Abstract
Since the emergence of SARS-CoV-2 at Wuhan in the Hubei province of China in 2019, the virus has accumulated various mutations, giving rise to many variants. According to the combinations of mutations acquired, these variants are classified into lineages and greatly differ in infectivity and transmissibility. In 2021 alone, a variant of interest (VoI) Mu (B.1.621), as well as, variants of concern (VoC) Delta (B.1.617.2) and Omicron (BA.1, BA.2) and later in 2022, BA.4, BA.5, and BA.2.12.1 have emerged. Since then, the world has seen prominent surges in the rate of infection during short periods of time. However, not all populations have suffered equally, which suggests a possible role of host genetic factors. Here, we investigated the strength of binding of the spike glycoprotein receptor-binding domain (RBD) of the SARS-CoV-2 variants: Mu, Delta, Delta Plus (AY.1), Omicron sub-variants BA.1, BA.2, BA.4, BA.5, and BA.2.12.1 with the human angiotensin-converting enzyme 2 (hACE2) missense variants prevalent in major populations. In this purpose, molecular docking analysis, as well as, molecular dynamics simulation was performed of the above-mentioned SARS-CoV-2 RBD variants with the hACE2 containing the single amino acid substitutions prevalent in African (E37K), Latin American (F40L), non-Finnish European (D355 N), and South Asian (P84T) populations, in order to predict the effects of the lineage-defining mutations of the viral variants on receptor binding. The effects of these mutations on protein stability were also explored. The protein-protein docking and molecular dynamics simulation analyses have revealed variable strength of attachment and exhibited altered interactions in the case of different hACE2-RBD complexes. In vitro studies are warranted to confirm these findings which may enable early prediction regarding the risk of transmissibility of newly emerging variants across different populations in the future.
Keywords: SARS-CoV-2, COVID-19, hACE2, Missense variants, Molecular docking, Molecular dynamics, MM/GBSA
1. Introduction
The novel coronavirus, named Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), also referred to as nCoV-19, was first identified in the Wuhan, Hubei province of China in late December 2019 as the causative agent of the Coronavirus Disease 2019 (COVID-19) [1]. Since then, the virus has spread all over the world, affecting more than 440 million people and causing more than 5.97 million deaths worldwide, as of March 2022. These numbers only represent the clinically diagnosed cases. The real number of infections is much higher because the virus can cause asymptomatic infections [2]. SARS-CoV-2 belongs to the Betacoronavirus genus under the Coronaviridae family. The single-stranded positive-sense RNA genome of the SARS-CoV-2 is approximately 29.9 kb in length. It is replicated by an RNA-dependent RNA polymerase (RdRP), encoded in the viral genome [3].
The virus spreads via respiratory droplets, targets the respiratory tract of humans, and infects the human angiotensin-converting enzyme 2 (hACE2) positive lung cells, which also express transmembrane protease, serine 2 (TMPRSS2) e.g: nasal and bronchial epithelial cells, also alveolar type II pneumocytes [4]. Upon entering the body it interacts with hACE2 of cells via its spike glycoprotein (S). The spike protein has two distinct subunits, S1 and S2, which are responsible for receptor binding and membrane fusion, respectively. Receptor-binding domain (RBD) of the spike protein interacts directly with the hACE2 and is a part of the S1 subunit, spanning residues 319–541. Cathepsin L and TMPRSS2 mediated priming of the spike protein also facilitates membrane fusion of the virus, through cleavage at the S1/S2 junction and S2’ site [5].
Many generic therapeutic regimens implemented in the case of previous outbreaks of coronaviruses, i.e. SARS-CoV and MERS-CoV have been repurposed for SARS-CoV-2 infection. Moreover, with advanced computer-aided drug design, many specific therapeutic components in the forms of small molecules, monoclonal antibodies, siRNA and miRNA have been proposed since the emergence of the novel coronavirus [[6], [7], [8]]. In this short period of time, several vaccines have been produced with considerable efficacy and many are currently in clinical trial [[9], [10], [11], [12]]. In addition, multi-epitope vaccine constructs have been designed through in silico approaches as potential prophylactics [13].
Like other RNA viruses, the SARS-CoV-2 has a high mutation rate, and this has caused the emergence of many variants of this virus since its identification [14]. The mutations that are necessarily present in all isolates of a particular variant of the virus are called lineage-defining mutations for that variant. An isolate is assigned to a variant clade based on the presence of certain lineage-defining mutations [15]. Based on the nature of the mutations, the variants may exhibit alteration to many different characteristics of the virus such as transmissibility, disease severity, immune escape, increase in virulence, change in chemical manifestations of disease, and diagnostic or therapeutic escape. These mutations may also contribute to community transmission and bring about detrimental changes in COVID-19 epidemiology.
According to the CoVariant portal of GISAID (https://covariants.org/), 27 variants have been identified so far. The World Health Organization (WHO) has declared 5 of them (Alpha, Beta, Gamma, Delta, Omicron) as Variant of Concern (VoC) and 2 of them (Mu, Lambda) as Variant of Interest (VoI). The VoCs are named according to their time of emergence with respect to the identification of the Wuhan strain, which is considered the reference for SARS-CoV-2. The Alpha variant emerged in the UK (reported in December 2020), Beta in South Africa (December 2020), Gamma in Brazil (January 2021), Delta and Delta plus in India (May 2021), and Omicron in South Africa (November 2021). The VoI Lambda emerged in Peru (December 2020) and Mu in Columbia (January 2021) [16]. The mutations cause changes in the respective protein's structures [17]. Mutations occurring in the spike protein have conferred the virus higher transmissibility and allow them to escape from the immune response generated by the monoclonal neutralizing antibody or vaccination [18]. There are nine mutations in the spike protein of Delta and Mu variant, 36 in Omicron BA.1, 31 in Omicron BA.2, 34 mutations shared by Omicron BA.4 and BA.5 and 33 in Omicron BA.2.12.1, compared to the Wuhan strain. Among these two are in the RBD for Delta variant, three for Mu, 15 for Omicron BA.1, 16 for Omicron BA.2, 17 for Omicron BA.4, BA.5, and 17 for Omicron BA.2.12.1.
The host receptor hACE2 harbours many polymorphisms, of which, some are missense variants that result in changes in the amino acid residue of the protein, and subsequent conformational change. These may alter the structure of the hACE2 which may affect the attachment with the spike protein of SARS-CoV-2. The polymorphisms are inherited within and can be attributed to a population in a region, where one may prevail in a given population, and others may prevail in other populations inhabiting different regions. The population frequency of a particular polymorphism might appear as a small number in percent value, but technically it includes a very large number of people. For example, the P84T missense variant in the hACE2 occurs at a frequency of 3.58% in the South Asian population, according to gnomAD exomes r2.1.1 dataset, and is estimated to be harboured by almost 56 million people in India and Bangladesh alone [19]. Therefore, it is important to take into consideration the effect of hACE2 polymorphism while analysing the virulence of SARS-CoV-2. The mutations occurring within the viral genome as well as natural variants within its receptor, hACE2 may contribute to the binding affinity followed by the entry and transmission of the virus in a population or region-specific manner [20]. Thus, the unique combinations of population-specific hACE2 polymorphisms and SARS-CoV-2 spike protein mutations may elucidate the region-dependent emergence and preference of each SARS-CoV-2 variant for one or more particular populations.
Molecular docking analysis provides a prediction of the binding affinity and stability of a complex. Therefore, it is possible to detect alterations in binding affinity between the reference and the variant complexes, resulting from the minute deviations of the interacting protein structures. An increase or decrease in binding affinity depends on the molecular interactions between the interacting molecules. Amino acid substitutions, depending on their physicochemical properties, often disrupt existing interactions and establish new interactions. Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) is end-point free energy calculation methodology which is vigorously used in the prediction of binding free energy (BFE) and identification of appropriate protein-protein complexes. The technique in conjunction with per-residue energy decomposition of the binding free energy of a protein-protein complex is particularly useful in highlighting the key residues present in the binding interface. Furthermore, Molecular dynamics (MD) simulation provides insights into the changes in a protein's structure and behaviour in physiological circumstances with respect to time. Initially, proteins were conceptualized to be rigid entities as the X-ray diffraction method produces structures that are fixed in a crystal lattice [21]. However, just like all other macromolecules in physiological systems, proteins exist in a dynamic equilibrium. Advances in computational technologies have enabled us to simulate a physiological system in a computer system and observe the dynamics of any biological macromolecule in that system [22]. Through MD simulation, the stability and dynamics of protein-protein interactions can be observed, thus providing insights into the interactions between proteins, and the behaviour of each of the protein's constituent amino acids during the course of simulation runtime.
Several in silico and experimental studies have shown promising associations of hACE2 polymorphisms with population-wise disease susceptibility and prevalence [[23], [24], [25], [26]]. Additionally, in silico and experimental studies have also been conducted to evaluate the binding affinity of the novel SARS-CoV-2 variants to hACE2 [[27], [28], [29]]. These approaches generate observations regarding variations in receptor-binding interactions based on either receptor hACE2 or the viral spike protein RBD. We aim to bridge the gap in the comparative assessment of binding interactions between polymorphic hACE2 and emerging SARS-CoV-2 variants.
This study, firstly, aims to evaluate the structural effects of the mutations occurring in the spike glycoproteins of the recently emerged SARS-CoV-2 variants. Secondly, considering the structural deviations of the emerging SARS-CoV-2 variants induced by these mutations, we sought to investigate the role of polymorphisms of hACE2 on the variable susceptibility of these variants among different populations.
2. Methods and materials
2.1. Retrieval of amino acid sequences and sequence variants of SARS-CoV-2 spike glycoprotein and human ACE2
The complete reference sequence of SARS-CoV-2 surface glycoprotein (accession: YP_009724390) was retrieved from the NCBI virus database (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/). Lineage defining mutations of the protein in the virus variants – Delta (21A), Delta Plus (21I), Mu (21H), and Omicron sub-variants BA.1 (21K), BA.2 (21L), BA.4 (22A), BA.2 (22B), and BA.2.12.1 (22C) were listed from the CoVariants portal on GISAID database (https://covariants.org/) (Supplementary Table 1).
The reference sequence of human angiotensin-converting enzyme 2 (hACE2) (accession: Q9BYF1) was obtained from the UniProt database (https://www.uniprot.org/uniprot/Q9BYF1). The interacting domain of hACE2 with the receptor-binding domain (RBD) of the viral spike glycoprotein encompasses the amino acid residues: 30–41, 82–84, and 353–357 of the protein, as documented in UniProt. Missense variants of hACE2 protein were then filtered using these amino acid coordinates from the variant table of hACE2 gene in the Ensembl database (available at https://asia.ensembl.org/Homo_sapiens/Gene/Variation_Gene/Table?db=core;g=ENSG00000130234;r=X:15494566-15607236). Population frequencies of each variant were obtained from the gnomAD v2.1.1 dataset (https://gnomad.broadinstitute.org/). The most highly prevalent missense variant within a population was chosen as the representative missense variant of that population. For more than one variant classified into the same population, co-occurrence was tested for the variant pair using the Variant Co-Occurrence tool (https://gnomad.broadinstitute.org/variant-cooccurrence). Between the variants occurring in different haplotypes, the one with the higher population frequency was selected, as the representative variant of the respective population. Four SNPs were thus listed for four populations: African (AFR), Latin American (AMR), non-Finnish European (NFE), and South Asian (SAS); as reported in the gnomAD v2.1.1 dataset.
2.2. Mutational analysis of spike glycoprotein variants
Structure-based analysis of the lineage-defining mutations of the spike protein of the SARS-CoV-2 variants: Delta, Delta Plus, Mu, Omicron subvariants BA.1, BA.2, BA.4, BA.5 and BA.2.12.1 was performed using MAESTROweb (https://pbwww.services.came.sbg.ac.at/maestro/web/maestro/) [30], INPS-3D (https://inpsmd.biocomp.unibo.it/inpsSuite/default/index3D) [31], CUPSAT (http://cupsat.tu-bs.de/) [32], PremPS (https://lilab.jysw.suda.edu.cn/research/PremPS/) [33], and SDM2 (http://marid.bioc.cam.ac.uk/sdm2/) [34] web servers. These tools predict ΔΔG, the difference in free energy of unfolding between wild-type and mutant proteins. Based on their algorithm, the tools interpret results differently. For MAESTROweb and PremPS, if the ΔΔG value is less than zero (negative), the mutation is inferred as destabilising [35,36]. For SDM2, CUPSAT, and INPS-3D, ΔΔG values greater than zero (positive) indicate a stabilising mutation and vice versa [35]. For a given mutation, the prediction provided by the majority of the tools (3 out 5) is considered and has been used for further analysis.
2.3. Construction and validation of mutant models of the viral spike glycoprotein receptor-binding domain (RBD) and human ACE2 protein using homology modelling
The three-dimensional structure of SARS-CoV-2 spike glycoprotein RBD bound with hACE2 (PDB ID: 6M0J) was retrieved from the Protein Data Bank (https://www.rcsb.org/structure/6m0j). The lineage-defining mutations were incorporated into the amino acid sequences of the respective spike glycoprotein variants (Delta, Mu, and Omicron). Chain E (RBD) of the hACE2-RBD complex (PDB ID: 6M0J) served as the template for the construction of the RBD models of the SARS-CoV-2 variants on the Swiss-Model server (https://swissmodel.expasy.org/) [37]. Chain A (hACE2) of the hACE2-RBD complex (PDB ID: 6M0J) was entered along with the single amino acid substitutions specified for the populations into the Missense 3D server (http://missense3d.bc.ic.ac.uk/missense3d/) [38] to generate the population-specific mutant models of hACE2 protein.
The modelled structures of both hACE2 and RBD variants were validated using Swiss-Model Structure Assessment (https://swissmodel.expasy.org/assess), ERRAT [39] & PROCHECK [40] (https://saves.mbi.ucla.edu/) and ProSA-web (https://prosa.services.came.sbg.ac.at/prosa.php) [41] servers.
2.4. Molecular docking assay and analysis of protein-protein interactions
Template-based protein-protein docking between the hACE2 and RBD was performed using the HDOCK server (http://hdock.phys.hust.edu.cn/) [42]. Protein-protein docking utilises the measure of intermolecular interactions to predict the strength of a molecular complex. The protein chains were prepared for docking experiments by removing water, heteroatoms, and ligand groups. The interacting residues for hACE2 were set as reported in the UniProt database as well as those detected by the PDBSum server (http://www.ebi.ac.uk/thornton-srv/databases/pdbsum) [43], using the hACE2-RBD complex (PDB ID: 6M0J) as reference. Using the same complex as the reference, the interacting residues of the RBD were set in a similar manner. The interacting residues of both hACE2 and RBD are presented in Table 1 . The docking energy scores were used for further analysis.
Table 1.
List of interacting residues for hACE2 and SARS-CoV-2 Spike glycoprotein receptor-binding domain (RBD).
| Protein | Interacting Residues |
|---|---|
| hACE2 | Gln24, Thr27, Phe28, Asp30, Lys31, Phe32, Asn33, His34, Glu35, Ala36, Glu37, Asp38, Lys39, Phe40, Tyr41, Gln42, Leu79, Met82, Tyr83, Pro84, Asn330, Lys353, Gly354, Asp355, Phe356, Arg357, Arg393 |
| Spike RBD | Lys417, Gly446, Tyr449, Tyr453, Leu455, Phe456, Ala475, Phe486, Asn487, Tyr489, Gln493, Gly496, Gln498, Thr500, Asn501, Gly502, Tyr505 |
Protein-protein interactions across hACE2 and RBD interfaces were visualised by BIOVIA Discovery Studio Visualizer v21.1.0.20298. Details regarding the two-dimensional interactions between the RBD variants and hACE2 (wildtype and selected missense variants) molecules were generated using the PDBsum server.
2.5. Prediction of binding energy and decomposition of free energy contributions
The binding energy of the reference and variant hACE2 – RBD complexes were calculated using an end-point free energy calculation methodology, Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) on the HawkDock server (http://cadd.zju.edu.cn/hawkdock/) [44]. The server analyses protein-protein interactions by integrating the ATTRACT docking algorithm, the HawkRank scoring function and the MM/GBSA free energy decomposition analysis. MM/GBSA is calculated based on the ff02 force field, the implicit solvent model and the GBOBC1 model (interior dielectric constant = 1). The whole system was minimized for 5000 steps with a cutoff distance of 12 Å for van der Waals interactions (2000 cycles of steepest descent and 3000 cycles of conjugate gradient minimizations).
In MM/GBSA analysis, the binding free energy (ΔGbind) of the system can be expressed as a change in free energy, which can be calculated by Equation (1)
| ΔGbind = ΔEinternal + ΔEelectrostatic + ΔEvdw + ΔGGB + ΔGSA – TΔS | (1) |
Here, ΔEinternal denotes change in internal MM energy, ΔEelectrostatic indicates change in electrostatic energy, ΔEvdw depicts change in van der Waals energy, ΔGGB denotes the electrostatic solvation energy, ΔGSA depicts the deviation of solvation free energy combined with non-electrostatic solvation, and TΔS is the total system binding entropy.
2.6. Molecular dynamics simulation
To evaluate the changes in stability and binding patterns of the docked protein-protein complexes with time, we performed a 125 ns molecular dynamics (MD) simulation of the Omicron BA.1 and BA.2 complexes, as well as, the highest and lowest scoring complexes of Delta and Mu variants. The simulation was run using the Desmond v3.6 program in the Schrödinger suite [45]. A single-point charge (SPC) water model was used to solvate the system and an orthorhombic periodic boundary box with the dimension of 10 Å * 10 Å * 10 Å was employed to maintain a stable volume. Na+ and Cl− ions were used at a concentration of 0.15 M to electronically neutralize the system. The OPLS3e force field was employed to optimize the system. Ensembles of the NPT (constant number of particles, pressure, and temperature) were maintained at 300.0 K temperature and 1.01325 bar pressure. After an initial relaxation, the system was recorded at 125 ps intervals with an energy of 1.2. The results of the MD simulation are presented as the evaluation of the Root Mean Square Deviation (RMSD) and Root Mean Square Fluctuation (RMSF) data.
2.7. RMSD analysis
RMSD is a measurement of the average displacement of selected atoms in a particular timeframe with respect to a reference frame [46]. During the initialization of the simulation, the protein structures were aligned with the backbone frame, and the RMSD was calculated using Equation (2).
| (2) |
Here, N is the number of selected atoms; tref is the reference time (usually the first frame is considered the reference frame, and time is taken as t = 0); Frame x is the location of the chosen atoms when superimposed with the reference frame. Every simulation frame underwent this calculation to provide the RMSD data.
2.8. RMSF analysis
RMSF is useful to observe the behavior of each amino acid residue during the simulation period. It is an indicator of local conformational change around the amino acid considered. The calculation is done by using Equation (3).
| (3) |
where T = trajectory time interval used to calculate the RMSF values, tref = the reference time interval, ri = the location of residue i, r’I = the location of the atoms in residue i after their overlap upon that reference, and the angle brackets (<>) indicates that the square distance is averaged across the residue's atoms.
3. Results
3.1. Retrieved high-frequency population-specific hACE2 missense variants within the hACE2-RBD binding interface
Six unique SNPs, specifying the hACE2-RBD binding interface residues, were initially retrieved. The African and the Non-Finnish European population each had two missense variants within the specified regions. When tested for co-occurrence, both of these variant pairs appeared to occur on different haplotypes. For the rest of the populations, each had only one missense variant. The four hACE2 SNPs thus obtained according to the highest population frequency are provided in Table 2 .
Table 2.
List of hACE2 SNPs representing the four populations according to their frequency along with their SNP IDs and resulting amino acid substitutions.
| Population | AFR | AMR | NFE | SAS |
|---|---|---|---|---|
| dbSNP ID | rs146676783 | rs924799658 | rs961360700 | rs759134032 |
| Amino acid substitution | E37K | F40L | D355 N | P84T |
| Population frequency(gnomAD) | 0.0512 | 0.0001 | 0.0175 | 0.0358 |
3.2. Validation of the predicted structures of RBD variants
The predicted RBD structures of the SARS-CoV-2 variants: Delta, Mu, Omicron BA.1, BA.2, BA.4 and BA.2.12.1 yielded favourable validation scores on Swiss-Model Structure Assessment, ERRAT, and ProSA-web servers (Table 3 ).
Table 3.
Validity scores of the predicted RBD structures of the SARS-CoV-2 variants.
| Variant | Substitutions | RC favoured | QMEAN | ERRAT | ProSA |
|---|---|---|---|---|---|
| Delta | 2 | 95.83% | −1.79 | 91.573 | −6.04 |
| Mu | 3 | 96.35% | −1.61 | 90.3409 | −5.83 |
| Omicron BA.1 | 15 | 94.27% | −2.01 | 91.4773 | −5.87 |
| Omicron BA.2 | 16 | 95.31% | −2.11 | 91.4286 | −5.68 |
| Omicron BA.4 | 17 | 94.79% | −2.06 | 91.4286 | −5.67 |
| Omicron BA.2.12.1 | 17 | 95.31% | −2.08 | 91.4286 | −5.7 |
All the modelled RBD structures produced QMEAN “Z-score” for absolute quality estimates of protein models well above −4.0 on Swiss-Model structure assessment indicating good quality models [47]. All RBD models scored an overall quality factor for non-bonded atomic interactions within a protein above 90 on ERRAT which satisfies the criteria for good quality model. The Ramachandran plots of the RBD models obtained from PROCHECK are presented in Supplementary Fig. 1. 90% or more residues of all the modelled RBD structures were in energetically most favoured region, hence, qualify as acceptable for further analyses [48]. The respective Z-scores of all RBD models, produced by ProSA-web server, were consistent with those determined experimentally as depicted in the Z-score plots (Supplementary Fig. 2).
3.3. Effects of spike protein mutations on protein stability and hACE2 binding affinity
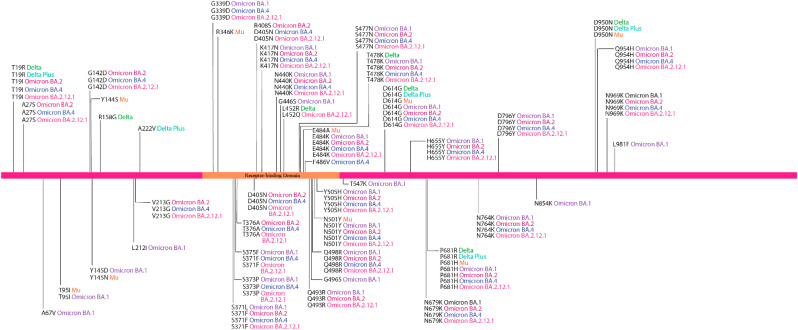
Lineage-defining amino acid substitutions of the SARS-CoV-2 variants are presented in Fig. 1 .
Fig. 1.
Lineage-defining amino acid substitutions of the SARS-CoV-2 variants.
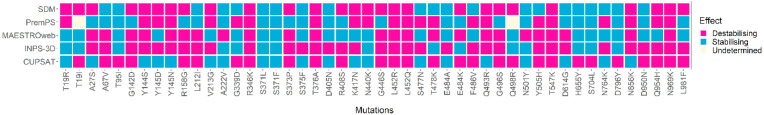
Predictions on the stability of 48 amino acid substitutions found in the spike protein of SARS-CoV-2 variants we investigated, provided by different tools, are presented in Fig. 2 .
Fig. 2.
Predictions on effects of mutations appearing in the spike protein of Delta, Delta plus, Mu, Omicron BA.1, BA.2 variants, BA.4 and BA.2.12.1 variants, as determined by five prediction tools. The columns represent single amino acid substitutions present in at least one SARS-CoV-2 variant mentioned above. The rows indicate the tools used for the prediction of the mutational effects. An amino acid substitution may have either of the effects: stabilising, destabilising or undetermined.
Six of these mutations (R346K, T376A, L452R, L452Q, T547K, N969K) were unanimously predicted to be destabilising, whereas, two of these (S371L, S371F) were unanimously – stabilising. 11 mutations (G142D, Y144S, Y145 N, R158G, S373P, G446S, Q493R, G496S, Y505H, N856K, Q954H) were passed as destabilising and nine mutations (T95I, A222V, S373F, D405 N, E484A, N501Y, H655Y, S704L, D796Y) were passed as stabilising by 80% of the prediction tools. 11 mutations (A27S, A67V, Y145D, L212I, V213G, R408S, K417 N, T478K, E484K, F486V, L981F) appeared to be destabilising, while, seven mutations (T19R, G339D, N440K, S477 N, D614G, N764K, D950 N) appeared as stabilising by more than half of the prediction tools employed. Two mutations (T19I and Q498R) were predicted to be destabilising by two of the five tools, undetermined by one tool, and stabilising by the rest of the tools, hence, undetermined.
For the spike glycoprotein of the Delta variant, three mutations were destabilising out of the six single amino acid substitutions analysed. Two of these mutations (L452R and T478K) occupy the receptor-binding domain. In addition to these three destabilising mutations found in the Delta variant, a stabilising mutation (A222V) was identified outside the RBD of the Delta plus variant. In the case of the Mu variant, eight mutations were analysed and four of them were destabilising. The RBD of the Mu variant contained two destabilising mutations (R346K and E484K) and one stabilising mutation (N501Y).
The Omicron sub-variants increased in both destabilising and stabilising mutations. For subvariant BA.1, 15 mutations were destabilising out of the 28 mutations analysed. These include the stabilising mutation N501Y, previously reported in Mu, and the destabilising mutation G496S. Omicron BA.2 had, however, 14 destabilising mutations out of 26 mutations analysed. This variant did not include destabilising mutation G496S. 13 out of 27 analysed mutations in Omicron BA.4, were shown to be destabilising, whereas, 12 were shown to be stabilising. For the most recent Omicron subvariant BA.2.12.1, 13 out of 27 mutations analysed were shown to be destabilising and 13 to be stabilising. Considering the emerging sequence of the variants, mutations are accumulating rapidly within the spike protein (Fig. 3 ), particularly, among RBD residues.
Fig. 3.
The pattern of stabilising and destabilising mutation accumulation for the emerging SARS-CoV-2 variants. The variants are presented sequentially, in order of their emergence in the virus's recent evolutionary timeline.
3.4. Docking energy score analysis of the hACE2-RBD complexes
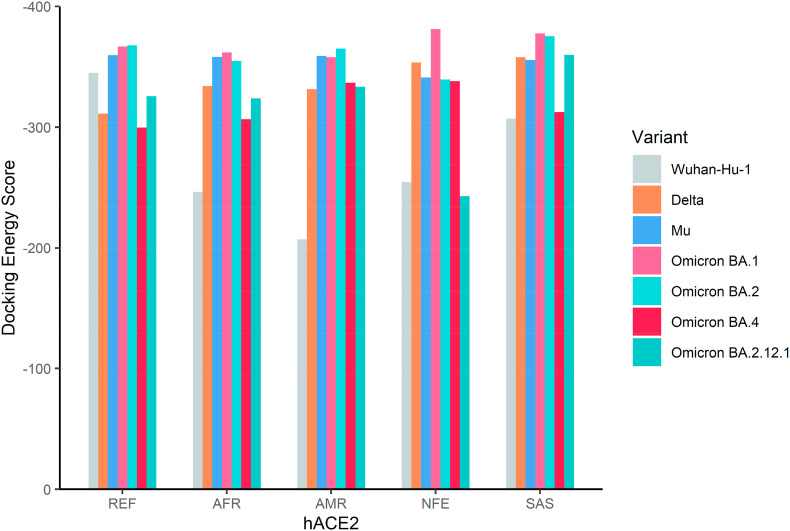
The docking energy scores of thirty-five complexes of spike RBD variants with the reference and missense hACE2 variants prevalent in four major populations are depicted in Fig. 4 . The numerical values are provided in Supplementary Table 2.
Fig. 4.
Docking energy scores of hACE2 reference and four singly mutated hACE2 variants (African, Latin American, Non-Finnish European, and South Asian) and the receptor-binding domains (RBD) of the spike glycoprotein from the Wuhan-Hu-1 (reference) and six emerging SARS-CoV-2 variants (Delta, Mu, Omicron BA.1, Omicron BA.2, Omicron BA.4, and Omicron BA.2.12.1).
Omicron BA.2 variant showed fairly the highest receptor-binding capacity (docking energy score = −367.74), surpassing the previous Omicron BA.1 variant (docking energy score = −366.84), in complex with the reference hACE2. The receptor-binding capacity of the Omicron BA.1 variant, however, remains high for the reference and all missense hACE2 variants we investigated. Especially, in the case of the NFE hACE2 containing the substitution D355 N, the Omicron BA.1 variant has the highest receptor-binding ability with the docking energy score of −381.24, compared to other missense hACE2 variants. However, the docking energy score was less negative for the Omicron BA.2 variant (−339.47) in complex with the NFE hACE2. For the AFR hACE2 containing the E37K substitution, the emerging variant spike RBDs exhibited a gradual increase in binding capacity, which slightly decreased in the case of the latest variant Omicron subvariant BA.2. Similar results were observed for the SAS hACE2 containing the P84T substitution, except for a more negative docking energy score in combination with the Delta RBD variant (−357.96). The receptor-binding ability of Omicron BA.1 and BA.2 was still very high with docking energy scores of −377.71 and −375.25, respectively. The reference and variant spike RBDs showed the most negative docking energy scores in the case of the AMR hACE2, containing the F40L substitution. This particular allele had a notably minor frequency (0.0001%). Nevertheless, the Omicron BA.2 manifested the highest receptor binding interaction with a docking energy score of −364.96 for AMR hACE2, compared with other populations. Among the most recently emerged Omicron sub-variants, Omicron BA.4 exhibits the least negative docking energy score −299.66 with the reference hACE2, compared with all variants analysed. However, the variant exhibits the most negative docking energy score (−338.16) with the NFE hACE2, compared with the other populations. Omicron BA.2.12.1, the closest relative to Omicron BA.2, produces a more negative docking energy score of −325.74. The variant shows the most negative docking energy score of −359.89 with SAS hACE2, much more towards the negative compared with Omicron BA.4 (docking energy score = −312.54). Interestingly, the recent Omicron sub-variant BA.4 exhibits a less negative docking energy score than the previous BA.1 and BA.2 overall.
3.5. Comparative analysis of the intermolecular interactions for different hACE2-RBD complexes
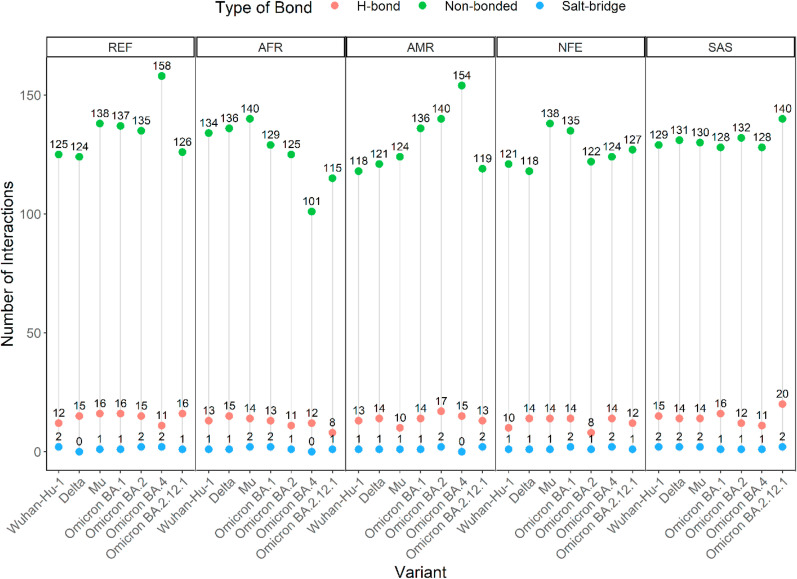
The summary of the interactions derived from the docked complexes of the hACE2 and RBD has been presented in Fig. 5 .
Fig. 5.
Comparison of the type and number of bonds formed between thirty-five different hACE2-RBD variant complexes. Types of bonds include salt-bridge (blue), H-bond (salmon), and non-bonded (green) interactions. Points of interaction vary with the SARS-CoV-2 variants among the AFR, AMR, and NFE groups of hACE2, whereas, consistent among SAS hACE2.
In the reference hACE2-RBD structural complex (6M0J), the following H-bonds were observed: Asp30-Lys417 [O⋯H–N], Glu35-Gln493 [O⋯H–N], Gln42-Gly446 [N–H⋯O], Lys353-Gly496 [N–H⋯O] and a salt bridge between Asp30 of hACE2 and Lys417 of RBD. The bond formation patterns of the thirty-five hACE2-RBD complexes are depicted in Fig. 6 and detailed in Supplementary Table 3.
Fig. 6.
Comparison of the salt bridges and H-bonds between the thirty-five hACE2 – RBD variant complexes, with an emphasis on the RBD residues mutated in at least one SARS-CoV-2 variant. Salt bridges and H-bonds are depicted as dashed and solid lines, respectively. RBD residues interacting with more than one hACE2 residue appear more than once.
The depiction compares the RBD residues which undergo mutations in at least one variant investigated and/or forms a salt bridge. The residues are Arg403, Lys417, Gly446, Glu484, Gln493, Gly496, Gln498, Asn501, Tyr505 of the spike protein RBD. The complex formed by the reference hACE2 and the spike RBD of the Wuhan-Hu-1 variant has been referred to as the ‘reference’ complex. The rest of the complexes are ‘variant’ complexes, in which either hACE2 or RBD is a variant.
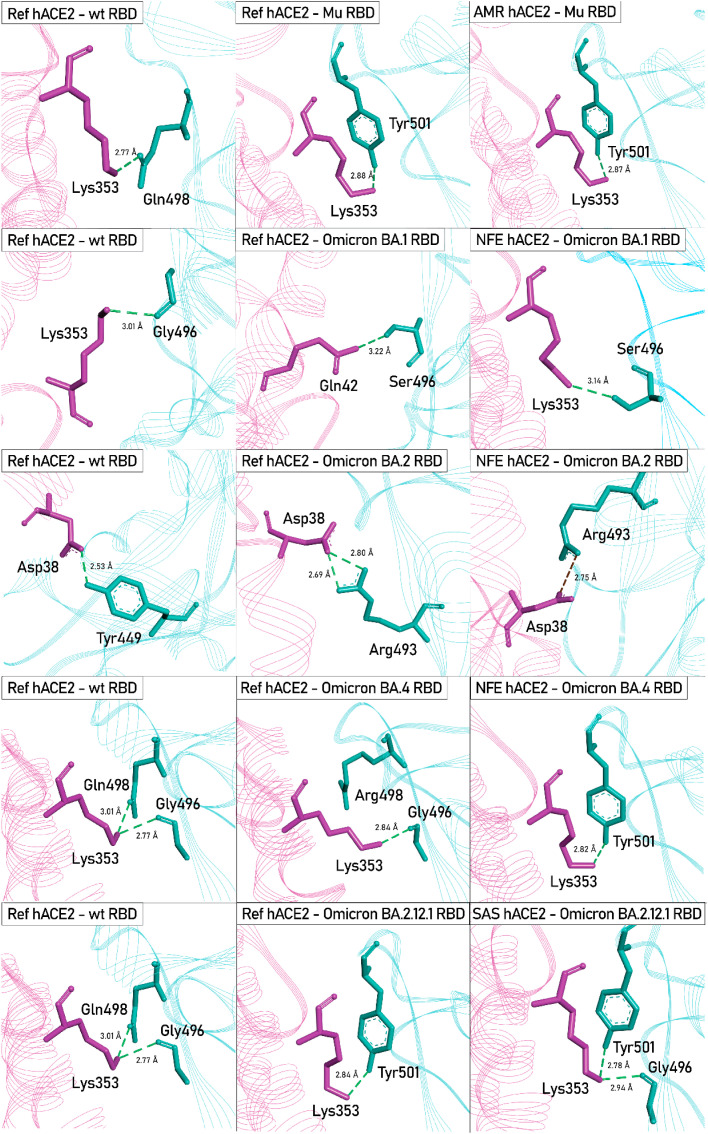
The notable H-bond and salt bridge interactions found within different variant hACE2-RBD complexes compared with the reference hACE2-RBD complex are depicted in Fig. 7 .
Fig. 7.
3D interaction diagrams of some unique interactions found in different hACE2 - RBD variant complexes, compared with the Ref hACE2 - Wuhan-Hu-1 RBD complex. hACE2 and RBD variants are depicted in pink and cyan, respectively. The Wuhan-Hu-1 strain of SARS-CoV-2 is denoted as wild-type (wt). H-Bonds are presented as green dashed lines and the salt bridge between Asp38-Arg493 in NFE hACE2-Omicron BA.1 RBD complex is presented in brick-red dashed line.
3.6. Prediction of free energy of protein-protein complexes and free energy decomposition of protein-protein interactions
The binding free energies predicted for the hACE2 – RBD variant complexes are summarized in Supplementary Table 4. The binding free energies of hACE2 variants in complex with Omicron BA.1, BA.2, BA.4, BA.2.12.1 and Delta are highlighted in Fig. 8 .
Fig. 8.
Comparison of binding free energy among Omicron BA.1, BA.2, BA.4, BA.2.12.1 and Delta RBD variant complexes with the reference and missense variants of hACE2. On the x-axis, VDW, ELE, GB, SA, and TOTAL denote the van der Waals Energy (kcal/mol), Electrostatic Energy (kcal/mol), Polar Solvation Energy (kcal/mol), Solvent Accessible Surface Area Energy (kcal/mol), Total Binding Energy (kcal/mol), respectively.
Omicron BA.1 sub-variant exhibited a more negative binding free energy in complex with AFR hACE2 (−79.11 kcal/mol) and NFE hACE2 (−77.53 kcal/mol), compared with reference (−74.51 kcal/mol). Whereas, Omicron BA.2 sub-variant depicted a significantly less negative binding free energy in complex with NFE hACE2 (−62.14 kcal/mol), compared with both SAS hACE2 (−74.36 kcal/mol) and the reference hACE2 (−76.95 kcal/mol). The Delta variant produced a significantly more negative binding free energy with SAS hACE2 (−74.03 kcal/mol), compared with the reference hACE2 (−68.77 kcal/mol). Omicron BA.4 manifests a much less negative binding energy in complex with both the reference (−66.78 kcal/mol) and NFE hACE2 (−66.72 kcal/mol), similar the pattern observed in case of docking energy score. In case of Omicron BA.2.12.1, highly negative binding free energy was observed in complex with both the reference (−78.55 kcal/mol) and NFE hACE2 (−76.7 kcal/mol).
The free energy contributions at the level of amino acid residues were calculated using free energy decomposition analyses. The differences in residual free energy contributions of wild type hACE2 residues and the mutated residues (E37K, F40L, D355 N, and P84T) caused by SNPs, highly prevalent in African (AFR), Latin American (AMR), Non-Finnish European (NFE), and South Asian (SAS) populations are depicted in Fig. 9 . The numerical values are provided in Supplementary Table 5.
Fig. 9.
The differences in residual free energy contributions (in kcal/mol) of wild type hACE2 residues and the mutated residues (A) E37K, (B) F40L, (C) D355 N, and (D) P84T, representing African (AFR), Latin American (AMR), Non-Finnish European (NFE), and South Asian (SAS) populations, respectively.
All mutated residues except F40L, were shown to shift the free energy towards a more positive value. Although it seems to decrease the binding affinity at a single residue level, the overall the binding free energy does appear to follow a similar trend.
3.7. RMSD analysis
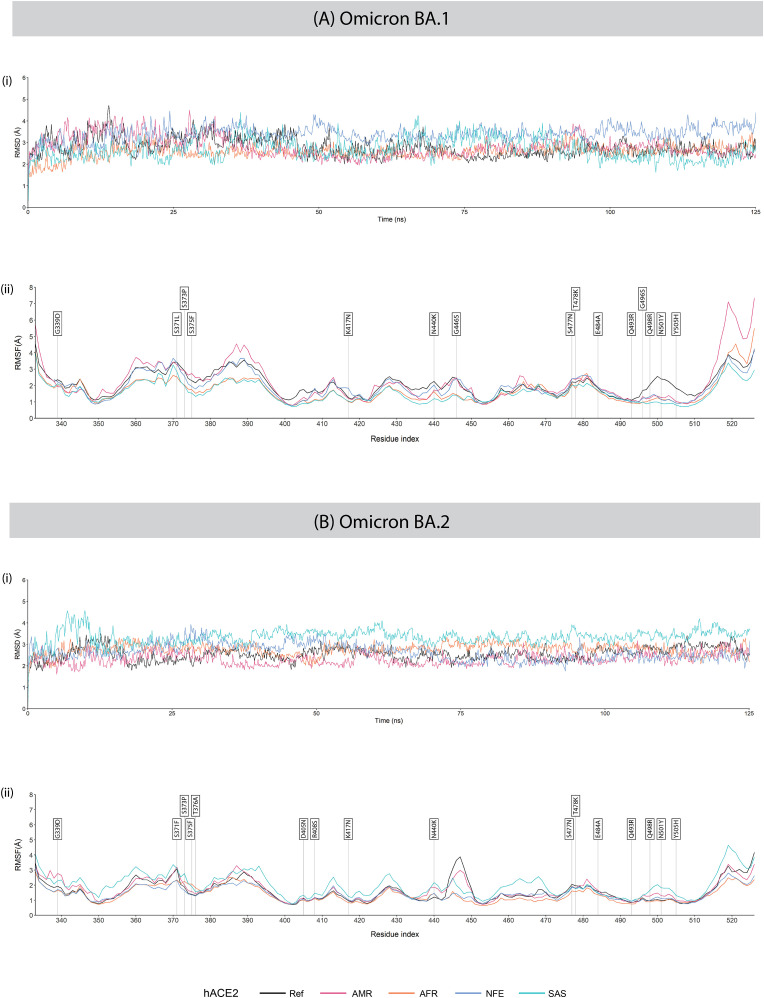
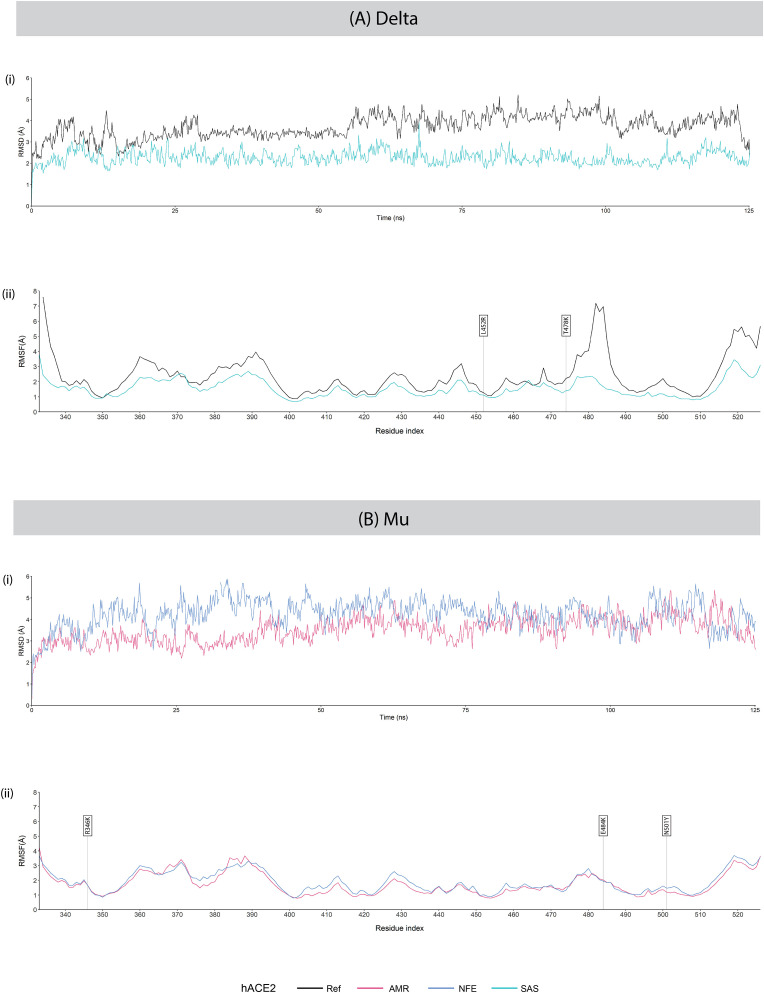
RMSD analysis of the backbones of RBD of Omicron BA.1 with different hACE2 variants revealed that Ref hACE2 – Omicron BA.1 RBD complex was initially unstable but later stabilised eventually with some momentary fluctuations during the simulation run time. RMSD values of the reference and Omicron sub-variant complexes of BA.1 and BA.2 during the 125 ns MD simulation are presented in Fig. 10 (A) (i), (B) (i); Delta and Mu variant complexes in Fig. 11 (A) (i), (B) (i) .
Fig. 10.
(i) RMSD and (ii) RMSF plots of hACE2 – RBD complexes of variants (A) Omicron BA.1 and (B) Omicron BA.2 during the 125 ns MD simulation. In the RMSF plots, RBD residues mutated in each SARS-CoV-2 variant are labelled with flags.
Fig. 11.
(i) RMSD and (ii) RMSF plots of hACE2 – RBD complexes of variants (A) Delta and (B) Mu during the 125 ns MD simulation. In the RMSF plots, RBD residues mutated ineach SARS-CoV-2 variant are labelled with flags.
There is less deviation on the MD trajectory of the AFR hACE2 – Omicron BA.1 RBD complex at the initial stage, indicating more stable binding that was maintained for the runtime. The SAS hACE2 – Omicron BA.1 RBD complex also showed stronger initial interaction as it had lower RMSD at the beginning compared to the Ref hACE2 – Omicron BA.1 RBD complex. For the hACE2 complexes with Omicron BA.2 RBD, an overall greater stability was observed in the Ref hACE2 – Omicron BA.2 RBD complex. The AFR hACE2 – Omicron BA.2 RBD and NFE hACE2 – Omicron BA.2 RBD complexes showed some fluctuations at the beginning. However, with time, the complexes stabilised and exhibited lower RMSD compared to the Ref hACE2 – Omicron BA.2 RBD complex. Interestingly, we observed opposite trends in RMSD in case of the NFE hACE2 and SAS hACE2 complexes with BA.1 and BA.2 variants. The NFE hACE2 – Omicron BA.2 RBD and SAS- hACE2 – Omicron BA.2 RBD are greatly stabilised compared to their complexes with Ref hACE2. The average RMSD values of the different hACE2 complexes with Omicron BA.1 and BA.2 RBD variants are given in Table 4 .
Table 4.
Average RMSD (Ǻ) of missense hACE2 variants in complexes with Omicron BA.1 and Omicron BA.2 RBD variants.
| Average RMSD (Ǻ) | ||
|---|---|---|
| hACE2 | Omicron BA.1 | Omicron BA.2 |
| Ref | 2.7918 | 2.5584 |
| AFR | 2.5755 | 2.7644 |
| AMR | 2.8106 | 2.3169 |
| NFE | 3.3771 | 2.6362 |
| SAS | 2.6804 | 3.3519 |
In case of SAS hACE2 – Delta RBD complex, very stable binding was observed compared with the Ref hACE2 – Delta RBD complex, although some minor fluctuations are observed momentarily. Mu RBD exhibited substantially higher RMSD values initially in complex with NFE hACE2, compared with that with AMR hACE2. The average RMSD values of the different hACE2 complexes with Delta and Mu RBD variants are given in Supplementary Table 6.
3.8. RMSF analysis
RMSF values of the RBD of the reference and Omicron sub-variantsBA.1 and BA.2 complexed with the reference hACE2 and different hACE2 variants obtained during the 125 ns MD simulation are presented in Fig. 10 (A) (ii), (B) (ii)); and RMSF values of the RBD of the Delta and Mu variants are presented in Fig. 11 (A) (ii), (B) (ii).
RMSF analysis of the protein backbone of the SARS-CoV-2 RBD variants revealed significant variations in RMSF values in the Omicron BA.1 RBD variants interacting with different hACE2 variants. The Asn360 was stable while interacting with AFR and SAS hACE2, however, exhibited fluctuation while interacting with AMR hACE2. The wild type Asn370 residue and the mutated Ser371Leu residue in the RBD were stabilised while interacting with AFR hACE2 but fluctuated higher in the rest of the cases. Leu387 and Asp389 showed high fluctuations while interacting with AMR hACE2 but were most stable in case of AFR and SAS hACE2. The mutated Gly446Ser residue exhibited greater stability while interacting with AFR and SAS hACE2 compared with the rest. The RBD region spanning residues 496–508 of Omicron BA.1 harbours four mutations and they have stabilised while interacting with all hACE2 variants compared with the reference hACE2.
In the Omicron BA.2 RBD variant complexes with different hACE2, greater fluctuations were observed at the RBD residues Asn360, Thr393, Gly413, Asp428, Asn460, Ile468, and Thr500 in interaction with the SAS hACE2 compared with the rest. The RBD residues of the AFR hACE2 – Omicron BA.2 RBD complex were most stabilised in terms of RMSF. Interestingly, we observed a high fluctuation in the Gly447 residue of the Omicron BA.2 RBD while interacting with reference hACE2. However, greatly reduced fluctuations were observed for interactions with SAS, NFE, and AFR hACE2. While interacting with AMR hACE2, the residue stabilised only moderately. On the other hand, the RBD residues of the Delta variant were observed to undergo major fluctuations in complex with reference hACE2, whereas, were substantially stabilised in complex with SAS hACE2.
The N-terminal and C-terminal residues of all complexes show higher RMSF value due to their positions at the ends of the protein molecule which fail to take part in any secondary structure formation, resulting in greater flexibility.
4. Discussion
Among the SARS-CoV-2 variants of concern, we especially investigated the Delta variant and Omicron sub-variants BA.1 and BA.2, as well as BA.4, BA.5 and BA.2.12.1, which emerged recently in 2022. A single amino acid substitution (A222V) distinguishes Delta plus variant from its predecessor Delta (B.1.617.2) which is located outside the RBD of the spike glycoprotein. The receptor-binding domains of the Delta and the Delta plus variants appear to be identical. Hence, the spike RBDs of both the Delta and the Delta plus variants are represented as the ‘Delta’ RBD variant in our results to avoid redundancy. Similarly, Omicron subvariants BA.4 and BA.5 share an identical spike protein, hence, the observations for ‘Omicron BA.4’ apply to both BA.4 and BA.5 variants. A variant of interest Mu was also included in this report as it showed limited spread within specific regions and despite acquiring noteworthy mutations, fewer studies have been conducted on the transmission pattern of this variant.
As hACE2 serves as the primary receptor for the entry of SARS-CoV-2, it is the first probable ‘genetic gateway’ that might determine disease progression [49]. The genetic polymorphisms of hACE2 are not evenly distributed across populations. Rather, the frequencies of these polymorphisms considerably differ among populations. Hence, a population may preferentially harbour one polymorphism at a much higher frequency, compared with the rest. Moreover, both experimental and in silico studies demonstrated that the binding interactions of SARS-CoV-2 vary in presence of hACE2 polymorphisms [50]. Furthermore, previous experiments have shown that the SARS-CoV-2 variants also interact differently with hACE2 [51]. The spread of the viral variants, thus, is likely to differ extensively among the host populations harbouring distinct hACE2 polymorphisms. However, none of the currently available studies have considered the RBD mutations and the hACE2 polymorphisms simultaneously. We combined both the recently emerged SARS-CoV-2 variants and the hACE2 single missense variants prevalent in major populations in a combinational approach to investigate the differences in the receptor-binding interactions between different RBD – hACE2 complexes. Our results indicate notable differences in receptor-binding interactions of the spike RBD of different SARS-CoV-2 variants within a population represented by a hACE2 single missense variant. The receptor-binding affinity of a particular SARS-CoV-2 variant also varies across populations carrying distinct single missense variants of hACE2. The findings may provide useful insights into the differential transmission rate of the emerging SARS-CoV-2 variants over individual populations.
This study focuses particularly on the missense mutations of hACE2 protein, present within its binding interface with the RBD of the spike glycoprotein of SARS-CoV-2. Substitutions of these residues are more likely to impact the binding affinity of the hACE2-RBD complex. Accordingly, instead of the whole spike glycoprotein, our focus remained on its RBD, which directly interacts with hACE2. Furthermore, the hACE2 missense variants with the highest frequencies within a population were preferred as they represent a larger target population, broadening the scope of this study. The hACE2 missense variants assigned to the African (AFR), Latin American (AMR), non-Finnish European (NFE), and the South Asian (SAS) populations represent 5.12%, 0.001%, 1.75%, and 3.58% of the respective populations.
Protein stability and protein-protein interactions are greatly affected by the accumulation of both stabilising and destabilising mutations. It is hence worthwhile to study the mutational pattern of the SARS-CoV-2 variants. This is particularly necessary for the evaluation of the efficacy of protective treatments against the disease, as well as, for predicting the fate of the ongoing pandemic. Stabilising mutations in the spike protein might increase its longevity under unfavourable conditions, whereas, destabilising mutations could significantly reduce the fitness of the protein. However, destabilising mutations tend to enhance protein flexibility, which often considerably affects the infectivity of a viral strain through an increase in receptor-binding affinity [52]. On the other hand, stabilising mutations render structural rigidity which may confer increased stability of the protein-protein complex [53]. In this study, the lineage-defining mutations of SARS-CoV-2 variants: Delta, Delta plus, Mu, Omicron sub-variants BA.1, BA.2, BA.4 and BA.2.12.1 were analysed. The destabilising mutation K417 N in the RBD region of the Omicron variants was reported to increase receptor-binding affinity [54]. This mutation along with five other notable mutations (N440K, G446S, E484A, Q493R and N501Y) have shown to markedly reduce the activity of monoclonal antibodies: bamlanivimab, etesevimab, casirivimab, and imdevimab [[55], [56], [57]] and moderate reduction in the activity in tixagevimab and sotrovimab [57]. Another destabilising mutation, L452R, present in the Delta variant was found to resist the activities of several monoclonal antibodies in therapeutic use [58]. The destabilising mutation E484K, present in the RBD of the spike glycoprotein of the Mu variant, exhibited an alteration in the complementarity of binding plasma polyclonal neutralizing antibodies [59]. The stabilising mutation N501Y, shared by the Mu, Omicron sub-variants BA.1, BA.2, BA.4 and BA.2.12.1 was found to increase the hACE2-binding affinity of the RBD [60]. This may also present a possible route of immune escape [61].
We sought to explore the effects of the SARS-CoV-2 variant spike glycoprotein RBD mutations on the binding with hACE2 containing the most prevalent single amino acid substitutions in the RBD binding regions for the African, Latin American, non-Finnish European and South Asian populations using molecular docking. A more negative docking energy score should indicate higher stability between receptor and ligand, hence, lower binding energy, increased affinity, and stronger interaction [62]. Furthermore, MM/GBSA calculations are theoretically more rigorous than scoring functions as commonly used in many of the docking algorithms, and can be used to validate the docking energy scores generated in this manner [63].
Previous studies have confirmed that Lys417, Tyr449, Phe456, Phe486, Gln493, Gly496, Gln498, Asn501 and Tyr505 are some key residues essential in the formation of a stronger complex with hACE2 [64,65]. Among which, a stable salt bridge between Lys417 of RBD and Asp30 of hACE2, along with three sustained H-bonds between RBD residues: Tyr449, Gln493 and Gln498 and hACE2 residues: Asp38, Glu35 and Lys353, distinguishes SARS-CoV-2 from SARS-CoV [66]. Hydrogen bonding plays a key role in hACE2-RBD binding among other intermolecular interactions. Mutations in the primary interface of hACE2-RBD induce stronger binding by increasing H-bonds as well as expanding buried solvent accessible surface [67]. Hence, we compared two types of interactions in terms of their number: salt-bridge, and hydrogen bonds between the reference and variant complexes. Increase and decrease in the quantity of these bonds fairly correlate with the strength, and to some extent, stability of the reference and variant complexes under investigation.
Molecular Dynamics simulation was performed to further evaluate the stability of interactions between the RBD and hACE2 variants over time using Root Mean Square Deviation (RMSD) values of the hACE2-RBD complexes. The stability of a protein-protein complex decreases as the RMSD value increases [68]. MD simulations also revealed differences in fluctuation among the RBD variant residues while in complex with different hACE2 variants through Root Mean Square Fluctuation (RMSF) values. The RMSF values represent the degree of fluctuation for each of the amino acid residues of the molecule of interest. High RMSF values may be correlated with the flexibility of the protein which may alter a protein-protein binding [69].
The N501Y substitution, first emerged in the Mu variant, had been reported to increase the affinity of the hACE2-RBD complex by forming new protein-protein interactions that modify internal structural dynamics [60,70]. Preferential selection of the mutation N501Y in more transmissible variants were also observed in an in vitro evolution study of the spike glycoprotein RBD [71] Our result is consistent with this finding as the binding free energy drastically moves towards the negative in case of the Mu variant in complex with the reference hACE2 (−80.43 kcal/mol) compared with that of the Delta variant (−68.77 kcal/mol) which lacks the given mutation, indicating higher binding affinity of the Mu variant for the reference hACE2. In comparison with the NFE hACE2, the Mu RBD manifests lower average RMSD (3.5072 Å) in complex with AMR-hACE2. Furthermore, key RBD residues of this variant, except Phe486, show lower RMSF while in complex with AMR hACE2 and much higher RMSF while in complex with NFE hACE2, when compared with reference hACE2. This finding aligns with the failure of the variant of South American origin to propagate into other populations, especially the Non-Finnish Europeans, as it revealed a less stable interaction with the NFE hACE2, compared with the AMR hACE2 [72].
The Lys353-Tyr501 [N–H⋯O] bond was also present in the NFE hACE2 – Omicron BA.1 RBD complex which exhibited a much higher affinity (−77.53 kcal/mol) compared with the variant complex with reference hACE2 (−74.51 kcal/mol). However, a substantially high RMSD (3.3771 Å) was observed in the NFE hACE2 - Omicron BA.1 RBD complex compared with the reference hACE2 (2.7918 Å), which indicates the binding was not maintained properly during the simulation runtime. The Gly496 residue is mutated in the RBD of the Omicron BA.1, but not in the Omicron BA.2 variant. The G496S substitution in the Omicron BA.1 variant has been reported to decrease the binding affinity of the complex [60]. The mutated Ser496 Omicron BA.1 RBD variant forms H-bonds with Asp38 and Gln42 of the reference hACE2 with distances 2.62 Å and 3.22 Å. Whereas, unmutated Gly496 forms a single H-bond with Lys353 of the reference hACE2 with a distance of 2.77 Å. Nevertheless, the AFR hACE2 – Omicron BA.1 RBD complex exhibited the most negative binding free energy (−79.11 kcal/mol). The complex also reported the lowest average RMSD (2.5755 Å). This may explain the origin and initial spread of the strain in the African population as the mutations accumulated in this strain appear to yield an advantage in maintaining stable binding to AFR hACE2, as demonstrated by the MD simulation data.
Our results suggest a higher affinity of the Omicron BA.2 variant, compared with its predecessor Omicron BA.1 variant. Furthermore, during the course of 125 ns MD simulation, Omicron BA.2 showed a lower average RMSD (2.5584 Å) in complex with the reference hACE2 compared with that of Omicron BA.1 (2.7918 Å). Hence, transmissibility is likely to increase for the Omicron BA.2 variant, which has caused a surge in the rate of infection. Since the WHO recognised Omicron as a Variant of Concern (VoC) on November 24, 2021, there have been 487,926 entries of complete genome sequences of Omicron BA.1 variant in the GISAID database [73] as of June 29, 2022. In contrast, there are 1,146,042 entries of complete genome of the Omicron BA.2 variant. This may represent the greater spread of Omicron BA.2 as the number of viral genome sequence deposition is representative of the circulating viral strain worldwide.
The higher RMSF at residues 496–508 in both Omicron BA.1 and BA.2 complexes may relate to previous reports of reduced antibody response [74]. Although a lower RMSF indicates a more rigid behaviour of the residue in concern, it does not guarantee a more stable protein-protein interactions as protein-protein interactions largely rely on the conformational flexibility [75].
The exceedingly low affinity of the reference hACE2 – Delta RBD variant complex, compared with a much higher affinity of the variant complex with the SAS hACE2 might be explained by comparing their patterns of interactions. The Delta RBD forms much fewer H-bonds with the reference hACE2, involving only four (Gln493, Gln498, Asn501, Phe486) of the key interacting residues mentioned above. Each of the residues exhibit high RMSF values in complex with the reference hACE2. Only Gln493 forms H-bonds with Lys31 and Gln35 of hACE2. Whereas, the Delta RBD residues manifest much lower RMSF values while interacting with SAS hACE2, of which Gln493 and Gln498 form H-bonds and Lys417 forms a salt bridge with hACE2 residues Lys31 and His34, Asp38, and Asp30, respectively. During the course of 125 ns MD simulation, the Delta variant maintained a much stable binding in complex with the SAS hACE2 compared with that of reference hACE2. This outcome is also notably consistent with the devastating spread of the variant in South-East Asia, as a large population of around 49.7 million in India alone would represent the SAS hACE2 harbouring the P84T substitution [76].
Although stronger binding to the receptor yields a higher probability of receptor endocytosis of the virus and subsequently successful replication, it is not the only determinant of the rate of transmissibility of the virus and the severity of the disease. A furin-like protease, transmembrane serine protease 2 (TMPRSS2), plays an equal role in the virus's ability to infect a permissive cell. The protease cleaves the viral spike protein at the S1/S2 junction, enabling its fusogenic activity and allowing subsequent fusion with the host cell [77]. An earlier report has suggested the Delta variant outcompetes the Omicron BA.1 variant in infecting TMPRSS2 overexpressing VeroE6 cells in culture [78]. In our result, the Omicron BA.1 and BA.2 RBD variant exhibited much higher receptor affinity, compared with the Delta RBD variant. However, the Delta variant may overcome the lower receptor-binding affinity with the heightened ability of infection using TMPRSS2 activity. Nevertheless, Omicron variants have been shown to employ a TMPRSS2-independent pathway for infection [79]. Hence, the higher receptor-binding capacity of the Omicron variants may still imply higher transmissibility of these emerging variants.
5. Conclusions
Our study suggests variation in the transmissibility of the emerging SARS-CoV-2 variants across populations harbouring specific hACE2 missense variants. Although viral susceptibility at the population level depends on many factors related to individual hosts, this study may provide insights into transmissibility of emerging SARS-CoV-2 variants among different populations. Furthermore, our results also reinforce the available evidence supporting that the Omicron subvariant BA.2 shows the highest transmissibility as well as the increasing incidences of Omicron BA.2.12.1, among the recently emerged SARS-CoV-2 variants. The severity and outcome of disease, however, can only be predicted upon considering host immune responses and related factors. Nevertheless, predictions on variant transmissibility with accurate interpretations may encourage effective implementation of preventive measures to contain community transmission and the rate of infection caused by the newly emerging variants.
Author contributions
AHMNN conceived the idea and supervised the research work. AT, RA and PB retrieved the sequences. AT and RA performed modelling of the proteins. MA, AAS and SC performed the mutational analyses. AT, RA and MA performed the docking analyses. AT performed the MMGBSA analyses. RA performed the MD simulations and subsequent data analysis. AHMNN, TY, AT and RA prepared the first draft. AT, RA, PB, MA and TY prepared the revised manuscript. AHMNN, AKMMH and TY then curated the revised draft critically. All authors read and approved the revised manuscript.
Declaration of competing interest
The authors declare that there is no conflict of interest.
Acknowledgement
Part of this research work was supported by the establishment developed from the special grants (Code#1280101-120008431-3631107) provided by the Information and Communication Technology Division under the Ministry of Posts, Telecommunications & Information Technology, Government of the People's Republic of Bangladesh (2019–2020), Centennial Research Grant of Dhaka University (2021) and United Graduate School of Agricultural Science, Gifu University, Japan.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.105903.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Abebe E.C., Dejenie T.A., Shiferaw M.Y., Malik T. The newly emerged COVID-19 disease: a systemic review. Virol. J. Jul. 2020;17(1) doi: 10.1186/S12985-020-01363-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johansson M.A., et al. SARS-CoV-2 transmission from people without COVID-19 symptoms. JAMA Netw. Open. Jan. 2021;4(1) doi: 10.1001/JAMANETWORKOPEN.2020.35057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham R.L., Sparks J.S., Eckerle L.D., Sims A.C., Denison M.R. SARS coronavirus replicase proteins in pathogenesis. Virus Res. 2008;133(1):88–100. doi: 10.1016/J.VIRUSRES.2007.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourgonje A.R., et al. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19) J. Pathol. Jul. 2020;251(3):228–248. doi: 10.1002/PATH.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/J.CELL.2020.02.058. e6, Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.al Saba A., Adiba M., Chakraborty S., Nabi A.N. Prediction of putative potential siRNAs for inhibiting SARS-CoV-2 strains, including variants of concern and interest. Future Microbiol. Apr. 2022;17(6):449–463. doi: 10.2217/FMB-2021-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang S., et al. The miRNA: a small but powerful RNA for COVID-19. Briefings Bioinf. 2021;22(2):1137–1149, Mar. doi: 10.1093/BIB/BBAB062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang S., et al. RNA-RNA interactions between SARS-CoV-2 and host benefit viral development and evolution during COVID-19 infection. Briefings Bioinf. Jan. 2022;23(1) doi: 10.1093/BIB/BBAB397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saeed U., Uppal S.R., Piracha Z.Z., Uppal R. SARS-CoV-2 spike antibody levels trend among sinopharm vaccinated people. Iran. J. Public Health. Jul. 2021;50(7):1486. doi: 10.18502/IJPH.V50I7.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahase E. Covid-19: pfizer and BioNTech submit vaccine for US authorisation. BMJ. Nov. 2020;371:m4552. doi: 10.1136/bmj.m4552. [DOI] [PubMed] [Google Scholar]
- 11.Mahase E. Covid-19: moderna vaccine is nearly 95% effective, trial involving high risk and elderly people shows. BMJ. Nov. 2020;371:m4471. doi: 10.1136/bmj.m4471. [DOI] [Google Scholar]
- 12.Knoll M.D., Wonodi C. Oxford–AstraZeneca COVID-19 vaccine efficacy. Lancet. Jan. 2021;397(10269):72–74. doi: 10.1016/S0140-6736(20)32623-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.al Saba A., Adiba M., Saha P., Hosen M.I., Chakraborty S., Nabi A.H.M.N. An in-depth in silico and immunoinformatics approach for designing a potential multi-epitope construct for the effective development of vaccine to combat against SARS-CoV-2 encompassing variants of concern and interest. Comput. Biol. Med. 2021;136(Sep) doi: 10.1016/j.compbiomed.2021.104703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao K., et al. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. Dec. 2021;22(12):757–773. doi: 10.1038/S41576-021-00408-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tegally H., et al. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat. Med. Mar. 2021;27(3):440–446. doi: 10.1038/S41591-021-01255-3. [DOI] [PubMed] [Google Scholar]
- 16.Lippi G., Mattiuzzi C., Henry B.M. Updated picture of SARS-CoV-2 variants and mutations. Diagnosis. Feb. 2022;9(1):11–17. doi: 10.1515/DX-2021-0149. [DOI] [PubMed] [Google Scholar]
- 17.Alabbas A.B., Alamri M.A. Analyzing the effect of mutations in SARS-CoV2 papain-like protease from Saudi isolates on protein structure and drug-protein binding: molecular modelling and dynamics studies. Saudi J. Biol. Sci. Jan. 2022;29(1):526–533. doi: 10.1016/J.SJBS.2021.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou W., Xu C., Wang P., Anashkina A.A., Jiang Q. Impact of mutations in SARS-COV-2 spike on viral infectivity and antigenicity. Briefings Bioinf. Jan. 2022;23(1) doi: 10.1093/BIB/BBAB375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karczewski K.J., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. May 2020;581(7809):434–443. doi: 10.1038/s41586-020-2308-7. 2020 581:7809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali F., Elserafy M., Alkordi M.H., Amin M. ACE2 coding variants in different populations and their potential impact on SARS-CoV-2 binding affinity. Biochemistry and Biophysics Reports. Dec. 2020;24:100798. doi: 10.1016/J.BBREP.2020.100798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkatakrishnan A.J., Flock T., Prado D.E., Oates M.E., Gough J., Madan Babu M. Structured and disordered facets of the GPCR fold. Curr. Opin. Struct. Biol. Aug. 2014;27(1):129–137. doi: 10.1016/J.SBI.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Hildebrand P.W., Rose A.S., Tiemann J.K.S. Bringing molecular dynamics simulation data into view. Trends Biochem. Sci. Jul. 2019;44(11):902–913. doi: 10.1016/J.TIBS.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Suryamohan K., et al. Human ACE2 receptor polymorphisms and altered susceptibility to SARS-CoV-2. Communications Biology. Apr. 2021;4(1):1–11. doi: 10.1038/s42003-021-02030-3. 2021 4:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou Y., et al. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. Jul. 2020;18(1):1–8. doi: 10.1186/s12916-020-01673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pouladi N., Abdolahi S. Investigating the ACE2 polymorphisms in COVID-19 susceptibility: an in silico analysis. Molecular Genetics & Genomic Medicine. Jun. 2021;9(6):e1672. doi: 10.1002/MGG3.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohebbi A., et al. Susceptibility of the Iranian population to severe acute respiratory syndrome coronavirus 2 infection based on variants of angiotensin i converting enzyme 2. Future Virol. Aug. 2020;15(8):507–514. doi: 10.2217/FVL-2020-0160. [DOI] [Google Scholar]
- 27.Han P., et al. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell. Feb. 2022;185(4):630–640. doi: 10.1016/J.CELL.2022.01.001. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lupala C.S., Ye Y., Chen H., Su X.D., Liu H. Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem. Biophys. Res. Commun. Jan. 2022;590:34–41. doi: 10.1016/J.BBRC.2021.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barton M.I., Macgowan S., Kutuzov M., Dushek O., Barton G.J., Anton Van Der Merwe P. Effects of common mutations in the sars-cov-2 spike rbd and its ligand the human ace2 receptor on binding affinity and kinetics. Elife. 2021;10(Aug) doi: 10.7554/ELIFE.70658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laimer J., Hiebl-Flach J., Lengauer D., Lackner P. MAESTROweb: a web server for structure-based protein stability prediction. Bioinformatics. 2016;32(9):1414–1416, Mar. doi: 10.1093/BIOINFORMATICS/BTV769. [DOI] [PubMed] [Google Scholar]
- 31.Savojardo C., Fariselli P., Martelli P.L., Casadio R. INPS-MD: a web server to predict stability of protein variants from sequence and structure. Bioinformatics. Mar. 2016;32(16):2542–2544. doi: 10.1093/BIOINFORMATICS/BTW192. [DOI] [PubMed] [Google Scholar]
- 32.Parthiban V., Gromiha M.M., Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. Mar. 2006;34 doi: 10.1093/NAR/GKL190. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y., Lu H., Zhang N., Zhu Z., Wang S., Li M. PremPS: predicting the impact of missense mutations on protein stability. PLoS Comput. Biol. Mar. 2020;16(12) doi: 10.1371/JOURNAL.PCBI.1008543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worth C.L., Preissner R., Blundell T.L. SDM—a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. Mar. 2011;39(2):W215–W222. doi: 10.1093/NAR/GKR363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marabotti A., del Prete E., Scafuri B., Facchiano A. Performance of Web tools for predicting changes in protein stability caused by mutations. BMC Bioinf. Jul. 2021;22(7):1–19. doi: 10.1186/s12859-021-04238-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das L., Shekhar S., Chandrani P., Varma A.K. In silico structural analysis of secretory clusterin to assess pathogenicity of mutations identified in the evolutionarily conserved regions. 2021. 10.1080/07391102.2021.2007791. [DOI] [PubMed]
- 37.Waterhouse A., et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. Mar. 2018;46(W1):W296–W303. doi: 10.1093/NAR/GKY427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ittisoponpisan S., Islam S.A., Khanna T., Alhuzimi E., David A., Sternberg M.J.E. Can predicted protein 3D structures provide reliable insights into whether missense variants are disease associated? J. Mol. Biol. Mar. 2019;431(11):2197–2212. doi: 10.1016/J.JMB.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colovos C., Yeates T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2(9):1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. Mar. 1993;26(2):283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 41.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. Jul. 2007;35(2) doi: 10.1093/NAR/GKM290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan Y., Zhang D., Zhou P., Li B., Huang S.Y. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. Jul. 2017;45(W1):W365. doi: 10.1093/NAR/GKX407. –W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laskowski R.A., Jabłońska J., Pravda L., Vařeková R.S., Thornton J.M. PDBsum: structural summaries of PDB entries. Protein Sci. Mar. 2018;27(1):129–134. doi: 10.1002/PRO.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weng G., et al. HawkDock: a web server to predict and analyze the protein-protein complex based on computational docking and MM/GBSA. Nucleic Acids Res. Jul. 2019;47:W322–W330. doi: 10.1093/NAR/GKZ397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bowers K.J., et al. Feb. 2007. Scalable algorithms for molecular dynamics simulations on commodity clusters. 43–43. [DOI] [Google Scholar]
- 46.Asseri A.H., et al. Toward the identification of natural antiviral drug candidates against merkel cell polyomavirus: computational drug design approaches. Pharmaceuticals. 2022;15(5) doi: 10.3390/ph15050501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benkert P., Biasini M., Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. Feb. 2011;27(3):343–350. doi: 10.1093/BIOINFORMATICS/BTQ662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. Apr. 1993;26(2):283–291. doi: 10.1107/S0021889892009944/FULL. [DOI] [Google Scholar]
- 49.Bao L., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. Mar. 2020;583(7818):830–833. doi: 10.1038/S41586-020-2312-Y. [DOI] [PubMed] [Google Scholar]
- 50.Möhlendick B., et al. ACE2 polymorphism and susceptibility for SARS-CoV-2 infection and severity of COVID-19. Pharmacogenetics Genom. 2021;31(8):165–171. doi: 10.1097/FPC.0000000000000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Celik I., et al. Interactions of the receptor binding domain of SARS-CoV-2 variants with hACE2: insights from molecular docking analysis and molecular dynamic simulation. Biology. Mar. 2021;10(9) doi: 10.3390/BIOLOGY10090880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rubayet Ul Alam S.M., et al. Dominant clade-featured SARS-CoV-2 Co-occurring mutations reveals plausible epistasis: an in silico based hypothetical model. medRxiv. Jul. 2021:2021. doi: 10.1101/2021.02.21.21252137. 02.21.21252137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ozono S., et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun. Feb. 2021;12(1):1–9. doi: 10.1038/s41467-021-21118-2. 2021 12:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shorthouse D., Hall B.A. SARS-CoV-2 variants are selecting for spike protein mutations that increase protein stability. J. Chem. Inf. Model. 2021 doi: 10.1021/ACS.JCIM.1C00990. [DOI] [PubMed] [Google Scholar]
- 55.Wilhelm A., et al. Reduced neutralization of SARS-CoV-2 omicron variant by vaccine sera and monoclonal antibodies. medRxiv. Mar. 2021:2021. doi: 10.1101/2021.12.07.21267432. 12.07.21267432. [DOI] [Google Scholar]
- 56.Falcone M., et al. Efficacy of bamlanivimab/etesevimab and casirivimab/imdevimab in preventing progression to severe COVID-19 and role of variants of concern. Infect. Dis. Ther. Dec. 2021;10(4):2479–2488. doi: 10.1007/s40121-021-00525-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quiros-Roldan E., et al. Monoclonal antibodies against sars-cov-2: current scenario and future perspectives. Pharmaceuticals. Dec. 01, 2021;14(12) doi: 10.3390/ph14121272. MDPI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baral P., et al. Mutation-induced changes in the receptor-binding interface of the SARS-CoV-2 Delta variant B.1.617.2 and implications for immune evasion. Biochem. Biophys. Res. Commun. Oct. 2021;574:14–19. doi: 10.1016/J.BBRC.2021.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jangra S., et al. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe. Jul. 2021;2(7):e283–e284. doi: 10.1016/S2666-5247(21)00068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu L., et al. SARS-CoV-2 Omicron RBD shows weaker binding affinity than the currently dominant Delta variant to human ACE2. Signal Transduct. Targeted Ther. Mar. 2022;7(1) doi: 10.1038/S41392-021-00863-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.S. Chakraborty, “E484K and N501Y SARS-CoV 2 spike mutants Increase ACE2 recognition but reduce affinity for neutralizing antibody,” Int. Immunopharm., vol. 102, Jan. 2022, doi: 10.1016/J.INTIMP.2021.108424. [DOI] [PMC free article] [PubMed]
- 62.Korb O., Stützle T., Exner T.E. Empirical scoring functions for advanced Protein-Ligand docking with PLANTS. J. Chem. Inf. Model. 2009;49(1):84–96. doi: 10.1021/CI800298Z. [DOI] [PubMed] [Google Scholar]
- 63.Genheden S., Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expet Opin. Drug Discov. May 2015;10(5):449–461. doi: 10.1517/17460441.2015.1032936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jawad B., Adhikari P., Podgornik R., Ching W.Y. Key interacting residues between RBD of SARS-CoV-2 and ACE2 receptor: combination of molecular dynamics simulation and density functional calculation. J. Chem. Inf. Model. 2021;61(9):4425–4441, Mar. doi: 10.1021/ACS.JCIM.1C00560. [DOI] [PubMed] [Google Scholar]
- 65.Andújar S.A., Gutiérrez L.J., Enriz R.D., Baldoni H.A. Vol. 47. 2021. pp. 1443–1454. (Structure, interface stability and hot-spots identification for RBD(SARS-CoV-2):hACE2 complex formation). 1979229. 17, 10.1080/08927022.2021.1979229. [DOI] [Google Scholar]
- 66.Ali A., Vijayan R. Dynamics of the ACE2–SARS-CoV-2/SARS-CoV spike protein interface reveal unique mechanisms. Sci. Rep. Aug. 2020;10(1):1–12. doi: 10.1038/s41598-020-71188-3. 2020 10:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lupala C.S., Ye Y., Chen H., Su X.D., Liu H. Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem. Biophys. Res. Commun. Jan. 2022;590:34–41. doi: 10.1016/J.BBRC.2021.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rehm S., Trodler P., Pleiss J. Solvent-induced lid opening in lipases: a molecular dynamics study. Protein Sci. 2010;19(11):2122–2130, Nov. doi: 10.1002/PRO.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li M., Bao F., Zhang Y., Song W., Chen C., Zhao J. Role of elemental carbon in the photochemical aging of soot. Proc. Natl. Acad. Sci. U. S. A. 2018;115(30):7717–7722, Jul. doi: 10.1073/PNAS.1804481115/SUPPL_FILE/PNAS.1804481115. SAPP.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khan A., et al. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J. Cell. Physiol. Oct. 2021;236(10):7045–7057. doi: 10.1002/JCP.30367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zahradník J., et al. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nature Microbiol. Aug. 2021;6(9):1188–1198. doi: 10.1038/s41564-021-00954-4. 2021 6:9. [DOI] [PubMed] [Google Scholar]
- 72.Halfmann P.J., et al. Characterization of the SARS-CoV-2 B.1.621 (Mu) variant. Sci. Transl. Med. May 2022 doi: 10.1126/SCITRANSLMED.ABM4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data - from vision to reality. Euro Surveill. : bulletin Europeen sur les maladies transmissibles = Eur. commun. dis. bullet. Mar. 2017;22(13) doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bhattacharya M., Sharma A.R., Dhama K., Agoramoorthy G., Chakraborty C. Omicron variant (B.1.1.529) of SARS-CoV-2: understanding mutations in the genome, S-glycoprotein, and antibody-binding regions. Geroscience. Apr. 2022;44(2):619–637. doi: 10.1007/S11357-022-00532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fayos R., Melacini G., Newlon M.G., Burns L., Scott J.D., Jennings P.A. Induction of flexibility through protein-protein interactions. J. Biol. Chem. May 2003;278(20):18581–18587. doi: 10.1074/JBC.M300866200. [DOI] [PubMed] [Google Scholar]
- 76.Yek C., et al. The pandemic experience in southeast Asia: interface between SARS-CoV-2, malaria, and dengue. Front. Tropical Dis. Nov. 2021;2 doi: 10.3389/FITD.2021.788590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bestle D., et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance. 2020;3(9) doi: 10.26508/LSA.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao H., et al. SARS-CoV-2 Omicron variant shows less efficient replication and fusion activity when compared with Delta variant in TMPRSS2-expressed cells. Emerg. Microb. Infect. Mar. 2022;11(1):277. doi: 10.1080/22221751.2021.2023329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.B. J. Willett et al., “The Hyper-Transmissible SARS-CoV-2 Omicron Variant Exhibits Significant Antigenic Change, Vaccine Escape and a Switch in Cell Entry Mechanism”, doi: 10.1101/2022.01.03.21268111.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.