of a chitin binding protein from Ginkgo biloba. FEBS Lett. 2000;478:123–126. doi: 10.1016/s0014-5793(00)01834-2. [DOI] [PubMed] [Google Scholar]
- 62.Huynh Q K, Borgmeyer J R, Zobel J F. Isolation and characterization of a 22 kDa protein with antifungal properties from maize seeds. Biochem Biophys Res Commun. 1992;182:1–5. doi: 10.1016/s0006-291x(05)80103-2. [DOI] [PubMed] [Google Scholar]
- 63.Huynh Q K, Hironaka C M, Levine E B, Smith C E, Borgmeyer J R, Shah D M. Antifungal proteins from plants. Purification, molecular cloning, and antifungal properties of chitinases from maize seed. J Biol Chem. 1992;267:6635–6640. [PubMed] [Google Scholar]
- 64.Huynh Q K, Borgmeyer J R, Smith C E, Bell L D, Shah D M. Isolation and characterization of a 30 kDa protein with antifungal activity from leaves of Engelmannia pinnatifida. Biochem J. 1996;15:723–727. doi: 10.1042/bj3160723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hwu L, Huang C C, Chen D T, Lin A. The action mode of the ribosome-inactivating protein α-sarcin. J Biomed Sci. 2000;7:420–428. doi: 10.1007/BF02255817. [DOI] [PubMed] [Google Scholar]
- 66.Ibeas J I, Lee H, Damsz B, Prasad D T, Pardo J M, Hasegawa P M, Bressan R A, Narasimhan M L. Fungal cell wall phosphomannans facilitate the toxic activity of a plant PR-5 protein. Plant J. 2000;23:375–383. doi: 10.1046/j.1365-313x.2000.00792.x. [DOI] [PubMed] [Google Scholar]
- 67.Iglesias R, Arias F J, Rojo M A, Escarmis C, Ferreras J M, Girbes T. Molecular action of the type 1 ribosome-inactivating protein saporin 5 on Vicia sativaribosomes. FEBS Lett. 1993;325:291–294. doi: 10.1016/0014-5793(93)81091-d. [DOI] [PubMed] [Google Scholar]
- 68.Iijima R, Kurata S, Natori S. Purification, characterization, and cDNA cloning of an antifungal protein from the hemolymph of Sarcophaga peregrina(flesh fly) larvae. J Biol Chem. 1993;268:12055–12061. [PubMed] [Google Scholar]
- 69.Iijima R, Kisugi J, Yamazaki M. Biopolymers from marine invertebrates. XIV. Antifungal property of Dolabellanin A, a putative self-defense molecule of the sea hare, Dolabella auricularia. Biol Pharm Bull. 1994;17:1144–1146. doi: 10.1248/bpb.17.1144. [DOI] [PubMed] [Google Scholar]
- 70.Izgu F, Altinbay D, Sagiroglu A K. Isolation and characterization of the K6 type yeast killer protein. Microbios. 1999;99:161–172. [PubMed] [Google Scholar]
- 71.Jach G, Gornhardt B, Mundy J, Logemann J, Pinsdorf E, Leah R, Schell J, Maas C. Enhanced quantitative resistance against fungal disease by combinatorial expression of different barley antifungal proteins in transgenic tobacco. Plant J. 1995;8:97–109. doi: 10.1046/j.1365-313x.1995.08010097.x. [DOI] [PubMed] [Google Scholar]
- 72.Joshi B N, Sainani M N, Bastawade K B, Gupta V S, Ranjekar P K. Cysteine protease inhibitor from pearl millet: a new class of antifungal protein. Biochem Biophys Res Commun. 1998;246:382–387. doi: 10.1006/bbrc.1998.8625. [DOI] [PubMed] [Google Scholar]
- 73.Kaminskyj S, Garrill A, Heath B. The relationship between turgor and tip growth in Saprolegnia ferax: turgor is necessary, but not sufficient to explain apical extension rates. Exp Mycol. 1992;16:64–75. [Google Scholar]
- 74.Kang S C, Park S, Lee D G. Purification and characterization of a novel chitinase from the entomopathogenic fungus, Metarhizium anisopliae. J Invert Pathol. 1999;73:276–281. doi: 10.1006/jipa.1999.4843. [DOI] [PubMed] [Google Scholar]
- 75.Kawabata S, Nagayama R, Hirata M, Shigenaga T, Agarwala S L, Saito T, Cho J, Nakajima H, Takagi T, Iwanaga S. Tachycitin, a small granular component in horseshoe crab hemocytes, is an antimicrobial protein with chitin-binding activity. J Biochem. 1996;120:1253–1260. doi: 10.1093/oxfordjournals.jbchem.a021549. [DOI] [PubMed] [Google Scholar]
- 76.Khardori N. Host-parasite interaction in fungal infections. Eur J Clin Microbiol Infect Dis. 1989;8:331–351. doi: 10.1007/BF01963468. [DOI] [PubMed] [Google Scholar]
- 77.Kim Y J, Hwang B K. Isolation of a basic 34 kilo Dalton β-1,3-glucanase with inhibitory activity against Phytophthora capsicifrom pepper stems. Physiol Mol Plant Pathol. 1997;50:103–115. [Google Scholar]
- 78.Kimura T, Kitamoto N, Kito Y, Iimura Y, Shirai T, Komiyama T, Furuichi Y, Ohmiya K. A novel yeast gene, RHK1, is involved in the synthesis of the cell wall receptor for the HM-1 killer toxin that inhibits β-1,3-glucan synthesis. Mol Gen Genet. 1997;254:139–147. doi: 10.1007/s004380050401. [DOI] [PubMed] [Google Scholar]
- 79.Kimura T, Komiyama T, Furuichi Y, Iimura Y, Karita S, Ohmiya K. N-glycosylation is involved inthe sensitivity of Saccharomyces cerevisiae to HM-1 killer toxin secreted from Hansenula mrakiiIFO 0895. Appl Microbiol Biotechnol. 1999;51:176–184. doi: 10.1007/s002530051379. [DOI] [PubMed] [Google Scholar]
- 80.Kitajima S, Sato F. Plant pathogenesis-related proteins: molecular mechanisms of gene expression and protein function. J Biochem. 1999;125:1–8. doi: 10.1093/oxfordjournals.jbchem.a022244. [DOI] [PubMed] [Google Scholar]
- 81.Koiwa H, Kato H, Nakatsu T, Oda J, Yamada Y, Sato F. Purification and characterization of tobacco pathogenesis-related protein PR-5d, an antifungal thaumatin-like protein. Plant Cell Physiol. 1997;38:783–791. doi: 10.1093/oxfordjournals.pcp.a029236. [DOI] [PubMed] [Google Scholar]
- 82.Koiwa H, Kato H, Nakatsu T, Oda J, Yamada Y, Sato F. Crystal structure of tobacco PR-5d protein at 1.8 Åresolution reveals a conserved acidic cleft structure in antifungal thaumatin-like proteins. J Mol Biol. 1998;286:1137–1145. doi: 10.1006/jmbi.1998.2540. [DOI] [PubMed] [Google Scholar]
- 83.Kolbe S, Fischer S, Becirevic A, Hinz P, Schrempf H. The Streptomyces reticulialpha-chitin-binding protein CHB2 and its gene. Microbiology. 1998;144:1291–1297. doi: 10.1099/00221287-144-5-1291. [DOI] [PubMed] [Google Scholar]
- 84.Komiyama T, Shirai T, Ohta T, Urakami H, Furuichi Y, Ohta Y, Tsukada Y. Action properties of HYI killer toxin from Williopsis saturnus var. saturnusand antibiotics, aculeacin A and papulacandin B. Biol Pharm Bull. 1998;21:1013–1019. doi: 10.1248/bpb.21.1013. [DOI] [PubMed] [Google Scholar]
- 85.Koo J C, Lee S Y, Chun H J, Cheong Y H, Choi J S, Kawabata S, Miyagi M, Tsunasawa S, Ha K S, Bae D W, Han C D, Lee B L, Cho M J. Two hevein homologs isolated from the seed of Pharbitis nilL. exhibit potent antifungal activity. Biochim Biophys Acta. 1998;1382:80–90. doi: 10.1016/s0167-4838(97)00148-9. [DOI] [PubMed] [Google Scholar]
- 86.Kouzuma Y, Inanaga H, Doi-Kawano K, Yamasaki N, Kimura M. Molecular cloning and functional expression of cDNA encoding the cysteine proteinase inhibitor with three cystatin domains from sunflower seeds. J Biochem. 2000;128:161–166. doi: 10.1093/oxfordjournals.jbchem.a022736. [DOI] [PubMed] [Google Scholar]
- 87.Kragh K M, Nielsen J E, Nielsen K K, Dreboldt S, Mikkelsen J D. Characterization and localization of new antifungal cysteine-rich proteins from Beta vulgaris. Mol Plant Microbe Interact. 1995;8:424–434. doi: 10.1094/mpmi-8-0424. [DOI] [PubMed] [Google Scholar]
- 88.Kristensen A K, Brunsted J, Nielsen J W, Mikkelsen J D, Roepstorff P, Nielsen K K. Processing, disulfide pattern, and biological activity of a sugar beet defensin, AX2, expressed in Pichia pastoris. Protein Expr Purif. 1999;16:377–387. doi: 10.1006/prep.1999.1085. [DOI] [PubMed] [Google Scholar]
- 89.Lacadena J, Martinez del Poxo A, Gasset M, Patino B, Campos-Olivas R, Vazquez C, Martinez-Ruiz A, Mancheno J M, Onaderra M, Gavilanes J G. Characterization of the antifungal protein secreted by the mould Aspergillus giganteus. Arch Biochem Biophys. 1995;324:273–281. doi: 10.1006/abbi.1995.0040. [DOI] [PubMed] [Google Scholar]
- 90.Lam S S, Wang H, Ng T B. Purification and characterization of novel ribosome inactivating proteins, α- and β-pisavins, from seeds of the garden pea Pisum sativum. Biochem Biophys Res Commun. 1998;253:135–142. doi: 10.1006/bbrc.1998.9764. [DOI] [PubMed] [Google Scholar]
- 91.Lamberty M, Ades S, Uttenweiler-Joseph S, Brookhart G, Bushey D, Hoffmann J A, Bulet P. Insect immunity. Isolation from the lepidopteran Heliothis virescensof a novel insect defensin with potent antifungal activity. J Biol Chem. 1999;274:9320–9326. doi: 10.1074/jbc.274.14.9320. [DOI] [PubMed] [Google Scholar]
- 92.Landon C, Sodano P, Hetru C, Hoffmann J, Ptak M. Solution structure of drosomycin, the first inducible antifungal protein from insects. Protein Sci. 1997;6:1878–1884. doi: 10.1002/pro.5560060908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Landon C, Pajon A, Vovelle F, Sodano P. The active site of drosomycin, a small insect antifungal protein, delineated by comparison with the modeled structure of Rs-AFP2, a plant antifungal protein. J Pept Res. 2000;56:231–238. doi: 10.1034/j.1399-3011.2000.00757.x. [DOI] [PubMed] [Google Scholar]
- 94.Langer M, Rothe M, Eck J, Mockel B, Zinke H. A nonradioactive assay for ribosome-inactivating proteins. Anal Biochem. 1996;243:150–153. doi: 10.1006/abio.1996.0493. [DOI] [PubMed] [Google Scholar]
- 95.Leah R, Tommerup H, Svendsen I, Mundy J. Biochemical and molecular characterization of three barley seed proteins with antifungal properties. J Biol Chem. 1991;266:1564–1573. [PubMed] [Google Scholar]
- 96.Lee K M, Kim D H, Lee Y H, Choi B S, Chung J H, Lee B L. Antifungal activities of recombinant antifungal protein by conjugation with polyethylene glycol. Mol Cells. 1999;9:410–416. [PubMed] [Google Scholar]
- 97.Lee S Y, Moon J H, Kurata S, Natori S, Lee B L. Purification and cDNA cloning of an antifungal protein from the hemolymph of Holotrichia diomphalialarvae. Biol Pharm Bull. 1995;18:1049–1052. doi: 10.1248/bpb.18.1049. [DOI] [PubMed] [Google Scholar]
- 98.Lee Y J, Chung T J, Park C W, Hahn Y, Chung J H, Lee B L, Han D M, Jung Y H, Kim S, Lee Y. Structure and expression of the tenecin 3 gene in Tenebrio molitor. Biochem Biophys Res Commun. 1996;218:6–11. doi: 10.1006/bbrc.1996.0002. . [Online.] [DOI] [PubMed] [Google Scholar]
- 99.Lee Y T, Kim K H, Suh J Y, Chung J H, Lee B L, Lee Y, Choi B S. Structural characteristics of tenecin 3, an insect antifungal protein. Biochem Mol Biol Int. 1999;47:369–376. doi: 10.1080/15216549900201393. [DOI] [PubMed] [Google Scholar]
- 100.Leung K C, Meng Z Q, Ho W K. Antigenic determination fragments of α-momorcharin. Biochim Biophys Acta. 1997;1336:419–424. doi: 10.1016/s0304-4165(97)00053-6. [DOI] [PubMed] [Google Scholar]
- 101.Levitz S. Overview of host defenses in fungal infections. Clin Infect Dis. 1992;14:37–42. doi: 10.1093/clinids/14.supplement_1.s37. [DOI] [PubMed] [Google Scholar]
- 102.Lin A, Lee T M, Rern J C. Tricholin, a new antifungal agent from Trichoderma viride, and its action in biological control of Rhizoctonia solani. J Antibiot. 1993;47:799–805. doi: 10.7164/antibiotics.47.799. [DOI] [PubMed] [Google Scholar]
- 103.Linthorst H J M, Melchers L S, Mayer A, Van Roekel J S C, Cornelissen B J C, Bol J F. Analysis of gene families encoding acidic and basic β-1,3 glucanases of tobacco. Proc Natl Acad Sci USA. 1990;87:8756–8760. doi: 10.1073/pnas.87.22.8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Linthorst H J M. Pathogenesis-related proteins of plants. Crit Rev Plant Sci. 1991;10:123–150. [Google Scholar]
- 105.Lipke P, Ovalle R. Yeast cell walls: new structures, new challenges. J Bacteriol. 1998;180:3735–3740. doi: 10.1128/jb.180.15.3735-3740.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu Y, Luo J, Xu C, Ren F, Peng C, Wu G, Zhao J. Purification, characterization, and molecular cloning of the gene of a seed-specific antimicrobial protein from pokeweed. Plant Physiol. 2000;122:1015–1024. doi: 10.1104/pp.122.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lyman C A, Walsh T J. Systemically administered antifungal agents: a review of their clinical pharmacology and therapeutic applications. Drugs. 1992;44:9–35. doi: 10.2165/00003495-199244010-00002. [DOI] [PubMed] [Google Scholar]
- 108.Magliani W, Conti S, Gerloni M, Bertolotti D, Polonelli L. Yeast killer systems. Clin Microbiol Rev. 1997;10:369–400. doi: 10.1128/cmr.10.3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Marcotte E M, Monzingo A F, Ernst S R, Brzezinski R, Robertus J D. X-ray structure of an anti-fungal chitosanase from StreptomycesN174. Nat Struct Biol. 1996;3:155–162. doi: 10.1038/nsb0296-155. [DOI] [PubMed] [Google Scholar]
- 110.Martinez-Ruix A, Martinez del Pozo A, Lacadena J, Mancheno J M, Onaderra M, Gavilanes J G. Characterization of a natural larger form of the antifungal protein (AFP) from Aspergillus giganteus. Biochim Biophys Acta. 1997;1340:81–87. doi: 10.1016/s0167-4838(97)00038-1. [DOI] [PubMed] [Google Scholar]
- 111.Marx F, Haas H, Reindl M, Stoffler G, Lottspeich F, Redl B. Cloning, structural organization and regulation of expression of the Penicillium chrysogenumpaf gene encoding an abundantly secreted protein with antifungal activity. Gene. 1995;167:167–171. doi: 10.1016/0378-1119(95)00701-6. [DOI] [PubMed] [Google Scholar]
- 112.Mathivanan N, Kabilan V, Murugesan K. Purification, characterization, and antifungal activity of chitinase from Fusarium chlamydosporum, a mycoparasite to groundut rust, Puccinia arachidis. Can J Microbiol. 1998;44:646–651. [PubMed] [Google Scholar]
- 113.Meins F, Neuhaus J M, Sperisen C, Ryals J. Characterization and analysis of thionin genes. In: Meins F, Boller T, editors. Genes involved in plant defense. Vienna, Austria: Springer-Verlag; 1992. pp. 245–282. [Google Scholar]
- 114.Melchers L S, Apotheker-de Groot M, van der Knaap J A, Ponstein A S, Sela-Buurlage M B, Bol J F, Cornelissen B J, van den Elzen P J, Linthorst H J. A new class of tobacco chitinases homologous to bacterial exo-chitinases displays antifungal activity. Plant J. 1994;5:469–480. doi: 10.1046/j.1365-313x.1994.5040469.x. [DOI] [PubMed] [Google Scholar]
- 115.Molina A, Segura A, Garcia-Olmedo F. Lipid transfer proteins (nsLTPs) from barley and maize leaves are potent inhibitors of bacterial and fungal plant pathogens. FEBS Lett. 1993;16:119–122. doi: 10.1016/0014-5793(93)81198-9. [DOI] [PubMed] [Google Scholar]
- 116.Molina A, Garcia-Olmedo F. Developmental and pathogen-induced expression of three barley genes encoding lipid transfer proteins. Plant J. 1993;4:983–991. doi: 10.1046/j.1365-313x.1993.04060983.x. [DOI] [PubMed] [Google Scholar]
- 117.Molina A, Gorlach J, Volrath S, Ryals J. Wheat genes encoding two types of PR-1 proteins are pathogen inducible, but do not respond to activators of systemic acquired resistance. Mol Plant Microbe Interact. 1999;12:53–58. doi: 10.1094/MPMI.1999.12.1.53. [DOI] [PubMed] [Google Scholar]
- 118.Money N. Measurement of hyphal turgor. Exp Mycol. 1990;14:416–425. [Google Scholar]
- 119.Money N, Harold F. Extension growth of the water mold Achyla: interplay of turgor and wall strength. Proc Natl Acad Sci USA. 1992;89:4245–4249. doi: 10.1073/pnas.89.10.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Money N, Harold F. Two water molds can grow without measurable turgor pressure. Planta. 1993;190:426–430. [Google Scholar]
- 121.Monzingo A F, Marcotte E M, Hart P J, Robertus J D. Chitinases, chitosanases, and lysozymes can be divided into procaryotic and eucaryotic families sharing a conserved core. Nat Struct Biol. 1996;3:133–140. doi: 10.1038/nsb0296-133. [DOI] [PubMed] [Google Scholar]
- 122.Moralejo F J, Cardoza R E, Gutierrez S, Martin J F. Thaumatin production in Aspergillus awamoriby use of expression cassettes with strong fungal promoters and high gene dosage. Appl Environ Microbiol. 1999;65:1168–1174. doi: 10.1128/aem.65.3.1168-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Morrisette J, Kratzschmar J, Haendler B, El-Hayek R, Mochca-Morales J, Martin B M, Patel J R, Moss R L, Schleuning W D, Coronado R, Possani L D. Primary structure and properties of helothermine, a peptide toxin that blocks ryanodine receptors. Biophys J. 1995;68:2280–2288. doi: 10.1016/S0006-3495(95)80410-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Müller E, Loeffler W. Mycology: an outline for science and medical students. G. Stuttgart, Germany: Thieme Publishers; 1976. [Google Scholar]
- 125.Muradov A, Petrasovits L, Davidson A, Scott K J. A cDNA clone for a pathogenesis-related protein 1 from barley. Plant Mol Biol. 1993;23:439–442. doi: 10.1007/BF00029021. [DOI] [PubMed] [Google Scholar]
- 126.Natori S. Function of antimicrobial proteins in insects. Ciba Found Symp. 1994;186:123–134. doi: 10.1002/9780470514658.ch8. [DOI] [PubMed] [Google Scholar]
- 127.Nawrath C, Metraux J. Salicylic acid induction-deficient mutants of Arabidopsisexpress PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Newman S L, Gootee L, Gabay J E, Selsted M E. Identification of constituents of human neutrophil azurophil granules that mediate fungistasis against Histoplasma capsulatum. Infect Immun. 2000;68:5668–5672. doi: 10.1128/iai.68.10.5668-5672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Niderman T, Genetet I, Buryere T, Gees R, Stintzi A, Legrand M, Fritig B, Mosinger E. Pathogenesis-related PR-1 proteins are antifungal. Isolation and characterization of three 14-kilodalton proteins of tomato and of a basic PR-1 of tobacco with inhibitory activity against Phytophthora infestans. Plant Physiol. 1995;108:17–27. doi: 10.1104/pp.108.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nielsen K K, Nielsen J E, Madrid S M, Mikkelsen J D. New antifungal proteins from sugar beet (Beta vulgarisL.) showing homology to non-specific lipid transfer proteins. Plant Mol Biol. 1996;31:539–552. doi: 10.1007/BF00042227. [DOI] [PubMed] [Google Scholar]
- 131.Nielsen K K, Nielsen J E, Madrid S M, Mikkelsen J D. Characterization of a new antifungal chitin-binding peptide from sugar beet leaves. Plant Physiol. 1997;113:83–91. doi: 10.1104/pp.113.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Oda T, Komatsu N, Muramatsu T. Cell lysis induced by ricin D and ricin E in various cell lines. Biosci Biotechnol Biochem. 1997;61:291–297. doi: 10.1271/bbb.61.291. [DOI] [PubMed] [Google Scholar]
- 133.Odani S, Koide T, Ono T. The complete amino acid sequence of barley trypsin inhibitor. J Biol Chem. 1983;258:7998–8003. [PubMed] [Google Scholar]
- 134.Ogata C M, Gordon P F, de Vos A M, Kim S H. Crystal structure of a sweet tasting protein Thaumatin I, at 1.65 Åresolution. J Mol Biol. 1992;228:893–908. doi: 10.1016/0022-2836(92)90873-i. [DOI] [PubMed] [Google Scholar]
- 135.Osaki T, Omotezako M, Nagayama R, Hirata M, Iwanaga S, Kasahara J, Hattori J, Ito I, Sugiyama H, Kawabata S. Horseshoe crab hemocyte-derived antimicrobial polypeptides, tachystatins, with sequence similarity to spider neurotoxins. J Biol Chem. 1999;274:26172–26178. doi: 10.1074/jbc.274.37.26172. [DOI] [PubMed] [Google Scholar]
- 136.Osborn R W, De Samblanx G W, Thevissen K, Goderis I, Torrekens S, Van Leuven F, Attenborough S, Rees S B, Broekaert W F. Isolation and characterization of plant defensins from seeds of Asteraceae, Fabaceae, Hippocastanaceae and Saxifragaceae. FEBS Lett. 1995;368:257–262. doi: 10.1016/0014-5793(95)00666-w. [DOI] [PubMed] [Google Scholar]
- 137.Ostoa-Saloma P, Carrero J C, Petrossian P, Herion P, Landa A, Laclette J P. Cloning, characterization and functional expression of a cyclophilin of Entamoeba histolytica. Mol Biochem Parasitol. 2000;107:219–225. doi: 10.1016/s0166-6851(00)00190-0. [DOI] [PubMed] [Google Scholar]
- 138.Parente A, De Luca P, Bolognesi A, Barbieri L, Batelli M G, Abbondanza A, Sande M J W, Gigliano G S, Tazzari P L, Stripe F. Purification and partial characterization of single-chain ribosome-inactivating proteins from the seeds of Phytolacca dioicaL. Biochim Biophys Acta. 1993;1216:43–49. doi: 10.1016/0167-4781(93)90035-c. [DOI] [PubMed] [Google Scholar]
- 139.Park K S, Cheong J J, Lee S J, Suh M C, Choi D. A novel proteinase inhibitor gene transiently induced by tobacco mosaic virus infection. Biochim Biophys Acta. 2000;1492:509–512. doi: 10.1016/s0167-4781(00)00126-3. [DOI] [PubMed] [Google Scholar]
- 140.Payne G, Ward E, Gaffney T, Goy P A, Moyer M, Harper A, Meins F, Jr, Ryals J. Evidence for a third structural class of β-1,3-glucanase in tobacco. Plant Mol Biol. 1990;15:797–808. doi: 10.1007/BF00039420. [DOI] [PubMed] [Google Scholar]
- 141.Pfaller M, Wenzel R. Impact of the changing epidemiology of fungal infections in the 1990's. Eur J Clin Microbiol Dis. 1992;11:287–291. doi: 10.1007/BF01962067. [DOI] [PubMed] [Google Scholar]
- 142.Pohi P, Antonenko Y N, Evtodienko V Y, Pohl E E, Saparov S M, Agapov I I, Tonevitsky A G. Membrane fusion mediated by ricin and viscumin. Biochim Biophys Acta. 1998;1371:11–16. doi: 10.1016/s0005-2736(98)00024-8. [DOI] [PubMed] [Google Scholar]
- 143.Ponstein A S, Bres-Vloemans S A, Sela-Buurlage M B, van den Elzen P J, Meichers L S, Cornelissen B J. A novel pathogen-and wound-inducible tobacco (Nicotiana tabacum) protein with antifungal activity. Plant Physiol. 1994;104:109–118. doi: 10.1104/pp.104.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pu Z, Lu B Y, Liu W Y, Jin S W. Characterization of the enzymatic mechanism of γ-momorcharin, a novel ribosome-inactivating protein with lower molecular weight of 11,500 purified from the seeds of bitter gourd (Momordica charantia) Biochem Biophys Res Commun. 1996;229:287–294. doi: 10.1006/bbrc.1996.1794. [DOI] [PubMed] [Google Scholar]
- 145.Rauscher M, Adam A L, Wirtz S, Guggenheim R, Mendgen K, Deising H B. PR-1 protein inhibits the differentiation of rust infection hyphae in leaves of acquired resistant broad bean. Plant J. 1999;19:625–633. doi: 10.1046/j.1365-313x.1999.00545.x. [DOI] [PubMed] [Google Scholar]
- 146.Rezzonico E, Flury N, Meins F, Jr, Beffa R. Transcriptional down-regulation by abscisic acid of pathogenesis-related β-1,3-glucanase genes in tobacco cell cultures. Plant Physiol. 1998;117:585–592. doi: 10.1104/pp.117.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Richardson M, Valdes-Rodriquez S, Blanco-Labra A. A possible function for thaumatin and a TMV-induced protein suggested by homology to a maize inhibitor. Nature. 1987;327:432–434. [Google Scholar]
- 148.Roberts W, Selitrennikoff C P. Zeamatin, an antifungal protein from maize with membrane-permeabilizing activity. J Gen Microbiol. 1990;136:1771–1778. [Google Scholar]
- 149.Ruiz-Herrera J. Fungal cell wall: structure, synthesis and assembly. Boca Raton, Fla: CRC Press; 1992. [Google Scholar]
- 150.Sakurada M, Morgavi D P, Komatani K, Tomita Y, Onodera R. Purification and characteristics of cytosolic chitinase from Piromyces communisOTS1. FEMS Microbiol Lett. 1996;137:75–78. doi: 10.1111/j.1574-6968.1996.tb08085.x. [DOI] [PubMed] [Google Scholar]
- 151.Salzman R A, Tikhonova I, Bordelon B P, Hasegawa P M, Bressan R A. Coordinate accumulation of antifungal proteins and hexoses constitutes a developmentally controlled defense response during fruit ripening in grape. Plant Physiol. 1998;117:465–472. doi: 10.1104/pp.117.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sanchez de la Hoz P, Castagnaro A, Carbonero P. Sharp divergence between wheat and barley at loci encoding novel members of the trypsin/alpha-amylase inhibitor family. Plant Mol Biol. 1994;26:1231–1236. doi: 10.1007/BF00040705. [DOI] [PubMed] [Google Scholar]
- 152a.Schimoler-O'Rourke R, Richardson M, Selitrennikoff C P. Zeamatin inhibits trypsin and α-amylase activities. Appl Environ Microbiol. 2001;67:2365–2366. doi: 10.1128/AEM.67.5.2365-2366.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Schmitt M J, Poravou O, Trenz K, Rehfeldt K. Unique double-stranded RNAs responsible for the anti-Candida activity of the yeast Hanseniaspora uvarum. J Virol. 1997;71:8852–8855. doi: 10.1128/jvi.71.11.8852-8855.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Segura A, Moreno M, Garcia-Olmedo F. Purification and antipathogenic activity of lipid transfer proteins (LTPS) from the leaves of Arabidopsisand spinach. FEBS Lett. 1993;332:243–246. doi: 10.1016/0014-5793(93)80641-7. [DOI] [PubMed] [Google Scholar]
- 155.Segura A, Moreno M, Molina A, Garcia-Olmedo F. Novel defensin subfamily from spinach (Spinacia oleracea) FEBS Lett. 1998;435:159–162. doi: 10.1016/s0014-5793(98)01060-6. [DOI] [PubMed] [Google Scholar]
- 156.Segura A, Moreno M, Madueno F, Molina A, Garcia-Olmedo F. Snakin-1, a peptide from potato that is active against plant pathogens. Mol Plant Microbe Interact. 1999;12:16–23. doi: 10.1094/MPMI.1999.12.1.16. [DOI] [PubMed] [Google Scholar]
- 157.Seguy N, Polonelli L, Dei-Cas E, Cailliez J C. Effect of a killer toxin of Pichia anomala on Pneumocystis. Perspectives in the control of pneumocytosis. FEMS Immunol Med Microbiol. 1998;22:145–149. doi: 10.1111/j.1574-695X.1998.tb01199.x. [DOI] [PubMed] [Google Scholar]
- 158.Selitrennikoff C P, Wilson S J, Clemons K V, Stevens D A. Zeamatin, an antifungal protein. Curr Opin Anti-Infect Investig Drugs. 2000;2:368–374. [Google Scholar]
- 159.Shao F, Xiong Y M, Huang Q Z, Wang C G, Zhu R H, Wang D C. A new antifungal peptide from the seeds of Phytolacca americana: characterization, amino acid sequence and cDNA cloning. Biochim Biophys Acta. 1999;1430:262–268. doi: 10.1016/s0167-4838(99)00013-8. [DOI] [PubMed] [Google Scholar]
- 160.Shivaraj B, Pattabiraman T N. Natural plant enzyme inhibitors. Characterization of an unusual α-amylase/trypsin inhibitor from ragi (Eleusine coracana) Biochem J. 1981;193:29–36. doi: 10.1042/bj1930029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Singh N K, Bracker C A, Hasegawa P M, Handa A K, Buckel S, Hermodson M A, Pfankoch E, Regnier F E, Bressan R A. Characterization of osmotin. Plant Physiol. 1987;85:529–536. doi: 10.1104/pp.85.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Smith M, Bruhn J, Anderson J. The fungus Armillaria bulbosais among the largest and oldest living organisms. Nature. 1992;356:428–431. [Google Scholar]
- 163.Stintzi A, Heitz T, Prasad V, Wiedemann-Merdioglu S, Kauffmann S, Geoffroy P, Legrand M, Fritig B. Plant ‘pathogenesis-related’ proteins and their role in defense against pathogens. Biochimie. 1993;75:687–706. doi: 10.1016/0300-9084(93)90100-7. [DOI] [PubMed] [Google Scholar]
- 164.Suzuki C, Shimma Y I. P-type ATPase spf1 mutants show a novel resistance mechanism to killer toxin SMKT. Mol Microbiol. 1999;32:813–823. doi: 10.1046/j.1365-2958.1999.01400.x. [DOI] [PubMed] [Google Scholar]
- 165.Tahiri-Alaoui A, Dumas-Gaudot E, Gianinazzi S. Immunocytochemical localization of pathogenesis-related PR-1 proteins in tobacco root tissues infected in vitro by the black root rot fungus Chalara elegans. Physiol Mol Plant Pathol. 1993;42:69–82. [Google Scholar]
- 166.Tassin S, Broekaert W, Marion D, Acland D, Ptak M, Vovelle F, Sodano P. Solution structure of Ace-AMP1, a potent antimicrobial protein extracted from onion seeds. Structural analogies with plant nonspecific lipid transfer proteins. Biochemistry. 1998;37:3623–3637. doi: 10.1021/bi9723515. [DOI] [PubMed] [Google Scholar]
- 167.Taylor B E, Irvin J D. Depurination of plant ribosomes by pokeweed antiviral protein. FEBS Lett. 1990;273:144–146. doi: 10.1016/0014-5793(90)81070-5. [DOI] [PubMed] [Google Scholar]
- 168.Terras F R, Schoofs H M, De Bolle M F, Van Leuven F, Rees S B, Vanderleyden J, Cammue B P, Broekaert W F. Analysis of two novel classes of plant antifungal proteins from radish (Raphanus sativus L.) seeds. J Biol Chem. 1992;267:15301–15309. [PubMed] [Google Scholar]
- 169.Terras F R, Torrekens S, Van Leuven F, Osborn R W, Vanderleyden J, Cammue B P, Broekaert W F. A new family of basic cysteine-rich plant antifungal proteins from Brassicaceaespecies. FEBS Lett. 1993;316:233–240. doi: 10.1016/0014-5793(93)81299-f. [DOI] [PubMed] [Google Scholar]
- 170.Terras F R, Eggermont K, Kovaleva V, Raikhel N V, Osborn R W, Kester A, Rees S B, Torrekens S, Van Leuven F, Vanderleyden J. Small cysteine-rich antifungal proteins from radish: their role in host defense. Plant Cell. 1995;7:573–588. doi: 10.1105/tpc.7.5.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Thevissen K, Ghazi A, De Samblanx G W, Brownlee C, Osborn R W, Broekaert W F. Fungal membrane responses induced by plant defensins and thionins. J Biol Chem. 1996;271:15018–15025. doi: 10.1074/jbc.271.25.15018. [DOI] [PubMed] [Google Scholar]
- 172.Thevissen K, Osborn R W, Acland D P, Broekaert W F. Specific, high affinity binding sites for an antifungal plant defensin on Neurospora crassahyphae and microsomal membranes. Biol Chem. 1997;272:32176–32181. doi: 10.1074/jbc.272.51.32176. [DOI] [PubMed] [Google Scholar]
- 173.Thevissen K, Terras F T, Broekaert W F. Permeabilization of fungal membranes by plant defensins inhibits fungal growth. Appl Environ Microbiol. 1999;65:5451–5458. doi: 10.1128/aem.65.12.5451-5458.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Thevissen K, Osborn R W, Acland D P, Broekaert W F. Specific binding sites for an antifungal plant defensin from Dahlia (Dahlia merckii) on fungal cells are required for antifungal activity. Mol Plant Microbe Interact. 2000;13:54–61. doi: 10.1094/MPMI.2000.13.1.54. [DOI] [PubMed] [Google Scholar]
- 175.Torres-Schumann S, Godoy J A, Pintor-Toro J A. A probable lipid transfer protein gene is induced by NaCl in stems of tomato plants. Plant Mol Biol. 1992;18:749–757. doi: 10.1007/BF00020016. [DOI] [PubMed] [Google Scholar]
- 176.Trudel J, Grenier J, Potvin C, Asselin A. Several thaumatin-like proteins bind to β-1,3-glucans. Plant Physiol. 1998;118:1431–1438. doi: 10.1104/pp.118.4.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tulasi R B, Nadimpalli S K. Purification of α-mannosidase activity from Indian lablab beans. Biochem Mol Biol Int. 1997;41:925–931. doi: 10.1080/15216549700201981. [DOI] [PubMed] [Google Scholar]
- 178.Turnay J, Olmo N, Jimenez A, Lizarbe M A, Gavilanes J G. Kinetic study of the cytotoxic effect of α-sarcin, a ribosome inactivating protein from Aspergillus giganteus, on tumour cell lines: protein biosynthesis inhibition and cell binding. Mol Cell Biochem. 1993;122:39–47. doi: 10.1007/BF00925735. [DOI] [PubMed] [Google Scholar]
- 179.Van Damme E J, Charels D, Roy S, Tierens K, Barre A, Martins J C, Rouge P, Van Leuven F, Does M, Peumans W J. A gene encoding a hevein-like protein from elderberry fruits is homologous to PR-4 and class V chitinase genes. Plant Physiol. 1999;119:1547–1556. doi: 10.1104/pp.119.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Van Loon L C. Pathogenesis-related proteins. Plant Mol Biol. 1985;116:111–116. doi: 10.1007/BF02418757. [DOI] [PubMed] [Google Scholar]
- 181.Vanderleyden, Cammue B P, Broekaert W F. Analysis of two novel classes of plant antifungal proteins from radish (Raphanus sativus L.) seeds. J Biol Chem. 1992;267:15301–15309. [PubMed] [Google Scholar]
- 182.Vigers A J, Wiedemann S, Roberts W K, Legrand M, Selitrennikoff C P, Fritig B. A new family of plant antifungal proteins. Mol Plant Microbe Interact. 1991;4:315–323. doi: 10.1094/mpmi-4-315. [DOI] [PubMed] [Google Scholar]
- 183.Vivanco J M, Savary B J, Flores H E. Characterization of two novel type I ribosome-inactivating proteins from the storage roots of the andean crop Mirabilis expansa. Plant Physiol. 1999;119:1447–1456. doi: 10.1104/pp.119.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Vu L, Huynh Q K. Isolation and characterization of a 27-kDa antifungal protein from the fruits of Diospyros texana. Biochem Biophys Res Commun. 1994;202:666–672. doi: 10.1006/bbrc.1994.1982. [DOI] [PubMed] [Google Scholar]
- 185.Wang H X, Ng T B. Quinqueginsin, a novel protein with anti-human immunodeficiency virus, antifungal, ribonuclease and cell-free translation-inhibitory activities from American ginseng roots. Biochem Biophys Res Commun. 2000;269:203–208. doi: 10.1006/bbrc.2000.2114. [DOI] [PubMed] [Google Scholar]
- 186.Wang X, Zafian P, Choudhary M, Lawton M. The PR5K receptor protein kinase from Arabidopsis thalianais structurally related to a family of plant defense proteins. Proc Natl Acad Sci USA. 1996;93:2598–2602. doi: 10.1073/pnas.93.6.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Watanabe T, Kanai R, Kawase T, Tanabe T, Mitsutomi M, Sakuda S, Miyashita K. Family 19 chitinases of Streptomycesspecies: characterization and distribution. Microbiology. 1999;145:3353–3363. doi: 10.1099/00221287-145-12-3353. [DOI] [PubMed] [Google Scholar]
- 188.Wnendt S, Felske-Zech H, Henze P-P C, Ulbrich N, Stahl U. Characterization of the gene encoding α-sarcin, a ribosome-inactivating protein secreted by Aspergillus giganteus. Gene. 1993;124:239–244. doi: 10.1016/0378-1119(93)90399-n. [DOI] [PubMed] [Google Scholar]
- 189.Wnendt S, Ulbrich N, Stahl U. Molecular cloning, sequence analysis and expression of the gene encoding an antifungal-protein from Aspergillus giganteus. Curr Genet. 1994;25:519–523. doi: 10.1007/BF00351672. [DOI] [PubMed] [Google Scholar]
- 190.Wong R N, Dong T X, Ng T B, Choi W T, Yeung H W. α-Kirilowin, a novel ribosome-inactivating protein from seeds of Trichosanthes kirilowii (family Cucurbitaceae): a comparison with β-kirilowin and other related proteins. Int J Pept Protein Res. 1996;47:103–109. doi: 10.1111/j.1399-3011.1996.tb00816.x. [DOI] [PubMed] [Google Scholar]
- 191.Xia X F, Sui S F. The membrane insertion of trichosanthin is membrane-surface-pH dependent. Biochem J. 2000;349:835–841. doi: 10.1042/bj3490835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Xu L, Lal K, Santarpia III R P, Pollock J J. Salivary proteolysis of histidine-rich polypeptides and the antifungal activity of peptide degradation products. Arch Oral Biol. 1993;38:277–283. doi: 10.1016/0003-9969(93)90133-7. [DOI] [PubMed] [Google Scholar]
- 193.Yamagami T, Taira T, Aso Y, Ishiguro M. Isolation and characterization of chitinase isoforms from the bulbs of four species of the genus Tulipa. Biosci Biotechnol Biochem. 1998;62:584–587. doi: 10.1271/bbb.62.584. [DOI] [PubMed] [Google Scholar]
- 194.Yang Y, Shah J, Klessig D F. Signal perception and transduction in plant defense responses. Genes Dev. 1997;11:1621–1639. doi: 10.1101/gad.11.13.1621. [DOI] [PubMed] [Google Scholar]
- 195.Yao Q Z, Yu M M, Ooi L S, Ng T B, Chang S T, Sun S S, Ooi V E. Isolation and characterization of a type 1 ribosome-inactivating protein from fruiting bodies of the edible mushroom (Volvariella volvacea) J Agric Food Chem. 1998;46:788–792. doi: 10.1021/jf970551h. [DOI] [PubMed] [Google Scholar]
- 196.Ye X Y, Wang H X, Ng T B. First chromatographic isolation of an antifungal thaumatin-like protein from French bean legumes and demonstration of its antifungal activity. Biochem Biophys Res Commun. 1999;263:130–134. doi: 10.1006/bbrc.1999.1166. [DOI] [PubMed] [Google Scholar]
- 197.Ye X Y, Wang H X, Ng T B. Sativin: a novel antifungal miraculin-like protein isolated from legumes of the sugar snap Pisum sativumvar. macrocarpon. Life Sci. 2000;67:775–781. doi: 10.1016/s0024-3205(00)00672-x. [DOI] [PubMed] [Google Scholar]
- 198.Ye X Y, Wang H X, Ng T B. Dolichin, a new chitinase-like antifungal protein isolated from field beans (Dolichos lablab) Biochem Biophys Res Commun. 2000;269:155–159. doi: 10.1006/bbrc.2000.2115. [DOI] [PubMed] [Google Scholar]
- 199.Ye X Y, Ng T B. Mungin, a novel cyclophilin-like antifungal protein from the mung bean. Biochem Biophys Res Commun. 2000;273:1111–1115. doi: 10.1006/bbrc.2000.3067. [DOI] [PubMed] [Google Scholar]
- 200.Yun D, Zhao Y, Pardo J, Narasimhan M L, Damsz B, Lee H, Abad L R, D'Urzo M P, Hasegawa P M, Bressan R A. Stress proteins on the yeast cell surface determine resistance to osmotin, a plant antifungal protein. Proc Natl Acad Sci USA. 1997;94:7082–7087. doi: 10.1073/pnas.94.13.7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Yun D J, Ibeas J I, Lee H, Coca M A, Narasimhan M L, Uesono Y, Hasegawa P M, Pardo J M, Bressan R A. Osmotin, a plant antifungal protein, subverts signal transduction to enhance fungal cell susceptibility. Mol Cell. 1998;1:807–817. doi: 10.1016/s1097-2765(00)80080-5. [DOI] [PubMed] [Google Scholar]
- 202.Zhang G P, Shi Y L, Wang W P, Liu W Y. Cation channel formed at lipid bilayer by Cinnamomin, a new type II ribosome-inactivating protein. Toxicon. 1999;37:1313–1322. doi: 10.1016/s0041-0101(99)00078-1. [DOI] [PubMed] [Google Scholar]