Abstract
We used denaturing gradient gel electrophoresis (DGGE) to study the diversity of picoeukaryotes in natural marine assemblages. Two eukaryote-specific primer sets targeting different regions of the 18S rRNA gene were tested. Both primer sets gave a single band when used with algal cultures and complex fingerprints when used with natural assemblages. The reproducibility of the fingerprints was estimated by quantifying the intensities of the same bands obtained in independent PCR and DGGE analyses, and the standard error of these estimates was less than 2% on average. DGGE fingerprints were then used to compare the picoeukaryotic diversity in samples obtained at different depths and on different dates from a station in the southwest Mediterranean Sea. Both primer sets revealed significant differences along the vertical profile, whereas temporal differences at the same depths were less marked. The phylogenetic composition of picoeukaryotes from one surface sample was investigated by excising and sequencing DGGE bands. The results were compared with an analysis of a clone library and a terminal restriction fragment length polymorphism fingerprint obtained from the same sample. The three PCR-based methods, performed with three different primer sets, revealed very similar assemblage compositions; the same main phylogenetic groups were present at similar relative levels. Thus, the prasinophyte group appeared to be the most abundant group in the surface Mediterranean samples as determined by our molecular analyses. DGGE bands corresponding to prasinophytes were always found in surface samples but were not present in deep samples. Other groups detected were prymnesiophytes, novel stramenopiles (distantly related to hyphochytrids or labyrinthulids), cryptophytes, dinophytes, and pelagophytes. In conclusion, the DGGE method described here provided a reasonably detailed view of marine picoeukaryotic assemblages and allowed tentative phylogenetic identification of the dominant members.
Small phototrophic and heterotrophic eukaryotes between 0.2 and 5 μm in diameter are found throughout the world's oceans at concentrations between 102 and 104 cells per ml in the upper photic zone (6, 23). They constitute an essential component of microbial food webs and play significant roles in global carbon and mineral cycles, especially in oligotrophic parts of the oceans (12, 23). However, the identities of the eukaryotic picoplankton have remained elusive due to their small size and the lack of distinctive taxonomic characteristics (43, 47). Conventional approaches based on morphological criteria, such as optical, epifluorescence, or electron microscopy (5, 36), can barely discriminate between these organisms, even at the class level. Although informative, analysis of diagnostic marker pigments by high-performance liquid chromatography (HPLC) provides information about the composition of photosynthetic picoplankton populations only at the class level (20) and appears to be a complementary method that is useful for gross characterization of populations. Culturing efforts have revealed the presence of novel lineages of heterotrophic (17, 50) and phototrophic (4, 18) picoeukaryotic organisms, but only a small percentage of marine picoeukaryotes have been grown in culture, and often the cultured organisms are not dominant in the plankton community (19, 24).
Molecular techniques based on rRNA genes obtained from natural assemblages have provided new insights into the diversity of marine microbial plankton (2, 41). In particular, cloning and sequencing of rRNA genes have been very useful for describing the compositions of marine bacterial (15) and archaeal (8, 13) assemblages. Similar studies have been performed recently with 18S rRNA eukaryotic genes (9, 27, 35), and the results suggest that the picoeukaryotic assemblage is very diverse and that there are many undiscovered taxa. However, analysis of clone libraries is time-consuming and not suitable when many different samples are analyzed. This is the case, for example, in studies focusing on changes in microbial assemblages exposed to a perturbation or on how the microbial composition changes along environmental gradients, such as depth in the water column, gradients across oceanographic features, or temporal changes with different time scales. A technique that allows processing of many samples simultaneously is necessary for such studies. Fingerprinting techniques, such as denaturing gradient gel electrophoresis (DGGE) (38, 39) or terminal restriction fragment length polymorphism (T-RFLP) analysis (25, 29, 34), offer the best compromise between the number of samples processed and the information obtained. DGGE, in particular, provides both rapid comparison data for many communities and specific phylogenetic information derived from excised bands (39).
DGGE has been widely used to investigate several patterns of distribution of marine bacterial assemblages (34, 37, 45, 46), but this technique has not been applied previously to the marine picoeukaryotic component. The first application of DGGE for detection of eukaryotic 18S rRNA genes was a study of fungal pathogens in coastal plants in which fungus-specific primers were used (21). A few recent studies have focused on the whole eukaryotic assemblage by using eukaryote-specific primers. These studies involved an analysis of temporal changes in an activated sludge bioreactor (30), a comparison of natural assemblages in different freshwater lagoons (51), and a description of the development of eukaryotic populations in a mesocosm experiment (52). As noted above, marine picoeukaryotic assemblages have not been investigated by DGGE previously.
In this study we examined the diversity of marine picoeukaryotes with DGGE by using two sets of primers specific for eukaryotic 18S rRNA genes. We optimized the conditions for both primer sets and tested their performance with cultures and environmental samples. The relative effectiveness of each set was evaluated by comparing community profiles obtained from different samples from the Mediterranean Sea. The most intense DGGE bands from a surface sample were sequenced, and tentative phylogenetic affiliations of organisms derived from eukaryotic picoplankton were determined. Finally, the DGGE results were compared with the results obtained by using two other molecular, PCR-based methods, gene cloning and T-RFLP analysis.
MATERIALS AND METHODS
Eukaryotic cultures.
Thalassiosira pseudonana (Bacillariophyceae), Heterosigma akashiwo (Raphydophyceae), Heterocapsa sp. (Dinophyceae), Platymonas suecica (Prasinophyceae), and Dunaliella primolecta (Chlorophyceae) were obtained from the culture collection of the Institut de Ciències del Mar, Barcelona, Spain. Pelagomonas calceolata (Pelagophyceae) and Nannochloropsis oculata (Eustigmatophyceae) were obtained from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton. Cultures were grown in f/2 medium (16) under continuous light conditions or under a daily regime consisting of 12 h of light and 12 h of darkness. When cultures reached sufficient biomass (after 7 to 10 days of growth), cells were harvested by filtration on 0.2-μm-pore-size Durapore filters. The filters were submerged in 2 ml of lysis buffer (40 mM EDTA, 50 mM Tris-HCl, 0.75 M sucrose), and nucleic acid extraction was performed immediately.
Marine samples.
Samples were collected during two MATER cruises of the B/O García del Cid and B.I.O. Hespérides. Samples were obtained from a frontal upwelling area (station ME-B) located at 36°14′N, 4°15′W across the Western Alborán Sea Gyre (southwest Mediterranean Sea) on 11 November 1997 (sample ME-B0) and four times in May 1998 (in this study we used only samples collected on 9 May [ME-B3] and 12 May [ME-B4]). Seawater from different depths was collected with Niskin bottles attached to a rosette. Microbial biomass was collected on a 0.2-μm-pore-size Sterivex unit (Durapore; Millipore) by filtering between 8 and 18 liters of seawater through a 5-μm-pore-size Durapore prefilter (diameter, 47 mm; Millipore) and the Sterivex unit in succession with a peristaltic pump, using filtration rates of 50 to 100 ml min−1. Each Sterivex unit was filled with lysis buffer and frozen at −70°C until nucleic acid extraction was performed in the laboratory.
Nucleic acid extraction.
Nucleic acid extraction was performed essentially as described by Massana et al. (32). For samples from cultures, 0.5-mm-diameter sterile glass beads were added to tubes containing filters, and the tubes were vortexed in order to physically disrupt the cells. For all samples, nucleic acid extraction started with addition of lysozyme (final concentration, 1 mg ml−1) and incubation of the filters at 37°C for 45 min. Then sodium dodecyl sulfate (final concentration, 1%) and proteinase K (final concentration, 0.2 mg ml−1) were added, and the filters were incubated at 55°C for 60 min. The lysates were purified twice by extraction with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1), and the residual phenol was removed by extraction with an equal volume of chloroform-isoamyl alcohol (24:1). Finally, nucleic acid extracts were further purified, desalted, and concentrated with a Centricon-100 concentrator (Millipore). The integrity of the total DNA was checked by agarose gel electrophoresis. The DNA yield was quantified by a Hoechst dye fluorescence assay (42). Nucleic acid extracts were stored at −70°C until analysis.
PCR.
About 10 ng of extracted DNA was used as the template in a PCR in which eukaryotic 18S ribosomal DNA (rDNA)-specific primers were used. For DGGE we tested two sets of primers (Table 1): set A (Euk1A and Euk516r-GC), which amplifies a fragment approximately 560 bp long, and set B (Euk1209f-GC and Uni1392r), which amplifies a fragment approximately 210 bp long. The PCR mixtures (50 μl) contained each deoxynucleoside triphosphate at a concentration of 200 μM, 1.5 mM MgCl2, each primer at a concentration of 0.3 μM, 2.5 U of Taq DNA polymerase (Gibco BRL), and the PCR buffer supplied with the enzyme. The PCR program for primer set A included an initial denaturation at 94°C for 130 s, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 45 s, and extension at 72°C for 130 s. The PCR program for primer set B included an initial denaturation at 94°C for 1 min and 10 touchdown cycles of denaturation at 94°C for 1 min, annealing at 65°C (with the temperature decreasing 1°C each cycle) for 1 min, and extension at 72°C for 3 min, followed by 20 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 3 min. During the last cycle of both programs, the length of the extension step was increased to 7 min. An aliquot of the PCR product was electrophoresed in a 0.8% agarose gel, stained with ethidium bromide, and quantified by using a standard (Low DNA Mass Ladder; Gibco BRL).
TABLE 1.
Oligonucleotide sequences used in this study
| Primer | Sequence (5′ to 3′) | Saccharomyces cerevisiae positions | Specificity | Reference |
|---|---|---|---|---|
| Set Aa | ||||
| Euk1A | CTGGTTGATCCTGCCAG | 4 to 20 | Eukarya | 48 |
| Euk516r-GCb | ACCAGACTTGCCCTCC | 563 to 548 | Eukarya | 2 |
| Set B | ||||
| Euk1209f-GCc | CAGGTCTGTGATGCCC | 1423 to 1438 | Eukarya | 14 |
| Uni1392r | ACGGGCGGTGTGTRC | 1641 to 1627 | Universal | 22 |
| Set C | ||||
| EukA | AACCTGGTTGATCCTGCCAGT | 1 to 21 | Eukarya | 33 |
| EukB | TGATCCTTCTGCAGGTTCACCTAC | 1795 to 1772 | Eukarya | 33 |
Set A was also used for T-RFLP analysis, using hexachlorofluorescein-labeled Euk1A and Euk516r without the GC clamp.
The GC clamp sequence is CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGG.
The GC clamp sequence is CGCGCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCG.
DGGE.
DGGE was performed with a DGGE-2000 system (CBS Scientific Company) as described previously (37, 39, 46). Electrophoresis was performed with 0.75-mm-thick 6% polyacrylamide gels (ratio of acrylamide to bisacrylamide, 37.5:1) submerged in 1× TAE buffer (40 mM Tris, 40 mM acetic acid, 1 mM EDTA; pH 7.4) at 60°C. Approximately 600 to 800 ng of PCR product from environmental samples and 100 ng of PCR product from cultures were applied to individual lanes in the gel. The following electrophoresis conditions were selected based on the results of perpendicular DGGE and time travel experiments (Fig. 1): 16 h at 100 V in a linear 40 to 65% denaturant agent gradient (100% denaturant agent was defined as 7 M urea and 40% deionized formamide) for primer set A; and 6 h at 200 V in a 40 to 80% denaturant agent gradient for primer set B. The gels were stained for 30 min in 1× TAE buffer with SybrGold nucleic acid stain (Molecular Probes) and visualized with UV radiation by using a Fluor-S MultiImager and the MultiAnalyst imaging software (Bio-Rad). The number of operational taxonomic units (OTUs) in each sample was defined as the number of DGGE bands.
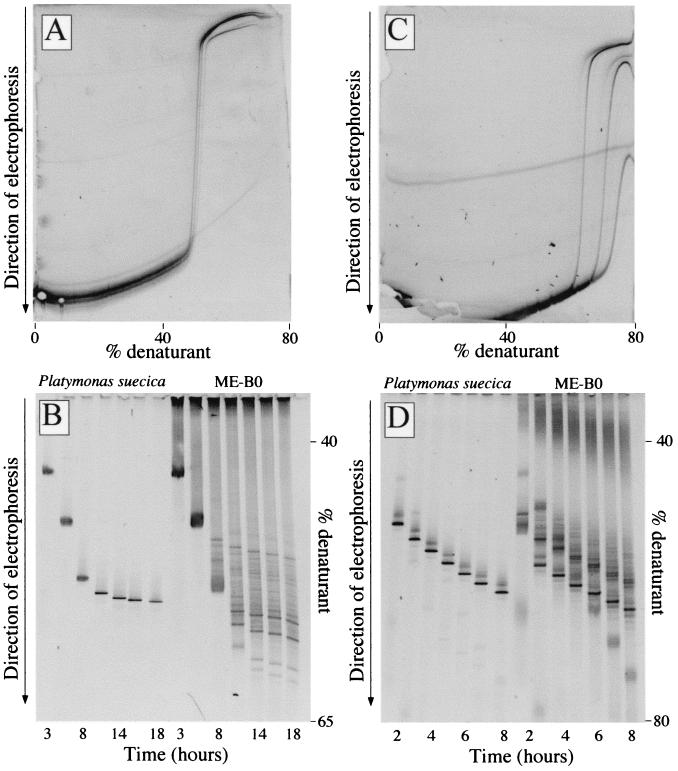
FIG. 1.
(A) Negative image of a perpendicular DGGE gel with PCR products obtained with primer set A from different algal cultures (P. calceolata, T. pseudonana, and P. suecica) and electrophoresed at 100 V for 16 h. (B) Time course separation of PCR products obtained with primer set A from an algal culture (P. suecica) and marine sample ME-B0. Samples were electrophoresed for 3, 5, 8, 11, 14, 16, and 18 h at 100 V. (C) Same as panel A but with PCR products amplified with primer set B. The electrophoresis conditions were 200 V for 6 h. (D) Same as panel B but with PCR products amplified with primer set B. Samples were electrophoresed for 2, 3, 4, 5, 6, 7, and 8 h at 200 V.
DGGE bands were sequenced after excision from the gel and reamplification. Briefly, bands were excised, resuspended in 20 μl of MilliQ water, and stored at 4°C overnight. An aliquot of supernatant was used for PCR reamplification with the original primer set. Between 30 and 50 ng of the reamplified PCR product was used for a sequencing reaction (with the corresponding forward primer) with a Thermo Sequenase v.2 kit (Amersham, U.S. Biochemicals) and an ABI PRISM model 377 (v. 3.3) automated sequencer. The sequences obtained (300 to 400 bases for primer set A and 100 to 200 bases for primer set B) were compared with public DNA database sequences by using BLAST (1).
Quantitative analysis of DGGE fingerprints.
Digitized DGGE images were analyzed with the Diversity Database software (Bio-Rad) as previously described (46). This software carries out a density profile analysis for each lane, detects the bands, and calculates the relative contribution of each band to the total band intensity in the lane after subtracting a rolling disk background value. Then the software identifies the bands occupying the same position in the different lanes of the gel. Two matrices were constructed; the first took into account the presence or absence of individual bands in all lanes (binary matrix), and the second incorporated the percentage of the intensity for each band based on the total intensity in the lane (intensity matrix). The binary matrix was used to calculate a similarity matrix with the Jaccard coefficient of similarity, and the intensity matrix was used to calculate a distance matrix with Euclidean distances. Both matrices were then used to construct a nonmetric multidimensional scaling (NMDS) diagram with the software SYSTAT 5.2.1. Such a diagram places each sample at a point in a plane (with dimensions of no special significance) so that very similar samples are plotted close together. By connecting consecutive data points (for instance, consecutive samples from a vertical profile), relative changes in the community structure can be visualized and interpreted (52).
T-RFLP analysis.
The PCR for T-RFLP analysis was performed with primer set A (Table 1) and the corresponding PCR program, except that primer Euk1A was 5′ labeled with hexachlorofluorescein (Operon Technologies) and primer Euk516r did not have the GC clamp. Fluorescently labeled PCR products were purified by using Wizard PCR purification columns (Promega). Purified PCR products were digested separately with restriction enzymes HhaI, MspI, and RsaI (Boehringer Mannheim Biochemicals). Terminal restriction fragments (TRFs) were resolved by electrophoresis at 3,000 V for 14 h in a denaturing 6% acrylamide gel (ratio of acrylamide to N,N-methylenebisacrylamide, 19:1) with an ABI PRISM model 373 automated sequencer. The sizes of TRFs were determined with the software GeneScan 2.1 at 1-bp resolution by using the size standard TAMRA-2500 (ABI), and the intensity of each TRF was measured using the peak area. The number of TRFs corresponded to the number of OTUs in each sample. TRF length predictions were made for most of the eukaryotic organisms by using complete sequences extracted from the Ribosomal Database Project (28) and the pattern-searching algorithm PatScan (10). The values obtained were used to identify the putative phylogenetic affiliations of the measured peaks. In cases in which the experimental fragment corresponded to several possible organisms, a most likely candidate was indicated when there were other supporting data.
Cloning and sequencing of 18S rDNA.
PCR was performed with primer set C (Table 1), which amplified almost the entire 18S rRNA gene. The PCR program involved an initial denaturation at 94°C for 3 min, 30 cycles of denaturation at 94°C for 45 s, annealing at 55°C for 1 min, and extension at 72°C for 3 min, and a final extension at 72°C for 5 min. The PCR product was used to construct a clone library with a TA cloning kit (Invitrogen). The presence of an 18S rDNA insert was confirmed by PCR reamplification with the same primers. Positive amplification products of the right size were digested with restriction enzyme HaeIII (Gibco BRL). The resulting restriction fragment length polymorphism (RFLP) products were separated by electrophoresis in a 2.5% low-melting-point agarose gel at 80 V for 2 to 3 h. Clones with the same RFLP pattern (same bands at the same positions) were considered members of the same OTU. At least one clone representative of each OTU was partially sequenced with an ABI PRISM model 377 (v. 3.3) automated sequencer.
RESULTS
DGGE optimization.
We optimized the use of two sets of eukaryotic 18S rDNA-specific primers. First, the four primers were checked against a database of about 4,000 eukaryotic sequences containing the whole 18S rDNA gene (more than 1,649 bases), and they gave very good results. The percentages of sequences having no mismatch and one mismatch with the primers were 79 and 93% for Euk1A, 87 and 96% for Euk516r, 90 and 99% for Euk1209f, and 96 and 98% for Uni1392r, respectively. Moreover, in most cases no consistent bias against any phylogenetic eukaryotic entity was identified; the only exceptions were the Cercomonas group with primer Euk1A and the Tetrahymena group with primer Euk516r. Second, the specificity of the primers was investigated by PCR by using as the templates DNA extracts of several marine algal cultures of organisms that belonged to the classes Pelagophyceae, Eustigmatophyceae, Bacillariophyceae, Chlorophyceae, Prasinophyceae, Raphydophyceae, and Dinophyceae. These cultures always yielded positive PCR amplification results with both sets of primers, whereas several bacterial and archaeal DNA extracts did not (data not shown). Third, a perpendicular DGGE analysis of a mixture of PCR products from three cultures (P. calceolata, T. pseudonana, and P. suecica) was performed to determine an appropriate gradient of denaturant concentrations for each primer set. At a denaturant concentration range of 50 to 55%, the fragments obtained with primer set A displayed reduced mobility (Fig. 1A), whereas with primer set B the three PCR products melted at 65 to 70% denaturant (Fig. 1C). We thus determined that the optimal denaturant gradient was 40 to 65% for primer set A and 40 to 80% for primer set B. Fourth, we performed time travel experiments with PCR products from a culture (P. suecica) and a natural sample (ME-B0) to determine the optimal electrophoresis time. PCR products obtained with primer set A were loaded into a gel every 2 to 3 h for 18 h and electrophoresed at 100 V (Fig. 1B). After 11 h bands were clearly defined and showed reduced mobility. PCR products obtained with primer set B were loaded into a gel every 1 to 2 h for 8 h and electrophoresed at 200 V (Fig. 1D). After 3 h the bands were clearly defined, but in this case the bands migrated continuously and did not show reduced mobility. Thus, the electrophoresis conditions used were 100 V for 16 h with primer set A and 200 V for 6 h with primer set B.
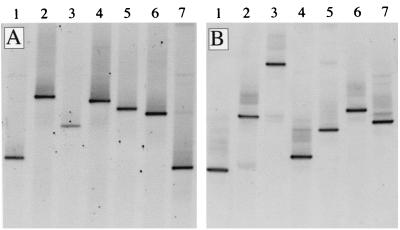
Once the optimal conditions for electrophoresis were defined for both primer sets, the performance of DGGE was tested further with the collection of algal cultures available (Fig. 2). The DGGE gel obtained indicated that each culture produced a single dominant band that appeared at a different position in the gel, indicating the potential of the primers to resolve different phylotypes. Some cultures produced additional bands, but the intensities of these bands were always much lower than the intensity of the dominant band.
FIG. 2.
DGGE fingerprints of algal cultures amplified with primer set A (A) and primer set B (B). The following cultures were tested: lane 1, P. calceolata; lane 2, N. oculata; lane 3, T. pseudonana; lane 4, D. primolecta; lane 5, P. suecica; lane 6, H. akashiwo; lane 7, Heterocapsa sp.
Fingerprinting of natural assemblages.
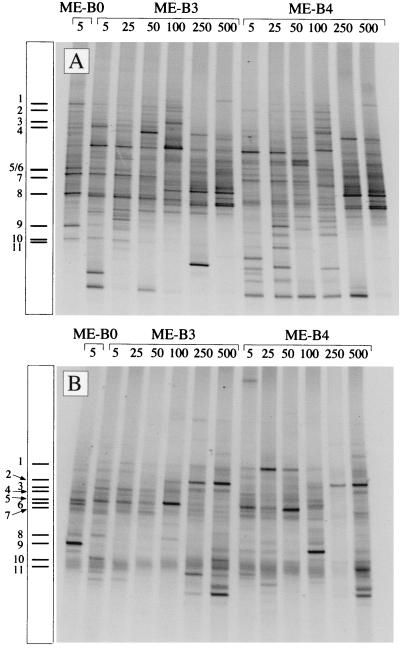
We used DGGE with primer set A (Fig. 3A) and primer set B (Fig. 3B) to compare the picoeukaryotic assemblages from 13 southwest Mediterranean Sea samples taken at the same station at different depths and on different dates. Each sample produced a complex fingerprint composed of a large number of bands; 33 to 45 bands at the surface (0 to 100 m) and 20 to 28 bands at depth (250 and 500 m) were obtained with primer set A, and 10 to 17 bands at the surface and approximately 20 bands at depth (250 to 500 m) were obtained with primer set B. Some bands were unique to surface samples, whereas other bands were obtained only with deep samples. With both sets of primers the fingerprints obtained for the surface samples were similar and the fingerprints obtained for the deep samples were similar, and there were clear differences when surface and deep fingerprints were compared.
FIG. 3.
DGGE fingerprints of picoeukaryotic assemblages obtained at station ME-B (southwest Mediterranean Sea) at different times (ME-B0, 11 November 1997; ME-B3, 9 May 1998; ME-B4, 12 May 1998) and at different depths (5 to 500 m). The fingerprints were obtained with primer set A (A) and primer set B (B). Bands from sample ME-B0 that were sequenced are indicated on the left side of each gel.
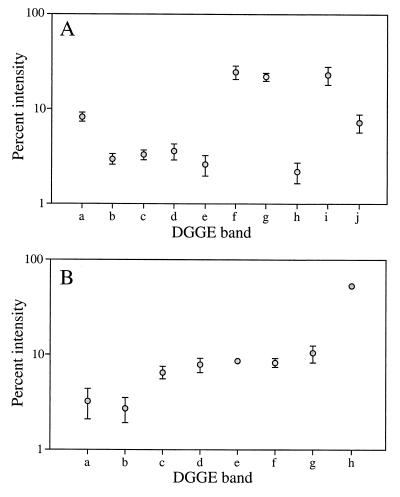
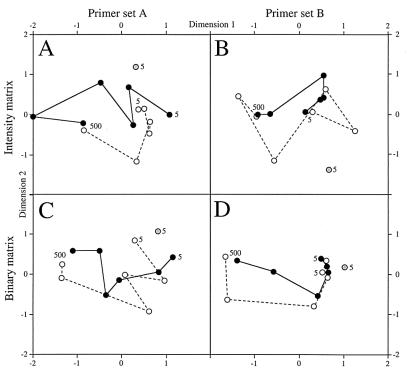
The data in the DGGE gels shown in Fig. 3 were extracted by image analysis, which resulted in a binary matrix (presence or absence of bands in different samples) and an intensity matrix (binary matrix information plus band intensity information). Using intensity matrices for comparative purposes requires reproducibility of band patterns. To test reproducibility, we selected the most intense bands from sample ME-B0 and determined the relative intensities of these bands in several DGGE fingerprints obtained after different PCRs and from different DGGE gels. As Fig. 4 shows, the intensities of these bands were always very reproducible; the average standard errors were 0.9% (n = 5) and 1.3% (n = 4) for bands obtained with primer sets A and B, respectively. The binary and intensity matrices were then used in an NMDS analysis for statistical comparison of the different samples (Fig. 5). The NMDS analysis showed that the samples grouped together primarily according to their positions in the water column. Thus, surface samples obtained on different dates appeared to be similar, and a cluster that included samples obtained in November 1997 and May 1998 was formed. Similarly, deep samples that were obtained at depths of 250 and 500 m formed another cluster that was clearly separated from surface samples. This distribution pattern was observed when we analyzed the data obtained with both primer sets and considered the two types of matrices. However, the results obtained when we used the binary matrix with both primer sets appeared to be more consistent with expectations of gradual change along a vertical profile; the differences between consecutive depths in the vertical profiles were more gradual, and both vertical profiles changed more in parallel.
FIG. 4.
Averages and standard errors of intensity values for DGGE bands of sample ME-B0 as quantified from separate PCR and DGGE analyses with primer set A (A) (n = 5) and primer set B (B) (n = 4). Where error bars are not visible, the error was smaller than the symbol.
FIG. 5.
NMDS diagrams relating the picoeukaryotic assemblages in ME-B samples on the basis of the DGGE gels shown in Fig. 3. NMDS diagrams were calculated from fingerprints obtained with primer set A using the intensity (A) and binary (C) matrices and from fingerprints obtained with primer set B using the intensity (B) and binary (D) matrices. On each diagram the grey circle corresponds to sample ME-B0, the solid circles correspond to ME-B3 samples, and the open circles correspond to ME-B4 samples. Solid and dashed lines join the data for the ME-B3 and ME-B4 samples, respectively, obtained from the surface (5 m) to a depth of 500 m. Only bands that accounted for at least 1% of the intensity in a lane were used in this analysis.
Identification of picoeukaryotic populations.
The potential of DGGE for identifying picoeukaryotic populations was addressed by sequencing DGGE bands of the ME-B0 sample. We sequenced 11 bands obtained with primer set A (Fig. 3A) that accounted for 70% of the total band intensity and 11 bands obtained with primer set B (Fig. 3B) that accounted for 84% of the total band intensity. The closest matches (and percentages of similarity) for the sequences retrieved were determined by a BLAST search (Table 2). The number of bases used to calculate each similarity value is also shown in Table 2 as an indication of the quality of the sequence. The most intense bands in the profiles obtained with both primer sets corresponded to the prasinophytes Mantoniella squamata (bands A7, B1, and B6 [Fig. 3 and Table 2]) and Ostreococcus tauri (bands A8, B3, B5, and B7) and to the appendicularian Oikopleura sp. (bands A9 and B9). Several other groups, such as prymnesiophytes (bands A5 and B4), cryptophytes (band A11), ciliates (bands A6, B10, and B11), dinophytes (band B2), and novel stramenopile groups closely related to labyrinthulids and hyphochytrids (bands A1 to A4), were also represented. Many of these groups are known to include organisms which are very small, and thus the sequences obtained probably belong to true picoeukaryotes. In other cases, such as ciliates and especially an appendicularian and a copepod, the presence of the organisms in the sample analyzed was obviously the result of inefficient prefiltration. Finally, only eukaryotic sequences were recovered, indicating that the two primer sets were very specific.
TABLE 2.
Sequence similarities of excised eukaryotic bands that appear in Fig. 3
| Band | Intensity (%) | Most closely related organism | % sequence similarity (no. of bases)a | Taxonomic group |
|---|---|---|---|---|
| Set A | ||||
| A1 | 6 | Thraustochytrium multirudimentale | 91 (484) | Stramenopiles |
| A2 | 3 | Thraustochytrium multirudimentale | 89 (424) | Stramenopiles |
| A3 | 1 | Thraustochytrium multirudimentale | 92 (270) | Stramenopiles |
| A4 | 3 | Thraustochytrium multirudimentale | 93 (432) | Stramenopiles |
| A5 | 3 | Unidentified prymnesiophyte | 89 (88) | Prymnesiophytes |
| A6 | 3 | Oxytricha sp. | 90 (67) | Ciliophora |
| A7 | 16 | Mantoniella squamata | 96 (269) | Prasinophytes |
| A8 | 14 | Ostreococcus tauri | 90 (407) | Prasinophytes |
| A9 | 19 | Oikopleura sp. | 98 (422) | Appendicularians |
| A10 | 9 | Calanus sp. | 95 (489) | Copepoda |
| A11 | 2 | Geminigera cryophila | 90 (285) | Cryptophytes |
| Set B | ||||
| B1 | 1 | Mantoniella squamata | 93 (90) | Prasinophytes |
| B2 | 3 | Prorocentrum sp. | 94 (114) | Dinophytes |
| B3 | 3 | Ostreococcus tauri | 86 (122) | Prasinophytes |
| B4 | 5 | Unidentified prymnesiophyte | 91 (33) | Prymnesiophytes |
| B5 | 11 | Ostreococcus tauri | 95 (172) | Prasinophytes |
| B6 | 11 | Mantoniella squamata | 97 (187) | Prasinophytes |
| B7 | 5 | Ostreococcus tauri | 89 (193) | Prasinophytes |
| B8 | 7 | Pelagomonas calceolata | 89 (169) | Pelagophytes |
| B9 | 34 | Oikopleura sp. | 96 (191) | Appendicularians |
| B10 | 2 | Oxytricha sp. | 91 (65) | Ciliophora |
| B11 | 2 | Strombidium sp. | 81 (141) | Ciliophora |
The numbers in parentheses are the numbers of bases used to calculate the levels of sequence similarity.
These results provided the identities of the most intense bands in the ME-B0 sample. This in turn permitted these phylotypes to be monitored along the vertical profiles in the Mediterranean Sea study (Fig. 3). Thus, bands corresponding to prasinophytes (especially bands A7, A8, B5, and B6) were detected at depths from the surface down to 50 m (sometimes down to 100 m) in both years, but they were absent at depths of 250 and 500 m. Some other bands, such as those corresponding to ciliates (bands A6, B10, and B11), seemed to be present at practically all depths. Something similar occurred with the band associated with a dinophyte (band B2), which was present at practically all depths. The intensity of this band increased with depth, and it was very intense at 250 to 500 m. Finally, the intense band corresponding to Oikopleura sp. (bands A9 and B9) was not found in the other samples, confirming that its presence was a prefiltration artifact that occurred only with the ME-B0 sample.
Comparison with other molecular techniques.
The phylogenetic composition of the picoeukaryotic assemblage in sample ME-B0 was also investigated by two other molecular techniques. The first technique was analysis of a clone library constructed with primer set C (Table 1), and the results are described in the accompanying paper (9). Ninety-nine eukaryotic clones in this library were analyzed by the RFLP method with HaeIII, and at least one representative of each of the 29 OTUs obtained was sequenced. The second technique was analysis of a T-RFLP fingerprint obtained with modified primer set A (Table 1) and assignment of the TRFs to phylogenetic entities by comparison with a computer-simulated restriction analysis of sequences in the database. Analyses of the results of three restriction digestions were performed, but only results obtained with HhaI are presented here since this enzyme gave more TRFs (19 different fragments) and the phylogenetic assignments were the least ambiguous. Whereas cloning and sequencing are very time-consuming but very informative, analysis of TRFs is fast but the phylogenetic assignments are only tentative. Similar numbers of OTUs (between 14 and 29 of OTUs) were detected with the three techniques (Table 3), which is remarkable since the definition of OTU was different for each technique. Moreover, the three techniques identified the same phylogenetic groups, and when the relative abundance of each group in the PCR pool was quantified (by estimating the percentage of total DGGE band intensity, the percentage of clonal representation, and the percentage of the total peak area for each TRF), the results were reasonably consistent (Table 3).
TABLE 3.
Relative levels of several eukaryotic groups in sample ME-B0 as estimated by different molecular methods, including DGGE band intensity determined with two primers sets, clonal representation in a genetic library, and TRF peak intensity
| Taxon | % of total in the following analyses:
|
|||
|---|---|---|---|---|
| DGGE analysis with primer set A (n = 29)a | DGGE analysis with primer set B (n = 14) | Library analysis with primer set C (n = 29) | HhaI T-RFLP analysis with primer set A (n = 19) | |
| Oikopleura sp. | 19 | 34 | 36 | 27 (195)b |
| Prasinophyceae | 30 | 31 | 16 | 24 (418, 420) |
| Prymnesiophyceae | 3 | 5 | 1 | 1 (280) |
| Pelagophyceae | 7 | 1 | 1 (442) | |
| Ciliophora | 3 | 4 | 5 | 0–8 (430)c |
| Eustigmatophyceae | 2 | 0–4 (433)c | ||
| Dinophyceae | 3 | 1 | ||
| Cryptophyceae | 2 | 3 | ||
| Diatoms | 6 | 0–12 (430, 433)c | ||
| Novel stramenopiles | 13 | 12 | ||
| Not assigned | 30 | 16 | 16 | 35 |
n is the number of OTUs identified.
The numbers in parentheses are the sizes of the corresponding TRFs (in base pairs).
The fragment(s) was produced by more than one phylogentic group.
All three techniques showed that the appendicularian Oikopleura sp. sequence was one of the most abundant 18S rDNA sequences in the sample; this sequence accounted for 19 and 34% of the total as estimated by DGGE with two primers sets, 36% of the total as estimated by clonal representation, and 27% of the total as estimated by the peak area percentage of the 195-bp fragment (Table 3). Two DGGE bands obtained with primer set A (accounting for 30% of the band intensity) and up to five DGGE bands obtained with primer set B (31% of the band intensity) were affiliated with the prasinophytes M. squamata and O. tauri. In the genetic library, 16% of the clones (distributed in three OTUs) were affiliated with the same two organisms. The T-RFLP analysis detected 418- and 420-bp TRFs, each representing 12% of the total fluorescence. The sizes of these TRFs were the sizes expected for O. tauri and M. squamata. Another important group in the clone library was a set of sequences (12% of the total) that formed novel lineages in the stramenopile group distantly related to the labyrinthulids and hyphochytrids (9). Similar sequences were detected by DGGE with primer set A (bands A1 to A4; 13% of the intensity) but were not detected by the other techniques. However, it must be noted that a significant fraction of the PCR product analyzed by the DGGE and T-RFLP techniques could not be identified (Table 3), and part of this fraction could account for these sequences. Several other groups, such as the dinophytes, prymnesiophytes, cryptophytes, ciliates, eustigmatophytes, diatoms, and pelagophytes, were detected by two or three of the techniques, and the level of these groups was always minor (Table 3). As an example of the conclusion that T-RFLP analysis results cannot be used for phylogenetic identification, the 430-bp TRF (8% of the intensity) was produced by the ciliate Oxytricha granulifera and the diatom Skeletonema costatum, and the 433-bp TRF (4% of the intensity) was produced by the eustigmatophyte Nannochloropsis sp. and the diatom Papiliocellulus elegans. This has been pointed out previously for prokaryotes (29, 31).
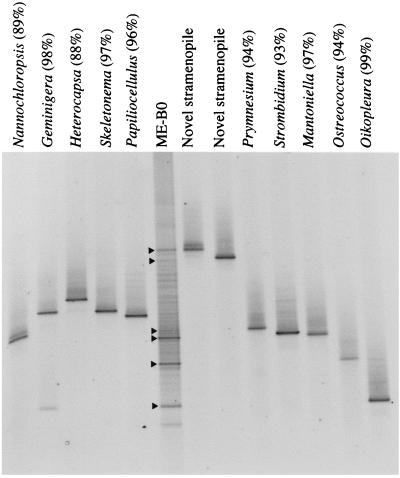
We performed a final check to directly compare the results obtained for the ME-B0 sample with DGGE and the clone library. Clones belonging to different OTUs and the ME-B0 sample were amplified by using primer set A and electrophoresed together in a DGGE gel (Fig. 6). The products from the amplified clones loaded to the right of the ME-B0 sample (novel stramenopiles, Prymnesium, Strombidium, Mantoniella, Ostreococcus, and Oikopleura) migrated to the same position in the gel as the community-derived DGGE bands having the same sequences (Fig. 6). This expected coincidence supported the results obtained by sequencing of the DGGE bands and the conclusion that different primers amplify the same sequences. On the other hand, clones loaded to the left of the ME-B0 sample (Nannochloropsis, Geminigera, Heterocapsa, Skeletonema, and Papiliocellulus) migrated to positions in the gel where there were several bands in the ME-B0 sample that could not be reamplified and/or sequenced.
FIG. 6.
DGGE fingerprints obtained with primer set A for the ME-B0 sample and several clones from the genetic library obtained from the same sample. The clone names are the names of the most closely related organisms in the database (levels of similarity are given in parentheses) found in a BLAST search. The lanes to the right of the ME-B0 sample contained clones representing phylotypes that have been retrieved by sequencing DGGE bands (indicated by arrowheads).
DISCUSSION
Planktonic picoeukaryotes are widely distributed in the photic zone in marine systems. They form a heterogeneous assemblage composed of small flagellated or coccoid algae and small heterotrophic flagellates. Direct identification of marine picoeukaryotes by microscopy is problematic because of their small sizes. Fortunately, diversity studies of picoeukaryotes can take advantage of the approaches used with marine prokaryotes (2, 41) since they can be similarly collected and processed by culture-independent techniques. The resulting environmental DNA extracts can be analyzed by an array of molecular techniques. Recent analysis of marine picoeukaryotes by gene cloning and sequencing (9, 27, 35) has indicated that the diversity of this assemblage is rather high and that this group includes organisms belonging to very different groups, some of which represent new and underscribed phylogenetic lineages. Here we describe the use of DGGE to compare the structures and compositions of different marine picoeukaryotic assemblages.
It is well known that quantification of organisms by PCR-based methods presents many uncertainties (53). Some biases may be due to differences in rRNA gene copy number (11), and this could be especially important for eukaryotic organisms that may contain up to several thousand copies of the rRNA gene (26). During PCR some phylotypes can be amplified preferentially due to preferential priming or differences in elongation rates between amplicons. Another bias can occur when the PCR includes many cycles; according to the kinetic model, when the number of cycles is increased, there is a tendency for the different amplicons to reach equimolarity (49). All of these potential biases can change the relative concentrations of PCR products so that the resulting profile of phylotypes no longer reflects the composition of the native community. In this study we attempted to quantify the relative levels of several picoeukaryote populations in one sample. In order to have a control for PCR biases, we compared the results obtained with three different approaches (DGGE, T-RFLP analysis, and gene cloning) using three different primer sets and different PCR protocols. The accidental presence of rDNA of Oikopleura in ME-B0 was used to illustrate relative quantitation of a single population with these different approaches. Thus, the relative levels of this rDNA were 19 and 34% as estimated by DGGE, 36% as estimated by clone library analysis, and 27% as estimated by T-RFLP analysis. These values were reasonably comparable considering the substantial technical differences among the three approaches. Note that the greatest difference occurred with the different primer sets used in the DGGE analysis. Moreover, a more exhaustive analysis of all the data revealed that the same phylotypes, at similar relative levels, were detected with the different techniques.
Fingerprinting techniques, such as DGGE and T-RFLP analysis, allow easy and quick comparison of profiles from related microbial assemblages and are now used in many ecological studies (25, 29, 34, 39). An advantage of DGGE is that selected bands can be sequenced, and thus, the presence of a particular phylotype can be monitored in the environmental samples studied. However, sequences obtained from DGGE bands are short (less than one-third the total length of small-subunit rRNA) and of variable quality. The shorter the sequence derived from DGGE fragments, the less refined the phylogenetic inference. Regarding the quality of the sequences, character ambiguities for directly sequenced PCR amplification products probably arise from amplification of different phylotypes with very similar electrophoretic mobilities. While these ambiguities do not prohibit identification with BLAST, the number of informative characters decreases in proportion to the number of ambiguities. One way to obtain cleaner sequences would be to clone excised bands and analyze several of the clones, but this would be prohibitively laborious when complex communities are analyzed. Therefore, as pointed out previously (7, 39), sequencing of DGGE bands is sufficient to determine broad phylogenetic affiliations but inadequate to perform a precise phylogenetic analysis.
We used two primer sets for DGGE in order to measure reliability. These primer sets amplify nonoverlapping regions of the 18S rRNA gene; set A amplifies a region between positions 4 and 563, including variable regions V1 to V3 (40), and set B amplifies a region between positions 1423 and 1641, including variable region V8. Primer set B amplifies the same region as a primer set described previously (51), but the primers are not the same. When tested with pure cultures, both sets were found to be specific for eukaryotic organisms and gave a single DGGE band, and when applied to environmental samples, they gave complex and reproducible fingerprints. The number of bands obtained with natural samples with set A (20 to 45 bands) was higher than the number of bands obtained with set B (10 to 22 bands). This could have been due to the fact that set B amplifies a smaller fragment with less sequence variability. Based on our results, there are at least two reasons to recommend using primer set A: it amplified a much larger DNA fragment and provided more phylogenetic information, and time travel experiments indicated that this primer set performed better.
The fingerprints obtained by the DGGE method were used to examine the similarity of a group of samples with NMDS diagrams calculated from binary and intensity matrices. The fact that the intensity of DGGE bands was reproducible argued in favor of using the intensity matrix for such analyses. In fact, this is what we proposed in a previous study in which marine bacterial assemblages were compared (46). However, the results for picoeukaryotes appeared to be better when the binary matrix was used (Fig. 5). This was due to the random presence in some samples of very intense bands corresponding to larger eukaryotic organisms, such as Oikopleura in sample ME-B0 or a copepod in sample ME-B3 obtained at 250 m (the lower, dominant band in the fingerprint [Fig. 3A]). The presence of these bands, which obviously did not correspond to picoeukaryotes, revealed that prefiltration did not always work perfectly. These bands could dominate the grouping of samples when the intensity matrix was used but were less influential when the binary matrix was used. This explains why the ME-B0 sample (which produced the intense Oikopleura band) always grouped better with the other surface samples when the binary matrix was used.
The picoeukaryotic diversity as measured by the different techniques appeared to be great; numerous OTUs and widely separated phylogenetic groups were detected (Table 3). The clone library provided a detailed list of the phylotypes present in sample ME-B0 that could be compared with the sequences obtained from DGGE bands and the database of terminal fragments. The prasinophyte group appeared to be the most abundant group, suggesting that these organisms are important components of marine picoplankton in Mediterranean waters. Significant levels of prasinophytes in other open ocean and coastal environments were detected in libraries of 18S rDNA genes (9, 35), in an analysis of plastidic clones of a bacterial 16S rDNA library (44), by HPLC pigment analysis (20), and by electron microscopy (5). Other groups detected in the clone library, such as prymnesiophytes, pelagophytes, and novel stramenopiles, were also identified by sequencing DGGE bands. In addition, clones belonging to groups not retrieved from DGGE bands, such as diatoms, cryptophytes, dinophytes, and eustigmatophytes, migrated to positions where several DGGE bands were not sequenced (Fig. 6), indicating that these groups could be represented in the unsequenced bands. Finally, we examined the ME-B0 sample by performing an HPLC analysis of pigments, a PCR-independent approach. This analysis attributed a high proportion of the phototrophic fraction to chlorophyll b-containing algae, including prasinophytes (M. Latasa, personal communication). Smaller amounts of pigments found in prymnesiophytes and cryptophytes were also detected by HPLC. Although the HPLC data were preliminary, they agreed with the molecular results.
In conclusion, we demonstrated that the combination of 18S rDNA community library sequencing and molecular fingerprinting is as revealing for picoeukaryotic communities as it is for prokaryotic communities. Similar phylogenetic groups at comparable relative levels were recovered by three different molecular approaches. Moreover, differences in community structure could be easily discerned with both DGGE and T-RFLP analysis. Direct application of these approaches to analysis and comparison of eukaryotic picoplankton assemblages should prove to be profitable.
ACKNOWLEDGMENTS
This work was funded by EU contracts MIDAS (MAS3-CT97-00154) and PICODIV (EVK3-CT1999-00021). Marine samples were collected on board B/O García del Cid and B.I.O. Hespérides during cruises funded by EU grant MATER (MAS3-CT96-0051). The T-RFLP analysis was funded in part by NSF grant NSF-DEB 8707224 to the Center for Microbial Ecology at Michigan State University.
We thank Mikel Latasa for sharing unpublished HPLC data, Lluïsa Cros for help with algal cultures, and Emilio O. Casamayor for helpful comments.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen R A, Saunders G W, Paskind M P, Sexton J P. Ultrastructure and 18S rRNA gene sequence for Pelagomonas calceolata gen. et sp. nov. and the description of a new algal class, the Pelagophyceae classis nov. J Phycol. 1993;29:701–715. [Google Scholar]
- 5.Andersen R A, Bidigare R R, Keller M D, Latasa M. A comparison of HPLC pigment signatures and electron microscopic observations for oligotrophic waters of the North Atlantic and Pacific oceans. Deep-Sea Res II. 1996;43:517–537. [Google Scholar]
- 6.Caron D A, Peele E R, Lim E L, Dennett M R. Picoplankton and nanoplankton and their trophic coupling in the surface waters of the Sargasso Sea south of Bermuda. Limnol Oceanogr. 1999;44:259–272. [Google Scholar]
- 7.Casamayor E O, Schäfer H, Bañeras L, Pedrós-Alió C, Muyzer G. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: a comparison of microscopy and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 2000;66:499–508. doi: 10.1128/aem.66.2.499-508.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Díez B, Pedrós-Alió C, Massana R. Genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl Environ Microbiol. 2001;67:2932–2941. doi: 10.1128/AEM.67.7.2932-2941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dsouza M, Larsen N, Overbeek R. Searching for patterns in genomic data. Trends Genet. 1997;13:497–498. doi: 10.1016/s0168-9525(97)01347-4. [DOI] [PubMed] [Google Scholar]
- 11.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fogg G E. Some comments on picoplankton and its importance in the pelagic ecosystem. Aquat Microb Ecol. 1995;9:33–39. [Google Scholar]
- 13.Fuhrman J A, McCallum K, Davis A A. Novel major archaebacterial group from marine plankton. Nature. 1992;356:148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- 14.Giovannoni S J, DeLong E F, Olsen G J, Pace N R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988;170:720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 16.Guillard R R L. Culture of phytoplankton for feeding marine invertebrates. In: Smith W L, Chanley M H, editors. Culture of marine invertebrate animals. New York, N. Y: Plenum Press; 1975. pp. 29–60. [Google Scholar]
- 17.Guillou L, Chrétiennot-Dinet M-J, Boulben S, Moon-van der Staay S Y, Vaulot D. Symbiomonas scintillans gen. et sp. nov. and Picophagus flagellatus gen. et sp. nov. (Heterokonta): two new heterothophic flagellates of picoplanktonic size. Protist. 1999;150:383–398. doi: 10.1016/S1434-4610(99)70040-4. [DOI] [PubMed] [Google Scholar]
- 18.Guillou L, Chrétiennot-Dinet M-J, Medlin L K, Claustre H, Loiseaux-de Goër S, Vaulot D. Bolidomonas: a new genus with two species belonging to a new algal class, the Bolidophyceae (Heterokonta) J Phycol. 1999;35:368–381. [Google Scholar]
- 19.Guillou L, Moon-van der Staay S Y, Claustre H, Partensky F, Vaulot D. Diversity and abundance of Bolidophyceae (Heterokonta) in two oceanic regions. Appl Environ Microbiol. 1999;65:4528–4536. doi: 10.1128/aem.65.10.4528-4536.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hooks C E, Bidigare R R, Keller M D, Guillard R R L. Coccoid eukaryotic marine ultraplankters with four different HPLC pigment signatures. J Phycol. 1988;24:571–580. [Google Scholar]
- 21.Kowalchuk G A, Gerards S, Woldendorp J W. Detection and characterization of fungal infections of Ammophila arenaria (marram grass) roots by denaturing gradient gel electrophoresis of specifically amplified 18S rDNA. Appl Environ Microbiol. 1997;63:3858–3865. doi: 10.1128/aem.63.10.3858-3865.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S RNA sequences for phylogenetic analysis. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W K W. Primary production of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol Oceanogr. 1994;39:169–175. [Google Scholar]
- 24.Lim E L, Dennet M R, Caron D A. The ecology of Paraphysomonas imperforata based on studies employing oligonucleotide probe identification in coastal water samples and enrichment cultures. Limnol Oceanogr. 1999;44:37–51. [Google Scholar]
- 25.Liu W L, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long E O, Dawid I B. Repeated genes in eukaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- 27.López-García P, Rodríguez-Valera F, Pedrós-Alió C, Moreira D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature. 2001;409:603–607. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- 28.Maidak B L, Cole J R, Lilburn T G, Parker C T, Jr, Saxman P R, Stredwick J M, Garrity G M, Li B, Olsen G J, Pramanik S, Schmidt T M, Tiedje J M. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 2000;28:173–174. doi: 10.1093/nar/28.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marsh T L. Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr Opin Microbiol. 1999;2:323–327. doi: 10.1016/S1369-5274(99)80056-3. [DOI] [PubMed] [Google Scholar]
- 30.Marsh T L, Liu W T, Forney L J, Cheng H. Beginning a molecular analysis of the eukaryal community in activated sludge. Water Sci Technol. 1998;37:455–460. [Google Scholar]
- 31.Marsh T L, Saxman P, Cole J, Tiedje J M. Terminal restriction fragment length polymorphism analysis program, a web-based research tool for microbial community analysis. Appl Environ Microbiol. 2000;66:3616–3620. doi: 10.1128/aem.66.8.3616-3620.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massana R, Murray A E, Preston C M, DeLong E F. Vertical distribution and phylogenetic characterization of marine planktonic Archaea in the Santa Barbara Channel. Appl Environ Microbiol. 1997;63:50–56. doi: 10.1128/aem.63.1.50-56.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medlin L, Elwood H J, Stickel S, Sogin M L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene. 1988;71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- 34.Moeseneder M M, Arrieta J M, Muyzer G, Winter C, Herndl G J. Optimization of terminal-restriction fragment length polymorphism analysis for complex marine bacterioplankton communities and comparison with denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1999;65:3518–3525. doi: 10.1128/aem.65.8.3518-3525.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moon-van der Staay S Y, De Wachter R, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature. 2001;409:607–610. doi: 10.1038/35054541. [DOI] [PubMed] [Google Scholar]
- 36.Murphy L S, Haugen E M. The distribution and abundance of phototrophic ultraplankton in the North Atlantic. Limnol Oceanogr. 1985;30:47–58. [Google Scholar]
- 37.Murray A E, Hollibaugh J T, Orrego C. Phylogenetic composition of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl Environ Microbiol. 1996;62:2676–2680. doi: 10.1128/aem.62.7.2676-2680.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muyzer G. DGGE/TGGE, a method for identifying genes from natural communities. Curr Opin Microbiol. 1999;2:317–322. doi: 10.1016/S1369-5274(99)80055-1. [DOI] [PubMed] [Google Scholar]
- 39.Muyzer G, Brinkhoff T, Nübel U, Santegoeds C, Schäfer H, Wawer C. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In: Akkermans A D L, van Elsas J D, de Bruijn F J, editors. Molecular microbial ecology manual. 3.4.4. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 1–27. [Google Scholar]
- 40.Neefs J-M, Van de Peer Y, De Rijk P, Chapelle S, De Wachter R. Compilation of small subunit RNA structures. Nucleic Acids Res. 1993;21:3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 42.Paul J H, Myers B. Fluorometric determination of DNA in aquatic microorganisms by use of Hoechst 33258. Appl Environ Microbiol. 1982;43:1393–1399. doi: 10.1128/aem.43.6.1393-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Potter D, Lajeunesse T C, Saunders G W, Andersen R A. Convergent evolution masks extensive biodiversity among marine coccoid picoplankton. Biodivers Conserv. 1997;6:99–107. [Google Scholar]
- 44.Rappé M S, Suzuki M T, Vergin K L, Giovannoni S J. Phylogenetic diversity of ultraplankton plastid small-subunit rRNA genes recovered in environmental nucleic acid samples from the Pacific and Atlantic coasts of the United States. Appl Environ Microbiol. 1998;64:294–303. doi: 10.1128/aem.64.1.294-303.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riemann L, Steward G F, Fandino L B, Campbell L, Landry M R, Azam F. Bacterial community composition during two consecutive NE Monsoon periods in the Arabian Sea studied by denaturing gradient gel electrophoresis (DGGE) of rRNA genes. Deep-Sea Res. 1999;46:1791–1811. [Google Scholar]
- 46.Schauer M, Massana R, Pedrós-Alió C. Spatial differences in bacterioplankton composition along the Catalan coast (NW Mediterranean) assessed by molecular fingerprinting. FEMS Microbiol Ecol. 2000;33:51–59. doi: 10.1111/j.1574-6941.2000.tb00726.x. [DOI] [PubMed] [Google Scholar]
- 47.Simon N, Barlow R G, Marie D, Partensky F, Vaulot D. Characterization of oceanic photosynthetic picoeukaryotes by flow cytometry. J Phycol. 1994;30:922–935. [Google Scholar]
- 48.Sogin M L, Gunderson J H. Structural diversity of eukaryotic small subunit ribosomal RNAs. Ann NY Acad Sci. 1987;503:125–139. doi: 10.1111/j.1749-6632.1987.tb40603.x. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki M T, Rappé M S, Giovannoni S J. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl Environ Microbiol. 1998;64:4522–4529. doi: 10.1128/aem.64.11.4522-4529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong S M. Developayella elegans nov. gen., nov. spec., a new type of heterotrophic flagellate from marine plankton. Eur J Protistol. 1995;31:24–31. [Google Scholar]
- 51.Van Hannen E J, van Agterveld M P, Gons H J, Laanbroek H J. Revealing genetic diversity of eukaryotic microorganisms in aquatic environments by denaturing gradient gel electrophoresis. J Phycol. 1998;34:206–213. [Google Scholar]
- 52.Van Hannen E J, Zwart G, van Agterveld M P, Gons H J, Ebert J, Laanbroek H J. Changes in bacterial and eukaryotic community structure after mass lysis of filamentous cyanobacteria associated with viruses. Appl Environ Microbiol. 1999;65:795–801. doi: 10.1128/aem.65.2.795-801.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Von Wintzingerode F, Goebel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]