Abstract
Macroalgae represent huge amounts of biomass worldwide, largely recycled by marine heterotrophic bacteria. We investigated the strategies of bacteria within the flavobacterial genus Zobellia to initiate the degradation of whole algal tissues, which has received little attention compared to the degradation of isolated polysaccharides. Zobellia galactanivorans DsijT has the capacity to use fresh brown macroalgae as a sole carbon source and extensively degrades algal tissues via the secretion of extracellular enzymes, even in the absence of physical contact with the algae. Co-cultures experiments with the non-degrading strain Tenacibaculum aestuarii SMK-4T showed that Z. galactanivorans can act as a pioneer that initiates algal breakdown and shares public goods with other bacteria. A comparison of eight Zobellia strains, and strong transcriptomic shifts in Z. galactanivorans cells using fresh macroalgae vs. isolated polysaccharides, revealed potential overlooked traits of pioneer bacteria. Besides brown algal polysaccharide degradation, they notably include oxidative stress resistance proteins, type IX secretion system proteins and novel uncharacterized polysaccharide utilization loci. Overall, this work highlights the relevance of studying fresh macroalga degradation to fully understand the metabolic and ecological strategies of pioneer microbial degraders, key players in macroalgal biomass remineralization.
Subject terms: Environmental microbiology, Marine microbiology, Microbial ecology
Introduction
Macroalgae are major primary producers in coastal zones, acting as a global carbon sink [1]. Specific polysaccharides dominate macroalgal extracellular matrices (ECM) and can represent up to 50% of the dry weight [2]. For example, brown algae produce alginates and fucose-containing sulfated polysaccharides (FCSPs). Alginates are linear polymers of β-D-mannuronic (M) and α-L-guluronic acids (G), representing between 10 and 45% of the algal dry weight [2]. FCSPs, accounting for 4–13% of the dry weight [3], refer to linear or highly branched polysaccharides containing α-linked L-fucose residues together with a variety of other neutral monosaccharides constituents (e.g. galactose, mannose, xylose, rhamnose) and uronic acids [4]. They hold many substitutions, mainly sulfate and acetyl groups. The structure of brown algal polysaccharides is consequently highly heterogeneous and varies according to species, seasons, geographical locations, thallus part, algal growth stages and environmental factors [3–7]. Within the ECM, these carbohydrates are cross-linked and associated with proteins (3–15%), minerals (7–36% such as iodine, calcium, iron, copper and magnesium), phenols (1–13%), vitamins, amino acids and small amounts of lipids (1–5%) to form a complex matrix [8–11]. Besides ECM polysaccharides, brown algae also produce laminarin (β-1,3-glucan) and mannitol [12] as storage carbohydrates.
Marine heterotrophic bacteria are crucial for algal biomass mineralization [13]. Macroalgae surfaces are constantly colonized by diverse bacterial communities with densities varying from 102 to 107 cells cm−2 of macroalgal tissue [14]. A fraction of these communities, mainly Bacteroidetes, Gammaproteobacteria, Verrucomicrobia and Planctomycetes, can degrade this complex biomass, showing abilities to hydrolyze purified high molecular weight algal compounds using a considerable enzymatic arsenal [15–18]. Over the last 20 years, many studies investigated the algal polysaccharide-processing capabilities of marine heterotrophic bacteria [19], deciphering new catabolic pathways and unraveling the role of carbohydrate active enzymes (CAZymes, http://www.cazy.org, [20]) including glycoside hydrolases (GHs), polysaccharide lyases (PLs) or carbohydrate esterases (CEs), and sulfatases (http://abims.sb-roscoff.fr/sulfatlas/, [21]). In Bacteroidetes, CAZymes are usually organized within clusters of co-regulated genes involved in carbohydrate binding, hydrolysis and transport, known as polysaccharide utilization loci (PULs). The regulations of these PULs during purified algal substrate degradation were recently studied in a few transcriptome-wide analyses, for both cultivated marine bacteria [22–26] and natural seawater bacterial communities [27]. However, using unique substrates does not reflect the complexity of the responses that might occur during the degradation of intact algal biomass. The term “pioneer” has been previously introduced to describe bacteria specialized in the breakdown of intact polysaccharides and release of degradation products that can fuel so-called scavenger bacteria [19, 28–30]. In nature, some pioneers should therefore be able to initiate algal tissue degradation and expose new substrate niches for other community members. Yet, the metabolic strategies of such algae-degrading pioneer bacteria have been seldom studied despite their crucial ecological relevance. To our knowledge, no previous work investigated the mechanisms involved in the degradation of fresh macroalgae, hindering our understanding of algal biomass recycling in coastal habitats. To date information on the mechanisms involved in raw algal material assimilation is scarce. “Bacillus weihaiensis” Alg07T and Bacillus sp. SYR4 grow with kelp and red algal powder, respectively [23, 31] and Microbulbifer CMC-5 grows with thallus pieces of the red alga Gracilaria corticata [32]. These studies suggested a successive use of the different brown algal polysaccharides contained in the algal ECM [23] and the release of degradation product in the medium [31, 32].
The genus Zobellia (Flavobacteriaceae family), frequently found associated with macroalgae [33–35], is composed of 15 validly described strains classified in 8 species [36–39]. Their genomes encode numerous CAZymes (263–336 genes representing from 6.4 to 7.6% of the coding sequences), and sulfatases [40–42]. Therefore, Zobellia spp. are considered as potent algal polysaccharide degraders. In particular, Zobellia galactanivorans DsijT, isolated from a red macroalga [36, 43], is a model strain to study macroalgal polysaccharide utilization [44]. It allowed the discovery of many novel CAZymes and the description of new PULs targeting alginates [45–47], carrageenans [25], agars [48, 49], laminarin [50, 51], mix-linked glucan [52] and mannitol [53]. Its complete transcriptome analysis revealed common regulations triggered by polysaccharides from the same algal phylum [24]. Z. galactanivorans DsijT is also well equipped to cope with algal defenses and can accumulate iodine [42, 54, 55]. Moreover, a previous study suggested that Z. galactanivorans DsijT might act as pioneer bacteria by initiating the breakdown of the kelp Laminaria digitata, and demonstrated the crucial role of the alginate lyase AlyA1 in this process [56].
In this study, we aim to better understand the mechanisms controlling fresh macroalgae degradation. To tackle this issue, (i) the capacity of Z. galactanivorans DsijT to act as a pioneer in the utilization of fresh algal tissue and to favor the growth of non-pioneer taxa was tested to shed light on social bacterial behavior, (ii) the complete transcriptome of Z. galactanivorans DsijT was analyzed during the degradation of three brown macroalgae with distinct chemical composition and compared with purified sugars to decipher key genes and mechanisms specifically triggered with fresh tissues and (iii) the ability of Z. galactanivorans DsijT to degrade fresh algae tissues was compared with other Zobellia spp. to assess its singular role and hypothesize on potential genetic determinant in fresh macroalgae breakdown.
Experimental procedure
Purified substrates
Maltose (Sigma-Aldrich, St. Louis, MO, USA), alginate from Laminaria digitata (Danisco [ref. Grindsted FD176], Landerneau, France) and FCSP-enriched fraction (hereafter FCSPs) from Ascophyllum nodosum (Algues & Mer [HMWFSA15424, fraction >100 kDa], Ouessant, France) were tested for growth. Treatment of this commercial FCSP extract with the alginate lyase AlyA1 [46] followed by Carbohydrate-PAGE [57] revealed it contained alginate impurities. Colorimetric assays [58, 59] showed that uronic acids accounted for ~24% (w/w) of the FCSP extract. Based on previous measurements of 9% uronic acid content in pure FCSPs from A. nodosum [60], we therefore estimated the alginate contamination in the FCSP-enriched fraction to be ca. 15%. Alginate, agar (Sigma-Aldrich), kappa- (Goëmar, St. Malo, France) and iota-carrageenans (Danisco) were used for enzymatic assays.
Strains
Eight Zobellia strains were used in this study (listed in Supplementary Table 1, together with previous results of their ability to use pure algal compounds [36–38]), as well as Tenacibaculum aestuarii SMK-4T [61]. They were first grown in Zobell 2216 medium [62] at room temperature before inoculation in marine minimum medium (MMM) complemented with antibiotics to which all the tested strains are resistant (see supplementary methods for composition) and amended with 4 g l−1 maltose as the sole carbon source. Pre-cultures were centrifuged (3200 g, 10 min) and pellets washed twice in 1X saline solution. Cells were inoculated in microcosms at OD600 0.05. For the co-culture experiment, Zobellia galactanivorans DsijT and Tenacibaculum aestuarii SMK-4T were pre-cultured in Zobell 2216 medium only, as T. aestuarii does not grow in maltose-amended MMM.
Macroalgae treatment
All algae were harvested at the Bloscon site (48°43′29.982″N, 03°58′8.27″W) in Roscoff (France) between May 2019 and March 2021 depending on the experiment. They were cut in pieces (ca. 2.5–3.5 cm2) with a sterile scalpel and immersed in 0.1% Triton in milli-Q water for 10 min followed by 1% iodine povidone in milli-Q water for 5 min to clean them from resident epibionts. Finally, algal pieces were rinsed in excess autoclaved seawater for 2 h, to minimize algal exudates and metabolites that could have been produced upon cutting.
Microcosm set up and sampling
All experiments were performed at 20 °C in MMM supplemented with antibiotics and strains inoculated at an initial OD600 of 0.05. Z. galactanivorans DsijT was grown in 50 ml with 10 macroalgal pieces, either young Laminaria digitata (<20 cm), Fucus serratus or Ascophyllum nodosum. For comparison it was also grown in the same conditions using 4 g l−1 maltose, alginate or FCSPs. All conditions were performed in triplicates, except for F. serratus in duplicates. During the exponential phase (at 65 h for fresh algae, 24 h for maltose and 72 h for alginate and FCSPs), culture medium (10 ml) was retrieved on ice for RNA extraction from the free-living bacteria. On ice, 0.5 volume of killing buffer (20 mM Tris-HCl pH 7.5, 5 mM MgCl2, 20 mM NaN3) was added to the liquid samples and cell pellets were frozen in liquid nitrogen after centrifugation (3200 g, 10 min, 4 °C). In parallel, algae-attached cells were also recovered for RNA extraction, as detailed in Supplementary Methods.
To assess Z. galactanivorans growth when cultivated in contact or physically separated from algal tissues, incubations were performed in two-compartment vessels (100 ml each) with round bottom and a 65 mm flat edge opening (Witeg [ref. 0861050], Wertheim, Germany), separated by a 0.2 μm filter. Each compartment was filled with 30 ml of MMM and ten L. digitata pieces (meristem part, i.e. <15 cm from the base) were immersed in one.
Co-culture experiments were carried out in duplicates by inoculating Z. galactanivorans DsijT and T. aestuarii SMK-4T in 50 ml with 15 pieces of the L. digitata meristem as the sole carbon source. Culture medium was collected during the degradation to monitor the growth of the two partners using sequential CARD-FISH.
For comparative physiology, the eight Zobellia strains were grown in 10 ml with three L. digitata pieces from the meristem part.
RNA extraction and sequencing
Details of the protocols are available in Supplementary Methods. Briefly, free-living bacterial cells were lysed by incubation 5 min at 65 °C in lysis buffer (400 µl) and phenol (500 μl). After phenol-chloroform extraction, RNA was treated 1 h at 37 °C with 2 units of Turbo DNAse (ThermoFisher Scientific, Waltham, MA, USA), purified using NucleoSpin RNA Clean-up (Macherey-Nagel, Hoerdt, France) and eluted in 50 μl of nuclease-free water. This protocol was modified to extract RNA from algae-attached cells (see Supplementary Methods).
DNA contamination was checked by PCR with primers S-D-Bact-0341-b-S-17 and S-D- Bact-0785-a-A-21 targeting the 16S rRNA gene [63]. RNA was quantified using the Qubit RNA HS assay kit (ThermoFisher Scientific) and its integrity assessed on a Bioanalyzer 2100 (Agilent Technology, Santa Clara, CA, USA) with the Agilent RNA 6000 Pico kit.
Paired-end RNA sequencing (RNA-seq) was performed by the I2BC platform (UMR9198, CNRS, Gif-sur-Yvette) on a NextSeq instrument (Illumina, San Diego, CA, USA) using the NextSeq 500/550 High Output Kit v2 (75 cycles) after a Ribo-Zero ribosomal RNA depletion step. A total of 24 samples were sequenced (Supplementary Table 2).
RNA-seq analysis
Demultiplexed and adapter-trimmed reads were processed with the Galaxy platform (https://galaxy.sb-roscoff.fr). After read quality filtering using Trimmomatic v0.38.0, transcripts were quantified using the pseudo-mapper Salmon v0.8.2 [64] with the Z. galactanivorans DsijT reference genome (retrieved from the MicroScope platform ([65], https://mage.genoscope.cns.fr), “zobellia_gal_DsiJT_v2”; Refseq NC_015844.1). Raw counts for individual samples were merged into a single expression matrix for downstream analysis. Raw and processed data were deposited under GEO accession number GSE189322. Cleaned reads were also mapped on the Z. galactanivorans DsijT reference genome using Bowtie2 ([66], Galaxy Version 2.3.2.2). The mean per nt coverage for the whole transcriptome was assessed using SAMtools v1.14 [67] (Supplementary Table 2). The mean per nt coverage and normalized read counts (after DESeq2 normalization) for three selected characterized house-keeping genes in Z. galactanivorans DsijT [68] are shown in Supplementary Table 3, and the coverage map for these three genes and for the well-characterized alginate PUL were visualized using the Integrative Genomics Viewer v2.11.9 [69] and shown in Supplementary Fig. 1. These data ensure the expression variability was not caused by low read coverage or promiscuous read mapping. Principal Component Analysis (PCA) and differential abundance analyses were performed on rlog-transformed data using DESeq2 v1.26.0 package [70] in R v3.6.2 [71]. Genes displaying a log2 fold-change |log2FC | > 2 and a Bonferroni-adjusted p value < 0.05 were considered to be significantly differentially expressed. The upset plot was created using the ComplexUpset package [72, 73]. Hierarchical clustering was performed using the Ward’s minimum variance method [74]. Graphics were prepared using ggplot2 [75].
Enzymatic assays
One volume of 0.2 μm filtered supernatant from the microcosms was incubated with 9 volumes of 0.2% polysaccharide substrate at 28 °C overnight. Controls were prepared with boiled supernatants. The amount of reducing ends released was quantified using the ferricyanide assay [76]. For each sample, the activity measured in controls was subtracted. Finally, the mean value (n = 3) measured for the non-inoculated microcosms was subtracted. Significant differences (p < 0.05) from 0 were tested using t-tests.
CARD-FISH
The Zobellia-specific probe ZOB137 was described in Brunet et al. 2021 [35] and the Tenacibaculum-specific probe TEN281 was designed in the same fashion (see Supplementary Methods).
Algal pieces and culture medium were fixed overnight at 4 °C with 2% paraformaldehyde. Free-living bacteria were harvested on a 0.2 μm polycarbonate membrane. Catalyzed reporter deposition-fluorescence in situ hybridization (CARD-FISH) was performed as described in [35] using the Zobellia-specific probe ZOB137 with helpers. For sequential CARD-FISH on co-culture medium, first hybridization and amplification were done using the probe ZOB137 and the fluorochrome Alexa546. HRPs were inactivated in 3% H2O2 (10 min), followed by a second hybridization and amplification with the probe TEN281 and the fluorochrome Alexa488. Cells on membrane were visualized with a Leica DMi8 epifluorescent microscope (oil objective 63X). Cells on algal tissues were detected with a Leica TCS SP8 confocal microscope (HC PL APO 63X/1.4 oil objective) using the 488 and 638 nm lasers to detect Alexa488 signal and algal autofluorescence signal, respectively. Z-stack images were collected using 1024 × 1024 scan format (0.29 μm thick layers, 400 Hz scan speed) and visualized using the surface channel mode of the 3D viewer module (Leica Las X software). Following sequential CARD-FISH, Zobellia and Tenacibaculum cell counts were processed manually from 5 different fields at each time.
Comparative genomics
Zobellia genomes were screened for GHs, PLs, CEs and sulfatases using dbCAN2 [77] on the MicroScope platform. Homologs (>50% identity and >80% alignment) were searched for genes of interest using synteny results on MicroScope.
Results
Zobellia galactanivorans DsijT degrades fresh brown macroalgae tissues and benefits non-degrading bacteria
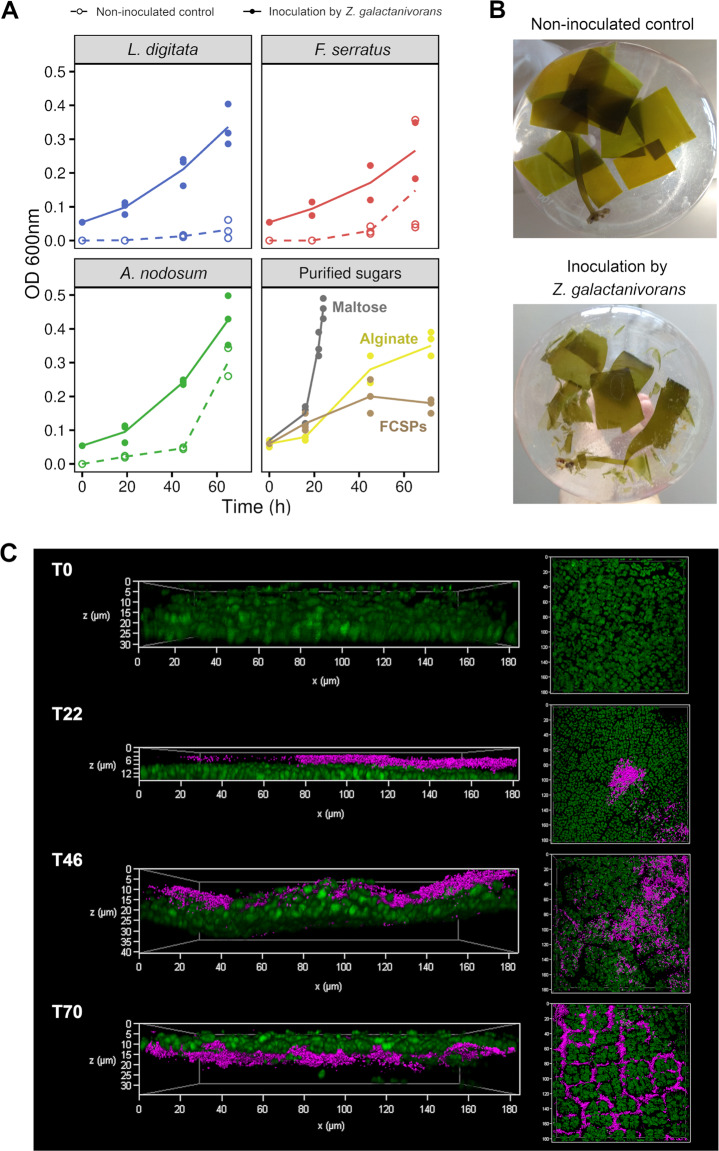
Z. galactanivorans growth was tested with three brown macroalgae from two different orders and with distinct chemical composition, Laminaria digitata (order Laminariales), Fucus serratus and Ascophyllum nodosum (order Fucales), as the sole carbon and energy source. Growth was detected with the three algal species (OD ≈ 0.2–0.5, Fig. 1A), with tissue bleaching and damages only visible on L. digitata pieces after 65 h (Fig. 1B). Zobellia-specific CARD-FISH assays revealed that even if antibiotic-resistant resident epibionts grew in the non-inoculated controls containing A. nodosum and F. serratus (one replicate), most of the bacterial biomass after 65 h in the Zobellia-inoculated microcosms was Zobellia cells (Supplementary Fig. 2).
Fig. 1. Z. galactanivorans DsijT is able to use different fresh brown macroalgae for its growth.
A Growth of Z. galactanivorans with either macroalgae pieces (Laminaria digitata, Fucus serratus and Ascophyllum nodosum) or purified sugars (maltose, alginate and FCSPs). Individual points for replicate experiments are shown. Lines are means of independent replicates (n = 2 or n = 3). B Photographs showing the integrity of the L. digitata tissues after 65 h. C L. digitata tissues colonization by Z. galactanivorans during the degradation. Micrographs are overlay of the CARD–FISH signal (magenta, Zobellia-specific probe with Alexa488 as the reporter signal) and the algal autofluorescence (green) and were obtained with the surface channel mode of the 3D viewer. For the different times, transversal views are shown on the left and top views on the right. The non-fluorescent gap between the bacterial cells and the algal cells likely represents the mucilage coat of L. digitata. The absence of algal autofluorescence signal below 25–30 µm is the result of its rapid decrease in intensity as we move away from the coverslip.
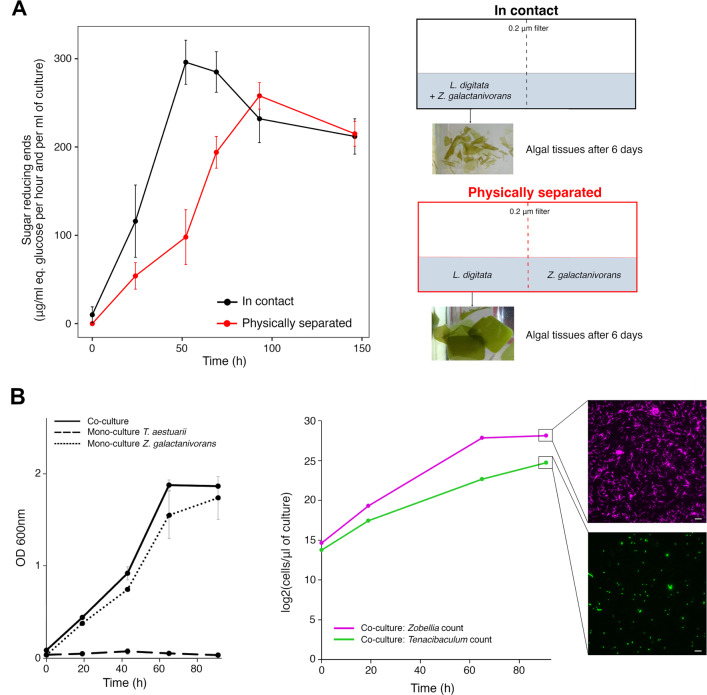
CARD-FISH assays on L. digitata tissues showed gradual tissue colonization by Z. galactanivorans, from cell patches at the surface of the L. digitata mucilage coat to deeper penetration within the tissue invading the intercellular space (Fig. 1C). To assess if algal degradation requires cell attachment, Z. galactanivorans was grown either in contact or physically separated from algal pieces (Fig. 2A). After 6 days, algal tissues were visually starting to decompose when bacteria were separated from algae, although to a lesser extent compared to the “contact” condition. Furthermore, extracellular alginolytic activity increased even without physical bacteria/algae contact and reached similar levels to that observed in the “contact” condition after 90 h. We further tested if this degradation behavior of Z. galactanivorans could lead to cooperative interactions with non-degrading bacteria. As a test case, we used the flavobacterium Tenacibaculum aestuarii that was not able to feed on fresh L. digitata pieces or initiate its degradation (Fig. 2B). When T. aestuarii was cultivated together with Z. galactanivorans, the optical density of the co-culture was between 7 and 23% higher than in the Z. galactanivorans monoculture. Sequential CARD-FISH analyses indicated an exponential growth of T. aestuarii during algal degradation in the co-culture microcosms, reaching 2.8 × 107 cells µl−1 after 91 h. T. aestuarii density was ~10 times lower and delayed compared to Z. galactanivorans. This delay suggests that T. aestuarii is able to grow after the accumulation of degradation products or secondary metabolites in the medium.
Fig. 2. Macroalgae degradation by Z. galactanivorans DsijT allows the growth of other taxa.
A Alginolytic activity of the enzymes secreted when Z. galactanivorans was grown in contact with L. digitata (black) or separated from L. digitata by a 0.2 µm filter (red). The activity was measured in each compartment (left and right) and summed. Values are mean ± s.d. (n = 3). B Cooperative interactions between Z. galactanivorans DsijT and T. aestuarii SMK-4T. Left: Co-culture and monoculture growth curve with L. digitata as sole carbon source. Values are replicate mean ± s.d. (n = 2). Right: Zobellia (magenta) and Tenacibaculum (green) cell concentration during the co-culture experiment. Cell counts were performed from 5 different microscopic fields of micrographs obtained after sequential CARD-FISH on one replicate. One microscopic field from the sample collected at 91 h is shown, showing either the Zobellia (magenta signal) or the Tenacibaculum (green signal) signal. Scale = 10 μm.
The transcriptomic profile of free-living bacteria shifts during fresh macroalgae degradation
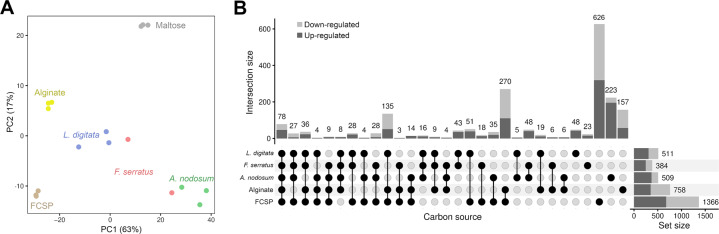
Z. galactanivorans DsijT transcriptome of free-living cells obtained during macroalgal degradation was compared to the responses occurring with a disaccharide, maltose, and with purified brown algal polysaccharides, alginate and FCSPs. Between 44 and 93% of the sequenced reads from free-living bacteria grown with macroalgae mapped on the genome of Z. galactanivorans DsijT (Supplementary Table 2). Multivariate analysis separated samples according to carbon source (Fig. 3A). Transcriptomes of cells grown with L. digitata were closer to that obtained with alginate or FCSPs compared to A. nodosum or F. serratus.
Fig. 3. General features of the transcriptomic responses occurring in free-living Z. galactanivorans DsijT during growth with macroalgae.
A Principal Component Analysis of the gene expression. B Upset plot of the differentially expressed genes with maltose as the control condition (Bonferroni-adjusted p value < 0.05 and |log2FC | > 2). Set size represents the total amount of genes regulated in each condition.
Differential abundance analysis revealed 1117 and 864 genes up- and down-regulated with at least one substrate, using maltose as control (Supplementary Table 4). Among them, 56% (628 upregulated genes) and 52% (449 downregulated genes) showed substrate-specific regulations (Fig. 3B). In particular, half of the genes regulated with A. nodosum and FCSPs were not differentially expressed in any other conditions. L. digitata was the algal species inducing the highest number of regulated genes shared with at least one polysaccharide (399, 254 and 217 genes with L. digitata, F. serratus and A. nodosum respectively). More regulations were shared between L. digitata and F. serratus (116 genes) than F. serratus and A. nodosum (89 genes) or L. digitata and A. nodosum (13 genes). Finally, a core set of 70 upregulated and 59 down-regulated genes responded to the three macroalgae.
Carbohydrate catabolism-related genes
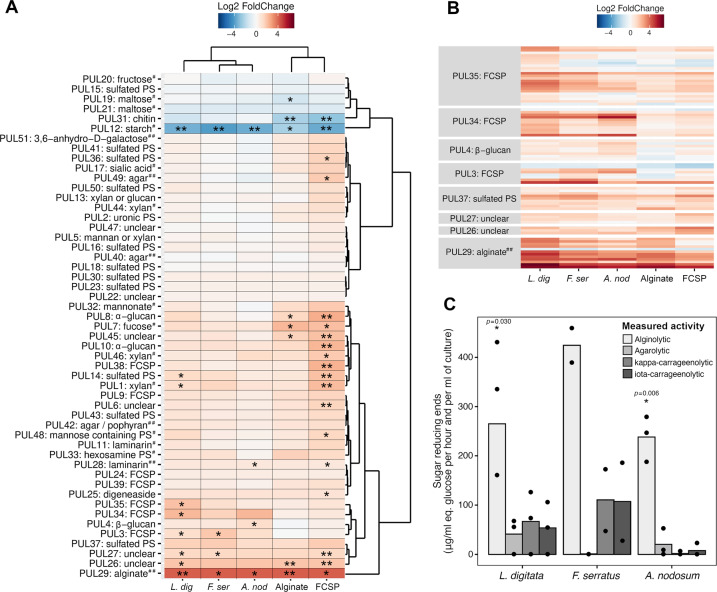
Hierarchical clustering of expression data of the 51 identified PULs in the Z. galactanivorans DsijT genome revealed that PULs predicted to target brown algal polysaccharides grouped together (Fig. 4A) and were significantly induced with macroalgae. In particular, the alginate-specific PUL29 was significantly overexpressed in all conditions compared with maltose (mean log2FC of 4) and the highest expression was observed with L. digitata (Fig. 4B, Supplementary Fig. 3). Some PULs were exclusively triggered by macroalgae: PUL34 and 35, likely targeting FCSPs (as they encode sulfatases and fucosidases), were significantly triggered by L. digitata, PUL4 targeting β-glucan responded to A. nodosum and the FCSP PUL3 was induced by both L. digitata and F. serratus. PUL26 and 27, whose function remains unclear, were both induced by L. digitata and FCSPs, as well as by alginate for PUL26 and F. serratus for PUL27. FCSPs also induced the expression of 14 PULs outside the described cluster, encompassing a large diversity of targeted substrate (notably β- and α-glucan, sulfated polysaccharides, xylan, unclear substrate). No PUL known to target red algal polysaccharides (e.g. PUL40, 42, 49 or 51) clustered with this set of overexpressed PULs, suggesting a specific induction of brown algal polysaccharide degradation mechanisms in the presence of brown algal tissues. The measured activity of secreted polysaccharidases corroborates this observation (Fig. 4C), as only the alginolytic activity was significantly higher when Z. galactanivorans was grown on macroalgae compared with the non-inoculated control (t-test, p < 0.05).
Fig. 4. Specific upregulation of pathways involved in brown algae carbohydrate catabolism during the utilization of fresh brown algal tissues.
A Heatmap of the 51 PULs identified in the genome of Z. galactanivorans DsijT. PUL 1 to 50 were identified during the annotation of the Z. galactanivorans DsijT genome by the presence of the susCD-like pair, their boundaries are based on bioinformatic predictions (Supplementary Table S3 in [42]). PUL51 targeting 3,6-anhydro-D-galactose and involved in carrageenan catabolism (but lacking the susCD-like pair) was further described [25]. For each PUL, the mean log2FC of all genes is represented, taking maltose as a control condition. Carbon sources and PULs were arranged according to a hierarchical clustering analysis (Ward’s method). A PUL was considered regulated (induced in red, repressed in blue) if more than 50% of the genes were significantly differentially expressed (*) and strongly regulated if more than 80% of the genes were significantly differentially expressed (**). Putative substrates targeted by the PULs are indicated. Hash signs denote PULs biochemically characterized previously in Z. galactanivorans (##) or in another organism (#). B Heatmap representing the log2FC of individual genes contained in the PULs induced with macroalgae and which clustered together in A. C Activity of extracellular polysaccharidases collected in the microcosms containing macroalgae after 65 h. The mean value measured in the uninoculated controls was subtracted from each value. Bars are means of independent replicates (n = 2 or 3) shown as individual points. Significant difference from zero was tested when n = 3 (t-test; *p < 0.05). L. dig: Laminaria digitata; F. ser: Fucus serratus; A. nod: Ascophyllum nodosum; FCSP: fucose-containing sulfated polysaccharide; PS: Polysaccharide.
On the other hand, PULs targeting simple sugars (maltose and fructose) or polysaccharides absent from brown algae (starch and chitin) were repressed with macroalgae and purified polysaccharides (Fig. 4A). The starch PUL12 was strongly under-expressed in all conditions while the chitin PUL31 showed a significant repression only with algal polysaccharides.
Specific induction with fresh algal tissues
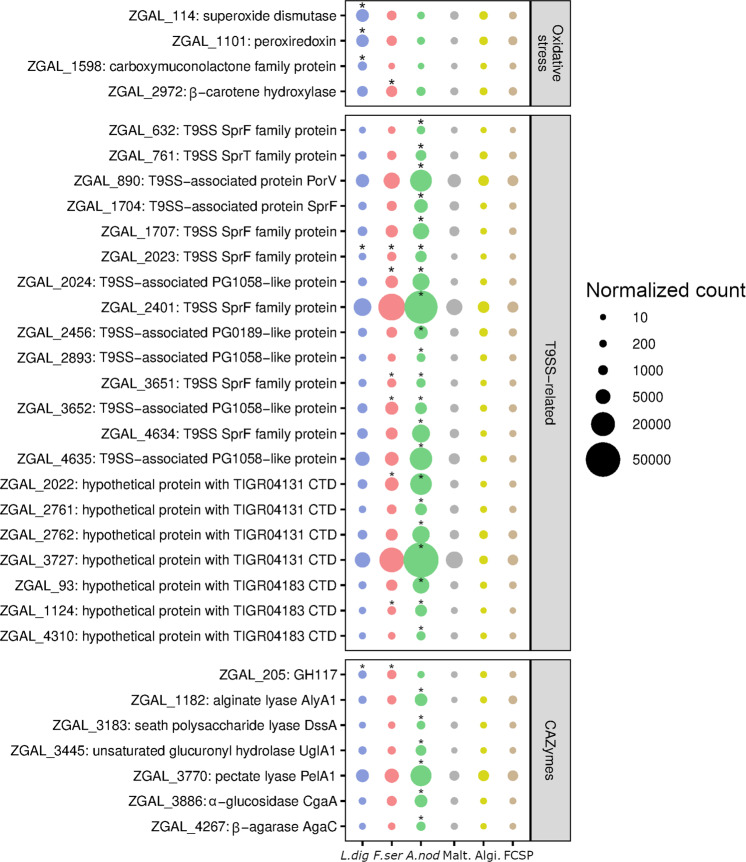
To unravel pathways specifically governing the degradation of fresh macroalgal biomass, we further focused on genes upregulated with at least one macroalgal species compared to maltose and purified polysaccharides. We detected 41, 59 and 189 genes following this pattern with L. digitata, F. serratus or A. nodosum, respectively (Supplementary Table 5). It included few CAZyme-encoding genes (Fig. 5), notably two genes within putative FCSP PULs (zgal_205 [GH117 in PUL3] and zgal_3445 [GH88 in PUL34]). Other polysaccharidase genes outside classical PUL structures were induced with A. nodosum, such as alyA1 (zgal_1182, alginate lyase PL7), cgaA (zgal_3886, glucan 1,4-α-glucosidase GH15), agaC (zgal_4267, β-agarase GH16), pelA1 (zgal_3770, pectate lyase PL1) and dssA (zgal_3183, sheath polysaccharide lyase PL9). GT2 (zgal_2991, 4154) and GT4 (zgal_2990, 3759) were also triggered with macroalgae. Additionally, many genes linked to oxidative stress responses and Type IX secretion systems (T9SS) were specifically induced with macroalgae (Fig. 5). A large gene cluster (zgal_1071-1105) notably encoding three oxidoreductases, a DNA topoisomerase and a peroxiredoxin was upregulated with L. digitata and F. serratus. Other genes encoding antioxidant proteins were triggered, especially on L. digitata, such as the superoxide dismutase SodC (ZGAL_114) or a β-carotene hydroxylase (ZGAL_2972), as well as a carboxymuconolactone decarboxylase family protein (ZGAL_1598) which includes enzyme involved in antioxidant defense [78]. Two catalases (ZGAL_1427 and ZGAL_3559) were induced in the presence of L. digitata and F. serratus in comparison to maltose and alginate (Supplementary Table 5). Despite the poor sequencing depth of RNA extracted from algae-attached cells (Supplementary Table 2), the induction of stress resistance mechanisms tended to be even more pronounced in algae-attached bacteria compared with the free-living ones, especially through the expression of chaperones (Supplementary Fig. 4).
Fig. 5. Genes related to oxidative stress response and T9SS are specifically induced by fresh tissue.
Mean expression values (n = 3, except for F. serratus n = 2) of selected genes significantly triggered (*) with at least one macroalgae compared to both the purified polysaccharides and maltose (see Supplementary Table 4). L. dig: Laminaria digitata; F. ser: Fucus serratus; A. nod: Ascophyllum nodosum; Malt.: Maltose; Algi.: Alginate; FCSP: fucose-containing sulfated polysaccharide.
Several genes predicted to encode T9SS components were significantly induced during macroalga degradation, in particular with A. nodosum (14 out of 33 genes identified in the genome, against 1 and 5 with L. digitata and F. serratus respectively) (Fig. 5). They include particularly genes encoding SprF family proteins and T9SS-associated PG1058-like proteins. In addition, 7 unknown proteins containing a conserved C-terminal domain (CTD) from families TIGR04131 (gliding motility - ZGAL_2022, 2761, 2762, 3727) and TIGR04183 (Por secretion system - ZGAL_93, 1124, 4310) were triggered. These CTDs are typical of cargo proteins secreted by the T9SS.
Comparative physiology and genomics of fresh macroalga degradation by Zobellia
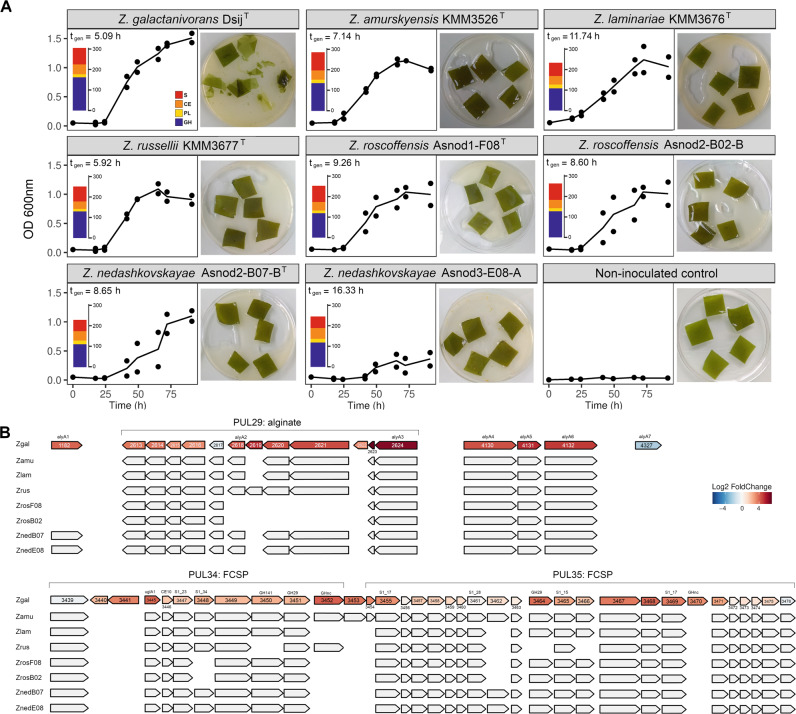
The degrading abilities of other members of the genus Zobellia were investigated (Fig. 6A). All tested Zobellia strains used fresh L. digitata tissues for their growth. Z. galactanivorans DsijT had the highest final cell density (OD600 = 1.5) and shortest generation time (5.09 h). Z. nedashkovskayae Asnod3-E08-A formed cell aggregates that biased OD600 readings, likely explaining the apparent limited growth (final OD600 = 0.4) and long generation time (tgen = 16.33 h). Other strains showed intermediate behaviors (OD600 ≈ 1, 5.92 < tgen < 11.74 h). These growth differences were reflected in the final aspect of macroalgal pieces. Only Z. galactanivorans DsijT completely broke down algal tissues after 91 h. Both Z. nedashkovskayae strains caused limited algal peeling and breakdown at the corners of the pieces., No visible trace of degradation was detected for other strains. A strong negative correlation was found between the number of GHs and the generation time (Spearman, rho = −0.90; p = 0.006) (Fig. 6A, Supplementary Table 6). Twenty-two out of the 305 genes upregulated by Z. galactanivorans DsijT with L. digitata compared to maltose had no homologs in the genome of the seven other Zobellia strains (Supplementary Table 7). They include two GHs, zgal_3349 (GH20 in PUL33) and zgal_3470 (GHnc in PUL35), and a susCD-like pair (zgal_3440, 3441) in PUL34. Other upregulated genes within the FCSP PUL34/35 are not conserved in all Zobellia strains (Fig. 6B). Likewise, several alginolytic genes were not conserved across the genus, especially in the two Z. roscoffensis strains that lack 7 of them. zgal_1182 and zgal_4327, encoding the extracellular endo-alginate lyases AlyA1 and AlyA7 respectively, were not conserved in the other strains (zgal_4327) or only found in the Z. nedashkovskayae strains (zgal_1182). Two other genes related to carbohydrate assimilation (zgal_334 and zgal_2296 encoding a GHnc and a lipoprotein with CBM22, respectively) are missing in five strains (Supplementary Table 7). zgal_334 neighbors genes encoding sulfatases, fucosidases and PLs and might belong to a FCSP-targeting cluster (absent from the 51 identified PULs as the pair susCD-like is absent).
Fig. 6. All Zobellia spp. use fresh L. digitata but Z. galactanivorans DsijT displays the highest tissue breakdown capacity.
A Growth of eight Zobellia strains with L. digitata pieces (meristem of adult individuals). The generation time tgen is indicated for each strain as well as the number of glycoside hydrolases (GH, blue), polysaccharides lyases (PL, yellow), carbohydrate esterases (CE, orange) and sulfatases (S, red) predicted in their genome (dbCAN search on the MaGe platform). Individual points for duplicate experiments are shown. Lines are means of independent replicates (n = 2 or n = 3). B Comparison of genomic loci among the eight Zobellia strains. For Z. galactanivorans, genes were colored according to their expression log2FC for the comparison L. digitata vs. maltose. Gene ID is indicated inside arrows and CAZymes and sulfatases are specified above. Top: genes involved in the alginate-utilization system. Bottom: genes contained in putative FCSP PUL34 and 35. Zgal: Z. galactanivorans DsijT; Zamu: Z. amurskyensis KMM 3526 T; Zlam: Z. laminariae KMM 3676 T; Zrus: Z. russellii KMM 3677 T; ZrosF08: Z. roscoffensis Asnod1-F08T; ZrosB02: Z. roscoffensis Asnod2-B02-B; ZnedB07: Z. nedashkovskayae Asnod2-B07-BT; ZnedE08: Z. nedashkovskayae Asnod3-E08-A.
Discussion
Zobellia galactanivorans DsijT: a sharing pioneer role during brown macroalgae utilization
By degrading macroalgae, marine heterotrophic bacteria are central to nutrient cycling in coastal habitats. The ecological strategies of different functional guilds not equally equipped to process biomass were recently conceptualized [19, 28, 29]. First, pioneer bacteria degrade complex organic matter by producing specific hydrolytic enzymes. The hydrolysate can then fuel other bacteria called exploiters or scavengers, which cannot feed on intact substrates. Such cooperative interactions were previously characterized during alginate [79] or chitin [80, 81] assimilation. Hence, in nature pioneer bacteria likely control the initial attack on fresh macroalgae, a hitherto rarely studied process that cannot be fully deciphered when using purified polysaccharides or crushed algae. Here, we showed that Z. galactanivorans DsijT uses fresh brown algal tissues for its growth, highlighting its pioneer role in algal biomass recycling. Similar growth rates were observed with three brown algal species, and Z. galactanivorans completely broke down L. digitata tissues. Transcriptomes obtained with L. digitata were closest to that with alginate and FCSP, suggesting a greater capacity to access and digest ECM polysaccharides within the L. digitata tissues compared to A. nodosum and F. serratus. The limited degradation of Fucales tissues might originate from their higher phlorotannin content [82], possibly inhibiting CAZymes [83]. In addition, A. nodosum induced a wider cellular response with many specific regulations. This might partly be due to the growth of antibiotic-resistant epiphytic bacteria that could have affected Z. galactanivorans behavior or to its much thicker and rigid thallus. Furthermore, A. nodosum is associated with various symbionts, especially the obligate endophytic fungus Mycophycias ascophylli [84] that secretes compounds potentially preventing tissue grazing and/or offering additional substrate niches.
We showed that although Z. galactanivorans can colonize L. digitata, it does not require a physical contact to initiate algal breakdown. This suggests a crucial role for secreted enzymes during the first stages of the degradation, in line with the measured extracellular alginolytic activity. Constitutively expressed extracellular enzymes, such as the alginate lyases AlyA1 and AlyA7 acting as sentry enzymes, would rapidly release diffusible degradation products, allowing remote substrate sensing [45]. We previously showed that when grown with purified alginate or algal tissues, Z. galactanivorans accumulates low molecular weight (LMW) alginate oligosaccharides that act as effectors for the expression of the alginolytic PUL [45, 56]. The release of degradation products could possibly govern cooperative interactions between Z. galactanivorans and other taxa. We specifically demonstrated here that Z. galactanivorans was able to support the growth of a Tenacibaculum species during algal tissue utilization, as first suggested in a previous study [56]. Therefore, we propose that Z. galactanivorans would behave as a “sharing” pioneer: by initiating algal breakdown, it would provide degradation products as public goods to opportunist taxa, contrary to “selfish” pioneers which sequester LMW products by producing essentially surface-associated hydrolytic enzymes with minor loss of hydrolysate to the medium [85, 86]. Tenacibaculum spp. were regularly reported on macroalgae [87], although their role in macroalgae degradation remains unclear. Members of the genus Tenacibaculum were identified as active incorporators of carbon from alginate within natural seawater communities [88]. The number of CAZymes is highly variable within the genus (between 34 and 102 for the 8 Tenacibaculum genomes available in the CAZy database) but is largely below the 221 CAZymes found in Z. galactanivorans DsijT [20]. Tenacibaculum spp. might then lack key genetic determinants to initiate the algal breakdown but are likely able to use processed algal compounds. On the other hand, the benefits of cooperative interactions for sharing pioneers remain elusive. Indeed, studies on chitin degradation by marine bacterial communities suggest that the presence of non-degrading organisms decreases degradation rates and overall productivity, possibly due to competition for space and/or nutrients [89, 90]. Yet, the benefits of cooperative interactions might be more relevant in limiting conditions, if scavengers produce one or several public goods (e.g. vitamins, siderophores) to be used by pioneers [91]. Additionally, scavengers might hinder the growth of other competing microorganisms that may have negative interactions with pioneers.
We further showed that the pioneer behavior can be strain-specific within the alga-associated genus Zobellia. All Zobellia spp. tested successfully grew with fresh L. digitata but without causing pronounced tissues damages as observed with Z. galactanivorans. Their catabolic profiles (Supplementary Table 1) indicate different growth capacities with purified brown algal sugars. For example, Z. roscoffensis strains and Z. laminariae KMM 3676 T display limited or no abilities to use alginate, FCSPs and laminarin for their growth. Hence, with macroalgae, they likely did not use these complex polysaccharides but rather fed on soluble algal exudates (e.g. mannitol). At least some of the tested Zobellia strains are known to co-occur in nature: Z. roscoffensis and Z. nedashkovskayae strains were isolated simultaneously from brown algae in Roscoff, and Z. galactanivorans DsijT was retrieved from the same location 30 years earlier. Therefore, different Zobellia species likely stably coexist at the surface of algae and their contrasted behavior towards fresh algal biomass degradation suggests they might not compete for the same resources, thus occupying different ecological niches. Some species, especially Z. galactanivorans DsijT, would be well adapted to breakdown fresh brown algae, while others would exploit soluble carbon sources such as dissolved polysaccharides, algal exudates (e.g. mannitol), degradation products (oligosaccharides, secondary metabolites) or non-carbohydrate cell wall compounds (e.g. proteins). Comparative genomics suggested that CAZyme content influences the strain capacity to use and break down fresh algal tissues. The highest number of CAZymes in Z. galactanivorans DsijT could explain its greater success at degrading algae. In particular, some Zobellia strains lack homologs of overexpressed genes contained in L. digitata-induced PULs targeting alginate or FCSPs. For example, alyA1 homologs were only found in the two other strains that caused visible algal damage (Z. nedashkovskayae Asnod2-B07-BT and Asnod3-E08-A). Accordingly, alyA1 is known to have a crucial role in initiating algae breakdown [56]. Such genes would therefore represent potential genetic determinants of pioneer bacteria. Yet, Z. galactanivorans DsijT lacks key genes to use some specific algal compounds, including particular FCSP structures (e.g. FCSP from Pelvetia canaliculata) or ulvans from green algae [42]. When faced with such compounds, Z. galactanivorans DsijT might therefore benefit from other highly specialized taxa possessing complete degradation pathways, such as members of the verrucomicrobium genus ‘Lentimonas’ [92] or the flavobacterium Formosa agariphila KMM 3901T [93] for FCSP and ulvan degradation respectively. This and other differences in gene content or regulatory programs between co-occurring strains could also explain the maintenance of a diverse pioneer bacteria population potentially acting as a consortium towards the complete breakdown of macroalgae.
Deciphering the metabolic mechanisms involved in fresh tissue breakdown, including new catabolic pathways
Regardless of the algal species, the well-characterized alginolytic PUL29 was the most induced among all PULs. Alginate is the most abundant polysaccharide in brown algal ECM and likely the most accessible as it embeds the cellulose-FCSP network [11]. This PUL was particularly triggered with L. digitata, likely reflecting the higher alginate content in this species [2] and/or easier substrate accessibility. Furthermore, several uncharacterized PULs were triggered with macroalgae. In particular, three out of the seven predicted FCSP PULs were significantly upregulated with macroalgae but not with extracted A. nodosum FCSPs, and to various degrees depending on algal species. This suggests different substrate specificities, consistent with the large structural diversity of FCSPs and cross-linkage to other compounds [4, 7, 94] that might not be equally extracted during purification. By preserving the original polysaccharide structure and environment, the study of fresh macroalga degradation may therefore be a more effective way to reveal specific genes crucial for macroalgae breakdown by pioneer bacteria but undetectable when using purified polysaccharides.
By contrast to alginate- and FCSP-targeting PULs, the characterized laminarin PUL11 and PUL28 were poorly regulated with the three algae. An uncharacterized β-glucan PUL4 was significantly induced only with A. nodosum, and also found triggered with purified laminarin in a previous study [24]. As raised above, the presence of endosymbionts in A. nodosum could result in specific laminarin structures that might be targeted by PUL4. The absence of induction of typical laminarin PULs with macroalgae might also indicate that Z. galactanivorans DsijT first uses ECM polysaccharides and later access intracellular storage polysaccharides. Such a prioritization of multiple substrates within algal material was previously observed for Bacillus weihaiensis Alg07T grown on algal powder [23]. Koch et al. [26] showed that Alteromonas macleodii 83-1 prioritized laminarin over alginate and pectin when grown on a mixture of purified polysaccharides. Thus, prioritization might differ between bacterial strains and whether substrates are under soluble form or within algal tissues, underlining the importance to consider intact macroalgae to understand the pioneer behavior. Furthermore, future time-resolved transcriptome analyses could inform on regulations at different degradation stages and help decipher prioritization effects.
Besides carbohydrate utilization, our approach unveiled several traits specifically induced upon macroalgal degradation and potentially linked to the pioneer behavior, including the resistance to algal defense and T9SS. One of the algal defense mechanisms is the production of reactive oxygen species (ROS), which in L. digitata is partly induced by endogenous elicitors (i.e. oligo-alginates) derived from the degradation of their own cell wall [95]. Breakdown of L. digitata tissues by Z. galactanivorans likely produced large amounts of elicitors, triggering a massive accumulation of ROS in the closed microcosm setup, in line with the strong induction of genes encoding ROS-detoxifying enzymes in this condition. In contrast, A. nodosum and F. serratus do not respond to the addition of endogenous elicitors [96], potentially explaining the lower induction of antioxidant pathways in Z. galactanivorans DsijT with these algae. To our knowledge, this is the first time ROS detoxification is shown as an important component of macroalgae degradation by marine bacteria. It is reminiscent of previous results showing the induction of oxidative stress responses in plant-associated terrestrial bacteria [97]. Another algal defense response is the emission of halogenated compounds. One vanadium-dependent iodoperoxidase (vIPO3) and a haloacid dehalogenase (HAD, [55]) were significantly upregulated with A. nodosum compared to alginate and maltose respectively. HAD expression was also 3-fold higher with L. digitata and F. serratus compared to maltose, although large variations precluded significance. Overall, our results suggest that pioneer bacteria might have evolved to cope with increasing stress levels upon algal degradation. Such metabolization of toxic compounds might also be a hitherto overlooked additional benefit that sharing pioneer bacteria provide to less stress-resistant scavengers. For example, Grigorian et al., 2021 [55] revealed that Tenacibaculum is one of the two genera within the Flavobacteriaceae family lacking Type II haloacid dehalogenase and that the growth of Tenacibaculum strains was inhibited by iodo and bromoacetic acid, while Z. galactanivorans DsijT was more tolerant. Hence, besides the sharing of degradation products, detoxification of algal defense compounds by Z. galactanivorans might partly explain the cooperative interaction we showed here with T. aestuarii. Further investigations must be pursued to characterize Z. galactanivorans interactions with its opportunistic partners and assess whether they rely on public good secretion, cross-feeding interactions and/or even physical mechanisms.
Specific to Bacteroidetes, T9SS is involved in biofilm formation, protease virulence factors delivery and secretion of polysaccharidases and cell-surface gliding motility adhesins [98, 99]. Here, we showed that growth with macroalgae strongly induced genes encoding T9SS components, T9SS-translocated proteins and several glycosyl transferases from families GT2 and GT4. Glycosyltransferases with a GT4_CapM-like domain were recently shown to N-glycosylate CTD in Cytophaga hutchinsonii, an essential step for the recognition of cargo proteins by T9SS [100]. Hence, our data suggest T9SS might be a key determinant of pioneer behavior for some members of the Bacteroidetes phylum, to secrete ECM-targeting CAZymes and/or attach to macroalgal surfaces. Only induced in the presence of algal tissues, this T9SS system might not be triggered by oligo-alginate or oligo-FCSP, but rather by other algal metabolites, such as ROS.
Conclusion
This study provides the first insights into the metabolic strategies of sharing pioneer bacteria during fresh macroalgae utilization and represents a source of potential genetic determinants for further functional characterization. Altogether, our results raised the relevance to consider the full complexity of whole macroalgae tissues in further degradation studies, as it would take a step forward in the understanding of the algal biomass recycling through the identification of new metabolic pathways or the characterization of bacterial cooperative interactions. Integrative time-series investigations would be particularly helpful to bring a more comprehensive view of the strategies taking place during algal breakdown.
Supplementary information
Acknowledgements
The authors thank Tatiana Rochat for advice during transcriptomic analyses, Sébastien Colin for guidance in confocal manipulation, Bernhard Fuchs and Rudolf Amann for guidance in designing and testing the TEN281 probe, Philippe Potin and Cécile Hervé for helpful discussions and Yan Jaszczyszyn from the I2BC sequencing platform. This work has benefited from the facilities of the Genomer platform and from the computational resources of the ABiMS bioinformatics platform (FR 2424, CNRS-Sorbonne Université, Roscoff), which are part of the Biogenouest core facility network. This work was funded by the French Government via the National Research Agency programs ALGAVOR (ANR-18-CE02-0001-01) and IDEALG (ANR-10-BTBR-04).
Author contributions
Author contributions following the CRediT taxonomy (https://casrai.org/credit/) are as follows: Conceptualization: FT, TB, MB; Formal analysis: FT, MB; Funding acquisition: FT; Investigation: FT, NLD, MB, TB; Project administration: FT; Visualization: FT, MB; Writing—original draft: MB; Writing—review and editing: FT, MB, TB.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-022-01251-6.
References
- 1.Duarte C, Middelburg JJ, Caraco N. Major role of marine vegetation on the oceanic carbon cycle. Biogeosciences. 2005;2:1–8. doi: 10.5194/bg-2-1-2005. [DOI] [Google Scholar]
- 2.Kloareg B, Quatrano RS. Structure of the cell walls of marine algae and ecophysiological functions of the matrix polysaccharides. Ocean Mar Biol Annu Rev. 1988;26:259–315. [Google Scholar]
- 3.Fletcher HR, Biller P, Ross AB, Adams JMM. The seasonal variation of fucoidan within three species of brown macroalgae. Algal Res. 2017;22:79–86. doi: 10.1016/j.algal.2016.10.015. [DOI] [Google Scholar]
- 4.Deniaud-Bouët E, Hardouin K, Potin P, Kloareg B, Hervé C. A review about brown algal cell walls and fucose-containing sulfated polysaccharides: Cell wall context, biomedical properties and key research challenges. Carbohydr Polym. 2017;175:395–408. doi: 10.1016/j.carbpol.2017.07.082. [DOI] [PubMed] [Google Scholar]
- 5.Haug A, Larsen B, Smidsrød O. Uronic acid sequence in alginate from different sources. Carbohydr Res. 1974;32:217–225. doi: 10.1016/S0008-6215(00)82100-X. [DOI] [Google Scholar]
- 6.Bruhn A, Janicek T, Manns D, Nielsen MM, Balsby TJS, Meyer AS, et al. Crude fucoidan content in two North Atlantic kelp species, Saccharina latissima and Laminaria digitata—seasonal variation and impact of environmental factors. J Appl Phycol. 2017;29:3121–3137. doi: 10.1007/s10811-017-1204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ponce NMA, Stortz CA. A comprehensive and comparative analysis of the fucoidan compositional data across the Phaeophyceae. Front Plant Sci. 2020;11:556312. doi: 10.3389/fpls.2020.556312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleurence J. The enzymatic degradation of algal cell walls: A useful approach for improving protein accessibility? J Appl Phycol. 1999;11:313–314. doi: 10.1023/A:1008183704389. [DOI] [Google Scholar]
- 9.Verhaeghe EF, Fraysse A, Guerquin-Kern JL, Wu TD, Devès G, Mioskowski C, et al. Microchemical imaging of iodine distribution in the brown alga Laminaria digitata suggests a new mechanism for its accumulation. J Biol Inorg Chem. 2008;13:257–269. doi: 10.1007/s00775-007-0319-6. [DOI] [PubMed] [Google Scholar]
- 10.Schiener P, Black KD, Stanley MS, Green DH. The seasonal variation in the chemical composition of the kelp species Laminaria digitata, Laminaria hyperborea, Saccharina latissima and Alaria esculenta. J Appl Phycol. 2015;27:363–373. doi: 10.1007/s10811-014-0327-1. [DOI] [Google Scholar]
- 11.Deniaud-Bouët E, Kervarec N, Michel G, Tonon T, Kloareg B, Hervé C. Chemical and enzymatic fractionation of cell walls from Fucales: Insights into the structure of the extracellular matrix of brown algae. Ann Bot. 2014;114:1203–1216. doi: 10.1093/aob/mcu096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michel G, Tonon T, Scornet D, Cock JM, Kloareg B. Central and storage carbon metabolism of the brown alga Ectocarpus siliculosus: Insights into the origin and evolution of storage carbohydrates in Eukaryotes. N. Phytol. 2010;188:67–81. doi: 10.1111/j.1469-8137.2010.03345.x. [DOI] [PubMed] [Google Scholar]
- 13.Mann K. Ecology of coastal waters—A systems approach, Berkeley: University of California Press; 1982.
- 14.Egan S, Harder T, Burke C, Steinberg P, Kjelleberg S, Thomas T. The seaweed holobiont: Understanding seaweed-bacteria interactions. FEMS Microbiol Rev. 2013;37:462–476. doi: 10.1111/1574-6976.12011. [DOI] [PubMed] [Google Scholar]
- 15.Kirchman DL. The ecology of Cytophaga-Flavobacteria in aquatic environments. FEMS Microbiol Ecol. 2002;39:91–100. doi: 10.1111/j.1574-6941.2002.tb00910.x. [DOI] [PubMed] [Google Scholar]
- 16.Thomas F, Hehemann JH, Rebuffet E, Czjzek M, Michel G. Environmental and gut Bacteroidetes: The food connection. Front Microbiol. 2011;2:93. doi: 10.3389/fmicb.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM, et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 2012;336:608–611. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 18.Wietz M, Wemheuer B, Simon H, Giebel HA, Seibt MA, Daniel R, et al. Bacterial community dynamics during polysaccharide degradation at contrasting sites in the Southern and Atlantic Oceans. Environ Microbiol. 2015;17:3822–3831. doi: 10.1111/1462-2920.12842. [DOI] [PubMed] [Google Scholar]
- 19.Arnosti C, Wietz M, Brinkhoff T, Hehemann J-H, Probant D, Zeugner L, et al. The biogeochemistry of marine polysaccharides: sources, inventories, and bacterial drivers of the carbohydrate cycle. Ann Rev Mar Sci. 2020;13:9.1–9.28. doi: 10.1146/annurev-marine-032020-012810. [DOI] [PubMed] [Google Scholar]
- 20.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barbeyron T, Brillet-Guéguen L, Carré W, Carrière C, Caron C, Czjzek M, et al. Matching the diversity of sulfated biomolecules: Creation of a classification database for sulfatases reflecting their substrate specificity. PLoS One. 2016;11:1–33. doi: 10.1371/journal.pone.0164846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang K, Lin Y, Han Y, Jiao N. Characterization of potential polysaccharide utilization systems in the marine Bacteroidetes Gramella flava JLT2011 using a multi-omics approach. Front Microbiol. 2017;8:220. doi: 10.3389/fmicb.2017.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Y, Chen P, Bao Y, Men Y, Zeng Y, Yang J, et al. Complete genome sequence and transcriptomic analysis of a novel marine strain Bacillus weihaiensis reveals the mechanism of brown algae degradation. Sci Rep. 2016;6:38248. doi: 10.1038/srep38248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas F, Bordron P, Eveillard D, Michel G. Gene expression analysis of Zobellia galactanivorans during the degradation of algal polysaccharides reveals both substrate-specific and shared transcriptome-wide responses. Front Microbiol. 2017;8:1808. doi: 10.3389/fmicb.2017.01808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ficko-Blean E, Préchoux A, Thomas F, Rochat T, Larocque R, Zhu Y, et al. Carrageenan catabolism is encoded by a complex regulon in marine heterotrophic bacteria. Nat Commun. 2017;8:1685. doi: 10.1038/s41467-017-01832-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koch H, Dürwald A, Schweder T, Noriega-Ortega B, Vidal-Melgosa S, Hehemann JH, et al. Biphasic cellular adaptations and ecological implications of Alteromonas macleodii degrading a mixture of algal polysaccharides. ISME J. 2019;13:92–103. doi: 10.1038/s41396-018-0252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bunse C, Koch H, Breider S, Simon M, Wietz M. Sweet spheres: succession and CAZyme expression of marine bacterial communities colonizing a mix of alginate and pectin particles. Environ Microbiol. 2021;23:3130–3148. doi: 10.1111/1462-2920.15536. [DOI] [PubMed] [Google Scholar]
- 28.Hehemann JH, Arevalo P, Datta MS, Yu X, Corzett CH, Henschel A, et al. Adaptive radiation by waves of gene transfer leads to fine-scale resource partitioning in marine microbes. Nat Commun. 2016;7:12860. doi: 10.1038/ncomms12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gralka M, Szabo R, Stocker R, Cordero OX. Trophic interactions and the drivers of microbial community assembly. Curr Biol. 2020;30:R1176–R1188. doi: 10.1016/j.cub.2020.08.007. [DOI] [PubMed] [Google Scholar]
- 30.Jiménez DJ, Dini-Andreote F, DeAngelis KM, Singer SW, Salles JF, van Elsas JD. Ecological insights into the dynamics of plant biomass-degrading microbial consortia. Trends Microbiol. 2017;25:788–796. doi: 10.1016/j.tim.2017.05.012. [DOI] [PubMed] [Google Scholar]
- 31.Kang S, Kim JK. Reuse of red seaweed waste by a novel bacterium, Bacillus sp. SYR4 isolated from a sandbar. World J Microbiol Biotechnol. 2015;31:209–217. doi: 10.1007/s11274-014-1778-x. [DOI] [PubMed] [Google Scholar]
- 32.Jonnadula R, Verma P, Shouche YS, Ghadi SC. Characterization of Microbulbifer strain CMC-5, a new biochemical variant of Microbulbifer elongatus type strain DSM6810T isolated from decomposing seaweeds. Curr Microbiol. 2009;59:600–607. doi: 10.1007/s00284-009-9480-1. [DOI] [PubMed] [Google Scholar]
- 33.Martin M, Barbeyron T, Martin R, Portetelle D, Michel G, Vandenbol M. The cultivable surface microbiota of the brown alga Ascophyllum nodosum is enriched in macroalgal-polysaccharide-degrading bacteria. Front Microbiol. 2015;6:1487. doi: 10.3389/fmicb.2015.01487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dogs M, Wemheuer B, Wolter L, Bergen N, Daniel R, Simon M, et al. Rhodobacteraceae on the marine brown alga Fucus spiralis are abundant and show physiological adaptation to an epiphytic lifestyle. Syst Appl Microbiol. 2017;40:370–382. doi: 10.1016/j.syapm.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 35.Brunet M, le Duff N, Fuchs B, Amann R, Barbeyron T, Thomas F. Specific detection and quantification of the marine flavobacterial genus Zobellia on macroalgae using novel qPCR and CARD-FISH assays. Syst Appl Microbiol. 2021;44:126269. doi: 10.1016/j.syapm.2021.126269. [DOI] [PubMed] [Google Scholar]
- 36.Barbeyron T, L’Haridon S, Corre E, Kloareg B, Potin P. Zobellia galactanovorans gen. nov., sp. nov., a marine species of Flavobacteriaceae isolated from a red alga, and classification of [Cytophaga] uliginosa (ZoBell and Upham 1944) Reichenbach 1989 as Zobellia uliginosa gen. nov., comb. nov. Int J Syst Evol Microbiol. 2001;51:985–997. doi: 10.1099/00207713-51-3-985. [DOI] [PubMed] [Google Scholar]
- 37.Barbeyron T, Thiébaud M, Le Duff N, Martin M, Corre E, Tanguy G, et al. Zobellia roscoffensis sp. nov. and Zobellia nedashkovskayae sp. nov., two flavobacteria from the epiphytic microbiota of the brown alga Ascophyllum nodosum, and emended description of the genus Zobellia. Int J Syst Evol Microbiol. 2021;71:004913. [DOI] [PubMed]
- 38.Nedashkovskaya OI, Suzuki M, Vancanneyt M, Cleenwerck I, Lysenko AM, Mikhailov VV, et al. Zobellia amurskyensis sp. nov., Zobellia laminariae sp. nov. and Zobellia russellii sp. nov., novel marine bacteria of the family Flavobacteriaceae. Int J Syst Evol Microbiol. 2004;54:1643–1648. doi: 10.1099/ijs.0.63091-0. [DOI] [PubMed] [Google Scholar]
- 39.Nedashkovskaya O, Otstavnykh N, Zhukova N, Guzev K, Chausova V, Tekutyeva L, et al. Zobellia barbeyronii sp. nov., a new member of the family Flavobacteriaceae, isolated from seaweed, and emended description of the species Z. amurskyensis, Z. laminariae, Z. russellii and Z. uliginosa. Diversity. 2021;13:520. doi: 10.3390/d13110520. [DOI] [Google Scholar]
- 40.Chernysheva N, Bystritskaya E, Stenkova A, Golovkin I. Comparative genomics and CAZyme genome repertoires of marine Zobellia amurskyensis KMM 3526T and Zobellia laminariae KMM 3676T. Mar Drugs. 2019;17:661. doi: 10.3390/md17120661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chernysheva N, Bystritskaya E, Likhatskaya G, Nedashkovskaya O, Isaeva M. Genome-wide analysis of PL7 alginate lyases in the genus Zobellia. Molecules. 2021;26:2387. doi: 10.3390/molecules26082387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbeyron T, Thomas F, Barbe V, Teeling H, Schenowitz C, Dossat C, et al. Habitat and taxon as driving forces of carbohydrate catabolism in marine heterotrophic bacteria: Example of the model algae-associated bacterium Zobellia galactanivorans DsijT. Environ Microbiol. 2016;18:4610–4627. doi: 10.1111/1462-2920.13584. [DOI] [PubMed] [Google Scholar]
- 43.Potin P, Sanseau A, Le Gall Y, Rochas C, Kloareg B. Purification and characterization of a new k‐carrageenase from a marine Cytophaga‐like bacterium. Eur J Biochem. 1991;201:241–247. doi: 10.1111/j.1432-1033.1991.tb16280.x. [DOI] [PubMed] [Google Scholar]
- 44.Lami R, Grimaud R, Sanchez-Brosseau S, Six C, Thomas F, West NJ, et al. Marine bacterial models for experimental biology. In: Boutet A, Schierwater B, editors. Handbook of Marine Model Organisms in Experimental Biology. London: Taylor & Francis Ltd; 2021.
- 45.Dudek M, Dieudonné A, Jouanneau D, Rochat T, Michel G, Sarels B, et al. Regulation of alginate catabolism involves a GntR family repressor in the marine flavobacterium Zobellia galactanivorans DsijT. Nucleic Acids Res. 2020;48:7786–7800. doi: 10.1093/nar/gkaa533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas F, Lundqvist LCE, Jam M, Jeudy A, Barbeyron T, Sandström C, et al. Comparative characterization of two marine alginate lyases from Zobellia galactanivorans reveals distinct modes of action and exquisite adaptation to their natural substrate. J Biol Chem. 2013;288:23021–23037. doi: 10.1074/jbc.M113.467217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas F, Barbeyron T, Tonon T, Génicot S, Czjzek M, Michel G. Characterization of the first alginolytic operons in a marine bacterium: from their emergence in marine Flavobacteriia to their independent transfers to marine Proteobacteria and human gut Bacteroides. Environ Microbiol. 2012;14:2379–94. doi: 10.1111/j.1462-2920.2012.02751.x. [DOI] [PubMed] [Google Scholar]
- 48.Jam M, Flament D, Allouch J, Potin P, Thion L, Kloareg B, et al. The endo-β-agarases AgaA and AgaB from the marine bacterium Zobellia galactanivorans: Two paralogue enzymes with different molecular organizations and catalytic behaviours. Biochem J. 2005;385:703–713. doi: 10.1042/BJ20041044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hehemann JH, Correc G, Thomas F, Bernard T, Barbeyron T, Jam M, et al. Biochemical and structural characterization of the complex agarolytic enzyme system from the marine bacterium Zobellia galactanivorans. J Biol Chem. 2012;287:30571–30584. doi: 10.1074/jbc.M112.377184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Labourel A, Jam M, Jeudy A, Hehemann JH, Czjzek M, Michel G. The β-glucanase ZgLamA from Zobellia galactanivorans evolved a bent active site adapted for efficient degradation of algal laminarin. J Biol Chem. 2014;289:2027–2042. doi: 10.1074/jbc.M113.538843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Labourel A, Jam M, Legentil L, Sylla B, Hehemann JH, Ferrières V, et al. Structural and biochemical characterization of the laminarinase ZgLamCGH16 from Zobellia galactanivorans suggests preferred recognition of branched laminarin. Acta Crystallogr. 2015;D71:173–184. doi: 10.1107/S139900471402450X. [DOI] [PubMed] [Google Scholar]
- 52.Dorival J, Ruppert S, Gunnoo M, Orłowski A, Chapelais-Baron M, Dabin J, et al. The laterally-acquired GH5 ZgEngAGH5_4 from the marine bacterium Zobellia galactanivorans is dedicated to hemicellulose hydrolysis. Biochem J. 2018;475:3609–3628. doi: 10.1042/BCJ20180486. [DOI] [PubMed] [Google Scholar]
- 53.Groisillier A, Labourel A, Michel G, Tonon T. The mannitol utilization system of the marine bacterium Zobellia galactanivorans. Appl Environ Microbiol. 2015;81:1799–1812. doi: 10.1128/AEM.02808-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fournier JB, Rebuffet E, Delage L, Grijol R, Meslet-Cladière L, Rzonca J, et al. The vanadium iodoperoxidase from the marine Flavobacteriaceae species Zobellia galactanivorans reveals novel molecular and evolutionary features of halide specificity in the vanadium haloperoxidase enzyme family. Appl Environ Microbiol. 2014;80:7561–7573. doi: 10.1128/AEM.02430-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grigorian E, Groisillier A, Thomas F, Leblanc C, Delage L. Functional characterization of a L-2-haloacid dehalogenase from Zobellia galactanivorans DsijT suggests a role in haloacetic acid catabolism and a wide distribution in marine environments. Front Microbiol. 2021;12:725997. doi: 10.3389/fmicb.2021.725997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu Y, Thomas F, Larocque R, Li N, Duffieux D, Cladière L, et al. Genetic analyses unravel the crucial role of a horizontally acquired alginate lyase for brown algal biomass degradation by Zobellia galactanivorans. Environ Microbiol. 2017;19:2164–2181. doi: 10.1111/1462-2920.13699. [DOI] [PubMed] [Google Scholar]
- 57.Zablackis E, Perez J. A partially pyruvated carrageenan from hawaiian Grateloupia filicina (Cryptonemiales, Rhodophyta) Bot Mar. 1990;33:273–276. doi: 10.1515/botm.1990.33.3.273. [DOI] [Google Scholar]
- 58.Filisetti-Cozzi T, Carpita N. Measurement of uronic acids without interference from neutral sugars. Anal Biochem. 1991;197:15162. doi: 10.1016/0003-2697(91)90372-Z. [DOI] [PubMed] [Google Scholar]
- 59.Blumenkrantz N, Asboe-Hansen G. New method for quantitative determination of uronic acids. Anal Biochem. 1973;54:484–489. doi: 10.1016/0003-2697(73)90377-1. [DOI] [PubMed] [Google Scholar]
- 60.Cumashi A, Ushakova NA, Preobrazhenskaya ME, D’Incecco A, Piccoli A, Totani L, et al. A comparative study of the anti-inflammatory, anticoagulant, antiangiogenic, and antiadhesive activities of nine different fucoidans from brown seaweeds. Glycobiology. 2007;17:541–552. doi: 10.1093/glycob/cwm014. [DOI] [PubMed] [Google Scholar]
- 61.Jung SY, Oh TK, Yoon JH. Tenacibaculum aestuarii sp. nov., isolated from a tidal flat sediment in Korea. Int J Syst Evol Microbiol. 2006;56:1577–1581. doi: 10.1099/ijs.0.64302-0. [DOI] [PubMed] [Google Scholar]
- 62.ZoBell C. Studies on marine bacteria. I. The cultural requirements of heterotrophic aerobes. J Mar Res. 1941;4:75. [Google Scholar]
- 63.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vallenet D, Calteau A, Dubois M, Amours P, Bazin A, Beuvin M, et al. MicroScope: An integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 2020;48:D579–D589. doi: 10.1093/nar/gkz926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thomas F, Barbeyron T, Michel G. Evaluation of reference genes for real-time quantitative PCR in the marine flavobacterium Zobellia galactanivorans. J Microbiol Methods. 2011;84:61–6. doi: 10.1016/j.mimet.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 69.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.R Core Team. R: A language and environment for statistical computing. 2018. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
- 72.Lex A, Gehlenborg N, Strobelt H. UpSet: Visualization of intersecting sets. IEEE Trans Vis Comput Graph. 2014;20:1983–1992. doi: 10.1109/TVCG.2014.2346248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krassowski M. krassowski/complex-upset. 2020. 10.5281/zenodo.3700590.
- 74.Murtagh F, Legendre P. Ward’s hierarchical clustering method: clustering criterion and agglomerative algorithm. J Classif. 2014;31:274–295. doi: 10.1007/s00357-014-9161-z. [DOI] [Google Scholar]
- 75.Wickham H Use R! ggplot2: Elegant graphics for data analysis. 2nd ed. London: Springer; 2016.
- 76.Kidby DK, Davidson DJ. Ferricyanide estimation of sugars in the nanomole range. Anal Biochem. 1973;55:321–325. doi: 10.1016/0003-2697(73)90323-0. [DOI] [PubMed] [Google Scholar]
- 77.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, et al. DbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen X, Hu Y, Yang B, Gong X, Zhang N, Niu L, et al. Structure of lpg0406, a carboxymuconolactone decarboxylase family protein possibly involved in antioxidative response from Legionella pneumophila. Protein Sci. 2015;24:2070–2075. doi: 10.1002/pro.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Enke TN, Datta MS, Schwartzman J, Cermak N, Schmitz D, Barrere J, et al. Modular assembly of polysaccharide-degrading marine microbial communities. Curr Biol. 2019;29:1528–1535.e6. doi: 10.1016/j.cub.2019.03.047. [DOI] [PubMed] [Google Scholar]
- 80.Pollak S, Gralka M, Sato Y, Schwartzman J, Lu L, Cordero OX. Public good exploitation in natural bacterioplankton communities. Sci Adv. 2021;7:eabi4717. [DOI] [PMC free article] [PubMed]
- 81.Pontrelli S, Szabo R, Pollak S, Schwartzman J, Ledezma D, Cordero OX, et al. Metabolic cross-feeding structures the assembly of polysaccharide degrading communities. Sci Adv. 2022;8:eabk3076. [DOI] [PMC free article] [PubMed]
- 82.Holdt SL, Kraan S. Bioactive compounds in seaweed: Functional food applications and legislation. J Appl Phycol. 2011;23:543–597. doi: 10.1007/s10811-010-9632-5. [DOI] [Google Scholar]
- 83.Kawamura-Konishi Y, Watanabe N, Saito M, Nakajima N, Sakaki T, Katayama T, et al. Isolation of a new phlorotannin, a potent inhibitor of carbohydrate-hydrolyzing enzymes, from the brown alga Sargassum patens. J Agric Food Chem. 2012;60:5565–5570. doi: 10.1021/jf300165j. [DOI] [PubMed] [Google Scholar]
- 84.Garbary DJ, Brown NE, MacDonell HJ, Toxopeux J. Ascophyllum and its symbionts — A complex symbiotic community on North Atlantic shores. Algal and Cyanobacteria Symbioses. 2017:547–572.
- 85.Pluvinage B, Grondin JM, Amundsen C, Klassen L, Moote PE, Xiao Y, et al. Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont. Nat Commun. 2018;9:1043. doi: 10.1038/s41467-018-03366-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reintjes G, Arnosti C, Fuchs BM, Amann R. An alternative polysaccharide uptake mechanism of marine bacteria. ISME J. 2017;11:1640–1650. doi: 10.1038/ismej.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hollants J, Leliaert F, de Clerck O, Willems A. What we can learn from sushi: A review on seaweed-bacterial associations. FEMS Microbiol Ecol. 2013;83:1–16. doi: 10.1111/j.1574-6941.2012.01446.x. [DOI] [PubMed] [Google Scholar]
- 88.Thomas F, Le Duff N, Wu TD, Cébron A, Uroz S, Riera P, et al. Isotopic tracing reveals single-cell assimilation of a macroalgal polysaccharide by a few marine Flavobacteria and Gammaproteobacteria. ISME J. 2021;15:3062–3075. doi: 10.1038/s41396-021-00987-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Datta MS, Sliwerska E, Gore J, Polz MF, Cordero OX. Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat Commun. 2016;7:11965. doi: 10.1038/ncomms11965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Enke TN, Leventhal GE, Metzger M, Saavedra JT, Cordero OX. Microscale ecology regulates particulate organic matter turnover in model marine microbial communities. Nat Commun. 2018;9:2743. doi: 10.1038/s41467-018-05159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sichert A, Cordero OX. Polysaccharide-bacteria Interactions from the lens of evolutionary ecology. Front Microbiol. 2021;12:705082. doi: 10.3389/fmicb.2021.705082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sichert A, Corzett CH, Schechter M, Unfried F, Markert S, Becher D, et al. Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan. Nat Microbiol. 2020;5:1026–1039. doi: 10.1038/s41564-020-0720-2. [DOI] [PubMed] [Google Scholar]
- 93.Reisky L, Préchoux A, Zühlke MK, Bäumgen M, Robb CS, Gerlach N, et al. A marine bacterial enzymatic cascade degrades the algal polysaccharide ulvan. Nat Chem Biol. 2019;15:803–812. doi: 10.1038/s41589-019-0311-9. [DOI] [PubMed] [Google Scholar]
- 94.Mabeau S, Kloareg B, Joseleau J-P. Fractionation and analysis of fucans from brown algae. Phytochemistry. 1990;29:2441–2445. doi: 10.1016/0031-9422(90)85163-A. [DOI] [Google Scholar]
- 95.Küpper FC, Kloareg B, Guern J, Potin P. Oligoguluronates elicit an oxidative burst in the brown algal kelp Laminaria digitata. Plant Physiol. 2001;125:278–291. doi: 10.1104/pp.125.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Küpper FC, Müller DG, Peters AF, Kloareg B, Potin P. Oligoalginate recognition and oxidative burst play a key role in natural and induced resistance of sporophytes of Laminariales. J Chem Ecol. 2002;28:2057–2081. doi: 10.1023/A:1020706129624. [DOI] [PubMed] [Google Scholar]
- 97.Leonard S, Hommais F, Nasser W, Reverchon S. Plant–phytopathogen interactions: bacterial responses to environmental and plant stimuli. Environ Microbiol. 2017;19:1689–1716. doi: 10.1111/1462-2920.13611. [DOI] [PubMed] [Google Scholar]
- 98.Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, McBride MJ, et al. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. PNAS. 2010;107:276–281. doi: 10.1073/pnas.0912010107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Eckroat TJ, Greguske C, Hunnicutt DW. The type 9 secretion system is required for Flavobacterium johnsoniae biofilm formation. Front Microbiol. 2021;12:660887. doi: 10.3389/fmicb.2021.660887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xie S, Tan Y, Song W, Zhang W, Qi Q, Lu X. N-glycosylation of a cargo protein C-terminal domain recognized by the type IX secretion system in Cytophaga hutchinsonii affects protein secretion and localization. Appl Environ Microbiol. 2022;88:e0160621. doi: 10.1128/AEM.01606-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.