Abstract
Oral cancer with high incidence rates is occurring in many countries including in India, Pakistan, Bangladesh, Sri Lanka and Taiwan. Smoking, alcoholism, and betel nut chewing are considered to be the main risk factors for oral cancer. Further, deaths from oral cancer have increased year by year. Although several oral cancer-associated biomarkers have been reported, very few useful biomarkers have been applied for early diagnosis. Therefore, the investigation of oral cancer-specific biomarkers is urgently needed. We previously investigated N-glycomes of oral cancer cells and patient plasma. We found that both mRNA levels of FUT8 and core-fucosylated glycoproteins increase in cases of oral cancer relative to normal cases. In this study we aim to discover novel core-fucosylated glycoprotein biomarkers for oral cancer diagnosis with glycoproteomic approaches. First, forty plasma samples obtained from the Human Bioinformation Bank of NCKUH were subjected to AAL (Aleuria aurantia lectin) affinity chromatography. Core-fucosylated proteins were collected and applied for LC-MS/MS followed by electrophoresis. Fourteen proteins were identified, and expression levels of proteins in plasma were verified by western blot. Expression levels of some glycoproteins were elevated in the oral cancer group, including ceruloplasmin, haptoglobin, and leucin-rich alpha-2-glycoprotein 1 (LRG1). However, levels of some glycoproteins decreased in the cancer group, including apolipoprotein A-I (apo A-I) and apolipoprotein A-IV (apo A-IV). Via ELISA analysis, we found that apo A-IV and apo A-IV/ total protein ratios were decreased in plasma accompanied with cancer stages. The LRG1/total protein ratio was found to increase while plasma levels of LRG1 were not found to differ between the oral cancer plasma and normal groups. An ROC curve analysis reveals strong diagnosis performance when combining apo A-IV levels and LRG1/total protein ratios. Taken together, apo A-IV and LRG1, given their strong performance in detecting oral cancer, can serve as useful biomarkers and may be used as a useful tool for oral cancer screening and early diagnosis.
Keywords: Apo A-IV, Core fucose, LRG1, Oral cancer
1. Introduction
Oral cancer, caused by malignancies occurring in the oral cavity and oropharynx, is a form of head and neck cancer accounting for roughly 90 percent of cases of squamous cell carcinoma [1]. Smoking, alcoholism, and betel nut chewing, cultural practices in Asian countries, are the main risk factors of oral cancers. In Asia, oral cancer is one of the six most frequent cancers with high incidence rates reported in many countries such as India, Pakistan, Bangladesh, Sri Lanka and Taiwan [2,3]. Oral cancer is believed to mainly occur in the elderly among those between 50 and 70 years of age. However, a shift toward younger cases has recently been observed, with the percentage of patients below 40 years of age increasing to 17%. In terms of gender, men present higher incidence rates than women in most of the countries. In Japan, the male to female ratio is 1.45, that for Pakistan is 1.5, and that for Yemen is 1.65, and Taiwan presents the highest ratio of 10.5 [4]. According to the Taiwan cancer registry, oral cancer is the fourth leading cause of mortality among men with prevalence and incidence rates increasing each year [5]. While the five-year survival rate of oral cancer can be improved with early diagnosis and treatment, physical examinations of oral cavities with biopsy confirmation remain as the main screening and diagnosis methods used, leading to delays in diagnosis and accounting for the majority of patients with late-stage presentation [6]. Although many screening tests have been developed, such as Toluidine blue and Fluorescence staining tests, Exfoliative cytology tests and Brush biopsies, varying sensitivity to these examinations makes it difficult to lower risks of dying from oral cancer [7]. Rather, by the time they are identified, most oral and oropharyngeal cancers have already migrated to lymph nodes or to other areas [8]. Thus, the development of effective screening tests that can detect oral cancer early on is urgently needed.
Glycosylation is an important regulatory mechanism involved in several biological processes, including those of cell proliferation, differentiation, inflammation, cell-cell interaction and carcinogenesis. The progression of malignancies such as cancer development and metastasis is considered to be associated with glycosylation alterations of cell surface proteins [9–11]. As aberrant glycosylation occurs, it can influence many physiological and pathological events, contributing to cancer progression as well as angiogenesis and metastasis. Glycans alterations of malignancies take multiple forms such as those of over or underexpression, truncation, modified branching, precursor accumulation and less commonly the appearance of novel structures [12]. In oral cancer, the dysregulation of the N-glycosylation-regulating gene, DPAGT1, which controls the N-glycosylation of E-cadherin and alters gene coexpression networks among DPAGT1, Wnt and E-cadherin, has been proven to be a major cause of oral cancer progression [13,14]. Wei-ling, Lin et al. also revealed that oral cancer migration can be promoted by the glycosylation of Lewisy antigen on epidermal growth factor receptors [15]. Moreover, sialylation and fucosylation, which are considered to be the two of most commonly occurring cancer-associated changes in N-glycosylation [16], have been found to be associated with oral cancer [17–19].
Glycoproteomics, different from proteomics and glycomics, involves the study of protein glycosylation, which is involved in several physiological functions and biological processes. Glycoprotein enrichment and proteomics technologies are integrated to systematically identify and quantify glycoproteins in sample mixtures. Mass spectrometry, which is commonly used to identify proteins and to characterize glycan structures, has played a crucial role in the determination of the mass of glycopeptide and glycoprotein sequencing [20]. Owing to the considerable complexity of biological samples, the constitution of glycans of a single glycoprotein is too limited to detect directly. Thus, lectins and hydrophilic affinity gel for glycoprotein enrichment are always used prior to mass spectrometry for the identification of glycan and glycopeptides [21]. In recent years, it has been of great interest to apply glycoproteomic technological strategies to clinical translational research and especially to glycoprotein biomarkers.
In our previous studies we found that fucosylation and especially core-fucosylation is enhanced in oral cancer cells relative to normal gingival keratinocytes [22]. We have also identified several serum N-glycans that can serve as valuable biomarkers for oral cancer diagnosis [23]. To further identify novel biomarkers for the early diagnosis of oral cancer, corefucosylated serum glycoproteins of normal volunteers and patients were purified by lectin chromatography, and the alteration of glycoproteins was analyzed by electrophoresis and mass spectrometry. The identified glycoproteins were then verified by western blotting and finally validated by ELISA.
2. Materials and methods
2.1. Plasma samples
This study was approved by National Cheng Kung University Hospital Institutional Review Board. Plasma samples of oral cancer patients (N = 40, test group; N = 71, validation group; Supplementary Table 1) were obtained from the National Cheng Kung University Hospital Human Bioinformation Bank. Cancer-free healthy volunteers (N = 40 of 40 years of age and above; N = 15 of under 40 years of age) were recruited and venous blood was collected into Becton Dickinson (BD) EDTA-treated tubes (lavender tops) in cooperation with the Health Examination Center over two months. Each volunteer subject to routine health examination within the normal physiological range fasted for 8–12 h and was sampled in the morning. Blood sampling was executed by a professional medical technologist. All blood samples were centrifuged at 3000 rpm for 10 min and upper liquid (plasma) was transferred into clean microcentrifuge tubes stored at −80 °C immediately after collection. Samples with hemolysis, lipemia and icteric that could have generated invalid data were excluded.
2.2. Aleuria aurantia lectin (AAL) affinity chromatography
To purify core fucosylated plasma proteins, 500 μl 50% slurry of A. aurantia lectin conjugated agarose beads (Vector Laboratory) was added to the columns. Before coupling with plasma samples, beads were rinsed 3 times throughout the column volume with TBS buffer (pH 7.4) to remove sugars added to stabilize lectins in the storage buffer. Pooled plasma samples of 500 μl were subjected to the lectin columns with TBS buffer diluted 2 times after the depletion of albumin and Immunoglobulin G. Mixtures in the columns were incubated overnight at 4 °C and were quickly mixed on the rotor. The separation of unbound plasma proteins was performed by rinsing column volumes with TBS wash buffer 5 times. To elute out the glycoproteins bound to the lectin beads, simple sugar (l-fucose 100 mM) was used as an elution buffer and was added to the column with 1 h of 4 °C incubation, and this was followed by 10 rounds of column volumes of elution buffer addition to the columns. Finally, the eluted target proteins were collected and concentrated through Amicon® Ultra centrifugal filters (Millipore).
2.3. In-gel digestion of proteins separated by SDS-PAGE for mass spectrometry (MS) analysis
Excised gel bands with targeted proteins were cut into small pieces and applied by enzyme digestion according to the following procedure. After the elimination of Coomassie blue dye via rinsing with 100 mM NH4HCO3/CH3CN (1:1 v/v), proteins in gel pieces were reduced through the addition of dithiothreitol (DTT) (10 mM final). Alkylation was performed by adding iodoacetic acid (20 mM final) and by incubating the samples for 30 min at room temperature in the dark followed by the addition of CH3CN to dehydrate the gel pieces. Trypsin solution (20 ng/μl) was then added to the gel samples at a 1:50 (w/w) ratio, and reactions were incubated at 37 °C for over 5 h. Enzymatic digestion was quenched through the addition of 1% formic acid/2% CH3CN, and digested peptides were dried in a speed-vac prior to mass analysis [24].
2.4. Liquid chromatography–mass spectrometry (nanoflow LC–MS/MS)
High-resolution, high-mass accuracy nanoflow LC–MS/MS was performed with a combined linear quadrupole ion trap (LTQ) and Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer (LTQ-FT system, Thermo Fisher Scientific, Waltham, MA) equipped with a nanoelectrospray ion source (New Objective, Woburn, MA), an Agilent 1100 Series binary high-performance liquid chromatography pump (Agilent Technologies, Palo Alto, CA), and a Famos autosampler (LC Packings, San Francisco, CA). Each digested peptide solution was injected onto a self-packed precolumn (150 μm ID × 20 mm, 5 μm, 200 Ä). Chromatographic separation was performed on a self-packed reverse-phase C18 nanocolumn (75 μm ID × 150 mm, 5 μm, 100 Ä) using 0.1% formic acid in water (mobile phase A) and 0.1% formic acid in 80% acetonitrile (mobile phase B). This was done by applying a linear gradient from 5% to 40% mobile phase B for 40 min at a flow rate of 300 nl/min provided across a flow splitter by the HPLC pumps. Electrospray voltage was applied at 2 kV, and the capillary temperature was set to 200 °C. A scan cycle was initiated with a full-scan survey MS spectrum (m/z 300–2000); this was performed with the FT-ICR mass spectrometer at a resolution of 100,000 (at m/z 400). The 10 most abundant ions detected in the scan were subjected to MS/MS analysis performed in the LTQ mass spectrometer. Ion accumulation (Auto Gain Control target number) and the maximal ion accumulation time for full scanning and MS/MS modes were set at 1 × 106 ions, 1000 ms and 5 × 104 ions, 200 ms, respectively. Parameters for collision-induced dissociation were set at 35% normalized collision energy, 0.3 activation Q, and 30 ms activation time [24].
2.5. Data evaluation and statistics
The identified protein data obtained via mass spectrometry were exported to a table form with information on the molecular weights, intensities and analyzed scores of the protein candidates. When comparing the duplicated results, proteins with the top three analyzed scores were chosen and we assessed potentiality based on previous studies. From western blot results, relative abundances of plasma proteins for two groups were first evaluated through a Mann–Whitney U test. For the ELISA results, the statistical significance of two groups was determined using through a student’s t test. The analysis described above was conducted using GraphPad Prism (version 5.0, GraphPad Software, Inc.) while considering a p-value of ≦ 0.05 as statistically significant. The receiver-operator characteristic (ROC) curve analysis was conducted with MedCalc (MedCalc Software bvba) and optimal cut-off values were set according to the Youden’s index (J).
3. Results
3.1. Purification and identification of core fucosylated glycoproteins from albumin/IgG depleted plasma
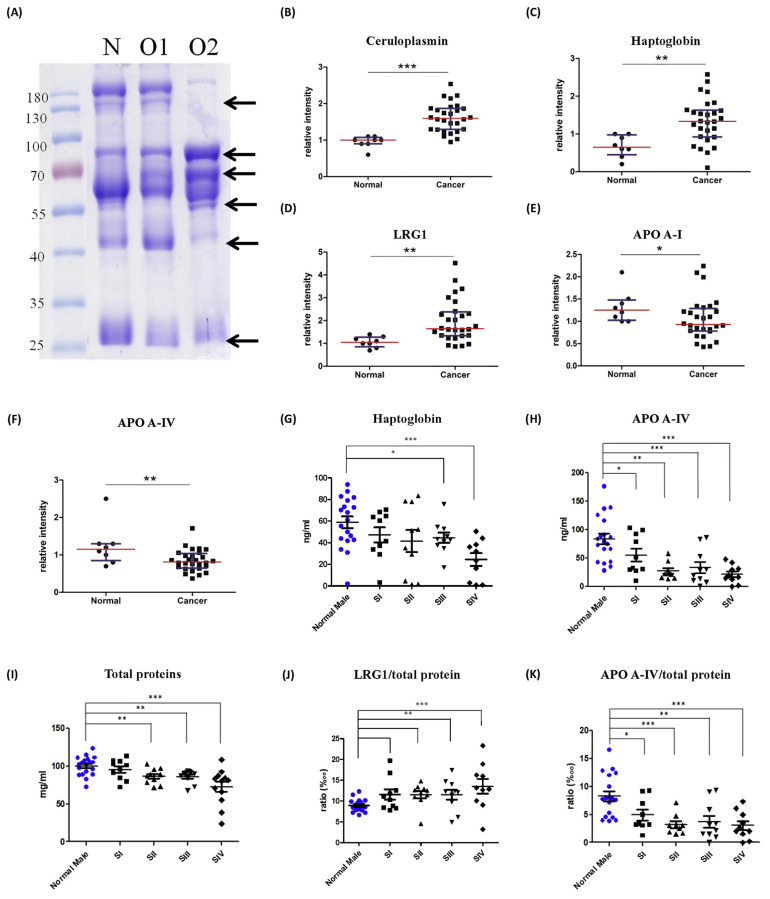
In this study, A. aurantia lectin (AAL), which specifically recognizes fucose residues linked to N-acetylglucosamine, was chosen to capture core fucosylated glycoproteins. To obtain a sufficient volume of plasma glycoproteins of a certain glycosylation type from one individual, pooled plasma was used in the enrichment step. Plasma samples were sorted into three groups: a normal group (N), an oral cancer group without regional lymph node metastasis (O1) and an oral cancer group with regional lymph node metastasis (O2). Since albumin and immunoglobulin G were the most abundant proteins that can interfere with lectin-glycoprotein binding efficiency, both were depleted prior to lectin chromatography. Equal amounts (20 μg) of core fucosylated protein were loaded into SDS-PAGE gel and glycoproteins were separated by molecular weight into multiple protein bands visualized by Coomassie Blue staining (Fig. 1A). In comparing the three groups we found that differences in patterns of the proportions of protein bands, and protein bands with significant differences between groups were then selected and identified. The experiments were done in duplicate. After in-gel digestion followed by mass spectrometry analysis, raw data with identified protein information were exported to the table forms. First, we eliminated laboratory contaminant proteins such as keratin and trypsin. Albumin, hemoglobin and immunoglobulin were excluded as well. Second, data collected from the duplicate experiments were compared and proteins overlapping in both datasets were selected. Then, selected proteins were filtered based on glycosylation features of the proteins with core fucose. Finally, 14 protein candidates were preliminarily assessed by reference consultation and are listed in Table 1. A further analysis was conducted on seven protein candidates through random selection (Table 2).
Fig. 1.
(A) SDS-PAGE analysis of core-fucosylated plasma proteins with Albumin/IgG depletion followed by AAL lectin chromatography with Coomassie staining. Core fucosylated protein purified from normal (N)/oral cancer without lymphatic metastasis (O1)/oral cancer for the lymphatic metastasis group (O2). Different proportions of proteins were compared and protein bands with strong variations among groups were excised (arrows) followed by MALDI-TOF MS identification. The relative abundance of (B) ceruloplasmin, (C) haptoglobin, (D) LRG1, (E) apolipoprotein A-I, and (F) apolipoprotein A-IV in normal (N) and oral cancer patients (P) analyzed by western blot. Statistical results for different cancer stages of (G) haptoglobin, (H) APO-AI, (I) total protein, (J) LRG1/total protein, and (K) APO-A IV/total protein levels in plasma. ***, p < 0.001; **, p < 0.01; *, p < 0.05 relative to normal results.
Table 1.
List of candidate proteins identified by glycoproteomic approaches and correlations with other diseases in literatures.
| Glycoprotein | MW(kD) | Score | Associated disease | Protein levels/ Altered glycoforms | Detection technique |
|---|---|---|---|---|---|
| Haptoglobulin | 45 | 997 | Pancreatic cancer | fucosylation sialylation | MALDI-TOF, immunoblot LC-MS/MS |
| Oral squamous cell carcinoma | up-regulated | ELISA | |||
| Ceruloplasmin | 122 | 101 | Hepatocellular carcinoma (HCC) | core fucosylation | LC-MS/MS |
| Pancreatic cancer | sLeX increase | LC-ESI-QTOF | |||
| Leucine-rich | 38 | 105 | Pancreatic cancer | up-regulated | MDLD |
| alpha-2-glycoprotein 1 (LRG1) | Ovarian cancer | up-regulated | ELISA, RT-PCR, WB, MS | ||
| Hemopexin | 51 | 355 | Hepatocellular carcinoma | fucosylated | HPLC, LC MS/MS, Lectin FLISA |
| Apolipoprotein-AI | 30 | 242 | Bladder cancer | increased expression | MS, Western blot |
| Liver fibrosis | down regulated | MALDI-TOF/TOF | |||
| Apolipoprotein A-II | 11 | 96 | Cholangiocarcinoma | up-regulated | SELDI-TOF-MS, ELISA, WB |
| Apolipoprotein A-IV | 45 | 333 | Ovarian cancer | decrease of conc. | ELISA |
| Zinc-alpha2-glycoprotein | 34 | 87 | Breast cancer | increase of conc. | IHC |
| Hepatocellular carcinoma (HCC) | up-regulated | MALDI-TOF-MS | |||
| CD5 antigen-like | 38 | 62 | Hepatocellular carcinoma (HCC) | increase of conc. | ELISA, LC-MS/MS |
| Alpha2-macroglobulin | 163 | 217 | Liver fibrosis | up-regulated | MALDI-TOF/TOF |
| Bone meta in prostate cancer | decrease of conc. | EIA | |||
| Galectin-3 binding protein | 65 | 95 | Behç et’s disease | increased expression galectin-mediated tumor cell aggregation | ELISA |
| Alpha-1-acid glycoprotein | 40 | 121 | Lymphoma; Ovarian tumor | fucosylation | MALDI-TOF, LDA |
| Lung adenocarcinoma | N-linked glycosylation, increased expression | MALDI-TOF/TOF | |||
| Hepatocellular carcinoma | increased expression | HPLC, Western blot | |||
| Alpha 2-HS Glycoprotein | 43 | 102 | Breast Cancer | up-regulated | MS, Immunoblot |
| Hypopharyngeal squamous cell carcinoma | up-regulated | MALDI-TOF/TOF MS, WB, ELISA | |||
| Apolipoprotein C-III | 10 | 108 | Papillary thyroid carcinoma | down-regulated | SELDI-TOF-MS, HPLC, LC-MS/MS |
| Gastric cancer | up-regulated | MS |
Table 2.
Seven protein candidates were further verified by western blot analysis.
| Protein name | Mascot score | Mol. wt | Peptide matches |
|---|---|---|---|
| Apolipoprotein A-I (Apo A-I) | 113 | 30,759 | 10 |
| Apolipoprotein A-II (Apo A-II) | 96 | 11,168 | 3 |
| Apolipoprotein A-IV (Apo A-IV) | 333 | 45,371 | 24 |
| Ceruloplasmin (Cp) | 228 | 122,128 | 22 |
| Haptoglobin (Hpt) | 336 | 45,177 | 23 |
| Hemopexin (HPX) | 55 | 51,643 | 8 |
| Leucine-rich | 105 | 38,154 | 8 |
| alpha-2-glycoprotein 1 (LRG1) |
3.2. Western blot analysis of protein candidates
To evaluate differences in candidate proteins in plasma among the normal and cancer groups, seven candidate proteins were further analyzed by western blot. The results show that protein levels of ceruloplasmin, haptoglobin and leucine-rich alpha-2-glycoprotein 1 were significantly higher in the oral cancer plasma than in the normal group with p-values of less than 0.001, 0.01 and 0.01, respectively (Fig. 1B–D). In contrast, apolipoprotein A-I and apolipoprotein A-IV levels in the oral cancer group were significantly lower than those of the normal group with p-values of less than 0.05 and 0.01, respectively (Fig. 1E, F). However, hemopexin and apolipoprotein A-II presented no significant differences in either group. To evaluate the expression changes of these plasma glycoproteins, volumes of candidate proteins between the normal and cancer groups were further analyzed by ELISA analysis.
3.3. ELISA analysis of protein candidates
To confirm levels of ceruloplasmin, haptoglobin, leucine-rich alpha-2-glycoprotein 1, apolipoprotein A-I and apolipoprotein A-IV observed through western blot analysis, 40 normal volunteers (19 males and 20 females) and 40 oral cancer plasmas were subjected to ELISA analysis. Haptoglobin (Hpt), which is used to define conditions of hemolysis, is also an acute-phase reactant (increasing levels of infection and inflammation). Haptoglobin levels in the plasma showed no discrepancies by gender. However, relative to the normal group, the oral cancer group showed significantly lower levels of haptoglobin. By comparison, among the cancer stages we found that only stage IV showed significantly lower levels, and no differences were observed in early stages (Fig. 1G). Apolipoprotein A-IV (Apo A-IV) is the main circulating apolipoprotein, and an assessment of plasma through chylomicrons reveals very low levels of low-density lipoproteins (VLDL) and HDL. No differences by gender were found. Compared to the normal group, oral cancer patients tended to present lower levels of apo A-IV in plasma. Further, a comparison of the cancerous and normal groups shows that with the exception of stage I, the other three stages show a significant decrease in apo A-IV levels with p-values of less than 0.001, 0.01, and 0.001 (Fig. 1H). Moreover, it has been noted that volumes of total proteins in plasma tend to be lower when malignancies occur. A significantly decreased total amount of plasma proteins was also found in oral cancer patients with decreasing trends following cancer progression (Fig. 1I). To eliminate the influence of reduced total protein levels found in the patients, ratios of candidate proteins to total plasma proteins were calculated and subjected to data analysis. We found that the proportion of haptoglobin in all proteins showed no significant differences while absolute quantities of haptoglobin were lower in oral cancer plasma than in the normal group. Although LRG1 presented no significant changes in absolute quantities across the two groups, ratios of LRG1/total protein gradually and significantly increased with cancer progression (Fig. 1J). Further, apolipoprotein A-IV with decreased absolute quantities in oral cancer plasma significantly decreased in proportions of proteins and gradually declined with cancer progression (Fig. 1K).
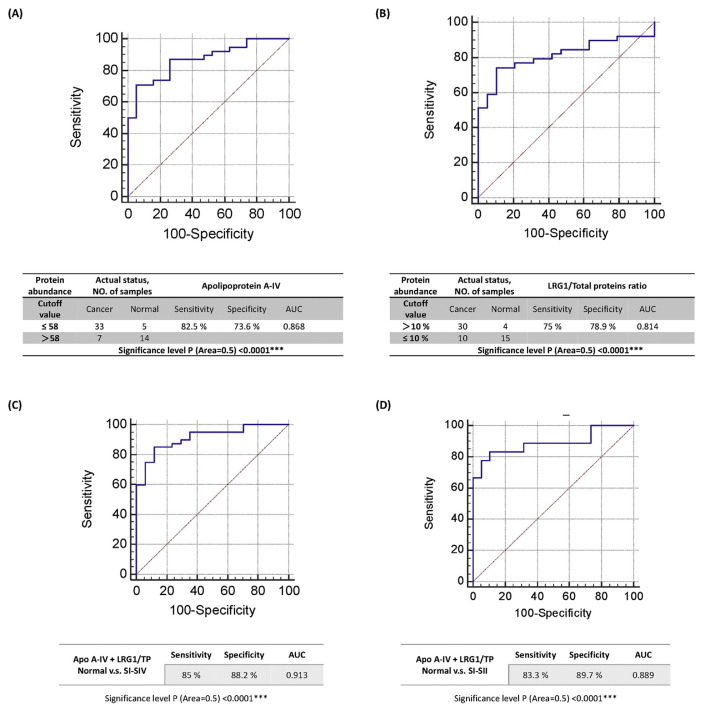
3.4. ROC curve analysis
According to the results of the ELISA analysis, apolipoprotein A-IV and LRG1 are two biomarkers that may cause significant alterations in plasma levels or in proportions of total proteins in oral cancer plasma. To evaluate the diagnostic performance of these two biomarkers for oral cancer diagnosis, an ROC curve analysis was performed. As is shown in Fig. 2A, the diagnostic performance of plasma levels of apolipoprotein A-IV in detecting oral cancer is supported by our results: AUC (area under the curve) = 0.868, sensitivity = 82.5%, and specificity = 73.6% with a significance level P (Area = 0.5) of <0.0001. An ROC curve analysis of LRG1/total protein ratios was also performed and diagnostic performance in detecting oral cancer is supported by the following results: AUC = 0.814, sensitivity = 75%, and specificity = 78.9% with a significance level P (Area = 0.5) of <0.0001 (Fig. 2B). An ROC curve analysis of the combination of apo A-IV/total protein and LRG1/total protein ratios was also conducted and the highest AUC of 0.913 with 85% sensitivity and 88.24% specificity was observed (Fig. 2C). To verify the performance of the combination for detecting early stages of oral cancer, stages I and II were isolated and compared to the normal group. The results show that the combination of apo A-IV and LRG1/total protein ratios presents an AUC of 0.889 with 83.3% sensitivity and 89.7% specificity for early stages of oral cancer (Fig. 2D).
Fig. 2.
The ROC curve and diagnostic performance of (A) apolipoprotein A-IV, (B) the LRG1 to total protein ratio, (C) the ROC curve and the diagnostic performance of combining apolipoprotein A-IV with LRG1 relative to the total proteins ratio, and (D) the ROC curve and the diagnostic performance of combining apolipoprotein A-IV and LRG1 relative to the total protein ratio in differentiating the normal group from early cancer stages.
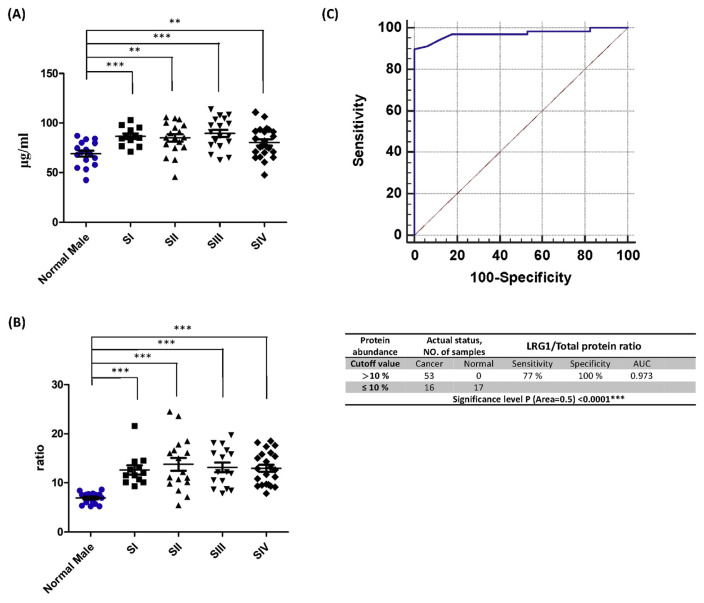
3.5. Validation of LRG1/total protein and apolipoprotein A-IV
To validate the role of LRG1/total protein and apolipoprotein A-IV in oral cancer diagnosis, an independent validation group with seventy-one OSCC patients (Supplementary Table 1) and seventeen normal volunteers was used to evaluate the sensitivity and specificity of apolipoprotein A-IV. Levels of LRG1 and LRG1/total protein increased with oral cancer stages (Fig. 3A, B). Based on the cutoff value set for the test group, the performance of the validation group exhibits 77.0% sensitivity with the detection of 53 of 69 cancer patients and 100% specificity with no normal volunteers falsely identified as cancer patients (Fig. 3C). The ROC curve of the validation group also shows strong performance with an AUC of 0.973.
Fig. 3.
Comparison of LRG1/total protein levels of the oral cancer validation group across normal cases and different cancer stages. (A) LRG1/total protein levels in plasma. (B) LRG1/total protein levels divided by levels of total protein in plasma. (C) The ROC curve and diagnostic performance of LRG1/total proteins in the validation group.
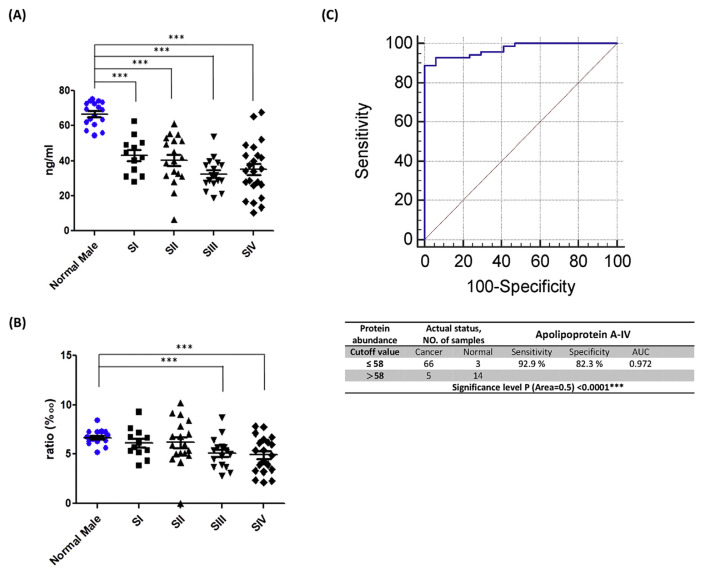
Although the apo A-IV to total proteins ratio also shows a decreasing trend, statistical significance was only observed in late stages (Fig. 4A, B). Compared to the test group, the ratio of apo A-IV to total proteins for the validation group exhibits poor performance in discriminating oral cancer. Based on the cutoff value set for the test group, the performance of the validation group exhibits 92.9% sensitivity with the detection of 66 of 71 cancer patients and 82.3% specificity with only 3 of 14 normal volunteers being falsely identified as cancer patients (Fig. 4C). The ROC curve of the validation group also shows good performance with an AUC of 0.972.
Fig. 4.
Comparison of apolipoprotein A-IV levels of the oral cancer validation group for normal cases and different cancer stages. (A) Apolipoprotein A-IV levels in plasma. (B) Apolipoprotein A-IV quantities divided by the amount of total protein in plasma. (C) The ROC curve and diagnostic performance of apolipoprotein A-IV in the validation group.
4. Discussion
Oral cancer the sixth most frequently diagnosed for a cancer with a high incidence rate reported in many countries and with prevalence rates increasing each year [2]. However, the diagnosis of oral cancer is currently only confirmed by open biopsy. Although biopsy is effectively used to diagnose oral cancer, it involves obvious defects such as invasiveness [25]. Therefore, effective biomarkers assisted by screening and diagnosing oral cancers are in urgent need of development. Biomarkers, which are defined as biological molecules in body fluids, are important and useful tools for distinguishing between health and the presence of disease. Although there is still no specific biomarker for oral cancer, many oral cancer-associated biomarkers have been identified in recent years. For example, ITGA3 (integrin α3) and IGTB4 (integrin β4) have been found to be useful genomic biomarkers associated with distant metastases and with the prognosis of oral cancer [4]. The cluster of differentiation factor 34 (CD34) serves as an angiogenetic marker that can importantly predict recurrent cases of oral cancer [26]. Salivary biomarkers are currently more appealing due to the simplicity and noninvasiveness of obtaining saliva samples. However, the complexity of oral cavity environments containing diverse substances from internal and external environments complicates the study and sample pretreatment of saliva and no criteria to apply before clinical laboratory tests have been developed. Therefore, biomarkers of serum/plasma samples that can be quickly adopted in clinical laboratories are still indispensable in addressing oral cancer. Many oral cancer-associated biomarkers have been found in serum/plasma, including Cyclin D1 [27], C-reactive protein [28], and matrix metalloproteinase enzymes-3 (MMP- 3) [29]. In our study, we used glycoproteomics to filtrate core fucosylated plasma glycoproteins as a first step to find oral cancer biomarkers based on different features.
Glycosylation, as one of the most abundant post-translational modifications, is believed to efficiently regulate protein activities. Aberrant glycosylation may lead to cancer development. Core fucosylation catalyzes by α1,6- Fucosyltransferase (Fut8), which transfers GDP-fucose to the innermost GlcNAc residue of hybrid and complex of N-glycans via α1,6-linkages. Kang Shao et al. found that lower levels of core fucosylation of E-cadherin are related to lung cancer progression by inducing an epithelial–mesenchymal transition-like process in lung cancer cells [30]. Xiangchun Wang et al. claimed that core fucose influences EGFR-mediated signal transduction, which regulates cell growth, differentiation, and migration [31]. In cases of oral cancer, correlations to core fucosylation have not been identified. Previously we found that core-fucosylated N-glycans are highly expressed on the cell surfaces of oral cancer cells relative to primary human oral keratinocytes (HOK) through an analysis of cell surface glycans [32]. Moreover, higher core fucosylation N-glycans have been obtained from plasma of oral cancer patients relative to normal volunteers. Therefore, core fucosylated glycoproteins contributing to the alteration of core fucosylation N-glycans levels in plasma are major targets as potential plasma protein biomarkers.
The detection of plasma protein biomarkers together with their protein glycans, which may take different glycosylation forms in different forms of cancer, may be more specific to a given disease. Asazawa H. et al. achieved a high levels of specificity in distinguishing chronic liver disease by detecting serum fucosylated haptoglobin (Fut-Hpt) [33]. LRG1, a secreted glycoprotein first isolated from plasma, has been identified from multiple malignancies with aberrant expressions. LRG1 has been reported to regulate endothelial TGFβ signaling, promoting angiogenesis [34]. It has also been noted that LRG1 is associated with cell proliferation, apoptosis inhibition and epithelial-mesenchymal transition in cases of colorectal cancer [35,36]. Although biological functions of oral cancer remains unknown, we found the proportion of LRG1 to proteins to increase significantly in oral cancer plasma. Interestingly, while men present lower levels of LRG1 than women, this cannot be verified since due to insufficient cancer plasma samples obtained from women, we cannot determine whether women present the same results as men. Indeed, it remains unclear whether alterations of LRG1 shown in our results are associated with the angiogenesis of oral cancer cells or with other molecular mechanisms. More studies must therefore be conducted on this point. In addition, LRG1 is a glycoprotein with five potential glycosylation sites that may take different glycosylation forms and which may be secreted by different cells [37]. Therefore, it is worth to further investigating different glycosylation forms of LRG1 that may present more specific results for oral cancers.
In this study we found that true amounts of apolipoprotein A-IV and relative amounts of LRG1 are significantly altered in the plasma of oral cancer patients. Previous studies have noted that decreased plasma levels of apo A-IV are related to ovarian cancer [38], but biological functions associated with cancer progression remain unknown. It has been reported that apo A-IV, which is synthesized primarily by the intestines with minor synthesis from the liver, is related to chylomicrons, a high-density lipoprotein (HDL) of the lipid transport system with a large lipoprotein-free fraction in plasma. Due to its anti-oxidative and anti-inflammatory properties and mediation of reverse-cholesterol transport, apo A-IV has been identified as a promising therapeutic target of metabolic disease [39]. It is interesting that though apo A-IV is mainly synthesized in the intestines, substantial alterations were found in the oral cancer group while only slight alterations were found in the colon cancer group. Both the test group and validation group exhibited high levels of sensitivity and specificity in differentiating normal cases from cases of oral cancer. Although it is not specific to oral cancer, apo A-IV may still serve as a biomarker for oral cancer screening and diagnosis.
Some discrepancies between the western blot and ELISA results were found in this study. While haptoglobin was found in higher volumes in the cancer group analyzed by western blot, a declining trend was found in the cancer group through ELISA testing. Ceruloplasmin was found at higher levels in cancer patients by western blot analysis, but no difference was found between normal and cancer groups by ELISA testing. Apolipoprotein A-I was downregulated in cancer plasma by western blot analysis, but no differences between the groups was found via ELISA testing. The above phenomena were observed for the following reasons. (1) Different quantification methods were applied for the two analyses. Ceruloplasmin studied by western blot, a relative quantification method, exhibited upregulation in the cancer group while no differences were found from the ELISA test by absolute quantification. However, after division by total proteins, the relative abundance derived from the ELISA results presents an increasing trend. (2) The sample size of the normal group used for western blot test was too small, generating inaccurate results. From western blot data for haptoglobin we observe high levels of dispersion among cancer patients but not for the normal group, which only included eight individuals while both groups with more samples presented high levels of dispersion in ELISA results. In apolipoprotein A-I, while higher levels of dispersion were found in cancer patients than in normal group in both analyses, the healthy and cancer groups were considered to be the same population through a statistical comparison with a larger sample size conducted via ELISA analysis.
Finally, core fucosylated glycoproteins in plasma were purified using a glycoproteomic approach and were identified by LD-MS/MS. After a comparative analysis by western blot and ELISA, two glycoproteins, apolipoprotein A-IV and LRG1, were found to be potential biomarkers of oral cancer. Plasma quantities of apolipoprotein A-IV and the proportion of LRG1 to total proteins were found to significantly differ between normal and oral cancer plasmas. The diagnostic performance of both shows high levels of sensitivity (83.3%) and specificity (89.7%) with an AUC of 0.889 for stages I and II. Hence, we suggest that alterations of these two plasma biomarkers may serve supportive tools of oral cancer screening, increasing the efficiency of early diagnosis.
Acknowledgement
We are grateful to the Human Biobank, Research Center of Clinical Medicine, National Cheng Kung University Hospital for providing the OSCC patient plasma.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jfda.2018.12.008.
Funding Statement
This work was supported by grants from the Ministry of Science and Technology, Taipei, Taiwan (MOST 106-2321-B-006-004 and MOST 107-2320-B-006-028).
Footnotes
Grant support
This work was supported by grants from the Ministry of Science and Technology, Taipei, Taiwan (MOST 106-2321-B-006-004 and MOST 107-2320-B-006-028).
Conflict of interest
The authors declare no conflicts of interest.
References
- 1. Sanderson RJ, Ironside JA. Squamous cell carcinomas of the head and neck. BMJ. 2002;325:822–7. doi: 10.1136/bmj.325.7368.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chiang CJ, Lo WC, Yang YW, You SL, Chen CJ, Lai MS. Incidence and survival of adult cancer patients in Taiwan, 2002–2012. J Formosan Med Assoc Taiwan yi zhi. 2016;115:1076–88. doi: 10.1016/j.jfma.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4. Krishna Rao SV, Mejia G, Roberts-Thomson K, Logan R. Epidemiology of oral cancer in Asia in the past decade–an update (2000–2012) Asian Pac J Cancer Prev. 2013;14:5567–77. doi: 10.7314/apjcp.2013.14.10.5567. [DOI] [PubMed] [Google Scholar]
- 5.Taiwan Cancer Registry. 癌症五年相對存活率. 2014. Available at: http://tcr.cph.ntu.edu.tw/main.php?Page=N2.
- 6.The Oral Cancer Foundation. Cancer screening protocols. 2016. Available at: http://oralcancerfoundation.org/discovery-diagnosis/cancer-screening-protocols/
- 7. Jitender S, Sarika G, Varada HR, Omprakash Y, Mohsin K. Screening for oral cancer. J Exp Ther Oncol. 2016;11:303–7. [PubMed] [Google Scholar]
- 8.Institute NC.Oral cavity and oropharyngeal cancer screening. 2016. Available at: https://www.cancer.gov/types/head-and-neck/patient/oral-screening-pdq#section/_24.
- 9. Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer. 2015;15:540–55. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 10. Xu C, Ng DT. Glycosylation-directed quality control of protein folding. Nat Rev Mol Cell Biol. 2015;16:742–52. doi: 10.1038/nrm4073. [DOI] [PubMed] [Google Scholar]
- 11.Varki A, Freeze HH, Vacquier VD. Glycans in development and systemic physiology. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, et al., editors. Essentials of glycobiology. 2nd ed. NY: Cold Spring Harbor; 2009. [PubMed] [Google Scholar]
- 12.Varki A, Kannagi R, Toole BP. Glycosylation changes in cancer. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, et al., editors. Essentials of glycobiology. 2nd ed. NY: Cold Spring Harbor; 2009. [PubMed] [Google Scholar]
- 13. Varelas X, Bouchie MP, Kukuruzinska MA. Protein N-glycosylation in oral cancer: dysregulated cellular networks among DPAGT1, E-cadherin adhesion and canonical Wnt signaling. Glycobiology. 2014;24:579–91. doi: 10.1093/glycob/cwu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nita-Lazar M, Noonan V, Rebustini I, Walker J, Menko AS, Kukuruzinska MA. Overexpression of DPAGT1 leads to aberrant N-glycosylation of E-cadherin and cellular discohesion in oral cancer. Cancer Res. 2009;69:5673–80. doi: 10.1158/0008-5472.CAN-08-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin WL, Lin YS, Shi GY, Chang CF, Wu HL. Lewisy promotes migration of oral cancer cells by glycosylation of epidermal growth factor receptor. PLoS One. 2015;10:e0120162. doi: 10.1371/journal.pone.0120162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Christiansen MN, Chik J, Lee L, Anugraham M, Abrahams JL, Packer NH. Cell surface protein glycosylation in cancer. Proteomics. 2014;14:525–46. doi: 10.1002/pmic.201300387. [DOI] [PubMed] [Google Scholar]
- 17. Shah M, Telang S, Raval G, Shah P, Patel PS. Serum fucosylation changes in oral cancer and oral precancerous conditions: alpha-L-fucosidase as a marker. Cancer. 2008;113:336–46. doi: 10.1002/cncr.23556. [In eng] [DOI] [PubMed] [Google Scholar]
- 18. Dadhich M, Prabhu V, Pai VR, D’Souza J, Harish S, Jose M. Serum and salivary sialic acid as a biomarker in oral potentially malignant disorders and oral cancer. Indian J Cancer. 2014;51:214–8. doi: 10.4103/0019-509X.146720. [DOI] [PubMed] [Google Scholar]
- 19. Vajaria BN, Patel KR, Begum R, Shah FD, Patel JB, Shukla SN, et al. Evaluation of serum and salivary total sialic acid and alpha-l-fucosidase in patients with oral precancerous conditions and oral cancer. Oral Surg Oral Med Oral Pathol Oral Radiol. 2013;115:764–71. doi: 10.1016/j.oooo.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 20. Tissot B, North SJ, Ceroni A, Pang PC, Panico M, Rosati F, et al. Glycoproteomics: past, present and future. FEBS Lett. 2009;583:1728–35. doi: 10.1016/j.febslet.2009.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pan S, Chen R, Aebersold R, Brentnall TA. Mass spectrometry based glycoproteomics–from a proteomics perspective. Mol Cell Proteomics MCP. 2011;10:R110003251. doi: 10.1074/mcp.R110.003251. [In eng] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen YT, Chong YM, Cheng CW, Ho CL, Tsai HW, Kasten FH, et al. Identification of novel tumor markers for oral squamous cell carcinoma using glycoproteomic analysis. Clinica Chimica Acta Int J Clin Chem. 2013;420:45–53. doi: 10.1016/j.cca.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 23. Guu SY, Lin TH, Chang SC, Wang RJ, Hung LY, Fang PJ, et al. Serum N-glycome characterization and anti-carbohydrate antibody profiling in oral squamous cell carcinoma patients. PLoS One. 2017;12:e0178927. doi: 10.1371/journal.pone.0178927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fang CY, Shen CH, Wang M, Chen PL, Chan MW, Hsu PH, et al. Global profiling of histone modifications in the polyomavirus BK virion minichromosome. Virology. 2015;483:1–12. doi: 10.1016/j.virol.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 25. Ishikawa S, Sugimoto M, Kitabatake K, Sugano A, Nakamura M, Kaneko M, et al. Identification of salivary metabolomic biomarkers for oral cancer screening. Sci Rep. 2016;6:31520. doi: 10.1038/srep31520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kademani D, Lewis JT, Lamb DH, Rallis DJ, Harrington JR. Angiogenesis and CD34 expression as a predictor of recurrence in oral squamous cell carcinoma. J Oral Maxillofac Surg. 2009;67:1800–5. doi: 10.1016/j.joms.2008.06.081. [DOI] [PubMed] [Google Scholar]
- 27. Feng XY, Li JH, Li JZ, Han ZX, Xing RD. Serum SCCA, Cyfra 21-1, EGFR and Cyclin D1 levels in patients with oral squamous cell carcinoma. Int J Biol Markers. 2010;25:93–8. doi: 10.1177/172460081002500206. [DOI] [PubMed] [Google Scholar]
- 28. Khandavilli SD, Ceallaigh P, Lloyd CJ, Whitaker R. Serum C-reactive protein as a prognostic indicator in patients with oral Squamous cell carcinoma. Oral Oncol. 2009;45:912–4. doi: 10.1016/j.oraloncology.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 29. Tadbir AA, Purshahidi S, Ebrahimi H, Khademi B, Malekzadeh M, Mardani M, et al. Serum level of MMP-3 in patients with oral squamous cell carcinoma–lack of association with clinico-pathological features. Asian Pac J Cancer Prev. 2012;13:4545–8. doi: 10.7314/apjcp.2012.13.9.4545. [DOI] [PubMed] [Google Scholar]
- 30. Shao K, Chen ZY, Gautam S, Deng NH, Zhou Y, Wu XZ. Posttranslational modification of E-cadherin by core fucosylation regulates Src activation and induces epithelial-mesenchymal transition-like process in lung cancer cells. Glycobiology. 2016;26:142–54. doi: 10.1093/glycob/cwv089. [DOI] [PubMed] [Google Scholar]
- 31. Wang X, Gu J, Ihara H, Miyoshi E, Honke K, Taniguchi N. Core fucosylation regulates epidermal growth factor receptor-mediated intracellular signaling. J Biol Chem. 2006;281:2572–7. doi: 10.1074/jbc.M510893200. [DOI] [PubMed] [Google Scholar]
- 32.Chong YM. Master’s Thesis. National Cheng Kung University; 2011. Site-specific N-glycan Analysis of Oral Cancer Cell (Unpublished Master’s Thesis) [Google Scholar]
- 33. Asazawa H, Kamada Y, Takeda Y, Takamatsu S, Shinzaki S, Kim Y, et al. Serum fucosylated haptoglobin in chronic liver diseases as a potential biomarker of hepatocellular carcinoma development. Clin Chem Lab Med. 2015;53:95–102. doi: 10.1515/cclm-2014-0427. [DOI] [PubMed] [Google Scholar]
- 34. Wang X, Abraham S, McKenzie JA, Jeffs N, Swire M, Tripathi VB, et al. LRG1 promotes angiogenesis by modulating endothelial TGF-beta signalling. Nature. 2013;499:306–11. doi: 10.1038/nature12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang J, Zhu L, Fang J, Ge Z, Li X. LRG1 modulates epithelial-mesenchymal transition and angiogenesis in colorectal cancer via HIF-1alpha activation. J Exp Clin Cancer Res. 2016;35:29. doi: 10.1186/s13046-016-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou Y, Zhang X, Zhang J, Fang J, Ge Z, Li X. LRG1 promotes proliferation and inhibits apoptosis in colorectal cancer cells via RUNX1 activation. PLoS One. 2017;12:e0175122. doi: 10.1371/journal.pone.0175122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Druhan LJ, Lance A, Li S, Price AE, Emerson JT, Baxter SA, et al. Leucine Rich alpha-2 Glycoprotein: a Novel Neutrophil Granule Protein and Modulator of Myelopoiesis. PLoS One. 2017;12:e0170261. doi: 10.1371/journal.pone.0170261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dieplinger H, Ankerst DP, Burges A, Lenhard M, Lingenhel A, Fineder L, et al. Afamin and apolipoprotein A-IV: novel protein markers for ovarian cancer. Cancer Epidem Biomar. 2009;18:1127–33. doi: 10.1158/1055-9965.EPI-08-0653. [In English] [DOI] [PubMed] [Google Scholar]
- 39. Wang F, Kohan AB, Lo CM, Liu M, Howles P, Tso P. Apolipoprotein A-IV: a protein intimately involved in metabolism. J Lipid Res. 2015;56:1403–18. doi: 10.1194/jlr.R052753. [DOI] [PMC free article] [PubMed] [Google Scholar]