Significance
Memory formation relies on a plethora of functions, including epigenetic modifications. Over recent years, multiple studies have indicated the potential of HDAC inhibitors (HDACis) as cognitive enhancers, but their mode of action is not fully understood. Here, we tested whether HDACi treatment improves memory formation via “cognitive epigenetic priming,” stipulating that HDACis—without inherent target specificity—specifically enhance naturally occurring plasticity processes. We found that combining HDACis with fear learning, but not either treatment alone, enhances synaptic plasticity as well as memory-promoting transcriptional signaling in the hippocampus, a brain area recruited by fear learning, but not in unrelated areas. These results lend experimental support to the theory of cognitive epigenetic priming.
Keywords: epigenetic priming, fear learning, HDACi, NGS, synaptic plasticity
Abstract
Long-term memory formation relies on synaptic plasticity, neuronal activity-dependent gene transcription, and epigenetic modifications. Multiple studies have shown that HDAC inhibitor (HDACi) treatments can enhance individual aspects of these processes and thereby act as putative cognitive enhancers. However, their mode of action is not fully understood. In particular, it is unclear how systemic application of HDACis, which are devoid of substrate specificity, can target pathways that promote memory formation. In this study, we explore the electrophysiological, transcriptional, and epigenetic responses that are induced by CI-994, a class I HDACi, combined with contextual fear conditioning (CFC) in mice. We show that CI-994–mediated improvement of memory formation is accompanied by enhanced long-term potentiation in the hippocampus, a brain region recruited by CFC, but not in the striatum, a brain region not primarily implicated in fear learning. Furthermore, using a combination of bulk and single-cell RNA-sequencing, we find that, when paired with CFC, HDACi treatment engages synaptic plasticity-promoting gene expression more strongly in the hippocampus, specifically in the dentate gyrus (DG). Finally, using chromatin immunoprecipitation-sequencing (ChIP-seq) of DG neurons, we show that the combined action of HDACi application and conditioning is required to elicit enhancer histone acetylation in pathways that underlie improved memory performance. Together, these results indicate that systemic HDACi administration amplifies brain region-specific processes that are naturally induced by learning.
Long-term memory is the result of facilitated synaptic communication as well as activity-dependent transcription that is regulated by epigenetic signaling (1–4). For example, memory-forming tasks, such as contextual fear conditioning (CFC), are paralleled by gene expression and histone acetylation changes in the hippocampus (5–7), while impaired cognition is coupled with a reduction in hippocampal histone acetylation and plasticity-related gene expression (8–12). Some of these epigenetic and transcriptional changes can be augmented by systemic HDAC inhibitor (HDACi) treatment, which improves memory in both healthy and cognitively impaired mice (9–11, 13, 14). Although the use of HDACis in these studies testifies to their suitability as pharmacological memory aids, the mechanisms by which HDACi enhances memory are not fully understood. In particular, it is unclear how systemic application of HDACis, most of which are devoid of substrate specificity, can enhance pathways that promote memory formation.
One proposed theoretical mode of action for HDACis as cognitive enhancers is called “cognitive epigenetic priming” (3, 15). This model is, on the one hand, inspired by evidence from cancer research, where HDACis have been shown to improve target efficacy of anticancer treatments (16, 17); and on the other hand, by addiction research, where chronic drug abuse durably enriches histone acetylation, which relaxes the chromatin structure into a primed state and thereby lowers the activation threshold for gene expression changes during subsequent drug exposures (18, 19). Analogously, for cognition, this theory stipulates that by broadly increasing histone acetylation, HDACi treatment leads to an overall primed state, while memory-induced neuronal activity, which is inherently characterized by high target specificity (2), would then further enrich such HDACi-induced histone acetylation in a selective manner, and thereby facilitate the transcription of synaptic plasticity-related genes.
In this study, we tested the concept of cognitive epigenetic priming in mice on three different levels. First, we investigated whether systemic HDACi treatment elicits brain region-specific electrophysiological and transcriptional responses after CFC, a hippocampus-dependent memory task. Second, we assessed whether and to which extent specific cell types are affected by the HDACi treatment in combination with CFC using single nuclear RNA-sequencing (snRNA-seq) of the hippocampus. Third, we determined which gene loci are epigenetically regulated by HDACi-supported CFC using chromatin immunoprecipitation (ChIP) followed by sequencing (ChIP-seq). These experiments were designed to better understand the underlying mechanisms of HDACis as potential cognitive enhancers.
Results
Systemic HDACi Treatment Enhances Memory Consolidation after Subthreshold CFC.
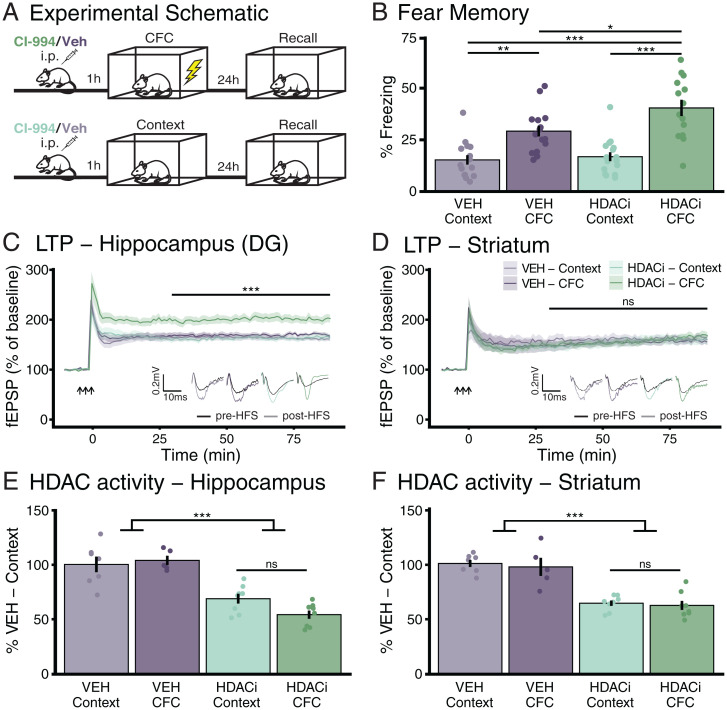
To investigate the mechanisms by which systemic HDACi treatment enhances fear memory, we treated mice with the HDACi CI-994 before subjecting them to a mild CFC task, a Pavlovian conditioning paradigm that, alone, only results in modest memory performance (20). CI-994 is a class I HDACi that selectively impedes HDACs 1 to 3 (21), promotes functional recovery after stroke (22), and has shown promise against cognitive dysfunctions in preclinical animal models (14, 23, 24). One hour prior to CFC or context-only exposure (Context), mice were intraperitoneally injected with 30 mg/kg of CI-994 or its vehicle (VEH) (Fig. 1A). One day later, freezing was measured during a 3-min context exposure. We found that pairing the CFC paradigm with the HDACi significantly improved memory retention compared to VEH-treated mice (P = 3.03e-02), and compared to HDACi treatment without CFC [Tukey’s honestly significant difference (HSD) following one-way ANOVA, F(3, 57) = 17.45, P = 3.72e-08] (Fig. 1B). In line with previous results (14), HDACi treatment had no effect on speed [F(3, 57) = 1.28, P = 0.29] or distance traveled [F(3, 57) = 1.26, P = 0.30], and did not alter time spent in the inner region of the behavioral apparatus [F(3, 57) = 0.493, P = 0.69] during habituation (SI Appendix, Fig. S1). These results indicate that the HDACi treatment facilitates contextual fear learning and leads to long-term memory retention.
Fig. 1.
HDACi treatment enhances contextual fear memory formation and hippocampal, but not striatal LTP, despite reducing HDAC activity in both brain regions. (A) Schematic of the experimental outline. (B) HDACi combined with CFC increases the percent of time spent freezing (>1 s) during 3-min reexposure to the conditioning chamber 24 h after conditioning. n (VEH-Context) = 15; n (VEH-CFC and HDACi-Context) = 16; n (HDACi-CFC) = 14. (C and D) HDACi combined with CFC enhances LTP in response to HFS in the perforant pathway of the DG (C) but not in cortico-striatal fibers (D) 1 h after conditioning. Statistical differences were calculated from 30 min (end of short-term potentiation) to end of recording. n = 8 animals per group. (E and F) HDAC activity was reduced after HDACi in both the hippocampus (E) and striatum (F) with no further reduction in HDAC activity in response to CFC. Hippocampus and striatum: n (VEH-Context) = 7; n (VEH-CFC) = 5; n (HDACi-Context and CFC) = 8. One or two-way ANOVA with Tukey’s HSD. Graphs represent mean ± SEM, *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant.
Systemic HDACi Treatment Regulates Long-Term Potentiation in an Activity-Specific Manner.
To explore whether HDACi treatment improves memory via cognitive epigenetic priming, we first assessed its mode of action on synaptic plasticity. To this end, we measured the effects of HDACi on long-term potentiation (LTP) 1 h after CFC in the hippocampus, a brain region activated by CFC (25), and the striatum, a brain region that is not directly involved (26). We found a significant increase in LTP at perforant path synapses of the dentate gyrus (DG) of the hippocampus when CFC was paired with the HDACi (Fig. 1C) [one-way ANOVA, F(3, 28) = 10.57, P = 8.09e-05]. Without CFC, the HDACi had no effect on LTP; similarly, CFC alone did not facilitate LTP. Conversely, at cortico-striatal fibers, the HDACi treatment had no effect regardless of the behavioral paradigm [F(3, 28) = 0.234, P = 0.872] (Fig. 1D). Neither paired-pulse facilitation (PPF) nor input/output (I/O) relationships were changed in either brain region (SI Appendix, Fig. S2). Importantly, combining CFC with HDACi also enhanced LTP at Schaffer collaterals of the CA1, another hippocampal subregion [one-way ANOVA, F(3, 28) = 5.213, P = 0.005] after subthreshold CFC, which, with the exception of the HDACi-Context sample, was not accompanied by any I/O changes (SI Appendix, Fig. S3). These findings indicate a brain area-specific effect of the HDACi treatment, with only brain areas engaged by CFC displaying enhanced synaptic plasticity.
This brain region-specific effect on synaptic plasticity occurred despite the same degree of HDAC activity inhibition in both brain regions. HDAC activity was reduced by about 50% in both the hippocampus [F(1, 24) = 60.15, P = 5.44e-08] (Fig. 1E) and the striatum [F(1, 24) = 68.96, P = 1.62e-08] (Fig. 1F) in response to HDACi, with no difference in HDAC activity induced by learning itself. Thus, despite the same extent of HDAC inhibition induced by the HDACi, synaptic plasticity was only altered in the brain area directly engaged by CFC.
To evaluate whether such brain region-specific HDACi-mediated enhancement of plasticity is restricted to hippocampus-dependent tasks, we also tested CI-994 treatment during rotarod training, a motor skill learning task known to depend on the cortico-striatal pathway (27). We found that HDACi-treated animals stayed on the apparatus for longer than their VEH-treated counterparts (SI Appendix, Fig. S4 A and B) [one-way ANOVA, F(1, 120) =12.155, P = 0.0007], indicating improved motor learning. Moreover, while neither training nor HDACi had an effect on hippocampal or striatal LTP (SI Appendix, Fig. S4 C and D), HDACi paired with rotarod training selectively increased striatal PPF (SI Appendix, Fig. S4 E and F) [two-way ANOVA, F(3, 192) = 12.217, P = 2.37e-07], known to underlie motor learning in the striatum (28). There were no major differences in I/O in either the striatum or the hippocampus (SI Appendix, Fig. S4 G and H). These electrophysiological data are thus in support of the cognitive epigenetic priming hypothesis at the level of these two brain areas, insofar as the HDACi application alone did not yield any measurable difference, but necessitated task-specific neuronal activity to reveal its potentiating effect.
HDACi Activates Different Transcriptional Cascades in Response to CFC in the Hippocampus and Striatum.
To further understand the molecular mechanisms by which cognitive epigenetic priming leads to improved memory performance, we next used bulk RNA-seq in the hippocampus and striatum to determine which genes are activated when CFC is combined with HDACi treatment. For this, we extracted and sequenced total mRNA from whole-tissue homogenates 1 h after behavior, using the same experimental set-up as for the electrophysiological recordings.
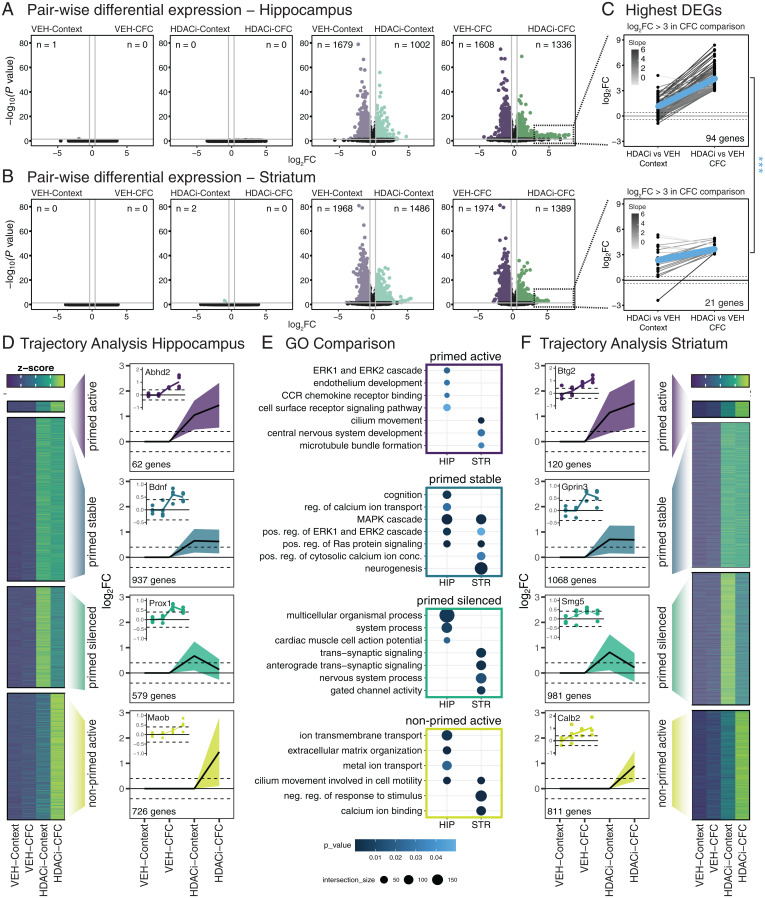
In the hippocampus, consistent with previous data (29), we found no differentially expressed genes (DEGs) (P ≤ 0.05; log2 fold-change [FC] ≥ 0.4) between CFC and context-only exposure in VEH-treated animals (Fig. 2 A, Far Left). Likewise, when comparing CFC with the context-only group in HDACi-treated animals, no DEGs were detected, indicating that CFC alone is not sufficient to induce detectable transcriptional changes (Fig. 2 A, Center Left). Conversely, when context exposure was compared with and without CI-994, we found 1,002 and 1,679 genes significantly up- and down-regulated, respectively, indicating that the addition of the HDACi per se can alter the transcriptional landscape (Fig. 2 A, Center Right, and SI Appendix, Fig. S5). Interestingly, when CFC training was compared with and without HDACi, we detected 1,336 up-regulated genes, a 25% increase to the context-only comparison, but a similar number (1,608) of down-regulated genes (Fig. 2 A, Far Right, and SI Appendix, Fig. S5).
Fig. 2.
HDACi elicits brain region-specific transcriptional responses to CFC. (A and B) Volcano plots of differential expression versus statistical significance of pairwise comparisons labeled above each plot in the hippocampus (A) and striatum (B). The n values represent number of DEGs (log2FC ≥ 0.4; P ≤ 0.05). (C) Juxtaposition of the DEGs with log2FC ≥ 3 in the HDACi-CFC vs. VEH-CFC (Right column) comparison to those same genes in the HDACi-Context vs. VEH-Context (Left column) comparison in the hippocampus (Upper) and striatum (Lower). Lines are colored by log2FC difference between the two comparisons. The blue line represents the average slope. Student’s t test compares slopes between hippocampus and striatum. ***P < 0.001. (D) Heat map of z-scores of average gene counts in the hippocampus (Left). Line graphs in trajectory plots represent significant log2FC values for each cluster (Right). Count in lower left corner indicates number of genes in each cluster. Line plots shown as mean ± SEM. Representative DEGs from each cluster in inset. Normalized counts for each replicate were compared to average normalized count for VEH-Context. (E) GO analysis of hippocampal (Left) and striatal (Right) trajectories. (F) Gene cluster analysis for striatum RNA-seq data as in D. n = 4 biological replicates.
In the striatum, there were no DEGs between the CFC and context-only exposure in either the VEH- or HDACi-treated animals (Fig. 2 B, Left panels). Similar to the hippocampus, when HDACi-Context was compared to VEH-Context, 1,486 and 1,968 genes were significantly up- and down-regulated (Fig. 2 B, Center Right). In contrast to the hippocampus, however, no further increase in the number of DEGs was detected when HDACi was paired with CFC (Fig. 2 B, Right, and SI Appendix, Fig. S5). Lists of pairwise differential expression for both brain regions can be found in Dataset S1.
When comparing strongly up-regulated genes (with a stringent log2FC ≥ 3 cutoff) after the combined HDACi and CFC treatment in both the hippocampus and the striatum, (Fig. 2C), we detected 4.5× more hippocampal DEGs, which also had a higher magnitude of activation than striatal DEGs (Student’s t test of slope values, P = 2.304e-05) (Fig. 2C). These hippocampal DEGs (Dataset S2) were enriched in ion-transport ontologies and included transthyretin, Ttr, which provides neuroprotection in aged mice and is associated with enhanced memory (30, 31). In contrast, highly expressed striatal genes were primarily predicted genes (Dataset S2). This indicates that, in the hippocampus, the expression of these DEGs is further enhanced when the HDACi is paired with CFC, while pairing HDACi with CFC had no such effect in the striatum.
Next, we set out to identify transcriptional patterns of CFC and HDACi treatment. To this end, all DEGs (P ≤ 0.05, log2FC ≥ 0.4) underwent decision tree clustering, in which they were grouped based on their FC expression pattern from the VEH-Context group (Fig. 2 D–F; see also details in Materials and Methods). Four major up-regulated clusters were identified as trajectories of interest (Fig. 2D). In the hippocampus, this analysis yielded: 1) genes that were up-regulated by the HDACi treatment alone (HDACi-VEH) and further increased when HDACi was paired with CFC (HDACi-CFC), which we termed “primed active” (n = 62); 2) genes that were increased by HDACi treatment but showed no further CFC-driven increase, which we termed “primed stable” (n = 937); 3) genes that were enriched by HDACi treatment, but were reduced when the HDACi was paired with CFC, which we termed “primed silenced” (n = 579); and 4) genes that were only activated when combining HDACi with CFC, but not by either condition alone, which we termed “non-primed active” (n = 726). In the striatum, the order of magnitude of DEGs was similar (Fig. 2F and Dataset S3). qPCR analysis confirmed the expression changes of these genes (SI Appendix, Fig. S6).
Gene ontology (GO) analysis revealed brain and trajectory-specific gene expression differences that are relevant for synaptic plasticity. We found that in the hippocampus, the primed active cluster was enriched for the ERK cascade, which has been implicated in synaptic plasticity as well as learning and memory (32, 33). Conversely, in the striatum, the primed active cluster was not enriched for any ontologies involved in MAPK/ERK signaling or learning and memory (Fig. 2E and Dataset S4). Furthermore, the primed stable cluster was characterized by learning and memory-related pathways, such as cognition and regulation of calcium ion transport in the hippocampus, but not the striatum. Hippocampal DEGs in this cluster included brain-derived neurotrophic factor (Bdnf) and the proto-oncogene Jun (Jun), both immediate-early genes (IEGs) induced by neuronal activity and implicated in synaptic plasticity as well as learning and memory (34–36). In the striatum, this cluster did not include memory-related IEGs. It did, however, contain pathways involved in intracellular signal transduction that also regulate learning and memory (5, 37) such as the MAPK cascade, Ras protein signal transduction, and Erk1 and Erk2 cascade. This comparison stipulates that HDACi similarly primes the MAPK pathway in both brain areas but further potentiates only the primed active genes in the hippocampus. In the primed silenced or non-primed active states, no ontologies associated with synaptic signal transduction were found. Finally, the hippocampal non-primed active cluster—representing genes that are only transcribed after combined HDACi-CFC—was enriched for “ion transmembrane transport” pathways, while in the striatum, it was enriched for genes involved in a “negative response to stimulus.” This could indicate that the combination of HDACi treatment and CFC increases inhibitory signaling in the striatum, possibly related to the decreased motor response following conditioning.
Of note, none of the clusters in which HDACi reduced transcription included pathways involved in learning and memory or synaptic plasticity in either brain area, despite a substantial number of down-regulated genes (SI Appendix, Fig. S7 A and B). Rather, down-regulated genes in both brain areas were primarily associated with cellular metabolism and genetic regulation (SI Appendix, Fig. S7C and Dataset S4).
Taken together, these results illustrate that HDACi treatment is a generic driver of differential transcription between the hippocampus and the striatum. It enhances MAPK signaling in both the hippocampus and the striatum, as seen in the comparisons of the primed stable groups. However, when paired with CFC, HDACi treatment results in a specific increase of learning-related genes in the primed active state selectively in the hippocampus.
HDACi Activates Different Transcriptional Cascades across Hippocampal Cell Types.
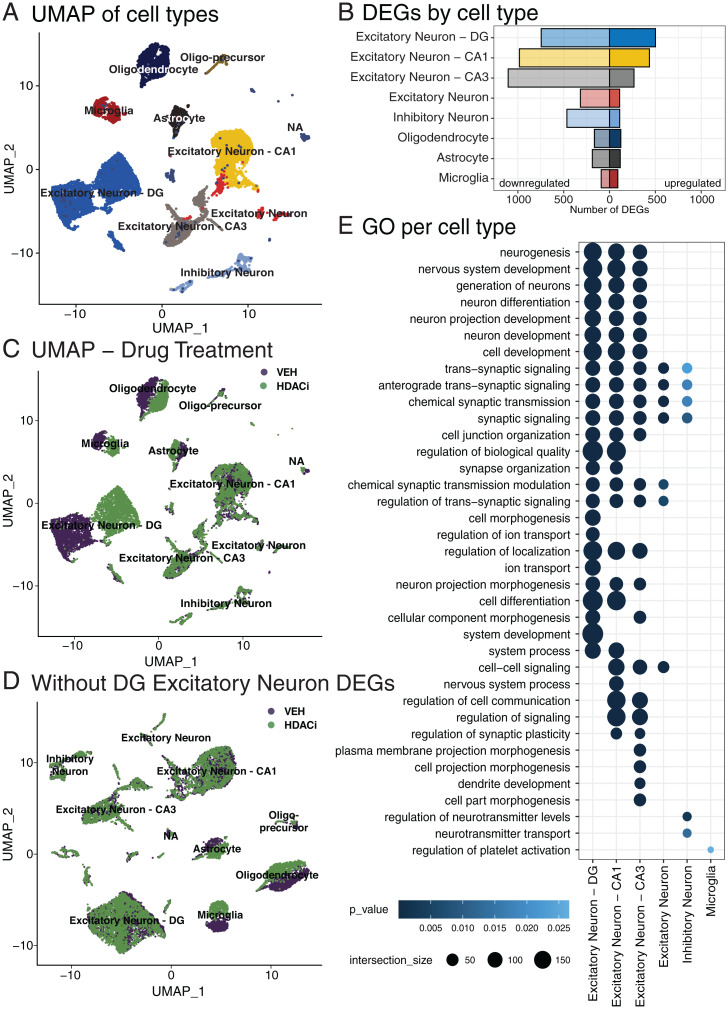
Next, we aimed to understand which cell types within the hippocampus are most responsive to HDACi treatment. To do so, we used snRNA-seq on isolated hippocampi from HDACi- or VEH-treated animals. Since transcriptional differences were most prominent in the HDACi-CFC vs. the VEH-CFC groups (Fig. 2C and SI Appendix, Fig. S5), we focused primarily on this comparison (for the HDACi-Context vs. VEH-Context comparison see SI Appendix, Fig. S8). We performed dimensionality reduction using uniform manifold approximation and projection (UMAP) and clustered nuclei by k-nearest neighbors, revealing 30 distinct clusters consisting of 15,339 total nuclei and expressing a total of 24,271 genes (SI Appendix, Fig. S9A). Clustering was similar across replicates (SI Appendix, Fig. S9B). These clusters were then assigned to known cell types by comparing expression of cell type-specific genes taken from previously published datasets (38–42) (SI Appendix, Fig. S9C). This identified 10 distinct cell types: 4 clusters of excitatory neurons that split based on hippocampal location (5,175 DG nuclei, 2,871 CA1 nuclei, 1,657 CA3 nuclei, and 507 nuclei with no location marker); 794 inhibitory neurons; 4 glial clusters (1,960 oligodendrocytes, 254 oligo-precursors, 763 astrocytes, and 880 microglia); and a cluster of 478 nuclei (NA), which could not be assigned to a single cell-type based on its expression profile (Fig. 3A and SI Appendix, Fig. S9D). In line with previous work (43), neuronal clusters had more expressed genes than glial clusters (SI Appendix, Fig. S9E) and the proportions of cell types were similar to those reported for the hippocampal region in the Blue Brain Atlas (44) (SI Appendix, Fig. S9F).
Fig. 3.
HDACi activates different transcriptional cascades within hippocampal cell types. (A) UMAP of 15,339 nuclei from the full hippocampus colored by 10 identified cell types. (B) Number of up-regulated (Right; log2FC ≥ 1; false-discovery rate [FDR] ≤ 0.05) and down-regulated (Left; log2FC ≤ −1; FDR ≤ 0.05) genes in each cell type that has significant GOs. (C) UMAP of nuclei colored by sample drug treatment. (D) UMAP, colored by drug treatment, after removing the 501 up-regulated genes in the DG excitatory neurons and reclustering. (E) Top 25 enriched GOs of genes up-regulated after HDACi treatment and CFC in each cell type. Cell types that have no ontologies are not represented. n = 2 biological replicates per group (HDACi-CFC and VEH-CFC).
We then explored whether pairing CFC with HDACi induces distinct responses across cell types. Augur, a tool prioritizing a population’s responsiveness to experimental perturbations (45), reported a similar global responsiveness for all clusters (SI Appendix, Fig. S10A), and with the exception of oligo-precursors, HDACi treatment did not change cluster cell-type composition (SI Appendix, Fig. S10B). However, HDACi treatment differentially regulated a distinct set of genes in each cell type (Fig. 3B), which were highly cluster-specific (SI Appendix, Fig. S10 C–F and Dataset S5), with excitatory neurons having the largest HDACi response for up-regulated genes (SI Appendix, Fig. S10C).
Interestingly, we found an HDACi-induced separation for excitatory neurons in the DG and for glia, but not for any other cluster (Fig. 3C). This drug-induced split was mainly mediated by the up-regulated genes within the DG, as removing those genes and rerunning the dimension reduction in silico remerged the split DG cluster (Fig. 3D). Conversely, there was no cluster remerging when removing up-regulated DEGs from CA1, glia, or from any other cell type (SI Appendix, Fig. S11A). Furthermore, DG cluster remerging was specific to the up-regulated genes, as removing only down-regulated genes had no effect either (SI Appendix, Fig. S11B).
Among the HDACi-induced up-regulated genes within each cluster, we found that excitatory neurons were predominatly enriched for genes involved in neuronal functioning and synaptic signaling (Fig. 3E and Dataset S6). Among the HDACi-induced down-regulated genes within each cluster, which were also cluster-specific (SI Appendix, Fig. S10F), there were only a few GOs significantly represented, which included regulation of metabolic and cellular processes in the excitatory neurons cluster (Dataset S6). Importantly, the differential expression patterns in the snRNA-seq dataset were in line with the bulk RNA-seq results (SI Appendix, Fig. S12), since most DEGs from both analyses were differentially expressed in the same direction (Dataset S7), with genes up-regulated in both experiments associated with cell signaling and communication, and genes down-regulated in both experiments mostly associated with RNA metabolism, DNA repair, and other nuclear processes important for cell maintenance (Dataset S8). Together, these results provide evidence that pairing CFC with HDACi treatment transcriptionally activates different gene sets across cell types, with a particularly strong response among up-regulated genes in the DG that is relevant for synaptic signaling.
HDACi Combined with CFC Enriches H3K27ac at Genes Involved in Synaptic Communication.
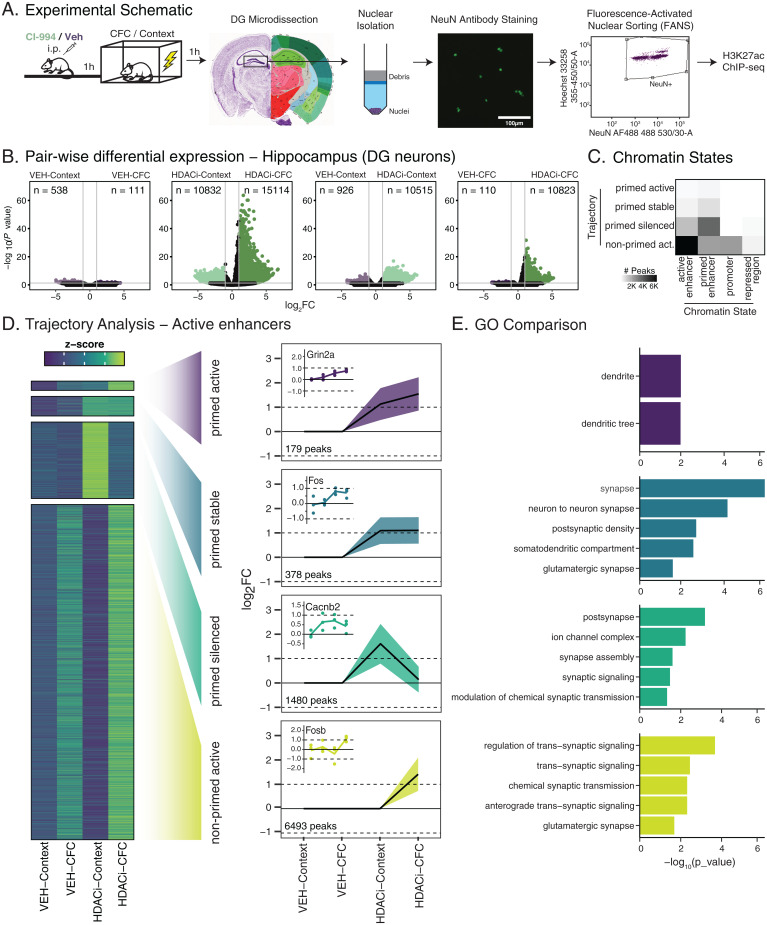
To better understand the HDACi-induced epigenetic regulation of the up-regulated genes in DG excitatory neurons, we next characterized histone acetylation in these cells using ChIP-seq. We focused on H3K27ac, a known marker of active enhancers that is enriched at activity-dependent regulatory elements after neuronal activation (29, 46–49), and correlates with gene transcription (29, 50). Furthermore, H3K27ac, alongside H3K9ac and H4K12ac, is directly influenced by CI-994, as revealed by Western blotting (SI Appendix, Fig. S13) [two-way ANOVA, F(3, 60) = 22.47, P = 1.11e-13]. For ChIP-seq, we used three biological replicates, each from pooled DG from five mice, following fluorescence activated nuclear sorting of NeuN+ nuclei (Fig. 4A).
Fig. 4.
HDACi enriches H3K27ac at genes involved in neuronal synaptic communication. (A) Schematic of experimental outline. (B) Volcano plots showing differential H3K27ac enrichment versus statistical significance for pairwise comparisons (labeled above each plot). n values represent the number of peaks that are enriched (log2FC ≥ 1; FDR ≤ 0.05). P values were calculated by the Wald test and corrected for multiple comparisons using FDR. (C) Heat map representing number of peaks in the trajectories per chromatin state. (D) Heat map of z-scores of the average normalized H3K27ac peak counts for all four trajectories of interest. Peak sets underwent decision tree clustering based on significant log2FC values for associated peaks in each group when compared to VEH-Context. Line graphs in trajectory plots represent significant log2FC values for each group in clusters of interest. Count in lower left corner indicates number of peaks. Line plots shown as mean ± SEM. Insets represent differentially enriched active enhancer peaks from each cluster. Normalized counts for each replicate were compared to average normalized count for VEH-Context. (E) GO analysis of genes associated with H3K27ac peaks. n = 3 biological replicates (pooled 5 animals).
Differential enrichment analysis (Diffbind, DEseq2: data in Dataset S9) revealed that CFC in VEH-treated animals led to only marginal changes in H3K27ac enrichment (Fig. 4 B, Left), in line with the transcriptional data (Fig. 2A). Conversely, when CFC occurred in the presence of HDACi, more than 10,000 and 15,000 regions were significantly down- and up-regulated (log2FC ≥ 1), respectively, indicating that in the presence of HDACi, the behavioral paradigm triggers substantial epigenetic changes. Furthermore, the HDACi treatment itself also enriched a significant number of regions, ∼10,500 regions in both context- and CFC-treated groups (Fig. 4 B, Right plots), suggesting that both CFC and HDACi treatment alter H3K27ac enrichment. This is in contrast with the Western blot comparisons (SI Appendix, Fig. S13), where no significant differences in H3K27ac levels were observed between the HDACi-Context and HDACi-CFC groups, but is likely attributed to the increased sensitivity of the ChIP-seq analysis.
Next, we performed a decision tree analysis for H3K27ac peaks at active enhancers, the chromatin state with the largest number of peaks (Fig. 4C and SI Appendix, Fig. S14; other chromatin states are included in SI Appendix, Fig. S15), focusing on the same four trajectories as in the RNA-seq (Fig. 2 D–F). The primed active cluster for active enhancers was the smallest, containing 179 peaks (Fig. 4D and Dataset S10). This cluster represented ontologies associated with dendritic locations (Fig. 4E and Dataset S11) and included peaks associated with NMDA receptor 2A (Grin2a) and the calcium voltage-gated channel subunit (Cacna1e). In the primed stable cluster, there were 378 active enhancers for which H3K27ac was increased after HDACi treatment but not further enriched after CFC. This cluster included ontologies specific to synaptic locations, and a previously described enhancer of cFos, whose regulation by histone acetylation was recently validated by targeted dCas9-p300 manipulations (47). The 1,480 enhancers of the primed silenced cluster were also associated with genes that are involved in synaptic assembly and signaling (Datasets S10 and S11). Finally, the non-primed active cluster was the largest and contained 6,493 active enhancer peaks, the ontologies for which were also associated with regulation of synaptic signaling. This cluster included enhancers for multiple plasticity and memory-related genes: for example, Fosb, Jun, Junb, and JunD, which are members of the AP-1 complex, known to be involved in neuronal plasticity processes during CFC (36, 49); calcium-dependent protein kinases, which are crucial for signaling at glutamatergic synapses (51); and genes in the MAPK/ERK signaling cascade, which regulates H3 acetylation during CFC and helps to establish the stabilization of long-term memory (5, 33, 37).
Taken together, these data show that HDACi-induced H3K27ac enrichment after context or CFC is highly specific to neuronal signaling processes. However, in contrast to the RNA-seq data, the H3K27ac enrichment appears to be most relevant in the non-primed active cluster, and thus most responsive to the combined HDACi and CFC treatment, which closely resembles the behavioral and electrophysiological results. This is interesting insofar as we would expect changes in acetylation, or the priming step, to be relevant in all HDACi-treated groups, but transcriptional activation to be more specific to the paired HDACi and CFC conditions. Thus, to better understand this disconnect between transcriptional activation and acetylation enrichment, we directly compared which genes are both enriched and transcriptionally activated and which genes are only enriched for H3K27ac at enhancer regions.
Transcriptional Activation and H3K27ac Accumulation Occur at Genes Involved in Synaptic Communication.
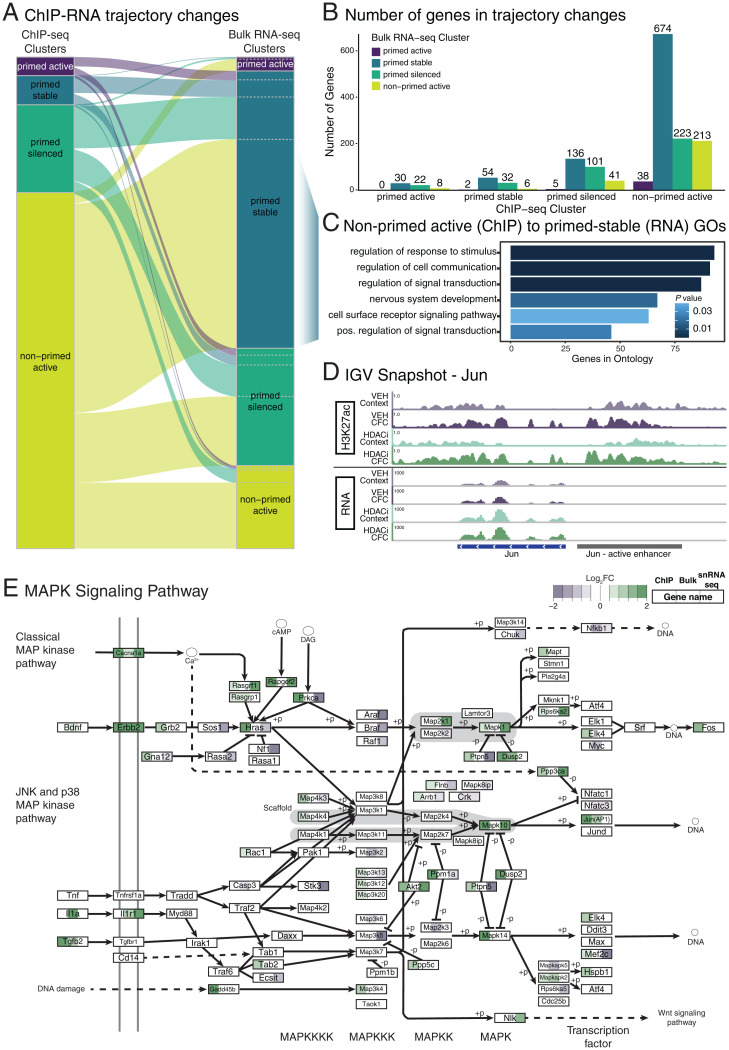
When we related H3K27ac accumulation at active enhancers to the expression changes of their associated genes, we found that multiple genes underwent trajectory changes (Fig. 5 A and B). The most pronounced trajectory change occurred for genes associated with active enhancers that were in the non-primed active cluster in the ChIP dataset, of which 58% changed to being transcriptionally activated by HDACi regardless of whether CFC had occurred (primed stable). This indicates that, while CFC was needed to drive their acetylation changes, it was no longer required for transcription. These genes were enriched for ontologies, including “positive regulation of signal transduction” and “nervous system development” (Fig. 5C), whereas ontology analysis for other cluster changes did not yield any significant hits. Genes in this group included voltage-gated potassium channels (such as Kcna1), a transcription factor required for fear learning (Neurod2) (52), and the IEG and AP-1 complex member, Jun (Fig. 5D). In addition, this cluster switch was enriched for various genes belonging to the MAPK signaling pathway, such as Mapk4 and Rapgef2 (Fig. 5E).
Fig. 5.
Overlap between HDACi-induced acetylation and transcriptional changes. (A) Sankey plot showing change in trajectory association for the 1,585 active enhancer-associated genes present in both the ChIP (Left) and bulk RNA-seq (Right). (B) Number of genes changing between ChIP-seq and the bulk RNA-seq clusters. (C) GOs for the 674 genes in the ChIP non-primed active cluster shifted to the RNA-seq primed stable cluster. (D) Example genome track of the H3K27ac and mRNA expression for Jun (active enhancer in gray and gene in blue). (E) KEGG visualization of the MAPK signaling pathway. Each box represents a gene for which the significant log2FC for the HDAC-CFC vs. VEH-CFC in the ChIP-seq (Left), bulk RNA-seq (Center), and snRNA-seq (Right) are color coded.
When comparing H3K27ac enrichment to the transcriptional activation in single nuclei of the DG (SI Appendix, Fig. S16A), we found that only 199 of the 4,594 genes were up-regulated in both analyses after combined HDACi-CFC treatment (SI Appendix, Fig. S16B). Despite this being a small subset—likely due to technical differences between the bulk and single nuclei preparations—these genes were also relevant to learning and memory in that they included NMDA receptors (Grin2a and Grin2b), a calcium voltage-gated ion channel (Cacna1e), and again, members of the MAPK pathway, including Mapk10 and Ras-guanine-nucleotide releasing factor 1(Rasgrf-1) (Fig. 5E), all of which contribute to glutamatergic synapse communication.
Finally, when comparing all three datasets together—namely enhancer acetylation, bulk and snRNA-seq transcriptional changes—we found the MAPK pathway to be predominantly activated (Fig. 5E). The MAPK/ERK pathway is necessary for memory consolidation (53, 54) and, once activated, ERK phosphorylates protein targets that are implicated in histone acetylation, gene transcription, protein synthesis, and synaptic plasticity (5, 33). Importantly, previous studies using different HDACis (i.e., sodium butyrate) have already demonstrated that HDACi-induced learning requires the activation of the MAPK pathway: knockout mice for mitogen- and stress-activated protein kinase 1 (Msk1), a member of the MAPK pathway, showed no HDACi-induced memory enhancement compared to wild-type mice (55, 56). In addition to the MAPK pathway, 18 more genes with H3K27ac enhancer enrichment were transcriptionally activated in all experiments (Dataset S12), of which two genes—autism susceptibility candidate 2 (Auts2) and cortactin binding protein 2 (Cttnbp2)—protect against autism-like behavior and regulate object recognition memory (57, 58). Taken together, these data suggest that genes involved in synaptic communication and MAPK signaling are the primary epigenetic and transcriptional targets of HDACi treatment, which in turn indicates these pathways to underlie HDACi-mediated memory enhancement.
Discussion
In this study, we aimed to determine the mechanisms by which HDACi application facilitates memory formation. We found that the HDACi CI-994 improves behavioral responses to contextual fear learning (Fig. 1B) and rotarod training (SI Appendix, Fig. S4B), regulated by the hippocampus and striatum, respectively. In both behavioral paradigms, CI-994 selectively enhanced unique aspects of synaptic communication within each brain region (Fig. 1 C and D and SI Appendix, Fig. S4F), despite these brain areas showing comparably reduced HDAC activity (Fig. 1 E and F). At the molecular level, HDACi treatment transcriptionally activated distinct gene subsets in each brain region (Fig. 2) and between different cell types within the hippocampus (Fig. 3). Finally, in DG neurons, HDACi treatment enriched H3K27ac at the enhancers of genes associated with synaptic function (Fig. 4), particularly at those involved in MAPK signaling (Fig. 5). Together, these findings indicate that CI-994, although applied systemically, results in brain region, cell-type, and pathway-specific effects.
These findings support the notion that CI-994 at least partly acts via cognitive epigenetic priming (3, 15). This model has originally been inspired by evidence from cancer research, where HDACi application—inherently devoid of target specificity—improves the efficacy of ongoing cancer treatments, while per se having no beneficial effects (16, 17). Analogously, here we found the HDACi application itself to have minimal effects; but when applied jointly with CFC, the HDACi treatment elicited electrophysiological, transcriptional, and epigenetic changes that paralleled the improved memory performance.
The brain region-specific effects likely occur because HDACi treatment reinforces behaviorally relevant cellular pathways in each brain area. When paired with CFC, HDACi specifically enhanced hippocampal LTP, which is known to underlie contextual fear learning (59–62), a finding similar to previous results in which CI-994 enhanced LTP in Schaffer collaterals when combined with extinction learning (14). Conversely, when paired with rotarod training, HDACi enhanced cortico-striatal PPF, which is known to underlie motor learning (28, 63, 64). This specialization is further supported by the differential transcriptional programs activated in these brain areas. While HDACi treatment enriched MAPK signaling in both brain regions irrespective of whether the animals were fear-conditioned or only context-exposed, the learning- and memory-related ERK1 and ERK2 cascade, as well as Bdnf and Jun, which are both involved in MAPK/ERK signaling (32, 47, 65, 66), were only enriched in the hippocampus in combination with learning. This suggests that HDACi generally targets the MAPK pathway but that, when paired with CFC, it leads to a further transcriptional enhancement thereof.
At the epigenetic level, we found a matching correlation between improved contextual memory formation, hippocampal LTP, and enhancer H3K27ac enrichment when HDACi treatment was paired with CFC (Fig. 4). But even after HDACi treatment alone, we observed H3K27ac-enriched pathways to be mainly associated with synaptic functions. Interestingly, past results have indicated that either HDACi (67) or CFC alone (29) enrich histone acetylation at regions that are already acetylated in baseline conditions. This suggests that the HDACi acts by reinforcing acetylated regions, which is likely, given that HDACs are known to be predominantly recruited to and act upon previously activated chromatin regions (68). Furthermore, H3K27ac enrichment also occurred at enhancers of the MAPK pathway (Fig. 5), which expands on previous findings linking HDACi treatment to this pathway (5, 55), and testifies to the importance of H3K27ac-induced epigenetic priming for improved memory performance.
At the same time, we observed that H3K27ac changes were not always translated into transcriptional changes (Fig. 5), which indicates some gene activation to be independent of H3K27ac priming at this time after learning. It is possible that this decoupling is driven by different cell populations in these experiments. Alternatively, the decoupling suggests that transcriptional changes likely rely on other epigenetic modifications, such as posttranslational histone modifications and DNA methylation, which are known to be important for memory formation (29, 36, 49, 69–73). However, since previous studies have also alluded to an epigenetic-transcriptional decoupling at similar time points after memory formation (29, 67), this is likely not the whole explanation. Indeed, our observation bears striking resemblance to a recent study that described an initial increase in enhancer accessibility following CFC, which was not yet paralleled by transcriptional changes, but only after several days postconditioning (73). Together, these findings suggest the following temporal order by which HDACi may improve memory formation. First, HATs and HDACs are recruited to regions of active gene transcription (68), which often occurs via the transcription machinery itself (74, 75). Second, during HDAC inhibition, these regions would remain open. Finally, during a subsequent neuronal activation, such as recall or high-frequency stimulation (HFS), genes primed in this way can then be more readily activated and thereby enhance memory retention. A precise experimental investigation of this time course is now required to test this order.
Given the multifactorial physiological and molecular underpinnings of learning and memory, there are several open questions emerging from this study. For example, while we only assessed DG-related histone acetylation changes, we cannot exclude the role of other hippocampal subregions, in particular CA1 (60), to be epigenetically altered by HDACi in response to CFC. Another limitation is the possibility that measuring mRNA and histone acetylation changes 1 h after CFC might be more representative of secondary-wave effects of HDACi application and CFC training, considering that many IEGs, acting as transcription factors themselves, are already up-regulated 30 min after CFC (46, 76). Of equal interest is the further study of HDACi-mediated down-regulated gene expression as observed here and previously (14, 67, 77). As down-regulated genes in both the bulk RNA-seq and snRNA-seq analyses were more associated with cellular metabolism and genetic regulation, this could indicate that as HDACi treatment enhances transcription of genes necessary for synaptic communication immediately following learning, it simultaneously silences processes involved in cellular maintenance. Finally, it remains to be determined whether similar molecular and physiological cascades are triggered by other HDACis or in bona fide learning-induced engram cells (78).
These open questions notwithstanding, the findings presented here shed light on the mechanisms by which systemic HDACi treatment can lead to specific memory-promoting effects. By enhancing neuronal activity-induced epigenetic and transcriptional cascades, HDACi application reaches a high level of target specificity despite being devoid of such specificity per se. Together with previous studies illustrating beneficial effects of HDACi-mediated regulation on synaptic genes in several neurodegenerative disorders (8–10, 79), these findings promote HDACis as a potential pharmacological intervention to improve cognitive capacities in both healthy and impaired conditions.
Materials and Methods
Animals.
Twelve-week-old male C57BL/6J mice were used for all experiments. Animals were housed in groups of four to five animals at 22 to 25 °C on a 12-h light/dark cycle with food and water ad libitum. All procedures, including animal experiments, were handled according to protocols approved by Swiss animal licenses VD2808/2808.1, VD2875/2875.1, VD3169, and VD3413 and according to the standard operating procedures of E-PHY-SCIENCE SAS [ENV/JM/MNO (2077)].
Drug Administration.
CI-994 (14) was dissolved in 10% dimethyl sulfoxide (Sigma-Aldrich), 30% Kolliphor (Sigma-Aldrich), and 60% 0.9% saline (Braun). Animals were interperitoneally injected with 30 mg/kg of CI-994 or a corresponding volume of VEH.
CFC.
CFC consisted of a 3-min habituation, two 1-s foot shocks (0.2 mA) separated by 29 s, and a final 15 s in the chamber (TSE Systems GmbH at École Polytechnique Fédérale de Lausanne; Imetronic for electrophysiology). For the HDACi assay, CFC consisted of three 2-s foot shocks (0.8 mA). Context groups were exposed to the conditioning chamber without shocks. To measure fear learning, animals were re-exposed to the chamber 24 h after training. Percentage of time spent freezing (>1 s) was calculated with an infrared beam detection system (MultiConditioning System, TSE Systems GmbH). For all molecular experiments, animals were left in their home cage for 1 h post-CFC before killing and dissection.
Electrophysiology.
One hour after behavioral experiments, mice were anesthetized (isoflurane), decapitated, and brains were extracted. Slices (350-μm thick) were prepared with a vibratome (VT 1000S; Leica Microsystems) in ice-cold gassed artificial cerebrospinal fluid (aCSF; 124 mM NaCl, 1.3 mM MgSO4, 4 mM KCl, 1.0 mM Na2HPO4, 2.0 mM CaCl2, 26 mM NaHCO3, 10 mM d-glucose). Sections were kept at room temperature for 1 h before recording.
For recordings, one slice was submerged in the recording chamber and superfused with gassed (95% O2, 5% CO2) aCSF at a constant rate (2 mL/min). Electrical stimulation (0.25 Hz, 100-µs duration) evoked field excitatory postsynaptic potential (fEPSPs) in the perforant path, Schaffer collaterals, or the cortico-striatal pathway. Downstream extracellular fEPSPs were recorded in the DG, CA1, and striatum, respectively, using a glass micropipette filled with aCSF. Stable fEPSPs were recorded by stimulating at 30% maximal field amplitude (single stimulation [0.25 Hz] every 30 s). HFS (three trains of 100-Hz stimulation; 2× 1 s, separated by 20 s), was delivered and a 90-min test period was recorded. Signals were amplified with an Axopatch 200B (Molecular Devices) digitized by a Digidata 1322A interface (Molecular Devices) and sampled at 10 kHz. Recordings were acquired using Clampex (Molecular Devices) and analyzed with Clampfit (Molecular Devices). LTP was measured as percent of baseline fEPSP slope recorded over a 10-min period before HFS delivery. This value was taken as 100% of the excitatory postsynaptic potential and all recorded values were normalized to this.
HDAC Activity Assay.
HDAC activity of hippocampal and striatal hemispheres was assessed using the Fluor de Lys HDAC fluorometric activity kit (Enzo Life Science, BML-AK500) (for details see SI Appendix).
RNA-Seq.
RNA extraction and library preparation.
Total RNA was isolated from hippocampal and striatal hemispheres using TRIzol Reagent (Life Technologies), and purified using the RNase-Free DNase Set (Qiagen, Cat# 79254) and the RNAeasy Mini Kit (Qiagen). Libraries were prepared using the TruSeq Stranded mRNA Preparation Kit (Illumina) then multiplexed and sequenced on the Illumina HiSEq. 4000, yielding 100-bp, paired-end reads.
RNA-seq analysis.
Truseq adapter sequences were trimmed from FASTQ files using bcl2fastq (v2.20.0, Illumina). STAR (v2.6) (80) aligned FASTQ reads to the mouse mm10 reference genome with annotations from Ensembl release 93 (81). Custom R scripts were used to count reads mapping to exonic regions. Differential expression was performed using DEseq2 (82) and DEGs were considered if they had P ≤ 0.05 and a log2FC ≥ 0.4, as described previously (14). For the trajectory analysis, genes were grouped using custom-written decision tree clustering in R.
Nuclear Extraction.
Nuclear extraction was performed for both ChIP and snRNA-seq experiments. Frozen brain tissue was homogenized in 6 mL Solution D (0.25 M Sucrose, 25 mM KCl, 5 mM MgCl2, 20 mM Tris⋅HCl, pH 7.5; plus 5 µg/mL actinomycin D [Sigma, Cat# A4262] for snRNA-seq). Samples were purified twice with Optiprep (Serumwerk Bernburg) and centrifuged for 10 min at 3,200 × g. Pellets were resuspended in PBS-T for the ChIP and N-PBS (PBS, 5% BSA, 5 µg/mL actinomycinD, and 0.2 U/µL RNase inhibitor [Thermo Fisher Scientific]) for the snRNA-seq then filtered into 5-mL polysterene tubes with filter (75 mm) snap-caps (Corning).
ChIP-Seq.
ChIP and library preparation.
Extracted nuclei were cross-linked in 1% formaldehyde (AppliChem) for 5 min and quenched with 125 mM glycine (VWR). Approximately 750,000 nuclei per sample were resuspended in 500 µL PBS-T. Nuclei were stained with 1:50 Alexa Fluor488 conjugated anti-NeuN (Millipore, MAB377X) for 30 min, then washed in PBS-T. Nuclei were stored in 200 µL PBS-T, passed through a 26-G needle, and incubated with Hoechst (1:1,000) before sorting on the FACSAriaIII (BD Bioscience). Sorted NeuN+ nuclei were pelleted (4 °C, 1,250 × g, 5 min) and lysed in 750 µL RIPA buffer for 10 min. Samples were sonicated on an E220 Focused-ultra-sonicator (Covaris) for 20 min (peak power = 140 W, duty = 5, cycle/burst = 200). Low-input ChIP was carried out using the Low Cell ChIP-Seq Kit (Active Motif) using H3K27ac (Abcam, ab4729). The Next Gen DNA Library and Indexing Kit (Active Motif) were used to prepare libraries (for details, see SI Appendix). Libraries were resuspended in 25 µL Low EDTA TE buffer then analyzed with the fragment analyzer (NGS High Sensitivity kit, Agilent) and sequenced, paired-end, on the Illumina NextSEq. 500.
ChIP-seq analysis.
Molecular identifier sequences were appended to FASTQ headers. Adapter sequences and low-quality regions were removed using Trimmomatic (v0.38) (83) (ILLUMINACLIP:Y2_adapter_seq.fa:0:6:6 SLIDINGWINDOW:10:20 MINLEN:36). Processed FASTQs were aligned to the mm10 genome using Bowtie2 (v2.3.5) (84). Differentially acetylated regions were identified using Diffbind (v2.16.2) (85) and DEseq2 (v1.28.1) (82). ChromHMM (v1.22) (86) was run on independently published ChIP-seq data (29). Five chromatin states were assigned (control regions, repressed regions, promoter regions, primed enhancers, active enhancers) based on their histone marks. We assigned enhancers to genes using HOMER (v4.11) annotatePeaks.pl (87). Trajectory analysis was performed for each chromatin state.
snRNA-Seq.
Library sequencing.
Hippocampi from five mice were pooled into two replicates per group. Nuclei were extracted and diluted to 1,000 nuclei per microliter. Libraries were constructed using Chromium SingleCell 3′Reagent Kit v3 chemistry (10X Genomics; CG000183 - Rev A), then pooled and sequenced across 2 NextSEq. 500 (v2.5) chips for 75 cycles.
snRNA-seq analysis.
Cellranger count (CellRanger v3.0.1) aligned FASTQ files to the mm10 pre-mRNA genome (expect-cells = 4,000, chemistry = SC3Pv3, r2-length = 56). Seurat (v4.0.3) (88) was used to calculate quality-control metrics. DoubletFinder (89) was used to find and remove doublets and normalization and was done using SCTransform (90). Seurat then performed UMAP clustering and defined clusters. Differential expression analysis was performed for each cell type using the logistic regression framework in Seurat’s FindMarkers.
All in-house analysis code can be found at https://github.com/allie-burns/2022_Burns_etal.
Accession Codes.
Raw data sets generated during this study are available in the Gene Expression Omnibus repository using accession no. GSE185455 (including subseries: GSE185452, GSE185453, and GSE185454)
Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Visualization.
The MAPK KEGG pathway (mmu04010) was downloaded and drawn using Pathview (91). Pathways were simplified for visualization purposes by only plotting MAPK subpathways containing at least one differentially acetylated or transcribed gene.
Supplementary Material
Acknowledgments
We thank all past and current members of the Laboratory of Neuroepigenetics for their support and discussion throughout this project, in particular, Paola Arguello and Diego Camacho for their contribution to molecular protocols and analysis; the École Polytechnique Fédérale de Lausanne (EPFL) Gene Expression Core Facility for their technical assistance with experiment planning and sequencing; the EPFL Flow Cytometry Core Facility for providing the nuclear sorting; and the EPFL Center of Phenogenomics for ensuring the welfare of the laboratory animals. The work in the laboratory of J.G. is supported by the European Research Council (ERC-2015-StG 678832), the Swiss National Science Foundation (SNSF, 31003A_155898), the National Competence Center for Research SYNAPSY (51NF40-185897), and the Floshield and Dragon Blue Foundations.
Footnotes
Competing interest statement: J.G. is coauthor of a patent (12/917,402) using CI-994 for the treatment of cognitive disorders.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116797119/-/DCSupplemental.
Data Availability
RNA-seq, snRNA-seq, ChIP-seq, and in-house analysis code data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE185455; including subseries: GSE185452, GSE185453, and GSE185454) (92) and GitHub (https://github.com/allie-burns/2022_Burns_etal) (93).
References
- 1.Kandel E. R., The molecular biology of memory storage: A dialogue between genes and synapses. Science 294, 1030–1038 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Lee Y. S., Silva A. J., The molecular and cellular biology of enhanced cognition. Nat. Rev. Neurosci. 10, 126–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gräff J., Tsai L. H., Histone acetylation: Molecular mnemonics on the chromatin. Nat. Rev. Neurosci. 14, 97–111 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Campbell R. R., Wood M. A., How the epigenome integrates information and reshapes the synapse. Nat. Rev. Neurosci. 20, 133–147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levenson J. M., et al. , Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 279, 40545–40559 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Villain H., Florian C., Roullet P., HDAC inhibition promotes both initial consolidation and reconsolidation of spatial memory in mice. Sci. Rep. 6, 27015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bousiges O., et al. , Spatial memory consolidation is associated with induction of several lysine-acetyltransferase (histone acetyltransferase) expression levels and H2B/H4 acetylation-dependent transcriptional events in the rat hippocampus. Neuropsychopharmacology 35, 2521–2537 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janczura K. J., et al. , Inhibition of HDAC3 reverses Alzheimer’s disease-related pathologies in vitro and in the 3xTg-AD mouse model. Proc. Natl. Acad. Sci. U.S.A. 115, E11148–E11157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Benito E., et al. , HDAC inhibitor-dependent transcriptome and memory reinstatement in cognitive decline models. J. Clin. Invest. 125, 3572–3584 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volmar C. H., et al. , M344 promotes nonamyloidogenic amyloid precursor protein processing while normalizing Alzheimer’s disease genes and improving memory. Proc. Natl. Acad. Sci. U.S.A. 114, E9135–E9144 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peleg S., et al. , Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 328, 753–756 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Gräff J., et al. , An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature 483, 222–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vecsey C. G., et al. , Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J. Neurosci. 27, 6128–6140 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gräff J., et al. , Epigenetic priming of memory updating during reconsolidation to attenuate remote fear memories. Cell 156, 261–276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burns A. M., Gräff J., Cognitive epigenetic priming: Leveraging histone acetylation for memory amelioration. Curr. Opin. Neurobiol. 67, 75–84 (2021). [DOI] [PubMed] [Google Scholar]
- 16.Scandura J. M., et al. , Phase 1 study of epigenetic priming with decitabine prior to standard induction chemotherapy for patients with AML. Blood 118, 1472–1480 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terranova-Barberio M., et al. , HDAC inhibition potentiates immunotherapy in triple negative breast cancer. Oncotarget 8, 114156–114172 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mews P., Walker D. M., Nestler E. J., Epigenetic priming in drug addiction. Cold Spring Harb. Symp. Quant. Biol. 83, 131–139 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robison A. J., Nestler E. J., Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci. 12, 623–637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parsons R. G., Davis M., A metaplasticity-like mechanism supports the selection of fear memories: Role of protein kinase a in the amygdala. J. Neurosci. 32, 7843–7851 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradner J. E., et al. , Chemical genetic strategy identifies histone deacetylase 1 (HDAC1) and HDAC2 as therapeutic targets in sickle cell disease. Proc. Natl. Acad. Sci. U.S.A. 107, 12617–12622 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li S., et al. , Early histone deacetylase inhibition mitigates ischemia/reperfusion brain injury by reducing microglia activation and modulating their phenotype. Front. Neurol. 10, 893 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sada N., et al. , Inhibition of HDAC increases BDNF expression and promotes neuronal rewiring and functional recovery after brain injury. Cell Death Dis. 11, 655 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper A., et al. , Inhibition of histone deacetylation rescues phenotype in a mouse model of Birk-Barel intellectual disability syndrome. Nat. Commun. 11, 480 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips R. G., LeDoux J. E., Differential contribution of amygdala and hippocampus to cued and contextual fear conditioning. Behav. Neurosci. 106, 274–285 (1992). [DOI] [PubMed] [Google Scholar]
- 26.Ferreira T. L., Moreira K. M., Ikeda D. C., Bueno O. F. A., Oliveira M. G. M., Effects of dorsal striatum lesions in tone fear conditioning and contextual fear conditioning. Brain Res. 987, 17–24 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Costa R. M., Cohen D., Nicolelis M. A. L., Differential corticostriatal plasticity during fast and slow motor skill learning in mice. Curr. Biol. 14, 1124–1134 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Assous M., et al. , Neuropilin 2 signaling mediates corticostriatal transmission, spine maintenance, and goal-directed learning in mice. J. Neurosci. 39, 8845–8859 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halder R., et al. , DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat. Neurosci. 19, 102–110 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Liz M. A., et al. , A narrative review of the role of transthyretin in health and disease. Neurol. Ther. 9, 395–402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brouillette J., Quirion R., Transthyretin: A key gene involved in the maintenance of memory capacities during aging. Neurobiol. Aging 29, 1721–1732 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Peng S., Zhang Y., Zhang J., Wang H., Ren B., ERK in learning and memory: A review of recent research. Int. J. Mol. Sci. 11, 222–232 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis S., Vanhoutte P., Pagès C., Caboche J., Laroche S., The MAPK/ERK cascade targets both Elk-1 and cAMP response element-binding protein to control long-term potentiation-dependent gene expression in the dentate gyrus in vivo. J. Neurosci. 20, 4563–4572 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J., Wu X., Zhang H., Pfeifer G. P., Lu Q., Dynamics of RNA polymerase II pausing and bivalent histone H3 mthylation during neuronal differentiation in brain development. Cell Rep. 20, 1307–1318 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang H. J., Schuman E. M., Neurotrophin-induced modulation of synaptic transmission in the adult hippocampus. J. Physiol. Paris 89, 11–22 (1995). [DOI] [PubMed] [Google Scholar]
- 36.Su Y., et al. , Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat. Neurosci. 20, 476–483 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adams J. P., Sweatt J. D., Molecular psychology: Roles for the ERK MAP kinase cascade in memory. Annu. Rev. Pharmacol. Toxicol. 42, 135–163 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Zalocusky K. A., et al. , Neuronal ApoE upregulates MHC-I expression to drive selective neurodegeneration in Alzheimer’s disease. Nat. Neurosci. 24, 786–798 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaeger B. N., et al. , A novel environment-evoked transcriptional signature predicts reactivity in single dentate granule neurons. Nat. Commun. 9, 3084 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hrvatin S., et al. , Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat. Neurosci. 21, 120–129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y., et al. , An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lein E. S., et al. , Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Zeisel A., et al. , Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Erö C., Gewaltig M. O., Keller D., Markram H., A cell atlas for the mouse brain. Front. Neuroinform. 12, 84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Squair J. W., Skinnider M. A., Gautier M., Foster L. J., Courtine G., Prioritization of cell types responsive to biological perturbations in single-cell data with Augur. Nat. Protoc. 16, 3836–3873 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Tyssowski K. M., et al. , Different neuronal activity patterns induce different gene expression programs. Neuron 98, 530–546.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L. F., et al. , Enhancer histone acetylation modulates transcriptional bursting dynamics of neuronal activity-inducible genes. Cell Rep. 26, 1174–1188.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malik A. N., et al. , Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat. Neurosci. 17, 1330–1339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandez-Albert J., et al. , Immediate and deferred epigenomic signatures of in vivo neuronal activation in mouse hippocampus. Nat. Neurosci. 22, 1718–1730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Creyghton M. P., et al. , Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U.S.A. 107, 21931–21936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hinds H. L., Goussakov I., Nakazawa K., Tonegawa S., Bolshakov V. Y., Essential function of α-calcium/calmodulin-dependent protein kinase II in neurotransmitter release at a glutamatergic central synapse. Proc. Natl. Acad. Sci. U.S.A. 100, 4275–4280 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin C. H., et al. , The dosage of the neuroD2 transcription factor regulates amygdala development and emotional learning. Proc. Natl. Acad. Sci. U.S.A. 102, 14877–14882 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Atkins C. M., Selcher J. C., Petraitis J. J., Trzaskos J. M., Sweatt J. D., The MAPK cascade is required for mammalian associative learning. Nat. Neurosci. 1, 602–609 (1998). [DOI] [PubMed] [Google Scholar]
- 54.Blum S., Moore A. N., Adams F., Dash P. K., A mitogen-activated protein kinase cascade in the CA1/CA2 subfield of the dorsal hippocampus is essential for long-term spatial memory. J. Neurosci. 19, 3535–3544 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chwang W. B., Arthur J. S., Schumacher A., Sweatt J. D., The nuclear kinase mitogen- and stress-activated protein kinase 1 regulates hippocampal chromatin remodeling in memory formation. J. Neurosci. 27, 12732–12742 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Monsey M. S., Ota K. T., Akingbade I. F., Hong E. S., Schafe G. E., Epigenetic alterations are critical for fear memory consolidation and synaptic plasticity in the lateral amygdala. PLoS One 6, e19958 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hori K., et al. , Heterozygous disruption of autism susceptibility candidate 2 causes impaired emotional control and cognitive memory. PLoS One 10, e0145979 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shih P. Y., et al. , Autism-linked mutations of CTTNBP2 reduce social interaction and impair dendritic spine formation via diverse mechanisms. Acta Neuropathol. Commun. 8, 185 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bliss T. V. P., Collingridge G. L., A synaptic model of memory: Long-term potentiation in the hippocampus. Nature 361, 31–39 (1993). [DOI] [PubMed] [Google Scholar]
- 60.Sacchetti B., et al. , Time-dependent inhibition of hippocampal LTP in vitro following contextual fear conditioning in the rat. Eur. J. Neurosci. 15, 143–150 (2002). [DOI] [PubMed] [Google Scholar]
- 61.Whitlock J. R., Heynen A. J., Shuler M. G., Bear M. F., Learning induces long-term potentiation in the hippocampus. Science (80-.) 313, 1093–1097 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Nabavi S., et al. , Engineering a memory with LTD and LTP. Nature 511, 348–352 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haber S. N., Corticostriatal circuitry. Dialogues Clin. Neurosci. 18, 7–21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cataldi S., Stanley A. T., Miniaci M. C., Sulzer D., Interpreting the role of the striatum during multiple phases of motor learning. FEBS J. 289, 2263–2281 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sütterlin P., et al. , The molecular basis of the cooperation between EGF, FGF and eCB receptors in the regulation of neural stem cell function. Mol. Cell. Neurosci. 52, 20–30 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Revest J. M., et al. , BDNF-TrkB signaling through Erk1/2 MAPK phosphorylation mediates the enhancement of fear memory induced by glucocorticoids. Mol. Psychiatry 19, 1001–1009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lopez-Atalaya J. P., Ito S., Valor L. M., Benito E., Barco A., Genomic targets, and histone acetylation and gene expression profiling of neural HDAC inhibition. Nucleic Acids Res. 41, 8072–8084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Z., et al. , Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138, 1019–1031 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miller C. A., Campbell S. L., Sweatt J. D., DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol. Learn. Mem. 89, 599–603 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gupta S., et al. , Histone methylation regulates memory formation. J. Neurosci. 30, 3589–3599 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Duke C. G., Kennedy A. J., Gavin C. F., Day J. J., Sweatt J. D., Experience-dependent epigenomic reorganization in the hippocampus. Learn. Mem. 24, 278–288 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pearce K., Cai D., Roberts A. C., Glanzman D. L., Role of protein synthesis and DNA methylation in the consolidation and maintenance of long-term memory in Aplysia. eLife 6, e18299 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marco A., et al. , Mapping the epigenomic and transcriptomic interplay during memory formation and recall in the hippocampal engram ensemble. Nat. Neurosci. 23, 1606–1617 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ali I., et al. , Crosstalk between RNA Pol II C-terminal domain acetylation and phosphorylation via RPRD proteins. Mol. Cell 74, 1164–1174.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carullo N. V. N., et al. , Enhancer RNAs predict enhancer-gene regulatory links and are critical for enhancer function in neuronal systems. Nucleic Acids Res. 48, 9550–9570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saha R. N., et al. , Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat. Neurosci. 14, 848–856 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Greer C. B., et al. , Histone deacetylases positively regulate transcription through the elongation machinery. Cell Rep. 13, 1444–1455 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Josselyn S. A., Köhler S., Frankland P. W., Finding the engram. Nat. Rev. Neurosci. 16, 521–534 (2015). [DOI] [PubMed] [Google Scholar]
- 79.Siebzehnrübl F. A., et al. , Early postnatal behavioral, cellular, and molecular changes in models of Huntington disease are reversible by HDAC inhibition. Proc. Natl. Acad. Sci. U.S.A. 115, E8765–E8774 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zerbino D. R., et al. , Ensembl 2018. Nucleic Acids Res. 46, D754–D761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stark R., Brown G., DiffBind: Differential binding analysis of ChIP-Seq peak data (2011). https://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf. Accessed 16 September 2020.
- 86.Ernst J., Kellis M., ChromHMM: Automating chromatin-state discovery and characterization. Nat. Methods 9, 215–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heinz S., et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hao Y., et al. , Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McGinnis C. S., Murrow L. M., Gartner Z. J., DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hafemeister C., Satija R., Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luo W., Brouwer C., Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29, 1830–1831 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.A. M. Burns, J. Gräff, GEO database release for Burns et al., PNAS (2022). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE185455. Deposited 6 October 2021. [Google Scholar]
- 93.A. M. Burns, allie-burns/2022_Burns_etal: Code Repository Release for Burns et al., 2022, PNAS (2022). Zenodo. 10.5281/zenodo.6543201. Deposited 12 May 2022. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq, snRNA-seq, ChIP-seq, and in-house analysis code data have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE185455; including subseries: GSE185452, GSE185453, and GSE185454) (92) and GitHub (https://github.com/allie-burns/2022_Burns_etal) (93).