Significance
Actin filament nucleation by Arp2/3 complex must be triggered by activators like WASP family proteins. Understanding how WASP proteins activate Arp2/3 complex has been a major challenge due to a lack of high-resolution structures of the complex in an activated state. We determined a high-resolution (∼3.9 Å) structure of the WASP-activated Arp2/3 complex at a branch junction and used biochemical, cell biological, and molecular dynamic simulations to understand the mechanism of WASP-mediated activation. This work shows in detail the contacts between the fully activated Arp2/3 complex, the nucleated daughter actin filament, and the mother actin filament and provides important insights into how conformational rearrangements in the Arp2/3 complex are stimulated during activation.
Keywords: Arp2/3, cryo-electron microscopy, nucleation, actin, cytoskeleton
Abstract
Arp2/3 complex nucleates branched actin filaments that provide pushing forces to drive cellular processes such as lamellipodial protrusion and endocytosis. Arp2/3 complex is intrinsically inactive, and multiple classes of nucleation promoting factors (NPFs) stimulate its nucleation activity. When activated by WASP family NPFs, the complex must bind to the side of a preexisting (mother) filament of actin to complete the nucleation process, ensuring that WASP-mediated activation creates branched rather than linear actin filaments. How actin filaments contribute to activation is currently not understood, largely due to the lack of high-resolution structures of activated Arp2/3 complex bound to the side of a filament. Here, we present the 3.9-Å cryo-electron microscopy structure of the Arp2/3 complex at a branch junction. The structure reveals contacts between Arp2/3 complex and the side of the mother actin filament that likely stimulate subunit flattening, a conformational change that allows the actin-related protein subunits in the complex (Arp2 and Arp3) to mimic filamentous actin subunits. In contrast, limited contact between the bottom half of the complex and the mother filament suggests that clamp twisting, a second major conformational change observed in the active state, is not stimulated by actin filaments, potentially explaining why actin filaments are required but insufficient to trigger nucleation during WASP-mediated activation. Along with biochemical and live-cell imaging data and molecular dynamics simulations, the structure reveals features critical for the interaction of Arp2/3 complex with actin filaments and regulated assembly of branched actin filament networks in cells.
To drive complex processes like motility and division, cells must precisely control the architecture, timing, and localization of the assembly of actin into filamentous networks. Polymerization of new actin filaments is limited by a slow nucleation step, so actin filament nucleator proteins provide a mechanism for cells to exert spatiotemporal control over actin filament assembly (1–3). Actin-related protein 2/3 (Arp2/3) complex, a seven-subunit ∼225-kDa protein assembly, was the first actin filament nucleator discovered and is the only one known to nucleate branched actin filaments (3), although depending on the identity of its activator, the complex can also nucleate linear actin filaments (4). Branched actin filaments nucleated by Arp2/3 complex provide pushing forces for processes ranging from lamellipodial protrusion to endocytic invagination, pathogen invasion, cellular differentiation, meiotic spindle positioning, and DNA repair (5–10).
Arp2/3 complex has little or no activity on its own; regulatory proteins called nucleation promoting factors (NPFs) must bind to the complex to trigger its actin filament nucleation activity (11). Structural and biochemical data have begun to reveal the molecular basis for nucleation. Two of the seven subunits in the complex, namely, Arp2 and Arp3, show close sequence and structural similarity to actin (12). In the inactive state, Arp3 and Arp2 are arranged in an end-to-end (splayed) conformation, an arrangement that is structurally distinct from that of two consecutive actin subunits within a filament (13). In addition, the Arps are in a twisted conformation, in which they resemble actin monomers rather than filamentous actin subunits, which have a flattened conformation (14, 15). A recent cryo-electron microscopy (cryo-EM) structure of Arp2/3 complex bound to a WISH/DIP/SPIN90 family NPF provided the first high-resolution structure of the complex in an activated state (15). It demonstrated that during activation, intra- and intersubunit conformational changes bring the complex into a conformation in which the two actin-related protein subunits Arp2 and Arp3 mimic the arrangement of two consecutive subunits along the short pitch helical axis of an actin filament (i.e., the short pitch conformation) and each Arp transitions from a twisted (monomer-like) to a flattened (filament-like) conformation (15). How activators of Arp2/3 complex drive these conformational changes has been an important open question.
WASP proteins compose the largest and best studied family of NPFs. They are characterized by the presence of a C-terminal Verprolin homology-Central-Acidic (VCA) segment, the minimal region of WASP sufficient to activate Arp2/3 complex (16, 17). When activated by WASP, Arp2/3 complex nucleates branched actin filaments (18, 19). Therefore, the coordinated action of WASP and Arp2/3 complex results in the assembly of highly dendritic actin networks that are optimal for pushing against broad, flat membrane surfaces (20, 21). Importantly, WASP family proteins alone are insufficient for activation of Arp2/3 complex. To complete activation, WASP must also bind and recruit actin monomers to the complex, and the complex must bind to the side of a preformed (mother) actin filament (16, 22). The requirement for all three of these factors (WASP, WASP-recruited actin monomer, and preformed actin filament) is thought to be critical for the proper function of the Arp2/3 complex in cells; WASP connects the Arp2/3 complex to cellular signaling pathways (23), the requirement for actin monomers helps regulate the density of branches created by the complex (24), and the requirement for preformed actin filaments ensures that WASP-mediated activation of the complex creates branches (22). Therefore, understanding the role of each activating factor—and why all three are required for activation—is critical to understanding how cells properly regulate branched actin assembly in cells.
Due to the efforts of multiple laboratories, there are now clear roles for WASP and WASP-recruited actin monomers in triggering activating conformational changes in Arp2/3 complex. For instance, several biochemical experiments, including engineered chemical crosslinking and FRET experiments, demonstrate that WASP and WASP-recruited actin monomers drive the two actin-related subunits into the short pitch conformation (25–28). Decreasing the ability of WASP (and WASP-recruited actin monomers) to stimulate the short pitch conformation through mutations or with small-molecule inhibitors blocks nucleation, demonstrating the importance of the short pitch conformational change (26, 28, 29). In contrast to WASP, little is known about the role of actin filaments in activation. While some models have postulated that actin filaments act cooperatively with WASP to trigger the short pitch conformational change, new data indicate that filaments do not stimulate the short pitch conformation and instead trigger distinct activating changes (30), although the nature of these changes are not yet known. Furthermore, while it is understood that actin filaments are required for WASP-mediated activation, how the activating function of preformed filaments is suppressed in the absence of WASP is poorly understood.
Our understanding of the role of actin filaments in activation has been hampered by insufficient structural information about the interactions between Arp2/3 complex and the sides of preexisting (mother) filaments. A 26-Å reconstruction of negatively stained branch junctions modeled all seven subunits of Arp2/3 complex in contact with the mother filament but showed that the main interactions were made by the clamp subunits in the complex, namely, ARPC2 and ARPC4, as had been indicated by mutational analysis (31, 32). A recent 9-Å reconstruction of branch junctions in isolated fibroblasts verified the general position of Arp2/3 complex on filaments but showed that the interface was far less extensive than predicted based on the 26-Å structure, with only five of the seven subunits contacting the mother filament (32, 33). Both structures provided important advances, but their relatively low resolution limited our understanding of the role of mother filaments in driving the Arp2/3 complex to a nucleation competent state. Furthermore, while a high-resolution (∼3.9 Å) cryo-EM structure of the Dip1-activated Arp2/3 complex was recently solved, Dip1 belongs to a class of NPFs that activates Arp2/3 complex without requiring a preformed actin filament (4, 15). Therefore, in that structure, the Arp2/3 complex is bound only to Dip1 and the pointed end of the new filament that it nucleated—no mother filament is present (15).
Here, we present a 3.9-Å cryo-EM structure of activated Bos taurus Arp2/3 complex at an actin filament branch junction. The structure shows how binding to the side of the actin filament can stimulate subunit flattening in Arp2/3 complex, thus providing a structural rationale for the requirement of actin filaments in WASP-mediated activation. The binding mode of the complex, in which only approximately its top half contacts the mother filament, likely prevents filaments from stimulating the short pitch conformation, potentially explaining why actin filaments alone are insufficient for activation. The branch junction model along with biochemical, live-cell imaging, and computational data reveal key functional and structural roles for conserved interactions of the complex with actin filaments and show that actin filament binding by the Arp2/3 complex is critical for proper endocytic actin assembly in yeast.
Results
Cryo-EM Reconstruction of Arp2/3 Complex at a Branched-Actin Junction.
Obtaining high-resolution structures of Arp2/3 complex at branched-actin junctions has been a major challenge. This is partly due to the difficulty of obtaining a sufficient density of branched junctions on the electron microscopy (EM) grid, which limits the number of particles that can be included in a high-resolution three-dimensional (3D) reconstruction. Additionally, the Y-shape of branches causes them to adopt limited orientations in ice (34). To overcome these obstacles, we developed a reconstitution strategy for generating a high density of branched actin junctions in vitro. Specifically, we created branched actin filaments by mixing the Bos taurus Arp2/3 complex, Oryctolagus cuniculus skeletal muscle actin, and the Arp2/3 complex activator N-WASP (SI Appendix, Fig. S1). We included the NPF protein SPIN90 to generate seed actin filaments to activate the WASP-bound Arp2/3 complex and capping protein to promote branching nucleation over elongation (4, 35). We also added the Type II NPF cortactin and phalloidin to the reaction mixes to stabilize the branches (36, 37). Furthermore, we optimized the data collection strategy by tilting the specimen, enabling us to obtain various projections of branched-junction particles in the micrographs, thus overcoming the limited orientation problem.
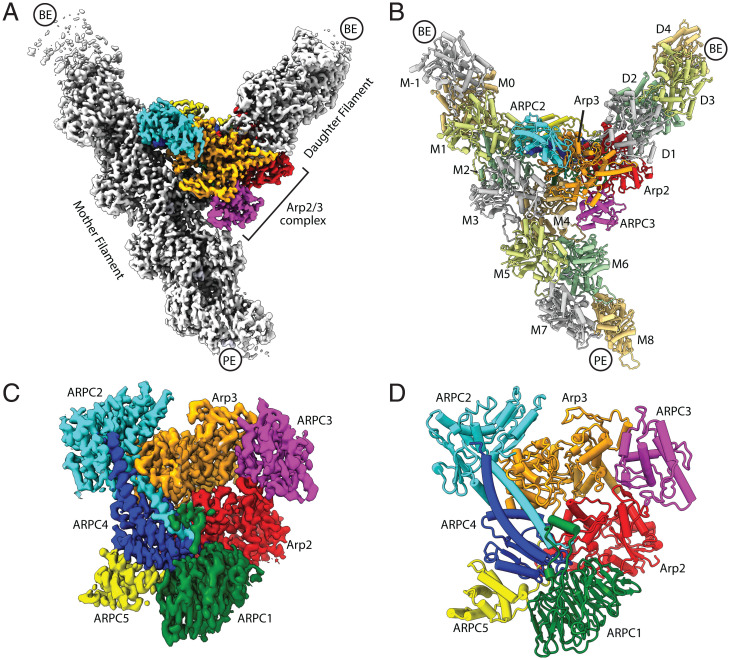
All seven subunits of Arp2/3 complex are visible in the reconstruction (Fig. 1 and SI Appendix, Fig. S3 and Table S1). The final map also includes 10 actin subunits within the mother filament and 4 actin subunits within the nucleated (daughter) filament (Fig. 1 A and B and SI Appendix, Fig. S3). The angle between the mother and daughter filament is 70 degrees, consistent with the recent in situ tomographic reconstruction of a branch junction at 9 Å (33). The reconstructed density is weakest for the peripheral actin subunits in the mother and daughter filaments and strongest for Arp2/3 complex subunits and actin subunits proximal to the complex (SI Appendix, Figs. S1 and S3).
Fig. 1.
Overview of Cryo-EM structure of the branch junction. (A) Reconstructed map of the branch junction. PE, pointed end; BE, barbed end. (B) Molecular model of the branch junction. (C) Reconstructed map of Arp2/3 complex at the branch junction. (D) Molecular model of Arp2/3 complex at a branch junction.
Actin subunits in the mother and daughter filaments showed density in their nucleotide pocket consistent with ADP (SI Appendix, Fig. S4), as expected based on the rate constants for ATP hydrolysis and phosphate release by actin (38). ADP also fits the density well in the active sites of both Arp2 and Arp3, suggesting that both actin-related subunits have also hydrolyzed their nucleotides (SI Appendix, Fig. S4). This observation is important because ATP hydrolysis by the complex helps destabilize branches during actin network disassembly (39, 40), and technical limitations have prevented biochemical confirmation that the Arp3 subunit catalyzes nucleotide hydrolysis (41, 42). We did not observe density attributable to WASP in the map, consistent with previous biochemical experiments showing that WASP is released before nucleation (43). Density for cortactin is also missing in the reconstruction, so we do not expect its presence in the reaction to have a significant influence on the branch junction structure. Five molecules of phalloidin are bound to the daughter filament and 10 are bound to the mother filament (SI Appendix, Fig. S5). While phalloidin binding is reported to influence the conformation of the D-loop in filamentous actin subunits under some conditions (44), the structures of the mother and daughter filaments presented here are nearly identical to that of ADP-bound actin filaments without phalloidin (45, 46) (SI Appendix, Fig. S5). Therefore, we do not expect that phalloidin significantly influences the interactions of actin subunits with Arp2/3 complex at the branch junction.
Interactions between the Mother Filament and Arp2/3 Complex.
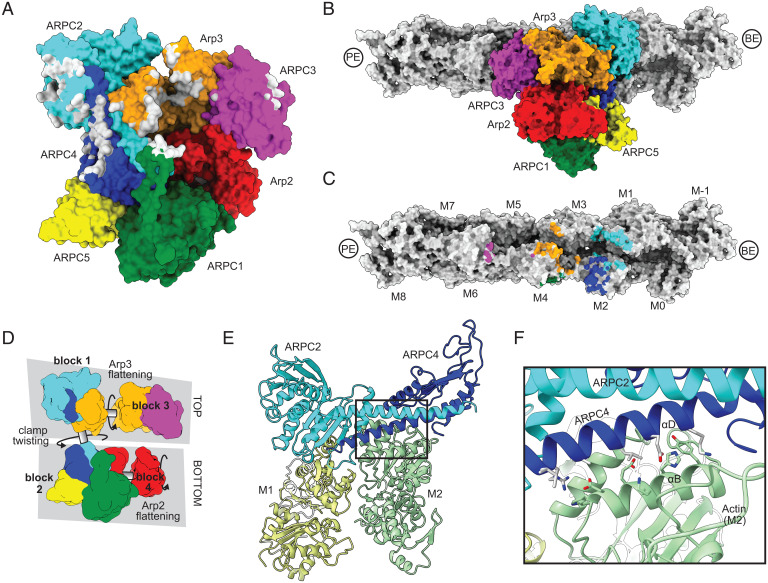
The arrangement of Arp2/3 complex and the geometries of the mother and daughter filament are overall very similar to the recent 9-Å reconstruction of actin filament branches in cells presented in Fäßler et al. (33). Arp2/3 complex bridges the two protofilament strands within the mother filament to bury a total of ∼3,500 Å2 at the interface (Fig. 2 A–C). The complex contacts a total of five actin subunits in the mother filament (M1, M2, M3, M4, and M6), but most of the contacts (∼87% of the buried surface area) are made with three adjacent actin subunits (M1, M2, and M4) toward the barbed end side of the interface (Fig. 2C). While a previous low-resolution EM reconstruction reported that actin subunits M4 and M2 are distorted at the branch junction (32), we find here that all actin subunits within the mother filament have conformations nearly identical to high-resolution cryo-EM structures of unbound actin filaments (SI Appendix, Fig. S6 A–C) (45, 46). Furthermore, the twist and rise of the subunits within the mother filament of actin are nearly identical to that observed in high-resolution cryo-EM structures of bare actin filaments (SI Appendix, Fig. S6D) (45, 46). Therefore, the mother filament does not undergo major conformational changes upon branching. These findings, which are in agreement with the reconstruction of actin filament branches in cells (33), indicate that the rate of branching nucleation is not limited by a major structural change in the mother filament, as some previous experiments have suggested (32, 47).
Fig. 2.
Contacts of Arp2/3 complex with the mother filament of actin. (A) Surface representation of the Arp2/3 complex at the branch junction with atoms contacting actin mother filament (within 5 Å) colored gray. (B) Surface representation of Arp2/3 complex bound to the mother filament. (C) Surface representation of actin mother filament with atoms contacting Arp2/3 complex (within 5 Å) colored the same color as the subunit they contact. Nomenclature for actin filament subunits is as defined in Rouiller et al. (31) (D) Schematic showing four blocks of subunits within Arp2/3 complex that move as rigid bodies during activation via rotations about three axes. (E) Ribbon diagram of mother filament subunits M1 and M2 showing their interaction with the clamp subunits ARPC2 and ARPC4. The filament is oriented with the viewer looking at the pointed end, (approximately) down the helical axis. (F) Close up of interactions between ARPC4 helix αD and actin subunit M2 helix αB. Zoomed in area is boxed in E.
The conformational changes that activate Arp2/3 complex, which we describe in detail below, occur via rotations of four separate rigid bodies (blocks 1 to 4) along three axes (Fig. 2D). The majority of the contacts (2,500 Å2 of the buried surface area) between Arp2/3 complex and the mother filament are made with the top half of the complex, which includes block 1 and 3. Block 1 includes most of ARPC2 and the long alpha helix in ARPC4, plus subdomains 1 and 2 of Arp3 (Fig. 2D). Block 1 packs against actin subunits M1 and M2 and is the dominant contributor to interactions with the mother filament (1,800 Å2 of the 2,500 Å2 buried by the top half of the complex) (Fig. 2 A, E, and F). Block 3 contributes an additional 700 Å2 to the interface, with interactions coming from subdomains 2 and 4 of Arp3 and from ARPC3. Block 3 interactions require subunit flattening in Arp3 (see below). The bottom half of the complex, which comprises blocks 2 and 4, protrudes out from the side of the actin filament and so contributes far less to the mother filament interactions (Fig. 2B). The minimal interactions between the bottom half of Arp2/3 complex and the mother filament have important implications for the activation mechanism, which we discuss below.
Contacts of Arp2/3 Complex with the Daughter Filament.
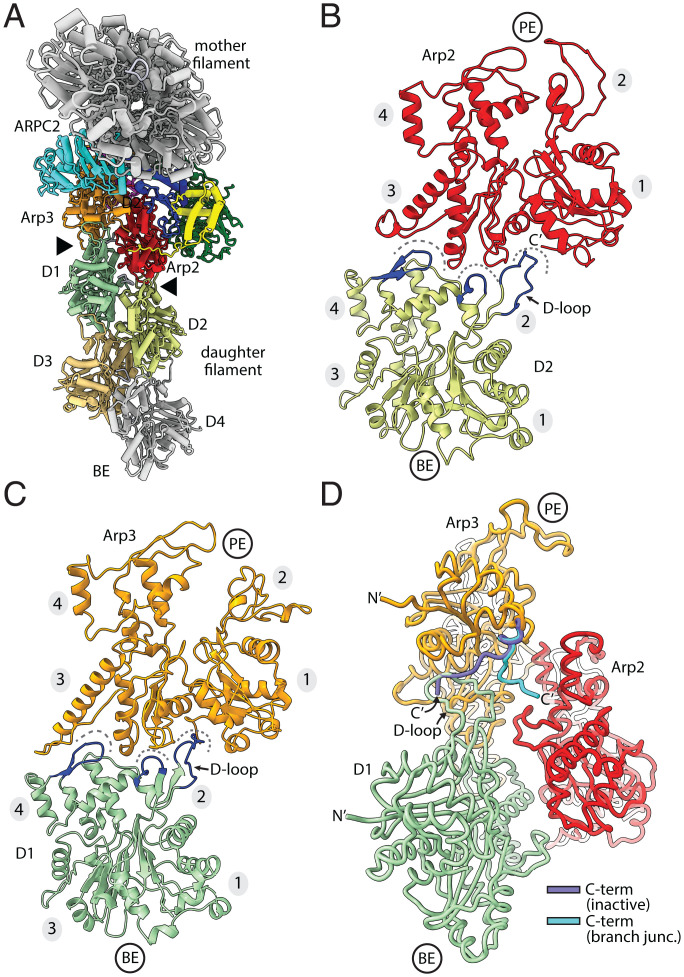
The daughter filament remains attached to Arp2/3 complex via interactions between the pointed ends of its first two actin subunits, namely, D1 and D2, with the barbed ends of Arp2 and Arp3 (Fig. 3 A–C). Actin subunits D1 and D2 insert three conserved features into three corresponding seams on the barbed ends of Arp2 and Arp3, as observed in the linear actin filament nucleated by the Dip1-activated Arp2/3 complex (15) (Fig. 3 B and C). The contacts at the Arp2/3 complex-daughter filament interface are very similar in the branch junction and linear filament structures (SI Appendix, Fig. S7) (15).
Fig. 3.
Contacts of Arp2/3 complex with the daughter filament of actin. (A) Ribbon representation of the branch junction looking down the helical axis of the mother filament. This view highlights the daughter filament-Arp2/3 complex interface (black arrowheads). (B and C) Long pitch (intrastrand) interactions between each Arp and actin subunits in the daughter filament. Dashes indicate three seams at the barbed end of each Arp that accept key structural protrusions (blue) from the pointed end of the actin subunits. Subdomains in actin and Arps are numbered 1 to 4 in this figure and throughout. (D) Diagram showing the position of the Arp3 C-terminal tail in the present branch junction model versus its position in the inactive Arp2/3 complex. The inactive C-terminal tail was modeled by overlaying Arp3 in 4JD2 onto subdomains 1 and 2 of Arp3 in the branch junction structure.
The C-terminal extension of Arp3 is an important autoinhibitory feature at the interface between Arp2/3 complex and the nucleated daughter filament. Biochemical and structural data indicate that this extension helps keep the complex inactive in the absence of WASP by blocking the association of actin monomers with the Arp3 barbed end and by stabilizing the splayed (inactive) conformation of Arp2 and Arp3 (15, 26, 27). Relief of autoinhibition is thought to require the release of the Arp3 C-terminal extension from the Arp3 barbed end groove (26, 27). Importantly, the Arp3 C-terminal extension is released from the barbed end groove in the branch junction structure we present here (Fig. 3D). It bends toward the helical axis, opening the Arp3 barbed end groove for interaction with the D-loop on the pointed end of the actin subunit D1 (Fig. 3D). The bent conformation appears to be stabilized by the packing of a conserved phenylalanine (F414Arp3) in the extension against two conserved leucine residues (L196 and L199) in Arp2 (SI Appendix, Fig. S8). However, these contacts are unlikely to be important in preventing the extension from engaging the groove in the activated state because 1) in the Dip1-activated Arp2/3 complex structure, the C-terminal end of the extension is disengaged but disordered and 2) the Arp3 C-terminal extension is also released from the groove in the recent 9-Å cryo-EM branch junction structure, but it adopts a different conformation that makes different contacts with Arp2 than we observe here (15, 33).
Two major conformational changes trigger branching nucleation.
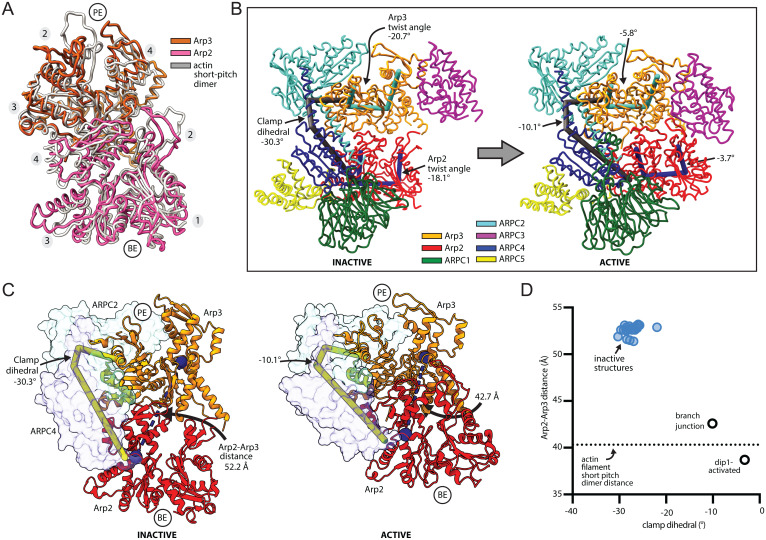
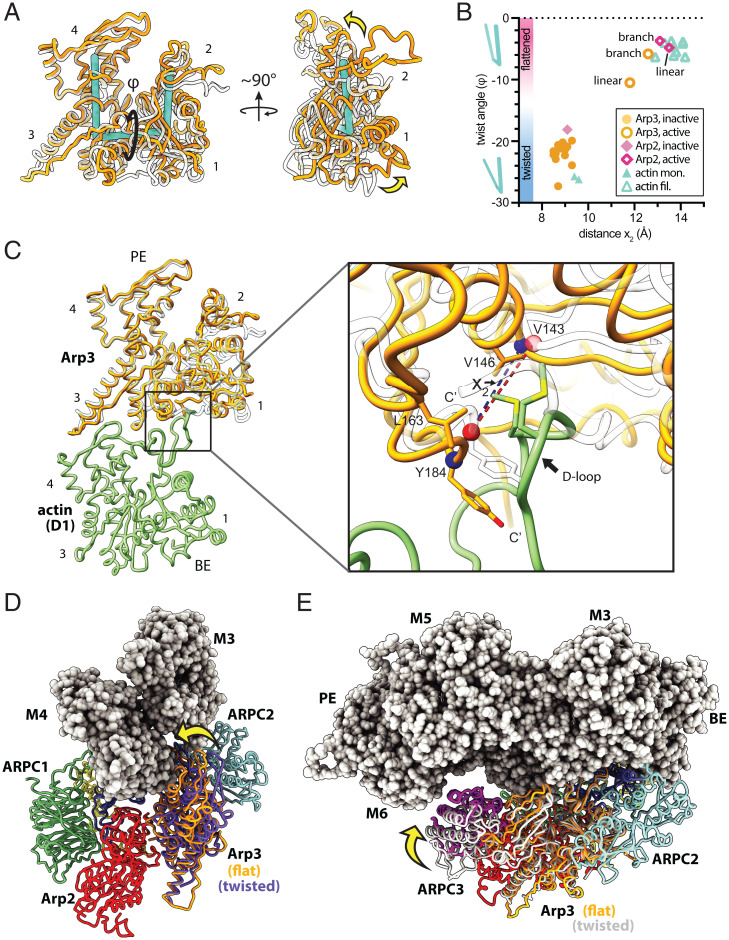
Examination of the structure reveals the conformational changes that allow the Arp subunits to mimic a filamentous actin dimer during branching nucleation (Fig. 4 A and B and SI Appendix, Fig. S9 and Movie S1). First, Arp2 moves into position next to Arp3 to create the Arp2-Arp3 short pitch dimer, as expected based on previous biochemical and structural data (Fig. 4 A and B) (15, 25, 26, 28, 33). Second, each Arp subunit transitions from a twisted (monomer-like) to a flattened (filament-like) conformation (SI Appendix, Movies S2 and S3). To adopt the short pitch conformation, Arp2 moves its center of mass ∼19 Å via a rigid body rotation of the entire bottom half of Arp2/3 complex (blocks 2 and 4), which includes ARPC1, ARPC5, Arp2, and the globular portion of ARPC4 (Fig. 4 C and D and SI Appendix, Movie S1). This rigid body motion, which was initially predicted based on the first inactive structure of Arp2/3 complex (13), is caused by twisting of the two clamp subunits ARPC2 and ARPC4 around an axis nearly parallel to the two long intertwined helices in the clamp (Fig. 4 B and C). Importantly, clamp twisting also moves Arp2 into the short pitch conformation in the activated Dip1-bound structure of Arp2/3 complex, indicating clamp twisting is a general mechanism used to reach this conformational state regardless of whether WASP or WISH/DIP/SPIN90 (WDS) proteins trigger activation (Fig. 4D) (15). However, clamp twisting and subsequent movement of Arp2 up the helical axis is less extensive in the branch junction than in the Dip1-activated Arp2/3 complex structure (Fig. 4D) (15). This conformational difference is likely due to increased flattening of the Arp3 subunit in the branch junction structure, which limits how far Arp2 can move up the helical axis without clashing with ARPC3 (see below).
Fig. 4.
Two major structural changes trigger branching nucleation. (A) Structural superposition of Cα atoms from Arp2 and Arp3 from the branch junction model with a filamentous actin dimer (PDB 6DJO, rmsd = 4.4 Å). (B) Comparison of the inactive (PDB 4JD2) Arp2/3 complex to activated Arp2/3 complex at the branch junction. The major conformational differences, clamp twisting and flattening of each Arp, can be measured via three different dihedral angles. (C) Comparison of the activated branch junction structure to inactive Arp2/3 complex (PDB 4JD2). The clamp (subunits ARPC2 and ARPC4, transparent surface) rotates to move Arp2 into the short pitch conformation, in which its center of mass (blue spheres) is closer to the Arp3 center of mass. Subdomains 3 and 4 were used for calculating the center of mass because subdomains 1 and 2 of Arp2 are disordered in most inactive structures. (D) Plot of the distance between the centers of mass of subdomains 3 and 4 of Arp2 and Arp3 versus the clamp dihedral angle. The equivalent distance between adjacent actin subunits along the short pitch helical axis in the mother filament is shown as a dotted line.
Subunit flattening, which occurs in both Arp2 and Arp3 subunits, involves a scissor-like motion that decreases the magnitude of the dihedral angle between the centers of mass of the four subdomains in each Arp (Fig. 5 A and B and SI Appendix, Movies S2 and S3). Flattening transforms each Arp from a conformation that approximately matches monomeric actin—the twisted conformation—to one that mimics filamentous actin subunits—the flattened conformation (Fig. 5B) (14). Interestingly, Arp3 flattens more in the branch junction structure than the Dip1-activated Arp2/3 complex structure (Fig. 5B). This structural plasticity of the nucleation-competent state may have important functional implications for the regulation of Arp2/3 complex by each class of NPF. For instance, flattening is thought to trigger ATP hydrolysis by the Arps (15, 48, 49), and ATP hydrolysis at branch junctions accelerates debranching (39, 40). On the other hand, structural data indicate that ATP is not hydrolyzed in the Dip1-activated Arp2/3 complex during linear actin filament nucleation (15). Therefore, the greater extent of subunit flattening in the branch junction structure may permit ATP hydrolysis after branching nucleation and could contribute to recycling of branched actin networks. In contrast, the partial flattening of Arp2/3 complex during linear filament nucleation triggered by WDS proteins may explain the stability of ATP in the Arp3 nucleotide binding cleft and of the Arp2/3 complex on the pointed end of the filament (15, 50). We note that the stability of the Arp2/3 complex on the filament end after activation by WDS proteins may be important for regulating the balance between branched and linear actin filaments. The strong association of the WDS protein Dip1 with Arp2/3 complex at the filament end prevents Dip1 from releasing and catalyzing multiple rounds of Arp2/3 complex activation, unlike WASP, which is released from the complex before nucleation (43, 50). This “single turnover” mechanism is hypothesized to limit Dip1-mediated linear filament nucleation relative to WASP-mediated branching nucleation during assembly of dendritic actin networks in cells (50).
Fig. 5.
Flattening of Arp3 increases contacts with the mother filament. (A) Structural superposition of subdomains 3 and 4 of Arp3 from the branch junction structure with Arp3 from the inactive Arp2/3 complex (PDB 4JD2). Flattening/twisting is measured as the dihedral angle (φ) defined by the center of mass of each subdomain. (B) Plot of the subunit twist angle versus the barbed end groove opening distance (see C) for structures of the active and inactive Arp2/3 complex and polymerized or monomeric actin. (C) Long pitch (intrastrand) interactions between Arp3 and actin subunit D1 in the daughter filament. Inactive Arp3 (PDB 4JD2, semitransparent rendering) is superposed onto Arp3 from the branch junction structure using subdomains 3 and 4. (D) Structural superposition of subdomains 1 and 2 of inactive Arp3 (4JD2, purple) with the same subdomains from Arp3 in the branch junction structure (orange). Yellow arrow shows the movement of subdomains 3 and 4 in Arp3 toward the mother filament caused by flattening. ARPC3 is omitted for clarity. (E) Reorientation of the structural superposition shown in D. The inactive (twisted) conformation of the complex is shown in white ribbon representation in this panel, and ARPC3 is rendered in magenta.
Actin Filaments Likely Stimulate Activation by Favoring Flattening in Arp3.
Both major structural changes that occur during activation are likely important for nucleation; by allowing Arp2 and Arp3 to mimic a filamentous actin dimer, they create the binding sites for the first actin monomers to be incorporated into the daughter filament, thereby templating filament assembly (15). Biochemical data have confirmed that the short pitch conformational change is required for activation (29). The requirement for subunit flattening has not been biochemically confirmed, but our data here and the Dip1-activated Arp2/3 complex structure show that flattening is correlated with widening of the barbed end groove of the Arps (Fig. 5 B and C) (15). Widening allows the D-loops of actin subunits D1 and D2 in the daughter filament to insert into the barbed end groove of each Arp (Fig. 5C). By permitting these interactions, flattening allows the Arps to make intrastrand (long pitch) contacts with the first actin subunits in the daughter filament.
Given its potential importance, we examined the contacts at the branch junction to determine if flattening could be stimulated by binding of Arp2/3 complex to the mother filament. Flattening of Arp3 occurs via rotation about a dihedral angle in Arp3 that rotates block 1 relative to block 3 (Figs. 2D and 5A and SI Appendix, Movie S3), bringing block 3—which includes half of Arp3 and all of ARPC3—closer to the mother filament. Specifically, flattening moves Arp3 subdomain 4 into position to contact actin subunit M4 and ARPC3 into position to contact actin subunit M6 (Fig. 5 D and E). These interactions increase the buried surface area between Arp2/3 complex and the mother filament by ∼640 Å2 compared to when Arp3 is in the twisted conformation. The increased interactions indicate that filaments help trigger nucleation by favoring the flattened conformation of Arp3. Most of the interactions at the block 3-mother filament interface are made by subdomain 4 of Arp3 (∼500 Å2 of buried surface area), including Arg209Arp3 with Glu125actin(M4), Pro217Arp3 with Pro367actin(M4), Glu218Arp3 with the backbone of Ala365actin(M4), and Gln205Arp3 with Asp363actin(M4) (SI Appendix, Fig. S10). ARPC3 buries 180 Å2 of the surface area against the mother filament and packs Tyr96ARPC3 against the side chain of Gln354actin(M4). These interactions may explain why Arp3 flattens by 5 degrees more in the branch-junction structure than the Dip1-activated Arp2/3 complex structure (Fig. 5B), which lacks these interactions because Dip1 activates Arp2/3 complex without a mother filament.
Interaction of Arp2/3 Complex with Actin Filaments May Not Stimulate Movement of Arp2 into the Short Pitch Conformation.
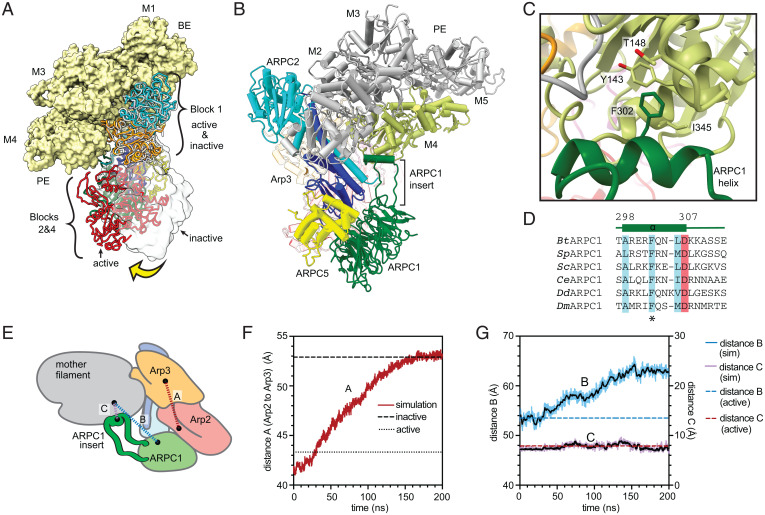
To determine if actin filament binding could stimulate clamp twisting and subsequent movement of Arp2 into the short pitch conformation, we compared the branch junction structure to a hypothetical model of an inactive (splayed) Arp2/3 complex bound to the side of an actin filament (Fig. 6A). To construct the model, we assumed that the inactive Arp2/3 complex (Protein Data Bank [PDB] 4JD2) binds filaments by inserting block 1 into the groove between actin subunits M1/M2 and M3/M4, as observed in the branch junction structure (Fig. 2). In this conformation, blocks 2 and 4 hang off the side of the filament, so they can rotate between inactive (splayed) and short pitch states without significantly changing the interactions of the complex with the mother filament of actin (Fig. 6A and SI Appendix, Fig. S11 and Movie S4). Therefore, while the top of the clamp (which is included in block 1) provides most of the contacts to the mother filament, its engagement with the filament may not influence clamp twisting.
Fig. 6.
Interaction of Arp2/3 complex with actin filaments may not stimulate movement of Arp2 into the short pitch conformation. (A) Comparison of mother filament binding contacts of the activated (flattened, short pitch) Arp2/3 complex to those of the inactive (splayed, twisted) Arp2/3 complex (PDB 4JD2, rendered in gray ribbon or transparent gray surface). The inactive Arp2/3 complex was modeled onto the mother filament by superposing block 1 onto block 1 in the branch junction model. Yellow arrow shows movement of blocks 2 and 4 stimulated by clamp twisting. Block 3 is omitted for clarity. (B) Ribbon diagram of a portion of the branch junction model highlighting the interaction between the ARPC1 protrusion helix and the mother filament. (C) Interaction of the ARPC1 protrusion helix with subunit M4 in the mother filament. (D) Sequence alignment showing conservation of a subset of hydrophobic (cyan) and acidic (red) residues in the ARPC1 protrusion helix. (E) Schematic showing key distances in molecular dynamics (MD) simulations plotted in F and G. (F) Plot of distance A, which measures the distance between the centers of mass of Arp2 and Arp3 (subdomains 3 and 4, see Materials and Methods), versus time during the simulation. The equilibrated branch junction structure is pulled to the splayed conformation over 150 ns via a biased MD simulation. The distance between Arp2 and Arp3 in the active (short pitch, flattened) conformation in the branch junction is shown with a dotted line, while the distance in the inactive (splayed, twisted) conformation is shown with a dashed line. (G) Plot of distances B and C, which measure the distance between the globular portion of ARPC1 (distance B) or the ARPC1 protrusion helix (distance C) and the mother filament versus time during the simulation.
Close examination of the model revealed only one obvious structural feature that could make substantially different interactions with the mother filament when the clamp twists, namely, the ARPC1 insert. This long insert extends away from the globular WD repeat domain of ARPC1 (part of block 2) and stretches back toward the mother filament (Fig. 6B). It inserts an alpha helix (the protrusion helix) into the barbed end groove of actin subunit M4 in the branch junction structure (Fig. 6 C and D), making interactions similar to those reported in the 9-Å cryo-EM branch junction reconstruction (33). We wondered if the protrusion helix interacts with filaments exclusively when Arp2/3 complex is in the short pitch conformation since ARPC1 is further away from the mother filament when the complex is in the splayed (inactive) conformation (Fig. 6 E–G). If so, this would provide a mechanism by which actin filaments could stimulate clamp twisting; the binding energy of the protrusion helix could help offset the energetic cost of the conformational change. However, because the ARPC1 protrusion helix is connected to the WD repeat domain of ARPC1 via long linker sequences, it may be able to stretch or retract to maintain contacts with the mother filament as the clamp twists/untwists. To test this, we computationally pulled Arp2/3 complex from the short pitch to the splayed conformation in molecular dynamics simulations and asked if the protrusion helix maintained contact with the filament. In three simulations (at three different pulling rates) the helix remained in contact during the transition even though the globular portion of ARPC1 swung further away from the mother filament as the complex moved into the splayed conformation (Fig. 6 E–G and SI Appendix, Fig. S12). Taken together, these observations suggest that the mother filament contacts made by splayed and short pitch conformations of Arp2/3 complex are similar or identical, so filament binding may not stimulate clamp twisting and the subsequent movement of Arp2 into the short pitch conformation. These observations are consistent with a recent study that used site-specific crosslinking of an engineered Arp2/3 complex to show that actin filaments do not stimulate the adoption of the short pitch conformation (30). Therefore, stimulation of the short pitch conformational change is likely stimulated solely by the other activating factors required for branching nucleation: WASP and WASP-recruited actin monomers. An inability of actin filaments to stimulate this conformational change could explain how branching nucleation is repressed in the absence of WASP; if actin filaments alone could stimulate both major activating conformational changes—subunit flattening and clamp twisting/short pitch adoption—WASP could become dispensable for activation.
Increasing Interactions with Actin Filaments Is a Primary Function of the ARPC1 Insert.
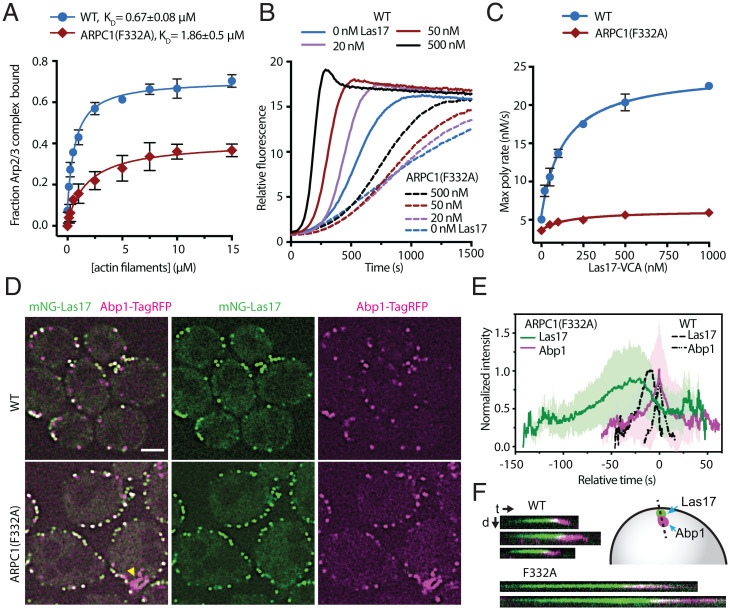
While our data suggest the ARPC1 insert does not play a role in stimulating clamp twisting, its interactions with the mother filament suggest that it may influence mother filament binding, as predicted based on the first crystal structure of the inactive Arp2/3 complex (13). Examination of the structure revealed that the ARPC1 protrusion helix inserts a conserved phenylalanine (F302) into the barbed end groove of subunit M4 in the mother filament (Fig. 6C). To test the importance of this interaction, we mutated the equivalent phenylalanine in budding yeast Arp2/3 complex to alanine (F332A) and tested the affinity of the mutant complex for actin filaments in a copelleting assay. The F332A mutation decreased the affinity of the complex for actin filaments by approximately threefold, and also decreased the fraction bound at saturating filament concentrations (Fig. 7A, see Materials and Methods for additional details). To determine if the ARPC1(F332A) mutation causes defects in the ability of Arp2/3 complex to nucleate actin filaments, we tested its activity in a pyrene actin polymerization assay. The mutant Arp2/3 complex was severely defective in these assays (Fig. 7 B and C and SI Appendix, Fig. S13). In reactions with 500 nM Las17, the Saccharomyces cerevisiae WASP family protein, the ARPC1(F332A) mutant nucleated 85% fewer barbed ends than the wild-type complex when polymerization was 75% complete (SI Appendix, Fig. S13). Together, these data suggest that the ARPC1 insert is important for Arp2/3 complex activation because it increases the affinity of the complex for actin filaments.
Fig. 7.
The ARPC1 insert is important for Arp2/3 complex function in vitro and in vivo. (A) Binding isotherm showing the fraction of 60 nM WT and mutant Arp2/3 complexes that copelleted with preformed actin filaments. Fraction bound was determined by measuring depletion of the Arp2/3 complex from the supernatant. Note that neither WT nor ARPC1(F332A) Arp2/3 complexes reach 100% bound even at saturating concentrations of actin filaments (see SI Appendix, Methods for more details). The F332 residue in S. cerevisiae is equivalent to F302 in B. taurus ARPC1. n = 3 for each concentration. Error bar: SD. (B) Time courses of pyrene actin polymerization for reactions containing 20 nM WT or ARPC1(F332A) mutant Arp2/3 complex and the indicated concentration of the VCA segment of Las17 (Las17-VCA). (C) Quantification of the maximum polymerization rate from the data in B. Data points are averages of three replicates. Error bars: SD. (D) Spinning disk confocal images of a central z-slice of WT or ARPC1(F332A) strains taken in optical pixel reassignment mode. Las17 is labeled at its N terminus with mNeonGreen (mNG), and Abp1 is labeled at its C terminus with TagRFP-T. Yellow arrowhead highlights a nonpunctate Abp1-TagRFP-T structure in the ARPC1(F332A) strain. Scale bar: 2.0 μm. (E) Plot of the average normalized intensity versus time for mNG-Las17 and Abp1-TagRFP-T patches in WT (n = 76 patches) and ARPC1(F332A) (n = 74 patches) cells. Paired trajectories were aligned at the maximum signal for Abp1-TagRFP-T. Error bars show the SD for all aligned trajectories. (F) Kymographs from widefield fluorescence images showing mNG-Las17 and Abp1-TagRFP-T signal over time for three events in WT and two events in the mutant cells. t, time; d, distance.
Previous reports indicated that the ARPC1 insert may contribute to activation of the Arp2/3 complex via mechanisms unrelated to filament binding. For instance, one study reported that the ARPC1 insert is important for Las17 binding to the Arp2/3 complex (51). However, we found the ARPC1(F332A) mutant bound to Las17 with similar affinity to wild-type Arp2/3 complex in a glutathione-S-transferase pulldown assay (SI Appendix, Fig. S14A). A separate study reported that the ARPC1 insert helps stabilize the short pitch arrangement of Arp2 and Arp3 (28). The branch junction model we present here does not reveal a structural basis by which the ARPC1 insert could stabilize the short pitch conformation, so we sought additional strategies to test which activation steps are influenced by the F332A mutation. Specifically, we asked if Dip1, a WDS protein, could activate the ARPC1(F332A) complex. WDS proteins activate the Arp2/3 complex without preexisting actin filaments (4). Consequently, if the F332A mutation affects only mother filament binding, it will not prevent Dip1 from activating Arp2/3 complex (SI Appendix, Fig. S14B). In contrast, if the F332A mutation blocks the adoption of the short pitch conformation—which is required during activation by both WASP and WDS family proteins (29, 52)—Dip1 will not activate the ARPC1(F332A) mutant. Pyrene actin polymerization assays showed that Dip1 activated the ARPC1(F332A) complex nearly as potently as wild-type Arp2/3 complex (SI Appendix, Fig. S14C). At 10 μM Dip1, the ARPC1(F332A) complex showed 84% of the wild-type activity, measured by the number of barbed ends created when the reaction was 75% complete. Therefore, we conclude that facilitating interactions of the Arp2/3 complex with actin filaments is the primary function of the ARPC1 insert. However, we note that Dip1 did not fully activate the ARPC1(F332A) mutant. Whether this small defect could be due to inefficient stabilization of the short pitch conformation remains an open question.
The ARPC1 Insert Is Critical for Proper Endocytic Actin Assembly.
Defects in actin filament binding in the ARPC1(F332A) mutation decreased the ability of the complex to nucleate filaments in vitro (Fig. 7). However, we wondered whether high local concentrations of actin filaments in cells could compensate for the decrease in actin filament binding affinity, thus alleviating the nucleation defect (53). In S. cerevisiae, the assembly of endocytic actin networks depend on the actin filament nucleation activity of Arp2/3 complex (54), so we created a budding yeast strain containing the ARPC1 F332A mutation together with mNeonGreen-tagged Las17 (mNG-Las17) and TagRFP-T labeled Abp1 (Fig. 7D). Abp1 binds actin filaments within endocytic actin networks, allowing us to monitor actin assembly defects that might result from mutations in the complex (55). In wild-type cells, Las17 accumulates in cortical patches for ∼18 s before actin filaments begin to assemble and recruit Abp1, consistent with previous reports (Fig. 7 E and F and SI Appendix, Fig. S15 and Movie S5) (55). In contrast, in the ARPC1(F332A) strain, Las17 puncta persist for ∼60 s before the Abp1 signal can be detected, indicating a significant delay in the initiation of actin assembly (Fig. 7 E and F and SI Appendix, Fig. S15 and Movie S6). Even after Abp1 assembly is initiated in the mutant, it accumulates more slowly than in the wild-type strain, reaching a peak signal over 24 s—∼2.5 times slower than in the wild-type strain (Fig. 7 E and F and SI Appendix, Fig. S15). Most Abp1 puncta failed to move off of the cortex and into the cytoplasm in the mutant strain, suggesting defective internalization of endocytic cargo (SI Appendix, Fig. S15 and Movie S6). Finally, some of the mutant cells showed long strand-like Abp1-marked features that were frequently connected to a cortical punctum of Abp1 (Fig. 7D and SI Appendix, Movie S6). Taken together, these data indicate that perturbations in the interaction of Arp2/3 complex with actin filaments cause significant defects in actin assembly in cells. Furthermore, they specifically demonstrate the importance of the ARPC1 insert in assembling functional endocytic actin networks.
Discussion
Here, we present a ∼3.9-Å cryo-EM reconstruction of Arp2/3 complex at a branch junction. This structure provides a snapshot of the final step in the branching nucleation reaction, in which the complex is fully activated and bound to mother and daughter actin filaments, but WASP has been released. The structure supports a model in which the mother filament contributes to activation by stimulating subunit flattening in Arp3, a conformational change that facilitates interactions of the complex with the first actin subunit in the daughter filament (Fig. 5) (15). The requirement for subunit flattening may explain why actin filaments are necessary for WASP-mediated activation of Arp2/3 complex and, consequently, how WASP triggers exclusively branched (and not linear) actin filament nucleation by the complex (18, 22). We note that while the branch junction structure revealed an obvious mechanism by which actin filament binding could stimulate flattening of the Arp3 subunit, how flattening is stimulated in Arp2 remains an important open question. One possibility is that Arp2 flattening may be weakly coupled to the other major activating conformational changes or, alternatively, it may be stimulated by contacts with WASP-recruited actin monomers.
We show here that—consistent with the recent 9-Å branch reconstruction (33)—contacts between Arp2/3 complex and the mother filament are far less extensive than observed in the 26-Å reconstruction by Roullier et al. (32), who reported that all subunits in the complex contact the mother filament. Instead, the complex uses only its top half to bind the mother filament, allowing clamp twisting/untwisting to toggle Arp2 and the bottom half of the complex between the splayed and short pitch conformations without substantially changing contacts with the mother filament. This result suggests that actin filaments do not trigger clamp twisting, thereby explaining recent biochemical data showing that actin filaments fail to stimulate movement of Arp2 and Arp3 into the short pitch conformation (30). Therefore, our data support a model in which WASP and WASP-recruited actin monomers—which were shown via biochemical and biophysical assays to stimulate the short pitch conformation (25–29)—are solely responsible for triggering clamp twisting and movement into the short pitch arrangement. Given that the short pitch conformational switch is one of two major conformational changes that allow Arp2 and Arp3 to mimic a filamentous actin dimer, the failure of filaments to stimulate this step may explain why they alone are not sufficient for activation and, therefore, how the requirement for WASP is programmed into the nucleation mechanism. However, it is important to note that while some experiments suggest that the sole activating function of WASP (and WASP-recruited actin monomers) is to simulate the short pitch conformation (26), other experiments suggest additional required activating functions for WASP (28). The precise role(s) of WASP and WASP-recruited actin monomers in activation remains an important open question.
While multiple studies have demonstrated a role for the ARPC1 insert in the activity of the Arp2/3 complex (28, 51), our results confirm that its primary role is to increase the binding affinity of the complex for the actin mother filament. The sensitivity of the complex to defects in actin filament binding is consistent with single-molecule visualization of the complex, which showed that branching nucleation is inefficient because of low affinity of Arp2/3 complex for actin filaments, with the complex releasing from filaments in 98% of the binding events instead of nucleating a branch (56). Importantly, while previous work demonstrated the importance of actin filament binding under the conditions of in vitro assays (31), it was unknown whether moderate defects in actin filament binding would disrupt actin assembly in cells. Here, we showed that the mutation of yeast ARPC1 residue F332 (equivalent of mammalian F302) to alanine potently disrupted actin assembly at endocytic sites in yeast. The sensitivity of Arp2/3 complex to perturbations in actin binding affinity underscores the importance of developing a detailed molecular understanding of how the activity of the complex is coordinated with actin regulators that influence its binding to filaments, including coronin, Abp1, and cortactin (57–59).
Together, our data provide a detailed look at the interactions of activated Arp2/3 complex at the branch junction, including key contacts it makes with the mother filament of actin. We anticipate that the structure presented here will provide a foundation for future experiments aimed at understanding the molecular basis of actin regulation by the complex.
Materials and Methods
Please see SI Appendix, Methods for a complete description of the methods. This description includes all protein purification procedures, including those for the Arp2/3 complex, WASP, actin, cortactin, and capping protein. SI Appendix, Methods also describes sample preparation for EM, details about how the EM data were acquired, how images were processed, and how the model was built. Details are also included about biochemical assays, including actin filament copelleting assays and pyrene actin polymerization assays. Details are also included about yeast strain construction and live-cell imaging and fluorescence image analysis for the Arp2/3 complex mutations in yeast. Finally, the SI Appendix, Methods describes in detail the molecular dynamics simulations and structural analysis procedures. The cryo-EM reconstructed map was deposited in the Electron Microscopy Data Bank (EMDB) with accession identifier (ID) EMD-26063, and the corresponding atomic model was deposited in the PDB (www.rcsb.org/pdb) with PDB accession ID 7TPT.
Supplementary Material
Acknowledgments
We thank Qing Luan for help with purification of the Bos taurus Arp2/3 complex, Maisie Topping for assistance with the preparation of the Arp2 D-loop triple mutant, Adam Fries and Mike Lynch for assistance with processing live-cell imaging data. We thank Pablo Guardado Calvo for sharing the rotation_axis.py script for the structural analysis. Cryo-EM data were collected at the Stony Brook University Cryo-EM center, which is supported by National Institutes of Health Grant number S10 OD012272. This work was supported by Stony Brook University start-up funds to S.C., by NIH Grant number R35-GM138312 to G.M.H., and NIH Grant number R35-GM136319 to B.J.N.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2202723119/-/DCSupplemental.
Data Availability
Reconstructed density map and coordinates have been deposited in EMDB (EMD-26063), and PDB (7TPT).
References
- 1.Chesarone M. A., Goode B. L., Actin nucleation and elongation factors: Mechanisms and interplay. Curr. Opin. Cell Biol. 21, 28–37 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollard T. D., Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 36, 451–477 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Skau C. T., Waterman C. M., Specification of architecture and function of actin structures by actin nucleation factors. Annu. Rev. Biophys. 44, 285–310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner A. R., Luan Q., Liu S.-L., Nolen B. J., Dip1 defines a class of Arp2/3 complex activators that function without preformed actin filaments. Curr. Biol. 23, 1990–1998 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goley E. D., Welch M. D., The ARP2/3 complex: An actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7, 713–726 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Schrank B. R., et al. , Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature 559, 61–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swaney K. F., Li R., Function and regulation of the Arp2/3 complex during cell migration in diverse environments. Curr. Opin. Cell Biol. 42, 63–72 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welch M. D., Way M., Arp2/3-mediated actin-based motility: A tail of pathogen abuse. Cell Host Microbe 14, 242–255 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yi K., et al. , Dynamic maintenance of asymmetric meiotic spindle position through Arp2/3-complex-driven cytoplasmic streaming in mouse oocytes. Nat. Cell Biol. 13, 1252–1258 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rotty J. D., Wu C., Bear J. E., New insights into the regulation and cellular functions of the ARP2/3 complex. Nat. Rev. Mol. Cell Biol. 14, 7–12 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Siton-Mendelson O., Bernheim-Groswasser A., Functional actin networks under construction: The cooperative action of actin nucleation and elongation factors. Trends Biochem. Sci. 42, 414–430 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Muller J., et al. , Sequence and comparative genomic analysis of actin-related proteins. Mol. Biol. Cell 16, 5736–5748 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson R. C., et al. , Crystal structure of Arp2/3 complex. Science 294, 1679–1684 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Oda T., Iwasa M., Aihara T., Maéda Y., Narita A., The nature of the globular- to fibrous-actin transition. Nature 457, 441–445 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Shaaban M., Chowdhury S., Nolen B. J., Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state. Nat. Struct. Mol. Biol. 27, 1009–1016 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marchand J. B., Kaiser D. A., Pollard T. D., Higgs H. N., Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat. Cell Biol. 3, 76–82 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Rohatgi R., et al. , The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell 97, 221–231 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Amann K. J., Pollard T. D., The Arp2/3 complex nucleates actin filament branches from the sides of pre-existing filaments. Nat. Cell Biol. 3, 306–310 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Blanchoin L., et al. , Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature 404, 1007–1011 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Mogilner A., Oster G., Cell motility driven by actin polymerization. Biophys. J. 71, 3030–3045 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullins R. D., Heuser J. A., Pollard T. D., The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc. Natl. Acad. Sci. U.S.A. 95, 6181–6186 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Achard V., et al. , A “primer”-based mechanism underlies branched actin filament network formation and motility. Curr. Biol. 20, 423–428 (2010). [DOI] [PubMed] [Google Scholar]
- 23.Burianek L. E., Soderling S. H., Under lock and key: Spatiotemporal regulation of WASP family proteins coordinates separate dynamic cellular processes. Semin. Cell Dev. Biol. 24, 258–266 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mullins R. D., Bieling P., Fletcher D. A., From solution to surface to filament: Actin flux into branched networks. Biophys. Rev. 10, 1537–1551 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Espinoza-Sanchez S., Metskas L. A., Chou S. Z., Rhoades E., Pollard T. D., Conformational changes in Arp2/3 complex induced by ATP, WASp-VCA, and actin filaments. Proc. Natl. Acad. Sci. U.S.A. 115, E8642–E8651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodnick-Smith M., Luan Q., Liu S.-L., Nolen B. J., Role and structural mechanism of WASP-triggered conformational changes in branched actin filament nucleation by Arp2/3 complex. Proc. Natl. Acad. Sci. U.S.A. 113, E3834–E3843 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodnick-Smith M., Liu S.-L., Balzer C. J., Luan Q., Nolen B. J., Identification of an ATP-controlled allosteric switch that controls actin filament nucleation by Arp2/3 complex. Nat. Commun. 7, 12226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zimmet A., et al. , Cryo-EM structure of NPF-bound human Arp2/3 complex and activation mechanism. Sci. Adv. 6, eaaz7651 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hetrick B., Han M. S., Helgeson L. A., Nolen B. J., Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem. Biol. 20, 701–712 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narvaez-Ortiz H. Y., Nolen B. J., Unconcerted conformational changes in Arp2/3 complex integrate multiple activating signals to assemble functional actin networks. Curr. Biol. 32, 975–987.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goley E. D., et al. , An actin-filament-binding interface on the Arp2/3 complex is critical for nucleation and branch stability. Proc. Natl. Acad. Sci. U.S.A. 107, 8159–8164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rouiller I., et al. , The structural basis of actin filament branching by the Arp2/3 complex. J. Cell Biol. 180, 887–895 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fäßler F., Dimchev G., Hodirnau V.-V., Wan W., Schur F. K. M., Cryo-electron tomography structure of Arp2/3 complex in cells reveals new insights into the branch junction. Nat. Commun. 11, 6437 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merino F., Pospich S., Raunser S., Towards a structural understanding of the remodeling of the actin cytoskeleton. Semin. Cell Dev. Biol. 102, 51–64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akin O., Mullins R. D., Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell 133, 841–851 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weaver A. M., et al. , Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 11, 370–374 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Mahaffy R. E., Pollard T. D., Influence of phalloidin on the formation of actin filament branches by Arp2/3 complex. Biochemistry 47, 6460–6467 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blanchoin L., Pollard T. D., Hydrolysis of ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry 41, 597–602 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Le Clainche C., Pantaloni D., Carlier M.-F., ATP hydrolysis on actin-related protein 2/3 complex causes debranching of dendritic actin arrays. Proc. Natl. Acad. Sci. U.S.A. 100, 6337–6342 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin A. C., Welch M. D., Drubin D. G., Arp2/3 ATP hydrolysis-catalysed branch dissociation is critical for endocytic force generation. Nat. Cell Biol. 8, 826–833 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Dayel M. J., Mullins R. D., Activation of Arp2/3 complex: Addition of the first subunit of the new filament by a WASP protein triggers rapid ATP hydrolysis on Arp2. PLoS Biol. 2, E91 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ingerman E., Hsiao J. Y., Mullins R. D., Arp2/3 complex ATP hydrolysis promotes lamellipodial actin network disassembly but is dispensable for assembly. J. Cell Biol. 200, 619–633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith B. A., et al. , Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. eLife 2, e01008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pospich S., Merino F., Raunser S., Structural effects and functional implications of Phalloidin and Jasplakinolide binding to actin filaments. Structure 28, 437–449.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Merino F., et al. , Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM. Nat. Struct. Mol. Biol. 25, 528–537 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Chou S. Z., Pollard T. D., Mechanism of actin polymerization revealed by cryo-EM structures of actin filaments with three different bound nucleotides. Proc. Natl. Acad. Sci. U.S.A. 116, 4265–4274 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ti S.-C., Jurgenson C. T., Nolen B. J., Pollard T. D., Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on Arp2/3 complex. Proc. Natl. Acad. Sci. U.S.A. 108, E463–E471 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murakami K., et al. , Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release. Cell 143, 275–287 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Vorobiev S., et al. , The structure of nonvertebrate actin: Implications for the ATP hydrolytic mechanism. Proc. Natl. Acad. Sci. U.S.A. 100, 5760–5765 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balzer C. J., Wagner A. R., Helgeson L. A., Nolen B. J., Single-turnover activation of Arp2/3 complex by Dip1 may balance nucleation of linear versus branched actin filaments. Curr. Biol. 29, 3331–3338.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balcer H. I., Daugherty-Clarke K., Goode B. L., The p40/ARPC1 subunit of Arp2/3 complex performs multiple essential roles in WASp-regulated actin nucleation. J. Biol. Chem. 285, 8481–8491 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luan Q., Liu S.-L., Helgeson L. A., Nolen B. J., Structure of the nucleation-promoting factor SPIN90 bound to the actin filament nucleator Arp2/3 complex. EMBO J. 37, e100005 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sirotkin V., Berro J., Macmillan K., Zhao L., Pollard T. D., Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast. Mol. Biol. Cell 21, 2894–2904 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winter D., Podtelejnikov A. V., Mann M., Li R., The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr. Biol. 7, 519–529 (1997). [DOI] [PubMed] [Google Scholar]
- 55.Sun Y., et al. , Direct comparison of clathrin-mediated endocytosis in budding and fission yeast reveals conserved and evolvable features. eLife 8, e50749 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith B. A., Daugherty-Clarke K., Goode B. L., Gelles J., Pathway of actin filament branch formation by Arp2/3 complex revealed by single-molecule imaging. Proc. Natl. Acad. Sci. U.S.A. 110, 1285–1290 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo S., et al. , Abp1 promotes Arp2/3 complex-dependent actin nucleation and stabilizes branch junctions by antagonizing GMF. Nat. Commun. 9, 2895 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helgeson L. A., Nolen B. J., Mechanism of synergistic activation of Arp2/3 complex by cortactin and N-WASP. eLife 2, e00884 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu S.-L., Needham K. M., May J. R., Nolen B. J., Mechanism of a concentration-dependent switch between activation and inhibition of Arp2/3 complex by coronin. J. Biol. Chem. 286, 17039–17046 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Reconstructed density map and coordinates have been deposited in EMDB (EMD-26063), and PDB (7TPT).