Abstract
The disulfide linkages of two etanercept products, Enbrel® (innovator drug) and TuNEX®, were characterized and compared using a multi-fragmentation approach consisting of electron transfer dissociation (ETD) and collision induced dissociation (CID) in combination with multi-enzyme digestion protocols (from Lys-C, trypsin, Glu-C, and PNGase F). Multi-fragmentation approach allowed multi-disulfide linkages contained in a peptide to be un-ambiguously assigned based on the cleavage of both the disulfide and the backbone linkages in a MS3 schedule. New insights gained using this approach were discussed. A total of 29 disulfides, Cys18-Cys31, Cys32-Cys45, Cys35-Cys53, Cys56-Cys71, Cys74-Cys88, Cys78-Cys-96, Cys98-Cys104, Cys112-Cys121, Cys115-Cys139, Cys-142-Cys157, Cys163-Cys178 in TNFR portion and Cys240-Cys240, Cys246-Cys246, Cys249-Cys249, Cys281-Cys341, Cys387-Cys445 in IgG1 Fc domain, were completely assigned with the demonstration of the same disulfide linkages between the Enbrel® and TuNEX® products. The data showed the higher order structure was preserved throughout the recombinant manufacturing processes and consistent between the two products.
Keywords: Enbrel®, TuNEX®, LC-MS, CID, ETD
1. Introduction
Disulfide linkages are directly involved in appropriate protein conformations and therefore have a significant influence on protein functions. As a result, regulatory agencies expect disulfide connectivity to be determined as part of product characterization [1,2]. Etanercept is a fusion protein that binds strongly to soluble tumor necrosis factor alpha (TNF-α). It consists of a recombinant human TNF receptor 2 bound to the Fc portion of an immunoglobulin, and was the first TNF-α antagonist approved in 1998 for moderate to severe plaque psoriasis, psoriatic arthritis, ankylosing spondylitis and rheumatoid arthritis [3,4]. Except complicated N/O-glycan forms in the hinge and Fc region [5,6], etanercept molecule contains thirteen intra-chain and three inter-chain disulfide bonds, the majority of which are located in the TNF receptor region of the molecule [2,7]. Such complicated disulfide bridging in etanercept represents a new quality attribute that is critical for biopharmaceutical functionality, and should thus be carefully monitored and controlled to guarantee patient safety [7].
Mapping highly complicated disulfide linkages such as etanercept is challenging. Currently, different strategies have been used to identify disulfide bonds like comparison of LC-MS/MS between a reduced and non-reduced protein digest [8–13]. Partial reduction and differential alkylation [14–17] or chemical labeling [18,19] have been applied in sample preparation to help the analysis of the disulfide bonds. Collision-induced dissociation (CID) in LC-MS is the most common means of fragmentation to derive polypeptide structure information [20–23]. However, modifications such as disulfide bonds are not typically fragmented by CID [22,24–26]. A triple digestion method has been utilized to map the disulfide linkages of etanercept by reducing the number of linkages of a native peptide digest such that the linkage could be assigned to the only two remaining cysteine residues on the peptide [26,27]. Nevertheless, with only limited or no peptide backbone sequence information gained by CID, it is hard to confirm the sequences that were linked by disulfides.
Alternatively, electron-transfer dissociation (ETD) has been shown to break disulfide linkages and allowed the linked sequences to be assigned unambiguously via MS3 [28,29]. Our goal is thus to use modern ETD technique coupled with ETD/CID method to characterize both the innovator etanercept drug (Enbrel®) and its biosimilar (TuNEX®). Results derived from this study using multiple fragmentations including ETD and CID were compared to that reported by using CID alone.
2. Experimental
2.1. Samples
TuNEX® was produced by Chinese Hamster Ovary (CHO) cells as a dimeric, secreted, soluble protein. After collection for several days, the expressed protein was purified by a sequential downstream process including several validated r-protein A chromatographic, ion-exchange chromatographic and ultrafiltration steps combined with viral filtration. The TuNEX® lot# 02F09 was provided as 1.0 mg/mL × 6 mL for characterization. Enbrel®, lot# 02S03, was obtained from Pfizer, and provided as 1.0 mg/mL × 2 mL. Each sample was aliquoted as 50 μL per vial (50 μg) and stored at −80 °C before analysis.
2.2. Reagents
Sequencing-grade trypsin was purchased from Promega (Madison, WI, USA). Achromobacter protease I (Lys-C) and Glu-C were obtained from Roche (Nutley, NJ, USA). PNGase F, ammonium bicarbonate (NH4HCO3), and formic acid (FA) were purchased from Sigma–Aldrich (St. Louis, MO, USA). LC-MS grade acetonitrile (ACN) was purchased from J. T Baker (Phillipsburg, NJ, USA). Amicon centrifugal filter (10 kDa MWCO) was obtained from Millipore (Bedford, MA, USA), and HPLC water was deionized to 18 MΩ by a Millipore Milli-Q system.
2.3. Enzymatic digestion
Protein solution (1.0 mg/mL of 100 μL) was buffer exchanged with 100 mM ammonium bicarbonate (pH 8 or pH 6.8) using a 10 kDa molecular weight cutoff filter to a concentration of 1 mg/mL (100 μL). In order to assess potential disulfide scrambling, in addition to pH 8, a slightly less than alkaline pH (pH 6.8) was also used in the enzymatic digestion protocol to examine whether any alternative disulfide linkages could be observed from the two different pH digestion conditions [20]. For trypsin digestion, trypsin (1:50, w/w) was added to the protein solution at room temperature for 8 h and then added a second time (1:50, w/w) for 12 h at room temperature. For Lys-C digestion, the endoproteinase Lys-C (1:50, w/w) was added to the protein solution for 4 h at 37 °C. For Lys-C plus trypsin digestion, the protein solution was added with endoproteinase Lys-C (1:50 w/w) and trypsin (1:50 w/w) for 24 h at room temperature. For Lys-C plus trypsin plus Glu-C digestion, the protein solution was added with endoproteinase Lys-C (1:50 w/w), trypsin (1:50 w/w), and Glu-C (1:20 w/w) for 24 h at room temperature. Similarly, for additional PNGase F digestion, PNGase F (10 units/mg) was added to the above enzyme solution for 24 h at room temperature. In all cases, digestion was terminated by addition of 1% formic acid. An aliquot of 2 μg of the enzyme digest was analyzed per LC-MS run.
2.4. LC-MS
An Agilent 1200 nano-HPLC system (Agilent, Santa Clara, CA, USA) with a Agilent pre-column (300 μm × 5 mm, 5 μm C18), and an analytical column (75 μm i.d. × 15 cm, 3 μm C18) was coupled online to an LTQ-Orbitrap-ETD XL mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA). Mobile phase A (0.1% formic acid in water) and mobile phase B (0.1% formic acid in acetonitrile) were used for the gradient consisted of (i) 5 min at 2% B for sample loading, (ii) linear from 2% to 40% B over 40 min, (iii) linear from 40% to 80% B over 10 min, and finally (iv) isocratic at 80% B for 10 min. Flow rate was maintained at 1 μL/min for the pre-column and 200 nL/min for the analytical column. The LTQ-Orbitrap-ETD XL mass spectrometer was operated in the data-dependent mode to switch automatically between MS (scan 1 at the Orbitrap), CID-MS2 (scan 2 at the LTQ), and ETD-MS2 (scan 3 at the LTQ). Briefly, after a survey MS spectrum from m/z 300 to m/z 2000, subsequent CID-MS2 and ETD-MS2 steps were performed on the same precursor ion with a ±2.5 m/z isolation width. Any inadequate information (assignment) in the CID-MS2 and ETD-MS2 spectra will be repeated by targeting the desired ions, e.g. the same precursor but with different charge state, in order to gain additional linkage information. These targeted approach will be repeated (e.g. targeting multiple charges of a precursor ion or the same disulfide-linked peptide but with different enzymatic cleavages or miscleavages) until the linkage information was adequate. In addition, a targeted CID-MS3 after ETD for the ions of interest was further performed as necessary.
2.5. Disulfide assignment
The anticipated disulfide-linked tryptic or multi-enzyme digested peptide masses with different charges were first calculated using free software (http://prospector.ucsf.edu/prospector/cgibin/msform.cgi?form=msproduct), and then matched to the observed masses in the LC-MS chromatogram. The matched masses (with <5 ppm mass accuracy) were further confirmed by the corresponding CID-MS2 and ETD-MS2 fragmentation manually, as well as by CID-MS3 fragmentation, as needed.
3. Results and discussion
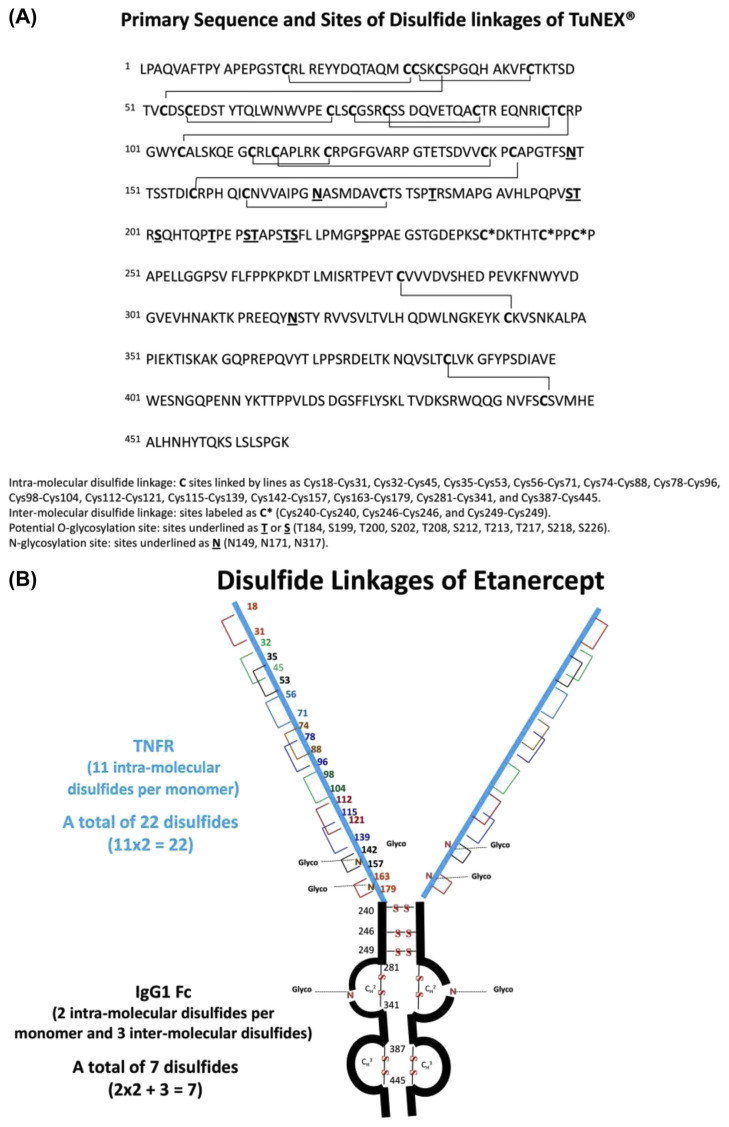
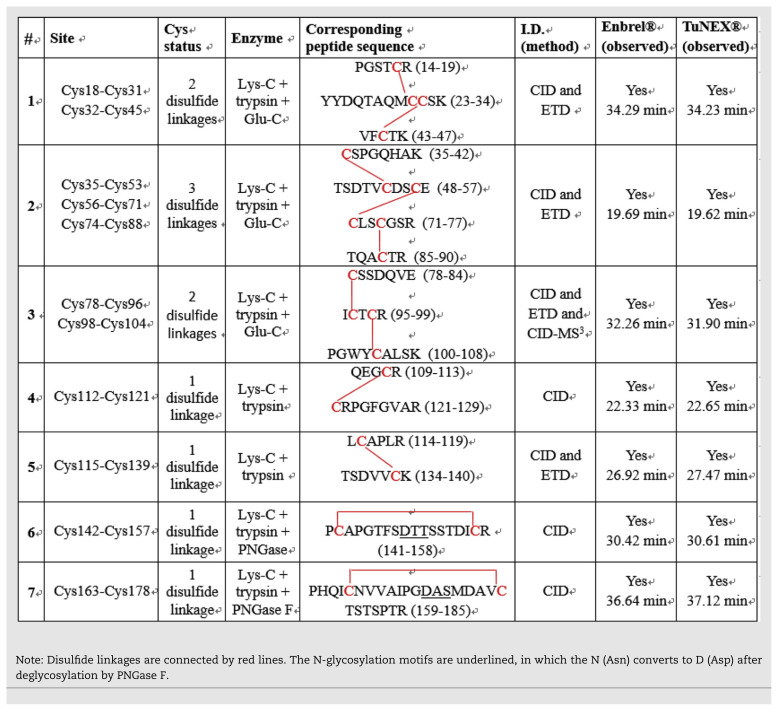
The fusion antibody (Enbrel® or TuNEX®) contains two identical pairs of monomers, which are connected by intermolecular and intra-molecular disulfides. The only obvious difference between the two products was a two-AA residue variance in the heavy chain (Fc) [30]. This, however, was not expected to affect the disulfide linkages. For illustration, the primary structure of TuNEX® with the sites of expected disulfide linkages is shown in Fig. 1A. For each monomer, there are two domains consisting of TNFR and IgG1 Fc (Fig. 1B). The TNFR domain contains only intra-molecular disulfides, a total of 22 disulfides, derived from the 11 intra-molecular disulfides multiply by 2 (2 identical monomers). The IgG1 Fc domain contains 4 intra-molecular disulfides (multiply by 2 equals 4) and 3 inter-molecular disulfides, a total of 7 disulfides. Thus, etanercept was expected to contain a total of 29 disulfides, as shown in Fig. 1B.
Fig. 1.
(A) Primary sequence and sites of disulfide linkages of TuNEX®. (B) Disulfide linkages of Etanercept.
For such a complicated disulfide structure, a proper enzyme digestion strategy is needed. In principle, identification of a single disulfide linkage is straight forward because there is usually only one possibility for connection. Consequently, enzymes, which can cut the protein to the peptide size containing only single disulfide, are desired. However, the intertwined disulfides or the protein’s backbone sequence may prevent enzyme digestion to the desired peptide size. In addition, peptide sizes are preferred to be 1–5 kDa. Recovery and electrospray ionization efficiency can be a problem for larger peptides, while smaller peptides may not retain well on a typical reversed phase column. Thus, selection of proper enzymes or multiple enzymes should be considered for the size adjustment. Also, for the disulfide-linked peptides containing N-glycosylation, additional PNGase F digestion needs to be considered to remove the N-linked glycans in order to reduce complexity. After surveying several enzyme combinations, a cocktail protocol, which combined Lys-C plus trypsin or Lys-C plus trypsin plus Glu-C, was found to work effectively for most of the disulfide analysis. With additional PNGase F, the disulfide analysis was successful for the peptides with N-glycosylation as well. It should be noted that enzymatic cleavages of the protein are the same by using either trypsin or Lys-C plus trypsin. Nevertheless, the use of Lys-C plus trypsin seems to yield slightly higher digestion efficiency than trypsin alone. Similarly, the use of PNGase F enzyme would not yield additional cleavages except the deglycosylation of N-glycosylated sites. The addition of PNGase F, even for the disulfides without N-glycosylation, was used to improve the digestion efficiency as needed.
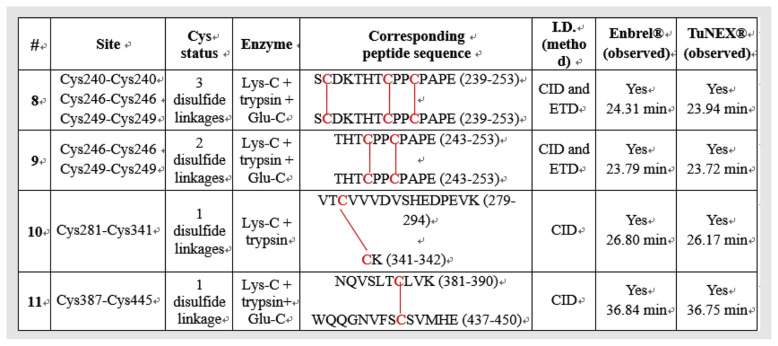
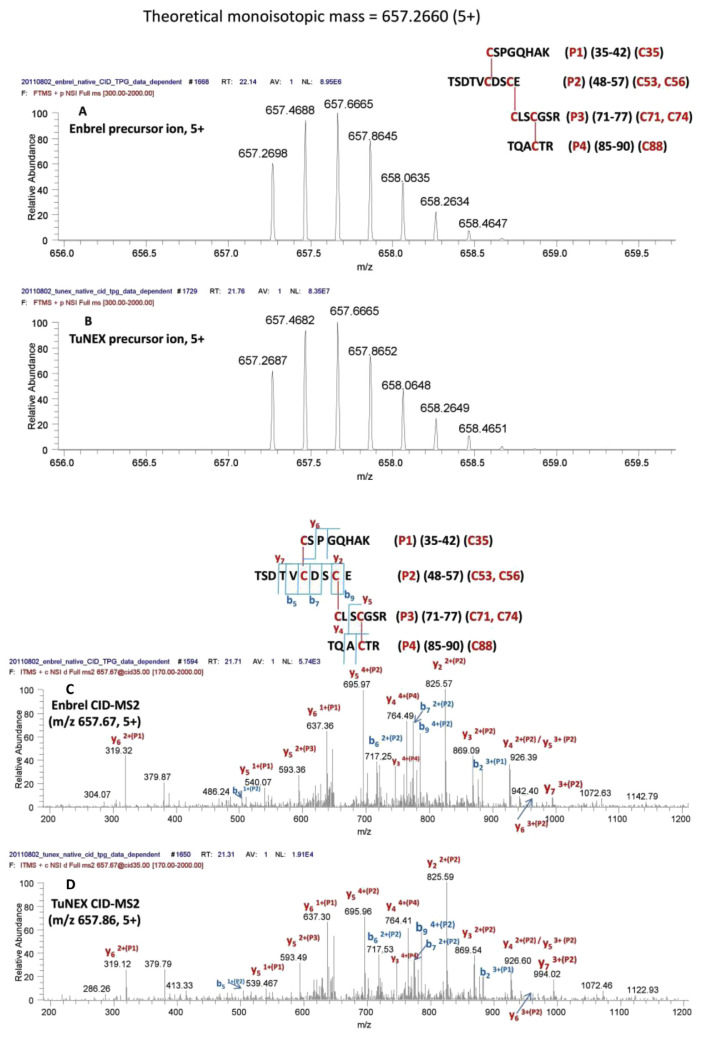
Table 1A lists the identified disulfide linkages of Enbrel® and TuNEX® from TNFR portion, respectively. Table 1B lists the identified disulfide linkages of Enbrel® and TuNEX® from IgG1 Fc domain, respectively. The linkage sites, corresponding peptide sequences, digest enzymes, identification methods by mass spectrometry, and observed positions in LC-MS analysis of Enbrel® and TuNEX® are summarized in the table. The detailed identification is described in the following text.
Table 1A.
Summary of disulfide linkages identification at TNFR of Etanercept for both Enbrel® and TuNEX®.
Table 1B.
Summary of disulfide linkages identification at IgG1 Fc of Etanercept for both Enbrel® and TuNEX®.
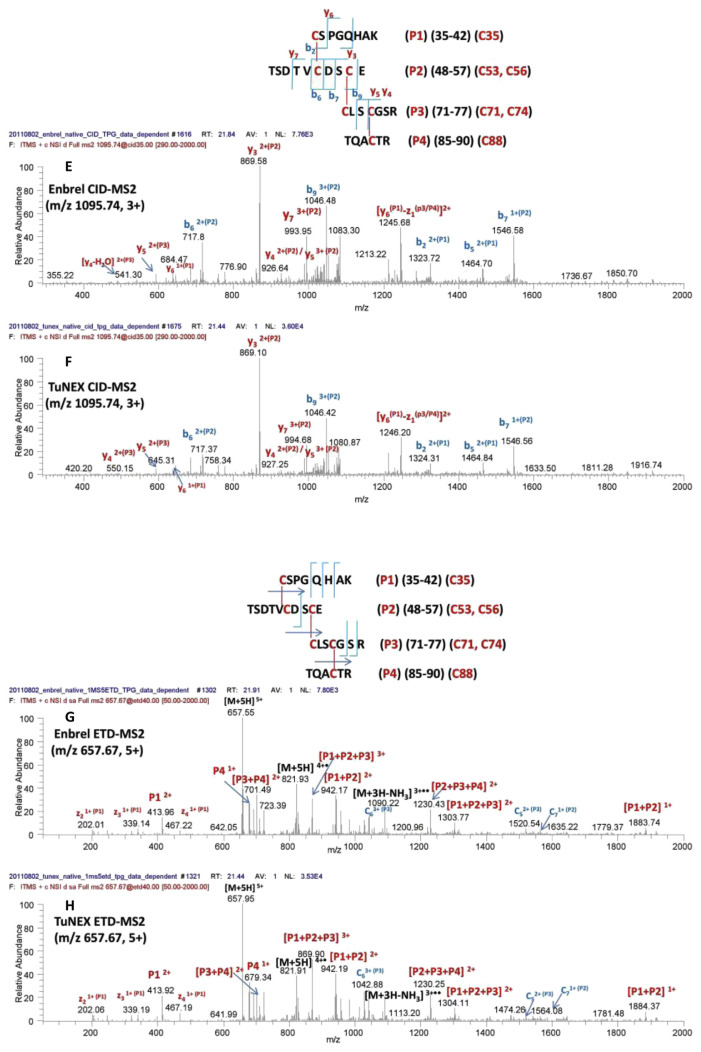
For the peptide with the three disulfides (Cys35-Cys53, Cys56-Cys71, and Cys74-Cys88, #2 in Table 1A), the same digestion protocol and identification methods were used. Again, indistinguishable mass (Fig. 2A,B) and fragmentation patterns (Fig. 2C–F) were observed for both Enbrel® and TuNEX® in this disulfide-linked peptide. For the linkage assignment, the CID fragmentation of the same disulfide-linked peptide but with different charge states were shown, i.e. m/z 657.67 of 5+ (Fig. 2C,D) and m/z 1095.74 of 3+ (Fig. 2E,F), with the identification of the same cleavage for disulfide linkage sites (i.e. y5 on P3 peptide). The fragmentation from the two different charge states was used to prove that the site assignment was correct not only with respect to the cleavages but also with the consistency. All these information provided strong evidence again that both products have the same disulfides as shown.
Fig. 2.
Mass and charge of Lys-C + trypsin + Glu-C-digested peptide with the three disulfides (Cys35-Cys53, Cys56-Cys71, and Cys74-Cys88), for Enbrel® (A) and TuNEX® (B). The sequence and theoretical mass of the peptide along with the observed monoisotopic (1st isotopic) mass are indicated in the insert. CID-MS2 spectrum of the precursor for Enbrel® (C) and TuNEX® (D). CID-MS2 spectrum of the precursor but with a different charge state (3+) from Figure 2A, B for Enbrel® (E) and TuNEX® (F). ETD-MS2 spectrum of the precursor for Enbrel® (G) and TuNEX®(H).
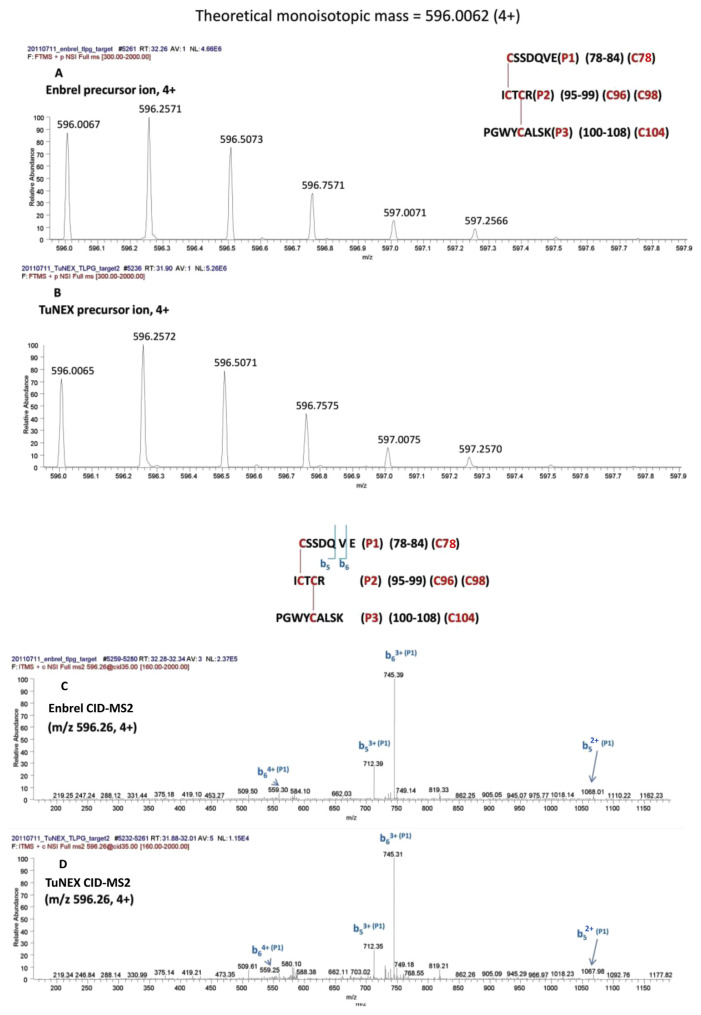
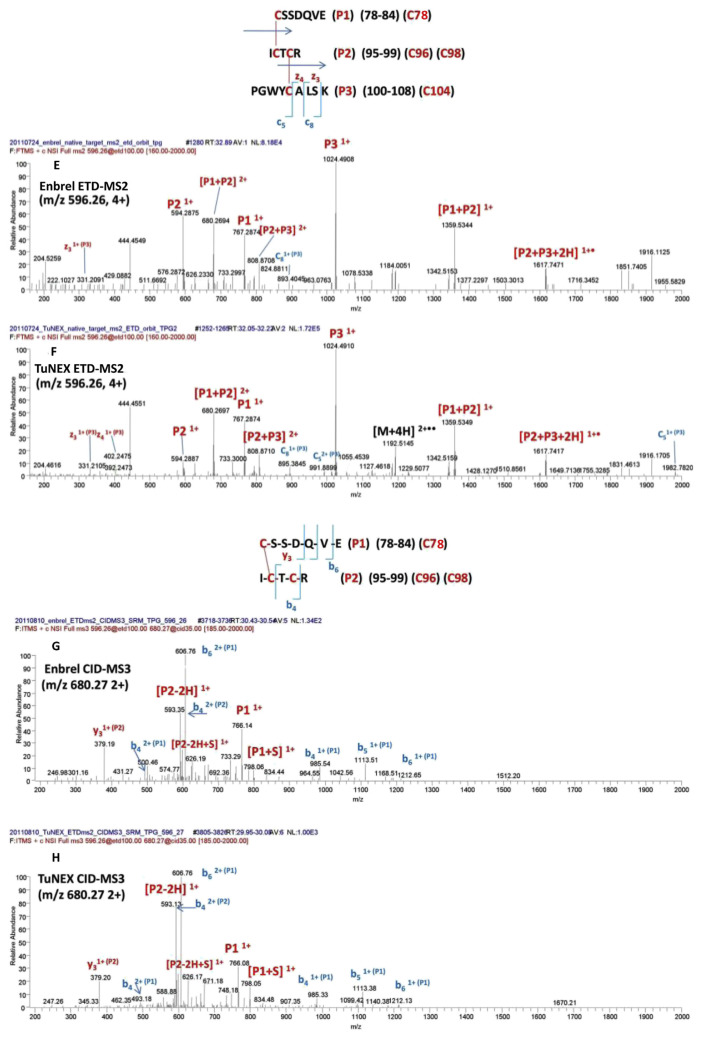
Similarly, for the peptide with the two disulfides (Cys78-Cys96 and Cys98-Cys104, #3 in Table 1A), the digestion protocol required 3 enzymes (the combination of Lys-C plus trypsin plus Glu-C). This cocktail digestion protocol cut the disulfide-linked peptide to a proper size effectively for fragmentation by mass spectrometry. As shown in Fig. 3A (Enbrel®) and 3B (TuNEX®), the corresponding mass and charge of the precursor ion from both products and were illustrated with the accurate monoisotopic mass within 1 ppm for both products (i.e. m/z 596.0067 for Enbrel® and m/z 596.0065 for TuNEX® at 4 + charge state). In the figure, both monoisotopic masses also accurately matched the theoretical monoisotopic mass (m/z 596.0062), which was the peptide with the two disulfides (a loss of 4H from the backbone sequence). The disulfide linkage information was obtained by CID-MS2 and ETD-MS2 of the precursor ion, as illustrated in Fig. 3C,E (Enbrel®), as well as Fig. 3D,F (TuNEX®), and by CID-MS3 of the precursor after ETD-MS2. In the linkage assignment, the corresponding CID-MS2 spectrum provided the sequence information (b ions). In addition to CID-MS2, the corresponding ETD-MS2 confirmed that the three peptides were linked together (the observation of P1, P2, and P3 peptides), and confirmed further by the partially disulfide-dissociated peptides (i.e. P1 + P2 and P2 + P3). As shown by ETD (Fig. 3E,F), the linked peptides were P1 with P2, and P2 with P3, but not P1 with P3. We chose m/z 680.27 of 2+ (Fig. 3E,F) to do CID-MS3 (Fig. 3G,H). The CID-MS3 provided us the information of disulfide linkage (y3 on P2 peptide). The fragmentation was used to prove Cys78-Cys96. And the remaining Cys98 should link Cys104. It should be noted that both Enbrel® and TuNEX® were demonstrated with indistinguishable precursor ion mass and fragmentation pattern by both CID and ETD spectra, which provided strong evidence that both products have the same disulfides in this linkage.
Fig. 3.
Mass and charge of Lys-C + trypsin + Glu-C-digested peptide with the two disulfides (Cys78-Cys96 and Cys98-Cys104), for Enbrel® (A) and TuNEX® (B). The sequence and theoretical mass of the peptide along with the observed monoisotopic (1st isotopic) mass are indicated in the insert. CID-MS2 spectrum of the precursor for Enbrel® (C) and TuNEX® (D). ETD-MS2 spectrum (using Orbitrap) of the precursor for Enbrel® (E) and TuNEX®(F). CID-MS3 spectrum of the precursor from Figure 3E (m/z 680.27) for Enbrel®(G) and Figure 3F (m/z 680.27) for TuNEX®(H).
Using the same approach, the rest of disulfide linkages were identified as shown from Figs. S1–S9, corresponding to the disulfides from #1 and #4 to #11 in Tables 1A and 1B, respectively. Again, indistinguishable mass and consistent fragmentation patterns were observed between the two products.
The peptides with a single disulfide, the assignment was straightforward (only one connection possible) and thus only CID-MS2 spectrum was sufficient and shown. The disulfides in the IgG1-Fc region, in which the linkages were known and conserved, the accurate mass and consistent pattern between Enbrel® and TuNEX® were shown for identification. In these case, we have successfully mapped disulfide linkages using online LC-MS with ETD based on the favorable breakage of the disulfide bond. The cleaved or partially cleaved disulfides were further fragmented by CID-MS3 to obtain specific linkage locations.
Compared to the previous report using HCD alone [7], three disulfide linkages appear to differ: Cys98-Cys104 differed from the reported Cys98-Cys115; Cys112-Cys121 differed from the reported Cys104-Cys112; Cys115-Cys139 differed from the reported Cys121-Cys139. Two of them, Cys104-Cys112 and Cys121-Cys139 linkages, were previously mapped by precursors with two or more disulfide linkages. Because of the lack of critical fragments to map multi-disulfide linkages using CID alone, their assignments were based on X-ray data derived from TNF binding protein rather than etanercept products. As shown in Table 1A, Cys98-Cys104 and Cys78-Cys96 were assigned here by the same precursor using ETD to break the disulfide bonds and further fragmented by CID-MS3 (Fig. 3G,H). Furthermore, the Cys112-Cys121 (Supplement Fig. S2B) and Cys115-Cys139 (Supplement Fig. S3B) were assigned here by peptides with single disulfide linkage (Table 1A) using our multi-enzyme protocol. We have further investigated our data based on manual extraction. The theoretical ions (m/z) with the previously reported Cys104-Cys112 linkage were not detected here from either Enbrel® or TuNEX®. Furthermore, previous method using HCD alone has revealed four disulfide scrambling, Cys18-Cys74, Cys71-Cys88, Cys71-Cys74, Cys78-Cys88 among which, the percentage of Cys78-Cys88 scrambling was reported to be as large as 10%. However, only a much lower and insignificant percentage (<1%) of scrambling on Cys71-Cys74 or Cys78-Cys88 was detected from Enbrel® or TuNEX® here. In addition, we have checked alternative linkages such as Cys71-Cys98/Cys96-Cys104 and could not find significant signals for fragments generated by such linkages.
4. Conclusions
In summary, a LC-MS protocol using multi-enzyme coupled with multi-fragmentation approach have been employed to assign the status of all entire disulfide linkages including TNFR portion (22 disulfides) and IgG1 Fc domain (7 disulfides) of two etanercept (Enbrel® and TuNEX®) products. Compared to previous reports using HCD fragmentation alone, our method provided more straight forwards and un-ambiguous assignments for disulfide linkages. Based on repetitive analyses of specific lots of the two etanercept products, our data showed the higher order structure of the two products was consistent.
Acknowledgements
This work was supported by Ministry of Science and Technology, Taiwan, Republic of China (MOST 105–2113-M-006–014-MY3).
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jfda.2018.11.007.
Funding Statement
This work was supported by Ministry of Science and Technology, Taiwan, Republic of China (MOST 105–2113-M-006–014-MY3).
REFERENCES
- 1. Liu H, May K. Disulfide bond structures of IgG molecules: structural variations, chemical modifications and possible impacts to stability and biological function. mAbs. 2012;4:17–23. doi: 10.4161/mabs.4.1.18347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cho IH, Lee N, Song DM, Jung SY, Bou-Assaf G, Sosic Z, et al. Evaluation of the structural, physicochemical, and biological characteristics of SB4, a biosimilar of etanercept. mAbs. 2016;8:1136–55. doi: 10.1080/19420862.2016.1193659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kohno T, Louie JS, Stevens SR. Differences in Fc receptor and C1q binding in tumor necrosis factor (TNF) antagonists may contribute to differences in mechanisms of action. J Invest Dermatol. 2005;124:A111–A. [Google Scholar]
- 4. Mukai Y, Nakamura T, Yoshikawa M, Yoshioka Y, Tsunoda S, Nakagawa S, et al. Solution of the structure of the TNFTNFR2 complex. Sci Signal. 2010;3 doi: 10.1126/scisignal.2000954. [DOI] [PubMed] [Google Scholar]
- 5. Houel S, Hilliard M, Yu YQ, McLoughlin N, Martin SM, Rudd PM, et al. N- and O-glycosylation analysis of etanercept using liquid chromatography and quadrupole time-of-flight mass spectrometry equipped with electron-transfer dissociation functionality. Anal Chem. 2014;86:576–84. doi: 10.1021/ac402726h. [DOI] [PubMed] [Google Scholar]
- 6. Huang LJ, Lin JH, Tsai JH, Chu YY, Chen YW, Chen SL, et al. Identification of protein O-glycosylation site and corresponding glycans using liquid chromatography-tandem mass spectrometry via mapping accurate mass and retention time shift. J Chromatogr A. 2014;1371:136–45. doi: 10.1016/j.chroma.2014.10.046. [DOI] [PubMed] [Google Scholar]
- 7. Lamanna WC, Mayer RE, Rupprechter A, Fuchs M, Higel F, Fritsch C, et al. The structure-function relationship of disulfide bonds in etanercept. Sci Rep. 2017;7 doi: 10.1038/s41598-017-04320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu SL, Kim J, Hancock WS, Karger B. Extended Range Proteomic Analysis (ERPA): a new and sensitive LC-MS platform for high sequence coverage of complex proteins with extensive post-translational modifications-comprehensive analysis of beta-casein and epidermal growth factor receptor (EGFR) J Proteome Res. 2005;4:1155–70. doi: 10.1021/pr050113n. [DOI] [PubMed] [Google Scholar]
- 9. Coon JJ, Syka JEP, Shabanowitz J, Hunt DF. Tandem mass spectrometry for peptide and protein sequence analysis. Biotechniques. 2005;38:519-+. doi: 10.2144/05384TE01. [DOI] [PubMed] [Google Scholar]
- 10. Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 11. Tsai PL, Chen SF, Huang SY. Mass spectrometry-based strategies for protein disulfide bond identification. Rev Anal Chem. 2013;32:257–68. [Google Scholar]
- 12. Srebalus Barnes CA, Lim A. Applications of mass spectrometry for the structural characterization of recombinant protein pharmaceuticals. Mass Spectrom Rev. 2007;26:370–88. doi: 10.1002/mas.20129. [DOI] [PubMed] [Google Scholar]
- 13. Yen TY, Joshi RK, Yan H, Seto NO, Palcic MM, Macher BA. Characterization of cysteine residues and disulfide bonds in proteins by liquid chromatography/electrospray ionization tandem mass spectrometry. J Mass Spectrom: JMS. 2000;35:990–1002. doi: 10.1002/1096-9888(200008)35:8<990::AID-JMS27>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 14. Gray WR. Disulfide structures of highly bridged peptides: a new strategy for analysis. Protein Sci. 1993;2:1732–48. doi: 10.1002/pro.5560021017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yen TY, Yan H, Macher BA. Characterizing closely spaced, complex disulfide bond patterns in peptides and proteins by liquid chromatography/electrospray ionization tandem mass spectrometry. J Mass Spectrom: JMS. 2002;37:15–30. doi: 10.1002/jms.241. [DOI] [PubMed] [Google Scholar]
- 16. Qi JF, Wu W, Borges CR, Hang DH, Rupp M, Torng E, et al. Automated data interpretation based on the concept of “negative signature mass” for mass-mapping disulfide structures of cystinyl proteins. J Am Soc Mass Spectrom. 2003;14:1032–8. doi: 10.1016/S1044-0305(03)00145-4. [DOI] [PubMed] [Google Scholar]
- 17. Jones MD, Hunt J, Liu JL, Patterson SD, Kohno T, Lu HS. Determination of tumor necrosis factor binding protein disulfide structure: deviation of the fourth domain structure from the TNFR/NGFR family cysteine-rich region signature. Biochemistry. 1997;36:14914–23. doi: 10.1021/bi971696k. [DOI] [PubMed] [Google Scholar]
- 18. Huang SY, Wen CH, Li DT, Hsu JL, Chen C, Shi FK, et al. Assignment of disulfide-linked peptides using automatic a(1) ion recognition. Anal Chem. 2008;80:9135–40. doi: 10.1021/ac8013725. [DOI] [PubMed] [Google Scholar]
- 19. Huang SY, Hsieh YT, Chen CH, Chen CC, Sung WC, Chou MY, et al. Automatic disulfide bond assignment using a(1) ion screening by mass spectrometry for structural characterization of protein pharmaceuticals. Anal Chem. 2012;84:4900–6. doi: 10.1021/ac3005007. [DOI] [PubMed] [Google Scholar]
- 20. Sung WC, Chang CW, Huang SY, Wei TY, Huang YL, Lin YH, et al. Evaluation of disulfide scrambling during the enzymatic digestion of bevacizumab at various pH values using mass spectrometry. Biochim et Biophys Acta. 2016;1864:1188–94. doi: 10.1016/j.bbapap.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 21. Wells JM, McLuckey SA. Collision-induced dissociation (CID) of peptides and proteins. Biol Mass Spectrom. 2005;402:148–85. doi: 10.1016/S0076-6879(05)02005-7. [DOI] [PubMed] [Google Scholar]
- 22. Zhang MX, Kaltashov IA. Mapping of protein disulfide bonds using negative ion fragmentation with a broadband precursor selection. Anal Chem. 2006;78:4820–9. doi: 10.1021/ac060132w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Janecki DJ, Nemeth JF. Application of MALDI TOF/TOF mass spectrometry and collision-induced dissociation for the identification of disulfide-bonded peptides. J Mass Spectrom. 2011;46:677–88. doi: 10.1002/jms.1938. [DOI] [PubMed] [Google Scholar]
- 24. Loo JA, Edmonds CG, Udseth HR, Smith RD. Effect of reducing disulfide-containing proteins on electrospray ionization mass-spectra. Anal Chem. 1990;62:693–8. doi: 10.1021/ac00206a009. [DOI] [PubMed] [Google Scholar]
- 25. Xu H, Zhang LW, Freitas MA. Identification and characterization of disulfide bonds in proteins and peptides from tandem MS data by use of the MassMatrix MS/MS search engine. J Proteome Res. 2008;7:138–44. doi: 10.1021/pr070363z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Clark DF, Go EP, Toumi ML, Desaire H. Collision induced dissociation products of disulfide-bonded peptides: ions result from the cleavage of more than one bond. J Am Soc Mass Spectrom. 2011;22:492–8. doi: 10.1007/s13361-010-0064-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wu SL, Jiang H, Hancock WS, Karger BL. Identification of the unpaired cysteine status and complete mapping of the 17 disulfides of recombinant tissue plasminogen activator using LC-MS with electron transfer dissociation/collision induced dissociation. Anal Chem. 2010;82:5296–303. doi: 10.1021/ac100766r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu SL, Jiang HT, Lu QZ, Dai SJ, Hancock WS, Karger BL. Mass spectrometric determination of disulfide linkages in recombinant therapeutic proteins using online LC-MS with electron-transfer dissociation. Anal Chem. 2009;81:112–22. doi: 10.1021/ac801560k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ni WQ, Lin M, Salinas P, Savickas P, Wu SL, Karger BL. Complete mapping of a cystine knot and nested disulfides of recombinant human arylsulfatase a by multi-enzyme digestion and LC-MS analysis using CID and ETD. J Am Soc Mass Spectrom. 2013;24:125–33. doi: 10.1007/s13361-012-0510-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tan Q, Guo Q, Fang C, Wang C, Li B, Wang H, et al. Characterization and comparison of commercially available TNF receptor 2-Fc fusion protein products. mAbs. 2012;4:761–74. doi: 10.4161/mabs.22276. [DOI] [PMC free article] [PubMed] [Google Scholar]