In this review, Rao et al. contrast mouse models of CHD, which maintain the anatomical arrangement of the heart, and human cellular models of CHD, which are more likely to capture human-specific biology but lack anatomical structure. They also discuss the recent development of cardiac organoids, which are a promising step toward more anatomically informative human models of CHD.
Keywords: congenital heart disease, genetics, heart
Abstract
Congenital heart defects (CHDs) are among the most common birth defects, but their etiology has long been mysterious. In recent decades, the development of a variety of experimental models has led to a greater understanding of the molecular basis of CHDs. In this review, we contrast mouse models of CHD, which maintain the anatomical arrangement of the heart, and human cellular models of CHD, which are more likely to capture human-specific biology but lack anatomical structure. We also discuss the recent development of cardiac organoids, which are a promising step toward more anatomically informative human models of CHD.
Cardiac development is achieved by the intricately coordinated actions of several transcription factors (TFs) and chromatin modifiers to guide timely activation of specific genes. This cascade of transcriptional events ensures that appropriate cellular and structural cues are relayed to generate a complex, four-chambered structure from the cardiac crescent. Congenital heart defects (CHDs) are malformations of the developing heart, and occur in at least one out of 100 live births (van der Linde et al. 2011). Some are very severe, resulting in neonatal death or requiring complex open heart surgery. Oftentimes, the repair only provides a temporary solution, and the patient requires heart transplantation a few years later. In addition, structural CHDs can be accompanied by physiological defects, such as abnormal impulse conduction or heart failure. Structural defects in CHD patients are often associated with mutations in the gene regulators that play a role in very early cardiogenesis, including TFs, chromatin remodeling factors, and signaling molecules (Nees and Chung 2019; Morton et al. 2022). However, the links between disease-causative variants and the resulting heart defect are not well understood. Therefore, there is a great need for disease modeling to better understand CHD pathophysiology and discover treatment options. Here, we review the current literature on modeling CHDs, exploring the relative benefits and limitations of mouse models and human cellular models.
Modeling CHDs in mice
The pathogenesis of CHDs is complex. A majority of CHDs lack a clearly identifiable heritable cause (for extensive review, see Nees and Chung 2019; Morton et al. 2022), although this may be attributable to myriad factors, including as yet unidentified genes, lowly penetrant multigenic disorders, or uterine environment. However, human genetics research has begun to unravel the genetic basis in some cases of CHDs, pointing to errors in common cellular mechanisms such as transcriptional regulation. Notably, many CHD variants have been identified in highly conserved cardiac TFs that are central to establishing cardiac morphology and progenitor identity. Apart from a few important exceptions, mouse heart development closely resembles human heart development, so mouse models of CHD can provide an excellent opportunity to understand how specific factors are involved in heart development and contribute to disease development. Here, we review mouse models that have provided insights into CHDs and the common themes that have emerged from disease modeling in mice. We particularly focus on some of the first TFs that were identified as CHD variants due to the extensive body of literature that has shed light on various aspects of their roles in CHD and cardiac development.
Roles of CHD genes in early cardiogenesis
Patterning of gene expression can provide clues about the potential function of CHD genes. TBX5 heterozygous loss-of-function mutations underlie Holt-Oram syndrome (HOS), an autosomal dominant developmental disorder characterized by skeletal abnormalities and, frequently, cardiac defects including atrial septal defects (ASDs), ventricular septal defects (VSDs), and conduction defects (Basson et al. 1994, 1997; Newbury-Ecob et al. 1996; Li et al. 1997). The pattern of Tbx5 expression in the developing embryo is consistent with the clinical characteristics of HOS (Chapman et al. 1996; Bruneau et al. 1999). In human embryos, TBX5 is highly expressed in the developing inflow tract, atrial wall, atrial septa, and atrioventricular (AV) endocardial cushions (Li et al. 1997). In mouse and chick embryos, where it has been characterized more extensively over the course of cardiac development, Tbx5 exhibits a unique asymmetric pattern of expression. It is initially expressed in the cardiac crescent, but becomes increasingly restricted to the atria and then the left ventricle following looping and septation (Bruneau et al. 1999). In the ventricles, Tbx5 expression is finely restricted to the boundary between the left and right sides of the interventricular septum (IVS). Indeed, disrupting the location of this boundary by either removing or extending the domain of Tbx5 expression leads to a lack of IVS. Lineage tracing further shows that Tbx5+ cells form what appears to be a lineage boundary at the junction of the left and right sides of the IVS (Devine et al. 2014). These data together suggest that VSDs caused by reduced TBX5 function might be due to mispatterning of this IVS boundary.
On the other hand, some genes, such as the cardiogenic TF Nkx2-5, are broadly expressed throughout embryonic heart development. In mice, Nkx2-5 is expressed in the earliest cardiac progenitors in the late primitive streak, before the onset of myogenic differentiation (Komuro and Izumo 1993; Lints et al. 1993). It is abundantly expressed throughout the atrial and ventricular myocardium, but not in the endocardium or epicardium. Heterozygous loss-of-function NKX2-5 mutations were initially identified in patients with secundum ASDs and AV conduction defects (Schott et al. 1998) Subsequent studies revealed that the spectrum of CHDs caused by NKX2-5 mutations is somewhat broader than initially recognized (Benson et al. 1999), but indicated that although the factor is widely expressed, only some regions of the heart are sensitive to reduced NKX2-5 function, and there is variation in which regions develop structural defects in its absence. This suggests that other spatially restricted cues might interact with NKX2-5 to confer dosage sensitivity.
During early cardiac development, the GATA transcription factor family homologs Gata4, Gata5, and Gata6 share very similar expression patterns, but they have overlapping yet distinct domains of expression during later stages (Arceci et al. 1993; Laverriere et al. 1994; Morrisey et al. 1996), suggesting that they may each contribute to fine spatial and temporal regulation differences in cardiac development. In contrast to GATA4 and GATA6, which are expressed broadly in the primitive streak mesoderm (Heikinheimo et al. 1994; Morrisey et al. 1996; Koutsourakis et al. 1999), GATA5 has a more restricted expression pattern in the precardiac mesoderm (Morrisey et al. 1997). GATA4 and GATA6 have overlapping expression patterns in the developing myocardium, vascular smooth muscles, and endocardial cushions (Heikinheimo et al. 1994; Morrisey et al. 1996; Koutsourakis et al. 1999), whereas GATA5 expression becomes progressively restricted to the atrial endocardial cells before its cardiac expression generally diminishes (Morrisey et al. 1997). GATA6 is highly expressed in the cardiac outflow tract (OFT) and neural crest cells that are required for proper OFT development (Lepore et al. 2006). GATA6 is also detected throughout the sinoatrial node by E14.5 in mice (Gharibeh et al. 2021) and, accordingly, cardiac conduction defects are observed in individuals with mutations in GATA6, whereas GATA4 mutations more often result in septal defects and endocardial cushion defects, but not issues with conduction (Garg et al. 2003). While compensation and genetic interaction of these factors have been studied in mouse knockout models, the extent to which each gene can compensate for the others in human CHDs is yet to be determined.
Haploinsufficiency in mouse models vs. human CHD
Virtually all genetically defined CHDs are due to reduced gene function, whether it be because of a gene deletion, nonsense mutation, or loss-of-function missense mutation. This indicates that a common molecular mechanism underlies many CHDs: exquisite sensitivity to the dosage of these proteins. Although humans have clear and often severe CHDs in the context of haploinsufficiency of the causative genes, in mice this is highly variable.
Both mice and humans display sensitivity to Tbx5 dosage (Bruneau et al. 2001; Mori et al. 2006; Moskowitz et al. 2007). Indeed, haploinsufficiency of Tbx5 in mice reasonably phenocopies the hallmarks of HOS, including the asymmetric bilateral upper limb, frequent ASDs and VSDs, and abnormal cardiac electrophysiology, possibly due to poor formation of the conduction system (Bruneau et al. 2001; Moskowitz et al. 2007), as well as impaired cardiac relaxation (Zhu et al. 2008). Notably, Tbx5 heterozygous null mice are not recovered at Mendelian ratios at birth, indicating perinatal lethality, and those that die in utero exhibit a complex range of cardiac defects including ASDs, muscular VSDs, and deformed left ventricles (Bruneau et al. 2001). Using mouse models with a gradation of Tbx5 dosage, Mori et al. (2006) found that the phenotype, including severity of ASDs, paralleled the expression level of Tbx5 regardless of genetic background. Changes in gene expression were also sensitive to Tbx5 dosage (Mori et al. 2006). However, humans with TBX5 mutations also have other cardiac defects including tetralogy of Fallot (ToF), and rarely hypoplastic left heart syndrome (HLHS) (Basson et al. 1994; Newbury-Ecob et al. 1996), but these defects are not observed in Tbx5 heterozygous mice.
Mutations in NKX2-5 have been identified in families with a high incidence of secundum ASDs and high penetrance of AV conduction defects (Benson et al. 1998; Schott et al. 1998). Affected individuals also have other structural abnormalities including VSDs, ToF, aortic stenosis, left ventricular noncompaction syndrome (LVNC), HLHS, and dilated cardiomyopathy (Schott et al. 1998; Elliott et al. 2003; Sarkozy et al. 2005; Costa et al. 2013). Initial studies examining the effects of Nkx2-5 loss of function in mice found that complete loss of Nkx2-5 resulted in embryonic lethality due to the inability of the heart tube to undergo rightward looping despite establishing proper myogenesis (Lyons et al. 1995). Surprisingly, mice that were heterozygous for a Nkx2-5-null allele were viable and seemingly normal. However, upon closer examination, it was determined that Nkx2-5 haploinsufficiency did in fact result in a low frequency of ASDs and an increased incidence of patent foramen ovale (PFO) (Biben et al. 2000; Tanaka et al. 2002). Mice with heterozygous loss of Nkx2-5 also exhibited mild conduction defects, with prolonged PR and QR intervals and extended atrial fibrillation (Tanaka et al. 2002), which was shown to be related to a change in the number of myocytes that become committed to the conduction system (Jay et al. 2004).
Despite the largely redundant expression patterns of the three cardiac-enriched GATA TFs (Laverriere et al. 1994), haploinsufficiency of the individual factors results in similar, yet distinct, phenotypes. Mouse embryos lacking Gata6 have defects in extraembryonic tissue development and die shortly after gastrulation (Koutsourakis et al. 1999), while its haploinsufficiency leads to defects in OFT septation and cardiac conduction (Lepore et al. 2006; Gharibeh et al. 2018, 2021), reflective of its expression pattern in these regions during development. Accordingly, human mutations in GATA6 have been identified in cases of patent ductus arteriosus (PDA) (Kodo et al. 2009) and atrial fibrillation (Yang et al. 2012; Gharibeh et al. 2021).
On the other hand, complete Gata4 loss of function, while also embryonic lethal, does permit development of cardiac lineage-committed progenitor cells in mice (Kuo et al. 1997; Molkentin et al. 1997). Interestingly, Gata6 expression was found to be increased in the absence of Gata4, suggesting at least a partial functional compensation between these two factors. In fact, loss of both factors resulted in acardia with a block in the progressive differentiation of cardiac progenitors to myocytes (Zhao et al. 2008). Although Gata4 heterozygous mice were initially believed to undergo normal cardiac development, a study of a gradation of GATA4 protein levels using various hypomorphic alleles revealed that small differences in Gata4 expression could profoundly affect embryonic viability (Pu et al. 2004). Whereas mice with hypomorphic alleles with 50% of normal GATA4 protein expression levels survived, a further reduction of 20% GATA4 expression resulted in embryonic death between E13.5 and E16.5. Thus, in mice there is a narrow window of tolerance for reduced Gata4 dosage, below which cardiomyocyte replication, endocardial cushion development, and proper chamber septation are compromised (Pu et al. 2004; Rajagopal et al. 2007). In humans, GATA4 mutations have been identified in two unrelated families with a history of CHDs. Two reported mutations in this study resulted in an autosomal dominant pattern of inheritance in which all affected individuals had ASDs (Garg et al. 2003). However, similar to other TFs implicated in CHDs, these mutations also resulted in a range of CHD phenotypes, including other septal defects and pulmonary valve thickening, but not cardiac conduction abnormalities. Other human mutations have also been identified in GATA4, some of which coincide with reported mouse haploinsufficiency phenotypes such as endocardial cushion defects, hypoplastic right ventricle, and secundum ASDs (Rajagopal et al. 2007). However, highly penetrant mouse phenotypes such as cardiomyopathy were not identified in the human patients.
These observations underscore the difference between mice and humans depending on the mutated gene. In addition, they reveal that physiological parameters such as the conduction system may be particularly sensitive to TF dosage and thus more likely to be recapitulated in mouse models than structural defects. Mouse models of these gene deletions have improved our understanding of the functional consequences of haploinsufficiency and how they might relate to disease phenotypes. However, mechanistic insight into how haploinsufficiency of a given TF affects the regulation of its direct targets is still an area of immense interest. Genomics-based approaches to interrogate the dose-dependent effects of haploinsufficiency are proving to be helpful in parsing these effects; however, mouse models can be limiting due to the challenges with generating sufficient material for such experiments.
Variability in phenotype
Aside from the primary clinical characteristics presented in each family, a wide range of other cardiac phenotypes are also frequently noted in CHD studies (Basson et al. 1994; Newbury-Ecob et al. 1996; Benson et al. 1998; Schott et al. 1998; Brassington et al. 2003; Elliott et al. 2003; Costa et al. 2013). There is variability in phenotype within families that share a common mutation, and this variability extends to the corresponding mouse knockout phenotype for each gene. For example, even mutations that are predicted to result in loss of function of TBX5 exhibit an array of phenotypes within the same family (Brassington et al. 2003).
One explanation for this variability is the contribution of genetic modifiers. Indeed, the remarkable concordance in clinical characteristics of monozygotic twins with a TBX5 mutation resulting in HOS argues for the direct effect of genotype on disease manifestation (Huang et al. 2002). In mice, this is evidenced by the fact that different strains of mice exhibit varying degrees of severity of their phenotype (Biben et al. 2000; Bruneau et al. 2001; Mori et al. 2006). This is also apparent in mouse models of Gata4 loss of function, where the genetic strain influences the degree of penetrance of cardiac abnormalities and the perinatal survival rates (Bisping et al. 2006; Rajagopal et al. 2007).
The combinatorial effect of multiple variants was nicely exemplified in a study of LVNC cardiomyopathy by Gifford et al. (2019), who used exome sequencing in a family with high penetrance of LVNC to discover damaging mutations in MYH7 and the TF MKL2 inherited from an affected father. However, the additional inheritance of a rare missense variant in NKX2-5 from their unaffected mother resulted in the three offspring having profoundly more severe disease and earlier age of onset. These conclusions were supported by modeling in mice, which showed that compound heterozygotes for all three genes exhibited LVNC-like phenotypes with hypertrabeculations in the left ventricular wall.
However, the fact that littermates within inbred lines can also show a wide range of variability suggests that genetic modifiers are not the only contributor to phenotypic variability. The discrepancy between human and mouse phenotypes in some cases may be attributable to the fact that the mutations are typically identified in families with a high incidence of disease. Thus, familial carriers of mutations with low disease severity are less likely to be identified, causing a skewed interpretation of the spectrum of phenotypes that these mutations might cause in humans.
Cooperativity of genes
Proper cardiac development is achieved through the convergent actions of multiple factors across different cell types. Through various studies of human and mouse genetics, a common theme that has emerged is that mutations in a single gene can result in a wide range of cardiac defects and, conversely, that mutations in multiple genes can result in very similar cardiac abnormalities. This is exemplified by the case of TBX5 and NKX2-5, as mutations of each result in ASDs, conduction defects, and other similar cardiac abnormalities. A physical interaction between these two factors was found to synergistically activate expression of common target genes such as ANF, Cx40, Nppa, and Id2 (Bruneau et al. 2001; Hiroi et al. 2001; Moskowitz et al. 2007). In fact, heterozygous loss of both Tbx5 and Nkx2-5 results in the lack of specification of the ventricular cardiac conduction system altogether (Moskowitz et al. 2007). Nkx2-5 was also found to interact with Gata4, a factor with which it shares a common expression pattern from early cardiogenesis (Durocher et al. 1997). A similar cooperative interaction between Gata4 and Tbx5 was identified and found to be sensitive to some patient variants of GATA4, suggesting one possible mechanism of action by which the mutations might affect downstream cardiac development (Garg et al. 2003).
Recent evidence has also drawn attention to the role of chromatin modifiers and epigenetic regulators in the regulation of cardiac development (Zaidi et al. 2013; Homsy et al. 2015). In fact, variants in several chromatin modifiers have been implicated in CHDs, and mouse models continue to shed light on the mechanisms by which mutations in these ubiquitous transcriptional regulators can result in tissue-specific abnormalities (Zaidi and Brueckner 2017). One mechanism by which they affect cardiac development is through cooperativity with TFs. For example, Smarcd3, encoding BAF60C, a subunit of the SWI/SNF-like BAF complex, is expressed specifically in the heart during early mouse development, and its loss results in severe cardiac abnormalities, including defects in development of the OFT and trabeculae of the myocardium (Lickert et al. 2004). Baf60c mediates the interaction between cardiac TFs like Tbx5, Nkx2-5, Gata4, and the core ATPase of the BAF complex, Brg1, as well as the Mef2 cofactor Myocardin (MYOCD) (Takeuchi et al. 2011; Sun et al. 2017). Brg1 itself displays strong dosage sensitivity in heart development that is further exacerbated by the compound loss of TFs Tbx5, Nkx2-5, and Tbx20 (Takeuchi et al. 2011).
Overall, mouse models have provided tremendous understanding into the molecular, structural, and phenotypic aspects of the contribution of these genes. However, often the consequences of heterozygous mutations in human CHD genes are not recapitulated in mouse models. This limitation, as well as challenges with phenotypic variability, argue for the complementary use of in vitro models of disease in which some of these issues can be circumvented. The combined insight from these approaches is likely to facilitate the discovery of better treatment options for CHD patients (Fig. 1).
Figure 1.
Advantages of mouse and iPSC-based models of CHD. Mouse models of CHDs enable investigation of the various cell types found in the heart that otherwise may not be generated using in vitro differentiation methods. Mouse models also provide spatial context, which is key to recapitulate structural defects found in humans. On the other hand, iPSC-based models allow studies to be carried out in the context of human genetic background and lend themselves to large-scale high-throughput assays. (Created with Biorender.com.)
Modeling CHDs in human iPSC-based systems
The combination of human pluripotent stem cells (hPSCs) and genome-editing tools has revolutionized human disease modeling. The isolation of human embryonic stem cells (hESCs) from human blastocysts (Thomson et al. 1998) and subsequent differentiation to cardiomyocytes (Kehat et al. 2001; Xu et al. 2002; He et al. 2003) opened the doors to cardiac disease modeling in human cells. More recently, the generation of induced pluripotent stem cells (iPSCs) from human adult somatic cells (Takahashi et al. 2007) has expanded the access to patient-specific cells for CHD studies. Genome-editing tools such as CRISPR have enabled the generation of hPSC lines harboring disease-relevant heterozygous or homozygous loss-of-function alleles (Hendriks et al. 2020). There are two major approaches to modeling CHD with these tools. The first involves reprogramming of somatic cells from CHD patients and healthy controls to generate iPSC lines, or isolation of patient iPSCs and the correction of the suspected mutation to create isogenic control lines. Alternatively, CRISPR technology can be used to introduce disease-causing mutations in publicly available “normal” iPSC lines such as WTC11 (Lee et al. 2009) or PGP1 (Miyaoka et al. 2014) and hESC lines such as H9 (Thomson et al.1998). These modified hPSC lines can be further differentiated into various cardiac cell types in vitro (Lian et al. 2012, 2013; Burridge et al. 2014) and used to study the mechanisms underlying cellular defects occurring in CHDs. In the following sections, we focus on a few hPSC-based CHD models to highlight the benefits of this system.
Controlling for variations in genetic background
CHD phenotypes in humans are often not recapitulated in mouse models, perhaps due to the differences between the human and mouse genomes. Modeling CHDs in hPSC-derived cells is an advantage because the human genetic background is preserved. However, in the case of patient-derived iPSC lines, variation in genetic background between patients and controls persists as a confounding factor. This issue can be circumvented by using CRISPR to correct the disease-causing mutation in the patient-derived iPSCs to generate isogenic control iPSC lines (Theodoris et al. 2015; Ang et al. 2016). Another approach is to show a direct link between the missing gene and CHD phenotype by introducing the candidate mutation via gene editing (Wang et al. 2014).
Mechanistic insights from iPSC-based CHD models
Cell types derived from diseased iPSCs often recapitulate phenotypes observed in patients, and therefore can provide invaluable insight into disease mechanisms. Despite growing in a dish, iPSC-derived cells are readily amenable to dissection of cellular pathways underlying CHDs via a wide variety of interventions including siRNA knockdown, treatment with inhibitory molecules, biochemical assays, and imaging modalities.
These advantages are exemplified by studies of Barth syndrome (BTHS) in iPSC-derived models. BTHS is a rare X-linked multisystem disorder encompassing dilated cardiomyopathy, skeletal muscle myopathy, growth delay, and neutropenia (Clarke et al. 2013; Saric et al. 2016). Studies of BTHS in human cellular models, including patient-derived fibroblasts (Barth et al. 1996) and lymphoblasts (Gonzalvez et al. 2008), were restricted by the limited supply of patient cells. Therefore, in order to study cardiomyocyte-specific disease pathogenesis in BTHS, Wang et al. (2014) generated two independent iPSC lines from unrelated Barth syndrome patients, which were then differentiated into cardiomyocytes (CMs) in vitro. BTHS is caused by mutations in the TAZ gene (Barth et al. 1983; Bione et al. 1996), which encodes tafazzin, an acyltransferase required for the acylation of a mitochondrial membrane phospholipid called cardiolipin (Neuwald 1997; Vreken et al. 2000). The patient-derived iPSC-CMs (referred to as BTHS iPSC-CMs) recapitulated a hallmark of BTHS: elevated ratios of monolysocardiolipin and mature cardiolipin. As BTHS is a metabolic disorder, the investigators leveraged various cell-based assays to demonstrate that BTHS iPSC-CMs displayed reduced ATP levels, higher oxygen consumption rates, and diminished respiratory capacity. The mitochondrial dysfunction in BTHS iPSC-CMs was further linked to irregularity in sarcomere spacing. The cardiomyopathy often observed in BTHS patients was reflected in the poor contractility of BTHS iPSC-CMs. Interestingly, TAZ gene replacement in BTHS iPSC-CMs returned cardiolipin levels to near normal and restored mitochondrial function, suggesting the potential for gene replacement therapy in the treatment of BTHS. Indeed, adenovirus-mediated TAZ gene replacement therapy in TAZ knockout mice led to reversal of cardiac dysfunction (Wang et al. 2020). These studies showcase the power of human iPSC-based models of CHD to elucidate disease mechanisms and serve as a preclinical testing ground for potential therapeutic approaches.
Scalability of hPSC-based models for ‘omics’ studies in CHDs
As discussed above, many CHDs are caused by mutations in early cardiac TFs such as GATA4, TBX5, and NKX2-5, which steer broad gene expression programs leading to changes in cell identity. Understanding how reduction in TF dosage can alter the transcriptional and epigenomic landscape requires multiomic approaches, such as single-cell RNA-seq, ChIP-seq, and ATAC-seq. Although cardiovascular cells from CHD patients are typically inaccessible for such studies, iPSC-derived cells are a renewable source. In vitro differentiation methods allow researchers to produce millions of diseased cells from hPSCs with relative ease, facilitating such large-scale assays.
A powerful example of such an approach involves the study of the G296S missense mutation in GATA4, which has been linked to fully penetrant atrial or ventricular septal defects, atrioventricular septal defects, and pulmonary stenosis (Garg et al. 2003). To study the effects of this mutation in human cells, Ang et al. (2016) generated iPSCs from four patients harboring G296S. Following cardiac differentiation, G296S CMs displayed defective calcium transients, disorganized sarcomeres, and decreased contractile force, mimicking the cardiomyopathy observed in patients. RNA-seq analysis in mutant iPSCs undergoing CM differentiation highlighted a broad down-regulation of the cardiac gene program, including genes involved in the Hedgehog signaling pathway, cardiac chamber morphogenesis, heart contraction, myofibril assembly, and cardiac progenitor differentiation. Instead, mutant cells showed an up-regulation of genes linked to the endocardial/endothelial development, with a concomitant increase in ATAC-seq signal at these sites, suggesting incomplete silencing of alternative gene programs during cardiac differentiation. In addition, the investigators characterized superenhancers (SEs)—large clusters of enhancers that are critical in controlling gene expression programs that drive cellular identity changes (Whyte et al. 2013). In G296S cells, TBX5 recruitment to SEs was reduced, especially near key cardiac genes such as RBM20, SMYD1, and SRF, suggesting that impaired GATA4–TBX5 interaction in mutant cells was a driving force in the mutant phenotype.
To explore the role of NKX2-5 in human cardiac development, Anderson et al. (2018) established a NKX2-5−/− human embryonic stem cell (hESC)-based model. Although NKX2-5−/− hESCs formed beating CMs, they lacked VCAM1, a marker of cardiac fate commitment, and sustained expression of PDGFRa, a cardiac progenitor marker, indicating stalled differentiation. NKX2-5−/− CMs displayed reduced asynchronous beating with a prolonged action potential duration, as well as reduced contractile force. A combination of gene expression profiling and NKX2-5 chromatin occupancy analysis revealed that the absence of NKX2-5 results in down-regulation of markers of ventricular development such as IRX4, HAND1, HEY2, and MYL2 and up-regulation of smooth muscle markers including MYH11 and TAGLN. Moreover, temporal expression of HEY2 in NKX2-5−/− CMs leads to restoration of VCAM1 expression and repression of MYH11 expression, thus implicating HEY2 as one of the mediators of NKX2-5 function.
Understanding how TF haploinsufficiency may lead to CHD requires identification of genes sensitive to TF dosage in a quantitative manner. As cardiac TFs may orchestrate the expression of a multitude of interconnected genes, building gene regulatory networks can be beneficial in this context. To study the effects of TBX5 dosage during human cardiac differentiation, our group developed a human iPSC-based allelic series consisting of heterozygous (TBX5in/+) and homozygous (TBX5in/del) mutant iPSC lines (Kathiriya et al. 2021). In this case, a mutation in TBX5 was introduced in a well-defined iPSC line, WTC11, by genome editing. As opposed to correcting a patient mutation, this approach does not carry the genetic background of a patient, which allows determination of the effect of a single introduced mutation on cellular phenotype. Single-cell RNA sequencing showed dysregulation of cardiac development genes, electrophysiology-related genes, and numerous human CHD genes, many of which changed in a stepwise fashion from wild type (WT) to TBX5in/+ to TBX5in/del in a TBX5 dose-dependent manner. Gene regulatory network analysis highlighted a human cardiac gene network controlled by TBX5, with changes in biological importance or “pagerank” of several genes in TBX5in/+ and TBX5in/del cells. The gene that showed the highest change in pagerank was MEF2C, and compound heterozygotes of Tbx5 and Mef2c in mice displayed ventricular septal defects. This study highlights the utility of human TF gene networks discovered in a two-dimensional iPSC model in uncovering genetic interactions underlying structural defects in an in vivo model.
In humans, the majority of mutations in GATA6 result in OFT abnormalities (Kodo et al. 2009). In a human iPSC-based model of GATA6 mutations, Sharma et al. (2020) used single-cell transcriptomics to show down-regulated expression of genes critical for OFT development, such as SMYD1, HAND2, and KDR. On the other hand, increased expression of fibroblast markers, epithelial-to-mesenchymal transition markers, and neural development genes indicated that GATA6+/− iPSCs may adopt alternative fates in the absence of normal cardiac signaling. Analysis of ChIP-seq and ATAC-seq data in WT cells showed 88% of GATA-bound peaks overlapped with closed chromatin, suggesting that GATA6 acts as a pioneer cardiac factor. Unlike open chromatin regions, GATA-bound peaks in closed chromatin regions were sensitive to reduction in GATA6 dosage in GATA6+/− cells.
Overall, these studies illustrate the potential for discovery of gene regulatory networks that might be disrupted in human CHDs. They also demonstrate the utility of in vitro iPSC models to understand physiological parameters that are independent of the structural defects found in patients.
iPSC-based disease models for drug discovery: NOTCH1 story
iPSC-derived cells allow high-throughput screening of candidate small molecules that can rescue disease phenotypes. Moreover, mass production of specific cell types from iPSCs enables drug screening to be performed in disease-relevant cell types. Hence, they serve as a promising preclinical platform to discover lifesaving drugs, which is especially beneficial for CHDs, where expensive surgeries are often the only available therapeutic intervention. Here, we discuss the contributions of iPSC models to the development of therapeutic drug candidates for valve disease (Fig. 2).
Figure 2.
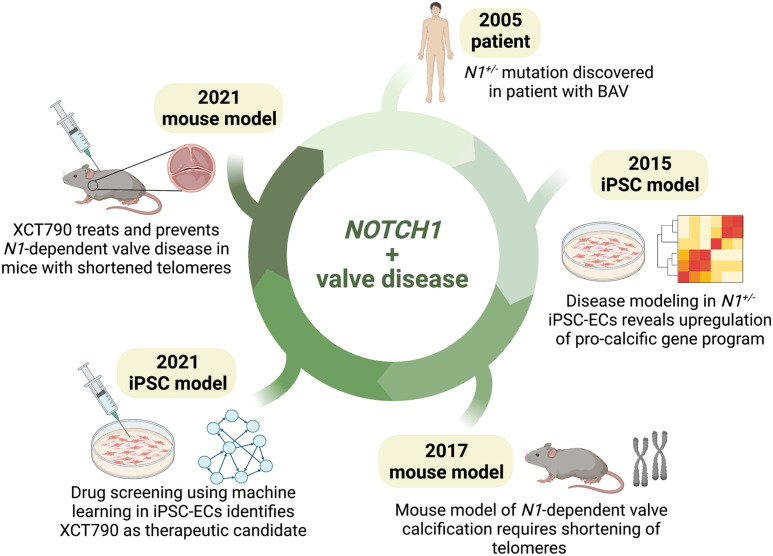
From bedside to bench and back: contributions of mouse and iPSC-based models to NOTCH1-related valve disease and drug development. A summary of discoveries made in mouse and human iPSC-based models demonstrates how contributions from both models have come full circle, leading to the development of therapy for valve disease. Since the discovery of N1+/− mutation in BAV patients, N1-related valve disease has been modeled in iPSC-derived endothelial cells (ECs), which demonstrated up-regulation of a procalcific gene program using single-cell transcriptomics. Modeling N1-dependent valve disease required shortening of telomeres, highlighting why some mouse models may not recapitulate human CHD phenotypes. A drug screen in the iPSC-based model was then used to discover XCT790 as a therapeutic candidate, which corrected aberrant gene networks in N1+/− ECs and treated valve calcification in the mouse model. (Created with Biorender.com.)
Heterozygous NOTCH1 mutations cause bicuspid aortic valve (BAV), a congenital heart defect that affects 1%–2% of the adult population (Garg et al. 2005). Mutations in NOTCH1 also lead to calcific aortic valve disease (CAVD) in humans (Garg et al. 2005), which is the most prevalent heart valve disease worldwide with no available therapies other than surgical valve replacement. Patient tissue samples are only available from individuals with severe aortic valve calcification, hindering studies of early disease progression. Hence, there was a pressing need for a model of NOTCH1 haploinsufficiency for mechanistic studies of CAVD.
Previous studies of valve calcification in mice implicated NOTCH1 signaling in endothelial cells (ECs) (Hofmann et al. 2012). Therefore, Theodoris et al. (2015) set out to establish a human iPSC-derived EC model of CAVD. To this end, they generated iPSCs from CAVD patients with heterozygous NOTCH1 mutations (N1+/−) and then differentiated them into ECs (Theodoris et al. 2015). To mimic the hemodynamic shear stress experienced by valves in the heart, N1+/− iPSC-ECs were subjected to shear stress in vitro. Transcriptomic profiling of WT iPSC-derived ECs subjected to shear stress showed activation of antiosteogenic and anti-inflammatory gene programs, whereas shear-stressed N1+/− ECs up-regulated pro-osteogenic genes such as BMP4 and proinflammatory genes such as IRF6. Furthermore, genes dysregulated in N1+/− ECs were associated with lower levels of bound N1, suggesting sensitivity to TF haploinsufficiency. Combining transcriptomic and epigenetic data in N1+/− ECs, a gene regulatory network was constructed, identifying several key downstream nodes, including procalcific genes SOX7 and TCF4.
Meanwhile, in mice, Theodoris et al. (2017) found that Notch1 heterozygosity did not lead to valve disease. Reasoning that calcification is an age-dependent phenomenon, these investigators bred the Notch1 heterozygous mice to mice lacking telomerase activity. Progressively shortening the length of the telomeres in Notch1 heterozygous mice caused valve stenosis, demonstrating that longer telomeres in mice may slow the progression of age-dependent valve disease. This provided a long-sought mouse model of valve calcification.
Next, Theodoris et al. (2020) used the N1+/− iPSC-derived EC model to perform a network-based drug screen. Wild-type or N1+/− ECs were treated with 1595 small molecules, followed by targeted RNA-seq of 119 genes, consisting of a subset of nodes identified from their previous NOTCH1 gene network (Theodoris et al. 2020). The investigators trained a machine learning algorithm to classify the gene networks of untreated wild-type and N1+/− ECs with 99.3% accuracy. When they used networks from treated N1+/− ECs as input, the algorithm identified 11 small molecules that corrected the aberrant N1+/− EC network to wild-type-like. Of these compounds, XCT790 corrected the most numbers of genes, including central players like SOX7 and TCF4. XCT790 was also effective in correcting the gene networks of primary valve ECs from patients with sporadic valve calcification in the absence of N1 haploinsufficiency, emphasizing XCT790 as a generalizable treatment for all forms of CAVD. Finally, the investigators tested the ability of XCT790 to correct gene networks in vivo in Notch1+/−/mTRG2 mice with valve calcification. Indeed, XCT790 not only restored gene networks in Notch1+/−/mTRG2 mice, but also significantly reduced aortic and pulmonary valve thickness in mice with valve disease. Notch1+/−/mTRG2 mice treated with XCT790 displayed a reduced risk of developing valve calcification, highlighting the use of XCT790 as a preventative medicine that could delay the onset of valve disease. The strength of this study lies in the strategy of using a gene network-level readout to test the efficacy of a drug molecule, rather than focusing on a limited number of genes or phenotypes. While bicuspid valves are irreversible, the use of XCT790 as a preventative and corrective therapy for valve calcification is highly promising and may reduce the clinical burden of valve replacements worldwide.
The series of experiments leading to the discovery of a potential therapeutic for CAVD illustrates the power of human iPSC-based modeling when combined with bioengineering and transcriptional and epigenomic analyses.
Modeling in 3D: the promise of cardiac organoids
Building on success in self-patterning of mouse ESCs into “gastruloids” with patterned early cardiac structures (Rossi et al. 2021), several groups have developed differentiating iPSCs cultured into three-dimensional (3D) structures that could be referred to as cardiac organoids. These have the potential to act as human organ-like models for the study of CHDs (Fig. 3).
Figure 3.
The promise of 3D in vitro models of CHDs. The disadvantages of mouse models include phenotypic variability from human disease. On the other hand, iPSC-based models suffer from the lack of spatial context. Engineered heart tissue, cardiac organoids, and gastruloids hold the promise of overcoming these limitations. (Created with Biorender.com.)
In an approach that used hESCs encapsulated in Matrigel and provided cardiac differentiation cues (e.g., CHIR), Drakhlis et al. (2021) were able to reproducibly derive patterned structures consisting of a noncardiac inner core surrounded by a layer of NKX2-5 and cTnT+ CMs. Immunostaining showed the presence of endothelial cells arranged in what appeared to be vasculature. An additional feature was the presence of foregut-like structures adjacent to the cardiac tissue. The CMs could beat and had electrophysiological properties similar to ventricular CMs. When the investigators used NKX2-5-null ESCs, the cardiac component was larger and exhibited disorganized sarcomeres and altered gene expression. These phenotypes were somewhat reminiscent of the effect of loss of Nkx2-5 in mice but did not indicate how loss of NKX2-5 might alter gene regulation in CHDs.
Drawing from known signaling pathways in vivo and established human pluripotent stem cell differentiation protocols (Kattman et al. 2011), Hofbauer et al. (2021) found that by withdrawing exogenous ECM and finely tuning BMP and WNT signaling, hESCs and iPSCs would reproducibly form beating spherical arrangements that they referred to as “cardioids.” The cardioid model has an interesting feature in that it has what resembles a chamber. Unlike normal heart development, however, the human cardioid “chamber” forms by cavitation. Nonetheless, it has features of a cardiac chamber in that its cellular arrangements are patterned from the inside out. Furthermore, depending on growth factors added, an endocardial lining could be visualized. In a proof of principle experiment, homozygous deletion of the chamber-promoting gene HAND1 led to a failure of cavitation, suggesting that this structure could be used to model CHDs that affect chamber formation.
A very different model involves the spontaneous coemergence of cardiac and gut tissue (Silva et al. 2021). Surrounded by a TBX18+ lining reminiscent of epicardium, these very large organoids develop a central core of beating myocardium, a vasculature, and structures that highly resemble intestinal tissue. Based on single-cell RNA-seq, a classifier trained on human fetal heart data predicted that the CMs in these organoids are atrial. This is an unusual finding, as most cardiac differentiation protocols yield mostly ventricular cardiomyocytes (Lee et al. 2017; Hofbauer et al. 2021; Kathiriya et al. 2021). The resulting CMs also had more mature phenotypes than those that arise in most two-dimensional cultures.
Limitations of CHD models
We have compared and contrasted the model systems used for studying CHDs and described major findings derived from each approach. No single model is a replacement for the others, and the insights from each model should be considered complementary toward our understanding of CHDs.
Mouse models faithfully represent many aspects of human cardiac development, and thus provide incredible insight into the role of specific genes and cell types that contribute to proper cardiogenesis. Despite the advantages that it presents, mutations modeled in mice often do not recapitulate many aspects of the human phenotype. While it is not always apparent what the underlying reasons for these discrepancies might be, additional genetic and environmental variants not captured by individual gene mutations might partially contribute to these differences. Thus, using hPSCs derived from patients may provide insight into these complex mechanisms.
While hPSC-based CHD models are highly scalable human models, there are currently no examples of CHDs that can be modeled only in hPSC-derived cells but not in mice. Rapidly evolving single-cell multiomics technologies now allow efficient capture of cell types from mouse hearts for genomics studies. Moreover, hPSC-based models are two-dimensional and lack the spatial context of the human heart, which is critical for studying structural defects of CHDs. On the other hand, spatial transcriptomics technologies are now facilitating transcriptomics assays in the heart while preserving spatial anatomy (Asp et al. 2019; Mantri et al. 2021).
Although cardiac organoids hold great promise as 3D models of CHD, some aspects of heart physiology such as blood flow cannot be mimicked in a dish. While there are only a few demonstrated examples of cardiac organoids, and they do not fully recapitulate cardiac anatomy, they illustrate that it is possible to generate 3D human structures.
Acknowledgements
We acknowledge support from Gladstone Institutes, Additional Ventures, and the Younger Family Fund.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.349678.122.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Anderson DJ, Kaplan DI, Bell KM, Koutsis K, Haynes JM, Mills RJ, Phelan DG, Qian EL, Leitoguinho AR, Arasaratnam D, et al. 2018. NKX2-5 regulates human cardiomyogenesis via a HEY2 dependent transcriptional network. Nat Commun 9: 1373. 10.1038/s41467-018-03714-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang Y-S, Rivas RN, Ribeiro AJS, Srivas R, Rivera J, Stone NR, Pratt K, Mohamed TMA, Fu J-D, Spencer CI, et al. 2016. Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell 167: 1734–1749.e22. 10.1016/j.cell.2016.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arceci RJ, King AA, Simon MC, Orkin SH, Wilson DB. 1993. Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol Cell Biol 13: 2235–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asp M, Giacomello S, Larsson L, Wu C, Furth D, Qian X, Wardell E, Custodio J, Reimegard J, Salmen F, et al. 2019. Spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell 179: 1647–1660.e19. 10.1016/j.cell.2019.11.025 [DOI] [PubMed] [Google Scholar]
- Barth PG, Scholte HR, Berden JA, Van Der Klei-Van Moorsel JM, Luyt-Houwen IEM, Van'T Veer-Korthof ETh, Van Der Harten JJ, Sobotka-Plojhar MA. 1983. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. J Neurol Sci 62: 327–355. 10.1016/0022-510X(83)90209-5 [DOI] [PubMed] [Google Scholar]
- Barth PG, den Bogert CV, Bolhuis PA, Scholte HR, Gennip AH, Schutgens RBH, Ketel AG. 1996. X-linked cardioskeletal myopathy and neutropenia (Barth syndrome): respiratory-chain abnormalities in cultured fibroblasts. J Inherit Metab Dis 19: 157–160. 10.1007/BF01799418 [DOI] [PubMed] [Google Scholar]
- Basson CT, Cowley GS, Solomon SD, Weissman B, Poznanski AK, Traill TA, Seidman JG, Seidman CE. 1994. The clinical and genetic spectrum of the Holt-Oram syndrome (heart-hand syndrome). N Engl J Med 330: 885–891. 10.1056/NEJM199403313301302 [DOI] [PubMed] [Google Scholar]
- Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, et al. 1997. Mutations in human cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet 15: 30–35. 10.1038/ng0197-30 [DOI] [PubMed] [Google Scholar]
- Benson DW, Sharkey A, Fatkin D, Lang P, Basson CT, McDonough B, Strauss AW, Seidman JG, Seidman CE. 1998. Reduced penetrance, variable expressivity, and genetic heterogeneity of familial atrial septal defects. Circulation 97: 2043–2048. 10.1161/01.CIR.97.20.2043 [DOI] [PubMed] [Google Scholar]
- Benson DW, Silberbach GM, Kavanaugh-McHugh A, Cottrill C, Zhang Y, Riggs S, Smalls O, Johnson MC, Watson MS, Seidman JG, et al. 1999. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J Clin Invest 104: 1567–1573. 10.1172/JCI8154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biben C, Weber R, Kesteven S, Stanley E, McDonald L, Elliott DA, Barnett L, Köentgen F, Robb L, Feneley M, et al. 2000. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2-5. Circ Res 87: 888–895. 10.1161/01.RES.87.10.888 [DOI] [PubMed] [Google Scholar]
- Bione S, D'Adamo P, Maestrini E, Gedeon AK, Bolhuis PA, Toniolo D. 1996. A novel X-linked gene, G4.5. is responsible for Barth syndrome. Nat Genet 12: 385–389. 10.1038/ng0496-385 [DOI] [PubMed] [Google Scholar]
- Bisping E, Ikeda S, Kong SW, Tarnavski O, Bodyak N, McMullen JR, Rajagopal S, Son JK, Ma Q, Springer Z, et al. 2006. Gata4 is required for maintenance of postnatal cardiac function and protection from pressure overload-induced heart failure. Proc Natl Acad Sci 103: 14471–14476. 10.1073/pnas.0602543103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brassington A-ME, Sung SS, Toydemir RM, Le T, Roeder AD, Rutherford AE, Whitby FG, Jorde LB, Bamshad MJ. 2003. Expressivity of Holt-Oram syndrome is not predicted by TBX5 genotype. Am J Hum Genet 73: 74–85. 10.1086/376436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG, Logan M, Davis N, Levi T, Tabin CJ, Seidman JG, Seidman CE. 1999. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev Biol 211: 100–108. 10.1006/dbio.1999.9298 [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, et al. 2001. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106: 709–721. 10.1016/S0092-8674(01)00493-7 [DOI] [PubMed] [Google Scholar]
- Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM, et al. 2014. Chemically defined generation of human cardiomyocytes. Nat Methods 11: 855–860. 10.1038/nmeth.2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman DL, Garvey N, Hancock S, Alexiou M, Agulnik SI, Gibson-Brown JJ, Cebra-Thomas J, Bollag RJ, Silver LM, Papaioannou VE. 1996. Expression of the T-box family genes, Tbx1–Tbx5, during early mouse development. Dev Dyn 206: 379–390. [DOI] [PubMed] [Google Scholar]
- Clarke SL, Bowron A, Gonzalez IL, Groves SJ, Newbury-Ecob R, Clayton N, Martin RP, Tsai-Goodman B, Garratt V, Ashworth M, et al. 2013. Barth syndrome. Orphanet J Rare Dis 8: 23. 10.1186/1750-1172-8-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MW, Guo G, Wolstein O, Vale M, Castro ML, Wang L, Otway R, Riek P, Cochrane N, Furtado M, et al. 2013. Functional characterization of a novel mutation in NKX2-5 associated with congenital heart disease and adult-onset cardiomyopathy. Circ Cardiovas Genet 6: 238–247. 10.1161/CIRCGENETICS.113.000057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine WP, Wythe JD, George M, Koshiba-Takeuchi K, Bruneau BG. 2014. Early patterning and specification of cardiac progenitors in gastrulating mesoderm. Elife 3: e03848. 10.7554/eLife.03848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakhlis L, Biswanath S, Farr C-M, Lupanow V, Teske J, Ritzenhoff K, Franke A, Manstein F, Bolesani E, Kempf H, et al. 2021. Human heart-forming organoids recapitulate early heart and foregut development. Nat Biotechnol 39: 737–746. 10.1038/s41587-021-00815-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durocher D, Charron F, Warren R, Schwartz RJ, Nemer M. 1997. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J 16: 5687–5696. 10.1093/emboj/16.18.5687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DA, Kirk EP, Yeoh T, Chandar S, McKenzie F, Taylor P, Grossfeld P, Fatkin D, Jones O, Hayes P, et al. 2003. Cardiac homeobox gene NKX2-5 mutations and congenital heart disease Associations with atrial septal defect and hypoplastic left heart syndrome. J Am Coll Cardiol 41: 2072–2076. 10.1016/S0735-1097(03)00420-0 [DOI] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, et al. 2003. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424: 443–447. 10.1038/nature01827 [DOI] [PubMed] [Google Scholar]
- Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. 2005. Mutations in NOTCH1 cause aortic valve disease. Nature 437: 270–274. 10.1038/nature03940 [DOI] [PubMed] [Google Scholar]
- Gharibeh L, Komati H, Bossé Y, Boodhwani M, Heydarpour M, Fortier M, Hassanzadeh R, Ngu J, Mathieu P, Body S, et al. 2018. GATA6 regulates aortic valve remodeling, and its haploinsufficiency leads to right-left type bicuspid aortic valve. Circulation 138: 1025–1038. 10.1161/CIRCULATIONAHA.117.029506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharibeh L, Yamak A, Whitcomb J, Lu A, Joyal M, Komati H, Liang W, Fiset C, Nemer M. 2021. GATA6 is a regulator of sinus node development and heart rhythm. Proc Natl Acad Sci 118: e2007322118. 10.1073/pnas.2007322118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford CA, Ranade SS, Samarakoon R, Salunga HT, de Soysa TY, Huang Y, Zhou P, Elfenbein A, Wyman SK, Bui YK, et al. 2019. Oligogenic inheritance of a human heart disease involving a genetic modifier. Science 364: 865–870. 10.1126/science.aat5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalvez F, Schug ZT, Houtkooper RH, MacKenzie ED, Brooks DG, Wanders RJA, Petit PX, Vaz FM, Gottlieb E. 2008. Cardiolipin provides an essential activating platform for caspase-8 on mitochondria. J Cell Biol 183: 681–696. 10.1083/jcb.200803129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JQ, Ma Y, Lee Y, Thomson JA, Kamp TJ. 2003. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ Res 93: 32–39. 10.1161/01.RES.0000080317.92718.99 [DOI] [PubMed] [Google Scholar]
- Heikinheimo M, Scandrett JM, Wilson DB. 1994. Localization of transcription factor GATA-4 to regions of the mouse embryo involved in cardiac development. Dev Biol 164: 361–373. 10.1006/dbio.1994.1206 [DOI] [PubMed] [Google Scholar]
- Hendriks D, Clevers H, Artegiani B. 2020. CRISPR–Cas tools and their application in genetic engineering of human stem cells and organoids. Cell Stem Cell 27: 705–731. 10.1016/j.stem.2020.10.014 [DOI] [PubMed] [Google Scholar]
- Hiroi Y, Kudoh S, Monzen K, Ikeda Y, Yazaki Y, Nagai R, Komuro I. 2001. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat Genet 28: 276–280. 10.1038/90123 [DOI] [PubMed] [Google Scholar]
- Hofbauer P, Jahnel SM, Papai N, Giesshammer M, Deyett A, Schmidt C, Penc M, Tavernini K, Grdseloff N, Meledeth C, et al. 2021. Cardioids reveal self-organizing principles of human cardiogenesis. Cell 184: 3299–3317.e22. 10.1016/j.cell.2021.04.034 [DOI] [PubMed] [Google Scholar]
- Hofmann JJ, Briot A, Enciso J, Zovein AC, Ren S, Zhang ZW, Radtke F, Simons M, Wang Y, Iruela-Arispe ML. 2012. Endothelial deletion of murine Jag1 leads to valve calcification and congenital heart defects associated with Alagille syndrome. Development 139: 4449–4460. 10.1242/dev.084871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, et al. 2015. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 350: 1262–1266. 10.1126/science.aac9396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Lock JE, Marshall AC, Basson C, Seidman JG, Seidman CE. 2002. Causes of clinical diversity in human TBX5 mutations. Cold Spring Harb Symp Quant Biol 67: 115–120. 10.1101/sqb.2002.67.115 [DOI] [PubMed] [Google Scholar]
- Jay PY, Harris BS, Maguire CT, Buerger A, Wakimoto H, Tanaka M, Kupershmidt S, Roden DM, Schultheiss TM, O'Brien TX, et al. 2004. Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J Clin Invest 113: 1130–1137. 10.1172/JCI19846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiriya IS, Rao KS, Iacono G, Devine WP, Blair AP, Hota SK, Lai MH, Garay BI, Thomas R, Gong HZ, et al. 2021. Modeling human TBX5 haploinsufficiency predicts regulatory networks for congenital heart disease. Dev Cell 56: 292–309.e9. 10.1016/j.devcel.2020.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. 2011. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8: 228–240. 10.1016/j.stem.2010.12.008 [DOI] [PubMed] [Google Scholar]
- Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A, Livne E, Binah O, Itskovitz-Eldor J, Gepstein L. 2001. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest 108: 407–414. 10.1172/JCI12131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodo K, Nishizawa T, Furutani M, Arai S, Yamamura E, Joo K, Takahashi T, Matsuoka R, Yamagishi H. 2009. GATA6 mutations cause human cardiac outflow tract defects by disrupting semaphorin-plexin signaling. Proc Natl Acad Sci 106: 13933–13938. 10.1073/pnas.0904744106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro I, Izumo S. 1993. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc Natl Acad Sci 90: 8145–8149. 10.1073/pnas.90.17.8145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsourakis M, Langeveld A, Patient R, Beddington R, Grosveld F. 1999. The transcription factor GATA6 is essential for early extraembryonic development. Development 126: 723–732. 10.1242/dev.126.4.723 [DOI] [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. 1997. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Gene Dev 11: 1048–1060. 10.1101/gad.11.8.1048 [DOI] [PubMed] [Google Scholar]
- Laverriere AC, MacNeill C, Mueller C, Poelmann RE, Burch JB, Evans T. 1994. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J Biol Chem 269: 23177–23184. 10.1016/S0021-9258(17)31636-8 [DOI] [PubMed] [Google Scholar]
- Lee J-H, Park I-H, Gao Y, Li JB, Li Z, Daley GQ, Zhang K, Church GM. 2009. A robust approach to identifying tissue-specific gene expression regulatory variants using personalized human induced pluripotent stem cells. PLoS Genet 5: e1000718. 10.1371/journal.pgen.1000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Protze SI, Laksman Z, Backx PH, Keller GM. 2017. Human pluripotent stem cell-derived atrial and ventricular cardiomyocytes develop from distinct mesoderm populations. Cell Stem Cell 21: 179–194.e4. 10.1016/j.stem.2017.07.003 [DOI] [PubMed] [Google Scholar]
- Lepore JJ, Mericko PA, Cheng L, Lu MM, Morrisey EE, Parmacek MS. 2006. GATA-6 regulates semaphorin 3C and is required in cardiac neural crest for cardiovascular morphogenesis. J Clin Invest 116: 929–939. 10.1172/JCI27363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QY, Newbury-Ecob RA, Terrett JA, Wilson DI, Curtis ARJ, Yi CH, Gebuhr T, Bullen PJ, Robson SC, Strachan T, et al. 1997. Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet 15: 21–29. 10.1038/ng0197-21 [DOI] [PubMed] [Google Scholar]
- Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. 2012. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci 109: E1848–E1857. 10.1073/pnas.1200250109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP. 2013. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc 8: 162–175. 10.1038/nprot.2012.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. 2004. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 432: 107–112. 10.1038/nature03071 [DOI] [PubMed] [Google Scholar]
- Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP. 1993. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 119: 419–431. [DOI] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. 1995. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeobox gene Nkx2-5. Gene Dev 9: 1654–1666. 10.1101/gad.9.13.1654 [DOI] [PubMed] [Google Scholar]
- Mantri M, Scuderi GJ, Abedini-Nassab R, Wang MFZ, McKellar D, Shi H, Grodner B, Butcher JT, De Vlaminck I. 2021. Spatiotemporal single-cell RNA sequencing of developing chicken hearts identifies interplay between cellular differentiation and morphogenesis. Nat Commun 12: 1771. 10.1038/s41467-021-21892-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y, Chan AH, Judge LM, Yoo J, Huang M, Nguyen TD, Lizarraga PP, So P-L, Conklin BR. 2014. Isolation of single-base genome-edited human iPS cells without antibiotic selection. Nat Methods 11: 291–293. 10.1038/nmeth.2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. 1997. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Gene Dev 11: 1061–1072. 10.1101/gad.11.8.1061 [DOI] [PubMed] [Google Scholar]
- Mori AD, Zhu Y, Vahora I, Nieman B, Koshiba-Takeuchi K, Davidson L, Pizard A, Seidman JG, Seidman CE, Chen XJ, et al. 2006. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev Biol 297: 566–586. 10.1016/j.ydbio.2006.05.023 [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Ip HS, Lu MM, Parmacek MS. 1996. GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev Biol 177: 309–322. 10.1006/dbio.1996.0165 [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Ip HS, Tang Z, Lu MM, Parmacek MS. 1997. GATA-5: a transcriptional activator expressed in a novel temporally and spatially-restricted pattern during embryonic development. Dev Biol 183: 21–36. 10.1006/dbio.1996.8485 [DOI] [PubMed] [Google Scholar]
- Morton SU, Quiat D, Seidman JG, Seidman CE. 2022. Genomic frontiers in congenital heart disease. Nat Rev Cardiol 19: 26–42. 10.1038/s41569-021-00587-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz IPG, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, Nobrega MA, Yokota Y, Berul C, Izumo S, et al. 2007. A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell 129: 1365–1376. 10.1016/j.cell.2007.04.036 [DOI] [PubMed] [Google Scholar]
- Nees SN, Chung WK. 2019. Genetic basis of human congenital heart disease. Cold Spring Harp Perspect Biol 12: a036749. 10.1101/cshperspect.a036749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF. 1997. Barth syndrome may be due to an acyltransferase deficiency. Curr Biol 7: R462–R466. 10.1016/S0960-9822(06)00237-5 [DOI] [PubMed] [Google Scholar]
- Newbury-Ecob RA, Leanage R, Raeburn JA, Young ID. 1996. Holt-Oram syndrome: a clinical genetic study. J Med Genet 33: 300–307. 10.1136/jmg.33.4.300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu WT, Ishiwata T, Juraszek AL, Ma Q, Izumo S. 2004. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev Biol 275: 235–244. 10.1016/j.ydbio.2004.08.008 [DOI] [PubMed] [Google Scholar]
- Rajagopal SK, Ma Q, Obler D, Shen J, Manichaikul A, Tomita-Mitchell A, Boardman K, Briggs C, Garg V, Srivastava D, et al. 2007. Spectrum of heart disease associated with murine and human GATA4 mutation. J Mol Cell Cardiol 43: 677–685. 10.1016/j.yjmcc.2007.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi G, Broguiere N, Miyamoto M, Boni A, Guiet R, Girgin M, Kelly RG, Kwon C, Lutolf MP. 2021. Capturing cardiogenesis in gastruloids. Cell Stem Cell 28: 230–240.e6. 10.1016/j.stem.2020.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saric A, Andreau K, Armand A-S, Møller IM, Petit PX. 2016. Barth syndrome: from mitochondrial dysfunctions associated with aberrant production of reactive oxygen species to pluripotent stem cell studies. Front Genet 6: 359. 10.3389/fgene.2015.00359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkozy A, Conti E, Neri C, D'Agostino R, Digilio MC, Esposito G, Toscano A, Marino B, Pizzuti A, Dallapiccola B. 2005. Spectrum of atrial septal defects associated with mutations of NKX2.5 and GATA4 transcription factors. J Med Genet 42: e16. 10.1136/jmg.2004.026740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott J-J, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. 1998. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science 281: 108–111. 10.1126/science.281.5373.108 [DOI] [PubMed] [Google Scholar]
- Sharma A, Wasson LK, Willcox JA, Morton SU, Gorham JM, DeLaughter DM, Neyazi M, Schmid M, Agarwal R, Jang MY, et al. 2020. GATA6 mutations in hiPSCs inform mechanisms for maldevelopment of the heart, pancreas, and diaphragm. Elife 9: e53278. 10.7554/eLife.53278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AC, Matthys OB, Joy DA, Kauss MA, Natarajan V, Lai MH, Turaga D, Blair AP, Alexanian M, Bruneau BG, et al. 2021. Co-emergence of cardiac and gut tissues promotes cardiomyocyte maturation within human iPSC-derived organoids. Cell Stem Cell 28: 2137–2152.e6. 10.1016/j.stem.2021.11.007 [DOI] [PubMed] [Google Scholar]
- Sun X, Hota SK, Zhou Y-Q, Novak S, Miguel-Perez D, Christodoulou D, Seidman CE, Seidman JG, Gregorio CC, Henkelman RM, et al. 2017. Cardiac-enriched BAF chromatin-remodeling complex subunit Baf60c regulates gene expression programs essential for heart development and function. Biol Open 7: bio029512. 10.1242/bio.029512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Lou X, Alexander JM, Sugizaki H, Delgado-Olguín P, Holloway AK, Mori AD, Wylie JN, Munson C, Zhu Y, et al. 2011. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat Commun 2: 187. 10.1038/ncomms1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Berul CI, Ishii M, Jay PY, Wakimoto H, Douglas P, Yamasaki N, Kawamoto T, Gehrmann J, Maguire CT, et al. 2002. A mouse model of congenital heart disease: cardiac arrhythmias and atrial septal defect caused by haploinsufficiency of the cardiac transcription factor Csx/Nkx2.5. Cold Spring Harb Symp Quant Biol 67: 317–326. 10.1101/sqb.2002.67.317 [DOI] [PubMed] [Google Scholar]
- Theodoris CV, Li M, White MP, Liu L, He D, Pollard KS, Bruneau BG, Srivastava D. 2015. Human disease modeling reveals integrated transcriptional and epigenetic mechanisms of NOTCH1 haploinsufficiency. Cell 160: 1072–1086. 10.1016/j.cell.2015.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoris CV, Mourkioti F, Huang Y, Ranade SS, Liu L, Blau HM, Srivastava D. 2017. Long telomeres protect against age-dependent cardiac disease caused by NOTCH1 haploinsufficiency. J Clin Invest 127: 1683–1688. 10.1172/JCI90338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoris CV, Zhou P, Liu L, Zhang Y, Nishino T, Huang Y, Kostina A, Ranade SS, Gifford CA, Uspenskiy V, et al. 2020. Network-based screen in iPSC-derived cells reveals therapeutic candidate for heart valve disease. Science 371: eabd0724. 10.1126/science.abd0724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282: 1145–1147. 10.1126/science.282.5391.1145 [DOI] [PubMed] [Google Scholar]
- van der Linde D, Konings EEM, Slager MA, Witsenburg M, Helbing WA, Takkenberg JJM, Roos-Hesselink JW. 2011. Birth prevalence of congenital heart disease worldwide a systematic review and meta-analysis. J Am Coll Cardiol 58: 2241–2247. 10.1016/j.jacc.2011.08.025 [DOI] [PubMed] [Google Scholar]
- Vreken P, Valianpour F, Nijtmans LG, Grivell LA, Plecko B, Wanders RJA, Barth PG. 2000. Defective remodeling of cardiolipin and phosphatidylglycerol in Barth syndrome. Biochem Biophys Res Commun 279: 378–382. 10.1006/bbrc.2000.3952 [DOI] [PubMed] [Google Scholar]
- Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L, et al. 2014. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med 20: 616–623. 10.1038/nm.3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Li Y, Xu Y, Ma Q, Lin Z, Schlame M, Bezzerides VJ, Strathdee D, Pu WT. 2020. AAV gene therapy prevents and reverses heart failure in a murine knockout model of Barth syndrome. Circ Res 126: 1024–1039. 10.1161/CIRCRESAHA.119.315956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. 10.1016/j.cell.2013.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Police S, Rao N, Carpenter MK. 2002. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ Res 91: 501–508. 10.1161/01.RES.0000035254.80718.91 [DOI] [PubMed] [Google Scholar]
- Yang Y-Q, Li L, Wang J, Zhang X-L, Li R-G, Xu Y-J, Tan H-W, Wang X-H, Jiang J-Q, Fang W-Y, et al. 2012. GATA6 loss-of-function mutation in atrial fibrillation. Eur J Med Genet 55: 520–526. 10.1016/j.ejmg.2012.06.007 [DOI] [PubMed] [Google Scholar]
- Zaidi S, Brueckner M. 2017. Genetics and genomics of congenital heart disease. Circ Res 120: 923–940. 10.1161/CIRCRESAHA.116.309140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, et al. 2013. De novo mutations in histone-modifying genes in congenital heart disease. Nature 498: 220–223. 10.1038/nature12141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Watt AJ, Battle MA, Li J, Bondow BJ, Duncan SA. 2008. Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Dev Biol 317: 614–619. 10.1016/j.ydbio.2008.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Gramolini AO, Walsh MA, Zhou Y-Q, Slorach C, Friedberg MK, Takeuchi JK, Sun H, Henkelman RM, Backx PH, et al. 2008. Tbx5-dependent pathway regulating diastolic function in congenital heart disease. Proc Natl Acad Sci 105: 5519–5524. 10.1073/pnas.0801779105 [DOI] [PMC free article] [PubMed] [Google Scholar]