Significance
Fibrin plays a vital role in biology as the fibrous network that stabilizes blood clots and also through interaction with numerous blood components. While much is known about fibrin mechanics, comparatively little is known about how fibrin’s mechanics influence its biochemistry. We show that structural changes in fibrin under mechanical tension reduces binding of tissue plasminogen activator, an enzyme that initiates lysis. Furthermore, these structural transitions also led to decreased platelet activation through suppressed binding between platelet integrins and fibrin. Our work shows that fibrin possesses an intrinsic mechano-chemical feedback loop that regulates its bioactivity via molecular structural rearrangements.
Keywords: fibrin, structure-function, mechanochemistry, extracellular matrix
Abstract
Fibrin is the fibrous protein network that comprises blood clots; it is uniquely capable of bearing very large tensile strains (up to 200%) due to multiscale force accommodation mechanisms. Fibrin is also a biochemical scaffold for numerous enzymes and blood factors. The biomechanics and biochemistry of fibrin have been independently studied. However, comparatively little is known about how fibrin biomechanics and biochemistry are coupled: how does fibrin deformation influence its biochemistry? In this study, we show that mechanically induced protein structural changes in fibrin affect fibrin biochemistry. We find that tensile deformation of fibrin leads to molecular structural transitions of α-helices to β-sheets, which reduced binding of tissue plasminogen activator (tPA), an enzyme that initiates fibrin lysis. Moreover, binding of tPA and Thioflavin T, a commonly used β-sheet marker, were mutually exclusive, further demonstrating the mechano-chemical control of fibrin biochemistry. Finally, we demonstrate that structural changes in fibrin suppressed the biological activity of platelets on mechanically strained fibrin due to reduced αIIbβ3 integrin binding. Our work shows that mechanical strain regulates fibrin molecular structure and biological activity in an elegant mechano-chemical feedback loop, which possibly extends to other fibrous biopolymers.
Fibrin is a mesh-like, three-dimensional fibrous protein network that is the primary component of blood clots, entrapping blood cells and platelets to prevent blood loss at wound sites (1). In its role as the fibrous network in blood clots, fibrin also acts as a biochemical scaffold, binding to and interacting with various molecules that participate in the wound healing response, as well as a mechanical stabilizer against forces from muscle contractions (tension) and blood flow (shear) (2, 3). Fibrin viscoelasticity, along with its strain stiffening behavior, has been shown to underlie hemostasis, blood clot stability, and supportive functions for wound healing in the dynamic in situ environment (4). These biomechanical properties of fibrin depend on its hierarchical structure, building up from the molecular to the fiber level (5) where applied forces can change the structure of monomers as well as fiber properties. Because of this hierarchical organization, fibrin mechanics and biochemistry are intertwined.
Several studies have concentrated on understanding and evaluating morphological changes in fibrin under mechanical deformation and how those changes correlate to clot biochemistry, in particular stability (4, 6–8). Stretching single fibrin fibers, for example, has been shown to both promote and hinder plasmin-mediated fibrinolysis. Bucay et al. (9) demonstrated that fiber lysis was faster for prestrained fibers whereas Li et al. (10) found that lysis was slower on strained fibrin fibers. Both studies hypothesized that plasmin bioactivity depends on fiber strain although with different mechanisms. At the network level, it has been shown that tensile forces on fibrin networks reduce the degradation rate of fibrin by plasmin (11, 12). Thus, the role of strain in fibrin enzymatic degradation is still debated. An important feature not explored in any of these studies is how fibrin degradability is sensitive to its molecular structure, which is the features that mediates enzyme-fibrin interaction.
In addition to degradation, variation in clot mechanical and morphological properties has been shown to affect several aspects of platelet-induced hemostasis and thrombosis (13, 14). Platelets sense and respond to their local environment based on fibrin mechanical properties (15). Qiu et al. (15) showed that changing fibrin network stiffness modulates platelet adhesion, spreading, and activation, highlighting how a heterogeneous fibrin distribution could potentially affect platelet response. Similarly, a correlation has also been found between fibrin fiber diameter and platelet aggregation. Fibrin networks with thinner diameter fibers not only showed prolonged (slower) lysis, but also resulted in substantial aggregation of platelets (16). Most of the literature to date focuses on how macroscale properties of fibrin (e.g., fiber density, fiber diameter, and fibrin network stiffness) affect platelet activity (15–18). The impact of the molecular properties of fibrin, specifically the conformation and molecular structure of fibrin proteins that constitute fibers, on fibrin biology and biochemistry are largely absent from the fibrin structure-function literature. This is despite evidence by many groups (8, 19, 20) showing that fibrin molecular structure changes from a dominant α-helix to a dominant β-sheet structure and that molecular distances in fibrin fibers shrink with increasing deformation (21–23).
In this study, we relate molecular structural changes in mechanically deformed fibrin with changes in fibrin biochemistry and platelet response. Unfolding of α-helices into β-sheets upon tensile deformation resulted in enhanced binding of Thioflavin T (ThT), a well-known β-sheet binding molecular rotor dye, and strongly reduced the binding of tissue plasminogen factor (tPA), an enzyme that converts plasminogen to plasmin upon fibrin binding. In addition, platelet activity was strongly suppressed on mechanically deformed fibrin due to structural alterations that decreased αIIbβ3 integrin binding to arginine glycine aspartic acid (RGD) motifs. This work provides a mechanistic perspective on how mechanical strain on fibrin regulates the biological activity of fibrin through changes in constituent protein structure.
Results
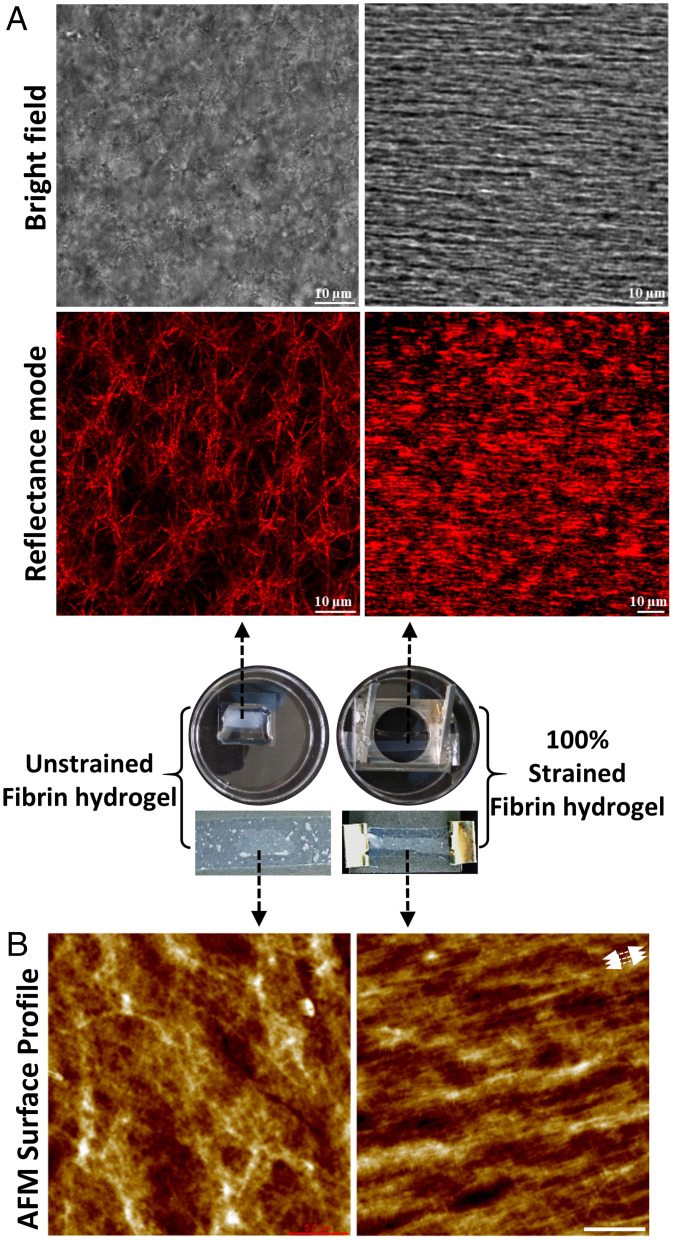
SI Appendix, Fig. S1 shows the preparation of the polydimethylsiloxane (PDMS)-fibrin construct that was used to apply tensile deformation to fibrin hydrogels (shown in Movies S1 and S2). We applied 100% strain to the flexible PDMS sheet, and this strain was partially transmitted through to the fibrin network via the adhesion between the network to the plasma-treated PDMS sheet (SI Appendix, Fig. S2). The free surface of the fibrin gel showed 73% strain while the surface attached to the PDMS was strained by 100%. Hereafter, we refer to strained condition as “100% strained” or “strained” fibrin. Tensile deformation of fibrin clearly showed distinct morphological changes in terms of fibrin fiber orientation, fiber diameter, and fiber packing density as shown by confocal reflectance images (Fig. 1A).
Fig. 1.
Morphology changes of fibrin under mechanical strain. (A) Transmitted light (Top) and reflectance (Bottom) confocal micrographs of unstrained (Left) and 100% strained fibrin networks (Right). Strain was applied in the horizontal laboratory frame. Images are taken ∼50 µm deep into network from the top surface, ∼1 mm away from the PDMS-fibrin interface where the strain was applied. (B) AFM topography images of unstrained and 100% strained fibrin (white arrows indicate the stretching direction), (Scale bar, 5 µm) on the top surface. The middle photos show samples corresponding to unstrained and 100% strained fibrin samples.
Unstrained (as prepared) fibrin hydrogels showed an isotropic fibrous network structure with an average fiber diameter of 0.4 ± 0.1 µm (mean ± SD, n > 50) as measured from the confocal reflectance images. Upon straining, the fibrin network expelled water and thinned; the fiber diameter in strained gels reduced to 0.25 ± 0.05 µm (n > 50), as similarly reported by others (11, 19). In addition, fibers in 100% strained fibrin aligned strongly in the stretching direction and came into the close contact with one another, forming a densely packed structure that nearly masked the borders of individual fibers. Studies have previously reported that elongation and alignment of fibers under mechanical strain resulted in a stiffer network due to strain-stiffening of fibrin fibers (11, 24). Like confocal reflectance, atomic force microscopy (AFM) images of the free (top) fibrin surface of unstrained fibrin hydrogels displayed randomly oriented fibers while the stretched fibrin showed elongated and aligned fibers along the stretching direction (indicated by white arrows) with densely packed structures (Fig. 1B). As the free surface is nominally 1 mm away from the flexible PDMS-fibrin interface, the AFM images of strained fibrin show the strains propagate through the entire network.
Mechanical deformation of fibrin is known to influence fiber morphology and molecular structure (4, 8). Fibrin molecular structural changes due to mechanical deformation have been studied by vibrational spectroscopy, which can provide information on protein secondary structure (α-helix, β-sheet, turn, and random coil structures) with variation in applied strain and showed a maximum structural change around 90% strain (25). As a result, in this work fibrin networks supported on thin PDMS sheets were stretched to 100% of their initial length to induce large molecular structural changes while not breaking the PDMS-fibrin attachment.
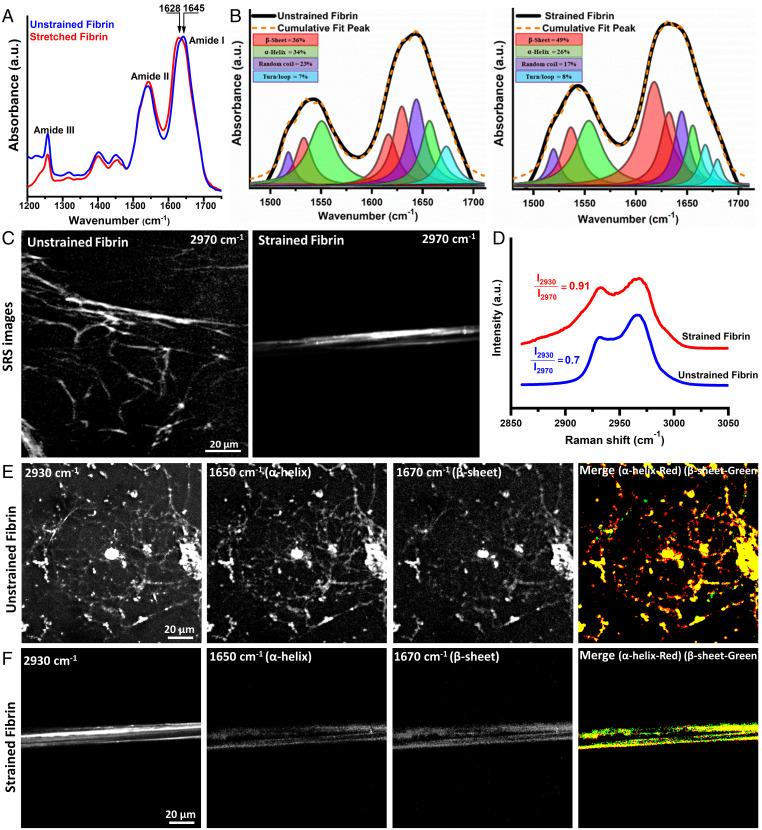
Fig. 2A shows attenuated total reflectance infrared (ATR-IR) spectra from 1,200 to 1,750 cm−1 for unstrained and 100% strained fibrin gel. The spectra show characteristic amide peaks (∼1,650 cm−1 for amide I, ∼1,550 cm−1 for amide II band, and ∼1250 cm−1 for amide III (26). Differences in the amide band shape were observed upon stretching, especially for the amide I peak. The fibrin amide I peak position for 100% strained fibrin shifted to lower frequency (1,628 cm−1) compared to unstrained fibrin (1,645 cm−1) (Fig. 2A), indicative of increased β-sheet content after stretching (20). We used second derivative analysis (SI Appendix, Fig. S3) and subsequent spectral decomposition of the amide I and II regions to provide quantitative information on the secondary structure before and after strain application (Fig. 2B, SI Appendix, Tables S1 and S2). This analysis showed that applying tensile forces to bulk fibrin networks resulted in a 36% increase in β-sheet content, whereas α-helix content decreased by 31% compared to the unstretched fibrin structure. These results are quantitatively consistent with our previous study and in good agreement with literature showing that fibrin tensile deformation is accompanied by α-helix to β-sheet structural transitions (8, 20, 27).
Fig. 2.
Tensile strain causes fibrin molecular and structural unfolding. (A) ATR-IR spectra of unstrained and 100% strained fibrin showing amide I to III bands with a clear amide I peak shift showing structural changes. (B) Decomposition of amide I and II modes from spectra in (A) into constituent peaks corresponding to different protein secondary structures shows the increased β-sheet content with strain. (C) SRS CH stretch (2,970 cm−1) image, (D) CH stretch spectra from 2,850 to 3,050 cm−1 of unstrained and strained fibrin. (E) and (F) SRS images from strained and unstrained fibrin acquired at the CH3 mode (2,930 cm−1), α-helix mode (1,650 cm−1), β-sheet mode (1,670 cm−1), and thresholded overlapped α-helix (red) and β-sheet (green) images. Imaging conditions were identical for all SRS images at the respective wavenumbers.
We extended these measurements to study molecular structural changes in sparse fibrin networks in which individual fibers could be observed using stimulated Raman scattering (SRS) vibrational microscopy (SI Appendix, Fig. S4). Fig. 2C shows SRS images of fibrin from 2,970 cm−1 in the CH region for sparse fibrin networks that are unstrained and strained. Fig. 2D shows the extracted spectral fingerprint of the CH region (2,800 to 3,050 cm−1), which has the characteristic CH3 symmetric and asymmetric CH3 stretching peaks at 2,935 cm−1 and 2,970 cm−1, respectively, that originate from side chains of alkyl amino acids in proteins (28). The ratio of the symmetric to asymmetric stretching peak was empirically observed to change with increasing strain on the fibrin network, and changes in CH peaks have been previously shown to reflect molecular scale packing of molecules (23). In the SRS spectra of sparse networks, we found the ratio was 0.7 ± 0.1 for unstrained fibrin fibers whereas strained fibrin fibers showed an increase in symmetric to asymmetric stretching ratio to 0.9 ± 0.2, which is consistent with our previous study. Combined with previous work showing increased protofibril packing in fibrin fibers using X-ray scattering (19, 21, 22), we surmise that the change in molecular distances between adjacent proto fibrils in fibrin fibers leads to increased steric hindrance for the CH3 asymmetric vibrational mode, making the lower energy CH3 symmetric mode more prevalent. Microscopically, the decreased molecular distance in fibers highlights the increased packing of protofibrils into tighter bundles prior to unfolding of coiled coil domains from helices into sheets. We next acquired SRS images at Raman shifts of 1,650 cm−1, and 1,670 cm−1, which are amide modes for α-helix and β-sheet motifs, respectively. Qualitatively looking at the merged images of the amide modes for unstrained and strain fibrin fibers in Fig. 2 E and F, unstrained fibers show more helical content. Quantitatively, unstrained fibers showed an Iα-helix:Iβ-sheet ratio of ∼1.5 compared to ∼0.8 for strained fibers after thresholding images and taking the ratio of intensities (SI Appendix, Fig. S5). Sparse fiber networks showed a 53% change in secondary structural transitions in proteins within single fibers. We note that, as in our previous work, we set the polarization of both lasers to preferentially sense the created β-sheet structures over α-helices, which will affect the absolute secondary structure percentages we obtain. However, this does not prohibit quantitative comparison between strained or unstrained fibrin networks as the laser polarization is fixed for all measurements.
Previous work has shown that natural, protein-based materials can undergo a reversable structural transitions (20, 29). Thus, we wanted to understand whether fibrin's strain-induced secondary structural changes are reversible. To evaluate structural reversibility, we took broadband coherent anti-Stokes Raman spectroscopy (CARS) images of unstrained (0%), strained (100%), and then strain-released (0%) fibrin gels as shown in SI Appendix, Fig. S6. At first, similar to SRS, CARS images and the accompanying secondary structure plot (SI Appendix, Fig. S6 A and B) showed a similar trend highlighting a decrease in α-helix and increase in β-sheet content upon straining (0 to 100%) fibrin gel. Next, upon releasing the strain and waiting 20 min, the fibrin gel showed more than 90% structural reversibility by decreasing β-sheet content and increasing α-helix, like the as-prepared fibrin hydrogels. This result is consistent with Fourier transform infrared spectroscopy studies from Litvinov et al. (20) on whole blood clots.
With a goal to investigate if the molecular structural changes in fibrin modulate its biochemical activity, we began by measuring how tPA, an enzyme involved in fibrinolysis, binding was affected by strain. In addition, we also quantified binding of ThT, a small molecule molecular rotor dye that is known to bind β-sheet rich protein structures (30, 31).
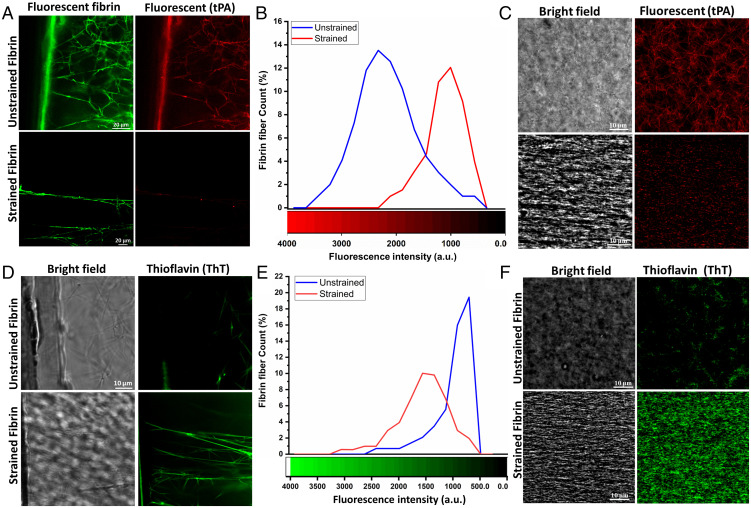
During the wound healing process, fibrin must also be enzymatically degraded by plasmin as the blood clot is slowly replaced with new tissue. To facilitate this process, fibrin serves as a biochemical scaffold that binds to tPA, which then activates plasminogen to plasmin. tPA binding to fibrin drastically increases its ability to activate plasminogen (32). As strained fibrin networks showed changes in secondary structure, we hypothesized that tPA binding could be sensitive to the tensile state, and by extension the structure, of fibrin. tPA binding to fibrin has been suggested to depend on fibrin structure via specific amino acids in coiled-coil region (residues 148 to 160 in the Aα chain) and in the D region of γ chain (residues 312 to 324) (33–35). As with the spectroscopy experiments, we used unsupported sparse and PDMS-supported bulk networks to measure tPA binding with and without strain (see SI Appendix, Fig. S7 for details on sparse network experiments). Incubation of tPA with unstrained fibrin showed effective binding to fibrin sparse and bulk and networks, with tPA essentially illuminating the fiber structure of the network. On the other hand, almost no binding was detected in strained sparse or bulk networks (Fig. 3 A and C). The mean tPA fluorescence intensity per fiber for spare networks shifted from 2,200 counts on unstrained to less than 1,000 counts on strained fibrin (Fig. 3B). Previous work has suggested that reduced network porosity may contribute to limited binding of enzymes to fibrin, but the sparse network measurements, where fiber density is substantially lower and diffusion to fibers is essentially free, demonstrates that diffusion is not limiting feature that reduced tPA binding. Rather, our data from Figs. 3 and 4 suggest that structural changes of fibrin are responsible for decreased tPA binding in line with previous reported literature (36, 37).
Fig. 3.
Differential binding of tPA and ThT on unstrained and strained fibrin. (A) Confocal micrographs of tPA bound to unstrained and strained sparse fibrin fibers, (B) tPA fluorescence intensity distribution for unstrained and strained sparse fibrin fibers (n ∼50), (C) Binding of tPA on unstrained and 100% strained bulk fibrin hydrogel, (D) Confocal micrographs of ThT bound to unstrained and strained sparse fibrin fibers and (E) ThT fluorescence intensity distribution for unstrained and strained sparse fibrin fibers (n ∼50) and (F) Binding of ThT on unstrained and 100% strained bulk fibrin hydrogel.
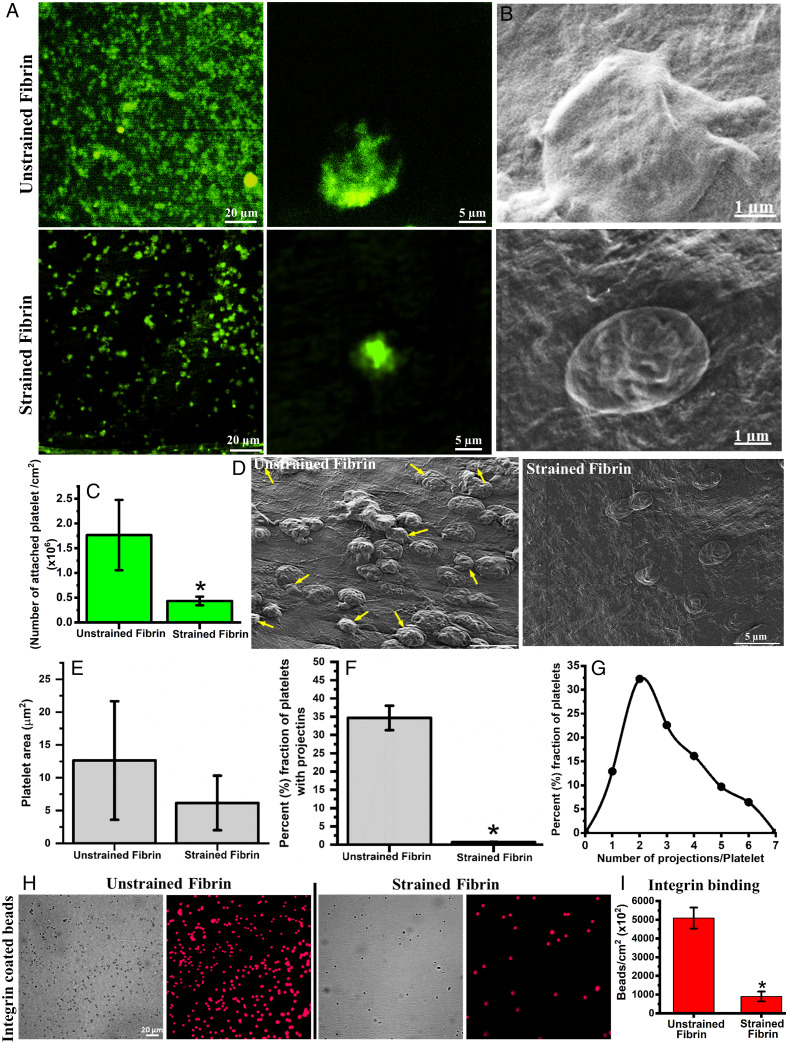
Fig. 4.
Attachment, morphology and αIIbβ3 integrin binding of platelets on different fibrin surfaces. (A) Fluorescence micrographs of attached platelets stained with Calcein on unstrained and strained fibrin gels. (B) SEM micrographs of platelets on unstrained and strained fibrin showing distinct morphology. (C) Quantification of attached platelets on unstrained and strained fibrin. (D) SEM micrographs of platelets on unstrained and strained fibrin surface (arrows indicate platelets with projections). (E) Quantification of platelet spreading (projected area) on unstrained and strained fibrin hydrogel. (F) and (G) Fraction of attached platelets with multiple projections and distribution profile of number of projections per platelets on unstrained fibrin. (H) Fluorescence micrograph of bound integrin-coated microbeads on unstrained and strained fibrin. (I) Quantitative plot of bound integrin coated microbeads on unstrained and strained fibrin. Statistically significant differences (P < 0.05) compared to unstrained fibrin is indicated by an asterisk.
As an alternative molecule to the 70 kDa tPA protein, we next probed if ThT, a small-molecule dye that binds to β-sheets showed preferential binding to unstrained or strained networks. As with tPA, we imaged ThT fluorescence in unstrained and strained fibrin hydrogels in both sparse and bulk fibrin samples (Fig. 3 D and F). In contrast to tPA, ThT bound significantly more to strained fibrin in both sparse and bulk networks compared to unstrained networks, further showing that fibrin is β-sheet rich after loading. ThT fluorescence was substantially less pronounced in relaxed fibrin gels under the same imaging conditions (Fig. 3D). The mean fluorescence intensity per fiber for spare networks shifted from 750 counts on unstrained to 1,600 counts on strained fibrin (Fig. 3E). We note that while unstrained fibrin is primarily α-helical, some parallel β-sheets exist in the D domains in the native state, based on the fibrinogen crystal structure, which is why ThT binding is nonzero in these networks.
To demonstrate the structural specificity of tPA and ThT binding, ThT and tPA solutions were added simultaneously and allowed to bind on unstrained or strained fibrin hydrogels. The z-stack projections in SI Appendix, Fig. S8 A and B and z-stack movies (Movies S3 and S4) showed depth profiles of bound tPA (green) and ThT (red) in unstrained and strained bulk fibrin hydrogel samples, respectively. ThT and tPA not only bound to the free surface but also were able to diffuse into the gel and bind to fibrin fibers at depth of at least 50 µm (our imaging depth). ThT and tPA bound to fibrin under opposite conditions, as shown by the data in Fig. 3. ThT binding was much more prominent on strained fibrin while tPA was more prominent on unstrained fibrin. Merged images of ThT and tPA showed very few spots of colocalization (indicated by arrows in SI Appendix, Fig. S8 C and D). These experiments showed that tPA and ThT bound primarily in mutual exclusion, where tPA binds to unstrained fibrin and ThT binds to strained. As the strained state on fibrin was already related to the secondary structure of fibrin, our results strongly suggest that tPA prefers to bind helical motifs whereas ThT prefers β-sheets. Given that we previously found that strain-induced secondary structure changes in fibrin were reversible from CARS imaging, we tested whether ThT binding showed similar behavior after strain reduction. SI Appendix, Fig. S9 shows that ThT binding to fibrin closely follows the results from CARS. Upon releasing the mechanical strain on fibrin networks, the mean fluorescence intensity of ThT decreased substantially, whereas stretching the same fibrin gel again showed an increase the mean fluorescence of ThT. As in the CARS experiment, the gel was allowed to relax after any strain changes to reduce the effects of viscoelasticity. This cyclic increase in the mean fluorescence of ThT on fibrin gel upon stretching illustrates that the viscoelastic relaxation does not affect the reversibility of the ThT binding to fibrin, which is consistent with the structural reversibility seen in SI Appendix, Fig. S6.
To resolve the question of whether limited mobility in the bulk fibrin network or the structural changes of fibrin were responsible for decreased tPA binding in bulk samples, we performed experiments to determine if bound tPA was able to unbind and diffuse out of the gel after strain application. tPA was initially bound to an unstrained fibrin hydrogel (SI Appendix, Fig. S8E), and upon stretching the gel, showed significantly decreased fluorescence after washing (SI Appendix, Fig. S8F and Movie S5 for more details), up to a depth of 50 µm into the network. We note that the surface of stretched fibrin, showing mostly broken or partially strained fibers retained some tPA binding (indicated by red circles in SI Appendix, Fig. S8F) whereas intact and stretched fibers in the gel interior showed near complete loss tPA fluorescence. Therefore, binding of tPA to unstrained was reversible by tensile deformation (as fibrin processes partial reversibility of secondary structure changes upon release of strain SI Appendix, Figs. S6 and S9), and the unbound tPA molecules were subsequently washed away. This experiment shows that tPA mobility in fibrin gels (with a fibrinogen concentration of 7.5 mg/mL) is not the rate-limiting step for tPA binding to fibrin, consistent with our sparse fibrin network experiments (Fig. 3D).
To further evaluate the biological activity of mechanically deformed fibrin, we studied platelet response on unstrained and strained fibrin gels. Freshly isolated platelets (SI Appendix, Fig. S10A) were deposited on fibrin samples and showed different features depending on whether they were incubated on strained or unstrained networks. Platelets showed nearly fourfold more attachment and more aggregation on unstrained fibrin compared to strained fibrin hydrogels (Fig. 4 A–C and SI Appendix, Fig. S10B). In addition, platelets on unstrained fibrin showed more spreading with nearly 35% of platelets having multiple cytoplasmic projections (on average ∼3 projections/platelet). Platelets on 100% strained fibrin, by comparison, were round with fewer cytoplasmic projections (Fig. 4 D–G). Increased cytoplasmic spreading, aggregation, and formation of multiple projections on unstrained fibrin indicated that platelets were in an activated state (38). We surmise that platelets adhered to strained fibrin were not activated as they showed limited spreading and no obvious projections.
Platelet adhesion and activation are regulated through the ligand-receptor interactions. Ligand binding to cell surface integrins, specifically αIIbβ3, mediates platelet adhesion, spreading, granule secretion, aggregation, and clot retraction (39). Given that we saw changes in platelet adhesion and activation with fibrin strain, where the structure of the coiled-coil regions was already shown to undergo structural transitions, we hypothesized that platelet integrin binding was modulated by stretching fibrin. To test this hypothesis, microbeads coated with the extracellular domain of integrin αIIbβ3, and verified to show integrin binding (SI Appendix, Fig. S11), were incubated on unstrained and strained fibrin hydrogels to test for integrin binding. After several washing steps, we imaged the attached beads and found that integrin αIIbβ3 coated beads attached greater than fivefold more to unstrained fibrin in comparison to strained fibrin (Fig. 4 H and I). Bovine serum albumin-coated beads were used as a control experiment, and these beads showed more than 10-fold reduced binding to unstrained fibrin and binding did not change with applied strain (SI Appendix, Fig. S12). This result shows that tensile deformation, and thereby secondary structure transitions in fibrin, directly affected integrin engagement, which ultimately influenced platelet adhesion and activation.
Discussion
Our results show that pulling on fibrin with near 100% tensile strain results in molecular unfolding of the constituent fibrin molecules that make up the network. This ultimately regulated various biochemical interactions, including platelet interactions and tPA enzyme binding. tPA binding to fibrin is critical for clot removal as it activates plasminogen much more efficiently when bound. tPA binds primarily to the coiled-coil region (and to a lesser extent to the nodule domains). Our data, combined with simulations (25), suggest that unfolding of coiled-coil from helices into β-sheets by tensile strain occludes the binding site, at least in the coiled-coil region. Together with previous work showing how fibrin degradation is reduced with tensile strain, we suggest a structure-function connection between the binding of tPA (and fibrin degradation) and fibrin blood clot stability. Urano et al. (37) have also suggested a similar connection between fibrin structure and fibrinolysis with increasing tensile strain. The structure-based binding of tPA and the structural transitions of fibrin under load forms an elegant mechano-chemical feedback loop that is intrinsically regulated, as we describe below. Only fibrin fibers that contain natively folded fibrin monomers, and thus not bearing large tensile loads, are subject to degradation. Loaded fibers in the gel are “marked” as load bearing by structural changes from helices to sheets, and the binding of tPA, and thus degradation rate, is drastically reduced. The ultimate outcome of such a mechanism is that parts of the fibrin clot that are actively working (load bearing) are retained while those not load-bearing are available for degradation and further processing via the wound healing cascade. This idea is consistent with the “use it or lose it” model proposed by Dingal and Discher for intracellular cytoskeletal networks (40, 41).
Studies have shown that platelets attach to fibrin through αIIbβ3 interaction with γAGDV (residues position at 409 in the γ chain) and αRGDF and αRGDS (residues positions 95 and 572, respectively, in the αA chain) (42, 43) in a mechano-sensitive way. Platelets have also been shown to sense anisotropic fiber orientation and network heterogeneity by applying piconewton forces (44, 45) via cytoplasmic, pseudopodia projections. The fact that platelet binding and activation on fibrin networks are also regulated by tensile loads, suggests a similar mechano-chemical feedback loop between platelets and fibrin as with tPA and fibrin. Activated platelets entrapped within a fibrin mesh exert significant force (ranging from 1.5 to 70 nN) on fibrin (45). According to recent work showing that platelets interact with and pull on 5 to 10 fibers in a fibrin clot (46), the force per fiber is easily above the level required to unfold the helical coiled-coil region of ∼75 pN (19, 45, 47). Thereby heterogenous platelet contraction of soft fibrin clots into highly packed and stiffer clots, could modify the protein structure of fibrin during contraction (by pulling against other contracting platelets or attachment points in the clot). As the RGD binding site of platelet integrins is in the coiled-coil region, structural changes, e.g., secondary structure unfolding, may reduce the binding efficiency and activation of additional platelets to the contracted clot. This idea consistent with experimental work from Lishko et al. (48), who showed that nascent fibrin gels supported platelet attachment, but the same gel, after removing attached platelets, showed substantially reduced platelet attachment. Their result suggests that the initial platelet adhesion altered fibrin in a way that reduced subsequent adhesion, possibly by causing structural transitions. Our results show that tensile mechanical deformation of fibrin alters the molecular structure of fibrin, including in the coiled-coil region, which drastically reduced αIIbβ3 integrin binding, platelet activation, and platelet binding. We suggest that platelet contraction of fibrin with sufficient force could alter the structure of the integrin binding domains (RGD motifs) on Aα chains, reducing platelet interaction on strained fibrin networks.
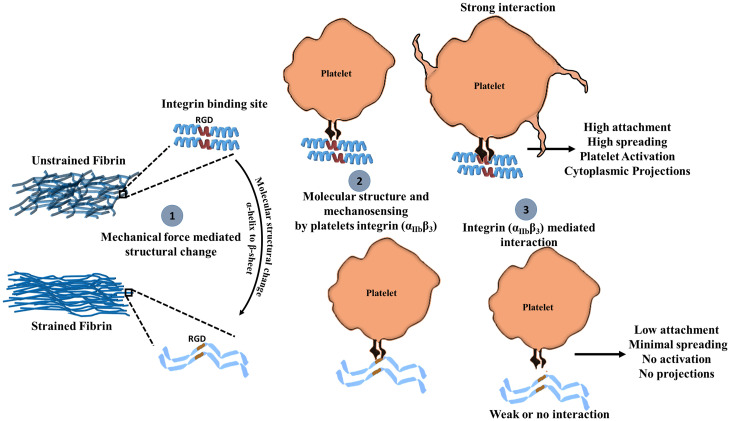
Our proposed mechano-chemical platelet fibrin interaction is summarized in Fig. 5. Sufficiently large tensile mechanical deformation of fibrin alters the structure of fibrin, resulting in densely packed, stiff, and aligned fibers (step 1). As the RGD binding site of platelets is in the Aα coiled-coil region, secondary structural changes can reduce its accessibility or binding compatibility. Thus, the interaction between αIIbβ3 the RGD binding site on fibrin is attenuated (step 2), and because the αIIbβ3-RGD interaction is mechanosensitive, platelets sense insufficient resistance from the fibrin network and do not form strong interactions or activate. On the other hand, platelets engaging with unstrained fibrin bind strongly to the RGD domain on natively folded coiled-coils of the Aα chain, which provides sufficient adhesion forces for attachment and results in more platelet activation (step 3). While our work shows strong evidence for molecular level structural transitions in fibrin as being responsible for modifying fibrin biological activity, we cannot completely exclude the possibility that squeezing water out of the intrafiber or interfiber space could alter tPA binding or platelet attachment. We note that Weisel and colleagues have shown that the average distance between the protofibrils is ∼5 nm under well-hydrated conditions. Given the size of macromolecules like tPA and plasminogen having diameters ∼6 nm (based on molecular weight), an intrafiber size of 5 nm would exclude these molecules even before stretching (49), which is not the case. Indeed, the diffusion coefficient of 40 kDa dextran, estimated to be 4.5 nm diameter, within similar networks (high thrombin low factor XIII, 5 mg/mL fibrinogen) was shown to be ∼85% of the value in saline and intrafiber transport was also suggested (50). While increasing intrafiber confinement and decreasing interfiber space may possibly contribute to reduced tPA binding in strained fibrin, the fact that tPA is able unbind and wash out from single fibrin fibers and bulk networks indicates that limited transport is not the primary explanation for the behavior we observe. Finally, we would also like to point out that mechanical deformation of fibrin hydrogel not only affects its molecular structure, but also influence topography. To distinguish the effects molecular structural changes on platelet response and tPA binding from changes in topography, further experiments are necessary. Future experiments are aimed at clarifying this issue using single fiber loading.
Fig. 5.
Proposed mechanism for mechano-chemical platelet activation on fibrin. (1) Mechanical force promotes molecular structural changes in fibrin. (2) Platelet integrin αIIbβ3 binds to unstrained fibrin RGD domains in the helical coiled-coil region of fibrin more effectively than to sheets in the same region, which leads to (3) increased platelet attachment, spreading, and activation.
Conclusions
The experiments presented here show that molecular structural changes in fibrin affect biochemical and biological activity, highlighting the unique mechano-chemical sensitivity of fibrin biomaterials. Fibrin strained to near 100% in tension showed densely packed and aligned fibers with secondary structural changes (α-helices to β-sheets) that prevented integrin-mediated platelet adhesion and activation as well as enzyme (tPA) binding. While our results demonstrate the concept of mechano-chemical regulation of by fibrin using two extreme cases, 0% and 100% tensile strain, clearly a subtler behavior exists. The features provided by such a mechano-chemical mechanism exemplify a structure-based response. The sensitivity of platelet attachment and activation on fibrin suggests that platelets sense fibrin molecular structure through their integrins, in addition to fiber anisotropy and mechanical stiffness. While we focused on fibrin, we note that other biomaterials, such as collagen, have been suggested to show similar effects (40), suggesting that mechano-chemical regulation of biomaterials is a broad phenomenon in fibrous biopolymers.
Materials and Methods
A detailed account of fibrin hydrogel preparation, flexible fibrin substrates, tensile deformation of fibrin gels, and other experimental protocols are given in SI Appendix, Materials and Methods. Briefly, for platelet isolation, blood from a de-identified, healthy volunteer was drawn by a licensed hematologist in accordance with the standard protocol approved by the institute with informed consent. All other details of molecular characterization, platelet isolation, imaging, and statistical analysis are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Florian Gericke and Marc-Jan van Zadel for technical support and Sabine Pütz for laboratory support. S.K. acknowledges Alexander von Humboldt Foundation Postdoctoral Fellowship and Science & Engineering Research Board, Startup Research Grant (SERB-SRG/2021/001886). S.H.P. acknowledges support from the Welch Foundation (F-2008-20190330), Texas 4000 funding, and the Human Frontiers in Science Program (RGP0045/2018). M.K.R. acknowledges support from the National Science Foundation Grant #2046148. M.K.R. and S.H.P. acknowledge support from the National Science Foundation Grant #2105175.
Footnotes
Competing interest statement: M.K.R. has a speaking agreement with Edwards Lifesciences.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117675119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Weisel J. W., Litvinov R. I., “Fibrin formation, structure and properties,” inFibrous Proteins: Structures and Mechanisms, Parry D.A.D., Squire J.M., Eds. (Springer, 2017), pp. 405–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tutwiler V., Wang H., Litvinov R. I., Weisel J. W., Shenoy V. B., Interplay of platelet contractility and elasticity of fibrin/erythrocytes in blood clot retraction. Biophys. J. 112, 714–723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rausch M. K., Parekh S. H., Dortdivanlioglu B., Rosales A. M., Synthetic hydrogels as blood clot mimicking wound healing materials. Prog. Biomed. Eng. 3, 042006 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Litvinov R. I., Weisel J. W., Fibrin mechanical properties and their structural origins. Matrix Biol. 60-61, 110–123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falvo M. R., Gorkun O. V., Lord S. T., The molecular origins of the mechanical properties of fibrin. Biophys. Chem. 152, 15–20 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim O. V., Litvinov R. I., Weisel J. W., Alber M. S., Structural basis for the nonlinear mechanics of fibrin networks under compression. Biomaterials 35, 6739–6749 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurniawan N. A., et al. , Fibrin networks support recurring mechanical loads by adapting their structure across multiple scales. Biophys. J. 111, 1026–1034 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleissner F., Bonn M., Parekh S. H., Microscale spatial heterogeneity of protein structural transitions in fibrin matrices. Sci. Adv. 2, e1501778 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bucay I., et al. , Physical determinants of fibrinolysis in single fibrin fibers. PLoS One 10, e0116350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W., et al. , Stretching single fibrin fibers hampers their lysis. Acta Biomater. 60, 264–274 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Varjú I., et al. , Hindered dissolution of fibrin formed under mechanical stress. J. Thromb. Haemost. 9, 979–986 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adhikari A. S., Mekhdjian A. H., Dunn A. R., Strain tunes proteolytic degradation and diffusive transport in fibrin networks. Biomacromolecules 13, 499–506 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weisel J. W., Biophysics. Enigmas of blood clot elasticity. Science 320, 456–457 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Weisel J. W., Biomechanics in hemostasis and thrombosis. J. Thromb. Haemost. 8, 1027–1029 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Qiu Y., et al. , Platelet mechanosensing of substrate stiffness during clot formation mediates adhesion, spreading, and activation. Proc. Natl. Acad. Sci. U.S.A. 111, 14430–14435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neergaard-Petersen S., et al. , Fibrin clot structure and platelet aggregation in patients with aspirin treatment failure. PLoS One 8, e71150 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swieringa F., et al. , Platelet control of fibrin distribution and microelasticity in thrombus formation under flow. Arterioscler. Thromb. Vasc. Biol. 36, 692–699 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Wufsus A. R., Macera N. E., Neeves K. B., The hydraulic permeability of blood clots as a function of fibrin and platelet density. Biophys. J. 104, 1812–1823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown A. E., Litvinov R. I., Discher D. E., Purohit P. K., Weisel J. W.. Multiscale mechanics of fibrin polymer: Gel stretching with protein unfolding and loss of water. Science 325, 741–744 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Litvinov R. I., Faizullin D. A., Zuev Y. F., Weisel J. W., The α-helix to β-sheet transition in stretched and compressed hydrated fibrin clots. Biophys. J. 103, 1020–1027 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jansen K. A.et al., . Molecular packing structure of fibrin fibers resolved by X-ray scattering and molecular modeling. Soft Matter 16, 8272–8283 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Vos B. E., Martinez-Torres C., Burla F., Weisel J. W., Koenderink G. H., Revealing the molecular origins of fibrin’s elastomeric properties by in situ X-ray scattering. Acta Biomater. 104, 39–52 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Wang Y., et al. , Probing fibrin’s molecular response to shear and tensile deformation with coherent Raman microscopy. Acta Biomater. 121, 383–392 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Piechocka I. K., Bacabac R. G., Potters M., Mackintosh F. C., Koenderink G. H., Structural hierarchy governs fibrin gel mechanics. Biophys. J. 98, 2281–2289 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barth A., Infrared spectroscopy of proteins. Biochim. Biophys. Acta. 1767, 1073–1101 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Lindon J. C., Tranter G. E., Koppenaal D., Encyclopedia of Spectroscopy and Spectrometry (Academic Press, 2016). [Google Scholar]
- 27.Purohit P. K., Litvinov R. I., Brown A. E., Discher D. E., Weisel J. W., Protein unfolding accounts for the unusual mechanical behavior of fibrin networks. Acta Biomater. 7, 2374–2383 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Howell N. K., Arteaga G., Nakai S., Li-Chan E. C., Raman spectral analysis in the C-H stretching region of proteins and amino acids for investigation of hydrophobic interactions. J. Agric. Food Chem. 47, 924–933 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Yu Y., Yang W., Wang B., Meyers M. A., Structure and mechanical behavior of human hair. Mater. Sci. Eng. C 73, 152–163 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Biancalana M., Koide S., Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim. Biophys. Acta. 1804, 1405–1412 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krebs M. R., Bromley E. H., Donald A. M., The binding of thioflavin-T to amyloid fibrils: Localisation and implications. J. Struct. Biol. 149, 30–37 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Longstaff C., Williams S., Thelwell C., Fibrin binding and the regulation of plasminogen activators during thrombolytic therapy. Cardiovasc. Hematol. 6, 212–223 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Nieuwenhuizen W., Fibrin-mediated plasminogen activation. Ann. N. Y. Acad. Sci. 936, 237–246 (2001). [DOI] [PubMed] [Google Scholar]
- 34.Kranenburg O., et al. , Tissue-type plasminogen activator is a multiligand cross-β structure receptor. Curr. Biol. 12, 1833–1839 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Schielen W., et al. , The sequence gamma-(312-324) is a fibrin-specific epitope. Blood 77, 2169–2173 (1991). [PubMed] [Google Scholar]
- 36.Longstaff C., et al. , The interplay between tissue plasminogen activator domains and fibrin structures in the regulation of fibrinolysis: Kinetic and microscopic studies. Blood 117, 661–668 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urano T., Castellino F. J., Suzuki Y., Regulation of plasminogen activation on cell surfaces and fibrin. J. Thromb. Haemost. 16, 1487–1497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Meijden P. E. J., Heemskerk J. W. M., Platelet biology and functions: New concepts and clinical perspectives. Nat. Rev. Cardiol. 16, 166–179 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Li Z., Delaney M. K., O’Brien K. A., Du X., Signaling during platelet adhesion and activation. Arterioscler. Thromb. Vasc. Biol. 30, 2341–2349 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saini K., Cho S., Dooling L. J., Discher D. E., Tension in fibrils suppresses their enzymatic degradation: A molecular mechanism for ‘use it or lose it’. Matrix Biol. 85-86, 34–46 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saini K., Discher D. E., Forced unfolding of proteins directs biochemical cascades. Biochemistry 58, 4893–4902 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Sun J. E., et al. , Bimolecular integrin-ligand interactions quantified using peptide-functionalized dextran-coated microparticles. Integr. Biol. 4, 84–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kloczewiak M., Timmons S., Lukas T. J., Hawiger J., Platelet receptor recognition site on human fibrinogen. Synthesis and structure-function relationship of peptides corresponding to the carboxy-terminal segment of the gamma chain. Biochemistry 23, 1767–1774 (1984). [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y., et al. , Platelet integrins exhibit anisotropic mechanosensing and harness piconewton forces to mediate platelet aggregation. Proc. Natl. Acad. Sci. U.S.A. 115, 325–330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lam W. A., et al. , Mechanics and contraction dynamics of single platelets and implications for clot stiffening. Nat. Mater. 10, 61–66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim O. V., Litvinov R. I., Alber M. S., Weisel J. W., Quantitative structural mechanobiology of platelet-driven blood clot contraction. Nat. Commun. 8, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown A. E., Litvinov R. I., Discher D. E., Weisel J. W., Forced unfolding of coiled-coils in fibrinogen by single-molecule AFM. Biophys. J. 92, L39–L41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lishko V. K., Yermolenko I. S., Ugarova T. P., Plasminogen on the surfaces of fibrin clots prevents adhesion of leukocytes and platelets. J. Thromb. Haemost. 8, 799–807 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bannish B. E., Chernysh I. N., Keener J. P., Fogelson A. L., Weisel J. W., Molecular and physical mechanisms of fibrinolysis and thrombolysis from mathematical modeling and experiments. Sci. Rep. 7, 1–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leonidakis K. A., et al. , Fibrin structural and diffusional analysis suggests that fibers are permeable to solute transport. Acta Biomater. 47, 25–39 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.