Abstract
Mass spectrometry (MS) is a type of analysis used to determine what molecules make up a sample, based on the mass spectrum that are created by the ions. Mass spectrometers are able to perform traditional target analyte identification and quantitation; however, they may also be used within a clinical setting for the rapid identification of bacteria. The causative agent in sepsis is changed over time, and clinical decisions affecting the management of infections are often based on the outcomes of bacterial identification. Therefore, it is essential that such identifications are performed quickly and interpreted correctly. Matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometer is one of the most popular MS instruments used in biology, due to its rapid and precise identification of genus and species of an extensive range of Gram-negative and-positive bacteria. Microorganism identification by Mass spectrometry is based on identifying a characteristic spectrum of each species and then matched with a large database within the instrument. The present review gives a contemporary perspective on the challenges and opportunities for bacterial identification as well as a written report of how technological innovation has advanced MS. Future clinical applications will also be addressed, particularly the use of MALDI-TOF MS in the field of microbiology for the identification and the analysis of antibiotic resistance.
Keywords: Antibiotic susceptibility testing, MALDI-TOF MS, Mass spectrometry, Microbiology, Microorganism identification
1. Introduction
Mass spectrometry (MS) identifies and quantifies molecules by analyzing the mass-to-charge ratios (m/z) of molecular ions. The mass spectrum within the mass spectrometer can be interpreted to determine the identity of different molecules within a sample. Mass spectrometers directly analyze any ionized susceptible biological molecule. The success of MS is only possible because of several important discoveries, including the electrospray ionization (ESI) discovered by John Fenn and matrix-assisted laser desorption/ionization (MALDI) discovered by Koicihi Tanaka [1,2].
MALDI has the benefit of requiring less pre-analysis workup, as samples are mixed with a chemical matrix followed by the generation of ions. MALDI time-of-flight (TOF) MS combines two technologies as the MALDI source and the TOF mass. Since its conception, this tool has revolutionized the method of microorganism identification in clinical microbiology laboratories, as it is a rapid, high throughput, low-cost and efficient system. One of the major advantages of using MALDI-TOF MS is the time saved, as bacterial identification may be performed in less than an hour as opposed to 24–48 h. This time is critical for patients with an underlying autoimmune disease or immunocompromised individuals [3].
The Antibiotic Sensitivity Test (AST) is another key piece of information that affects clinical treatment. It is also desirable to develop a fast and reliable method for detecting drug sensitivity using MALDI-TOF MS. Several AST detection principles are being developed and tested, such as (1) the detection of drug-resistant strain biomarkers; (2) mass spectrometric detection of antibiotic degradation; and (3) the detection of stable (non-radioactive) isotope-labeled amino acids etc. Nonetheless, these methods need further improvement to have the opportunity to apply to routine testing [4–6].
The present review discusses the background of the MALDI-TOF MS technique in detail, including sample preparation and workflow. It then be presented the applications of MALDI-TOF MS microbial identification in the clinic, the production of virulence factors for subtyping, and the presence of antimicrobial resistance. In addition, the current limitations and potential usage of MALDI-TOF MS in future daily clinical practice are discussed.
2. MALDI-TOF MS based microorganism identification and sample preparation
2.1. MALDI-TOF MS overview
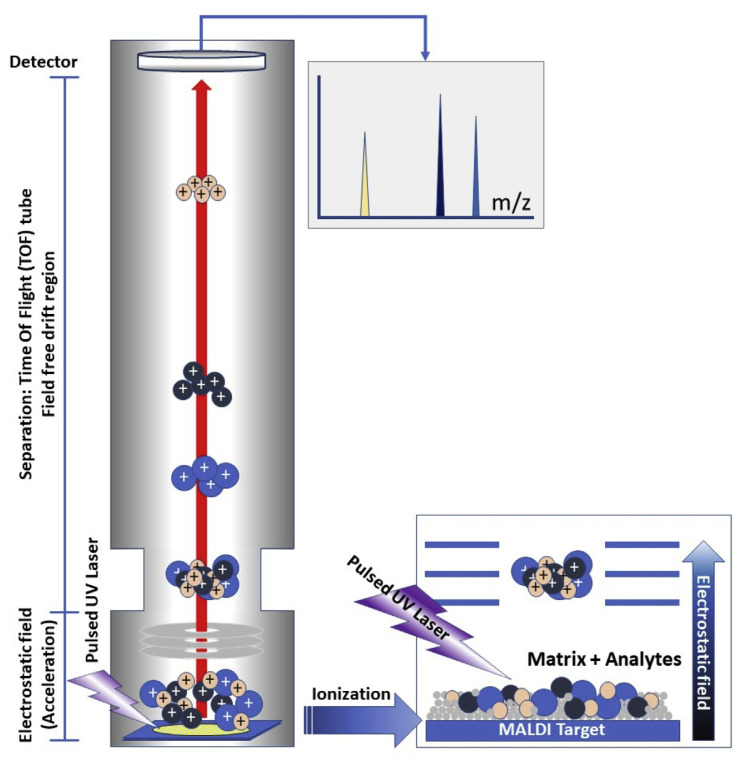
MS is used to detect the m/z ratio and MALDI-TOF MS provides a rapid, accurate, and sensitive spectra of the bio-analytes within a sample. MALDI is an ionization technique in which a matrix absorbs energy from ultraviolet lasers to create ions from large molecules with minimal fragmentation. The m/z ratio can be determined by the TOF of the ions which the detector measure TOF of ions to calculating masses of ions (Fig. 1). The α-Cyano-4-hydroxycinnamic acid (HCCA) and 2,5-dihydroxybenzoic acid are MALDI matrices used for various microbial analysis. For microorganism identification, positively charged peptides/proteins with a molecular weight between 2000 and 20,000 m/z are used. The sequence and size of ribosomal proteins are highly conserved among different bacterial species, and therefore be used to identify individual types of bacteria. Individual mass peaks are used for microorganism identification and provide valuable information for the fingerprinting of bacteria. To enable rapid, accurate, easy and reliable microorganism identification, the new MS system was designed with a highly automatic operational workflow and analysis process [3,7,8].
Fig. 1.
MALDI-TOF MS methodology. The sample is co-crystallized with the matrix on the sample target and to be desorbed and ionized by the MALDI ion source (e.g. ultraviolet laser). The ion molecules, including the microbial peptides/proteins, are accelerated by the electric field into the TOF analyzer. All the ions are separated by TOF in accordance with the m/z ratio and a mass spectrum.
2.2. Sample preparation
The majority of bacterial samples are separated and grown on a solid agar medium, and a single colony is then selected and smeared directly as a thin film on a MALDI target. However, Mycobacterium spp. and filamentous fungi usually require multi-step extraction methods to ensure the quality of biological proteome, which improves the spectrum and identification confidence. For cases with increased clinical urgency, direct pathogen identification may be performed from biological samples. Many types of body fluids may be required further purification to separate the pathogen from the host cells, peptides, and proteins. Preparation methods and identification workflows for different biological samples are needed to be optimized for the MALDI-TOF MS analysis (Fig. 2).
Fig. 2.
MALDI-TOF MS microorganism identification workflow in a clinical laboratory. All the bacterial colonies from a standard culture in a solid agar plate are applied directly via the standard smear method. Some liquid samples of urgent specimens, such as blood culture positive bottles and body fluids (CSF or urine), can be applied for direct identification after a multi-step sample preparation and extraction protocol. Mycobacterium spp. are applied following a specific sample preparation protocol for their rapid identification.
2.2.1. General preparation within a clinic
Ideally, an easy, rapid and robust sample preparation method would be the best for use within a clinical setting. Intact-cell MS, also known as whole-cell MS (WCMS), is a method of acquiring microorganism protein profile data by MALDI-TOF MS [7]. Microorganisms from a single colony of bacteria or yeast are smeared as a thin film directly onto a MALDI target plate. A total of 1 μl HCCA matrix solution (prepared with 50% acetonitrile and 2.5% trifluoroacetic acid in pure water) is then dropped onto the smear. This solvent is able to extract proteins from the most of bacteria for MALDI-TOF MS measurement.
Some bacteria are encased by glycocalyx or a capsule layer, including Mycobacterium spp. and certain yeast species; this means their internal proteins cannot be easily extracted by the matrix solution. The ethanol-formic acid (FA) extraction method is suggested to use for these samples to generate high-quality spectra for identification and further analysis [9]. One loop of bacteria is resuspended in 300 μl pure water and mixed with 900 μl pure ethanol to inactivate the microorganisms by vortex well in seconds. High-speed centrifugation is performed for 2 min and the supernatant is carefully removed. A total of 20–50 μl 70% FA is added and mixed well, then an equal amount of pure acetonitrile is added and mixed by vortex for several seconds. A bacteria pellet is formed by high-speed centrifugation for 2 min and 1 μl of the supernatant is applied to the MALDI target. After air dry, the sample was covered with 1 μl HCCA matrix solution for measuring.
2.2.2. Liquid samples
Positive blood cultures (BC) and various other body fluids are primarily liquid samples. Positive BCs may include blood cells, culture materials and resin or charcoal. Removal of blood cells and proteins derived from the host and culture media is key to prepare sample for MALDI-TOF MS. There are several approaches for direct bacterial identification from positive BC by MS. Low speed centrifugation for 10 min is used for the removal of blood cells, then the supernatant, which includes the bacteria, is taken for further high-speed centrifugation [10,11]. Detergents, such as saponin and sodium dodecyl sulfate (SDS), have also been used for the lysis of blood cells. Eukaryotic cells may be lysed, but prokaryotic cells are not affected by low concentrations of detergents. These treatments reduce the duration of low-speed centrifugation step required to remove host cells [11,12]. A CE-IVD (Conformité Européene In Vitro Diagnostics) labeled blood culture sample preparation kit can also be used for direct microorganism identification from 1.5 ml BC positive broth [13]. The purified bacteria would then be further processed by the FA extraction protocol prior to MALD-TOF MS analysis.
The sensitivity of MALDI-TOF MS for bacterial identification is ~104–105 CFU (colony-forming unit) of Escherichia coli [14,15]. The sensitivity and pure culture requirement of this method depends on the type of specimen. A urine sample could be used directly for bacteria identification. The urine sample undergoes flow cytometry to confirm the presence of bacteria. Then 4 ml of the positive urine sample undergoes low-speed centrifugation (2000×g) for 30 sec to remove leucocytes and epithelium cells. The supernatant then undergoes high-speed centrifugation (15,500×g) for 5 min and the bacteria are collected. The pellet is treated with FA for the extraction of bacterial protein prior to MALDI-TOF MS analysis [15]. There are previous reports of using cerebrospinal fluid (CSF) for rapid pathogen identification in patients with bacterial meningitis. A total of 500 μl CSF was used for the microbiological diagnosis; 100 μl 13% SDS was added to the CSF and vortexed for 10 sec, then centrifuged at 13,000×g for 2 min. The pellet was resuspended in 1 ml pure water and centrifuged at 13,000×g. The washed bacterial pellet was treated with FA for protein extraction prior to MALDI-TOF MS analysis [16–18].
2.2.3. Mycobacterium
In recent decades, the mycobacterium genus has undergone a large taxonomic reversion and there are currently more than 190 recognized species (bacterio.net/mycobacterium.html; Dec. 2018). The pathogenic Mycobacterium tuberculosis complex (MTC) causes tuberculosis (TB) and presents a serious public health issue worldwide. In 2016, 10.4 million people contracted TB, and 1.7 million died from the disease [19]. A number of non-TB mycobacteria (NTM) are also well-established pathogens and there is an increased risk of infection in immunocompromised individuals [20]. Identification of mycobacteria could be improved by the MALDI-TOF MS method as it has increased speed, accuracy, and is more economic compared with conventional methods. Regarding the slow growth time of Mycobacterium spp., the conventional biochemical identification, depending on the species, usually takes 7–21 days for the biochemical reaction completely after the colony was formed. In contrast, MALDI-TOF MS technology can get the identical result in hours after the colony applied for the sample preparation. Both the Bruker MALDI Biotyper (MBT, Bruker Daltoniks GmbH; Billerica, MA, USA) and Vitek MS (bioMérieux Inc., NC, USA) have their own protocol for mycobacteria identification. Inactivation of the mycobacterium species is one of the critical points in sample handling. Bruker uses heat inactivation for 30 min in a 95 °C heat block, while bioMérieux applied vortex mixing at 3000 rpm (Vortex Genie 2 [Scientific Industries] with MoBio Vortex Adaptor) for 10–15 min with silica beads in 70% ethanol. The bacteria mass is vortexed with silica beads for 1 min in pure acetonitrile or 10–15 min in 70% ethanol. After the mechanical disruption caused by the vortex step, the sample is sequentially treated with an equal amount of 70% FA and pure acetonitrile. A total of 1 μl of the extract is dropped onto the MALDI target, dried and covered with 1 μl HCCA matrix [21].
2.2.4. Filamentary fungi
A number of different cultivation conditions and extraction methods have been reported for filamentary fungi. The MALDI Biotyper workflow suggests that Sabouraud liquid broth should be cultivated with continuous rotation. Fungi cultivated on Sabouraud gentamicin-chloramphenicol agar plates are also widely used within laboratories, where they are usually incubated for 72 h at 27 °C. Part of the mycelium is harvested and subjected to modified FA extraction. A total of 300 μl pure water and 900 μl pure ethanol are added to the fungal samples and mixed well in a 1.5 ml tube. After centrifugation at 13,000×g for 10 min, the supernatant is removed and air dried for 1 h. The dried samples were incubated for 5 min in 20–50 μl 70% FA. Then equal amounts of pure acetonitrile were added for 10 min incubation before the samples were dropped onto the MALDI target for analysis [22,23].
3. Classification and identification of microorganisms
Clinical microbiology laboratories serve as a key partner to clinicians in the treatment of infectious disease, particularly in the identification of microorganisms from clinical specimens. The identification of bacterial species can guide the selection of appropriate antibiotics. With the aid of antimicrobial susceptibility testing, physicians can determine which antibiotics are effective and which enables the successful control of infection. A wide range of clinical microbial isolates with different culture conditions will be efficiently and accurately identified by MALDI-TOF MS technology (Table 1). Therefore, a rapid and accurate methodology for antimicrobial identification is vital for managing antibiotic treatment.
Table 1.
References of the evaluation of MALDI-TOF MS identification in clinical microorganism isolates.
| Study | Percentage (No.) organisms correctly detected by MALDI-TOF MS | Description of isolates of study | |
|---|---|---|---|
|
| |||
| Genus level | Species Level | ||
| Gram negative bacteria | |||
| Faron et al., 2015 [29] | 99.8% (2258/2263) | 98.2% (2222/2263) | 2263 clinical isolates of aerobic Gram-negative bacteria. |
| Gamer et al., 2013 [30] | 92.5% (357/386) | 91.7% (351/386) | 386 isolates of anaerobic Gram-negative bacteria. |
| Gram positive bacteria | |||
| Rychert et al., 2013 [31] | 95.5% (1094/1146) | 92.8% (1063/1146) | 1146 isolates of aerobic Gram-positive from multicenter. |
| Gamer et al., 2013 [30] | 92.5% (245/265) | 91.7% (243/265) | 265 isolates of anaerobic gram Positive bacteria. |
| Anaerobic bacteria | |||
| Garner et al., 2013 [30] | 92.5% (602/651) | 91.2% (591/651) | 651 isolates anaerobic bacteria. |
| Yeast and mold | |||
| Wang et al., 2016 [46] | n.a.a | 98.8% (2651/2683) | 2683 clinical isolates of yeast. |
| Chao et al., 2014 [47] | 92.5% (185/200) | 200 clinical isolates of yeast. | |
| Chen et al., 2013 [45] | 94.9% (93/98) | 74.5% (73/98) | 98 Clinical isolates of yeast. |
| Becker et al., 2014 [49] | n.a. | 95.4% (372/390) | 390 clinical mold isolates. 760 customized reference spectra. |
| Gautier et al., 2014 [50] | n.a. | 98.8% (1094/1107) | 1107 clinical mold isolates (107 distinct species). 2832 customized reference spectra. |
| Mycobacterium | |||
| Wilen et al., 2015 [21] | n.a. | 89.2% (140/157) | 157 mycobacterial isolates (including 16 isolates of M. tuberculosis). |
| Rodrigues-Sanchez et al., 2015 [44] | n.a. | 88.8% (111/125) | 125 non-toberculosis mycobacterial (NTM) isolates. |
| Chen et al., 2013 [45] | 87.3% (89/102) | 62.8% (64/102) | 102 mycobacterial isolates. |
| Blood culture positive bottles | |||
| Chien et al., 2016 [36] | 405 blood culture (BC) positive samples. | ||
| 89.6% (327/365) | 72.1% (263/365) | 365/405 monomicrobial growth BC samples. | |
| 92.5% (37/40) | 82.5% (33/40) | 40/405 polymicrobial growth BC positive samples. | |
| Arroyo et al., 2017 [13] | 94.0% (188/200) | 91.5% (181/200) | 200 isolates of Gram-negative bacilli from monomicrobial growth of BC positive samples. |
| Chen et al., 2013 [11] | 97.8% (177/181) | 81.8% (148/181) | 181 monomicrobial growth BC positive samples. |
| Kok et al., 2011 [35] | 100% (358/358) | 78.5% (281/358) | 358 monomicrobial growth BC positive samples. |
| 100% (195/195) | 67.7% (132/195) | 195 Gram-positive isolates | |
| 100% (163/163) | 91.4% (149/163) | 163 Gram-negative isolates | |
Not applicable.
3.1. Identification from culture plates
The clinical application of MALDI-TOF MS in the identification of microorganisms primarily comes from culture plates. Using this method, a small amount of microbial biomass (104–105 bacteria) can yield the bacterial identification of a large variety of species in a few minutes. Bacterial colonies are handpicked from agar culture plates with a sterile tip, then smeared onto a ground steel MALDI target plate, which has a highly regular and fine structure for facilitating homogenous preparations. The samples are dried and a small amount of matrix, such as HCCA, is added to co-crystallize with the bacterial sample on MALDI target, then the sample is introduced into the mass spectrometer for data acquisition.
Comprehensive comparison studies have evaluated the reproducibility and accuracy of MALDI-TOF MS. In 2010, a study compared a MALDI-TOF MS device with traditional biochemical tests routinely used for the identification of bacterial species. A high-confidence correct identification was found in 99.1% of cases [24]. Another study evaluated the overall correct identification rate in coagulase-negative staphylococci as 93.2% [25]. In 2011, a study demonstrated that MALDI-TOF MS is reliable (accuracy, 99.3%) in identifying various staphylococci species, and may be considered equivalent to the standard method of rpoB sequence-based identification [26]. In 2012, a study reported the correct identification rates of MALDI-TOF MS are 94.9% and 83,4% at genus (39 genera) and species (102 species) level, respectively from 296 strains covering Gram-positive and -negative bacteria [27]. Another study suggested that MALDI-TOFMS is the method of choice for the identification of Campylobacter and related microorganisms compared with other commercial systems [28]. For Gram-negative bacteria evaluation, there are 2263 clinical significant species can be analyzed by MALDI-TOF MS identification. Faron et al. showed that MALDI-TOF MS system correctly identified 99.8% (2258/2263) to genus and 98.2% (2222/2263) to species level of all isolates from multiple centers [29]. In addition, Garner et al. showed the identification of anaerobic Gram-negative isolates was 91.7% to the species level and 92.5% of the isolates to the genus level [30]. For Gram-positive bacteria evaluations, a multi-center evaluation of MALDI-TOF MS was used to identify 1146 isolates of Gram-positive aerobic bacteria, including 13 genera and 42 species. Rychert et al. showed 92.8% (1063/1.146) correct identification to species level and 95.5% (1094/1146) to the genus level [31]. Furthermore, Garner et al. showed that the identification of anaerobic Gram-negative isolates was 90.9% to the species level and 92.5% to the genus level [30]. In the group of anaerobic bacteria identification, MALDI-TOF MS identification showed 91.2% (591/651) exhibited the correct identification at the species level and 92.5% were correctly identified at the genus level of the 651 analyzed isolates [30]. MALDI-TOF MS has been used for the rapid identification of bacteria from positive BC bottles after a short incubation time of 5.5 h on solid media, with an accuracy rate of 82.3% [32]. Not only the high accuracy but the time saving is improved in MALD-TOF MS technology compared with conventional methods.
Therefore, MALDI-TOF MS is considered a highly accurate, appropriate method for high-throughput microbial identification from culture plates, and is a powerful clinical technique that supersedes the previous conventional biochemical or molecular identification systems.
3.2. Identification from positive blood culture (BC)
MALDI-TOF MS can be performed directly for identifying bacterial isolates in BC [33]. In a previous study, 584 positive BCs were tested and 562 were found to contain unique bacterial species [34]. Another study using the MALDI Biotyper revealed that the positive identification at genus and species level was 97.8% in 181 mono-microbial cultures [11]. However, some major problems remain. When there is a mixture of species in BC, only one bacterial species is identified and false identification can occur. In addition, it does not allow for the identification of certain microorganisms, including Viridans Streptococci and encapsulated microorganisms, such as Klebsiella pneumoniae and Haemophilus influenza. The components of human blood can cause interference or generate noisy spectra when BC broth is analyzed directly. Therefore, preprocessing of the BC broth is important to decrease interference and to concentrate the bacteria. To solve these problems, the MBT Sepsityper™ kit was developed. In one study investigating the Sepsityper™ kit, more Gram-negative organisms (100% for genus and 91.4% for species level) were accurately identified than Gram-positive organisms (100% for genus and 67.7% for species level). Difficulties for anaerobic bacteria, alpha-hemolytic streptococci, and polymicrobial were informed [35]. Arroyo et al. showed 200 isolates of Gram-negative bacilli from monomicrobial growth positive blood cultures were identified by MALDI-TOF MS directly. It identified 94% (188/200) and 91% (181/200) to the genus and species levels [13]. In addition, Chien et al. demonstrated MALDI-TOF MS successfully identified bacteria and fungi in 405 positively flagged blood culture bottles directly. 365 monomicrobial growth were correctly identified to the species (72.1%) or genus (89.6%) level. The remaining 40 polymicrobial growth showed 82.5% (33/40) of the isolates were correctly identified to the species level and 92.5% (37/40) to the genus level [36]. Pretreatments to remove non-bacterial proteins are a prerequisite for the successful application of MALDI-TOF MS in patient-derived BCs. For the timing from positive blood culture alarm to microorganism identification result only take a half hour and more than 80% of the BC positive sample can get a confidence microorganism identification result. By the traditional workflow, the positive blood culture sample needs to be separated on an agar plate and culture overnight or longer, with different microbial species/strains. When getting the bacterial mass, the biochemical method was used for the identification. The new workflow can use 1ml of blood culture positive broth directly, without the bacteria culture step via a 30 min sample preparation and MALDI-TOF MS identification. Compared with the time of conventional and MALDI-TOF MS identification of positive blood culture sample, it saves the time of biochemical reaction around 24–48 h or longer [37]. A CE-IVD certificated sample preparation kits and software are available for medical laboratory application.
3.3. Identification directly from patient urine
Urinary tract infections (UTIs), including cystitis (infection of the bladder/lower urinary tract) and pyelonephritis (infection of the kidney/upper urinary tract) are among the most common types of clinical bacterial infections. Prompt diagnosis and treatment are important as patients with acute, complicated UTIs. Therefore, reducing the time taken for microorganism identification can be important, and this may be achieved by direct analysis of urine samples. One study showed that utilization of 4 ml urine sample for centrifuging at low speed to remove the leukocytes, and then at a high speed to collect the bacteria. The pellet was washed and applied directly to the MALDI-TOF MS plate. Microorganism identification was 91.8% and 92.7% at the species and genus level, respectively [15]. Another study revealed that MALDI-TOF MS was able to reliably identify bacteria directly in urine samples at concentrations as low as 103 colony forming units/ml [38]. These results indicate that MALDI-TOF MS is an accurate method of urinary bacterial identification and may be used more expansively in the future.
3.4. Mycobacterium identification
Mycobacteria are one of the primary types of pathogens causing a serious public health problem globally [39]. Mycobacterium is a genus of Actinobacteria, which comprises over 190 known species. M. tuberculosis is the pathogen which causes the most morbidity in humans. In recent years, the incidence of diseases due to NTM infection has escalated, this is due to the increasing numbers of patients with autoimmune diseases and immunocompromised individuals [40].
The gold standard for M. tuberculosis diagnosis is a culture of bacterium isolated from human secretions, such as sputum, bronchoalveolar lavage, or pleural fluid and confirmation by smear microscopy. However, the identification of the Mycobacterium species is challenging due to their long incubation time for each step of culture and biochemical reaction (7 to more than 21 days). Other diagnostic tools, including sputum acid-fast bacilli smear stains, are less sensitive than culture. Thus, faster and more accurate methods of Mycobacterium identification are required to improve patient outcomes. Recently, sequencing and probing methods have been used for bacterial identification. Nucleic acid hybridization (NAH) or nucleic acid amplification testing are helpful for the rapid diagnosis (24–48 h) of organisms belonging to the MTC in patients with suspected TB.
In one study comparing the use of MALDI-TOF MS with NAH, it was observed that MALDI-TOF MS identified 100% of the M. tuberculosis strains correctly; however, only 38.5% of the NTM strains were identified by NAH [41]. MALDI-TOF MS has been demonstrated to have a high sensitivity for the identification of MTC in a clinical laboratory, and reduces the identification time (45 min) with fewer costs [41]. Although MALDI-TOF MS is the most cost-effective and fastest method of MTC identification, NTM are becoming increasingly important clinically, and the identification of NTM using MALDI-TOF MS should be investigated further. A method proposed from the study might be able to reduce the incidence of misidentification of Mycobacterium avium complex (MAC) by MALDI TOF MS. Some closely related species have similar spectra and cannot be separated by MALDI-TOF MS technology. For example, Mycobacterium chimaera and Mycobacterium intracellulare are often difficult to be distinguished from each other. However, M. intracellulare are significantly more likely to be associated with a pulmonary infection compared with M. chimaera. The precise identification between M. intracellulare and M. chimaera within MAC can be helpful in clinical significance. This study performed subtype processing by extending the MALDI spectra analysis to enable the accurate identification of M. intracellulare and M. chimaera [42]. It is also difficult to distinguish bacteria within the Mycobacterium abscessus complex such as Mycobacterium massiliense (M. abscessus subspecies bolletti) and M. abscessus (Sensu Stricto). The different bacterial subtypes present different antibiotic resistance phenotypes and their precise, rapid identification can be useful for the administration of the correct antibiotics. A report showed that M. massiliense can be accurately separate from M. abscessus with using the genetic algorithms [43]. MALDI Biotyper has an updated mycobacterium library in 2017, which includes 164 species; however, a wide range of NTM details remain unreported. For the evaluation of Mycobacterium species, Wilen et al. evaluated the efficiency of MALDI-TOF identification by 157 mycobacterial isolates (including 16 Mycobacterium tuberculosis isolates) and showed the 89.2% (140/157) reliable identification. Another study, Rodrigues-Sanchez et al. used 125 non-tuberculosis mycobacterial (NTM) isolates to evaluate the MALDI-TOF MS identification [44]. There are 88.8% (111/125) correct identification at the species level with a score above 1.7 [21]. Chen et al. MALDI-TOF MS generated reliable identification for 87.3% (89/102) and 62.8% (64/102) clinical mycobacterial isolates at the genus and species levels from the solid culture media, respectively [45].
3.5. Yeast and fungi identification
Yeast and fungal infections are common complications in immunosuppressed patients and therefore they are associated with therapies for autoimmune diseases, hematologic malignancies, hematopoietic cell transplantation and solid organ transplantation. Invasive candidiasis is a common central venous catheter-associated infection, and invasive pulmonary Aspergillosis can result in pneumonia, fever, pleuritic chest pain and hemoptysis symptoms. Due to the high-cost and time-consuming nature of conventional phenotypic methods used for yeast and fungi identification, the application of MALDI-TOF MS in routine clinical examination has been recently supported. For example, Wang et al. showed that 41 distinct species were identified by MALDI-TOF MS from 2683 clinical yeast isolates. The correct identification of all the yeast isolates is 98.8% (2651/2683) [46]. Chao et al. showed 92.5% (185/200) identification rate with 200 clinical yeast isolates [47]. Furthermore, Chen et al. applied 98 clinical isolates of yeast to evaluate the identification rate of MALDI-TOF MS, and results showed that 74.5% (73/98) was identified to species level and 94.9% (93/98) to genus level [45]. Therefore, MALDI-TOF MS is less expensive and simpler than the current DNA based gold standard method of identification [9,48].
For filamentous fungi, Becker et al. showed MALDI-TOF MS correctly identified 95.4% of isolates at the species level [49]. Gautier et al. examined the identification of clinical mold isolates from patients with aspergillosis, fusariosis and mucormycosis. The study created 2832 reference spectra, corresponding to 347 species of mold to improve the identification of mold. Next, this workflow identified 1094 of 1107 (98.8%) clinical mold isolates, corresponding to 107 distinct species, superior to morphological identification (78.2%) [50]. Furthermore, in one study using an extended in-house database, MALDI-TOF MS appeared to obtain more accurate identifications of filamentous fungi [51]. Compared with biochemical techniques, MS is easier, faster, cheaper, more accurate and allows for the identification of closely related species. Continuous extension of the library is essential for the maintenance of enhanced reliability of MS in the future.
4. Antimicrobial susceptibility testing
MALDI-TOF MS microbial identification technology is frequently used within clinical microbiology laboratories. Furthermore, MALDI-TOF MS is also used for antimicrobial susceptibility testing (AST). Many scientists have designed various methods to analyze AST using MALDI-TOF MS. The majority follow one of the three approaches listed below.
4.1. Detection of antibiotic degradation
This approach primarily determines the β-lactam antibiotic resistance. Resistance to β-lactam antibiotics is mainly due to β-lactamase expression, which hydrolyses the β-lactam ring and inactivates the antibiotic’s activity. The hydrolysis of a βlactam ring by β-lactamase corresponds to the addition of a Water molecular, and this reaction leads to an 18 Da mass change, which can be easily detected by MALDI-TOF MS. Clinical isolates of model E. coli strains were used to develop the method. The model strains were treated with many different types of β-lactam antibiotics [52], then the clinical species were tested to determine the AST result of each strain. A study showed that MALDI-TOF MS can detect the carbapenem hydrolysis product, which created with carbapenemase. This demonstrates the potential of rapid β-lactam antibiotic resistance testing by MALDI-TOF MS [53]. The method was also applied to BC positive samples for direct detection of carbapenemase activity. This enables a faster decision to be made on which antibiotic treatment to administer and speeds up the implementation of infection control measures [54]. An important study showed that in certain cases low enzymatic activity or delayed enzyme expression could lead to a false-negative result. The extension of incubation time to 4 h could help avoid these mistakes [55].
4.2. Specific biomarker discovery for detecting antibiotic resistant strains
Samples prepared in the same way as for microorganism identification by MALDI-TOF WCMS (whole cell mass spectrometry) may be used for biomarker detection of antibiotic resistant strains. Some specific signals can be detected in cfiA-positive Bacteroides fragilis strains. cfiA is encoded by a class B metallo-β-lactamase, which confers resistance in B. fragilis to all β-lactam antibiotics, including carbapenems; almost 100% of carbapenem-resistance Bacteroides strains are cfiA positive [56,57]. Methicillin-resistant Staphylococcus aureus (MRSA) is a critical issue in public health and hospital hygiene in most countries worldwide. The ability to rapidly identify MRSA would be a critical improvement within healthcare. Many studies have shown a signal at 2415 m/z is a small peptide PSM-mec encoded on three SCCmec cassettes, is highly associated with MRSA [58]. There are many different mechanisms of methicillin-resistance in S. aureus, and the 2415 m/z signal is shown in 37% of MRSA. Although the sensitivity is low, the specificity is high (≥98%), which indicates that this peak still has value in clinical applications without any additional sample preparation and cost [59].
4.3. Phenotypic antibiotic resistance analysis of bacterial strains
This method is similar to the phenotypic observation of antibiotic resistance in microorganisms. The advantage of this method is that it is faster than conventional technology, as it only requires 2–3 h incubation. In principle, it can be used to detect any mechanism of antibiotic resistance by optimizing the growth of microorganisms in the presence of antibiotics [60]. When microorganisms are incubated with antibiotics, they are grown in medium containing isotopically labeled amino acids. Only resistant, growing microorganisms are able to incorporate the labeled amino acids. The de novo biosynthesis of proteins by resistant isolates with labeled, heavy amino acids reveals different protein profile spectra for susceptible and resistant isolates. By comparing the resistant profile spectra with the standard spectra, the AST results can be determined in hours [61,62].
5. Subtyping strategy by MALDI-TOF MS peptide/protein profile
The conventional biochemical bacteria identification provides only the results of the species. MALDI-TOF MS can be used to obtain the protein profile of the strain. Using the protein profile, there is a chance to detect phenotype-related biomarkers, such as drug-resistant strains, or similar species identification. The Subtyping’s strategy is to use the protein profile to obtain the subtype information using the same protein profile after the strain identification results. It can be applied to the identification of drug-resistant strains or the accurate identification of a bacterial species complex.
The preclinical antibiotics-resistance strain detection can be applied to methicillin-resistant S. aureus (MRSA) strains [5]. Most of the cfiA positive B. fragilis strains are carbapenem resistant [63]. A blaKPC positive related signal is highly correlated with K. pneumoniae carbapenem resistance strain, which produces K. pneumoniae carbapenemase (KPC) [6]. The accurate identification of a bacterial species complex could be used to Listeria monocytogenes group (including L. monocytogenes, Listeria ivanovii, Listeria innocua, Listeria welshimeri, Listeria seeligeri and Listeria grayi) show only slight differences in genetic and proteomic profile levels [64].
6. Current limitations of microorganism identification by MALDI-TOF MS
At present MALDI-TOF MS is unsuitable for the differentiation of Shigella and E. coli [65], Bordetella pertussis and B. bronchioseptica, Achromobacter xylosoxidans and A. ruhlandii, as well as Bacteroides nordii and B. salyersiae. The Enterobacter cloacae complex is a group of six very closely related species (E. asburiae, E. cloacae, E. hormaechei, E. kobei, E. ludwigii, and E. nimipressuralis) with similar resistance patterns, which are not yet differentiated [66]. In addition, pneumococci, viridans streptococci and encapsulated microorganisms, such as K. pneumoniae and H. influenza can be misidentified by MALDI-TOF MS. Most of these bacterial species can be identified with the utilization of the reference spectra. However, a small number of these strains that have a very similar genetic background or wide diversity inter-species cannot be distinguished by the mass spectrometry signal and to obtain high-confidence identification results. Furthermore, the precise identification of NTM is not feasible at present. For filamentous fungi, the primary limitation of MALDI TOF MS identification is limited by the reference spectrum. Continuous upgrades of spectral databases and optimal sample enrichment within the MALDI-TOF MS system will increase its power and potential applications.
7. Conclusion
MALDI-TOF MS is an emerging technology and has been recently used for the identification of microorganisms. It is a high-throughput, cost-effective method with a high degree of accuracy. This is especially important for routine clinical microbiology, as the results could come directly from positive BCs, urine or other biological fluids, as well as subcultures on agar plates and broth media. The reliability and accuracy of MALDI-TOF MS have been verified in a number of studies, and the precision of microorganism identification is critically dependent on the number of database entries. Therefore, continuous updating of the reference database is indispensable for the advancement of performance. Further database refinements in the MALDI-TOF MS system allow for the rapid identification of antibiotic resistance characteristics and reliability, as well as the direct identification of pathogens in urine samples and BCs. This technique has significant potential in diagnostic laboratories in the future.
REFERENCES
- 1. Koichi Tanaka HW, Yutaka Ido, Satoshi Akita, Yoshikazu Yoshida, Tamio Yoshida, Matsuo T. Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 1988;2:151–3. [Google Scholar]
- 2. Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 3. Croxatto A, Prod’hom G, Greub G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol Rev. 2012;36:380–407. doi: 10.1111/j.1574-6976.2011.00298.x. [DOI] [PubMed] [Google Scholar]
- 4. Vrioni G, Tsiamis C, Oikonomidis G, Theodoridou K, Kapsimali V, Tsakris A. MALDI-TOF mass spectrometry technology for detecting biomarkers of antimicrobial resistance: current achievements and future perspectives. Ann Transl Med. 2018;6:240. doi: 10.21037/atm.2018.06.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Camoez M, Sierra JM, Dominguez MA, Ferrer-Navarro M, Vila J, Roca I. Automated categorization of methicillin-resistant Staphylococcus aureus clinical isolates into different clonal complexes by MALDI-TOF mass spectrometry. Clin Microbiol Infect. 2016;22:161 e1–e7. doi: 10.1016/j.cmi.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 6. Gaibani P, Galea A, Fagioni M, Ambretti S, Sambri V, Landini MP. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of KPC-producing Klebsiella pneumoniae. J Clin Microbiol. 2016;54:2609–13. doi: 10.1128/JCM.01242-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hsieh SY, Tseng CL, Lee YS, Kuo AJ, Sun CF, Lin YH, et al. Highly efficient classification and identification of human pathogenic bacteria by MALDI-TOF MS. Mol Cell Proteomics. 2008;7:448–56. doi: 10.1074/mcp.M700339-MCP200. [DOI] [PubMed] [Google Scholar]
- 8. Mellmann A, Cloud J, Maier T, Keckevoet U, Ramminger I, Iwen P, et al. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J Clin Microbiol. 2008;46:1946–54. doi: 10.1128/JCM.00157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marklein G, Josten M, Klanke U, Muller E, Horre R, Maier T, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J Clin Microbiol. 2009;47:2912–7. doi: 10.1128/JCM.00389-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sharma M, Gautam V, Mahajan M, Rana S, Majumdar M, Ray P. Direct identification by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) from positive blood culture bottles: an opportunity to customize growth conditions for fastidious organisms causing bloodstream infections. Indian J Med Res. 2017;146:541–4. doi: 10.4103/ijmr.IJMR_823_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen JH, Ho PL, Kwan GS, She KK, Siu GK, Cheng VC, et al. Direct bacterial identification in positive blood cultures by use of two commercial matrix-assisted laser desorption ionization-time of flight mass spectrometry systems. J Clin Microbiol. 2013;51:1733–9. doi: 10.1128/JCM.03259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Meex C, Neuville F, Descy J, Huynen P, Hayette MP, De Mol P, et al. Direct identification of bacteria from BacT/ALERT anaerobic positive blood cultures by MALDI-TOF MS: MALDI Sepsityper kit versus an in-house saponin method for bacterial extraction. J Med Microbiol. 2012;61:1511–6. doi: 10.1099/jmm.0.044750-0. [DOI] [PubMed] [Google Scholar]
- 13. Arroyo MA, Denys GA. Parallel evaluation of the MALDI sepsityper and verigene BC-GN assays for rapid identification of gram-negative bacilli from positive blood cultures. J Clin Microbiol. 2017;55:2708–18. doi: 10.1128/JCM.00692-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Conway GC, Smole SC, Sarracino DA, Arbeit RD, Leopold PE. Phyloproteomics: species identification of Enterobacteriaceae using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Mol Microbiol Biotechnol. 2001;3:103–12. [PubMed] [Google Scholar]
- 15. Ferreira L, Sanchez-Juanes F, Gonzalez-Avila M, Cembrero-Fucinos D, Herrero-Hernandez A, Gonzalez-Buitrago JM, et al. Direct identification of urinary tract pathogens from urine samples by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2010;48:2110–5. doi: 10.1128/JCM.02215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Segawa S, Sawai S, Murata S, Nishimura M, Beppu M, Sogawa K, et al. Direct application of MALDI-TOF mass spectrometry to cerebrospinal fluid for rapid pathogen identification in a patient with bacterial meningitis. Clin Chim Acta. 2014;435:59–61. doi: 10.1016/j.cca.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 17. Brunetti G, Ceccarelli G, Giordano A, Navazio AS, Vittozzi P, Venditti M, et al. Fast and reliable diagnosis of XDR Acinetobacter baumannii meningitis by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. New Microbiol. 2018;41:77–9. [PubMed] [Google Scholar]
- 18. Arca-Suarez J, Galan-Sanchez F, Marin-Casanova P, Rodriguez-Iglesias MA. Direct identification of microorganisms from thioglycolate broth by MALDI-TOF MS. PLoS One. 2017;12:e0185229. doi: 10.1371/journal.pone.0185229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.WHO. WHO global tuberculosis report 2017. Geneva, Switzerland: WHO; 2017. [Google Scholar]
- 20. Management of opportunist mycobacterial infections: joint tuberculosis committee Guidelines 1999. Subcommittee of the joint tuberculosis committee of the British thoracic society. Thorax. 2000;55:210–8. doi: 10.1136/thorax.55.3.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilen CB, McMullen AR, Burnham CA. Comparison of sample preparation methods, instrumentation platforms, and contemporary commercial databases for identification of clinically relevant Mycobacteria by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2015;53:2308–15. doi: 10.1128/JCM.00567-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chalupova J, Raus M, Sedlarova M, Sebela M. Identification of fungal microorganisms by MALDI-TOF mass spectrometry. Biotechnol Adv. 2014;32:230–41. doi: 10.1016/j.biotechadv.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 23. Cassagne C, Ranque S, Normand AC, Fourquet P, Thiebault S, Planard C, et al. Mould routine identification in the clinical laboratory by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. PLoS One. 2011;6:e28425. doi: 10.1371/journal.pone.0028425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cherkaoui A, Hibbs J, Emonet S, Tangomo M, Girard M, Francois P, et al. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J Clin Microbiol. 2010;48:1169–75. doi: 10.1128/JCM.01881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dupont C, Sivadon-Tardy V, Bille E, Dauphin B, Beretti JL, Alvarez AS, et al. Identification of clinical coagulase-negative staphylococci, isolated in microbiology laboratories, by matrix-assisted laser desorption/ionization-time of flight mass spectrometry and two automated systems. Clin Microbiol Infect. 2010;16:998–1004. doi: 10.1111/j.1469-0691.2009.03036.x. [DOI] [PubMed] [Google Scholar]
- 26. Spanu T, De Carolis E, Fiori B, Sanguinetti M, D’Inzeo T, Fadda G, et al. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to rpoB gene sequencing for species identification of bloodstream infection staphylococcal isolates. Clin Microbiol Infect. 2011;17:44–9. doi: 10.1111/j.1469-0691.2010.03181.x. [DOI] [PubMed] [Google Scholar]
- 27. Carbonnelle E, Grohs P, Jacquier H, Day N, Tenza S, Dewailly A, et al. Robustness of two MALDI-TOF mass spectrometry systems for bacterial identification. J Microbiol Methods. 2012;89:133–6. doi: 10.1016/j.mimet.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 28. Martiny D, Dediste A, Debruyne L, Vlaes L, Haddou NB, Vandamme P, et al. Accuracy of the API Campy system, the Vitek 2 Neisseria-Haemophilus card and matrix-assisted laser desorption ionization time-of-flight mass spectrometry for the identification of Campylobacter and related organisms. Clin Microbiol Infect. 2011;17:1001–6. doi: 10.1111/j.1469-0691.2010.03328.x. [DOI] [PubMed] [Google Scholar]
- 29. Faron ML, Buchan BW, Hyke J, Madisen N, Lillie JL, Granato PA, et al. Multicenter evaluation of the Bruker MALDI biotyper CA system for the identification of clinical aerobic gram-negative bacterial isolates. PLoS One. 2015;10:e0141350. doi: 10.1371/journal.pone.0141350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garner O, Mochon A, Branda J, Burnham CA, Bythrow M, Ferraro M, et al. Multi-centre evaluation of mass spectrometric identification of anaerobic bacteria using the VITEK(R) MS system. Clin Microbiol Infect. 2014;20:335–9. doi: 10.1111/1469-0691.12317. [DOI] [PubMed] [Google Scholar]
- 31. Rychert J, Burnham CA, Bythrow M, Garner OB, Ginocchio CC, Jennemann R, et al. Multicenter evaluation of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for identification of Gram-positive aerobic bacteria. J Clin Microbiol. 2013;51:2225–31. doi: 10.1128/JCM.00682-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Altun O, Botero-Kleiven S, Carlsson S, Ullberg M, Ozenci V. Rapid identification of bacteria from positive blood culture bottles by MALDI-TOF MS following short-term incubation on solid media. J Med Microbiol. 2015;64:1346–52. doi: 10.1099/jmm.0.000168. [DOI] [PubMed] [Google Scholar]
- 33. Lagace-Wiens PR, Adam HJ, Karlowsky JA, Nichol KA, Pang PF, Guenther J, et al. Identification of blood culture isolates directly from positive blood cultures by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry and a commercial extraction system: analysis of performance, cost, and turnaround time. J Clin Microbiol. 2012;50:3324–8. doi: 10.1128/JCM.01479-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. La Scola B, Raoult D. Direct identification of bacteria in positive blood culture bottles by matrix-assisted laser desorption ionisation time-of-flight mass spectrometry. PLoS One. 2009;4:e8041. doi: 10.1371/journal.pone.0008041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kok J, Thomas LC, Olma T, Chen SC, Iredell JR. Identification of bacteria in blood culture broths using matrix-assisted laser desorption-ionization sepsityper and time of flight mass spectrometry. PLoS One. 2011;6:e23285. doi: 10.1371/journal.pone.0023285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chien JY, Lee TF, Du SH, Teng SH, Liao CH, Sheng WH, et al. Applicability of an in-house saponin-based extraction method in Bruker biotyper matrix-assisted laser desorption/ionization time-of-flight mass spectrometry system for identification of bacterial and fungal species in positively flagged blood cultures. Front Microbiol. 2016;7:1432. doi: 10.3389/fmicb.2016.01432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yonetani S, Ohnishi H, Ohkusu K, Matsumoto T, Watanabe T. Direct identification of microorganisms from positive blood cultures by MALDI-TOF MS using an in-house saponin method. Int J Infect Dis. 2016;52:37–42. doi: 10.1016/j.ijid.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 38. Kohling HL, Bittner A, Muller KD, Buer J, Becker M, Rubben H, et al. Direct identification of bacteria in urine samples by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and relevance of defensins as interfering factors. J Med Microbiol. 2012;61:339–44. doi: 10.1099/jmm.0.032284-0. [DOI] [PubMed] [Google Scholar]
- 39. Bar-On O, Mussaffi H, Mei-Zahav M, Prais D, Steuer G, Stafler P, et al. Increasing nontuberculous Mycobacteria infection in cystic fibrosis. J Cyst Fibros. 2015;14:53–62. doi: 10.1016/j.jcf.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 40. Donohue MJ. Increasing nontuberculous Mycobacteria reporting rates and species diversity identified in clinical laboratory reports. BMC Infect Dis. 2018;18:163. doi: 10.1186/s12879-018-3043-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Samli A, Ilki A. Comparison of MALDI-TOF MS, nucleic acid hybridization and the MPT64 immunochromatographic test for the identification of M. tuberculosis and non-tuberculosis Mycobacterium species. New Microbiol. 2016;39:259–63. [PubMed] [Google Scholar]
- 42. Pranada AB, Witt E, Bienia M, Kostrzewa M, Timke M. Accurate differentiation of Mycobacterium chimaera from Mycobacterium intracellulare by MALDI-TOF MS analysis. J Med Microbiol. 2017;66:670–7. doi: 10.1099/jmm.0.000469. [DOI] [PubMed] [Google Scholar]
- 43. Teng SH, Chen CM, Lee MR, Lee TF, Chien KY, Teng LJ, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry can accurately differentiate between Mycobacterium masilliense (M. abscessus subspecies bolletti) and M. abscessus (sensu stricto) J Clin Microbiol. 2013;51:3113–6. doi: 10.1128/JCM.01239-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rodriguez-Sanchez B, Ruiz-Serrano MJ, Marin M, Lopez Roa P, Rodriguez-Creixems M, Bouza E. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of nontuberculous Mycobacteria from clinical isolates. J Clin Microbiol. 2015;53:2737–40. doi: 10.1128/JCM.01380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen JH, Yam WC, Ngan AH, Fung AM, Woo WL, Yan MK, et al. Advantages of using matrix-assisted laser desorption ionization-time of flight mass spectrometry as a rapid diagnostic tool for identification of yeasts and mycobacteria in the clinical microbiological laboratory. J Clin Microbiol. 2013;51:3981–7. doi: 10.1128/JCM.01437-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang H, Fan YY, Kudinha T, Xu ZP, Xiao M, Zhang L, et al. A comprehensive evaluation of the Bruker biotyper MS and Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry systems for identification of yeasts, part of the National China hospital invasive fungal surveillance net (CHIF-NET) study, 2012 to 2013. J Clin Microbiol. 2016;54:1376–80. doi: 10.1128/JCM.00162-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chao QT, Lee TF, Teng SH, Peng LY, Chen PH, Teng LJ, et al. Comparison of the accuracy of two conventional phenotypic methods and two MALDI-TOF MS systems with that of DNA sequencing analysis for correctly identifying clinically encountered yeasts. PLoS One. 2014;9:e109376. doi: 10.1371/journal.pone.0109376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bizzini A, Durussel C, Bille J, Greub G, Prod’hom G. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of bacterial strains routinely isolated in a clinical microbiology laboratory. J Clin Microbiol. 2010;48:1549–54. doi: 10.1128/JCM.01794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Becker PT, de Bel A, Martiny D, Ranque S, Piarroux R, Cassagne C, et al. Identification of filamentous fungi isolates by MALDI-TOF mass spectrometry: clinical evaluation of an extended reference spectra library. Med Mycol. 2014;52:826–34. doi: 10.1093/mmy/myu064. [DOI] [PubMed] [Google Scholar]
- 50. Gautier M, Ranque S, Normand AC, Becker P, Packeu A, Cassagne C, et al. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry: revolutionizing clinical laboratory diagnosis of mould infections. Clin Microbiol Infect. 2014;20:1366–71. doi: 10.1111/1469-0691.12750. [DOI] [PubMed] [Google Scholar]
- 51. Normand AC, Cassagne C, Gautier M, Becker P, Ranque S, Hendrickx M, et al. Decision criteria for MALDI-TOF MS-based identification of filamentous fungi using commercial and in-house reference databases. BMC Microbiol. 2017;17:25. doi: 10.1186/s12866-017-0937-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sparbier K, Schubert S, Weller U, Boogen C, Kostrzewa M. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based functional assay for rapid detection of resistance against beta-lactam antibiotics. J Clin Microbiol. 2012;50:927–37. doi: 10.1128/JCM.05737-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hrabak J, Studentova V, Walkova R, Zemlickova H, Jakubu V, Chudackova E, et al. Detection of NDM-1, VIM-1, KPC, OXA-48, and OXA-162 carbapenemases by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2012;50:2441–3. doi: 10.1128/JCM.01002-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Carvalhaes CG, Cayo R, Visconde MF, Barone T, Frigatto EA, Okamoto D, et al. Detection of carbapenemase activity directly from blood culture vials using MALDI-TOF MS: a quick answer for the right decision. J Antimicrob Chemother. 2014;69:2132–6. doi: 10.1093/jac/dku094. [DOI] [PubMed] [Google Scholar]
- 55. Chong PM, McCorrister SJ, Unger MS, Boyd DA, Mulvey MR, Westmacott GR. MALDI-TOF MS detection of carbapenemase activity in clinical isolates of Enterobacteriaceae spp., Pseudomonas aeruginosa, and Acinetobacter baumannii compared against the Carba-NP assay. J Microbiol Methods. 2015;111:21–3. doi: 10.1016/j.mimet.2015.01.024. [DOI] [PubMed] [Google Scholar]
- 56. Nagy E, Becker S, Soki J, Urban E, Kostrzewa M. Differentiation of division I (cfiA-negative) and division II (cfiA-positive) Bacteroides fragilis strains by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Med Microbiol. 2011;60:1584–90. doi: 10.1099/jmm.0.031336-0. [DOI] [PubMed] [Google Scholar]
- 57. Fenyvesi VS, Urban E, Bartha N, Abrok M, Kostrzewa M, Nagy E, et al. Use of MALDI-TOF/MS for routine detection of cfiA gene-positive Bacteroides fragilis strains. Int J Antimicrob Agents. 2014;44:474–5. doi: 10.1016/j.ijantimicag.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 58. Josten M, Dischinger J, Szekat C, Reif M, Al-Sabti N, Sahl HG, et al. Identification of agr-positive methicillin-resistant Staphylococcus aureus harbouring the class A mec complex by MALDI-TOF mass spectrometry. Int J Med Microbiol. 2014;304:1018–23. doi: 10.1016/j.ijmm.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 59. Rhoads DD, Wang H, Karichu J, Richter SS. The presence of a single MALDI-TOF mass spectral peak predicts methicillin resistance in staphylococci. Diagn Microbiol Infect Dis. 2016;86:257–61. doi: 10.1016/j.diagmicrobio.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 60. van Belkum A, Welker M, Pincus D, Charrier JP, Girard V. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry in clinical microbiology: what are the current issues? Ann Lab Med. 2017;37:475–83. doi: 10.3343/alm.2017.37.6.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sparbier K, Lange C, Jung J, Wieser A, Schubert S, Kostrzewa M. MALDI biotyper-based rapid resistance detection by stable-isotope labeling. J Clin Microbiol. 2013;51:3741–8. doi: 10.1128/JCM.01536-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jung JS, Hamacher C, Gross B, Sparbier K, Lange C, Kostrzewa M, et al. Evaluation of a semiquantitative matrix-assisted laser desorptionionization-time of flight mass spectrometry method for rapid antimicrobial susceptibility testing of positive blood cultures. J Clin Microbiol. 2016;54:2820–4. doi: 10.1128/JCM.01131-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wybo I, De Bel A, Soetens O, Echahidi F, Vandoorslaer K, Van Cauwenbergh M, et al. Differentiation of cfiA-negative and cfiA-positive Bacteroides fragilis isolates by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2011;49:1961–4. doi: 10.1128/JCM.02321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Barbuddhe SB, Maier T, Schwarz G, Kostrzewa M, Hof H, Domann E, et al. Rapid identification and typing of listeria species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl Environ Microbiol. 2008;74:5402–7. doi: 10.1128/AEM.02689-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pupo GM, Lan R, Reeves PR. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci U S A. 2000;97:10567–72. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Paauw A, Caspers MP, Schuren FH, Leverstein-van Hall MA, Deletoile A, Montijn RC, et al. Genomic diversity within the Enterobacter cloacae complex. PLoS One. 2008;3:e3018. doi: 10.1371/journal.pone.0003018. [DOI] [PMC free article] [PubMed] [Google Scholar]