Significance
Ribosomes translate the genetic codes of messenger RNA (mRNA) to make proteins. Translation must begin at the correct initiation site; otherwise, abnormal proteins will be produced. Here, we show that a short ribosome-specific sequence in the upstream followed by an unstructured downstream sequence is a favorable initiation site. Those mRNAs lacking either of these two characteristics do not associate tightly with the ribosome. Initiator transfer RNA (tRNA) and initiation factors facilitate the binding. However, when the downstream site forms structures, initiation factor 3 triggers the dissociation of the accommodated initiator tRNA and the subsequent disassembly of the ribosome–mRNA complex. Thus, initiation factors help the ribosome distinguish unfavorable structured sequences that may not act as the mRNA translation initiation site.
Keywords: translation initiation, initiation factors, single-molecule, smFRET, optical tweezers
Abstract
Initiation of protein synthesis from the correct start codon of messenger RNA (mRNA) is crucial to translation fidelity. In bacteria, the start codon is usually preceded by a 4- to 6-mer adenosine/guanosine-rich Shine–Dalgarno (SD) sequence. Both the SD sequence and the start codon comprise the core ribosome-binding site (RBS), to which the 30S ribosomal subunit binds to initiate translation. How the rather short and degenerate information inside the RBS can be correctly accommodated by the ribosome is not well understood. Here, we used single-molecule techniques to tackle this long-standing issue. We found that the 30S subunit initially binds to mRNA through the SD sequence, whereas the downstream RBS undergoes dynamic motions, especially when it forms structures. The mRNA is either dissociated or stabilized by initiation factors, such as initiation factor 3 (IF3). The initiator transfer RNA (tRNA) further helps the 30S subunit accommodate mRNA and unwind up to 3 base pairs of the RBS structure. Meanwhile, the formed complex of the 30S subunit with structured mRNA is not stable and tends to disassociate. IF3 promotes dissociation by dismissing the bound initiator tRNA. Thus, initiation factors may accelerate the dynamic assembly–disassembly process of 30S–mRNA complexes such that the correct RBS can be preferentially selected. Our study provides insights into how the bacterial ribosome identifies a typical translation initiation site from mRNA.
Translation is initiated by recruitment of the ribosome to messenger RNA (mRNA) at the start codon to assume the correct reading frame. In bacteria, the small (30S) ribosomal subunit first binds to the ribosome-binding site (RBS), which usually contains the Shine–Dalgarno (SD) sequence (1), to form the 30S preinitiation complex (30S PIC). The SD sequence is complementary to the 3′ tail (named anti-SD) of 16S ribosomal RNA (rRNA) of the 30S subunit. The 30S initiation complex (30S IC) forms when initiator transfer RNA (tRNA) is accommodated onto the peptidyl-tRNA–binding site (P site). Roughly 30 nucleotides (nt) of the mRNA wrap around the neck region of the 30S subunit (2) and outline the ribosome footprint. Finally, the elongation-competent 70S IC forms after assembly of the large (50S) ribosomal subunit (3–5).
As key factors for the efficiency of translation, initiation factors (IF1, IF2, and IF3) can join the 30S subunit before or after mRNA association (6, 7) and are essential for Escherichia coli cell viability (8–10). IF1 binds to the aminoacyl-tRNA–binding site (A site) of the 30S subunit (11) and ensures the decoding position of initiator tRNA. IF1 also assists IF2-associated initiator tRNA accommodation (12, 13). IF2, a GTP-binding protein, not only helps the accommodation of the initiator tRNA but also facilitates the association of the 50S subunit (6, 14, 15).
IF3 is suggested to help selection of a cognate start codon and initiator tRNA, as its conformation between the N- and C-terminal domains (NTD and CTD, respectively) is sensitive to whether the 30S IC is correctly assembled (16). This function of IF3 is supported by a cryoelectron microscopy study showing that in the absence of initiator tRNA, the IF3 CTD, together with 16S rRNA, makes several specific contacts with the start codon in or near the P site (13). When initiator tRNA joins, the IF3 NTD binds to the elbow of the tRNA, followed by a series of conformational changes to accommodate the tRNA in the P site (13). Accommodation of the initiator tRNA results in closure of the A site mRNA latch (formed between helices h18 and h34 of 16S rRNA) (13), potentially stabilizing the downstream mRNA on the channel.
During the initiation process, the 30S subunit may encounter various mRNA structures that are used to regulate translation (17–19). A genome-wide analysis showed that, in polycistronic mRNAs, each open reading frame forms distinct structures, which are separated by less-structured regions centered at the start codon (roughly from positions −25 to +25; the first nucleotide of the start codon is designated as +1) (20). The exposed single-stranded initiation site may facilitate ribosome association. Consistent with this finding, several studies have systematically varied the sequence of the 5′ untranslated region (5′ UTR) (21), the hairpin harboring the start codon (18), or the synonymous codons of a reporter protein (22) and have shown that secondary structures formed near the RBS tend to down-regulate translation. A moderate mRNA structure containing the SD sequence can interact rapidly with the 30S subunit through a neighboring single strand, but further stabilization of the complex requires the participation of initiation factors and initiator tRNA (7). The downstream mRNA that enters the 30S subunit is located at position +13 to +15 and is surrounded by ribosomal proteins uS3, uS4, and uS5 (2, 23). The entrance site possesses helicase activity that can unwind mRNA structures during elongation (23–26), but whether it is also involved in initiation is unknown. Overall, how mRNA structures affect initiation depends on the structure’s position, stability, and flanking sequences, among others, yet the detailed molecular mechanisms remain elusive.
To explore the accommodation of mRNA with various sequences and structures, we measured the interaction between mRNA and the 30S subunit at the initiation stage using single-molecule Förster resonance energy transfer (smFRET) (27) and optical tweezers (28). We found that mRNA was initially recruited to the 30S subunit through the SD sequence, while the downstream RBS was only dynamically associated, especially when it formed a structure. Initiation factors could facilitate further accommodation of the mRNA. Finite unwinding of the downstream RBS structure was observed only when initiator tRNA was present. However, the formed 30S ICs with structured mRNA were not stable. IF3 (and likely IF2 as well) promoted dissociation of the initiator tRNA from the complex, followed by disassembly of the 30S–mRNA complex. Thus, initiation factors may accelerate the assembly–disassembly kinetics of 30S ICs such that a correct RBS can be quickly identified by the 30S subunit during initiation.
Results
Accommodation of Unstructured mRNA on 30S Subunits Is Facilitated by Initiator tRNA and Initiation Factors.
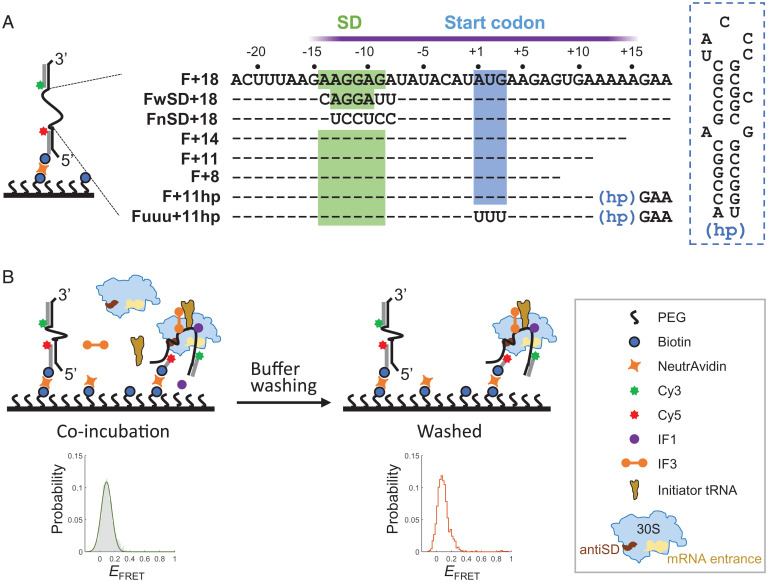
To study the real-time mRNA accommodation on the 30S subunit by smFRET, we designed a single-stranded mRNA flanked by a Cy3 dye (donor) and a Cy5 dye (acceptor) through complementary DNA handles; the construct was named F+18, according to the downstream boundary at position +18 (Fig. 1A). The 5′ UTR of F+18 (−22 ∼ +3) was derived from the gene of T7 major capsid protein (accession number LR745710), and the coding region was changed to minimize the potential of forming secondary structures. The mRNA construct was immobilized onto the surface of a slide chamber, the 30S subunit and other indicated factors were added and incubated (Fig. 1B, Left), and time traces of FRET efficiency (EFRET) of the mRNA were recorded. Then, the chamber was washed to remove free and weakly bound components (Fig. 1B, Right). This buffer washing step also allowed us to distinguish the conformational changes (FRET dynamics) of mRNA caused by repeated dissociation–reassociation of the 30S subunit from those caused by transitioning among various accommodation states on the tightly bound 30S subunit. The former would be more affected by washing than the latter.
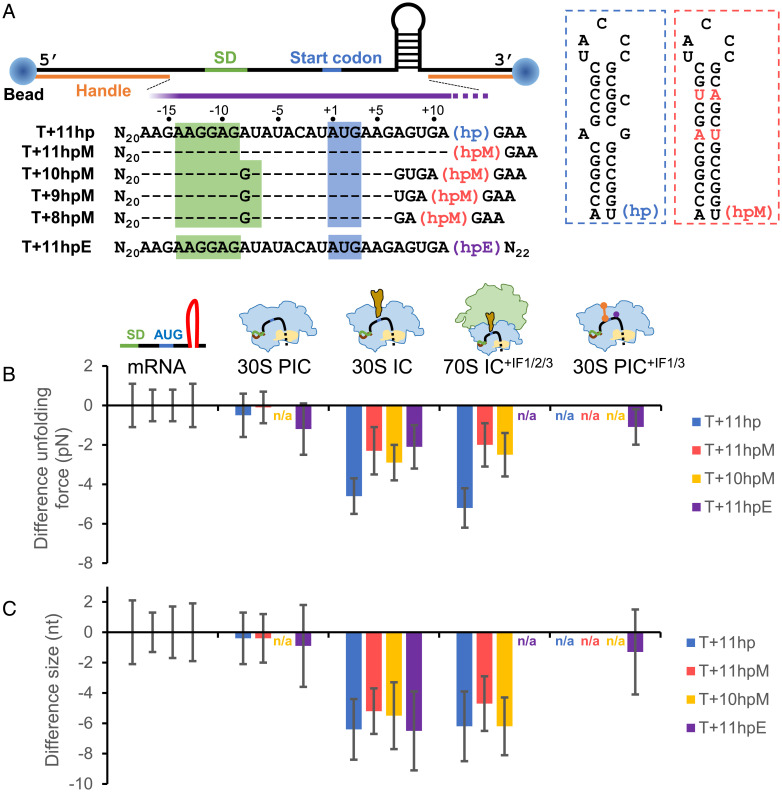
Fig. 1.
smFRET experimental setup. (A) Design of mRNA constructs. Left, mRNA was labeled with Cy3 and Cy5 dyes through complementary DNA handles at the 3′ and 5′ ends, respectively. The 5′ handle also contained a biotin tag for immobilization onto the slide surface. Middle, The mRNA sequences (5′ to 3′) between the two handles are shown. Sequences identical to F+18 are denoted as dashes for clarity. The approximate ribosome footprint is indicated by the purple bar. Right, the secondary structure of the hairpin (hp). (B) Schematic of a typical experimental procedure. Left, The 30S subunits and indicated components were incubated with immobilized, dye pair–labeled mRNA. FRET time traces were recorded and used to construct the ensemble EFRET distribution (gray histogram), which usually could be fit to a Gaussian function (green curve). Right, after recording, buffer was injected to remove unbound components. FRET time traces were recorded again, and the ensemble EFRET was constructed (orange line); PEG, polyethylene glycol.
When present alone, most F+18 molecules exhibited a stable and high FRET efficiency (EFRET; Fig. 2A, Inset). The ensemble EFRET histogram showed a narrow distribution, which was fit to a Gaussian function centered at 0.93 (Fig. 2A). The high EFRET was caused by compaction of the single-stranded RNA in the presence of Mg2+ (7 mM; SI Appendix, Fig. S1A) (29). Similar results were observed in our previous experiments using the same smFRET design with different RNA sequences (30).
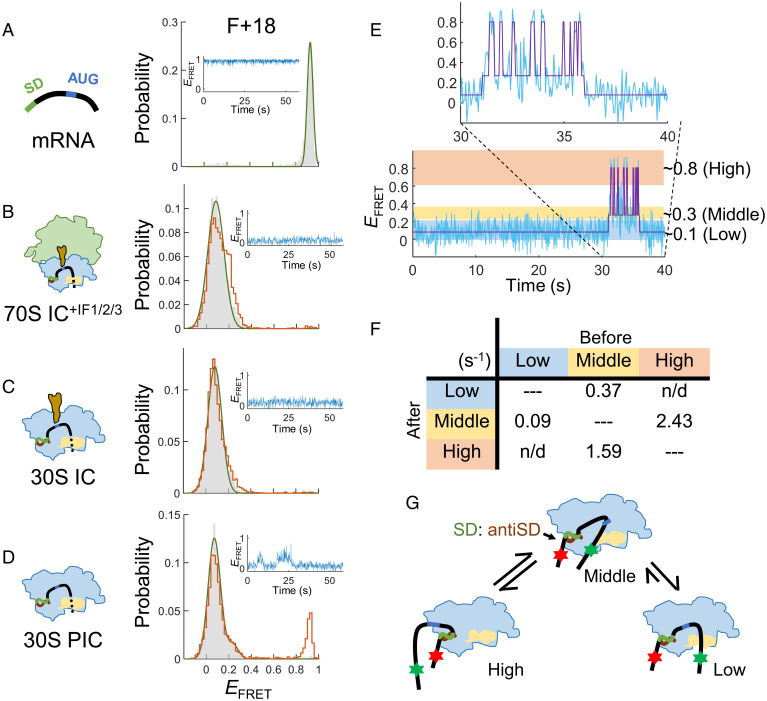
Fig. 2.
Accommodation of unstructured mRNA on 30S subunits is facilitated by initiator tRNA and initiation factors. The F+18 construct was used to measure the conformational changes of unstructured mRNA at various ribosome-binding states. (A) mRNA alone. (B) 70S IC formed in the presence of IF1, IF2, and IF3. (C) 30S IC. (D) 30S PIC. EFRET distributions before (gray histograms) and after (orange lines) buffer washing and a representative FRET time trace are shown. (E) Hidden Markov modeling for the 30S PIC. A FRET time trace (20 Hz) was fit to three EFRET states, centered at around 0.1, 0.3, and 0.8, and referred to as the low-, middle-, and high-FRET states, respectively. (F) Summary of transition rates from one state (Top) to another (Left) from the hidden Markov modeling analysis. The rates are in s−1; n/d, not detectable. (G) A model to illustrate a possible conformational change of mRNA in the 30S PIC.
We then determined how the mRNA conformation was changed when forming 70S IC. As shown in Fig. 2B (gray histogram), 99.2% of the EFRET signals appeared in a single peak at 0.08, a dramatic shift from 0.93. Note that SI Appendix, Table S1, shows the details of the major EFRET peaks of mRNA in the ribosome-bound states for the constructs measured in this study. The distribution was essentially not changed when the chamber was washed with buffer (Fig. 2B, orange line), indicating that the complexes were stable. The distance between the bacterial ribosome’s mRNA entrance and exit sites is ∼80 Å (Protein Data Bank [PDB] ID: 2HGP) (31), consistent with the low EFRET measured. To confirm that the low EFRET was caused by separation of the FRET dye pair after ribosome binding, we annealed a 30-nt DNA oligomer complementary to the RBS of mRNA as mimicry of ribosome binding. The result showed a similar distribution of EFRET at 0.12 (SI Appendix, Fig. S1B).
Initiation factors are not components of the 70S IC, but their involvement can greatly facilitate the formation of stable 70S IC. To further explore their role in the initiation process, we omitted these factors and examined how the formation of 30S IC was affected. The results showed that the EFRET distributions of 30S IC with tRNAfMet (unaminoacylated; Fig. 2C) or with fMet-tRNAfMet (SI Appendix, Fig. S1C) were indistinguishable from those observed for 30S IC+IF2 (SI Appendix, Fig. S1D; the superscript indicates the initiation factor[s] included in the reaction) and 70S IC+IF1/2/3 (Fig. 2B), indicating that the unstructured F+18 mRNA can be efficiently and stably accommodated onto 30S subunits without the assistance of initiation factors, as long as the initiator tRNA was present.
Then, we further excluded the initiator tRNA from the reaction and measured the formation of 30S PIC (we use “30S PIC” to denote the 30S IC devoid of initiator tRNA). Like in 30S IC, the EFRET distribution appeared at low values (Fig. 2D, gray), but a minor 0.92 EFRET peak reappeared after buffer washing (Fig. 2D, orange), and the time trace showed occasional FRET fluctuations (Fig. 2D, Inset). These results suggest that part of the mRNA strand bound in 30S PIC was not adequately accommodated, and thus the complex may dissociate. After quantifying the FRET dynamics with an empirical Bayesian method for hidden Markov models (32), we found that the conformation of mRNA transited among three apparent EFRET states, ∼0.1, ∼0.3, and ∼0.8, referred to as the low-, middle-, and high-FRET states, respectively (Fig. 2E). Interestingly, interchanges between the low- and the high-FRET states were not found (Fig. 2F), suggesting that the middle-FRET state was an intermediate mRNA conformation prior to 30S accommodation (Fig. 2G).
The above data showed that the initiator tRNA, in the absence of initiation factors, could stabilize the interaction between F+18 and the 30S subunit. Conversely, we asked whether initiation factors alone could have a similar effect. In the presence of both IF1 and IF3, 30S PIC+IF1/3 showed a similar low EFRET distribution (SI Appendix, Fig. S1F) as in 30S IC. The same low EFRET peak was dominant in 30S IC+IF2 (90%), and the population remained unchanged after buffer washing (SI Appendix, Fig. S1E). These results support that initiation factors (IF1 + IF3 or IF2) could further promote the accommodation of unstructured mRNA on the 30S subunit.
mRNAs with Weak or Null SD Sequences Are Compromised in Recruiting 30S Subunits.
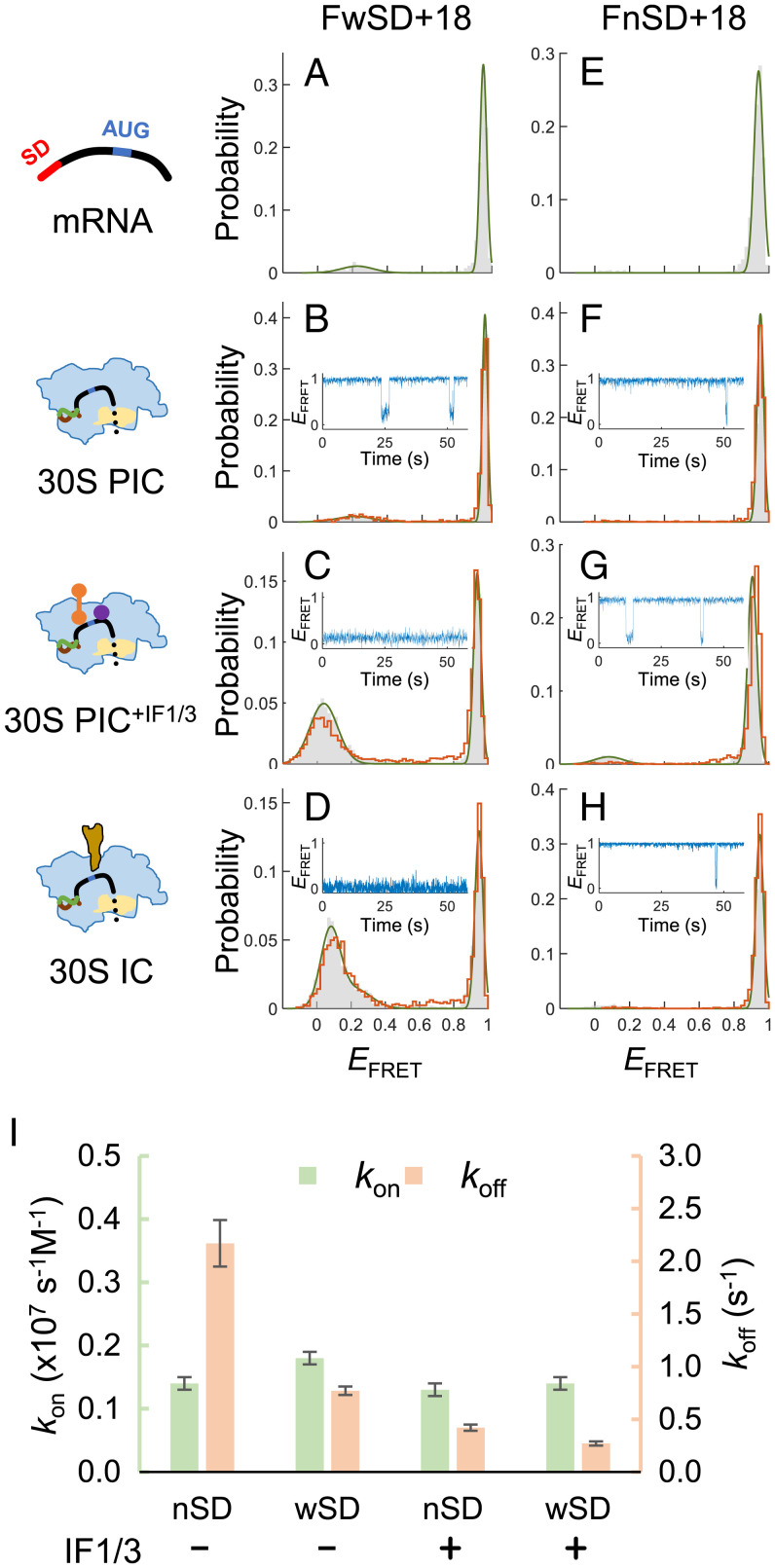
F+18 contained a 6-nt SD sequence (AAGGAG), which was relatively longer (and stronger) than most SD sequences (4 to 5 nt) found in E. coli (33). We then investigated whether the strength of SD sequences affected the interaction between mRNA and 30S subunits. In contrast to F+18, when the SD sequence was changed to a weaker one (AGGA; FwSD+18; Fig. 1A), a stable 30S PIC with low EFRET was barely formed (Fig. 3 A and B); only short-lived complexes were detected (Fig. 3B, Inset). Stable 30S–mRNA complexes could form in the presence of IF1 and IF3 (Fig. 3C) or initiator tRNA (Fig. 3D), but the efficiency was compromised. Moreover, the SD-free mRNA (FnSD+18; Fig. 1A) did not form any apparent complexes with 30S subunits, even in the presence of IF1/IF3 or initiator tRNA (Fig. 3 E–H). Kinetic analysis of the time traces showed that the dissociation rates of 30S subunits (koff) decreased from 2.17 s−1 to 0.77 s−1 when the SD sequence was changed from the null to the weak ones (Fig. 3I). Dissociation events for F+18 (with a strong SD) were rare; on average, less than one was observed per time trace (recorded for 60 s), and thus we could estimate an upper limit of koff, 0.017 (=1/60) s−1. In the presence of IF1/IF3, koff was further decreased and still SD dependent (Fig. 3I). By contrast, the association rates of 30S subunits (kon) did not change apparently with the SD strengths and initiation factors (Fig. 3I). These data support that the SD sequence, but not the start codon, of mRNA was the primary site to recruit 30S subunits; a strong SD sequence and initiation factors lengthen the lifetime of the formed complex, which increases the efficiency of start codon recognition and accommodation of the downstream RBS. Thus, the whole complex can be further stabilized.
Fig. 3.
mRNAs with weak or null SD sequences are compromised in recruiting 30S subunits. (A–D) EFRET histograms of FwSD+18 (with a weak SD sequence AGGA) in the states of mRNA alone (A), 30S PIC (B), 30S PIC+IF1/3 (C), and 30S IC (D). Gray bars and orange lines represent histograms before and after buffer washing, respectively. A representative FRET time trace is shown in B–D. (E–H) Same as in A–D, except that the SD-free construct FnSD+18 was measured. (I) Association (kon) and dissociation (koff) rate constants of 30S PIC with FwSD+18 (wSD) or FnSD+18 (nSD), measured in the presence (+) or absence (−) of both IF1 and IF3. Errors were estimated by a bootstrapping method (SI Appendix, Methods).
Binding of Initiator tRNA Stabilizes the Weak 30S PIC Formed with Structured mRNA.
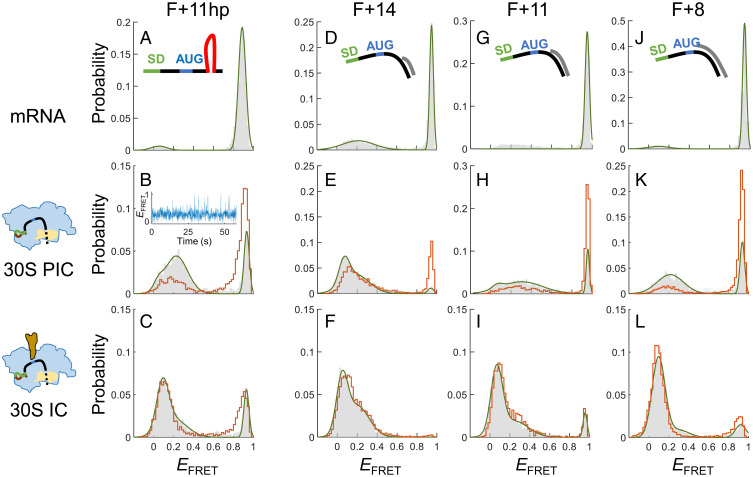
Naturally occurring mRNA may form structures around the RBS that can interfere with the initiation reaction and serve as a regulatory strategy (17, 18). To investigate accommodation of structured mRNA, we made a construct identical to F+18 except that a hairpin derived from the dnaX transcript of E. coli was inserted after position +11 (F+11hp; Fig. 1A). Given the ribosome footprint, approximately the first 3 base pairs (bp) of the stem must be opened before the mRNA is accommodated completely onto the ribosome.
F+11hp showed an EFRET of 0.89 (Fig. 4A). Binding of 30S subunits predominantly shifted the EFRET distribution to a widespread of low values with a major peak at 0.23 and a shoulder below 0.1 (Fig. 4B, gray), and substantial EFRET fluctuations at around 0.3 with frequent excursions to the higher and lower FRET states were observed (Fig. 4B, Inset). Similar FRET dynamics were also observed in F+18, but the major conformation with F+11hp was the middle-FRET state as opposed to the low-FRET state with F+18 (Fig. 2 D and E), suggesting that F+11hp was not adequately accommodated on the 30S subunit. Accordingly, most bound 30S subunits fell off after buffer washing, shifting the major EFRET peak back to 0.91 (Fig. 4B, orange). Thus, the downstream mRNA structure can severely destabilize the 30S–mRNA interactions.
Fig. 4.
Binding of initiator tRNA stabilizes the 30S PIC formed with structured mRNA. (A–C) EFRET histograms of F+11hp in the states of mRNA alone (A), 30S PIC (B), and 30S IC (C). Gray bars and orange lines represent histograms before and after buffer washing, respectively. A representative FRET time trace is shown for the 30S PIC. (D–F) Corresponding to A–C, except that F+14 was measured. (G–I) Corresponding to A–C, except that F+11 was measured. (J–L) Corresponding to A–C, except that F+8 was measured.
The frequent EFRET excursions described above were not caused by repeated dissociation–reassociation of the 30S subunit, because similar time traces were also observed after removing free 30S subunits. To further confirm this argument, we used biotinylated 30S subunits (34) for surface immobilization and incubated them with a low concentration of dye-labeled F+11hp (50 pM; SI Appendix, Fig. S2A). The results showed that the EFRET distributions were at low values (SI Appendix, Fig. S2B), and most time traces exhibited uninterrupted fluorescence and FRET fluctuations (SI Appendix, Fig. S2C), similar to the pattern shown in the Inset of Fig. 4B. Thus, these observed FRET signals were mostly caused by conformational changes of mRNA on the associated 30S subunit.
When the initiator tRNA was coincubated, the bipartite EFRET peaks of F+11hp were merged into the lower one (< 0.1; Fig. 4C, gray), which was retained after buffer washing (Fig. 4C, orange). The data indicate that binding of initiator tRNA could stabilize the low FRET conformation, analogous to the 30S IC with F+18. To confirm if the formation of stable complexes resulted from correct codon–anticodon recognition at the P site of the 30S subunit, we changed the AUG start codon to UUU, a cognate codon for tRNAPhe. Again, the new construct Fuuu+11hp (Fig. 1A) showed a major EFRET peak at 0.11 in the presence of tRNAPhe, and this peak largely remained after buffer washing (SI Appendix, Fig. S3A). By contrast, none of the 30S-bound complexes survived buffer washing when the noncognate tRNAfMet was substituted for tRNAPhe (SI Appendix, Fig. S3B). All these data support that cognate codon–anticodon interaction in the P site helped form stable 30S IC with structured mRNA. We note that after buffer washing, the 30S-bound fraction of Fuuu+11hp/tRNAPhe (22.4%) was smaller than that of F+11hp/tRNAfMet (62.0%). On this basis, the canonical AUG start codon was more potent than the UUU codon in forming ICs.
To explore the interaction between the 30S subunit and the downstream mRNA structure in more detail, we designed the following mRNA constructs for smFRET experiments: the downstream single-stranded sequence of F+18 was trimmed such that the annealed Cy3-containing DNA handle was placed immediately after positions +14, +11, and +8 (corresponding to F+14, F+11, and F+8; Fig. 1A). In this design, the handle (DNA/RNA duplex) would mimic the hairpin, and the Cy3 dye could directly reflect the response of the structure when interacting with the ribosome. Like F+18, these mRNA constructs exhibited a dominant EFRET peak at high values (Fig. 4 D, G, and J).
The downstream duplex of F+14 was expected to be located exactly outside the ribosome footprint. Compared with F+18, F+14 bound less tightly to the 30S subunit, as fewer complexes were retained after buffer washing (57.3% for F+14 and 82.0% for F+18; Figs. 2D and 4E, respectively), indicating that a spare single-stranded sequence to span the ribosome footprint was important for the complex formation. Nevertheless, initiator tRNA significantly promoted the formation of 30S IC (Fig. 4F). With the downstream duplexes further extending into the ribosome footprint, both F+11 and F+8 could barely bind 30S subunits after buffer washing (Fig. 4 H and K). However, stable 30S IC was formed in the presence of initiator tRNA (Fig. 4 I and L).
The above results of the 30S IC with structured mRNAs can be explained by 1) that the downstream structure overlapping the ribosome footprint was unwound by the 30S subunit after accommodating the initiator tRNA or 2) that base pairing between the initiator tRNA and the AUG start codon further stabilized the association of the downstream strand (either structured or unstructured) with the 30S subunit. In the next section, we used optical tweezers to test the first possibility.
The Ribosome Can Unwind Up to 3 bp of the Downstream mRNA Structure after the Binding of Initiator tRNA.
To facilitate measurements with optical tweezers, the two flanking handles of F+11hp were lengthened and respectively attached to two polystyrene beads. The construct was renamed T+11hp (Fig. 5A), whereby the prefixes “T” and “F” are used to denote the constructs designed for measurements from optical tweezers and smFRET, respectively. On optical tweezers, the construct was pulled through the two polystyrene beads, and the RNA structure was unfolded by gradually increasing the force. The stability and size of the structure were respectively determined by the unfolding force and the extension change after structural unfolding.
Fig. 5.
The ribosome can unwind up to 3 bp of the downstream mRNA structure after the binding of initiator tRNA. (A) Experimental setup for optical tweezers. An anti-digoxigenin antibody–coated polystyrene bead (Left) and a streptavidin-coated bead (Right) were attached to the ends of mRNA through the digoxigenin- and biotin-labeled DNA handles, respectively. The mRNA sequences (5′ to 3′) between the two handles are shown. Sequences identical to T+11hp are denoted as dashes for clarity. The approximate ribosome footprint is indicated by the purple bar, which overlaps part of the downstream hairpins (hp, hpM, and hpE). In hpM (red boxed), the mismatched and bulged bases of hp (blue boxed) were changed such that the whole stem was correctly base paired. For hpE, see SI Appendix, Fig. S4A for detail. (B and C) Difference in unfolding force (B) and size (C) of the downstream hairpins. For each of the indicated mRNA constructs, the unfolding force and size of the hairpin in the designated ribosome-binding states (30S PIC, 30S IC, 70S IC, and 30S PIC+IF1/3) were plotted as the difference from that of the mRNA alone. Error bars represent SDs; n/a, not available.
T+11hp showed an unfolding force of 19.1 pN and a size of 30.6 nt (SI Appendix, Table S2; note that this table summarizes the results of optical tweezer measurements described in this report). The measured size matched the actual length of the downstream hairpin (30 nt). In the presence of the 30S subunit, the hairpin was only slightly affected, with a marginal decrease in force (0.5 pN; Fig. 5B, “30S PIC”, blue) and size (0.4 nt; Fig. 4C). To further confirm that the RNA molecule under measurements was bound by a 30S subunit, we designed another construct, T+11hpE, which would form an additional short stem covering the SD sequence (Fig. 5A and SI Appendix, Fig. S4A for the predicted structure). The extra structure could be identified from the measured force extension curves of the RNA without 30S binding (SI Appendix, Fig. S4B, Left), but it disappeared when the 30S subunit had bound to the SD sequence (SI Appendix, Fig. S4B, Right). The result showed that the decrease in force (1.2 pN; Fig. 5B, “30S PIC”, purple) became more apparent, supporting that the 30S subunit bound at the SD sequence could partially destabilize the downstream RNA structure.
By contrast, after forming 30S IC or 70S IC+IF1/2/3, the hairpin size of T+11hp was significantly decreased by 6.2–6.4 nt (Fig. 5C, blue), and the force was dropped by 4.6–5.2 pN (Fig. 5B, blue), indicating that 3 bp of the hairpin were unwound in these complexes. A consistent result was obtained when a more stable hairpin was used (T+11hpM; Fig. 5A), although the unwinding size was slightly decreased (4.7–5.2 nt; Fig. 5C, red), and the force did not drop to the same level (2.0–2.3 pN; Fig. 5B, red). These results confirm that, after accommodation of initiator tRNA, the 30S subunit could unwind the mRNA downstream structure steadily to the position at approximately +14, matching the footprint size of the ribosome (2, 23).
Furthermore, to determine the maximal unwinding capacity of the 30S subunit during the initiation stage, we made three constructs (derived from T+11hpM), of which the hairpin was located immediately after positions +10 (T+10hpM), +9 (T+9hpM), and +8 (T+8hpM), corresponding to 4 bp, 5 bp, and 6 bp, respectively, of unwinding (Fig. 5A). We found that partial opening of the hairpin was detectable only in 30S IC with T+10hpM but not the other two (SI Appendix, Table S2), and the complex formation efficiency of T+10hpM was low (14% compared with 72% for T+11hpM). Moreover, the hairpin of T+10hpM in 30S IC and in 70S IC+IF1/2/3 was unwound by only 2.8 bp (5.5 nt) and 3.1 bp (6.2 nt; Fig. 5C, yellow), respectively, both smaller than the expected 4 bp. This could probably result from the putative flexibility of the mRNA within the downstream ribosome footprint. We have recently found that the mRNA strand inside the ribosome can be stretched when the ribosome translocated forward yet the encountered RNA structure resisted unwinding (35). These data demonstrate that the downstream mRNA structure overlapping the ribosome footprint by no more than 3 bp can effectively be opened during the formation of 30S IC, whereas the ribosome complexes are not formed properly when the overlapping structure is longer.
Given the finite structure-unwinding capability of the 30S subunits, the downstream mRNA structure overlapping the RBS by more than 3 bp (such as for F+8) should remain folded inside the 30S IC. Thus, the stable 30S IC found in F+8 (Fig. 4L) could be attributed to the initiator tRNA-dependent stabilization, which was not highly sensitive to the mRNA structure. However, such formed 30S complexes with unwound mRNA structures buried inside would not serve as an appropriate substrate for the translation initiation to proceed and should be circumvented. Here, we expect that initiation factors may play a role in coordinating the interaction between the 30S subunit and structured mRNA. This potential function was investigated below.
Initiation Factors Accelerate Association–Dissociation Kinetics of 30S Subunits with Structured mRNA.
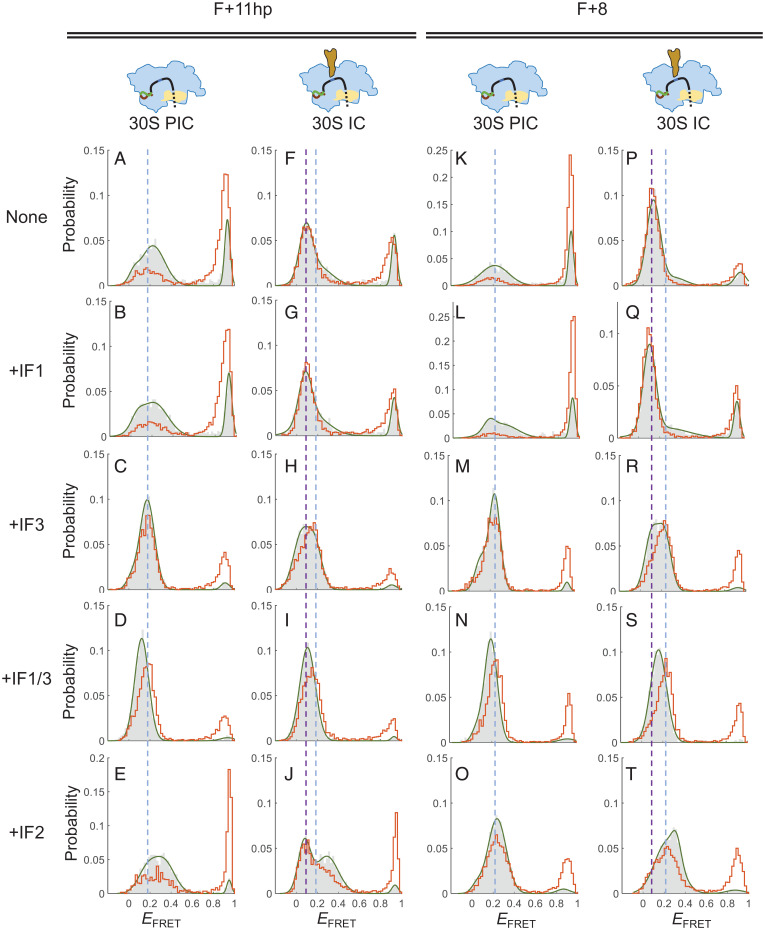
We used smFRET to explore the influence of initiation factors on the interaction between 30S subunits and structured mRNA. First, F+11hp was used as a model, whose downstream RBS-overlapping structure could be unwound by 30S subunits. In the presence of IF1, the overall EFRET distributions of 30S PIC (Fig. 6B) and 30S IC (Fig. 6G) were similar to those without IF1 (Fig. 6 A and F, respectively), indicating that IF1 did not affect the binding of 30S subunits to F+11hp. Similar results were observed for IF2 (Fig. 6 E and J), except that a higher population of 30S PIC was formed with IF2 (89.2%) than without IF2 (65.6%) before buffer washing (compare the gray histograms between Fig. 6 A and E). The data suggest that IF2 may promote the formation but not the stability of 30S PIC with F+11hp.
Fig. 6.
Initiation factors facilitate the association of 30S subunits with structured mRNA. (A–E) EFRET histograms of F+11hp in the state of 30S PIC without initiation factors (A), with IF1 (B), with IF3 (C), with both IF1 and IF3 (D), or with IF2 + 1 mM GTP (E). Gray bars and orange lines represent histograms before and after buffer washing, respectively. The blue dashed line is shown as a reference across panels to indicate the major EFRET peak (0.17) of 30S PIC+IF3 (C). (F–J) Corresponding to A–E, except that the state was 30S IC (with initiator tRNA). The purple dashed line is a reference indicating the major EFRET peak (0.11) of 30S IC (F). The same blue dashed line as in A–E is also shown for comparison. (K–O) Corresponding to A–E, except that F+8 was measured. The blue dashed line is a reference indicating the major EFRET peak (0.24) of 30S PIC+IF3 (M). (P–T) Corresponding to K–O, except that the state was 30S IC (with initiator tRNA). The purple dashed line is a reference indicating the major EFRET peak (0.09) of 30S IC (P). The same blue dashed line as in K–O is also shown for comparison.
By contrast, IF3 greatly stabilized the association between F+11hp and 30S subunits, and the major EFRET distribution appeared in a narrower peak at 0.17 (Fig. 6C). This result could be explained as IF3 CTD is located near the P site and makes some specific contacts with the start codon (13). The combination of IF1 and IF3 also showed a comparable result (Fig. 6D). This highly stable conformation raised a possibility that the downstream RBS-overlapping structure was unwound and the mRNA was well accommodated on the 30S subunit. To test this, we used optical tweezers to measure. The results showed that the T+11hpE hairpin in 30S PIC+IF1/3 was destabilized by only 1.1 pN and unwound by ∼0.6 bp, both comparable to those measured in 30S PIC (without initiation factors; Fig. 5 B and C, purple). Thus, this observation did not support unwinding of the mRNA hairpin in the IF1/IF3-bound 30S PIC.
In the formation of 30S IC (containing initiator tRNA), the overall stability was also increased in the presence of IF3 (Fig. 6H) or IF3 plus IF1 (Fig. 6I). Interestingly, the major EFRET peak (after washing) was slightly shifted from ∼0.11 without IF3 (Fig. 6 F and G, purple dashed line) to ∼0.17 with IF3 (Fig. 6 H and I, blue dashed line). This 0.17 EFRET peak was equivalent to the major peak in the 30S PIC containing IF3 (Fig. 6 C and 6D, blue dashed line). Although this FRET shift was modest, it indeed reflected a significant IF3-dependent mRNA conformational change in the 30S IC. This argument was supported by the following experiments.
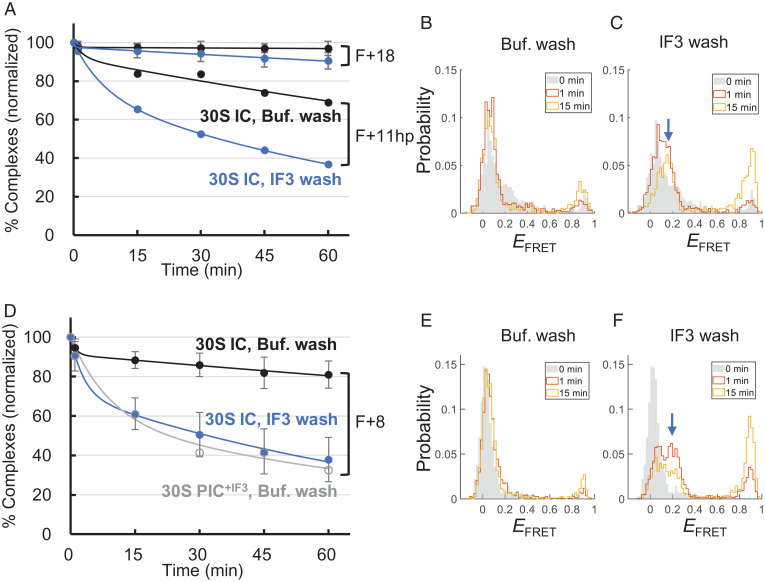
We conducted time course measurements to determine the retention of 30S IC after buffer washing. In a parallel experiment, 0.5 μM IF3 was included in the buffer such that IF3 was incubated inside the chamber after washing (termed “IF3 wash”). As shown in Fig. 7A, the 30S IC with F+11hp exhibited time-dependent disassembly, which was faster with IF3 wash than with buffer wash. The data could be fitted well to a kinetic model containing two rate constants. The major rate constant (k1) from the IF3 wash was 2.0 × 10−4 s−1, about 2.5-fold faster than that from the buffer wash (7.7 × 10−5 s−1; SI Appendix, Table S3). In addition, the EFRET distribution was split from a major single peak (∼0.1) to a bipartite (with a shoulder at ∼0.2) within 1 min after IF3 wash, and this 0.2 EFRET peak became dominant at a later time (Fig. 7C, blue arrow), whereas the EFRET splitting was not observed in the buffer wash (Fig. 7B). These results suggest that, after binding of IF3, the conformation of 30S IC was converted from the 0.1 EFRET state (initiator tRNA dependent) into the 0.2 EFRET state (IF3 dependent), and the latter was prone to disassembly. By contrast, the IF3-dependent disassembly of 30S IC was not apparent for the unstructured F+18 (Fig. 7A).
Fig. 7.
IF3 facilitates the disassembly of 30S IC. (A) Time-dependent disassembly of F+18 and F+11hp in the 30S IC states. Normalized percentages of F+18 and F+11hp retained in the 30S complexes were measured after buffer washing (Buf. wash; black filled circles) or IF3 washing (blue filled circles). Each data set was fitted to a kinetic model containing two rate constants (SI Appendix, Table S3). (B and C) Time-dependent EFRET histograms of F+11hp in the 30S IC state after buffer (B) and IF3 (C) washing. The blue arrow indicates the IF3-dependent emergency of the middle FRET state. (D) Time-dependent disassembly of F+8 in the 30S IC state after buffer washing (black filled circles) or IF3 washing (blue filled circles) and F+8 in the 30S PIC+IF3 state after buffer washing (gray open circle). (E and F) Corresponding to B and C, except that F+8 in the 30S IC state was measured. In A and D, shown are data averaged from three (with error bars) or two (without error bars) independent experiments. The error bars represent SDs.
Then, we did similar measurements for the highly structured F+8, whose downstream duplex overlapped the RBS by 6 bp and would remain closed upon 30S binding. Overall, the influence of IF1 and/or IF3 to the formation of 30S PIC (Fig. 6 K–N) and 30S IC (Fig. 6 P–S) were analogous to the corresponding results with F+11hp. Likewise, the initiator tRNA-dependent EFRET peak (0.09; Fig. 6P, purple dashed line) was shifted to ∼0.25 in the presence of IF3 (Fig. 6 R and S), matching the IF3-dependent position (blue dashed line). Interestingly, unlike F+11hp, F+8 was greatly promoted to form stable complexes with 30S subunits by IF2 (compare Fig. 6O with Fig. 6E). Moreover, the 30S IC+IF2 exhibited a major EFRET peak at 0.25 (Fig. 6T), suggesting that IF2 disfavored the highly structured F+8 to adopt the initiator tRNA-dependent conformation, although IF2 is known to facilitate the binding of initiator tRNA to 30S IC (36).
Further kinetic analysis showed that IF2 and IF3 increased the “wrapping” rate (high-to-middle FRET transition) and concurrently decreased the “unwrapping” rate (middle-to-high FRET transition) of F+8 on the 30S subunit, resulting in a three- to fourfold increase of the equilibrium constant toward the middle-FRET state (SI Appendix, Fig. S5). The results account for the enhanced stability of 30S PIC by IF2 and IF3. Nevertheless, the complex in the middle-FRET state was prone to dissociation with time. As shown in Fig. 7D, 30S PIC+IF3 (gray curve; k1 = 1.5 × 10−4 s−1; SI Appendix, Table S3) was dissociated about 4.3-fold faster than 30S IC (black curve; k1 = 3.5 × 10−5 s−1), where the 30S PIC+IF3 was mainly in the middle-FRET state (Fig. 6M) and the 30S IC in the low-FRET state (Fig. 6P). In addition, the low EFRET peak of 30S IC was retained substantially after buffer wash (Fig. 7E), but it was quickly split into another FRET population at ∼0.2 after IF3 wash (Fig. 7F), and the dissociation of the complex was accelerated (Fig. 7D, blue curve). Thus, the IF3-dependent conformational changes of 30S IC with F+8 were analogous to those found in F+11hp (Fig. 7 A–C).
In summary, IF3 and IF2 catalyzed the initiation reaction of structured mRNA to form a metastable, intermediate complex (middle-FRET state) from a primarily assembled 30S–mRNA complex (high-FRET state) or from an initiator tRNA-stabilized 30S IC (low-FRET state). The intermediate complex alone would disassemble gradually.
Discussion
Deciphering how bacterial ribosomes identify the RBS from a long mRNA template is important in understanding the mechanism of translation initiation. The largest ribosomal protein bS1 on the 30S subunit may act as the mRNA catching arm (36–38) to establish the primary association with the mRNA, where a single-stranded standby site at the 5′ UTR is a preferred target, especially when the RBS forms a structure (39, 40). bS1 was found to bind to the region preceding the SD sequence (41, 42) and could help the 30S subunit unfold structured RBS for the positioning of the SD sequence and start codon (43). Thus, the SD sequence appears to function as the docking site after the initial recruitment of the 30S subunit. Indeed, the 30S subunit binding affinity for SD-containing mRNA is more than an order of magnitude higher than that for SD-free mRNA (44), and mRNA containing the SD sequence was enriched from a randomized library for 30S subunit binding (45). In addition, we recently found that stronger SD sequences having more Gs exhibited lower dissociation rates with the 30S subunit in vitro and resulted in higher protein expression levels in vivo (30). Here, we also demonstrated that the formation of 30S PIC was highly dependent on the strength of the SD sequence (Figs. 2D and 3). These findings support the role of SD sequences in the initial recruitment of 30S subunits.
After the initial binding through SD sequences, the downstream mRNA strand was found to interact with the 30S subunit dynamically. An unstructured sequence was mostly accommodated on the 30S subunit (low-FRET state; Fig. 2E) but occasionally transited to other conformations (middle- and high-FRET states). The middle- and high-FRET states were interchanged quickly, but the low state was achieved only through the middle state (Fig. 2F). We inferred that the middle-FRET state was an intermediate mRNA conformation before full accommodation onto its binding channel of the 30S subunit (Fig. 2G). A previous study showed that, in the IC, nucleotide C1397 of 16S rRNA interacted with mRNA at positions +6 and +7, and Gln162 of ribosomal protein uS3 formed a hydrogen bond with mRNA at position +9 (46). These features shall give a structural account for our proposed intermediate mRNA conformation. Consistently, the middle-FRET state became dominant when the downstream RBS (after position +11) formed a hairpin structure (Fig. 4B), which hindered itself from fitting into the narrow mRNA entrance site of the 30S subunit (2). Thus, mRNA with unstructured RBS can be efficiently accommodated onto the 30S subunit without being trapped in the intermediate conformation, resulting in increased translation rates (22, 47–49).
It has been shown that the 30S–mRNA complexes were stabilized after binding of initiator tRNA (6, 13, 50–52). We also found that, in the presence of initiator tRNA, both unstructured and structured mRNAs were stabilized at the low-FRET state (Figs. 2C and 4C, respectively). This stabilization was attributed to not only the codon–anticodon base pairing at the P site but also the closure of the A site mRNA latch (13). The mRNA entrance site was probably also closed to lock the single-stranded sequence at around position +14, as we found that 3 bp of the downstream mRNA hairpin overlapping the ribosome footprint were persistently open in the 30S IC (Fig. 5C). This ribosomal helicase activity was initiator tRNA dependent; the downstream mRNA hairpin was only marginally affected by 30S subunits in the absence of initiator tRNA (Fig. 5 B and C, “30S PIC”). Between the two known RNA unwinding mechanisms of the ribosome during elongation, that is, open-state stabilization and mechanical unwinding (23), the 30S subunit is likely to use the open-state stabilization mechanism in the initiation process. When the structure junction fluctuates (“breathes”) thermally, the transiently released single strand can be further accommodated through the entrance site, which is then closed if initiator tRNA is present. In this model, the initiator tRNA plays a passive role in helping accommodate structured mRNA.
Like initiator tRNA, IF3 also facilitated the association between the 30S subunit and mRNA but did not lead to unwinding of structured mRNA (Fig. 5C, “30S PIC+IF1/3”). A structural study showed that, in the absence of initiator tRNA, the CTD of IF3 made several contacts with the +1, +2, and +4 nt of mRNA at around the P site (13). These extra interactions from IF3 could further anchor unstructured mRNA on the binding channel of the 30S subunit. However, when the downstream mRNA sequence formed a structure, it would clash with the entrance site and not be accommodated completely, resulting in the formation of an intermediate conformation of 30S PIC+IF3 observed in F+11hp (Fig. 6C) and F+8 (Fig. 6M). Further kinetic analysis showed that the 30S PIC+IF3 with F+8 was disassembled faster than the corresponding 30S IC (Fig. 7D), indicating that the 30S PIC+IF3 was a metastable complex. Interestingly, when IF3 was added, the disassembly rate of the 30S IC was greatly accelerated and matched that of 30S PIC+IF3. This result suggests that the initiator tRNA was triggered to fall off from the complex when IF3 was bound to the 30S subunit, consistent with known function of IF3 that destabilizes the binding of initiator tRNA to the 30S subunit (53, 54).
IF2 is another initiation factor that helps the 30S subunit recruit mRNA. For the unstructured F+18, highly stable 30S–mRNA complexes were formed in the presence of IF2 (SI Appendix, Fig. S1E), initiator tRNA (Fig. 2C), or both (SI Appendix, Fig. S1D). For the structured F+8, however, the conformation of 30S–mRNA complexes with initiator tRNA (low-FRET state; Fig. 6P) appeared to be different from that with IF2 (middle-FRET; Fig. 6O), and IF2 was dominant when both were included (Fig. 6T). In this regard, IF2 functioned in a similar way as IF3; when an mRNA structure hindered it from proper accommodation (such as F+8), IF2 would dislodge the bound initiator tRNA from the 30S subunit. On the other hand, when the structure can be partially unfolded to fit the mRNA channel in the presence of initiator tRNA (such as F+11hp), the 30S IC was largely retained in a stable, low-FRET state with IF2 (Fig. 6J); yet, it was transitioned to a metastable, middle-FRET state by IF3 (Fig. 6H) and gradually dissociated (Fig. 7A, bottom blue curve). Thus, IF2 and IF3 may counteract in recruiting the mRNA harboring an initiator tRNA-dependent, resolvable structure.
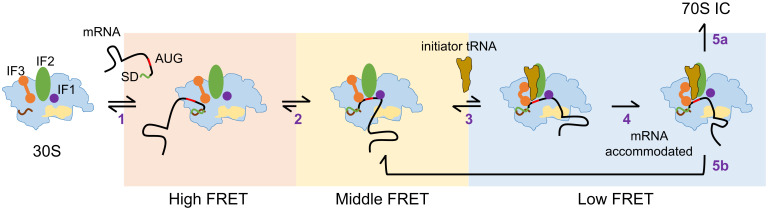
It has been shown that initiation factors play a role in accelerating the kinetics, instead of equilibrium, of the interaction between the 30S subunit and initiator tRNA (53). These features may help the ribosome quickly and correctly select the cognate initiator tRNA during initiation. Here, as shown in Fig. 8, we propose that the selection of RBS by the 30S subunit may also follow a similar dynamic strategy. The initial selection of RBS is based on the SD sequence (step 1); mRNA with a weak SD sequence is retained only briefly on the 30S subunit. IF2 and IF3 further prolong the binding of mRNA by quickly moving it from the high-FRET state to the middle-FRET state, where the start codon is placed near the P site (step 2). Accommodation of initiator tRNA to the P site further anchors the mRNA in the binding channel, resulting in a low-FRET state (step 3). The reaction will not proceed forward if the mRNA contains a structure not fitting into the entrance site (e.g., F+8), and the initiator tRNA will be rejected by IF3 (and probably IF2), and the reaction will be reverted. By contrast, the mRNA will be accommodated completely (step 4) if its downstream RBS contains no structures (e.g., F+18) or weak structures (e.g., F+11hp). The former will proceed to form 70S IC (step 5a), whereas the latter may revert to the middle-FRET state (step 5b), depending on the outcome of counteracting between IF2 and IF3.
Fig. 8.
A model for the dynamic RBS selection by the 30S subunit during initiation. The initiation factors (IF1, IF2, and IF3) may bind to the 30S subunit before or after its binding to mRNA. Step 1, association of the 30S subunit to mRNA through SD:anti-SD base pairing. Step 2, movement of the start codon to the proximity of the P site, which is assisted by initiation factors. Step 3, binding of the initiator tRNA to the P site, such that the downstream RBS is placed at around the mRNA entrance site. Step 4, closure of the entrance site if the RBS can be accommodated completely. Step 5, stable 70S IC is formed in the presence of 50S subunits (5a), or the initiator tRNA is dissociated and the reaction is reverted to a previous step (5b). The corresponding FRET states are shown in each step.
Overall, our study suggests that the formation of 30S PIC and 30S IC may involve a repeated assembly–disassembly process, which is accelerated by initiation factors. Through this process, an unstructured RBS containing the SD sequence and the AUG start codon will be preferentially selected over the others. This sequence feature may represent a typical RBS, as similar features of the translation initiation sites have been identified in a genome-wide analysis (20). In addition, initiation factors, especially IF2 and IF3, not only ensure the efficiency but also sustain the fidelity in the formation of 30S ICs.
Materials and Methods
Sample Preparations.
Ribosomes were purified from E. coli. IF1, IF2, and IF3 were overexpressed in BL21 cells (Agilent #230280) and purified through column chromatography. RNAs were in vitro transcribed and then annealed with specific tag-labeled DNA handles for single-molecule measurements. Further details are described in SI Appendix, Methods.
Single-Molecule Experiments.
We used an objective-type total internal reflection fluorescence (TIRF) microscope for smFRET measurements. A 532-nm laser (CL532-075-L, CrystaLaser) and a 638-nm laser (DL638-035, CrystaLaser) were used for excitation. On optical tweezers, a micropipette and an optical trap were used to manipulate a pair of micron-sized beads for force measurements. Further details are described in SI Appendix, Methods.
Data Analysis.
The single-molecule data were analyzed by custom-written MATLAB programs, as described previously for optical tweezers (55) and for smFRET (56). Briefly, on optical tweezers, the measured distance change (x) of an RNA structure was converted to the corresponding number of nucleotides by the worm-like chain model (57):
where F is the force, kB is the Boltzmann constant, T is the absolute temperature, P is the persistent length, and L is the contour length of the sequence that forms the structure. For single-stranded RNA, P = 1 nm and L = 0.59 nm per nt (58). The diameter of the hairpin (2 nm) was calibrated for the conversion.
For the time traces recorded from smFRET experiments, EFRET values of frames 3 to 12 (0.1 to 0.6 s) from each trace were averaged, and the distribution was plotted as a histogram.
A hidden Markov model based on the empirical Bayesian method, named ebFRET (32), was used to analyze the time traces of FRET to obtain kinetic data. The maximum number of states for fitting was determined by the apparent features of the traces; usually two or three states were assigned.
Supplementary Material
Acknowledgments
We thank Dr. Harry Noller (University of California Santa Cruz) for the plasmids of initiation factors (IF1, IF2, and IF3) and for the E. coli KLF203 strain used in the preparation of the biotinylated 30S ribosomal subunits and Dr. Jin Chen (then at University of California San Francisco) for the protocols for purification of ribosomal subunits. We thank Dr. Kai-Chun Chang for the MATLAB script package for analyzing smFRET data and other lab members for materials and suggestions. We thank the Technology Commons of National Taiwan University College of Life Science for the core facility. This work was supported by the Ministry of Science and Technology (106-2311-B-002-012-MY2, 109-2311-B-002-009-MY3).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2118099119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Shine J., Dalgarno L., The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: Complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. U.S.A. 71, 1342–1346 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yusupova G. Z., Yusupov M. M., Cate J. H., Noller H. F., The path of messenger RNA through the ribosome. Cell 106, 233–241 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Rodnina M. V., Translation in prokaryotes. Cold Spring Harb. Perspect. Biol. 10, a032664 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaledhonkar S., et al. , Late steps in bacterial translation initiation visualized using time-resolved cryo-EM. Nature 570, 400–404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wen J. D., Kuo S. T., Chou D., . The diversity of Shine-Dalgarno sequences sheds light on the evolution of translation initiation. RNA Biol. 18, 1489–1500 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milón P., Maracci C., Filonava L., Gualerzi C. O., Rodnina M. V., Real-time assembly landscape of bacterial 30S translation initiation complex. Nat. Struct. Mol. Biol. 19, 609–615 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Studer S. M., Joseph S., Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol. Cell 22, 105–115 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Cummings H. S., Hershey J. W., Translation initiation factor IF1 is essential for cell viability in Escherichia coli. J. Bacteriol. 176, 198–205 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olsson C. L., Graffe M., Springer M., Hershey J. W., Physiological effects of translation initiation factor IF3 and ribosomal protein L20 limitation in Escherichia coli. Mol. Gen. Genet. 250, 705–714 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Laalami S., Putzer H., Plumbridge J. A., Grunberg-Manago M., A severely truncated form of translational initiation factor 2 supports growth of Escherichia coli. J. Mol. Biol. 220, 335–349 (1991). [DOI] [PubMed] [Google Scholar]
- 11.Moazed D., Samaha R. R., Gualerzi C., Noller H. F., Specific protection of 16 S rRNA by translational initiation factors. J. Mol. Biol. 248, 207–210 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Stringer E. A., Sarkar P., Maitra U., Function of initiation factor 1 in the binding and release of initiation factor 2 from ribosomal initiation complexes in Escherichia coli. J. Biol. Chem. 252, 1739–1744 (1977). [PubMed] [Google Scholar]
- 13.Hussain T., Llácer J. L., Wimberly B. T., Kieft J. S., Ramakrishnan V., Large-scale movements of IF3 and tRNA during bacterial translation initiation. Cell 167, 133–144 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simonetti A., et al. , Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor. Proc. Natl. Acad. Sci. U.S.A. 110, 15656–15661 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caban K., Gonzalez R. L. Jr., The emerging role of rectified thermal fluctuations in initiator aa-tRNA- and start codon selection during translation initiation. Biochimie 114, 30–38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elvekrog M. M., Gonzalez R. L. Jr., Conformational selection of translation initiation factor 3 signals proper substrate selection. Nat. Struct. Mol. Biol. 20, 628–633 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozak M., Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361, 13–37 (2005). [DOI] [PubMed] [Google Scholar]
- 18.de Smit M. H., van Duin J., Control of prokaryotic translational initiation by mRNA secondary structure. Prog. Nucleic Acid Res. Mol. Biol. 38, 1–35 (1990). [DOI] [PubMed] [Google Scholar]
- 19.Duval M., Simonetti A., Caldelari I., Marzi S., Multiple ways to regulate translation initiation in bacteria: Mechanisms, regulatory circuits, dynamics. Biochimie 114, 18–29 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Burkhardt D. H., et al. , Operon mRNAs are organized into ORF-centric structures that predict translation efficiency. eLife 6, e22037 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evfratov S. A., et al. , Application of sorting and next generation sequencing to study 5′-UTR influence on translation efficiency in Escherichia coli. Nucleic Acids Res. 45, 3487–3502 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kudla G., Murray A. W., Tollervey D., Plotkin J. B., Coding-sequence determinants of gene expression in Escherichia coli. Science 324, 255–258 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qu X., et al. , The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature 475, 118–121 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takyar S., Hickerson R. P., Noller H. F., mRNA helicase activity of the ribosome. Cell 120, 49–58 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Wen J. D., et al. , Following translation by single ribosomes one codon at a time. Nature 452, 598–603 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang K. C., Salawu E. O., Chang Y. Y., Wen J. D., Yang L. W., Resolution-exchanged structural modeling and simulations jointly unravel that subunit rolling underlies the mechanism of programmed ribosomal frameshifting. Bioinformatics 35, 945–952 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Prabhakar A., Puglisi E. V., Puglisi J. D., Single-molecule fluorescence applied to translation. Cold Spring Harb. Perspect. Biol. 11, a032714 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaiser C. M., Tinoco I. Jr., Probing the mechanisms of translation with force. Chem. Rev. 114, 3266–3280 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newby Lambert M., et al. , Mg2+-induced compaction of single RNA molecules monitored by tethered particle microscopy. Biophys. J. 90, 3672–3685 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo S. T., et al. , Global fitness landscapes of the Shine-Dalgarno sequence. Genome Res. 30, 711–723 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yusupova G., Jenner L., Rees B., Moras D., Yusupov M., Structural basis for messenger RNA movement on the ribosome. Nature 444, 391–394 (2006). [DOI] [PubMed] [Google Scholar]
- 32.van de Meent J. W., Bronson J. E., Wiggins C. H., Gonzalez R. L. Jr., Empirical Bayes methods enable advanced population-level analyses of single-molecule FRET experiments. Biophys. J. 106, 1327–1337 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osterman I. A., Evfratov S. A., Sergiev P. V., Dontsova O. A., Comparison of mRNA features affecting translation initiation and reinitiation. Nucleic Acids Res. 41, 474–486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu T., et al. , Direct measurement of the mechanical work during translocation by the ribosome. eLife 3, e03406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu C. F., et al. , Formation of frameshift-stimulating RNA pseudoknots is facilitated by remodeling of their folding intermediates. Nucleic Acids Res. 49, 6941–6957 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramanian A. R., Structure and functions of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol. 28, 101–142 (1983). [DOI] [PubMed] [Google Scholar]
- 37.Lauber M. A., Rappsilber J., Reilly J. P., Dynamics of ribosomal protein S1 on a bacterial ribosome with cross-linking and mass spectrometry. Mol. Cell. Proteomics 11, 1965–1976 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loveland A. B., Korostelev A. A., Structural dynamics of protein S1 on the 70S ribosome visualized by ensemble cryo-EM. Methods 137, 55–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sterk M., Romilly C., Wagner E. G. H., Unstructured 5′-tails act through ribosome standby to override inhibitory structure at ribosome binding sites. Nucleic Acids Res. 46, 4188–4199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Romilly C., Deindl S., Wagner E. G. H., The ribosomal protein S1-dependent standby site in tisB mRNA consists of a single-stranded region and a 5′ structure element. Proc. Natl. Acad. Sci. U.S.A. 116, 15901–15906 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sengupta J., Agrawal R. K., Frank J., Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc. Natl. Acad. Sci. U.S.A. 98, 11991–11996 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boni I. V., Isaeva D. M., Musychenko M. L., Tzareva N. V., Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 19, 155–162 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duval M., et al. , Escherichia coli ribosomal protein S1 unfolds structured mRNAs onto the ribosome for active translation initiation. PLoS Biol. 11, e1001731 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calogero R. A., Pon C. L., Canonaco M. A., Gualerzi C. O., Selection of the mRNA translation initiation region by Escherichia coli ribosomes. Proc. Natl. Acad. Sci. U.S.A. 85, 6427–6431 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao R., et al. , Deep sequencing reveals global patterns of mRNA recruitment during translation initiation. Sci. Rep. 6, 30170 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jenner L. B., Demeshkina N., Yusupova G., Yusupov M., Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 17, 555–560 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Goodman D. B., Church G. M., Kosuri S., Causes and effects of N-terminal codon bias in bacterial genes. Science 342, 475–479 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Bentele K., Saffert P., Rauscher R., Ignatova Z., Blüthgen N., Efficient translation initiation dictates codon usage at gene start. Mol. Syst. Biol. 9, 675 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voges D., Watzele M., Nemetz C., Wizemann S., Buchberger B., Analyzing and enhancing mRNA translational efficiency in an Escherichia coli in vitro expression system. Biochem. Biophys. Res. Commun. 318, 601–614 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Masuda T., et al. , Initiation factor 2, tRNA, and 50S subunits cooperatively stabilize mRNAs on the ribosome during initiation. Proc. Natl. Acad. Sci. U.S.A. 109, 4881–4885 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Milon P., et al. , The ribosome-bound initiation factor 2 recruits initiator tRNA to the 30S initiation complex. EMBO Rep. 11, 312–316 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaminishi T., et al. , A snapshot of the 30S ribosomal subunit capturing mRNA via the Shine-Dalgarno interaction. Structure 15, 289–297 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Antoun A., Pavlov M. Y., Lovmar M., Ehrenberg M., How initiation factors tune the rate of initiation of protein synthesis in bacteria. EMBO J. 25, 2539–2550 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Antoun A., Pavlov M. Y., Lovmar M., Ehrenberg M., How initiation factors maximize the accuracy of tRNA selection in initiation of bacterial protein synthesis. Mol. Cell 23, 183–193 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Wu Y. J., Wu C. H., Yeh A. Y., Wen J. D., Folding a stable RNA pseudoknot through rearrangement of two hairpin structures. Nucleic Acids Res. 42, 4505–4515 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen Y. T., et al. , Coordination among tertiary base pairs results in an efficient frameshift-stimulating RNA pseudoknot. Nucleic Acids Res. 45, 6011–6022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bustamante C., Marko J. F., Siggia E. D., Smith S., Entropic elasticity of lambda-phage DNA. Science 265, 1599–1600 (1994). [DOI] [PubMed] [Google Scholar]
- 58.Liphardt J., Onoa B., Smith S. B., Tinoco I. Jr., Bustamante C., Reversible unfolding of single RNA molecules by mechanical force. Science 292, 733–737 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.