Abstract
Background and Aims
In eukaryotes, the total kinetochore size (defined as a chromosomal region containing CENH3-positive nucleosomes) per nucleus strongly correlates with genome size, a relationship that has been hypothesized to stem from general intracellular scaling principles. However, if larger chromosomes within a karyotype required larger kinetochores to move properly, it could also be derived from the mechanics of cell division.
Methods
We selected seven species of the plant subfamily Agavoideae whose karyotypes are characterized by the presence of small and very large chromosomes. We visualized the kinetochore regions and chromosomes by immunolabelling with an anti-CENH3 antibody and DAPI (6′-diamidino-2-phenylindole) staining. We then employed 2D widefield and 3D super-resolution microscopy to measure chromosome and kinetochore areas and volumes, respectively. To assess the scaling relationship of kinetochore size to chromosome size inside a karyotype, we log-transformed the data and analysed them with linear mixed models which allowed us to control for the inherent hierarchical structure of the dataset (metaphases within slides and species).
Key Results
We found a positive intra-karyotype relationship between kinetochore and chromosome size. The slope of the regression line of the observed relationship (0.277 for areas, 0.247 for volumes) was very close to the theoretical slope of 0.25 for chromosome width based on the expected physics of chromosome passage through the cytoplasm during cell division. We obtained similar results by reanalysing available data from human and maize.
Conclusions
Our findings suggest that the total kinetochore size to genome size scaling observed across eukaryotes may also originate from the mechanics of cell division. Moreover, the potential causal link between kinetochore and chromosome size indicates that evolutionary mechanisms capable of leading kinetochore size changes to fixation, such as centromere drive, could promote the size evolution of entire chromosomes and genomes.
Keywords: Asparagaceae, cell division, centromere, chromosome size evolution, genome size evolution, intracellular scaling, linear mixed models, structured illumination microscopy
Introduction
The centromere is a critical chromosomal region for eukaryotic cell divisions because it is the assembly site of the kinetochore, a protein complex mediating chromosome attachment to spindle microtubules (Talbert and Henikoff, 2020). The cornerstone of the kinetochore in most eukaryotes is centromeric histone H3 (CENH3 or CENP-A), which recruits other kinetochore proteins (Murillo-Pineda and Jansen, 2020).
It has been shown in grasses (Poaceae) that the total kinetochore size, defined as the sum of the CENH3-binding domain sizes within a genome, is strongly correlated with genome size (Zhang and Dawe, 2012). Recently, we have shown that this strong relationship is universal across eukaryotes (Plačková et al., 2021). It has been suggested that a mechanism maintaining the stable proportion of total centromere size to the genome size stems from general intracellular scaling principles (Wang et al., 2021) that maintain the size ratio of intracellular components to ensure their proper function (Levy and Heald, 2012). However, it could also come from the mechanics of cell division, if larger chromosomes inside a karyotype possessed larger kinetochores. In such a case, a positive intra-karyotype scaling relationship between the kinetochore and chromosome sizes would be expected.
One can imagine that a larger chromosome as a larger cargo simply needs to be pulled by more microtubules, which requires a larger kinetochore. The original calculations of the cytoplasm viscosity based on Brownian motion suggested that the force needed to move a chromosome was approx. 0.1 picoNewton (pN; Nicklas, 1965; Taylor, 1965). This was puzzling as the pulling power that the anaphase spindle could exert was approx. 700 pN (Nicklas, 1983; Anjur-Dietrich et al., 2021). However, it appears that the microtubule network of the spindle itself exerts a force on the chromosomes that is orders of magnitude higher than previously thought (Houchmandzadeh et al., 1997; Shimamoto et al., 2011; Kramer et al., 2021). As the number of attached microtubules depends on kinetochore size (McEwen et al., 1998; Zhang and Dawe, 2012; Drpic et al., 2018), some authors speculated that a minimal number of microtubules reflecting chromosome size is required for proper chromosomal segregation (Moens, 1979; Cherry et al., 1989; McEwen et al., 1998). Some evidence supporting this notion was found in unstable barley hybrids (Hordeum vulgare × H. bulbosum) (Schwarzacher et al., 1992; Heslop-Harrison and Schwarzacher, 1993). In these unstable hybrids, chromosomes with smaller centromeres were lagging during embryogenesis and underwent elimination (Schwarzacher et al., 1992). Furthermore, microtubule staining revealed that the centromeres of lagging chromosomes interacted with fewer microtubules (Heslop-Harrison and Schwarzacher, 1993). Subsequent studies on haploid plant formation showed that the failure to load enough centromeric CENH3 leads to the formation of no or small kinetochores, and such chromosomes are lost (reviewed in Wang and Dawe, 2018). Also, monocentric chromosomes with kinetochores which are too large are susceptible to missegregation (Drpic et al., 2018). Reports of moderate correlations between kinetochore size and chromosome size in humans (Irvine et al., 2004) and maize (Wang et al., 2021) indeed suggest possible restrictions on the range of kinetochore size concerning chromosome size. However, the intra-karyotype relationships between kinetochore and chromosome size and their scaling laws remain poorly understood.
A model system suitable for exploring the within-karyotype correlation between kinetochore and chromosome size should exhibit large differences in chromosome size. Therefore, in the present study, we focused on species from the plant subfamily Agavoideae (Asparagales) containing so-called bimodal karyotypes characterized by small as well as very large chromosomes displaying 5- to 8-fold (e.g. Agave), and even up to 10-fold (e.g. Hosta and Yucca) size differences (Watkins, 1936; Kaneko, 1966; Guadalupe et al., 2008). We exploited the large intra-karyotype differences in chromosome size in Agavoideae and analysed the kinetochores of seven species from this subfamily to explore the intra-karyotype relationship between kinetochore and chromosome size.
MATERIALS AND METHODS
Plant materials
Seeds of agavoid species used in this study – Hesperaloe funifera (K.Koch) Trel., H. parviflora (Torr.) J.M.Coult., Hesperoyucca whipplei (Torr.) Trel., Yucca carnerosana (Trel.) McKelvey, Y. constricta Buckley and Y. elata (Engelm.) Engelm. – were germinated on Petri dishes with water and gibberellic acid (1 g L–1). Some seedlings were transplanted into pots. These plants and adult plants of Agave tequilana F. A. C. Weber were grown under greenhouse conditions in the Department of Botany and Zoology, Masaryk University, Czech Republic.
Generation of agavoid CENH3-specific antibodies
To identify putative CENH3 sequences of Agavoideae, we performed BLAST searches in the GenBank (NCBI Resource Coordinators, 2018) and 1000 plant (1KP) transcriptomes (Carpenter et al., 2019; One Thousand Plant Transcriptomes Initiative, 2019) databases. Purified polyclonal rabbit IgG antibodies recognizing a peptide corresponding to the N-terminus of agavoid CENH3 (MARVKHKPQPQPRRRLVLNEA; amino acids 1–21) were generated by LifeTein (USA).
Chromosome preparation and immunostaining
For chromosome preparation and immunostaining, we followed a previously published protocol (Houben et al., 2007). Root tips from germinated seeds or plants were either untreated or were pre-treated for up to 26 h in ice-cold distilled water and fixed in an ice-cold 4% paraformaldehyde solution (PFA) dissolved in 1× phosphate-buffered saline (1×PBS; pH 7.4) for 25 min. After washing in 1×PBS, root tips were digested in 0.7% (w/v) cellulase R-10 (Duchefa Biochemie), cellulysin cellulase (Merck Millipore), 1% (w/v) pectolyase Y23 (Duchefa Biochemie) and cytohelicase (Sigma-Aldrich) in 1× PBS at 37°C for 1 h. Chromosome spreads were prepared by squashing and freezing in liquid nitrogen. Slides were incubated with blocking solution [4% (w/v) bovine serum albumin (BSA) and 0.01% (v/v) Tween-20 in 1×PBS] at room temperature for 1 h and then incubated with primary CENH3 antibodies used at a dilution of 1:200 to 1:500 at 4°C overnight. After washing in 1×PBS, samples were incubated with Alexa Fluor 488-conjugated secondary anti-rabbit antibodies (Jackson Immuno; diluted 1:500) at 37°C for 1 h. Finally, slides were washed in 1×PBS, dehydrated using a graded ethanol series (70, 85 and 100%), and chromosomes were counterstained with 6′-diamidino-2-phenylindole (DAPI; 1.5 µg mL–1) in Vectashield (Vector Laboratories).
Microscopy and measurement
Chromosomes prepared by the above-described protocol were used for both 2D and 3D measurements (see below). First, we obtained 2D images and then 3D-SIM image stacks from the same slides.
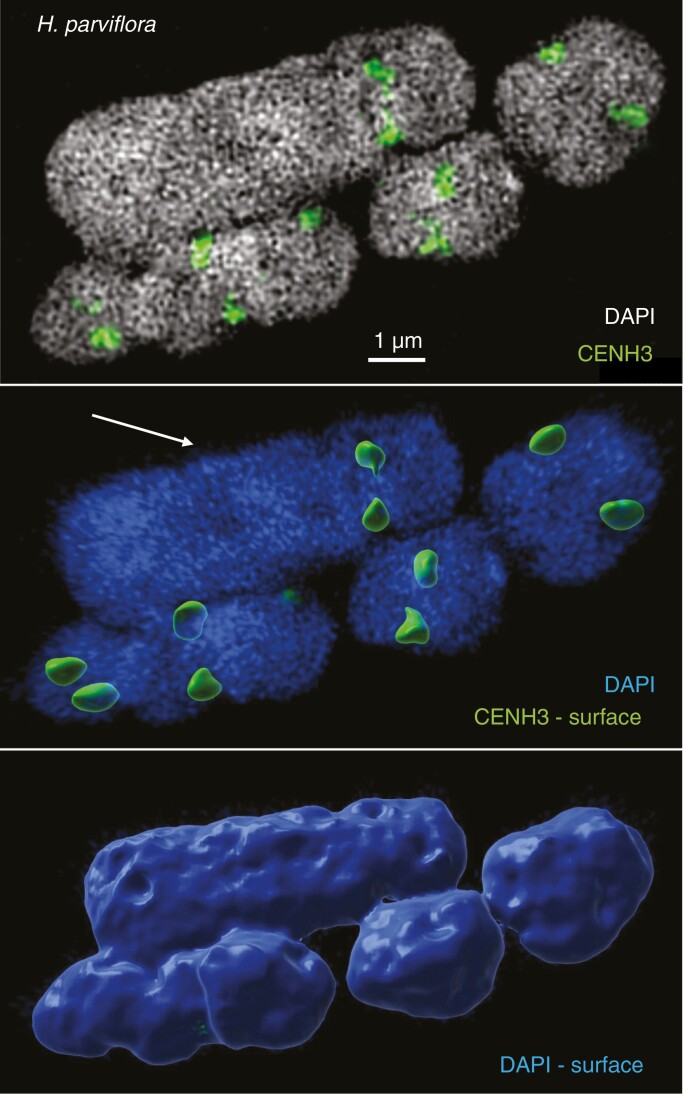
Images were captured using Olympus BX51 and BX61 microscopes equipped with a CCD camera (ORCA-ER, Hamamatsu) and a ColorView II camera (Olympus), respectively, and the Cell^F software. Areas of chromosomes and their CENH3-labelled kinetochores were measured in wide-field photographs using the ImageJ software (Supplementary data Fig. S1). To identify the boundaries of CENH3 labelling, we enlarged the CENH3-labelled photographs to 400% and applied the ‘Elliptical selections’ function. To identify boundaries of DAPI-stained chromosomes, the photographs were enlarged to 300% and the boundaries were set using the ‘Freehand selections’ option (Supplementary data Fig. S1). To measure the volumes of metaphase cells at the super-resolution level, spatial structured illumination microscopy (3D-SIM) using an Elyra PS.1 microscope system equipped with a 63×/1.40 objective and the software ZENBlack (Carl Zeiss GmbH) was applied (Weisshart et al., 2016; Kubalová et al., 2021a). The Imaris 9.7 (Bitplane) software was used to measure chromosome and CENH3 signal volumes after surface rendering of 3D-SIM image stacks (Fig. 1; Supplementary data Movie S1).
Fig. 1.
One large (arrow) and four small chromosomes selected from the bimodal Hesperaloe parviflora karyotype. Slices of 3D-SIM image stacks (top) were used for surface rendering to measure the volumes of the chromosomes (bottom) and CENH3-labelled centromere signals (middle). See also Supplementary data Movie S1.
Altogether, we performed 1447 chromosome and kinetochore size measurements in seven Agavoideae species. Of these, 1229 were area measurements based on 2D fluorescence microscopy and 218 were volume quantifications based on 3D-SIM (Supplementary data Tables S1 and S2). For five species, we had both volumes and area measurements. For Yucca elata, we had only area measurements. For Y. constricta, we had only volume measurements.
In each metaphase, we measured only those chromosomes and their kinetochores that were clearly separated from other chromosomes, which allowed a reliable measurement (see Supplementary data Fig. S1). Because each metaphase chromosome had two kinetochores (one for each chromatid), we measured both and used their mean size in all the analyses. To avoid bias in the data caused, for instance, by the chromatin condensation differences between metaphases, we applied a hierarchical statistical approach that allows controlling for such noise in the data (see below).
Statistical analyses
To test the relationship between kinetochore and genome size, we applied a linear mixed model using the ‘lme’ function implemented in the package nlme (Pinheiro et al., 2021) in R v4.0.2 (R Core Team, 2020). We set the kinetochore size as the response variable and chromosome size as the explanatory variable. To account for random variation caused by differences between different metaphases, slides, plants and species, we used these variables as nested random effects as follows: metaphases nested within slides nested within plants nested within species. Because we are interested in the proportional scaling relationship between kinetochore and chromosome size, we log-transformed (base 10) both the kinetochore size and chromosome size before the regression analyses (Glazier, 2013). Log transformation (base 10) also increases the homoscedasticity and normality of the residuals, which are the assumptions of linear regression models used for the analyses.
RESULTS
We measured chromosome size (from DAPI-stained areas and volumes) and kinetochore size using the immunolabelled CENH3 region as a proxy (see the Materials and Methods). We choose CENH3 because this histone variant is the essential kinetochore protein upon which the kinetochore structure is built in most eukaryotes (Wang and Dawe, 2018; Murillo-Pineda and Jansen, 2020), and thus the amount of CENH3 reflects the kinetochore size.
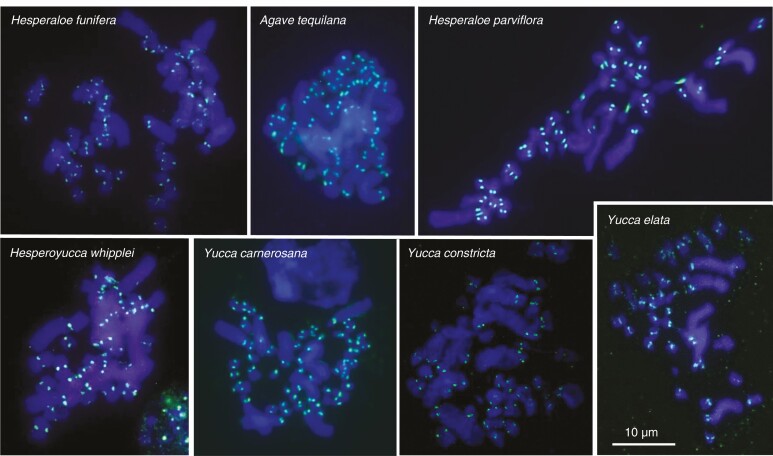
Using BLAST searches, we identified two isoforms of CENH3 in Agavoideae (Supplementary data Fig. S2). Corresponding CENH3 antibodies were raised against the N-terminal region (see the Materials and Methods), which is conserved across both isoforms (Supplementary data Fig. S2). After indirect immunostaining, these antibodies labelled the centromeric regions of all analysed Agavoideae species (Figs. 1 and 2).
Fig. 2.
CENH3 antibodies label (in green) the centromeres of seven Agavoideae species.
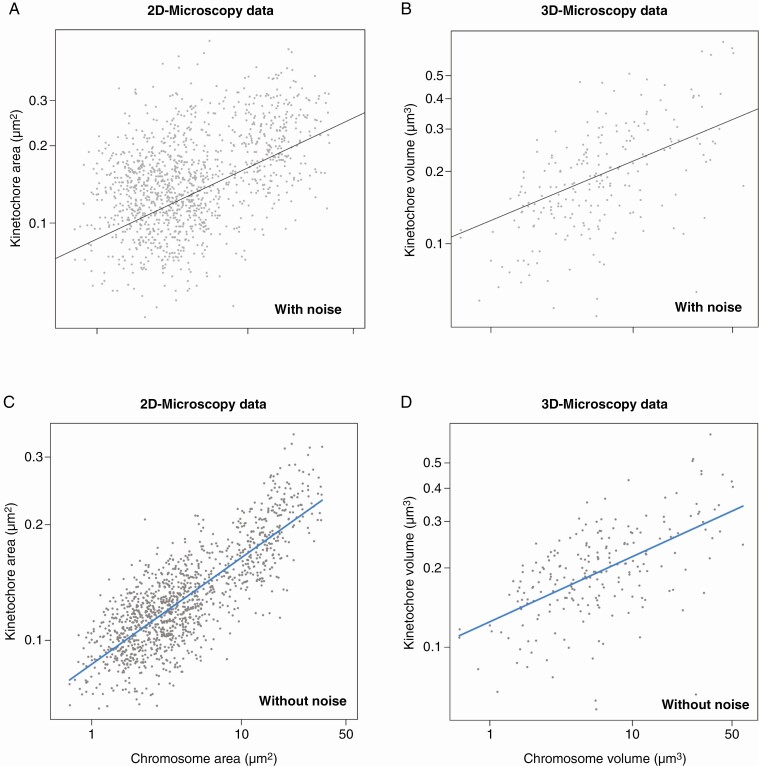
Although the visual inspection of the immunostained chromosomes, as shown in Figs. 1 and 2, does not reveal obvious kinetochore size differences between small and large chromosomes, we found after the analysis of 1447 chromosomes that, on average, the kinetochore size is positively related to the chromosome size in Agavoideae (Fig. 3), regardless of whether we analysed kinetochore/chromosome areas (Fig. 3A) or volumes (Fig. 3B). The slope of the regression line fit for log-transformed data is mathematically equivalent to the scaling exponent of non-transformed data (Glazier, 2013). Thus, if the slope is 1, kinetochore size would be in direct proportion to chromosome size. If the slope is higher than 1, kinetochore size increases faster with chromosome size. If the slope is lower than 1, kinetochore size increases more slowly than chromosome size. In our case, the observed slope of the regression line of the kinetochore and chromosome size relationship was 0.277 (P = 0) for areas and 0.247 (P = 0) for volumes (Fig. 3A–D; Table 1). The relationships between chromosome and kinetochore size for each analysed species based on 2D and 3D measurements are shown in Supplementary data Figs. S3 and S4, respectively. When the differences between the analysed metaphases, slides and individual plants were accounted for (see the Materials and Methods) (Figs. 3C, D), the chromosome size explained 83 and 51% of the variance in kinetochore size for areas and volumes, respectively (Table 1).
Fig. 3.
Scaling relationship of kinetochore and chromosome size in Agavoideae based on microscopic area (A, C) and volume (B, D) measurements. The equation of the regression line is y = 0.277x – 1.062 for areas (A, C) and y = 0.247x – 0.903 for volumes. (A and B) The original data including noise introduced by the hierarchical structure of the data (metaphases within slides and species). (C and D) Partial residual plots of the relationship between chromosome and kinetochore size after accounting for the noise introduced by differences between metaphases, slides, and species. Note that the regression lines (their slope and intercept) are identical in both plots with original data and partial residual plots. The partial residual plots were constructed in the R package ‘visreg’ (Breheny and Burchett, 2017).
Table 1.
Outcome of the regression model showing the effect of chromosome size on kinetochore size
| Analysis based on | Model term | b i | 95% CI | t | P | R 2 |
|---|---|---|---|---|---|---|
| 2D microscopy | Intercept | –1.062 | –1.133, –0.994 | –32.034 | 0 | 83.04 |
| Chromosome size | 0.277 | 0.263, 0.291 | 39.593 | 0 | ||
| 3D-SIM | Intercept | –0.903 | –0.983, –0.820 | –22.272 | 0 | 51.47 |
| Chromosome size | 0.247 | 0.186, 0.309 | 7.880 | 0 |
To test the relationship between kinetochore and genome size, we applied a linear mixed model. We set the kinetochore size as the response variable and chromosome size as the explanatory variable. To account for random variation caused by differences between different metaphases, slides, plants and species, we used these variables as nested random effects as follows: metaphases nested within slides nested within plants nested within species.
b i, parameters estimates; 95% CI, lower and upper bounds of the 95% confidence interval of the parameter estimate; t, t statistics; P, significance of the estimate difference from zero; R2, conditional R-squared that indicates the percentage of explained variance in kinetochore size by the chromosome size after accounting for random effects (i.e. the differences between metaphases, slides and species) used in the linear mixed model.
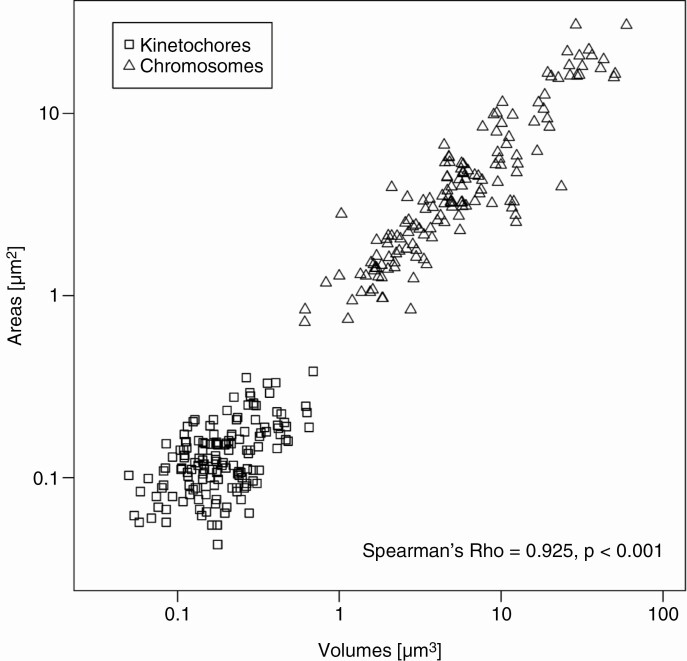
We further tested the correlation between the 2D and 3D quantifications by comparing the measurements of the same kinetochores and chromosomes. We found a very strong correlation (Fig. 4; Spearman’s rho = 0.925, P < 0.0001), indicating the reliability of both approaches.
Fig. 4.
Comparison of 2-D wide-field microscopy and 3-D-SIM measurements on a logarithmic scale. Spearman’s rho correlation was used.
Discussion
We showed that chromosome size is a strong predictor of kinetochore size in the karyotypes of Agavoideae species, suggesting that a larger chromosome needs a larger kinetochore. This is in accordance with the notion that for chromosomes of a specific size, a limited range of kinetochore size exists, ensuring the attachment of an adequate number of spindle fibres for proper chromosome movement and, consequently, separation of the sister chromatid during cell division (Plačková et al., 2021). Because both 2D and 3D approaches delivered similar results (Figs. 3A, B), the simpler and less time-consuming 2D approach may be used effectively in future studies.
Our data are in agreement with the idea that larger chromosomes need more sites for microtubule attachment, and thus a larger kinetochore. However, it is not yet clear what the physical mechanisms of the cell division might predict for the slope of the observed relationship. On average, the slopes of kinetochore and chromosome size relationships (from both 2D and 3D analyses) are very close to 0.25 (Fig. 3), which is a theoretical value proposed by Kramer et al. (2021) based on their across-species analysis of chromosome’s DNA content relative to its width (see below). The only available data on intra-karyotype relationships between kinetochore and chromosome size except those for this study were published for human (Irvine et al., 2004) and maize (Wang et al., 2021). When reanalysing the human data, we obtained a slope of 0.24 [95% confidence interval (CI) 0.11–0.38] (Supplementary data Fig. S5). Reanalysis of the data from maize (Wang et al., 2021) revealed a slope of 0.33 (95% CI 0.20–0.46) (Supplementary data Fig. S6). Again, these values are close to the theoretical slope of 0.25 (Kramer et al., 2021). The same value characterizes the relationship of a chromosome’s DNA content to its width, which probably stems from the physics of the elastic response of a chromatid to the forces that act on it during cell division (Kramer et al., 2021). The longer the chromosome, the more difficulties it can encounter during cell division because, in a high-viscosity environment, the drag force on elongated bodies is approximately proportional to their length (Nicklas, 1965; Ui et al., 1984). To prevent the formation of chromosomes which are too long and which are not stable during cell division if the size of the cell is limited (Schubert and Oud, 1997), they should have a proportionally larger cross-section area, i.e. they need to be wider (Kramer et al., 2021). This seems to be realized via increasing the helical turn size of the chromonemata forming the chromatids of large chromosomes (Kubalová et al., 2021b). The DNA content of a chromosome corresponds to the degree of chromatin compaction and is the product of its length and the square of its width (Kramer et al., 2021). Thus, if the kinetochore size is indeed related to chromosome width, perhaps because the kinetochore needs to cover the width of the chromosome to serve as an effective surface for microtubule capture, then its relatively small change may be related to a relatively large change in the DNA content of the chromosome. A factor that could also contribute to such a proportional relationship between kinetochore and chromosome size may be the size-dependent position of chromosomes in the metaphase plate. Even though we did not observe any such patterns, pioneering studies in Agavoideae reported that the large chromosomes tend to be placed at the edges of metaphase plates and small chromosomes in the middle (McKelvey and Sax, 1933; Watkins, 1936; Granick, 1944). If the density of the microtubule network decreased towards the edge of the metaphase plate (which is currently unknown), large chromosomes would face lower resistance and friction than in the centre and their kinetochores could thus be smaller than would normally be appropriate for their size.
If there is a causal link between kinetochore and chromosome size, what is the direction? Is it the chromosome size change that requires a change in kinetochore size? This possibility would mean that chromosome size changes would exert pressure on the kinetochore size to increase with chromosome size. This could happen relatively easily in small populations where genetic drift rather than selection affects the fate of new variants. In larger populations, such a chromosome would be selected against, unless its change in size would not be linked to some advantage. If the causal direction was from kinetochores to chromosomes, then any evolutionary mechanism capable of leading kinetochore size changes to fixation could thus promote the size evolution of entire chromosomes and genomes. Such chromosomes could then grow within the size limits permitted by their kinetochores. Kinetochore size change could become fixed either by chance via genetic drift or by deterministic processes such as centromere drive. Centromere drive would be expected to increase the rate of chromosome size evolution in lineages with asymmetric meiosis (usually female meiosis, where only one of the meiotic products – the egg or megaspore – survives) (Henikoff et al., 2001; Malik, 2009; Kursel and Malik, 2018; Chang and Malik, 2021), but not in lineages with symmetric meiosis (in both male and female meioses, all four products survive) (Zedek and Bureš, 2016). Confirming this expectation, chromosome size in ferns and lycophytes showing exclusively symmetric meiosis remains remarkably conserved compared with the high variability present in ‘asymmetric’ angiosperms and gymnosperms (Nakazato et al., 2008; Clark et al., 2016; Fujiwara et al., 2021).
We found that kinetochore size scales with chromosome size within metaphase cells of bimodal Agavoideae species. This finding suggests that the total centromere size to genome size scaling observed across eukaryotes (Zhang and Dawe, 2012; Plačková et al., 2021) may also originate from the mechanics of cell division. This conclusion is further supported by the conformity of our results with the theoretical predictions for chromosome width based on the physics of chromosome passage through the cytoplasm.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Figure S1: Example of area measurements in Hesperaloe parviflora (metaphase code Hepa30s1). Figure S2: Protein alignment of Agavoideae CENH3 sequences showing the two isoforms and the region used for antibody production. Figure S3: Partial residual plots show the relationship between chromosome and kinetochore size in Agavoideae species after accounting for differences between metaphases and slides. Figure S4: Partial residual plots show the relationship between chromosome and kinetochore size in Agavoideae species after accounting for differences between metaphases and slides. Figure S5: Regression model applied on the data on chromosome and kinetochore size from human. Figure S6: Linear mixed regression model applied on the data on chromosome and kinetochore size from maize. Table S1: Measurements of chromosome and kinetochore areas in Agavoideae species using 2D fluorescence wide-field microscopy. Table S2: Measurements of chromosome and kinetochore volumes in Agavoideae species using 3D-SIM. Movie S1: Centromeres of Hesperaloe parviflora chromosomes labelled by CENH3.
ACKNOWLEDGEMENTS
We thank Kelly Dawe and Na Wang for kindly providing data on kinetochore and chromosome size from maize lines reported in Wang et al. (2021). We are indebted to Eric Kramer for fruitful discussions and insightful comments on the manuscript. We also thank the editor Trude Schwarzacher and all the anonymous reviewers for their constructive comments and suggestions that helped to improve the manuscript. We acknowledge the core facility CELLIM of CEITEC supported by the Czech-BioImaging large RI project (LM2018129 funded by MEYS CR) for their support in obtaining scientific data presented in this paper. The data that support the findings of this study are available in the supplementary data of this article. K.P. and V.S. performed karyological analyses and the measurements. A.H. initiated the study. F.Z. and P.B. conceived the study. K.P. and F.Z. analysed the data. All authors contributed to the text and the final form of the manuscript.
Contributor Information
Klára Plačková, Department of Botany and Zoology, Faculty of Science, Masaryk University, Kotlářská 2, 611 37 Brno, Czech Republic.
František Zedek, Department of Botany and Zoology, Faculty of Science, Masaryk University, Kotlářská 2, 611 37 Brno, Czech Republic.
Veit Schubert, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) Gatersleben, D-06466 Seeland, Germany.
Andreas Houben, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) Gatersleben, D-06466 Seeland, Germany.
Petr Bureš, Department of Botany and Zoology, Faculty of Science, Masaryk University, Kotlářská 2, 611 37 Brno, Czech Republic.
Funding
This work was financially supported by the Czech Science Foundation, grant no. GA20-15989S, and the Grant Agency of Masaryk University, project MUNI/C/1660/2019. A.H. was supported by the Deutsche Forschungsgemeinschaft, grant no. HO1779/32-1.
LITERATURE CITED
- Anjur-Dietrich MI, Kelleher CP, Needleman DJ. 2021. Mechanical mechanisms of chromosome segregation. Cells 10: 465. doi: 10.3390/cells10020465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breheny P, Burchett W. 2017. Visualization of regression models using visreg. R Journal 9: 56–71. [Google Scholar]
- Carpenter EJ, Matasci N, Ayyampalayam S, et al. 2019. Access to RNA-sequencing data from 1,173 plant species: the 1000 Plant transcriptomes initiative (1KP). Gigascience 8: giz126. doi: 10.1093/gigascience/giz126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Malik HS. 2021. Putting the brakes on centromere drive in Mimulus. PLoS Genetics 17: e1009494. doi: 10.1371/journal.pgen.1009494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry LM, Faulkner AJ, Grossberg LA, Balczon R. 1989. Kinetochore size variation in mammalian chromosomes: an image analysis study with evolutionary implications. Journal of Cell Science 92: 281–289. [DOI] [PubMed] [Google Scholar]
- Clark J, Hidalgo O, Pellicer J, et al. 2016. Genome evolution of ferns: evidence for relative stasis of genome size across the fern phylogeny. New Phytologist 210: 1072–1082. doi: 10.1111/nph.13833. [DOI] [PubMed] [Google Scholar]
- Drpic D, Almeida AC, Aguiar P, et al. 2018. Chromosome segregation is biased by kinetochore size. Current Biology 28: 1344–1356. doi: 10.1016/j.cub.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Liu H, Meza-Torres EI, et al. 2021. Evolution of genome space occupation in ferns: linking genome diversity and species richness. Annals of Botany 389. doi: 10.1093/aob/mcab094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazier DS. 2013. Log-transformation is useful for examining proportional relationships in allometric scaling. Journal of Theoretical Biology 334: 200–203. doi: 10.1016/j.jtbi.2013.06.017 [DOI] [PubMed] [Google Scholar]
- Granick EB. 1944. A karyosystematic study of the genus Agave. American Journal of Botany 31: 283–298. [Google Scholar]
- Guadalupe P, Martínez J, Méndez I. 2008. Karyotype studies in cultivars of Agave tequilana Weber. Caryologia 61: 144–153. doi: 10.1080/00087114.2008.10589622. [DOI] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. 2001. The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293: 1098–1102. doi: 10.1126/science.1062939 [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison JS, Schwarzacher T. 1993. Molecular cytogenetics – biology and applications in plant breeding. In: Sumner AT, Chandley AC, eds. Chromosomes today. Dordrecht: Springer, 191–198. [Google Scholar]
- Houben A, Schroeder-Reiter E, Nagaki K, et al. 2007. CENH3 interacts with the centromeric retrotransposon cereba and GC-rich satellites and locates to centromeric substructures in barley. Chromosoma 116: 275–283. doi: 10.1007/s00412-007-0102-z [DOI] [PubMed] [Google Scholar]
- Houchmandzadeh B, Marko JF, Chatenay D, Libchaber A. 1997. Elasticity and structure of eukaryote chromosomes studied by micromanipulation and micropipette aspiration. Journal of Cell Biology 139: 1–12. doi: 10.1083/jcb.139.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine DV, Amor DJ, Perry J, Sirvent N, Pedeutour F, Choo KHA, Saffery R. 2004. Chromosome size and origin as determinants of the level of CENP-A incorporation into human centromeres. Chromosome Research 12: 805–815. doi: 10.1007/s10577-005-5377-4. [DOI] [PubMed] [Google Scholar]
- Kaneko K. 1966. Cytological studies on some species of Hosta I. Karyotypes of H. montana, H. lancifolia, H. chibai and H. capitate. Botanical Magazine Tokyo 79: 131– 137. doi: 10.15281/jplantres1887.79.131. [DOI] [Google Scholar]
- Kramer EM, Tayjasanant PA, Cordone B. 2021. Scaling laws for mitotic chromosomes. Frontiers in Cell and Developmental Biology 9: 684278. doi: 10.3389/fcell.2021.684278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubalová I, Němečková A, Weisshart K, Hřibová E, Schubert V. 2021a. Comparing super-resolution microscopy techniques to analyze chromosomes. International Journal of Molecular Sciences 22: 1903. doi: 10.3390/ijms22041903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubalová I, Câmara AS, Cápal P, et al. 2021. b. Helical metaphase chromatid coiling is conserved. bioRxiv doi: 10.1101/2021.09.16.460607. Preprint. [DOI] [Google Scholar]
- Kursel LE, Malik HS. 2018. The cellular mechanisms and consequences of centromere drive. Current Opinion in Cell Biology 52: 58–65. doi: 10.1016/j.ceb.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DL, Heald R. 2012. Mechanisms of intracellular scaling. Annual Review of Cell and Developmental Biology 28: 113–135. doi: 10.1146/annurev-cellbio-092910-154158. [DOI] [PubMed] [Google Scholar]
- Malik HS. 2009. The centromere-drive hypothesis: a simple basis for centromere complexity. Progress in Molecular and Subcellular Biology 48: 33–52. doi: 10.1007/978-3-642-00182-6_2 [DOI] [PubMed] [Google Scholar]
- McEwen BF, Ding Y, Heagle AB. 1998. Relevance of kinetochore size and microtubule-binding capacity for stable chromosome attachment during mitosis in PtK1 cells. Chromosome Research 6: 123–132. doi: 10.1023/a:1009239013215. [DOI] [PubMed] [Google Scholar]
- McKelvey SD, Sax K. 1933. Taxonomic and cytological relationships of Yucca and Agave. Journal of the Arnold Arboretum 14: 76–81. [Google Scholar]
- Moens PB. 1979. Kinetochore microtubule numbers of different sized chromosomes. Journal of Cell Biology 83: 556–561. doi: 10.1083/jcb.83.3.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo-Pineda M, Jansen LET. 2020. Genetics, epigenetics and back again: lessons learned from neocentromeres. Experimental Cell Research 389: 111909. doi: 10.1016/j.yexcr.2020.111909. [DOI] [PubMed] [Google Scholar]
- Nakazato T, Barker MS, Rieseberg LH, Gastony GJ. 2008. Evolution of the nuclear genome of ferns and lycophytes. In: Ranker TA, Haufler CH, eds. Biology and evolution of ferns and lycophytes. Cambridge: Cambridge University Press, 175–198. [Google Scholar]
- NCBI Resource Coordinators. 2018. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research 46: D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. 1965. Chromosome velocity during mitosis as a function of chromosome size and position. Journal of Cell Biology 25: 119– 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. 1983. Measurements of the force produced by the mitotic spindle in anaphase. Journal of Cell Biology 97: 542–548. doi: 10.1083/jcb.97.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- One Thousand Plant Transcriptomes Initiative. 2019. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574: 679–685. doi: 10.1038/s41586-019-1693-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D, R Core Team. 2021. nlme: linear and nonlinear mixed effects models. R package version 3.1-152. https://cran.r-project.org/web/packages/nlme/.
- Plačková K, Bureš P, Zedek F. 2021. Centromere size scales with genome size across Eukaryotes. Scientific Reports 11: 19811. doi: 10.1038/s41598-021-99386-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2020. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. http://www.R-project.org/. [Google Scholar]
- Schubert I, Oud JL. 1997. There is an upper limit of chromosome size for normal development of an organism. Cell 88: 515–520. doi: 10.1016/s0092-8674(00)81891-7 [DOI] [PubMed] [Google Scholar]
- Schwarzacher T, Heslop-Harrison JS, Anamthawat-Jonsson K, Finch RA, Bennett MD. 1992. Parental genome separation in reconstructions of somatic and premeiotic metaphases of Hordeum vulgare × H. bulbosum. Journal of Cell Science 101: 13–24. [Google Scholar]
- Shimamoto Y, Maeda YT, Ishiwata S, Libchaber AJ, Kapoor TM. 2011. Insights into the micromechanical properties of the metaphase spindle. Cell 145: 1062–1074. doi: 10.1016/j.cell.2011.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. 2020. What makes a centromere? Experimental Cell Research 389: 111895. doi: 10.1016/j.yexcr.2020.111895. [DOI] [PubMed] [Google Scholar]
- Taylor EW. 1965. Brownian and saltatory movements of cytoplasmic granules and the movement of anaphase chromosomes. Proceedings of the International Congress on Rheology 4: 175–191. [Google Scholar]
- Ui TJ, Hussey RG, Roger RP. 1984. Stokes drag on a cylinder in axial motion. Physics of Fluids 27: 787–795. doi: 10.1063/1.864706. [DOI] [Google Scholar]
- Wang N, Dawe RK. 2018. Centromere size and its relationship to haploid formation in plants. Molecular Plant 11: 398–406. doi: 10.1016/j.molp.2017.12.009 [DOI] [PubMed] [Google Scholar]
- Wang N, Liu J, Ricci WA, Gent JI, Dawe RK. 2021. Maize centromeric chromatin scales with changes in genome size. Genetics 217: iyab020. doi: 10.1093/genetics/iyab020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins GM. 1936. Chromosome numbers and species characters in Yucca. American Journal of Botany 23: 328–333. [Google Scholar]
- Weisshart K, Fuchs J, Schubert V. 2016. Structured illumination microscopy (SIM) and photoactivated localization microscopy (PALM) to analyze the abundance and distribution of RNA polymerase II molecules in flow-sorted Arabidopsis nuclei. Bio-protocol 6: e1725. http://www.bio-protocol.org/e1725 [Google Scholar]
- Zedek F, Bureš P. 2016. CenH3 evolution reflects meiotic symmetry as predicted by the centromere drive model. Scientific Reports 6: 33308. doi: 10.1038/srep33308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Dawe RK. 2012. Total centromere size and genome size are strongly correlated in ten grass species. Chromosome Research 20: 403–412. doi: 10.1007/s10577-012-9284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.