Abstract
Genome analyses predict that the cofactor cobalamin (vitamin B12, called B12 herein) is produced by only one-third of all prokaryotes but almost all encode at least one B12-dependent enzyme, in most cases methionine synthase. This implies that the majority of prokaryotes relies on exogenous B12 supply and interacts with producers. B12 consists of a corrin ring centred around a cobalt ion and the lower ligand 5’6-dimethylbenzimidazole (DMB). It has never been tested whether availability of this pivotal cofactor, DMB or its intermediate α-ribazole affect growth and composition of prokaryotic microbial communities. Here we show that in the subtropical, equatorial and polar frontal Pacific Ocean supply of B12 and α-ribazole enhances heterotrophic prokaryotic production and alters the composition of prokaryotic and heterotrophic protist communities. In the polar frontal Pacific, the SAR11 clade and Oceanospirillales increased their relative abundances upon B12 supply. In the subtropical Pacific, Oceanospirillales increased their relative abundance upon B12 supply as well but also downregulated the transcription of the btuB gene, encoding the outer membrane permease for B12. Surprisingly, Prochlorococcus, known to produce pseudo-B12 and not B12, exhibited significant upregulation of genes encoding key proteins of photosystem I + II, carbon fixation and nitrate reduction upon B12 supply in the subtropical Pacific. These findings show that availability of B12 and α-ribazole affect growth and composition of prokaryotic and protist communities in oceanic systems thus revealing far-reaching consequences of methionine biosynthesis and other B12-dependent enzymatic reactions on a community level.
Subject terms: Microbial communities, Microbial ecology
Introduction
The cofactor vitamin B12 (cobalamin, called B12 herein) is widely used among prokaryotes and eukaryotes for numerous metabolic functions. Based on genomic sequences to-date, only about one third of prokaryotes including Thaumarchaeota, Cyanobacteria and alpha- and gammaproteobacterial lineages are predicted to produce B12 [1–4]. Among these prokaryotic groups biosynthesis of cobalamin varies greatly [3, 4]. In oceanic systems Rhodobacterales and Rhizobiales are the main alphaproteobacterial B12 producers, whereas the abundant alphaproteobacterial SAR11 clade, 99% of Flavo- and Sphingobacteria and the euryarchaeotal Marine Group II are B12 auxotrophic [1, 5–7]. In contrast to the rather limited proportion of prokaryotes predicted or shown to produce B12, almost all marine prokaryotic and more than half of all marine eukaryotic microbes are known or genomically predicted to possess B12-dependent enzymes [1, 3, 4, 8]. Thus, all marine B12-dependent eukaryotic and the majority of marine prokaryotic organisms rely on exogenous supply of this pivotal cofactor. This disparity between dependency and supply of B12 results in close microbial interactions [8–12].
In pelagic marine ecosystems, concentrations of B12 greatly vary, ranging from highest concentrations of ~90 pM to below the detection limit of <1 pM in surface waters [11, 13–15]. B12 has been shown to be growth-limiting for distinct phytoplankton groups such as diatoms and dinoflagellates and thus crucial in shaping phytoplankton community composition [12, 16–19]. However, despite the fact that many marine bacteria are B12 auxotrophic no information exists whether growth and composition of prokaryotic microbial communities in pelagic marine systems are also affected by the availability of B12.
Cobamides summarise a group of coenzymes of the B12-family, consisting of a corrin ring centred around a cobalt ion to which a lower axial ligand, consisting of a phenolic, purine, or benzimidazole derivative, is attached. The chemical form of the lower ligand alters the cobamide structure and thereby affects the cofactor binding and consequently the coenzyme catalysis [4, 20–23]. Availabilities of dissolved exogenous cobamide variants differ greatly, depending on the ambient prokaryotic communities [3, 4, 12, 24]. Cobalamin, the most common and bioavailable cobamide cofactor in near-surface marine ecosystems, comprises 5,6-dimethylbenzimidazole (DMB) as lower ligand (Fig. 1A), whereas pseudocobalamin encompasses adenine as lower ligand and is synthesised by Cyanobacteria and less bioavailable to most other organisms [20, 25]. Notably, some prokaryotic as well as eukaryotic microorganisms acquired an adaptive strategy to remodel unusable cobamides in the presence of suitable lower ligands [23, 25]. In particular, α-ribazole, the activated form of DMB, has been shown to be taken up by a bacterium, enabling the synthesis of cobalamin, thus complementing the genetic deficiency of the biosynthesis of this lower ligand intermediate [26]. Based on comparative genome analyses, 17% of all bacterial B12 auxotrophs feature partial biosynthetic pathways of cobamides and thus presumably possess the capability to salvage B12 building blocks [3, 27]. Lower ligand salvaging towards the most common and bioactive cobalamin is mostly tested by targeting DMB which has been detected in soil, a freshwater creek and the intestine of animals [24, 28]. Nothing is known about its presence in marine waters. However, there is some information available that the lower ligand intermediate α-ribazole is present in marine waters. A molecular formula matching that of α-ribazole has been detected in marine dissolved organic matter (DOM) and within the exometabolome of several marine prokaryotes [29–32]. Life of the majority of marine microorganisms depends on the availability of B12, however only a minority of marine prokaryotes holds the genetic apparatus for its de novo synthesis. Hence we hypothesise that uptake of α-ribazole, thus salvaging its biosynthetic deficiency, is a pathway to overcome B12 auxotrophy and to function as crucial element in microbial vitamin networks, especially for prokaryotic communities. To test whether growth and composition of pelagic marine prokaryotic communities are affected by the availability of B12 or α-ribazole we supplemented mesocosms in the Pacific Ocean with these compounds and monitored growth activity and composition of the prokaryotic communities over several days.
Fig. 1. Cobalamin (vitamin B12) molecular structure and location of the mesocosm experiments in the Pacific Ocean.
A Molecular structure of B12 and its lower axial ligand 5,6-dimethylbenzimidazole (DMB) and intermediate α-ribazole. B Location of stations where mesocosm experiments M1, M2 and M3 were conducted. The graphic was created using ocean data view. For further details of stations see Table 1.
Material and methods
Location, sampling, hydrography and set-up of mesocosm experiments
Mesocosm experiments were carried out in three different biogeographic regions of the Pacific Ocean, the South Pacific Subtropical Gyre (SPSG; station 2, M1), the Pacific North Equatorial Current (PNEQ; station 7, M2) and the North Pacific Polar Frontal region (NPPF, station 14, M3). At these stations, temperature and salinity profiles and concentrations of chlorophyll a were determined. They are part of a transect between 30°S and 59°N investigated for hydrographic, biogeochemical and microbial variables during a cruise wit RV Sonne (SO248) from May 3rd to June 1st, 2016. Further details are described elsewhere [33, 34]. Here we present data of the depth of sampling for the mesocosm experiments, 20 m at stations 2 and 14, and 75 m at station 7 (Table 1). We collected water from 75 m at station 7 in PNEQ to reach the less nutrient- and phytoplankton-depleted deeper water with a higher chlorophyll fluorescence reading closer to the deep chlorophyll maximum. Water was collected using 20 L-Niskin bottles mounted on a Sea-Bird Electronics (SBE, Bellevue, WA, USA) 32 Carousel Water Sampler containing 24 ×20 L-Niskin bottles (Ocean Test Equipment Inc., Fort Lauderale, FL, USA). The carousel included the CTD system SBE 911 plus a probe with double-sensors for temperature (SBE 3), conductivity (SBE 4), pressure (Digiquartz), Chl a-fluorescence combined with turbidity (FluoroWetlab ECO_AFL FL, WET Labs Inc., Philomath, OR, USA) and dissolved Oxygen (Optode 4831F, Aanderaa, Bergen, Norway) and an altimeter (Teledyne Benthos, North Falmouth, MA, USA). After retrieval, water was transferred to 25 l Nalgene polycarbonate bottles.
Table 1.
Location, hydrographic and microbial features at the mesocosm stations.
| Province, mesocosm ID Station no. | Depth (m) | Latitude | Longitude | Date (2016) | Temperature (°C) | Salinity | Chl a (µg l−1) | Prokaryote cells (105 ml−1) | HPP (ng C l−1 h−1) | Growth rate (day−1) | DFAA turnover (day−1) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SPSG (M1) Station 2 | 20 | 26.99°S | 178.21°E | 04 May | 25.22 | 35.64 | 0.27 | 4.5 | 9.8 | 0.02 | 0.03 |
| PNEQ (M2) Station 7 | 60 | 04.66°N | 179.40°E | 14 May | 28.71 | 34.32 | 0.10 | 4.4 | 382.1 | 0.71 | 0.14 |
| NPPF (M3) Station 14 | 20 | 45.00°N | 178.75°E | 24 May | 5.86 | 33.31 | 0.72 | 9.7 | 93.0 | 0.13 | 0.14 |
Given are the biogeographic provinces (SPSG: South Pacific Subtropical Gyre; PNEQ: Pacific North Equatorial Current; NPPF: North Pacific Polar Frontal Region), ID of the mesocosm experiments, station number, latitude, longitude, date of sampling, temperature, salinity, concentrations of chlorophyll a (Chl a), prokaryote cell abundance, heterotrophic prokaryotic production (HPP), bulk growth rate of the prokaryotic community and turnover rates of dissolved free amino acids (DFAA) at 20 m at stations, 2 and 14, the depths of sampling for the mesocosms, and at 60 m depth at station 7. At station 7, respective data of the depth of sampling, 75 m, are not available. For further details of provinces see Giebel et al. [33].
Mesocosm experiments
The Nalgene bottles were first washed twice with acidified ultrapure water (MilliQ, pH 2) and then rinsed with the respective mesocosm water. The bottles were filled to a volume of 23 l and amended with FeSO4 (5 nM), NaH2PO4 (1 µM), NaSiO3 (32 µM), NaNO3 (16 µM) and CoCl2 (500 pM) to avoid a B12 co-limitation (Supplementary Table S1) as observed by previous studies [13, 14, 16]. All experiments were conducted in triplicates with supplements of B12 (cyanocobalamin; 100 pM; Sigma-Aldrich, St. Louis, MO, USA) or α-ribazole (100 pM; own preparation, see below) and a negative control. Incubation for six days was performed at the respective in situ temperature in a temperature controlled room (Table 1) and illuminated in a day:night rhythm of 12:12 hours (15–20 μmol photons m−2 s−1). Subsamples were withdrawn periodically to assess prokaryotic cell numbers, chlorophyll autofluorescence and heterotrophic prokaryotic production (HPP). Subsamples for analysing the composition of the pro- and eukaryotic microbial communities and metatranscriptomic analyses were withdrawn throughout the experiment. Metatranscriptomic analyses were only conducted for experiments M1 and M2.
Preparation and purity verification of α-ribazole
Alpha-ribazole is not commercially available. Therefore it was prepared by alkaline hydrolysis of B12 and purified as described in detail in the Supplementary Methods (see also Supplementary Fig. S1).
Prokaryotic abundance, chlorophyll autofluorescence and HPP
Prokaryotic abundance, chlorophyll autofluorescence of eukaryotic pico- and nanophytoplankton were measured immediately after sampling on board using a BD Accuri C6 cytometer (BD Biosciences, Heidelberg, Germany) as described elsewhere [33, 35]. Briefly, abundance of heterotrophic prokaryotes was determined after staining with SybrGreen I and that of autofluorescent pico- and nanophytoplankton by re-gating data from a plot of fluorescence FL3 (red, >670 nm) vs. FL2 (orange, 585 ± 20 nm) to a cytogram of FL4 (red, 675 ± 12.5 nm) vs. forward scattered light (FSC) and translation into cell numbers. Volume calibration was done by using TruCount tube controls (BD Biosciences). Rates of HPP and turnover of dissolved free amino acids (DFAA) were determined immediately after sampling by incorporation of 14C-leucine and a mixture of 3H-amino acids as described elsewhere [33, 34]. Samples were incubated for 2–6 h thus adjusting for varying ambient temperature and growth of the prokaryotic communities in the mesocosms. Variables characterising the biogeographic provinces, inorganic nitrate and phosphate, chlorophyll a and particulate organic carbon (POC) were also determined as described previously [33, 34].
Microbial community analysis
The composition of the prokaryotic and eukaryotic microbial communities was analysed in all experiments three hours after the onset and at days 3 and 6. Five hundred ml of water were withdrawn from the mesocosms and concentrated by vacuum filtration on a 0.2 µm polycarbonate filter (Millipore, Burlington, MA, USA), immediately deep-frozen in liquid nitrogen and stored at − 80 °C. DNA and RNA were extracted simultaneously as described elsewhere [36].
Prokaryotic and eukaryotic microbial communities were analysed using taxonomic marker gene sequencing. For prokaryotes the V3-V4 variable region of the 16S rRNA gene was targeted and for eukaryotes the V9 variable region of the 18S rRNA gene using the primer sets 341F (CCTAYGGGRBGCASCAG) and 806R (GGACTACNNGGGTATCTAAT) [37], and 1389F (TTGTACACACCGCCC) and 1510R (CCTTCYGCAGGTTCACCTAC) [38], respectively. PCR amplification of the extracted DNA and sequencing of the purified amplicon libraries on a MiSeq instrument (Illumina, San Diego, USA) were carried out as described in the Supplementary Methods. Additionally, for targeting the active prokaryotic communities, cDNA of the V3-V4 region of the 16S rRNA was sequenced after preparation from RNA extracts as described in the Supplementary Methods.
DNA reads in the demultiplexed sequencing files were trimmed for quality using Trimmomatic v. 0.32 [39] with the settings SLIDINGWINDOW:5:3 and MINLEN: 275, and reads were merged using FLASH v. 1.2.7 [40] with the settings -m 350 -M 525. The trimmed reads were dereplicated and formatted for use in the UPARSE workflow [41]. The dereplicated reads were clustered using the usearch v. 7.0.1090 -cluster_otus command with default settings, and the resulting OTU abundances were estimated using the usearch v. 7.0.1090 -usearch_global command with -id 0.97 -maxaccepts 0 -maxrejects 0. Taxonomy was assigned using the RDP classifier [42] as implemented in the parallel_assign_taxonomy_rdp.py script in QIIME [43], using –confidence 0.8 and the SILVA database, release 132 [44].
The results were analysed in R v. 3.5.0 (R Core Team, 2017) through the Rstudio IDE using the ampvis package v.2.3.16 [45]. Alpha diversities were estimated after rarefaction using the R-packages ‘drc,’ ‘picante,’ ‘Cairo,’ ‘vegan,’ and ‘ape,’. Rarefaction curves and statistical evaluation data are presented in Supplementary Fig. S2 and Supplementary Data 1. The community structure based on OTUs was normalised through Hellinger transformation, fitted under an NMDS based on Bray–Curtis dissimilarities calculated with the function vegdist in the R-package ‘vegan’. Permanova and homogeneity of multivariate dispersion are listed in Supplementary Data 1. The 14 most abundant OTUs, from their mean relative abundance in a given mesocosm, are presented as Box-Whisker plots and significant differences between the treatments compared to the control were calculated running two-sample Student’s t test (see Supplementary Data 1). Rarefaction curves showed that sampling efforts in the mesocosm experiments varied and that in M3 saturation was almost reached (Supplementary Fig. S2). All sequencing and bioinformatic analyses were carried out by DNASense (Aalborg, Denmark).
Metagenome and metatranscriptome analyses
Metagenome and metatranscriptome library preparation and sequencing was carried out as described in detail in the Supplementary Methods on a HiSeq and NovaSeq system (Illumina), respectively by DNASense (Aalborg, Denmark). Illumina reads were quality checked and low-quality regions as well as adaptor sequences were trimmed using Trimmomatic 0.36 [39] (ADAPTER:2:30:10 SLIDINGWINDOW:4:25 MINLEN:100). The high quality (HQ) reads were assembled using metaSPAdes 3.11.1 [46] using k-mer sizes 21, 33, 55 and 77. Contigs smaller than 210 bp and average coverage <2 were discarded. Gene-coding sequences of the assembled contigs were predicted using Prodigal 2.6.2 in meta-mode [47]. Genes shorter than 210 bp and longer than 4500 bp were discarded resulting in 6.83 M unique gene sequences.
Gene catalogue generation
The gene catalogue was generated as previously described [48] clustering at 95% identity, resulting in 1.89 Mio cluster centroids. These were taxonomically classified using Kaiju 1.6 ([49]) against NCBI nr database (downloaded 2018-05-29) and the proGenomes database [50] (downloaded 2019-07-26). Genes were functionally classified against the Kyoto Encyclopedia of Genes and Genomes (KEGG) as previously described [48]. 39% of all sequences were assigned to a KEGG orthologue (KO). Both mesocosm M1 and M2 sequencing efforts were near saturation as shown by rarefaction (Supplementary Fig. S3).
Transcriptome read abundance and normalisation
Reads were filtered for ribosomal RNA using SortMeRNA version 4.0.0 [51] matching against concatenated SILVA 90% database (16S, 18S and 23S rRNA) [44] (release 132) and the rfam 5s database [52]. HQ Illumina reads longer than 75 bp were mapped to the representative gene sequences using bowtie2 [53] 2.3.5 (--very-sensitive-local mode). SAMtools [54] version 1.9-58-gbd1a409 was used to convert the SAM alignment file to read abundance tables, discarding reads that did not map to any non-redundant sequence. Normalisation and scaling factors were calculated and applied as previously described [48, 55]. Analysed transcribed genes were normalised to the gene abundance of the taxonomic group under investigation.
Metatranscriptome evaluation
For each prokaryotic annotated gene transcript, the log2-fold change and p value was calculated, treatments (B12 or α-ribazole addition) with negative control on each sampling point (mesocosm 1 and 2, 3 h, days 1 and 3). Volcano plots were generated using the R-package ‘cairo’. The majority of gene transcripts with log2-fold change >2 and p < 0.05 were associated to key proteins of photosystem I and II and carbon fixation of Prochlorococcus (Supplementary Fig. S4), ammonium and amino acid transport and metabolism of Pelagibacteraceae as well as ammonium transport, B12 transport and cell motility of Gammaproteobacteria. Therefore, we grouped protein coding transcripts involved in cellular functions of respective taxonomic groups. The considered genes of the investigated cellular functions are listed in Supplementary Data 1. In order to understand the effect of B12 and α-ribazole supply on the regulation of the B12 pathway and the B12 outer membrane permease, encoded by btuB in the prokaryotic community, we grouped respective gene transcripts. Differentially expressed genes between treatments and negative control were tested by two-sample Student’s t test (p < 0.05; Supplementary Data 1).
Data deposition
Sequence data of the microbial community, metagenome and metatranscriptome analyses were deposited in the European Nucleotide Archive (ENA) at EMBL-EBI with accession numbers PRJEB43936, PRJEB43939, PRJEB43941, PRJEB43944 and PRJEB43946, using the data brokerage service of the German Federation for Biological Data (GFBio, [56]), in compliance with the Minimal Information about any (X) Sequence (MIxS) standard [57].
Results
During a cruise across the Pacific Ocean from New Zealand to Alaska we carried out mesocosm experiments in three biogeographic provinces, SPSG, PNEQ and NPPF, to test for the effect of B12 and α-ribazole on growth and related changes in microbial community composition (Fig. 1B). These biogeographic provinces differ greatly regarding hydrographic, biogeochemical, biotic and microbial features [33, 34]. SPSG encompasses the most oligotrophic and permanently stratified region of the Pacific Ocean with water temperatures of 26°–30 °C. In the upper 100 m, we encountered nitrate concentrations close to or below the detection limit (0.3 µM), concentrations of chlorophyll a below 0.35 µg l−1, except at the deep chlorophyll maximum (DCM) around 90 m depth, and rates of HPP not exceeding 30 ng C l−1 h−1. PNEQ adjacent to the equatorial upwelling is more productive. The upper 100 m are as warm as SPSG and concentrations of inorganic nutrients, chlorophyll a, POC and rates of HPP were also low in the upper 40 m but increased at 60 m and below. At these depths, inorganic phosphate concentrations reached 0.23 µM, well above the detection limit of 0.04 µM, and HPP 382 ng C l−1 h−1. Chlorophyll a concentrations remained as low as in SPSG with a maximum at the DCM at 105 m. Prokaryotic cell numbers were also as low as in SPSG, but bulk growth rates were much higher and reached 0.71 d−1, reflecting the metabolically highly active heterotrophic prokaryotic communities. The cold and nutrient-rich NPPF exhibited highest concentrations of chlorophyll a of 0.7–1.1 µg l−1 and highest numbers of prokaryotes (up to 1.8 106 cells ml−1) but lower rates of HPP than in PNEQ, 90–140 µg C l−1 h−1. Highest values consistently occurred at 20 m depth and continuously decreased below. Further details of the biogeochemical and microbial features of these provinces, stations and depths of sampling are published elsewhere [33, 34] and shown in Table 1.
Microbial abundance and HPP
In experiment M1 in SPSG abundance of heterotrophic prokaryotes increased continuously and similarly in both treatments and the control (Fig. 2A). In experiment M2 in PNEQ abundance of heterotrophic prokaryotes increased only until day 3 and then remained constant (Fig. 2B). In the B12 treatment, numbers remained lower onwards from day 1 than in the α-ribazole treatment and the control. This reduced prokaryotic abundance is consistent with the hypothesis of enhanced protist grazing in the B12 treatment. Relative proportions of the uncultured marine stramenopiles (MAST) group 1,and “other” protist groups, comprising more than 45% of total eukaryotic reads, were enhanced in this treatment relative to the control and the α-ribazole treatment on days 3 and/or 6 (Supplementary Fig. S5). As these MAST lineages comprise heterotrophic protists which can graze on nanoplankton and bacteria [58, 59] we hypothesise that an enhanced grazing pressure in the B12 treatment led to reduced numbers of prokaryotes. In experiment M3 in NPPF with the highest initial prokaryotic abundance, cell numbers decreased slightly from the beginning until day 3 and more so thereafter. Those in the B12 treatment were lower than in the control and partially also than in the α-ribazole treatment (Fig. 2C). As “other” protist and the MAST 1 groups in M3 constituted more than 50% of total eukaryotic reads and significantly higher values in the B12 treatment relative to the control at day 6 these data are also consistent with the protist grazing hypothesis (Fig. 2C; Supplementary Fig. S6). Initial abundances of eukaryotic pico- and nanophytoplankton were highest in M3, in line with the highest concentrations of chlorophyll a (Table 1). Abundances decreased in all experiments continuously after day 1 whereas initially in experiment M2 they increased and in M3 remained constant (Supplementary Fig. S7). In M1 data for hour 3 were not recorded.
Fig. 2. Prokaryotic abundance and heterotrophic prokaryotic production (HPP) in the course of the mesocosm experiments.
A–C Prokaryotic abundance of the treatments with additions of α-ribazole and B12 and an untreated control. D–F HPP of the treatments and an untreated control. Shown are means of triplicates and standard deviations. Significant differences of the means of a treatment and control (T-test, *p < 0.05, **p < 0.01, ***p < 0.001).
Rates of HPP exhibited positive responses to the supply of B12 and α-ribazole in all three mesocosm experiments. In experiment M1, HPP increased until day 3 and exhibited twofold higher values in the B12 treatment at days 3 and 6 and 1.5-fold higher values in the α-ribazole treatment at day 3 relative to the control (Fig. 2D). In experiment M2, rates of HPP in the α-ribazole and B12 treatments were two- and threefold higher than in the control already at the initial sampling at hour 3 (Fig. 2E). At this station and in this experiment, prokaryotic growth activities were very high, also in the control. This is evident from the very high rates of HPP and prokaryotic growth at 60 m depth, in fact the highest value at this depth over the entire transect (Table 1 and reference [34]). At hour 3 we observed in both treatments and at day 2 in the B12 treatment significantly higher rates of HPP than in the control and recorded an up and down pattern throughout the experiment (Fig. 2E and Supplementary Data 1). In experiment M3 in NPPF, rates of HPP were lowest (Fig. 2F). Highest values occurred on day 3 and decreased thereafter, in line with decreasing prokaryotic abundance. Addition of B12 yielded a significantly higher HPP rate at most sampling points throughout this experiment. Similarly, α-ribazole supply yielded significantly higher rates at days 4 and 5 than the control (Supplementary Data 1).
Composition of microbial communities
In experiments M1 and M2, the initial prokaryotic communities were largely dominated by Prochlorococcus and two sub-lineages of the SAR11 clade. In M2, the SAR86 clade affiliated to Oceanospirillales, Rhodospirillaceae and euryarchaeotal Marine Group II (Thermoplasmatales) constituted 5–7%. Other lineages, including SAR116, Flavobacteriaceae, Rhodobacteraceae, Alteromonas and Marine Group III, constituted less than 3% each at the beginning of both experiments (Supplementary Figs. S8 and S9). In experiment M3, the prokaryotic community initially was largely dominated by Alphaproteobacteria, predominantly various lineages of the SAR11 clade, and Flavobacteriia, mostly sublineages of the NS2, NS4 and NS5 groups, constituting 58% and 21% of the total. Gammaproteobacteria, predominantly the SAR86 clade, constituted 11% and other lineages including Cyanobacteria and Thermoplasmatales < 3% each (Supplementary Fig. S10). The initial eukaryotic communities in M1 and M2 were dominated by Dinoflagellata, Protalveolata and Retaria (Supplementary Figs. S11 and S5) whereas in M3 it was more diverse and dominated by Ochrophyta, including diatoms, and MAST-1, representing 21% and 10% of the total, respectively (Supplementary Fig. S6). In all initial communities, “other” eukaryotic groups, representing accumulated lineages of <3% each, constituted large proportions. The initial composition of the mesocosm communities was very similar to that of the ambient communities at the stations and depth ranges at which the water for the experiments was collected (Supplementary Fig. S12).
During the course of all experiments and as response to the B12 or α-ribazole supply the composition of the prokaryotic as well as eukaryotic communities changed substantially. The global effects of these observations are visualised by a non-metric multidimensional scaling (NMDS) analysis based on the V3-V4 fragment of the 16S rRNA gene and its transcript (cDNA) and of the V9 fragment of the 18S rRNA gene (Fig. 3). The clustering exhibits differences not only over time but also reflecting the effect of the B12 and α-ribazole supply on the composition of the prokaryotic and eukaryotic communities. Details of the temporal differences of the major prokaryotic groups including Prochlorococcus, the SAR11 and SAR86 clades, Rhodospirillaceae, Pseudoalteromonas, Flavobacteriaceae and Marine Group II are shown in Supplementary Figs. S8, S9 and S10. The different treatments led also to pronounced and significant changes in single prokaryotic groups. In M1, the accumulated OTUs affiliated to Oceanospirillales increased in the B12 treatment relative to the other treatment and control at day 3 and 6 with a highly significant increase on the latter day (Fig. 4C). In M2, the percentages of Marine Group III (Thermoplasmatales), Alteromonas, one family of the SAR86 clade and Rhodospirillaceae increased in either the B12 or α-ribazole treatment or both relative to the control on days 3 and/or 6 (Fig. 4F, Supplementary Fig. S9). In M3, significant changes of the prokaryotic community were predominantly caused by enhanced proportions in the B12 treatment of the SAR11 clade and Oceanospirillales at day 6, similar to M1, and reduced proportions of flavobacteriial lineages (Fig. 4G, Supplementary Fig. S10).
Fig. 3. Compositional changes of the microbial communities during the mesocosm experiments.
Non-metric multi-dimensional scaling (NMDS) analysis of the prokaryotic and eukaryotic communities. A–C NMDS plots of the prokaryotic communities of mesocosms M1, M2 and M3 and the treatments with B12, α-ribazole, and the control at hour 3, days 3 and 6; D–F similar plot based on the transcript (rRNA) level; G–I NMDS plots of the eukaryotic communities of mesocosms M1, M2 and M3. The NMDS analysis was calculated as a Bray–Curtis similarity based on Hellinger transformed microbial community data.
Fig. 4. Prokaryotic lineages exhibiting the most pronounced changes in the communities during the mesocosm experiments.
A–C M1, SAR11 clade (predominantly Candidatus Pelagibacter), Cyanobacteria (predominantly Prochlorococcus) and Oceanospirillales (predominantly SAR86 clade); D–F M2, SAR11 clade (predominantly Candidatus Pelagibacter), Oceanospirillales (predominantly SAR86 clade) and Thermoplasmatales (Marine Group I and III); G–I M3, SAR11 clade (predominantly Candidatus Pelagibacter), Oceanospirillales (predominantly SAR86 clade and Oceanospirillaceae) and Flavobacteriales (predominantly Flavobacteriaceae). Given are relative proportions of the lineages in the treatments with supply of B12 (red), α-ribazole (a-R, pink) and a control (C, black) at hour 3 and days 3 and 6. Significant differences of the means of a treatment and control (T-test, *p < 0.05, **p < 0.01, ***p < 0.001).
The composition of the eukaryotic communities also changed as a result of the B12 and α-ribazole supply. In M1, the community composition among the two treatments and the control remained rather similar except for reduced fractions of Ochrophyta in the B12 treatment (Supplementary Fig. S11). In M2, lineages MAST 1 and “other” significantly increased on day 6 in the B12 treatment relative to the control whereas Protalveolata decreased (Supplementary Fig. S5 and Supplementary Data 1). In M3, shifts in the eukaryotic community correlated with increased proportions of the heterotrophic lineages MAST-1, MAST-7 and Picomonadidae and reduced proportions of Ochrophyta and Prymnesiales at days 3 and/or 6 in the B12 treatment relative to the control (Supplementary Fig. S6). The strong relative increases of the heterotrophic flagellates MAST-1 and MAST-7 in the B12 treatment are consistent with the hypothesis of a grazing-induced decrease of prokaryotic cell numbers in this treatment.
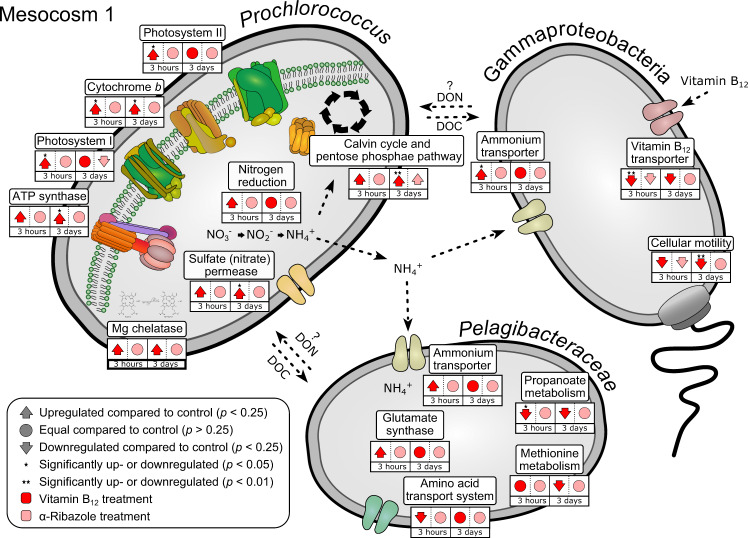
The major effects of the B12 and α-ribazole supply on prokaryotic abundance, HPP, responsive prokaryotic and eukaryotic lineages are summarised in Fig. 5.
Fig. 5. Environmental and biotic features of the locations of mesocosm experiments M1, M2 and M3 and major effects of the supply of B12 and α-ribazole in the course of the experiments.
Treatment effects include changes in the composition of prokaryotic and eukaryotic communities and changes in transcription of genes encoding different proteins in Gammaproteobacteria (mainly SAR86 clade), Pelagibacteraceae and Prochlorococcus. -: decrease; +: increase. d1, d2, d3, d4, d6: day 1, 2, 3, 4 and 6; h3: hour 3. ++ or -- indicate strong changes. When no + or no – is given no significant change between a treatment and the control was detected. M3 was not analysed for metatranscriptomics (accentuated in grey). For further details and specific responses of individual lineages see Figs. 4 and 6 and Supplementary Figs. S5 and S7 to S12. For other abbreviations see text.
Metatranscriptomic responses
The prokaryotic communities exhibited transcriptional responses predominantly to the B12 treatment in experiments M1 and M2 in which metatranscriptomic analyses were conducted. We focused our analyses on identified genes affiliated to the major prokaryotic groups in both experiments which exhibited high transcriptional responses and/or encode proteins which are potentially affected by B12. As in experiment M1 transcriptional responses were much more pronounced than in M2 we focus mainly on the former. For results of M2 see Supplementary Fig. S13. Transcription of btuB, encoding the outer membrane permease for B12 for subsequent import into the cell [27], was downregulated in the B12 treatment relative to the control at hour 3 and day 3 (Fig. 6). Transcripts affiliated almost exclusively to Oceanospirillales of Gammaproteobacteria, indicating that this order directly took advantage of the B12 supply by reducing energetic costs for the biosynthesis of its importer. We did not find a transcriptional downregulation of genes associated to B12 biosynthesis pathways in both treatments relative to the control. Similarly, we did not find a transcriptional upregulation of genes encoding enzymes requiring B12 as cofactor such as the B12-dependent methionine synthase metH. As often few transcripts of a single gene and taxon were detected as significantly upregulated at one sampling point we pooled transcripts of individual taxa to major prokaryotic groups. Significant metabolic responses were found for Prochlorococcus, Oceanospirillales and Pelagibacteraceae. Transcripts affiliated to Prochlorococcus constituted 23.3 ± 4.4% in M1 and 14.2 ± 2.3% in M2, respectively. Prochlorococcus also constituted proportions of 15-45% of the prokaryotic communities in both experiments until day 3 (Fig. 4, Supplementary Fig. S9). In M1, already three hours after the B12 supply, the number of transcripts of genes encoding various functions related to the light and dark reaction of photosynthesis significantly increased relative to the control and the α-ribazole treatment (Fig. 6). Upregulated transcripts included genes encoding key features of photosystem I and II, cytochrome b, the magnesium chelatase, a key enzyme of chlorophyll biosynthesis, ATP synthase, CO2-fixation via the Calvin Benson cycle and monosaccharide metabolism via the pentose phosphate pathway. This pathway is closely linked to the Calvin Benson cycle in Prochlorococcus [60]. Further, a gene encoding a sulfate permease which mediates also the uptake of nitrate [61] and genes encoding the reduction of nitrate to nitrite (narB) and further to ammonium (nirA) were upregulated (Fig. 6). Nitrate reduction was recently shown to be an important trait of several lineages of Prochlorococcus [62, 63]. Some of these transcriptional upregulations were still present at day 3 (Fig. 6). For Oceanospirillales, in addition to the transcriptional downregulation of B12 import, the gene encoding ammonium transport (amt) was upregulated at hour 3 whereas genes encoding motility were downregulated at this time point and also at day 3 (Fig. 6). For Pelagibacteraceae, the genes encoding ammonium transport (amt) and binding to glutamate (gltB, gltD) were also upregulated at hour 3 whereas others encoding transport of branched chain amino acids (livK), propanoate (adh1, acuL) and methionine metabolism were downregulated at this time point and/or at day 3 (Fig. 6). For a complete list of regulated genes see Supplementary Data 1. The major effects of the B12 and α-ribazole supply on the transcriptional response of genes or gene families are summarised in Fig. 5.
Fig. 6. Up- and downregulated transcripts of genes in Gammaproteobacteria, Pelagibacteraceae and Prochlorococcus encoding proteins of different functions in experiment M1.
Gammaproteobacteria (mainly SAR86 clade of Oceanospirillales), transporters of B12 and ammonium and cellular motility; Pelagibacteraceae, transporters of ammonium and amino acids, glutamate synthase, propanoate and methionine metabolism; Prochlorococcus, ATP synthase, photosystem I and II, cytochrome b, Magnesium chelatase, Calvin Benson cycle, pentose phosphate pathway, sulfate/nitrate permease, nitrate reduction. Genes associated to respective cellular functions were pooled and are shown in Supplementary Data 1. Dotted arrows indicate putative compounds released and exchanged among the three groups of organisms. Circles show no difference in the gene regulation between treatment and control, whereas arrows illustrate up- or downregulation (T-test, p < 0.25). Significant differences between treatment vs. control are highlighted by * (T-test, p < 0.05) and (T-test, p < 0.01). Red compares vitamin B12 treatment vs. control and pink compares α-ribazole vs. control.
Discussion
Supply of B12 and α-ribazole enhanced HPP and prokaryotic growth and led to changes in the composition of prokaryotic and eukaryotic communities, visualised globally by NMDS analyses (Fig. 3), in all three mesocosm experiments in distinctly different biogeographic provinces of the Pacific Ocean. Effects of B12 were greater than of α-ribazole. Supply in particular of B12 also affected expression of genes of the major groups of the prokaryotic community predominantly in experiment M1 and to a lesser degree in M2. Surprisingly, Prochlorococcus yielded the most pronounced transcriptional response even though most known cyanobacteria, including Synechococcaceae, produce and use pseudo-B12 [12, 25]. These findings add important information to previous observations that supply and availability of B12 affects growth of various eukaryotic phytoplankton groups in different oceanic regions [8, 16–19].
Effects of B12 and α-ribazole on heterotrophic prokaryotes
Genomic and metatranscriptomic analyses predict that B12 is produced only by about one-third of prokaryotes, mainly comprising Thaumarchaeota, Cyanobacteria and alpha- and gammaproteobacterial lineages including Rhodobacterales, Rhizobiales and 80% of Rickettsiales [1, 3, 5, 12]. These analyses further predict B12 auxotrophy for the alphaproteobacterial lineage SAR11, marine Flavobacteriia and Sphingobacteriia and for 54% of Oceanospirillales [1, 3, 5]. For the SAR86 clade, affiliated also to Oceanospirillales but not included in the mentioned analyses, genomic information on B12-related metabolism is scarce. In the only sequenced genome of subclade B, B12 biosynthesis is encoded whereas in the only sequenced genome of subclade A it is not and for subclades C and D no information is available [64]. Genomic data further show that most Alteromonadales are B12 auxotrophic [1] but a few lineages such as Alteromonas macleodii encode a B12-dependent (metH) and a B12-independent methionine synthase (metE) [65]. For E. coli it was shown that the former is >100-fold more efficient than the latter [66] and for Vibrio cholerae metH was shown to be operational when B12 is available and metE knocked out [67]. These findings indicate that the simple division into B12-auxo- and prototrophic organisms may not be sufficient to provide a comprehensive understanding of the B12 requirements in a given ecosystem. Thus, it is also conceivable that A. macleodii uses the metH-encoded methionine synthase when B12 is available and that Alteromonadales are responsive to B12 supply irrespective of auxo- or prototrophy. Our results corroborate the genomic predictions because in all three experiments one or several of these lineages or sublineages responded to the addition of B12 by enhancing their proportions in the course of the experiment (Fig. 4). Most pronounced were the positive responses of Oceanospirillales, mainly represented by different lineages of the SAR86 clade, in experiments M1 and M3, of Thermoplasmatales (Marine Group III) in M2 and the SAR11 clade in M3. The positive effect of B12 supply on Oceanospirillales in experiment M1 is reflected by two observations. i) relative proportions of Oceanospirillales significantly increased; ii) downregulation of btuB transcription (Fig. 6). The biosynthesis of the btuB encoded protein in prokaryotes is often regulated by the availability of B12 via a B12-dependent riboswitch [27, 68] and thus consistent with our findings. This suggest that Oceanospirillales benefitted from available B12 resulting in an enhanced abundance in experiment M1. This order presumably outcompeted the SAR11 clade in the uptake of B12, as we identified no downregulated transcripts of btuB affiliated to the SAR11 clade. It is unknown, though, whether transcription of btuB in the genome-streamlined SAR11 clade is regulated by B12. Downregulated gene transcripts encoding enzymes of methionine metabolism, as observed at day 3 (Fig. 6), also suggest a lower availability of B12 to SAR11. Oceanospirillales and the SAR11 clade presumably initially benefitted from ammonium released by Prochlorococcus. Evidence for this is provided by the upregulated nitrate metabolism in this primary producer and the upregulation of amt transcription in both heterotrophic lineages and the downregulated expression of genes encoding transporters of branched-chain amino acids. Further, transcription of genes encoding motility were downregulated in Oceanospirillales. We are not aware of any direct functional link between motility and B12 availability and speculate that this downregulation may have been an indirect consequence of the altered supply by organic compounds during the incubation. The relative increase of the abundance of the SAR11 clade in experiment M3 is consistent with its genomically encoded B12 auxotrophy [1, 64]. However, another possible positive effect on the SAR11 clade in M3 as a result of B12 supply, about which we can only speculate, could be an increased availability of methionine. Bacteria of the SAR11 clade I require for their growth exogenous reduced sulphur compounds, which can be partly obtained from methionine [69]. Its synthesis in turn depends on B12 availability in most bacteria [1]. Hence, an enhanced supply of methionine by other bacteria and eukaryotes to SAR11 due to a relaxed control of methionine synthesis and release into the surrounding water could also explain the relatively enhanced proportion of SAR11. Oceanospirillales also responded in this experiment to the B12 supply but much less than in experiment M1. These differences between both experiments may be explained by the presence of different SAR86 subclades in both biogeographic provinces diverging in their B12-biosynthesis traits. In experiment M2, Marine Group III responded most significantly to B12 supply by enhancing its proportions. This euryarchaeotal group has been studied little so far and there is no genomic information on its potential for vitamin biosynthesis [70]. The only genomic information on Thermoplasmatales is available from a metagenome assembled genome of Marine Group II indicating that the biosynthetic pathway for B12 is not encoded whereas proteins for transport of B12 are encoded [6]. If these traits are a general genomic feature of Thermoplasmatales this may explain why Marine Group III responded positively to the B12 supply in experiment M2. In this case, growth of Marine Group II, however, would have been controlled differently as it did not respond to B12 supply (Supplementary Fig. S9). Hence, these observations provide some evidence for the B12 auxotrophy of Marine Group III.
Our findings indicate that growth of the major marine prokaryotic lineages or sublineages including the SAR86 clade of Oceanospirillales, SAR11 and possibly Marine Group III is limited by the availability of B12. Responses of individual groups vary in the different oceanic regions. The environmental and biotic conditions and the interacting microbial communities may also affect the responses to B12 availability. In the only other study, which examined a response of B12 supply to natural prokaryotic communities in a pelagic system, the Southern Ocean, and applied metatranscriptomic analyses, only one unidentified Gammaproteobacterium exhibited a downregulation of a transcript of a single gene, cobC, encoding the protein which catalyses the final step in the B12 biosynthesis [17].
As indicated by increasing or constant prokaryotic cell numbers in experiments M1 and M2 the responding groups raised their proportions of the prokaryotic communities by enhanced growth. In experiment M3 with generally decreasing cell numbers the responding groups increased their proportions by a relatively slower temporal decrease than the other groups by a more sustained growth due to supply of B12. In this experiment heterotrophic protists such as MAST-1, “other” protists and Protalveolata increased from ~10% to >35% thus covarying inversely with prokaryotic abundance. This inverse correlation is consistent with the hypothesis that grazing by these protozoans led to the continuous reduction of prokaryotic cell numbers (see above, Supplementary Fig. S6). A further indication of the relatively enhanced proportions of the mentioned prokaryotic lineages in the B12 treatment of the M3 experiment were the lower cell numbers relative to the other treatment and the control, also consistent with the hypothesis of enhanced grazing on the actively growing prokaryotes in this treatment. Our findings indicate that availability and use of B12 needs to be explored much better as a growth-controlling factor of oceanic prokaryotic communities.
In addition to the effect of B12 our results also showed that supply of its lower ligand intermediate α-ribazole affected the composition of prokaryotic communities in the course of the mesocosm experiment. These global effects were evident from the NMDS analyses (Fig. 3) but effects on growth of individual lineages such as the SAR11 and SAR86 clades and of an Alteromonas sublineage were non-significant. The findings imply that if α-ribazole or possibly also DMB are available in oceanic marine systems these compounds should be considered as another factor affecting growth of pelagic prokaryotic communities.
B12 occurrence and producing and remodelling prokaryotes
Concentrations of B12 in the mixed layer of the oceans range between <1 and about 20 pM and >30 pM at 200 m and below [14, 15, 71]. Our B12 additions of 100 pM thus raised the ambient concentrations by an unknown factor. All other experiments, which tested the effect of B12 supply on pelagic prokaryotic and eukaryotic microbial communities, applied concentration of 90–200 pM [16–19]. So the concentration of our B12 supply is well in the range of those even though this concentration is well above in situ concentrations. In situ concentrations of α-ribazole or lower ligand derivates are unknown. However, a mass formula matching that of α-ribazole has been detected in the exometabolome of various Rhodobacteraceae, Nitrosopumilus maritimus and in marine DOM [29–32] and a gene enabling transport of α-ribazole into a bacterium was identified [26]. Hence, it is highly probable that lower ligands occur in one or the other form in oceanic systems as a release product of prokaryotes and are available to and used by prokaryotes.
Looking for the supply of B12 to auxotrophic members of oceanic microbial communities Rhodobacteraceae, Rhizobiales and sublineages of Rhodospirillales, Oceanospirillales and Pseudomonadales are the most important candidates as genomic analyses of the great majority of the members of the two former and about 50% of the latter lineages predict them to be B12 prototrophic [1, 12]. In fact, a metatranscriptomic and a metaproteomic study in neritic ecosystems identified active B12 producers, including Rhodobacterales, Rhizobiales and also Cyanobacteria [5, 7]. Several Rhodobacterales are well known to produce and provide phytoplankton algae with B12 and constitute 2 to 15% of pelagic prokaryotic communities [72–75]. Usually, also at the stations of our experiments, members of these lineages constitute small fractions of the prokaryotic communities [34]. Hence it is reasonable to speculate that these members act as black queens for supply of B12 [76] to auxotrophic prokaryotes and possibly to grazing protists. Another phylum of cobamide suppliers are Cyanobacteria but most of them including Prochlorococcus and Synechococcus produce pseudo-B12 which differs from B12 in that adenine is the lower ligand instead of DMB. Some heterotrophic prokaryotes also produce and use pseudo-B12 [77, 78]. Whereas pseudo-B12 can be used by several prokaryotes its availability to other pro- and eukaryotes may require its remodelling to B12 [12, 25, 67]. Remodelling depends on the availability of the lower ligand DMB as has been shown for Vibrio cholerae [67]. Hence, when DMB, or possibly α-ribazole, and appropriate prokaryotes are present remodelling of pseudo-B12 is possible and this pathway may contribute to salvaging B12 requirements by prokaryotes as well as eukaryotes. A compound with a mass formula identical to that of α-ribazole was identified as an exometabolite of various prokaryotes and in marine DOM (see above) but no information is available on DMB in marine pelagic systems. DMB has been detected in soil, freshwater and the intestine of animals [24, 28]. This provides some evidence that remodelling of pseudo-B12 by using α-ribazole could be another yet unexplored pathway to salvage B12 requirements of marine microbes.
Effects of B12 and α-ribazole on Prochlorococcus
Unexpectedly, Prochlorococcus responded to the B12 addition in experiment M1 by enhancing transcription of genes encoding key proteins of photosynthesis, carbon fixation and nitrate uptake and reduction (Fig. 6). Principally there are two possible explanations for this response; i) a direct response of B12 on the functions encoded by these genes; ii) an indirect response via a direct effect of B12 supply on other prokaryotes favouring photosynthesis and carbon fixation of Prochlorococcus by other factors. Even though it is well established that Cyanobacteria including Prochlorococcus produce, use and release pseudo-B12 as cofactor [12, 25, 79], a direct effect by B12 cannot be ruled out. It has been shown that Salmonella enterica synthesises pseudo-B12 but can readily use pseudo-B12 as well as B12 [80]. If B12 can be used in a similar way by Prochlorococcus, its strong transcriptional response could be a direct result of the B12 supply. As considered for the effect of B12 on the photosynthesis of a diatom [81], a B12 deficiency could lead to reduced availability of phylloquinone (vitamin K1), an electron carrier of photosystem I, because its synthesis requires B12-dependent S-adenosyl methionine [82]. Phylloquinone also serves as electron carrier in photosystem I of cyanobacteria. Hence, the enhanced supply of B12 could explain the upregulation of gene transcripts encoding photosystem I and the energy metabolism of Prochlorococcus we observed.
There is, however, also experimental evidence for the second explanation. It has been reported that when Prochlorococcus was growing in co-culture with an A. macleodii strain in particular transcripts of genes encoding pseudo-B12 biosynthesis, photosystem I and biosynthetic pathways were upregulated even though growth was not enhanced relative to an axenic control culture [65]. The authors speculate that supply of pseudo-B12 to A. macleodii (and subsequent remodelling to B12) favoured growth of this organism which responded in supplying Prochlorococcus with unknown goods favouring energy conservation via photosystem I, excessive release of DOM and possibly biosynthesis of pseudo-B12. The elevated nitrate concentration in our experiments may have further enhanced these effects as genes encoding proteins for nitrate uptake and reduction were upregulated. Nitrate uptake can be mediated via a sulfate permease [61] and the gene encoding this permease was upregulated in experiment M1. Nitrate assimilation and reduction is encoded in quite a few Prochlorococcus sublineages [63] and growth yield of Prochlorococcus is significantly reduced when using nitrate as nitrogen source as compared to ammonium [62].
We are unable to decide which of both explanations better support our observations on the transcriptional effects of B12 on Prochlorococcus and these explanations are even not mutually exclusive. In any case, our findings indicate that availability of B12 can effect photosynthesis and energy acquisition of Prochlorococcus and should be considered as a yet unexplored factor regulating growth of this globally important cyanobacterium. Future in-depth studies need to elucidate the details of how B12 availability effects photosynthesis and growth of Prochlorococcus.
Effects of B12 and α-ribazole on eukaryotes
The eukaryotic communities in all three experiments were also affected by supply of B12 and α-ribazole as shown by the NMDS analyses (Fig. 3) and the shifts in the community composition with enhanced proportions mainly of heterotrophic protists (Supplementary Figs. S5 and S6). In previous studies, a positive effect of B12 on growth of phytoplankton algae has been shown in subpolar and polar seas [16–18]. Our experiments provide good evidence that growth of heterotrophic protists in oceanic microbial communities is also stimulated by supply of B12. In particular, the uncultured lineages MAST-1 and MAST-7 were stimulated even though to varying extent in the different experiments and regions. Lineages of these groups have been shown to be abundant in the Pacific Ocean and to graze on bacteria [58, 83]. So far only scarce information is available on the effect of B12 on growth of heterotrophic protists [84, 85] and none from marine environments. It has been shown that an amoeba requires a B12 prototrophic bacterium for growth and that bacterial remodelling of pseudo-B12 can also meet the B12 requirements of this amoeba [85]. From our experiments, we cannot disentangle whether B12 had a direct or indirect effect via ingested B12-enriched prokaryotic cells on growth of the protists or by directly taking up B12. Based on the scarce available information we assume that protists benefitted from grazing on B12-enriched prokaryotic cells. However, irrespective of the mode of action of B12, our findings shed new light on the significance this growth factor has for growth of heterotrophic protists in marine ecosystems. The availability of this growth factor does not only affect growth of phytoplankton algae, as shown in previous studies, but also of heterotrophic protists and prokaryotes in marine pelagic systems. The different effects of B12 availability on protists in the experiments in different oceanic regions may reflect the different partitioning of prokaryotes, pico- and nano-eukaryotes in the food for the heterotrophic protists.
Conclusion
Often the effect of B12 is considered in a rather narrow biochemical context of methionine biosynthesis in prokaryotic species, despite its known role as a cofactor also in other biochemical reactions. We have shown that effects of B12 and α-ribazole go far beyond these species-specific biochemical reactions: Availability of B12 and α-ribazole affects growth and related changes in the composition of prokaryotic and eukaryotic microbial communities in oceanic regions of different trophic state. Both growth factors but more so B12 favour growth of members of the major lineages of pelagic marine prokaryotes such as SAR11, Oceanospirillales/SAR86 and the euryarchaeotal Marine Group III, and several heterotrophic protist groups, in particular of the uncultured MAST-lineages. Surprisingly also Prochlorococcus responded positively to supply of B12, suggesting that it also benefits from availability of this growth factor. Our findings show in particular the significance of B12 for controlling growth of and complex interactions among hetero- and autotrophic prokaryotes and eukaryotes in oceanic systems. The positive effect of α-ribazole on altering the composition of prokaryotic communities, together with the detection of a mass formula similar to this compound in previous studies, suggests that this lower ligand intermediate is available in oceanic systems and plays a role in salvaging B12 requirements by distinct prokaryotic lineages.
Supplementary information
Supplementary Methods, Tables and Figures
Acknowledgements
We thank the master L. Mallon and the crew of RV Sonne for their most valuable support on shipboard, T. Badewien and R. Henkel for the hydrographic data and technical assistance in sampling. We are grateful to F. Milke, M. Wolterink, and B. Kuerzel for their support in sampling and sample analysis on shipboard and to M. Wolterink also for his contribution to the logistics of this cruise. Sequencing was carried out by DNASense (Aalborg, Denmark), with professional support in data processing by Mads Albertsen and his team. The study was supported by Deutsche Forschungsgemeinschaft within the Collaborative Research Center TRR51 Roseobacter and by grant BacGeoPac 03G0248A awarded to MS from the German Ministry of Education and Research (BMBF).
Author contributions
GW designed the experiment, carried out most of the data analysis and wrote large parts of the manuscript; LD carried out the basic bioinformatic evaluation of the metatranscriptomic analyses; RH synthesised α-ribazole; HW supervised the synthesis of α-ribazole; HAG carried out the flow cytometric analysis and enumeration of the prokaryotic and eukaryotic microbial communities; MS contributed to designing the experiments, carried out the HPP measurements, advised data evaluation and wrote parts of and finalised the manuscript. All authors revised carefully the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01250-7.
References
- 1.Sañudo-Wilhelmy SA, Gómez-Consarnau L, Suffridge C, Webb EA. The role of B vitamins in marine biogeochemistry. Annu Rev Mar Sci. 2014;6:339–67. doi: 10.1146/annurev-marine-120710-100912. [DOI] [PubMed] [Google Scholar]
- 2.Doxey AC, Kurtz DA, Lynch MD, Sauder LA, Neufeld JD. Aquatic metagenomes implicate Thaumarchaeota in global cobalamin production. ISME J. 2015;9:461–71. doi: 10.1038/ismej.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shelton AN, Seth EC, Mok KC, Han AW, Jackson SN, Haft DR, et al. Uneven distribution of cobamide biosynthesis and dependence in bacteria predicted by comparative genomics. ISME J. 2019;13:789–804. doi: 10.1038/s41396-018-0304-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sokolovskaya OM, Shelton AN, Taga ME. Sharing vitamins: cobamides unveil microbial interactions. Science. 2020;369:eaba0165. doi: 10.1126/science.aba0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gómez-Consarnau L, Sachdeva R, Gifford SM, Cutter LS, Fuhrman JA, Sañudo-Wilhelmy SA, et al. Mosaic patterns of B-vitamin synthesis and utilization in a natural marine microbial community. Environ Microbiol. 2018;20:2809–23. doi: 10.1111/1462-2920.14133. [DOI] [PubMed] [Google Scholar]
- 6.Iverson V, Morris RM, Frazar CD, Berthiaume CT, Morales RL, Armbrust EV. Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science. 2012;335:587–90. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- 7.Isaac A, Francis B, Amann RI, Amin SA. Tight Adherence (Tad) Pilus genes indicate putative niche differentiation in phytoplankton bloom associated Rhodobacterales. Front Microbiol. 2021;12:718297. doi: 10.3389/fmicb.2021.718297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Croft MT, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- 9.Cruz-López R, Maske H. The vitamin B1 and B12 required by the marine dinoflagellate Lingulodinium polyedrum can be provided by its associated bacterial community in culture. Front Microbiol. 2016;7:560. doi: 10.3389/fmicb.2016.00560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grant MA, Kazamia E, Cicuta P, Smith AG. Direct exchange of vitamin B12 is demonstrated by modelling the growth dynamics of algal–bacterial cocultures. ISME J. 2014;8:1418–27. doi: 10.1038/ismej.2014.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sañudo‐Wilhelmy SA, Gobler CJ, Okbamichael M, Taylor GT. Regulation of phytoplankton dynamics by vitamin B12. Geophys Res Lett. 2006;33:L04604. doi: 10.1029/2005GL025046. [DOI] [Google Scholar]
- 12.Heal KR, Qin W, Ribalet F, Bertagnolli AD, Coyote-Maestas W, Hmelo LR, et al. Two distinct pools of B12 analogs reveal community interdependencies in the ocean. Proc Natl Acad Sci USA. 2017;114:364–69. doi: 10.1073/pnas.1608462114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menzel DW, Spaeth JP. Occurrence of vitamin B12 in the Sargasso Sea. Limnol Oceanogr. 1962;7:151–54. doi: 10.4319/lo.1962.7.2.0151. [DOI] [Google Scholar]
- 14.Suffridge CP, Gómez‐Consarnau L, Monteverde DR, Cutter L, Arístegui J, Alvarez‐Salgado XA, et al. B Vitamins and their congeners as potential drivers of microbial community composition in an oligotrophic marine ecosystem. J Geophys Res Biogeosciences. 2018;123:2890–907. doi: 10.1029/2018JG004554. [DOI] [Google Scholar]
- 15.Suffridge C, Cutter L, Sañudo-Wilhelmy SA. A new analytical method for direct measurement of particulate and dissolved B-vitamins and their congeners in seawater. Front Mar Sci. 2017;4:11. doi: 10.3389/fmars.2017.00011. [DOI] [Google Scholar]
- 16.Bertrand EM, Saito MA, Rose JM, Riesselman CR, Lohan MC, Noble AE, et al. Vitamin B12 and iron colimitation of phytoplankton growth in the Ross Sea. Limnol Oceanogr. 2007;52:1079–93. doi: 10.4319/lo.2007.52.3.1079. [DOI] [Google Scholar]
- 17.Bertrand EM, McCrow JP, Moustafa A, Zheng H, McQuaid JB, Delmont TO, et al. Phytoplankton–bacterial interactions mediate micronutrient colimitation at the coastal Antarctic sea ice edge. Proc Natl Acad Sci USA. 2015;112:9938–43. doi: 10.1073/pnas.1501615112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koch F, Marcoval MA, Panzeca C, Bruland KW, Sañudo-Wilhelmy SA, Gobler CJ. The effect of vitamin B12 on phytoplankton growth and community structure in the Gulf of Alaska. Limnol Oceanogr. 2011;56:1023–34. doi: 10.4319/lo.2011.56.3.1023. [DOI] [Google Scholar]
- 19.Browning TJ, Achterberg EP, Rapp I, Engel A, Bertrand EM, Tagliabue A, et al. Nutrient co-limitation at the boundary of an oceanic gyre. Nature. 2017;551:242–46. doi: 10.1038/nature24063. [DOI] [PubMed] [Google Scholar]
- 20.Crofts TS, Seth EC, Hazra AB, Taga ME. Cobamide structure depends on both lower lgand availability and cobTsubstrate specificity. Chem Biol. 2013;20:1265–74. doi: 10.1016/j.chembiol.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Mok KC, Taga ME. Growth inhibition of Sporomusa ovata by incorporation of benzimidazole bases into cobamides. J Bacteriol. 2013;195:1902–11. doi: 10.1128/JB.01282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stupperich E, Steiner I, Eisinger HJ. Substitution of Co alpha-(5-hydroxybenzimidazolyl)cobamide (factor III) by vitamin B12 in Methanobacterium thermoautotrophicum. J Bacteriol. 1987;169:3076–81. doi: 10.1128/jb.169.7.3076-3081.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yi S, Seth EC, Men Y-J, Stabler SP, Allen RH, Alvarez-Cohen L, et al. Versatility in corrinoid salvaging and remodeling pathways supports corrinoid-dependent metabolism in Dehalococcoides mccartyi. Appl Environ Microbiol. 2012;78:7745–52. doi: 10.1128/AEM.02150-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu X, Heal KR, Ingalls AE, Doxey AC, Neufeld JD. Metagenomic and chemical characterization of soil cobalamin production. ISME J. 2020;14:53–66. doi: 10.1038/s41396-019-0502-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Helliwell KE, Lawrence AD, Holzer A, Kudahl UJ, Sasso S, Kräutler B, et al. Cyanobacteria and eukaryotic algae use different chemical variants of vitamin B12. Curr Biol. 2016;26:999–1008. doi: 10.1016/j.cub.2016.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray MJ, Escalante-Semerena JC. A New Pathway for the synthesis of α-ribazole-phosphate in Listeria innocua. Mol Microbiol. 2010;77:1429–38. doi: 10.1111/j.1365-2958.2010.07294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundrigan MD, Köster W, Kadner RJ. Transcribed sequences of the Escherichia coli btuB gene control its expression and regulation by vitamin B12. Proc Natl Acad Sci USA. 1991;88:1479–83. doi: 10.1073/pnas.88.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crofts TS, Men Y, Alvarez-Cohen L, Taga ME. A bioassay for the detection of benzimidazoles reveals their presence in a range of environmental samples. Front Microbiol. 2014;5:592. doi: 10.3389/fmicb.2014.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson WM, Kido Soule MC, Kujawinski EB. Evidence for quorum sensing and differential metabolite production by a marine bacterium in response to DMSP. ISME J. 2016;10:2304–16. doi: 10.1038/ismej.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romano S, Dittmar T, Bondarev V, Weber RJM, Viant MR, Schulz-Vogt HN. Exo-metabolome of Pseudovibrio sp. FO-BEG1 analyzed by ultra-high resolution mass spectrometry and the effect of phosphate limitation. PLoS One. 2014;9:e96038. doi: 10.1371/journal.pone.0096038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wienhausen G, Noriega-Ortega BE, Niggemann J, Dittmar T, Simon M. The exometabolome of two model strains of the Roseobacter group: a marketplace of microbial metabolites. Front Microbiol. 2017;8:1985. doi: 10.3389/fmicb.2017.01985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Law KP, He W, Tao J, Zhang C. Characterization of the exometabolome of Nitrosopumilus maritimus SCM1 by liquid chromatography–ion mobility mass spectrometry. Front Microbiol. 2021;12:1355. doi: 10.3389/fmicb.2021.658781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balmonte JP, Simon M, Giebel H-A, Arnosti C. A sea change in microbial enzymes: heterogeneous latitudinal and depth-related gradients in bulk water and particle-associated enzymatic activities from 30°S to 59°N in the Pacific Ocean. Limnol Oceanogr. 2021;66:3489–507. doi: 10.1002/lno.11894. [DOI] [Google Scholar]
- 34.Giebel H-A, Arnosti C, Badewien TH, Bakenhus I, Balmonte JP, Billerbeck S, et al. Microbial growth and organic matter cycling in the Pacific Ocean along a latitudinal transect between subarctic and subantarctic waters. Front Mar Sci. 2021;8:1888. doi: 10.3389/fmars.2021.764383. [DOI] [Google Scholar]
- 35.Giebel H-A, Wolterink M, Brinkhoff T, Simon M. Complementary energy acquisition via aerobic anoxygenic photosynthesis and carbon monoxide oxidation by Planktomarina temperata of the Roseobacter group. FEMS Microbiol Ecol. 2019;95:fiz050. doi: 10.1093/femsec/fiz050. [DOI] [PubMed] [Google Scholar]
- 36.Schneider D, Wemheuer F, Pfeiffer B, Wemheuer B. Extraction of total DNA and RNA from marine filter samples and generation of a cDNA as universal template for marker gene studies. In: Streit WR, Daniel R, editors. Metagenomics: methods and protocols. 2017. New York, NY: Springer; 2017. p. 13–22. [DOI] [PubMed]
- 37.Sundberg C, Al-Soud WA, Larsson M, Alm E, Yekta SS, Svensson BH, et al. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol Ecol. 2013;85:612–26. doi: 10.1111/1574-6941.12148. [DOI] [PubMed] [Google Scholar]
- 38.Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One. 2009;4:e6372. doi: 10.1371/journal.pone.0006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinforma Oxf Engl. 2011;27:2957–63. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–98. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 42.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–67. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–96. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Albertsen M, Karst SM, Ziegler AS, Kirkegaard RH, Nielsen PH. Back to basics – the influence of DNA extraction and primer choice on phylogenetic analysis of activated sludge communities. PLoS One. 2015;10:e0132783. doi: 10.1371/journal.pone.0132783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and Its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dlugosch L, Poehlein A, Wemheuer B, Pfeiffer B, Badewien TH, Daniel R, et al. Significance of gene variants for the functional biogeography of the near-surface Atlantic Ocean microbiome. Nat Comm. 2022;13:456. doi: 10.1038/s41467-022-28128-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Menzel P, Ng KL, Krogh A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun. 2016;7:11257. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mende DR, Letunic I, Huerta-Cepas J, Li SS, Forslund K, Sunagawa S, et al. proGenomes: a resource for consistent functional and taxonomic annotations of prokaryotic genomes. Nucleic Acids Res. 2017;45:D529–34. doi: 10.1093/nar/gkw989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinforma Oxf Engl. 2012;28:3211–17. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 52.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–41. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–59. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinforma Oxf Engl. 2009;25:2078–79. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci Theor Den Biowissenschaften. 2012;131:281–85. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 56.Diepenbroek M, Glöckner FO, Grobe P, Güntsch A, Huber R, König-Ries B, et al. Towards an integrated biodiversity and ecological research data management and archiving platform: the German federation for the curation of biological data (GFBio). In: Plödereder E, Grunske L, Schneider E, Ull D (eds.). Informatik. Gesellschaft für Informatik e.V. Bonn 2014. pp. 1711–l21.
- 57.Yilmaz P, Kottmann R, Field D, Knight R, Cole JR, Amaral-Zettler L, et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat Biotechnol. 2011;29:415–20. doi: 10.1038/nbt.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin Y-C, Campbell T, Chung C-C, Gong G-C, Chiang K-P, Worden AZ. Distribution patterns and phylogeny of marine Stramenopiles in the north Pacific Ocean. Appl Environ Microbiol. 2012;78:3387–99. doi: 10.1128/AEM.06952-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Massana R, Unrein F, Rodríguez-Martínez R, Forn I, Lefort T, Pinhassi J, et al. Grazing rates and functional diversity of uncultured heterotrophic flagellates. ISME J. 2009;3:588–96. doi: 10.1038/ismej.2008.130. [DOI] [PubMed] [Google Scholar]
- 60.Waldbauer JR, Rodrigue S, Coleman ML, Chisholm SW. Transcriptome and proteome dynamics of a light-dark synchronized bacterial cell cycle. PLoS One. 2012;7:e43432. doi: 10.1371/journal.pone.0043432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maeda S, Sugita C, Sugita M, Omata T. Latent nitrate transport activity of a novel sulfate permease-like protein of the cyanobacterium Synechococcus elongatus. J Biol Chem. 2006;281:5869–76. doi: 10.1074/jbc.M513196200. [DOI] [PubMed] [Google Scholar]
- 62.Berube PM, Biller SJ, Kent AG, Berta-Thompson JW, Roggensack SE, Roache-Johnson KH, et al. Physiology and evolution of nitrate acquisition in Prochlorococcus. ISME J. 2015;9:1195–07. doi: 10.1038/ismej.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berube PM, Rasmussen A, Braakman R, Stepanauskas R, Chisholm SW. Emergence of trait variability through the lens of nitrogen assimilation in Prochlorococcus. eLife. 2019;8:e41043. doi: 10.7554/eLife.41043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dupont CL, Rusch DB, Yooseph S, Lombardo M-J, Alexander Richter R, Valas R, et al. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J. 2012;6:1186–99. doi: 10.1038/ismej.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Biller SJ, Coe A, Chisholm SW. Torn apart and reunited: impact of a heterotroph on the transcriptome of Prochlorococcus. ISME J. 2016;10:2831–43. doi: 10.1038/ismej.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.González JC, Banerjee RV, Huang S, Sumner JS, Matthews RG. Comparison of cobalamin-independent and cobalamin-dependent methionine synthases from Escherichia coli: two solutions to the same chemical problem. Biochemistry. 1992;31:6045–56. doi: 10.1021/bi00141a013. [DOI] [PubMed] [Google Scholar]
- 67.Ma AT, Tyrell B, Beld J. Specificity of cobamide remodeling, uptake and utilization in Vibrio cholerae. Mol Microbiol. 2020;113:9–102. doi: 10.1111/mmi.14402. [DOI] [PubMed] [Google Scholar]
- 68.Nahvi A, Barrick JE, Breaker RR. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 2004;32:143–50. doi: 10.1093/nar/gkh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tripp HJ, Kitner JB, Schwalbach MS, Dacey JWH, Wilhelm LJ, Giovannoni SJ. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature. 2008;452:741–44. doi: 10.1038/nature06776. [DOI] [PubMed] [Google Scholar]
- 70.Haro-Moreno JM, Rodriguez-Valera F, López-García P, Moreira D, Martin-Cuadrado A-B. New insights into marine group III Euryarchaeota, from dark to light. ISME J. 2017;11:1102–17. doi: 10.1038/ismej.2016.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sañudo-Wilhelmy SA, Cutter LS, Durazo R, Smail EA, Gómez-Consarnau L, Webb EA, et al. Multiple B-vitamin depletion in large areas of the coastal ocean. Proc Natl Acad Sci USA. 2012;109:14041–45. doi: 10.1073/pnas.1208755109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wagner-Döbler I, Ballhausen B, Berger M, Brinkhoff T, Buchholz I, Bunk B, et al. The complete genome sequence of the algal symbiont Dinoroseobacter shibae: a hitchhiker’s guide to life in the sea. ISME J. 2010;4:61–77. doi: 10.1038/ismej.2009.94. [DOI] [PubMed] [Google Scholar]
- 73.Segev E, Wyche TP, Kim KH, Petersen J, Ellebrandt C, Vlamakis H, et al. Dynamic metabolic exchange governs a marine algal-bacterial interaction. eLife. 2016;5:e17473. doi: 10.7554/eLife.17473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Voget S, Wemheuer B, Brinkhoff T, Vollmers J, Dietrich S, Giebel H-A, et al. Adaptation of an abundant Roseobacter RCA organism to pelagic systems revealed by genomic and transcriptomic analyses. ISME J. 2015;9:371–84. doi: 10.1038/ismej.2014.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Billerbeck S, Wemheuer B, Voget S, Poehlein A, Giebel H-A, Brinkhoff T, et al. Biogeography and environmental genomics of the Roseobacter-affiliated pelagic CHAB-I-5 lineage. Nat Microbiol. 2016;1:1–8. doi: 10.1038/nmicrobiol.2016.63. [DOI] [PubMed] [Google Scholar]
- 76.Morris JJ, Lenski RE, Zinser ER. The Black Queen hypothesis: evolution of dependencies through adaptive gene loss. mBio. 2012;3:e00036–12. doi: 10.1128/mBio.00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stupperich E, Kräutler B. Pseudo vitamin B12 or 5-hydroxybenzimidazolyl-cobamide are the corrinoids found in methanogenic bacteria. Arch Microbiol. 1988;149:268–71. doi: 10.1007/BF00422016. [DOI] [Google Scholar]
- 78.Hoffmann B, Oberhuber M, Stupperich E, Bothe H, Buckel W, Konrat R, et al. Native corrinoids from Clostridium cochlearium are adeninylcobamides: spectroscopic analysis and identification of pseudovitamin B12 and factor A. J Bacteriol. 2000;182:4773–82. doi: 10.1128/JB.182.17.4773-4782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bonnet S, Webb EA, Panzeca C, Karl DM, Capone DG, Wilhelmy SAS. Vitamin B12 excretion by cultures of the marine cyanobacteria Crocosphaera and Synechococcus. Limnol Oceanogr. 2010;55:1959–64. doi: 10.4319/lo.2010.55.5.1959. [DOI] [Google Scholar]
- 80.Anderson PJ, Lango J, Carkeet C, Britten A, Kräutler B, Hammock BD, et al. One pathway can incorporate either adenine or dimethylbenzimidazole as an α-axial ligand of B12 cofactors in Salmonella enterica. J Bacteriol. 2008;190:1160–71. doi: 10.1128/JB.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.King AL, Sañudo-Wilhelmy SA, Leblanc K, Hutchins DA, Fu F. CO2 and vitamin B12 interactions determine bioactive trace metal requirements of a subarctic Pacific diatom. ISME J. 2011;5:1388–96. doi: 10.1038/ismej.2010.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lohmann A, Schöttler MA, Bréhélin C, Kessler F, Bock R, Cahoon EB, et al. Deficiency in phylloquinone (vitamin K1) methylation affects prenyl quinone distribution, photosystem I abundance, and anthocyanin accumulation in the Arabidopsis AtmenG mutant. J Biol Chem. 2006;281:40461–72. doi: 10.1074/jbc.M609412200. [DOI] [PubMed] [Google Scholar]
- 83.Orsi W, Wilken S, del Campo J, Heger T, James E, Richards T, et al. Identifying protist consumers of photosynthetic picoeukaryotes in the surface ocean using stable isotope probing. Environ Microbiol. 2017;20:815–27. doi: 10.1111/1462-2920.14018. [DOI] [PubMed] [Google Scholar]
- 84.Bonhomme A, Durand M, Quintana C, Halpern S. Effect of cobalt and vitamin B12 on growth and survival of rumen ciliates in vitro as a function of bacterial population. Reprod Nutr Dev. 1982;22:107–22. doi: 10.1051/rnd:19820109. [DOI] [PubMed] [Google Scholar]
- 85.Ma AT, Beld J, Brahamsha B. An amoebal grazer of cyanobacteria requires cobalamin produced by heterotrophic bacteria. Appl Environ Microbiol. 2017;83:e00035–17. doi: 10.1128/AEM.00035-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods, Tables and Figures