Abstract
Metabolomics is considered an effective approach for understanding metabolic responses in complex biological systems. Accordingly, it has attracted increasing attention for biomarker discovery, especially in cancer. In this study, we used a non-invasive method to evaluate four urine metabolite biomarker candidates—o-phosphoethanolamine, 3-amio-2-piperidone, uridine and 5-hydroxyindoleactic acid—for their potential as bladder cancer diagnostic biomarkers. To analyze these targeted amine- and phenol-containing metabolites, we used differential 12C2-/13C2-dansylation labeling coupled with liquid chromatography/tandem mass spectrometry, which has previously been demonstrated to exhibit high sensitivity and reproducibility. Specifically, we used ultra-performance liquid chromatography (UPLC) coupled with high-resolution Fourier transform ion-cyclotron resonance MS system (LC-FT/MS) and an ion trap MS with MRM function (LC-HCT/MS) for targeted quantification. The urinary metabolites of interest were well separated and quantified using this approach. To apply this approach to clinical urine specimens, we spiked samples with 13C2-dansylatedsynthetic compounds, which served as standards for targeted quantification of 12C2-dansylated urinary endogenous metabolites using LC-FT/MS as well as LC-HCT/MS with MRM mode. These analyses revealed significant differences in two of the four metabolites of interest—o-phosphoethanolamine and uridine—between bladder cancer and non-cancer groups. O-phosphoethanolamine was the most promising single biomarker, with an area-under-the-curve (AUC) value of 0.709 for bladder cancer diagnosis. Diagnostic performance was improved by combining uridine and o-phosphoe-thanolamine in a marker panel, yielding an AUC value of 0.726. This study confirmed discovery-phase features of the urine metabolome of bladder cancer patients and verified their importance for further study.
Keywords: Bladder cancer, Biomarker, Phosphoethanolamine, Uridine, Metabolites
1. Introduction
It has been estimated that bladder cancer will account for 81,990 new cases and 17,240 bladder cancer-estimated deaths worldwide during 2018 [1]. Bladder cancer progression and death rates are highly related to tumor stage and cancer grade; thus, these metrics are crucial indicators of clinic prognosis and optimal treatments [2]. The progression rate for bladder cancer patients with low-grade tumors is approximately 6%, whereas that for patients with high-grade lesions is approximately 17% [3]. Hence, early detection and precise prognosis of cancer severity are critical for clinical management and assessment of progression risk.
Among the current methods for diagnosing disease, predicting progression and monitoring recurrence are cystoscopy, biopsy, urine cytology, and imaging [4]. However, these prognostic tools are not without drawbacks. Both cystoscopies and biopsies are invasive processes, and cystoscopy is expensive, painful and less sensitive for detecting high-grade tumors [4]. Although urine cytology is a highly sensitive, specific, cost-effective and convenient screening technique, it lacks the ability to detect low-grade bladder cancer [2]. An ideal screening test would retain these desirable characteristics (inexpensiveness, high sensitivity, specificity and non-invasiveness), while minimizing drawbacks.
Metabolomics, one of the “omics” technologies, is a comprehensive technique for global metabolite screening and profiling. Metabolomics is capable of investigating relatively small molecules in biofluids (blood, urine, plasma) or tissue, and is therefore complementary to genomics, transcriptomics, and proteomics. Moreover, because metabolism-related proteins are subject to modification through a series of complex biological processes at transcriptional, translational and post-translational levels [5], metabolomics reflects the dynamic responses of living systems to path physiological stimuli or genetic influences [6]. Similarly, specific metabolites secreted into biofluids reflect the operation of molecular pathways involving cellular ontogenesis, providing information that cannot be obtained directly using other omics approaches [6].
The bladder is the organ responsible for the storage of urine. Therefore, screening for cancer-associated metabolites in urine is a potentially on-invasive approach for disease prognosis and examination of bladder cancer recurrence. Accordingly, metabolomics is a promising approach for the discovery of bladder cancer biomarkers. Despite these potential advantages, current research on metabolic biomarker discovery faces certain limits. After applying metabolomics as a platform for biomarker discovery, candidate metabolic biomarker candidates must be confirmed and accurately quantified, a requirement made more difficult by the small, but significant, differences in levels of metabolites in biological specimens; even efforts to subsequently confirm metabolites identified in the discovery phase have met with limited success [7]. The lack of synthetic standards and limited availability of clinical specimens for novel metabolite identification are the main reasons for difficulties in verifying metabolite biomarker candidates.
One approach for addressing these issues is ultra-performance liquid chromatography (UPLC) coupled with mass spectrometry (LC-MS), which provides a good platform for metabolic biomarker discovery. Advantages of LC-MS include the ability to separate, ionize and detect a wide range of analytes and identify chemical features of molecules, such as retention time and mass-to-charge ratio (m/z). Multiple reaction monitoring (MRM)-MS provides additional information on fragments of targeted metabolites [7] and can be used for absolute quantification. Nevertheless, urinary metabolites are highly diverse [8] and some have properties that limit their retention on a reverse phase column, resulting in difficulties in analyzing the entire metabolome at once. Kuo et al. [8] demonstrated a solution to this challenging problem, showing that the chemical behaviors of amine- and phenol-containing metabolites could be altered by dansyl chloride labeling. This study [8] also demonstrated several important characteristics of differential isotope dansylation labeling, including high sensitivity, an expanded detection range for highly diverse molecules, and impressive reproducibility. Dansylated metabolites, even polar or ionic metabolites that are normally not retained on a reverse-phase (RP) column, can be separated by RPLC, increasing metabolite signal intensity and producing a better signal-to-noise ratio [8]. In addition, 12C-/13C-isotope dansylation labeling offers the characteristic of signal pairs on a spectrum that facilitates the confidence in identification and quantification of endogenous metabolites. Because it uses isotope-labeled synthetic compounds as a quantitative reference standard, 12C-/13C-isotope dansylation labeling coupled with LC-MS/MS is considered as a promising technique for metabolic quantification and biomarker discovery [8–11].
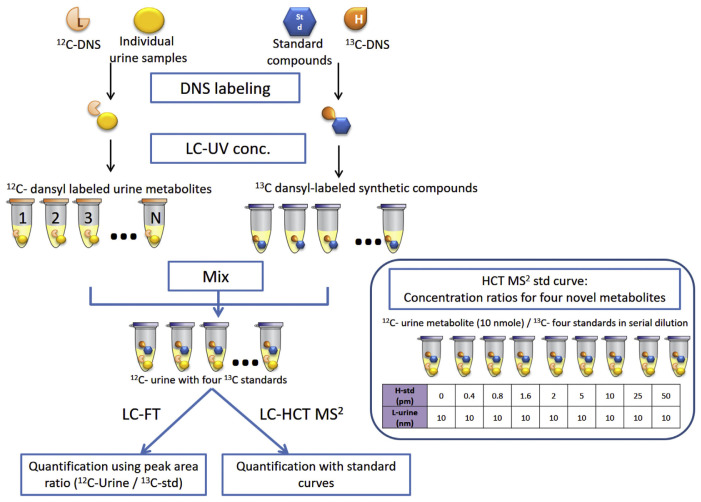
Our previous work [12] identified numerous putative markers for bladder cancer using isotope dansylation labeling in conjunction with HPLC-MS (Supplemental Table 1). However, analytical methods for assessing these potential markers have not been established for metabolite identification and large-scale quantification for clinical applications. The identification of these previous potential metabolite biomarker candidates was based on matching their molecular weight with compounds from metabolome databases. An alternative identification and quantification strategy is to perform analyses after danslyation labeling utilizing highly purified commercial compounds. In this study, we sought to apply HPLC coupled with two types of mass spectrometry—Fourier transform ion-cyclotron resonance MS (FT/MS) and ion trap MS with MRM function (HCT/MS)—to verify the clinical utility of five potential urinary metabolite biomarkers discovered in our previous work [12] using the five commercially-available compounds as standards. The standard compounds were used to compare features (e.g., m/z, fragmentation masses, and method specific retention time) to confirm the identification of candidate metabolite. As indicated above, stage and grade of bladder cancer are crucial for clinical prognosis. Accordingly, three clinical subgroups of bladder cancer patients were recruited for the current study: patients with low-grade, early-stage (LgEs) cancer; patients with high-grade, early-stage (HgEs) cancer; and patients with high-grade, advanced-stage (HgAs) cancer. Age-matched hernia patients were selected as the control group for targeted quantification of these five metabolites and evaluation of their utility as bladder cancer biomarkers. Fig. 1 shows the workflow for targeting amine- and phenol-containing metabolites in bladder cancer urine using dansylation isotope labeling and LC-MS.
Fig. 1.
Workflow for targeting amine- and phenol-containing metabolites in bladder cancer urine using dansylation isotope labeling and LC-MS. Commercial standard compounds were labeled with 13C2-DnsCl, spiked with 12C2-DnsCl–labeled urinary metabolites, and then analyzed by LC-FT/MS and LC-HCT/MS. The blue block on the right represents serial dilutions used to prepare standard curves in LC-HCT/MS.
2. Material and methods
2.1. Reagents
Compounds used for dansylation, including sodium carbonate (NaHCO3), sodium bicarbonate (Na2CO3), sodium hydroxide (NaOH), formic acid, and the authentic reference standards of the previously annotated putative urine biomarkers, 3-amino-2-piperidone, pyroglutamic acid, o-phosphoethanolamine, 5-hydroxyindoleacetic acid and uridine, and amino acid standards (#AAS 18) were purchased from Sigma–Aldrich (St. Louis, MO, USA)MS grade water, acetonitrile (ACN) containing 0.1% formic acid, and water containing 0.1% formic acid were also obtained from Sigma–Aldrich. Can and formic acid used as elution buffers for LC-UV were purchased from J.T. Baker-Avantor (Easton, PA, USA) and Sigma–Aldrich, respectively. Ultrapure water was obtained by purification using a Milli-Q system (Merck Millipore, Darmstadt, Germany). 12C2-Dansyl chloride(12C2-DnsCl) was from Sigma–Aldrich, and 13C2-dansyl chloride( 13C2-DnsCl) was obtained from Wuxi Beita Pharmatech (Jiangsu, China).
2.2. Samples and preparation
2.2.1. Urine sample collection and preparation of quality control (QC) urine
All urine specimens were collected followed a previously established protocol [12]. In brief, the first morning urine was collected from each subject and kept on ice; thereafter, samples were filtered twice with 0.22 nm filters (Millipore) and stored at −80 °C before dansylation labeling. All human urine samples were collected at Chang Gung Memorial Hospital in Taiwan with approval of the local Institutional Review Board.
In this study, metabolites discovered as bladder cancer biomarker candidates in our previous study were subjected to new quantitative analytical methods. Our original statistical analysis showed that these biomarkers exhibited differences in concentration between cancer and control specimens [12]. Clinical urine specimens included those from hernia patients (control group) and bladder cancer patients, the latter of which was further divided into three subgroups: LgEs, HgEs, and HgAs. An external quality control sample (QC urine) was prepared by pooling 150 human urine specimens followed by dansylation labeling for analysis by LC-MS. QC urine samples were analyzed daily to ensure the performance stability and reproducibility of the LC-MS system.
2.2.2. Dansylation labeling
The dansylation method used was based on a previous study by Peng and coworkers [12]. In brief, clinical urine samples were thawed at 4 °C, and authentic reference standard compounds were dissolved in MS grade water (Fluka). Sample solutions were centrifuged at 10,000 rpm at 4 °C for 10 min using an Eppendorf 5417R centrifuge to remove cells and debris before labeling.
Urine and standard compounds were labeled with 12C2-DnsCl and 13C2-DnsCl, respectively, by first adding 25 μl of 0.5 M NaHCO3/Na2CO3 and 75 μl of dansyl chloride solution (1 mg in 80.5 μl of ACN) into 50 μl of each individual sample or 50 μl of the standard compounds (2 mM). Urine specimens were diluted with water if the concentrations of metabolites in original urines were high [13,14]. Dansylation reactions were performed by incubating sample mixtures at 40 °C for 45 min on an orbital shaker at 200 rpm, and reactions were stopped by immediately transferring reaction mixtures to ice. The reaction was quenched by adding 10 μl of 250 mM NaOH, followed by shaking at 200 rpm at 40 °C for 10 min. Samples on ice were prepped for LC-UV concentration determination by adding 50 μl of 425 mM formic acid in 50% ACN to each specimen.
2.2.3. Concentration normalization of dansylated metabolites by LC-UV
Since the efficiencies of dansylation of all metabolites from different urine samples may vary, a concentration normalization process is essential. To this end, we used an Acquity UPLC with ultra violet detection (Acquity TUV) system (Waters, Milford, MA, USA) to determinate the total concentration of dansylated metabolites after dansylation labeling.
A standard curve for quantification of dansylated metabolites was prepared by dansyl chloride labeling of a serially diluted (0.52, 1.3, 2.6, 5.2, and 7.8 mM) 18-amino-acid standard mixture, followed by LC with UV detection of absorption at 338 nm. LC-UV concentration normalization was performed by injecting 2 μl of labeled samples onto an ACQUITY LC-UV system (Waters) at a flow rate of 0.45 ml/min and eluting with the following gradient: 1% ACN (J.T. Baker) from 0 to 1 min, 95% ACN from 1 to 3 min; and 1% ACN from 3 to 6 min. A fixed amount (45 nmol) of each individual of 12C2-Dns–labeled urine sample was spiked with the mixture of 13C2-Dns–labeled standards (45 pmole for each compound). Samples were dried, re-dissolved in 30 μl 50% ACN containing 0.1% formic acid, and subsequently analyzed by LC/MS.
2.3. Sample analysis
2.3.1. MS analysis
Two mass specters, an ion trap MS (HCT/MS; Bruker Daltonics, Bremen, Germany), and a Fourier transform ion cyclotron resonance MS (FT/MS) system (Apex-Qe-SHEDS FTICR, 9.4 T; Bruker Daltonics, Bremen, Germany) were utilized for quantification of metabolites. Both MS systems were linked to an ACQUITY UPLC system (Waters), eluted with MS grade solvents (solvent A: 5% ACN in water containing 0.1% formic acid; solvent B: 99% ACN containing 0.1% formic acid) according to the following program: 0–2 min, 5% B; 2–3 min, 5% B to 15% B; 3–13 min, 15% B to 35% B; 13–25 min, 35% B to 70% B; 25–28 min, 70% B to 99% B; 28–30 min, 99% B; 30–30.1 min, 99% B to 5% B; 30.1–32.5 min, 5% B. All MS spectra were obtained in positive ion mode.
All samples for the two systems were prepared as described above. The optimized injection amount was 10 nmole (6.6 μl) of 12C2-Dns–labeled endogenous urinary metabolites and 10 pmole of each 13C2-Dns–labeled standard compound per LC-MS run. A QC sample was analyzed daily to ensure the stability of the system. Measurements of control group and study group samples were randomized to minimize instrument-and environment-related variations. Each metabolite sample was analyzed in triplicate in consecutive runs, with a wash run in between; an additional blank run (5% ACN containing 0.1% formic acid) was performed to prevent signal interference by carry-over between different clinical samples.
2.3.2. Confirmation of synthetic standards detection and relative quantification using LC-FT/MS system
An LC-FT/MS system with Waters binary management was configured by connecting an Acquity UPLC Waters system with an Acquity UPLC HSS T3 1.8 μmcolumn (Waters) to an FT/MS system (Bruker). To investigate ionization abilities of these reference compounds without labeling, the compounds were dissolved at a concentration of 2 mM in LC-MS grade water (Fluka), then diluted to yield 10, 100, and 500 pmol/μl samples, which were injected separately by direct infusion at a flow rate of 180 μl/min apart from this, ionization and detectability of dansyl-labeled standard compounds (5, 10, and 50 pmol/run) were evaluated by injecting standards onto the LC-FT/MS system. The LC gradient used for LC-FT/MS analysis of individual specimen was the same as that used for LC-HCT/MS.
2.3.3. Precise quantification by LC-HCT/MS analysis: MS2 fragmentation and standard curve
A high-capacity spherical trap (HCT) ultra MS (Bruker) coupled with an Acquity UPLC BEH C18 (1.7 μm) column was employed for quantification of metabolites. To ensure that the commercial standards were detectable after labeling, we injected 50, 100 and 500 pmol/μl of the labeled standard compounds onto the LC-HCT system by direct infusion at a flow rate of 180 μl/min and acquired MS2 fragmentation spectra. For quantification, fragment ions of m/z 252.1, 234.1, 366.1, and 381.2 were acquired in the MS2 spectrum of the parent ions m/z 375.1 (12C2-o-phosphoethanolamine), m/z 348.2 (12C2-Dns-3-amino-2-piperidone), m/z 478.1 (12C2-uridine), and m/z 425.1 (12C2-5-hydroxyindoleacetic acid. For the real sample analysis, we increased the number of data points by splitting the time course into four segments with the following MS2 fragmentation settings: 0–5 min with no MS2 fragmentation; 5–9.5 min with fragmentation of m/z 375.1 and m/z 377.1; 9.6–17.1 min with parent ions of m/z 348.2, m/z 350.1, m/z 478.1, and m/z 480.1; and 17.2–32 min with m/z 425.1 and m/z 427.1 as the precursor ions. The time segments were determined based on the retention times of standard compounds. The corresponding ions of parent and fragment ions of 12C-labeled endogenous metabolites and corresponding internal standards are summarized as Table 1A. The mixture of 13C2-Dns standard compounds was serially diluted (0, 0.4, 0.8, 1.6, 2, 5, 10, 25, and 50 pmol), and a standard curve was prepared by spiking each concentration point with 10 nmol of 12C2-Dns–labeled endogenous urinary metabolites.
Table 1A.
Fold-changes in targeted metabolites in bladder cancer urine specimens compared with specimens from hernia patients. Fold-changes in metabolites in the total bladder cancer group and three bladder cancer subgroups compared with that in the hernia group are shown, together with their retention times in FT/MS and HCT/MS. Information on putative biomarkers reported by Jun Peng et al. [12] is also shown. O-phosphoethanolamine and uridine concentrations were significantly different between urine samples from bladder cancer and hernia patients.
| Metabolomic Analysis Peng [12] | Targeted Analysis FT-MS | Q1 (m/z) | Q3 (m/z) | Time segments (min) | Targeted Analysis HCT-MS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||||
| Fold change | Fold change | ||||||||||||||
|
|
|
|
|||||||||||||
| RT (min) | Fold change BC/hernia | RT (min) | BC | LgEs | HgEs | HgAs | RT (min) | BC | LgEs | HgEs | HgAs | ||||
| O-phosphoethanolamine | 2.6 | 1.53 | 6 | 1.63 | 1.34 | 1.47 | 2.08 | 375.1 | 252.1 | 5.0–9.5 | 5.1 | 1.57 | 1.31 | 1.5 | 1.9 |
| 377.1 | 254.1 | ||||||||||||||
| 3-Amino-2-piperidone | 9.19 | 1.39 | 11.7 | 1.15 | 1.30 | 1.19 | 0.95 | 348.2 | 234.1 | 9.6–17.1 | 9.6 | 1.19 | 1.33 | 1.27 | 0.95 |
| 350.1 | 236.1 | ||||||||||||||
| Uridine | 7.5 | –1.25 | 12.6 | 1.78 | 1.70 | 1.71 | 1.94 | 478.1 | 366.1 | 9.6–17.1 | 10.6 | 1.80 | 1.73 | 1.78 | 1.89 |
| 480.1 | 368.2 | ||||||||||||||
| 5-Hydroxyindoleacetic acid | 16.7 | 1.32 | 19.9 | 1.10 | 1.10 | 1.15 | 1.04 | 425.1 | 381.2 | 47.2–32.0 | 17.9 | 1.08 | 1.09 | 1.13 | 1.02 |
| 427.1 | 383.2 | ||||||||||||||
2.4. Data analysis and statistics
Metabolites were quantified by obtaining a specific MS2 fragment of the corresponding precursor ion using extract ion chromatography (EIC) and exporting the results to an Excel file for analysis. Peak area data were subsequently converted into urinary metabolite concentrations based on the ratio of 12C2-Dns-metabolite to 13C2-Dns-reference for FT/MS data or the standard curve for HCT/MS data. The levels of individual targeted metabolites were then obtained. Therefore, fold change (Table 1A) was determinate by the ratio of average targeted metabolite amount of disease group (bladder cancer) to which of control group (hernia). Similarly, fold changed of bladder cancer subgroups (LgEs, HgEs and HgAs) were calculated with the same method. Prism or SPSS (v12.0, SPSS Inc.) software was used for additional statistical analyses, including receiver operating characteristic (ROC) curve and chi square analyses, as well as preparation of bar charts and dot plots.
3. Results
To establish targeted, quantitative methods for verifying previously discovered dansylated amine- and phenol-compounds as bladder cancer-associated urinary proteins, we collected urine specimens from 59 hernia patients without hematuria (HU) or urinary tract infection (UTI) conditions as a control group, and a total of 60 bladder cancer patients divided into three subgroups (n = 20/subgroup). Commercially available standard compounds were labeled with 13C2-Dns, and individual urine samples were labeled with 12C2-Dns. The endogenous concentration of targeted metabolites in urine samples was then quantified based on the signals of known concentrations of standard compounds using LC-FT/MS and LC-HCT/MS. The workflow for targeting amine- and phenol-containing metabolites in bladder cancer urine using dansylation isotope labeling and LC-MS is shown as Fig. 1.
To ensure the detectability of dansylated commercial standards, we injected 5, 10, and 50 pmole of dansylated standard compounds into the LC-FT/MS system. This analysis showed that four of the five labeled commercial standards (Supplementary Table 2)—O-phosphoethanolamine, 3-amino-2-piperidone, uridine, and 5-hydroxyindolectic acid—but not dansylated pyroglutamic acid, were detectable by UV and LC-MS. Therefore, only these four 13C-standard compounds were targeted for the development of quantification methods based on optimization of MS2 fragmentation using the LC-HCT/MS system.
Complex compounds in urine samples were analyzed by applying a LC system to separate metabolites of interest. The accuracy of quantitative results largely depends on establishing LC conditions under which the four targeting metabolites can be well separated. Accordingly, we investigated different LC conditions to determine optimal elution conditions for separation of commercial standards. Using these conditions (see Materials and methods), we found that, of the four dansylated standard compounds of interest, 3-amino-2-piperidone and uridine eluted with very similar retention times in LC-MS spectra, but the peaks of the remaining two metabolites were well separated.
3.1. Selection of MS2 fragments using LC-HCT/MS and application for quantification
Using dansyl chloride-labeled standard compounds, we examined MRM fragments of the corresponding parent ions to evaluate suitable daughter ions that would not be susceptible to detect ion interference, ion suppression, or matrix effects contributed by the biological samples. As described above, the four detectable standard compounds were analyzed by HCT/MS by direct injection at a flow rate of 180 μl/min, followed by MS/MS fragmentation at the appropriate ion mass. The resulting MS2 spectra from direct infusion in HCT MRM mode yielded the following daughter ions of the four labeled metabolites: 186.2, 188.2, 234.1 and 236.1 for dansylated 3-amino-2-piperidone; 252.1, 254.1277.1 and 279.1 for dansylated o-phosphoethanolamine; 366.1 and 368.2 for dansylated uridine; and 381.2 and 383.2 for dansylated 5-hydroxyindolectic acid.
Given the relatively short interval separating Dns-3-amino-2-piperidone (~11.7 min) and Dns-uridine (~12.6 min), we sought to collect specific daughter ions between these time frames for these particular metabolites. Hence, for MRM ionization on the LC-HCT system, we defined three time segments with appropriate ionized ions: (1) ionization of o-phosphoethanolamine (m/z 375.1or m/z 377.1) from 5 to 9.5 min, (2) ionization of 3-amino-2-piperidone (m/z 348.2 or m/z 350.1) and uridine (m/z 478.1 or m/z 480.1) from 9.5 to 17.1 min, and (3) ionization of 5-hydroxyindolectic acid (m/z 425.1 or m/z 427.1) from 17.1 to 30 min. After comparison of the LC-MS experimental conditions of metabolomics profiling [12] and targeted analyses by two LC-MS systems in this work, Eclipse plus C18 column (2.1 mm × 100 mm, 1.8 μm), an Waters Acquity UPLC BEH C18 (1.0 mm × 100 mm,1.7 μm) as well as Waters Acquity UPLC HSS T3 (1.0 mm × 100 mm, 1.8 μm) were utilized for the metabolomic analysis [12], LC-HCT/MS and LC-FT/MS, respectively. To optimize each elution condition, LC gradient of elution solvents were different in these studies. The difference in elution order of 3-amino-2-piperidone and uridine in the previous work (Table 1A) could be caused by the different columns and gradient of LC systems.
3.2. Development of a method for measuring the urinary concentration of four targeted metabolites
Because we applied two mass systems in this study for analysis of biological samples, we needed to apply two different quantification methods. Although LC-FT is a high-resolution chromatographic technique, it may be susceptible to isobaric inference. Because peak area is based on parent ions under full-scan mode, there is a concern that other compounds with the same m/z value as the isobaric interference could contribute to peak areas. By contrast, the LC-HCT system is able to fragment parent ions into MS2 or MS3, and thus is able to more reliably identify parent/daughter ion pairs as the correct compounds. However, the resolution of HCT/MS is lower than that of FT/MS. Accordingly, using both of these systems provides increased confidence in both the identification (LC-HCT/MS) and quantification (LC-HCT/MS) of the metabolites of interests.
Only five out of the ten putative metabolites were commercial availability for the following work. Among the five metabolites, dimethylated-pyroglutamic acid standard could not be accurately quantified due to poor detectability by UV and MS. The identification of the metabolites was based on accurate mass of the MS result for searching metabolite databases to gain one or more putative identifications. The putative identification will need further validation experiment to confirm its true chemical identification. In this study, commercially available pyroglutamic acid standard after dimethylation was not detected on UV nor on MS. We speculated the reasons were possibly due to poor yield of dansylation during sample preparation, or low-ionization efficiency in MS analysis, or wrong putative identification in the database searching of the discovery phase of metabolite biomarker discovery. Metabolites were quantified based on the isotopic ratio of 12C2 (endogenous metabolites) and known concentrations of 13C2-DnsCl–labeled metabolite standards. The first step in this process was identification of peaks of endogenous and standards of four of the five previously discovered metabolite biomarker candidates using isotopic pairs with accurate m/z and co-eluted retention times. For quantification by high-resolution FT/MS, EIC isotopic pairs for the four labeled analytes of interest were acquired according to the molecular weights listed in Supplementary Table 2. For quantification by HCT/MS, MS2 signals in Table 1A were acquired in MRM mode, which has lower background interference. The defined peaks of the designated m/z values were extracted, and their areas were integrated and converted to measures of the concentrations of the corresponding labeled metabolites by applying the arithmetic operation described below.
3.2.1. FT/MS – peak area ratio
The concentrations of metabolites of interest in individual urine specimens were calculated from the ratio of the corresponding 12C2-DnsCl-labeled metabolites to 13C2-DnsCl–labeled standards. To avoid possible interference from the matrix background, we first used high-resolution FT/MS to quantify these metabolite biomarker candidates. Since the amounts of 13C2-DnsCl–labeled standards are known (10 pmol), the quantities of labeled urinary metabolites in all individual urine specimens could be obtained by conversion of peak area ratios of 12C2-DnsCl–labeled metabolites to their corresponding 13C2-DnsCl–labeled standards. The relative quantification was obtained according to the formula below:
3.2.2. HCT/MS – standard curve
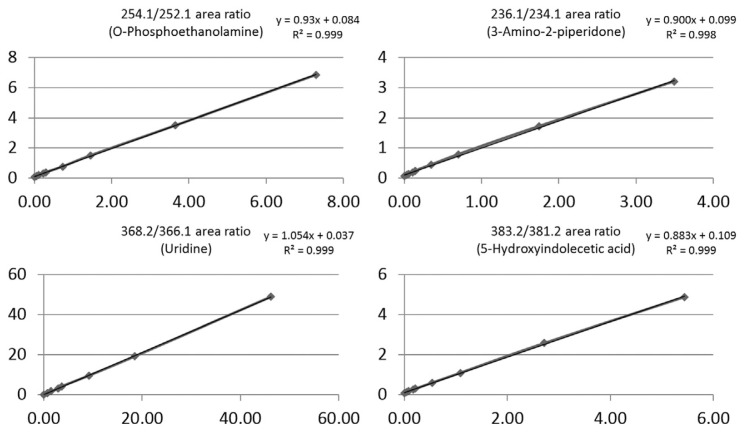
To evaluate the dynamic range for quantification of the four endogenous metabolites, we established a response curve using HCT/MS in MRM scanning mode. A fixed amount of labeled, total endogenous urine metabolites was separately added into different concentrations of 13C2-DnsCl–labeled standards. Because the amount of the four metabolites in urine varied, the level of each 12C2-DnsCl–labeled metabolite in urine was calculated based on the peak areas of corresponding serially diluted 13C2-DnsCl–labeled standards using the average peak areas (n = 3) of isotopically labeled metabolite pairs. The concentrations of the four detectable 12C2-DnsCl–labeled metabolites in urine samples were applied separately to the serially diluted 13C2-DnsCl–labeled standards (0, 0.4, 0.8, 1.6, 2, 5, 10, 25 and 50 pmol), yielding individual regression formulas for ratio-to-ratio plots for the different analytes. Each of the four response curves showed a linear response with good R2 values, as shown in Fig. 2. Uridine (m/z 366.1) showed are remarkable drop in peak area for the 50 pmol 13C2-DnsCl–labeled standard; thus, this point was excluded from the standard curve. Using the resulting standard curves, we calculated the concentrations of the four urinary metabolites—o-phosphoethanolamine, 3-amino-2-piperidone, uridine and 5-hydroxyindoleacetic acid—in all individual clinical urine samples.
Fig. 2.
Response curves of dansylated standard compounds. Response curves are presented as the ratio of the dansylated standards to the corresponding dansylated endogenous urinary metabolites. Mass-to-charge (m/z) corresponds to MS2 fragment ions obtained by LC-HCT/MS in MRM mode. Serial dilutions of 13C2-DnsCl–labeled standard concentrations (0, 0.4, 0.8, 1.6, 2, 5, 10, 25 and 50 pmole) were spiked with a fixed amount (10 nmole) of 12C2-DnsCl–labeled urinary metabolites. The x-axis is the concentration ratio and y-axis is the EIC peak area ratio (13C2-DnsCl–labeled standards/12C2-DnsCl–labeled metabolites).
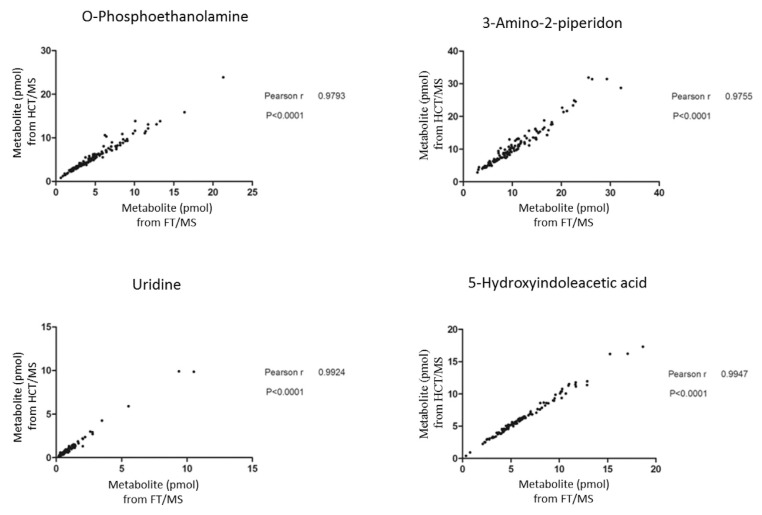
3.2.3. Correlation between HCT and FT quantification of metabolites
The performance of FT/MS and HCT/MS in terms of mass resolution and scan rates differs. Thus, taking into account signal interference, mass-to-charge accuracy and quantitative dynamic range, using a single system for quantification might be problematic. We thus next examined the correlation between metabolite quantification results obtained by HCT/MS and FT/MS. Concentration estimates for all four compounds were well correlated between the two methods, with Pearson r-values greater than 0.9; notably, the Pearson r-values for uridine and 5-hydroxyindoleacetic acid reached 0.99. Correlation plots of o-phosphoethanolamine and 3-amino-2-piperidone in urine exhibited some dispersion of points at higher levels of metabolites, resulting in a slight reduction in Pearson r-values (0.97). We then conducted an additional experiment to validate the precision and accuracy of the quantification methods used in the study. The standard curves of the four metabolites were established and quantification of a spiked sample based on the methods described in the method section (n = 3). A mixture of 10 pmol of each 13C2-dansylated standard compounds (as internal standards for quantification) with 12C2-dansylated endogenous urinary metabolites were measured first to quantify the endogenous concentration of four targeted metabolites based the peak area ratios of 12C2- to 13C2-dansylated signals. Then, comparison of quantitative results in triplicates of spiked 12C2-dansylated standard compounds to their certified values (5 pmol) in the above mixture samples provided a measure of method accuracy and precision for the targeted metabolites. The results showed that the spike recoveries fell within the range of 80–120% for all the four compounds which is used for evaluation of accuracy. Among them, spike recoveries were within 90–110% for o-phosphoethanolamine and 5-hydroxyindolecetic acid which showing good accuracy (Supplementary Table 4). The coefficient of variation (CV) values were all below 10% which were also satisfactory.
3.3. Comparison of concentrations of urinary metabolites between control and bladder cancer patients
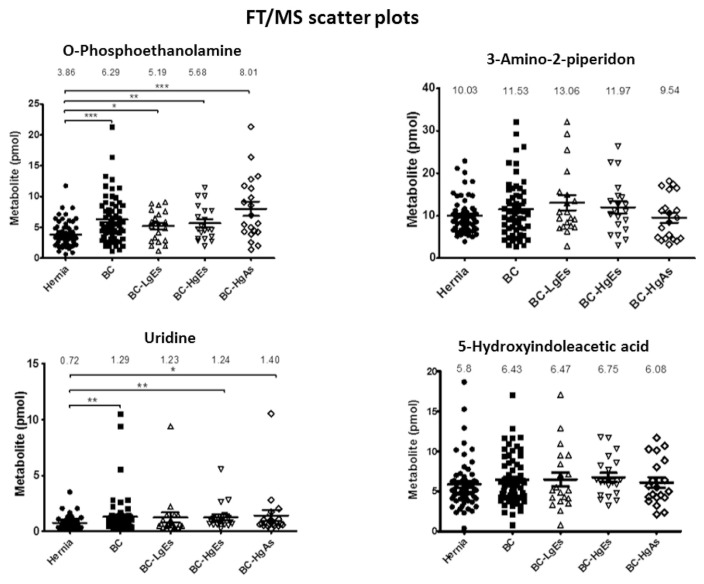
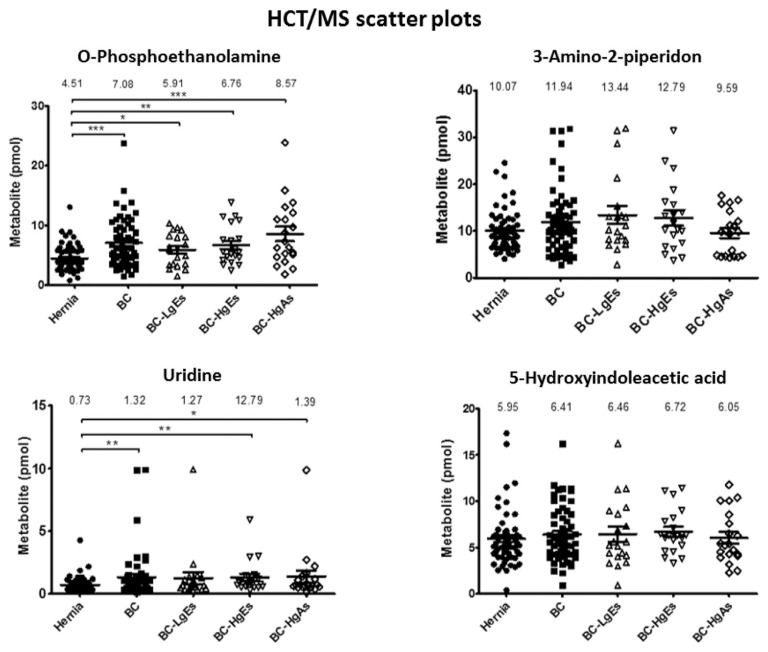
To evaluate concentration differences between control and bladder cancer groups, we performed statistical analyses on urinary quantification data for the four metabolites in 119 clinical urine specimens. The clinical information of samples used in this study are listed as Table 1C. These analyses showed that urinary concentrations of two of the four urinary metabolites—o-phosphoethanolamine and uridine—were significant different between bladder cancer and control groups by both FT/MS (Fig. 3) and HCT/MS (Fig. 4). As shown in Tables 1B and C, the o-phosphoethanolamine concentration in urine samples from the bladder cancer group was ~1.6-times higher than that in the control group, based on HCT/MS. The urinary concentration of uridine was also higher significantly in bladder cancer patients than in the control group. Notably, o-phosphoethanolamine levels in bladder cancer patients, determined by HCT/MS, were correlated with cancer stage severity, exhibiting a 1.3-, 1.5-, and 1.9-fold increase compared with controls in LgEs, HgEs and HgAs subgroups, respectively. Results obtained using FT/MS were similar to those of HCT/MS (Table 1B, Fig. 5 and Supplementary Fig. 1). Furthermore, the small values of absolute differences between the two LC-MS systems indicated well correspondence in quantification of four target metabolites as shown in Supplementary Table 5. In contrast, there was no significant difference in the urinary concentration of 3-amino-2-piperidone or 5-hydroindoleacetic acid between hernia control and bladder cancer patients using either FT/MS or HCT/MS.
Table 1C.
Clinical information of samples used in this study.
| Classification | Diagnosis Status | Number of male Patients | Age |
|---|---|---|---|
| Hernia | Hernia | 59 | 67.2 ± 7.6 |
| Low grade/Early stage (LgEs) – BC | pTa | 20 | 66.6 ± 12.1 |
| High grade/Early stage (HgEs) – BC | Tis, pTa, pT1 | 20 | 67.3 ± 9.5 |
| High grade/Advanced stage (HgAs) – BC | pT2, pT3, pT3b, pT4 | 20 | 65.8 ± 10.5 |
Fig. 3.
Scatter plots of urinary metabolite concentrations (pmole) in different clinical groups obtained by LC-FT/MS. Each dot represents the concentration of a urinary metabolite from the analysis of LC-FT/MS data, expressed as means ± SEM (error bars). Labels: Hernia, control group (filled circles); BC, bladder cancer (total study group; filled squares); BC-LgEs, low-grade early-stage subgroup (open triangles); BC-HgEs, high-grade early-stage subgroup (open inverted triangles); BC-HgAs, high-grade advanced-stage subgroup (open diamonds). P < 0.05, **P < 0.01, **P < 0.001; ns, not significant. Y-axis values are pmole of individual urinary metabolites in a total of 10 nmole.
Fig. 4.
Scatter dot plots of urinary metabolic concentrations (pmole) of LC-HCT/MS results in different clinical groups. Each dot represents the concentration of a urinary metabolite from the analysis of LC-HCT/MS data, expressed as means ± SEM (error bars). Labels: Hernia, control group (filled circles); BC, bladder cancer (total study group; filled squares); BC-LgEs, low-grade early-stage subgroup (open triangles); BC-HgEs, high-grade early-stage subgroup (open inverted triangles); BC-HgAs, high-grade advanced-stage subgroup (open diamonds). P < 0.05, **P < 0.01, **P < 0.001; ns, not significant. Y-axis values are pmole of individual urinary metabolites in a total of 10 nmole.
Table 1B.
The amount average (pmole) of the fore urinary metabolites in total amount of 10 nmol dansyled urine metabolites.
| FT/MS | HCT/MS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Mean (pmol) ± SEM | Mean (pmol) ± SEM | |||||||||
|
|
|
|||||||||
| Hernia | BC | LgEs | HgEs | HgAs | Hernia | BC | LgEs | HgEs | HgAs | |
| O-Phosphoethanolamine | 3.86 ± 0.27 | 6.29 ± 0.5 | 5.19 ± 0.56 | 5.68 ± 0.62 | 8.01 ± 1.17 | 4.51 ± 0.29 | 7.08 ± 0.52 | 5.91 ± 0.61 | 6.76 ± 0.73 | 8.57 ± 1.21 |
| 3-Amino-2-piperidone | 10.03 ± 0.54 | 11.53 ± 0.86 | 13.06 ± 1.78 | 11.97 ± 1.4 | 9.54 ± 1.17 | 10.07 ± 0.57 | 11.94 ± 0.91 | 13.44 ± 1.88 | 12.79 ± 1.64 | 9.59 ± 1.05 |
| Uridine | 0.72 ± 0.07 | 1.29 ± 0.24 | 1.23 ± 0.44 | 1.24 ± 0.27 | 1.40 ± 0.5 | 0.73 ± 0.07 | 1.32 ± 0.24 | 1.27 ± 0.47 | 12.79 ± 0.29 | 1.39 ± 0.47 |
| 5-Hydroxyindoleacetic acid | 5.8 ± 0.4 | 6.43 ± 0.4 | 6.47 ± 0.88 | 6.75 ± 0.57 | 6.08 ± 0.64 | 5.95 ± 0.39 | 6.41 ± 0.38 | 6.46 ± 0.81 | 6.72 ± 0.54 | 6.05 ± 0.62 |
Fig. 5.
Correlation of quantitative results obtained by LC-HCT/MS and LC-FT/MS. Quantitative results obtained by LC-FT/MS are shown in the x-axis and results of LC-HCT/MS are shown in the y-axis.
3.4. Diagnostic efficacies of four dansylated amine- and phenol-containing metabolites in detecting bladder cancer
It is conceivable that changes in the concentrations of urinary metabolites could result from factors other than diseases, such as age. Accordingly, we performed Chi square tests to investigate the relationship between age and metabolite levels in urines. Similar tests for effects of biological sex were not performed because all study subjects were male. For HCT/MS results on o-phosphoethanolamine, Pearson r-values for disease stage and age were 0.005 and 0.583, respectively (Supplementary Table 6), indicating that changes in o-phosphoethanolamine concentration were correlated with disease status rather than patients’ age.
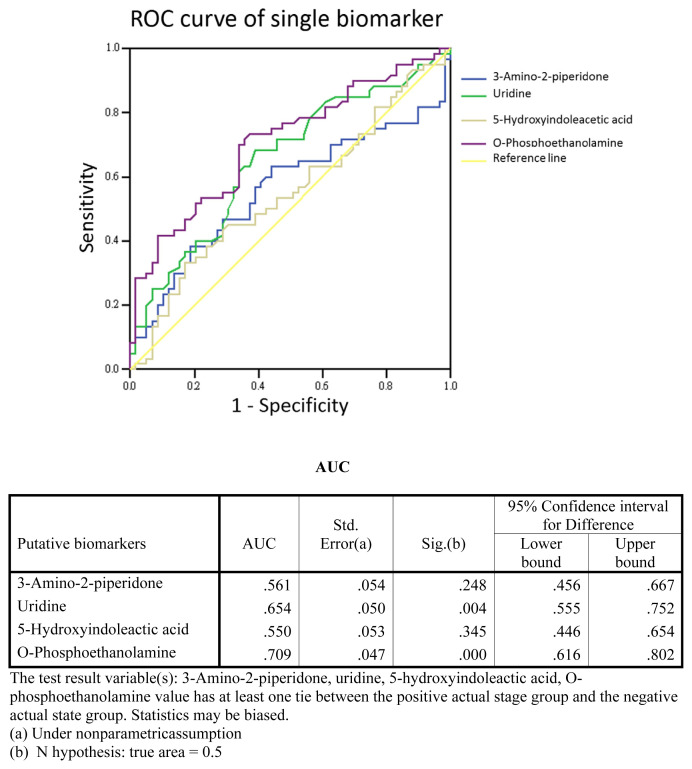
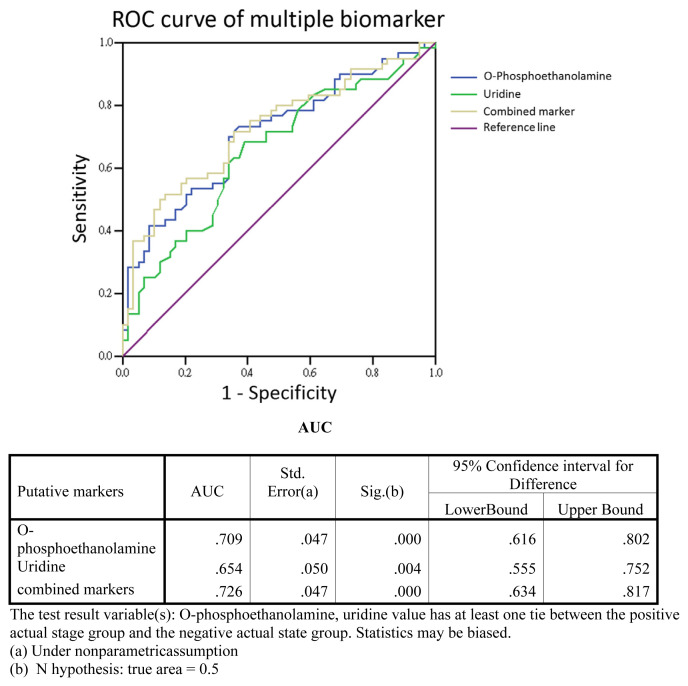
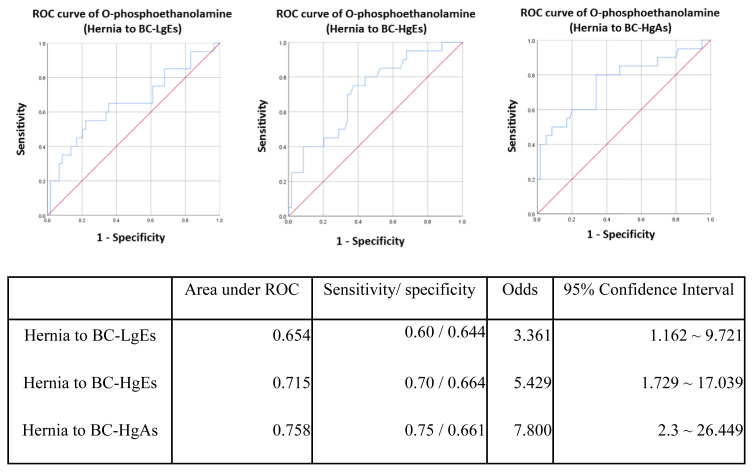
A previous study [12] reported 10 urinary metabolites as potential biomarkers for bladder cancer, five of which were examined in the current study. To test the performance of these metabolites as biomarkers, we carried out a receiver operating characteristic (ROC) curve analysis. In an analysis of single-marker performance (Fig. 6), we found that o-phosphoethanolamine (m/z 252.1) in the HCT/MS system was moderately predictive, with an AUC (area under the curve) of 0.709; the AUC for uridine was 0.654. Combining these two biomarkers improved the AUC to 0.726 (Fig. 7), indicating that this two-biomarker panel has better predictive value than either single biomarker alone. Odds-ratio analysis was also performed to estimate that the different degrees of bladder cancer response to o-phosphoethanolamine in this study (Fig. 8). The cut off values of the analysis were determinate by the sensitivity and specificity of ROC curve, which were equal to or greater than 0.6 with lower false-possibility. The odd-ratios with 95% confidence interval (CI) of hernia versus BC LgEs, HgEs and HgAs were 3.361 (1.162–9.721), 5.429 (1.729–17.039) and 7.8 (2.3–26.449), respectively. The odd ratios were greater than 1 and increased along the severity of bladder cancer in the study indicating a positive correlation between the metabolite and the severity of bladder cancer subgroups.
Fig. 6.
Single-biomarker performance. The performance of four urinary metabolites, quantified by LC-HCT/MS, as single markers of bladder cancer was evaluated using ROC curve analysis. The AUC for O-phosphoethanolamine was 0.709.
Fig. 7.
Two-biomarker panel performance. The four urinary metabolites, quantified by LC-HCT/MS, were combined by logistic regression, and the performance of multi-biomarker panels in detecting bladder cancer was assessed by ROC curve analysis. The performance of O-phosphoethanolamine and uridine as single markers is shown for comparison. The AUC for the performance of the o-phosphoethanolamine and uridine two-biomarker panel in detecting bladder cancer was increased to 0.726.
Fig. 8.
The ROC curves of hernia group to BC-LgEs, BC-HgEs and BC-HgAs separately. The cut offs were defined as where the points on curves were close to the left top point (sensitivity = 1) and with lowest false-possibility.
4. Discussion
The field of metabolomics has become increasingly important for exploring markers of changes in biological reactions that are associated with pathophysiology, environmental toxicity, changes in biological mechanisms, or disease [15,16]. This technology is considered as a complementary tool for genomics, transcriptomics, and proteomics approaches, given that the genome-scale changes detected by these latter approaches occur at a higher, upstream level, and might not be reflected in subsequent changes in regulation or components [16]. Additionally, analyzing metabolites in urine is a non-invasive means for biomarker discovery—an advantage for clinical practice and diagnosis. Difficulties in analyzing metabolites in biological fluids include the high complexity of biological matrices, detection limits, and lower confidence in metabolite identification. Moreover, many hydrophilic metabolites are not likely to be retained on the reverse-phase columns common employed, resulting in difficulties in separating and analyzing complex mixtures. Dansylation labeling [8] represents a promising approach for increasing sensitivity and detection probability of compounds with specific functional groups, namely primary amines, secondary amines, or phenolic hydroxyl group(s) [8]. In the current study, clinical samples used for verification were collected, prepared, and analyzed according to the method described above. Using differential 12C2-/13C2-isotope dansylation labeling, we were able to explore analytes by identifying correct peak pairs of 12C2-labeled endogenous metabolites and corresponding 13C2-labeled commercial metabolite standards, an approach that correctly and precisely targets metabolites with high confidence.
Because of clinical or biological variability, candidate biomarkers identified in the discovery stage need to be validated across a different set of individual samples. There are several reasons that part of the biomarker candidates that discovered in the previously work have not been verified successfully in this study. This could be the causes of different patient enrollment or LC systems between pervious work and this study. Besides, the identification of the metabolites in the previous work was potential biomarkers obtained from database according to accurate molecular weight, yet their true identification required further validation. The comparison of patient enrollment between discovery and verification phases is shown in Supplementary Table 3. Only low percentage (5.5%) of patients were used in both metabolomics profiling and targeted quantification which was good for evaluation of biological variation. A biomarker candidate verification phase eliminates this bottleneck in biomarker development pipeline by ensuring that only the most promising putative biomarkers found in discovery go on to the validation stage.
The reason that we did not use the same LC methods in two LC-MS systems was due to the routine setting of two LC systems. In this work, the main purpose was targeted quantification of metabolite for verification the clinical performance of potential metabolite biomarkers. The metabolite identity could be confidently confirmed using the commercial standards although the LC methods of two MS systems were different. The first issue encountered in establishing a quantitative workflow for novel metabolite biomarkers is confirmation of the correctness of metabolite identification, determined by reference to a metabolite database based on accurate molecular masses. Therefore, a high-resolution MS system is required to confirm detected m/z signals and retention times in an LC-MS run. Fourier transform mass spectrometry (FT/MS) is known for its high accuracy and resolution, which are beneficial for metabolite identification. However, the ion capture mode in FT/MS has a limited ability to accurately quantify metabolites owing to its relative slow scan speed, which limits the number of data points during signal acquisition. Despite its accuracy, FT/MS is susceptible to effects of the complexity of analytes in urine sample, which may result in other molecules with the same mass-to-charge signal after ionization contributing to peak areas. In this case, signal interference caused by matrix complexity results in imprecise quantification. To overcome this issue, we implemented an additional MS system—HCT/MS—which has compensatory properties relative to FT/MS. Specifically, HCT/MS is capable of multiple-reaction monitoring (MRM), which gives information about chemical fragments, meaning that it is complementary to FT/MS for molecular quantification.
The second quantification method, LC-HCT/MS in MRM mode—an ion trap system that is robust and easy to use, and delivers a rapid MSn function—was used to establish response curves for quantifying the four targeted amine- and phenol-metabolite in clinical urine specimens. The response curves also provided insight into ion suppression by matrix effects of the complex biological mixture. Moreover, based on scatter plots (Figs. 3 and 4) and response curves (Fig. 2), quantified urinary metabolites fell within reasonable measurement ranges, despite some outliers. In summary, the targeted metabolites could be quantified based on standard curves and provided accurate estimates of concentration using a simple ion-trap MS system. Notably, quantitative results obtained for the four amine- and phenol-metabolites using the two MS systems (Table 1B, Figs. 3 and 4) were highly correlated.
There was a slight, but significant, increase in the concentrations of the metabolites, uridine and o-phosphoethanolamine, in the bladder cancer group compared with the control group, whereas the other two metabolites showed no significant difference between groups. Of these two metabolites, o-phosphoethanolamine concentrations in urine correlated with disease stage severity, increasing from low-grade early stage (LgEs), to high-grade early stage (HgEs) and high-grade advanced stage (HgAs). This suggests that o-phosphoethanolamine could be an indicator of bladder cancer generally as well as a marker of disease stage. However, the fold-changes in both uridine and o-phosphoethanolamine concentrations in urine samples from the bladder cancer group compared with those from the hernias group were not remarkable. There for, because changes in metabolite levels can be related to other factors (e.g., age, biological sex, hematuria), we performed Chi square tests on o-phosphoethanolamine levels to test the possible role of age in addition to disease stage (Supplementary Tables 2 and 3). These analyses demonstrated that o-phosphoethanolamine levels were related to bladder cancer, but did not likely reflect age effects.
Metabolites are the final products of a series of complex biological processes, and reflect the involvement of numerous gene-scale regulatory events, including transcription, as well as post-transcriptional and post-translational mechanisms. Moreover, various molecules are likely associated with complex interactions within a single pathway and the multiple regulatory cross-links among molecular and biological modifications involved in the development of cancer [17]. Given this, using a single metabolite as a disease biomarker may not provide a powerful, or even valid, clinical approach for diagnosing or predicting a disease. Accordingly, it is important to consider the use of multiple biomarkers in the form of a biomarker panel using ROC curve analysis together with logistic regression. To this end, we analyzed ROC curves to assess the performance of single and combined biomarkers (Fig. 7). As a single biomarker (Fig. 6), o-phosphoethanolamine was confirmed to be capable of discriminating bladder cancer urine from control urine specimens. When combined with uridine, the resulting two-biomarker panel showed improved ability to discriminate between these two groups. Of these two urinary panel members, o-phosphoethanolamine is an important metabolite that is involved in phospholipid metabolism and is associated with several key cellular functions, including cell membrane turnover and lipid signaling pathways that regulate cell proliferation and survival; it may also exert therapeutic effects in animal melanoma models [18]. However, detailed mechanisms underlying the potential role of o-phosphoethanolamine in bladder cancer biology remain unclear and will require further study. In summary, we verified that urinary phosphoethanolamine is a potential non-invasive biomarker for use in bladder cancer screening that shows improved discriminating power when combined with uridine in a two-biomarker panel.
5. Conclusions
In this study, 12C-/13C-isotope dansylation labeling was utilized as a platform to quantify four targeted phenol- and amine-containing metabolites in urine samples and evaluate the performance of these metabolites as non-invasive biomarkers of bladder cancer. Using dansyl chloride-labeled synthetic standards, we verified the identity and quantify ability of four previously discovered novel urinary metabolites, and assessed their performance in differentiating bladder cancer specimens from control specimens. Two different MS systems—HCT/MS and FT/MS—were used for quantification, and the results obtained with the two systems were well correlated. This indicates that the platform is practical and accessible for use in precisely and accurately evaluating urinary metabolite levels. Although FT/MS offers high accuracy of m/z analysis, it has limitations including high cost and intense maintenance. On the other hand, HCT/MS is relative cost-effective and the MRM method established in the study is practical for clinical screening, such as newborn blood screening using tandem mass spectrometric platform. Therefore, as the consideration of accessibility and applicability, targeted analysis of patient specimens performed with an LC-HCT/MS or similar instrumentation is more practical for clinical screening. The results of quantification showed that o-phosphoethanolamine increased in the urine of bladder cancer patients, independent of the age of patients. Therefore, o-phosphoethanolamine is a potential biomarker for bladder cancer diagnosis. Diagnostic performance was further improved by combining uridine and o-phosphoethanolamine in a two-biomarker panel.
Acknowledgement
This research was supported by grants from the Ministry of Science and Technology of Taiwan, Republic of China (106-2113-M-182-001-MY2), and Chang Gung Memorial Hospital (CMRPD1G0131, CMRPD1G0132, CMRPD3E0251, CMRPD3E0252, and BMRPD78). The instrumental and data analysis resources were supported by the proteomic core lab (CLRPD190017) at Chang Gung University, Taoyuan, Taiwan. This work was financially supported by the “Molecular Medicine Research Center, Chang Gung University” from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jfda.2018.11.008.
Funding Statement
This research was supported by grants from the Ministry of Science and Technology of Taiwan, Republic of China (106-2113-M-182-001-MY2), and Chang Gung Memorial Hospital (CMRPD1G0131, CMRPD1G0132, CMRPD3E0251, CMRPD3E0252, and BMRPD78).
Footnotes
Conflicts of interest
The authors declare no conflict of interest.
REFERENCES
- 1. Society AC. Cancer facts & figures 2018. 2018 [Google Scholar]
- 2. Kirkali Z, Chan T, Manoharan M, Algaba F, Busch C, Cheng L, et al. Bladder cancer: epidemiology, staging and grading, and diagnosis. Urology. 2005;66:4–34. doi: 10.1016/j.urology.2005.07.062. [DOI] [PubMed] [Google Scholar]
- 3. Chang SS, Boorjian SA, Chou R, Clark PE, Daneshmand S, Konety BR, et al. Diagnosis and treatment of non-muscle invasive bladder cancer: AUA/SUO guideline. J Urol. 2016;196:1021–9. doi: 10.1016/j.juro.2016.06.049. [DOI] [PubMed] [Google Scholar]
- 4. Alberice JV, Amaral AF, Armitage EG, Lorente JA, Algaba F, Carrilho E, et al. Searching for urine biomarkers of bladder cancer recurrence using a liquid chromatography-mass spectrometry and capillary electrophoresis-mass spectrometry metabolomics approach. J Chromatogr A. 2013;1318:163–70. doi: 10.1016/j.chroma.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 5. Ganti S, Weiss RH. Urine metabolomics for kidney cancer detection and biomarker discovery. Urol Oncol. 2011;29:551–7. doi: 10.1016/j.urolonc.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang Z, Lin L, Gao Y, Chen Y, Yan X, Xing J, et al. Bladder cancer determination via two urinary metabolites: a biomarker pattern approach. Mol Cell Proteomics. 2011;10:M111 007922. doi: 10.1074/mcp.M111.007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–67. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 8. Guo K, Li L. Differential 12C-/13C-isotope dansylation labeling and fast liquid chromatography/mass spectrometry for absolute and relative quantification of the metabolome. Anal Chem. 2009;81:3919–32. doi: 10.1021/ac900166a. [DOI] [PubMed] [Google Scholar]
- 9. Guo K, Bamforth F, Li L. Qualitative metabolome analysis of human cerebrospinal fluid by 13C-/12C-isotope dansylation labeling combined with liquid chromatography Fourier transform ion cyclotron resonance mass spectrometry. J Am Soc Mass Spectrom. 2011;22:339–47. doi: 10.1007/s13361-010-0033-4. [DOI] [PubMed] [Google Scholar]
- 10. Wu Y, Li L. Dansylation metabolite assay: a simple and rapid method for sample amount normalization in metabolomics. Anal Chem. 2014;86:9428–33. doi: 10.1021/ac503359v. [DOI] [PubMed] [Google Scholar]
- 11. Zhou R, Li L. Quantitative metabolomic profiling using dansylation isotope labeling and liquid chromatography mass spectrometry. Methods Mol Biol. 2014;1198:127–36. doi: 10.1007/978-1-4939-1258-2_9. [DOI] [PubMed] [Google Scholar]
- 12. Peng J, Chen YT, Chen CL, Li L. Development of a universal metabolome-standard method for long-term LC-MS metabolome profiling and its application for bladder cancer urine-metabolite-biomarker discovery. Anal Chem. 2014;86:6540–7. doi: 10.1021/ac5011684. [DOI] [PubMed] [Google Scholar]
- 13. Han W, Li L. Matrix effect on chemical isotope labeling and its implication in metabolomic sample preparation for quantitative metabolomics. Metabolomics. 2015;11:1733–42. [Google Scholar]
- 14. Panuwet P, Hunter RE, Jr, D’Souza PE, Chen X, Radford SA, Cohen JR, et al. Biological matrix effects in quantitative tandem mass spectrometry-based analytical methods: advancing biomonitoring. Crit Rev Anal Chem. 2016;46:93–105. doi: 10.1080/10408347.2014.980775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Timbrell JA. Biomarkers in toxicology. Toxicology. 1998;129:1–12. doi: 10.1016/s0300-483x(98)00058-4. [DOI] [PubMed] [Google Scholar]
- 16. van Ravenzwaay B, Cunha GC, Leibold E, Looser R, Mellert W, Prokoudine A, et al. The use of metabolomics for the discovery of new biomarkers of effect. Toxicol Lett. 2007;172:21–8. doi: 10.1016/j.toxlet.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 17. Urquidi V, Rosser CJ, Goodison S. Molecular diagnostic trends in urological cancer: biomarkers for non-invasive diagnosis. Curr Med Chem. 2012;19:3653–63. doi: 10.2174/092986712801661103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mambelli LI, Teixeira SF, Jorge SD, Kawamura B, Meneguelo R, Barbuto JAM, et al. Phosphoethanolamine induces caspase-independent cell death by reducing the expression of C-RAF and inhibits tumor growth in human melanoma model. Biomed Pharmacother. 2018;103:18–28. doi: 10.1016/j.biopha.2018.03.135. [DOI] [PubMed] [Google Scholar]