SUMMARY
The kinetochore links chromosomes to spindle microtubules to drive chromosome segregation at cell division. While we know nearly all mammalian kinetochore proteins, how these give rise to the strong yet dynamic microtubule attachments required for function remains poorly understood. Here, we focus on the Astrin-SKAP complex, which localizes to bioriented kinetochores and is essential for chromosome segregation, but whose mechanical role is unclear. Live imaging reveals that SKAP depletion dampens movement and decreases coordination of metaphase sister kinetochores, and increases tension between them. Using laser ablation to isolate kinetochores bound to polymerizing vs depolymerizing microtubules, we show that without SKAP kinetochores move slower on both polymerizing and depolymerizing microtubules, and that more force is needed to rescue microtubules to polymerize. Thus, in contrast to previously described kinetochore proteins that increase grip on microtubules under force, Astrin-SKAP reduces grip, increasing attachment dynamics and force responsiveness and reducing friction. Together, our findings suggest a model where the Astrin-SKAP complex effectively “lubricates” correct, bioriented attachments to help preserve them.
Graphical Abstract
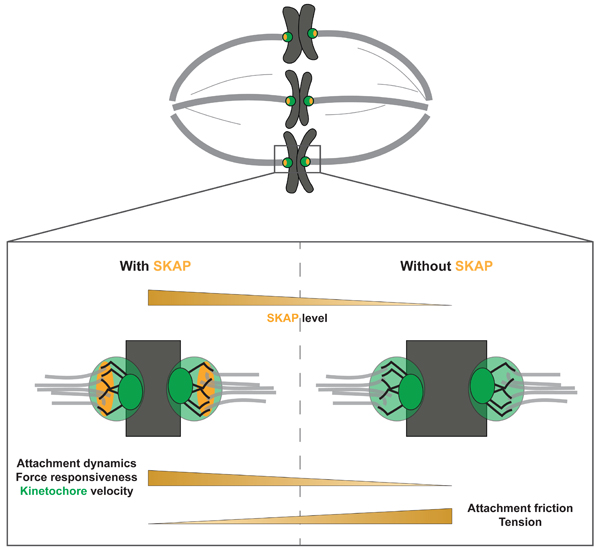
eTOC Bulrb
Rosas-Salvans et al. show that the Astrin-SKAP complex reduces friction at the mammalian kinetochore-microtubule interface, in contrast to other kinetochore proteins thought to increase friction. Astrin-SKAP increases interface dynamics and force responsiveness, effectively lubricating attachments as they stabilize, which may help preserve them.
INTRODUCTION
The kinetochore links each chromosome to spindle microtubules at cell division, transmitting spindle forces to move chromosomes. To perform its function, the kinetochore must not only bind microtubules strongly enough to resist cellular forces, but also slide on them to move and segregate chromosomes. While we now have a detailed map of mammalian kinetochore components, and are uncovering their structure, biochemistry and biophysics, how these components together give rise to the mechanics of the kinetochore-microtubule interface remains poorly understood. Indeed, we cannot as yet reconstitute mammalian kinetochores or the microtubule bundles they bind to in vitro, and applying precise mechanical perturbations to mammalian kinetochores remains challenging in vivo. How mammalian kinetochore-microtubule attachments can be robust and strong yet dynamic remains an open question. Answering this question is central to understanding how cells accurately segregate their chromosomes.
To perform its function, the kinetochore-microtubule interface both generates and responds to force. In mammalian cells, kinetochores bind to the 15–25 microtubules that form a kinetochore-fiber (k-fiber)1,2, and that both polymerize and depolymerize. When sister kinetochores oscillate together at metaphase, active (energy consuming) force generation from microtubule depolymerization at the “front” kinetochore (moving towards the pole) largely drives movement of the pair; in turn, passive, frictional force at the “back” kinetochore (moving away from the pole) is generated as kinetochore proteins slide on microtubules and oppose movement (Figure 1A)3–8. The kinetochore-microtubule interface also responds to force. For example force coordinates microtubule dynamics at both sister kinetochores as chromosomes move7,9,10, and helps maintain chromosomes in the spindle center through spatially regulated polar ejection forces11,12. Key to the interface’s ability to generate and respond to force, it is dynamic: this allows kinetochore mobility on microtubules and microtubule growth and shrinkage, and as such perturbing microtubule dynamics causes segregation defects13. While we now know different kinetochore molecules that generate force and increase grip at the microtubule interface14–21, the mechanisms that make this interface dynamic and able to respond to force despite this grip remain poorly defined.
Figure 1. SKAP increases kinetochore mobility and is essential for sister kinetochore coordination in metaphase.
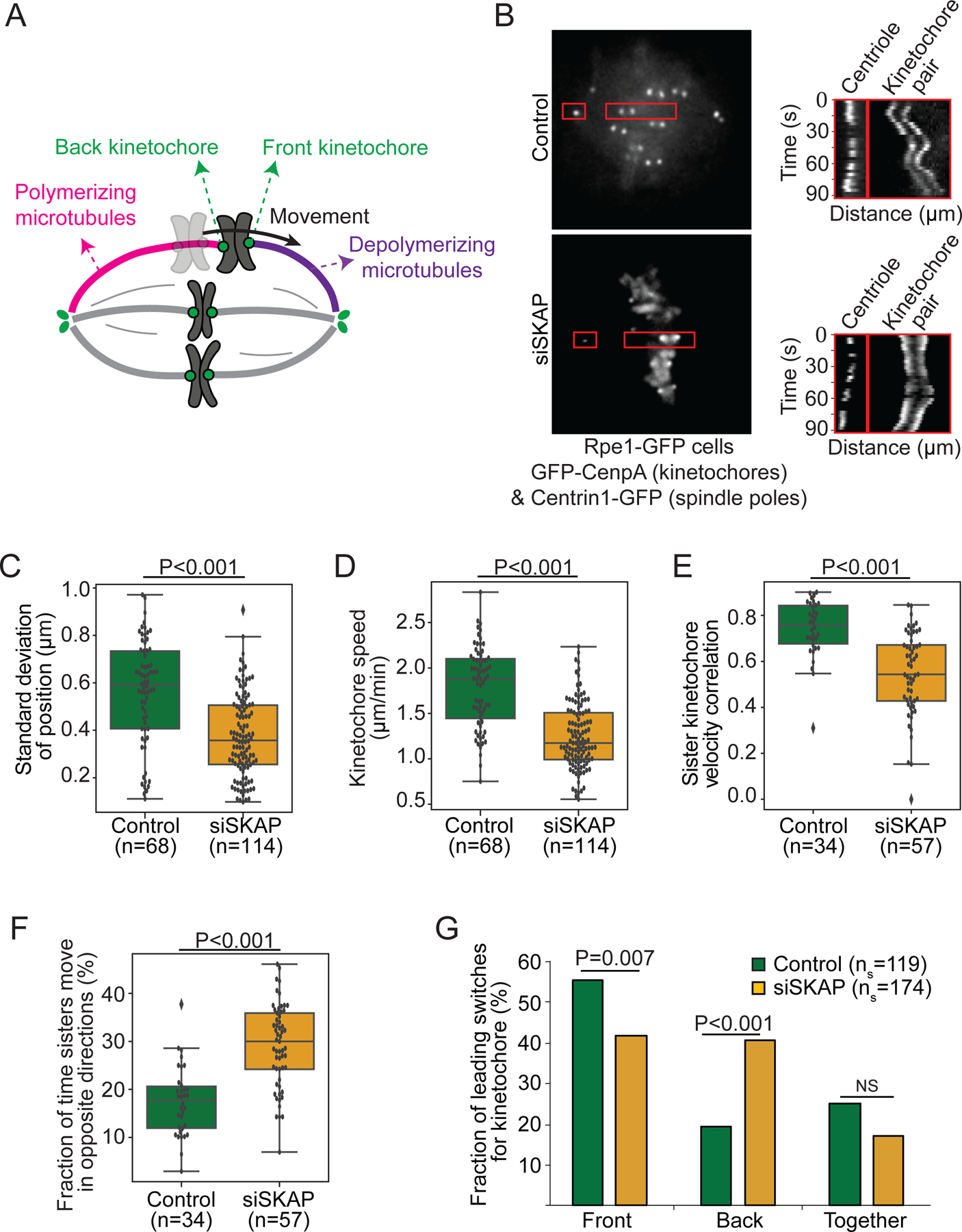
(A) Simplified representation of metaphase chromosome oscillations. Force from depolymerizing microtubules (purple) at the front kinetochore drives movement of both sisters. Frictional force at the back kinetochore, bound to polymerizing microtubules (pink), opposes movement. (B) Representative live images (left) of a control and siSKAP metaphase Rpe1-GFP cell (GFP-CenpA and centrin1-GFP) with red boxes highlighting regions used for kymographs (right) of centriole and kinetochore movements for those cells. (C) Standard deviation of the position of individual control and siSKAP metaphase kinetochores over time (Mann-Whitney test). (D) Average speed of individual control and siSKAP metaphase kinetochores (Mann-Whitney test). (E) Velocity correlation between metaphase sister kinetochores (Mann-Whitney test). (F) Fraction of time individual metaphase sister kinetochores move in opposite directions (Mann-Whitney test). (G) Fraction of metaphase directional switches in which the front or back kinetochore switches first, or both switch together (Fisher’s exact test) (ns=number of switches). (C-G) from individual kinetochore tracks obtained from the dataset as (B) (n=number of kinetochore pairs, 1–4 kinetochore pairs per analyzed cell from 18 control and 20 siSKAP cells). See also Figure S1, Videos S1-2.
Ndc80 and Ska kinetochore complexes play central roles in microtubule attachment in mammalian cells. The Ndc80 complex is essential to the formation of kinetochore-microtubule attachments in vivo14 and directly binds microtubules forming load-bearing attachments in vitro22,23. During mitosis, progressive dephosphorylation of the Ndc80 component Hec1 increases its microtubule affinity24, load-bearing ability20, and grip on polymerizing microtubules17,18. Thus, Ndc80 binds more stably to microtubules as attachments mature, with one phosphorylation site (S69 on protein Hec1) maintaining basal interface dynamics25. In turn, the Ska complex is essential for proper chromosome alignment and mitotic progression, but is only loaded at kinetochores once these biorient, and in a Ndc80-dependent manner26. In vivo, Ska increases attachment stability to depolymerizing microtubules under force19. In vitro, Ska directly binds microtubules27, and increases Ndc80’s affinity for microtubules, and microtubule tracking and load-bearing ability on depolymerizing microtubules16,20,28. Thus, Ska is thought to be a “locking” factor increasing grip on microtubules and stabilizing mature attachments. In addition to Ndc80 and Ska, the Astrin-SKAP (SKAP for short thereafter) complex has been proposed to contribute to microtubule attachment. Like Ska, SKAP is essential for chromosome alignment and mitotic progression, and is only loaded at bioriented kinetochores in a Ndc80-dependent manner29–31. Strikingly, similar to Ndc80 dephosphorylation18,32, SKAP depletion decreases k-fiber poleward flux33, suggesting that SKAP’s presence at the kinetochore may not increase grip on microtubules. Yet, SKAP directly interacts with microtubules in vitro34,35, synergistically with Ndc8035. Similar to Ndc80 and Ska36,37, mutations or changes in SKAP expression are highly frequent in some cancers and increase aneuploidy38,39. Yet, SKAP’s mechanical role at the kinetochore-microtubule interface, if any, is not known. More broadly, whether all microtubule-binding kinetochore proteins increase microtubule grip, as Ndc80 and Ska complexes do, or whether the presence of some proteins instead reduce grip to “lubricate” the attachment, remains an open question.
Here, we show that SKAP decreases grip at the kinetochore-microtubule interface, effectively “lubricating” it. We use live imaging to show that SKAP increases the magnitude of metaphase kinetochore movements and the coordination between sisters, and yet that it decreases the tension sisters are under. Using laser ablation, we show that SKAP increases the velocity with which kinetochores move on both polymerizing and depolymerizing microtubules, and that it makes the dynamics of attached microtubules more force-responsive. Thus, not all kinetochore proteins increase the grip on microtubules: SKAP does exactly the opposite, reducing friction at the interface. We propose that SKAP promotes accurate segregation by “lubricating” correct, mature attachments, ensuring that they can smoothly slide despite mechanisms that stabilize them late in mitosis. More broadly, our work suggests that maintaining a strong yet dynamic kinetochore-microtubule interface not only requires components that grip – which are being actively studied – but components that help slide, distinct from those that grip.
RESULTS
SKAP increases kinetochore mobility and is essential for sister kinetochore coordination in metaphase
To probe the mechanical role of the Astrin-SKAP complex at the kinetochore-microtubule interface, we live imaged metaphase chromosome movements in human Rpe1 (human retinal pigment epithelial-1) GFP-CenpA (kinetochore) and centrin1-GFP (centriole) cells (Rpe1-GFP40 cells thereafter). Upon SKAP depletion by RNAi (siSKAP, Figure S1A-B), kinetochore pairs oscillated less far about their mean position than control (0.4±0.2 µm vs 0.6±0.2 µm standard deviation, Figure 1B-C, Video S1-2), as expected33. siSKAP kinetochores moved at slower velocity than control (1.2±0.4 µm/min vs 1.8±0.4 µm/min, Figures 1B, D, Videos S1-2), indicating that SKAP increases kinetochore mobility. This suggests that SKAP increases kinetochore-microtubule interface dynamics. Additionally, sister kinetochore movement coordination decreased without SKAP. siSKAP sister kinetochore pairs showed lower velocity correlation than control sister pairs (0.54±0.18 vs 0.74±0.12, Figure 1E), with siSKAP sister kinetochores moving in opposite directions a higher fraction of time than control (29±7% vs 17±7%, Figure 1F). SKAP silencing using an alternative siRNA sequence confirmed the specificity of the oscillation phenotypes associated with SKAP silencing (Figure S1C-G). Further, while in control sister kinetochores the front kinetochore usually reverses direction before the back kinetochore (Figure 1G and Figure S1H7), in siSKAP sisters this preference was lost and the back kinetochore switched first more often than control (41% vs 19%, Figure 1G). Together, these findings indicate that SKAP is essential for sister kinetochore mobility and coordination at metaphase.
SKAP decreases tension at the kinetochore-microtubule interface
Mechanical force from the sister kinetochore and from the spindle on chromosome arms is thought to coordinate sister kinetochore movement and inform directional kinetochore switching3,7,12. To probe if miscoordination in siSKAP sister kinetochore movement (Figure 1E-G) could be due to defects in force generation or in how the kinetochore respond to force, we measured the interkinetochore (K-K) distance in siSKAP cells. Decreasing the activity of microtubule-binding kinetochore proteins (such as Hec1 phosphorylation or Ska depletion) typically reduces the kinetochore’s grip or ability to sustain force before sliding on microtubules, and thus leads to a lower K-K distance17,41,42 (Figure 2A). In contrast, we found that in live Rpe1-GFP cells siSKAP kinetochore pairs had a higher K-K distance than control (1.4±0.2 µm vs 1.1±0.1 µm, Figure 2B), consistent with some30 but not other29,43 previous reports. We used an alternative siRNA sequence to silence SKAP and confirmed that the K-K distance increase was a specific effect of SKAP silencing, rather than an off-target effect of the siRNA used in this study (Figure S2A).
Figure 2. SKAP decreases tension at the kinetochore-microtubule interface.
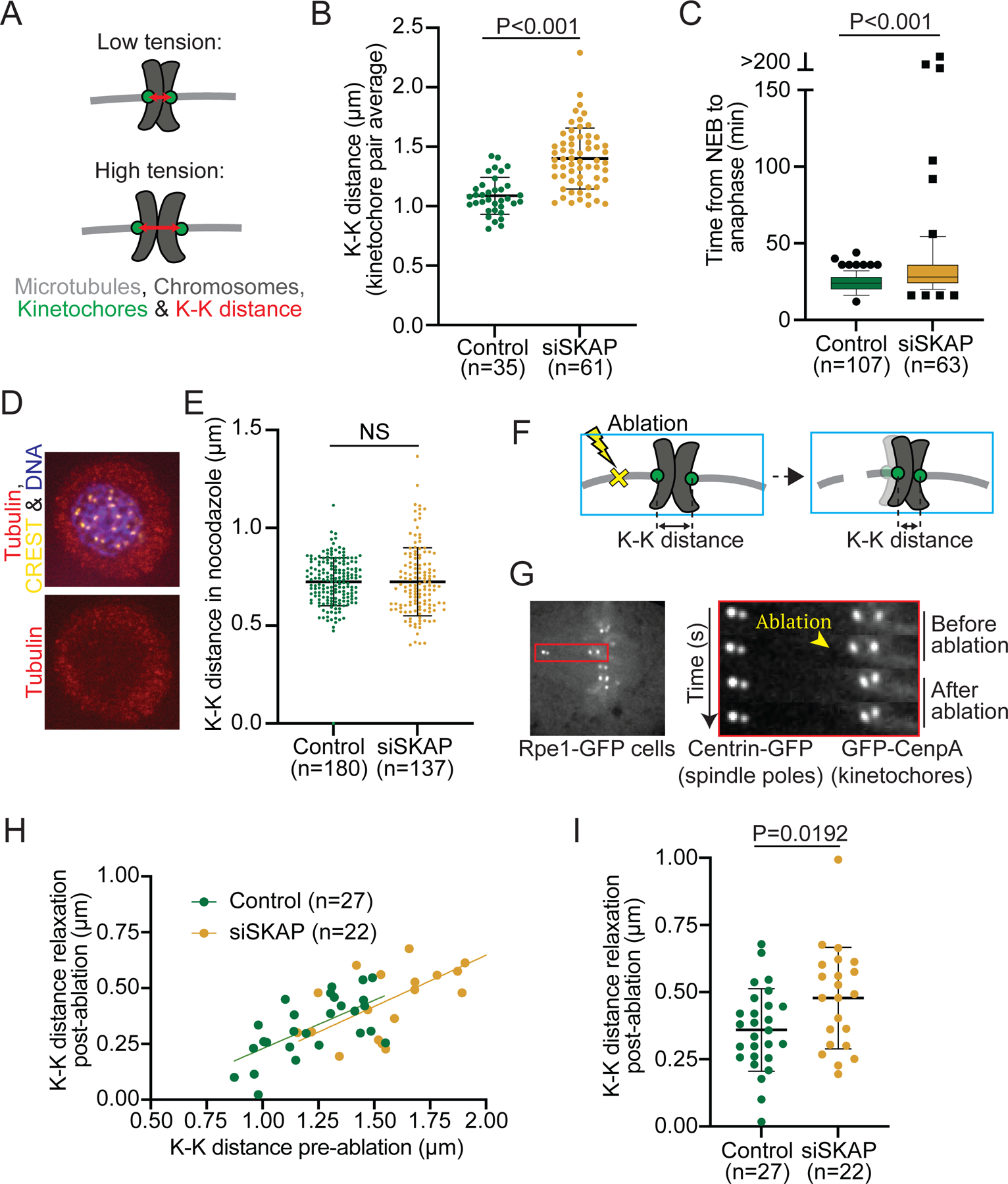
(A) High tension between sister kinetochores leads to a high K-K distance (red double arrow). High tension can stem from high spindles forces, a tighter grip of kinetochores on spindle microtubules, or both. (B) K-K distance average over time for individual kinetochore pairs in control and siSKAP Rpe1-GFP cells from the dataset in Figure 1 (studentś t-test) (n=number of kinetochore pairs, 1–4 kinetochore pairs per analyzed cell from 18 control and 20 siSKAP cells). (C) Time that individual control or siSKAP cells spend from nuclear envelope breakdown (NEB) to anaphase onset (Mann-Whitney test) (n=number of cells). Box and whiskers graph (quartiles including 5–95%) (D) Representative immunofluorescence images in control and siSKAP nocodazolde treated Rpe1-GFP cells (2µM nocodazole, 3 h) stained for CREST (yellow), chromosomes (purple) and tubulin (red). (E) K-K distance for individual sister pairs in control and siSKAP cells treated with nocodazole (Mann-Whitney test) (n=number of kinetochore pairs). (F-G) Laser ablation (yellow X) of k-fiber near a kinetochore releases tension, if present, across a sister pair (schematic cartoon) (F), as shown in representative kymograph images of K-K distance relaxation upon k-fiber ablation (yellow arrowhead) in Rpe1-GFP cells (G). (H-I) K-K distance relaxation (decrease) post-ablation as a function of K-K distance pre-ablation (linear regression lines for each condition) (H) or as a direct comparison (I) (studentś t-test) in control vs siSKAP cells (n=number of ablations, one ablation per cell). See also Figure S2.
In principle, an increase in K-K distance could stem from increased tension at the kinetochore-microtubule interface, or decreased centromere stiffness7,44. To test whether siSKAP cells could arrest at mitosis, and thereby indirectly undergo cohesion fatigue and centromere softening44, we imaged and measured mitotic duration (nuclear envelope breakdown to anaphase onset) in Rpe1-GFP cells. Mitotic duration was ~24 min in control and ~8 min longer in siSKAP cells (23.9±5.8 min vs 31.4±15.0 min, Figure 2C). However, when we artificially induced mitotic arrest using the proteasome inhibitor MG132 (10µM), the K-K distance only detectably increased starting 106 min post-MG132 (Figure S2A-B44). Considering that just 6% of siSKAP cells had a mitotic duration longer than 100 min (Figure 2C), cohesion fatigue is unlikely responsible for increasing K-K distance in siSKAP cells, as the mitotic delay observed in siSKAP cells is not long enough to induce this effect. To test whether SKAP could more directly affect centromere stiffness, we compared chromosome movements in these MG132-treated cells to siSKAP cells (Figure S2). MG132-treated cells selected for their high mean K-K distance (Figure S2B), indicative of low centromere stiffness, had poor sister kinetochore velocity correlation, indistinguishable from siSKAP cells, but kinetochore velocity indistinguishable from control (Figure S2D-E). Thus, although an effect on centromere stiffness cannot be fully excluded, SKAP cannot simply increase centromere stiffness. Consistent with SKAP not affecting centromere stiffness, treating Rpe1-GFP cells with nocodazole to remove microtubules and spindle forces led to reduced K-K distances that were indistinguishable between siSKAP and control (0.7±0.2 µm siSKAP vs 0.7±0.1µm control, Figure 2D-E). Thus, the increased K-K distance in siSKAP kinetochores depends on microtubules. Supporting this idea, laser ablating k-fibers in Rpe1-GFP cells to release spindle forces (Figure 2F-G) indicates that siSKAP kinetochores are under more tension than control. As expected for a spring, sister pairs with higher K-K distances pre-ablation relaxed more post-ablation than those with lower K-K distances in both control and siSKAP cells (Figure 2H). Notably, K-K distance relaxed more post-ablation in siSKAP kinetochores than control (0.5±0.2 µm vs 0.4±0.1 µm, Figure 2I). This difference indicates that siSKAP kinetochores are under higher tension at the kinetochore-microtubule interface, and is consistent with sister kinetochores coordinating movement more poorly (Figure 1E-G). Thus, SKAP reduces tension at the kinetochore-microtubule interface, and not all microtubule couplers at the kinetochore increase tension at this interface.
SKAP decreases kinetochore friction on polymerizing microtubules
To decrease tension at the kinetochore-microtubule interface, SKAP could either reduce passive, frictional force, or active, energy consuming force generated at this interface, or both. At metaphase, the back sister kinetochore is typically bound to polymerizing microtubules through a largely passive interface, and the front sister to depolymerizing microtubules through an interface that is both active and passive6. Thus, decoupling the roles of SKAP at passive versus active interfaces requires uncoupling the effect of SKAP depletion at kinetochores in polymerizing and depolymerizing microtubules.
Since both sisters are attached together, exerting force on each other and holding on to microtubules in opposite polymerization states, we turned to laser ablation to decouple their responses. To probe kinetochores bound to polymerizing microtubules and generating passive force, we ablated k-fibers to trigger spindle-based force generation on a sister pair18. K-fiber ablation generates new microtubule minus-ends that are recognized by dynein and pulled to the spindle pole (Figure 3A), which exerts an “external” force on the attached sister pair and induces microtubule polymerization at the back (away from the pole) kinetochore45,46. Assuming that this dynein pulling force is comparable between conditions, the velocity of this back kinetochore reports on the passive, frictional force at its microtubule interface18. We ablated k-fibers in Rpe1-GFP cells with and without SKAP (Figure 3B, Video S3-4), and tracked both sisters. The velocity of the front kinetochore was indistinguishable in siSKAP and control cells (3.2±1.5 µm/min siSKAP vs 3.0±1.4 µm/min in controls, Figure 3C, E). This is consistent with the dynein-generated force not being affected in siSKAP cells. In contrast, the back kinetochore moved slower in siSKAP cells than control (2.0±0.9 vs 2.8±1.2 µm/min, Figure 3D-E). While front and back kinetochores moved at indistinguishable velocities in control cells (3.0±1.4 µm/min in front vs 2.8±1.2 µm/min in back kinetochores), back kinetochores moved at lower velocities than front ones in siSKAP cells (2.0±0.9 µm/min vs 3.2±1.5 µm/min) (Figure 3E). This led to a persistent increase in the K-K distance during this sister kinetochore movement in siSKAP kinetochore pairs (Figure S3). Notably, the K-K distance at the time of back kinetochore directional switching (starting to move away from the pole) was higher in siSKAP kinetochores than control (1.4±0.5 µm vs 1.0±0.2 µm, Figure 3F), again suggesting that siSKAP kinetochores are less sensitive to force changes. Together, these findings indicate that SKAP decreases friction between kinetochores and polymerizing microtubules.
Figure 3. SKAP decreases kinetochore friction on polymerizing microtubules.
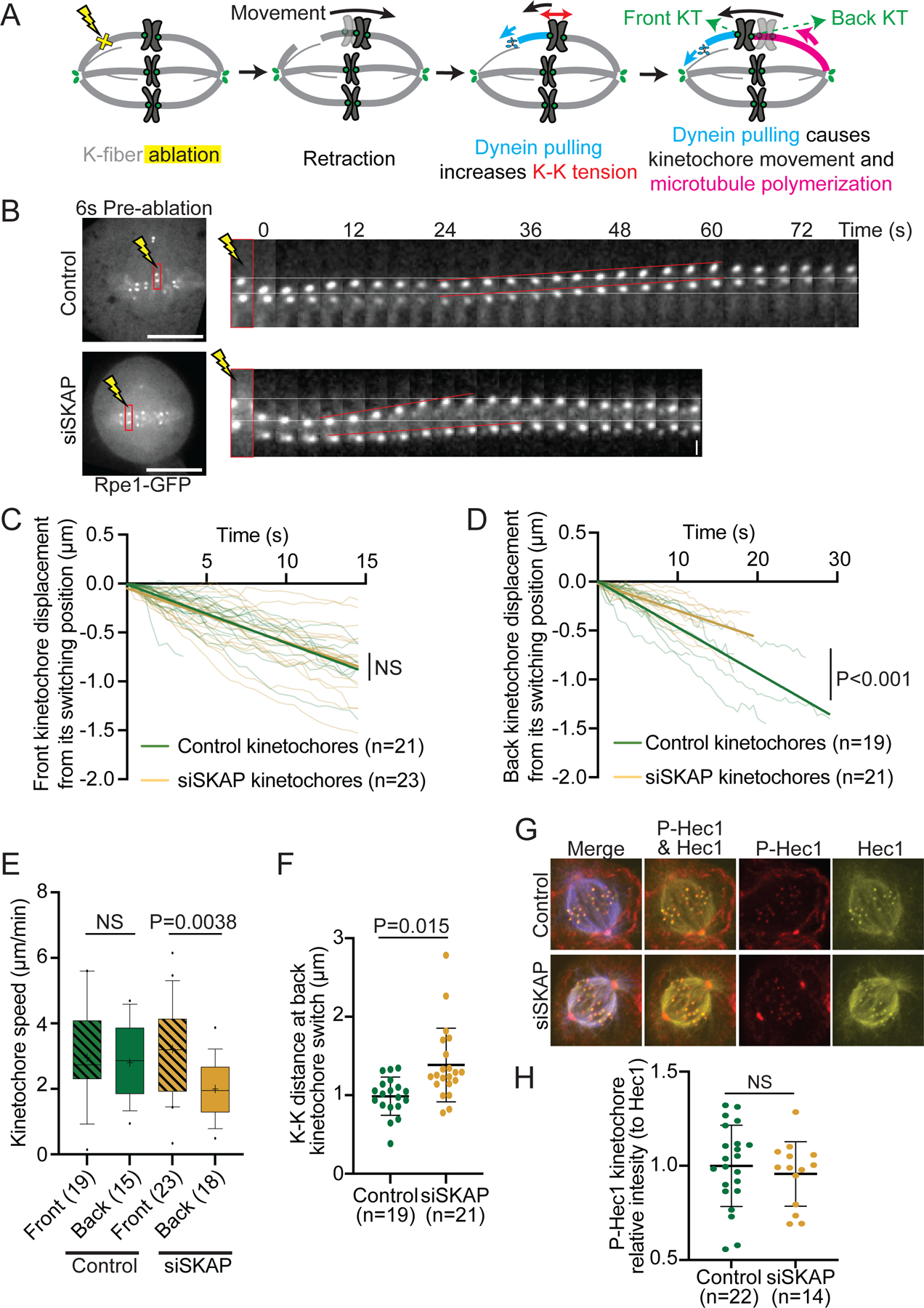
(A) K-fiber ablation assay to isolate kinetochores associated to polymerizing microtubules: laser ablation (yellow X) leads to transient retraction of the kinetochore pair, the creation of new minus-ends leads to the recruitment of dynein (light blue) which pulls on a kinetochore and leads the microtubules attached to its sister to polymerize (pink). (B) Representative time lapse of k-fiber ablation in Rpe1-GFP control and siSKAP cells. White lines mark the kinetochore positions pre-ablation at the top, red lines represent the movement of sister kinetochores. (C) Individual tracks from control and siSKAP front kinetochores moving post-ablation, showing kinetochore displacement from its switching position after dynein pulling (time=0 corresponds to the timepoint of switch to poleward movement), with linear regression fits (straight lines) (analysis of covariance test, ANCOVA). (D) Individual tracks from control and siSKAP back kinetochores moving post-ablation, showing kinetochore displacement from its switching position after dynein pulling (time=0 corresponds to the timepoint of switch to away from the pole movement), with linear regression fits (straight lines) (analysis of covariance test, ANCOVA). (E) Front (lined boxes) and back (clear boxes) kinetochore speed distribution post-ablation in control and siSKAP kinetochores (studentś t-test) from data in (C) and (D). (F) K-K distance at the time the back kinetochore switched to away-from-pole movement post-ablation in control and siSKAP kinetochore pairs (Mann-Whitney test). In (C-F), n=number of ablations, one ablation per cell. (G) Representative immunofluorescence images of Rpe1-GFP control and siSKAP cells stained for Hec1-S69 phosphorylation (red), tubulin (blue) and Hec1 (yellow). (H) p-Hec1-S69 kinetochore intensity relative to Hec1 kinetochore intensity in control and siSKAP cells (studentś t-test; n=number of cells). See also Figure S3, Videos S3-4.
Hec1 dephosphorylation is known to increase friction at the attachment interface17,18. Hec1-S69 phosphorylation (by Aurora A) is maintained during mitosis, and is required and sufficient to preserve kinetochore movement dynamics at metaphase25. Therefore, we tested whether SKAP depletion affected the level of Hec1-S69 phosphorylation. Immunofluorescence quantification showed that SKAP depletion did not affect kinetochore levels of Hec1-S69 phosphorylation (Figure 3G-H), indicating that friction regulation by SKAP occurs independently of Hec1-S69 phosphorylation. Together, these findings indicate that SKAP, in contrast to the proposed role of other microtubule binding proteins at the kinetochore17,18,20,28, decreases friction between kinetochores and polymerizing microtubules, and does so through a mechanism independent of that setting Hec1’s basal dynamic state25. SKAP could either do so by increasing microtubule tip polymerization dynamics, producing an apparent change in friction, or by directly reducing kinetochore friction on the microtubule lattice.
SKAP increases k-fiber depolymerization velocity and kinetochore force-responsiveness
Given that SKAP regulates the kinetochore’s frictional interface with polymerizing microtubules (Figure 3), we asked whether it also regulates its interface with depolymerizing ones. To decouple both attached sister kinetochores and isolate SKAP’s role on depolymerizing microtubules, we laser ablated one kinetochore in a sister pair in Rpe1-GFP cells, and tracked the movement of the remaining sister. As expected, the remaining sister was pulled poleward by depolymerizing microtubules, and reversed direction near the pole, pushed by polar ejection forces3,9,12 (Figure 4A-B, Video S5). The poleward velocity of siSKAP kinetochores was slower than that in control (2.1±0.6 µm/min vs 3.5±0.8 µm/min, Figure 4B-D, Video S5-6). This indicates that SKAP increases the velocity of microtubule depolymerization at the kinetochore interface; here too, it could do so either by lowering friction on the microtubule lattice or by increasing depolymerization dynamics and thereby decreasing apparent friction. Consistent with SKAP acting directly at the interface, SKAP depletion did not detectably change kinetochore levels of key microtubule plus-end depolymerases involved in metaphase kinetochore movements, MCAK and Kif18A47,48 (Figure S4). Further, in the same kinetochore ablation experiments siSKAP kinetochores switched direction (to away from pole movement) closer to the pole than control kinetochores as they got pushed by polar ejection forces (3.2±1.2 µm vs 4.0±1.8 µm (Figure 4E)). Thus, SKAP increases the kinetochore’s force sensitivity when it is bound to depolymerizing microtubules, favoring a switch to the polymerization state, as observed after k-fiber ablation (Figure 3F) and during metaphase oscillations (Figure 1G). Together, these findings indicate that Astrin-SKAP does not increase the kinetochore’s grip on microtubules as other microtubule-binding proteins are thought to do, but instead reduces grip, lowering friction and effectively lubricating the interface, making it more dynamic and force responsive.
Figure 4. SKAP increases k-fiber depolymerization velocity and kinetochore force-responsiveness.
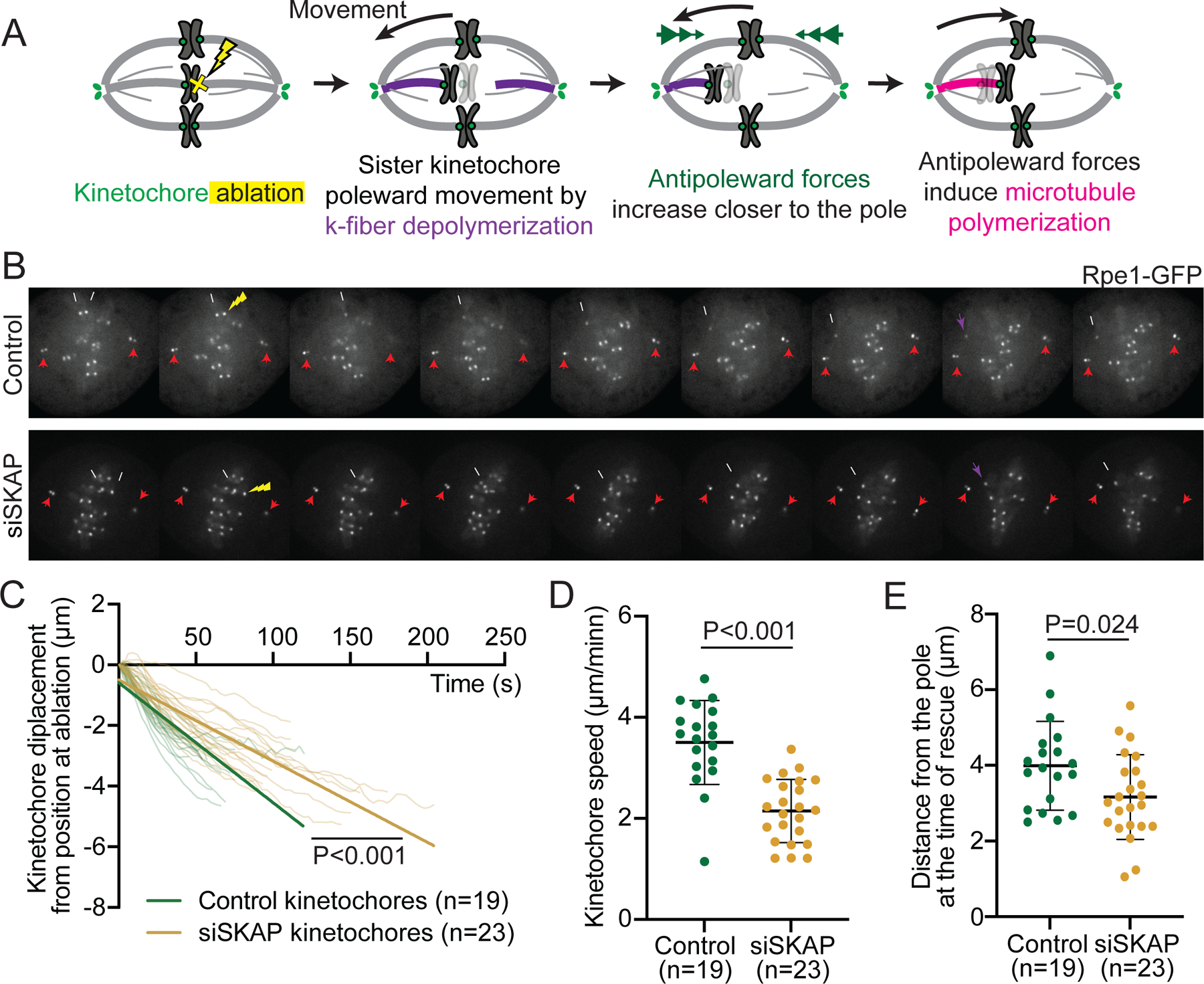
(A) Kinetochore ablation assay to isolate kinetochores associated to depolymerizing microtubules: laser ablation (yellow X) of one sister leads to the other sister moving poleward as its microtubules depolymerize (purple), and to later move away-from-the pole as polar ejection forces (green arrowheads) increase and microtubules rescue and polymerize (pink). (B) Representative time lapse images of kinetochore ablation (yellow arrow) in control (top) and siSKAP (bottom) Rpe1-GFP cells, with remaining sister kinetochore (white arrow) and centrioles (red arrows) marked, and kinetochore directional switch marked (purple arrow). (C) Distance to position at ablation as a function of time for individual kinetochores post-ablation (t=0 corresponds to the first timepoint post-ablation) in control and siSKAP cells, with linear regression fits (straight lines) (analysis of covariance test, ANCOVA). (D) Average speed of individual kinetochores post sister ablation in control and siSKAP cells (Mann-Whitney test). (E) Kinetochore distance from the spindle pole of individual control and siSKAP cells at the time of direction switch from poleward to away-from-pole movement (rescue) post-ablation (studentś t-test), with a smaller distance typically reflecting a higher force at rescue. In (C-E), n=number of ablations, one ablation per cell. See also Figure S4, Videos S5-6.
DISCUSSION
Faithful chromosome segregation requires kinetochores to hold on to dynamic microtubules as they grow and shrink. Here we ask: How does the mammalian kinetochore-microtubule interface stay dynamic and force responsive while maintaining a strong grip on microtubules? Mechanisms increasing grip as attachments mature are being actively studied, including Ndc80 dephosphorylation17,18 and Ska recruitment19,20. Here, we combine molecular and mechanical perturbations to define the mechanical role of SKAP at this interface, and we show that it in turn decreases grip at the interface. We demonstrate that SKAP increases sister kinetochore mobility and coordination in metaphase (Figure 1), and decreases tension at the kinetochore-microtubule interface (Figure 2). We show that SKAP increases the velocity at which kinetochores move on both polymerizing (Figure 3) and depolymerizing (Figure 4) microtubules, and that it makes the attachment more responsive to force changes (Figures 3–4). Together, our findings indicate that SKAP reduces friction at the kinetochore-microtubule interface, effectively lubricating it. As such, and given SKAP’s arrival as kinetochores biorient29,30, we propose that SKAP keeps correct kinetochore-microtubule attachments dynamic to preserve them as they stabilize and mature17,19. The association of SKAP mutations and changes in expression with some cancers and aneuploidy38,39,49 is consistent with SKAP’s lubrication function being key for faithful chromosome segregation.
Our work indicates that SKAP lowers friction at the kinetochore-microtubule interface, raising the question of what function a lower friction could serve. Basal levels of attachment dynamics are essential for accurate chromosome segregation25,50. Ska recruitment and gradual dephosphorylation of Hec1 increases kinetochore grip on microtubules during mitosis17–20. In contrast to other Hec1 phosphosites, S69 is persistently phosphorylated during mitosis, maintaining basal dynamics25. Yet, these dynamics are lost and friction increases upon SKAP depletion (Figure 1, 3-4), although phosphorylation levels of Hec1-S69 are not affected by this depletion (Figure 3G-H). Thus, SKAP acts as a lubricant independently of Hec1-S69 phosphorylation. By reducing friction, SKAP may increase microtubule dynamics, increasing microtubule depolymerization (Figure 4B-D) and poleward flux33. To our knowledge, SKAP is the first microtubule binder proposed to decrease friction at the kinetochore-microtubule interface. We propose that using SKAP to lower friction, instead of loosening Ndc80-Ska’s grip to lower friction, allows the cell to keep a dynamic kinetochore-microtubule interface without losing grip of microtubules and attachment stability, by having one molecule specialized for each activity. Indeed, having distinct mechanisms to tune grip and dynamics may lead to finer regulatory control “knobs” to preserve stable attachments.
By lubricating correct, bioriented attachments, SKAP could in principle help preserve them and prevent their increased microtubule affinity from making them less mobile and less responsive to force. The presence of more dynamic, lower friction interactions with microtubules could help stabilize attachments under force by increasing their adaptability to force changes. The force responsiveness of attachments is essential for accurate chromosome movement, alignment and sister kinetochore coordination7,12,51, and SKAP depleted cells have chromosome alignment and segregation defects29–31. Indeed, we show that SKAP makes kinetochores more mobile (Figure 1C-D), and the kinetochore-microtubule interface more sensitive to force changes (Figure 1E-G, 3F, 4E). This could explain why siSKAP attachments are under higher tension (Figure 2), inefficient at dissipating force (Figure 3B-E) and at responding to sister movement (Figure 1E-G, 3F), and it suggests that a function of lowering friction could be to make kinetochores responsive to force. While additional work will be needed to reveal how SKAP increases force responsiveness, in principle lowering friction would be sufficient to do so.
While SKAP reduces friction at the kinetochore-microtubule interface (Figure 3A-E), the mechanism by which it does so is not clear. SKAP depletion could in principle increase friction by reducing microtubule dynamics. However, lowering microtubule dynamics globally with drugs (as taxol or eribulin) decreases interkinetochore tension (or K-K distance), rather than increase it as we see for siSKAP (Figure 2)52,53. Thus, decreasing microtubule dynamics is not sufficient to recapitulate the effects of SKAP depletion. Further, we could not detect any change in kinetochore recruitment of key microtubule dynamics regulators (MCAK, Kif18A) in siSKAP cells (Figure S4) and changing their activity is not consistent with our findings: MCAK depletion decreases both interkinetochore tension and microtubule depolymerization47, and Kif18A depletion increases kinetochore velocity and oscillations amplitudes and decreases switching rates48,51, and we do not see these observables change as such (Figures 1C-D, 2, 3F, 4C-E). Alternatively, SKAP might affect microtubule dynamics through its described interaction with the plus end factor EB133,54. Contradicting this possibility, SKAP’s interaction with EB1 is not needed for SKAP’s role in chromosome alignment and spindle assembly, suggesting that SKAP’s role at the kinetochore-microtubule interface is independent of its interaction with EB155. Thus, while we cannot exclude the possibility that SKAP regulates the kinetochore activity of these or other microtubule dynamics regulators, or directly affect microtubule dynamics, a change in microtubule dynamics alone is not sufficient to recapitulate all the behaviors of siSKAP cells. Together, these findings are consistent with SKAP playing a mechanical role at the kinetochore-microtubule interface, and not simply regulating microtubule dynamics.
Mechanistically, SKAP could regulate kinetochore-microtubule friction in different ways. One way it could do so is by changing the sliding friction of other kinetochore proteins on microtubules. SKAP could induce conformational changes in Ndc80 or Ska complexes which lower their friction on microtubules (Model A, Figure 5), independently of Hec1-S69 phoshoregulation (Figure 3G-H). Alternatively, SKAP could directly bind microtubules and act as a kinetochore-microtubule coupler, as suggested by in vitro work30,34,35. Accordingly, the microtubule binding domain of SKAP is essential for its role in chromosome alignment and segregation55. As such, SKAP could then compete out other microtubule binders that have higher friction on microtubules (Model B, Figure 5), as microtubule affinity (binding energy) and friction coefficient (transition state energy in moving between lattice binding sites) are not strictly coupled56. The affinity of Ndc80-SKAP on microtubules35 is similar, though lower, than that of Ndc80-Ska16, suggesting that both complexes could compete for microtubule binding; if Ndc80-SKAP had lower friction on microtubules than Ncd80-Ska, SKAP kinetochore localization could lower friction at the kinetochore-microtubule interface. In principle, the above models hold independently of how SKAP is recruited to the kinetochore, for example through Ndc8030,35 or Mis1333. Whether SKAP lowers other kinetochore proteins’ (Ndc80, Ska) friction on microtubules (Model A) and whether SKAP and other kinetochore proteins compete for microtubule binding and generate different friction (Model B) are non-exclusive models and a rich area of future study.
Figure 5. Models for the Astrin-SKAP complex decreasing friction at the mammalian kinetochore-microtubule interface.
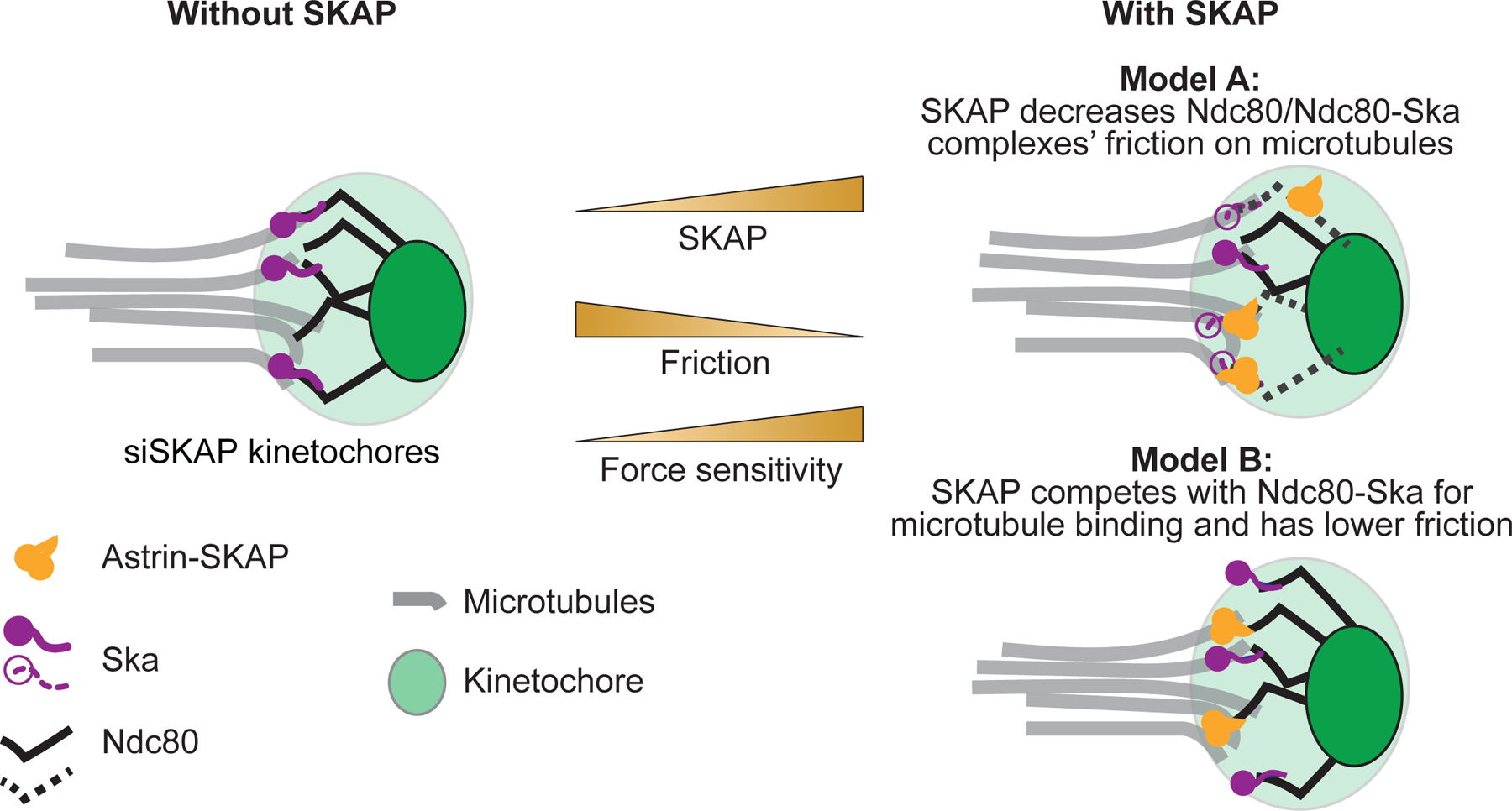
Representation of the kinetochore-microtubule interface in the absence (left) and presence (right) of SKAP. Two models for how Astrin-SKAP (yellow) could increase friction at the kinetochore-microtubule interface (right). In Model A (top right), SKAP affects how Ndc80 (black) or Ndc80-Ska (purple) complexes bind microtubules (dashed black and purple), decreasing their friction on microtubules and indirectly reducing attachment friction and increasing dynamics. In Model B (bottom right), SKAP directly binds microtubules, with Ndc80-SKAP and Ndc80-Ska competing for microtubule binding with similar affinities (binding energy) but with Ndc80-SKAP moving on microtubules with lower friction (lower transition state energy in moving between lattice binding sites). In both models, the more SKAP molecules are at the kinetochore, the lower the friction at the kinetochore-microtubule interface and the higher the sensitivity to force (center).
Across scales in biology, diverse interfaces need to be both strong and dynamic. For example, motor proteins must walk on their tracks without letting them go, and cell-cell junctions must robustly persist yet remodel. Understanding the mechanics of such interfaces will require not only understanding how strong, robust interactions are achieved, but how the interface can remain dynamic. The latter may require looking beyond mechanisms that grip, as is the case for SKAP which makes the human kinetochore-microtubule interface more dynamic.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Sophie Dumont (sophie.dumont@ucsf.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data reported in this paper has been deposited at Mendeley Data and is publicly available as of the date of publication (DOI: 10.17632/9gwpmj4hpr.1).
All original code has been deposited at github.com and is publicly available as of the date of publication (https://github.com/miquelrosassalvans/kinetochore-oscillations-analysis.git).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Rpe1-GFP cells (gift from A. Khodjakov, Wadsworth Center40) were cultured in DMEM/F12 (Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12, Thermo Fisher Scientific, 11320082) supplemented with 10% qualified and heat-inactivated fetal bovine serum (FBS) (10438–026, Gibco) and penicillin/streptomycin and maintained at 37°C and 5% CO2. Cells used in this study were not authenticated by the authors.
METHOD DETAILS
Cell culture, siRNA transfection and drug treatments in Rpe1 cells
Cells were plated in 35 mm glass-bottom dishes (poly-D-lysine coated; MatTek Corporation) for live imaging experiments or in six wells plates after addition on #1.5 25 mm coverslips (acid cleaned and poly-L-lysine coated) for immunofluorescence experiments. For knockdown experiments, siRNA targeting SKAP (siSKAP: 5′-AGGCUACAAACCACUGAGUAA-3′ 31; siSKAP2: 5′-GAAAGAGUCCGAUUCCUAG-3′ 30) or luciferase siRNA (5’ CGUACGCGGAAUACUUCGA 3’, control, WD01818022) were transfected in Rpe1-GFP cells and, using 4 µl of lipofectamine siRNAmax (Thermo Fisher Scientific, 13778075) and 10 µM of siRNA for 2 ml of cell culture media. Cells were incubated at 37 °C for 6–8 h before media wash, and imaged or processed 24 h after transfection. SiR-tubulin (Cytoskeleton, CY-SC002) was used at 1 µM concentration and added to the cells 40 min before imaging in experiments for Figure 2C. Nocodazole (Sigma-Aldrich, M1404–50MG) was used at 2 µM concentration and added 3 h before fixation in experiments for Figure 2 D-E. MG132 (carbobenzoxy-Leu-Leu-leucinal, EMB Millipore, 474790–5MG) was used at 10uM concentration in experiments for Figure S2.
Immunofluorescence and immunoblotting
To validate the SKAP siRNA (Figure S1A-B), cells were seeded in six well plates, transfected with luciferase (control) or SKAP siRNA 24 h after transfection and processed for immunoblotting. Cells were collected in PBS1X (Phoshpate Buffered Saline solution) using a cell scrapper and lysed in PBS1X + 1% NP40 on ice for 30 min. Samples were run in a 4–12% Bis-Tris gel (Invitrogen, NPO335BOX) and transferred to a nitrocellulose membrane (Thermo Scientific, 88018). The following primary and secondary antibodies and dyes where used (incubated in TBS1X (Tris-buffered saline), 3% milk, 0.1 % Tween for 1 h or 45 min, respectively): anti-SKAP (1ug/ml, rabbit, Origene, TA333584), anti-α-tubulin (DM1A, 1:1000, mouse, Sigma, T6199), goat anti-mouse IgG-HRP (1:1000, sc-2005, Santa Cruz Biotechnology) and mouse anti-rabbit IgG-HRP (1:1000, sc-2357, Santa Cruz Biotechnology). Blots were exposed with SuperSignal West Pico Substrate (Thermo Scientific) and imaged with a Bio-Rad ChemiDoc XRS+ system.
For immunofluorescence experiments cells were fixed in 99.8% methanol for 10 min at −20 °C, 24 h after siRNA transfection, and permeabilized in PBS1x, 0.5% BSA, 0.1% Triton (IF buffer thereafter) for 30 min. The following primary antibodies were incubated for 1 h in IF buffer: SKAP (1ug/ml, rabbit, gift from I. Cheeseman55), Kif18A (2ug/ml, rabbit, Bethyl, A301–080A-M), MCAK (1:500, rabbit, cytoskeleton, AKIN05), α-Tubulin DM1A (1:1000, mouse, Sigma, T6199), p-Hec1-s69 (1:3000, rabbit, gift from J. DeLuca25), CREST (1:100, human, Antibodies Incorporated, 15–234-0001), Hec1 (1:200, mouse, Abcam, ab3613), tubulin (1:2000, rat, Bio-rad, MCA77G). There washes in IF buffer (10 min each) were done before incubation with the following secondary antibodies (1:1000 in IF buffer, 45 min): goat anti-mouse IgG Alexa Fluor 405, 488 and 568 (A31553, A11001 and A11004, Invitrogen), goat anti-rabbit IgG Alexa Fluor 405 and 568 (A31556 and A11011, Invitrogen), goat anti-rat IgG Alexa Fluor 488 (A11006, Invitrogen) and goat anti-human IgG Alexa Fluor 568 (A21090, Invitrogen). Samples were washed once in IF buffer and twice in PBS1X (10 min each) before mounting in ProLong Gold Antifade reagent (P36934, Thermo Fisher).
Microscopy and laser ablation
Samples were imaged using an inverted microscope (Eclipse Ti-E; Nikon) with a spinning disk confocal (CSU-X1; Yokogawa Electric Corporation), head dichroic Semrock Di01-T405/488/568/647 for multicolor imaging, equipped with 405 nm (100 mW), 488 nm (120mW), 561 nm (150mW), and 642 nm (100mW) diode lasers, emission filters ET455/50M, ET525/50M, ET630/75M and ET690/50M for multicolor imaging, and an iXon3 camera (Andor Technology) operated by MetaMorph (7.7.8.0; Molecular Devices)46. Cells were imaged through a 100X 1.45 Ph3 oil objective and 1.5X lens.
For live imaging and laser ablation experiments, cells were maintained in a stage-top incubation chamber (Tokai Hit) at 37 °C and 5 % CO2. Metaphase oscillations were imaged every 3 s (Figures 1, 2B and S2). Laser ablation (30–40 pulses of 3 ns at 20 Hz) with 514 nm light was performed using the MicroPoint Laser System (Andor). For laser ablation experiments, images were acquired more slowly prior to ablation and then acquired more rapidly after ablation (3 s prior and 0.5 s after k-fiber ablation, and 6 s prior and 3 s after kinetochore ablation (Figures 3 and 4, respectively)). Successful k-fiber ablation was verified by immediate K-K distance relaxation (Figure 2G-I, 3 and S3) and posterior front kinetochore movement poleward by dynein pulling. Successful kinetochore ablation was verified by immediate poleward movement of the remaining sister kinetochore (Figure 4). For long term imaging experiments (Figure 2C), Rpe1-GFP cells treated with 100 nM SiR-DNA were imaged every 4 min for 18–20 h using a 20X objective.
Study design and data inclusion criteria
Two general criteria for inclusion of cells in metaphase oscillation and laser ablation experiments were applied. First, cells must express detectable levels of GFP-CenpA at kinetochores, but not so high as to completely label chromosome arms. Second, cells must be in metaphase, with a defined metaphase plate. For oscillation experiments (Figure 1–2B and S2), 2–4 kinetochore pairs per cell were analyzed, and the two kinetochores from the pair must stay in focus for a minimum of 90 s. For MG132 treatment experiments (Figure S2), cells with low centromere stiffness where selected if their mean K-K distance over time was higher than two standard deviations over the mean control K-K distance. For kinetochore speed calculations in ablation experiments (Figure 3E-4D), kinetochore tracks shorter than 5 timepoints were excluded due to the absence of a consistent directional movement.
We did not pre-estimate a required sample size before performing experiments nor did we blind or randomize samples during experimentation or analysis. The ablation experiments in this study are low throughput by nature, which does not enable us to report averages from multiple independent replicate experiments. Instead, we pool cells from across different independent experiments (with at least three independent experiments per condition per assay).
QUANTIFICATION AND STATISTICAL ANALYSIS
Analyzing fluorescence intensity
Images from immunofluorescence experiments were processed and fluorescence intensity was quantified using FIJI (Version 2.3.0/1.53f)57. For protein intensity measurements, a color threshold mask (Yen method) was applied using the CREST or Hec1 signal (for kinetochore selection) or DM1A signal (for spindle microtubule selection) to define the areas in which the fluorescence intensity would be measured for each protein of interest. Fluorescence intensity was measured for the proteins of interest (Hec1-S69, MCAK, Kif18A) and for the reference (CREST, Hec1 or Tubulin). Intensity of the protein of interest was normalized by the kinetochore or microtubule marker intensity by dividing the total intensity of the protein of interested by the intensity of the reference in the selected area. Figures 3H and S4C-D showing data from individual representative experiments, three independent experiments were performed for each quantification, obtaining comparable results.
Analyzing kinetochore behaviors
Kinetochores and centrioles were manually tracked from GFP-CenpA /centrin1-GFP videos using the MtrackJ plugin from FIJI58. GFP-centriole position was used as a marker for the spindle pole position. Kinetochore position was calculated as the distance from the spindle pole position. In metaphase oscillations experiments, all quantifications and statistical analyses were performed using home-written Phyton code (https://github.com/miquelrosassalvans/kinetochore-oscillations-analysis.git). Kinetochore speed at each timepoint was calculated as the difference in kinetochore position between two consecutive timepoints. Kinetochore speed was calculated by obtaining the slope of the best fitting regression line of individual kinetochore tracks (Figures 3E and 4D) or of entire tracks together (Figures 3C, D and 4C). Sister kinetochore movement coordination was obtained by calculating the correlation of sister kinetochore velocity over time or by the percentage of timepoints in which sister kinetochore movement direction was opposite. Kinetochore directional switch was determined by the consistent movement of a kinetochore in a the new direction for 3 consecutive timepoints. In ablation experiments, time after ablation was measured from the fist timepoint immediately after ablation. In all experiments, a kinetochore directional switch was defined by the consistent movement of the kinetochore in the opposite direction for a period of 3 consecutive timepoints (9 s in oscillations and kinetochore ablations and 1.5 s for k-fiber ablation experiments). K-K distance was calculated by subtracting the position of sister kinetochore A from sister kinetochore B, obtaining the length of the vector, and calculating the K-K distance average over time for each kinetochore pair. K-K distance relaxation was obtained by subtracting the K-K distance at the first timepoint after ablation from the last timepoint before ablation.
Statistical analysis
Statistical analysis was performed in Phyton or Graphpad (Prism 9). The Fisher’s exact test was used in Figure 1G, Student’s T test was used for parametric datasets (Figures 2B,I, 3E,H, 4E ad S4C-D), Mann-Whitney test for non-parametric datasets (Figures 1C-F, 2C,E, 3F, 4D and S2A,C-D) and analysis of covariance test, ANCOVA, for linear regression slopes comparison (Figures 3C,D-4C and S3).
Supplementary Material
Video S1. Metaphase kinetochore oscillations in control cell. Related to Figures 1–2. Representative kinetochore oscillations in a metaphase control Rpe1-GFP cell. Kinetochores (GFP-CenpA) and centrioles (centrin1-GFP) in grey. Time in min:s. The video was collected using a spinning disk confocal microscope at 1 frame every 3 s and adjusted to play at 75x real time. Video corresponds to cell shown in Figure 1B.
Video S2. Metaphase kinetochore oscillations in siSKAP cell. Related to Figures 1–2. Representative kinetochore oscillations in a metaphase siSKAP Rpe1-GFP cell. Kinetochores (GFP-CenpA) and centrioles (centrin1-GFP) in grey. Time in min:s. The video was collected using a spinning disk confocal microscope at 1 frame every 3s and adjusted to play at 75x real time. Video corresponds to cell shown in Figure 1B.
Video S3. K-Fiber ablation in control cell. Related to Figure 3. Laser ablation of a k-fiber in a metaphase control Rpe1-GFP cell. Kinetochores (GFP-CenpA) and Centrioles (centrin1-GFP) in grey, time in seconds. After k-fiber ablation (yellow arrow, time = 0) and immediate K-K distance relaxation, the front kinetochore (sister kinetochore on the ablation side) is pulled poleward by dynein activity at the new k-fiber minus-ends (white arrows point to the kinetochores of interest during movement). K-K distance increases as a result of this force, triggering microtubule polymerization at the back k-fiber and back kinetochore (away-from-pole) movement (following the front one). The whole cell is shown on the right and a magnified area containing the kinetochore pair of interest and the spindle pole is shown on the left. The video was collected using a spinning disk confocal microscope at 1 frame every 3 s before ablation and 0.5 s after ablation and adjusted to play at 8 frames/s. Video corresponds to cell shown in Figure 3B.
Video S4. K-fiber ablation in siSKAP cell. Related to figure 3. Laser ablation of a k-fiber in a metaphase siSKAP Rpe1-GFP cell. Kinetochores (GFP-CenpA) and centrioles (centrin1-GFP) in grey, time in seconds. After k-fiber ablation (yellow arrow, time = 0) and immediate K-K distance relaxation, the front kinetochore (sister kinetochore on the ablation side) is pulled poleward by dynein activity at the new k-fiber minus-ends (white arrows point to the kinetochores of interest during movement). K-K distance increases as a result of this force, triggering microtubule polymerization at the back k-Fiber and back kinetochore away-from-pole movement (following the front one). The whole cell is shown on the right and a magnified area containing the kinetochore pair of interest and the spindle pole is shown on the left. The video was collected using a spinning disk confocal microscope at 1 frame every 3 s before ablation and 0.5 s after ablation and adjusted to play at 8 frames/s. Video corresponds to cell shown in Figure 3B.
Video S5. Kinetochore ablation in control cell. Related to Figure 4. Laser ablation of a kinetochore in a metaphase control Rpe1-GFP cell. Kinetochores (GFP-CenpA) and Centrioles (centrin1-GFP) in grey, time in seconds. After individual kinetochore ablation (yellow arrow, time = 0), the sister kinetochore moves poleward, driven by microtubule depolymerization at the associated k-fiber (white arrow points to the kinetochore of interest). Kinetochore movement switches to away-from-pole direction (purple arrow) near the spindle pole due to polar ejection forces that trigger microtubule rescue. The whole cell is shown on the right and a magnified area containing the kinetochore of interest and the spindle pole is shown on the left. The video was collected using a spinning disk confocal microscope at 1 frame every 6 s before ablation and 3 s after ablation and adjusted to play at 10 frames/s. Video corresponds to cell shown in Figure 4B.
Video S6. Kinetochore ablation in siSKAP cell. Related to Figure 4. Laser ablation of a kinetochore in a metaphase siSKAP Rpe1-GFP cell. Kinetochores (GFP-CenpA) and Centrioles (centrin1-GFP) in grey, time in seconds. After individual kinetochore ablation (yellow arrow, time = 0), the sister kinetochore moves poleward, driven by microtubule depolymerization at the associated k-fiber (white arrow points to the kinetochore of interest). Kinetochore movement switches to away-from-pole direction (purple arrow) near the spindle pole, due to polar ejection forces that trigger microtubule rescue. The whole cell is shown on the right and a magnified area containing the kinetochore of interest and the spindle pole is shown on the left. The video was collected using a spinning disk confocal microscope at 1 frame every 6 s before ablation and 3 s after ablation and adjusted to play at 10 frames/s. Video corresponds to cell shown in Figure 4B.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-SKAP | Origene | TA333584 |
| Rabbit anti-SKAP | I. Cheeseman lab | 55 |
| Mouse anti-α-tubulin (DM1A) | Sigma-Aldrich | T6199 |
| Rabbit anti-Kif18A | Bethyl | A301–080A-M |
| Rabbit anti-MCAK | cytoskeleton | AKIN05 |
| Rabbit anti-p-Hec1-S69 | J. DeLuca lab | 25 |
| Human anti-CREST | Antibodies Incorporated | 15–234-0001 |
| Mouse anti-Hec1 | Abcam | ab3613 |
| Rat anti-α-tubulin | Bio-rad | MCA77G |
| Goat anti-mouse IgG Alexa Fluor 405, 488 and 568 | Invitrogen | A31553, A11001 and A11004 |
| Goat anti-rabbit IgG Alexa Fluor 405 and 568 | Invitrogen | A31556 and A11011 |
| Goat anti-rat IgG Alexa Fluor 488 | Invitrogen | A11006 |
| Goat anti-human IgG Alexa Fluor 568 | Invitrogen | A21090 |
| Chemicals, peptides, and recombinant proteins | ||
| Nocodazole | Sigma-Aldrich | M1404–50MG |
| MG132 | EMB Millipore | 474790–5MG |
| SiR-tubulin | Cytoskeleton | CY-SC002 |
| Experimental models: Cell lines | ||
| Rpe1-GFP cells | A. Khodjakov lab | 40 |
| Oligonucleotides | ||
| siRNA SKAP: 5′-AGGCUACAAACCACUGAGUAA-3′ | Sigma-Aldrich | 31 |
| siRNA SKAP2: 5′-GAAAGAGUCCGAUUCCUAG-3′ | Sigma-Aldrich | 30 |
| siRNA luciferase: 5’ CGUACGCGGAAUACUUCGA 3’ | Sigma-Aldrich | WD01818022 |
| Software and algorithms | ||
| FIJI (Version 2.3.0/1.53f) | ImageJ | 57 |
| MtrackJ | FIJI | 58 |
| Pyton code for kinetochore oscillations analysis | This paper | https://github.com/miquelrosassalvans/kinetochore-oscillations-analysis.git |
| Other | ||
| lipofectamine siRNAmax | Thermo Fisher Scientific | 13778075 |
| ProLong Gold Antifade reagent | Thermo Fisher Scientific | P36934 |
Highlights.
SKAP increases kinetochore motility during mammalian metaphase
SKAP decreases friction at the kinetochore-microtubule interface, lubricating it
SKAP increases kinetochore-microtubule interface’s dynamics and force responsiveness
Reducing interface friction independently of grip may be key to robust attachments
ACKNOWLEDGMENTS
We thank Iain Cheeseman for anti-SKAP antibody and discussion, Jennifer DeLuca for Hec1-pS69 antibody, Alexey Khodjakov for Rpe1-GFP (GFP-CenpA and centrin1-GFP) cells and Charles Asbury and Rob Phillips for discussions. We thank the members of the Dumont Lab for discussions and critical reading of the manuscript. This work was supported by NIH DP2GM119177, NIH R01GM134132, NIH R35GM136420, NSF CAREER 1554139, NSF 1548297 Center for Cellular Construction, the Rita Allen Foundation and the Chan Zuckerberg Biohub (S.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTEREST
The authors declare no conflict of interest.
Video preparation
Videos were prepared using FIJI. Brightness and contrast were linearly adjusted to clearly visualize the kinetochores and centrioles.
REFERENCES
- 1.Wendell KL, Wilson L, and Jordan MA (1993). Mitotic block in HeLa cells by vinblastine: Ultrastructural changes in kinetochore-mcirotubule attachment and in centrosomes. J. Cell Sci 104, 261–274. [DOI] [PubMed] [Google Scholar]
- 2.McEwen BF, Heagle AB, Cassels GO, Buttle KF, and Rieder CL (1997). Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J. Cell Biol 137, 1567–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khodjakov A, and Rieder CL (1996). Kinetochores moving away from their associated pole do not exert a significant pushing force on the chromosome. J. Cell Biol 135, 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grishchuk EL, and McIntosh JR (2006). Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J 25, 4888–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joglekar AP, Bloom KS, and Salmon ED (2010). Mechanisms of force generation by end-on kinetochore-microtubule attachments. Curr. Opin. Cell Biol 22, 57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumont S, Salmon ED, and Mitchison TJ (2012). Deformations within moving kinetochores reveal different sites of active and passive force generation. Science (80-. ) 337, 355–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan X, Cimini D, Cameron LA, and Salmon ED (2012). The coupling between sister kinetochore directional instability and oscillations in centromere stretch in metaphase PtK1 cells. Mol. Biol. Cell 23, 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rago F, and Cheeseman IM (2013). The functions and consequences of force at kinetochores. J. Cell Biol 200, 557–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNeill PA, and Berns MW (1981). Chromosome behavior after laser microirradiation of a single kinetochore in mitotic PtK2 cells. J. Cell Biol 88, 543–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skibbens RV, Rieder CL, and Salmon ED (1995). Kinetochore motility after severing between sister centromeres using laser microsurgery: evidence that kinetochore directional instability and position is regulated by tension. J. Cell Sci 108, 2537–2548. [DOI] [PubMed] [Google Scholar]
- 11.Rieder CL, and Salmon ED (1994). Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J. Cell Biol 124, 223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ke K, Cheng J, and Hunt AJ (2009). The Distribution of Polar Ejection Forces Determines the Amplitude of Chromosome Directional Instability. Curr. Biol 19, 807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bakhoum SF, and Compton DA (2012). Kinetochores and disease: Keeping microtubule dynamics in check! Curr. Opin. Cell Biol 24, 64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLuca JG, Dong Y, Hergert P, Strauss J, Hickey JM, Salmon ED, and McEwen BF (2004). Hec1 and Nuf2 Are Core Components of the Kinetochore Outer Plate Essential for Organizing Microtubule Attachment Sites. Mol. Biol. Cell 16, 519–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheeseman IM, and Desai A (2008). Molecular architecture of the kinetochore–microtubule interface. Nat. Rev. Mol. Cell Biol 9, 33–46. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt JC, Arthanari H, Boeszoermenyi A, Dashkevich NM, Wilson-Kubalek EM, Monnier N, Markus M, Oberer M, Milligan RA, Bathe M, et al. (2012). The Kinetochore-Bound Ska1 Complex Tracks Depolymerizing Microtubules and Binds to Curved Protofilaments. Dev. Cell 23, 968–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaytsev AV, Sundin LJR, DeLuca KF, Grishchuk EL, and DeLuca JG (2014). Accurate phosphoregulation of kinetochore-microtubule affinity requires unconstrained molecular interactions. J. Cell Biol 206, 45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long AF, Udy DB, and Dumont S (2017). Hec1 Tail Phosphorylation Differentially Regulates Mammalian Kinetochore Coupling to Polymerizing and Depolymerizing Microtubules. Curr. Biol 27, 1692–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auckland P, Clarke NI, Royle SJ, and McAinsh AD (2017). Congressing kinetochores progressively load Ska complexes to prevent force-dependent detachment. J. Cell Biol 216, 1623–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huis In ‘T Veld PJ, Volkov VA, Stender ID, Musacchio A, Dogterom M, Huis PJ, Volkov VA, Stender ID, Musacchio A, and Dogterom M (2019). Molecular determinants of the Ska-Ndc80 interaction and their influence on microtubule tracking and force-coupling. Elife 8, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long AF, Kuhn J, and Dumont S (2019). The mammalian kinetochore–microtubule interface: robust mechanics and computation with many microtubules. Curr. Opin. Cell Biol 60, 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, and Desai A (2006). The Conserved KMN Network Constitutes the Core Microtubule-Binding Site of the Kinetochore. Cell 127, 983–997. [DOI] [PubMed] [Google Scholar]
- 23.Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, and Asbury CL (2009). The Ndc80 Kinetochore Complex Forms Load-Bearing Attachments to Dynamic Microtubule Tips via Biased Diffusion. Cell 136, 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaytsev AV, Mick JE, Maslennikov E, Nikashin B, DeLuca JG, and Grishchuk EL (2015). Multisite phosphorylation of the NDC80 complex gradually tunes its microtubule-binding affinity. Mol. Biol. Cell 26, 1829–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeLuca KF, Meppelink A, Broad AJ, Mick JE, Peersen OB, Pektas S, Lens SMA, and DeLuca JG (2018). Aurora A kinase phosphorylates Hec1 to regulate metaphase kinetochore-microtubule dynamics. J. Cell Biol 217, 163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanisch A, Silljé HH, and Nigg EA (2006). Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J 25, 5504–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welburn JPI, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR, and Cheeseman IM (2009). The Human Kinetochore Ska1 Complex Facilitates Microtubule Depolymerization-Coupled Motility. Dev. Cell 16, 374–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helgeson LA, Zelter A, Riffle M, Maccoss MJ, Asbury CL, and Davis TN (2018). Human Ska complex and Ndc80 complex interact to form a load-bearing assembly that strengthens kinetochore-microtubule attachments 115, 2740–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang L, Seki A, and Fang G (2009). SKAP associates with kinetochores and promotes the metaphase-to-anaphase transition. Cell Cycle 8, 2819–2827. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt JC, Kiyomitsu T, Hori T, Backer CB, Fukagawa T, and Cheeseman IM (2010). Aurora B kinase controls the targeting of the Astrin-SKAP complex to bioriented kinetochores. J. Cell Biol 191, 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunsch AK, Linnane E, Barr FA, and Gruneberg U (2011). The astrin-kinastrin/SKAP complex localizes to microtubule plus ends and facilitates chromosome alignment. J. Cell Biol 192, 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cimini D, Wan X, Hirel CB, and Salmon ED (2006). Aurora Kinase Promotes Turnover of Kinetochore Microtubules to Reduce Chromosome Segregation Errors. Curr. Biol 16, 1711–1718. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Zhuang X, Cao D, Chu Y, Yao P, Liu W, Liu L, Adams G, Fang G, Dou Z, et al. (2012). Mitotic Regulator SKAP Forms a Link between Kinetochore Core Complex KMN and Dynamic Spindle Microtubules *. J. Biol. Chem 287, 39380–39390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friese A, Faesen AC, Huis In ‘t Veld PJ, Fischböck J, Prumbaum D, Petrovic A, Raunser S, Herzog F, and Musacchio A (2016). Molecular requirements for the inter-subunit interaction and kinetochore recruitment of SKAP and Astrin. Nat. Commun [DOI] [PMC free article] [PubMed]
- 35.Kern DM, Monda JK, Su K-C, Wilson-Kubalek EM, and Cheeseman IM (2017). Astrin-SKAP complex reconstitution reveals its kinetochore interaction with microtubule-bound Ndc80. Elife 6:e26866, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao H, Pan Q, Wang Y, and Chen Q (2021). Impact of KMN network genes on progression and prognosis of non-small cell lung cancer. Anticancer. Drugs Publish Ah, 1–11. [DOI] [PubMed]
- 37.Yu DC, Chen XY, Li X, Zhou HY, Yu DQ, Yu XL, Hu YC, Zhang RH, Zhang XB, Zhang K, et al. (2021). Transcript levels of spindle and kinetochore-associated complex 1/3 as prognostic biomarkers correlated with immune infiltrates in hepatocellular carcinoma. Sci. Rep 11, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siprashvili Z, Bhaduri A, Ungewickell A, Lee CS, Rios EJ, Straight A, Khavari PA, Aasi SZ, Kim J, Mah A, et al. (2014). Recurrent point mutations in the kinetochore gene KNSTRN in cutaneous squamous cell carcinoma. Nat. Genet 46, 1060–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng P, Zhou R, Zhang J, and Cao L (2021). Increased Expression of KNSTRN in Lung Adenocarcinoma Predicts Poor Prognosis: A Bioinformatics Analysis Based on TCGA Data. J. Cancer 12, 3239–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paul R, Mogilner A, Magidson V, Connell CBO, Lon J, O’Connell CB, Lončarek J, Paul R, Mogilner A, and Khodjakov A (2011). The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell 146, 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, and Salmon ED (2006). Kinetochore Microtubule Dynamics and Attachment Stability Are Regulated by Hec1. Cell 127, 969–982. [DOI] [PubMed] [Google Scholar]
- 42.Raaijmakers JA, Tanenbaum ME, Maia AF, and Medema RH (2009). RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J. Cell Sci 122, 2436–2445. [DOI] [PubMed] [Google Scholar]
- 43.Huang Y, Wang W, Yao P, Wang X, Liu X, Zhuang X, Yan F, Zhou J, Du J, Ward T, et al. (2011). CENP-E Kinesin Interacts with SKAP Protein to Orchestrate Accurate Chromosome Segregation in Mitosis *. J. Biol. Chem 287, 1500–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daum JR, Potapova TA, Sivakumar S, Daniel JJ, Flynn JN, Rankin S, and Gorbsky GJ (2011). Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr. Biol 21, 1018–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sikirzhytski V, Magidson V, Steinman JB, He J, Le Berre M, Tikhonenko I, Ault JG, McEwen BF, Chen JK, Sui H, et al. (2014). Direct kinetochore-spindle pole connections are not required for chromosome segregation. J. Cell Biol 206, 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elting MW, Hueschen CL, Udy DB, and Dumont S (2014). Force on spindle microtubule minus ends moves chromosomes. J. Cell Biol 206, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wordeman L, Wagenbach M, and Dassow G. Von (2007). MCAK facilitates chromosome movement by promoting kinetochore microtubule turnover 179, 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stumpff J, Dassow G. Von, Wagenbach M, Asbury C, and Wordeman L (2008). The Kinesin-8 Motor Kif18A Suppresses Kinetochore Movements to Control Mitotic Chromosome Alignment 252–262. [DOI] [PMC free article] [PubMed]
- 49.Jaju PD, Nguyen CB, Mah AM, Atwood SX, Li J, Zia A, Chang ALS, Oro AE, Tang JY, Lee CS, et al. (2015). Mutations in the kinetochore gene KNSTRN in basal cell carcinoma. J. Invest. Dermatol 135, 3197–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bakhoum SF, Thompson SL, Manning AL, and Compton D a (2009). Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat. Cell Biol 11, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stumpff J, Wagenbach M, Franck A, Asbury CL, and Wordeman L (2012). Kif18A and Chromokinesins Confine Centromere Movements via Microtubule Growth Suppression and Spatial Control of Kinetochore Tension. Dev. Cell 22, 1017–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waters JC, Chen RH, Murray AW, and Salmon ED (1998). Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol 141, 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okouneva T, Azarenko O, Wilson L, Littlefield BA, and Jordan MA (2008). Inhibition of centromere dynamics by eribulin (E7389) during mitotic metaphase. Mol. Cancer Ther 7, 2003–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tamura Naoka, Simon JE, Nayak A, Shenoy R, Hiroi N, Boilot V, Funahashi A, and Draviam VM (2015). A proteomic study of mitotic phase-specific interactors of EB1 reveals a role for SXIP-mediated protein interactions in anaphase onset. Biol. Open 4, 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kern DM, Nicholls PK, Page DC, and Cheeseman IM (2016). A mitotic SKAP isoform regulates spindle positioning at astral microtubule plus ends. J. Cell Biol 213, 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forth S, Hsia KC, Shimamoto Y, and Kapoor TM (2014). Asymmetric friction of nonmotor MAPs can lead to their directional motion in active microtubule networks. Cell 157, 420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meijering E, Dzyubachyk O, and Smal I (2012). Methods for cell and particle tracking 1st ed. (Elsevier Inc.). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Metaphase kinetochore oscillations in control cell. Related to Figures 1–2. Representative kinetochore oscillations in a metaphase control Rpe1-GFP cell. Kinetochores (GFP-CenpA) and centrioles (centrin1-GFP) in grey. Time in min:s. The video was collected using a spinning disk confocal microscope at 1 frame every 3 s and adjusted to play at 75x real time. Video corresponds to cell shown in Figure 1B.
Video S2. Metaphase kinetochore oscillations in siSKAP cell. Related to Figures 1–2. Representative kinetochore oscillations in a metaphase siSKAP Rpe1-GFP cell. Kinetochores (GFP-CenpA) and centrioles (centrin1-GFP) in grey. Time in min:s. The video was collected using a spinning disk confocal microscope at 1 frame every 3s and adjusted to play at 75x real time. Video corresponds to cell shown in Figure 1B.
Video S3. K-Fiber ablation in control cell. Related to Figure 3. Laser ablation of a k-fiber in a metaphase control Rpe1-GFP cell. Kinetochores (GFP-CenpA) and Centrioles (centrin1-GFP) in grey, time in seconds. After k-fiber ablation (yellow arrow, time = 0) and immediate K-K distance relaxation, the front kinetochore (sister kinetochore on the ablation side) is pulled poleward by dynein activity at the new k-fiber minus-ends (white arrows point to the kinetochores of interest during movement). K-K distance increases as a result of this force, triggering microtubule polymerization at the back k-fiber and back kinetochore (away-from-pole) movement (following the front one). The whole cell is shown on the right and a magnified area containing the kinetochore pair of interest and the spindle pole is shown on the left. The video was collected using a spinning disk confocal microscope at 1 frame every 3 s before ablation and 0.5 s after ablation and adjusted to play at 8 frames/s. Video corresponds to cell shown in Figure 3B.
Video S4. K-fiber ablation in siSKAP cell. Related to figure 3. Laser ablation of a k-fiber in a metaphase siSKAP Rpe1-GFP cell. Kinetochores (GFP-CenpA) and centrioles (centrin1-GFP) in grey, time in seconds. After k-fiber ablation (yellow arrow, time = 0) and immediate K-K distance relaxation, the front kinetochore (sister kinetochore on the ablation side) is pulled poleward by dynein activity at the new k-fiber minus-ends (white arrows point to the kinetochores of interest during movement). K-K distance increases as a result of this force, triggering microtubule polymerization at the back k-Fiber and back kinetochore away-from-pole movement (following the front one). The whole cell is shown on the right and a magnified area containing the kinetochore pair of interest and the spindle pole is shown on the left. The video was collected using a spinning disk confocal microscope at 1 frame every 3 s before ablation and 0.5 s after ablation and adjusted to play at 8 frames/s. Video corresponds to cell shown in Figure 3B.
Video S5. Kinetochore ablation in control cell. Related to Figure 4. Laser ablation of a kinetochore in a metaphase control Rpe1-GFP cell. Kinetochores (GFP-CenpA) and Centrioles (centrin1-GFP) in grey, time in seconds. After individual kinetochore ablation (yellow arrow, time = 0), the sister kinetochore moves poleward, driven by microtubule depolymerization at the associated k-fiber (white arrow points to the kinetochore of interest). Kinetochore movement switches to away-from-pole direction (purple arrow) near the spindle pole due to polar ejection forces that trigger microtubule rescue. The whole cell is shown on the right and a magnified area containing the kinetochore of interest and the spindle pole is shown on the left. The video was collected using a spinning disk confocal microscope at 1 frame every 6 s before ablation and 3 s after ablation and adjusted to play at 10 frames/s. Video corresponds to cell shown in Figure 4B.
Video S6. Kinetochore ablation in siSKAP cell. Related to Figure 4. Laser ablation of a kinetochore in a metaphase siSKAP Rpe1-GFP cell. Kinetochores (GFP-CenpA) and Centrioles (centrin1-GFP) in grey, time in seconds. After individual kinetochore ablation (yellow arrow, time = 0), the sister kinetochore moves poleward, driven by microtubule depolymerization at the associated k-fiber (white arrow points to the kinetochore of interest). Kinetochore movement switches to away-from-pole direction (purple arrow) near the spindle pole, due to polar ejection forces that trigger microtubule rescue. The whole cell is shown on the right and a magnified area containing the kinetochore of interest and the spindle pole is shown on the left. The video was collected using a spinning disk confocal microscope at 1 frame every 6 s before ablation and 3 s after ablation and adjusted to play at 10 frames/s. Video corresponds to cell shown in Figure 4B.
Data Availability Statement
All data reported in this paper has been deposited at Mendeley Data and is publicly available as of the date of publication (DOI: 10.17632/9gwpmj4hpr.1).
All original code has been deposited at github.com and is publicly available as of the date of publication (https://github.com/miquelrosassalvans/kinetochore-oscillations-analysis.git).


