Abstract
Background:
Many issues, such as severity assessment and antibody responses, remain to be answered eagerly for evaluation and understanding of COVID-19. Immune lesion is one of key pathogenesis of the disease. It would be helpful to understand the disease if an investigation on antigenemia and association was conducted in the patients with SARS-CoV-2 infection.
Methods:
A total of 156 patients admitted to the First People's Hospital of Hefei or Anhui Provincial Hospital on January to February 2020 were involved in this study. SARS-CoV-2 nucleocapsid (NP) antigen, specific IgM/IgG antibodies, and RNA were detected in sequential sera from three COVID-19 patients, and additional 153 COVID-19 patients by means of NP-antigen capture enzyme-linked immunosorbent assay, colloidal gold quick diagnosis, and real-time RT-PCR, respectively. The clinical types of COVID-19 patients were classified into asymptomatic, mild, moderate, severe, and critical, following on the Chinese guideline of COVID-19 diagnosis and treatment. The demographic and clinical data of patients were obtained for comparable analysis.
Results:
NP antigen was detected in 5 of 20 sequential sera collected from three COVID-19 patients with typically clinical symptoms, and 60.13% (92/153) expanded samples collected within 17 days after illness onset. No SARS-CoV-2 RNA segment was detected in these sera. The NP positive proportion reached a peak (84.85%, 28/33) on 6 to 8 days after illness onset. Both NP concentration and positive proportion were increased with the increase of clinical severity of COVID-19. Compared to NP negative patients, NP positive patients had older age [years, medians (interquartile ranges (IQR)), 49 (6) vs. 31 (11)], lower positive proportion of NP specific IgM [27.17% (25/92) vs. 59.02% (36/61)], and IgG [21.74% (20/92) vs. 59.02% (36/61)] antibodies, and longer duration [days, medians (IQR), 24 (10) vs. 21 (13)] from illness to recovery.
Conclusions:
SARS-CoV-2 NP antigenemia occurred in COVID-19, and presented highly prevalent at early stage of the disease. The antigenemia was related to clinical severity of the disease, and may be responsible for the delay of detectable SARS-Cov-2 IgM.
Keywords: COVID-19, Antibody response, Antigenemia, Clinical severity, SARS-CoV-2
Introduction
Since the emergence of the novel coronavirus (SARS-CoV-2) causing the Coronavirus Disease 2019 (COVID-19), global concerns are being raised because of its core with high transmissibility, high mobility, and motility.[1,2] The virus has caused global pandemic after undergoing only 3 months around since it was first detected in the end of December 2019. Patients with COVID-19 present a broad spectrum of clinical presentation from asymptomatic to severe life-threatening disease with major complications like severe pneumonia, ARDS, acute cardiac injury, and septic shock.[3,4] The reports in 2020 showed that 19% of 72,314 symptomatic patients in China progressed to severe and critical illness with an estimated 1.4% symptomatic case fatality risk.[5,6]
SARS-CoV-2 is the first one causing pandemic in coronavirus family in traceable human history. Many issues associated with the disease, such as severity assessment and antibody responses, remain to be answered eagerly for evaluation and understanding of the disease. In this study, we tested SARS-CoV-2 nucleocapsid (NP) antigen in serial serum samples collected from the first three COVID-19 patients with typically clinical features, and expanded tests to evaluate the seroprevalence of NP antigenemia in 153 COVID-19 patients, and analyzed the demographic, clinical, and antibody response features in these patients.
Methods
Ethical approval
Data collection and analysis of cases and close contacts were determined by the National Health Commission of China to be part of a continuing public health outbreak investigation and were thus considered exempt from institutional review board approval.
Patients
A total of 156 (3 initially tested patients whom sequential serum samples were collected from, and additional 153 COVID-19 patients) admitted to the First People's Hospital of Hefei or Anhui Provincial Hospital on January to February 2020 were involved in this study. Those patients were confirmed with SARS-Cov-2 infection by real-time RT-PCR (rRT-PCR) based on the Chinese novel coronavirus pneumonia prevention and control program (3rd–4th ed.).[7] Serum samples were collected from these patients for detections of viral antigen, rRT-PCR, and SARS-Cov-2 NP specific IgM/IgG antibodies.
Data sources
Demographic and clinical characteristics data were obtained with standardized data collection forms through interviews of infected persons, relatives, close contacts, and health care workers.
Clinical typing of COVID-19
Clinical typing of COVID-19 follows on the Chinese guideline of COVID-19 diagnosis and treatment:[8] (1) Asymptomatic, SARS-CoV-2 RNA detection is positive but no clinical symptoms; (2) Mild, the clinical symptoms are mild, and there was no signs of pneumonia on radiograph; (3) Moderate, patients have pneumonia on radiograph with fever respiratory tract and other symptoms; (4) Severe, patients have at least one of the following: dyspnea, respiratory rate ≥30 times/min; oxygen saturation ≤93%; the pulmonary imaging showed a progressed lesions more than 50% within 24 to 48 hours; (5) Critical, patients have at least one of the following: respiratory failure occurred, and mechanical ventilation is required; shock is occurred; patients with extra pulmonary organ failure need to be admitted to Intensive Care Unit for treatment. To analyze the association of the antigen concentration in sera with disease progress, the duration after illness onset were grouped into 0 to 2, 3 to 5, 6 to 8, 9 to 11, 12 to 14, and 15 to 17 days.
Real-time PCR detection
According to the surveillance scheme of pneumonia cases with SARS-Cov-2 infection and the guideline of laboratory detection for COVID-19, RNA was extracted from throat swabs and/or sputum specimens collected from COVID-19 suspected patients. Specific rRT-PCR assays were performed to identify gene open reading frame 1ab (ORF) and nucleocapsid protein (NP) of SARS-Cov-2 using commercial kits qualified by Chinese FDA. The collected serum samples were also tested by rRT-PCR with same protocol.
NP antigen-capture enzyme-linked immunosorbent assay (ELISA)
SARS-COV-2 NP antigen was quantified in sera of COVID-19 patients using a SARS-CoV-2 NP Antigen-capture Quantitative Determination ELISA Kit (Catalog No.: 208.01.25.01) according to the manufacturer's instructions (BIOHIT Healthcare (Hefei), China).
SARS-CoV-2 NP specific IgM and IgG detection
SARS-CoV-2 NP specific IgM and IgG was detected in sera of COVID-19 patients by colloidal gold quick diagnosis kit (Catalog No.: 207.01.25.01) according to the manufacturer's instructions (INNOVITA Biotech, China; BIOHIT Healthcare (Hefei), China). In brief, 10 μL serum was dropped on to the sample placement region of the kit. Three drops (100–150 μL) of the mixed solution were then dropped on to the solution placement region of the kit. Results were visualized after 5 to 10 minutes of incubation at room temperature.
The sample was designated to be IgM or IgG positive when visualized results were observed in both detection kits.
Statistical analysis
Normally distributed continuous variables were presented as means ± standard deviation and non-normally distributed continuous variables as medians [interquartile ranges (IQR)]. The sera concentration of SARS-CoV-2 NP antigen, rRT-PCR Ct values, patient ages, and days from illness onset to recovery were compared by Mann-Whitney U test. The categorical variables were expressed as number (%) and compared by Fisher's exact test. Differences were considered significant at P < 0.05 with a two-tailed test. All analysis was performed using Instat software (Vision 5.0, GraphPad Prism San Diego, CA, USA).
Results
SARS-CoV-2 NP antigenemia in 3 COVID-19 patients
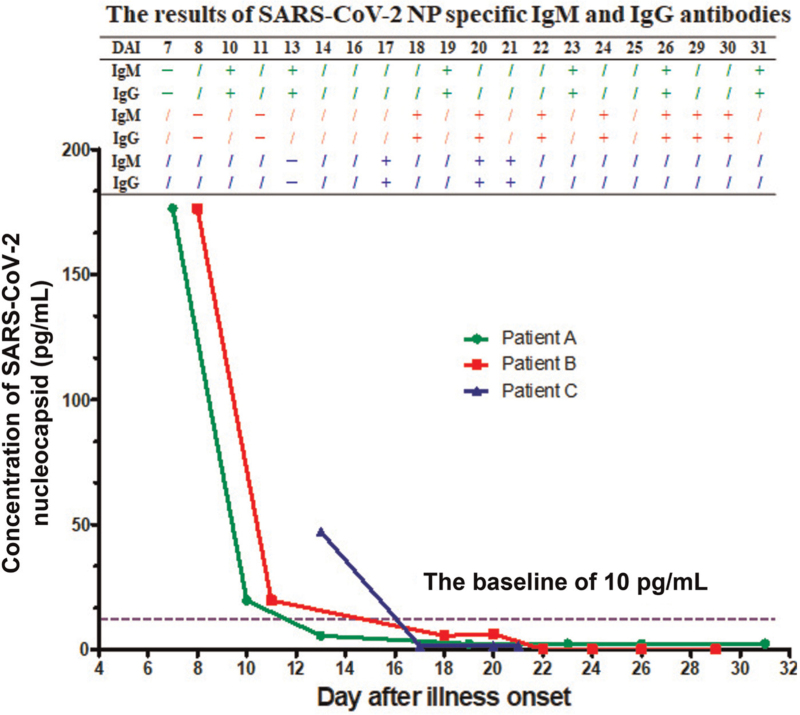
SARS-CoV-2 NP antigen was detected in 5 of 20 sequential sera collected from three COVID-19 patients by NP antigen-capture ELISA. The cutoff value of the assay was 10 pg/mL which was determined through assessing the results of 633 sera collected from patients with other respiratory infection (n = 369), pregnant women (n = 100), people with increased level of rheumatoid factor (n = 119), and hemolytic samples of healthy donors (n = 45) by the kit (data not shown). Of the five positive sera, there were two collected from patient A on day 7 and 10 after illness onset respectively, two collected from patient B on days 8 and 11 after illness onset respectively, and one collected from patient C on day 13 after illness onset [Figure 1]. SARS-CoV-2 NP specific IgM and IgG antibodies were detected in only one out of the five antigen positive sera samples whereas IgM and IgG antibodies were detected in the other 15 antigen negative sera samples [Figure 1].
Figure 1.
The kinetics of SARS-CoV-2 NP antigen concentration in sera collected from three COVID-19 patients on 7 to 31 days after illness onset, and the corresponding results of SARS-CoV-2 NP specific IgM and IgG antibodies in those sera. DAI: Day after illness onset; NP: Nucleocapsid.
As shown in Table 1, the three patients were 35 years old male (patient A), 67 years old female (patient B), and 44 years old male (patient C), respectively. They presented illness onset on the end of January or early of February. When typing clinically, both patient A and C were moderate, and patient B was severe. They showed typically clinical features of COVID-19 reported previously,[3,9,10] including fever with or without respiratory symptom (cough with or without expectoration), bilateral pneumonia imaged by ground-glass opacity, acute liver damage, and/or failure of respiratory function. To be similar to previous report,[9] laboratory abnormalities of the patients’ blood presented in increased level of aspartate aminotransferase, alanine aminotransferase, lactate dehydrogenase, γ-glutamyl transpeptidase, C reactive protein, and/or IL-6 [Supplementary Table 1]. In addition, the prothrombin time was prolonged in all three patients [Supplementary Table 1]. They all recovered after treated with antiviral therapy, antibiotic therapy, and other symptomatic supports [Table 1].
Table 1.
Characteristics of COVID-19 patient A, B, and C.
| Characteristics | Patient A | Patient B | Patient C |
|---|---|---|---|
| Age (years) | 35 | 67 | 44 |
| Gender | Male | Female | Male |
| Underlying medical condition | Chronic HBV infection with abnormal liver function | Hypothyroidism, angiosclerotic heart disease | No |
| Clinical typing | Moderate | Severe | Moderate |
| Date of illness onset | 31-Jan, 2020 | 25-Jan, 2020 | 1-Feb, 2020 |
| Day after illness onset on admission | 1 | 5 | 3 |
| Fever | Yes | Yes | Yes |
| The highest temperature | 38.4°C | 38.0°C | 38.0°C |
| Cough | Yes | Yes | No |
| Expectoration | No | Yes | No |
| Chest radiograph findings on admission | Bilateral pneumonia, ground-glass opacity | Bilateral pneumonia, multiple mottling, and ground-glass opacity | Bilateral pneumonia, ground-glass opacity in left upper and lower lobe and right middle lobe |
| Complications | |||
| Failure of respiratory function | No | Yes | No |
| Acute liver damage | Abnormal liver function before illness onset | Yes | Yes |
| Antiviral therapy | Lopinavir and ritonavir tablets (500 mg bid, started at February 2 till 20), Oseltamivir (started at February 2 till 5) | Lopinavir and ritonavir tablets, arbidol hydrochloride granules, aerosol therapy with α-interferon | Lopinavir and ritonavir tablets, arbidol hydrochloride granules, chloroquine phosphate, aerosol therapy with α-interferon |
| Antibiotic therapy | Yes | Yes | Yes |
| Outcome | Recovered | Recovered | Recovered |
The prevalence of SARS-CoV-2 NP antigenemia in COVID-19 patients
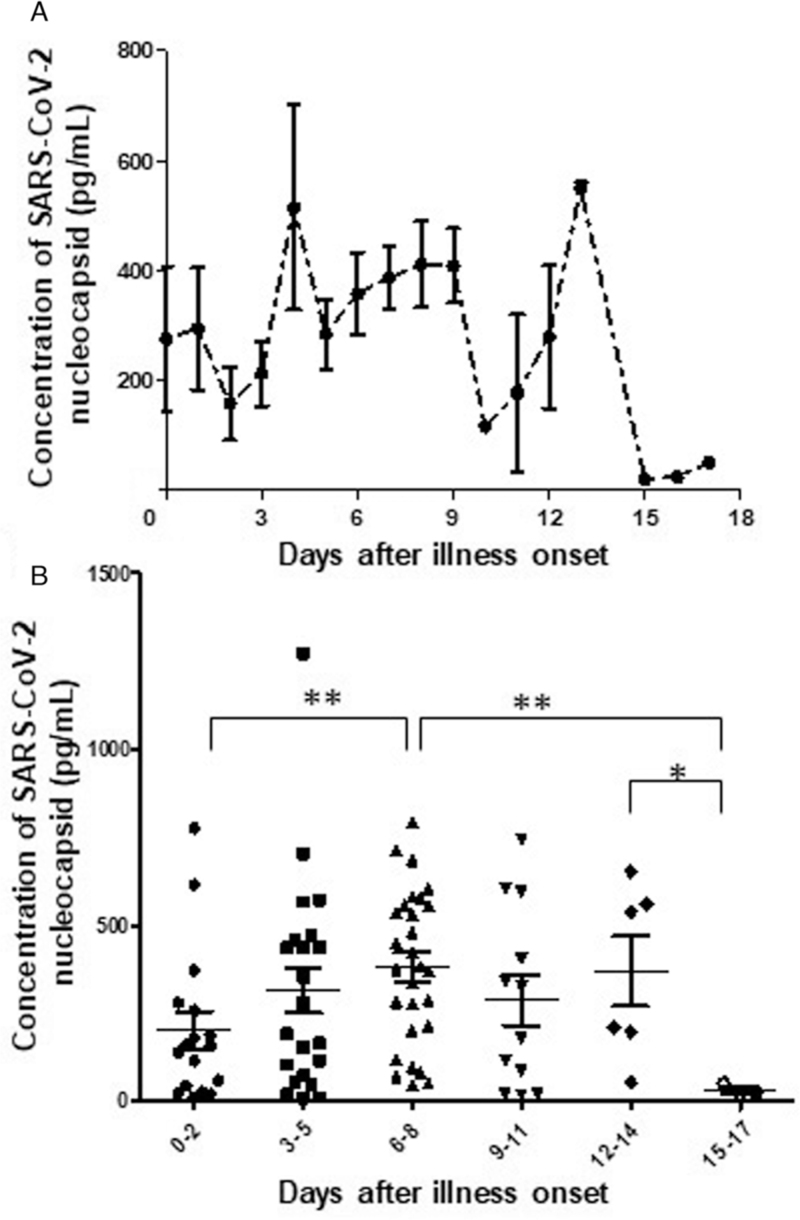
To determine the seroprevalence of SARS-CoV-2 NP antigenemia in COVID-19 patients, additional 153 COVID-19 patients were conducted detection of SARS-CoV-2 NP antigen. The results showed that, of the 153 COVID-19 patients, 92 (60.13%) presented SARS-CoV-2 NP antigen positive. But the kinetic curve of NP concentration did not indicate an obvious peak within 17 days after illness onset [Figure 2A]. However, when grouping the duration, the concentration of the NP antigen presented significantly higher level in sera collected on 6 to 8 days (381.3 ± 217.2 pg/mL) after illness onset than on 0 to 2 or 15 to 17 days after illness onset (P < 0.01), and the concentration was significantly higher in sera collected on 12 to 14 days than 15 to 17 days (369.2 ± 245.4 pg/mL vs. 32.0 ± 16.7 pg/mL) after illness onset (P < 0.05) [Figure 2B]. Meanwhile, if comparing the positive proportion, NP antigen presented the highest proportion (84.85%, 28/33) on 6 to 8 days after illness onset but the lowest ratio (3/13) on 15 to 17 days after illness onset [Table 2]. And compared to those sera sampled on 15 to 17 days after illness onset, except to group 12 to 14 days after illness onset (6/18), NP antigen had a significantly higher positive proportion in group 0 to 2 (62.96%, 17/27), 3 to 5 (75.00%, 24/32), or 9 to 11 (12/17) days after illness onset [Table 2]. Whether NP antigen positive or not, however, both SARS-CoV-2 ORF and NP gene segments were not detected by rRT-PCR in these sera.
Figure 2.
The kinetics of SARS-CoV-2 NP antigen concentration and its distribution in different duration of the disease. (A) The kinetics of SARS-CoV-2 NP antigen concentration in sera collected from 92 expanded COVID-19 patients within 17 days after illness onset. (B) The NP antigen concentration in sera collected during six grouped duration of the disease (0–2, 3–5, 6–8, 9–11, 12–14, and 15–17 days after illness onset). Statistical significance was analyzed by Mann-Whitney U test. ∗P < 0.05, ∗∗P < 0.01. NP: Nucleocapsid.
Table 2.
Characteristics of COVID-19 patients with or without SARS-CoV-2 nucleocapsid antigenemia.
| Variable | With antigenemia (n = 92) | Without antigenemia (n = 61) | P |
|---|---|---|---|
| Age [years, M (IQR)] | 49 (16) | 31 (21) | <0.001 |
| Females [n (%)] | 40 (43.48) | 29 (47.54) | 0.740 |
| IgM positive [n (%)] | 25 (27.17) | 36 (59.02) | 0.001 |
| IgG positive [n (%)] | 20 (21.74) | 36 (59.02) | <0.001 |
| Clinical typing [n (%)] | |||
| Asymptomatic | 2 (15.38) | 11 (84.62) | <0.001 |
| Mild | 5 (45.45) | 6 (54.54) | 0.002 |
| Moderate | 69 (61.06) | 44 (38.94) | 0.001 |
| Severe & critical | 16 (100.00) | 0 (0) | Reference |
| Days after illness onset [n (%)]∗ | |||
| 0–2 | 17 (62.96) | 10 (37.04) | 0.040 |
| 3–5 | 24 (75.00) | 8 (25.00) | 0.002 |
| 6–8 | 28 (84.85) | 5 (15.15) | 0.001 |
| 9–11 | 12 (70.59) | 5 (29.41) | 0.030 |
| 12–14 | 6 (33.33) | 12 (66.66) | 0.700 |
| 15–17 | 3 (23.08) | 10 (76.92) | Reference |
| Days from illness onset to recovery [M (IQR)]∗ | 24 (10)† | 21 (13) | 0.020 |
| Mild | 34 (10) | 30 (9) | 0.360 |
| Moderate | 24 (10) | 21 (12) | 0.030 |
| Severe & critical | 23 (8)† | 0 (0) | Reference |
M (IQR): median (interquartile rang); –: Not applicable.
Asymptomatic patients were not involved because of no illness onset.
Excluded one patient who died on day 8 after illness onset.
The association of serum NP concentration with viral load in respiratory tract or clinical severity of the disease
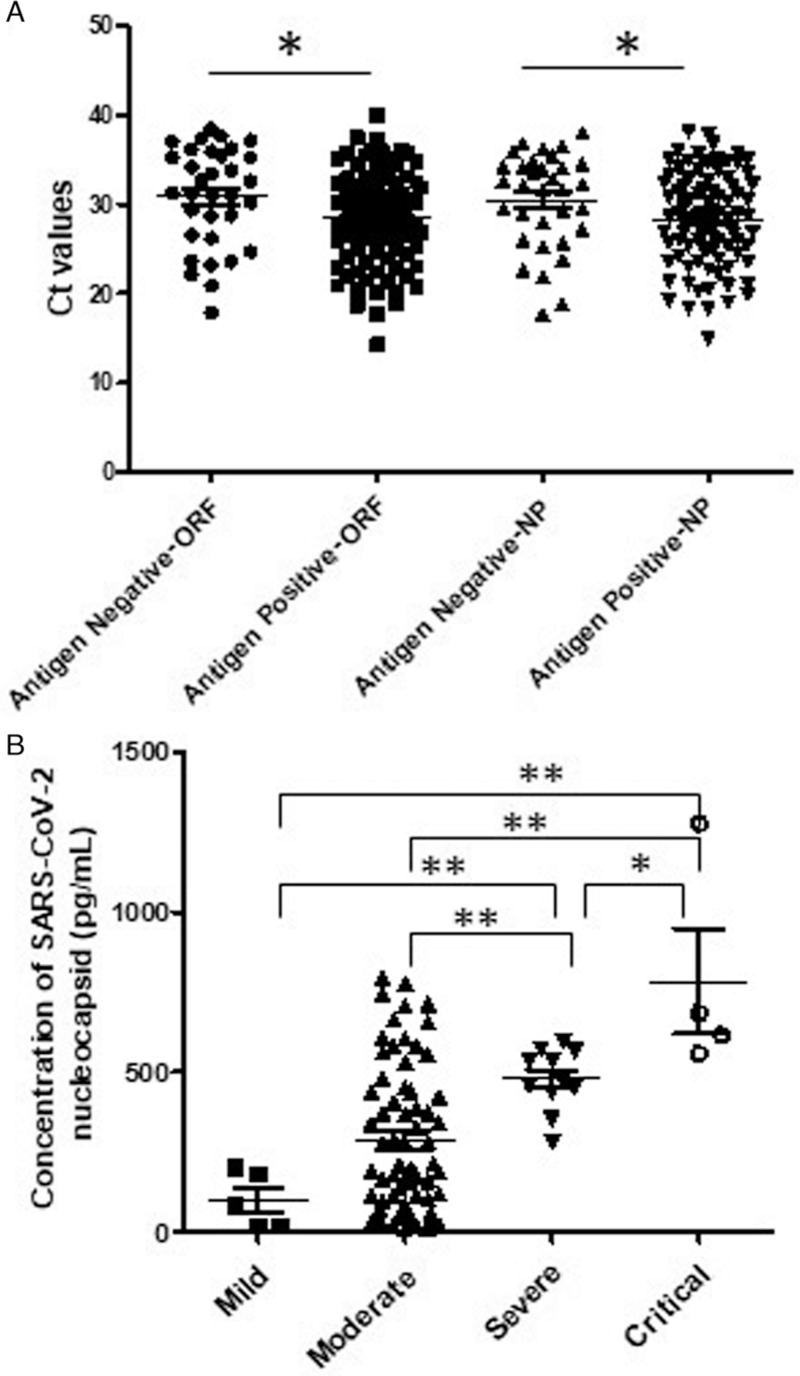
To analyze possible association serum concentration of NP antigen with viral load in respiratory tract of COVID-19 patients or clinical severity of the disease, we compared the rRT-PCR Ct values of throat swabs or sputum samples between NP antigen positive patients (NAPP) and NP antigen negative patients (NANP), and serum concentration of NP antigen among mild, moderate, severe, or critical NAPP. The results showed that, compared to NANP, NAPP presented significantly lower rRT-PCR Ct values of both gene ORF and NP in throat swabs or sputum samples [Figure 3A], indicating that NAPP may have a higher viral load in infected respiratory tract than NANP. In addition, critical (783.2 ± 331.2 pg/mL) patients had the highest serum concentration of NP antigen, then severe (478.0 ± 97.06 pg/mL) and moderate (285.4 ± 231.0 pg/mL) patients [Figure 3B]. The mild patients had the lowest level (100.5 ± 86.03 pg/mL) of NP antigen in sera although no significant difference was observed between mild patients and moderate patients (P > 0.05).
Figure 3.
rRT-PCR Ct values of respiratory samples from NP antigen positive or negative COVID-19 patients (A), and the sera concentration of NP antigen in mild, moderate, severe, or critical COVID-19 patients by clinical typing (B). Statistical significance was analyzed by Mann-Whitney U test. ∗P < 0.05, ∗∗P < 0.01. NP: Nucleocapsid; rRT-PCR: real-time RT-PCR.
Clinical characteristics of COVID-19 patients with or without SARS-CoV-2 NP antigenemia
We further compared the clinical characteristics between NAPP and NANP. As shown in Table 2, NAPP had much older age (median: 49 years, IQR: 16 years) than NANP (median: 31 years, IQR: 21 years) while the percentage of sex had no significant difference between them. Both positive proportions of IgM and IgG were significantly much lower in NAPP [27.17% (25/92), 21.74% (20/92)] than NANP [59.02% (36/61), 59.02% (36/61)]. NP antigen was not detected in sera collected from the other 66 COVID-19 patients on 18 to 110 days after illness onset while both IgM and IgG were detected in these patients except three (data not shown). NP antigen was detected in 2 of 13 asymptomatic patients, 5 of 11 mild patients, 69 of 113 (61.06%) moderate patients, and all 16 severe and critical patients. And significantly statistical differences were observed between asymptomatic, mild, or moderate patients and severe & critical patients. In addition, it took much longer time from illness onset to recovery in NAPP (median: 24 days, IQR: 10 days) than NANP (median: 21 days, IQR: 13 days). When typed patients clinically, the time from illness onset to recovery is longer in mild, moderate, or severe & critical patients with antigenemia than ones without antigenemia although no significantly statistical difference was observed between mild patients with antigenemia and mild patients without antigenemia.
Discussion
The SARS-CoV-2 sustained human-to-human transmission making it a world concerning and serious public health threat.[11] A number of important characteristics of SARS-CoV-2 infection have already been identified, but there are many unclear issues, such as severity assessment and antibody responses, which are very crucial to understand COVID-19 or impose improvement of associated managements on the disease. Here, we provide an investigation on seroprevalence of SARS-CoV-2 antigen and associated characteristics in COVID-19.
Our study strongly suggested that SARS-CoV-2 NP antigenemia was induced, and presented highly prevalent in COVID-19 at early stage of the disease. SARS-CoV-2 NP antigen was detected in 5 of 20 sequential sera collected from three COVID-19 patients with typically clinical symptom, as well as in 92 of 153 (60.13%) expanded COVID-19 patients. Most patients (90%, 81/90) with NP antigenemia were detected within 11 days after illness onset [Table 2]. The positive ratio of NP antigenemia reached a peak on 6 to 8 days after illness onset while the concentration of NP antigen also presented a high level on 6 to 8 days after illness onset in COVID-19 patients. To be our knowledge, antigenemia is very rare in respiratory virus infection. An individual report showed that antigenemia was detected in individual patients with SARS.[12] In addition, our results suggested that appearance of IgM or IgG was associated with the disappearance of the antigenemia. Correspondingly, both positive ratios of specific NP IgM and IgG were significantly much lower in NAPP than NANP within 17 days after illness onset. And no antigenemia was detected in sera collected from COVID-19 patients on more than 17 days after illness onset while almost all patients had IgM and IgG antibodies. Previous studies had demonstrated that IgM can recognize non-self particles,[13] and plays an important role on small particle clearance through macrophage.[14] It may explain why IgM of most COVID-19 cannot be detected at the early stage of COVID-19 by colloidal gold quick diagnosis kit or IgG and IgM occurred simultaneously as reported in previous study.[15] It also indicated that the colloidal gold quick detection of SARS-CoV-2 IgM antibody may not be suitable for early diagnosis of COVID-19.
SARS-CoV-2 NP antigenemia was not related to viremia in COVID-19. Previous studies suggested that SARS-CoV-2 may result in viremia in COVID-19 because of appearance of viral RNA or live virus.[16,17] In April 2003, Drosten et al. reported that viral RNA was detected at extremely low concentrations in plasma during the acute symptomatic phase of SARS-CoV infection.[18] In 2004, Singapore researchers reported that SARS-CoV can be detected in the blood of infected patients with SARS.[19] However, NP antigenemia was not related to viremia in COVID-19 because no viral RNA segments were detected in all serum samples in this study. But, as indicated by results of rRT-PCR, the viral load may be much higher in respiratory tract of NAPP than one of NANP. Intracytoplasmic inclusion bodies, which are commonly observed in RNA viral infection, were abundantly observed in the autopsied lung tissue of COVID-19 patients.[20] In addition, autopsy pathology suggested that exudative diffuse alveolar damage with massive capillary congestion was the main feature of histological lesion in COVID-19,[21] and coagulopathy may play an important role on the disease progressing.[22] Coincidentally, the prothrombin time was also extended in the all first investigated three patients in this study. Hence, possibly, antigenemia may be associated with the infusion of viral protein from intracytoplasmic inclusion bodies through damaged capillary in lung.
SARS-COV-2 NP antigenemia may be used as a biomarker for evaluation of COVID-19 severity. Our results showed that both the serum concentration of NP antigen and the ratio of NP antigenemia were increased with the increase of clinical severity of the disease. This phenomenon was reported in some other viral infections (eg, dengue,[23] CMV[24]). Previous studies have demonstrated that dengue virus fatal cases had higher NS1 antigenemia than the non-fatal cases,[23] and the degree of NS1 antigenemia correlated positively with hematocrit and liver transaminases in dengue patients.[25] Liver transaminases were highly frequently increased in COVID-19. And, similarly, immunopathology were believed to be an important pathogenesis in both COVID-19 and dengue.[26,27] In addition, our results showed that NAPP had much older age than NANP, and NAPP have longer duration from illness to recovery than NANP. Correspondingly, previous report has demonstrated that the increased risk of in-hospital death was associated with older age in COVID-19.[28]
In conclusion, we first reported SARS-CoV-2 antigenemia in COVID-19, and the NP antigenemia presented highly prevalent at early stage of COVID-19. And the antigenemia was not related to viremia but did to clinical severity of the disease. In addition, the antigenemia may be responsible for the delay of detectable SARS-Cov-2 IgM. The study would be helpful to understand COVID-19, impose improvement of associated managements on the disease although we did not detect all antigens of SARS-CoV-2 in sera of patients in this study because of the limitation of unavailable detection kits.
Acknowledgments
The authors would like to thank all the hospitals in Hefei for the collection of clinical specimens, and great appreciate to BIOHIT Healthcare (Hefei) Co. for the supply of corona diagnostic kits.
Funding
This study was supported by National Mega-projects for Infectious Diseases (2017ZX10304402-001-019), National Natural Scientific Foundation of China (81971946), and Hefei Municipal Health Commission (hwk2018zd001).
Author Contributors
Rongbao Gao and Wenyan Zhang designed the study. Kefu Zhao, Enqing You, Xuxiang Liu, Liwei Zhu, Lili Chen, Wenjing Wang, Lijie Wang, and Jinju Wu gathered data. Lei Zhang and Xiuzhen Wang joined into the clinical treatment. Jing Jin, Renshu Tang, and Wei Liu transferred samples. Wei Zhou, Wei Liu, Jiawang Lin, Jing Jin, Renshu Tang, Yindi Zhu, and Qiang Zhang did experimental tests. Rongbao Gao and Wenyan Zhang did analyses and wrote the report. Jinju Wu joined in statistical analysis and text checking. All authors contributed to review and revision and have seen and approved the final version.
Conflicts of Interest
None.
Supplementary Material
References
- [1].Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 2020;382(13):1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen T, Wu D, Chen H, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ 2020;368:m1091. doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mizumoto K, Kagaya K, Zarebski A, et al. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill 2020;25(10):2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wu JT, Leung K, Bushman M, et al. Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China. Nat Med 2020;26(4):506–510. doi: 10.1038/s41591-020-0822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- [7].Novel coronavirus pneumonia prevention and control program (3rd ed.) (in Chinese). Available from: http://wwwnhcgovcn/xcs/zhengcwj/202001/470b128513fe46f086d79667db9f76a5/files/8faa1b85841f42e8a0febbea3d8b9cb2pdf). Accessed December 20, 2021. [Google Scholar]
- [8].Novel coronavirus pneumonia diagnosis and treatment program (6th ed.) (in Chinese). Available from: http://wwwnhcgovcn/xcs/zhengcwj/202002/8334a8326dd94d329df351d7da8aefc2/files/b218cfeb1bc54639af227f922bf6b817pdf. Accessed December 20, 2021. [Google Scholar]
- [9].Zhao D, Yao F, Wang L, et al. A comparative study on the clinical features of COVID-19 pneumonia to other pneumonias. Clin Infect Dis 2020;71(15):756–761. doi: 10.1093/cid/ciaa247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Patel A, Jernigan DB. 2019-nCoV CDC Response Team. Initial public health response and interim clinical guidance for the 2019 novel coronavirus outbreak - United States, December 31, 2019–February 4, 2020. MMWR Morb Mortal Wkly Rep 2020;69(5):140–146. doi: 10.15585/mmwr.mm6905e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Che XY, Di B, Zhao GP, et al. A patient with asymptomatic severe acute respiratory syndrome (SARS) and antigenemia from the 2003-2004 community outbreak of SARS in Guangzhou, China. Clin Infect Dis 2006;43(1):e1–e5. doi: 10.1086/504943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang M, Carroll MC. Natural antibody mediated innate autoimmune response. Mol Immunol 2007;44(1–3):103–110. doi: 10.1016/j.molimm.2006.06.022. [DOI] [PubMed] [Google Scholar]
- [14].Litvack ML, Post M, Palaniyar N. IgM promotes the clearance of small particles and apoptotic microparticles by macrophages. PLoS One 2011;6(3):e17223. doi: 10.1371/journal.pone.0017223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- [16].Chen W, Lan Y, Yuan X, et al. Detectable 2019-nCoV viral RNA in blood is a strong indicator for the further clinical severity. Emerg Microbes Infect 2020;9(1):469–473. doi: 10.1080/22221751.2020.1732837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cho HJ, Koo JW, Roh SK, et al. COVID-19 transmission and blood transfusion: a case report. J Infect Public Health 2020;13(11):1678–1679. doi: 10.1016/j.jiph.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- [19].Ng LF, Wong M, Koh S, et al. Detection of severe acute respiratory syndrome coronavirus in blood of infected patients. J Clin Microbiol 2004;42(1):347–350. doi: 10.1128/JCM.42.1.347-350.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zeng Z, Xu L, Xie XY, et al. Pulmonary pathology of early phase COVID-19 pneumonia in a patient with a benign lung lesion. Histopathology 2020;77(5):823–831. doi: 10.1111/his.14138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Menter T, Haslbauer JD, Nienhold R, et al. Post-mortem examination of COVID-19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings of lungs and other organs suggesting vascular dysfunction. Histopathology 2020;77(2):198–209. doi: 10.1111/his.14134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wichmann D. Autopsy findings and venous thromboembolism in patients with COVID-19. Ann Intern Med 2020;173(12):1030. doi: 10.7326/L20-1206. [DOI] [PubMed] [Google Scholar]
- [23].Nunes PCG, Nogueira RMR, Heringer M, et al. NS1 antigenemia and viraemia load: potential markers of progression to dengue fatal outcome? Viruses 2018;10(6):326. doi: 10.3390/v10060326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].The TH, van der Bij W, van den Berg AP, et al. Cytomegalovirus antigenemia. Rev Infect Dis 1990;12 Suppl 7:S734–S744. [PubMed] [Google Scholar]
- [25].Erra EO, Korhonen EM, Voutilainen L, et al. Dengue in travelers: kinetics of viremia and NS1 antigenemia and their associations with clinical parameters. PLoS One 2013;8(6):e65900. doi: 10.1371/journal.pone.0065900. Print 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Henderson LA, Canna SW, Schulert GS, et al. On the alert for cytokine storm: immunopathology in COVID-19. Arthritis Rheumatol 2020;72(7):1059–1063. doi: 10.1002/art.41285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Screaton G, Mongkolsapaya J, Yacoub S, et al. New insights into the immunopathology and control of dengue virus infection. Nat Rev Immunol 2015;15(12):745–759. doi: 10.1038/nri3916. [DOI] [PubMed] [Google Scholar]
- [28].Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.