Abstract
Primer sets were designed to target specific 16S ribosomal DNA (rDNA) sequences of photosynthetic bacteria, including the green sulfur bacteria, the green nonsulfur bacteria, and the members of the Heliobacteriaceae (a gram-positive phylum). Due to the phylogenetic diversity of purple sulfur and purple nonsulfur phototrophs, the 16S rDNA gene was not an appropriate target for phylogenetic rDNA primers. Thus, a primer set was designed that targets the pufM gene, encoding the M subunit of the photosynthetic reaction center, which is universally distributed among purple phototrophic bacteria. The pufM primer set amplified DNAs not only from purple sulfur and purple nonsulfur phototrophs but also from Chloroflexus species, which also produce a reaction center like that of the purple bacteria. Although the purple bacterial reaction center structurally resembles green plant photosystem II, the pufM primers did not amplify cyanobacterial DNA, further indicating their specificity for purple anoxyphototrophs. This combination of phylogenetic- and photosynthesis-specific primers covers all groups of known anoxygenic phototrophs and as such shows promise as a molecular tool for the rapid assessment of natural samples in ecological studies of these organisms.
Photosynthetic ability is widely distributed among microorganisms. Anoxygenic photosynthetic bacteria, or anoxyphototrophs, are prokaryotes capable of utilizing light as an energy source, but, unlike cyanobacteria, they do not evolve molecular oxygen. Anoxyphototrophs typically use sulfide or other reduced sulfur compounds as well as molecular hydrogen or a variety of small organic molecules as electron donors in photosynthesis (14). There are four known phylogenetic groups of anoxygenic phototrophs: the green sulfur bacteria, the green nonsulfur bacteria, the heliobacteria, and the purple bacteria (31, 36).
Green sulfur bacteria contain bacteriochlorophyll a and either bacteriochlorophyll c, d, or e and require sulfide for growth (14). The green nonsulfur bacteria (Chloroflexus group) contain bacteriochlorophyll c or d and chlorosomes, as do certain green sulfur bacteria, but produce a photosynthetic reaction center of the purple bacterial type (28). Heliobacteria are gram positive and produce a unique photosynthetic pigment, bacteriochlorophyll g (18, 19). Phototrophic purple bacteria are the most phylogenetically diverse anoxyphototrophs, with members in the α, β, and γ subclasses of the Proteobacteria (36). Purple bacteria contain bacteriochlorophyll a or b, and both purple sulfur and purple nonsulfur phototrophs are known (14).
The ecological and biogeochemical importance of anoxyphototrophs has long been recognized. These organisms supply fixed carbon to chemotrophic organisms and, because they are also excellent nitrogen-fixing bacteria (17), contribute to the fertility of the ecosystems in which they reside (16, 27). Anoxyphototrophs also play significant roles in the carbon and sulfur cycles (27, 33). In aquatic ecosystems, the oxidation of sulfide to sulfur and then to sulfate is a major biogeochemical activity, and anoxyphototrophs are often responsible for these activities in stratified lakes (27, 29).
The microbial ecology and diversity of anoxygenic phototrophs have been investigated using a variety of techniques. While the traditional approach of liquid enrichment or establishing a most-probable-number series leads to the isolation of a pure isolate, enrichment culture bias (1) must be taken into consideration. Although whole-cell detection of green sulfur bacteria using labeled oligonucleotides (32) and antibodies (4) has recently been described, most studies utilize a PCR technique for the sensitive detection of specific genes from bacterial populations (10, 12, 24, 25). This molecular approach overcomes the problem of enrichment bias and also allows for the detection of organisms present in small numbers that may nevertheless play significant ecological roles in the community. Through these types of molecular analyses, purple and green anoxyphototrophs have been identified from natural samples using nonspecific 16S ribosomal DNA (rDNA) primers followed by denaturing gradient gel electrophoresis (DGGE) and sequencing (6, 8). In addition, methods have been developed for the identification of purple bacteria from analyses of their low-molecular-weight RNA (5S RNA plus tRNA) banding patterns (5). Although this method is of some use in identifying pure cultures of purple phototrophs, its use in natural environments is doubtful.
Here we report the design and application of PCR primer sets targeted to phylogenetic genes for the detection of green sulfur bacteria, green nonsulfur bacteria, and heliobacteria and primers specific for a conserved photosynthesis gene for the detection of purple phototrophic bacteria. We also show the usefulness of these primers both with pure isolates of phototrophic bacteria and for environmental analyses.
MATERIALS AND METHODS
DNA extraction and purification.
Genomic DNAs from bacterial strains used as positive and negative controls were extracted using the PureGene DNA isolation kit (Gentra Systems, Inc., Minneapolis, Minn.). Alternatively, 1.5 ml of liquid culture was centrifuged, the cell pellet was resuspended in 40 μl of sterile water and 5 μl of chloroform, and the mixture was boiled at 95°C for 10 min to lyse the cells. Total community DNA was extracted from environmental samples using a FastDNA SPIN Kit for soil (Bio 101, Vista, Calif.) with cell disruption by ceramic beads.
Primer design and PCR amplification.
Four sets of primers targeted to specific groups of anoxygenic phototrophs were designed for this study (Table 1). Primers targeting 16S rDNA were used for detecting green sulfur bacteria, green nonsulfur bacteria, and heliobacteria. The primers were designed by downloading16S rDNA sequences from target organisms and their close relatives from the Ribosomal Database Project (20) and GenBank (2) and constructing an alignment based on secondary structure using the computer program SeqApp (11). Regions of 16 to 20 bases that were unique to the target organisms were selected as priming sites. The 16S rDNA primers were checked against Ribosomal Database Project II using the Probe Match program (20), primarily to identify small-subunit rDNAs from other organisms that each primer might target. Except for the heliobacterial primer HB.418F, each 16S rDNA primer was specific to the target group. In the case of HB.418F, although this primer could potentially bind to 16S rDNAs from organisms other than heliobacteria (e.g., Nitrospira and Dietzia spp.), the high specificity of the reverse primer HB.1159R would ensure that only DNAs from heliobacterial species would be amplified.
TABLE 1.
Photosynthetic and phylogenetic primers used in this study
| Primer | Target group | Sequence (5′→3′) |
|---|---|---|
| pufM.557F | Purple phototrophic bacteria | CGCACCTGGACTGGAC |
| pufM.750R | Purple phototrophic bacteria | CCCATGGTCCAGCGCCAGAA |
| GS.619F | Green sulfur bacteria | GGGGTTAAATCCATGTGCT |
| GS.1144R | Green sulfur bacteria | CAGTTCARTTAGAGTCC |
| CFX.856F | Green nonsulfur bacteria | TGCCTTAGCTCACGCGGTAA |
| CFX.1240R | Green nonsulfur bacteria | GCAACGCATTGTCGTGGCCA |
| HB.418F | Heliobacteria | TCTTCGGATTGTAAACCC |
| HB.1159R | Heliobacteria | CCGGTCGTCCCGGGCA |
Amplimer sizes for the different primer sets are as follows: pufM, 229 bp; GS, 525 bp; CFX, 384 bp; HB, 741 bp. All 16S rDNA primer names are based on E. coli numbering.
Because of the diverse phylogeny of purple anoxygenic phototrophs (36), a single set of 16S rDNA primers that would target all of these bacteria could not be designed (14). Thus, primers were designed to target the pufM gene, encoding a protein for the M subunit of the photosynthetic reaction center in purple sulfur and purple nonsulfur bacteria (9). An alignment of 29 pufM sequences was used to identify appropriate conserved priming sites in the pufM gene (the alignment is available on request). The pufM.750R primer sequence is a modification of a previously published pufM primer (23). All oligonucleotide primers were synthesized by Life Technologies (Grand Island, N.Y.).
The custom primer sets were used in PCR amplification mixtures containing 37.75 μl of tissue culture water, 5 μl of 10× PCR buffer without MgCl2, 3 μl of 25 mM MgCl2, 1 μl of each deoxynucleoside triphosphate (dATP, dCTP, dGTP, and dTTP) at 10 mM, 0.5 μl of each forward and reverse primer at 125 μg/ml, 1 μl of DNA template (ca. 100 ng), and 1 U of Taq DNA polymerase. All reaction components were from Promega (Madison, Wis.). Reactions were cycled in a Perkin-Elmer GeneAmp 2400 thermocycler at the following parameters: 94°C for 3 min, followed by 30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, with a final extension step at 72°C for 10 min. The resulting amplification products were electrophoresed in 0.7% (wt/vol) Tris-acetate-EDTA agarose gels and photographed.
DGGE analysis.
DGGE was used to separate DNA fragments of the same size that differed in base composition (22). A DCode Universal Mutation Detection System (Bio-Rad, Hercules, Calif.) was used to prepare and electrophorese all DGGE gels. The pufM amplification products were electrophoresed on a 10% acrylamide-bisacrylamide gel prepared from 40% acrylamide-bisacrylamide (38.93 g of acrylamide and 1.07 g of bisacrylamide dissolved in 100 ml of double-distilled water). The high and low concentrations of denaturant were varied in different experiments, but initially a 20 to 80% gradient was employed. Improved resolution of specific bands was achieved by narrowing the field of denaturant. Although a GC clamp is typically attached to one of the PCR primers for subsequent use in DGGE (21), high resolution of the DGGE bands was consistently obtained without a GC clamp.
Cloning and sequencing.
DGGE bands selected for sequence analysis were excised from the gel and placed in sterile water at 4°C overnight to elute the DNA (26). The eluate was purified and concentrated into 10 μl using a Geneclean Spin Kit (Bio 101). Following reamplification, 1 μl of the PCR product was cloned into the pCR 2.1-TOPO vector (Invitrogen, Carlsbad, Calif.) and sequenced using vector primers with the ThermoSequenase cycle sequencing kit (Amersham Life Science, Cleveland, Ohio).
RESULTS
Primers for green sulfur bacteria.
Although primers for the detection of green sulfur bacteria have previously been described (24), these primers do not match target sites for several green sulfur bacteria. Therefore, we designed a new set of primers targeting the 16S rDNA of green sulfur bacteria that is more inclusive and will detect six additional known members of the green sulfur bacteria according to the Probe Match program available through Ribosomal Database Project II.
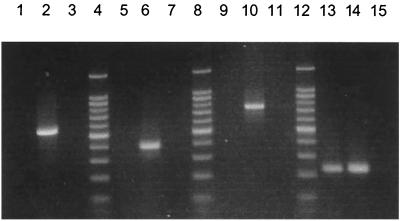
The primer set GS.619F and GS.1144R (Table 1) was tested using the green sulfur bacteria Chlorobium tepidum, Chlorobium limicola, and Chlorobium phaeobacteroides as positive controls. Predicted negative controls included Myroides (formerly Flavobacterium) odoratus, a close relative of the Chlorobiaceae (25, 36, 37), along with a number of purple bacteria and green nonsulfur bacteria, including Rhodospirillum rubrum, Thermochromatium (Chromatium) tepidum, Rhodobacter capsulatus, Klebsiella pneumoniae, Escherichia coli, and Chloroflexus aurantiacus strain OK-70-f1. Amplifications yielded a product of 525 bp only from DNAs from Chlorobium species (Fig. 1).
FIG. 1.
Agarose gel of amplification products obtained using the phototroph-specific primer sets. LF 10 m is DNA from an environmental sample of the water column of Lake Fryxell, Antarctica, collected at a depth of 10 m; only the pufM primer set resulted in a positive amplification using this sample (lane 13). The negative controls contained no DNA template. Lanes 4, 8, and 12, 100-bp molecular size marker; lanes 1 to 3, green sulfur primer set (LF 10 m, Chlorobium tepidum, and negative control, respectively); lanes 5 to 7, green nonsulfur primer set (LF 10 m, Chloroflexus aurantiacus, and negative control, respectively); lanes 9 to 11, heliobacterial primer set (LF 10 m, Heliorestis daurensis, and negative control, respectively); lanes 13 to 15, pufM primer set (LF 10 m, Rhodocyclus purpureus, and negative control, respectively).
Environmental samples and enrichment cultures were also tested using the green sulfur bacterial primer set. Four New Zealand hot spring enrichment samples from the Rotorua region, where Chlorobium tepidum was first discovered (7, 34), were amplified, and all were positive for green sulfur bacteria. In addition, water samples from six permanently frozen and stratified lakes in the Vestfold Hills of East Antarctica all revealed the presence of green sulfur bacteria (Fig. 2A). By contrast, two microbial mat samples from Yellowstone thermal springs (Bath Lake mat, 55°C; New Pit Spring mat, 48°C) (both located in the Upper Terraces region of Mammoth Hot Springs) tested negative for green sulfur bacteria.
FIG. 2.
Agarose gels of amplification products from environmental samples using the specific primer sets. (A) PCR using the green sulfur bacterial primer set (amplified fragments are 525 bp). Lane 1, Pendant Lake, Antarctica; lane 2, New Zealand hot spring JP-1; lane 3, 1-kb ladder. (B) PCR using the green nonsulfur bacterial primer set (amplified fragments are 384 bp). Lane 1, Yellowstone hot spring New Pit; lane 2, Yellowstone hot spring Bath Lake; lane 3, 1-kb ladder. (C) PCR using the pufM primer set (amplified fragments are 229 bp). Lane 1, Yellowstone hot spring New Pit; lane 2, Lake Fryxell, Antarctica; lane 3, 100-bp ladder.
Primers for green nonsulfur bacteria.
The green nonsulfur bacteria primers, CFX.856F and CFX.1240R, were designed for detecting Chloroflexus species; therefore, three different strains of Chloroflexus aurantiacus, the most common and well studied of the green nonsulfur bacteria (28), were used as positive controls. The strains included ones from Yellowstone (strain Y-400-fl), Oregon (strain OK-70-fl), and Japanese (strain J-10-fl) hot springs. Negative controls included Chlorobium tepidum, Chlorobium phaeobacteroides, Chlorobium limicola, Rhodobacter capsulatus, K. pneumoniae, and E. coli. A 384-bp amplification product was obtained only from DNAs from the three Chloroflexus strains (Fig. 1).
Using the CFX.856F and CFX.1240R primer set, green nonsulfur bacteria were detected in the two hot spring samples from Yellowstone that were tested (Fig. 2B) but not in the four water samples from the Vestfold Hills of East Antarctica that were tested.
Primers for heliobacteria.
Because the Heliobacteriaceae form a sister group with the endospore-forming, low-GC, gram-positive bacteria (3, 18), it was necessary to test several genera from this group as negative controls. Using primers HB.418F and HB.1159R, no amplification was observed from the low-GC gram-positive bacteria Bacillus subtilis, Bacillus cereus, Bacillus stearothermophilus, Bacillus megaterium, Sporosarcina ureae, Clostridium perfringens, and Clostridium pasteurianum. In addition and as expected, neither the purple bacteria Rhodobacter capsulatus, Rhodobacter blasticus, or Rhodobacter sphaeroides; the green nonsulfur bacteria Chloroflexus aurantiacus strains OK-70-fl and J-10-fl; nor the green sulfur bacteria Chlorobium tepidum and Chlorobium limicola yielded amplification products using the heliobacterial primer set. However, amplification products of 741 bp resulted from PCR using the HB.418F and HB.1159R primer set on five species of heliobacteria, including Heliobacterium gestii, Heliobacterium chlorum, Heliobacillus mobilis, the thermophile Heliobacterium modesticaldum (15), and the alkaliphile Heliorestis daurensis (3) (Fig. 1).
Photosynthetic pufM primers.
Because of the broad phylogenetic diversity of purple anoxyphototrophs, the primer set pufM.557F-pufM.750R was tested against a wide variety of both phototrophic and nonphototrophic Proteobacteria. A number of purple bacteria, both purple sulfur and purple nonsulfur species, were used as positive controls, including Rhodobacter capsulatus, Rhodobacter blasticus, Rhodobacter sphaeroides, Rhodobaca bogoriensis, Rhodopseudomonas palustris, Rhodomicrobium vannielii, and Rhodospirillum rubrum (all α Proteobacteria); Rhodocyclus tenuis, Rhodocyclus purpureus, and Rhodoferax antarcticus (all β Proteobacteria); and Thermochromatium tepidum from the γ subclass. Chloroflexus aurantiacus, a green nonsulfur bacterium that contains a purple bacterial-type reaction center (35), was also used as a positive control. In all cases, the expected 229-bp amplification product was observed (see data for Rhodocyclus purpureus in Fig. 1).
Nonphototrophic Proteobacteria, most notably those closely related to species of anoxyphototrophs, proved negative in PCRs with the pufM primer set. These included Bradyrhizobium japonicum (α); Dechloromonas agitata, Dechlorosoma suillum, and Nitrosomonas europaea (β); E. coli, K. pneumoniae, Salmonella enterica serovar Typhimurium, Pseudomonas aeruginosa, Pseudomonas fluorescens, Methylobacterium extorquens, Serratia marcesans, Azotobacter choococcum, Thiobacillus neapolitanus, and Shewanella putrefaciens (γ); Pelobacter carbinolicus (δ); and Campylobacter foetus (ɛ). As additional negative controls, the gram-positive species Staphylococcus aureus, B. stearothermophilus, and Sporosarcina ureae were tested along with the green sulfur bacterium Chlorobium tepidum and the cyanobacterium Fremyella deplosiphon; using the pufM primers, no amplification products were obtained from any of these organisms.
Because the pufM primer set was designed specifically for use in environmental population studies, several experiments were performed to ensure its usefulness and application in this arena. One such experiment involved spiking environmental soil samples with cells from pure cultures of anoxyphototrophs as positive controls for a specific primer set, extracting total genomic DNA from the sample, and amplifying to confirm that the chosen primer set would successfully detect the target cells in the sample (data not shown). Amplifications from environmental samples known from enrichment studies to contain certain species of anoxyphototrophs were also performed to confirm the utility of the primer sets for environmental detection of purple bacteria. For example, using the pufM primer set, purple anoxyphototrophs were readily detected in the New Pit Spring sample from Yellowstone and in all water and microbial mat samples tested from Lakes Fryxell and Hoare, Antarctica (Fig. 1 and 2C).
DGGE of pure bacterial cultures.
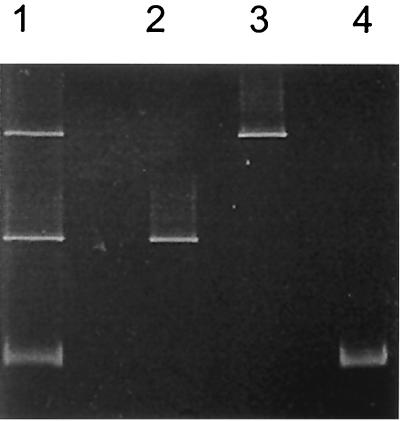
To test the utility of the pufM primers for detecting different species of purple anoxyphototrophs in the environment, DGGE was performed on pufM amplification products obtained from a known mixture of purple bacteria. Three anoxygenic phototrophic species were tested, including Thermochromatium tepidum, Rhodopseudomonas palustris, and Rhodobacter sphaeroides. Genomic DNA from each of the pure cultures was amplified with the pufM.557F-pufM.750R primer set. In addition, a mixture of the three genomic DNAs was also amplified with the same primer set. All of the PCR products were visualized on an agarose gel to verify that each amplification resulted in a single 229-bp product (data not shown). The amplification products were then electrophoresed on a DGGE gel, and, following electrophoresis, the mobilities of the single bands from the pure cultures were compared with those in the mixed culture lane. The pufM products from the three species were easily resolved by DGGE, and each single band corresponded to a band in the mixed lane (Fig. 3). DNA from each of the bands on the DGGE gel was then extracted, cloned, and sequenced. As expected, the pufM sequences from the pure-culture DGGE bands were identical to the sequences of the corresponding DGGE bands from the mixed amplification (data not shown).
FIG. 3.
DGGE gel of pufM amplification products from Rhodobacter sphaeroides (lane 2), Rhodobacter palustris (lane 3), Thermochromatium tepidum (lane 4), and a mixture of all three bacterial species (lane 1).
DISCUSSION
Using the suite of phylogenetic and photosynthetic primers described here, a molecular analysis of an environmental sample can be made for the presence and diversity of all prokaryotes that constitute an ecophysiological group: the anoxygenic phototrophic bacteria. These primer sets should greatly benefit studies of the ecology of these organisms and perhaps even reveal new habitats containing new species of these interesting and ecologically significant phototrophs.
The broad phylogenetic diversity of purple anoxyphototrophs made it impossible to design a single phylogenetically based primer set that would exclusively amplify the target organisms. In addition, the close evolutionary relationships of many of the purple anoxyphototrophs to their nonphotosynthetic counterparts resulted in a lack of appropriate priming sites that were unique to the phototrophic members of the group. Therefore, a metabolic gene, pufM, was chosen because it is unique to purple phototrophic bacteria and universally distributed across the group (9). Thus, because the pufM primers are based on a gene unique to organisms possessing a purple bacterial light-harvesting reaction center, they can be used to detect any photosynthetic member of the Proteobacteria regardless of phylogenetic affiliation. The pufM primer set will also detect Chloroflexus aurantiacus, a green nonsulfur bacterium that synthesizes a purple bacterial-type reaction center (35). However, use of both the green nonsulfur bacterial 16S rDNA and pufM primer sets on the same sample should allow for some resolution as to whether the detected organisms are green nonsulfur bacteria or purple bacteria (that is, a positive pufM and negative green nonsulfur bacterial result would conclusively indicate the presence of purple bacteria).
The vast database of available small-subunit rDNA sequences has resulted in the use of 16S rDNA probes in many ecological studies designed to assess bacterial populations (13, 30). The advantage of employing a 16S rDNA PCR primer set in such a setting is that the results are phylogenetically informative; sequence data gleaned from 16S rDNA can be analyzed in an evolutionary framework (36). Our contribution here is the deployment for the first time of specific primers for two phylogenetically distinct groups of anoxygenic phototrophs, the green nonsulfur bacteria and the heliobacteria, and alternative primers for green sulfur bacteria, a group for which primer sets have previously been described (32).
Metabolic gene probes, such as pufM, have several advantages over rDNA-based probes. The pufM primers provide a link to a physiology unique to a phylogenetically diverse group of organisms. Furthermore, if one were to analyze functional gene expression of pufM using mRNA detection techniques, it would be possible to determine whether or not photosynthetic metabolism was occurring in a specific environment. Thus, using reverse transcriptase PCR with the pufM primer set, it should be possible not only to track purple bacteria in a given habitat but also to determine which phylotypes are most metabolically active.
DGGE is superior to cloning and sequencing of PCR products in that it provides an immediate and quantitative assessment of the components of a population in a less time-consuming manner (21). While amplification with the group-specific primers in this study indicates the presence of anoxyphototrophs in environmental samples, amplification alone does not reveal how many different phylotypes (organisms with unique sequences that may or may not represent different species) of the target group might be present. However, DGGE can reveal many of the phylotypes in a PCR mixture by separating DNA fragments of the same size which differ in sequence (22).
In the case of pufM, the use of DGGE is particularly advantageous. Besides resolving PCR products of different sequence through DGGE, the small size of the pufM fragments (229 bp) allows for rapid sequencing and database analysis to yield candidate organisms from the environmental sample. Because the pufM gene is subject to horizontal transfer among the purple bacteria (23), pufM sequences obtained in this manner will not provide reliable phylogenetic information. However, using the pufM primer set, one could track the distribution of purple anoxyphototrophs through a stratified water column, for example, and, using quantitative PCR techniques, determine the relative abundances of different phylotypes. Moreover, the use of pufM to guide the progress of enrichment cultures for purple anoxyphototrophs, comparing the banding patterns with that of the environmental sample, should yield information on whether the major phylotypes present in the environment are being successfully enriched in the laboratory.
ACKNOWLEDGMENTS
We thank John Bowman for the Vestfold Hills lake samples, David M. Ward for the Yellowstone mat samples, Mary Lidstrom for the Methylobacterium DNA, and Jess Shively for the Thiobacillus DNA. We also thank Carl Bauer for suggesting pufM as a biomarker for purple anoxyphototrophs.
This work was funded by grant OPP0989195 from the National Science Foundation to L.A.A. and M.T.M.
REFERENCES
- 1.Amann R I, Ludwig W, Schileifer K. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson D A, Karsch-Mizrachi I, Lipman D J, Ostell J, Rapp B A, Wheeler D L. GenBank. Nucleic Acids Res. 2000;28:8–15. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryantseva I A, Gorlenko V M, Kompantseva E I, Achenbach L A, Madigan M T. Heliorestis daurensis gen. nov. sp. nov., an alkaliphilic coiled to rod-shaped phototrophic heliobacterium from an alkaline Siberian soda lake. Arch Microbiol. 1999;172:167–174. doi: 10.1007/s002030050756. [DOI] [PubMed] [Google Scholar]
- 4.Cahill A D, Stolz J F. Polyclonal antibodies to chlorosome proteins as probes for green sulfur bacteria. Appl Environ Microbiol. 1995;61:784–787. doi: 10.1128/aem.61.2.784-787.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casamayor E O, Calderón-Paz J I, Mas J, Pedrós-Alió C. Identification of phototrophic sulfur bacteria through the analysis of lmwRNA band patterns. Arch Microbiol. 1998;170:269–278. doi: 10.1007/s002030050642. [DOI] [PubMed] [Google Scholar]
- 6.Casamayor E O, Schäfer H, Bañeras L, Pedrós-Alió C, Muyzer G. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2000;66:499–508. doi: 10.1128/aem.66.2.499-508.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castenholz R W, Bauld J, Jørgensen B B. Anoxygenic microbial mats of hot springs: thermophilic Chlorobium sp. FEMS Microbiol Ecol. 1990;74:325–336. [Google Scholar]
- 8.Coolen M J L, Overmann J. Analysis of subfossil molecular remains of purple sulfur bacteria in a lake sediment. Appl Environ Microbiol. 1998;64:4513–4521. doi: 10.1128/aem.64.11.4513-4521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corson G E, Nagashima K V P, Matsuura K, Sakuragi Y, Wettasinghe R, Qin H, Allen R, Knaff D B. Genes encoding light-harvesting and reaction center proteins from Chromatium vinosum. Photosyn Res. 1999;59:39–52. [Google Scholar]
- 10.Fröstl J M, Overmann J. Arch. Microbiol. 174:50–58. 2000. Phylogenetic affiliation of the bacteria that constitute phototrophic consortia. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert D G. SeqApp. 1.9a157 ed. Bloomington: Biocomputing Office, Biology Dept., Indiana University; 1993. [Google Scholar]
- 12.Giovannoni S J, Rappe M S, Vergin K L, Adair N L. 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the green non-sulfur bacteria. Proc Natl Acad Sci USA. 1996;93:7979–7984. doi: 10.1073/pnas.93.15.7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gordon D A, Priscu J, Giovannoni S. Origin and phylogeny of microbes living in permanent Antarctic lake ice. Microb Ecol. 2000;39:197–202. doi: 10.1007/s002480000016. [DOI] [PubMed] [Google Scholar]
- 14.Imhoff J F. Taxonomy and physiology of phototrophic purple bacteria and green sulfur bacteria. In: Blakenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–15. [Google Scholar]
- 15.Kimble L K, Mandelco L, Woese C R, Madigan M T. Heliobacterium modesticaldum, sp. nov., a thermophilic heliobacterium of hot springs and volcanic soils. Arch Microbiol. 1995;163:259–267. [Google Scholar]
- 16.Madigan M T. Microbiology, physiology, and ecology of phototrophic bacteria. In: Zehner A J B, editor. Biology of anaerobic microorganisms. New York, N.Y: John Wiley & Sons; 1988. pp. 39–111. [Google Scholar]
- 17.Madigan M T. Microbiology of nitrogen fixation in photosynthetic bacteria. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 915–928. [Google Scholar]
- 18.Madigan M T. The family Heliobacteriaceae. In: Garrity G, editor. Bergey's manual of systematic bacteriology. Vol. 1. New York, N.Y: Springer; 2000. pp. 615–620. [Google Scholar]
- 19.Madigan M T, Ormerod J G. Taxonomy, physiology and ecology of heliobacteria. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 17–30. [Google Scholar]
- 20.Maidak B L, Cole J R, Parker C T, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muyzer G, der Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers R M, Maniatis T, Lerman L S. Detection and localization of single base changes by denaturing gradient gel electrophoresis. Methods Enzymol. 1987;155:501–527. doi: 10.1016/0076-6879(87)55033-9. [DOI] [PubMed] [Google Scholar]
- 23.Nagashima K V P, Hiraishi A, Shimada K, Matsuura K. Horizontal transfer of genes coding for the photosynthetic reaction centers of purple bacteria. J Mol Evol. 1997;45:131–136. doi: 10.1007/pl00006212. [DOI] [PubMed] [Google Scholar]
- 24.Overmann J, Coolen M J L, Tuschak C. Specific detection of different phylogenetic groups of chemocline bacteria based on PCR and denaturing gradient gel electrophoresis of 16S rRNA gene fragments. Arch Microbiol. 1999;172:83–94. doi: 10.1007/s002030050744. [DOI] [PubMed] [Google Scholar]
- 25.Overmann J, Tuschak C. Phylogeny and molecular fingerprinting of green sulfur bacteria. Arch Microbiol. 1997;167:302–309. doi: 10.1007/s002030050448. [DOI] [PubMed] [Google Scholar]
- 26.Øvreås L, Forney L, Daae F L, Torsvik V. Distribution of bacterioplankton in meromictic Lake Sælenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol. 1997;63:3367–3373. doi: 10.1128/aem.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfennig N. Ecology of phototrophic purple and green sulfur bacteria. In: Schlegel H G, Bowien B, editors. Autotrophic bacteria. New York, N.Y: Springer-Verlag; 1989. pp. 81–96. [Google Scholar]
- 28.Pierson B K, Castenholz R W. Taxonomy and physiology of filamentous anoxygenic phototrophs. In: Blakenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 31–47. [Google Scholar]
- 29.Pierson B K, Olson J M. Photosynthetic bacteria. In: Amesz J, editor. Photosynthesis. Amsterdam, The Netherlands: Elsevier Science Publishers, B.V.; 1987. pp. 21–42. [Google Scholar]
- 30.Skirnisdottir S, Hreggvidsson G O, Hjorleifsdottir S, Marteinsson V T, Petursdottir S K, Holst O, Kristjansson J K. Influence of sulfide and temperature on species composition and community structure of hot spring microbial mats. Appl Environ Microbiol. 2000;66:2835–2841. doi: 10.1128/aem.66.7.2835-2841.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trüper H G, Pfennig N. Characterization and identification of the anoxygenic phototrophic bacteria. In: Starr M P, Stolp H, Trüper H G, Balows A, Schlegel H G, editors. The prokaryotes. New York, N.Y: Springer-Verlag; 1981. pp. 299–312. [Google Scholar]
- 32.Tuschak C, Glaeser J, Overmann J. Specific detection of green sulfur bacteria by in situ hybridization with a fluorescently labeled oligonucleotide probe. Arch Microbiol. 1999;171:265–272. doi: 10.1007/s002030050709. [DOI] [PubMed] [Google Scholar]
- 33.Van Gemerden H, Beeftink H H. Ecology of phototrophic bacteria. In: Ormerod J G, editor. The phototrophic bacteria. Berkeley: University of California Press; 1983. pp. 146–185. [Google Scholar]
- 34.Wahlund T, Castenholz R W, Woese C R, Madigan M T. A thermophilic green sulfur bacterium from New Zealand hot springs. Chlorobium tepidum, nov. sp. Arch Microbiol. 1991;156:81–91. [Google Scholar]
- 35.Watanabe Y, Feick R G, Shiozawa J A. Cloning and sequencing of the genes encoding the light-harvesting B806-866 polypeptides and initial studies on the transcriptional organization of puf2B, puf2A and puf2C in Chloroflexus aurantiacus. Arch Microbiol. 1995;163:124–130. doi: 10.1007/BF00381786. [DOI] [PubMed] [Google Scholar]
- 36.Woese C R. Prokaryote systematics: the evolution of a science. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1992. pp. 3–18. [Google Scholar]
- 37.Woese C R, Mandelco L, Yang D, Gherna R, Madigan M T. The case for relationship of the flavobacteria and their relatives to the green sulfur bacteria. Syst Appl Microbiol. 1990;13:258–262. doi: 10.1016/s0723-2020(11)80196-7. [DOI] [PubMed] [Google Scholar]